Abstract
Tumor heterogeneity is prevalent in both treatment-naïve and end-stage metastatic castration-resistant prostate cancer (PCa), and may contribute to the broad range of clinical presentation, treatment response, and disease progression. To characterize molecular heterogeneity associated with de novo metastatic PCa, multiplatform single cell profiling was performed using high definition single cell analysis (HD-SCA). HD-SCA enabled morphoproteomic and morphogenomic profiling of single cells from touch preparations of tissue cores (prostate and bone marrow biopsies) as well as liquid samples (peripheral blood and bone marrow aspirate). Morphology, nuclear features, copy number alterations, and protein expression were analyzed. Tumor cells isolated from prostate tissue touch preparation (PTTP) and bone marrow touch preparation (BMTP) as well as metastatic tumor cells (MTCs) isolated from bone marrow aspirate were characterized by morphology and cytokeratin expression. Although peripheral blood was examined, circulating tumor cells were not definitively observed. Targeted proteomics of PTTP, BMTP, and MTCs revealed cell lineage and luminal prostate epithelial differentiation associated with PCa, including co-expression of EpCAM, PSA, and PSMA. Androgen receptor expression was highest in MTCs. Hallmark PCa copy number alterations, including PTEN and ETV6 deletions and NCOA2 amplification, were observed in cells within the primary tumor and bone marrow biopsy samples. Genomic landscape of MTCs revealed to be a mix of both primary and bone metastatic tissue. This multiplatform analysis of single cells reveals several clonal origins of metastatic PCa in a newly diagnosed, untreated patient with polymetastatic disease. This case demonstrates that real-time molecular profiling of cells collected through prostate and bone marrow biopsies is feasible and has the potential to elucidate the origin and evolution of metastatic tumor cells. Altogether, biological and genomic data obtained through longitudinal biopsies can be used to reveal the properties of PCa and can impact clinical management.
Keywords: prostate cancer, tumor heterogeneity, metastatic tumor cells, liquid biopsies, circulating tumor cells, high definition single cell analysis (HD-SCA)
Introduction
Despite early detection and aggressive intervention, prostate cancer (PCa) is the third-leading cause of death among men in United States, with an estimated 160 000 new cases and approximately 26 000 annual deaths in 2017 [1, 2]. Multiple therapeutic agents have been shown to improve overall survival in end-stage metastatic castrate-resistant PCa (mCRPC). Additionally, recent clinical trials have provided evidence that use of combination therapies, including docetaxel, with first-line androgen deprivation therapy (ADT), significantly increases overall survival in de novo metastatic patients [3, 4]. However, not all patients respond to combination therapy, and it is unknown whether or not the efficacy of combinatorial approaches can be optimized based upon biological and genomic features of the tumor.
Intra-patient spatiotemporal molecular profiling has the potential to provide treatment response signatures, insight into heterogeneity, and prognostic information for patients with metastatic PCa. While primary tumor and metastasis biopsies provide information about tumor type, grade, and pathological features, information on heterogeneity, clonality, and the likelihood of treatment response is limited. In contrast, liquid biopsies provide access to circulating tumor cells (CTCs) from routine peripheral blood (PB) samples, disseminated tumor cells (DTCs) or metastatic tumor cells (MTCs) from the bone marrow aspirates (BMA), and cell-free DNA (cfDNA) [5]. DTCs and MTCs describe two different clinical stages in PCa. DTCs describe tumor cells present in bone marrow of a patient without clinical bone metastatic whereas MTCs are tumor cells found in bone marrow of a patient with clinical bone metastasis [5]. Comprehensive analysis of these specimens will provide an in depth assessment of tumor heterogeneity and clonality and may lead to improved prognostication and prediction of treatment efficacy. Many new technological advances allow for high-resolution single cell analysis and when combined with rare cell detection, provide clinical insights that may impact clinical management.
We have previously developed and technically validated the high definition single cell analysis (HD-SCA) workflow for enumeration, morphoproteomic, and morphogenomic characterization of rare cells in order to identify and quantify cellular heterogeneity [5–9, 18]. The workflow utilizes single-cell profiling to characterize CTCs, DTCs, and MTCs followed by genomic and proteomic characterization that can be correlated with morphology data [6, 7, 10]. CfDNA extraction from a liquid biopsy prior to HD-SCA sample processing allows for genomic assessment of circulating tumor DNA (ctDNA) found in both PB and BMA. The same multiplatform single-cell analysis can be used not only for liquid biopsy samples, but also for genomic and proteomic characterization of primary and metastatic tissue preparation [9]. Additionally, as demonstrated by our recently published data, the HD-SCA platform can be adapted to the fluid form of BMA, allowing characterization and comparison of circulatory and bone marrow cancer cells [5]. Carlsson et al demonstrated the value of non-guided BMA as a feasible and cost-effective procedure for longitudinal sampling during cancer progression and treatment similar to current clinical practice in the liquid malignancies [11].
In this report, we used the HD-SCA workflow to characterize cancer cells in a patient with newly diagnosed de novo polymetastatic PCa. Single-cell morphoproteomic and morphogenomic analysis allowed direct comparison of solid phase primary and metastatic tissue tumor cells with those extracted from liquid biopsies. The clinical utility of liquid biopsy (CTC, DTC, MTC, and/or ctDNA) lies in the ability to illuminate indicators of treatment response/resistance that may guide therapeutic selection. This report demonstrates that genomic and proteomic data collected from the HD-SCA workflow provides information on heterogeneity, clonality, and marker expression that may influence prognostication and treatment response in patients harboring lethal PCa.
Material and methods
Specimen collection and HD-SCA sample preparation
The patient is a 73 year-old African–American male diagnosed with de novo polymetastatic PCa via prostate needle biopsy (PNBX) within the Greater Los Angeles Veterans’ Affairs Healthcare System. PB and bone marrow aspiration/biopsy were collected at the time of diagnostic PNBX and placed into 10 ml cfDNA BCT Streck tubes (STRECK, Omaha, USA, Cat#62790315). Approximately 12 prostate tissue cores were obtained in a random fashion from the right and left base, mid-gland, and apex under transrectal ultrasound guidance. Prostate tissue touch preparation (PTTP) was performed with each biopsy core by gentle rolling onto glass slides prior to formalin fixation and paraffin embedding (FFPE). A standard percutaneous bone marrow biopsy and aspiration were performed at the right posterior iliac crest. The bone marrow core was gently rolled onto a glass slide to prepare bone marrow touch preparation (BMTP). Samples were shipped overnight to the Kuhn laboratory at the University of Southern California, Los Angeles, CA, USA. Upon arrival of PB and BMA samples in Streck tubes, red blood cells were lysed, and remaining cells were plated on a custom-made adhesive glass slide (Marienfeld, Lauda-Königshofen, Germany) as a monolayer of 3.0 × 106 nucleated cells [5]. Unstained slides were covered with coverslips and stored at −80 °C before analysis (figure 1). All touch preparation slides were air dried, blocked with 7% BSA in PBS, and stored in −80 °C until processing.
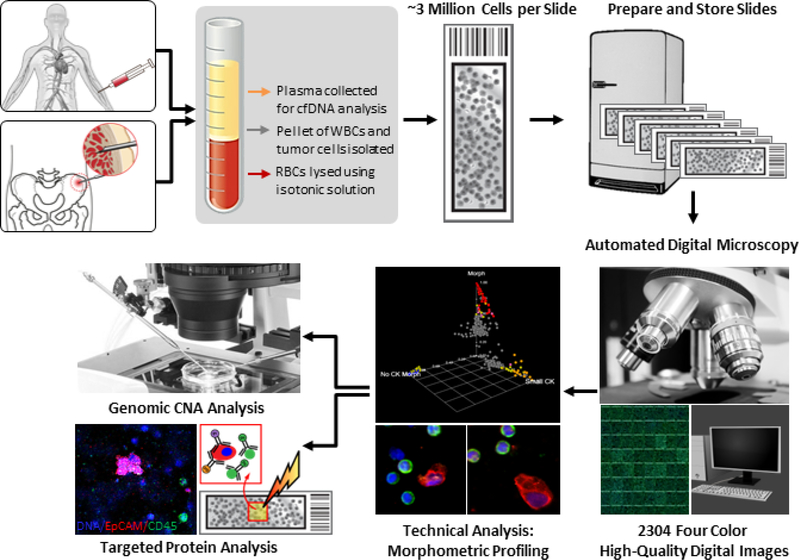
Figure 1.
