ABSTRACT
Approximately 3 billion people around the world have gone into some form of social separation to mitigate the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. The uncontrolled influx of patients in need of emergency care has rapidly brought several national health systems to near-collapse with deadly consequences to those afflicted by Coronavirus Disease 2019 (COVID-19) and other critical diseases associated with COVID-19. Solid scientific evidence regarding SARS-CoV-2/COVID-19 remains scarce; there is an urgent need to expand our understanding of the SARS-CoV-2 pathophysiology to facilitate precise and targeted treatments. The capacity for rapid information dissemination has emerged as a double-edged sword; the existing gap of high-quality data is frequently filled by anecdotal reports, contradictory statements, and misinformation. This review addresses several important aspects unique to the SARS-CoV-2/COVID-19 pandemic highlighting the most relevant knowledge gaps and existing windows-of-opportunity. Specifically, focus is given on SARS-CoV-2 immunopathogenesis in the context of experimental therapies and preclinical evidence and their applicability in supporting efficacious clinical trial planning. The review discusses the existing challenges of SARS-CoV-2 diagnostics and the potential application of translational technology for epidemiological predictions, patient monitoring, and treatment decision-making in COVID-19. Furthermore, solutions for enhancing international strategies in translational research, cooperative networks, and regulatory partnerships are contemplated.
Keywords: Acute respiratory distress syndrome, animal models, clinical trials, immuno-modulation, pandemic, pneumonia
INTRODUCTION
In late 2019, a novel human coronavirus, termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in Wuhan, China. SARS-CoV-2 quickly spread in several Chinese provinces producing a high incidence of acute respiratory illness (1). Following its further spread, the World Health Organization (WHO) has coined this global illness Coronavirus Disease 2019 (COVID-19) and has declared the outbreak to be a Public Health Emergency of International Concern on January 30, 2020. SARS-CoV-2 infections quickly escalated to a pandemic with virtually all continents reporting COVID-19 cases (2). The SARS-CoV-2 infections appeared to have spread in China over several weeks prior to the first delayed reports of patients suffering from severe pneumonia in late December 2019. Since then, the number of confirmed cases has exponentially increased, first in China, subsequently across other continents with almost five million people affected (3). The countries currently most affected by this pandemic are the United States (US) as well as Russia, the UK, Spain, Italy and France in Europe, and the UK in Europe accounting for more than 70% of the entire global death toll (3). According to the WHO, the April spike of SARS-CoV-2 infections in Africa positions this continent as the next epicenter with dire consequences for its population. In May 2020, the US recorded the highest number of known coronavirus cases (with new cases continuously increasing) compared to any other country with one-and-half million cases. However, an enormous variability in testing and data reporting (deaths usually not confirmed through autopsy) exist, which impedes precise estimations. The yet known global propagation pathways of SARS-CoV-2 based on its genetic fingerprinting are displayed in Figure 1(4).
Fig. 1.
Phylogeny, evolutionary relationships, global propagation pathways, and timeline of SARS-CoV-2 viruses from the ongoing COVID-19 pandemic.
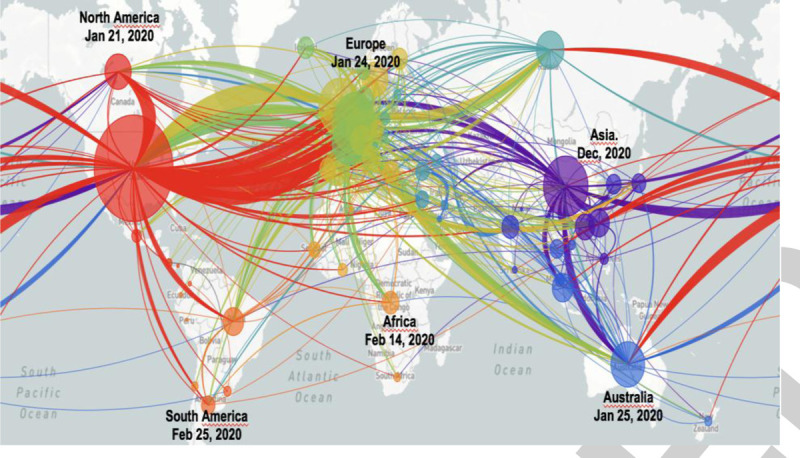
Despite relatively clear genetic relationships among sampled viruses, an uncertainty for specific transmission dates and the reconstruction of the geographic spread remains. Note that the specific inferred transmission patterns (connecting lines) are only hypothetical (4). Thousands of complete genomes are available and increase on a daily basis. The visualization is based upon sub-sampled available genome data (see more under: https://nextstrain.org/ncov). As the pathogen replicates and spreads, its genome is replicated and random mutations/errors accumulate in the genome. Such random mutations allow tracking of the SARS-CoV-2 spread inferences regarding its transmission routes and dynamics. The colors indicate the origin/source of the various viral strains, while circle diameters reflect the size of the transmission clusters. The initial SARS-CoV-2/COVID-19 emergence occurred in Wuhan, China, in November and December 2019, with the first (officially announced) COVID-19-related death on January 11, 2020. The phylogeny is rooted relative to early samples obtained from Wuhan, China. Thereafter, a sustained human-to-human transmission with the first case outside of China (Thailand) was confirmed on January 13, 2020. On January 21, 2020 the first case was confirmed in North America (Wash, USA) and 4 days later in Australia (Victoria). The first three cases in Europe were reported in France on January 24, 2020 (first death on February 15, 2020 France). COVID-19 surveillance was implemented by the European CDC and WHO in the European Region on January 27, 2020, 3 days later the WHO declared SARS-CoV-2 a global emergency. On February 14, 2020 the first case in Africa (Egypt) was confirmed. On February 21, 2020 nine European countries (Belgium, Finland, France, Germany, Italy, Russia, Spain, Sweden, and UK) reported SARS-CoV-2/COVID-19 cases. On February 25, 2020 SARS-CoV-2 reached South America (São Paulo, Brazil). On March 11, 2020 the WHO declared SARS-CoV-2 a pandemic and 2 days later Europe was announced the active pandemic center; on 17 March 2020, all European countries reported confirmed SARS-CoV-2/COVID-19 cases. On March 31, 2020 the number of COVID-19-related deaths (>3,500) in the US surpassed those (officially reported) in China. The highest worldwide daily death toll of 10,761 was recorded on April 27, 2020; as of May 18, 2020, 4,727,625 confirmed cases in 213 countries/territories and two international conveyances with 315,389 deaths were reported (www.worldometers.info). Graphic modified based on https://nextstrain.org/ncov (accessed on April 27, 2020). COVID-19 indicates coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Given that the initial containment was often delayed and insufficient (5, 6), the most affected countries have introduced a range of restrictive mitigation mechanisms such as social distancing, border closures, and travel/business restrictions. On March 18, 2020, more than 250 million people went into lockdown in Europe. The enormous and uncontrolled influx of patients in need of specialized medical care has rapidly brought several national health systems to near-collapse. Adhering to the WHO call (7), the healthcare systems worldwide rapidly increased hospital capacities and adapted to the specific needs of COVID-19 patients as a fundamental response measure. These adaptation mechanisms, however, are difficult (impossible) to achieve in many underprivileged regions/countries with an under-developed health infrastructure putting those populations at much higher risk.
SARS-CoV-2 is assumed to be mainly spread via small droplets produced at coughing/sneezing in close contact (up to 2 m) although longer distances cannot be ruled out (8). Experimental evidence shows that SARS-CoV-2 may remain viable in aerosols up to 3 h and up to 72 h on various surfaces such as plastic, steel, copper, and cardboard (9). Of note, the American Centers for Disease Control and Prevention (CDC) reported that SARS-CoV-2 RNA was identified on cabin surfaces of a cruise ship 17 days after cabins had been vacated; it is unclear whether the material remained infectious (10). High viral load and active shedding in the upper respiratory tract that peaks during the first week of symptoms, suggests that SARS-CoV-2 is most contagious in already symptomatic subjects although some spread is likely before occurrence of symptoms (11). The SARS-CoV-2 infection may be asymptomatic in some people; analysis of the “Diamond Princess” cruise ship cohort indicated that approximately 19% of the infected passengers remained clinically healthy (12). The viral load in asymptomatic patients may reach a level comparable to the one seen in symptomatic patients; preliminary evidence demonstrates that asymptomatic patients may transmit the virus but the transmission pathways and timing are yet to be identified (13, 14). The percentage of patients who remain truly asymptomatic for the course of their infection is unknown; it is likewise unclear what percentage of individuals who initially present with an asymptomatic infection subsequently progress into clinical disease.
Clinical features of COVID-19 are nonspecific and are hardly distinguishable from other causes of severe community and hospital-acquired pneumonia. While approximately 80% of cases follow a relatively mild trajectory, the elderly and/or patients with comorbidities (e.g., chronic lung conditions, hypertension, diabetes, and obesity) are at risk for severe COVID-19 course with pneumonia as the typical manifestation (Fig. 2) (15). Not infrequently, patients may show a disproportionate extent of radiographic pulmonary involvement compared to the mild level of hypoxemia. Some of these patients suddenly deteriorate to severe respiratory failure followed by intubation and mechanical ventilation requirement. Deaths appear to be dominated by severe respiratory failure, fulminant myocarditis (leading to heart failure), thrombo-embolic events (stroke, infarcts, embolism), and late secondary sepsis with severe single or multiorgan dysfunction (typically involving the liver and kidneys) (16–18). Renal dysfunction may be an early sign for later deterioration. Emerging data suggest that severe COVID-19 phenotypes are associated with a significant (hyper-) coagulopathy that correlates with disease severity (19–21). Direct viral infection of the endothelial cells and diffuse endothelial inflammation with a shift of the vascular equilibrium toward enhanced vasoconstriction (with subsequent organ ischemia), inflammation with an associated tissue edema, and a pro-coagulant state may constitute the main underpinnings of the severe clinical phenotypes (22, 23). Studies confirm the high rate of comorbidities among deceased SARS-CoV-2/COVID-19 patients, but serial (and better powered) studies are needed to precisely identify the cause(s) of death in the most severe cases (24).
Fig. 2.
A pulmonary presentation of SARS-CoV-2 infection in a severely ill, intubated, and mechanically ventilated COVID-19 patient by computed tomography (CT; panel A) and plain chest x-ray imaging (panel B).
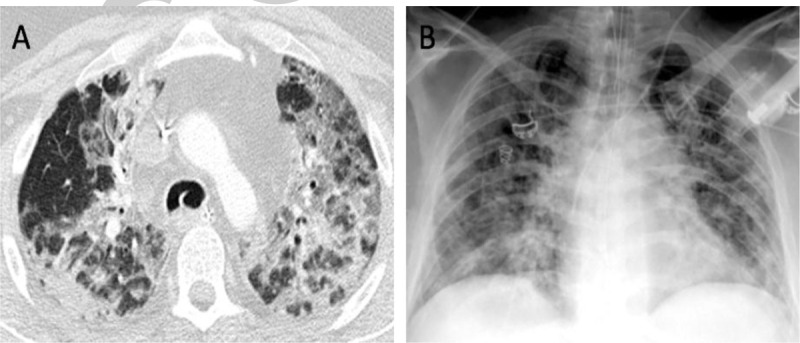
The CT shows characteristic milk-glass like opacities with consolidations in both upper lobes (A). CT findings may be unspecific and the primary diagnosis of SARS-CoV-2 remains laboratory-based. However, if indicated, imaging studies are helpful in assessing the severity and the course of COVID-19 pneumonia. A CT score can be used to evaluate the severity of the disease (15). The risks of an in-hospital transfer and potential contamination need to be considered. (Source: Axel Gossmann (MD), Department of Radiology, Cologne-Merheim Medical Center (CMMC), Cologne, Germany). COVID-19 indicates coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
With most of the world in lockdown, an ongoing SARS-CoV-2 spread and new infection waves expected, the pandemic will continue to represent a major global threat (25). As solid scientific evidence remains scarce, there is an urgent need to expand our understanding of the SARS-CoV-2 evolving epidemiology, its infectivity and body site-specific replication as well as COVID-19 immuno-inflammatory characteristics and treatment strategies against it (25). The existing gap of high-quality, reproducible evidence-based data is frequently filled by anecdotal reports, contradictory statements (26), and misinformation. The present review addresses several important aspects unique to the SARS-CoV-2/COVID-19 pandemic highlighting the newest evidence, most relevant knowledge gaps, and windows-of-opportunity.
COMPARISON OF COVID-19 TO THE RECENT VIRAL PANDEMICS
COVID-19 is the fourth viral pandemic of the last two decades following the SARS virus in 2002/2003, the influenza A virus H1N1 in 2009, and the Middle East Respiratory Syndrome (MERS) virus in 2012 (Table 1). All these viruses are enveloped by a host cell-derived membrane and contain a single RNA as genome. Many of the RNA viruses possess the capacity to induce zoonotic infections. Wild birds are a reservoir for influenza A viruses that may be transmitted to swine, human, and other mammals (27). SARS-CoV and SARS-CoV-2 have high similarity to coronaviruses (CoV) in bats and likely have been transmitted from bats to humans via an intermediate host (civet for SARS-CoV; possibly pangolin for SARS-CoV-2) (28–30). MERS-CoV was transmitted from camels to humans, with limited human-to-human transmission capacity (but high pathogenicity; (31)). Due to the tight contact between camels and humans, in some communities, MERS-CoV continues to circulate and temporal disease clusters arise.
Table 1.
