Abstract
Core binding factor beta (Cbfβ), the partner protein of Runx family transcription factors, enhances Runx function by increasing the binding of Runx to DNA. Null mutations of Cbfb result in embryonic death, which can be rescued by restoring fetal hematopoiesis but only until birth where bone formation is still nearly absent. Here we address a direct role of Cbfβ in skeletal homeostasis by generating osteoblast-specific Cbfβ-deficient mice (CbfbΔob/Δob) from Cbfb-floxed mice crossed with mice expressing Cre from the Col1a1 promoter. CbfbΔob/Δob mice showed normal growth and development, but exhibited reduced bone mass, particularly of cortical bone. The reduction of bone mass in CbfbΔob/Δob mice is similar to the phenotype of mice with haploinsufficiency of Runx2. Although the number of osteoblasts remained unchanged, the number of active osteoblasts decreased in CbfbΔob/Δob mice and resulted in lower mineral apposition rate. Immunohistochemical and quantitative real-time PCR analyses showed that the expression of osteogenic markers, including Runx2, osterix, osteocalcin and osteopontin, was significantly repressed in CbfbΔob/Δob mice compared to wild type mice. Cbfβ deficiency also reduced Runx2 protein levels in osteoblasts. The mechanism was revealed by forced expression of Cbfβ which increased Runx2 protein levels in vitro by inhibiting polyubiquitination-mediated proteosomal degradation. Collectively, these findings indicate that Cbfβ stabilizes Runx2 in osteoblasts by forming a complex, and thus facilitates the proper maintenance of bone mass, particularly cortical bone.
Keywords: Cbfβ, osteoblasts, bone mass, polyubiquitination, Runx2
INTRODUCTION
Runx2 is an essential regulator of bone formation, which is identified by the absence of mineralized skeleton in Runx2 null (Runx2−/−) mice and mice expressing a Runx2 mutant protein that truncates the functional C-terminus (1–3). Haploinsufficiency of Runx2 and deletion of one isoform of Runx2 result in a lack of cranial suture closure and low bone mass in mice (4–6). In vitro cell studies established that Runx2 is the “master” transcription regulator for osteoblast differentiation (7, 8). Together these findings indicate that the level of cellular Runx2 is crucial for the regulation of bone formation and maintenance of bone homeostasis in skeletal tissues.
Many Runx2 partner proteins regulate the function and protein levels of Runx2 during osteogenesis and osteoblast differentiation (9). Core binding factor β (Cbfβ), a key cofactor of Runx transcription factors that enhances their DNA binding, is expressed ubiquitously (10–12). Cbfβ cannot bind to DNA directly, but it facilitates the transcriptional activities of Runx transcription factors through forming a Runx/Cbfβ complex (13, 14). Cbfβ knockout (Cbfb−/−) mice die before bone formation occurs due to failure of fetal liver hematopoiesis and brain hemorrhage (15, 16). Cbfb−/− mice rescued from ablation of fetal hematopoiesis were viable but exhibited severely compromised bone formation, similar to Runx2−/− mice (17, 18), pointing to a possible essential function of Cbfβ in bone formation. Interestingly, the hypomorphic expression of Cbfβ delays endochondral bone formation in relations with Runx2 (19). However, the specific mechanism of how Cbfβ supports skeletal homeostasis is not known.
To directly address the function of Cbfβ on bone formation and investigate the relationship between Cbfβ, Runx2, and postnatal bone formation in vivo, we deleted Cbfβ from mature osteoblasts using Col1a1-Cre and Cbfb-floxed mice. Both 5 week and 12 week-old mice were assessed to observe the role of osteoblast-specific Cbfβ during both growth and maturity.
MATERIALS AND METHODS
Materials
Alkaline phosphatase (ALP) kits, silver nitrate, Alizarin red S, methyl green, toluidine blue, calcein, β-glycerophosphate, ascorbic acid, and anti-Flag monoclonal antibody were purchased from Sigma-Aldrich (St. Louis, MO). ECL western blotting detection reagents and Protein A Sepharose were provided by GE Healthcare (Bucks, UK). Polyclonal antibodies of Osterix (Abcam, Cambridge, UK) and Myc (Abcam), mouse monoclonal antibody for Myc (Invitrogen, Carlsbad, CA) were purchased. Other antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), including rabbit polyclonal antibodies for Cbfβ, Runx2, and Osteocalcin, goat polyclonal antibodies for Lamin B1, goat anti-mouse or rabbit IgG and donkey anti-goat IgG conjugated with horseradish peroxidase (HRP), and mouse or rabbit normal IgG.
Animal models
Mice harboring an osteoblast-specific deletion of Cbfb (CbfbΔob/Δob) were generated by crossing Col1a1-Cre transgenic mice (CreTg/+) (20) with Cbfbfl/fl mice (21), both maintained on C57BL/6N background. Cbfbfl/fl and Cbfbfl/+ mice served as wild type control (WT). Five and 12 week-old male mice were used for in vivo assessments to investigate the role of Cbfβ in mature osteoblasts during growth and maturity, respectively. . All animal procedures were carried out in accordance with the guidelines issued by the Institutional Animal Care and Use Committee of Kyungpook National University (KNU-201079).
Micro-computed tomography (μCT) analysis
Micro-CT scanning was accomplished using eXplore Locus SP (GE Healthcare, London, Ontario, Canada) with 8 μM resolution. All bone morphometric parameters were calculated three-dimensionally with eXplore MicroView version 2.2 (GE Healthcare). Bone parameters and density were analyzed at the region between 0.7 mm and 2.3 mm below the growth plate of the distal femur. All bone μCT nomenclature follows the guidelines of the American Society for Bone and Mineral Research (ASBMR) (22).