HD-SCA platform for morphoproteogenomic profiling of liquid biopsy. PB and BMA samples are initially spun down for plasma extraction. Next, they undergo red blood cell lysis before plating approximately 3 million nucleated cells on each slide. Prepared slides are stored at −80 °C until needed for fluorescent antibody staining. Stained slides are first morphometrically profiled using automated digital microscopy at 10 × magnification, followed by classification by a technical analyst. Identified tumor cells are then re-imaged at 40 × magnification and proceed for genomic CNA or targeted protein analysis via imaging mass cytometry.
Detailed sample processing procedures have been described previously [7]. Briefly, two slides per sample were stained using four fluorescent markers. Cell nuclei were identified using DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride, Cat#D1306, Invitrogen, Waltham, MA). Epithelial cells were identified using a mix of cytokeratin (CK) 19 (1:100; Dako, Carpinteria, USA, Cat#M0888) and pan-CK antibodies (1:100; Sigma-Aldrich, St. Louis, USA, Cat#C2562) with an Alexa Fluor 555 secondary antibody (Invitrogen, Carlsbad, USA, Cat#A21127). An anti-CD45 Alexa Fluor 647–conjugated antibody (1:125; Biorad, Hercules, USA, Cat#MCA87A647X) was used as a leukocyte exclusion marker. An androgen receptor (AR) rabbit monoclonal antibody (1:250; Cell Signaling Technology, Danvers, USA, Cat#5153) and Alexa Fluor 488 secondary antibody (Invitrogen, Carlsbad, USA, Cat#A11034) were used for evaluation of AR levels in cells [5, 10] (figure 1).
Candidate cell imaging and morphological analysis
PB and BMA slides were imaged at 10× magnification using automated high-throughput microscopy. Candidate cells were computationally identified and semi-manually classified using morphology and DAPI+/CK+/CD45− expression criteria as previously established (figure 1) [12, 13]. PTTP and BMTP were manually imaged at 10× and 40× magnification. AR expression and localization were reviewed and quantified using average fluorescent intensity within a fixed-size circle centered around the cell [14]. The threshold for AR positivity was defined as a signal more than 6 standard deviations over the mean signal intensity (SDOM) observed in the surroundings leukocytes (background). For morphometric analysis of candidate cells, nuclear area and nuclear circularity was measured using DAPI intensity within a fixed-size circle centered around the cell using image object features from the EBImage R package [15].
Single-cell next-generation sequencing and analysis
Single cells from PB, BMA, PTTP, and BMTP were isolated and underwent genomic amplification as previously described [16, 17] (figure 2(a)). In short, tumor cells were isolated off the slides using a robotic micromanipulator system and placed in individual tubes for whole genome amplification (Sigma-Aldrich, St. Louis, USA, Cat#WGA4). Prior to cell capture, PTTP and BMTP slides were incubated with dispase type II (1:1000; ThermoFisher, Waltham, USA, Cat#17105041) and collagenase (1:1000; ThermoFisher, Waltham, USA, Cat#17018029) in PBS at 37 °C for 30 min. PTTP and BMTP slides were washed with PBS twice for 3 min, and individual cells or cell clusters from PTTP and BMTP were isolated and extracted in the same manner. Following DNA purification, 50 ng of DNA was sonicated to 200 bp fragments in AFA fiber pre-slit snap-cap microtubes (Covaris, Woburn, USA, Cat# 520077) with the Covaris S2 using the following setting: intensity of 5, 10% duty cycle, 200 cycles per burst, 3 min treatment time, and temperature less than 7 °C. Sonicated DNA was used for library construction with the DNA Ultra Library Prep Kit and Multiplex Oligos for Illumina (New England Biolabs, Ipswich, USA, Cat#E7370 and E7600). Copy number alteration (CNA) profiles and heatmaps were created as previously described, with R unsupervised hierarchical clustering using the Ward method and Euclidian distance to distinguish subclones [16, 17].
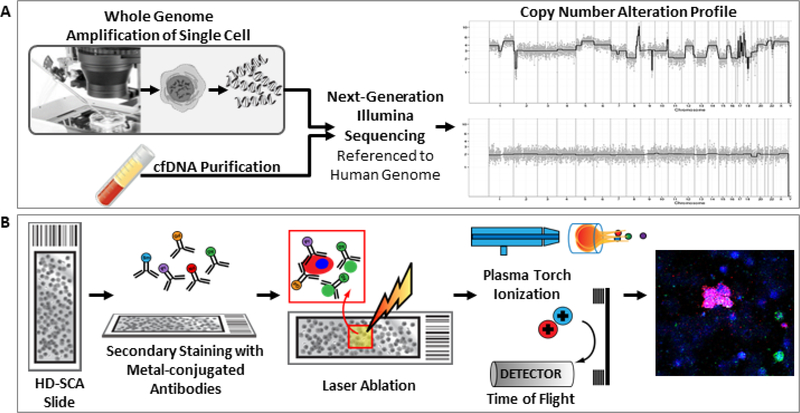
Figure 2.
(a) Single cell and cfDNA genomic preparation for next-generation sequencing and CNA analysis. Single cells are extracted from slides with a robotic micromanipulator prior to whole genome amplification and DNA purification followed by Illumina DNA library preparation for sequencing. CfDNA is extracted from plasma and Illumina DNA libraries are constructed similarly to single cells. CNA profiles are created using the human genome as reference where copy number is calculated then displayed as the ratio to the median. (b) Targeted proteomic analysis via IMC. Slides previously labeled with fluorescent antibodies are stained with 21 metal-conjugated antibodies. Regions of interest are laser ablated with plasma ionization and ions are detection using Cytometry by Time of Flight (CyTOF) technology. Rasterized images are generated from ion count data, and protein expression on tumor cells are scored for expression levels.
CfDNA next-generation sequencing and analysis
PB and BMA were fractionated by centrifugation at 2000 g for 10 min at RT, 2 ml of plasma were collected from each sample, and the removed plasma volume was reconstituted to its original concentration with 1 × PBS. Plasma was spun at 14 000 g for 10 min at RT and supernatant was stored at −80 °C for future analysis. CfDNA was extracted with the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany, Cat# 55114) according to the manufacturer’s instructions. Illumina libraries were constructed from 5 ng cfDNA using the NEBNext Ultra II DNA Library Prep Kit and Multiplex Oligos for Illumina according to the manufacturer’s instructions (New England Biolabs, Ipswich, USA, Cat#E7370 and E7600) as seen in figure 2(a). CNA profiles were created as described under single-cell next-generation sequencing and analysis.
Single-cell targeted proteomic analysis
Previously identified candidate cells from BMA, PTTP, and BMTP slides were subjected to protein analysis using Fluidigm Hyperion Imaging System (IMC) (Fluidigm, San Francisco, USA). Slides were washed in PBS to remove cell media before secondary staining. Slides were stained with 21 markers out of the 40 available channels to detect the expression of leukocyte, epithelial, endothelial, and prostate cell protein markers (table 2). Slides were blocked using 1% BSA with 0.2 mg ml−1 mouse IgG Fc fragment (Thermofisher, Waltham, USA, Cat#31205) in PBS for 60 min at 37 °C. The MaxPar™ metal-labeled antibody cocktail was prepared in 0.1% Tween and 1% BSA in PBS with antibodies from Fluidigm according to the manufacturer’s dilutions. Antibody cocktail was added to each slide for 90 min at room temperature and washed with PBS twice for 3 min. IR-193 DNA intercalator (Fluidigm, San Francisco, USA, Cat#201192 A) was added to slides for 30 min at room temperature. Slides were washed with PBS twice for 3 min and dipped in ddH2O for 5 s to remove salt. Slides were dried for 2 h and stored at room temperature until IMC runs [18].
