Abstract
DNA lesions can severely compromise transcription and block RNA synthesis by RNA polymerase (RNAP), leading to subsequent recruitment of DNA repair factors to the stalled transcription complex. Recent structural studies have uncovered molecular interactions of several DNA lesions within the transcription elongation complex. However, little is known about the role of key elements of the RNAP active site in translesion transcription. Here, using recombinantly expressed proteins, in vitro transcription, kinetic analyses, and in vivo cell viability assays, we report that point amino acid substitutions in the trigger loop, a flexible element of the active site involved in nucleotide addition, can stimulate translesion RNA synthesis by Escherichia coli RNAP without altering the fidelity of nucleotide incorporation. We show that these substitutions also decrease transcriptional pausing and strongly affect the nucleotide addition cycle of RNAP by increasing the rate of nucleotide addition but also decreasing the rate of translocation. The secondary channel factors DksA and GreA modulated translesion transcription by RNAP, depending on changes in the trigger loop structure. We observed that although the mutant RNAPs stimulate translesion synthesis, their expression is toxic in vivo, especially under stress conditions. We conclude that the efficiency of translesion transcription can be significantly modulated by mutations affecting the conformational dynamics of the active site of RNAP, with potential effects on cellular stress responses and survival.
Keywords: RNA polymerase (RNAP), translesion transcription, DNA repair, trigger loop, DksA, GreA, nucleotide excision repair (NER), transcription-coupled repair, secondary channel factor, RNA, RNA polymerase, transcription, DNA damage
RNA extension by RNA polymerase (RNAP), consisting of alternating steps of nucleotide addition and RNAP translocation, is a nonuniform process that can be interrupted by transient pausing and permanent stalling induced by nucleotide misincorporation, recognition of specific regulatory sequences, roadblock proteins, or DNA lesions. In both bacteria and eukaryotes, RNAP serves as a major sensor of DNA lesions and recruits nucleotide excision repair (NER) factors to the sites of DNA damage during transcription-coupled repair (1–3).
Experiments in vitro and in vivo demonstrated that various lesions can significantly decrease transcription fidelity and impair RNA extension by both bacterial and eukaryotic RNAPs. In particular, it was shown that bulky lesions (such as thymine dimers: cyclobutane pyrimidine dimers (CPDs)) or adducts that disrupt complementary pairing (such as 1,N6-ethenoadenine (εA)) can effectively block RNA synthesis (4–12). Similarly, apurinic/apyrimidinic (AP) sites that lack the templating base in the transcribed DNA strongly inhibit transcription and can be slowly bypassed through nontemplated nucleotide incorporation (preferably adenine) by both Escherichia coli RNAP and Saccharomyces cerevisiae RNAP II (11–18). Other nucleotide modifications, such as 8-oxoguanine or O6-methylguanine, can be bypassed by RNAP with nucleotide misincorporation, resulting in transcriptional mutagenesis (12, 19–24).
Transient stalling of the transcription elongation complex (TEC) can lead to RNAP backtracking and disengagement of the RNA 3′-end from the active site (reviewed in Ref. 25). Such complexes can be reactivated by transcript cleavage factors (Gre factors in bacteria and TFIIS in eukaryotic RNAP II) that bind within the secondary channel/pore of RNAP and stimulate endonucleolytic cleavage of nascent RNA (26–29). Surprisingly, however, TFIIS could stimulate transcription only at nonbulky lesions (8-oxoguanine) (23, 30), whereas in several other studies, GreA and TFIIS did not stimulate or even inhibited translesion transcription by RNAP (11, 31, 32), suggesting that the effects of secondary channel factors may strongly depend on the type of the lesion.
Previous studies identified few mutations in RNAP that can affect translesion RNA synthesis. In particular, certain substitutions in the secondary channel in S. cerevisiae RNAP II (G740D) and in the bridge helix in the active site in E. coli RNAP (T790A), which decreased the elongation rate of RNAP, expectedly increased RNAP stalling at DNA lesions (7, 10, 12). In contrast, substitutions at the base of the trigger loop (TL), directly involved in catalysis (E1103G and T1095G in the largest subunit of RNAP II), stimulated translesion RNA synthesis likely by facilitating TL closure during nucleotide addition (7, 10).
RNAP stalled at DNA lesions can be directly recognized by transcription-repair coupling factors (Mfd and/or UvrD in bacteria, and CSA and CSB in eukaryotes), which further displace the TEC and bring other components of the NER system to damaged DNA (33–40). The efficiency of DNA lesion recognition by RNAP (the efficiency of TEC stalling at the damaged site) can have significant effects on subsequent DNA repair. In particular, substitutions in RNAP II that increase or decrease translesion RNA synthesis by affecting TL folding respectively decreased or increased the efficiency of transcription-coupled repair (TCR) and affected yeast cell survival after UV irradiation (7, 41).
These studies suggested that conformational changes of the TL in the active site of RNAP might be a key factor in translesion RNA synthesis on damaged DNA templates. However, the role of the TL dynamics in translesion transcription by bacterial RNAPs and its possible interplay with secondary channel factors have not been studied. Here, we demonstrate that point amino acid substitutions at nonconserved positions in the TL can have significant effects on transcription on damaged DNA by bacterial RNAP and modulate the action of secondary channel factors GreA and DksA on translesion RNA synthesis.
Results
Mutations in the TL stimulate translesion RNA synthesis
During our previous analysis of chimeric variants of E. coli RNAP containing parts from Deinococcus radiodurans, we discovered that certain substitutions significantly changed its catalytic properties (42). In particular, it was found that replacement of the whole TL stimulated translesion RNA synthesis on various damaged DNA templates.3 In this work, we demonstrate that these effects can be largely explained by substitution of just two amino acid residues in the TL, G1136M and I937T. In E. coli RNAP, these residues are located in the two α helixes when the TL is fully folded (Fig. 1, A and B) and are separated by a nonconserved 188-residue insertion (SI3, sequence insertion 3) (43), which is absent in most bacteria, including D. radiodurans (Fig. 1). Thus, further experiments were performed with E. coli RNAP variants lacking this insertion (ΔSI3) and bearing point amino acid substitutions in the TL (G1136M or I937T), in parallel with WT RNAP. In addition, we included in our analysis RNAP with the G1136Q substitution. Glutamine at this position is found in RNAPs from many bacteria lacking the SI3 insertion, including Thermus thermophilus (Fig. 1) and cyanobacteria (see Fig. S9 in Ref. 42). Recently, E. coli RNAP with the G1136Q substitution was shown to have an increased level of the intrinsic RNA cleavage activity (44), similarly to the G1136M substitution (42). Below, we present a detailed analysis of the transcription properties of these RNAP variants during RNA synthesis on normal and damaged DNA templates.
Figure 1.
Structure of the RNAP active site and conformational changes in the TL during nucleotide addition. A, structure of the TEC of T. thermophilus RNAP with partially folded TL and NTP bound in the preinsertion state (complex with streptolydigin, not shown here; Protein Data Bank code 2PPB) (75). B, structure of the TEC with fully folded TL and NTP bound in the catalytically competent conformation (Protein Data Bank code 2O5J) (75). Catalytic Mg2+ ions are shown as pink spheres; the TL and bridge helix (BH) are blue and turquoise, respectively. The positions of the G1136M and I937T substitutions are indicated (correspond to Gln1254 and Thr1243 in the T. thermophilus RNAP structure). The position of the SI3 insertion in E. coli RNAP is shown with a gray sphere (located in a disordered TL region in the preinsertion complex structure).
To reveal the effects of the mutant RNAPs on translesion RNA synthesis, we performed transcription on control and damaged DNA templates containing three modifications that were previously shown to strongly inhibit nucleotide addition by RNAP (11, 12, 16): thymine–thymine dimers (CPD), εA, and the AP site (Fig. 2). TECs containing damaged template DNA strands and a short RNA transcript, whose 3′-end was located immediately upstream of the lesions, were reconstituted in vitro from synthetic oligonucleotides with either WT or mutant core enzymes of E. coli RNAP. Transcription was performed at 37 °C for CPD and εA or at 25 °C for the AP site to increase RNAP stalling at the latter lesion. Only three NTPs were added to the reaction, resulting in the addition of up to six nucleotides to the starting RNA transcript, which allowed visualization of both RNAP stalling at the sites of lesions and read-through RNA synthesis (Fig. 2, top panels).
