A TAF1 mutation that lowers levels of the transcript causes X-linked dystonia parkinsonism. Hernández et al. show reduced levels of TAF1, and of the RNA-binding protein SREK1 which targets TAF1, also in Huntington’s disease. SREK1 overexpression restores TAF1 levels in the R6/1 mouse model, and reduces motor deficits.
Keywords: SREK1, TAF1, RNA-binding proteins (RBP), splicing, SR proteins
Abstract
Huntington’s disease and X-linked dystonia parkinsonism are two monogenic basal ganglia model diseases. Huntington’s disease is caused by a polyglutamine-encoding CAG repeat expansion in the Huntingtin (HTT) gene leading to several toxic interactions of both the expanded CAG-containing mRNA and the polyglutamine-containing protein, while X-linked dystonia parkinsonism is caused by a retrotransposon insertion in the TAF1 gene, which decreases expression of this core scaffold of the basal transcription factor complex TFIID. SRSF6 is an RNA-binding protein of the serine and arginine-rich (SR) protein family that interacts with expanded CAG mRNA and is sequestered into the characteristic polyglutamine-containing inclusion bodies of Huntington’s disease brains. Here we report decreased levels of the SRSF6 interactor and regulator SREK1—another SR protein involved in RNA processing—which includes TAF1 as one of its targets. This led us to hypothesize that Huntington’s disease and X-linked dystonia parkinsonism pathogeneses converge in TAF1 alteration. We show that diminishing SRSF6 through RNA interference in human neuroblastoma cells leads to a decrease in SREK1 levels, which, in turn, suffices to cause diminished TAF1 levels. We also observed decreased SREK1 and TAF1 levels in striatum of Huntington’s disease patients and transgenic model mice. We then generated mice with neuronal transgenic expression of SREK1 (TgSREK1 mice) that, interestingly, showed transcriptomic alterations complementary to those in Huntington’s disease mice. Most importantly, by combining Huntington’s disease and TgSREK1 mice we verify that SREK1 overexpression corrects TAF1 deficiency and attenuates striatal atrophy and motor phenotype of Huntington’s disease mice. Our results therefore demonstrate that altered RNA processing upon SREK1 dysregulation plays a key role in Huntington’s disease pathogenesis and pinpoint TAF1 as a likely general determinant of selective vulnerability of the striatum in multiple neurological disorders.
Introduction
Huntington’s disease is a fatal neurological disorder characterized by prominent motor symptoms and marked atrophy of the nucleus striatum (Walker, 2007). Huntington’s disease is caused by a CAG repeat expansion in the Huntingtin (HTT) gene encoding a polyglutamine (polyQ) tract in the N-terminal region of the HTT protein (Huntington’s Disease Collaborative Research Group, 1993). Huntington’s disease thus belongs, together with inherited dominant ataxias, spinal and bulbar muscular atrophy (SBMA) and myotonic dystrophy type 1 (DM1), to the group of neurological disorders caused by expansions of unstable triplet repeats (Orr and Zoghbi, 2007). Despite the well documented pathogenic role of expanded polyQ, growing evidence supports toxicity of expanded triplet containing mRNA (Ranum and Cooper, 2006; Li et al., 2008), mainly due to sequestration of specific RNA binding proteins (RBPs), such as MBNL1—in the case of DM1—which binds to the expanded CUG mRNA repeat (Ranum and Cooper, 2006).
Serine and arginine-rich (SR) proteins are RBPs that contain one or two RNA recognition motifs (RRM) and a domain rich in serine and arginine dipeptides (RS domain) (Long and Caceres, 2009). The RRMs impart substrate specificity through sequence-specific RNA binding while the RS domain participates in protein-protein interactions (Birney et al., 1993). SR proteins play significant roles in constitutive and alternative splicing of pre-mRNAs as well as in nuclear export, nonsense-mediated decay and translation of mRNAs (Long and Caceres, 2009). SRSF6 (also known as SRp55) is an SR protein that was bioinformatically predicted (Sathasivam et al., 2013) and biochemically confirmed (Schilling et al., 2019) to bind mutant huntingtin (mHTT) CAG repeat RNA, thus contributing to two pathogenic mis-splicing events in Huntington’s disease. Namely, incomplete splicing of the mHTT gene giving rise to a highly toxic N-terminal mHTT fragment (Neueder et al., 2018) and increased inclusion of tau (MAPT) exon 10 leading to a pathogenic increase in tau isoforms with four tubulin binding repeats (4R-tau) (Fernandez-Nogales et al., 2014). Besides, aberrant MAP2 splicing and levels in Huntington’s disease also correlate with SRSF6 dysregulation (Cabrera and Lucas, 2017) and alteration of SRSF6 activity in striatum of Huntington’s disease patients and mouse models is evidenced by its sequestration into polyQ inclusion bodies and hyperphosphorylation (Fernandez-Nogales et al., 2014), the latter leading to dissociation from nuclear speckles (Yin et al., 2012; Naro and Sette, 2013).
SR proteins act cooperatively in functional networks (Pandit et al., 2013) and their reciprocal physical interaction may modulate their phosphorylation, levels and activity (Wu and Maniatis, 1993; Kohtz et al., 1994; Xiao and Manley, 1997). SREK1 is a member of the SR protein superfamily with an additional glutamic acid-lysine (EK)-rich domain (Barnard and Patton, 2000) that regulates RNA processing by modulating multiple SR proteins. In fact, SREK1 is a direct interactor and regulator of SRSF6 (Barnard and Patton, 2000) and is particularly interesting because one of its target mRNAs is TAF1 (TATA-box binding protein associated factor 1) (Solis and Patton, 2010). TAF1 encodes the largest component and core scaffold of the TFIID basal transcription factor complex (Albright and Tjian, 2000) and is relevant to neurodegeneration because a retrotransposon insertion mutation leading to decreased TAF1 levels causes X-linked dystonia parkinsonism (XDP, also known as Lubag syndrome or X-linked dystonia of Panay) (Aneichyk et al., 2018), a rare neurodegenerative motor disorder that, like Huntington’s disease, is characterized by a marked striatal atrophy (OMIM #314250).
Here, we aimed to explore whether the SRSF6-interacting SR protein SREK1 is altered upon SRSF6 alteration in Huntington’s disease and whether this would correlate with altered levels of its target TAF1. This has been corroborated experimentally here indicating that molecular pathogenesis of Huntington’s disease and XDP converge in decreased TAF1 expression and pinpoint a new general mechanism for neurodegeneration and for therapeutic intervention.
Materials and methods
Cell culture, lentiviral transduction and actinomycin D treatment
Lentiviral packaging
293T cells were plated at 60% of confluence in P100 tissue culture dishes with 10% foetal bovine serum (FBS) (Gibco) supplemented Dulbecco’s modified Eagle medium (DMEM) and penicillin–streptomycin (Sigma) at 37°C with 5% CO2 and 24 h later were co-transfected with 4 µg of the corresponding lentivector plasmid, 2 µg of the packaging plasmid pCMVdR8.74 (Addgene plasmid 22036) and 2 µg of the VSV G envelope protein plasmid pMD2G (Addgene plasmid 12259) using LipofectamineTM 2000 (Thermo Fisher), as per instructions from the supplier. The lentivectors used (Mission, Merck) encode shRNA sequences for either SRSF6 or SREK1 interference. The shRNA sequences were: shSRSF6, CCGGCGAACAAATGAGGGTGTAATTCTCGAGAATTACACCCTCATTTGTTCGTTTTTG; and shSREK1, CCGGGAAGAAGTAATGAAGCGAGTACTCGAGTACTCGCTTCATTACTTCTTCTTTTTG.