Table 2.
Targets and descriptions of 21 metal-conjugated antibodies for IMC analysis. Markers are subdivided into epithelial, endothelial, leukocyte, and prostate-specific panels for classification of cell type and origin.
| Target | Description |
|---|---|
| ALDH1 | ALDH1 belongs to the aldehyde dehydrogenase family. Nineteen ALDHs are present in humans, expressed in a variety of organelles, and having different substrate preferences. It is also expressed in the epithelium of many organs |
| Caspase 3 | Caspase-3 is activated during the early stages of apoptosis. Caspase-3 is either partially or totally responsible for the proteolytic cleavage of many key proteins, such as the nuclear enzyme polymerase. Caspase-3 is widely distributed including high expression in cells of lymphoid origin and active caspase-3 is a marker for cells undergoing apoptosis |
| CD14 | CD14 is a glycosylphosphatidylinositol (GPI)-linked membrane glycoprotein. CD14 is expressed at high levels on antigen presenting innate immune cells including monocytes and macrophages and at lower levels of granulocytes. It has also been reported to be expressed on some dendritic cell sub-populations |
| CD24 | CD24 is expressed on the surface of B cells, granulocytes, follicular dendritic cells, and epithelial cells and may play a role in the regulation of B-cell proliferation and maturation |
| CD3 | CD3 is a transmembrane subunit of the T Cell receptor complex. It is a member of the immunoglobulin superfamily that plays a role in antigen recognition, signal transduction, and T cell activation. CD3 is expressed by T lymphocytes (thymocytes) at the highest levels on mature cell types |
| CD31 | CD31, also known as platelet endothelial cell adhesion molecule-1, is a type I transmembrane glycoprotein. It binds CD38 and plays a role in wound healing, angiogenesis, removing aged neutrophils and cellular migration in an inflammatory situation |
| CD38 | CD38 is a transmembrane glycoprotein. It is expressed at variable levels across most human hematopoietic cells and in other tissues (i.e. Brain, muscle, and kidney). It is expressed at the highest levels on plasma cells (memory B cells) as well as activated T and B cells |
| CD44 | CD44 is a cell surface glycoprotein. It is involved in a variety of cellular functions including: cell-to-cell interactions, adhesion, homing and migration, as well as being a receptor for hyaluronic acid. CD44 is expressed on a number of cell types including leukocytes, endothelial cells, hepatocytes, and mesenchymal cells and has been reported to be a marker for memory cell subsets |
| CD45 | CD45 is a type I transmembrane glycoprotein. It is expressed on the plasma membrane of all hematopoietic cells, except mature red blood cells and platelets. Its intracellular domain is a tyrosine phosphatase that serves to regulate signal transduction in most hematopoietic cells. |
| CD45 RA | CD45A is a type I transmembrane glycoprotein. It is a specific splice variant of the transmembrane tyrosine phosphatase CD45. The CD45RA isoform is most highly expressed on resting/naïve T cells, B cells and monocytes. Like CD45, CD45A’s intracellular domain is a tyrosine phosphatase that servers to regulate signal transduction event—in particular, enhancing both T cell receptor and B cell receptor signalling |
| CD61 | CD62 is a type I transmembrane glycoprotein. CD61 is a member of integrin family and is expressed on platelets and megakaryocytes playing a role in platelet activation and aggregation. CD61 in complex with CD51 has also been reported on osteoclasts, fibroblast, macrophages and some tumour cells |
| CD66 | CD66a is a glycoprotein belonging to the immunoglobulin superfamily. It plays a role in multiple cellular activities such as differentiation and arrangement of tissue 3D structure, angiogenesis, apoptosis, tumour suppression, metastasis and the modulation of innate and adaptive immune responses. It can be found expressed on the surface of endothelial/epithelial cells, neutrophils and monocytes and can be induced on T cells, B cells, and CD16-negative NK cells |
| CD8a | CD8 is a type I membrane glycoprotein. CD8 is a member of the immunoglobulin superfamily found on the majority of thymocytes, a subset of PB T cells, and natural killer cells (which express almost exclusively CD8a homodimers). CD8 acts as a co-receptor with MHC class I-restricted T cell receptors in antigen recognition and T cell activation and has been shown to play a role in thymic differentiation |
| CK8/18 | CytoKeratin 8 and 18 are intermediate filaments that provides mechanical support and serves variety of functions in epithelial cells. They are part of the cytoskeleton of the cell. CK8/18 is used by pathologist to evaluate pathogenesis of biopsy tissue in breast cancer |
| E-Cadherin | E-Cadherin (Epithelial cadherin) is a member of the cadherin superfamily. E-cadherin is a calcium-dependent, transmembrane cell–cell adnesion glycoprotein composed of 4 extracellular cadherin repeats and a highly conserved cytoplasmic tail region. It functions as cell adhesion molecule involved in development, bacterial, pathogenesis, and tumour invasion |
| EpCAM | Epithelial cell adhesion molecule (EpCAM) is a type I transmembrane protein. EpCAM functions as homotypic calcium-independent cell adhesion molecule and believed to be involved in carcinogenesis by inducing genes involved in cellular metabolism and proliferation. It is highly expressed in bone marrow, colon, lung, most epithelial cells and on carcinomas of gastrointestinal origin |
| HLA-DR | HLA-DR or human leukocyte antigen DR is an MHC class II cell surface receptor that is a cell surface glycoprotein. HLA-DR is expressed on B cells, activated T cells, monocytes/macrophages, dendritic cells, and other non-professional antigen presenting cells (APCs). HLA-DR is critical for efficient antigen presentation to CD4 + T cells |
| KI-67 | Ki-67 protein is a cellular marker for proliferation. It is strictly associated with cell proliferation. During interphase, the Ki-67 antigen can exclusively detected within the cell nucleus, whereas in mitosis most of the protein is relocated to the surface of the chromosomes |
| PSA | Prostate-specific antigen (PSA) is a glycoprotein enzyme encoded in humans by the KLK3 gene. PSA is a member of the kallikrein-related peptidase family and is secreted by the epithelial cells of the prostate gland |
| PSMA | Human prostate-specific membrane antigen (PSMA) is a type II transmembrane zinc metallopeptidase. As a tumour marker in PCa, PSMA expression has been shown to correlate with disease progression |
| Vimentin | Vimentin are class-III intermediate filaments found in various non-epithelial cells, especially mesenchymal cells. It is highly expressed in fibroblasts, low in T- and B-lymphocytes. Vimentin has dynamic structural changes and spatial re-organization in response to extracellular stimuli help to coordinate various signalling pathways. Remodelling of Vimentin and other intermediate filaments is important during lymphocyte adhesion and migration through the endothelium |
Epithelial Markers | Endothelial Markers | Leukocyte Markers | Prostate-specific Markers
Laser ablation with time-of-flight detection and analysis was performed using the IMC as seen in figure 2(b). A 400 μm × 400 μm region of interest around each cell of interest was ablated aerosolizing a 1 μm2 area/pulse (200 Hz), followed by ionization and quantification in the CyTOF Helios instrument. Ion mass data were collected, resulting in the construction of 1 μm2 resolution images depicting the tumor cells of interest and surrounding white blood cells (WBC) as reference. For BMA, this section encompasses the MTC of interest and the surrounding WBCs totaling approximately 500 cells. For BMTP and PTTP, the ablated area contained hundreds to thousands of cells, depending on cell density and the size of the imprint.
A four-level scoring system was developed where 0 is below limit of detection (LOD), 1 is at LOD, and 2–3 is above. The LOD for each marker was set as equal to signal to noise ratio (S/N) ⩾ 3 or standard deviation of mean (SDOM) > 3.3. A score of 1 was given to cells exceeding the LOD, a score of 2 was given to signals with S/N of 7–20 or SDOM > 6, and finally a score of 3 was assigned to signals of S/N > 20 or SDOM > 12.
