Abstract
Human leukocyte immunoglobulin-like receptors (LILRs) typically regulate immune activation by binding to the human leukocyte antigen class I molecules. LILRA2, a member of the LILR family, was recently reported to bind to other unique ligands, the bacterially degraded Igs (N-truncated Igs), for the activation of immune cells. Therefore, LILRA2 is currently attracting significant attention as a novel innate immune receptor. However, the detailed recognition mechanisms required for this interaction remain unclear. In this study, using several biophysical techniques, we uncovered the molecular mechanism of N-truncated Ig recognition by LILRA2. Surface plasmon resonance analysis disclosed that LILRA2 specifically binds to N-truncated Ig with weak affinity (Kd = 4.8 μm) and fast kinetics. However, immobilized LILRA2 exhibited a significantly enhanced interaction with N-truncated Ig due to avidity effects. This suggests that cell surface-bound LILRA2 rapidly monitors and identifies bi- or multivalent abnormal N-truncated Igs through specific cross-linking to induce immune activation. Van't Hoff analysis revealed that this interaction is enthalpy-driven, with a small entropy loss, and results from differential scanning calorimetry indicated the instability of the putative LILRA2-binding site, the Fab region of the N-truncated Ig. Atomic force microscopy revealed that N truncation does not cause significant structural changes in Ig. Furthermore, mutagenesis analysis identified the hydrophobic region of LILRA2 domain 2 as the N-truncated Ig-binding site, representing a novel ligand-binding site for the LILR family. These results provide detailed insights into the molecular regulation of LILR-mediated immune responses targeting ligands that have been modified by bacteria.
Keywords: cell surface receptor, immunoglobulin G (IgG), protein-protein interaction, surface plasmon resonance (SPR), atomic force microscopy (AFM), immune regulatory receptor, bacterially cleaved immunoglobulin, immune regulation, leukocyte immunoglobulin-like receptor (LILR), LILRA2, immunology
Leukocyte immunoglobulin (Ig)-like receptors (LILRs or LIRs), also known as Ig-like transcripts (ILTs) or the CD85 family, are expressed on the surface of immune cells, such as monocytes, macrophages, and dendritic cells (1). LILRBs (LILR subfamily B) are considered immune checkpoint molecules, like PD-1 and CTLA-4 (2, 3). LILRs belong to a paired receptor family that has homologous extracellular domains but opposite intracellular signaling motifs. The extracellular domains of most LILRs are composed of four Ig-like domains (D1, D2, D3, and D4). LILRBs (LILRB1, -B2, -B3, -B4, and -B5) have an immunoreceptor tyrosine-based inhibitory motif (ITIM). For example, LILRB1 and -B2 on immune cells bind to the human leukocyte antigen (HLA) on target cells to inhibit cellular activation (4–7). In contrast, LILRAs (LILRA1, -A2, -A4, -A5, and -A6) have a positively charged Arg residue in the transmembrane domain, which facilitates coupling with molecules harboring the immunoreceptor tyrosine-based activation motif (ITAM), such as the FcRγ chain. The activating receptor LILRA1 binds to HLA-B27, which is a well-known factor for diseases like ankylosing spondylitis (8). LILRA1 and LILRA3 preferentially bind to β2 microglobulin-free HLA-C, which is associated with symptoms of HIV infection (9). The stimulation of immune cells, such as monocytes, macrophages, eosinophils, and basophils, by anti-LILRA2 leads to the secretion of inflammation cytokines (10–13). Recently, Hirayasu et al. reported that LILRA2 ligands are cleaved Igs whose VH domain is cleaved by proteases secreted by bacteria, such as Legionella pneumophila, Haemophilus influenzae, Streptococcus pneumoniae, and mycoplasma (14). The cleaved Igs were found in L. pneumophilia-infected mice and pus fluid from S. pneumoniae-infected patients (14). Moreover, Mycoplasma Ig binding protein has been reported to recruit Mycoplasma Ig protease to cleave the heavy chain of Ig (15). Therefore, the cleavage of Igs is considered a highly conserved immune evasion mechanism for bacteria (14). In contrast, Hirayasu et al. also found that LILRA2 ingeniously bound to the cleaved Igs and stimulated the immune system to inhibit bacterial growth. For example, the growth of L. pneumophila was inhibited by stimulating primary monocytes with LILRA2 (14). This is a novel immune activation mechanism for sensing immune evasion by bacteria; however, the detailed molecular recognition mechanism remains unknown.
Here, we report the biophysical characteristics of the interaction between LILRA2 and N-truncated Ig. We purified WT LILRA2 and N-truncated Ig and determined the binding affinity, kinetics, and thermodynamic characteristics of the LILRA2/N-truncated Ig interaction using surface plasmon resonance (SPR). Furthermore, we identified the N-truncated Ig binding site on LILRA2 using mutagenesis studies. We also observed N-truncated Ig and full-length Ig structure by high-speed atomic force microscopy (HS-AFM). These results provided important molecular insights into microbially cleaved Ig recognition by LILRA2 as well as strategies for suitable regulation of host immune systems.
Results
Expression and purification of LILRA2 and N-truncated Ig
The constructs used for the expression of proteins in this study are summarized in Fig. 1, A and B, and Fig. S1A. The N-terminal domains 1 and 2 (D1D2) of the extracellular region of human LILRA2 receptor (residues 1–195, here designated LILRA2D1D2) were expressed in Escherichia coli as inclusion bodies and refolded in vitro by a dilution method (4). The refolded LILRA2D1D2 was purified by size exclusion chromatography and eluted as a monomer based on analysis using calibration standards (Fig. S1B). SDS-PAGE analysis indicated that the protein obtained was highly purified and suitable for further experiments (Fig. S1C). The domains 1 to 4 (D1D4) of the extracellular region of LILRA2 (residues 1–409, here designated LILRA2D1D4) was fused to a His tag and expressed in HEK293S GnTI(−) cells. The recombinant protein was purified by Ni-affinity chromatography. The subsequent size exclusion chromatogram showed that LILRA2D1D4 also eluted as a monomer (Fig. S1, D and E). Previous reports showed an unusual refolding of the LILRA2 mutant (R142C) forming a domain-swapped dimer (16, 17). However, the ectodomain of the WT LILRA2 exists as a monomer. LILRA2 D1D4 fused with the Fc region (residues 1–409 of LILRA2 fused with Fc, here designated Fc fusion LILRA2D1D4) was transiently expressed by HEK293T cells as a secreted protein, as described previously (14). The protein was purified from the culture supernatant by protein G affinity chromatography followed by size exclusion chromatography.
Figure 1.
