Abstract
Bacillus thuringiensis subsp. israelensis produces crystal inclusions composed of three-domain Cry proteins and cytolytic Cyt toxins, which are toxic to different mosquito larvae. A key component is the Cyt toxin, which synergizes the activity of the other Cry toxins, thereby resulting in high toxicity. The precise mechanism of action of Cyt toxins is still debated, and two models have been proposed: the pore formation model and the detergent effect. Here, we performed a systematic structural characterization of the Cyt toxin interaction with different membranes, including in Aedes aegypti larval brush border membrane vesicles, small unilamellar vesicle liposomes, and rabbit erythrocytes. We examined Cyt1Aa insertion into these membranes by analyzing fluorescence quenching in solution and in the membrane-bound state. For this purpose, we constructed several Cyt1Aa variants having substitutions with a single cysteine residue in different secondary structures, enabling Cys labeling with Alexa Fluor 488 for quenching analysis using I-soluble quencher in solution and in the membrane-bound state. We identified the Cyt1Aa residues exposed to the solvent upon membrane insertion, predicting a possible topology of the membrane-inserted toxin in the different membranes. Moreover, toxicity assays with these variants revealed that Cyt1Aa exerts its insecticidal activity and hemolysis through different mechanisms. We found that Cyt1Aa exhibits variable interactions with each membrane system, with deeper insertion into mosquito larva membranes, supporting the pore formation model, whereas in the case of erythrocytes and small unilamellar vesicles, Cyt1Aa's insertion was more superficial, supporting the notion that a detergent effect underlies its hemolytic activity.
Keywords: Cyt1Aa toxin, Bacillus thuringiensis, membrane insertion, fluorescence quenching, pore formation, detergent effect, insecticide, larvicidal agent, mosquito control, Bti bacterium
Bacillus thuringiensis subsp. israelensis (Bti) is the active component of many larvicidal products used worldwide for mosquito control such as Aedes aegypti, Anopheles spp., and Culex spp. Nowadays, the control of A. aegypti is especially important because it is the vector of several important human diseases such as dengue, chikungunya, yellow fever, West Nile fever, and Zika, which are increasing worldwide (1–3). The advantage of using Bti bacterium for mosquito control is its high specificity against mosquito larvae and the absence of resistance evolution by the mosquito (4). The larvicidal activity of Bti is due to the insecticidal toxins found in parasporal inclusions that are formed during the sporulation growth phase. These parasporal crystals are composed of different kinds of δ-endotoxins such as the three-domain Cry proteins (Cry4Aa, Cry4Ba, Cry10Aa, and Cry11Aa) and cytolytic toxins (Cyt1Aa, Cyt2Ba, and Cyt1Ca) (5). The absence of Bti-resistant mosquitoes is principally due to the multiple mechanisms of action displayed by these toxins and to their synergism (6, 7). The toxicity of the whole Bti crystal inclusion is much higher than the sum of the individual toxicities of each toxin found in the crystal (8). The key component is the Cyt toxin that synergizes the activity of the Cry toxins present in the crystal, and it is able to overcome the resistance of mosquito populations against individual Cry or multiple Cry toxins (6, 8). Cry and Cyt protoxins are solubilized in insect gut and activated by midgut proteases, resulting in activated toxins that finally insert into the membrane of their hosts, forming pores that kill the larvae (7). It was proposed that Cyt1Aa synergizes the toxicity of Cry toxins such as Cry11Aa or Cry4Ba, by functioning as a receptor that facilitates their oligomerization. These Cry oligomeric structures are prone to interact with additional receptors and insert into the membrane killing the mosquito larvae (9–11).
The Cyt1Aa is a protein composed of a single α–β domain, where two outer layers of α-helix hairpins wrap around a β-sheet (12). The mechanism of action of Cyt1Aa toxin is not dependent on receptor proteins, because this toxin interacts with unsaturated membrane lipids present in brush border membranes from the mosquito larvae (13). Cyt1Aa toxin is toxic to dipteran insects, but toxicity against coleopteran larvae and pea aphid has also been reported, as well as cytolytic effect against some mammalian cell lines and erythrocytes (7, 14–17).
Two distinct models explaining the mechanism of action of Cyt toxins have been proposed (18). First, the pore formation model suggests that after binding to the cell membrane, the Cyt toxin forms a cation-selective channel, leading to colloid-osmotic lysis of the cell (19, 20). The second model suggests a detergent effect, whereby nonspecific aggregation of the toxin on the surface of the membrane leads to lipid bilayer disassembly and cell death (21).
The precise mechanism by which the Cyt toxin inserts into the membrane is not known. In the pore formation model, it was proposed that the β-sheet region of Cyt1Aa (composed of strands β-6, β-7, and β-8; Fig. 1) is involved in membrane insertion of the toxin, whereas helices α-1 and α-3 are important for membrane recognition and oligomerization (12, 17, 20, 22, 23) to finally form high-molecular-weight oligomers, which insert into the membrane and form the lytic pores composed of 16 monomers (19, 24, 25). It has been shown that Cyt1Aa oligomerization is needed for membrane insertion and pore formation into insect brush border membrane vesicles (BBMV) because Cyt1Aa helix α-3 mutants were unable to oligomerize and lost toxicity against mosquitoes and hemolytic activity, suggesting that oligomerization is needed for membrane insertion (23). In the case of Cyt2Aa, mutagenesis of helix α-1 showed that this helix could also be involved in toxin oligomerization and toxicity (26). However, mutations in helix α-1 in the Cyt1Aa toxin were not affected in oligomer formation, insecticidal activity, and synergism with Cry11Aa but were severely affected in their hemolytic activity, suggesting that insecticidal and hemolytic activities of Cyt1Aa could involve different mechanisms (17).
Figure 1.
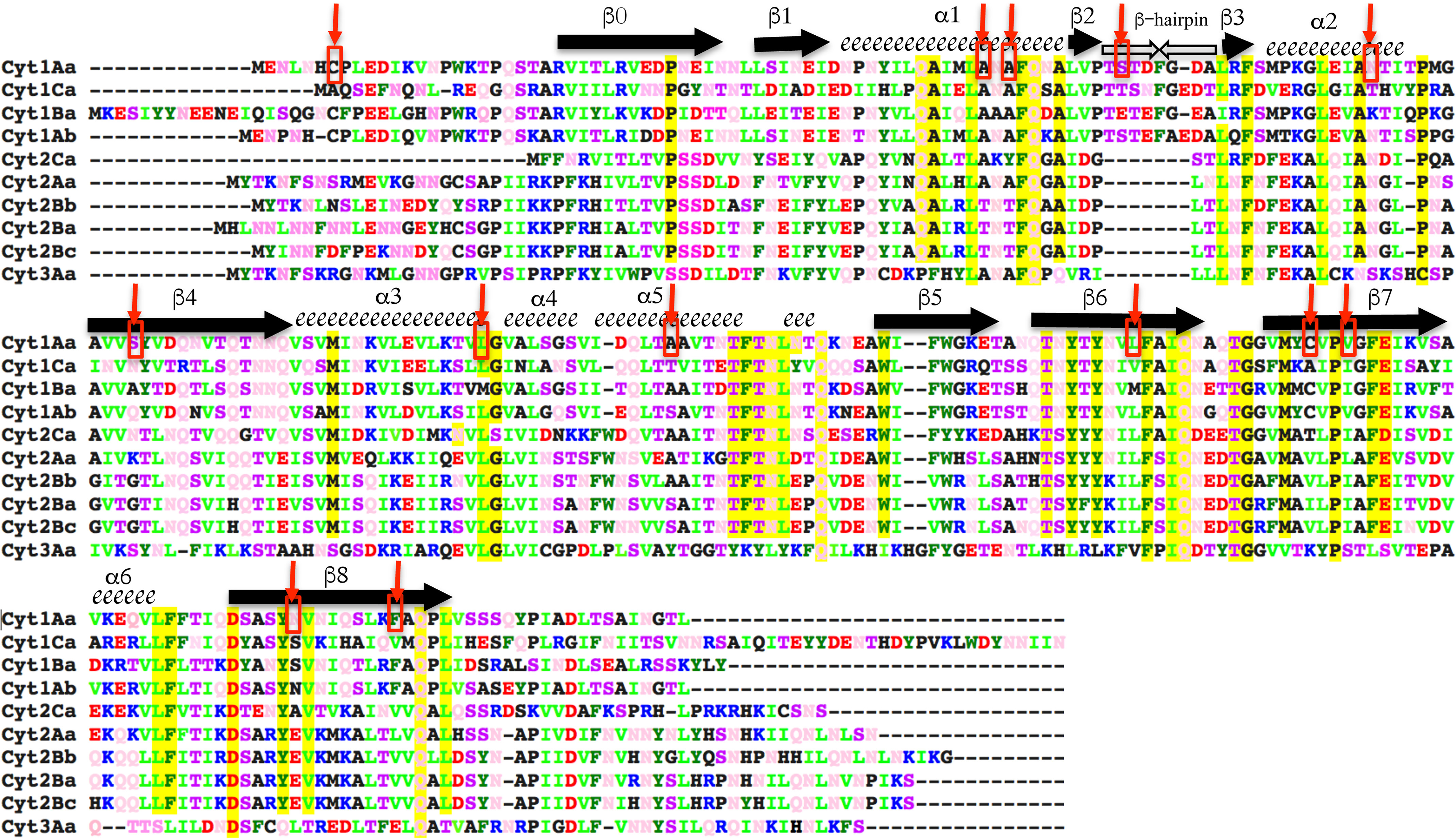
Sequence alignment of Cyt1Aa with other members of Cyt family. Cyt1Aa secondary structure elements are labeled above the corresponding sequence; α-helices are depicted as spirals, and β-strands are arrows. A color code was used to show the different residues. Blue is used for positively charged residues (Arg and Lys); red is for negatively charged residues (Asp and Glu); dark green is for aromatic residues (Trp, Phe, and Tyr); light green is for basic residues (Leu, Val, and Iso); pink is for polar neutral amino acids (Gln and Asn); and polar residues (Thr and Ser) are in purple. Highly conserved residues in the Cyt family of proteins are highlighted in yellow. Residues selected for mutagenesis in Cyt1Aa are labeled with red arrows and within red squares.