Bone histological and morphological analyses
Mice were sacrificed at 5 and 12 weeks and fixed in 4% paraformaldehyde (Merck, Darmstadt, Germany) at 4°C overnight. To estimate dynamic histomorphometry, 30 mg/kg body weight of calcein was injected twice before sacrifice intraperitoneally separated by 3 or 5 days. For bone histological analysis, tibiae were embedded in paraffin after decalcification with 10% EDTA and sectioned at a thickness of 3 μm. For osteocyte analysis, we used modified Bodian staining methods (23). For von Kossa staining, undecalcified bones were embedded in methyl-methacrylate (Sigma) and sectioned at a thickness of 6 μm as previously described (24). Histomorphometrical analyses were performed with the Bioquant Osteo II program (Bio-Quant. Inc., Nashville, TN). All bone histomorphometry nomenclature follows the guidelines of the ASBMR (25). OxiPOHT POL model polarized microscope (Zeiss) was used to assess collagen fiber.
Immunohistochemistry
Endogenous peroxidase activity of sections was quenched with 3% H2O2 and then antigens were retrieved by boiling in TEG-buffer (1.211 g of Tris and 0.190 g of EGTA in 1L MilliQ-water, pH 9.0). After blocking with 1% bovine serum albumin for 1 hour at room temperature, sections were incubated with anti-Cbfβ, anti-Runx2, anti-Osterix, and anti-Osteocalcin antibodies for 16 hours at 4°C and bound antibodies were detected with goat anti-rabbit IgG conjugated with HRP. Signals were visualized by developing with a DAB substrate-chromogen system (Dakocytomation, Denmark).
Culture and differentiation of bone marrow stromal cells (BMSCs)
BMSCs were isolated from femur and tibia of 6-8 week-old mice and cultured as previously described (4). BMSCs were plated into 24-well culture dishes at a density of 2 × 104 cells/well and differentiated in vitro in osteogenic medium with 10 mM β-glycerophosphate and 50 μg/ml ascorbic acid for 4 weeks. To evaluate osteoblast differentiation, we performed ALP staining using ALP kits according to the manufacturer’s instructions and Alizarin red S staining.
Quantitative real-time PCR (Q-PCR) analyses
Total RNA from bone tissues of 4 week-old and 12 week-old mice and BMSCs differentiated in vitro for 2 weeks was isolated using the easy-BLUE Total RNA Extraction Kit (iNtRON Biotechnology, Seongnam-si, Gyeonggi-do, Korea) and cDNA was synthesized from 2 μg of total RNA using SuperScript II Reverse Transcriptase (Invitrogen). Q-PCR was performed using the Power SYBR green master mixture (Applied Biosystems, Foster city, CA). All primers used for Q-PCR analyses were designed using Primer Express software (Applied Biosystems). Primer descriptions are located in Table S1.
Primary osteoblast culture
Primary osteoblasts were isolated from the calvaria of E17.5 or E18.5 mice by serial digestion with 0.1% type II collagenase (Gibco, Grand Island, NY) and cultured in α-MEM medium (Welgene, Daegu, Korea) supplemented with 15% fetal bovine serum (FBS; Gibco) and penicillin/streptomycin (Lonza, Walkersville, MD).
Ubiquitination assay
MC3T3E1 cells and HEK293T cells were transfected with expression constructs of 3xFlag-Runx2, HA-Ubiqutin with or without Myc-Cbfβ and cells were incubated with 20 μM MG132 for 6 hours. Total cell lysates were isolated with a lysis buffer (20 mM HEPES, pH 7.9, 300mM KCl, 10% glycerol, 10% NP-40, 1mM DTT and protease inhibitors), precleared with protein A Sepharose beads (GE healthcare) for 30 minutes, and immunoprecipitated with anti-Flag antibody overnight at 4°C on a shaker. Protein A Sepharose beads were then added for 2 hours at 4°C on a shaker and mixtures were washed three times with immunoprecipitation (IP) buffer (20 mM HEPES, pH 7.9, 150 mM KCl, 10% glycerol, 0.1% NP-40, 1 mM DTT and protease inhibitors). Bound proteins were eluted by boiling the beads with 20 μl of gel loading buffer. Ubiquitination levels were checked by immunoblotting with anti-HA antibody. 3xFlag-Runx2 and Myc-Cbfβ were detected with anti-Flag antibody (Sigma) and anti-Myc antibody (Invitrogen), respectively.
Co-immunoprecipitation (Co-IP) assay
Expression constructs of 3xFlag-Runx2, Myc-Cbfβ, or co-expression of 3xFlag-Runx2 and Myc-Cbfβ were transfected into HEK293T cells. After 24 hours, the cells were treated with 100 ug/ml of dithiobis (succinimidyl propionate) (DSP, Pierce, Rockford, IL, USA) at 37°C for 20 minutes. Nuclear extracts were obtained with the ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit according to the manufacturer’s instructions (Fermentas LIFE SCIENCES). Nuclear extracts were used for Co-IP as previously described (26) using antibodies of anti-Myc (Invitrogen) and anti-IgG (Santa Cruz Biotechnology).
Statistical Analysis
Statistical differences were analyzed by Student’s t test using Sigma Plot 10.0 (Systat Software Inc, Chicago, IL). Results are presented as mean ± SDs and statistical significance was accepted when p < 0.05.