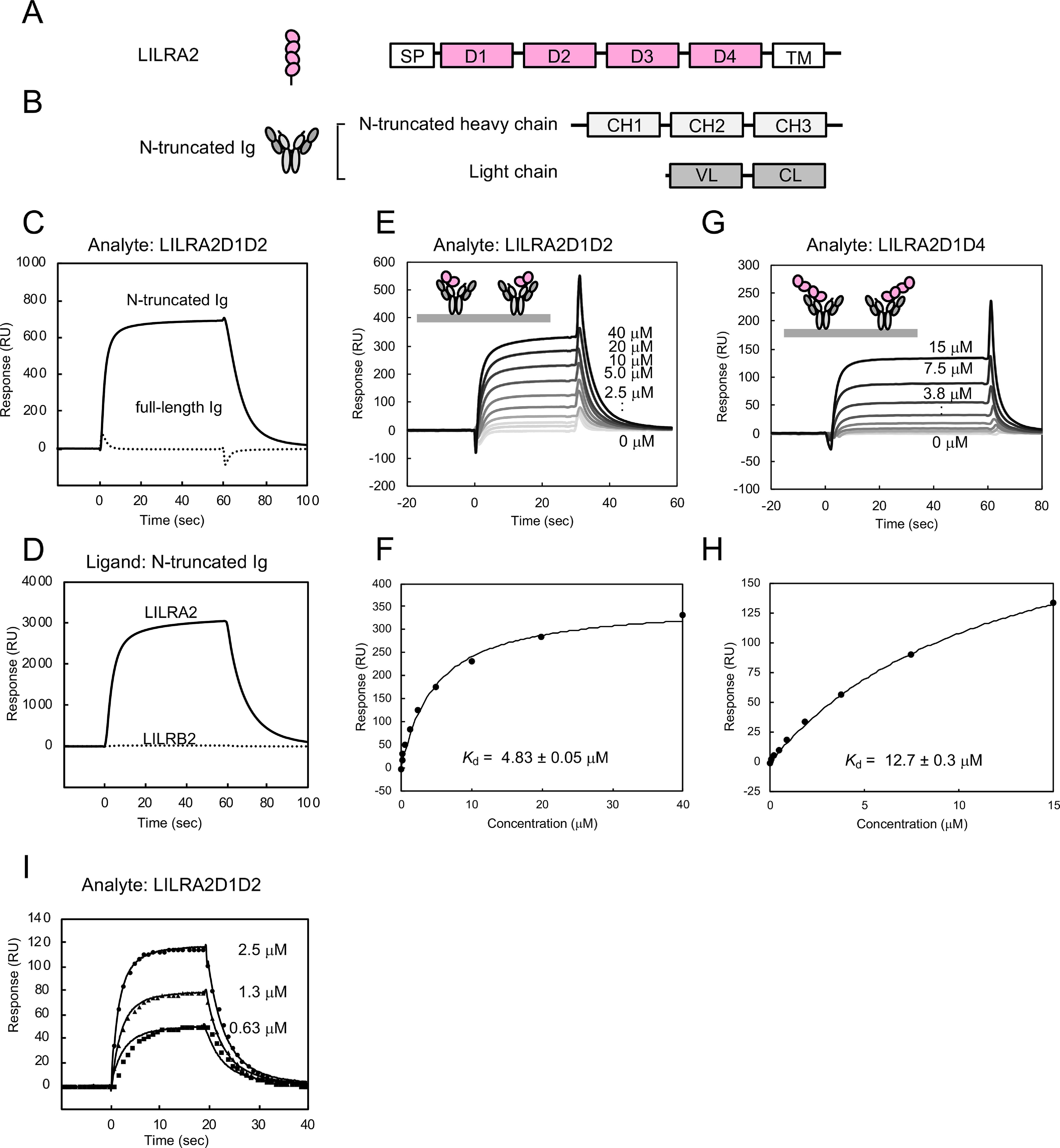
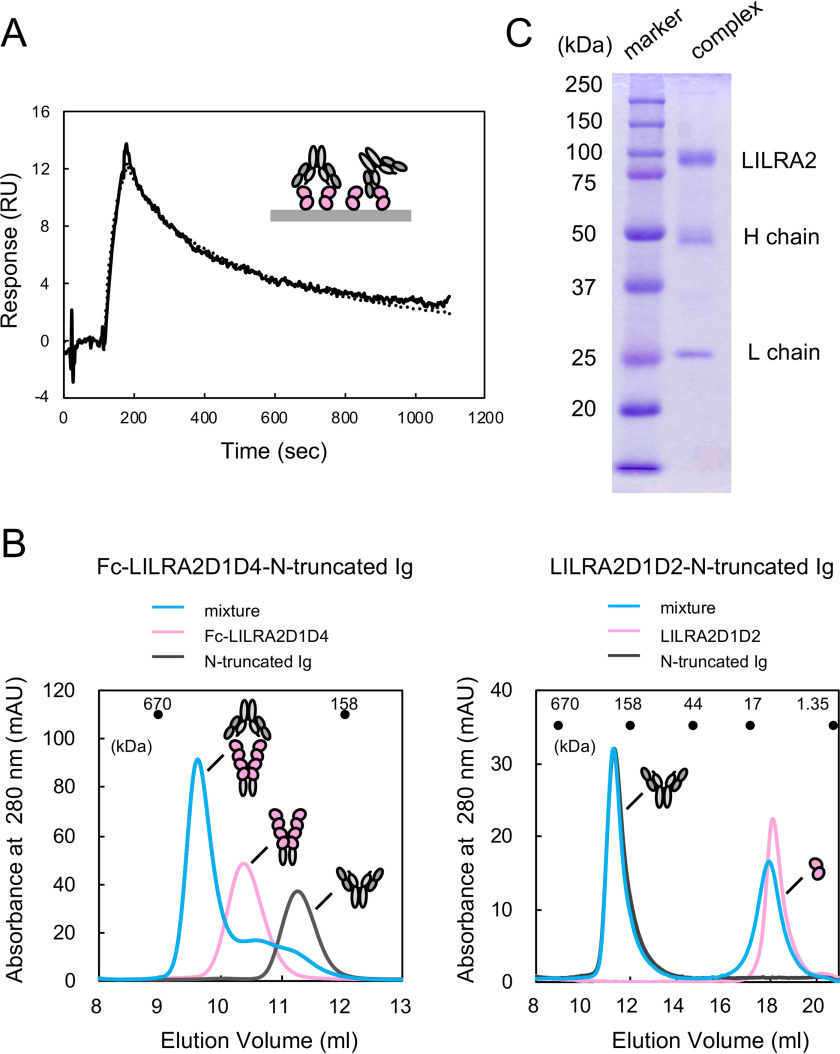
Binding analysis between LILRA2 and N-truncated Ig. A, Schematic structures of LILRA2. SP, signal peptide; D, domain; TM, transmembrane domain. B, Schematic structures of N-truncated Ig. C, The SPR sensorgrams of injection of LILRA2D1D2 over N-truncated Ig (solid lines) or full-length Ig (dotted lines) are shown. D, Interaction analysis of LILRA2D1D2 or LILRB2D1D2 with N-truncated Ig. N-truncated Ig was immobilized on a sensor chip at 20,000 RU. 9 μm LILRA2D1D2 or LILRB2D1D2 was injected as the analyte. E, SPR sensorgram of continuous injection of LILRA2D1D2 at various concentrations, from 0.16 μm to 40 μm, over N-truncated Ig immobilized sensor chip. F, Equilibrium analysis of panel E. The black line represents nonlinear fits of the 1:1 Langmuir binding fitting. Kd value is presented as mean ± standard deviation from three experiments. G, SPR sensorgram of continuous injection of LILRA2D1D4 at various concentrations, from 0.12 μm to 15 μm, over N-truncated Ig immobilized sensor chip. H, Equilibrium analysis of panel G. The black line represents nonlinear fits of the 1:1 Langmuir binding fitting. Kd value is presented as mean ± range from two experiments. I, The kinetic analysis of LILRA2 binding to N-truncated Ig. SPR sensorgram of LILRA2 at various concentrations, from 0.63 μm to 2.5 μm, over N-truncated Ig immobilized sensor chip, is shown. Circle, triangle, and square show the fitting curve based on the global fitting analysis using the 1:1 Langmuir binding model.
Full-length Ig (amino acid residues 1–495 of heavy chain and amino acid residues 1–217 of light chain) and N-truncated Ig (corresponding to amino acid residues 106–495 of the heavy chain of full-length IgG3 and amino acid residues 1–217 of the light chain) were also expressed and purified by the same method as the Fc fusion LILRA2D1D4 described above. The purity of the protein was confirmed by SDS-PAGE analysis (Fig. S1F).
SPR analysis of the LILRA2–N-truncated Ig interaction
SPR analysis was performed to elucidate the molecular recognition mechanism of LILRA2 by N-truncated Ig, using the purified recombinant proteins obtained above. Soluble LILRA2D1D2 protein was injected onto a CM5 sensor chip where N-truncated Ig or full-length Ig (as a negative control) was directly immobilized by the amine coupling method. Specific binding of LILRA2 to N-truncated Ig was observed but not to the full-length Ig (Fig. 1C). LILRB2D1D2 did not show specific binding to N-truncated Ig by SPR analysis, despite having a sequence similarity of almost 79% to LILRA2D1D2 (Fig. 1D). This result was consistent with previous flow cytometric analysis, which indicated that Fc-LILRB2 did not bind to N-truncated Ig, using the coexpression system with CD79α/β on HEK293T cells (14). We injected LILRA2D1D2 (from 0.16 μm to 40 μm) or LILRA2D1D4 (from 0.12 μm to 15 μm) to N-truncated and full-length Igs immobilized on the chip to determine the dissociation constants (Kd) by equilibrium binding analysis (Fig. 1, E–H). The dissociation constants of the interaction between LILRA2D1D2 or LILRA2D1D4 and N-truncated Ig were determined to be 4.83 ± 0.05 μm and 12.7 ± 0.3 μm, respectively. These data indicate that D1-D2 domains, not D3-D4 domains, of LILRA2 play a pivotal role in the N-truncated Ig binding. LILRA2D1D2 (0.63, 1.3, and 2.5 μm) was injected onto an N-truncated Ig immobilized chip to determine the kinetic parameters by global fitting analysis, using a 1:1 Langmuir binding model (Fig. 1I). The association and dissociation rate constants were 1.1 ± 0.0 (105 M−1·s−1) and 0.23 ± 0.00 (s−1), respectively. The Kd derived from the kinetic parameters was 2.0 ± 0.0 μm. Similar Kd values obtained from the kinetic and equilibrium analysis provide further evidence of the accuracy of these kinetic parameters.