Forty-eight hours after transfection supernatant was collected and filtered to remove cell debris. Viral media was immediately used for transduction or aliquoted in cryotubes for long-term storage at −80°C.
Transduction
Human SH-SY5Y neuroblastoma cells were cultured in 10% FBS (Gibco) supplemented DMEM with penicillin–streptomycin (Sigma) at 37°C in a 5% CO2 atmosphere. The day before transduction 3 × 105 cells per condition were placed in a six-well plate (Falcon). SH-SY5Y was transduced using 293T supernatant containing either scramble, shSRSF6 or shSREK1 lentiviral particles with polybrene (final concentration, 8 µg/ml, Santa Cruz). After 6 h, transduction medium was replaced with 10% FBS-supplemented DMEM. Finally, samples were collected for analysis after 48 h of transduction.
Actinomycin D treatment
For transcription inhibition assays we plated 5 × 105 Neuro-2a neuroblastoma cells in postnatal Day 35 tissue culture dishes for each condition. Twenty-four hours after plating, 5 µg/ml actinomycin D (Thermo Fisher) was added and cells were harvested after 0, 1, 3 or 8 h of treatment for RNA analysis.
Human brain tissue samples
Brain specimens from Huntington’s disease patients and control subjects were provided by the Institute of Neuropathology Brain Bank (HUB-ICO-IDIBELL, Hospitalet de Llobregat, Spain), the Neurological Tissue Bank of the IDIBAPS Biobank (Barcelona, Spain), the Banco de Tejidos Fundación Cien (BT-CIEN, Madrid, Spain) and the Netherlands Brain Bank (Amsterdam, The Netherlands). Written informed consent for brain removal after death for diagnostic and research purposes was obtained from brain donors and/or next of kin. Procedures, information and consent forms were approved by the corresponding Bioethics Committees.
Mice
R6/1 transgenic mice (JAX-Mice 002809) for the human HTT-exon 1 sequence were maintained in B6CBAF1 background; HdhQ111 mice (JAX-Mice 003456) were maintained as homozygotes; CamKII-tTA line (Mayford et al., 1996) was maintained in a pure C57BL/6J background. Conditional mice expressing human SREK1 (TgSREK1) were generated for this study (see the ‘Generation of TgSREK1 mice’ section below) and were maintained in C57BL/6J background.
All animals were housed at the Centro de Biología Molecular ‘Severo Ochoa’ animal facility. Mice were housed maximum four per cage with food and water available ad libitum and maintained in a temperature-controlled environment on a 12/12-h light-dark cycle with light onset at 8:00 am. Animal housing and maintenance protocols followed the guidelines of the Council of Europe Convention ETS123, revised as indicated in the Directive 86/609/EEC. Animal experiments were performed under protocols (P15/P16/P18/P22) approved by the Centro de Biología Molecular Severo Ochoa Institutional Animal Care and Utilization Committee (Comité de Ética de Experimentación Animal del CBM, CEEA-CBM), and Comunidad de Madrid PROEX 293/15.
Generation of TgSREK1 mice
Human SREK1 cDNA was cloned into a plasmid containing a bidirectional TetO sequence to also express the β-Gal reporter with a nuclear localization signal (pBI‐G, Clontech, 631004). The construct was microinjected into single‐cell C57BL/6JxCBA embryos and resulting β-Gal-BiTetO-SREK1 founder mice were backcrossed with wild-type C57BL/6J mice for maintenance. These mice were then crossed with CamKII-tTA mice to obtain conditional double transgenic mice with forebrain neuronal expression of SREK1 (TgSREK1 mice).
Rotarod test
Motor coordination was assessed with an accelerating rotarod apparatus (Ugo Basile). Similar proportions of male and female mice of each genotype were tested. Mice were first trained in two sessions at fixed speed over two consecutive days. On the first day, the mice were placed in the rotarod four times, fixed at 4 rpm for 1 min each. On the second day, the mice were placed in the rotarod four times for 2 min each (the first minute at 4 rpm and the second minute at 8 rpm). The mice were then tested on a third day with the rotarod set to accelerate from 4 to 40 rpm over 5 min and the mice were tested in four trials. The latency to fall from the rotarod was measured as a mean of the four accelerating trials.
Western blot
Samples from human brain were stored at −80°C and ground with a mortar in a freezing environment with liquid nitrogen to prevent thawing, resulting in tissue powder. Mouse brains were quickly dissected on an ice-cold plate and the different structures stored at −80°C. Human and mouse protein extracts were prepared by homogenizing brain structures in ice-cold extraction buffer [20 mM HEPES pH 7.4, 100 mM NaCl, 20 mM NaF, 1% TritonTM X-100, 1 mM sodium orthovanadate, 1 μM okadaic acid, 5 mM sodium pyrophosphate, 30 mM β-glycerophosphate, 5 mM EDTA, protease inhibitors (Complete, Roche)]. Homogenates were centrifuged at 15 000 rpm for 15 min at 4°C. The resulting supernatant was collected, and protein content determined by Quick StartTM Bradford Protein Assay (Bio-Rad, 500-0203). Total protein (30 μg) was electrophoresed on 10% SDS-polyacrylamide gel, transferred to a nitrocellulose blotting membrane (Amersham Protran® 0.45 μm, GE Healthcare Life Sciences) and blocked in TBS-T (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.1% Tween 20) supplemented with 5% non-fat dry milk. Membranes were incubated overnight at 4°C with either mouse anti-SRSF6 (1:1000; Millipore, MABE152), rabbit anti-SREK1 (1:500, Abnova, H00140890-B01P), rabbit anti-TAF1 (1:500; Bethyl, A303-504A), mouse anti-β-ACTIN (1:50 000; Sigma, A2066) or rabbit anti-VINCULIN (1:20 000; Abcam, ab129002) in TBS-T supplemented with 5% non-fat dry milk. Membranes were washed with TBS-T and then incubated with HRP-conjugated anti-rabbit IgG (1:2000; Dako, P0448) or anti-mouse IgG (1:2000; Dako, P0447) and developed using the ECL detection kit (PerkinElmer, NEL105001EA).
Tissue preparation for immunohistochemistry
For human samples, formalin-fixed (4%, 24 h), paraffin-embedded tissue from striatum were used. Sections (5-μm thick) were mounted on SuperFrost™ Plus tissue slides (Menzel-Gläser) and deparaffinized. Peroxidase activity was quenched with 0.3% H2O2 in methanol for 30 min, followed by antigen retrieval with 10 mM pH 6.0 citrate buffer heated in a microwave for 15 min.
For mouse samples, mice were euthanized in a CO2 chamber. Brains were immediately removed and the left hemispheres placed in 4% paraformaldehyde prepared in Sorensen’s phosphate buffer overnight followed by three phosphate-buffered saline (PBS) washes and, finally, 30% sucrose in PBS for 72 h for cryoprotection. Samples were then added to optimum cutting temperature (OCT) compound (Tissue-Tek®), frozen and stored at −80°C until use. Sagittal sections (30 µm) were cut on a cryostat (Thermo Scientific), collected and stored free floating in glycol-containing buffer (30% glycerol, 30% ethylene glycol in 0.02 M phosphate buffer) at −20°C.