Figure 2.

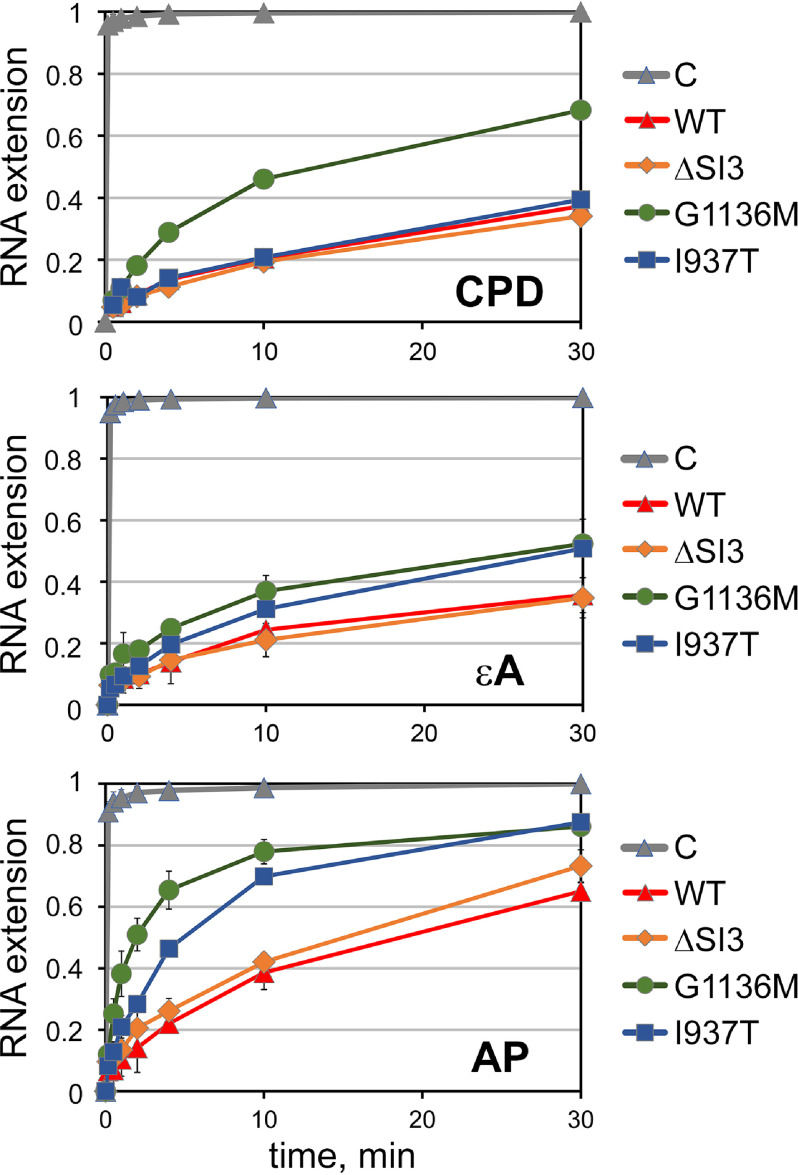
Transcription of damaged and control DNA templates by the WT and mutant RNAP variants. The structures of nucleic-acid scaffolds used for the TEC assembly are shown in the top panels. Positions of damaged nucleotides and read-through RNA transcripts are indicated. A, CPD and control TT templates (see the second replica in Fig. S2). B, εA and control A templates. C, AP site and control T templates. Transcription was performed in the presence of ATP, GTP, and UTP (100 μm each) at either 37 °C (CPD and εA) or 25 °C (AP site). The reaction times were 10 s, 30 s, 1 min, 2 min, 4 min, 10 min, and 30 min for the AP site and 30 s, 1 min, 2 min, 4 min, 10 min, 30 min, and 60 min for CPD and εA.
In the case of the control templates corresponding to the CPD and εA lesions, all analyzed RNAPs—WT, ΔSI3, I937T(ΔSI3), G1136M(ΔSI3), and G1136Q(ΔSI3)—readily extended RNA to the expected position (Fig. 2, A and B, left panels). In the case of the control reactions corresponding to the AP site, some RNAP stalling was observed after the addition of just three nucleotides together with synthesis of full-length RNA, likely as a result of lower reaction temperature in this case. However, almost all starting RNA was rapidly extended for all the templates (Fig. 2C, left panels, and 3; and Fig. S1).
Figure 3.
Kinetics of RNA extension by the WT and mutant RNAP variants opposite various DNA lesions. The efficiency of RNA extension at each time point is calculated as the ratio of all extended RNA products to the sum of all RNAs, including the starting 12-nt RNA oligonucleotide. The gray curves show the kinetics of RNA extension on control undamaged templates by WT RNAP. For both control and damaged templates, the data were normalized to the maximum RNA extension observed for corresponding undamaged templates at the 30-min time point. Means and standard deviations from two or three independent experiments are shown. For the CPD template, the calculation is shown only for the experiment from Fig. 2 (see Fig. S2 for the second replica). All kinetic plots are presented separately in Fig. S1.
DNA lesions greatly impaired RNA extension by WT E. coli RNAP, in agreement with previous reports (Fig. 2, A–C) (12, 16). For most reactions, we performed two or three independent measurements that were reproduced well (see all data points in Fig. S1). For unknown reasons, the two experiments for the CPD lesion produced different reaction kinetics. The results of these two experiments for the CPD template are therefore presented separately (Figs. 2 and 3 and Figs. S1 and S2). A single measurement was performed for the G1136Q(ΔSI3) RNAP.
As can be seen, although almost all RNA (≥80%) is already extended at the first time point (10 s) for all control templates, the extension efficiency for the damaged templates varies between <5 and 10% at 10 s and 40–60% at 30 min (Fig. 3 and Fig. S1). For all three tested lesions, strong RNAP stalling is observed in the starting TEC, just before nucleotide addition opposite the lesion. For CPD and the AP site, transcription is also blocked after first nucleotide addition, resulting in accumulation of RNA products exactly corresponding to the sites of lesions (Fig. 2).
Changes in the TL significantly affected translesion transcription. The ΔSI3 deletion did not change RNAP activity on all tested templates compared with WT RNAP (Figs. 2 and 3). The I937T(ΔSI3) substitution significantly stimulated transcription on the εA and AP templates. The G1136M(ΔSI3) and G1136Q(ΔSI3) substitutions strongly stimulated translesion RNA synthesis on the CPD and AP templates, with some stimulatory effect also observed for the εA template (Figs. 2 and 3 and Fig. S1). Although the overall rate of translesion synthesis on the CPD template by all analyzed polymerases was different in the two replicate experiments, the same stimulation of translesion synthesis was observed for the G1136M substitution (compare Figs. 2 and 3 with Fig. S2). For all mutant RNAPs, both initial nucleotide incorporation and further RNA extension were increased. This suggested that substitutions in the trigger loop allow RNAP to tolerate changes in the DNA template during nucleotide addition and also changes in the conformation of the DNA–RNA hybrid during further RNA extension.
Stimulation of translesion transcription is not associated with decreased fidelity of RNA synthesis by mutant RNAPs
Previous studies demonstrated that various DNA lesions can significantly change the fidelity of nucleotide incorporation, which may in turn affect the efficiency of further RNA extension (7, 10). We therefore analyzed whether stimulation of translesion RNA synthesis by mutations might be associated with changes in the specificity of nucleotide incorporation opposite the lesions. For each template variant, we performed transcription in the presence of each single NTP and analyzed the extension products. It was shown that WT RNAP incorporated predominantly AMP opposite first thymine of CPD (with little further extension) and the AP site and AMP or GMP opposite εA (Fig. 4). This agrees with previously published data for both bacterial RNAP and eukaryotic RNAP II (4, 5, 7, 11, 12, 14, 16, 45, 46). The ΔSI3 RNAP had similar specificity. Importantly, both G1136M(ΔSI3) and I937T(ΔSI3) RNAPs also revealed similar patterns of NTP addition, with higher overall efficiency of RNA extension by the G1136M RNAP. In particular, incorporation of the second thymine opposite CPD was markedly increased in the case of this RNAP (Fig. 4).