In contrast, the model of detergent action proposed that the interaction of Cyt1Aa with lipids triggers Cyt structural changes, and the toxin is spread in the surface of the membrane affecting lipid organization, resulting in breakdown of the liposome and releasing its content into the media. It was shown that small and high-molecular-weight polymers could be released from the vesicles by Cyt action, and it was estimated that at least 140 toxin molecules must bind to PC large unilamellar vesicles for inducing their breakdown (27).
These data indicate that Cyt toxin may interact with different types of membrane systems and probably the conformational changes in this toxin that are needed to affect membrane integrity could be different. Recently some key steps in the mode of action of Cyt1Aa have been further studied in insect cell lines and large unilamellar vesicles (28). The authors proposed that hairpin of helices α3/α4 moves away from the β-sheet upon activation. The toxin aggregates in the surface of the bilayer and inserts forming large pores with more than 56 monomers (28).
To understand the Cyt1Aa mechanism involved in membrane insertion, we analyzed the insertion of Cyt1Aa toxin into different lipid membranes, such as A. aegypti BBMV isolated from midgut tissue, erythrocyte membranes from rabbit red blood cells (RBC), and small unilamellar vesicle (SUV) liposome membranes. We constructed Cyt1Aa mutants with a single cysteine residue in the different secondary structures to label them with Alexa Fluor 488 for quenching analysis using a KI-soluble quencher that allowed us to identify residues exposed to the solvent upon membrane insertion. Our data indicate that Cyt1Aa insertion is different in the three membrane systems analyzed.
Results
Insecticidal and hemolytic activities of Cyt1Aa mutants
We constructed a collection of single cysteine mutants with the purpose of labeling them with Alexa Fluor 488 maleimide and determining their conformational changes upon membrane insertion, by analyzing the fluorescence quenching with the soluble quencher KI when the protein is in solution and in the membrane-inserted state. Fig. 1 shows the selected residues labeled with red arrows in the sequence of Cyt1Aa, and Fig. S1 shows that all these residues are exposed to the solvent in the 3D structure of the toxin after analyzing their accessibility by using Swiss PDB Viewer.
The WT Cyt1Aa protein has two Cys residues; one is located in the N-terminal region that is cleaved out during activation (Cys7), and the second is located in the β-7 strand (Cys190). These two residues were changed to Ser to provide a Cyt1Aa background that could be used to introduce single Cys residues in other secondary structures of this protein. Point mutations in these Cys7 and Cyt190 residues did not affect the toxicity to A. aegypti larvae (Table 1). Also, the toxicity against A. aegypti larvae of the rest of the single-point Cys mutants was not affected, with exception of the Cyt1Aa-N89C mutant located in helix α-2. This was the only mutant affected in toxicity against A. aegypti larvae (Table 1). Because the Cyt1Aa-N89C mutant lost toxicity, it was not further analyzed in this work, although it was used for other analyses elsewhere (29). Some mutants showed increased insecticidal activity, especially mutants Cyt1Aa-L176C and Cyt1Aa-N218C with 13- and 15-fold higher toxicity when compared with the Cyt1Aa protein.
Table 1.
A. aegypti larval toxicity of different single-point mutations in the Cyt1Aa toxin
| Mutation | Location | Insecticidal activity LC50 (fiducial limit) |
|---|---|---|
| ng/ml | ||
| Cyt1Aa | 897.55 (731–1053.6) | |
| C7S | N-terminal | 721.5 (484.5–970.7) |
| C190S | β-7 | 389.3 (231–589.2) |
| A59C | Helix α-1 | 2419 (1861–3653) |
| A61C | Helix α-1 | 223.8 (165.2–307.7) |
| S70C | β-Hairpin | 1119.8 (869.3–1427.1) |
| N89C | Helix α-2 | >5000 |
| L120C | Helix α-3 | 964.7 (675.3–1294.6) |
| A141C | Helix α-5 | 254.4 (185.9–359.1) |
| L176C | β-6 | 68.4 (51.1–89.1) |
| V193C | β-7 | 196.6 (120.9–285.6) |
| N218C | β-8 | 57.8 (30.8–86.2) |
| C7S/C190S | N-terminal β-7 | 523.4 (386.8–720.9) |
Because single-point mutants Cyt1Aa-C7 and Cyt1Aa-C190 did not affect toxicity, the double mutant Cyt1Aa-C7S-C190S was constructed. This double mutant was also not affected in toxicity against A. aegypti larvae (Table 1). We used the Cyt1Aa-C7S-C190S mutant as background for construction of all other mutants with a single Cys residue in different locations of the toxin (i.e. triple mutants). Table 2 shows the insecticidal activity of these triple mutants. Mutant Cyt1AaTriple-A59C was slightly affected in insecticidal activity when compared with the WT Cyt1Aa toxin. Some mutants such as Cyt1AaTriple-A61C, Cyt1AaTriple-S99C, Cyt1Aatriple-A141C, Cyt1AaTriple-V193C, and Cyt1Aatriple-N218C showed 5-, 11-, 3-, 4-, and 5-fold higher toxicity, respectively, than Cyt1Aa. The fact that all the triple mutants retained toxicity indicated that the structures of these mutants were not severely affected by these point mutations and could be used for labeling with the fluorescent dyes to perform structural analysis with BBMV and compare with other membrane systems.
Table 2.
Toxicity of different triple mutations in the Cyt1Aa toxin against A. aegypti larvae
| Mutations | Location of the single Cys | Insecticidal activity LC50 (fiducial limit) |
|---|---|---|
| ng/ml | ||
| Cyt1AaTriple-A59C | Helix α-1 | 3595.9 (2695.6–5084.7) |
| Cyt1AaTriple-A61C | Helix α-1 | 175.06 (53.3–312) |
| Cyt1AaTriple-S70C | β-Hairpin | 1459.1 (1154.6–1964.8) |
| Cyt1AaTriple-S99C | β-4 | 80.56 (−89.6–144.1) |
| Cyt1AaTriple-L120C | Helix α-3 | 1207.6 (978.9–1472.3) |
| Cyt1AaTriple-A141C | Helix α-5 | 289.15 (211.4–399.9) |
| Cyt1AaTriple-L176C | β-6 | 453.9 (204–773.3) |
| Cyt1AaTriple-V193C | β-7 | 242.1 (187.6–319.1) |
| Cyt1AaTriple-N218C | β-8 | 175.7 (86.8–278.5) |
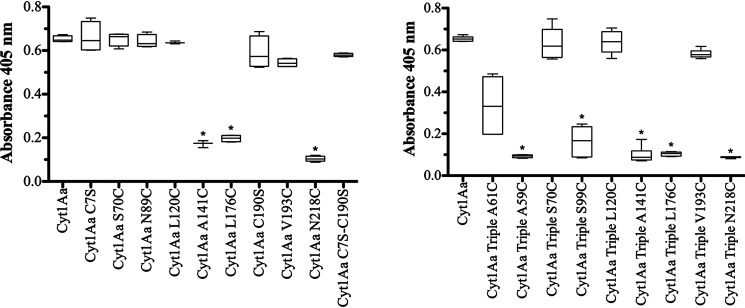
The hemolytic activity of these mutants was analyzed (Fig. 2). Several single mutants such as Cyt1Aa-A141C, Cyt1Aa-L176C, and Cyt1AaN218C were affected in hemolysis (Fig. 2, left panel). Cyt1Aa-A59C and Cyt1Aa-A61C were not analyzed because their effect in hemolytic activity was previously reported, showing that Cyt1Aa-A59C was severely affected, and Cyt1Aa-A61C showed reduced hemolytic activity (17). The double mutant Cyt1Aa-C7S-C190S showed a similar hemolytic activity as the Cyt1Aa toxin. The triple mutants Cyt1AaTriple-A59C, Cyt1Atriple-A61C, Cyt1Atriple-S99C, Cyt1AaTriple-A141C, Cyt1AaTriple-L176C, and Cyt1AaTriple-N218C also showed reduced hemolytic activity (Fig. 2, right panel). It is important to make clear that the point mutant Cyt1Aa-C7S has a single Cys residue that corresponds to Cys190; this mutant is not affected in hemolysis (Fig. 2, left panel).
Figure 2.
Hemolytic activity of soluble Cyt1Aa proteins. The hemolytic activity of Cyt1Aa toxins was analyzed against rabbit red blood cells. Left panel, hemolytic activity of Cyt1Aa single-point mutations. Right panel, hemolytic activity of Cyt1Aa-Triple mutants. Positive control showing 100% hemolysis was defined after incubation of the same volume of rabbit red blood cells with dechlorinated H2O. Negative controls were red blood cells incubated with buffer A. These assays were performed four to six times in triplicate each time. The data are presented as box-and-whisker plots. A t test was performed using the statistical program GraphPad Prism with statistical significance set at P < 0.01. Asterisks indicate significant differences from the WT Cyt1Aa.