Immunohistochemistry
Sections were first washed in PBS and then immersed in 0.3% H2O2 in PBS for 45 min to quench endogenous peroxidase activity. Following PBS washes, sections were immersed for 1 h in blocking solution [PBS containing 0.5% FBS, 0.3% TritonTM X-100 and 1% bovine serum albumin (BSA)] and incubated overnight at 4°C with either rabbit anti-SREK1 (1:500, Abnova, H00140890-B01P), anti-TAF1 (1:1000, Sigma, HPA001075), anti-β-Gal (1:2000, Thermo Fisher, A-11132), anti-DARPP32 (1:5000, Chemicon, AB1656) or anti-cleaved caspase 3 (1:50, Cell Signaling, 9664) diluted in blocking solution. After washing, brain sections were incubated first with biotinylated goat anti-rabbit secondary antibody and then with avidin-biotin complex using the Elite Vectastain kit (Vector Laboratories, PK-6101-2). Chromogen reactions were performed with diaminobenzidine (SIGMAFAST™ DAB, Sigma, D4293) for 10 min. Mouse sections were mounted on glass slides and coverslipped with Mowiol® (Calbiochem) while human sections where first dehydrated and then mounted with DePeX (SERVA). Images were captured using an Olympus BX41 microscope with an Olympus camera DP-70 (Olympus Denmark A/S).
Quantification of cleaved caspase-3-positive cells
Wild-type, R6/1 and R6/1:TgSREK1 mice (n = 5) were analysed at the age of 3.5 months. The total number of immunopositive cells in three sagittal sections (lateral coordinates 0.72, 1.92 and 3.00 mm) was quantified per each animal using an Olympus BX41 microscope with an Olympus camera DP-70 (Olympus Denmark A/S). Means per genotype were used for statistical comparison.
Striatal volumetry
Mouse sagittal sections (30-µm thick) were cut on a cryostat and every sixth section was counterstained with toluidine blue pH 4.0 (1 g/l toluidine blue, Sigma, 198161 in 0.8 M glacial acetic acid). Digital images were captured at 2.5× magnification (Canon EOS 450D digital camera) and striatal areas were calculated using ImageJ software.
RNA sequencing
Total RNA was isolated from striatum of wild-type (n = 3) and TgSREK1 (n = 3) mice using the Maxwell® 16 LEV simplyRNA Tissue Kit (Promega, AS1280). Total RNA was quantified by Qubit® RNA BR Assay kit (Thermo Fisher Scientific) and the RNA integrity number (RIN) was estimated using the RNA 6000 Nano Bioanalyzer 2100 Assay (Agilent) detecting, on average, a RIN value over 9. The RNA-seq libraries were prepared with KAPA Stranded mRNA-Seq Illumina® Platforms Kit (Roche) following the manufacturer’s recommendations. Briefly, 500 ng of total RNA was used as the input material, the poly-A fraction was enriched with oligo-dT magnetic beads and the mRNA was fragmented. The strand specificity was achieved during the second strand synthesis performed in the presence of dUTP instead of dTTP. The blunt-ended double-stranded cDNA was 3′ adenylated and Illumina platform compatible adaptors with unique dual indexes and unique molecular identifiers (Integrated DNA Technologies) were ligated. The ligation product was enriched with 15 PCR cycles and the final library was validated on an Agilent 2100 Bioanalyzer with the DNA 7500 assay.
The libraries were sequenced on HiSeq 4000 (Illumina, Inc) with a read length of 2 × 101 bp using HiSeq 4000 SBS kit in a fraction of a HiSeq 4000 PE Cluster kit sequencing flow cell lane generating >70 million paired-end reads per sample. Image analysis, base calling and quality scoring of the run were processed using the manufacturer’s software Real Time Analysis (RTA 2.7.7).
RNA-seq data processing and analysis
Reads in fastq format were aligned to the Mus musculus reference genome (GRCm38) and analysed using VAST-TOOLS (Tapial et al., 2017). We then prepared a pre-ranked file with the average fold-change of each gene in TgSREK1 with respect to wild-type mice ordered from negative to positive and used GSEA for enrichment analysis (Mootha et al., 2003; Subramanian et al., 2005). For enriched signature detection we used 186 Kyoto Encyclopedia of Genes and Genomes (KEGG) curated gene sets from MSigDB collections and in particular, the Huntington’s disease KEGG gene set containing 156 genes. For comparison with transcriptomic changes in the Huntington’s disease model mice, we used RNA-seq data from the striatum of R6/1 mice (Elorza et al., 2020). For differential alternative splicing analysis we applied the function ‘compare’ of VAST-TOOLS to the three wild-type and the three TgSREK1 samples. From the resulting table, we selected those events with an absolute dPSI (change in per cent spliced-in) >10 between wild-type and TgSREK1 mice.
Real-time polymerase chain reaction
Relative quantification was carried out for mRNA analysis in N2a cells transduced with either scramble RNA or shRNA targeting SRSF6, and in striatum of R6/1 and R6/1:TgSREK1 mice. The housekeeping genes Actb, Gapdh and Tubb were used for normalization. Qunatitative RT-PCR (CFX 384 Bio-Rad) was carried out with 5 ng of cDNA in a volume of 4 µl with 1 µl of 5 µM forward and reverse primers mix (Supplementary Table 1), and 5 µl of SsoFastTM EvaGreen® Supermix premix (Bio-Rad). Triplicate reactions were carried out for each mRNA. The following amplification protocol was used: initial denaturation of 5 s at 95°C + 40 cycles × (5 s at 95°C + 5 s at 60°C) + (5 s at 60°C + 5 s at 95°C). Fluorescence was taken at the end of the elongation step.
Data analysis
Statistical analysis was performed with SPSS 21.0 (SPSS® Statistic IBM®). Data are represented as mean ± SEM (standard error of the mean). The normality of the data was analysed by Shapiro-Wilk or Kolmogorov-Smirnov tests. For two-group comparison, two-tailed Student’s t-test was performed. For multiple comparisons, data were analysed by one-way ANOVA test followed by a Games-Howell post hoc test. A critical value for significance of P < 0.05 was used throughout the study. Benjamini-Hochberg correction was applied for multiple testing in RNA-seq analysis.
Data availability
Data are available from the corresponding author upon reasonable request.