Bivalent effect on LILRA2-N-truncated Ig interaction
Antibodies typically have two or more antigen binding sites and, therefore, demonstrate strong binding potential against antigens expressed on the cell surface due to an avidity effect. The N-truncated Ig presumably has two potential receptor binding sites and, thus, may also exhibit the avidity effect on LILRA2 expressed on the cell surface. To examine this avidity effect, N-truncated Ig was injected onto the LILRA2D1D2 immobilized sensor chip. It showed a slower dissociation sensorgram than the opposite orientation (Fig. 2A). Furthermore, to verify the slow dissociation in solution, mixtures of either LILRA2D1D2 (one ligand binding site) or Fc fusion LILRA2D1D4 (two ligand binding sites) and N-truncated Ig were analyzed by size exclusion chromatography. The size exclusion chromatogram of the mixture of LILRA2D1D2 and N-truncated Ig showed separate elution peaks for each component protein, while that of Fc fusion LILRA2D1D4 and N-truncated Ig showed an earlier elution peak, which was not observed for each component protein (Fig. 2B). SDS-PAGE analysis revealed that this peak contained Fc fusion LILRA2D1D4 and N-truncated Ig (Fig. 2C). These data indicated that N-truncated Ig has two LILRA2 binding sites and a significant avidity effect for LILRA2 binding.
Figure 2.
Bivalent effect of the interaction between LILRA2 and N-truncated Ig. A, 47 nm N-truncated Ig was injected over the LILRA2D1D2 immobilized (100 RU) sensor chip. The black solid line represents raw data. The dotted line represents the fitting curve based on the bivalent analyte model. The ka1, kd1, ka2, and kd2 of the interaction were determined by bivalent fitting to be 1.6 (105 M−1·s−1), 0.080 (s−1), 0.35 (RU−1·s−1), and 0.094 (s−1), respectively. B, The size exclusion chromatogram of the mixture of Fc fusion LILRA2D1D4 or LILAR2D1D2 and N-truncated Ig (blue line). The chromatogram of free Fc fusion LILRA2D1D4 or LILRA2D1D2 and N-truncated Ig are also shown as a pink line and gray line, respectively. Approximate positions of molecular weight standards are shown above the chromatogram. C, SDS-PAGE of the eluted fraction of the complex of Fc fusion LILRA2D1D4 and N-truncated Ig from size exclusion chromatography of panel B.
Thermodynamic properties of the LILRA2–N-truncated Ig interaction
We investigated the thermal stability of N-truncated Ig by differential scanning calorimetry (DSC) analysis to further understand its thermodynamic properties (Fig. 3A). Deconvolution analysis of the N-truncated Ig shows two peaks, with melting temperatures (Tms) of 57.8 and 66.3 °C. To clarify the assignment of these transitions, DSC analysis of the full-length Ig was also performed (Fig. 3B). It showed two major peaks, with Tms of 69.0 and 73.5 °C, corresponding to the unfolding temperatures of Fc and Fab, respectively, as previously described by Garber et al. (18). Since the N-truncated Ig was purified by protein G affinity chromatography, the Fc portion of the N-truncated Ig presumably has a structure similar to that of full-length Ig. Therefore, the Tm of 66.3 °C in N-truncated Ig was assigned to the unfolding temperature of the Fc of N-truncated Ig. The other Tm of 57.8 °C in N-truncated Ig was assigned to the unfolding temperature of the truncated Fab of N-truncated Ig. The Tms of N-truncated Ig, especially that of the truncated Fab, were lower than that of full-length Ig, suggesting that Fab truncation reduces the stability of N-truncated Ig, especially affecting the stability of the Fab region.
Figure 3.
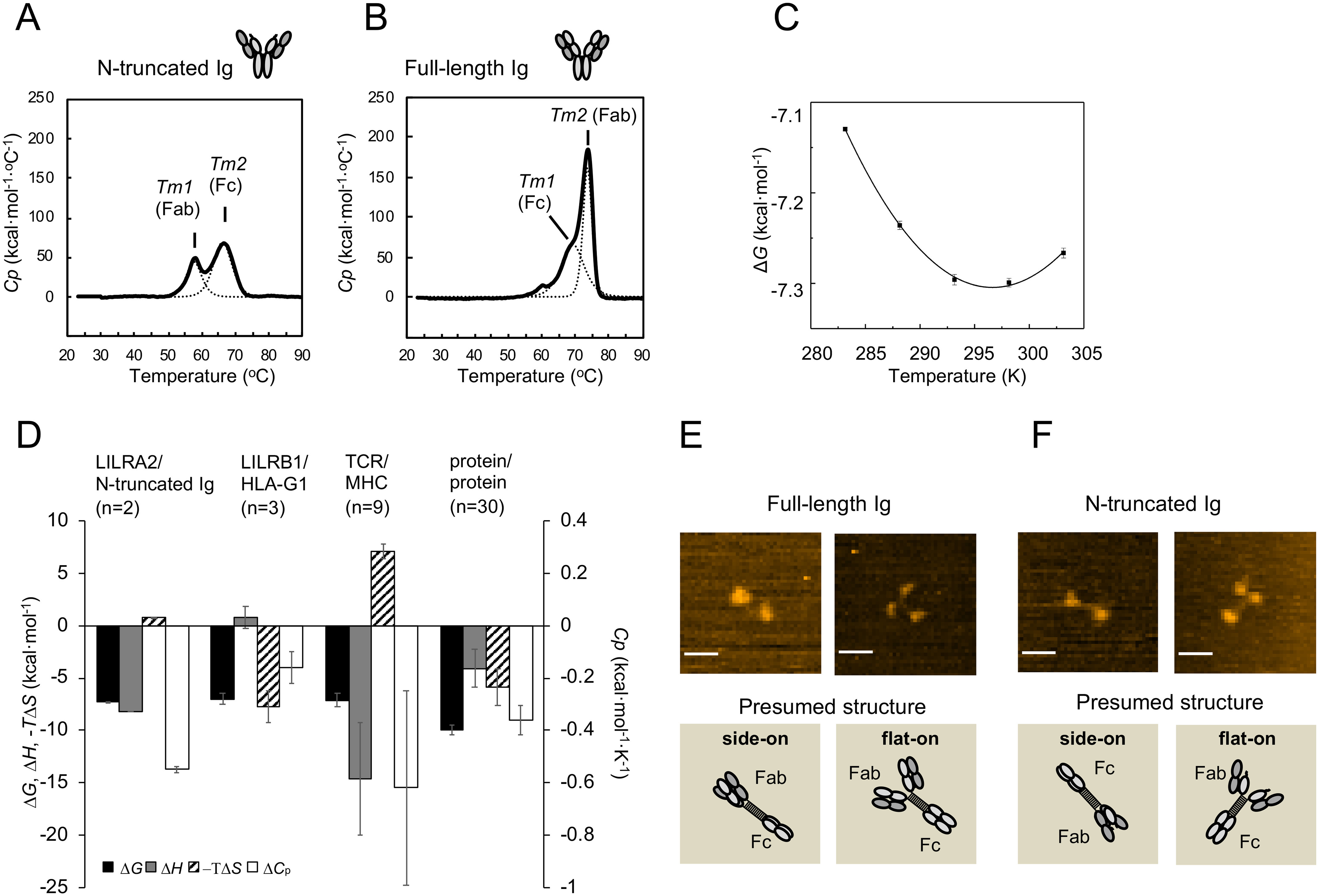
Thermodynamic features of LILRA2/N-truncated Ig interaction and structural characteristics of N-truncated Ig. A–B, DSC analyses of N-truncated Ig (A) and full-length Ig (B) are shown. Solid lines represent raw data. Dotted lines represent fitting curves. C, Thermodynamic analysis of LILRA2 binding to N-truncated Ig. The interaction was measured at several temperatures (10–30 °C) and converted into the standard free energy of binding (ΔG). Values for the enthalpic (ΔH) and standard entropic (–TΔS) changes (at 25 °C) and the specific heat capacity (ΔCp) were calculated by fitting the nonlinear form of the van't Hoff equation to these data (see Experimental procedures). Error bars show the range from two independent experiments. D, Comparison of thermodynamic properties (at 25 °C) of several cell surface intermolecular interactions. Data for LILRB1/HLA-G1 and TCR/peptide–MHC interactions are from references 37 and 26, 38–42, respectively. Data for protein–protein interactions (excluding antibody–antigen interactions) are from reference 28. The mean and standard error of 30 types of protein–protein interactions are depicted. E–F, Representative HS-AFM images of full-length Ig (E) and N-truncated Ig (F) (scale bars, 25 nm). The images show full-length Ig and N-truncated Ig absorbed on mica in different orientations. Schematic images corresponding to the above HS-AFM images show presumed absorbed protein structure. The upper right image of panel F was selected from Fig. S3 as a representative of the flat-on structure.