The crystal inclusions of the triple mutants were purified and trypsin-activated, showing a 25-kDa size similar to the WT Cyt1Aa toxin (Fig. 3A). The 25-kDa band of these proteins were all recognized with the anti-Cyt1Aa polyclonal antibody (Fig. 3B), suggesting that mutations did not induce degradation of the proteins during their purification, because a single band that corresponds to the size of the protein was detected in the Western blotting assays. Table 2 shows that these mutants retain insecticidal activity, supporting that their structures were not affected by these point mutations.
Figure 3.
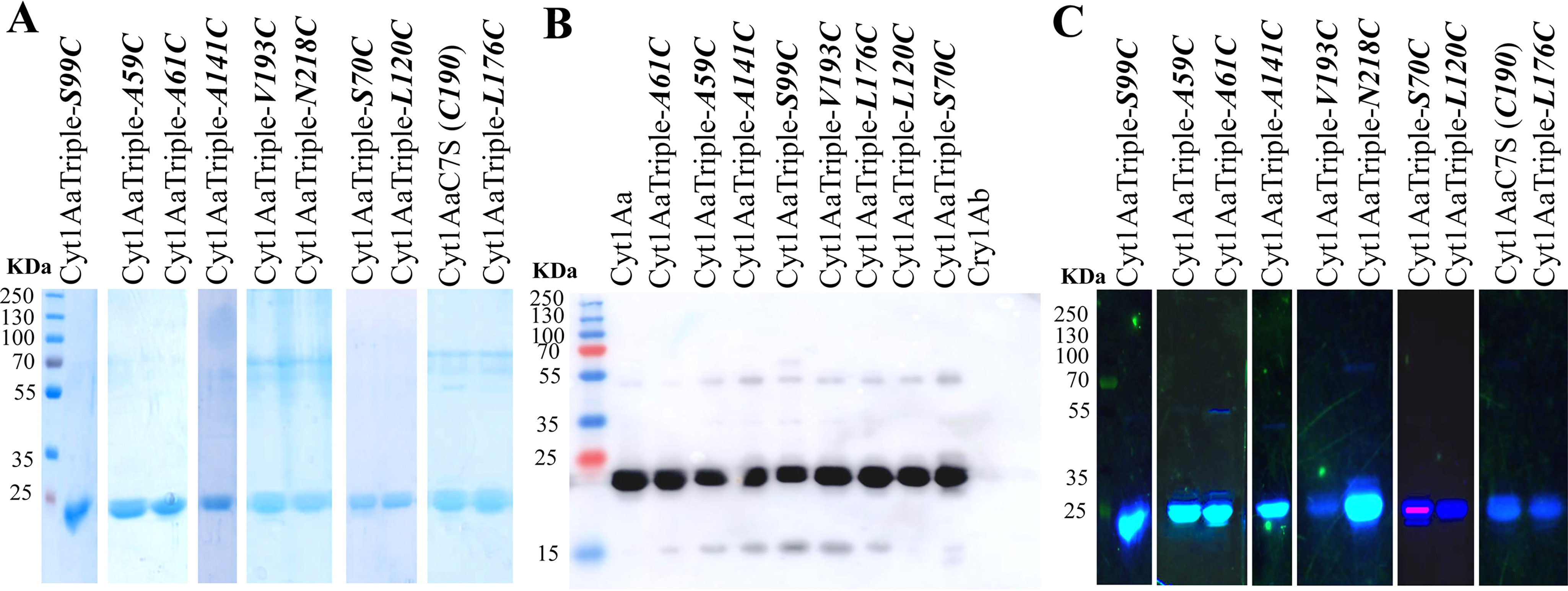
Cyt1Aa mutant stability to trypsin and labeling with Alexa Fluor 488. Cyt1Aa and labeled proteins were activated with trypsin and purified using anion-exchange chromatography. A, purified Cyt1Aa proteins analyzed in SDS-PAGE stained with Coomassie Brilliant Blue. B, Western blotting using anti-Cyt1Aa polyclonal antibody. C, visualization of the labeled proteins with Alexa Fluor 488 in the SDS-PAGE by excitation with UV light transilluminator. Molecular mass markers used were precision prestained plus standards all blue (Bio-Rad).
Evaluation of synergism between Cyt1Aa and Cry11Aa proteins
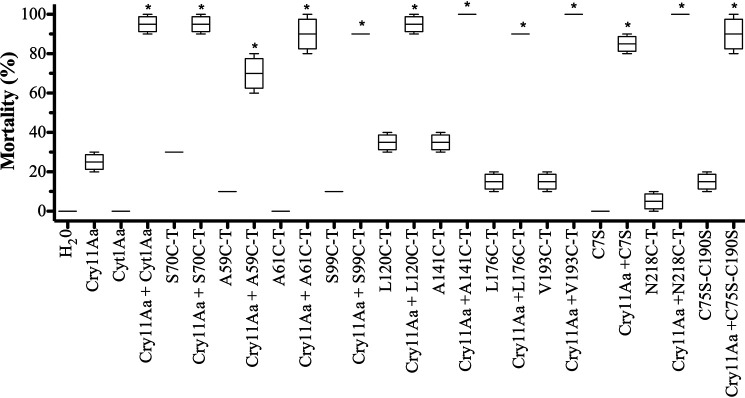
To determine whether the triple mutations introduced in Cyt1Aa affect their synergism with Cry11Aa toxin, qualitative estimation of synergism was obtained by mixing a Cyt1Aa amount that would produce an estimated 10% mortality with 95 ng/ml of Cry11Aa that was estimated to produce 20% mortality. Fig. 4 shows that all the triple mutants retained the capacity to synergize Cry11Aa toxin because the mixtures with Cry11Aa resulted in toxicities up to 70–80%. Also, the Cyt1A C75SC190S double mutant synergized Cry11Aa toxicity (Fig. 4). The SF values were calculated as described under “Experimental procedures” using a 1:1 mixture of Cyt1Aa proteins with Cry11Aa. Table S1 shows that all Cyt1Aa triple mutants synergize Cry11Aa toxicity with SF values higher than 1.
Figure 4.
Analysis of synergism of Cyt1Aa or Cyt1Aa-mutant toxins with Cry11Aa toxin.
Cry11Aa toxin was used at 95 ng/ml that kills 10% of A. aegypti larvae, and Cyt1Aa or Cyt1Aa mutant toxins were used at concentration that kills 10–20% of these larvae. Negative control (dechlorinated water) was included in the bioassay. Larvae mortality was examined 24 h after treatment. These assays were performed four times in triplicate each time. The data are presented as box-and-whisker plots. The data with low variation did not show a box. Asterisks indicate significant differences of the mixtures of proteins compared with single proteins by analysis of variance with significant differences P < 0.01 by using GraphPad Prism.
Analysis of the insertion of Cry1Ab-labeled mutants into different membrane systems
Cyt1Aa mutants were labeled with Alexa Fluor 488, and the efficiency of labeling was determined with respect to its molar extinction coefficient. Five mutants (Cyt1AaTriple-S70C, Cyt1AaTriple-L120C, Cyt1AaTriple-L176C, Cyt1AaTriple-V193C, and the single mutant Cyt1AaC7S toxin, which retains original Cys190 residue) showed lower efficiency of labeling (0.2–0.3 mol dye/mol toxin). The rest of the mutants showed that efficiency of labeling was 0.6–0.8 mol of dye/mol Cyt1Aa protein. The labeling of the toxins was also visualized directly on the SDS-PAGE excited with UV light transilluminator (Fig. 3C). The Cyt1AaTriple-S70C, Cyt1AaTriple-L120C, Cyt1AaTriple-L176C, and Cyt1AaTriple-V193C mutants and the single mutant Cyt1AaC7S-C190 toxin showed low fluorescence in the SDS-PAGE because of their lower efficiency of labeling.
The interaction of activated Cyt1Aa with the different membrane systems was analyzed by determining the fluorescence of the Alexa Fluor 488–labeled single-Cys proteins in its soluble state and then compared with the membrane-associated proteins. As mentioned above, three different membrane systems were analyzed: A. aegypti BBMV, RBC, and synthetic PC–Ch–S SUV liposomes. The degree of solvent exposure of each labeled residue was determined in the presence of different concentrations of the soluble I− quencher (from 0 to 0.5 M). Fig. S2 shows all the Stern–Volmer plots (Fig. S2). The values of the apparent dynamic quenching constant KSV derived from the slope of these plots are presented in Table 3.
Table 3.