Results
Interference of SRSF6 in neuroblastoma cells leads to decreased SREK1 and subsequent TAF1 deficiency
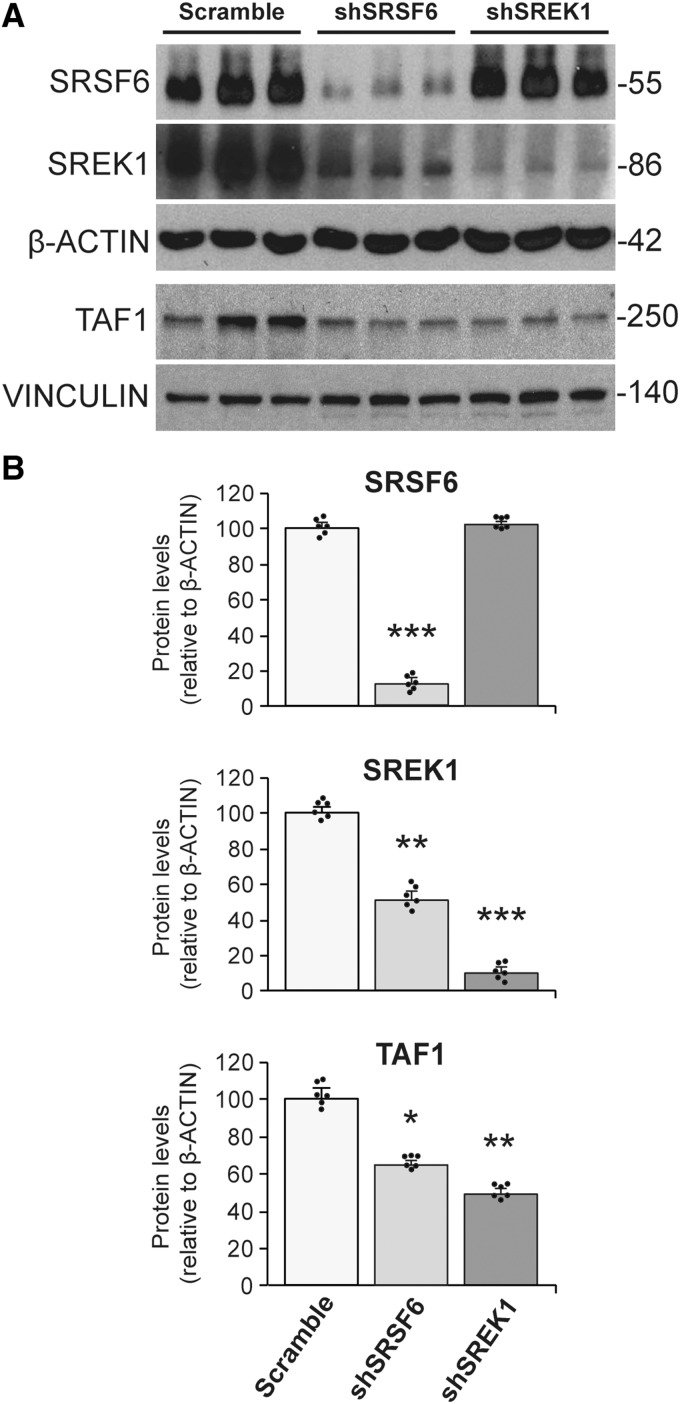
To obtain possible mechanistic evidence of altered SRSF6 levels and/or activity leading to the hypothesized deranged SREK1 levels and subsequent TAF1 alteration, we performed RNA-interference experiments on SH-SY5Y human neuroblastoma cells. This revealed that diminishing SRSF6 levels through shRNA transduction leads to decreased levels of both SREK1 (53% decrease, P = 0.008) and TAF1 (37% decrease, P = 0.017) (Fig. 1). As we did not observe by RT-PCR any difference in the rate of inclusion of the only evolutionary conserved alternative splicing (AS) event in SREK1 transcript (skipping of the 124-nt exon 3, http://vastdb.crg.eu) nor in total SREK1 transcript levels (Supplementary Fig. 1A), decreased SREK1 protein levels upon interference of SRSF6 do not seem to have originated at the pre-mRNA level and might simply reflect diminished SREK1 protein stability due to decreased levels of its direct interactor SFSF6 (Barnard and Patton, 2000). Besides, by performing the equivalent experiment with an shRNA targeting SREK1, we could then demonstrate that diminishing SREK1 levels is sufficient to induce decreased TAF1 protein levels (53% decrease, P = 0.001) (Fig. 1) without altering total TAF1 transcript levels (Supplementary Fig. 1B). These data therefore demonstrate that diminished SRSF6 is sufficient to induce a decrease in SREK1 levels which, in turn, suffices to cause diminished TAF1 levels. Together with the previously reported sequestration of SRSF6 into immunoblots of Huntington’s disease brains (Fernandez-Nogales et al., 2014), these results suggest that SREK1 and TAF1 might be diminished in brains of patients with Huntington’s disease.
Figure 1.
Knockdown of SRSF6 in SH-SY5Y cells leads to decreased SREK1 levels with a subsequent decrease in TAF1. (A) Representative western blots of SRSF6, SREK1 and TAF1 proteins in SH-SY5Y cells upon transduction with scramble RNA (n = 6), shRNA targeting SRSF6 (shSRSF6) (n = 6), or shRNA targeting SREK1 (shSREK1) (n = 6). (B) Bar charts show quantification of SRSF6, SREK1 and TAF1 protein levels (normalized to β-actin) after interference of SRSF6 or SREK1 versus the scramble condition. ANOVA followed by Games-Howell, *P < 0.05; **P < 0.01; ***P < 0.001. Data represent mean + SEM.
Decreased SREK1 and TAF1 in Huntington’s disease brains
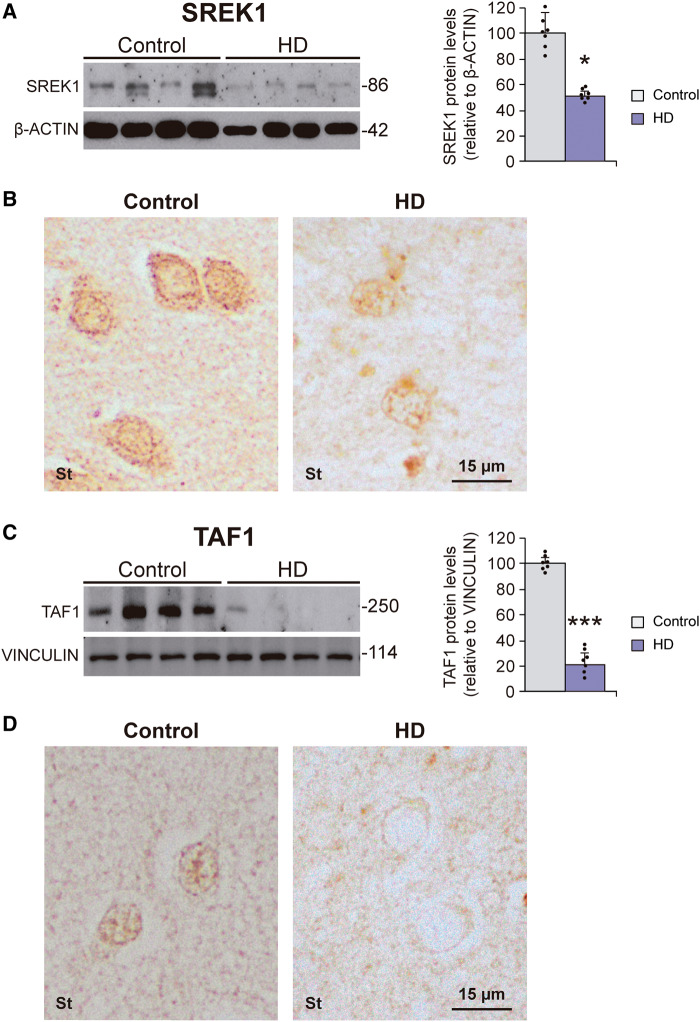
We then carried out western blot analysis of the levels of SREK1 in striatum of Huntington’s disease patients. This revealed a 48% decrease (P = 0.017) in SREK1 protein levels in striatum of Huntington’s disease subjects compared to control subjects (Fig. 2A). This decrease in SREK1 was also evident by immunohistochemistry (Fig. 2B). In view of the decreased TAF1 levels achieved upon SREK1 silencing in neuroblastoma cells, we reasoned that the observed SREK1 decrease in striatum of Huntington’s disease patients could be mirrored by a decrease in TAF1 levels. This was confirmed by western blot analysis on striatal homogenates from Huntington’s disease patients and control subjects that revealed a 79% decrease (P < 0.001) of TAF1 protein levels in Huntington’s disease compared to control samples (Fig. 2C) without changes in total TAF1 transcript levels (Supplementary Fig. 2A). As expected, a marked TAF1 decrease was also observed by immunohistochemistry (Fig. 2D).
Figure 2.
Decreased levels of SREK1 and its target TAF1 in striatum of Huntington’s disease patients. Representative western blots of SREK1 (A) and TAF1 (C) proteins in striatum of Huntington’s disease (HD) patients (n = 7) and controls (n = 7) and their quantifications normalized to β-actin or vinculin (Student’s t-test; *P < 0.05; ***P < 0.001). Data represent mean + SEM. (B) SREK1- and (D) TAF1-immunohistochemistry staining in striatum (St) of Huntington’s disease patients and control samples.
