Abstract
Cyclic changes of the human endometrium, such as proliferation, secretion, and decidualization, occur during regular menstrual cycles. Heart– and neural crest derivatives–expressed transcript 2 (HAND2) is a key transcription factor in progestin-induced decidualization of human endometrial stromal cells (ESCs). It has been suggested that HAND2 regulates interleukin 15 (IL15), a key immune factor required for the activation and survival of uterine natural killer (uNK) cells. Activated uNK cells can promote spiral artery remodeling and secrete cytokines to induce immunotolerance. To date, no studies have evaluated the transcription factors that regulate IL15 expression in human ESCs. In the present study, we examined whether HAND2 controls IL15 transcriptional regulation in human ESCs. Quantitative RT-PCR and histological analyses revealed that HAND2 and IL15 levels increase considerably in the secretory phase of human endometrium tissues. Results from ChIP-quantitative PCR suggested that HAND2 binds to a putative HAND2 motif, which we identified in the upstream region of the human IL15 gene through in silico analysis. Using a luciferase reporter assay, we found that the upstream region of the human IL15 gene up-regulates reporter gene activities in response to estradiol and a progestin representative (medroxyprogesterone) in ESCs. The upstream region of the human IL15 gene also exhibited increasing responsiveness to transfection with a HAND2 expression vector. Of note, deletion and substitution variants of the putative HAND2 motif in the upstream region of IL15 did not respond to HAND2 transfection. These findings confirm that HAND2 directly up-regulates human IL15 transcription in ESCs.
Keywords: gene transcription, basic helix-loop-helix transcription factor (bHLH), reproduction, interleukin, stromal cell, heart- and neural crest derivatives-expressed transcript 2 (HAND2), gene regulation, interleukin 15 (IL15), endometrium, decidualization, Forkhead box protein O1 (FOXO1), gene expression, FOXO
The human endometrium undergoes growth and regeneration during each menstrual cycle. In the absence of an implantation, the endometrium sheds, is repaired, proliferates, and terminally differentiates to prepare for subsequent embryo implantation (1, 2). These sequential events are controlled by the interplay of ovarian steroid hormones, estradiol (E2) and progesterone (P4) (3).
During the proliferative phase, human endometrial epithelial cells and stromal cells (ESCs) increase in response to E2, leading to a thickening of the stratum functionalis of the endometrium. After ovulation, under P4 dominance, the endometrium, which is regulated by autocrine, paracrine, and transcription factors, transitions from the proliferative to the secretory phase (4). The endometrial transition leads to spontaneous decidualization of human ESCs. Decidualization co-occurs with functional and morphological changes in human ESCs (5).
Uterine natural killer (uNK) cells are the predominant leukocyte in normal human endometrium (6). Approximately 70–80% of uNK cells are characterized as CD56brightCD16− (7). Activated uNK cells can produce angiogenic factors, which promote spiral artery remodeling, and secrete cytokines that direct the migration and invasion of the trophoblast by interaction with trophoblast surface antigens (8–10). Importantly, the uNK cells and other leukocytes in the endometrium do not express P4 receptors (11). The activation and survival of uNK cells have been associated with the decidual-specific factor, interleukin-15 (IL15), in the human endometrium (12).
Previously, we showed that one transcription factor, heart and neural crest derivatives-expressed transcript 2 (HAND2), was up-regulated by E2 and a representative progestin, medroxyprogesterone (MPA), in a time- and dose-dependent manner for ESC decidualization (13). Originally, HAND2 was identified as a basic helix-loop-helix transcription factor for embryonic right ventricles (14, 15). In human ESCs, silencing of HAND2 reduced both the morphological differentiation and the decidual-specific factors, including prolactin, forkhead box O1A (FOXO1A), interleukin-15 (IL15), fibulin-1, and tissue inhibitor of metalloproteinase-3 (16). Therefore, it is conceivable that IL15 transcription is directly or indirectly regulated by HAND2.
To the best of our knowledge, no studies have evaluated transcription factors directly affecting IL15 in human ESCs. We have applied in silico analysis here to identify HAND2 motifs (CHTCTGG) (17) and found candidate sequences in the upstream and intronic region of the human IL15 locus. The present study aimed to examine whether HAND2 participates in IL15 transcriptional regulation in human ESCs.
Results
Up-regulation in HAND2 and IL15 during the secretory phase of the human endometrium
First, we examined whether HAND2 and IL15 were up-regulated in the human endometrium as well as cultured ESCs. During the secretory phase (days 15–28 from previous menstruation, n = 12), HAND2 and IL15 were significantly increased compared with the proliferative phase (days 6–11 from previous menstruation, n = 8) (HAND2: 6.08 ± 5.40 (mean ± S.D.) versus 2.00 ± 0.73, p < 0.05; IL15: 6.86 ± 4.63 versus 2.47 ± 1.34, p < 0.05) (Fig. 1). There is a significant correlation between HAND2 and IL15 in the human endometrium (r2 = 0.4239, p < 0.05) (Fig. 1).
Figure 1.
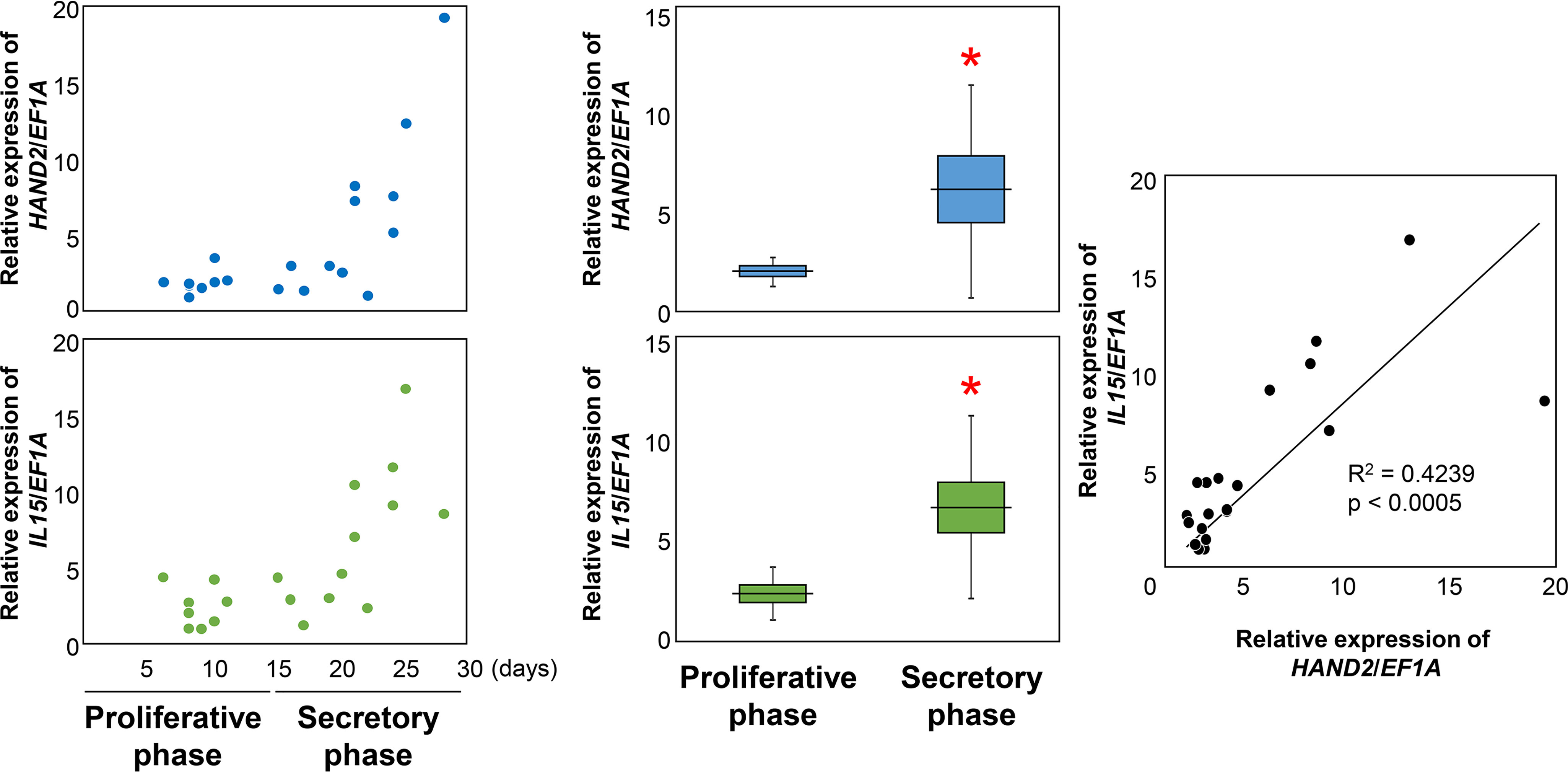
Relative expression of HAND2 and IL15 in the human endometrium. HAND2 and IL15 were evaluated using RT-qPCR using human endometrium cDNA from the proliferative phase and the secretory phase. Each dot indicates the relative expression level from each subject. Changes in transcription levels between the proliferative phase and the secretory phase were analyzed using Student's t test. *, p < 0.05 versus the proliferative phase. Correlations between HAND2 and IL15 in all phases were analyzed using Spearman's rank correlation coefficient. Horizontal lines and squares indicate the mean ± S.E. Error bars, S.D. Each dot indicates relative expression data from each patient.