We performed van't Hoff analysis using the Kd determined by SPR, at different temperatures (10–30 °C), to further clarify the thermodynamic aspect of this interaction. The binding enthalpy (ΔH), entropy (ΔS), and heat capacity (ΔCp) were determined by using the nonlinear van't Hoff equation as described in the Experimental procedures (Fig. 3C). −TΔS was found to be 0.85 ± 0.03 kcal·mol−1 and ΔH was −8.2 ± 0.0 kcal·mol−1, indicating that this interaction is enthalpically driven with a slight entropy loss at 25 °C (Fig. 3D and Table S1). ΔCp was calculated as −0.56 ± 0.02 kcal·mol−1·K−1 (Fig. 3D and Table S1).
AFM analysis of N-truncated Ig and full-length Ig
High-speed atomic force microscopy (HS-AFM) was utilized to examine the structural features of N-truncated Ig. AFM images of full-length Igs in Fig. 3E and Fig. S2 show that full-length Ig exhibits two representative images, Y-shaped (flat-on) and dumbbell-like (side-on) structures. The Fab and Fc regions are relatively distant (the average distance is 21 ± 3 nm, mean ± S.D., n = 10) compared with IgG1 structures previously reported (19). Moreover, Movie S1 shows that the connection between Fab and Fc regions (two or three spots) of Ig particles are highly mobile, while the longest distances are 26 nm. Since the full-length Ig is IgG3 type and has a long linker region, these structural and dynamic features are reasonable for IgG3-type antibodies. On the other hand, N-truncated Ig (Fig. 3F, Fig. S3, and Movie S2) exhibited Y-shaped and dumbbell-like structures with highly mobile features, similar to full-length Ig (the average distance is 20 ± 4 nm, mean ± S.D., n = 10). These results indicate that N truncation does not cause any significant changes in overall structure and that the movement of Fab is independent of that of the Fc region (Fig. 3, E and F, Fig. S2 and S3, and Movies S1 and S2).
Identification of amino acid residues of LILRA2 critical for N-truncated Ig binding
We designed LILRA2 mutants based on a comparison of the primary sequence of LILRA2 and group 1 LILRs to determine the N-truncated Ig binding site on LILRA2 (Fig. 4A). Unique residues in LILRA2 with completely conserved corresponding amino acid residues in other group 1 LILRs were identified to prepare Q31E, E33Q, N41K, R48T, P53L, G54V, L112S, R142S, and W154R mutants (Fig. S4). We also focused on residues that are not universally conserved among group 1 LILRs and constructed the following LILRA2D1D2 mutants: I50_Q51insR/Q51P, H73G/Y85L, and H79R/N80A/H81R/S82W (Fig. S4). CD spectra of all mutants were generally similar to that of the WT protein (Fig. S5). SPR analysis of LILRA2 mutants showed that all domain 1 mutants (Q31E, E33Q, N41K, R48T, P53L, G54V, I50_Q51insR/Q51P, H73G/Y85L, and H79R/N80A/H81R/S82W) and two domain 2 mutants (L112S and R142S) bound to the N-truncated Ig with Kd similar to that of the WT. However, one domain 2 mutant, W154R, did not bind to the N-truncated Ig (Fig. 4B, Fig. S6, S7, and S8A, Table 1, and Table S2). These results suggest that the region surrounding Trp154 on LILRA2 domain 2 is involved in N-truncated Ig binding. We constructed an LILRA2D1-LILRB2D2 chimeric protein to test this hypothesis (Fig. S4). CD spectra of the chimera were similar to that of the WT protein (Fig. S5). SPR analysis showed that the chimeric protein does not bind to the N-truncated Ig (Fig. 4B). More LILRA2 mutants (V121E, D127G, H146Q, S147P, and W152S) were designed to further clarify the N-truncated Ig binding region surrounding Trp154, based on the structures of the LILR family members and prepared by the refolding method. SPR analysis showed that LILRA2 D127G, H146Q, and S147P bound to N-truncated Ig but LILRA2 V121E and W152S did not (Fig. S8B). Mapping of these residues on the LILRB2 structure (PDB entry 2GW5) showed that these residues are located on the same surface of domain 2 (Fig. 4C). Based on the mutagenesis study, we propose a model of the LILRA2/N-truncated Ig complex (Fig. 4D).
Figure 4.
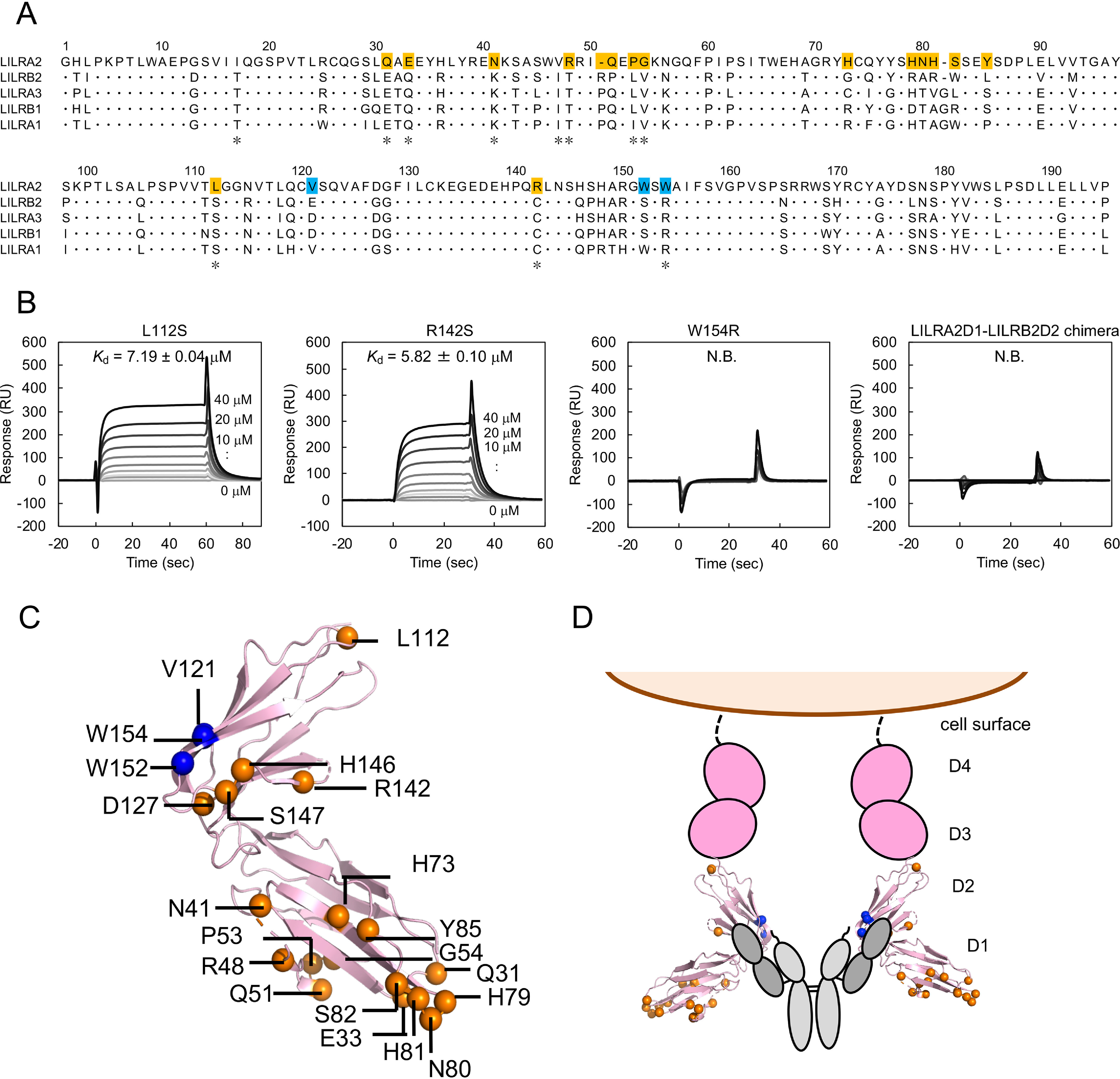
Mutagenesis analysis for LILRA2 binding mode toward N-truncated Ig. A, Alignment of group 1 LILRs. The asterisks below the sequence indicate the LILRA2 unique residues among group 1 LILRs. The mutated residues that did not affect N-truncated Ig binding are shown with orange background. The identified residues involved in ligand binding are shown with blue background. B, Mutagenesis analysis using SPR. LILRA2 mutants were injected onto N-truncated Ig immobilized chip. Kd value is presented as mean ± range from two experiments or mean ± standard deviation from three experiments. N.B. means no detectable binding. C, Mapping of mutated amino acid residues of LILRA2. Blue spheres represent the residues that are involved in N-truncated Ig binding. Orange spheres represent the residues that are not involved in N-truncated Ig binding. The structure was based on LILRB2 (PDB entry 2GW5). D, Proposed model of N-truncated Ig recognition by LILRA2.