Main differences between COVID-19 and previous viral pandemics
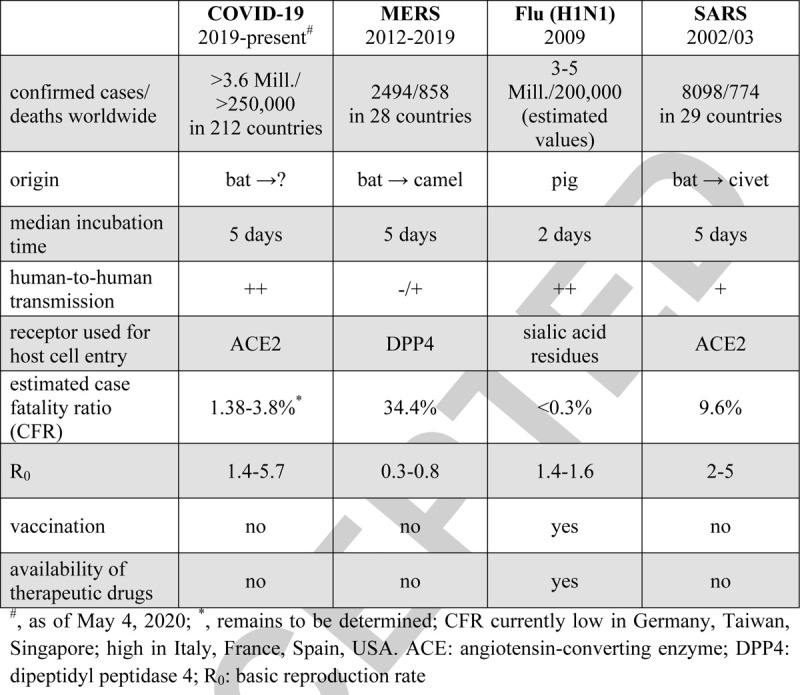
H1N1 (and other influenza viruses) belong to the Orthomyxoviridae and induce upper respiratory tract infections. Influenza A virus enters the host via the viral hemagglutinin attaching to sialic acid residues present on upper respiratory tract cells with an average incubation time of 2 days (32). The 2009 pandemic H1N1 virus integrated gene segments from multiple avian and mammalian strains thereby forming a novel virus unrecognizable by any pre-existing immunity (27). Despite relatively efficient vaccines and antiviral drugs, the flu continues to kill >200,000 people worldwide per year (33).
SARS, MERS, and the novel COVID-19 are caused by CoV that are widely distributed, highly infectious and responsible for the symptoms associated with common cold in humans, i.e., the strains NL63, OC43, 229E, and HKU1. For their entry into the human cells, SARS-CoV and SARS-CoV-2 use a spike protein that binds to the angiotensin-converting enzyme (ACE) 2 on alveolar type II cells in the lung (also airways mucosa) (34). The expression of ACE2 in the liver, heart, and gastrointestinal tract (Table 2) may explain why some infected patients also develop liver injury, fulminant myocarditis with subsequent heart failure, and diarrhea in addition to severe pneumonia (35–38). In contrast to SARS-CoV, MERS-CoV uses the enzyme dipeptidyl peptidase (DPP) 4 as receptor for host cell entry (39). The median incubation time for CoV infections is 5 days but up to 2 weeks incubation time is not uncommon.
Table 2.
Comparison of selected aspects of immune response to SARS-CoV, MERS, and SARS-CoV-2∗
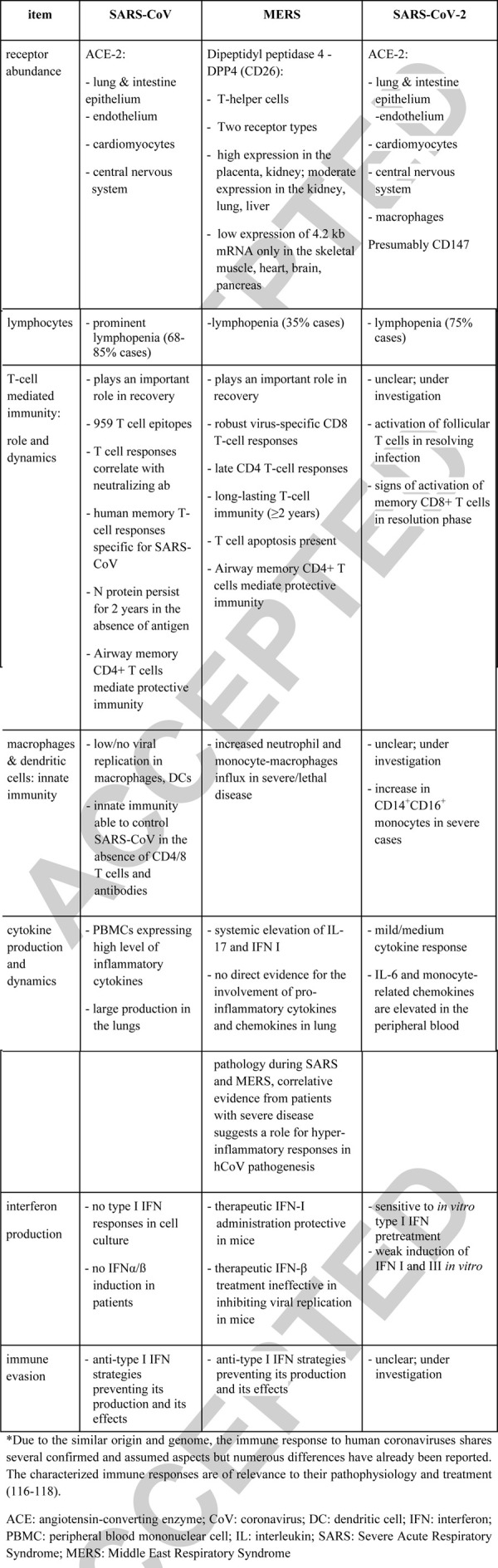
The extraordinarily high spread and the number of SARS-CoV-2 infections indicate that its infectivity is higher compared to SARS-CoV, with a basic reproductive number (Ro) at approximately 3 (40, 41); a recent estimate by the CDC defines the SARS-CoV-2 Ro at approximately 5.7 (42). This may be explained by an improved virus entry due to a molecular change of the receptor-binding domain and the insertion of a furin-cleavage site in the spike protein of SARS-CoV-2; this enables a 10-fold increased binding affinity to ACE2 and fusion with the host cell membrane (43). SARS-CoV-2 may additionally infect epithelial cells in the upper respiratory tract, thereby facilitating transmission of the shed virus via respiratory droplets. Presently, it appears that SARS-CoV-2 is more pathogenic than the influenza A virus but less pathogenic than SARS-CoV (44); the reported overall case fatality ratio of SARS-CoV-2 ranges between 1.38 and 3.8 (1, 25, 45). Notably, wide testing and identification of asymptomatic carriers versus focal testing restricted to the hospitalized (symptomatic) patients may either underestimate or overestimate the CRF.
IMMUNOPATHOGENESIS OF SARS-CoV-2
Given the vague COVID-19 pathogenesis, references to the earlier SARS/MERS-CoV pandemics are inevitable. While useful given the similar CoV origin, this is not ideal as several significant immunological differences among the three diseases are apparent (Table 2).
Upon infection, virus internalization typically evokes intracellular pattern-recognition receptors signaling likely via RIG-I, OAS (46), and TLR-7 inducing interferons (IFN) I/III (and IFN-stimulated genes; IFGs) subsequently triggering a local immune response. In uncomplicated COVID-19, an increase in circulating follicular helper T cells and antibody secreting B cells was observed (47) concurrent with an upregulation of activation markers on CD14+ and CD8+-T cells. In contrast, there was a reduction of circulating CD14+CD16+ monocytes. Interestingly, the systemic cytokine response has been typically negligible in mild COVID-19, while rarely soaring in severe COVID-19 cases (48–50). Such a scenario reflects an optimal orchestration of the immune system and a balance between the inflammatory response and disease tolerance leading to uneventful pathogen eradication. Unfortunately, in a subgroup of patients developing life-threatening COVID-19 phenotype this balance is deranged. In the following sections, Table 2 and Figure 3 summarize the rudimentarily understood dynamics of the immuno-inflammatory processes in patients with varying COVID-19 severity and phenotype.
Fig. 3.
Summary of potentially protective and harmful host responses during the SARS-CoV2 infection based on the currently available data.
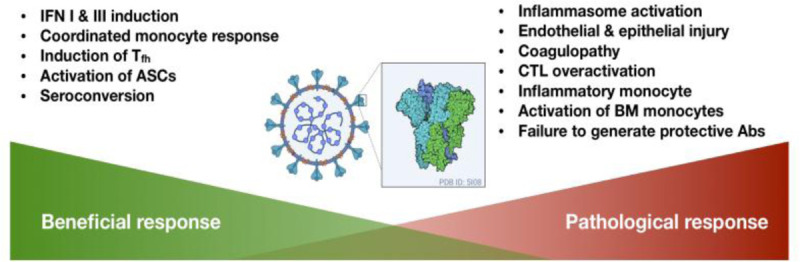
ASC indicates antibody secreting cells; BM, bone marrow; CTL, cytotoxic T-cells; IFN, interferon; Tfh, follicular helper T-cells; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Viral load
In adults, viral load was found to correlate with COVID-19 severity and has been suggested as a potential mechanism responsible for the disease progression (51). However, the role of the viral load is unclear since it may reflect failure of the immune response and a high viral load can also be the result of immuno-evasion (SARS-CoV-2 poorly upregulates IFN I and III) (46). Endocytosis of the virions bounded to ACE2 decreases activity of this enzyme which can skew the angiotensin II/angiotensin (1–3, 5–8) balance by increasing ACE1 activity (52). It remains to be verified whether the increase in angiotensin II indeed contributes to propagation of the inflammation and impaired hypoxic vasoconstriction in COVID-19 patients. In human alveolar epithelial cells, SARS-CoV-2 induces cytopathic effects (53). SARS-CoV-2 appears to be very cytotoxic; infection of epithelial Vero-E6 cells with a patient-isolated SARS-CoV-2 strain was highly cytopathic after 48 h (M. Ranawak, personal communication). It can only be hypothesized that, similarly to SARS-CoV, SARS-CoV-2 activates the NLPR3 inflammasome leading to pyroptotic cell death (54); an abundant serum lactate dehydrogenase release in severe COVID-19 supports this notion (55). Interestingly, in bats, MERS-CoV poorly activates the inflammasome which contributes to their disease tolerance (56).
Innate immunity
Even in severe COVID-19, a mild neutrophilia is typically observed (48, 49), while circulating monocyte counts typically remain unaltered (49). An increased fraction of CD14+CD16+ cells was reported in severe cases (57) and several groups found blood monocytes IL-1β+, IL-6+ with IFGs expression (57, 58). Monocytes in COVID-19 were found to reduce their Human Leukocyte Antigen – DR isotype expression but the magnitude and time course of that reduction remains to be determined (59, 60). This suggests a pathological role of inflammatory monocytes in the immune response to SARS-CoV-2. Moreover, transcriptomic analysis of peripheral blood mononuclear cells (PBMCs) revealed upregulation of complement-related genes and increase in C-C motif ligand (CCL)2, C-X-C motif ligand10, CCL3, and CCL4 chemokines expression (61). Importantly, the CD169+ macrophages can be stimulated and infected by the virus up-regulating IL-6 (62). In some severe COVID-19 patients, high circulating ferritin was reported suggestive of hemophagocytosis by activated macrophages (48, 60).
Adaptive immunity
Severe cases of COVID-19 are characterized by peripheral lymphopenia attributable to the loss of T-cells, while B and NK cells remain only marginally affected (48, 49). Mechanisms of the T-cell lymphopenia remain mostly hypothetical but upregulation of pro-apoptotic genes expression has been suggested (61). Hallmarks of apoptosis and necrosis were present in lymph nodes and spleens in three autopsied COVID-19 patients (62). Alternatively, lymphopenia may be due to a robust lung infiltration and tissue homing (of circulating lymphocytes) which needs to be verified in further autopsy studies. Differentiating between these two mechanisms is key for understanding the pathophysiology and potential treatments. The circulating CD4+ and CD8+ T-cells are activated, but there is discrepancy in the available reports regarding their ability to produce IFN-γ(49, 63). Notably, in aged mice, CD4+ but not CD8+ T-cells were crucial in controlling interstitial inflammation and clearing SARS-CoV-infection (64). One group reported an increased frequency of central memory with a decrease in naive T-cells subpopulations and clonal expansion of cytotoxic CD8+ T cells (58), while another group showed a rise in the frequency of naive CD4+ T-cells (49). It can be hypothesized that an uncontrolled “bystander activation” (in a cytokine-dependent manner) of the cytotoxic T-cells and their tissue sequestration are among the major pathogenic events which is supported by activation markers in severe cases (63). Interestingly, only in the ICU-admitted patients the pathogenic GM-CSF+IFN-γ+ CD4+-T cells were found (57). Simultaneously, the regulatory T cells decrease (49) suggesting an impairment of the immune regulation. A single-cell analysis revealed an increase in memory B and plasma cells, which maintained activation and antibody production (with IgA overrepresentation) (58). Most antibody-related findings indicate that specific anti-SARS-CoV-2 immunoglobulins possess a neutralizing potential in vitro(65). However, the role of antispike protein antibodies should be further evaluated as these molecules may aggravate COVID-19 by promoting proinflammatory monocyte activation (66) and enable infection of immune cells (67). Those cellular changes were accompanied by seroconversion and appearance of Immunoglobulin G (IgG) and Immunoglobulin M (IgM) immunoglobulins. However, seroconversion takes from 5 to 9 days (68–70). Whether this humoral immunity is fully protective or could also contribute to antibody-dependent immunopathology (71, 72) remains to be established. Nevertheless, when studied in vitro, a correlation was reported between antibody titers and neutralization properties. At present, several studies investigate convalescent plasma-containing antibodies for treatment (73, 74).
Cytokines
Despite evidence of the so-called “cytokine storm” in SARS and MERS (75, 76), recent data suggest that this is not necessarily the case in SARS-CoV-2 infection (48, 51). Conflicting viewpoints backed by yet insufficient data have emerged fueling controversy about this concept (77, 78). Circulating cytokines (both pro- and anti-inflammatory) increase in COVID-19 patients; however, only some of them (e.g., IL-2, IL-7, IL-10, G-CSF, MCP1, TNFα) are relatively robustly increased in severely (ICU-admitted) ill patients when compared with moderate/mild cases (48, 51). Compared with hyperreactive responses recorded in bacterial septic patients (79) and after noninfectious triggers (80), the cytokine upregulation in COVID-19 appears to be at least one log of magnitude lower. Many controversies arise regarding IL-6 as this cytokine was shown to be slightly increased in some severely ill (51), while it was also markedly up-regulated in non-survivors (81) and critically ill COVID-19 patients (82). Others found IL-6 predictive of mechanical ventilation requirement at a cutoff of 80 pg/mL (83). Of note, the expression of IL-6R in bronchoalveolar lavage fluid cells was downregulated and unchanged in PBMCs, suggesting that this pathway may not be crucial for COVID-19 pathophysiology (61). The current rationale to inhibit IL-6 is also controversial as IL-6 promotes antibodies formation (84), regeneration of airway ciliated cells from basal stem cells (85) and protects against H1N1 influenza (86). An indiscriminate application of IL-6 (and other) blockers has potential for harm, especially when done in a “blind” fashion in a heterogenous population of COVID-19 patients.