RESULTS
Reduced cortical bone thickness in CbfbΔob/Δobmice
Genomic deletion of exon5 of Cbfb was confirmed in long bone of CbfbΔob/Δob mice using genomic DNA PCR, which was obtained from long bone of WT and CbfbΔob/Δob mice (Fig. 1A). Cbfb mRNA expression in CbfbΔob/Δob bone tissue was reduced by 95% compared to WT (Fig. 1B). In addition, reduction of Cbfβ protein in tibiae was confirmed in Cbfb∆ob/∆ob mice by immunohistochemistry (Fig. 1C). Col1a1-Cre activity was characterized using Rosa26 reporter (R26R) mice (27). Mature osteoblasts of R26R; CreTg/+mice at E18.5 showed Cre activity in the ulna and metacarpal bones (Fig. S1).
Fig. 1. Osteoblast-specific deletion of Cbfb by Col1a1-Cre.
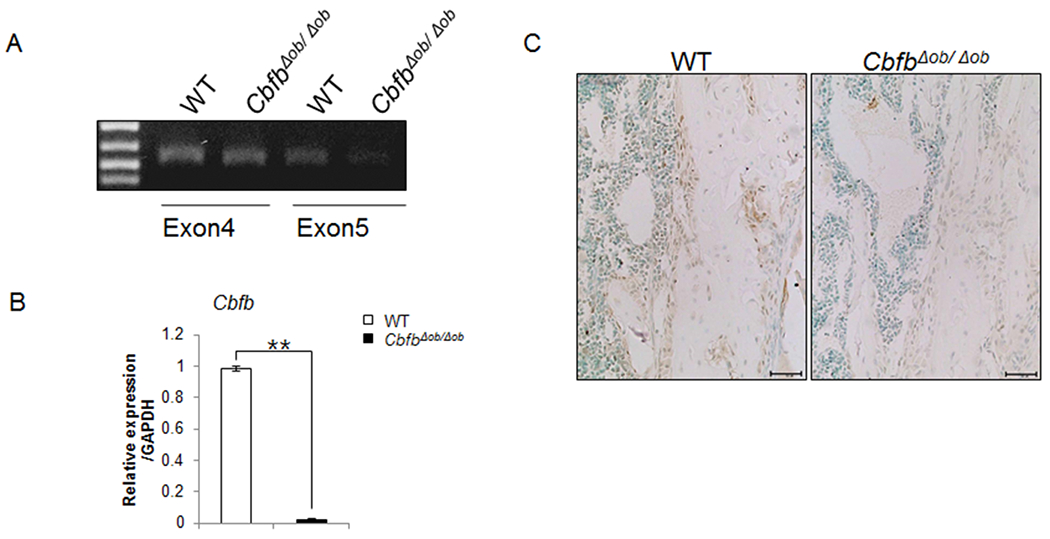
The osteoblast-specific deletion of Cbfb was determined by PCR using genomic DNA obtained from tibia (A). Reduction of Cbfβ was confirmed by Q-PCR using RNA isolated from bone tissue of 4-week-old mice (B). Immunohistochemistry of tibia shows deletion of Cbfβ in Cbfb∆ob/∆ob mice (C). The tibiae of 12-week-old CbfbΔob/Δob mice were used (A and C). WT mice (□, n = 3), CbfbΔob/Δob mice (■, n = 3). ** p<0.01. Results are mean ± SD.
CbfbΔob/Δob mice exhibited lower bone volume compared to WT at 12 weeks of age despite normal development (Fig. 2A). Reduction of bone volume was evident in µCT analysis. Femoral BV/TV was reduced by about 10 % in CbfbΔob/Δob mice compared to WT (Fig. 2B). Higher BS/TV and Tb.N and lower Tb.Th indicated more fragmented bone in CbfbΔob/Δob mice compared with WT mice. However, there was no significant difference in Tb.Sp. BMD of cortical bone was lower in CbfbΔob/Δob mice compared to WT mice. Ct.Th of CbfbΔob/Δob mice was reduced in the diaphysis (Fig. 2B) and humeri (Fig. 2C) compared to WT at 12 week-old age. Similar but more subtle phenotypes were observed in 5 week-old CbfbΔob/Δob mice. Bone mass of 5 week-old CbfbΔob/Δob was lower than WT and diaphyseal cortical thickness (Ct.Th) was 26% thinner compared to WT (Fig. S2). Interestingly, no significant difference in vertebral bone volume was detected at either age (data not shown). These results indicate that Cbfβ in mature osteoblasts is essential in maintaining Ct.Th.
Fig. 2. Lower BV/TV and Ct.Th in 12 week-old CbfbΔob/Δob mice.

Less tibiae bone in 12-week-old CbfbΔob/Δob mice compared to WT is observed by H&E staining (A) and μCT (B). Lower Ct.Th in humeri of CbfbΔob/Δob mice was confirmed by von Kossa staining(C). WT mice (□, n = 3–5), CbfbΔob/Δob mice (■, n = 3–7), *p<0.05 **p<0.01. Results are mean ± SD.
Reduced MAR and active osteoblast numbers in CbfbΔob/Δobmice
The endocortical MAR and mineralized surface to bone surface ratio (MS/BS) were compromised in 12 week-old CbfbΔob/Δob mice (Fig. 3A). Although the number of endocortical osteoblasts was similar, the number of active cuboidal-shaped osteoblasts was less in these mice compared to WT mice (Fig. 3B). These results were similar in 5 week-old CbfbΔob/Δob mice (data not shown). No significant differences in osteoblast proliferation and apoptosis were observed in 12 week-old CbfbΔob/Δob mice (Fig. S3). Interestingly, osteocyte processes appeared abnormal in 12 week-old CbfbΔob/Δob mice compared to WT mice, though the number of osteocytes was similar (Fig. S4A–B). We also confirmed the reduced expression of Phex and Dmp1 without change of Fgf23 by Q-PCR in bone tissues (Fig. S4C). These abnormal expressions of osteocytes markers may be associated with the woven bone-like appearance of cortical bone observed through polarized microscope (Fig. S4D). Though osteoblasts regulate osteoclastogenesis (28), osteoclast number (N.Oc), osteoclast surface/bone surface (Oc.S/BS) and activity (Fig. 3C, Fig. S5A) were not affected by osteoblast-specific deletion of Cbfβ. While the ratio of Rankl/Opg was increased, Rank and M-Csf were decreased in bone tissue of CbfbΔob/Δob mice as shown by Q-PCR (Fig. S5B). These data suggest that the lower Ct.Th in CbfbΔob/Δob mice is due to compromised osteoblast function and not osteoblast number or osteoclast activity.