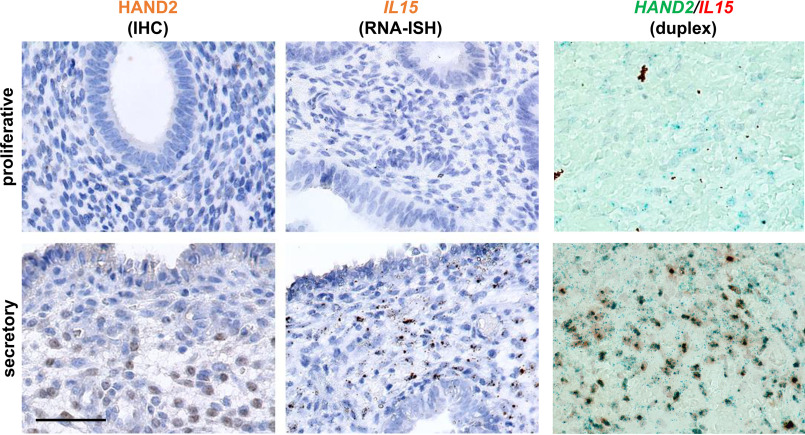
In the endometrium sections (Fig. 2), pseudostratification of the nuclei in the glandular epithelium was observed as the proliferative phase unfolded. In the secretory phase, glandular epithelium transformed to a secretory form in the glands as monolayer cubic epithelium. ESCs in the endometrium transformed from fibroblast-like in the proliferative phase to epithelium-like with cytoplasmic expansion, large pale nuclei, and rounding of cells in the secretory phase as predecidua. Similar to quantitative PCR (qPCR) results, histological analysis indicated that HAND2 protein and IL15 mRNA increased in the secretory phase in the endometrium compared with the proliferative phase (Fig. 2). Although both are estimated to be expressed in ESCs, HAND2 and IL15 mRNA were observed in many, but not all, ESCs in the endometrium during the secretory phase (Fig. 2). HALO analysis shows that during the secretory phase, there are 390.7 ± 50.3 (312–464) HAND2-positive cells/0.1 mm2 (Fig. S1). Cells with IL15 signals exhibited 100% merge with HAND2 signals in the secretory phase. Conversely, cells with HAND2 signals did not always merge with IL15 signals; the results demonstrated that 60.3 ± 6.4 (48.9–70.3)% of HAND2-positive cells were merged with the IL15, suggesting a time lag for IL15 transcription start by HAND2.
Figure 2.
Localization of HAND2 and IL15 in the human endometrium. The top panels show the stained sections in the proliferative phase, whereas the bottom panels show them in the secretory phase. HAND2 protein was detected using immunohistochemical (IHC) staining (left panels). HAND2 was localized in the nuclei of ESCs, especially in the secretory phase, as brown signals. IL15 was detected using RNA in situ hybridization (ISH) (middle panels). IL15 was predominantly expressed in ESCs, especially in the secretory phase of the endometrium. One brown dot signal represents one mRNA copy in the RNAscope results. Double staining with HAND2 and IL15 was conducted by RNAscope duplex (right panels). Almost all cells with HAND2 in the secretory phase merged with IL15. One red and green dot represent one mRNA copy of IL15 and HAND2 in duplex, respectively. Black dots in IL15 staining of the proliferative phase section and brown debris in duplex of the proliferative phase section are regarded as the artifacts of tissue fragments. Bar, 50 μm.
HAND2 binds to the upstream region of human IL15 gene in ESCs
Similar regions within 2 kbp upstream regions from transcription start sites between human IL15 and mouse, rat, or monkey IL15 gene were analyzed using VISTA (18, 19) (Fig. S2). IL15 upstream regions between humans and mice or humans and rats are not conserved, but regions between humans and monkeys are well-conserved. Among them, the positions from −439 to −1 in the human IL15 upstream region are not conserved (Fig. S2). Next, a homology search among Homo sapiens, Macaca fascicularis, and Mus musculus was performed using ClustalW version 2.1 (Fig. 3A). We found the putative consensus sequence for HAND2 binding (17) in the well-conserved upstream region and intronic regions of human IL15 locus using in silico analysis. Putative HAND2 motifs are denoted as CCTCTGG at position −1628/−1622, relative to the transcription start site, in the upstream region of the human IL15 gene (Fig. 3, A and B). One putative HAND2 motif was also found in the first intron of the human IL15 locus and is denoted as CATCTGG at position intervening sequences (IVS)1_16239/16245 (Fig. 3B). Introns have been shown to affect gene expression by a number of known and unknown mechanisms, collectively known as intron-mediated enhancement (IME) (20). However, introns must be within 1 kbp of the transcription start to exert an effect via IME. Because the length of exon 1 in the human IL15 gene is 153 nucleotides, it has been suggested that the putative HAND2 motif at IVS1_16239/16245 is involved in IME. Therefore, ChIP-PCR analysis was performed using primers specific to the human IL15 upstream region around the upstream putative motif, the putative motif in intron 1, negative control primers (pair −1110F and −933R and pair −1937F and −1761R) (Fig. 3B), and immunoprecipitated purified DNA. ChIP-PCR analysis using the −1687F and −1515R primer pair revealed a positive reaction for the human IL15 upstream region around the putative HAND2 motif (Fig. 3C). An additional positive reaction was found in the experiment using the −1649F and −1405R primer pair (Fig. 3C). PCR against the putative HAND2 motif included in intron 1 with the IVS1_16153F and IVS1_16346R primer pair or the IVS1_16002F and IVS1_16253R primer pair showed no positive amplifications. No amplification was detected from purified DNA incubated with normal rabbit IgG (Fig. 3C) or water, which were used as negative control templates, or negative control experiments with the −1110F and −933R primer pair and the −1937F and −1761R primer pair (Fig. 3C). We also confirmed the binding of HAND2 to the HAND2 motifs at −1628/−1622 using ChIP-qPCR with ESC immunoprecipitated chromatin samples from three independent patients (Fig. 3D). Even in ChIP-qPCR, HAND2 recognized the CCTCTGG sequence at position −1628/−1622 of the IL15 upstream region without the CATCTGG at position IVS1_16239/16245 and the negative control region (Fig. 3D).
Figure 3.