Apparent dynamic quenching constant in Cyt1Aa toxin mutants
Apparent dynamic quenching constant (KSV) observed in Cyt1Aa toxin mutants labeled with Alexa Fluor 448 and quenched with KI when the toxin is in solution compared with after insertion into different membrane systems. The bold letters in the table were included to indicate the degree of solvent exposure of these residues arbitrarily based in their KSV value as follows: HB, highly buried KSV value between 1 and 3.5; B, buried KSV value between 3.5 and 5; PB, partially buried KSV value between 5 and 8; and E, exposed KSV value between 8 and 31.
| Mutation | Localization of Cys residue |
KSV value |
|||
|---|---|---|---|---|---|
| In solution | In BBMV | In RBC | In liposomes | ||
| Cyt1AaTriple-A59C | Helix α-1 | 14.39 ± 1.76 E | 2.38 ± 0.74 HB | NDa | 6.92 ± 0.52 PB |
| Cyt1AaTriple-A61C | Helix α-1 | 13.92 ± 0.95 E | 3.02 ± 0.61 HB | 2.5 ± 0.42 HB | 7.06 ± 0.44 PB |
| Cyt1AaTriple-S70C | β-Hairpin | 23.67 ± 2.32 E | 7.39 ± 0.31 PB | 3.8 ± 0.29 B | 13.62 ± 0.09 E |
| Cyt1AaTriple-S99C | β-4 | 30.4 ± 1.81 E | 17.76 ± 1.69 E | 9.17 ± 1.87 E | 27.4 ± 2.11 E |
| Cyt1AaTriple-L120C | Helix α-3 | 28.05 ± 1.02 E | 4.73 ± 0.49 B | 8.23 ± 0.49 E | 4.8 ± 1.12 B |
| Cyt1AaTriple-A141C | Helix α-5 | 31.66 ± 1.01 E | 5.62 ± 0.87 PB | 9.1 ± 0.14 E | 12.82 ± 1.77 E |
| Cyt1AaTriple-L176C | β-6 | 15.63 ± 1.33 E | 2.14 ± 0.47 HB | 4.36 ± 0.42 B | 11.57 ± 1.27 E |
| Cyt1AaC7S-C190 | β-7 | 13.08 ± 1.59 E | 2.91 ± 0.89 HB | 7.69 ± 0.52 PB | 5.74 ± 1.25 PB |
| Cyt1AaTriple-V193C | β-7 | 25.16 ± 4.5 E | 0.95 ± 1.46 HB | 2.40 ± 1.63 HB | 5.59 ± 0.42 PB |
| Cyt1AaTriple-N218C | β-8 | 23.98± 3.4 E | 4.46 ± 1.00 B | 5.91 ± 0.75 PB | 6.07 ± 0.65 PB |
aND, not determined.
Quenching analysis of the Alexa Fluor 488–labeled mutant toxins performed in solution showed that all residues were exposed to the solvent, based in their KSV values, although these data also indicate that residues S70C (β-hairpin), S99C (β-4), L120C (α-3), A141C (α-5), V193C (β-7), and N218C (β-8) were more exposed to the solvent than residues A59C (α-1), A61C (α-1), L176C (β-6), and C190 (β-7) (Table 3).
When the Cyt1Aa mutants were incubated with BBMV, all labeled residues showed a drastic reduction in their susceptibility to be quenched by I− when compared with proteins in solution. The only exception was residue S99C (β-4), which remains exposed to the solvent. Specifically, residues located in helix α-1, A59C (α-1), A61C (α-1), and those located in the long β-strands, L176C (β-6), Cys190, and V193C (β-7), were shown to be deeply buried because their KSV values were below 3.5 (Table 3).
When Cyt1Aa mutants were incubated with RBC, residues S99C (β-4), L120C (α-3), and A141C (α-5) were more exposed to the solvent that the rest of the labeled residues. Residues A61C (α-1) and V193C (β-7) showed KSV values similar to those obtained when the protein was interacting with BBMV (below 3.5). The residue S70C (β-hairpin) showed a more buried localization than BBMV interaction (Table 3). Overall, the positions of the other residues when the toxin interacts with RBC showed more exposed locations compared with the same mutants after interaction with BBMV. Finally, the interaction of Cyt1Aa with synthetic SUV liposomes showed that Cyt1Aa has a more superficial interaction because residues S70C (β-hairpin), S99C (β-4), A141C (α-5), and L176C (β-6) were exposed to the solvent with KSV values higher than 11.5, and none of the residues were highly buried because none of them showed KSV values lower than 4.8 (Table 3).
Discussion
Bti Cyt1Aa toxin is a versatile protein that has proven to be toxic to different insect orders and also to erythrocytes and some mammalian cell lines (7, 14–17). For more than 20 years, different groups have studied the mechanism of action of Cyt toxins. Two models have been proposed: pore formation and detergent action (18). However, a strong controversy remains, because solid data have been presented supporting both models (20, 21). The main problem in most of these studies is that the model membranes characterized were only lipid liposomes or erythrocytes membranes, and studies that characterize Cyt toxin interaction with BBMV isolated from mosquito larvae are missing. The only exception is a study showing Cyt2Aa oligomerization in liposomes, erythrocytes, and A. aegypti BBMV showing that this protein forms similar high-molecular-weight aggregates in the three membrane systems (30). Here, we compared the insertion of Cyt1Aa in these three membrane systems and show that interaction with each membrane system resulted in different conformational changes of Cyt1Aa.
A collection of 23 Cyt1Aa single, double, or triple mutants were used and characterized. Analysis of insecticidal and hemolytic activities of these mutants allowed us to conclude that Cyt1Aa toxin exerts toxicity by different mechanisms. Some point mutations such as Cyt1Aa-L176C and Cyt1Aa-N218C mutants resulted in a 13–15-fold increase in insecticidal activity and at the same time these two mutants showed reduced hemolytic activity. Other single-point mutants were severely affected in hemolysis (Cyt1Aa-A59C and Cyt1Aa-A141C) but retained an insecticidal activity similar to that of the Cyt1Aa toxin. Finally, Cyt1Aa-N89C lost insecticidal activity but retained its hemolytic activity. Further characterization of Cyt1Aa-N89C and other mutations of Cyt1Aa helices α-1 and α-2 showed that oligomerization of Cyt1Aa is required for its insecticidal activity but not for hemolysis, indicating that the mode of action of Cyt1Aa is different in the two systems (29). Interestingly, these data indicate that multiple mutations can affect the hemolytic activity of the toxin. Similarly, the mutation T144A in Cyt2Aa (located in the loop between helix α-4 and strand β-5; Fig. 1) showed lower hemolytic activity but retained larvicidal activity against mosquitoes (30). These results also indicate that hemolytic activity is not restricted to a single region of the toxin because mutations in different regions of the toxin such as helices α-1 and α-5, the loop between α-4 and β-5, and β-strands β-6 and β-8 resulted in significant reduction of the hemolytic activity.
Because double point mutant Cyt1Aa-C7S-C190S, eliminating the two Cys residues of the Cyt1Aa, was toxic to the mosquito larvae and showed similar hemolytic activity as the WT Cyt1Aa, we used this double mutant as background to construct a collection of triple mutants containing single Cys residue located in different structures of the toxin (Fig. 1). All triple mutants were active against mosquitoes (Table 2), and some of these mutants showed reduced hemolytic activity (Fig. 2). The triple mutants were labeled with Alexa Fluor 488, and the degree of solvent exposure of each labeled residue was determined by quenching analysis in its soluble state, as well as after interaction with BBMV, RBC, and SUV (PC–Ch–S) liposomes.
Previously, Promdonkoy and Ellar (25) analyzed acrylodan-labeled mutants of Cyt2Aa inserted in erythrocytes. Acrylodan is a fluorescent dye that is highly sensitive to the solvent polarity, showing distinct emission characteristics in different polar environments. Residues that are located in the long β-strands of Cyt2Aa (β-6, β-7, and β-8) were labeled, but unfortunately, they did not label any other region of the toxin, so no comparison was provided with residues exposed to the solvent. Their data showed that all the residues analyzed were in contact with the lipids and that the emission spectra of dilapidated proteins after membrane interaction recovered the emission spectra similar to that of the soluble state of the toxin. In addition, the deepness of the insertion of these residues into the bilayer was not analyzed. Unfortunately, BBMV or synthetic liposomes were not analyzed (20). Here, similar residues as those discussed above in Cyt2Aa were analyzed, because Cyt2Aa residues Leu172, Val186, Leu189, and Glu214 corresponds to Cyt1Aa Leu176, Cys190, Val193, and Asn218. Our data show that effective KSV for I− quenching of all mutant toxins was much lower in the membrane-inserted state than in solution, indicating that Cyt1Aa interacts with the three membrane models analyzed. However, when the Cyt1Aa mutants were incubated with BBMV, several residues reported extremely low KSV values (below 3.5 value) showing a drastic reduction in their susceptibility to be quenched by I− than in solution. These data indicate that these residues could be deeply buried into the membrane environment or in the oligomeric structure of the toxin. These residues correspond to the residues located in the middle of β-strands, β-6 and β-7, that were previously suggested to be inserted into the membrane (20) and in helix α-1, suggesting that this helix may be also interacting with the bilayer or is involved in oligomer formation as was previously reported (29). The Cyt1Aa Asn218 residue that is located in the initial part of the β-8 strand showed a KSV value slightly higher (KSV = 4.46) than those located in the middle part of β-6 and β-7, indicating that it could be located close to the lipid water interphase.
As mentioned above, our data indicated that helix α-1 of Cyt1Aa is located in a position deeply buried to KI quenching when the Cyt1Aa interacts with BBMV and RBC, suggesting that this region could be located inside the membrane environment or in protein–protein contacts involved in oligomerization of the toxin. Du et al. (31) analyzed the proteolysis profile of Cyt2Aa and Cyt1Aa after interaction with PC–Ch–S liposomes, showing that a fragment of Cyt2Aa and Cyt1Aa was protected from cleavage with different proteases. Under these conditions the long β-strands were protected from degradation, suggesting that the conformational change after interaction with the membrane exposes the N-terminal half of the toxin, resulting in its degradation and supporting that β-strands were inserted into the bilayer. However, analysis of N-terminal sequence of the membrane protected fragments of Cyt1Aa, as well as MS/MS sequence of membrane-associated fragments of this toxin, showed that helix α-1 of Cyt1Aa was also protected from degradation, suggesting that it also may be located in the interior of the membrane (31). These data and our quenching data support the model proposed by Li et al. (32) that suggested that the hairpin composed of helices α-1 and α-2 might be package against the long β-sheet during insertion into the membrane (32). Mutations in helix α-1 of Cyt1Aa showed that helix α-1 may not be involved in oligomerization because the mutants in this region were still able to oligomerize, showing an important increase in insecticidal activity against mosquito and coleopteran larvae (17). However, analysis of other Cyt1Aa mutations in helices α-1 and α-2 identified mutations that affect toxin oligomerization and toxicity to A. aegypti (29).