R6/1 mice mimic the decreased SREK1 and TAF1 levels seen in Huntington’s disease
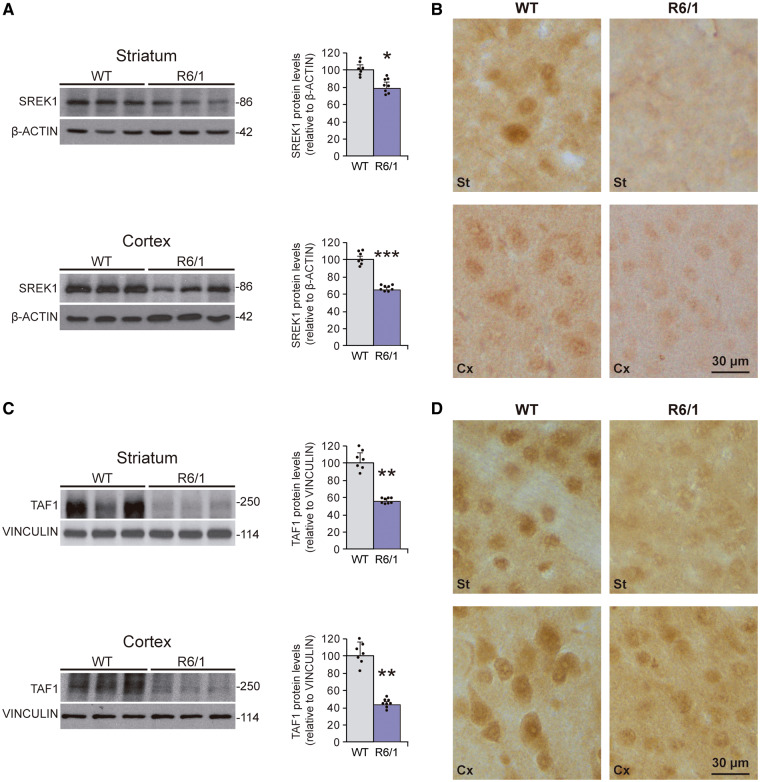
Transgenic mouse models of Huntington’s disease are an excellent tool to explore, through mouse genetics, the pathogenic relevance of the biochemical abnormalities observed in human post-mortem tissue. For this reason, we explored whether the commonly used R6/1 mouse model of Huntington’s disease recapitulates the decreased SREK1 and TAF1 levels seen in Huntington’s disease brains. Remarkably, early symptomatic (3.5-month-old) R6/1 mice show decreased SREK1 protein levels both in striatum (22%, P = 0.023) and cortex (37%, P < 0.001) as evidenced by western blot (Fig. 3A). As for human tissue, the decrease in SREK1 levels observed by western blot was mirrored by decreased SREK1 immunohistochemistry staining in both striatal and cortical sections (Fig. 3B).
Figure 3.
Decreased SREK1 and TAF1 levels in striatum and cortex of Huntington’s disease mouse model. Representative immunoblots of SREK1 (A) and TAF1 (C) protein levels in striatum and cortex of 3.5-month-age R6/1 mice (n = 8) and wild-type (WT) mice (n = 7) and their quantification normalized to β-actin or vinculin (Student’s t-test; *P < 0.05; ***P < 0.001). Data represent mean + SEM. (B) SREK1- and (D) TAF1-immunohistochemistry staining in striatum (St) and cortex (Cx) of R6/1 and wild-type mice.
In good agreement with the data from Huntington’s disease brains, R6/1 mice also showed decreased striatal (45%, P = 0.004) and cortical (56%, P = 0.004) TAF1 levels as evidenced by western blot (Fig. 3C) and by immunohistochemistry (Fig. 3D) without changes in total TAF1 transcript levels (Supplementary Fig. 2B). Interestingly, we also found decreased SREK1 and TAF1 protein levels in brains of a full-length HTT mouse model, namely homozygous HdhQ111 mice (Supplementary Fig. 3). Together, the results shown here indicate that the altered levels and/or activity of SRSF6 previously observed in brains of Huntington’s disease patients and mice (Fernandez-Nogales et al., 2014) correlate with a decrease in SREK1 levels that can then become detrimental by contributing to cause a decrease in TAF1 levels similar to that underlying XDP.
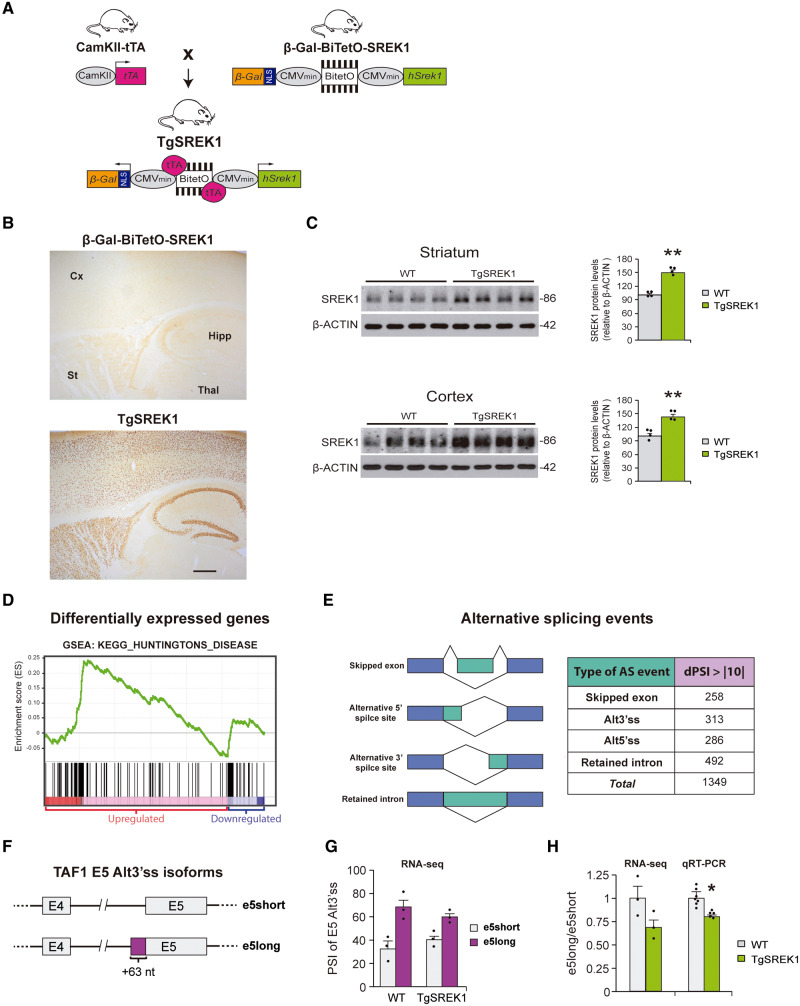
SREK1 overexpressing mice display transcriptomic alterations complementary to those of Huntington’s disease mice
To test in vivo if the observed decrease in SREK1 level is pathogenic—at least in part due to the subsequent decrease in TAF1—we generated transgenic mice with neuronal SREK1 overexpression to see whether this counteracts the deficit of TAF1 (along with other underexpressed genes) of Huntington’s disease mice and, most importantly, their neuropathological and motor symptoms. To restrict transgene expression to postnatal forebrain neurons, we used a conditional double transgenic approach with CamKII-tTA driver mice (Fig. 4A). The responder mice (β-Gal-BiTetO-SREK1) harbour a bidirectional cassette with the hSREK1 sequence in one direction and the reporter β-Gal in the other (Fig. 4A). As shown in Fig. 4B, the resulting TgSREK1 mice show transgenic expression restricted to neurons of forebrain structures such as striatum, cortex and hippocampus. We then verified increased SREK1 levels in striatum and cortex of TgSREK1 mice by western blot (Fig. 4C). To verify that the observed SREK1 overexpression has an impact on global mRNA stability and processing, we performed RNA-seq analysis on striatal tissue of 3.5-month-old TgSREK1 mice. Regarding transcript levels, we observed that 3.2% of genes show significantly increased transcript levels, while 4.4% showed decreased transcript levels (FDR < 0.05, Supplementary Table 2). Interestingly, the majority (87.5%) of genes with maximally increased transcript levels (fold change > 1.6) in TgSREK1 correspond to genes with decreased transcript levels in R6/1 mice (Table 1). In line with the complementarity with R6/1 mice, gene set enrichment analysis (GSEA) showed that most upregulated transcripts in the striatum of TgSREK1 mice present a significant (FDR < 0.001) enrichment in Huntington’s disease-related genes (Fig. 4D). Particularly interesting among top upregulated genes in TgSREK1 is Hes5 which, apart from being decreased in R6/1 mice, has recently been shown to be protective in a CAG/polyQ disease, namely SBMA (Kondo et al., 2019). Other top upregulated genes in TgSREK1 and decreased in R6/1 include Mpp7, Dsp14, Id3 and Pvalb (Table 1). As expected, many of the latter also show decreased transcript levels in shSREK1 SHSY-5Y cells (Supplementary Fig. 4A).