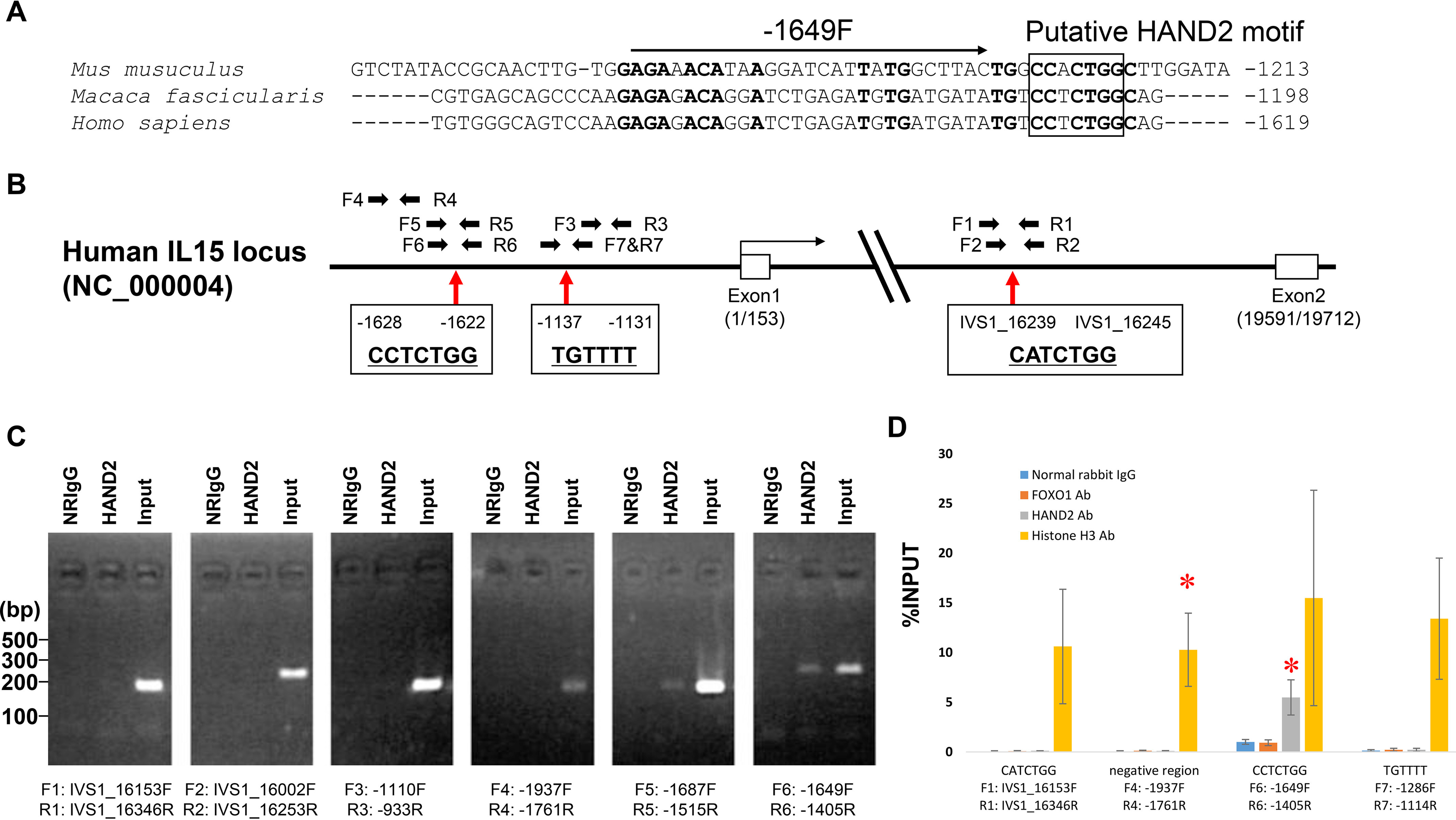
Schematic representation of the human IL15 locus and gene regulatory region. A, positions from −439 to −1 in the human IL15 upstream region are not conserved among species and are specific for human genome. The putative HAND2 binding motif is shown in the box. Conserved sequences among species are shown in boldface type. Numbers on the right indicate the positions from transcriptional start sites. B, boxes are exons. Arrows indicate primers for Chip-PCR. C, ChIP-PCR. NRIgG, normal rabbit IgG; Ab, antibody. HAND2, chromatin was immunoprecipitated with HAND2 antibody. Input, 10% input of total chromatin. D, ESCs were treated with E2 (10−8 mol/liter) and MPA (10−7 mol/liter) for 12 days. After that, chromatin was immunoprecipitated with corresponding antibodies. The transcription factors HAND2 and FOXO1 to the 1.8-kbp upstream region of the IL15 locus were analyzed using a ChIP-qPCR assay. Histone H3 antibody and normal rabbit IgG were used as positive and negative controls, respectively. The relative recruitment levels were analyzed and plotted as the ratio of immunoprecipitated DNA to the total input DNA sample (%INPUT). Data are represented as mean ± S.D. Multiple comparisons were performed using Student's t test with a Bonferroni correction. *, p < 0.01666 versus normal rabbit IgG in each region.
IL15 up-regulation in ESCs treated with E2 and MPA is controlled by IL15 upstream region alone
Subsequently, to clarify the significance of the 1.8-kbp upstream region, including the position at −1628/−1622 of the IL15 gene on IL15 transcription in ESCs, we performed a luciferase reporter assay with IL15ups/pGL4.10 (Fig. 4A) under treatment with E2 and MPA. No significant responses were observed between pGL4.10 with/without E2 and MPA treatment (1.19 ± 0.09/1.00 ± 0.38) (Fig. 4B), indicating an absence of confounding ESCs endogenous transcriptional regulatory elements in the pGL4.10 plasmid. Additionally, there are no significant responses between pGL4.10 and IL15ups/pGL4.10 without treatment (1.35 ± 0.35), but E2 and MPA treatment significantly increased luciferase activity of IL15ups/pGL4.10 (6.78 ± 1.06, p < 0.05) (Fig. 4B) compared with no treatment. Even in ESCs, from a different patient with the estimated secretory phase, E2 and MPA stimulation significantly increased luciferase activity (18.79 ± 2.20, p < 0.05; Fig. S3) compared with no stimulation. It is suggested that the upstream region of the IL15 gene can regulate IL15 transcription in ESCs under E2 and MPA stimulation in isolation, without intronic sequence. Alternatively, in this different patient's ESCs, IL15ups/pGL4.10 showed significant elevation in luciferase activities without E2 and MPA stimulation (5.47 ± 0.67, p < 0.05; Fig. S3) compared with pGL4.10 without E2 and MPA stimulation (1.00 ± 0.06) (Fig. S3), suggesting the presence of an endogenous transcription factor for IL15 prior to E2 and MPA stimulation in this ESCs.
Figure 4.