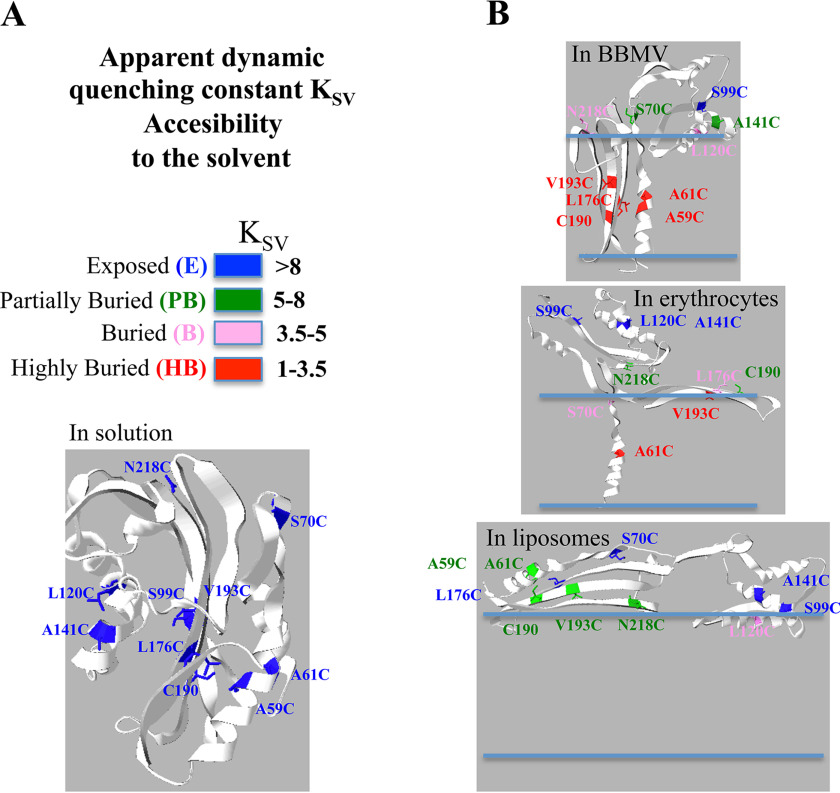
The only residue that remained highly exposed to the solvent after interaction of Cyt1Aa with BBMV was S99C. This residue is found in strand β-4, which is located between the two hairpins of α-helices (α-1 and α-2 in the first hairpin and α-3 and α-5 in the second hairpin; see Fig. 1). Based on the quenching constant KSV values presented in Table 3, we displayed an exposure color code and used this color code to construct graphical models of the possible Cyt1Aa structures that could integrate all our data regarding the conformational changes of Cyt1Aa after interaction with the different membranes (Fig. 5). The models constructed show that Cyt1Aa inserts deeply into the membrane in the case of BBMV, where helix α-1 and half of the β-hairpin that is attached to helix α-1 are buried into the bilayer together with the long β-strands, whereas the rest of the toxin is located in the surface of the membrane (Fig. 5). This model is compatible with other two structural models previously proposed supporting oligomerization and membrane insertion to perforate the membrane bilayer (28, 32, 33). Nevertheless, the final structure of Cyt1Aa oligomers in the membrane remain to be precisely defined by other means as high resolution microscopy and modeling, cryoEM data, AFM data, or X-ray 3D structure analysis, among other techniques.
Figure 5.
Models representing the possible structures of Cry1Aa in the different membrane systems accordingly to the KSV values obtained in the quenching assays. A, color code used to label the different residues accordingly to their different KSV values and 3D structure of Cyt1Aa showing the analyzed residues in solution. B, model representations of the possible structural changes upon membrane interaction of the Cyt1Aa with the different membrane systems. Structural models were constructed with Swiss PDB viewer program drawing each α-helix and β-strand according to the data of the Table 3 that indicated the possibility of each selected residue to be exposed to the solvent.
When Cyt1Aa was incubated with RBC, only two residues showed a KSV value below 3.5; one of these residues is A61C located in helix α-1, suggesting that this residue may be located in the interior of the membrane as explained above. The other residue is V193C that is located in the middle part of the β-7. However, a very nearby residue in the same β-strand (Cys190) is not deeply buried because the KSV value was 7.69. These mutations did not affect the hemolytic activity, suggesting that hemolysis does not involve a deep insertion of β-7 strand into the bilayer. Other mutants that conserved hemolytic activity were the Cyt1AaTriple-S70C and Cyt1AaTriple-L120C. The residue L120C (α-3) showed a higher KSV value (KSV =8.23) in RBC than when the protein inserted into BBMV, indicating that it would be more accessible to the solvent. In contrast, S70C (β-hairpin) is located in a position less accessible to the solvent when compared with insertion into BBMV. These data indicated that the topology of the toxin after interaction with RBC membrane is different from when the toxin interacts with the insect BBMV. Our data indicate that in RBC the Cyt1Aa has a different conformation of β-strands compared with BBMV that represent a more superficial arrangement in RBC compared with interaction with BBMV. The β-strands may not be deeply inserted into the erythrocytes bilayer, and the overall protein may be more exposed to the solvent (Fig. 5). The electrophysiological and AFM data presented by Tetraeu et al. (28), also supported our data because it was proposed that residues 154–234 are membrane-associated, forming the hydrophobic part of the pore. Nevertheless, the exact structure of the Cyt1Aa when inserted into BBMV still remains to be determined. It is highly interesting that the cryoEM data reported indicated that Cyt1Aa form oligomers when interacting with large unilamellar vesicle liposomes (28). It will be interesting to see whether similar structures are observed in erythrocytes or BBMV membrane systems.
Finally, regarding the interactions of Cyt1Aa toxin with synthetic liposomes SUV, our data show important differences from that with RBC and BBMV. With SUV the β-strands β-4 and β-6, the β-hairpin, and helix α-5 are highly exposed to the solvent. The other long β-strands, β-7 and β-8, showed KSV values of >5.59 that could indicate they are partially buried. Helix α-1 is also partially buried (KSV values > 5.88). These data indicate that interaction with synthetic liposomes resulted in a more superficial topology than with the other two membrane systems analyzed (BBMV and RBC), which could be compatible with the detergent effect (34) (Fig. 5). It was proposed that the toxin is adsorbed into the membrane surface like a carpet structure, destabilizing lipid packaging and breaking the membrane organization. The detergent action model of Cyt1Aa has received support from multiple studies performed in liposomes especially with PC lipids. Analysis of lifetime of Trp residues in the presence and absence of lipids have similar values, suggesting that these residues, located in strand β-5 (Fig. 1), have a rather superficial topology (21).
Quenching analysis performed with brominated phospholipids or spin-labeled lipids showed they could not quench Trp fluorescence, confirming that Trp residues do not insert deeply into the bilayer (35). Analysis of proton–deuteron exchange showed that in the presence of PC liposomes, the toxin showed a loose structure, suggesting that an important part of the toxin remains in the surface of the membrane (35). It was proposed that the tertiary structure of the protein is affected after lipid interaction, whereas the secondary structure is maintained (35). These data are compatible with the proposed tertiary structure changes of Cyt2Aa after interaction with lipid/Ch bilayer membranes, where it was stated that this toxin binds to the lipid membrane, forming a softer protein-lipid layer covering almost fully the membrane surface (36). Recently it was clearly shown that Cyt1Aa does not form large aggregates when bound to SUV made with l-α-phosphatidylcholine lipids and that this protein is only able to form aggregates after interaction with l-α-phosphatidylcholine–large unilamellar vesicles (28). Nevertheless, oligomerization assays of Cyt1Aa with PC:Ch:S–SUV showed formation of high-molecular-weight oligomers (23). One possible explanation could be the type of lipids or the toxin concentration used in these assays. Here, we worked only with PC:Ch:S–SUV, and we concluded that interaction with these membranes is highly superficial, suggesting that oligomers formed in the presence of SUV do not insert into the membrane. The proposed model for Cyt1Aa interaction with synthetic liposomes (Fig. 5B) is compatible with all data on Cyt toxin interaction with membranes previously obtained with synthetic liposomes.
Our data support that in BBMV the Cyt1Aa forms a structured pore where β-strand regions may be forming a β-barrel, and α-1 may also be inserted also into the membrane. However, it is still possible that helix α-1 may be buried within the oligomers of Cyt1Aa, while the rest of the protein remains in the lipid surface. When the toxin interacts with RBC, the conformational changes are different, with a lower insertion of the β-strands into the bilayer and higher exposure of the toxin to the solvent. In SUV liposomes the toxin seems to spread in the surface of the lipids. Thus, a detergent mode of action could be more compatible for the hemolytic activity of Cyt1Aa and with the Cyt1Aa interaction with SUV liposomes, whereas pore formation could be responsible for its insecticidal activity. Our data help to elucidate the initial events in the membrane interaction process of Cyt1Aa toxin that could significantly contribute to our understanding of other β-barrel cytolytic toxins such as Staphylococcus aureus α-hemolysin (37), Streptococcus streptolysin O (38), and Aeromonas hydrophila aerolysin (39), among others.