Fig. 3. Lower osteoblast activity due to diminished population of cuboid osteoblasts in 12 week old CbfbΔob/Δob mice.
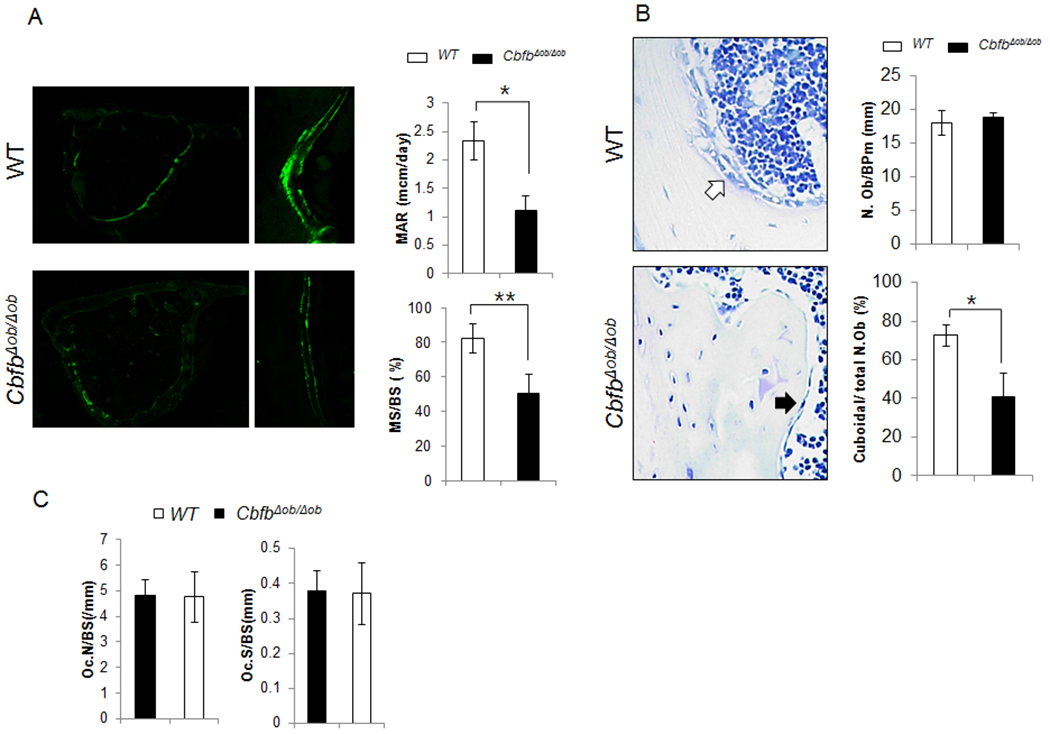
Endocortical MAR and MS/BS were lower in femurs of 12-week-old CbfbΔob/Δob mice compared to WT mice (A). The population of active cuboidal osteoblasts (Cuboidal N.Ob/total N.Ob) was lower in CbfbΔob/Δob mice compared to WT (B). Osteoclast number (N.Oc/T.A) and osteoclast surface (Oc.S/BS) were measured in both WT and CbfbΔob/Δob mice (C). WT (□, n=4–5), CbfbΔob/Δob mice (■, n=4–7). *p<0.05, ** p<0.01. Results are presented as mean ± SD.
Diminished osteogenic markers and delayed osteoblast differentiation of bone marrow stromal cells (BMSCs) in CbfbΔob/Δobmice
Osteoblast-specific deletion of Cbfb resulted in reduced Cbfβ and Runx2 levels in osteoblasts on the bone surface (Fig. 4A). Moreover, Runx2 downstream targets osterix and osteocalcin, were also diminished in CbfbΔob/Δob mice on the bone surface. The mRNA expressions of Cbfb and many osteogenic markers, such as Runx2 and its target genes that encode osterix, collagen type I, osteocalcin, osteopontin, ALP, and osteonectin, were already lower in CbfbΔob/Δob bone tissue compared to those from WT mice at 4 weeks (Fig. 4B). The reduced levels of osteogenic markers may be due in part to the compromised function of osteoblasts in the cortical bone of CbfbΔob/Δob mice.
Fig. 4. Lower expression of osteogenic markers in CbfbΔob/Δob mice compared to WT mice at 12 weeks.