The interferons
The precise role of IFNs in SARS-CoV-2 infection is yet unclear and requires further investigation. It is unclear whether patients by default produce amounts of interferon like certain mammals (87) or low IFN concentrations may reflect the coronaviruses’ capacity to prevent/reduce its production and action (88–95). The latter may be plausible given that CoV are well capable of preventing NF-κB activation (96) and protein translation (97). It is also important to define the specific role played by IFN-ε in the mucosa; this may explain differences between humans versus bats/pangolins regarding their response to a CoV infection (98–100). Better understanding of the crosstalk between SARS-CoV-2 and IFNs will offer insights into the COVID-19 immunopathogenesis. Some data suggest that the strategy of a very early administration of type I IFN could be considered (101–103). However, the negative effect of a late exposure to IFNs must also be considered; a study combining single-cell RNA-sequencing data and in vitro analysis showed that ACE2 is upregulated by type I and II IFNs in human and primate airway epithelial cells (104). Furthermore, IFN-inducible genes are highly expressed in cells from broncho-alveolar lavage fluid of COVID-19 patients and exhibited pathogenic potential with overrepresentation of genes involved in inflammation (105). Of note, the National Institute of Health (NIH) recommends against the use of IFNs for the treatment of COVID-19 outside of clinical trials (https://www.covid19treatmentguidelines.nih.gov/therapeutic-options-under-investigation/host-modifiers-immunotherapy/). Carefully designed trials will hopefully inform on the potential of the type I interferon application (106).
Coagulopathy and endotheliopathy
Coagulopathy appears to be a critical element in the context of severe COVID-19 courses. Elevation of D-dimers (fibrin degradation products) has been frequently observed in severe cases and identified as a significant risk factor (107). One study revealed presence of a procoagulant state even during the early COVID-19 stage (21) and disseminated intravascular coagulation has been diagnosed in most critically ill patients (108). Moreover, the incidence of thromboembolic events is high (comparable to sepsis) and most likely underdiagnosed (109). It can be hypothesized that there is dominant local pulmonary vessel microthrombosis which correlates with the severity of hypoxemia and high compliance (110) and stays in accordance with fibrin accumulation found in the lungs (110, 111). Presumably, there are several pathways which may contribute to the clinical coagulopathy observed. Direct infection of the endothelium, thereby triggering endothelial injury, inflammation, and cell death (22), can directly activate the coagulation cascade. Pyroptosis and inflammasome-released mediators are other potent coagulation cascade activators (112, 113). Histopathological examination revealed deposition of activated complement complexes that may propel microvascular injury and subsequent activation of the clotting pathway (114). Neutrophilic infiltrates can also activate coagulation through the generation of neutrophil extracellular traps (115) and a recent anecdotal study confirmed the presence of antiphospholipid antibodies in three COVID-19 patients with severe coagulopathy (116). An analysis of over 3,500 genes (differentially expressed in the lungs) following murine SARS-CoV infection identified various procoagulatory factors (especially urokinase) to be strongly associated with mortality (116). Urokinase activity results in the generation of plasmin and in turn in fibrinolysis with elevated D-dimers manifested by alveolar coagulopathy and pulmonary hemorrhage. Furthermore, serpin1 knockout mice confirmed an enhanced pulmonary expression of procoagulatory and profibrinolytic proteins and clinical susceptibility to SARS-CoV (117). Apart from cytokine effects on the pulmonary endothelium, it has also been proposed that disruptions of the kallikrein-bradykine axis can increase microvascular permeability and cause angioedema (118).
EXPERIMENTAL THERAPIES AND PRECLINICAL EVIDENCE
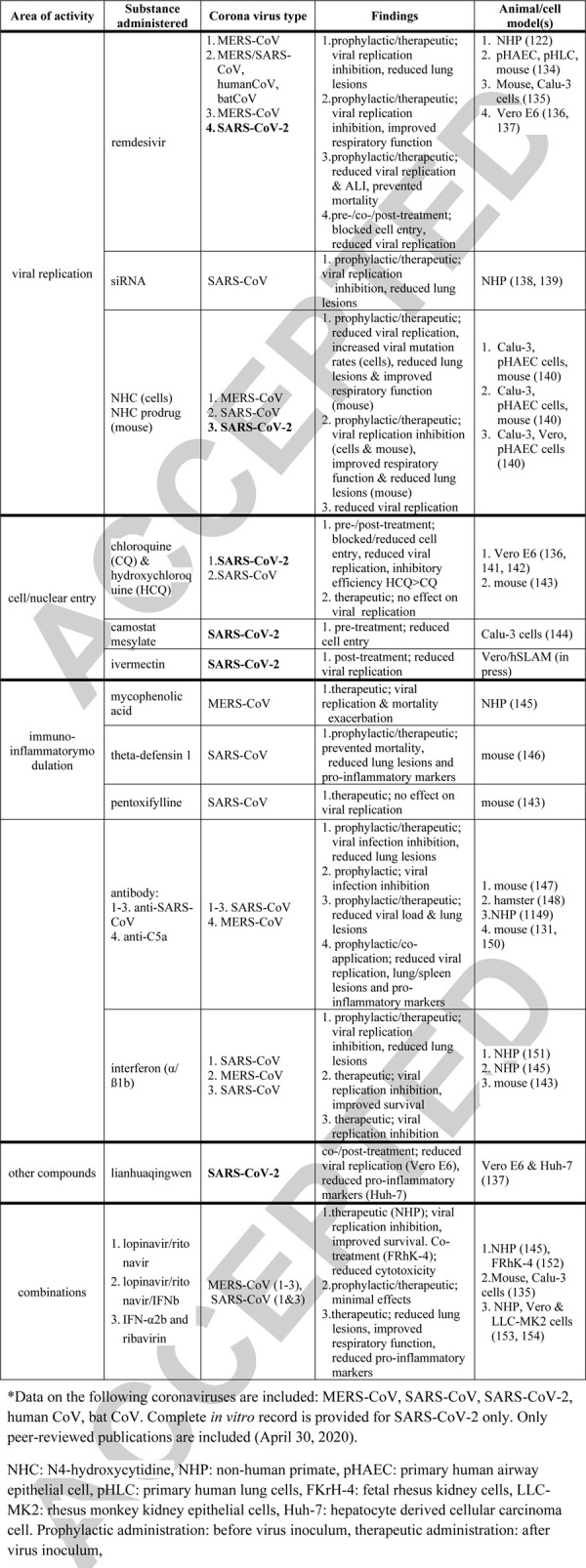
The number of proposed therapeutic strategies against COVID-19 grows weekly; presently, there are over 50 substances considered as potential remedies including brand new (Small interfering RNA) (119)) as well as well-known/repurposed chemicals (chloroquin, interferons, remdesivir). Yet, the currently available pre- and clinical evidence supportive of the experimental therapies is suggestive at best. Several substances with different targets have been proposed based on sparse peer-reviewed publications (Table 3) (120–143); some based on non-peer-reviewed (medRxiv and bioRxiv) preprints and/or anecdotal evidence only. As of May 19, 2020, there is only one peer-reviewed study (140) that tested an anti-COVID-19 candidate drug in a relevant SARS-CoV-2 animal model (Table 3); many drugs have not yet been directly tested against SARS-CoV-2 in vitro. Except a single case (140), the only available animal model-based evidence stems from MERS/SARS-CoV studies (Table 3), but these diseases are not identical to COVID-19 (Table 2).
Table 3.
Preclinically tested therapeutics against SARS-CoV, MERS-CoV, and SARS-CoV-2 and related illness∗
Mechanistically, we are only at the very beginning to understand how SARS-CoV-2 infects, targets the lungs (and other organs), and causes severe vascular and tissue damage. Despite the aforementioned limitations, the existing experimental findings warrant a well-organized, collaborative verification of therapeutics aiming at clearing the virus load, modulating the inflammatory response, protecting/repairing damaged tissues (e.g., endothelium), and ameliorating vascular coagulopathy. The mechanisms of pulmonary and remote organ response to SARS-CoV-2 present an additional level of complexity; any pathophysiological (and subsequently therapeutic) leads remain mostly speculative and require adequate modeling and experimental verification. This is currently difficult to achieve given that (mostly non-peer-reviewed) evidence suggests the only animal models suitable for studying SARS-CoV-2/COVID-19 include non-human primates, ferrets, and transgenic mice. The WHO asserts high reproducibility of SARS-CoV-2 infection in Rhesus macaques and ferrets (144). Macaca mulatta was the most susceptible to SARS-CoV-2 (compared with M fascicularis and C jacchus) displaying a wide array of clinical-like COVID-19 symptoms (145). In another study, six macaques developed infection and pneumonia but remained asymptomatic (120). Kim et al. (146) demonstrated ferrets as an apt model both for SARS-CoV-2 infection and transmission; animals were symptomatic (e.g., fever, cough), displayed high viral RNA in the lungs and upper respiratory tract and shedded virus via multiple routes. The disease phenotype in ferrets was reproduced by another study (147) and there is unpublished evidence from the Australian Centre for Disease Preparedness, Geelong, Victoria.
However, small laboratory animals remain the most accessible and cost-effective model to study. Transgenic human ACE2 mice (both sexes) infected with the HB-01 strain developed COVID-19-like interstitial pneumonia, high viral load, and produced high specific IgG titer but the disease was generally mild (148). The most recent study largely reproduced this phenotype in hACE mice (149). Syrian hamsters constitute another potential option; two groups recently recapitulated a mild but widely symptomatic COVID-19 phenotype (150, 151). In contrast, species such as pigs, chickens, and ducks were virus-free when either inoculated or exposed to the virus, cats were asymptomatic despite postexposure infection whereas dogs were minimally susceptible to SARS-CoV-2 exposure (152). Experimental drugs will soon be tested in the newly emerging COVID-19 models. While modeling of SARS-CoV-2 infection in healthy and young animals will be informative, it is important to consider that the most severe COVID-19 phenotypes appear in aged patients with comorbidities—the animal models should reflect this to maximize their translational capability for pathophysiology studies and drug testing. The most recent macaque experiment attests to that; 15-year-old animals developed an exacerbated COVID-19 phenotype while the young ones did not (153). Another important element in preclinical modeling is to promote study designs that allow drug testing in divergent, precisely defined and relatively homogeneous COVID-19 phenotypes (provided they can be recapitulated in animals). It is likely that a given therapeutic may be either beneficial or detrimental contingent upon the timing of its administration and/or specific COVID-19 pathophysiological characteristic. Compared with patients, animal studies present a relatively cheap and safe platform to establish such relationships.
The yet limited evidence derived from preclinical studies coupled with the emerging clinical data as discussed previously indicates that SARS-CoV-2-induced coagulopathy may be one of the key interests for experimental studies (116). In this context, another less apparent but interesting target for potential COVID-19 therapy is the complement activation pathway. Genetic absence of the complement C3 component was associated with reduced pulmonary/systemic inflammation and improved lung function (154). MERS-CoV-infected mice displayed elevated complement component C5a in the lungs and blood and blockade of the C5aR receptor attenuated inflammation in the lungs and spleen and reduced pulmonary viral replication (137). As aforementioned, adaptive immunity cells activation is prerequisite for eradication of the virus. Whether specific cell types of adaptive immune cells may help to induce pulmonary healing processes in SARS-CoV is currently unclear. Mechanistically, the large surface area of the alveolar capillary endothelium appears as a key target organ; protection and repair of the dysfunctional air–blood barriers (through reduction of endothelial swelling, damage, boosting epithelial regeneration) and endothelium-derived coagulopathy (e.g., formation of microthrombi) could be life-saving in COVID-19 patients, especially in those with advanced pulmonary damage and severe respiratory failure. Unfortunately, these postulates remain largely speculative and valid SARS-CoV-2/COVID-19 models are needed for verification.
An application of experimental (unlicensed) therapeutics via the compassionate use protocol (CUP) while justified as last resort (155) should not be reflexively overexpanded in the context of SARS-CoV-2/COVID-19 infection; CUP rarely provides reliable efficacy data given multiple confounding factors and inobjectivity. Whereas the magnitude of pandemia may justify extraordinary measures to save patients’ lives, these measures must be counterbalanced by an extraordinary analytical rigor and resistance to overoptimistic interpretation of the daily-emerging in vitro/silico, animal and clinical data. In a disease with a relatively low mortality and rudimentary understanding of its pathophysiology, potential risks for life-threatening side effects by unproven therapeutics must not be ignored in a pursuit of the desired benefits. The hazards of adverse effects are additionally aggravated by the typically advanced age of COVID-19 patients, who frequently present with various comorbidities and comedications (156). Properly designed animal experiments and clinical studies will gradually reveal the cons and pros of the experimental therapeutics.
CLINICAL MANAGEMENT AND TRIALS
With the exception of remdesivir, which has received an emergency use authorization by the US Food and Drug Administration (FDA) for the treatment of hospitalized COVID-19 patients on May 1, 2020, (European Medical Agency (EMA) announced on May 18, an upcoming conditional marketing authorization), there are no drugs yet approved by any professional authority to prevent and treat COVID-19; results from any large-scale clinical trials are not yet available. The WHO, EMA, and CDC strongly discourage the application of any unlicensed therapeutics outside of adequately designed clinical trials (https://www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html#r8; https://apps.who.int/iris/bitstream/handle/10665/331446/WHO-2019-nCoV-clinical-2020.4-eng.pdf?sequence=1&isAllowed=y; https://apps.who.int/iris/handle/10665/330680). The most reliable strategies continue to rest on supportive ICU care practices including supplemental oxygen, mechanical ventilation, and, in extremis, extracorporeal membrane oxygenation. However, traditional ventilation protocols used in acute respiratory distress syndrome (ARDS) may not be adequate for all COVID-19 patients. Interim guidelines to inform clinicians how to care for patients with COVID-19 have been released by the NIH (https://covid19treatmentguidelines.nih.gov/introduction/) and by the Surviving Sepsis Campaign as an initiative supported by the Society of Critical Care Medicine and the European Society of Intensive Medicine (SCCM/ESICM, (157)). Health professionals from the European Respiratory Society partner societies have compiled an international directory of guidelines and best practice recommendations documents intended to help share COVID-19 expertise around the world (https://www.ersnet.org/covid-19-guidelines-and-recommendations-directory). However, the guidelines are based upon current limited evidence and will require rapid updates to account for any new and compelling evidence; the NIH guidelines are conceptualized as living guidelines (158). Some of the more prominent, currently investigated therapies include antimalarials, antivirals, immunotherapeutics, and corticosteroids (158).