Experimental procedures
Site-directed mutagenesis
Plasmid pWF45 containing cyt1Aa gene (40) was used as template for site-directed mutagenesis with a QuikChange mutagenesis kit from Thermo Fisher Scientific, following the manufacturer's instructions. Mutagenic oligonucleotides were synthesized at the Instituto de Biotecnologia Universidad Nacional Autónoma de México facilities, and their sequences are presented in Table 4. The mutated plasmids were transformed in Escherichia coli X-L1 blue strain, and mutated colonies were selected in LB-Ampicillin 100 μg/ml at 25 °C for 2 days. Plasmid DNA was extracted from selected colonies using a DNA extraction Wizard®PLUS SV kit (Promega) and sequenced at the Institute of Biotechnology Universidad Nacional Autónoma de México facilities. These plasmids were transformed into the acrystalliferous Bt 407 strain as described (41). Bt transformants were selected in LB-erythromycin 10 μg/ml at 30 °C. Single-point mutations in helix α-1 were previously described (17).
Table 4.
Sequence of mutagenic oligonucleotides
The sequences of the mutated residue are bold and underlined.
| Mutated residue | Location in Cyt1Aa toxin | Sequence of mutagenic oligonucleotide |
|---|---|---|
| C7S | N-terminal | 5′-GAAAATTTAAATCATTCTCCATTAGAAGATATAAAGGTAAATCC-3′ |
| S70C | β-Hairpin | 5′-CAAAATGCATTAGTTCCCACTTGTACAGATTTTGGTGATGCCCTAC-3 |
| N89C | Helix α-2 | 5′-CAAAAGGTTTAGAAATCGCATGCACAATTACACCGATGGGTGC-3 |
| S99C | β-4 | 5′-CACCGATGGGTGCTGTAGTGTGTTATGTTGATCAAAATGTAAC-3′ |
| L120C | Helix α-3 | 5′-GTGTTATGATTAATAAAGTCTGCGAAGTGTTAAAAACTGTATTAG-3 |
| A141C | Helix α-5 | 5′-CTGTAATAGATCAATTAACTTGCGCAGTTACAAATACGTTTAC-3′ |
| L176C | β-6 | 5′-CAAATTACACATACAATGTCTGCTTTGCAATCCAAAATGCCCAAAC-3 |
| C190C | β-7 | 5′-CAAACTGGTGGCGTTATGTATTCTGTACCAGTTGGTTTTGAAATTAAAG-3′ |
| V193C | β-7 | 5′-GCGTTATGTATTGTGTACCATGCGGTTTTGAAATTAAAGTATC-3′ |
| N218C | β-8 | 5′-CAAGATTCTGCGAGCTACTGTGTTAACATCCAATCTTTGAAATTTG-3′ |
| F226C | β-8 | 5′-GTTAACATCCAATCTTTGAAATGTGCACAACCATTAGTTAGCTC-3′ |
Production of Cyt1Aa and Cry11Aa proteins
Cyt1Aa and Cry11Aa protoxins were produced in B. thuringiensis 407− acrystalliferous strain transformed with WT or mutated plasmids. Plasmid pWF45 express cyt1Aa gene (40) and plasmid pCG6 express cry11Aa gene (42). Bt transformant strains were grown for 4 days at 30 °C in HCT sporulation medium (43) supplemented with 10 μg/ml Erm for Cyt1Aa or 25 μg/ml Erm for Cry11Aa. Spores and crystals were washed four times with 0.3 m NaCl, 0.01 m EDTA, pH 8, and four times with 1 mm phenylmethylsulfonyl fluoride by centrifugation for 10 min at 10,000 rpm at 4 °C, and the pellet was stored at 4 °C. Cry11Aa crystal inclusions were purified by centrifugation in sucrose gradients as described (16), whereas those of Cyt1Aa were purified by the aqueous two-phase system as previously described (44). This aqueous two-phase system includes 40% phosphate buffer (15 g of K2HPO4, 5 g of KH2PO4, 30 g of H2O) and 40% PEG (20 g of PEG 4000, 30 g of H2O). Briefly, each spore/Cyt1Aa-crystal suspension was suspended in 0.1% Triton X-100 (0.2 g spore/crystal, 0.2 ml of 0.1% Triton X-100 v/v), 1 ml of H2O, and 1.6 g of PEG at 40% were added and mixed in vortex. 1 g of KHPO4 at 40% was added, and suspension was mixed again by vortex. The samples were centrifuged 1 min at 500 rpm. The crystals in the interphase were recovered and stored at 4 °C. The Cyt1A proteins were solubilized 1 h at 37 °C in 50 mm Na2CO3, 0.2% of β-mercaptoethanol, pH 10.5, with slight shaking (350 rpm). Soluble proteins were recovered in the supernatant after centrifugation for 10 min at 14,000 rpm 4 °C. Cyt1Aa protoxin was activated with trypsin 1:50 ratio (trypsin: Cyt1Aa ratio) (Sigma–Aldrich) (w/w) at pH 8.5 for 2 h at 37 °C with agitation 350 rpm. After this, incubation 1 mm of phenylmethylsulfonyl fluoride was added to stop proteolysis. Protein concentration was determined by the Bradford assay and by UV absorbance on a Nanodrop 2000 (Thermo Scientific), and the protein profile was analyzed in SDS-PAGE with 15% acrylamide. Activated Cyt1Aa toxins were purified by anion-exchange chromatography on a GE Healthcare HiTrapTM Q HP column (Uppsala, Sweden) in an AKTA FPLC system from Amsterdam Biosciences by using a 50 mm Na2CO3, pH 8.5 buffer and a linear NaCl concentration gradient from 50 to 600 mm. Elution of protein fractions were analyzed by Coomassie Blue staining in SDS-PAGE with 15% acrylamide.
Insect bioassays
A. aegypti insects were reared at Instituto de Biotecnologia, Universidad Nacional Autónoma de México facilities at 28 °C, 75% humidity, with 12 h:12 h light:dark photoperiods. Bioassays were performed with different concentrations (25–5000 ng/ml) of spore/crystal suspensions of Cyt1Aa against 10 early fourth-instar larvae in 100 ml of dechlorinated water. Negative control (dechlorinated water) was included in the bioassay. Larvae viability was examined after 24 h. The mean lethal concentration (LC50) was determined by Probit analysis (Polo-Plus LeOra Software) using statistical parameters using data obtained in triplicate from three independent assays.
Qualitative estimation of synergism was determined by the analysis of a Cyt1Aa toxin amount that would produce 10% mortality (different for each Cyt1Aa mutant depending on its LC50 value) and a Cry11Aa concentration that would produce 20% mortality (95 ng/ml). Additive toxicities will produce 30% mortality, whereas synergistic interactions would produce much higher toxicity. The quantitative estimation of synergism (synergism factor, SF), between Cyt1Aa and Cry11Aa proteins was done as previously described according to a Tabashnik equation assuming a simple additive effect (45). For each ratio of toxin mixtures, the theoretical LC50 value is the harmonic mean of the intrinsic LC50 values of each component weighted by the ratio used in the mixture,
| (Eq. 1) |
where rCyt1A and rCry11A are the Cyt1A and Cry11A protein proportions used in the final ratio of the mixture, and LC50(Cyt1A) and LC50(Cry11A) are the LC50 values for each individual toxin. SF was calculated by dividing the theoretical toxicity by the observed toxicity of the mixture in bioassays. SF values greater than 1 indicate synergism.
Hemolysis assays
These assays were done as previously described (46). Rabbit RBC were washed three times in buffer A (0.1 m dextrose, 0.07 m NaCl, 0.02 m sodium citrate, 0.002 m citrate, pH 7.4) and diluted to 2 × 108 cells/ml in buffer A. The samples containing 20 μl of washed RBC and 1000 ng of Cyt1Aa toxin in a final volume of 200 μl of buffer A were incubated at 37 °C for 30 min in 96-well microtiter plates. The supernatants were collected in a new microtiter plate after centrifugation at 2,500 × g for 5 min at 4 °C, and hemolytic activity was quantified by measuring the absorbance of the supernatant at 405 nm. Positive control showing 100% hemolysis was defined after incubation of the same volume of RBC with dechlorinated H2O. Negative control was RBC incubated with buffer A. These assays were performed four to six times in triplicate each time. The data are presented as box-and-whisker plots constructed with GraphPad Prism program. A t test was performed using the statistical program GraphPad Prism.
Labeling mutant proteins with fluorescent dyes
Pure Cyt1Aa protein samples were incubated with 3 mm DTT, 1 mm EDTA for 15 min at room temperature to improve the labeling of the Cys residues. The DTT was removed by dialyzing exhaustively with PBS buffer, pH 7.2, and centrifugation for 15 min at 14,000 rpm at 4°C. The soluble proteins were quantified by UV absorbance on a Nanodrop 2000. Proteins (200–300 μg) were then incubated overnight in the dark at 4 °C with 20-fold molar excess of the probe. Alexa Fluor 488 from Molecular Probes was used to label the Cyt1Aa mutants. Unbound label was removed by dialyzing exhaustively with PBS, pH 7.2. The efficiency of labeling was measured using the molar extinction coefficient of each probe and the following equation,
| (Eq. 2) |
where Ax corresponds to the maximum wavelength of dye absorbance, 495 nm for Alexa Fluor 488, ε corresponds to the molar extinction coefficient of Alexa Fluor 488, which is 72,000 m−1 cm−1 at 495 nm, and MW is the molecular weight.