Figure 4.
Transgenic mice with conditional overexpression of SREK1 in forebrain neurons display global alteration of Huntington’s disease-related transcripts. (A) Double transgenic mouse line TgSREK1 was generated by crossing tTA-expressing mice under the control of CamKII promoter (CamKII-tTA mice) with mice carrying a tTA-responsive SREK1 construct (β-Gal-BiTetO-SREK1 mice). (B) Immunohistochemistry with anti-β-Gal antibody in sagittal sections from a 1.5-month-old TgSREK1 mouse and a single-transgenic β-Gal-BiTetO-SREK1 control mouse showing transgenic expression in cortex (Cx), striatum (St) and hippocampus (Hipp) of TgSREK1 mice, but not in non-forebrain structures like thalamus (Thal). Scale bar = 500 μm. (C) Immunoblot of SREK1 protein levels in striatum and cortex of 1.5-month-old TgSREK1 and wild-type (WT) mice, and its quantification with respect to β-actin (Student’s t-test; **P < 0.01). (D) GSEA analysis showing landscape of enrichment score (green line) for the KEGG molecular signature corresponding to ‘Huntington’s disease’. Black vertical bars above the red and blue regions correspond to upregulated and downregulated transcripts, respectively in TgSREK1 mice with respect to wild-type. (E) The different types of alternative splicing events and the abundance of those with an absolute dPSI > 10 between wild-type and TgSREK1 mice. (F) Diagram of the short (e5short) and long (e5long) versions of exon 5 (E5) of TAF1 generated by an alternative 3′ splice site (Alt3′ ss). The e5long version carries 63 additional nucleotides (+63 nt) on its 5′ site. (G) Histogram shows PSI of the e5long and e5short events observed by RNA-seq in 3.5-month-old wild-type (n = 3) and TgSREK1 (n = 3) mice. (H) Ratios in 3.5-month-old wild-type (n = 7) and TgSREK1 (n = 5) mice of the PSI values of e5long over e5short obtained by RNA-seq and of e5long over e5short transcript isoforms by qRT-PCR. Data represent mean + SEM. AS = alternative splicing.
Table I.
Top upregulated transcripts in TgSREK1 versus wild-type mice
| Gene symbol | Gene name | Upregulated in TgSREK1 (FC) | Downregulated in R6/1 mice |
|---|---|---|---|
| Chrna7 | Cholinergic receptor nicotinic alpha 7 subunit | 2.76 | No |
| Mpp7 | Membrane palmitoylated protein 7 | 2.07 | Yes |
| Dusp14 | Dual specificity phosphatase 14 | 1.83 | Yes |
| Hes5 | Hes family BHLH transcription factor 5 | 1.76 | Yes |
| Id3 | Inhibitor of DNA binding 3, HLH protein | 1.71 | Yes |
| Pvalb | Parvalbumin | 1.70 | Yes |
| Serpinf1 | Serpin family F member 1 | 1.65 | Yes |
| Plk5 | Polo like kinase 5 | 1.61 | Yes |
FC = fold change.
Regarding global splicing alteration in TgSREK1 mice, we analysed the RNA-seq data with Vast-Tools (Fig. 4E and Supplementary Table 3). This revealed 1349 differential AS events with an absolute dPSI >10 in TgSREK1 mice with respect to wild-type mice (19.1% skipped exons, 23.2% alternative 3′ splice sites, 21.2% alternative 5′ splice sites, and 36.5% retained introns). Focusing on TAF1, TgSREK1 mice displayed no alteration in its total transcript levels, but they showed an imbalance in usage of the alternative 3′ splicing site of exon 5 which adds 63 nt to it (Fig. 4F). Such imbalance results in a decrease of the e5long/e5short ratio (Fig. 4G and H), which happens to be opposite to the imbalance observed in R6/1 mice (Supplementary Fig. 4B). Together, the RNA-seq data strongly suggest that SREK1 has a significant impact on Huntington’s disease altered gene expression.
SREK1 overexpression corrects TAF1 deficit and eases striatal atrophy and motor phenotype of Huntington’s disease mice
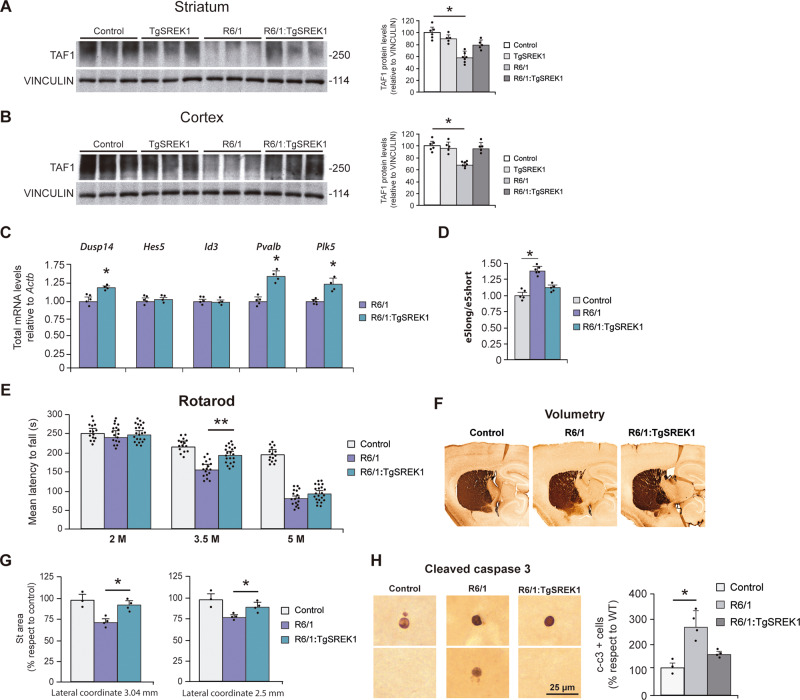
We then combined R6/1 and TgSREK1 mice to see whether SREK1 overexpression attenuates the Huntington’s disease-like biochemical, anatomical and behavioural abnormalities of R6/1 mice. First, we verified that the levels of SREK1 are normalized in 3.5-month-old R6/1:TgSREK1 mice (data not shown). Next, as decreased TAF1 expression is sufficient to cause a neurodegenerative motor disease (Aneichyk et al., 2018) and we have shown that diminished SREK1 leads to decreased TAF1 (Fig. 1), we checked whether transgenic SREK1 overexpression corrects the R6/1 deficit of TAF1, as a key determinant of toxicity. As expected, transgenic overexpression of SREK1 markedly attenuated the TAF1 deficit in striatum of 3.5-month-old R6/1 mice, as evidenced by western blot analysis (Fig. 5A) and such recovery was even more pronounced in the cortex (Fig. 5B). At the mRNA level, SREK1 overexpression in R6/1:TgSREK1 mice increased the transcript levels of multiple genes downregulated in R6/1 mice (Fig. 5C) and, importantly, it reversed the imbalance of e5long- and e5short-TAF1 splicing variants (Fig. 5D). Then, to explore whether AS of exon 5 might affect stability of TAF1 transcript, which in turn would account for the observed decreased TAF1 protein levels, we analysed mRNA stability of e5short- and e5long-TAF1 transcripts in neuroblastoma cells treated with a transcription inhibitor (Supplementary Fig. 5A). We found no difference between e5long- and e5short-TAF1 transcripts (despite a tendency to decreased stability of the e5long transcript). This fits with the fact that R6/1 brains, which show transcript isoform imbalance, do not show decreased total TAF1 transcript levels. Interestingly, the additional 21 amino acids present in the long version of exon 5 provide a degradation motif (DEG_OPDH, Supplementary Fig. 5B) missing in the e5short-TAF1 isoform. Thus offering a possible explanation at the protein level for the decrease observed in R6/1 mice and for its reversal in R6/1:TgSREK1 mice upon normalization of TAF1 splicing isoform ratios.