Table 1.
Summary of binding affinities (Kd) of LILRA2 mutants for N-truncated immunoglobulinsa
| LILRA2D1D2 | n | Kd (μm) |
|---|---|---|
| Wild type | 3 | 4.83 ± 0.05 |
| Q31E | 3 | 4.01 ± 0.02 |
| E33Q | 2 | 3.23 ± 0.01 |
| N41K | 3 | 3.38 ± 0.03 |
| R48T | 3 | 3.98 ± 0.05 |
| P53L | 2 | 2.87 ± 0.21 |
| G54V | 3 | 2.74 ± 0.05 |
| I50_Q51insR/Q51P | 2 | 6.10 ± 0.04 |
| H73G/Y85L | 2 | 4.81 ± 0.03 |
| H79R/N80A/H81R/S82W | 2 | 6.72 ± 0.05 |
| L112S | 2 | 7.19 ± 0.04 |
| V121E | 3 | N.B. |
| D127G | 2 | 1.03 ± 0.09 |
| R142S | 3 | 5.82 ± 0.10 |
| H146Q | 3 | 4.41 ± 0.04 |
| S147P | 3 | 4.93 ± 0.22 |
| W152S | 3 | N.B. |
| W154R | 3 | N.B. |
aNumbers of experiments are shown as n. Kd values are presented as means ± standard deviations from three experiments or means ± ranges from two experiments. N.B. means no detectable binding.
Discussion
In this study, we performed kinetic analysis using SPR to reveal the mechanistic details of the interaction between LILRA2 and N-truncated Ig. LILRA2 binds to N-truncated Ig with a relatively fast association rate constant (1.1 × 105 M−1·s−1) and dissociation rate constant (0.23 s−1) compared with those of general protein–protein interactions, suggesting that LILRA2 can rapidly detect N-truncated Ig in blood. The weak affinity of LILRA2/N-truncated Ig interaction (Kd of ∼4.8 μm) is typically observed in interactions which control cell–cell recognition (Table S3).
A strong avidity effect was observed in SPR when N-truncated Ig was injected onto an LILRA2 immobilized chip (Fig. 2A). In addition, the size exclusion chromatogram of the mixture of Fc fusion LILRA2D1D4 and N-truncated Ig showed an earlier elution peak, which contained the Fc fusion LILRA2D1D4 and N-truncated Ig, whereas that of LILRA2D1D2 and N-truncated Ig showed separate elution peaks for each component protein (Fig. 2B). These data suggest that LILRA2 binds to N-truncated Ig strongly by avidity effect. The avidity effect is observed in other ITAM/ITIM-harboring molecules, such as FcαR and LILRB1 and -B2 (20–24). We propose that the dimerization, which occurs by cross-linking upon the binding of N-truncated Ig, triggers immune activation through LILRA2. Thus, the LILRA2 expression level is also an immune-regulating factor.
Considering the characteristics of the LILRA2 ligand, IgM-expressing cells cultured with bacteria secreted not only the cleaved heavy chain but also the intact heavy chain (14). A previous study showed antibodies with one cleaved heavy chain and another intact heavy chain in Western blotting under nonreducing conditions (15). Our monovalent binding analysis in SPR indicated that these antibodies bind to LILRA2 but are rapidly dissociated. In contrast, the antibodies that contain two cleaved heavy chains have stronger binding affinity at nanomolar range and activate immune cells through LILRA2. Therefore, when LILRA2 encounters Igs that have both cleaved Fab domains, it is able to bind to them with strong affinity due to avidity effect and initiate effective signal transduction to activate the immune system.
LILRA2 specifically binds to the N-truncated Ig but not to the full-length Ig, suggesting that LILRA2 recognizes the exposed VL and the N-terminal peptide of cleaved VH of N-truncated Ig. DSC analysis showed the instability of the truncated Fab. Generally, the VL domain interacts with the VH domain (25). Therefore, instability is considered to arise from the flexibility of the exposed VL and N-terminus peptide of the cleaved VH of N-truncated Ig. LILRA2–N-truncated Ig interaction was characterized by enthalpically driven binding with small entropy loss (Table S1). Initially, we proposed that LILRA2 binding to N-truncated Ig causes the loss of dynamic flexibility of the N-truncated Ig and results in binding with entropy loss similar to that of T-cell receptor (TCR)/major histocompatibility complex (MHC) binding (26, 27). However, the entropy loss was too low, with a −TΔS of 0.85 ± 0.03 kcal·mol−1, which is lower than that of the TCR–MHC interaction (7.1 ± 5.7 kcal·mol−1) and larger than the average −TΔS of protein–protein interactions (−5.91 kcal·mol−1) (Fig. 3D) (28). In addition, we determined the heat capacity change upon binding, which provided information about conformational changes reflecting a change in the hydrated state (29, 30). For example, gp120–CD4 interaction, which requires structural changes for ligand binding, shows a large negative heat capacity change (∼−1.8 kcal·mol−1·K−1) (31). The ΔCp of LILRA2/N-truncated Ig was moderate (−0.56 ± 0.02 kcal·mol−1·K−1) compared with the heat capacity change of cell surface receptor–ligand interactions, such as gp120–CD4 and LILRB1–HLA-G interactions, suggesting a change in protein flexibility upon binding. The interaction shows fast association and dissociation, with moderate heat capacity change and small entropy loss, using the hydrophobic site composed of V121, W152, and W154 of LILRA2. These results suggest a hydrophobic effect and/or maintenance of conformational flexibility upon complex formation.
The AFM observations of N-truncated and full-length Igs revealed that both of them exhibit not only Y-shaped but also dumbbell-like structures. These different images likely arise from the absorbed orientation of the proteins toward mica, as similarly pointed out for the IgG structure before (32). We also observed that interdomain Fab-Fc connections for both N-truncated and full-length Igs are highly mobile due to the long hinge region. Therefore, the structure and dynamics of N-truncated and full-length Ig were similar to each other, suggesting that N truncation does not affect overall structure and mobility (Fig. 3, E and F, and Movies S1 and S2). We further performed the preliminary AFM experiments of a mixture of Fc-fusion LILRA2D1D4 and N-truncated Ig, which is mainly eluted as a complex by size exclusion chromatography, as shown in Fig. 2B (the details will be published elsewhere). Many particles presumably considered to be dimers and multimers were found, while such dimer/multimer particles were rarely observed in each AFM image of Fc-fusion LILRA2D1D4 or N-truncated Ig only, suggesting the direct recognition of LILRA2 toward N-truncated Ig.