Figure 4.
Fidelity of nucleotide incorporation by WT and mutant RNAPs on damaged DNA templates. A, CPD template. B, εA template. C, AP template. For each template, the TECs were incubated with either all four (N) or each single NTP (A, G, U, and C; 100 μm) at 37 °C (CPD and εA) or 25 °C (AP site) for 3 min. The positions of the lesions in each template are indicated with arrowheads. The overall efficiency of RNA extension, calculated as the ratio of extended RNA products to the sum of all RNAs in the reaction normalized by RNA extension in the presence of all four NTPs, is shown below each lane (means from three independent experiments).
We further compared the fidelity of NTP addition on a control undamaged template containing adenine at the +1 position by WT and G1136M(ΔSI3) RNAPs, by measuring the kinetics of RNA extension in the presence of high concentrations of each single NTP. This allowed us to observe both correct and incorrect nucleotide incorporation, as well as extension of RNA transcripts with mismatched 3′-ends, if the first nucleotide was incorrectly incorporated. The two RNAPs demonstrated very similar patterns of RNA extension with each of the four NTPs (Fig. S3). Therefore, changes in the TL associated with increased translesion activity do not dramatically change the fidelity of nucleotide incorporation by RNAP.
Mutations in the TL suppress transcriptional pausing
Previous studies demonstrated that changes in the TL can significantly modulate the catalytic properties of RNAP and its ability to recognize various regulatory signals. In particular, the TL is important for the regulation of hairpin-dependent pausing, which is stabilized by RNA hairpin formation in the RNA exit channel of RNAP (47). This is associated with swiveling of several structural modules of RNAP and changes in the conformation of the TL and the SI3 insertion, thus preventing nucleotide addition (48, 49). Accordingly, deletion of the SI3 domain suppresses hairpin-dependent pauses (43).
To reveal possible effects of the analyzed TL mutations on transcriptional pausing, we measured t1/2 times of the well-characterized hairpin-dependent his pause for WT, ΔSI3, and G1136M(ΔSI3) RNAPs. The experiments were performed in reconstituted TECs, in which RNA hairpin formation was mimicked by the addition of a short RNA oligonucleotide complementary to the RNA transcript (Fig. 5A) (50). It was shown that the SI3 deletion reduced the pause t1/2 ∼2.8-fold (t1/2 of 20 s compared with 55 s for the WT RNAP) (Fig. 5, B and C), in line with previous reports (43). At the same time, the G1136M substitution further decreased the pause t1/2 ∼3-fold to ∼7 s (Fig. 5, B and C). Thus, this substitution can likely change the catalytic properties of the TEC and modulate its ability to pause at DNA lesions or at regulatory pause signals.
Figure 5.
Analysis of hairpin-dependent pausing by the WT, ΔSI3, and G1136M(ΔSI3) RNAPs. A, structure of the nucleic-acid scaffold used for analysis of his pausing. The positions of the starting 17-nt transcript, the paused 19-nt transcript, and the read-through 21-nt transcript are indicated. B, kinetics of RNA extension in TECs reconstituted at the his pause site. C, Quantification of the pausing kinetics. The pause t1/2 times for each RNAP are shown on the right (means from two or three independent experiments).
Effects of changes in the TL on the nucleotide addition cycle of RNAP
To assess the effects of the SI3 deletion and the G1136M substitution on individual steps in the nucleotide addition cycle, we performed parallel, time-resolved measurements of nucleotide incorporation and postcatalytic relaxation (the pretranslocated TEC formed after nucleotide incorporation relaxes into the equilibrium mixture of the pre- and post-translocated states) by ΔSI3 and G1136M(ΔSI3) RNAPs (Fig. 6 and Fig. S4). The experiments were performed using a complete synthetic nucleic-acid scaffold containing fluorescent labels at the RNA 5′-end and in the template DNA (Fig. 6A). RNA extension was monitored by a rapid chemical quench-flow method, whereas the postcatalytic relaxation was monitored by measuring the increase in fluorescence of the 6-methyl-isoxanthopterin (6-MI) base incorporated in the template DNA strand in a stopped-flow instrument (51). Combined kinetic analysis of the data revealed that ΔSI3 RNAP was only marginally faster in GMP addition and marginally slower in reaching the translocation equilibrium than the WT RNAP (Fig. 6, B and D). In contrast, G1136M(ΔSI3) RNAP was 2-fold faster than the WT enzyme in adding GMP and approached the translocation equilibrium with nearly 6-fold slower rate.
Figure 6.

Analysis of the catalytic properties of the ΔSI3 and G1136M(ΔSI3) RNAPs. A, the nucleic-acid scaffold employed in the nucleotide addition/translocation assay. The guanine analog 6MI was initially positioned in the RNA:DNA hybrid eight nucleotides upstream of the RNA 3′-end. The 6-MI fluorescence was quenched by the neighboring base pairs in the initial TEC (state 1) and the pretranslocated TEC that formed following the nucleotide incorporation (state 2) but increases when the 6-MI relocates to the edge of the RNA:DNA hybrid upon translocation (state 3). The bridge helix (BH) and the lid loop (LL) are two structural elements of the β′ subunit that flank the RNA:DNA hybrid in the multisubunit RNAPs. B, the pretranslocated state was generated by rapid GMP addition, and the apparent translocation rate (the rate of relaxation to the equilibrium) was determined from the delay between the GMP addition (discrete time points) and the translocation curve (continuous time trace) using the irreversible two step model (schematics below the graph and also D). C, the completeness of translocation was assessed by extending the TEC with 2′-dGTP or 3′-dGTP in the presence of 0.5 mm CTP. The individual forward and backward translocation rates were then determined from the relaxation rate using the relationships presented below the graph. D, the kinetic parameters of the nucleotide addition cycle determined from the data in B and C.
To assess the completeness of translocation, we extended the TECs with 3′-dGMP (RNA chain terminator) and forward-biased the 3′-dGMP-extended TECs with CTP (the next incoming NTP in our system). The extension of the ΔSI3 TEC with 3′-dGMP in the presence of 0.5 mm CTP resulted in the same fluorescence intensity as that following the extension with GMP (Fig. 6C). Furthermore, the same fluorescence intensity was observed after the extension of the TEC with 2'-dGMP, which promotes forward translocation because the 2′-OH group at the RNA 3′-end is essential for stabilizing the pretranslocated state (52). These observations collectively suggested that the fraction of the pretranslocated state was below the detection threshold (∼10%) in the ΔSI3 TEC, similarly to the situation in the WT RNAP (42, 52, 53). In contrast, the fluorescence intensity of the G1136M(ΔSI3) TEC extended with GMP was ∼40% lower than that observed upon the extension of the TEC by 2'dGMP or by 3′-dGMP in the presence of 0.5 mm CTP (Fig. 6C). These observations suggested that ∼40% of the β′G1136M(ΔSI3) TEC was in the pretranslocated state and ∼60% was in the post-translocated sate after the equilibrium between the translocation states was established.
Next, the knowledge of the relaxation rates toward the equilibrium (Fig. 6B) and the equilibrium fractions of the pre- and post-translocated states (Fig. 6C) allowed for the inference of the forward and backward translocation rates. Following the rules of the formal kinetics, the relaxation rate is the sum of the forward and backward translocation rates, whereas the ratio of the forward and backward translocation rates equals the ratio of the fractions of the post- and pretranslocated states. In the case of G1136M(ΔSI3) RNAP, 60% of the relaxation rate corresponds to the forward (8 s−1), and 40% corresponds to the backward (5 s−1) translocation rate (Fig. 6D). Our failure to detect the pretranslocation state in the ΔSI3 (Fig. 6C) or the WT (42, 52, 53) TECs suggests that the relaxation rate approximately corresponds to the forward translocation rate for those RNAPs. We additionally assumed that the WT, ΔSI3, and G1136M(ΔSI3) TECs all have the same backward translocation rate (Fig. 6D). In doing so we reasoned that the backward translocation rate is the property of the post-translocated state in which the RNA 3′-end is positioned far away from the TL irrespective of the TL conformation. Therefore, the backward translocation rate is unlikely affected by the alterations in the TL. This assumption is further supported by the observation that we obtained the same estimate for the backward translocation rate (5 s−1) for three different RNAPs when using the same GMP incorporating TEC (Fig. 6A): β′G1136M(ΔSI3) (this work), β′F773V, and β′P750L (53).