Figure 5.
SREK1 overexpression rescues TAF1 levels and improves striatal atrophy and motor coordination of Huntington’s disease mice. (A and B) Representative western blots of TAF1 protein levels in striatum (A) and cortex (B) of 3.5-month-old Control (n = 6), TgSREK1 (n = 5), R6/1 (n = 7) and combined R6/1:TgSREK1 (n = 5) mice. Histograms show quantification of TAF1 normalized to VINCULIN and represented in % respect to Control levels. (C) Quantification of genes analysed by RT-PCR in 3.5-month-old R6/1 (n = 5) and R6/1:TgSREK1 (n = 4) normalized to Actb. (D) Quantification of e5long/e5short transcript isoform ratio in Control (n = 5), R6/1 (n = 6) and R6/1:TgSREK1 (n = 5) mice. (E) Rotarod test. Evolution of the mean latency to fall from the rod in the four accelerating testing trials of Control (eight males and seven females); R6/1 (10 males and seven females); and R6/1:TgSREK1 (12 males and 10 females) mice at 2, 3.5 and 5 months of age. Analysis of variance (ANOVA), followed by Games-Howell, **P < 0.01. (F) Representative images of DARPP-32-stained sagittal sections from Control, R6/1 and R6/1:TgSREK1 mice at 3.5 months of age. (G) Quantification of the striatal area measured in sagittal sections at two different lateral coordinates of Control (n = 3), R6/1 (n = 4) and R6/1:TgSREK1 (n = 4) mice. ANOVA, followed by Games-Howell, *P < 0.05. (E) Representative fields showing immunostaining of cleaved caspase-3 (left) and quantification apoptotic (cleaved caspase-3-positive) cells in Control (n = 3), R6/1 (n = 4) and R6/1:TgSREK1 (n = 4) mice. ANOVA followed by Games-Howell. Data represent mean ± SEM.
We then explored whether SREK1 overexpression with the subsequent normalization of TAF1 levels attenuate the motor coordination deficit of R6/1 mice. This is the case, as evidenced in the Rotarod test, in early symptomatic (3.5 months old) R6/1 mice. More precisely, R6/1:TgSREK1 mice showed a 33% recovery in mean latency to fall with respect to R6/1 mice in the accelerating paradigm (Fig. 5E, P = 0.008). However, this behavioural rescue was no longer evident at the age of 5 months and this prompted us to test whether the biochemical rescue of decreased TAF1 levels was also transient. For this, we performed western blot analysis in 6.5-month-old mice of the four relevant genotypes (Control, TgSREK1, R6/1 and R6/1:TgSREK1). This revealed that aged TgSREK1 mice no longer show increased levels of SREK1 and, accordingly, that the decreased TAF1 levels in striatum and cortex of R6/1 mice are no longer rescued in R6/1:TgSREK1 mice at older age (Supplementary Fig. 6).
We then tested whether the normalization of TAF1 levels and the amelioration of motor phenotype seen in 3.5-month-old R6/1:TgSREK1 mice was accompanied by an attenuation of the striatal atrophy evident at this age in lateral striatum of R6/1 mice with respect to wild-type mice. This is the case, as shown in Fig. 5F and G (P < 0.05). Finally, as TAF1 has been reported to play a role in apoptosis regulation (Kimura et al., 2008), we reasoned that this might be one of the mechanisms by which restoring SREK1 and TAF1 levels attenuate motor phenotype and striatal atrophy in R6/1:TgSREK1 mice. In agreement, when we scored the incidence of apoptosis by immunohistochemistry with an antibody against cleaved caspase-3 in forebrain of Control, R6/1 and R6/1:TgSREK1 mice, we observed that the increased incidence of apoptosis observed in R6/1 mice is fully prevented in R6/1:TgSREK1 mice (Fig. 5H). Together, the biochemical recovery of TAF1 levels and the attenuation of Huntington’s disease-like neuropathology and motor symptoms in R6/1:TgSREK1 mice demonstrate that the observed decrease of SREK1 in brains of Huntington’s disease mouse models and patients is pathogenic and strongly suggest that subsequent TAF1 decrease is one of its key determinants.
Discussion
By exploring the status of the SRSF6-interacting SR protein SREK1 in Huntington’s disease brains, we detected a marked decrease in its protein levels, which led us to analyse its XDP-causing target TAF1, which we also found markedly decreased. Interestingly, gene silencing of SRSF6 in human neuroblastoma cells decreased both SREK1 and TAF1 levels and we also found that SREK1 decrease, per se, suffices to diminish TAF1 levels. To explore in vivo the pathogenic relevance of the observed SREK1 decrease in Huntington’s disease brains and demonstrate that the XDP-like deficiency of TAF1 is secondary to such SREK1 decrease, we also generated mice with transgenic SREK1 overexpression (TgSREK1 mice). Interestingly, RNA-seq analysis of TgSREK1 mice revealed a transcriptomic alteration complementary to that of Huntington’s disease mice. Most importantly, SREK1 overexpression corrects TAF1 deficiency and attenuates striatal atrophy and motor phenotype of Huntington’s disease mice, thus demonstrating convergence of Huntington’s disease and XDP pathogenic pathways in TAF1 through a pathogenic SREK1 decrease.
Thus, here we provide a molecular link between the Huntington’s disease-causing mutation and TAF1, the effector of XDP. In this mechanistic model, SRSF6 would participate immediately downstream of the Huntington’s disease mutation as it is known to directly interact with the expanded CAG mRNA (Schilling et al., 2019) and to get sequestered into the characteristic polyQ-containing inclusion bodies as well as hyperphosphorylated (Fernandez-Nogales et al., 2014). This, in turn, affects its direct interactor and regulator SREK1 whose levels get decreased. As a consequence, there is a transcriptomic rearrangement that, apart from many Huntington’s disease-related transcripts, affects TAF1, whose levels also get diminished, as in XDP.