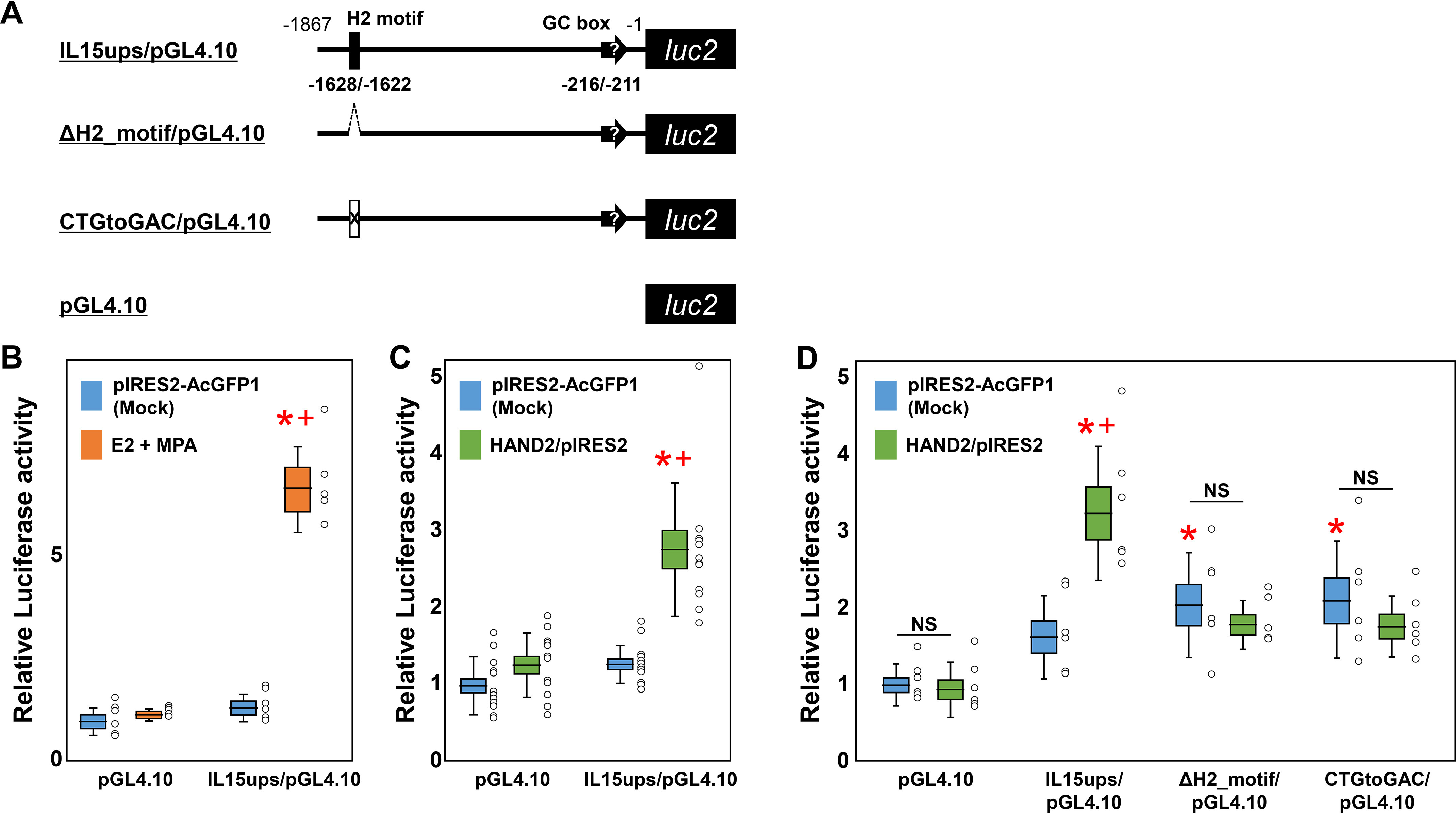
Luciferase activity in the 1.8-kbp human IL15 upstream region of human ESCs. A, schematic representation of the reporter plasmid containing the 1.8-kbp human IL15 upstream region with/without mutants. ΔH2_motif/pGL4.10 reporter has a deletion of the putative HAND2 motif (H2 motif) at −1628/−1622. CTGtoGAC/pGL4.10 reporter has three nucleotide substitutions at −1623/−1625. The 1.8-kbp upstream region includes a putative GC box promoter at −216/−211, as shown by black arrows with quotation marks. B, luciferase activity in the 1.8-kbp human IL15 upstream region of human ESCs against E2 and MPA treatment. ESCs were treated with E2 (10−8 mol/liter) and MPA (10−7 mol/liter) 1 day prior to transfection. 2 days after transfection, ESCs were lysed, and luciferase activity was measured. Multiple comparisons were performed using ANOVA with a Steel–Dwass test. *, p < 0.05 versus pGL4.10 with mock; †, p < 0.05 versus mock in each condition. n ≥ 5 in each condition. C, luciferase activity in the 1.8-kbp human IL15 upstream region of human ESCs against HAND2. 2 days after transfection with HAND2 expression vectors, ESCs were lysed, and luciferase activity in the human IL15 upstream region was measured. Multiple comparisons were performed using ANOVA with a Steel–Dwass test. *, p < 0.05 versus pGL4.10 with mock; †, p < 0.05 versus mock in each condition. n ≥ 6 in each condition. D, effects of mutants against luciferase activity in the 1.8-kbp human IL15 upstream region. 2 days after transfection, ESCs were lysed, and luciferase activity was measured. Multiple comparisons were performed using ANOVA with Tukey's test. *, p < 0.05 versus pGL4.10 with mock; †, p < 0.05 versus IL15ups/pGL4.10. n ≥ 5 in each condition. NS, no significance. The deletion mutant ΔH2_motif/pGL4.10 contains an internal deletion at −1628/−1622, and nucleotide substitution reporter CTGtoGAC/pGL4.10 has three substitutions at −1623/−1625 within the IL15ups/pGL4.10. Horizontal lines and squares indicate the mean ± S.E. Error bars, S.D. Each dot indicates relative luciferase activity from each well.
HAND2 regulates IL15 transcription via putative HAND2 motif in the upstream region of IL15 gene in ESCs
Thereafter, we examined whether HAND2 regulates IL15 transcription via the 1.8-kbp upstream region of IL15, including the position at −1628/−1622, in ESCs. Co-transfection of the pGL4.10 plasmid with the pIRES2-AcGFP1 (1.00 ± 0.36) or HAND2/pIRES2 (1.26 ± 0.42) vectors exhibited similar luciferase expression in ESCs (Fig. 4C). Luciferase activity from IL15ups/pGL4.10 with the pIRES2-AcGFP1 vector (mock) exhibited no significant increase compared with the pGL4.10 and pIRES2-AcGFP1 vector (mock) (1.27 ± 0.26) in ESCs (Fig. 4C). Activity in the IL15ups/pGL4.10 group increased significantly via co-transfection with the HAND2/pIRES2 vector compared with mock in ESCs (2.76 ± 0.88; p < 0.05; Fig. 4C).
Subsequently, the luciferase activity of deletion (ΔH2_motif/pGL4.10) (Fig. 4A) and substitution (CTGtoGAC/pGL4.10) (Fig. 4A) mutants of the putative HAND2 motif were examined. The luciferase activity from IL15ups/pGL4.10 with HAND2/pIRES2 (3.25 ± 0.85) was significantly up-regulated in estimated secretory phase ESCs from different patients compared with pIRES2-AcGFP1 (1.64 ± 0.51, p < 0.05) (Fig. 4D). These results indicate that there is no individual difference in HAND2-induced IL15 up-regulation in each ESC series. Conversely, the ΔH2_motif/pGL4.10 (2.05 ± 0.65, p < 0.05) or CTGtoGAC/pGL4.10 (2.08 ± 0.74, p < 0.05) with pIRES2-AcGFP1 showed significant promoter activity compared with pGL4.10 with pIRES2-AcGFP1 (1.00 ± 0.25) (Fig. 4D). There are no responses against HAND2/pIRES2 in the ΔH2_motif/pGL4.10 (1.80 ± 0.30) and CTGtoGAC/pGL4.10 (1.74 ± 0.39).
To examine whether HAND2 effect against IL15 transcription is specific in ESCs, we conducted a reporter assay using IL15ups/pGL4.10 in HEK293T cells (Fig. S4). Although the IL15ups/pGL4.10 showed higher luciferase activities in HEK293T cells than in ESCs, no reactivity was observed in the 1.8-kbp IL15 upstream region against HAND2 in HEK293T cells (Fig. S4). This suggests that HAND2 effect against IL15 transcription might be specific in ESCs, or the 1.8-kbp IL15 upstream region might be suppressed, and this suppression might be released by HAND2 in ESCs.
No FOXO1 effect observed on the 1.8-kbp upstream region of the IL15 gene in ESCs
In our previous siRNA experiments against HAND2 (16), FOXO1 expression was synchronized with HAND2 expression. This may suggest that FOXO1 transcription is regulated by HAND2 and that FOXO1 coordinately regulates IL15 transcription in ESCs. We have also identified putative FOXO1 motifs (TGTTTT) (21–24) in the upstream region of human IL15 gene (positions at −1131/−1137 and −1993/−1998). Therefore, we examined whether IL15 was indeed regulated by FOXO1 in ESCs. ChIP-qPCR analysis was performed using primers to the putative FOXO1 motif at −1131/−1137 (Fig. 3B) and at −1993/−1998 (Fig. S5) and immunoprecipitated purified DNA. ChIP-qPCR analysis using the −1286F and −1114R, −2065F to −1916R, and −2051F to −1907R primer pairs showed no significant difference between the immunoprecipitated-DNA with normal rabbit IgG and FOXO1 antibody (Fig. 3D and Fig. S5), indicating no recruitment of FOXO1 to the IL15 upstream region in our ESCs as reported in previous ChIP-sequencing analysis with FOXO1 antibody and decidualizing ESCs (25).