Antimalarials
The trial-based evidence has been fluctuating very dynamically since the dawn of the SARS-CoV-2 pandemic. Initially, it was proposed to treat severely ill Chinese COVID-19 patients with chloroquine phosphate (159). Following these observations, a combination of oral hydroxychloroquine and azithromycin was proposed in patients with signs of lower respiratory tract involvement. This was based on a small-scale open-label clinical trial, in which 36 patients were allocated to treatment with hydroxychloroquine (n = 20) or left untreated (n = 16). Hydroxychloroquine was associated with virological cure on days 3, 4, and 5 in 50%, 60%, and 70%, respectively, compared with 6.3%, 25%, and 12.5% of untreated controls (azithromycin co-administration in six patients further improved it to 83%) (160). This French study was further confirmed by the same team on 80 patients (161). These studies were heavily criticized by the MRC-NIHR Trials Methodology Research Partnership for numerous shortcomings (160). The two subsequent studies with hydroxychloroquine indicated no improvement (162, 163). The most recent reports emphasize risks associated with chloroquine-based trials; due to adverse cardiac reactions, chloroquine treatment (with and without azithromycin) was stopped in Brazilian (164) and French (165) trials; and several hospitals in Sweden stopped administering hydroxychloroquine to COVID-19 patients based on similar findings (166). A recent preprint article that retrospectively analyzed hydroxychloroquine use in hospitalized US veterans found an association of increased overall mortality with its use (167).
Antivirals
Other suggested alternatives have been antiviral treatments with remdesivir and lopinavir-ritonavir. The most recent remdesivir CUP study was performed in 53 patients with severe COVID-19 and the median follow-up showed an improvement in oxygen support in 36 patients (68%) including 17 of 30 patients (57%) with mechanical ventilation that were successfully extubated (168). However, the study has been widely questioned as no comparator group was involved (https://www.sciencemediacentre.org/expert-reaction-to-a-study-about-compassionate-use-of-remdesivir-for-patients-with-severe-covid-19/). Another open-label randomized trial in 199 COVID-19 patients showed that lopinavir-ritonavir failed to accelerate clinical improvement, reduce mortality, and diminish viral RNA detectability (169). Nearly 14% of patients in the lopinavir-ritonavir arm could not complete the full 14-day treatment course due to gastrointestinal adverse events. Remdesivir and (hydroxy) chloroquine are both under evaluation in the largest international trial in severe COVID-19 patients launched by the WHO and partners. The “SOLIDARITY” trial compares four options: local standard of care (LSC), or LSC plus either remdesivir, chloroquine/hydroxychloroquine, lopinavir with ritonavir, or lopinavir with ritonavir plus IFNß-1a. As of May 18, 2020, over 90 countries are active participants to this trial (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments). As the most recent development, results of two different randomized, placebo-controlled, multicenter studies testing remdesivir were released on April 29, 2020: the Chinese trial (NCT04257656; 237 patients; (170)) demonstrated no statistical benefit, whereas an official announcement (apnews.com) stated that an NIH-led trial (NCT04280705; 460 patients at interim analysis) shortened the time to recovery to 31%. Two days later, on May 1, 2020, the FDA approved remdesivir for emergency use to treat COVID-19 patients based upon the belief that “the known and potential benefits of [remdesivir] outweigh the known and potential risks of the drug for the treatment of patients hospitalized with severe COVID-19” (https://www.fda.gov/media/137564/download).
Immuno-inflammatory modulation
Various strategies aiming to modulate the immune response in critically ill COVID-19 patients have been proposed. Specific examples include the transfusion of convalescent plasma and targeted anti-inflammatory treatments. In two case-series critically ill COVID-19 patients (171, 172) received compatible transfusions of 250 mL to 400 mL of convalescent plasma. In the first case-series (171), all five patients were under co-administration of methylprednisolone and antivirals (mainly lopinavir/ritonavir). After 12 days from transfusion, an improvement was noted in the Sequential Organ Failure Assessment score, an increase of the partial oxygen to fraction of inspired oxygen (pO2/FiO2) ratio along with virological cure was observed (172). In the second case-series, clinical improvement was not observed in the 10 enrolled patients but all patients experienced virological cure (172). Three trials testing convalescent plasma in COVID-19 patients are currently recruiting patients (NCT04321421; NCT04343755; NCT04355897).
The idea of modulating the immune response of the host originates from the observed alterations of various cytokines in the blood of COVID-19 patients (49, 60, 172). For example, circulating cytokines were compared between 286 severe and 166 non-severe COVID-19 patients; TNFα, IL-2 receptor, IL-6, IL-8, and IL-10 were significantly higher in the severe versus non-severe patients. Another recent study analyzed immune characteristics of 54 COVID-19 patients with (n = 28) or without (n = 26) mechanical ventilation requirement suggesting two divergent dysregulation patterns: a generalized immune hyperactivation and dysregulation associated with a decreased Human Leukocyte Antigen – DR isotype expression (60). Several clinical trials are currently testing a range of inflammatory modulators; four medium-size trials investigate the efficacy of biologically targeting the IL-1 and the IL-6 pathway: recombinant IL-1 receptor antagonist anakinra (NCT04330638/COV-AID, 342 patients), IL-6 receptor antagonists tocilizumab (NCT04330638/COV-AID, 342 patients; NCT04320615/COVACTA, 330 patients), and sarilumab (NCT04320615/CORIMUNO-SARI, 240 patients). Mortality, ventilator-free days, and the change of the respiratory ratio are the most common endpoints. Smaller-scale trials also aim (not yet recruiting) to investigate the impact of immunostimulants like PD-1 blockers thymosine (NCT04268537; 120 patients) and novilumab (NCT04343144/CORIMUNO-NIVO, 92 patients), and Treg stimulant human recombinant IL-2 (NCT04357444/LILIADE-COVID, 30 patients). However, these strategies are not controversy-free given that there is no clear consensus as to what extent the magnitude of the inflammatory response generated by SARS-CoV-2 is detrimental to COVID-19 patients (hence favoring cytokine inhibition) or necessary for eradication of the virus and host defense (hence discouraging their blockage and/or favoring immuno-supportive interventions).
Corticosteroids
The use of corticosteroids in the acute respiratory distress syndrome (ARDS) of COVID-19 remains also a matter of debate as no evidence of their efficacy currently exists. In a recent report in 84 COVID-19 patients with ARDS, the administration of methylprednisolone (dosage similar to the SCCM/ESICM recommendations (173)) was associated with a reduced risk of death (0.38; 95% CI, 0.20–0.72) (107). Although opinions vary on the administration of corticosteroids in COVID-19 patients, the two largest studies on H1N1 and SARS (n = 7568) were supportive to their use (174, 175). However, the WHO interim (https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected) and NIH (https://covid19treatmentguidelines.nih.gov/introduction/) guidelines do not recommend corticosteroids in viral pneumonia outside of clinical trials. There is preliminary information from China and Italy, suggesting that the use of corticosteroids in COVID-19-associated ARDS could be considered (176) but such anecdotal evidence is burdened by bias and should not be used as reference for altering treatment practices. The WHO has prioritized the evaluation of corticosteroids in COVID-19 with five running randomized controlled trials. At present, no definite conclusion can be drawn from the available experimental and clinical data to assess the acute and long-term effects of corticosteroids for the resolution of the local and systemic inflammatory response in COVID-19 and/or the development of fibrotic complications.
Coordinating clinical trials
An overwhelming number of clinical trials has emerged (Fig. 4) aiming to assess therapies ranging from antivirals, immune-therapeutics, and host-directed therapies, vitamins, gases, mesenchymal stem cells to Traditional Chinese Medicine —all in the bid to save lives of COVID-19 patients. As of May 18, 2020, the cumulative number of registered COVID-19 clinical trials according to the WHO Clinical Trials Portal was 2356 (https://www.who.int/ictrp/en/). The trial count grows daily and is frequently updated. The deluge of trials scheduled to recruit simultaneously may create a realistic “traffic jam” risk for under-enrolment and is perceived as chaotic (177). An additional major concern of many ongoing COVID-19 trials is their relatively low power to detect significant differences in meaningful outcomes. Low-powered trials rarely provide unequivocal evidence justifying the use of tested therapeutics. In this respect, pandemics provide a unique window of opportunity for large-scale collaborative initiatives, thus, enabling networks to jointly generate and address common goals as well as to standardize data collection in large patient volumes. The WHO recommends the development of “Master Protocols” for high-powered, adequately-designed trials as a tool for the most rapid and reliable evaluation of investigational therapeutics and vaccines (https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus/en/). The five ongoing global clinical trials SOLIDARITY (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments), REMAP-CAP (A Randomised, Embedded, Multifactorial, Adaptive Platform Trial for Community-Acquired Pneumonia; (NCT02735707; remapcap.org (178), RECOVER (Randomized Evaluation of COVID-19 Therapy; https://www.recoverytrial.net/; https://doi.org/10.1186/ISRCTN50189673) and PRINCIPLE (Platform Randomized trial of Interventions against COVID-19 in older people; ISRCTN86534580 (179), and Trial of Treatments for COVID-19 in Hospitalized Adults (DisCoVeRy) (https://clinicaltrials.gov/ct2/show/NCT04315948(180) exemplify such an approach.
Fig. 4.
Global clinical research activities on SARS-CoV2/COVID-19 based upon trial registration data.
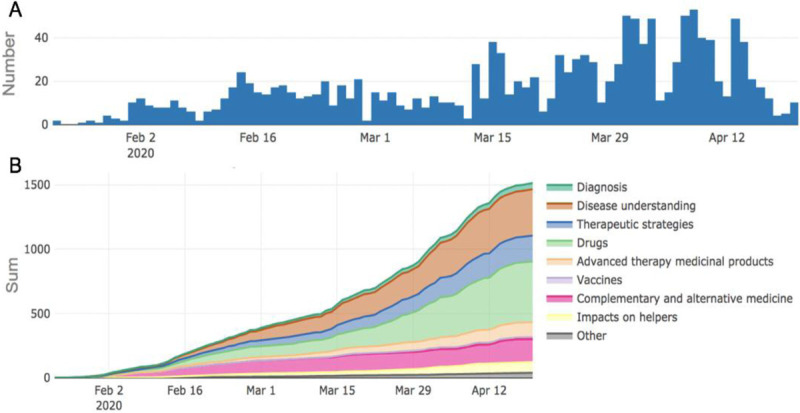
A, Registration of COVID-19 clinical trials for each day. B, The cumulative number of registered COVID-19 clinical trials exceeds 1,500. Information based on data from the WHO Clinical Trials Search Portal (https://apps.who.int/trialsearch/) for COVID-19-related clinical trials as of May 18, 2020. This portal allows access to a central database containing the trial registration data sets provided by international registries. The WHO portal is updated every Friday by six important registries and every 4 weeks by additional registries (https://covid-19.heigit.org/clinical_trials.html; COVID-19-Karte der Hoffnung; Universität Heidelberg; May 18, 2020). COVID-19 indicates coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Vaccines
Finding a safe and effective vaccine to prevent infection with SARS-CoV-2 has become an urgent public health priority and a number of legacy drug-makers and startups have begun to develop vaccines (and treatments) that target SARS-CoV-2. At present, the global COVID-19 vaccine R&D landscape includes 115 vaccine candidates, of which 78 are confirmed as active and 37 are unconfirmed ((181), https://clinicaltrials.gov/). Of the 78 confirmed active projects, 73 are currently at exploratory or preclinical stages. The most advanced candidates have recently been moved into clinical development, including mRNA-1273, Ad5-nCoV, INO-4800, LV-SMENP-DC, and pathogen-specific aAPC (181). On March 16, 2020, the first-ever injection of an investigational vaccine for SARS-CoV-2 (mRNA-1273) was administered to four volunteers participating in an open-label phase 1 clinical trial in the United States. The study is expected to conclude June 1, 2021. Multiple other vaccine developers have communicated plans to initiate human testing within this year.
SARS-CoV-2 DIAGNOSIS AND OPTIMIZATION OF TESTING
The differentiation of the SARS-CoV-2-positive cases from the healthy is among the main clinical challenges. Many asymptomatic persons are a source of infection despite being considered healthy prior to a positive test result (182). The prevalence of asymptomatic (SARS-CoV-2 positive) patients ranges between 1.5% and 30% (182–184). Non-test-based COVID-19 diagnosis is difficult given that the most common clinical symptoms, e.g., fever, fatigue, dry cough, myalgia, and dyspnea (51, 185) are unspecific and overlap with other viral diseases (185). Several hematologic and immuno-inflammatory abnormalities observed in SARS-CoV-2-positive patients resemble MERS/SARS-CoV infection symptoms (185). To reduce the transmission risk, aggressive containment, mitigation, and treatment strategies must be combined and rapid and accurate testing is key. A few countries/territories such as Taiwan, Hong Kong, Singapore, and South Korea have rapidly implemented aggressive testing (186); South Korea has performed >300,000 tests (5,829 tests/million population) within the 9 weeks after the first SARS-CoV-2 case was identified, effectively containing the SARS-CoV-2 spread (187, 188). The majority of other countries have been struggling with tests approval, availability, and/or operationalization of high-throughput testing, which was accompanied by substantial differences in reported numbers.
The current recommendation of many professional radiological associations and societies is that imaging should not be employed as a screening nor diagnostic tool for COVID-19 but reserved for the evaluation of complications. Although used frequently in early reports with some characteristic features described ((4), Fig. 2), imaging has limited sensitivity for COVID-19 and the definitive test for SARS-CoV-2 remains laboratory-based. A wider evaluation of the CT value in COVID-19 patients is further complicated by its different use among different countries (e.g., frequent in China, infrequent in Europe, sporadic in the US).