Mutations in the TL decrease Km for nucleotide substrates on damaged templates
Our analysis revealed that the catalytic properties of the G1136M(ΔSI3) RNAP are significantly different from the control WT or ΔSI3 enzymes, with a higher rate of nucleotide incorporation, a reduced rate of forward translocation, and, as a result, an increased fraction of the pretranslocated state observed for the mutant RNAP (Fig. 6) (42). This suggested that the mutations in the TL might affect its folding and, possibly, incoming NTP binding during transcription of damaged DNA templates (see “Discussion”). To test this possibility, we compared apparent Km values for nucleotide incorporation on the CPD and AP site templates for ΔSI3 and G1136M(ΔSI3) RNAPs. We also performed this analysis for the I937T(ΔSI3) RNAP that also stimulated translesion synthesis (see above).
For the ΔSI3 RNAP, the Km,app value for nucleotide incorporation on the CPD template was 560 ± 110 μM (Fig. 7), significantly higher than previously reported Km values for undamaged templates (54, 55). For the G1136M(ΔSI3) RNAP, this value was decreased to 120 ± 15 μM (Fig. 7), significantly lower than in the case of the control RNAP (p < 0.05). The I937T substitution also decreased the Km,app, but the effect was not significant (385 ± 40 μm, p = 0.10).
Figure 7.

Determination of the apparent Km values for NTP substrates on the CPD (A) and AP site (B) templates. The reactions were performed in reconstituted TECs formed with control or damaged templates with ΔSI3 or mutant G1137M(ΔSI3) and I937T(ΔSI3) RNAPs at increasing NTP concentrations. The efficiency of RNA extension was calculated and normalized to the maximum efficiency observed at the highest NTP concentration, and the data were fitted to a hyperbolic equation (means and standard deviations from three independent experiments).
For the AP site template, the Km,app for the ΔSI3 RNAP was 360 ± 20 μM and was decreased more than 3-fold for the G1136M mutant (100 ± 15 μm, p < 0.05). A similar but nonsignificant effect was observed for the I937T RNAP (250 ± 35 μm, p = 0.12). This suggested that increased translesion synthesis by the mutant RNAPs may at least in part be explained by stimulation of the incoming nucleotide binding in the active site of RNAP.
Secondary channel factors inhibit translesion RNA synthesis depending on mutations in the TL
Recently, we observed that secondary channel factors of the extremophilic bacterium D. radiodurans, GreA and Gfh1, can significantly increase TEC stalling at the sites of DNA lesions, acting on D. radiodurans RNAP (11). In comparison, E. coli GreA had a smaller inhibitory effect on translesion transcription by E. coli RNAP (11). We showed that E. coli GreA did not stimulate but inhibited RNA extension also by the ΔSI3 RNAP (Fig. S5). Similarly, the G1136M(ΔSI3) RNAP was not stimulated by GreA but still had a higher level of translesion activity compared with the control ΔSI3 RNAP (Fig. S5).
In addition to Gre factors, E. coli contains another secondary channel regulator, DksA, that together with small alarmone ppGpp regulates transcription initiation by destabilizing promoter complexes (56). DksA and ppGpp were also proposed to regulate transcription elongation, in particular, by decreasing nucleotide misincorporation and preventing RNAP conflicts with DNA replication (57, 58). The ability of DksA to interact with actively transcribing RNAP, however, can be inhibited by the SI3 insertion in the TL, which partially occupies the binding site of DksA in the secondary channel. It was therefore proposed that DksA might be involved in the regulation of RNAP activity at specific sites during RNA elongation, possibly including DNA lesions (59, 60).
To test this hypothesis directly, we compared translesion transcription by the WT, ΔSI3, and G1136M(ΔSI3) RNAPs in the absence and in the presence of DksA and ppGpp, using the CPD template (Fig. 8). It was found that the addition of DksA/ppGpp indeed decreased translesion transcription by the ΔSI3 RNAP; in particular, the efficiency of overall RNA extension after 30 min was decreased from ∼60% to ∼30% (Fig. 8, A and B; the fraction of nonextended RNA is shown in blue). Similarly, DksA alone could inhibit translesion synthesis by the ΔSI3 RNAP but not WT RNAP (Fig. S6), suggesting that the SI3 domain indeed “gates” the secondary RNAP channel against binding of DksA (60). The inhibitory effect of DksA/ppGpp on RNA extension was partially suppressed by the G1136M(ΔSI3) substitution (Fig. 8B and Fig. S6, bottom panels). Thus, DksA/ppGpp can modulate the efficiency of translesion transcription, depending on changes in the structure of the TL in the active site of RNAP.
Figure 8.
Effects of DksA and ppGpp on RNA synthesis on the CPD template. A, analysis of the RNA extension products. The reactions were performed with either ΔSI3 or G1136M(ΔSI3) RNAPs for 10 s 1 min, 5 min, or 30 min at 37 °C; DksA and ppGpp were added to 2 and 200 μm, respectively. Positions of damaged nucleotides are indicated with arrowheads. B, quantification of the RNA extension efficiencies. For each time point, the relative amounts of starting RNA, RNA transcripts corresponding to the sites of lesion, and read-through RNAs are shown. The data are the means and standard deviations from three independent experiments.
Effects of RNAP mutations on cell viability
Previous studies of eukaryotic RNAP II performed in S. cerevisiae demonstrated that RNAP mutations affecting translesion transcription in vitro also change cell sensitivity to DNA damage in vivo, leading to changes in cell survival after UV irradiation (7, 41). Because the G1136M (and, similarly, G1136Q) substitution significantly affected translesion RNA synthesis, we tested whether it could also have any effects on cell viability in E. coli strains transformed with plasmids encoding different β′ subunit variants.
In the first experiment, the ΔSI3 or G1136M(ΔSI3) β′ variants were expressed from pBAD vectors in E. coli MG1655 in the presence of arabinose; control experiments were performed without the addition of arabinose. After the cells were grown in liquid medium for identical times, they were or were not UV-irradiated, and the number of viable cells in each experiment was determined by serial dilutions on LB agar plates (Fig. 9). Expression of the ΔSI3 β′ subunit decreased bacterial titer by 1–2 orders of magnitude in the absence of UV irradiation (Fig. 9, compare first and third rows) and had even a stronger effect after UV irradiation (Fig. 9, fifth and seventh rows), suggesting that the SI3 domain is important for cell viability, in agreement with published data (43). Expression of the β′ subunit containing the G1136M(ΔSI3) substitution had even stronger effects on cell survival. Without UV irradiation, the numbers of cells were significantly reduced (by 1–2 orders of magnitude) compared with the ΔSI3 variant with no substitution (Fig. 9, fourth row). No cell growth was detected for the G1136M(ΔSI3) RNAP after UV exposure (Fig. 9, eighth row). After UV irradiation, the presence of the G1136M(ΔSI3)-encoding plasmid also decreased cell viability compared with the ΔSI3 plasmid even without the addition of arabinose, suggesting that the leaky RNAP expression in the absence of the inductor is sufficient for toxicity under stress conditions (Fig. 9, compare the fifth and sixth rows). Therefore, the G1136M(ΔSI3) substitution appears to be toxic for cells when the mutant β′ subunit is expressed in either normal or stress conditions, even despite the presence of the WT rpoC gene constantly expressed in the same strains.
Figure 9.
Effects of the mutant rpoC alleles on cell viability in the MG1655 strain. Expression of the β′ subunit variants, ΔSI3 and G1136M(ΔSI3), was induced from a pBAD-based vector by arabinose, and the cells were grown under identical conditions and plated with serial dilutions, either without or with UV irradiation.