The purity of these proteins was analyzed on SDS-PAGE with 15% acrylamide. The labeling of the proteins was visualized by excitation of the SDS-PAGE gel with UV light transilluminator and analyzed in an Amersham Biosciences Imager 600 (GE Healthcare).
Preparation of BBMV
Insect midgut tissue of fourth-instar A. aegypti larvae were dissected and used to prepare BBMV by the differential precipitation method described by Wolfersberger et al. (47) using MgCl2 in absence of protease inhibitors. The BBMV were finally suspended in 50 mm Na2CO3, pH 9, and stored at −70 °C until used. BBMV protein concentration was analyzed using the Lowry DC protein assay (Bio-Rad).
Preparation of synthetic liposomes
SUVs were prepared as previously described (23). We used a mixture in a 10:3:1 proportion (PC:Ch:S) (Avanti Polar Lipids, Alabaster, AL, USA, and Sigma–Aldrich) from chloroform stocks. This lipid mixture was dried by nitrogen flow evaporation, followed by overnight storage under vacuum to remove residual chloroform. The lipid mixture was finally hydrated in 10 mm CHES, 150 mm KCl, pH 9, for 1 h at room temperature. The final concentration of the total lipid mixture after hydration was 1 mm. The samples were extensively mixed by vortex and sonicated three to five times for 20 s each in a Branson-1200 bath sonicator (Danbury, CT). SUVs were used the same day of preparation.
Steady-state fluorescence quenching measurements
Experiments were carried out in a Tecan Infinite M1000 Pro microplate reader. The excitation wavelength was 495 nm for Alexa Fluor 488, and the emission spectrum was recorded from 450 to 550 nm. The spectra show the averages of three to four scans and were also corrected for background and dilution.
Alexa Fluor 488–labeled proteins were incorporated in the three selected membrane systems. For incorporation into BBMV, 1–5 µg of Alexa Fluor 488–labeled proteins were incubated with 10 μg of A. aegypti BBMV in a final volume of 50 μl of 50 mm Na2CO3, pH 10.5 buffer for 1 h at 30°C with shaking at 350 rpm. For incorporation into synthetic SUV liposomes 0.2–1 µg of labeled proteins were mixed with 90 μl of synthetic SUV liposomes in 50 mm Na2CO3, pH 10.5 buffer for 1 h at 30°C with shaking at 350 rpm. For incorporation into RBC 0.4–2 µg of labeled proteins were mixed with 95 μl of buffer A (reported above in hemolysis assays) containing 2× 108 RBC/ml prepared as described above with shaking at 350 rpm for 1 h at 37 °C. After incubation with these membranes, the samples were ultracentrifuged for 30 min at 55,000 rpm at 4 °C. The supernatant was discarded, and the pellets were washed once. The BBMV and SUV were suspended in 50 mm Na2CO3 buffer, pH 10.5, whereas RBC were suspended in buffer A. The membranes without Alexa Fluor 488–labeled proteins were included as controls.
Fluorescent quenching experiments were performed with KI for Alexa Fluor 488–labeled proteins. KI was added to samples at final concentrations of 0, 50, 100, 250, and 500 mm. KCl was used to maintain ionic strength at 500 mm in all samples. Na2S2O3 was added at 8 mm to the sample to avoid production iodine ions. The volumes were adjusted to 200 μl with sample buffer. The emission spectra were recorded. Equivalent samples without labeled Cyt1Aa but with KI were used as negative controls.
Effective Stern–Volmer constants (KSV) were obtained from the fluorescent data according to the Stern–Volmer equation for dynamic quenching (48),
| (Eq. 3) |
where Fo and F are the florescence intensities in the absence and presence of the quencher, respectively. The value for KSV was obtained from Stern–Volmer plots, calculating the slope of Fo/F versus concentration of quencher. The value for KSV is considered to be a reliable reflection of the bimolecular collisional constant for collisional quenching, because KSV = kq·to, where kq is the bimolecular collisional constant, and to is the lifetime constant in the absence of quencher.
Structural models
Structural models were constructed with Swiss PDB viewer program drawing each α-helix and β-strand accordingly to the data of the Table 3 that indicated the possibility of each labeled residue to be exposed the solvent.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We are grateful to Lizbeth Cabrera and Jorge Sánchez for technical assistance.
This article contains supporting information.
Funding and additional information—This work was supported by National Institutes of Health Grant 5R01AI066014 (to S. S. G., M. S., and A. B.) and Dirección general de asuntos del personal académico, Universidad Nacional Autónoma de México Grant IN202718 (to A. B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare no conflicts of interest in regards to this manuscript.
- Bti
- B. thuringiensis subsp. israelensis
- BBMV
- brush border membrane vesicles
- SUV
- small unilamellar vesicles
- PC
- egg-yolk phosphatidyl choline
- Ch
- cholesterol
- S
- stearylamine
- KI
- potassium iodide
- RBC
- red blood cells
- SF
- synergism factor
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid.
References
- 1. Staples J. E., Breiman R. F., and Powers A. M. (2009) Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 49, 942–948 10.1086/605496 [DOI] [PubMed] [Google Scholar]
- 2. Kyle J. L., and Harris E. (2008) Global spread and persistence of dengue. Annu. Rev. Microbiol. 62, 71–92 10.1146/annurev.micro.62.081307.163005 [DOI] [PubMed] [Google Scholar]
- 3. Enserink M. (2015) Infectious diseases: an obscure mosquito-borne disease goes global. Science 350, 1012–1013 10.1126/science.350.6264.1012 [DOI] [PubMed] [Google Scholar]
- 4. Lacey L. A. (2007) Bacillus thuringiensis serovar israelensis and Bacillus sphaericus for mosquito control. J. Am. Mosq. Control. Assoc. 23, 133–163 10.2987/8756-971X(2007)23[133:BTSIAB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 5. Berry C., O'Neil S., Ben-Dov E., Jones A. F., Murphy L., Quail M. A., Holden M. T., Harris D., Zaritsky A., and Parkhill J. (2002) Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68, 5082–5095 10.1128/AEM.68.10.5082-5095.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wirth M. C., Georghiou G. P., and Federici B. A. (1997) CytA enables CryIV endotoxins of Bacillus thuringiensis to overcome high levels of CryIV resistance in the mosquito, Culex. Proc. Natl. Acad. Sci. U.S.A. 94, 10536–10540 10.1073/pnas.94.20.10536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bravo A., Gill S. S., and Soberón M. (2007) Mode of action of Bacillus thuringiensis toxins and their potential for insect control. Toxicon 49, 423–435 10.1016/j.toxicon.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khasdan V., Ben-Dov E., Manasherob R., Boussiba S., and Zaritsky A. (2001) Toxicity and synergism in transgenic Escherichia coli expressing four genes from Bacillus thuringiensis subsp. israelensis. Environ. Microbiol. 3, 798–806 10.1046/j.1462-2920.2001.00253.x [DOI] [PubMed] [Google Scholar]
- 9. Cantón P. E., Zanicthe Reyes E. Z., Ruiz de Escudero I., Bravo A., and Soberón M. (2011) Binding of Bacillus thuringiensis subsp. israelensis Cry4Ba to Cyt1Aa has an important role in synergism. Peptides 32, 595–600 10.1016/j.peptides.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pérez C., Fernández L. E., Sun J., Folch J. L., Gill S. S., Soberón M., and Bravo A. (2005) Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc. Natl. Acad. Sci. U.S.A. 102, 18303–18308 10.1073/pnas.0505494102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pérez C., Muñoz-Garay C., Portugal L. C., Sánchez J., Gill S. S., Soberón M., and Bravo A. (2007) Bacillus thuringiensis ssp. israelensis Cyt1Aa enhances activity of Cry11Aa toxin by facilitating the formation of a pre-pore oligomeric structure. Cell. Microbiol. 9, 2931–2937 10.1111/j.1462-5822.2007.01007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen S., Albeck S., Ben-Dov E., Cahan R., Firer M., Zaritsky A., and Dym O. (2011) Cyt1Aa toxin: crystal structure reveals implications for its membrane-perforating function. J. Mol. Biol. 413, 804–814 10.1016/j.jmb.2011.09.021 [DOI] [PubMed] [Google Scholar]
- 13. Gill S. S., Singh G. J. P., and Hornung J. M. (1987) Cell membrane interaction of Bacillus thuringiensis subsp. israelensis cytolytic toxins. Infect. Immun. 55, 1300–1308 10.1128/IAI.55.5.1300-1308.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Federici B. A., and Bauer L. S. (1998) Cyt1Aa protein of Bacillus thuringiensis is toxic to the cottonwood leaf beetle, Chrysomela scripta, and suppresses high levels of resistance to Cry3Aa. Appl. Environ. Microbiol. 64, 4368–4371 10.1128/AEM.64.11.4368-4371.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porcar M., Grenier A.-M., Federici B., and Rahbe Y. (2009) Effects of Bacillus thuringiensis δ-endotoxins on the pea aphid (Acyrthosiphon pisum). Appl. Environm. Microbiol. 75, 4897–4900 10.1128/AEM.00686-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas W. E., and Ellar D. J. (1983) Bacillus thuringiensis var. israelensis crystal δ endotoxin: effects on insect and mammalian cells in vitro and in vivo. J. Cell Sci. 60, 181–197 [DOI] [PubMed] [Google Scholar]