Results and discussion
Single cell morphometric analysis of identified tumor cells
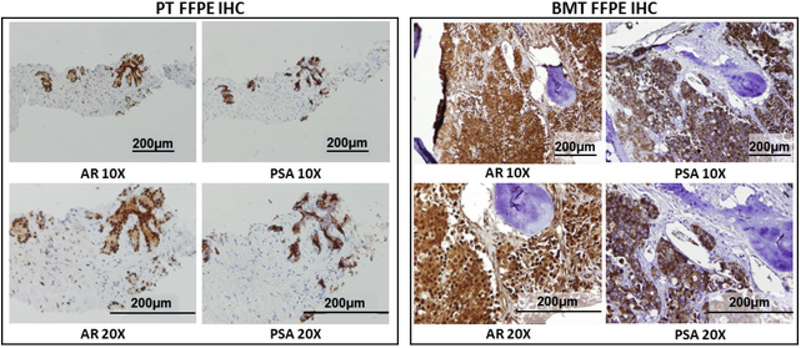
To examine intra-patient heterogeneity at time of diagnosis, samples were collected from a patient with de novo polymetastatic PCa. During a routine examination in 2016, the patient was found to have an elevated serum prostate specific antigen (PSA) level of 234 ng ml−1, which triggered a diagnostic workup for PCa. A bone scan demonstrated widespread metastatic disease involving skull, bilateral ribs, multilevel lumbar spine, and pelvic bone. Diagnostic PNBX revealed highgrade PCa with a Gleason score of 8 (4 + 4) in 9 of 12 biopsy cores. Immunohistochemistry (IHC) staining of FFPE PNBX and bone marrow biopsy core revealed adenocarcinoma that expressed AR and PSA (figure 3).
Figure 3.
IHC staining of FFPE PT from diagnostic PNBX and BMT from bone marrow biopsy. IHC staining for AR and PSA were performed on both PT and BMT samples. Samples were imaged using a light microscope at 10 and 20× objective. Staining results were reported as positive or negative for each marker.
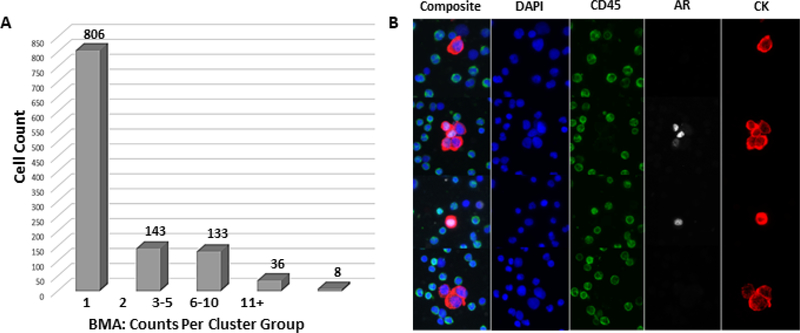
For the HD-SCA workflow, PB, BMA, and bone marrow tissue (BMT) were collected at the time of PNBX. Following processing and staining as described in Materials and Methods, we identified and categorized candidate cells from PB and BMA using a semi-automated reporting system. Candidate cells were evaluated based on DAPI+/CK+/CD45− criteria and marked as CTCs in PB and MTCs in BMA [6, 7]. Enumeration of tumor cells revealed a high count of MTCs and lack of CTCs. Counts for CTCs in PB and MTCs in BMA were 0 cells ml−1 and 3673 cells ml−1, respectively (figure 4(a)). The HD-SCA threshold for a positive sample is ⩾1 cell per case based on previous studies, making BMA positive for this patient [23, 24]. MTCs were detected as both single cells and clusters of 2–5 cells (25%), 6–10 cells (3%), and 11 + cells (1%) (figure 4(a)).
Figure 4.
(a) MTC enumeration in BMA. Total counts are reflected by cluster group size. The y-axis reflects the number of cells within each cluster group and the x-axis reflects each cluster group category including single cells, cluster of 2 cells, cluster of 3–5 cells, cluster of 6–10 cells, and clusters of 11 and more cells. Overall, 3673 cells ml−1 were detected in BMA. (b) AR expression heterogeneity in MTCs. Each row shows both composite and individual DAPI, CK, CD45, and AR channel for each cell. DAPI is shown in blue, CK in red, AR in white, and CD45 in green. AR expression was located in the nucleus in both single cells and cell clusters despite differences in expression levels. Other single cells or cell clusters had no AR at all, showing heterogeneity in MTCs.
AR expression was observed in both BMTP and PTTP via IHC analysis (figure 3). Single-cell morphometric analysis of MTCs revealed AR expression localized to the nucleus (figure 4(b)). Despite observed heterogeneity in AR expression, AR staining was confined to the nucleus as expected, regardless of expression levels (figure 4(b)). To further confirm nuclear localization, detailed confocal images were acquired (figure 5(a)). We observed marked heterogeneity across MTCs in nuclear area, nuclear circularity, CK stain intensity SDOM, and AR expression SDOM (figure 5(b)), revealing several MTC subclones. Collectively, these data demonstrate that tumor heterogeneity can be detected at a morphometric level using measurements such as protein expression and nuclear features.
Figure 5.
(a) Confocal images of MTCs. Nuclear localization and heterogeneity of AR expression are shown in three different clusters of MTCs. DAPI is shown in blue, CK in red, AR in white, and CD45 in green. Some clusters were entirely AR-positive or AR-negative while others presented mixed expression within the cluster. (b) Morphometric analysis of MTCs in BMA. A wide range of nuclear area, nuclear circularity, CK intensity SDOM, and AR intensity SDOM was displayed, highlighting the ability to detect tumor heterogeneity through morphometric measurements. The distribution of cells is shown along the y-axis for nuclear area, nuclear circularity, CK intensity SDOM, and AR intensity SDOM.
Single cell and cfDNA genomic analysis using CNA
To explore tumor heterogeneity on a molecular level, we selected a total of 73 single cells for CNA analysis from 3 sample types: BMA (n = 22), BMTP (n = 17), and PTTP (n = 32). This analysis included WBC as controls (2 from BMA). CNA profiles of tumor cells in prostate tissue (PT) and BMT showed prostate-specific alterations across the genome as well as clonality, which we analyzed further (figure 6). Table 1 summarizes all observed alterations with their frequency in MTCs, BMTP, and PTTP and their function and implications in cancer. We define clonality as two or more cells that share two or more genomic alterations. Alterations of tumor suppressor genes such as PTEN, RB1, and TP53 have been shown to contribute to tumor and metastatic development as well as invasiveness [19, 20]. PTEN deletion was observed in 77% (17/22) and 34% (11/32) of MTCs and PTTP, respectively. Partial chromosome 13 loss was observed in 23% (4/17) of BMTP, resulting in at least hemizygous loss of RB1. TP53 loss was observed in 68% (15/22) of MTCs and 82% (14/17) of BMTP. No TP53 deletion was observed in PT; thus, TP53 loss may have been acquired after the cancer cells left the PT and settled in the BM. Research has shown that in the absence of or dysregulation of TP53, DNA damage and spindle damage occur, resulting in polyploidy in tumor cells [21]. 64% (14/22) of MTCs have four copies of each chromosome and are thus polyploid. Regardless of this increase in DNA content, they remain clonal and share common alterations with the rest of the tumor cells.
Figure 6.
Heatmap and phylogenic tree of CNAs across the entire population of cells from MTC, PTTP, and BMTP. Sample type and clones are identified using color key. Three clones were identified: clone 1 consisting of prostate cells with hallmark alterations, clone 2 with few CNAs, and clone 3 of bone marrow specimens with additional alterations from that of clone 1. Key genes such as MYC, NCOA2, PTEN, and TP53 are highlighted by chromosome location across clone and sample type.
Table 1.