In the second experiment, we compared the effects on cell growth of WT or mutant RNAP variants expressed from pVS10-based vectors encoding all core RNAP subunits (61). E. coli BL21(DE3) was transformed with plasmids encoding WT, ΔSI3, G1136M(ΔSI3), or G1136Q(ΔSI3) RNAP variants or a control ampicillin-resistance plasmid lacking RNAP genes (pBR322) and grown at 30 or 42 °C. In the case of the WT RNAP, no effects on cell growth were observed at either temperature (Fig. S7, compare first and second rows). For the ΔSI3, G1136M(ΔSI3), or G1136Q(ΔSI3) RNAPs, bacterial titer was decreased ∼10-fold at 30 °C (Fig. S7, left panel, third, fourth, and fifth rows). The effects of the G1136M and G1136Q substitutions on cell titer became stronger at 42 °C (right panel), further suggesting that the mutations have detrimental effects on cell growth under stress conditions.
Discussion
RNAP is the key participant in NER in the transcribed genomic regions. During transcription, it directly senses DNA lesions, stalls, and recruits repair enzymes to the sites of DNA damage (1–3, 33, 37). On the other hand, the ability of RNAP to transcribe through DNA lesions—translesion RNA synthesis (TLS)—may be important for cell survival under stress conditions, as shown for yeast RNAP II mutants with changed TLS activities (7, 41). The balance between RNAP stalling and readthrough synthesis is therefore essential for both efficient DNA repair and transcription, producing full-length RNA products and avoiding conflicts with DNA replication (58, 62). However, only a handful of studies aimed at understanding of these relationships have been performed to date, primarily in eukaryotic models, whereas the features of bacterial RNAP that are important for transcription of damaged DNA and may determine the balance between TLS and TCR have remained unknown.
Here, we show that changes in the key element of the active site—the TL that encloses the incoming NTP substrate during catalysis (Fig. 1)—have profound effects on the nucleotide addition cycle and the TLS activity of bacterial RNAP. In particular, the G1136M substitution in the C-terminal α-helix of the TL changes the rates of nucleotide addition and RNAP translocation, decreases RNAP pausing, stimulates translesion synthesis, and decreases RNAP sensitivity to the secondary channel factors. Similarly, translesion synthesis is stimulated by the G1136Q substitution at the same position. At the same time, deletion of the large SI3 domain in the TL, characteristic for E. coli and related bacteria, has much milder effects on transcription, suggesting that SI3 functions may be restricted to specific regulatory signals and transcription factors. Below, we discuss these findings in light of available biochemical and structural data.
Comparison of nucleotide addition and translocation by the WT, ΔSI3, and G1136M(ΔSI3) RNAPs revealed that the latter enzyme is markedly different from the former two. The G1136M(ΔSI3) RNAP has a 2-fold higher rate of nucleotide incorporation, a 7-fold reduced rate of forward translocation, and a markedly elevated fraction of the pretranslocated state under equilibrium conditions. As a result, processive transcript elongation by the G1136M(ΔSI3) RNAP is likely limited by the rate of forward translocation, similarly to previously characterized pause-resistant β′F773V, β′P750L E. coli RNAPs (53), and Rpb1-E1103G S. cerevisiae RNAPII (63–65). We further argue that the intrinsically stabilized helical conformation of the TL is likely behind the slow forward translocation rate and the insensitivity to pauses of G1136M(ΔSI3) RNAP, as has been proposed earlier for β′F773V, β′P750L, and Rpb1-E1103G RNAPs.
Interestingly, the decrease in the rates of postcatalytic relaxation and forward translocation observed for the G1136M(ΔSI3) RNAP does not result in a net decrease in the average rate of elongation for this RNAP on either normal (42) or damaged DNA templates (this work). Thus, other factors, such as improved nucleotide binding in the active site and decreased pausing, likely compensate for the observed changes in the catalytic cycle in the G1136M(ΔSI3) RNAP. The effects of several other mutations in the TL, including substitution G1136S in E. coli RNAP, on the rate of nucleotide incorporation were shown to correlate with their effects on transcriptional pausing (47, 66); however, their influence on the postcatalytic relaxation and RNAP translocation remains to be tested.
In contrast to the G1136M substitution, deletion of the SI3 domain only marginally affects the rates of nucleotide addition and translocation, despite its location in the flexible part of the TL that changes its conformation during catalysis (Fig. 1). The largely unaffected forward translocation suggests that the helical conformation of TL is not intrinsically stabilized by the SI3 deletion. The insensitivity of the ΔSI3 RNAP to some pause sites (43, 67–69) thus likely originates from stabilization of the helical TL only at a subset of sequence positions where RNAP adopts a swiveled conformation and the TL folding is inhibited because of the clash of the SI3 domain with the β-lobe domain (48). Overall, the insertion of the SI3 domain in the TL has relatively little effect on the catalytic activity of RNAP, and its primary function may be in gating secondary channel factors toward the active site in various types of TECs and in stimulation of hairpin-dependent pausing (48, 49, 59, 60).
We further show that mutations in the TL in bacterial RNAP can stimulate translesion transcription, likely by affecting the folding dynamics of the TL during nucleotide addition and translocation. In particular, stabilization of the helical TL by the analyzed amino acid substitutions may facilitate NTP binding in the active site on damaged DNA templates. Indeed, the G1136M (and, to some extent, I937T) substitution was shown to decrease Km,app for NTPs on the CPD and AP site templates severalfold. Furthermore, the ability of the G1136M substitution to also decrease hairpin-dependent pausing suggests a functional connection between transcriptional pausing at site-specific pause signals and at DNA lesions.
No structural information on transcription of damaged DNA by bacterial RNAP is available to date. However, analysis of the structures of TECs of eukaryotic RNAP II stalled at several DNA lesions demonstrated that the lesion can either result in nucleotide mispairing (8-oxoguanine or 5-carboxycytosine) (21, 70), impair translocation of the damaged nucleotide into the active site (cisplatin, CPD) (5–7, 10), or induce RNAP stalling prior to lesion by forming unfavorable interactions within the downstream DNA-binding channel (bulky minor groove adducts) (71). In particular, CPD in the template strand prevents its translocation in the active site, resulting in nontemplated incorporation of A against the first T and preferable misincorporation of U against the second thymine (5, 7). Similarly to our findings, mutations in the active site of eukaryotic RNAP II that favor TL folding (E1103G and T1095G in Rpb1) also increase bypass of CPD, possibly by increasing NTP retention in the active site and thus promoting its incorporation into the nascent RNA, even in the absence of the templating base. In contrast, the G730D substitution in RNAP II, which decreases the rate of transcription, dramatically increases transcriptional stalling at CPD, likely by impairing nucleotide addition (7, 10). Similar effects of substitutions that stabilize the folded TL conformation on the ability of E. coli and S. cerevisiae RNAPs to transcribe damaged DNA suggest a common mechanism of TLS in the bacterial and eukaryotic systems.
In eukaryotic RNAP II, changes in the efficiency of lesion bypass strongly correlate with the efficiency of TCR and cell survival in vivo, with stronger stalling associated with stronger TCR but decreased viability (7, 41). Interestingly, the expression of G1136M (and G1136Q) RNAP in E. coli had toxic effects on cell survival, which were exacerbated under UV irradiation or at elevated temperature. This could in part result from the SI3 domain deletion in the mutant RNAP variants, which by itself inhibits cell growth (Fig. 9 and Fig. S7 (43). At the same time, the G1136M substitution was more toxic than the deletion of the SI3 domain, even under DNA damaging conditions, suggesting that its positive effects on TLS could not compensate for its possible negative effects on the recognition of regulatory pause signals and RNAP interactions with regulatory factors in vivo. It remains to be tested whether the I937T substitution could also affect cell survival, whether the in vivo effects of the mutations in the TL might be associated with changes in the level of RNAP expression, and how these effects could be modulated by the SI3 domain insertion.
Finally, we show that secondary channel factors, GreA and DksA/ppGpp, can inhibit translesion synthesis by E. coli RNAP, depending on the TL structure. Previously, the G1136M substitution was found to increase the rate of intrinsic RNA cleavage by E. coli RNAP (42); this potentially might facilitate translesion RNA synthesis by increasing the efficiency of RNA proofreading at the sites of lesions. However, the inability of GreA to stimulate translesion synthesis by both control and mutant RNAPs, as well as the absence of additional cleavage products during transcription of damaged templates by the G1136M(ΔSI3) RNAP, argues against this possibility.