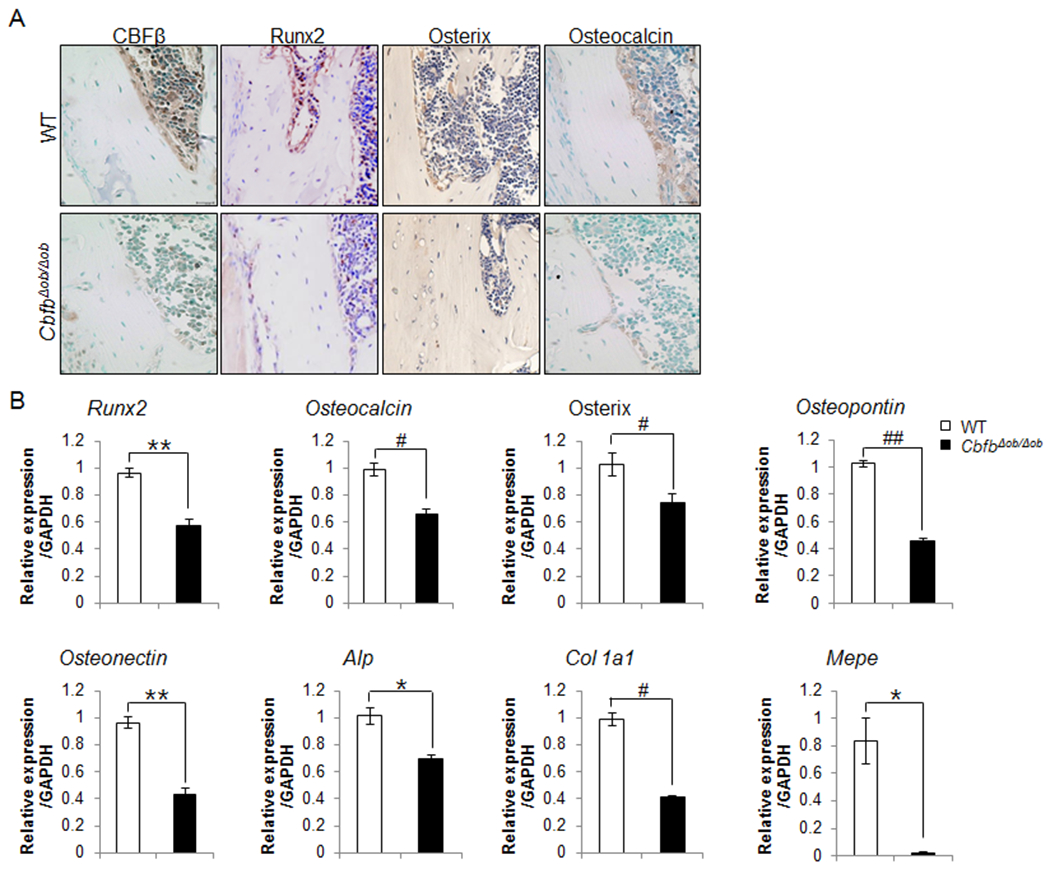
The levels of Runx2 and its downstream proteins, such as, osterix and osteocalcin were reduced in the tibiae of 12-week-old CbfbΔob/Δob mice, as determined by immunohistochemistry (A). The mRNA expression level of several osteogenic markers in bone tissue of CbfbΔob/Δob mice were reduced from bone tissues of 4-week-old mice (B). WT (□, n=3), CbfbΔob/Δob (■, n=3), *p<0.05, **p<0.01. Results are presented as means ± SD.
Initial histochemistry of in vitro analyses showed diminished osteoblast differentiation of CbfbΔob/Δob BMSCs compared to WT BMSCs at day 14 and 28 (Fig. S6A). The mRNA expressions of Cbfb and many osteogenic markers, such as Runx2 and its target genes that encode osterix, collagen type I, osteocalcin, osteopontin, ALP, and osteonectin, were lower in differentiated BMSCs derived from CbfbΔob/Δob compared to those from WT mice by day 14. (Fig. S6B). These results suggest that the osteoblast-specific deletion of Cbfb decreases Runx2 protein and mRNA levels and, consequently, the expression of downstream osteogenic markers which result in decreased synthesis of bone matrix in vitro and inhibit bone formation in vivo.
Effect of Cbfβ on the stability and nuclear localization of Runx2
Runx2 expression was severely diminished in primary osteoblasts derived from CbfbΔob/Δob mice (Fig. 5A). On the other hand, Runx2 expression was increased when Runx2 and Cbfβ were co-expressed in primary osteoblasts as compared with Runx2 expression alone (Fig. 5B). Co-expression of Cbfβ with Runx2 in HEK293T cells (Fig 5C) and MC3T3E1 cells (Fig. 5D) resulted in an increase in Runx2 levels and a dramatic decrease in polyubiquitination of Runx2 compared to when Runx2 was expressed alone. After transient transfection of Runx2 and Cbfb, Co-IP showed that Cbfβ formed a heterodimer with Runx2 (Fig. 5E upper panel) and localization of Runx2 and Cbfβ in the nucleus was increased when they were co-expressed in HEK293 cells (Fig. 5E, lower panel), suggesting that Cbfβ binds with Runx2 and increases the stability of Runx2 protein by protecting it from proteasomal degradation.
Fig. 5. Cbfβ protected Runx2 from polyubiquitination.

Runx2 protein levels were lower in cell extracts of calvarial osteoblasts from CbfbΔob/Δob mice than that of WT mice (A). Overexpression of Cbfβ in primary osteoblasts increased Runx2 (B). Polyubiquitination of Runx2 was lower when Runx2 was co-expressed with Cbfβ in HEK293 cells (C) and MC3T3E1 cells (D) compared to when Runx2 was expressed alone. Runx2/Cbfβ complex was detected in co-transfected cells (E, upper panel). Expression of Runx2 and Cbfβ were detected by anti-Flag and anti-Myc antibodies, respectively (E, lower panel). Both Runx2 and Cbfβ were higher in co-transfected HEK293 cell nuclear extracts (NE) compared to single transfected cells. Lamin B (LB) was shown as an internal control. WB, western blot; TL, total lysates.