As of March 9, 2020, the WHO recommends testing based on clinical and epidemiological factors contingent on the likelihood of SARS-CoV-2 infection and/or exposure (189). Similarly, the CDC has established a priority system for diagnostic testing for SARS-CoV-2 infection based on the availability of tests; the CDC testing guidance is updated periodically (https://covid19treatmentguidelines.nih.gov/overview/). In asymptomatic or mildly symptomatic subjects who were in contact with SARS-CoV-2/COVID-19, virus nucleic acid amplification tests (NAAT), such as RT-PCR, is typically recommended (189). Given that NAAT recommendations are difficult to meet in many countries due to limited testing resources and/or excessive demand, the WHO suggests adopting screening protocols tailored to the local situation and provides reference laboratories for confirmatory testing (https://www.ecdc.europa.eu/en/novel-coronavirus/laboratory-support) as well as shipment instructions if local testing is unavailable (190). As co-infections and false-negative diagnoses occur, testing for other respiratory pathogens should be also performed utilizing routine laboratory procedures (189). Currently, two types of diagnostic kits for SARS-CoV-2 are available: genomic (RNA-based, long, precise) and serological (IgM/IgG-based; fast, relatively imprecise).
Nucleic acid amplification test (NAAT)
The WHO recommends a RT-PCR test for SARS-CoV-2 detection (185, 189); RT-PCR was the first established and rapidly commercialized diagnostic method in the SARS-CoV-2 pandemic (191, 192). The WHO testing recommendations differ depending on the SARS-CoV-2 prevalence. In the virus-free areas, a positive NAAT for at least two target SARS-CoV-2 genes is considered reliable; in areas with SARS-CoV-2 presence, a confirmation of a single discriminatory target is sufficient (185, 189). The Chinese National Institute for Viral Disease Control and Prevention recommends a cycle threshold value (Ct-value) below 37 as positive, Ct-value >40 as negative test (193), and (40) <Ct-value < 40 requiring retesting. The intermediate values have frequently been associated with reported false-negative rates (194). Therefore, a negative RT-PCR result does not necessarily rule out the possibility of SARS-CoV-2 infection (194). The WHO quotes several culprit factors including a poor specimen quality, timing of specimen collection, specimen handling, and/or shipping and various technical problems (189). A small Chinese study demonstrated that some COVID-19 patients who met criteria for hospital discharge and/or quarantine discontinuation (free of clinical symptoms, radiological abnormalities and with two negative RT-PCR tests) were tested positive (by RT-PCR) 5 to 13 days later (195). A South Korean study reported that two out of 10 negative COVID-19 cases by RT-PCR were later confirmed as positive (196). In a large study, the second RT-PCR test was positive in 12.5% of initially negative 384 patients (197). Thus, the detection of SARS-CoV-2 is highly specific (no false positives) but the sensitivity is not ideal (198). These findings reveal limitations of RT-PCR testing and indicate that some recovered patients may continue to be virus carriers (195). This phenomenon, if confirmed on a larger scale, may force changes in the current criteria for hospital discharge and quarantine discontinuation. Furthermore, the SARS-CoV-2 diagnostics should account for clinical findings; preliminary data from China showed that isolated patients (with presumed COVID-19) with initially negative RT-PCR but with typical clinical and radiological COVID-19 symptoms were confirmed as SARS-CoV-2-positive after repeated swab testing (194). Thus, a combination of repeated RT-PCR testing and radiological imaging may be helpful in determining suspected false-negative cases.
Although the RT-PCR constitutes the current diagnostic standard, the test kits suffer considerable limitations. In 610 patients, in the first all-patient test, 27.5% cases were positive, 0.2% weakly positive, 9.3% dubious, and 63% were negative (197). Among the patients with initial negative results, the second test was positive in 12.5%, dubious in 7% patients, negative in 73% patients, and results were not available for 7.6% patients (197). The false-negative readouts are also contingent on the workflow for molecular detection (e.g., and isolation method with/without commercial kit) (194, 195, 199). Additionally, RT-PCR has long turnaround times, requires certified laboratories, expensive equipment, and trained personnel. Due to these limitations, RT-PCR may not be practical as a rapid and simple diagnostic and/or screening tool. There is an urgent need for a fast, sensitive, accurate, and simple test to process a large number of suspected and asymptomatic carriers. Moreover, there is a pressing need for rapid, digital analyses of viral load as the field moves forward.
Serological testing
Serologic detection of specific IgM/IgG antibodies constitutes a rapid, simple, and cost-effective complementation to NAAT (170). The WHO continues to evaluate and update all available immunodiagnostic tests for COVID-19 (200). One of the newest point-of-care tools is the lateral flow immunoassay test able to detect IgM and IgG in the blood sample within 15 min (201). Since the IgG concentration begins to rise as IgM levels start to drop, the dynamic pattern in COVID-19 patients is consistent with an acute viral infection (202).
The serological SARS-CoV-2 testing with an average sensitivity of 88.7% and a specificity of 90.6% faces the risks of both false-positive and false-negative readouts (201). The key testing limitations include low antibody concentrations, differences in individual immune responses and antibody production, and quick changes in the IgM/IgG ratio (201); possible cross-reactivity with other corona and flu viruses cannot be excluded (203, 204). The above shortcomings practically preclude defining the exact infection time-point and the length of infection. Over 500 commercial SARS-CoV-2 tests are currently available or under development ((205); https://www.finddx.org/covid-19/pipeline/); however, their efficacy remains limited and the validation of their clinical performance and quality constitutes a global priority (200). As of April 8, 2020, the WHO does not recommend the use of antibody-detecting rapid diagnostic tests for direct patient assessment; their application is encouraged for COVID-19 surveillance and epidemiological research (200).
MODELING AND TRANSLATIONAL TECHNOLOGY FOR MONITORING AND THERAPEUTIC STRATEGIES
The knowledge gaps in our understanding of COVID-19 pathophysiology and the complexity of treatment validation are major limitations for the current standard of care. One possible strategy to overcome this is to engage complex statistical and physiological modeling. In silico models devoted to big data analysis and encompassing systems biology and precision medicine concepts have been introduced to inflammation, trauma, and sepsis research as a new complementary tool supporting translational applications (206–208). Similarly, advanced data analysis techniques constitute a potential support in the interpretation of different clinical COVID-19 presentations, in the timely identification of infected subjects prone to ARDS, and in the prediction of patient trajectories and their complication risks while on the ICU.
Artificial intelligence (AI) has been already introduced into the fight against SARS-CoV-2 (209). Despite the poor quality and inhomogeneity of the available data (that affect the modeling robustness and accuracy), AI may hold potential for the prediction and monitoring of SARS-CoV-2 outbreak hot-spots, transmission, and spread. For example, the analysis of big data from the commercial aviation industry was used to predict patterns of the international SARS-CoV-2 spread (210). Given the suboptimally coordinated response to the original outbreak, it is advisable that transnational organizations such as the WHO as well as national governments and local health systems integrate AI-based analyses to support their decision-making in: the design and implementation of lockdown and post-lockdown strategies and the early identification and prevention of second infection waves. Such applications have already been proposed to predict the preparedness and vulnerability to SARS-CoV-2 in Africa (211) and could be expanded to other continents/regions around the globe. Artificial intelligence can be also used in a cluster fashion in individual clinics. For example, AI-powered automated analysis of chest X-rays and CT scans effectively reduced the workload of radiologists (212–215); AI identified COVID-19-induced airway injury and pneumonia of varying magnitude as accurately as human operators. Thus, AI applications appear as a time- and resource-saving alternative in diagnosing COVID-19 in mildly symptomatic individuals. Recently, a multidisciplinary global initiative to leverage AI in the fight against COVID-19 was launched in Belgium together with hospitals and organizations worldwide (icometrix/icolung, Leuven (Belgium), Chicago). This multinational collaboration resulted in the development of an AI solution for rapid and objective quantification of lung pathology on chest CT scans in admitted COVID-19 patients (Fig. 5).
Fig. 5.
Example of a fully automated assessment of the total and lobar disease burden in COVID-19 pneumonia based upon an AI algorithm.
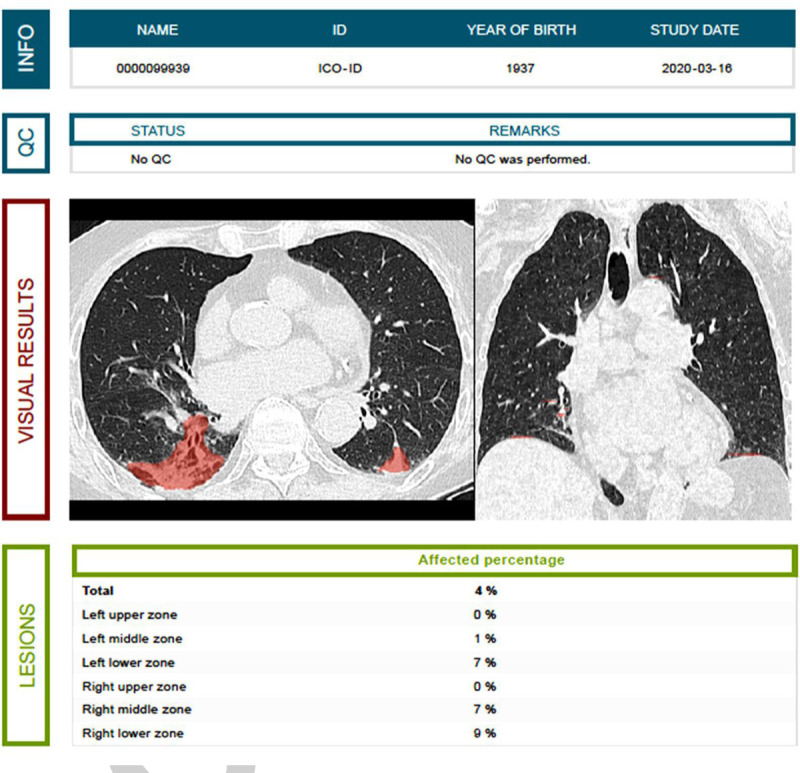
Evaluating the type, pattern, and extent of lung pathology on chest CT can help in the assessment, triage, and follow-up of COVID-19 patients. (Source: Jan Verheyden, icometrix (icolung), Leuven, Belgium). COVID-19 indicates coronavirus disease 2019; CT, computed tomography; AI, artificial intelligence.
Despite its epidemiological and diagnostic potential, it is premature to expect AI to be a game-changer in real-time patient monitoring (216). It is unclear whether AI can precisely prognosticate, and its usefulness has been modest in the context of therapy design. Even though AI algorithms have been used to predict the efficacy of existing drugs repurposed to treat COVID-19 (217, 218), the candidate drugs (and pharmacological targets) still require experimental validation before translating them to bedside and into clinical practice. Interestingly, promising results were obtained from the analysis of high-throughput data on the interaction between SARS-CoV-2 and ACE2 receptors encouraging research on interventions preventing the internalization of SARS-CoV-2 (131, 219).
The contribution of data-driven models in the improvement of patient monitoring and in the design of new therapeutic approaches critically depends on the quality of the available data; it is key to ensure the best possible consistency and homogeneity across databases. Initiatives such as the COVID-19 data portal by the European Molecular Biology Laboratory—European Bioinformatics Institute (EMBL-EBI) (https://www.covid19dataportal.org/about) and the CORD-19 (COVID-19 Open Research Dataset) by the Allen Institute for AI (Seattle, Wash) and collaborators (https://allenai.org/data/cord-19) exemplify this paradigm and support the development of knowledge-based models to formulate and test hypotheses on the desired pathological mechanisms of interest, and characterize and interpret the dynamics of the disease progression in real time. Physiological models will likely benefit from the integration of data from different levels of the biological scale. In particular, high-throughput, multi-omics data have been increasingly used in acute illness (220–226) to investigate disease pathways and their molecular signatures, and to search for novel diagnostic biomarkers and therapeutic targets. The use of multiscale models in COVID-19 research could shed light on the complexity of phenotypes and enhance the design of combinatorial treatments not limited to a single drug or intervention. A recent study (227) used a multi-omic approach and machine learning algorithms to analyze COVID-19 signatures; the data showed that COVID-19 severity was associated with distinct profiles in the serum proteome and metabolome, implicating massive metabolic suppression, macrophage dysregulation, platelet degranulation, and complement system imbalance. Multilevel data integration, including the -omics, hemodynamic, biochemical, and clinical readouts, could further advance our understanding of the relationships among various COVID-19 phenotypes such as bilateral pneumonia/ARDS requiring mechanical ventilation (228), shock requiring vasopressor support (229, 230), cardiomyopathy (231), and neurological manifestations (232).
Retrospective studies will be necessary to build and validate physiological multiscale models. The success of this effort will rely on the availability of well-annotated, cohesive, and consistent databases and corresponding biobanks. The integration of statistical (data-driven) modeling and physiological (knowledge-based) modeling should be pursued to enhance the translational potential of computational analytics for: diagnostics and monitoring, the design of novel, more effective treatments, the guidance of therapy by decision support systems.
DISSEMINATION OF DATA AND LARGE-SCALE TRANSLATIONAL RESEARCH NETWORKS
The SARS-CoV-2 outbreak has triggered an unprecedented cooperative activity worldwide, yet with varying perception and outcomes. Major funding agencies including the European Commission with dedicated emergency funding action under the Horizon 2020 Framework Program, the National Institutes of Health and other funding agencies such as the Department of Defense among others, and numerous national agencies have mobilized funds to facilitate rapid basic and clinical research on the SARS-CoV-2 pathogenesis and COVID-19 treatments. While this greatly supports COVID-19-oriented research, it simultaneously puts other important research areas at risk of slowdown and under-funding. To enable an uninhibited and rapid exchange of emerging knowledge, major publishers such as Elsevier (https://www.elsevier.com/connect/coronavirus-information-center), Springer Nature (https://www.springernature.com/gp/researchers/campaigns/coronavirus) JAMA, (https://ja.ma/covidyt), The Lancet, The New England Journal of Medicine, and Science, among others, have gone open-access covering multiple SARS-CoV-2/COVID-19-related topics. The video/audio knowledge exchange has been at its finest; the JAMA Update podcast with Dr. M Cecconi (March 3, 2020) from the Italian COVID-19 frontline was followed by over million viewers (233) and the 7 h-long COVID-19 Marathon Webinar organized by ESICM (March 23, 2020) gathered expert-speakers from 15 countries and four continents (https://esicm-tv.org/covid19/). However, perception of the rapid data dissemination by the preprint servers (e.g., bioRxiv.org; medRxiv.org) is less favorable. There has been a growing concern that such a steep growth of unscrutinized scientific evidence (3718 bio/medRxiv.org articles on SARS-CoV-2/COVID-19; accessed May 19, 2020) rather provokes more confusion than meritorious guidance and indiscriminately selected pieces of the preprinted data are recurrently sensationalized by the lay press.