In agreement with previous data obtained with undamaged DNA templates (59, 60), the inhibitory effect of DksA on translesion synthesis is increased when the SI3 domain is deleted. Furthermore, the increased activity of the G1136M(ΔSI3) RNAP in the presence of GreA and DksA/ppGpp, which both inhibit translesion synthesis by control RNAP more efficiently, suggest that in the case of the mutant RNAP, stabilized TL folding may compete with binding of these factors within the secondary channel. Transcription factors and regulatory signals might therefore modulate the translesion activity of RNAP by affecting the position and conformation of the TL and the SI3 domain and changing their interactions with GreA and DksA (26, 72). Thus, regulatory factors and inhibitors affecting TL dynamics may significantly change the properties of transcribing RNAP and its sensitivity to DNA lesions, providing an opportunity for development of novel antibacterial compounds that would modulate translesion RNA synthesis by RNAP.
Experimental procedures
Proteins
WT E. coli core RNAP and its mutant variants with deletion of the SI3 domain (β′Δ945-1132) and with the G1136M, G1136Q, and I937T substitutions were obtained expressed from pVS10-based plasmids, encoding all core RNAP subunits, by autoinduction as described previously (42, 61). GreA and DksA were expressed and purified from E. coli (42, 73).
Transcription in vitro
Synthetic DNA oligonucleotides containing modified bases were purchased from TriLink BioTechnologies (San Diego, CA). TECs containing control or modified DNA templates were reconstituted from DNA and RNA oligonucleotides (Fig. 2A) and E. coli core RNAP as described previously (11, 12). For the CPD and εA templates, 5′-P32-labeled RNA was mixed with the template and nontemplate DNA oligonucleotides (final concentrations, 0.5, 1, and 5 μm) in transcription buffer (40 mm Tris-HCl, pH 7.9, 40 mm NaCl, 10 mm MgCl2) at 65 °C and cooled down to 20 °C at 1 °C/min. For the AP site, the complex was assembled for 10 min at 37 °C. The annealed templates were diluted with the transcription buffer to 10 nm RNA concentration, and core RNAP was added to 25 nm. Then the samples were incubated for 10 min at 37 °C for TEC formation and transferred to the required temperature, and NTPs were added to 100 μM. When indicated, GreA, DksA (final concentration, 2 μm) and ppGpp (200 μm) were added 5 min prior to NTP addition. The reactions were stopped after the indicated time intervals, and RNA products were analyzed by 23% denaturing PAGE, followed by phosphorimaging (Typhoon 9500, GE Healthcare).
For analysis of hairpin-dependent pausing, TECs were reconstituted from synthetic RNA (5′-P32-labeled) and DNA oligonucleotides shown in Fig. 5A as previously described (50, 74). NTP substrates (2 μm GTP and 100 μm UTP and CTP) were added at 30 °C, and the reactions were terminated after increasing time intervals (4, 10, 20, 40, 90, and 180 s), followed by PAGE analysis of RNA products. Pause efficiencies at each time point were calculated as the ratio of the paused RNA product (minus the background value) to the sum of the paused and read-through products. Observed rate constants (kobs) for the pause escape were calculated by fitting the data to a single-exponential equation (P = Pmax × exp(−kobs × t), where P is the pausing efficiency), and the reaction half-times were determined (t1/2 = ln2/kobs).
For measurements of the apparent Km values for nucleotide substrates, the reactions were performed in reconstituted TECs formed with CPD or AP site templates at increasing NTP concentrations (ATP, GTP, and UTP) for 1 min at 37 °C in the case of the CPD template and for 10 s at 25 °C in the case of the AP template. The efficiency of RNA extension was calculated for each reaction point and normalized to the maximum efficiency observed at the highest NTP concentration. To determine apparent Km values, the data were fitted to a hyperbolic equation.
Kinetics of nucleotide addition and RNA elongation
TECs for analysis of nucleotide addition and translocation were assembled on a scaffold containing fluorescently labeled RNA and DNA oligonucleotides (Fig. 6). DNA and RNA oligonucleotides were purchased from IBA Biotech (Göttingen, Germany), Fidelity Systems (Gaithersburg, MD, USA), and Eurofins Genomics (Ebersberg, Germany). The nucleotide addition rates were measured after mixing the TEC with 400 μm GTP (final concentration, 200 μm) in a rapid quench-flow instrument (RQF 3, KinTek Corporation) at 25 °C as described previously (42). The reactions were quenched with 0.5 m HCl, and the RNA products were separated by 16% denaturing PAGE and analyzed by fluorimetry. Equilibrium levels of fluorescence were determined by recording emission spectra of 6-MI (excitation at 340 nm) with an LS-55 spectrofluorometer (PerkinElmer Life Sciences) at 25 °C. The fluorescence at peak emission wavelength (420 nm) was used for data analysis and representation. Time-resolved translocation measurements were performed in an Applied Photophysics (Leatherhead, UK) SX.18 MV stopped-flow instrument at 25 °C as described previously (42). The reaction was initiated by mixing the TEC with 400 μm GTP (final concentration, 200 μm). The detailed descriptions of the experimental conditions and the analysis routines are presented in the supporting text.
In vivo assays of cell viability
For experiments in Fig. 9, E. coli MZ1655 Z1 was transformed with pBAD-based plasmids containing either WT (ΔSI3) or mutant G1136M(ΔSI3) alleles of the rpoC gene under the control of arabinose-inducible promoter. The cells were grown under identical conditions from overnight cultures in liquid LB with ampicillin (100 μg/ml) and spectinomycin (25 μg/ml), either without or with 0.1% arabinose, for 9 h at 37 °C and plated with serial dilutions onto LB plates (300 μg/ml ampicillin, 100 μg/ml spectinomycin, either without or with 0.1% arabinose). The plates were or were not irradiated with UV (0.007 J) and incubated in the dark overnight at 37 °C.
For experiments in Fig. S7, E. coli BL21(DE3) was transformed with pBR322 or pVS10-based plasmids containing all core RNAP subunits, with WT or mutant rpoC alleles. Night cultures were grown from fresh colonies overnight in liquid LB with ampicillin (100 μg/ml), inoculated into fresh LB with ampicillin, and grown for 8 h. Serial dilutions were prepared and plated onto LB agar plates with ampicillin, and the plates were incubated at 30 or 42 °C for 24 and 16 h, respectively.
Data availability
All data presented are contained in the article and are available upon request from A. Kulbachinskiy (akulb@img.ras.ru) and D. Esyunina (es_dar@inbox.ru).
Supplementary Material
Acknowledgments
We thank Irina Artsimovitch for plasmids and modified DNA oligonucleotides.
This article contains supporting information.
Author contributions—A. A., A. I., M. T., G. B., D. E., and A. K. formal analysis; A. A., A. I., M. T., and D. E. investigation; A. A. and G. B. methodology; G. B., D. E., and A. K. conceptualization; G. B., D. E., and A. K. supervision; G. B., D. E., and A. K. writing-review and editing; A. K. funding acquisition; A. K. writing-original draft.
Funding and additional information—This work was supported by Russian Science Foundation Grant 17-14-01393 (analysis of translesion transcription) and Academy of Finland Grant 286205 (analysis of nucleotide addition cycle).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
D. Esyunina, A. Ignatov, and A. Kulbachinskiy, unpublished observations.
- RNAP
- RNA polymerase
- NER
- nucleotide excision repair
- TCR
- transcription-coupled repair
- TEC
- transcription elongation complex
- TL
- trigger loop
- CPD
- cyclobutane pyrimidine dimer
- AP
- apurinic/apyrimidinic site
- εA
- 1,N6-ethenoadenine
- 6-MI
- 6-methyl-isoxanthopterin
- TLS
- translesion RNA synthesis
- nt
- nucleotide.