DISCUSSION
Osteoblast-specific deletion of Cbfb decreased bone mass, particularly in cortical bone. The flat morphology of these osteoblasts, the thinner Ct.Th, and reduced MAR and MS/BS in CbfbΔob/Δob mice suggests that osteoblast activity is diminished. This is further supported by delayed osteoblast differentiation in BMSCs derived from CbfbΔob/Δob mice. The Runx2/Cbfβ complex in MC3T3-E1 mouse osteoblastic cell line inhibited polyubiquitination of Runx2, thereby maintaining adequate levels of cellular Runx2 and activating its osteogenic target genes for osteoblasts to function normally. On the other hand, osteoclastogenesis was not affected by the absence of Cbfβ in osteoblasts. These results demonstrate that Cbfβ plays an essential role in mature osteoblasts by modulating Runx2 protein stability in vivo during both growth and adulthood.
Runx2 is a rate-limiting factor for osteoblastic maturation and activity but the amount of Runx2/Cbfβ is important for normal skeletal development. While Cbfβ is an essential co-regulatory partner of Runx factors for DNA binding to target genes, overexpression of Cbfβ does not affect bone (29). However, overexpression of Runx2 results in osteopenia, which worsens with the overexpression of both Runx2 and Cbfβ (29). The bone phenotypes we observe in CbfbΔob/Δob mice are similar to the recently reported osteoblast-specific Runx2 deleted mice (30) and to earlier mouse models of Runx2 haploinsufficiency, such as low bone mass (5,6). Importantly, the differences we observed between CbfbΔob/Δob mice and WT mice were similar but more drastic in 12 week-old mice compared to 5 week-old mice, indicating a requirement of Cbfβ to maintain bone mass during aging. The results also demonstrate that abnormal amounts, either high or low, of Runx2/Cbfβ impact osteoblast maturation and activity. These findings, combined with ubiquitination results, indicate that Cbfβ is required to prevent proteosomal degradation of Runx2, which controls osteoblast maturation and activity.
Runx2 has a well characterized PPXY site for interacting with many WW domain proteins including Smurf1 and WWP1 which are E3 ubiquitin ligases and promote Runx2 ubiquitination and proteosomal degradation (31–33). The Runx2/Cbfβ complex may prevent Runx2 ubiquitination by suppressing the binding of WWP1 to Runx2. WWP1 promotes Runx2 ubiquitination by interacting with the Runt domain of Runx2 with Shunurri-3 (31–33). As a result, Shunurri-3 null mice show an increase of osteoblast function (33). The Runx2/Cbfβ may also modulate other posttranslational modifications, such as phosphorylation (34), acetylation (35), and isomerization (36) of Runx2. Abnormal amounts of Runx2 and Cbfβ affect osteoblast activity and osteocyte formation. Overexpression of osteoblastic Runx2 and Cbfβ inhibits osteoblast maturation and transition to osteocytes (29). On the other hand, deletion of Cbfb impaired osteoblast activity by affecting its shape and further resulted in abnormal canaliculi of osteocytes. Thus, a threshold level of both Runx2 and Cbfβ to form adequate amounts of the Runx2/Cbfβ complex is required for bone health.
In addition to the role of Cbfβ increasing Runx2 stability, Cbfβ may also affect Runx2-DNA binding affinity. It is well-known that co-expression of Cbfβ and Runx2 results in a higher Runx2-DNA binding affinity compared to whenRunx2 is expressed alone regarding skeletal development (18, 19). Therefore it is necessary to investigate how the Runx2-DNA binding affinity is affected in osteoblasts of CbfbΔob/Δob mice.
The more prominent CbfbΔob/Δob effect on cortical bone compared to trabecular bone is possibly due to the high expression of Runx2 in the perichondrium and periosteum. Cortical bone has layers and thus higher numbers of mature osteoblasts compared to trabecular bone. Our results support the concept that Runx2 is essential for commitment of progenitor cells to osteoblasts, thereby having a greater influence on cortical bone surrounded by periosteum. Thus higher ubiquitination of Runx2 in cortical bone of CbfbΔob/Δob mice may have resulted in less formation of cortical bone in these mice.
The first demonstration of in vivo functional significance of the Runx2/Cbfβ complex in osteoblasts is further supported by some cases of human cleidocranial dysplaysia (CCD). RUNX2 mutations that do not affect DNA binding activity but inhibit RUNX2/CBFβ heterodimerization in CCD patients, such as S118R, T200A, and Q209R mutations, indicate the importance of RUNX2/CBFβ complex formation (37, 38). Under osteogenic conditions, large co-regulatory factors may be involved in the RUNX2/CBFβ complex formation, which exerts osteogenic conversion from preosteoblasts into osteoblasts (35, 36, 39).
Collectively, our findings indicate that Cbfβ regulates Runx2 stability by inhibiting Runx2 proteasomal degradation in mature osteoblasts, and that this has a positive effect on the postnatal maintenance of cortical bone thickness.
Supplementary Material
Fig. S2. Lower BV/TV and Ct.Th in 5 week-old CbfbΔob/Δob mice by μCT analysis
BV/TV of 5 week-old CbfbΔob/Δob mice was reduced compared to WT mice (A). Ct. Th was lower in the distal femoral metaphyses of CbfbΔob/Δob mice versus WT mice (B). WT mice (□, n = 6), CbfbΔob/Δob mice (■, n = 7). *p<0.05. Results are mean ± SD.
Fig. S3. Normal proliferation and apoptosis of osteoblasts derived from CbfbΔob/Δob mice
In vivo osteoblast proliferation and apoptosis in 12 week-old mice were assessed by BrdU and TUNEL assays, respectively. Positive signals of BrdU and TUNEL assays in trabecular bones were not different between WT and CbfbΔob/Δob mice (n=3-4 per group).