Similar Kd values were observed for the interaction between LILRA2D1D2 or LILRA2D1D4 and N-truncated Ig (Fig. 1, E–H). In addition, LILRA2D1-LILRB2D2 chimeric protein did not bind to the N-truncated Ig (Fig. 4B). These results suggest that domain 2 is crucial for N-truncated Ig binding. Our mutational study further revealed that V121 on strand B and W152 and W154 on the loop between strands C′ and E of LILRA2 domain 2 contribute to N-truncated Ig binding. Mapping of LILRA2 V121, W152, and W154 on the LILRB2 structure (PDB entry 2GW5) showed that these amino acid residues are located in the proximal region of the hinge region (Fig. 4C). The corresponding residues, E123 (V121 in LILRA2), S154 (W152), and R156 (W154), on the LILRB2 structure (PDB entry 2GW5) are located on the surface of the protein and are almost exposed (Fig. S9). We propose that the hydrophobic surface composed of V121, W152, and W154 is important for the interaction with the exposed VL and N-terminus peptide of the cleaved VH of the N-truncated Ig. Several kinds of N-truncated Igs can be recognized by LILRA2, with the hydrophobic feature being common among N-truncated Igs. Therefore, hydrophobic interaction might enable LILRA2 to recognize a broad range of N-truncated Igs specifically. Hirayasu et al. found that LILRA2 bound to N-truncated Ig, whose VH domain was cleaved at the amino acid positions 11, 12, 13, or 14 of the J region but not at position 15 or 16 (14). This result suggests that the section of the exposed J region (11 residues at the C-terminal side) of the VH domain after cleavage is important for binding. In addition, the mutagenesis analysis reveals that residues crucial for N-truncated Ig are located around positions 10 and 95, close to the tip of the C domain and the side of the VL domain, respectively, exposed after the cleavage. On the other hand, the present mutagenesis data on LILRA2 suggests that the binding site for N-truncated Ig is a small hydrophobic patch on the surface of the side of the LILRA2 D2 domain. Therefore, the hydrophobic residues of the LILRA2 D2 domain would interact with the exposed hydrophobic surface of N-truncated Ig, the left VH domain, the side of the VL domain, and/or the tip of the C domain. Other LILR or KIR members (Fig. S10) have larger ligand binding sites. For example, LILRB2 binds to multiple Ig domains of HLA proteins, both β2 microglobulin and the α3 domain (33). Thus, future investigation is warranted to confirm whether LILRA2 utilizes a wider surface for the binding. The hydrophobicity of LILRA2 V121, W152, and W154 is a distinct feature among group 1 LILRs. For example, the corresponding amino acid residue of LILRA2 W154 is typically arginine (Fig. 4A). This could be the reason for LILRA2 not binding to HLAs, which are common ligands for group 1 LILRs, except LILRA2. The N-truncated Ig binding site on LILRA2 is also a novel ligand recognition site among group 1 LILR family members and other immunoglobulin-like receptors, such as KIR2DS2, which bind to HLA. The hinge region between domain 1 and domain 2 of LILRB1, LILRB2, and KIR2DS2 is reported as the ligand recognition site (Fig. S10) (33–35). LILR genes supposedly have evolved rapidly by pathogen-driven evolution (36). Comparison of the ligand recognition sites suggests that LILRA2 evolved to become a specialized receptor to sense foreign antigens by sacrificing HLA recognition ability.
Conclusions
In this study, we report that LILRA2/N-truncated Ig binding shows relatively fast association and dissociation rates, as typically observed in cell-cell recognition mechanisms. However, the affinity with immobilized LILRA2 becomes much stronger due to the avidity effect, suggesting that strong affinity achieved by the cross-linking of LILRA2 on the cell surface is crucial for efficient signal transduction. Furthermore, DSC analysis showed the instability of N-truncated Ig compared with full-length Ig. Thermodynamic analysis showed that the interaction is enthalpy-driven, with low entropy loss and moderate heat capacity change. The AFM results suggested that the structure and mobility of N-truncated Ig were similar to those of full-length Ig. In addition, a mutagenesis study revealed that the N-truncated Ig binding site on LILRA2 was the hydrophobic region on domain 2, suggesting that LILRA2 binds to the exposed VL domain and left heavy chain, which is likely hydrophobic. The fast kinetics, moderate heat capacity change, and slight entropy loss could be explained by a hydrophobic effect and/or residual conformational flexibility upon binding. The understanding of the N-truncated Ig recognition mechanism by LILRA2 in this study provides insights into efficient strategies employed in effective regulation of host immune systems. Furthermore, our study could form the basis for the rational design of antibacterial drugs that activate LILRA2 in addition to innate N-truncated Ig.
Experimental procedures
Expression and purification of LILRA2D1D2, LILRA2D1D4, and Fc fusion LILRA2D1D4
Expression and purification of LILRA2D1D2 was performed as described previously for LILRB1 (24). Briefly, the extracellular domain of LILRA2D1D2 (residues 1–195) was cloned into a pGMT7 vector. The resulting plasmid was transformed into Escherichia coli BL21(DE3)pLysS and the LILRA2D1D2 protein expressed as inclusion bodies. The inclusion bodies were washed, solubilized with guanidine hydrochloride containing buffer, and refolded using the dilution method previously reported (4). The refolded LILRA2 was purified by size exclusion chromatography. The extracellular domain of LILRA2D1D4 (residues 1–409) was cloned into a pHLsec vector. Recombinant protein fused with a 6× His tag was transiently expressed in the culture supernatant of 293S GnTI(−) cells and purified using HisTrap FF (GE Healthcare), followed by size exclusion chromatography. Expression and purification of Fc fusion LILRA2 D1D4 was performed using a modified version of a previously described method (14). Briefly, Fc fusion LILRA2D1D4 transiently expressed in the culture supernatant of 293T cells was purified using affinity chromatography with protein G Sepharose resin (GE Healthcare), followed by size exclusion chromatography.
LILRA2 mutant preparation
LILRA2 mutants (Q31E, E33Q, N41K, R48T, P53L, G54V, I50_Q51insR/Q51P, H73G, H79R/N80A/H81R/S82W, L112S, V121E, D127G, R142S, H146Q, S147P, W152S, and W154R) were generated using the primer set listed in Table S4, using a plasmid encoding LILRA2D1D2 as the template. In addition, LILRA2 Y85L mutagenesis was performed using LILRA2 H73G plasmid as the template. The expression plasmid for the LILRA2D1-LILRB2D2 chimeric protein (residues 1–97 of LILRA2 fused with residues 99–196 of LILRB2) was constructed using the Gibson Assembly master mix (New England BioLabs) with LILRA2D1D2 and LILRB2D1D2 expression plasmids as templates. All mutants were prepared by the same method as the WT LILRA2 D1D2.
Expression and purification of N-truncated Ig and full-length Ig
Expression and purification of N-truncated Ig and full-length Ig were performed using a modified version of a previously described method (14). Briefly, recombinant proteins were produced by cotransfection of plasmids encoding the heavy chain and the light chain. Recombinant proteins transiently expressed in the culture supernatant of 293T cells were purified using affinity chromatography with protein G Sepharose resin (GE Healthcare), followed by size exclusion chromatography (GE Healthcare).
Interaction analysis using SPR
SPR experiments were performed using a BIAcore3000 (GE Healthcare). N-truncated Ig and full-length Ig (control protein) were immobilized on a research-grade CM5 chip (GE Healthcare) at 1400 and 1300 response units (RU). The purified WT LILRA2D1D2 and mutants in HBS-EP (GE Healthcare) were injected onto an immobilized N-truncated Ig, at a flow rate of 10 μl/min for 30 to 60 s, for the equilibrium analysis. The binding response at each concentration (from 0.16 μm to 40 μm) was calculated by subtracting the equilibrium response measured by the full-length Ig immobilized flow cell from the response in the N-truncated Ig immobilized flow cell. The data were analyzed using BIAevaluation software (v4.1) and Origin 7.0 software (OriginLab). Affinity constants (Kd) were derived by nonlinear curve fitting of the standard Langmuir binding isotherm. The purified LILRA2D1D4 (from 0.12 μm to 15 μm) in HBS-EP (GE Healthcare) was injected over the immobilized N-truncated Ig, at a flow rate of 10 μl/min for 60 s, for the equilibrium analysis. Kd values were determined as described above.
For kinetics analysis, LILRA2D1D2 was injected onto an N-truncated Ig immobilized chip (1400 RU) at a flow rate of 30 μl/min for 20 s. The global fitting analysis using the 1:1 Langmuir binding model was simultaneously performed with the raw data for the association and dissociation phases at different concentrations of the LILRA2 protein. Curve fitting was performed using the BIAevaluation software (v4.1).
The avidity effect was evaluated by sequential injection of 47 nm N-truncated Ig onto an LILRA2D1D2 immobilized chip (100 RU) for 1 min. The curve-fitting analysis was performed by using a bivalent analyte model as described above.
Binding analysis using size exclusion chromatography
For binding analysis, a mixture of Fc fusion LILRA2D1D4 (216 pmol) or LILRA2D1D2 (432 pmol) and N-truncated Ig (205 pmol) in 250 μl 20 mm Tris-HCl, 100 mm NaCl, pH 8.0, was incubated at room temperature for 1 h, followed by fractionation on a Superdex 200 10/300 Increase size exclusion column.
Thermal stability of N-truncated Ig by DSC
The thermal stability of N-truncated Ig and full-length Ig was determined by microCal VP-capillary DSC (Malvern). Briefly, 0.3 mg/ml of N-truncated Ig and full-length Ig were prepared in 10 mm HEPES and 150 mm NaCl, pH 7.4, for DSC analysis. Thermograms were collected at scan rates of 1 °C/min from 20 to 95 °C. The data were analyzed using Origin 7.0 software (OriginLab). The buffer reference scan was subtracted from the thermograms of N-truncated Ig and full-length Ig. The base line of the thermograms was determined by connecting the pre- and posttransition state base lines. The transition temperature (Tm) was calculated by fitting data to a non-two-state thermal transition model.