References
- 1. Spivak G. (2016) Transcription-coupled repair: an update. Arch. Toxicol. 90, 2583–2594 10.1007/s00204-016-1820-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strick T. R., and Portman J. R. (2019) Transcription-coupled repair: from cells to single molecules and back again. J. Mol. Biol. 431, 4093–4102 10.1016/j.jmb.2019.05.040 [DOI] [PubMed] [Google Scholar]
- 3. Pani B., and Nudler E. (2017) Mechanistic insights into transcription coupled DNA repair. DNA Repair 56, 42–50 10.1016/j.dnarep.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donahue B. A., Yin S., Taylor J. S., Reines D., and Hanawalt P. C. (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl. Acad. Sci. U.S.A. 91, 8502–8506 10.1073/pnas.91.18.8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brueckner F., Hennecke U., Carell T., and Cramer P. (2007) CPD damage recognition by transcribing RNA polymerase II. Science 315, 859–862 10.1126/science.1135400 [DOI] [PubMed] [Google Scholar]
- 6. Damsma G. E., Alt A., Brueckner F., Carell T., and Cramer P. (2007) Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat. Struct. Mol. Biol. 14, 1127–1133 10.1038/nsmb1314 [DOI] [PubMed] [Google Scholar]
- 7. Walmacq C., Cheung A. C., Kireeva M. L., Lubkowska L., Ye C., Gotte D., Strathern J. N., Carell T., Cramer P., and Kashlev M. (2012) Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol. Cell 46, 18–29 10.1016/j.molcel.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen S. E., Lewis C. A., Mooney R. A., Kohanski M. A., Collins J. J., Landick R., and Walker G. C. (2010) Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 15517–15522 10.1073/pnas.1005203107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dimitri A., Goodenough A. K., Guengerich F. P., Broyde S., and Scicchitano D. A. (2008) Transcription processing at 1,N2-ethenoguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. J. Mol. Biol. 375, 353–366 10.1016/j.jmb.2007.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walmacq C., Wang L., Chong J., Scibelli K., Lubkowska L., Gnatt A., Brooks P. J., Wang D., and Kashlev M. (2015) Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions. Proc. Natl. Acad. Sci. U.S.A. 112, E410–E419 10.1073/pnas.1415186112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agapov A., Esyunina D., and Kulbachinskiy A. (2019) Gre-family factors modulate DNA damage sensing by Deinococcus radiodurans RNA polymerase. RNA Biol. 16, 1711–1720 10.1080/15476286.2019.1656027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pupov D., Ignatov A., Agapov A., and Kulbachinskiy A. (2019) Distinct effects of DNA lesions on RNA synthesis by Escherichia coli RNA polymerase. Biochem. Biophys. Res. Commun. 510, 122–127 10.1016/j.bbrc.2019.01.062 [DOI] [PubMed] [Google Scholar]
- 13. Clauson C. L., Oestreich K. J., Austin J. W., and Doetsch P. W. (2010) Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 3657–3662 10.1073/pnas.0913191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuraoka I., Endou M., Yamaguchi Y., Wada T., Handa H., and Tanaka K. (2003) Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 278, 7294–7299 10.1074/jbc.M208102200 [DOI] [PubMed] [Google Scholar]
- 15. Tornaletti S., Maeda L. S., and Hanawalt P. C. (2006) Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 19, 1215–1220 10.1021/tx060103g [DOI] [PubMed] [Google Scholar]
- 16. Zhou W., and Doetsch P. W. (1993) Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 90, 6601–6605 10.1073/pnas.90.14.6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou W., and Doetsch P. W. (1994) Efficient bypass and base misinsertions at abasic sites by prokaryotic RNA polymerases. Ann. N.Y. Acad. Sci. 726, 351–354 10.1111/j.1749-6632.1994.tb52849.x [DOI] [PubMed] [Google Scholar]
- 18. Wang W., Walmacq C., Chong J., Kashlev M., and Wang D. (2018) Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 115, E2538–E2545 10.1073/pnas.1722050115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brégeon D., Doddridge Z. A., You H. J., Weiss B., and Doetsch P. W. (2003) Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol. Cell 12, 959–970 10.1016/S1097-2765(03)00360-5 [DOI] [PubMed] [Google Scholar]
- 20. Brégeon D., Peignon P. A., and Sarasin A. (2009) Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 5, e1000577 10.1371/journal.pgen.1000577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Damsma G. E., and Cramer P. (2009) Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J. Biol. Chem. 284, 31658–31663 10.1074/jbc.M109.022764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dimitri A., Burns J. A., Broyde S., and Scicchitano D. A. (2008) Transcription elongation past O6-methylguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. Nucleic Acids Res. 36, 6459–6471 10.1093/nar/gkn657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuraoka I., Suzuki K., Ito S., Hayashida M., Kwei J. S., Ikegami T., Handa H., Nakabeppu Y., and Tanaka K. (2007) RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair 6, 841–851 10.1016/j.dnarep.2007.01.014 [DOI] [PubMed] [Google Scholar]
- 24. Viswanathan A., and Doetsch P. W. (1998) Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem. 273, 21276–21281 10.1074/jbc.273.33.21276 [DOI] [PubMed] [Google Scholar]
- 25. Nudler E. (2012) RNA polymerase backtracking in gene regulation and genome instability. Cell 149, 1438–1445 10.1016/j.cell.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdelkareem M., Saint-Andre C., Takacs M., Papai G., Crucifix C., Guo X., Ortiz J., and Weixlbaumer A. (2019) Structural basis of transcription: RNA polymerase backtracking and its reactivation. Mol. Cell 75, 298–309 10.1016/j.molcel.2019.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheung A. C., and Cramer P. (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253 10.1038/nature09785 [DOI] [PubMed] [Google Scholar]
- 28. Laptenko O., Lee J., Lomakin I., and Borukhov S. (2003) Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 22, 6322–6334 10.1093/emboj/cdg610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sekine S., Murayama Y., Svetlov V., Nudler E., and Yokoyama S. (2015) The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol. Cell 57, 408–421 10.1016/j.molcel.2014.12.014 [DOI] [PubMed] [Google Scholar]
- 30. Charlet-Berguerand N., Feuerhahn S., Kong S. E., Ziserman H., Conaway J. W., Conaway R., and Egly J. M. (2006) RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 25, 5481–5491 10.1038/sj.emboj.7601403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng T. F., Hu X., Gnatt A., and Brooks P. J. (2008) Differential blocking effects of the acetaldehyde-derived DNA lesion N2-ethyl-2′-deoxyguanosine on transcription by multisubunit and single subunit RNA polymerases. J. Biol. Chem. 283, 27820–27828 10.1074/jbc.M804086200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu L., Wang W., Wu J., Shin J. H., Wang P., Unarta I. C., Chong J., Wang Y., and Wang D. (2017) Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 114, E7082–E7091 10.1073/pnas.1708748114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park J. S., Marr M. T., and Roberts J. W. (2002) E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767 10.1016/S0092-8674(02)00769-9 [DOI] [PubMed] [Google Scholar]
- 34. Deaconescu A. M., Chambers A. L., Smith A. J., Nickels B. E., Hochschild A., Savery N. J., and Darst S. A. (2006) Structural basis for bacterial transcription-coupled DNA repair. Cell 124, 507–520 10.1016/j.cell.2005.11.045 [DOI] [PubMed] [Google Scholar]
- 35. Smith A. J., and Savery N. J. (2008) Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair 7, 1670–1679 10.1016/j.dnarep.2008.06.020 [DOI] [PubMed] [Google Scholar]
- 36. Le T. T., Yang Y., Tan C., Suhanovsky M. M., Fulbright R. M. Jr., Inman J. T., Li M., Lee J., Perelman S., Roberts J. W., Deaconescu A. M., and Wang M. D. (2018) Mfd dynamically regulates transcription via a release and catch-up mechanism. Cell 172, 344–357.e15 10.1016/j.cell.2017.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Epshtein V., Kamarthapu V., McGary K., Svetlov V., Ueberheide B., Proshkin S., Mironov A., and Nudler E. (2014) UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature 505, 372–377 10.1038/nature12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamarthapu V., Epshtein V., Benjamin B., Proshkin S., Mironov A., Cashel M., and Nudler E. (2016) ppGpp couples transcription to DNA repair in E. coli. Science 352, 993–996 10.1126/science.aad6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fousteri M., and Mullenders L. H. (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18, 73–84 10.1038/cr.2008.6 [DOI] [PubMed] [Google Scholar]