Because it has been reported that FOXO1 binds to human IGFBP1 enhancer region in ESCs with decidualization (26), as a control experiment, we conducted ChIP-qPCR with a primer pair for the IGFBP1 enhancer region (Table S1). FOXO1 antibody significantly immunoprecipitated the human IGFBP1 enhancer region, including in our ESCs (Fig. S5), indicating the effect of FOXO1 antibody. FOXO1 protein was indeed expressed in this study and was recruited to IGFBP1 upstream region under our E2 and MPA treatment.
To further confirm the FOXO1 effects against 1.8 kbp of the IL15 upstream region, we performed a reporter assay. HAND2 significantly increased the luciferase activities even in the 1.8-kbp IL15 upstream region in these new ESCs obtained from a distinct patient; however, there is no reactivity found against FOXO1 (Fig. S6). We also examined the possibility of FOXO1 functioning as a repressor in the IL15 upstream region but found no effect of FOXO1 as the variants in the FOXO1 motif had no reactivity against FOXO1 (Fig. S6). Unlike HAND2, FOXO1 is unlikely to be involved in the transcriptional modulation of the 1.8-kbp upstream region of IL15 following E2 and MPA stimulation.
Discussion
This report is the first to clearly demonstrate that HAND2 is a transcription factor that directly regulates IL15 expression in human ESCs. We have identified a CCTCTGG sequence in the proximal region of the human IL15 gene as a putative HAND2 motif using ChIP-PCR analysis and a luciferase reporter assay, using a human IL15 upstream region and its mutants in ESCs. Our results suggest a functional connection between HAND2 and this putative HAND2 motif (CCTCTGG) in regulating human IL15 transcription. We did not find any functional association between FOXO1 and IL15 expression as reported previously (25).
VISTA analysis showed no significant similar region between rodents and primates with a criterion of 70% per 100 nucleotides. Unlike the human endometrium, rodent endometrium does not induce thickening of the stratum functionalis without embryo implantation. In actuality, decidualization of the endometrium is under maternal control in a handful of species, which includes higher primates (humans, apes, and Old World monkeys), some bats, and the elephant shrew (5). Therefore, the upstream regions are not conserved between rodents and primates, and whole transcriptional regulation of IL15 might be distinct among them. However, because ClustalW multiple alignment indicated that the HAND2 motif in the upstream region of the IL15 gene is well-conserved (Fig. 3C), the relationship between HAND2 and IL15 transcription in ESCs might be conserved even in rodents.
A chimeric motif is supposed to have been generated by connecting forward and backward sequences around the putative HAND2 motif in the deletion mutant. Further, the deletion mutant altered positional relations of all known and unknown motifs in the upstream region of the IL15 gene. For instance, we previously found that nuclear receptor subfamily 6, group A, member 1 (NR6A1), a DNA-binding factor, down-regulates hypocretin expression through an 18-bp nuclear receptor response element (NurRE) in the upstream region of the hypocretin gene (27). We then cloned a single copy or three copies of NurRE into the upstream region of the TATA box in pTAL-luc reporter plasmid to investigate whether the NurRE exerted transcriptional modulator activity (enhancer, silencer, or insulator). In the condition with the NR6A1 expression vector, three copies were surprisingly activated, although a single copy was repressed. This switching mode might be induced by a generated chimeric binding site connecting each NurRE to other motifs or the change with proximity of NurRE to the promoter. Therefore, we have examined the activities of not only a deletion mutant, but also a substitution mutant, to confirm that the putative HAND2 motif in the human IL15 transcription functions appropriately in ESCs. CTG changed to GAC at −1623/−1625 (purine base to pyrimidine base or pyrimidine base to purine base), but the length from the transcription start site and locations of the other putative motifs might not differ in this substitution mutant. Among these examinations, both mutants showed no response against HAND2. Therefore, HAND2 could regulate IL15 transcription via the putative HAND2 motif at −1628/−1622 in the upstream region of the IL15 gene in ESCs.
A recent single-cell analysis reported that a subclass of ESCs express IL15, ruling out IL15 expression by all ESCs (28). Therefore, two possibilities exist—either HAND2 transcription is different in each ESC, or HAND2 is expressed at a similar level in all ESCs, but the susceptibility (epigenetic alterations, such as methylation) of the IL15 locus against HAND2 is different for each ESC. Our immunohistochemical results show that HAND2 is not expressed in all ESCs; therefore, HAND2 may be regulated by different mechanisms in each ESC. Interestingly, lack of expression of IL15 might indicate no requirement of uNK around them, suggesting that a local prevalence of uNK cells could trigger IL15 expression. HAVCR2 (also known as TIM3)-LGALS9, a factor associated with immune checkpoint, influences the uNK-ESC cell interaction (28). HAVCR2, which is expressed on the surface of uNK cells, might affect ESCs through interacting with LGALS9 and various receptors on the surface of ESCs as seen in macrophage activation by TH1 cells by HAVCR2-LGALS9 interactions (29). We speculate that this interaction might affect IL15 transcription directly or indirectly via HAND2 in ESCs. Therefore, in future studies, it is necessary to perform experiments in which the HAVCR2 peptide is supplemented to culture medium or using a HAVCR2-coated culture dish to investigate the response of ESC, HAND2, and IL15 following E2 and MPA treatment.
We found the putative HAND2 motif, like the E-box motif, at position IVS1_16239/16245 in intron 1 of the IL15 locus. Introns within 1 kbp from the transcription start site have been shown to affect gene expression as IME (20, 30). The IMEter algorithm showed that many introns stimulate gene expression near the 5′-end of a gene (21, 31–34). However, our ChIP-PCR showed no binding of HAND2 at IVS1_16239/16245. Thus, the putative HAND2 motif at IVS1_16239/16245 found in an intron of the IL15 locus might not play roles in IME in ESCs.
Intriguingly, it is well-known that HAND2 up-regulates the T-box family of transcription factors, such as Tbx2 and Tbx3, in the early limb bud mesenchyme (22). Because the IL15 receptor is known to be regulated by T-box transcription factors (23), we speculated that HAND2 might regulate IL15 responsiveness from both ligand and receptor via T-box families.
As a limitation of this study, primary human ESCs cannot be maintained over prolonged periods in cell culture while retaining the primary ESC phenotype. Therefore, CRISPR-Cas9 editing (24) of primary human ESCs has been challenging for researchers, as it is difficult to generate genome-edited cell lines with primary ESC phenotypes and evaluate whether identified results are based on primary ESCs or other differentiated cell types. Alternatively, immortalized human ESCs have been generated via oncogenic transformation (35) or prolongation of cell division by introducing human telomerase reverse transcriptase (36–39). These cell lines are very appealing, but the decidualization capacity and response to steroids have not been fully reported. Recently, a novel immortalized human ESC line, KC02-44D, has been established. This cell line is able to fulfill characteristics, such as decidualization capacity, similar to primary human ESCs (40). Therefore, the KC02-44D cell line would provide a breakthrough in this research area.
In conclusion, we found that human IL15 transcription is directly regulated by HAND2 via a putative HAND2 motif in the upstream region of the human IL15 gene. Because up-regulation in HAND2 stimulation is relatively lower than that with E2 and MPA stimulation, HAND2 might regulate IL15 transcription in collaboration with other factors.
Experimental procedures
Ethics statement
This study was approved by the institutional review board of Kansai Medical University, and informed consent was obtained from each patient (protocol number 2006101). This study also abides by the Declaration of Helsinki principles.
Tissue collection
Human endometrial tissues were collected from 30 patients of 32–48 years of age with regular menstrual cycles. They underwent hysterectomies without preoperative hormonal therapy. Histological examination of the uteri showed benign tumors, such as myoma. ESCs were immediately purified from endometrial tissues as described below.
qPCR
Total RNA was isolated from each human endometrium (8 from proliferation phase, 12 from secretory phase) using the RNeasy Mini kit (Qiagen, Venlo, The Netherlands). Single-strand cDNA was synthesized with a ReverTra Ace qPCR RT master mix with gDNA remover (TOYOBO, Osaka, Japan). The mRNA expression levels were determined using qPCR on a Roter-Gene Q platform (Qiagen) using Thunderbird qPCR mix (TOYOBO) and the gene-specific primers listed in Table S1. Relative target gene expression levels were evaluated by the 2−ΔΔCt method (41) using EF1a as an internal control (13).