Summary of observed CNAs. Gene functions, alteration types, and implications are displayed with the frequencies of occurrence in MTCs, BMTP, and PTTP
| Gene | Function | Type of alteration | MTCs | BMTP | PTTP | Implications |
|---|---|---|---|---|---|---|
| PTEN | Tumor suppressor | Deletion | 77% (17/22) | 0% (0/17) | 34% (11/32) | Increased cell proliferation and reduced cell death due to dysfunction in cell cycle regulation |
| RB1 | Tumor suppressor | Hemizygous Deletion | 0% (0/22) | 23% (4/17) | 0% (0/32) | Excessive cell growth due to dysregulation of cell cycle inhibition |
| TP53 | Tumor suppressor | Deletion | 68% (15/22) | 82% (14/17) | 0% (0/32) | Dysfunction in DNA repair activation, cell cycle inhibition, activation of apoptosis and senescence response to short telomeres |
| ATM | Coordinates DNA repair | Deletion | 77% (17/22) | 94% (16/17) | 47% (15/32) | Dysfunction in recognition of DNA damage and activation of DNA repair pathways |
| BRCA1 | Tumor suppressor DNA repair | Deletion | 9% (2/22) | 29% (5/17) | 9% (3/32) | Dysfunction in repairing damaged DNA properly. Cell can no longer sense DNA damage and sent appropriate signals |
| BRCA2 | Tumor suppressor DNA repair | Hemizygous deletion | 0% (0/22) | 23% (4/17) | 0% (0/32) | Dysfunction in repairing damaged DNA properly. BRCA2 directly binds DNA and interact with other protein to initiation strand invasion during repair |
| CDK2 | Cell cycle regulator | Deletion | 36% (8/22) | 23% (4/17) | 9% (3/32) | Dysfunction in cell cycle regulation specifically G1-S transition |
| MLH1 | DNA mismatch repair | Deletion | 0% (0/22) | 2% (3/17) | 19% (6/32) | Dysfunction in DNA specifically in recognition and initiation of mismatch repair, therefore, elevating spontaneous mutation rate |
| NCOA2 | Transcription co-regulator | Amplification | 68% (15/22) | 100% (17/17) | 28% (9/32) | Upregulation of DNA expression for nuclear hormone receptors such as AR and ESR1 |
| MYC | Transcription factor | Amplification | 0% (0/22) | 12% (2/17) | 3% (1/32) | Upregulation of many genes involved in cell proliferation and cellular transformation via DNA over-replication |
| NKX3.1 | Tumor suppressor | Deletion | 73% (16/22) | 94% (16/17) | 50% (16/32) | As a transcription factor with critical function in prostate development NKX3.1 deletion results in increase prostate epithelial cell growth |
| AR | Transcription factor | Amplification | 0% (0/22) | 0% (0/17) | 0% (0/32) | As a transcription factor, AR amplification results in DNA over expression of genes such as IGFR and PSA that promote cell proliferation |
| ETV6 | Transcription factor | Deletion | 45% (10/22) | 41% (7/17) | 6% (2/32) | As a transcription factor it interacts with DNA to mostly inhibit transcription of its target genes that regulate both differentiation and cell growth |
| polyploidy | — | Numerical change in a whole set of chromosomes | 64% (14/22) | 0% (0/17) | 0% (0/32) | Polyploidy occurs due to abnormal cell division and have associated with TP53 deletion |
Genomic alterations affecting DNA repair pathways like ATM, BRCA1, BRCA2, CDK2, and MLH1 have been shown to affect sensitivity to platinum-based treatment in metastatic PCa patients [22]. Investigation of these alterations in this patient revealed genomic changes in DNA repair pathway genes. Deletions affecting ATM were found in 77% (17/22), 94% (16/17), and 47% (15/32) of MTCs, BMTP, and PTTP, respectively. Next, we investigated BRCA1 and detected a deletion in 9% (2/22), 29% (5/17), and 9% (3/32) of MTCs, BMTP, and PTTP, respectively. We found partial chromosome 13 loss in 23% (4/17) of BMTP, resulting in at least hemizygous loss of BRCA2. CDK2 gene deletions were also detected at frequencies of 36% (8/22), 23% (4/17), and 9% (3/32) of MTCs, BMTP, and PTTP, respectively. MLH1 deletion was only detected in BMTP and PTTP at frequencies of 2% (3/17) and 19% (6/32), respectively. Collectively, these data demonstrate multiple genomic alterations associated with DNA damage repair. These alterations were identified in a subset of tumor cells and required single-cell analysis for detection. Furthermore, we found the frequency of tumor cells harboring genomic aberrations to be increased in bone marrow tumor cells relative to prostate resident cells. This genomic heterogeneity and low frequency of alterations would have been overlooked in bulk analysis, demonstrating the power of single-cell sequencing.
Next, we focused on PCa specific alterations, such as in chromosome Xq12 where the AR gene resides. We did not observe focal amplification of the AR gene locus in any sample but noted a whole-chromosome gain of the X chromosome in 6% (1/17) and 3% (1/32) of BMTP and PTTP, respectively. In addition, NCOA2 (AR co-regulator) amplification was detected in 68% (15/22), 100% (17/17), and 28% (9/32) of MTCs, BMTP, and PTTP, respectively [20]. MYC amplification was observed in 12% (2/17) and 3% (1/32) of BMTP and PTTP, respectively [20]. All cells with MYC amplification have NCOA2 amplification as well. Consistent with published data showing that NCOA2 and MYC on 8q13 and 8q24 function as driver oncogenes, we observed an increased frequency of amplifications of these genes in MTCs compared to primary tumors [20, 23]. Thus, our observation of AR expression in MTCs and BMTP is likely linked to NCOA2 and MYC gene amplification in a majority of these cells. AR plays a major role in disease initiation as well as progression and is a target of first-line therapy in PCa. AR overexpression in PCa is well documented, and AR gene amplification or mutation are linked to castration-resistant PCa, which is believed to be a result of treatment pressure as opposed to natural disease evolution [20, 24]. Consequently, samples collected from a metastatic hormone-naïve PCa patient may present a higher expression of AR but not a genomic amplification, as seen here.
Other alterations observed include a loss in ERCC3, a DNA helicase that is involved in DNA damage repair. Studies have shown that an age-related decline in this gene leads to declining repair capacity in cells and may lead to development of cancer [25]. The gain in NFκB2 observed in this patient has been demonstrated to contribute to tumorigenesis by uncoupling the normal mode of regulation in immune regulation and inflammation [26]. We also detected deletion of transcription factor ETV6 in 45% (10/22), 41% (7/17), and 6% (2/32) of MTCs, BMTP, and PTTP, respectively. Studies have shown that ETV6 is deleted in 25% of PCa and is involved in tumor development and proliferation as an oncogene [27]. One of the most common forms of alterations in PCa is a loss of chromosome 8p encoding NKX3.1, a transcription factor suppressing cell growth in PT [28]. In this patient, NKX3.1 loss was observed in 73% (16/22), 94% (16/17), and 50% (16/32) of MTCs, BMTP, and PTTPs, respectively. Other observed gene deletions important for PCa initiation and progression include MEEK1, KRAS, and BCL2. Amplifications of the MET tyrosine kinase, BRAF threonine/serine kinase, and the methyltransferase and transcription suppressor EZH2 were also observed across MTCs, BMTP, and PTTP.
Comparison of PTTP, BMA, and BMTP CNAs across sample types enabled construction of a phylogenetic tree demonstrating heterogeneity as well as common patterns of CNAs between tumor cells in prostate and bone marrow (figure 6). Clones of tumor cells can be divided into three groups: clonal cells consisting of PCa hallmark genomic alterations residing in the PT (clone 1), tumor cells with a minimal number of CNAs (clone 2), and tumor cells similar to clone 1 and with additional CNAs residing only in the bone marrow (clone 3).
Amplification of chromosome 10p and loss of chromosome 16p distinguish two subclones contained in clone 3. Within the primary and MTCs additional sub-clonal populations were identified: a subclone of cells with a deletion in chromosome 2, a subclone with focal deletions in chromosome 5, a subclone with an amplification in chromosome 9, and a subclone with a deletion in chromosome 16. This heatmap also demonstrates the development of subclones just within prostate cells and those exclusive to the metastatic site. CNAs only in BM (MTCs and BMTP) included a deletion in chromosome 1 and amplifications in chromosomes 4, 10, and 14. An alteration unique to PTTP included the deletion in chromosome 4.