Fig. S1. The Col1α1-Cre transgene is active in osteoblasts
X-gal stained whole embryo and paraffin sections of the ulna and digit from R26R;Col1α1-Cre and WT mice at E18.5. Bars: 200 μm. Positive LaxZ signals were observed in the bone collar area.
Fig. S5. TRAP staining in CbfbΔob/Δob mice
Number of osteoclasts did not differ between genotype, which was determined by the BioquantOsteoII program after Tartrate-resistant acid phosphatase (TRAP) staining (A). Expression of several osteoclastogenesis related genes were decreased in 12 week old mice (B). WT mice (□, n = 3-4), CbfbΔob/Δob mice (■, n = 3-4). *p<0.05, **p<0.01. Results are mean ± SD.
Fig. S4. Abnormal canaliculi formation in CbfbΔob/Δob mice
Abnormal canaliculi appeared in CbfbΔob/Δob mice compared to WT mice as shown by bodian staining (A). There was no difference in osteocyte number between WT and CbfbΔob/Δob mice (B). Bone tissue mRNA of Dmp1 and Phex, osteocyte-specific genes, were decreased in CbfbΔob/Δob mice compared to WT mice while Fgf23 did not differ as shown by Q-PCR (C). Direction of collagen fiber was normal as observed under polarized light (D). WT mice (□, n = 3), CbfbΔob/Δob mice (■, n = 3). **p<0.01. Results are mean ± SD.
Fig. S6. Reduction of osteogenic markers in BMSC of CbfbΔob/Δob mice
Using BMSCs, ALP and Alizarin red S staining was performed at day 14 and day 28, respectively (A). Q-PCR of osteogenic markers was performed using total RNA isolated from BMSCs after 14 days in vitro osteoblast differentiation culture (B). *p<0.05, **p<0.01
ACKNOWLEDGEMENTS
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare (A111487) and the National Research Foundation of Korea grant (2010-0026741), Republic of Korea. This study also supported in part by NIH grants R01 AR039588 (GSS) R37DE012528 ARRA Merit Award (JBL).
Footnotes
DISCLOSURES
The authors declare no conflict of interest.
REFERENCES
- 1.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–64. Epub 1997/05/30. [DOI] [PubMed] [Google Scholar]
- 2.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–71. Epub 1997/05/30. [DOI] [PubMed] [Google Scholar]
- 3.Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci U S A. 2001;98(15):8650–5. Epub 2001/07/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong JH, Jung YK, Kim HJ, Jin JS, Kim HN, Kang SM, et al. The gene for aromatase, a rate-limiting enzyme for local estrogen biosynthesis, is a downstream target gene of Runx2 in skeletal tissues. Molecular and cellular biology. 2010;30(10):2365–75. Epub 2010/03/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JC, Lengner CJ, Gaur T, Lou Y, Hussain S, Jones MD, et al. Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. The Journal of biological chemistry. 2011;286(34):30057–70. Epub 2011/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou Y, Javed A, Hussain S, Colby J, Frederick D, Pratap J, et al. A Runx2 threshold for the cleidocranial dysplasia phenotype. Human molecular genetics. 2009;18(3):556–68. Epub 2008/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee C, McCabe LR, Choi JY, Hiebert SW, Stein JL, Stein GS, et al. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. Journal of cellular biochemistry. 1997;66(1):1–8. Epub 1997/07/01. [DOI] [PubMed] [Google Scholar]
- 8.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–54. Epub 1997/05/30. [DOI] [PubMed] [Google Scholar]
- 9.Westendorf JJ. Transcriptional co-repressors of Runx2. Journal of cellular biochemistry. 2006;98(1):54–64. Epub 2006/01/28. [DOI] [PubMed] [Google Scholar]
- 10.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, et al. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. Journal of virology. 1990;64(10):4808–19. Epub 1990/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(14):6859–63. Epub 1993/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Molecular and cellular biology. 1993;13(6):3324–39. Epub 1993/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speck NA, Terryl S. A new transcription factor family associated with human leukemias. Critical reviews in eukaryotic gene expression. 1995;5(3–4):337–64. Epub 1995/01/01. [DOI] [PubMed] [Google Scholar]
- 14.Bae SC, Ito Y. Regulation mechanisms for the heterodimeric transcription factor, PEBP2/CBF. Histol Histopathol. 1999;14(4):1213–21. Epub 1999/10/03. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, et al. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93(22):12359–63. Epub 1996/10/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. Epub 1996/11/15. [DOI] [PubMed] [Google Scholar]
- 17.Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, et al. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nature genetics. 2002;32(4):645–9. Epub 2002/11/16. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nature genetics. 2002;32(4):633–8. Epub 2002/11/16. [DOI] [PubMed] [Google Scholar]
- 19.Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nature genetics. 2002;32(4):639–44. Epub 2002/11/16. [DOI] [PubMed] [Google Scholar]
- 20.Baek WY, Lee MA, Jung JW, Kim SY, Akiyama H, de Crombrugghe B, et al. Positive regulation of adult bone formation by osteoblast-specific transcription factor osterix. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2009;24(6):1055–65. Epub 2008/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204(8):1749–55. Epub 2007/07/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25(7):1468–86. Epub 2010/06/10. [DOI] [PubMed] [Google Scholar]