Thermodynamic analyses using SPR
Equilibrium analyses of LILRA2D1D2 were performed at five temperatures (10–30 °C, every 5 °C). The amount of immobilized N-truncated Ig was 1400 RU. The standard state Gibbs energy change (ΔG) upon binding was obtained from equation 1:
| (1) |
where Kd is the dissociation constant, expressed in units mol/L, and R is the gas constant. ΔG of each data set was plotted against the different temperatures and fitted with the nonlinear van't Hoff equation 2:
| (2) |
where ΔH and ΔS are the binding enthalpy and entropy at 298.15 K, respectively, and ΔCp is the heat capacity, which is assumed to be temperature-independent.
Structural analysis using HS-AFM
The HS-AFM experiments were performed using a Nano Explorer high-speed atomic force microscope (Research Institute of Biomolecule Metrology Co., Ltd.) and a cantilever (BL-AC10DS-A2, Olympus). N-truncated Ig and full-length Ig were absorbed on a mica surface for 3 min and washed with Milli-Q water. AFM images were obtained at a scan area of 500 by 500 nm2 with 200 by 200 pixels and a scan rate of 1 frame/s. All of the images were analyzed by using Eagle and ImageJ software. A flatten filter was applied for each AFM image. The distances between Fab and Fc were calculated as those between the center of mass of each domain.
Data availability
Data are available from the corresponding author upon reasonable request.
Supplementary Material
This article contains supporting information.
Author contributions—R. Y., A. F., K. Y., and H. F. formal analysis; R. Y., A. F., and K. M. investigation; R. Y., A. F., and K. M. writing-original draft; R. Y., A. F., and K. M. writing-review and editing; A. F. and K. M. conceptualization; K. H. and H. A. resources; K. M. supervision; K.M. funding acquisition; K.M. project administration.
Funding and additional information—This research was partially supported by the “Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers” (no. S2701), by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), the Japan Agency for Medical Research and Development (AMED) under grant number 16am0101007j0005, the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED under grant numbers 17am0101093j0001, 18am0101093j0002, and 19am0101093j0003, by Hokkaido University, Global Facility Center (GFC), Pharma Science Open Unit (PSOU), funded by MEXT under “Support Program for Implementation of New Equipment Sharing System,” by the Hokkaido University Biosurface Project, by Takeda Science Foundation, and by Nitobe School Advanced Program Research Grant 2017 from the Hokkaido University's Frontier Foundation (R. Y.). H. A. is supported by JSPS HAKENHI grant numbers JP18H05279 and 18K19450, MEXT KAKENHI grant number JP190H04808, and AMED under grant numbers JP17fm0208004 and JP19ek0410053. K. H. is supported by JSPS HAKENHI, grant number JP19H03468. K. M. and H. A. are supported by the Japan Society for the Promotion of Science Research and CREST, JST.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- LILRs
- leukocyte Ig-like receptors
- ILTs
- Ig-like transcripts
- ITIM
- immunoreceptor tyrosine-based inhibitory motif
- ITAM
- immunoreceptor tyrosine-based activation motif
- SPR
- surface plasmon resonance
- HS-AFM
- high-speed atomic force microscopy
- TCR
- T-cell receptor
- MHC
- major histocompatibility complex
- RU
- response units.
References
- 1. Kuroki K., Furukawa A., and Maenaka K. (2012) Molecular recognition of paired receptors in the immune system. Front. Microbiol. 3, 429 10.3389/fmicb.2012.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sharma P., and Allison J. P. (2015) Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 10.1016/j.cell.2015.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kang X., Kim J., Deng M., John S., Chen H., Wu G., Phan H., and Zhang C. C. (2016) Inhibitory leukocyte immunoglobulin-like receptors: immune checkpoint proteins and tumor sustaining factors. Cell Cycle 15, 25–40 10.1080/15384101.2015.1121324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shiroishi M., Tsumoto K., Amano K., Shirakihara Y., Colonna M., Braud V. M., Allan D. S., Makadzange A., Rowland-Jones S., Willcox B., Jones E. Y., van der Merwe P. A., Kumagai I., and Maenaka K. (2003) Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. U S A 100, 8856–8861 10.1073/pnas.1431057100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chapman T. L., Heikeman A. P., and Bjorkman P. J. (1999) The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11, 603–613 10.1016/S1074-7613(00)80135-1 [DOI] [PubMed] [Google Scholar]
- 6. Borges L., Hsu M. L., Fanger N., Kubin M., and Cosman D. (1997) A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J. Immunol. 159, 5192–5196 [PubMed] [Google Scholar]
- 7. Cosman D., Fanger N., Borges L., Kubin M., Chin W., Peterson L., and Hsu M. L. (1997) A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 7, 273–282 10.1016/S1074-7613(00)80529-4 [DOI] [PubMed] [Google Scholar]
- 8. Allen R. L., Raine T., Haude A., Trowsdale J., and Wilson M. J. (2001) Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J. Immunol. 167, 5543–5547 10.4049/jimmunol.167.10.5543 [DOI] [PubMed] [Google Scholar]
- 9. Jones D. C., Kosmoliaptsis V., Apps R., Lapaque N., Smith I., Kono A., Chang C., Boyle L. H., Taylor C. J., Trowsdale J., and Allen R. L. (2011) HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J. Immunol. 186, 2990–2997 10.4049/jimmunol.1003078 [DOI] [PubMed] [Google Scholar]
- 10. Tedla N., Bandeira-Melo C., Tassinari P., Sloane D. E., Samplaski M., Cosman D., Borges L., Weller P. F., and Arm J. P. (2003) Activation of human eosinophils through leukocyte immunoglobulin-like receptor 7. Proc. Natl. Acad. Sci. U S A 100, 1174–1179 10.1073/pnas.0337567100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee D. J., Sieling P. A., Ochoa M. T., Krutzik S. R., Guo B., Hernandez M., Rea T. H., Cheng G., Colonna M., and Modlin R. L. (2007) LILRA2 activation inhibits dendritic cell differentiation and antigen presentation to T cells. J. Immunol. 179, 8128–8136 10.4049/jimmunol.179.12.8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huynh O. A., Hampartzoumian T., Arm J. P., Hunt J., Borges L., Ahern M., Smith M., Geczy C. L., McNeil H. P., and Tedla N. (2007) Down-regulation of leucocyte immunoglobulin-like receptor expression in the synovium of rheumatoid arthritis patients after treatment with disease-modifying anti-rheumatic drugs. Rheumatology (Oxford) 46, 742–751 10.1093/rheumatology/kel405 [DOI] [PubMed] [Google Scholar]
- 13. Sloane D. E., Tedla N., Awoniyi M., Macglashan D. W., Borges L., Austen K. F., and Arm J. P. (2004) Leukocyte immunoglobulin-like receptors: novel innate receptors for human basophil activation and inhibition. Blood 104, 2832–2839 10.1182/blood-2004-01-0268 [DOI] [PubMed] [Google Scholar]
- 14. Hirayasu K., Saito F., Suenaga T., Shida K., Arase N., Oikawa K., Yamaoka T., Murota H., Chibana H., Nakagawa I., Kubori T., Nagai H., Nakamaru Y., Katayama I., Colonna M., et al. (2016) Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat. Microbiol. 1, 16054 10.1038/nmicrobiol.2016.54 [DOI] [PubMed] [Google Scholar]
- 15. Arfi Y., Minder L., Di Primo C., Le Roy A., Ebel C., Coquet L., Claverol S., Vashee S., Jores J., Blanchard A., and Sirand-Pugnet P. (2016) MIB-MIP is a mycoplasma system that captures and cleaves immunoglobulin G. Proc. Natl. Acad. Sci. U S A 113, 5406–5411 10.1073/pnas.1600546113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y., Chu F., Gao F., Zhou B., and Gao G. F. (2007) Stability engineering, biophysical, and biological characterization of the myeloid activating receptor immunoglobulin-like transcript 1 (ILT1/LIR-7/LILRA2). Protein Expr. Purif. 56, 253–260 10.1016/j.pep.2007.08.010 [DOI] [PubMed] [Google Scholar]
- 17. Chen Y., Gao F., Chu F., Peng H., Zong L., Liu Y., Tien P., and Gao G. F. (2009) Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: molecular basis for its non-binding to MHC complexes. J. Mol. Biol. 386, 841–853 10.1016/j.jmb.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 18. Garber E., and Demarest S. J. (2007) A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem. Biophys. Res. Commun. 355, 751–757 10.1016/j.bbrc.2007.02.042 [DOI] [PubMed] [Google Scholar]