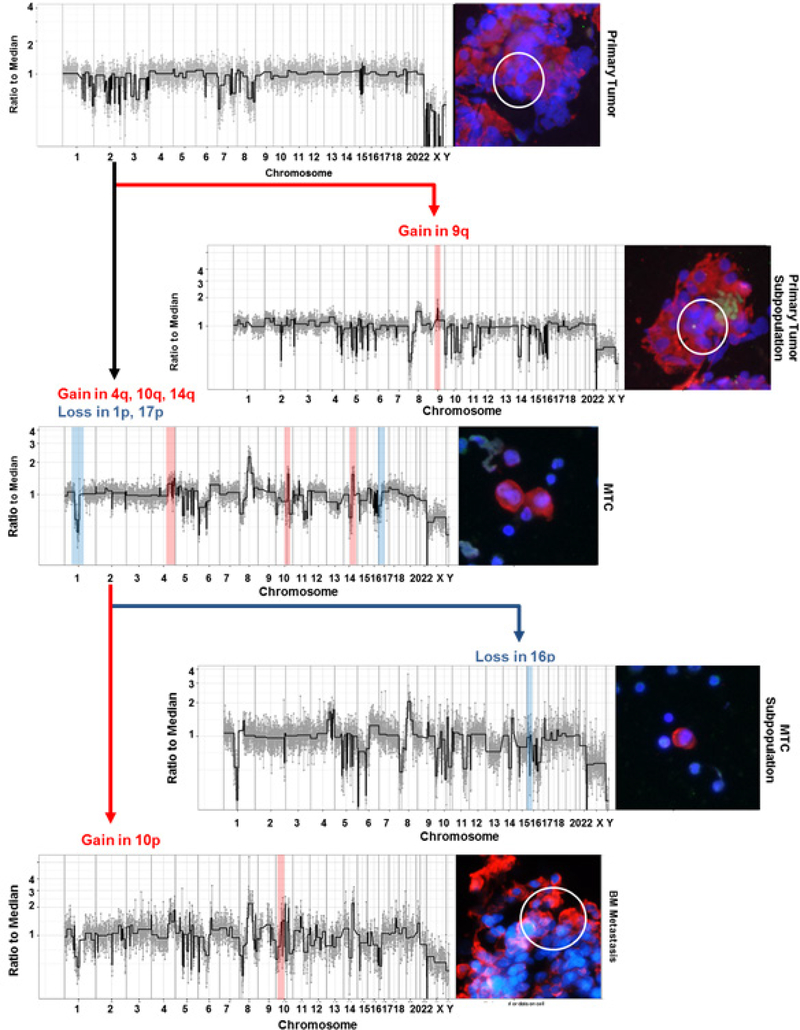
A single dominant clone was identified in all three compartments (BMA, BMTP, and PTTP) and was traced back to a few cells (9/32) in the primary tumor (figure 7). Based on genomic profiles, these cells did not share all of the CNAs of the clonal BM population (figure 6). At its origin in the primary tumor, the clonal population included losses in chromosomes 8p, 10, 11q, and 14q. MTCs in the BM descending from this clone gained additional alterations such as amplifications in 4q, 10q, and 14q as well as losses in 1p and 17p. In addition, MTCs from the BM aspirate gained additional alterations seen in the BM biopsy, such as gain in 10p. These changes can be tracked from PT to BMT. Further examination revealed subclones in the PTTP cells that have developed independently; for example, cells in PTTP with a gain in chromosome 9q were not found in the BM cells. Similarly, subclones in MTCs with a loss in chromosome 16p were observed that were not otherwise found in BMTP cells.
Figure 7.
Cell lineage of dominant clone starting from the primary tumor. Cells in the bone marrow gained additional alterations, such as losses in chromosomes 1 and 17 as well as amplifications in 4q, 10q, and 14q as they evolved from PT. A subpopulation within the MTC was distinguished by the addition of a loss in 16p, and BMTP cell exhibited a 10p gain. Deviations from the original dominant population are highlighted with amplifications in red and deletions in blue. Through this analysis, the lineage of the cancer cells can be tracked from PT to MTCs to BMT. DAPI is shown in blue, CK in red, AR in white, and CD45 in green.
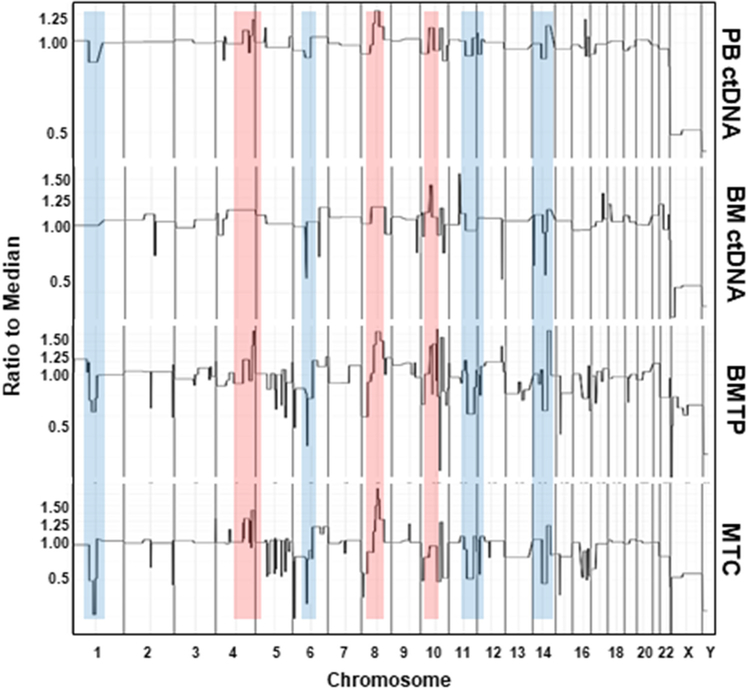
The HD-SCA genomic analysis extends beyond rare cells. Plasma was isolated from both PB and BMA for CNA analysis of cfDNA. Analysis of cfDNA from PB plasma finds shared genomic alterations of cfDNA with both MTCs and BMTP (figure 8). A detailed analysis revealed that the PB plasma profile is more closely related to tumor cells in the MTC than the BMTP sample. Deletions in chromosomes 1, 6, 10, 11, and 14 as well as amplifications in chromosomes 4, 8, and 10 in PB ctDNA matched with those found in the clonal population (figure 8). For ctDNA from BMA, CNA comparison revealed features present in both BMTP and MTCs. Such features included deletions in chromosomes 6 and amplifications in chromosomes 8, 10, and 11. Using in-house software, the fraction of tumor DNA to normal DNA in plasma can be estimated by comparing the amplitude of amplification and deletions of cfDNA to those observed in a single tumor cell where the gains and losses fall in integer steps. PB cfDNA consisted of 75% (3:1 ratio of tumor: normal) ctDNA whereas BMA cfDNA consisted of 50% ctDNA (1:1 ratio of tumor: normal). Thus, the data demonstrate that analysis of cfDNA in the liquid biopsy enables detection of genomic alterations that are present in both primary and metastatic tissue.
Figure 8.
CNA profile comparison of PB and BMA ctDNA to MTCs and BMTP cells. Genomic alterations in cfDNA analysis match CNA of domianant clonal poplation in BMTP and MTCs. Common amplifications across profile types are highlighted in red while common deletions are shown in blue.