While the majority of above activities are effective (and impressive), there is an important gap: the lack of a functional translational-research collaborative network on the SARS-CoV-2/COVID-19 pandemic. Although clinical studies are costly and organizationally challenging, clinical centers can join them with relative ease and without a need for additional and/or specialized core facilities. This is in sharp contrast to basic and translational studies. While the progress in modern basic science can be made using online resources like sequencing and structural data, e.g., enabling in silico modeling (234), translational studies require work with live SARS viruses and animal models at biosafety level (BSL)-3 facilities (https://www.cdc.gov/sars/guidance/f-lab/app5.html). The availability of SARS-CoV-2-susceptible animals and BSL-3 labs is limited. Therefore, efforts should be undertaken to enhance cooperation among existing centers by creating international collaborative research platforms able to simultaneously investigate multiple hypotheses/aims. This can be achieved by implementing three key objectives: to galvanize an open-access scientific exchange and tools (e.g., models, protocols) among laboratories, to facilitate a multilateral data flow among basic science researchers in an organized, standardized fashion, and to improve the validity of preclinical treatment studies by introduction of multicenter animal testing. Encouraging examples of such strategies exist; beginning with China (2020/01/21), hCoV-19 genome sequences have been rapidly and freely shared by the international scientific community (virological.org; GenBank; www.gisaid.org/), some SARS-CoV-2-related findings are published patent-free (235) and first multicenter animal studies have shown a superior reproducibility and validity (236). A few of such preclinical collaborative networks have also been found in the area of critical care medicine; Operation Brain Trauma Therapy is a fully operational US-based multicenter drug and biomarker screening consortium for traumatic brain injury (237), whereas the international Wiggers-Bernard Initiative is in the process of forming a multicenter preclinical RCT in mouse sepsis (https://wiggers-bernard.org/2019-topic/). In the context of COVID-19, especially the trials performed in non-human primates would highly benefit from a multicenter coordinated approach given that single-center studies are typically limited to very low numbers (n = 4–6) of animals (120, 238, 239). The translational research community should also learn from the clinical examples that employ additional, more advanced designs: for example, the REMAP-CAP trial enables multicenter large-scale and adaptive-design treatment testing. Adaptation and utilization of the above strategies in preclinical research will likely save time and resources, increase rigor of the findings, and enhance the bidirectional flow of data between the laboratories and clinic (and vice versa) given their tighter alignment.
Another key element is to strengthen regulatory partnerships among institutions and countries that would directly complement large-scale multicenter preclinical and clinical studies, through initiatives such as creation of shared data repositories and associated biobanks with patient or animal samples for future retrospective studies. A Dutch-based (private-government-academic) Molecular Diagnosis and Risk Stratification of Sepsis consortium (https://consortiapedia.fastercures.org/consortia/mars/) exemplifies such an approach; organized in four complementary working packages brings together several international partners and integrates clinical, discovery, and technology data-sharing platforms. Similarly, the largest existing military registry for combat-related trauma patients (with data on combat casualty care epidemiology, treatments, and longitudinal outcomes) has been created by the US Department of Defense Joint Trauma System (https://jts.amedd.army.mil/). On April 20, 2020 the European Commission, in cooperation with the EMBL-EBI, deployed a data platform specifically dedicated to COVID-19 to collect and share scientific evidence (https://www.covid19dataportal.org). The availability of diverse data types from different clinical centers can enhance the validation of both data-driven and knowledge-based models, leading to a faster introduction of diagnostic and patient monitoring tools as well as decision support systems.
CONCLUSIONS
COVID-19 represents a major global health challenge for the up-coming months and years. SARS-CoV-2 infection likely presents in various phenotypes reflected by differential immunopathogenesis. While many derangements contribute to the pathophysiology of COVID-19, particularly damage to the endothelium along with altered hemostasis appear to be the key driver and link between the different phenotypes. Despite many similarities, the SARS-CoV2/COVID-19 is not identical to MERS and SARS-CoV and should be contemplated as a novel and independent disease entity. Reliable scientific evidence regarding SARS-CoV-2/COVID-19 epidemiology, infectivity, host response, as well as clinical assessment and treatments needs to be generated through a verified, orchestrated, large-scale transnational, and interdisciplinary effort rather than anecdotal, idiosyncratic preclinical studies and multiple small-scale, low-powered clinical trials. It is imperative that the currently contemplated therapeutic strategies adhere to the theranostic-like principle to avoid the past failed treatments that indiscriminately targeted heterogenous patients (with sepsis as an example). Consensus-based “Master Protocols” for data reporting and clinical trials constitute the basis for uniform contingency protocols, rapid and coordinated implementation of emergency measures, and reliable evaluation of therapeutics and vaccines. Advanced data analysis techniques can help in the interpretation of different clinical COVID-19 presentations, rapid risk stratification, and prediction of outcomes. In the absence of approved drugs to prevent and/or treat COVID-19, the magnitude of the current pandemia may justify extraordinary measures to save patient lives but these have to be balanced against ethical and scientific safeguards. Any review of an ongoing pandemic is highly fluid and the dynamic nature of the research on the disease, its progression and our understanding of its epidemiology and pathogenesis may already be partly outdated on the day of publication. Thus, the complete, hopefully inspiring, story of the SARS-CoV-2/COVID-19 pandemic will be written in years from now.
Acknowledgment
The authors thank Michaela Steiner for the critical assessment and the suggestions to the section “SARS-CoV-2 diagnosis and optimization of testing.”
Footnotes
EJG-B: honoraria from AbbVie USA, Abbott CH, InflaRx GmbH, MSD Greece, XBiotech Inc. and Angelini Italy; independent educational grants from AbbVie, Abbott, Astellas Pharma Europe, AxisShield, bioMérieux Inc, InflaRx GmbH, and XBiotech Inc.; and funding from the FrameWork 7 program HemoSpec (granted to the National and Kapodistrian University of Athens), the Horizon2020 Marie-Curie Project European Sepsis Academy (granted to the National and Kapodistrian University of Athens), and the Horizon 2020 European Grant ImmunoSep (granted to the Hellenic Institute for the Study of Sepsis).
All other authors report no conflicts of interest.
REFERENCES
- 1.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, et al. China Medical Treatment Expert Group for Covid-19: clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) situation reports. 2020; Geneva:Worls Health Organization, Available at: https://who.int/emergencies/diseases/novel-coronavirus-2019/ situation-reports. Accessed May 17, 2020. [Google Scholar]
- 3.John Hopkins University & Medicine. Corona Research Center. Available at: https://coronavirus.jhu.edu/map.html (Retrieved May 19, 2020).
- 4.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34 (23):4121–4123, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Lancet: COVID-19: too little, too late? Lancet 395:755, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman SJ, Silverberg SL. Delays in global disease outbreak responses: lessons from H1N1, Ebola, and Zika. Am J Public Health 108 (3):329–333, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hospital readiness checklist for COVID-19. Available at: www.euro.who.int (Retrieved April 19, 2020).
- 8.Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. JAMA 2020; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 9.van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 382:1564–1567, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriarty LF, Plucinski MM, Marston BJ, Kurbatova EV, Knust B, Murray EL, Pesik N, Rose D, Fitter D, Kobayashi M, et al. CDC Cruise Ship Response Team; California Department of Public Health COVID-19 Team; Solano County COVID-19 Team: Public Health Responses to COVID-19 Outbreaks on Cruise Ships — Worldwide, February–March 2020. MMWR 69 (12):347–352, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill 2020; 25 (10): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 382:1177–1179, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Yang H. The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID-19): a Chinese perspective. J Med Virol 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z, Guo D, Li C, Fang F, Chen L, Yang R, Li X, Zeng W. Coronavirus disease 2019: initial chest CT findings. Eur Radiol 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan S. Likelihood of survival of coronavirus disease 2019. Lancet Infect Dis 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 41 (2):145–151, 2020.32064853 [Google Scholar]
- 18.Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, Yi F, Yang HC, Fogo AB, Nie X, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int 2020; pii: S0085-2538(20)30369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fogarty H, Townsend L, Ni Cheallaigh C, Bergin C, Martin-Loeches I, Browne P, Bacon CL, Gaule R, Gillett A, Byrne M, et al. COVID-19 coagulopathy in Caucasian patients. Br J Haematol 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiezia L, Boscolo A, Poletto F, Cerruti L, Tiberio I, Campello E, Navalesi P, Simioni P. COVID-19-related severe hypercoagulability in patients admitted to intensive care unit for acute respiratory failure. Thromb Haemost 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranucci M, Ballotta A, Di Dedda U, Bayshnikova E, Dei Poli M, Resta M, Falco M, Albano G, Menicanti L. The procoagulant pattern of patients with COVID-19 acute respiratory distress syndrome. J Thromb Haemost 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 395:1417–1418, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonetti PO, Lerman LO, Lerman A. Endothelial dysfunction: a marker of atherosclerotic risk. Arterioscler Thromb Vasc Biol 23 (2):168–175, 2003. [DOI] [PubMed] [Google Scholar]
- 24.Hanley B, Lucas SB, Youd E, Swift B, Osborn M. Autopsy in suspected COVID-19 cases. J Clin Pathol 73 (5):239–242, 2020. [DOI] [PubMed] [Google Scholar]
- 25.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, Cuomo-Dannenburg G, Thompson H, Walker PGT, Fu H, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis 2020; pii: S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Available at: https://www.ema.europa.eu/en/documents/press-release/ema-gives-advice-use-non-steroidal-anti-inflammatories-covid-19_en.pdf. Accessed May 19, 2020.
- 27.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325 (5937):197–201, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395 (10224):565–574, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Yang Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res 19:1351–1360, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 Outbreak. Curr Biol 30 (7):1346–1351, 2020. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Available at: www.who.int/en/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov). Accessed May 19, 2020.
- 32.Fujioka Y, Nishide S, Ose T, Suzuki T, Kato I, Fukuhara H, Fujioka M, Horiuchi K, Satoh AO, Nepal P, et al. A sialylated voltage-dependent Ca2+ channel binds hemagglutinin and mediates Influenza A Virus entry into mammalian cells. Cell Host Microbe 23 (6):809–818, 2018. e5. [DOI] [PubMed] [Google Scholar]
- 33.Simonsen L, Spreeuwenberg P, Lustig R, Taylor RJ, Fleming DM, Kroneman M, Van Kerkhove MD, Mounts AW, Paget WJ. GLaMOR collaborating teams: global mortality estimates for the 2009 Influenza Pandemic from the GLaMOR project: a modeling study. PLoS Med 10 (11):e1001558, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581 (7807):215–220, 2020. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, Liu J, Lu M, Yang D, Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int 40:998–1004, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, de Wit E, Munster VJ, Hensley LE, Zalmout IS, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5 (2):e00884–14, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemida MG, Chu DK, Poon LL, Perera RA, Alhammadi MA, Ng HY, Siu LY, Guan Y, Alnaeem A, Peiris M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis 20 (7):1231–1234, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reghunathan R, Jayapal M, Hsu LY, Chng HH, Tai D, Leung BP, Melendez AJ. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol 6:2, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495 (7440):251–254, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, Riley S, Abu-Raddad LJ, Ho LM, Thach TQ, Chau P, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 361 (9371):1761–1766, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, Gopalakrishna G, Chew SK, Tan CC, Samore MH, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 300 (5627):1966–1970, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome Coronavirus 2. Emerg Infect Dis 2020; 26 (7): [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 (6483):1260–1263, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu JT, Leung K, Bushman M, Kishore N, Niehus R, de Salazar PM, Cowling BJ, Lipsitch M, Leung GM. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 26 (4):506–510, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Available at: https://www.who.int/publications-detail/report-of-the-who-china-joint-mission-on-coronavirus-disease-2019-(covid-19). Accessed May 19, 2020.
- 46.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Møller R, Panis M, Sachs D, Albrecht RA, Benjamin R. tenOever: SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. 2020.
- 47.Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, Jia X, Nicholson S, Catton M, Cowie B, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med 26 (4):453–455, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest 130 (5):2620–2629, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis 2020; pii: ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Tan M, Chen X, Liu X, Huang J, Ou J, Deng X. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 51.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, Peiris M, Poon LLM, Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020; pii: S1473-3099(20)30232-2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crowley SD, Rudemiller NP. Immunologic Effects of the Renin-Angiotensin System. Immunologic Effects of the Renin-Angiotensin System. J Am Soc Nephrol 28 (5):1350–1361, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. China novel coronavirus investigating and research team: a novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382 (8):727–733, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus Viroporin 3a activates the NLRP3 inflammasome. Front Microbiol 10:50, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Y, Zhang H, Mu S, Wei W, Jin C, Xue Y, Tong C, Zha Y, Song Z, Gu G. Lactate dehydrogenase, a Risk Factor of Severe COVID-19 Patients. medRxiv. [DOI] [PMC free article] [PubMed]
- 56.Ahn M, Anderson DE, Zhang Q, Tan CW, Lim BL, Luko K, Wen M, Chia WN, Mani S, Wang LC, et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol 4 (5):789–799, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, Sun R, Tian Z, Xu X, Wei H. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv. 2020.
- 58.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J et al. Immune Cell Profiling of COVID-19 Patients in the Recovery Stage by Single-Cell Sequencing medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 59.Kox M, Frenzel T, Schouten J, van de Veerdonk F, Koenen HJPM, Pickkers P, RCI-COVID-19 study group: COVID-19 patients exhibit less pronounced immune suppression compared with bacterial septic shock patients. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 60.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami M-E, Katsaounou P, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. CellPress Journal pre-proof. Cell Host Microbe pii: S1931-3128(20)30236-5, 2020. [DOI] [PMC free article] [PubMed]
- 61.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 9 (1):761–770, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yongwen C, Feng Z, Diao B, Wang R, Wang G, Wang C, Wang C, Tan Y, Liu L, Wang C, et al. The novel severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv 2020. [Google Scholar]
- 63.Zheng H-Y, Zhang M, Yang C-X, Zhang N, Wang XC, Yang XP, Dong XQ, Zheng YT. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 17:541–543, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol 84 (3):1289–1301, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ju B, Zhang Q, Ge X, Wang R, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J et al. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. bioRxiv. 2020. [DOI] [PubMed]
- 66.Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 2020; 4 (4): pii: 123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, Chen KH, Liu FT, Liu WT, Chen YM, et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun 451 (2):208–214, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perera RA, Mok CK, Tsang OT, Lv H, Ko RL, Wu NC, Yuan M, Leung WS, Chan JMC, Chik TSH, et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill 25:2000421, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick L, Rattigan SM, Borgert B, Moreno C, Solomon BD, Rodriguez-Barraquer I, et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. April 14, 2020. [DOI] [PMC free article] [PubMed]
- 70.Grzelak L, Temmam S, Planchais C, Demeret C, Huon C, Guivel F, Staropoli I, Chazal M, Dufloo J, Planas D et al. SARS-CoV-2 serological analysis of COVID-19 hospitalized patients, pauci-symptomatic individuals and blood donors. medRxiv. 2020.