- 40. Xu J., Lahiri I., Wang W., Wier A., Cianfrocco M. A., Chong J., Hare A. A., Dervan P. B., DiMaio F., Leschziner A. E., and Wang D. (2017) Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657 10.1038/nature24658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W., Selvam K., Ko T., and Li S. (2014) Transcription bypass of DNA lesions enhances cell survival but attenuates transcription coupled DNA repair. Nucleic Acids Res. 42, 13242–13253 10.1093/nar/gku1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Esyunina D., Turtola M., Pupov D., Bass I., Klimasauskas S., Belogurov G., and Kulbachinskiy A. (2016) Lineage-specific variations in the trigger loop modulate RNA proofreading by bacterial RNA polymerases. Nucleic Acids Res. 44, 1298–1308 10.1093/nar/gkv1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Artsimovitch I., Svetlov V., Murakami K. S., and Landick R. (2003) Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 278, 12344–12355 10.1074/jbc.11214200 [DOI] [PubMed] [Google Scholar]
- 44. Riaz-Bradley A., James K., and Yuzenkova Y. (2020) High intrinsic hydrolytic activity of cyanobacterial RNA polymerase compensates for the absence of transcription proofreading factors. Nucleic Acids Res. 48, 1341–1352 10.1093/nar/gkz1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Selby C. P., Drapkin R., Reinberg D., and Sancar A. (1997) RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25, 787–793 10.1093/nar/25.4.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kalogeraki V. S., Tornaletti S., Cooper P. K., and Hanawalt P. C. (2005) Comparative TFIIS-mediated transcript cleavage by mammalian RNA polymerase II arrested at a lesion in different transcription systems. DNA Repair 4, 1075–1087 10.1016/j.dnarep.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 47. Zhang J., Palangat M., and Landick R. (2010) Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 17, 99–104 10.1038/nsmb.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kang J. Y., Mishanina T. V., Bellecourt M. J., Mooney R. A., Darst S. A., and Landick R. (2018) RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815 10.1016/j.molcel.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang J. Y., Mishanina T. V., Landick R., and Darst S. A. (2019) Mechanisms of transcriptional pausing in bacteria. J. Mol. Biol. 431, 4007–4029 10.1016/j.jmb.2019.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kolb K. E., Hein P. P., and Landick R. (2014) Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 289, 1151–1163 10.1074/jbc.M113.521393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malinen A. M., Turtola M., and Belogurov G. A. (2015) Monitoring translocation of multisubunit RNA polymerase along the DNA with fluorescent base analogues. Methods Mol. Biol. 1276, 31–51 10.1007/978-1-4939-2392-2_3 [DOI] [PubMed] [Google Scholar]
- 52. Malinen A. M., Turtola M., Parthiban M., Vainonen L., Johnson M. S., and Belogurov G. A. (2012) Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 40, 7442–7451 10.1093/nar/gks383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Malinen A. M., Nandymazumdar M., Turtola M., Malmi H., Grocholski T., Artsimovitch I., and Belogurov G. A. (2014) CBR antimicrobials alter coupling between the bridge helix and the β subunit in RNA polymerase. Nat. Commun. 5, 3408 10.1038/ncomms4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prajapati R. K., Rosenqvist P., Palmu K., Mäkinen J. J., Malinen A. M., Virta P., Metsä-Ketelä M., and Belogurov G. A. (2019) Oxazinomycin arrests RNA polymerase at the polythymidine sequences. Nucleic Acids Res. 47, 10296–10312 10.1093/nar/gkz782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yuzenkova Y., Bochkareva A., Tadigotla V. R., Roghanian M., Zorov S., Severinov K., and Zenkin N. (2010) Stepwise mechanism for transcription fidelity. BMC Biol. 8, 54 10.1186/1741-7007-8-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gourse R. L., Chen A. Y., Gopalkrishnan S., Sanchez-Vazquez P., Myers A., and Ross W. (2018) Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184 10.1146/annurev-micro-090817-062444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roghanian M., Zenkin N., and Yuzenkova Y. (2015) Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res. 43, 1529–1536 10.1093/nar/gkv003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tehranchi A. K., Blankschien M. D., Zhang Y., Halliday J. A., Srivatsan A., Peng J., Herman C., and Wang J. D. (2010) The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141, 595–605 10.1016/j.cell.2010.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Furman R., Sevostyanova A., and Artsimovitch I. (2012) Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 40, 3392–3402 10.1093/nar/gkr1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Furman R., Tsodikov O. V., Wolf Y. I., and Artsimovitch I. (2013) An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J. Mol. Biol. 425, 82–93 10.1016/j.jmb.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Svetlov V., and Artsimovitch I. (2015) Purification of bacterial RNA polymerase: tools and protocols. Methods Mol. Biol. 1276, 13–29 10.1007/978-1-4939-2392-2_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McGlynn P., Savery N. J., and Dillingham M. S. (2012) The conflict between DNA replication and transcription. Mol. Microbiol. 85, 12–20 10.1111/j.1365-2958.2012.08102.x [DOI] [PubMed] [Google Scholar]
- 63. Dangkulwanich M., Ishibashi T., Liu S., Kireeva M. L., Lubkowska L., Kashlev M., and Bustamante C. J. (2013) Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. eLife 2, e00971 10.7554/eLife.00971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kireeva M. L., Nedialkov Y. A., Cremona G. H., Purtov Y. A., Lubkowska L., Malagon F., Burton Z. F., Strathern J. N., and Kashlev M. (2008) Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell 30, 557–566 10.1016/j.molcel.2008.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Larson M. H., Zhou J., Kaplan C. D., Palangat M., Kornberg R. D., Landick R., and Block S. M. (2012) Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 109, 6555–6560 10.1073/pnas.1200939109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bar-Nahum G., Epshtein V., Ruckenstein A. E., Rafikov R., Mustaev A., and Nudler E. (2005) A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 10.1016/j.cell.2004.11.045 [DOI] [PubMed] [Google Scholar]
- 67. Conrad T. M., Frazier M., Joyce A. R., Cho B. K., Knight E. M., Lewis N. E., Landick R., and Palsson B. O. (2010) RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. U.S.A. 107, 20500–20505 10.1073/pnas.0911253107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Windgassen T. A., Mooney R. A., Nayak D., Palangat M., Zhang J., and Landick R. (2014) Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 42, 12707–12721 10.1093/nar/gku997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Svetlov V., Belogurov G. A., Shabrova E., Vassylyev D. G., and Artsimovitch I. (2007) Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 35, 5694–5705 10.1093/nar/gkm600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang L., Zhou Y., Xu L., Xiao R., Lu X., Chen L., Chong J., Li H., He C., Fu X. D., and Wang D. (2015) Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 523, 621–625 10.1038/nature14482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu L., Wang W., Gotte D., Yang F., Hare A. A., Welch T. R., Li B. C., Shin J. H., Chong J., Strathern J. N., Dervan P. B., and Wang D. (2016) RNA polymerase II senses obstruction in the DNA minor groove via a conserved sensor motif. Proc. Natl. Acad. Sci. U.S.A. 113, 12426–12431 10.1073/pnas.1612745113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Molodtsov V., Sineva E., Zhang L., Huang X., Cashel M., Ades S. E., and Murakami K. S. (2018) Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Mol. Cell 69, 828–839 10.1016/j.molcel.2018.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pupov D., Petushkov I., Esyunina D., Murakami K. S., and Kulbachinskiy A. (2018) Region 3.2 of the σ factor controls the stability of rRNA promoter complexes and potentiates their repression by DksA. Nucleic Acids Res. 46, 11477–11487 10.1093/nar/gky919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Esyunina D., Agapov A., and Kulbachinskiy A. (2016) Regulation of transcriptional pausing through the secondary channel of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 113, 8699–8704 10.1073/pnas.1603531113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vassylyev D. G., Vassylyeva M. N., Zhang J., Palangat M., Artsimovitch I., and Landick R. (2007) Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 10.1038/nature05931 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented are contained in the article and are available upon request from A. Kulbachinskiy (akulb@img.ras.ru) and D. Esyunina (es_dar@inbox.ru).