Targeted proteomics reveals origin of cells in BM
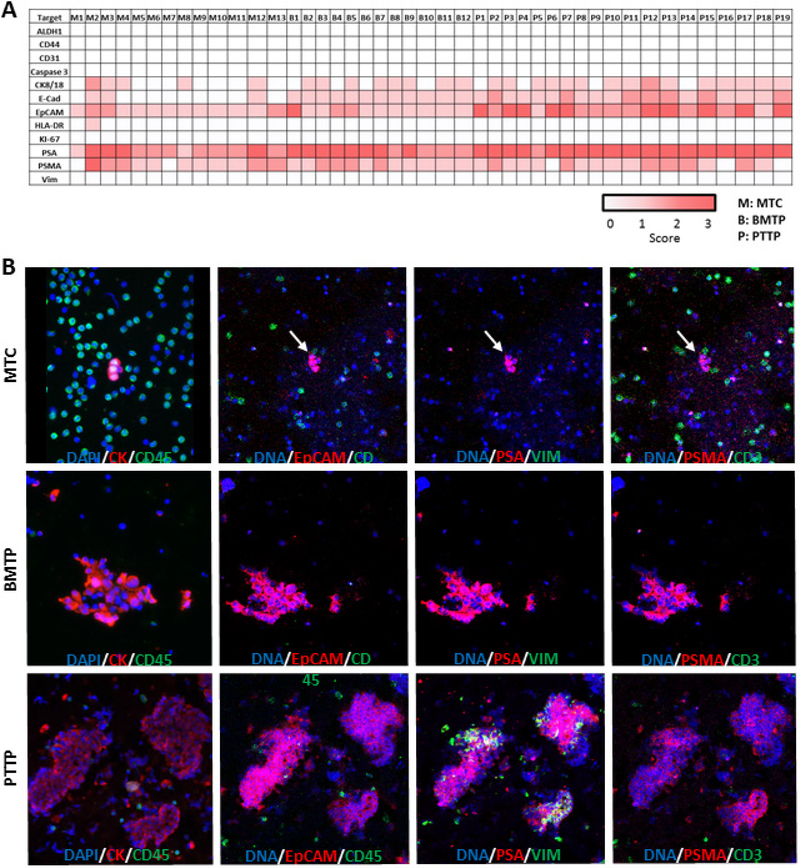
To generate single-cell targeted proteomic data we selected MTCs, PTTP, and BMTP tissue imprints for analysis. We have previously validated a panel of 21 protein markers including a comprehensive leukocyte, epithelial, endothelial, and prostate protein panel for analysis (table 2). A total of 13 MTCs immobilized on a glass slide, 12 BMTP tissue imprints, and 19 PTTP tissue imprints were stained and laser ablated for analysis. Each region of interest was scanned for ablation by the Fluidigm Hyperion Imaging System using a highly focused, pulsed laser that atomizes and ionizes a 1 μm2 region and the resulting ions are introduced into the inductively coupled plasma time-of-flight mass spectrometer [18, 29, 30]. The ion count for each pulse is reconstructed into a 1 μm2 images, and ion count for each protein is scored. The scores for each single MTC as well as BMTP and PTTP tissue imprints are demonstrated as a heatmap in figure 9(a).
Figure 9.
(a) Four-level scoring heatmap for IMC analysis of single MTC and tissue imprints of BMTP and PTTP. Each protein is scored by ion count, and the LOD is determined by a signal-to-noise (S/N) ratio ⩾ 3 or a standard deviation of the mean (SDOM) above 3.3. For each marker, below the LOD is a 0, above the LOD is a 1, a S/N ratio between 7 and 20 or SDOM > 6 is a 2, and a S/N ratio above 20 or SDOM above 12 is a 3. (b) Composite images comparing fluorescent imaging to IMC analysis of all three sample types. The first panel on the left shows fluorescent images with DAPI in blue, CK in red, and CD45 in green. All subsequent panels are IMC-generated composite images showing DAPI in blue followed by two different markers in red and green. EpCAM confirmed epithelial character of most of the cells while PSA and PSMA verified the prostatic source of the samples. Vimentin (VIM) expression in regions of PTTP highlighted the stromal cells of the prostate.
Targeted proteomics of MTCs, BMTP, and PTTP revealed co-expression of epithelial cellular adhesion molecule (EpCAM), PSA, and prostate specific membrane antigen (PSMA) in a majority of cells. E-Cadherin was expressed in 23% (3/13), 67% (8/12), and 100% (19/19) of MTCs, BMTP, and PTTP, respectively. CK8/18 was expressed in 38% (5/13), 100% (12/12), and 84% (16/19) of MTCs, BMTP, and PTTP, respectively. Evaluation of these markers in MTCs, PTTP, and BMTP revealed epithelial and prostatic cellular origin consistent with luminal PCa. Expression level of PSA and PSMA varied both within and between tissue sections. Expression of EpCAM, PSA, and PSMA was consistent across all 3 sample types, but expression of CK8/18 and E-Cadherin was mainly observed in BMTP and PTTP. Such observation may be due to microenvironmental pressures. Once MTCs reach their final destination in the bone marrow, that new microenvironment may lead to higher expression of CK8/18 and E-Cadherin. Examination of EpCAM also revealed higher expression in PTTP than BMTP, suggesting that once MTCs have established a BM metastasis, moderate EpCAM expression may be sufficient [31].
The prostate consists of both epithelial and stromal cells where the stromal cells express mesenchymal markers such as Vimentin [32]. We observed a mix of stromal and epithelial cells in some of the PTTP sections as seen by both EpCAM and Vimentin expression in different parts of the tissue (figure 9(b)). Expansion of our proteomic panel to include proteins such as NKX3.1, ERG, ATM, PTEN, and other markers could help further stratify the subclones of tumor cells. Heterogeneity of protein expression between each cell or cell cluster was visible even within each sample type in the same patient, revealing the power of single-cell data to study variability.
Conclusion
Assessment of liquid and solid biopsies from a patient with de novo polymetastatic PCa using single-cell technology revealed tumor heterogeneity, protein expression variability, and genomic clonality at the intra and inter-biopsy level. Such high-content extraction is feasible with a single cell biology research workflow that preserves morphology and molecular integrity following preservation of cells, identification, and enumeration. This preservation and maintenance of identity through the workflow enables genomic and proteomic analysis using CNA and protein expression, which can be correlated through the morphology. A detailed study of the dominant clonal population revealed modifications such as amplifications in NCOA2 and MYC and deletions in ETV6, PTEN, and TP53, among others. Genomic clonality was demonstrated both within and between each biopsy source (MTCs, BMTP, and PTTP). Tumor heterogeneity via protein expression has also been demonstrated in this patient. Proteomic analysis data shows heterogeneity in expression level of EpCAM, PSA, and PSMA within and across all three sample types.
Deletions in genes such as BRCA1, BRCA2, CDK2, and MLH1 revealed dysfunction in the DNA repair pathway. Many of these genes are candidate targets for therapeutics, and prior knowledge of such gene alterations may be relevant when it comes to choosing the optimal combination therapy. For other therapeutic targets such as PTEN and MYC, having knowledge of both genomic and proteomic status of these genes may lead to optimal combination therapy and personalized medicine for each patient.
We also identified specific gene alterations that are found only in BM while some are shared across all three compartments (BMA, BMTP, PTTP). Similar to Klein et al, our results show that once cells leave the primary tumor and enter the circulation and BM, they acquire independent alterations [33]. Using our single-cell analysis, we were able to demonstrate the same independent evolution in MTCs and BMTP. Such details can be revealed through single-cell study approaches where non-dominant molecular features are detected rather than masked by the dominant population, as is the case in bulk sampling. Information on such detailed features of each compartment (PT versus BM) may influence therapeutic recommendations in the future.
We used liquid biopsy as a source for both cellular and acellular tumor associated components. PB and BMA ctDNA analysis revealed a CNA profile similar to that of the dominant clonal population in BMT. CfDNA analysis has the potential for evaluation and close monitoring of tumor evolution and progression of the dominant clone when CTCs and/or MTCs are not available for analysis.
Cell lineage of MTCs and BMTP cells can be traced back to the primary tumor based on their genomic profile [34, 35]. Interestingly, we observed genomic heterogeneity not only between but also within each compartment. Detailed evaluation of individual CNAs revealed the rise of several small sub-clonal populations within each compartment, demonstrating independent evolution at each site. Tumor lineage of the dominant clonal population revealed that this population started in PT, gained additional alterations as a MTC, and evolved with additional genomic modifications once metastasized. This lineage resulted in a vast genomic diversity with sub-clonal populations within each compartment in this patient case study.