The fact that correcting SREK1 decrease in Huntington’s disease mouse models normalizes TAF1 levels and attenuates Huntington’s disease-like neuropathology and motor symptoms, clearly demonstrates that decreased SREK1 plays a central role in the above described mechanism. On the other hand, since TAF1 alteration suffices to cause a basal ganglia disorder that shares many features with Huntington’s disease (Aneichyk et al., 2018), it is therefore compelling to reason that TAF1 decrease also plays a key role in Huntington’s disease pathogenesis. We should, however, keep in mind that TAF1 might not be the only SREK1-target relevant to Huntington’s disease pathogenesis. Other interesting targets include Hes5, which plays an important role in SBMA (Kondo et al., 2019). Similarly, expanded CAG mRNA and polyQ have other toxic interactors—apart from the one explored here—that also significantly contribute to Huntington’s disease.
As the main function of TAF1 is to act as the largest and core subunit of the basal transcription factor TFIID (Albright and Tjian, 2000), it is conceivable that the detrimental consequences of decreased TAF1 levels in XDP and Huntington’s disease might be due to decreased global efficiency of transcription, in line with earlier observations in brain tissue of Huntington’s disease patients and mouse models (Cha, 2000). However, it should be noted that TAF1 also has kinase activity and that this is known to regulate apoptosis, in part through its ability to phosphorylate p53 (Li et al., 2004; Cai and Liu, 2008). This might be related to the apoptosis and brain atrophy modulated by SREK1, and presumably TAF1, levels in Huntington’s disease mice. In line with this, we have unpublished results indicating decreased phosphorylation of Thr-55 p53 in brains of Huntington’s disease patients and such phosphorylation has been shown to correlate with increased apoptosis (Li et al., 2004).
In summary, here we demonstrate a pathogenic SREK1 decrease in Huntington’s disease, which leads to a deficit in TAF1. This is evidence of a mechanistic convergence with XDP and identifies a possible general mechanism for striatal neurodegeneration as well as new targets for therapeutic intervention.
Supplementary Material
Acknowledgements
Human tissue was obtained from Institute of Neuropathology (HUB-ICO-IDIBELL) Brain Bank, the Neurological Tissue Bank of the IDIBAPS Biobank, the Banco de Tejidos Fundación CIEN, and the Netherlands Brain Bank. We thank Wilfried Meijer for advice on molecular cloning and Marcos Casado for advice on bioinformatics analysis of degradation motifs. We also thank excellent technical assistance by Miriam Lucas and by the following core facilities: CBMSO-Genomics & Massive Sequencing and CBMSO-Animal Facility. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Funding
This work was supported by CIBERNED-ISCIII collaborative grants PI2015-2/06-3 and PI2018/06-1 and by grants from Spanish Ministry of Economy and Competitiveness/Ministry of Science, Innovation and Universities (MINECO/MCIU/AEI/FEDER, UE): SAF2015-65371-R and RTI2018-096322-B-I00 to J.J.L. and PI18/00263 from the Instituto de Salud Carlos III (Ministry of Economy, Industry and Competitiveness)—cofunded by the European Regional Development Fund—to R.G.-E. and by institutional grant from Fundación Ramón Areces to CBMSO.
Competing interests
The authors report no competing interests.
Glossary
- polyQ =
polyglutamine;
- XDP =
X-linked dystonia parkinsonism
References
- Albright SR, Tjian R.. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 2000; 242: 1–13. [DOI] [PubMed] [Google Scholar]
- Aneichyk T, Hendriks WT, Yadav R, Shin D, Gao D, Vaine CA, et al. Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 2018; 172: 897–909.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Patton JG.. Identification and characterization of a novel serine-arginine-rich splicing regulatory protein. Mol Cell Biol 2000; 20: 3049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR.. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucl Acids Res 1993; 21: 5803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JR, Lucas JJ.. MAP2 splicing is altered in Huntington’s disease. Brain Pathol 2017; 27: 181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Liu X.. Inhibition of Thr-55 phosphorylation restores p53 nuclear localization and sensitizes cancer cells to DNA damage. Proc Natl Acad Sci USA 2008; 105: 16958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH. Transcriptional dysregulation in Huntington’s disease. Trends Neurosci 2000; 23: 387–92. [DOI] [PubMed] [Google Scholar]
- Elorza A, Márquez Y, Cabrera JR, Sánchez-Trincado JL, Santos-Galindo M, Hernández IH, et al. Huntington’s disease-specific mis-splicing captured by human-mouse intersect-RNA-seq unveils pathogenic effectors and reduced splicing factors. BIORXIV/2020/086017, 2020. [Google Scholar]
- Fernandez-Nogales M, Cabrera JR, Santos-Galindo M, Hoozemans JJ, Ferrer I, Rozemuller AJ, et al. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat Med 2014; 20: 881–5. [DOI] [PubMed] [Google Scholar]
- Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993; 72: 971–83. [DOI] [PubMed] [Google Scholar]
- Kimura J, Nguyen ST, Liu H, Taira N, Miki Y, Yoshida K.. A functional genome-wide RNAi screen identifies TAF1 as a regulator for apoptosis in response to genotoxic stress. Nucleic Acids Res 2008; 36: 5250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, et al. Protein-protein interactions and 5’-splice-site recognition in mammalian mRNA precursors. Nature 1994; 368: 119–24. [DOI] [PubMed] [Google Scholar]
- Kondo N, Tohnai G, Sahashi K, Iida M, Kataoka M, Nakatsuji H, et al. DNA methylation inhibitor attenuates polyglutamine-induced neurodegeneration by regulating Hes5. EMBO Mol Med 2019; 11: e8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Li AG, Sheppard HM, Liu X.. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: a role for TAF1 in cell G1 progression. Mol Cell 2004; 13: 867–78. [DOI] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM.. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 2008; 453: 1107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF.. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 2009; 417: 15–27. [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER.. Control of memory formation through regulated expression of a CaMKII transgene. Science 1996; 274: 1678–83. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–73. [DOI] [PubMed] [Google Scholar]
- Naro C, Sette C.. Phosphorylation-mediated regulation of alternative splicing in cancer. Int J Cell Biol 2013; 2013: 151839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neueder A, Dumas AA, Benjamin AC, Bates GP.. Regulatory mechanisms of incomplete huntingtin mRNA splicing. Nat Commun 2018; 9: 3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY.. Trinucleotide repeat disorders. Annu Rev Neurosci 2007; 30: 575–621. [DOI] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 2013; 50: 223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA.. RNA-mediated neuromuscular disorders. Annu Rev Neurosci 2006; 29: 259–77. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci USA 2013; 110: 2366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling J, Broemer M, Atanassov I, Duernberger Y, Vorberg I, Dieterich C, et al. Deregulated splicing is a major mechanism of RNA-induced toxicity in Huntington’s disease. J Mol Biol 2019; 431: 1869–77. [DOI] [PubMed] [Google Scholar]
- Solis AS, Patton JG.. Analysis of SRrp86-regulated alternative splicing: control of c-Jun and IkappaBbeta activity. RNA Biol 2010; 7: 486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapial J, Ha KCH, Sterne-Weiler T, Gohr A, Braunschweig U, Hermoso-Pulido A, et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res 2017; 27: 1759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker FO. Huntington’s disease. Lancet 2007; 369: 218–28. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T.. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 1993; 75: 1061–70. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL.. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev 1997; 11: 334–44. [DOI] [PubMed] [Google Scholar]
- Yin X, Jin N, Gu J, Shi J, Zhou J, Gong CX, et al. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J Biol Chem 2012; 287: 30497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding author upon reasonable request.