- 19. Umakoshi T., Udaka H., Uchihashi T., Ando T., Suzuki M., and Fukuda T. (2018) Quantum-dot antibody conjugation visualized at the single-molecule scale with high-speed atomic force microscopy. Colloids Surf. B Biointerfaces 167, 267–274 10.1016/j.colsurfb.2018.04.015 [DOI] [PubMed] [Google Scholar]
- 20. Herr A. B., Ballister E. R., and Bjorkman P. J. (2003) Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature 423, 614–620 10.1038/nature01685 [DOI] [PubMed] [Google Scholar]
- 21. Herr A. B., White C. L., Milburn C., Wu C., and Bjorkman P. J. (2003) Bivalent binding of IgA1 to FcalphaRI suggests a mechanism for cytokine activation of IgA phagocytosis. J. Mol. Biol. 327, 645–657 10.1016/S0022-2836(03)00149-9 [DOI] [PubMed] [Google Scholar]
- 22. Appel H., Gauthier L., Pyrdol J., and Wucherpfennig K. W. (2000) Kinetics of T-cell receptor binding by bivalent HLA-DR. Peptide complexes that activate antigen-specific human T-cells. J. Biol. Chem. 275, 312–321 10.1074/jbc.275.1.312 [DOI] [PubMed] [Google Scholar]
- 23. De Crescenzo G., Pham P. L., Durocher Y., and O'Connor-McCourt M. D. (2003) Transforming growth factor-beta (TGF-beta) binding to the extracellular domain of the type II TGF-beta receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high affinity binding. J. Mol. Biol. 328, 1173–1183 10.1016/S0022-2836(03)00360-7 [DOI] [PubMed] [Google Scholar]
- 24. Shiroishi M., Kuroki K., Ose T., Rasubala L., Shiratori I., Arase H., Tsumoto K., Kumagai I., Kohda D., and Maenaka K. (2006) Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J. Biol. Chem. 281, 10439–10447 10.1074/jbc.M512305200 [DOI] [PubMed] [Google Scholar]
- 25. Padlan E. A. (1994) Anatomy of the antibody molecule. Mol. Immunol. 31, 169–217 10.1016/0161-5890(94)90001-9 [DOI] [PubMed] [Google Scholar]
- 26. Willcox B. E., Gao G. F., Wyer J. R., Ladbury J. E., Bell J. I., Jakobsen B. K., and van der Merwe P. A. (1999) TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity 10, 357–365 10.1016/S1074-7613(00)80035-7 [DOI] [PubMed] [Google Scholar]
- 27. Ding Y. H., Baker B. M., Garboczi D. N., Biddison W. E., and Wiley D. C. (1999) Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11, 45–56 10.1016/S1074-7613(00)80080-1 [DOI] [PubMed] [Google Scholar]
- 28. Stites W. E. (1997) Proteinminus signProtein Interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 97, 1233–1250 10.1021/cr960387h [DOI] [PubMed] [Google Scholar]
- 29. Vega S., Abian O., and Velazquez-Campoy A. (2016) On the link between conformational changes, ligand binding and heat capacity. Biochim. Biophys. Acta 1860, 868–878 10.1016/j.bbagen.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 30. Lo Conte L., Chothia C., and Janin J. (1999) The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285, 2177–2198 10.1006/jmbi.1998.2439 [DOI] [PubMed] [Google Scholar]
- 31. Myszka D. G., Sweet R. W., Hensley P., Brigham-Burke M., Kwong P. D., Hendrickson W. A., Wyatt R., Sodroski J., and Doyle M. L. (2000) Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U S A 97, 9026–9031 10.1073/pnas.97.16.9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vilhena J. G., Dumitru A. C., Herruzo E. T., Mendieta-Moreno J. I., Garcia R., Serena P. A., and Pérez R. (2016) Adsorption orientations and immunological recognition of antibodies on graphene. Nanoscale 8, 13463–13475 10.1039/c5nr07612a [DOI] [PubMed] [Google Scholar]
- 33. Shiroishi M., Kuroki K., Rasubala L., Tsumoto K., Kumagai I., Kurimoto E., Kato K., Kohda D., and Maenaka K. (2006) Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc. Natl. Acad. Sci. U S A 103, 16412–16417 10.1073/pnas.0605228103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willcox B. E., Thomas L. M., and Bjorkman P. J. (2003) Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat. Immunol. 4, 913–919 10.1038/ni961 [DOI] [PubMed] [Google Scholar]
- 35. Liu J., Xiao Z., Ko H. L., Shen M., and Ren E. C. (2014) Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc. Natl. Acad. Sci. U S A 111, 2662–2667 10.1073/pnas.1322052111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Canavez F., Young N. T., Guethlein L. A., Rajalingam R., Khakoo S. I., Shum B. P., and Parham P. (2001) Comparison of chimpanzee and human leukocyte Ig-like receptor genes reveals framework and rapidly evolving genes. J. Immunol. 167, 5786–5794 10.4049/jimmunol.167.10.5786 [DOI] [PubMed] [Google Scholar]
- 37. Shiroishi M., Kuroki K., Tsumoto K., Yokota A., Sasaki T., Amano K., Shimojima T., Shirakihara Y., Rasubala L., van der Merwe P. A., Kumagai I., Kohda D., and Maenaka K. (2006) Entropically driven MHC class I recognition by human inhibitory receptor leukocyte Ig-like receptor B1 (LILRB1/ILT2/CD85j). J. Mol. Biol. 355, 237–248 10.1016/j.jmb.2005.10.057 [DOI] [PubMed] [Google Scholar]
- 38. Boniface J. J., Reich Z., Lyons D. S., and Davis M. M. (1999) Thermodynamics of T cell receptor binding to peptide-MHC: evidence for a general mechanism of molecular scanning. Proc. Natl. Acad. Sci. U S A 96, 11446–11451 10.1073/pnas.96.20.11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anikeeva N., Lebedeva T., Krogsgaard M., Tetin S. Y., Martinez-Hackert E., Kalams S. A., Davis M. M., and Sykulev Y. (2003) Distinct molecular mechanisms account for the specificity of two different T-cell receptors. Biochemistry 42, 4709–4716 10.1021/bi026864+ [DOI] [PubMed] [Google Scholar]
- 40. Garcia K. C., Radu C. G., Ho J., Ober R. J., and Ward E. S. (2001) Kinetics and thermodynamics of T cell receptor-autoantigen interactions in murine experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U S A 98, 6818–6823 10.1073/pnas.111161198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J. K., Stewart-Jones G., Dong T., Harlos K., Di Gleria K., Dorrell L., Douek D. C., van der Merwe P. A., Jones E. Y., and McMichael A. J. (2004) T cell cross-reactivity and conformational changes during TCR engagement. J. Exp. Med. 200, 1455–1466 10.1084/jem.20041251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davis-Harrison R. L., Armstrong K. M., and Baker B. M. (2005) Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J. Mol. Biol. 346, 533–550 10.1016/j.jmb.2004.11.063 [DOI] [PubMed] [Google Scholar]
- 43. Tabata S., Kuroki K., Wang J., Kajikawa M., Shiratori I., Kohda D., Arase H., and Maenaka K. (2008) Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J. Biol. Chem. 283, 8893–8901 10.1074/jbc.M709793200 [DOI] [PubMed] [Google Scholar]
- 44. Maenaka K., van der Merwe P. A., Stuart D. I., Jones E. Y., and Sondermann P. (2001) The human low affinity Fcgamma receptors IIa, IIb, and III bind IgG with fast kinetics and distinct thermodynamic properties. J. Biol. Chem. 276, 44898–44904 10.1074/jbc.M106819200 [DOI] [PubMed] [Google Scholar]
- 45. Maenaka K., Juji T., Nakayama T., Wyer J. R., Gao G. F., Maenaka T., Zaccai N. R., Kikuchi A., Yabe T., Tokunaga K., Tadokoro K., Stuart D. I., Jones E. Y., and van der Merwe P. A. (1999) Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J. Biol. Chem. 274, 28329–28334 10.1074/jbc.274.40.28329 [DOI] [PubMed] [Google Scholar]
- 46. Gao G. F., Willcox B. E., Wyer J. R., Boulter J. M., O'Callaghan C. A., Maenaka K., Stuart D. I., Jones E. Y., Van Der Merwe P. A., Bell J. I., and Jakobsen B. K. (2000) Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8alpha. J. Biol. Chem. 275, 15232–15238 10.1074/jbc.275.20.15232 [DOI] [PubMed] [Google Scholar]
- 47. Bakker T. R., Piperi C., Davies E. A., and Merwe P. A. (2002) Comparison of CD22 binding to native CD45 and synthetic oligosaccharide. Eur. J. Immunol. 32, 1924–1932 [DOI] [PubMed] [Google Scholar]
- 48. van der Merwe P. A., Bodian D. L., Daenke S., Linsley P., and Davis S. J. (1997) CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J. Exp. Med. 185, 393–403 10.1084/jem.185.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding author upon reasonable request.