Immunohistochemical staining for HAND2
The formalin-fixed paraffin-embedded tissues were cut into 3-µm-thick sections. The sections were then deparaffinized, rehydrated, and boiled for 10 min in antigen retrieval buffer (10 mm sodium citrate, pH 6.0). After cooling for 25 min at room temperature, the tissues were incubated with 3% hydrogen peroxidase for 10 min to inactivate endogenous peroxidases. After washing with distilled water, the sections were incubated with goat dHAND antibody (M-19) (catalog no. sc-9409, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) (17) (1:500 in TBS, 0.1% Tween 20; sc-9409) overnight at 4 °C and were then incubated with N-Histofine Simple Stain MAX PO (G) (Nichirei Bioscience Inc., Tokyo, Japan) for 30 min according to the manufacturer's protocol. After staining with DAB chromogen for 10 min, the sections were counterstained with hematoxylin. Bright-field images were captured with an Eclipse E-1000M digital camera (Nikon, Tokyo, Japan).
RNAscope for IL15 and HAND2
RNA in situ hybridization for IL15 and HAND2 was performed using an RNAscope 2.5 HD Reagent Kit-BROWN or RNAscope 2-plex Reagent Kit (Advanced Cell Diagnostics, Inc., Newark, CA, USA). Briefly, 5-μm-thick formalin-fixed paraffin-embedded sections were mounted onto Superfrost Plus microscope slides (Thermo Fisher Scientific, Waltham, MA, USA), baked, and deparaffinized. After incubation with hydrogen peroxidase, the sections were boiled in the Target Retrieval Reagents solution for 15 min and were treated with Protease Plus. They were then hybridized with four RNAscope probes, namely RNAscope Probe-Hs-IL15-C1, RNAscope Probe-Hs-HAND2-C2, RNAscope Positive Control Probe-Hs-PPIB, and RNAscope Negative Control Probe-DapB (Advanced Cell Diagnostics). A sequential signal amplification was performed with six or 10 serial amplifications. After the final amplification, DAB, FastRed, or FastGreen chromogenic detection was performed. They were counterstained with 50% Gill's hematoxylin. Bright-field images were captured with an Eclipse E-1000M digital camera.
Culture of human ESCs
The standard enzyme digestion method described previously was used to purify human ESCs. Purified ESCs were cultured in phenol red–free Dulbecco's modified Eagle's medium/F-12 (Thermo Fisher Scientific) containing 10% dextran-coated charcoal-stripped fetal calf serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mmol/liter GlutaMAX (Thermo Fisher Scientific), to remove the effect of endogenous steroid hormones, at 37 °C under a humidified atmosphere of 5% CO2 in air. Culture medium was changed every 3 days. ESCs were used for each experiment when the cells became nearly confluent. Vimentin-positive cells in confluent ESCs were detected at a rate of >99% by immunohistochemical analysis, as described previously (42).
Expression vectors and reporter plasmids
The firefly luciferase–encoding reporter plasmids pGL4.10[luc2] and pGL4.74[hRluc/TK] were purchased from Promega (Madison, WI, USA). The pGL4.74[hRluc/TK], which encodes Renilla luciferase, was used as an internal control for transfection efficiency. The 1,867-bp upstream region of the human IL15 transcription start site was amplified with KOD Fx, hIL15_−1867F_KpnI (5′-CGGGGTACCtgccttaagttcaccctacaagt-3′; the KpnI site is underlined) and hIL15_+1R_XhoI (5′-CCGCTCGAGcccctggcgaaaagaaaagtg-3′; the XhoI site is underlined). The PCR products were cloned into pGL4.10[luc2] (IL15ups/pGL4.10). The deletion mutant ΔH2_motif/pGL4.10, which contains an internal deletion at −1628/−1622 within the IL15ups/pGL4.10, was generated using site-directed mutagenesis and hIL15_−1629R_P and hIL15_−1621F_forDEL_P phosphorylated primers (Table S1). The deletion mutant ΔF1_motif/pGL4.10, which contains an internal deletion at −1131/−1137 within the IL15ups/pGL4.10, was generated using site-directed mutagenesis and hIL15_−1138R+P and hIL15_−1130FforDEL+P phosphorylated primers (Table S1). Nucleotide substitution reporter CTGtoGAC/pGL4.10 at −1623/−1625 was generated using site-directed mutagenesis with hIL15_−1629R_P and hIL15_−1628F (CTG>GAC)_P phosphorylated primers, and F1_substitution/pGL4.10 at −1132/−1137 was generated using site-directed mutagenesis with hIL15_−1138R+P and hIL15_−1137FforSubsti+P (Table S1). Substituted nucleotides are indicated as red characters in Table S1.
The expression vector, pIRES2-AcGFP1, was purchased from Takara Bio USA, Inc. (Mountain View, CA, USA). Human HAND2 open reading frame was subcloned into pIRES2-AcGFP1 at EcoRI and BamHI sites (HAND2/pIRES2). Human FOXO1 cDNA was subcloned into pcDNA3-FLAG vector (FOXO1/pcDNA3).
All constructs were confirmed to have no substitutional mutation, no insertion, and no deletion by sequencing analysis except for mutagenesis regions with a BigDye terminator cycle sequencing reaction kit (Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Both strands were read with sequence primers.
ChIP-PCR
ChIP-PCR was conducted according to a method reported previously (27, 43). Briefly, ESCs were treated with E2 (10−8 mol/liter) and MPA (10−7 mol/liter) for 12 days. ESCs were then cross-linked with 1% formaldehyde for 10 min at room temperature; a final concentration of 0.125 m glycine was added to block further cross-linking. Tissues were washed twice with ice-cold PBS, and a crude nuclear extract was prepared. To obtain digested genomic DNA, ranging between 150 and 900 bp, the nuclear extract was incubated with micrococcal nuclease (Cell Signaling Technology, Inc., Tokyo, Japan) for 20 min at 37 °C. The size of the DNA fragments was confirmed by gel electrophoresis following treatment with RNase I for 30 min at 37 °C and Proteinase K for 2 h at 65 °C. The lysate was sonicated at 43 kHz 10 times (20 s/sonication) at 4 °C and centrifuged, and the obtained supernatant was diluted in ChIP buffer (Cell Signaling Technology) overnight at 4 °C on a wheel rotator with either 2 μg of goat dHAND antibody (M-19) (catalog no. sc-9409, Santa Cruz Biotechnology, Inc.) (17) to capture the HAND2 protein, 1:100 FoxO1 (C29H4) rabbit mAb (catalog no. 2880, Cell Signaling Technology) to capture the FOXO1 protein, 1:50 histone H3 (D2B12) XP® rabbit mAb (catalog no. 4620, Cell Signaling Technology) as a positive control, or 2 μg of normal rabbit IgG (catalog no. 2729, Cell Signaling Technology) as a negative control. Samples were then incubated with Protein G–agarose beads for 2 h at 4 °C. Beads were washed with low- and high-salt wash buffers, and DNA templates for PCR were purified from the resulting DNA-protein complexes. ChIP-PCR analysis was performed using KOD plus DNA polymerase (Toyobo Life Sciences), primers (Table S1), and a thermal cycler, with the following settings: one cycle at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 68 °C for 30 s. Further, ChIP-qPCR was used to determine the relative levels of transcription factor recruitments, and the ratio of IP DNA to the 2% input DNA sample (%INPUT) was calculated.