- 71.de Alwis R, Chen S, Gan ES, Ooia EE: Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine 55:102768, 2020. [DOI] [PMC free article] [PubMed]
- 72.Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B, Liu S, Tan T, Huang W, Dong Y, Chen L, Chen Q, Zhang L, Zhong Q, Zhang X, et al. Treatment with convalescent plasma for critically ill patients with SARS-CoV-2 infection. Chest 2020; pii: S0012-3692(20)30571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bao L, Deng W, Gao H, Xiao C, Liu J, Xue J, Lv Q, Liu J, Yu P, Xu Y et al. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv. 2020.
- 75.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 39 (5):529–539, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang KJ, Su IJ, Theron M, Wu YC, Lai SK, Liu CC, Lei HY. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol 75 (2):185–194, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leisman DE, Deutschman CS, Legrand M: Facing COVID-19 in ICU: vascular dysfunction, thrombosis, and dysregulated inflammation. Intensive Care Med 2020 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 78.Hirano T, Murakami M: COVID-19: a new virus, but an old cytokine release syndrome. Int J Mol Sci 21(9). pii: E3330, 2020.
- 79.Cavaillon JM, Adib-Conquy M, Fitting C, Adrie C, Payen D. Cytokine cascade in sepsis. Scand J Infect Dis 35 (9):535–544, 2003. [DOI] [PubMed] [Google Scholar]
- 80.Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, Panoskaltsis N. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med 355 (10):1018–1028, 2006. [DOI] [PubMed] [Google Scholar]
- 81.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 46 (5):846–848, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, Men D, Huang Q, Liu Y, Yang B, et al: Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely associated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 83.Herold T, Jurinovic V, Arnreich C, Hellmuth JC, von Bergwelt-Baildon M, Klein M, Weinberger T: Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. medRxiv. 2020.
- 84.Hirano T, Yasukawa K, Harada H, Taga T, Watanabe Y, Matsuda T, Kashiwamura S, Nakajima K, Koyama K, Iwamatsu A, et al. Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature 324 (6092):73–76, 1986. [DOI] [PubMed] [Google Scholar]
- 85.Tadokoro T, Wang Y, Barak LS, Bai Y, Randell SH, Hogan BL. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc Natl Acad Sci U S A 111 (35):E3641–E3649, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dienz O, Rud JG, Eaton SM, Lanthier PA, Burg E, Drew A, Bunn J, Suratt BT, Haynes L, Rincon M. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol 5 (3):258–266, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cameron MJ, Kelvin AA, Leon AJ, Cameron CM, Ran L, Xu L, Chu YK, Danesh A, Fang Y, Li Q, et al. Lack of innate interferon responses during SARS coronavirus infection in a vaccination and reinfection ferret model. PLoS One 7 (9):e45842, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Y, Ye F, Zhu N, Wang W, Deng Y, Zhao Z, Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep 5:17554, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zinzula L, Tramontano E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antiviral Res 100 (3):615–635, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deng X, Hackbart M, Mettelman RC, O’Brien A, Mielech AM, Yi G, Kao CC, Baker SC. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114 (21):E4251–E4260, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits Type i interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 2017; 291 (8): e02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spiegel M, Pichlmair A, Martínez-Sobrido L, Cros J, García-Sastre A, Haller O, Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 79 (4):2079–2086, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matthews K, Schäfer A, Pham A, Frieman M. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol J 11:209, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signalling: role of nsp1 and rational design of an attenuated strain. J Virol 81 (21):11620–11633, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee JY, Kim SJ, Myoung J. Middle east respiratory syndrome coronavirus-encoded ORF8b inhibits RIG-I-like receptors in a differential mechanism. J Microbiol Biotechnol 29 (12):2014–2021, 2019. [DOI] [PubMed] [Google Scholar]
- 96.Lee JY, Bae S, Myoung J. Middle east respiratory syndrome coronavirus-encoded accessory proteins impair MDA5-and TBK1-mediated activation of NF-κB. J Microbiol Biotechnol 29 (8):1316–1323, 2019. [DOI] [PubMed] [Google Scholar]
- 97.Xiao H, Xu LH, Yamada Y, Liu DX. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One 3 (1):e1494, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xi Y, Day SL, Jackson RJ, Ranasinghe C. Role of novel type I interferon epsilon in viral infection and mucosal immunity. Mucosal Immunol 5 (6):610–622, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choo SW, Rayko M, Tan TK, Hari R, Komissarov A, Wee WY, Yurchenko AA, Kliver S, Tamazian G, Antunes A, et al. Pangolin genomes and the evolution of mammalian scales and immunity. Genome Res 26 (10):1312–1322, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Demers A, Kang G, Ma F, Lu W, Yuan Z, Li Y, Lewis M, Kraiselburd EN, Montaner L, Li Q. The mucosal expression pattern of interferon-ε in rhesus macaques. J Leukoc Biol 96 (6):1101–1107, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB, Jr, Meyerholz DK, Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 130:3625–3639, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cinatl J, Jr, Michaelis M, Scholz M, Doerr HW. Role of interferons in the treatment of severe acute respiratory syndrome. Expert Opin Biol Ther 4 (6):827–836, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ströher U, DiCaro A, Li Y, Strong JE, Aoki F, Plummer F, Jones SM, Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon- alpha. J Infect Dis 189 (7):1164–1167, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM et al. Lung Biological Network: SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell pii: S0092-8674(20)30500-6, 2020. [DOI] [PMC free article] [PubMed]
- 105.Zhou Z, Lili Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe pii: S1931-3128(20)30244-4, 2020. [DOI] [PMC free article] [PubMed]
- 106.Sallard E, Lescure FX, Yazdanpanah Y, Mentre F, Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res 178:104791, 2020[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18 (4):844–847, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lodigiani C, Iapichino G, Carenzo L, Cecconi M, Ferrazzi P, Sebastian T, Kucherd N, Studte J-D, Saccoa C, Alexiaf B, et al. On behalf of the humanitas COVID-19 Task Force: Venous and arterial thromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb Res 191:9–14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gattinoni L, Chiumello D, Caironi P, Busana M, Romitti F, Brazzi L, Camporota L. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fox SE, Akmatbekov A, Harbert JL, Li G, Brown JQ, Vander Heide RS. Pulmonary and Cardiac Pathology in Covid-19: The First Autopsy Series from New Orleans. medRxiv. April 6, 2020. [DOI] [PMC free article] [PubMed]
- 112.Lv B, Wang H, Tang Y, Fan Z, Xiao X, Chen F. High-mobility group box 1 protein induces tissue factor expression in vascular endothelial cells via activation of NF-kappaB and Egr-1. Thromb Haemost 102 (2):352–359, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu C, Lu W, Zhang Y, Zhang G, Shi X, Hisada Y, Grover SP, Zhang X, Li L, Xiang B, et al. Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity 50 (6):1401–1411, 2019. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Magro C, Mulvey JJ, Berlin D, Nuovo G, Salvatore S, Harp J, Baxter-Stoltzfus A, Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res 2020; pii: S1931-5244(20)30070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, Blair CN, Weber A, Barnes BJ, Egeblad M, et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020; pii: 138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Y, Xiao M, Zhang S, Xia P, Cao W, Jiang W, Chen H, Ding X, Zhao H, Zhang H, et al. Coagulopathy and antiphospholipid antibodies in patients with Covid-19. N Engl J Med e38:2020, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gralinski LE, Bankhead A, 3rd, Jeng S, Menachery VD, Proll S, Belisle SE, Matzke M, Webb-Robertson BJ, Luna ML, Shukla AK, et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. mBio 2013; 4 (4): e00271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van de Veerdonk F, Netea MG, van Deuren M, van der Meer JWM, Mast Q, Bruggemann RJ, van der Hoeven H: Kinins and Cytokines in COVID-19: A Comprehensive Pathophysiological Approach. Preprints. April 3, 2020.
- 119.Hodgson J. The pandemic pipeline. Nat Biotechnol 38 (5):523–532, 2020. [DOI] [PubMed] [Google Scholar]
- 120.de Wit E, Feldmann F, Cronin J, Jordan R, Okumura A, Thomas T, Scott D, Cihlar T, Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A 117 (12):6771–6776, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sheahan TP, Sims A, Graham R, Menachery V, Gralinski L, Case J, Leist S, Pyrc K, Feng J, Trantcheva I, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9 (396):eaal3653, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun 11 (1):222, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30 (3):269–271, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Runfeng L, Yunlong H, Jicheng H, Weiqi P, Qinhai M, Yongxia S, Chufang L, Jin Z, Zhenhua J, Haiming J, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res 2020; 104761.[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, Xu J, Liu Y, Zheng BJ, Woodle MC, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med 11 (9):944–951, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tang Q, Li B, Woodle M, Lu PY. Application of siRNA against SARS in the rhesus macaque model. Methods Mol Biol 442:139–158, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, 3rd, Stevens LJ, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12 (541):pii: eabb5883, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, Li Y, Hu Z, Zhong W, Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 6:16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020; pii: ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barnard DL, Day CW, Bailey K, Heiner M, Montgomery R, Lauridsen L, Chan PK, Sidwell RW. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir Chem Chemother 17 (5):275–284, 2006. [DOI] [PubMed] [Google Scholar]
- 131.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; pii: S0092-8674(20)30229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chan JF, Yao Y, Yeung ML, Deng W, Bao L, Jia L, Li F, Xiao C, Gao H, Yu P, et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis 212 (12):1904–1913, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wohlford-Lenane CL, Meyerholz DK, Perlman S, Zhou H, Tran D, Selsted ME, McCray PB., Jr Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J Virol 83 (21):11385–11390, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou L, Ni B, Luo D, Zhao G, Jia Z, Zhang L, Lin Z, Wang L, Zhang S, Xing L, et al. Inhibition of infection caused by severe acute respiratory syndrome-associated coronavirus by equine neutralizing antibody in aged mice. Int Immunopharmacol 2007; (3):392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhao G, Ni B, Jiang H, Luo D, Pacal M, Zhou L, Zhang L, Xing L, Zhang L, Jia Z, et al. Inhibition of severe acute respiratory syndrome-associated coronavirus infection by equine neutralizing antibody in golden Syrian hamsters. Viral Immunol 20 (1):197–205, 2007. [DOI] [PubMed] [Google Scholar]
- 136.Miyoshi-Akiyama T, Ishida I, Fukushi M, Yamaguchi K, Matsuoka Y, Ishihara T, Tsukahara M, Hatakeyama S, Itoh N, Morisawa A, et al. Fully human monoclonal antibody directed to proteolytic cleavage site in severe acute respiratory syndrome (SARS) coronavirus S protein neutralizes the virus in a rhesus macaque SARS model. J Infect Dis 203 (11):1574–1581, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jiang Y, Zhao G, Song N, Li P, Chen Y, Guo Y, Li J, Du L, Jiang S, Guo R, et al. Blockade of the C5a-C5aR axis alleviates lung damage in hDPP4-transgenic mice infected with MERS-CoV. Emerg Microbes Infect 7 (1):77, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jiang Y, Li J, Teng Y, Sun H, Tian G, He L, Li P, Chen Y, Guo Y, Li J, et al. Complement receptor C5aR1 inhibition reduces pyroptosis in hDPP4-transgenic mice infected with MERS-CoV. Viruses 11 (1):E39, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, van Amerongen G, van Riel D, de Jong T, Itamura S, Chan KH, et al. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med 10 (3):290–293, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Deng W, Xu Y, Kong Q, Xue J, Yu P, Liu J, Lv Q, Li F, Wei Q, Bao L: Therapeutic efficacy of Pudilan Xiaoyan Oral Liquid (PDL) for COVID-19 in vitro and in vivo. Signal Transduct Target Ther 8;5(1):66, 2020. [DOI] [PMC free article] [PubMed]
- 141.Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, Chan KS, Kao RY, Poon LL, Wong CL, Guan Y, et al. HKU/UCH SARS Study Group: role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59 (3):252–256, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med 19 (10):1313–1317, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep 3:1686, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.https://www.who.int/blueprint/priority-diseases/key-action/WHO-ad-hoc-Animal-Model-Working-Group_Summary.pdf.
- 145.Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, Kuang D, Yang M, Yang Y, Ma C et al. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. April 8, 2020.
- 146.Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 27 (5):704.e2–709.e2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shi J, Wen Z, Zhong G, Yang H, Wang C, Liu R, He X, Shuai L, Sun Z, Zhao Y, et al: Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. bioRxiv. March 30, 2020. [DOI] [PMC free article] [PubMed]
- 148.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F et al. The Pathogenicity of SARS-CoV-2 in hACE2 Transgenic Mice. bioRxiv. February 27, 2020. [DOI] [PubMed]
- 149.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020 [Epub ahead of print]. [DOI] [PubMed]
- 150.Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020; pii: ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 152.Shi J, Wen Z, Zhong G, Yang H, Wang C, Liu R, He X, Shuai L, Sun Z, Zhao Y et al. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. bioRxiv. March 30, 2020. [DOI] [PMC free article] [PubMed]
- 153.Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med 3 (1):93–97, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gralinski LE, Sheahan TP, Morrison TE, Menachery VD, Jensen K, Leist SR, Whitmore A, Heise MT, Baric RS. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio 2018; 9 (5): e01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Available at: https://www.who.int/emergencies/ebola/MEURI-Ebola.pdf. Accessed on May 19, 2020.