- 17. Bravo A., López-Diaz J. A., Yamamoto T., Harding K., Zhao J. J., Mendoza G., Onofre J., Torres M. C., Nelson M. E., Wu G., Sethi A., and Soberón M. (2018) Susceptible and mCry3A resistant corn rootworm larvae killed by a non-hemolytic Bacillus thuringiensis Cyt1Aa mutant. Sci. Rep. 8, 17805 10.1038/s41598-018-36205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soberón M., López-Díaz J. A., and Bravo A. (2013) Cyt toxins produced by Bacillus thuringiensis: a protein fold conserved in several pathogenic microorganims. Peptides 41, 87–93 10.1016/j.peptides.2012.05.023 [DOI] [PubMed] [Google Scholar]
- 19. Knowles B. H., Blatt M. R., Tester M., Horsnell J. M., Carroll J., Menestrina G., and Ellar D. J. (1989) A cytolytic δ-endotoxin from Bacillus thuringiensis var. israelensis forms cation-selective channels in planar lipid bilayers. FEBS Lett. 244, 259–262 10.1016/0014-5793(89)80540-X [DOI] [PubMed] [Google Scholar]
- 20. Promdonkoy B., and Ellar D. J. (2000) Membrane pore architecture of a cytolytic toxin from Bacillus thuringiensis. Biochem. J. 350, 275–282 10.1042/0264-6021:3500275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manceva S. D., Pusztai-Carey M., Russo P. S., and Butko P. (2005) A detergent-like mechanism of action of the cytolytic toxin Cyt1A from Bacillus thuringiensis var. israelensis. Biochemistry 44, 589–597 10.1021/bi048493y [DOI] [PubMed] [Google Scholar]
- 22. Li J., Derbyshire D. J., Promdonkoy B., and Ellar D. J. (2001) Structural implications for the transformation of the Bacillus thuringiensis δ-endotoxins from water-soluble to membrane-inserted forms. Biochem. Soc. Trans. 29, 571–577 10.1042/bst0290571 [DOI] [PubMed] [Google Scholar]
- 23. López-Diaz J. A., Cantón P. E., Gill S. S., Soberón M., and Bravo A. (2013) Oligomerization is a key step in Cyt1Aa membrane insertion and toxicity but not necessary to synergize Cry11Aa toxicity in Aedes aegypti larvae. Environ. Microbiol. 15, 3030–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chow E., Singh G. J., and Gill S. S. (1989) Binding and aggregation of the 25 kDa toxin of Bacillus thuringiensis subsp. israelensis to cell membranes and alteration by monoclonal antibodies and amino acid modifiers. Appl. Environ. Microbiol. 55, 2779–2788 10.1128/AEM.55.11.2779-2788.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Promdonkoy B., and Ellar D. J. (2003) Investigation of the pore-forming mechanism of a cytolytic δ-endotoxin from Bacillus thuringiensis. Biochem. J. 374, 255–259 10.1042/bj20030437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Promdonkoy B., Rungrod A., Promdonkoy P., Pathaichindachote W., Krittanai C., and Panyim S. (2008) Amino acid substitutions in αA and αC of Cyt2Aa2 alter hemolytic activity and mosquito-larvicidal specificity. J. Biotechnol. 133, 287–293 10.1016/j.jbiotec.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 27. Butko P., Huang F., Pusztai-Carey M., and Surewicz W. K. (1996) Membrane permeabilization induced by cytolytic δ-endotoxin CytA from Bacillus thuringiensis var. israelensis. Biochemistry 35, 11355–11360 10.1021/bi960970s [DOI] [PubMed] [Google Scholar]
- 28. Tetreau G., Banneville A.-S., Andreeva E. A., Brewster A. S., Hunter M. S., Sierra R. G., Teulon J.-M., Young I. D., Burke N., Grünewald T. A., Beaudouin J., Snigireva I., Fernandez-Luna M. T., Burt A., Park H.-W., et al. (2020) Serial femtosecond crystallography on in vivo–grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade. Nat. Commun. 11, 1153 10.1038/s41467-020-14894-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anaya P., Onofre J., Torres-Quintero M. C., Sánchez J., Gill S. S., Bravo A., and Soberón M. (2020) Oligomerization is a key step for Bacillus thuringiensis Cyt1Aa insecticidal activity but not for toxicity against red blood cells. Insect Biochem. Mol. Biol. 119, 103317 10.1016/j.ibmb.2020.103317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suktham K., Pathaichindachote W., Promdonkoy B., and Krittanai C. (2013) Essential role of amino acids in αD-β4 loop of a Bacillus thuringiensis Cyt2Aa2 toxin in binding a complex formation on lipid membrane. Toxicon 74, 130–137 10.1016/j.toxicon.2013.08.053 [DOI] [PubMed] [Google Scholar]
- 31. Du J., Knowles B. H., Li J., and Ellar D. J. (1999) Biochemical characterization of Bacillus thuringiensis cytolytic toxins in association with a phospholipid bilayer. Biochem. J. 338, 185–193 10.1042/0264-6021:3380185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J., Koni P. A., and Ellar D. J. (1996) Structure of the mosquitocidal δ-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J. Mol. Biol. 257, 129–152 10.1006/jmbi.1996.0152 [DOI] [PubMed] [Google Scholar]
- 33. Promdonkoy B., and Ellar D. J. (2005) Structure function relationships of a membrane pore forming toxin revealed by reversion mutagenesis. Mol. Membr. Biol. 22, 327–337 10.1080/09687860500166192 [DOI] [PubMed] [Google Scholar]
- 34. Butko P. (2003) Cytolytic toxin Cyt1Aa and its mechanism of membrane damage: data and hypotheses. Appl. Environ. Microbiol. 69, 2415–2422 10.1128/AEM.69.5.2415-2422.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Butko P., Huang F., Pusztai-Carey M., and Surewicz W. K. (1997) Interaction of the δ-endotoxin CytA from Bacillus thuringiensis var. israelensis with lipid membranes. Biochemistry 36, 12862–12868 10.1021/bi9702389 [DOI] [PubMed] [Google Scholar]
- 36. Tharad S., Moreno-Cencerrado A., Üzulmez Ö., Promdonkoy B., and Toca-Herrera J. L. (2017) Bacillus thuringiensis Cyt2Aa binding on lipid/cholesterol bilayer depends on protein concentration and time. Biochem. Biophys. Res. Commun. 492, 212–217 10.1016/j.bbrc.2017.08.051 [DOI] [PubMed] [Google Scholar]
- 37. Berube B. J., and Bubeck-Wardenburg J. (2013) Staphylococcus aureus α-toxin: nearly a century of intrigue. Toxins (Basel) 5, 1140–1166 10.3390/toxins5061140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilbert R. J. (2010) Cholesterol-dependent cytolysins. Adv. Exp. Med. Biol. 677, 56–66 10.1007/978-1-4419-6327-7_5 [DOI] [PubMed] [Google Scholar]
- 39. Abrami L., Fivaz M., and van der Goot F. G. (2000) Adventures of a pore-forming toxin at the target cell surface. Trends Microbiol. 8, 168–172 10.1016/S0966-842X(00)01722-4 [DOI] [PubMed] [Google Scholar]
- 40. Wu D., Johnson J. J., and Federici B. A. (1994) Synergism of mosquitocidal toxicity between CytA and CryIV proteins using inclusions produced from cloned genes of Bacillus thuringiensis subsp. israelensis. Mol. Microbiol. 13, 965–972 10.1111/j.1365-2958.1994.tb00488.x [DOI] [PubMed] [Google Scholar]
- 41. Lereclus D., Arantes O., Chaufaux J., and Lecadet M. M. (1989) Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 60, 211–217 10.1016/0378-1097(89)90511-9 [DOI] [PubMed] [Google Scholar]
- 42. Chang C., Yu Y. M., Dai S. M., Law S. K., and Gill S. S. (1993) High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl. Environ. Microbiol. 59, 815–821 10.1128/AEM.59.3.815-821.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muñóz-Garay C., Rodríguez-Almazán C. R., Aguilar J. N., Portugal L., Gómez I., Saab-Rincon G., Soberón M., and Bravo A. (2009) Oligomerization of Cry11Aa from Bacillus thuringiensis has an important role in toxicity against Aedes aegypti. Appl. Environ. Microbiol. 75, 7548–7550 10.1128/AEM.01303-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Güereca L., Bravo A., and Quintero R. (1994) Design of an aqueous two-phase system for the purification of ICP from Bacillus thuringiensis. Process Biochem. 29, 181–185 10.1016/0032-9592(94)85002-X [DOI] [Google Scholar]
- 45. Tabashnik B. E. (1992) Evaluation of synergism among Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 58, 3343–3346 10.1128/AEM.58.10.3343-3346.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rodriguez-Almazan C., Ruiz de Escudero I., Cantón P. E., Muñoz-Garay C., Pérez C., Gill S. S., Soberón M., and Bravo A., (2011) The amino- and carboxyl-terminal fragments of the Bacillus thuringiensis Cyt1Aa toxin have differential roles on toxin oligomerization and pore formation. Biochemistry 50, 388–396 10.1021/bi101239r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolfersberger M., Luethy P., Maurer A., Parenti P., Sacchi F. V., Giordana B., and Hanozet G. M. (1987) Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86, 301–308 10.1016/0300-9629(87)90334-3 [DOI] [Google Scholar]
- 48. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, 3rd Ed., Springer Science, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.