Luciferase reporter assay
Human ESCs were seeded into 24-well cell culture plates at a density of 2.5 × 105/well. Two types of luciferase plasmids were co-transfected with LipofectamineTM 3000 transfection reagent (Thermo Fisher Scientific) after pretreatment with E2 (10−8 mol/liter) and MPA (10−7 mol/liter) the day before, or two types of luciferase plasmids and one expression vector were co-transfected according to the manufacturer's protocol. The following amounts of co-transfected plasmids and vectors were placed in each well: 200 ng of firefly luciferase–encoding reporter plasmid (pGL4.10, IL15ups/pGL4.10 or IL15ups_mutants/pGL4.10), 20 ng of Renilla luciferase-encoding internal control plasmid (pGL4.74), and 200 ng of expression vectors (pIRES2-AcGFP1, HAND2/pIRES2, pcDNA3-FLAG, or FOXO1/pcDNA3). Approximately 48 h after transfection, ESCs were lysed with lysis buffer (100 µl/well), and 20 µl of cell lysate was transferred to an OptiPlate-96 96-well microplate (SUMILON, Tokyo, Japan). Thereafter, firefly luciferase luminescence from the pGL4.10 plasmids and Renilla luciferase luminescence from the pGL4.74 plasmids were sequentially measured in duplicate using a PicaGene dual sea pansy luminescence kit (TOYO INK CO. Ltd., Chuo-ku, Tokyo, Japan) and the 2030 ARVO X multilabel reader (PerkinElmer Japan Co. Ltd., Yokohama, Japan) according to the manufacturer's protocol. Relative luciferase activity per well was calculated by dividing firefly luciferase luminescence by Renilla luciferase luminescence. Relative luciferase activity was standardized using the corresponding control condition, which was transfected with pGL4.10 plasmid alone or co-transfected with a pGL4.10 plasmid and a mock vector (pIRES2-AcGFP1 or FOXO1/pcDNA3). Activity levels were expressed as the mean of at least six independent experiments ± S.D. and S.E.
Immunoblot analysis
HAND2 and FOXO1 proteins were confirmed in ESCs using immunoblot analysis after transfection. Total soluble proteins were extracted from total cell lysates in lysis buffer containing mammalian protein extraction reagent (Thermo Fisher Scientific) and protease inhibitor mixture (Nacalai Tesque, Osaka, Japan). Protein samples, heated for 5 min in SDS sample buffer, were loaded onto 7.5% Mini-PROTEAN TGX precast gel (Bio-Rad). They were then electrotransferred to immunoblot polyvinylidene difluoride membranes (Bio-Rad), and blocked with Blocking One (Nacalai Tesque) for 1 h and then incubated with goat dHAND antibody (M-19) (1:200), FoxO1 rabbit mAb (C29H4) (1:1,000), or mouse monoclonal β-actin antibody (1:5,000; AC-74, Sigma–Aldrich, Tokyo, Japan) in TBS, 0.1% Tween 20, containing 5% Blocking One at 4 °C overnight. Donkey anti-goat IgG peroxidase–linked antibody (1:5,000; sc-2020, Santa Cruz Biotechnology, Inc.), goat peroxidase-labeled anti-rabbit IgG (H + L) (1:5,000; PI-1000, Vector Laboratories), or sheep anti-mouse IgG peroxidase–linked antibody (1:10,000; NA931, GE Healthcare) were respectively used as secondary antibodies. Immune complexes were visualized using ECL Prime Western blotting detection reagent (GE Healthcare). The bands were detected using LAS 4000 (GE Healthcare). After FOXO1 transfection, we also detected FOXO1 proteins among the immunoprecipitated proteins by using FoxO1 rabbit mAb (C29H4) and immunoprecipitation kit (Protein G) (Sigma–Aldrich) according to the manufacturer's instructions.
Statistical analyses
Normal distribution was determined using the Shapiro–Wilk normality test. Comparisons between HAND2 and IL15 expression in the proliferative and secretory phase were performed using Student's t test. Correlation between HAND2 and IL15 expression was analyzed using Spearman's rank correlation coefficient. Certain groups in the luciferase reporter assay were not normally distributed. Therefore, we employed an ANOVA with Tukey's multiple-comparison test for normal distribution or a nonparametric Steel–Dwass multiple-comparison test for nonnormal distribution. All values were two-sided with statistical significance set at 0.05. Statistical analyses were performed using IBM SPSS Statistics version 21.0 (IBM Corp., Armonk, NY, USA).
Data availability
The data sets and images used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We are grateful to our colleagues in the Department of Obstetrics and Gynecology, Kansai Medical University for patient recruitment.
This article contains supporting information.
Author contributions—H. M., S. T., and H. O. conceptualization; H. M., S. T., T. T.-N., T. K., M. K.-K., N. K., Y. Hisamatsu, H. T., Y. Hashimoto, and H. O. resources; H. M. and S. T. data curation; H. M. and S. T. formal analysis; H. M., S. T., and H. O. validation; H. M. and S. T. investigation; H. M. and S. T. visualization; H. M. and S. T. methodology; H. M. and S. T. writing-original draft; H. M., S. T., and H. O. project administration; S. T. software; S. T. and H. O. funding acquisition; S. T., T. T.-N., T. K., M. K.-K., N. K., Y. Hisamatsu, H. T., Y. Hashimoto, M. K., and H. O. writing-review and editing; M. K. and H. O. supervision.
Funding and additional information—This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 17K11260 (to H. O.) and 19K06891 (to S. T.) and the Takeda Science Foundation (to S. T.). The agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- E2
- estradiol
- P4
- progesterone
- ESC
- endometrial stromal cell
- uNK
- uterine natural killer
- IL15
- interleukin 15
- HAND2
- heart– and neural crest derivatives–expressed transcript 2
- MPA
- medroxyprogesterone
- qPCR
- quantitative PCR
- IVS
- intervening sequence
- IME
- intron-mediated enhancement
- NurRE
- nuclear receptor response element
- ANOVA
- analysis of variance.
References
- 1. Cha J., Sun X., and Dey S. K. (2012) Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 18, 1754–1767 10.1038/nm.3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans J., Salamonsen L. A., Winship A., Menkhorst E., Nie G., Gargett C. E., and Dimitriadis E. (2016) Fertile ground: human endometrial programming and lessons in health and disease. Nat. Rev. Endocrinol. 12, 654–667 10.1038/nrendo.2016.116 [DOI] [PubMed] [Google Scholar]
- 3. Fox C., Morin S., Jeong J. W., Scott R. T. Jr., and Lessey B. A. (2016) Local and systemic factors and implantation: what is the evidence? Fertil. Steril. 105, 873–884 10.1016/j.fertnstert.2016.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel B., Elguero S., Thakore S., Dahoud W., Bedaiwy M., and Mesiano S. (2015) Role of nuclear progesterone receptor isoforms in uterine pathophysiology. Hum. Reprod. Update 21, 155–173 10.1093/humupd/dmu056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gellersen B., and Brosens J. J. (2014) Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 35, 851–905 10.1210/er.2014-1045 [DOI] [PubMed] [Google Scholar]
- 6. Nagler A., Lanier L. L., Cwirla S., and Phillips J. H. (1989) Comparative studies of human FcRIII-positive and negative natural killer cells. J. Immunol. 143, 3183–3191 [PubMed] [Google Scholar]
- 7. King A., and Loke Y. W. (1991) On the nature and function of human uterine granular lymphocytes. Immunol. Today 12, 432–435 10.1016/0167-5699(91)90014-K [DOI] [PubMed] [Google Scholar]
- 8. Jokhi P. P., King A., Sharkey A. M., Smith S. K., and Loke Y. W. (1994) Screening for cytokine messenger ribonucleic acids in purified human decidual lymphocyte populations by the reverse-transcriptase polymerase chain reaction. J. Immunol. 153, 4427–4435 [PubMed] [Google Scholar]
- 9. Dosiou C., and Giudice L. C. (2005) Natural killer cells in pregnancy and recurrent pregnancy loss: endocrine and immunologic perspectives. Endocr. Rev. 26, 44–62 10.1210/er.2003-0021 [DOI] [PubMed] [Google Scholar]
- 10. Kalkunte S. S., Mselle T. F., Norris W. E., Wira C. R., Sentman C. L., and Sharma S. (2009) Vascular endothelial growth factor C facilitates immune tolerance and endovascular activity of human uterine NK cells at the maternal-fetal interface. J. Immunol. 182, 4085–4092 10.4049/jimmunol.0803769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henderson T. A., Saunders P. T., Moffett-King A., Groome N. P., and Critchley H. O. (2003) Steroid receptor expression in uterine natural killer cells. J. Clin. Endocrinol. Metab. 88, 440–449 10.1210/jc.2002-021174 [DOI] [PubMed] [Google Scholar]