- 156.Lea M, Mowe M, Mathiesen L, Kvernrød K, Skovlund E, Molden E. Prevalence and risk factors of drug-related hospitalizations in multimorbid patients admitted to an internal medicine ward. PLoS One 14 (7):e0220071, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alhazzani W, Møller MH, Arabi YM, Loeb M, Gong MN, Fan E, Oczkowski S, Levy MM, Derde L, Dzierba A, et al. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med 46 (5):854–887, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Phua J, Weng L, Ling L, Egi M, Lim CM, Divatia JV, Shrestha BR, Arabi YM, Ng J, Gomersall CD, et al. Asian Critical Care Clinical Trials Group: Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir Med 8 (5):506–517, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14 (1):72–73, 2020. [DOI] [PubMed] [Google Scholar]
- 160.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020:105949, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 161.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Sevestre J, Mailhe M, Doudier B, Aubry C, Amrane S, et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med Infect Dis 34:101663, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Molina JM, Delaugerre C, Le Goff J, Mela-Lima B, Ponscarme D, Goldwirt L, de Castro N: No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med Mal Infect 50(4):384, 2020. [DOI] [PMC free article] [PubMed]
- 163.Mahevas M, Tran V-T, Roumier M, Chabrol A, Paule R, Guillaud C, Gallien S, Lepeule R, Szwebel T-A, Lescure X et al. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID-19 infection with oxygen requirement: results of a study using routinely collected data to emulate a target trial. medRxiv. April 10, 2020.
- 164.Borba MGS, Val FFA, Sampaio VS, Alexandre MAA, Melo GC, Brito M, Mourão MPG, Brito-Sousa JD, Baía-da-Silva D, Guerra MVF, et al. CloroCovid-19 team: effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw Open 3 (4.23):e208857, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Available at: https://www.newsweek.com/hydroxychloroquine-coronavirus-france-heart-cardiac-1496810. Accessed on May 19, 2020.
- 166.Available at: https://www.newsweek.com/swedish-hospitals-chloroquine-covid-19-side-effects-1496368. Accessed on May 19, 2020.
- 167.Magagnoli J, Narendran S, Pereira F, Cummings T, Hardin JW, Sutton SS, Ambati J: Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. medRxiv. April 16, 2020. [DOI] [PMC free article] [PubMed]
- 168.Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 382 (19):1787–1799, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395:1569–1578, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A 117:9490–9496, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Annane D, Pastores SM, Rochwerg B, Arlt W, Balk RA, Beishuizen A, Briegel J, Carcillo J, Christ-Crain M, Cooper MS, et al. Correction to: Guidelines for the diagnosis and management of critical illness-related corticosteroid insufficiency (CIRCI) in critically ill patients (Part I): Society of Critical Care Medicine (SCCM) and European Society of Intensive Care Medicine (ESICM) 2017. Intensive Care Med 44 (3):401–402, 2018. [DOI] [PubMed] [Google Scholar]
- 174.Li H, Yang SG, Gu L, Zhang Y, Yan XX, Liang ZA, Zhang W, Jia HY, Chen W, Liu M, et al. National Influenza A(H1N1)pdm09 Clinical Investigation Group of China: Effect of low-to-moderate-dose corticosteroids on mortality of hospitalized adolescents and adults with influenza A(H1N1)pdm09 viral pneumonia. Influenza Other Respir Viruses 11 (4):345–354, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chen RC, Tang XP, Tan SY, Liang BL, Wan ZY, Fang JQ, Zhong N. Treatment of severe acute respiratory syndrome with glucosteroids: the Guangzhou experience. Chest 129 (6):1441–1452, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Villar J, Confalonieri M, Pastores S, Meduri GU. Rationale for prolonged corticosteroid treatment in the acute respiratory distress syndrome caused by coronavirus disease 2019. Crit Care Explor (Commentary) 2 (4):e0111, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Angus DC. Optimizing the trade-off between learning and doing in a pandemic. JAMA 2020; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 178.Angus DC, Berry S, Lewis RJ, Al-Beidh F, Arabi Y, van Bentum-Puijk W, Bhimani Z, Bonten M, Broglio K, Brunkhorst F, et al. The randomized embedded multifactorial adaptive platform for community-acquired pneumonia (REMAP-CAP) Study: rationale and design. Ann Am Thorac Soc 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.A trial evaluating treatments for suspected coronavirus infection in people aged 50 years and above with pre-existing conditions and those aged 65 years and above; Available at: 10.1186/ISRCTN86534580. Accessed on May 19, 2020. [DOI]
- 180.Trial of Treatments for COVID-19 in Hospitalized Adults (DisCoVeRy); Available at: https://clinicaltrials.gov/ct2/show/NCT04315948. Accessed on May 19, 2020.
- 181.Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov 19:305–306, 2020. [DOI] [PubMed] [Google Scholar]
- 182.Wang Y, Liu Y, Liu L, Wang X, Luo N, Ling L. Clinical outcome of 55 asymptomatic cases at the time of hospital admission infected with SARS-Coronavirus-2 in Shenzhen, China. J Infect Dis 221 (11):1770–1774, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tao Y, Cheng P, Chen W, Wan P, Chen Y, Yuan G, Chen J, Huo D, Guan G, Sun D et al. High incidence of asymptomatic SARS-CoV-2 infection, Chongqing, China. medRxiv. March 16, 2020.
- 184.Qian G, Yang N, Ma AHY, Wang 1, Li G, Chen X, Chen X. A COVID-19 Transmission within a family cluster by presymptomatic infectors in China. Clin Infect Dis 2020; pii: ciaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang C, Yu H, Horby PW, Cao B, Wu P, Yang S, Gao H, Li H, Tsang TK, Liao Q, et al. Comparison of patients hospitalized with influenza A subtypes H7N9, H5N1, and 2009 pandemic H1N1. Clin Infect Dis 58 (8):1095–1103, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol 38 (5):515–518, 2020. [DOI] [PubMed] [Google Scholar]
- 188.Korean Society of Infectious Diseases; Korean Society of Pediatric Infectious Diseases; Korean Society of Epidemiology; Korean Society for Antimicrobial Therapy; Korean Society for Healthcare-associated Infection Control and Prevention; Korea Centers for Disease Control and Prevention: Report on the Epidemiological Features of Coronavirus Disease 2019 (COVID-19) Outbreak in the Republic of Korea from January 19 to March 2, 2020. J Korean Med Sci 35 (10):e112, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Available at: https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117. Accessed on May 19, 2020.
- 190.Available at: https://apps.who.int/iris/bitstream/handle/10665/331639/WHO-2019-nCoV-laboratory_shipment-2020.3-eng.pdf. Accessed on May 19, 2020.
- 191.Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta 505:172–175, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, Park JS, Kim GJ, Sung H, Roh KH, et al. Korean Society for Laboratory Medicine, COVID-19 Task Force and the Center for Laboratory Control of Infectious Diseases, the Korea Centers for Disease Control and Prevention. Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann Lab Med 40 (5):351–360, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Specific primers and probes for detection 2019 novel coronavirus Available at: http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html. Assessed on May 19, 2020.
- 194.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology 2020; 200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li D, Wang D, Dong J, Wang N, Huang H, Xu H, Xia C. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J Radiol 21 (4):505–508, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol 2020; doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25 (3): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fomsgaard AS, Rosenstierne MW: An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage. medRxiv. March 27, 2020. [DOI] [PMC free article] [PubMed]
- 200.Available at: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19. Assessed on May 19, 2020.
- 201.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (7798):270–273, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Infantino M, Damiani A, Gobbi FL, Grossi V, Lari B, Macchia D, Casprini P, Veneziani F, Villalta D, Bizzaro N, et al. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr Med Assoc J 22 (4):203–210, 2020. [PubMed] [Google Scholar]
- 204.Wan WY, Lim SH, Seng EH: Cross-reaction of sera from COVID-19 patients with SARS-CoV assays. medRxiv. March 17, 2020. [PubMed]
- 205.Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, Kim B1, Kim SJ. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J Microbiol Biotechnol 30 (3):313–324, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vodovotz Y, An G. Agent-based models of inflammation in translational systems biology: a decade later. Wiley Interdiscip Rev Syst Biol Med 11 (6):e1460, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vodovotz Y, Billiar TR. In silico modeling: methods and applications to trauma and sepsis. Crit Care Med 41:2008–2014, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chase JG, Preiser JC, Dickson JL, Pironet A, Chiew YS, Pretty CG, Shaw GM, Benyo B, Moeller K, Safaei S, et al. Next-generation, personalised, model-based critical care medicine: a state-of-the art review of in silico virtual patient models, methods, and cohorts, and how to validation them. Biomed Eng Online 17 (1):24, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Naudé W: Artificial Intelligence against COVID-19: An Early Review, IZA Institute of Labor Economics, Discussion Paper Series DP 13110.
- 210.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med 27 (2):pii: taaa008, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gilbert M, Pullano G, Pinotti F, Valdano E, Poletto C, Boëlle PY, D’Ortenzio E, Yazdanpanah Y, Eholie SP, Altmann M, et al. Preparedness and vulnerability of African countries against importations of COVID-19: a modelling study. Lancet 395 (10227):871–877, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bullock J, Luccioni A, Pham KH, Lam CSN, Luengo-Oroz M. Mapping the landscape of artificial intelligence applications against COVID-19. arXiv 2020; arXiv:2003.11336 [cs.CY]. [Google Scholar]
- 213.Apostolopoulos ID, Aznaouridis S, Tzani M. Extracting possibly representative COVID-19 Biomarkers from X-Ray images with Deep Learning approach and image data related to Pulmonary Diseases. arXiv 2020; [6 Apr 2020 v2.] arXiv:2004.00338[eess.IV]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hemdan EED, Shouman MA, Karar ME. COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv 2020; arXv:2003.11055 [eess.IV]. [Google Scholar]
- 215.Wang L, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv 2020; v3.] arXiv:2003.09871 [eess.IV]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wynants L, Van Calster B, Bonten MMJ, Collins GS, Debray TPA, De Vos M, Haller MC, Heinze G, Moons KGM, Riley RD, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ 369:m1328, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J 18:784–790, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stebbing J, Phelan A, Griffin I, Tucker C, Oechsle O, Smith D, Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis 20 (4):400–402, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Alimadadi A, Aryal S, Manandhar I, Munroe PB, Joe B, Cheng X. Artificial intelligence and machine learning to fight COVID-19. Physiol Genomics 52 (4):200–202, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, Chen B, Carin L, Suarez A, Mohney RP, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 5 (195):195ra95, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wong HR. Genome-wide expression profiling in pediatric septic shock. Pediatr Res 273 (4 pt 2):564–569, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Raymond SL, López MC, Baker HV, Larson SD, Efron PA, Sweeney TE, Khatri P, Moldawer LL, Wynn JL. Unique transcriptomic response to sepsis is observed among patients of different age groups. PLoS One 12 (9):e0184159, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bauzá-Martinez J, Aletti F, Pinto BB, Ribas V, Odena MA, Díaz R, Romay E, Ferrer R, Kistler EB, Tedeschi G, et al. Proteolysis in septic shock patients: plasma peptidomic patterns are associated with mortality. Br J Anaesth 121 (5):1065–1074, 2018. [DOI] [PubMed] [Google Scholar]
- 224.Cambiaghi A, Pinto BB, Brunelli L, Falcetta F, Aletti F, Bendjelid K, Pastorelli R, Ferrario M. Characterization of a metabolomic profile associated with responsiveness to therapy in the acute phase of septic shock. Sci Rep 7 (1):9748, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cambiaghi A, Díaz R, Martinez JB, Odena A, Brunelli L, Caironi P, Masson S, Baselli G, Ristagno G, Gattinoni L, et al. An innovative approach for the integration of proteomics and metabolomics data in severe septic shock patients stratified for mortality. Sci Rep 8 (1):6681, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Braga D, Barcella M, Herpain A, Aletti F, Kistler EB, Bollen Pinto B, Bendjelid K, Barlassina C. A longitudinal study highlights shared aspects of the transcriptomic response to cardiogenic and septic shock. Crit Care 23 (1):414, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. medRxiv. April 7, 2020. [DOI] [PMC free article] [PubMed]
- 228.Arentz M, Yim E, Klaff L, Lokhandwala S, Riedo FX, Chong M, Lee M. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK, Greninger AL, Pipavath S, Wurfel MM, Evans L, et al. Covid-19 in critically ill patients in the seattle region—case series. N Engl J Med 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ling L, So C, Shum HP, Chan PKS, Lai CKC, Kandamby DH, Ho E, So D, Yan WW, Lui G, et al. Critically ill patients with COVID-19 in Hong Kong: a multicentre retrospective observational cohort study. Crit Care Resusc 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bonow RO, Fonarow GC, O’Gara PT, Yancy CW. Association of Coronavirus Disease 2019 (COVID-19) with myocardial injury and mortality. JAMA Cardiol 2020; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 232.Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Available at: https://edhub.ama-assn.org/jn-learning/audio-player/18317079. Accessed on May 19, 2020.
- 234.Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput Biol Med 119:103670, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Rut W, Groborz K, Zhang L, Sun X, Zmudzinski M, Hilgenfeld R, Drag M: Substrate specificity profiling of SARS-CoV-2 Mpro protease provides basis for anti-COVID-19 drug design. bioRxiv. March 7, 2020.
- 236.Llovera G, Hofmann K, Roth S, Salas-Pérdomo A, Ferrer-Ferrer M, Perego C, Zanier ER, Mamrak U, Rex A, Party H, et al. Results of a preclinical randomized controlled multicenter trial (pRCT): anti-CD49d treatment for acute brain ischemia. Sci Transl Med 27 (299):299ra121, 2015. [DOI] [PubMed] [Google Scholar]
- 237.Kochanek PM, Bramlett H, Dixon C, Dietrich W, Mondello S, Wang KK, Hayes R, Lafrenaye A, Povlishock JT, Tortella FC, et al. Operation brain trauma therapy: 2016 update. Mil Med 183: Suppl_1: 303–312, 2018. [DOI] [PubMed] [Google Scholar]
- 238.Wang Q, Zhang L, Kuwahara K, Li L, Liu Z, Li T, Zhu H, Liu J, Xu Y, Xie J, et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect Dis 2 (5):361–376, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Luo F, Liao FL, Wang H, Tang HB, Yang ZQ, Hou W. Evaluation of antibody-dependent enhancement of SARS-CoV infection in rhesus macaques immunized with an inactivated SARS-CoV vaccine. Virol Sin 33 (2):201–204, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]