In conclusion, these results demonstrate the high content characterization that can be obtained using the HD-SCA workflow for profiling of liquid and solid biopsies. Using previously published data as validation, BMA was shown to be a feasible and applicable approach and provides additional opportunities for analysis in advanced PCa [5]. BMAs are easier and more cost-effective to obtain compared to image-guided biopsies and can be collected repeatedly during treatment cycles and progression. Longitudinal sampling and assessment provides critical insights into treatment response and allow monitoring of disease progression. Using morphology to integrate genomics and proteomics of single cells throughout the workflow maintains the single cell identity of CTCs, MTCs, and cells isolated from solid tissues. This viable, longitudinal approach has the potential to support treatment decisions throughout the disease course, an important precision medicine objective. Future studies include comprehensive investigation of a broader cohort of both treatment-naïve organ-confined PCa patients as well as additional metastatic cases to further characterize the biology of metastatic tumors.
Acknowledgments
We would like to thank all of our patients and their families who participate in our studies. We would like to thank Xiomara Villasenor, Tayebeh Hosseinzadegan, and the rest of the Kuhn Lab team for their support every day.
The authors would like to acknowledge funding for this project has been made in whole or in part by the Prostate Cancer Foundation Challenge Award 17CHAL04 (IPG, BSK, JH, PK); Prostate Cancer Foundation-Movember Foundation 17CHAL01 USA Award (AA, PK); Prostate Cancer Foundation-Movember Foundation 16CHAL04 (KP, PK); DOD CDMRP PC130244 (BSK); Federal funds from the National Cancer Institute PO1CA098912 (BSK); National Cancer Institute, National Institutes of Health, Leidos Biomedical Research under contract number HHSN261200800001E (JH, PK); the Jean Perkins Foundation (BSK); David and Janet Polak Foundation Fellowship in Convergent Science (PM, PK); and the Vicky Joseph Research Fellow and the Vassiliadis Research Fellow (LW, PK); Schlegel Family Endowed Fellowship Fund (PK); Kalayil and Leela Chacko, MD Fellowship (PK);. The content of this article is solely the responsibility of the authors and does not necessarily reflect the views of the foundations nor the Department of Health and Human Services, National Cancer Institute or the National Institutes of Health nor does mention of trade names, commercial products, or organizations imply endorsement.
Competing Financial Interests are PK: Advisor to Epic Sciences, royalty recipient from Epics Sciences, shareholder at Epic Sciences; JBH: On the Clinical Advisory Boards of Epic Sciences, Inc., La Jolla, CA and CelMatix, Inc. of NY, NY.
References
- [1].Howlader NNA et al. (ed) 2017. SEER Cancer Statistics Review, 1975–2014 National Cancer Institute, Bethesda, MD: (http://seer.cancer.gov/csr/1975_2014/, based on November 2016 SEER data submission, posted to the SEER web site, April 2017) [Google Scholar]
- [2].Bernard B, Muralidhar V, Chen YH, Sridhar SS, Mitchell EP, Pettaway CA, Carducci MA, Nguyen PL and Sweeney CJ 2017. Impact of ethnicity on the outcome of men with metastatic, hormone-sensitive prostate cancer Cancer 123 1536–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].James ND et al. 2017. Abiraterone for prostate cancer not previously treated with hormone therapy New Engl. J. Med. 377 338–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].van Soest RJ and de Wit R 2015. Irrefutable evidence for the use of docetaxel in newly diagnosed metastatic prostate cancer: results from the STAMPEDE and CHAARTED trials BMC Med. 13 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Carlsson A et al. 2017. Paired high-content analysis of prostate cancer cells in bone marrow and blood characterizes increased androgen receptor expression in tumor cell clusters Clin. Cancer Res. 23 1722–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marrinucci D et al. 2007. Case study of the morphologic variation of circulating tumor cells Hum. Pathol. 38 514–9 [DOI] [PubMed] [Google Scholar]
- [7].Marrinucci D et al. 2012. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers Phys. Biol. 9 016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J and Kuhn P 2009. Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type Arch. Pathol. Lab Med. 133 1468–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nieva J et al. 2012. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: a longitudinal analysis Phys. Biol. 9 016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dago AE. et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS One. 2014;9:e101777. doi: 10.1371/journal.pone.0101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Riley RS, Hogan TF, Pavot DR, Forysthe R, Massey D, Smith E, Wright L Jr and Ben-Ezra JM 2004. A pathologist’s perspective on bone marrow aspiration and biopsy: I. Performing a bone marrow examination J. Clin. Lab Anal. 18 70–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marrinucci D, Bethel K, Lazar D, Fisher J, Huynh E, Clark P, Bruce R, Nieva J and Kuhn P 2010. Cytomorphology of circulating colorectal tumor cells: a small case series J. Oncol. 2010 861341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hsieh HB et al. 2006. High speed detection of circulating tumor cells Biosens. Bioelectron. 21 1893–9 [DOI] [PubMed] [Google Scholar]
- [14].Lazar DC, Cho EH, Luttgen MS, Metzner TJ, Uson ML, Torrey M, Gross ME and Kuhn P 2012. Cytometric comparisons between circulating tumor cells from prostate cancer patients and the prostate-tumor-derived LNCaP cell line Phys. Biol. 9 016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pau G, Fuchs F, Sklyar O, Boutros M and Huber W 2010. EBImage—an R package for image processing with applications to cellular phenotypes Bioinformatics 26 979–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Navin N et al. 2011. Tumour evolution inferred by single-cell sequencing Nature 472 90–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baslan T et al. 2012. Genome-wide copy number analysis of single cells Nat. Protocols 7 1024–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gerdtsson E et al. 2017. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry Converg. Sci. Phys. Oncol. 4 015002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Krohn A et al. 2012. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer Am. J. Pathol. 181 401–12 [DOI] [PubMed] [Google Scholar]
- [20].Taylor BS et al. 2010. Integrative genomic profiling of human prostate cancer Cancer Cell 18 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Storchova Z and Pellman D 2004. From polyploidy to aneuploidy, genome instability and cancer Nat. Rev. Mol. Cell Biol. 5 45–54 [DOI] [PubMed] [Google Scholar]
- [22].Mateo J et al. 2015. DNA-repair defects and olaparib in metastatic prostate cancer New Engl. J. Med. 2015 1697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Agoulnik IU, Vaid A, Bingman WE III, Erdeme H, Frolov A, Smith CL, Avala G, Ittmann MM and Weigel NL 2005. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression Cancer Res. 65 7959–67 [DOI] [PubMed] [Google Scholar]
- [24].Liu Y 2017. The context of prostate cancer genomics in personalized medicine Oncol Lett 13 3347–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].DePinho RA. The age of cancer. Nature. 2000;408:248. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- [26].Haefner B 2002. NF-κB: arresting a major culprit in cancer Drug Discovery Today 7 653–63 [DOI] [PubMed] [Google Scholar]
- [27].Demichelis F et al. 2009. Distinct genomic aberrations associated with ERG rearranged prostate cancer Genes Chromosomes Cancer 48 366–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He WW et al. 1997. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3. 1) that maps to 8p21, a region frequently deleted in prostate cancer Genomics 43 69–77 [DOI] [PubMed] [Google Scholar]
- [29].Di Palma S and Bodenmiller B 2015. Unraveling cell populations in tumors by single-cell mass cytometry Curr. Opin. Biotechnol. 31 122–9 [DOI] [PubMed] [Google Scholar]
- [30].Giesen C et al. 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry Nat. Methods 11 417–22 [DOI] [PubMed] [Google Scholar]
- [31].Plaks V, Koopman CD and Werb Z 2013. Circulating tumor cells Science 341 1186–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gregg JL, Brown KE, Mintz EM, Piontkivska H and Fraizer GC 2010. Analysis of gene expression in prostate cancer epithelial and interstitial stromal cells using laser capture microdissection BMC Cancer 10 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Klein CA 2009. Parallel progression of primary tumours and metastases Nat. Rev. Cancer 9 302–12 [DOI] [PubMed] [Google Scholar]
- [34].Jiang R. et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6:44781. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Heitzer E et al. 2013. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing Cancer Res. 73 2965–75 [DOI] [PubMed] [Google Scholar]