- 12. Kitaya K., Yamaguchi T., and Honjo H. (2005) Central role of interleukin-15 in postovulatory recruitment of peripheral blood CD16(−) natural killer cells into human endometrium. J. Clin. Endocrinol. Metab. 90, 2932–2940 10.1210/jc.2004-2447 [DOI] [PubMed] [Google Scholar]
- 13. Cho H., Okada H., Tsuzuki T., Nishigaki A., Yasuda K., and Kanzaki H. (2013) Progestin-induced heart and neural crest derivatives expressed transcript 2 is associated with fibulin-1 expression in human endometrial stromal cells. Fertil. Steril. 99, 248–255 10.1016/j.fertnstert.2012.08.056 [DOI] [PubMed] [Google Scholar]
- 14. Srivastava D., Thomas T., Lin Q., Kirby M. L., Brown D., and Olson E. N. (1997) Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 16, 154–160 10.1038/ng0697-154 [DOI] [PubMed] [Google Scholar]
- 15. Yamagishi H., Olson E. N., and Srivastava D. (2000) The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J. Clin. Invest. 105, 261–270 10.1172/JCI8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shindoh H., Okada H., Tsuzuki T., Nishigaki A., and Kanzaki H. (2014) Requirement of heart and neural crest derivatives-expressed transcript 2 during decidualization of human endometrial stromal cells in vitro. Fertil. Steril. 101, 1781–1790.e1-5 10.1016/j.fertnstert.2014.03.013 [DOI] [PubMed] [Google Scholar]
- 17. Boeva V., Louis-Brennetot C., Peltier A., Durand S., Pierre-Eugène C., Raynal V., Etchevers H. C., Thomas S., Lermine A., Daudigeos-Dubus E., Geoerger B., Orth M. F., Grünewald T. G. P., Diaz E., Ducos B., et al. (2017) Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413 10.1038/ng.3921 [DOI] [PubMed] [Google Scholar]
- 18. Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S., and Dubchak I. (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- 19. Frazer K. A., Pachter L., Poliakov A., Rubin E. M., and Dubchak I. (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mascarenhas D., Mettler I. J., Pierce D. A., and Lowe H. W. (1990) Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol. Biol. 15, 913–920 10.1007/BF00039430 [DOI] [PubMed] [Google Scholar]
- 21. Rose A. B., Elfersi T., Parra G., and Korf I. (2008) Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20, 543–551 10.1105/tpc.107.057190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osterwalder M., Speziale D., Shoukry M., Mohan R., Ivanek R., Kohler M., Beisel C., Wen X., Scales S. J., Christoffels V. M., Visel A., Lopez-Rios J., and Zeller R. (2014) HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev. Cell 31, 345–357 10.1016/j.devcel.2014.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mackay L. K., Wynne-Jones E., Freestone D., Pellicci D. G., Mielke L. A., Newman D. M., Braun A., Masson F., Kallies A., Belz G. T., and Carbone F. R. (2015) T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 10.1016/j.immuni.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 24. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasquez Y. M., Mazur E. C., Li X., Kommagani R., Jiang L., Chen R., Lanz R. B., Kovanci E., Gibbons W. E., and DeMayo F. J. (2015) FOXO1 is required for binding of PR on IRF4, novel transcriptional regulator of endometrial stromal decidualization. Mol. Endocrinol. 29, 421–433 10.1210/me.2014-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura I., Jozaki K., Sato S., Shirafuta Y., Shinagawa M., Maekawa R., Taketani T., Asada H., Tamura H., and Sugino N. (2018) The distal upstream region of insulin-like growth factor-binding protein-1 enhances its expression in endometrial stromal cells during decidualization. J. Biol. Chem. 293, 5270–5280 10.1074/jbc.RA117.000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanaka S., Kodama T., Nonaka T., Toyoda H., Arai M., Fukazawa M., Honda Y., Honda M., and Mignot E. (2010) Transcriptional regulation of the hypocretin/orexin gene by NR6A1.) Biochem. Biophys. Res. Commun. 403, 178–183 10.1016/j.bbrc.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vento-Tormo R., Efremova M., Botting R. A., Turco M. Y., Vento-Tormo M., Meyer K. B., Park J.-E., Stephenson E., Polański K., Goncalves A., Gardner L., Holmqvist S., Henriksson J., Zou A., Sharkey A. M., et al. (2018) Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jayaraman P., Sada-Ovalle I., Beladi S., Anderson A. C., Dardalhon V., Hotta C., Kuchroo V. K., and Behar S. M. (2010) Tim3 binding to galectin-9 stimulates antimicrobial immunity. J. Exp. Med. 207, 2343–2354 10.1084/jem.20100687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rose A. B. (2008) Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 326, 277–290 10.1007/978-3-540-76776-3_15 [DOI] [PubMed] [Google Scholar]
- 31. Rose A. B. (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 40, 744–751 10.1111/j.1365-313X.2004.02247.x [DOI] [PubMed] [Google Scholar]
- 32. Callis J., Fromm M., and Walbot V. (1987) Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200 10.1101/gad.1.10.1183 [DOI] [PubMed] [Google Scholar]
- 33. Jeong Y. M., Mun J. H., Lee I., Woo J. C., Hong C. B., and Kim S. G. (2006) Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 140, 196–209 10.1104/pp.105.071316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Snowden K. C., Buchhholz W. G., and Hall T. C. (1996) Intron position affects expression from the tpi promoter in rice. Plant Mol. Biol. 31, 689–692 10.1007/BF00042241 [DOI] [PubMed] [Google Scholar]
- 35. Chapdelaine P., Kang J., Boucher-Kovalik S., Caron N., Tremblay J. P., and Fortier M. A. (2006) Decidualization and maintenance of a functional prostaglandin system in human endometrial cell lines following transformation with SV40 large T antigen. Mol. Hum. Reprod. 12, 309–319 10.1093/molehr/gal034 [DOI] [PubMed] [Google Scholar]
- 36. Barbier C. S., Becker K. A., Troester M. A., and Kaufman D. G. (2005) Expression of exogenous human telomerase in cultures of endometrial stromal cells does not alter their hormone responsiveness. Biol. Reprod. 73, 106–114 10.1095/biolreprod.104.035063 [DOI] [PubMed] [Google Scholar]
- 37. Krikun G., Mor G., Alvero A., Guller S., Schatz F., Sapi E., Rahman M., Caze R., Qumsiyeh M., and Lockwood C. J. (2004) A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology 145, 2291–2296 10.1210/en.2003-1606 [DOI] [PubMed] [Google Scholar]
- 38. Samalecos A., Reimann K., Wittmann S., Schulte H. M., Brosens J. J., Bamberger A. M., and Gellersen B. (2009) Characterization of a novel telomerase-immortalized human endometrial stromal cell line, St-T1b. Reprod. Biol. Endocrinol. 7, 76 10.1186/1477-7827-7-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura K., Yoshie M., Hara T., Isaka K., and Kogo H. (2007) Involvement of stathmin in proliferation and differentiation of immortalized human endometrial stromal cells. J. Reprod. Dev. 53, 525–533 10.1262/jrd.18129 [DOI] [PubMed] [Google Scholar]
- 40. Yuhki M., Kajitani T., Mizuno T., Aoki Y., and Maruyama T. (2011) Establishment of an immortalized human endometrial stromal cell line with functional responses to ovarian stimuli. Reprod. Biol. Endocrinol. 9, 104 10.1186/1477-7827-9-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 42. Okada H., Nakajima T., Yoshimura T., Yasuda K., and Kanzaki H. (2001) The inhibitory effect of dienogest, a synthetic steroid, on the growth of human endometrial stromal cells in vitro. Mol. Hum. Reprod. 7, 341–347 10.1093/molehr/7.4.341 [DOI] [PubMed] [Google Scholar]
- 43. Tanaka S., Honda Y., Takaku S., Koike T., Oe S., Hirahara Y., Yoshida T., Takizawa N., Takamori Y., Kurokawa K., Kodama T., and Yamada H. (2019) Involvement of PLAGL1/ZAC1 in hypocretin/orexin transcription. Int. J. Mol. Med. 43, 2164–2176 10.3892/ijmm.2019.4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets and images used and/or analyzed during the current study are available from the corresponding author on reasonable request.