- 23.Kusuzaki K, Kageyama N, Shinjo H, Takeshita H, Murata H, Hashiguchi S, et al. Development of bone canaliculi during bone repair. Bone. 2000;27(5):655–9. Epub 2000/11/04. [DOI] [PubMed] [Google Scholar]
- 24.Erben RG. Embedding of bone samples in methylmethacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. J Histochem Cytochem. 1997;45(2):307–13. Epub 1997/02/01. [DOI] [PubMed] [Google Scholar]
- 25.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2013;28(1):2–17. Epub 2012/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han MS, Kim HJ, Wee HJ, Lim KE, Park NR, Bae SC, et al. The cleidocranial dysplasia-related R131G mutation in the Runt-related transcription factor RUNX2 disrupts binding to DNA but not CBF-beta. Journal of cellular biochemistry. 2010;110(1):97–103. Epub 2010/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soriano P Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21(1):70–1. Epub 1999/01/23. [DOI] [PubMed] [Google Scholar]
- 28.Khosla S Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142(12):5050–5. [DOI] [PubMed] [Google Scholar]
- 29.Kanatani N, Fujita T, Fukuyama R, Liu W, Yoshida CA, Moriishi T, et al. Cbf beta regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296(1):48–61. Epub 2006/06/27. [DOI] [PubMed] [Google Scholar]
- 30.Takarada T, Hinoi E, Nakazato R, Ochi H, Xu C, Tsuchikane A, et al. An analysis of skeletal development in osteoblast- and chondrocyte-specific Runx2 knockout mice. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2013. Epub 2013/04/05. [DOI] [PubMed] [Google Scholar]
- 31.Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. The Journal of biological chemistry. 2003;278(30):27939–44. Epub 2003/05/10. [DOI] [PubMed] [Google Scholar]
- 32.Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. The Journal of biological chemistry. 2004;279(13):12854–9. Epub 2004/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312(5777):1223–7. Epub 2006/05/27. [DOI] [PubMed] [Google Scholar]
- 34.Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Cells, tissues, organs. 2009;189(1–4):144–52. Epub 2008/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon EJ, Lee KY, Choi NS, Lee MH, Kim HN, Jin YH, et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. The Journal of biological chemistry. 2006;281(24):16502–11. Epub 2006/04/15. [DOI] [PubMed] [Google Scholar]
- 36.Yoon WJ, Islam R, Cho YD, Woo KM, Baek JH, Uchida T, et al. Pin1-mediated Runx2 modification is critical for skeletal development. Journal of cellular physiology. 2013;228(12):2377–85. Epub 2013/05/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou G, Chen Y, Zhou L, Thirunavukkarasu K, Hecht J, Chitayat D, et al. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Human molecular genetics. 1999;8(12):2311–6. Epub 1999/11/05. [DOI] [PubMed] [Google Scholar]
- 38.Bravo J, Li Z, Speck NA, Warren AJ. The leukemia-associated AML1 (Runx1)--CBF beta complex functions as a DNA-induced molecular clamp. Nat Struct Biol. 2001;8(4):371–8. Epub 2001/03/29. [DOI] [PubMed] [Google Scholar]
- 39.Ohba S, Ikeda T, Kugimiya F, Yano F, Lichtler AC, Nakamura K, et al. Identification of a potent combination of osteogenic genes for bone regeneration using embryonic stem (ES) cell-based sensor. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21(8):1777–87. Epub 2007/02/24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S2. Lower BV/TV and Ct.Th in 5 week-old CbfbΔob/Δob mice by μCT analysis
BV/TV of 5 week-old CbfbΔob/Δob mice was reduced compared to WT mice (A). Ct. Th was lower in the distal femoral metaphyses of CbfbΔob/Δob mice versus WT mice (B). WT mice (□, n = 6), CbfbΔob/Δob mice (■, n = 7). *p<0.05. Results are mean ± SD.
Fig. S3. Normal proliferation and apoptosis of osteoblasts derived from CbfbΔob/Δob mice
In vivo osteoblast proliferation and apoptosis in 12 week-old mice were assessed by BrdU and TUNEL assays, respectively. Positive signals of BrdU and TUNEL assays in trabecular bones were not different between WT and CbfbΔob/Δob mice (n=3-4 per group).
Fig. S1. The Col1α1-Cre transgene is active in osteoblasts
X-gal stained whole embryo and paraffin sections of the ulna and digit from R26R;Col1α1-Cre and WT mice at E18.5. Bars: 200 μm. Positive LaxZ signals were observed in the bone collar area.
Fig. S5. TRAP staining in CbfbΔob/Δob mice
Number of osteoclasts did not differ between genotype, which was determined by the BioquantOsteoII program after Tartrate-resistant acid phosphatase (TRAP) staining (A). Expression of several osteoclastogenesis related genes were decreased in 12 week old mice (B). WT mice (□, n = 3-4), CbfbΔob/Δob mice (■, n = 3-4). *p<0.05, **p<0.01. Results are mean ± SD.
Fig. S4. Abnormal canaliculi formation in CbfbΔob/Δob mice
Abnormal canaliculi appeared in CbfbΔob/Δob mice compared to WT mice as shown by bodian staining (A). There was no difference in osteocyte number between WT and CbfbΔob/Δob mice (B). Bone tissue mRNA of Dmp1 and Phex, osteocyte-specific genes, were decreased in CbfbΔob/Δob mice compared to WT mice while Fgf23 did not differ as shown by Q-PCR (C). Direction of collagen fiber was normal as observed under polarized light (D). WT mice (□, n = 3), CbfbΔob/Δob mice (■, n = 3). **p<0.01. Results are mean ± SD.
Fig. S6. Reduction of osteogenic markers in BMSC of CbfbΔob/Δob mice
Using BMSCs, ALP and Alizarin red S staining was performed at day 14 and day 28, respectively (A). Q-PCR of osteogenic markers was performed using total RNA isolated from BMSCs after 14 days in vitro osteoblast differentiation culture (B). *p<0.05, **p<0.01


