Summary
Cardiac neural crest cells (cNCCs) are required for normal heart development. cNCCs are a multipotent and migratory cell lineage that differentiates into multiple cell types. cNCCs migrate into the developing heart to contribute to the septation of the cardiac outflow tract (OFT). Foxc1 and Foxc2 are closely related members of the FOX (Forkhead box) transcription factor family and are expressed in cNCC during heart development. However, the precise role of Foxc1 and Foxc2 in cNCCs has yet to be fully described. We found that compound NCC-specific Foxc1;Foxc2 mutant embryos exhibited persistent truncus arteriosus (PTA), ventricular septal defects (VSDs), and thinning of the ventricular myocardium. Loss of Foxc1/c2 expression in cNCCs resulted in abnormal patterns of cNCC migration into the OFT without the formation of the aorticopulmonary septum. Further, loss of Foxc1 expression in cNCCs resulted in normal OFT development but abnormal ventricular septal formation. In contrast, loss of Foxc2 expression in NCCs led to no obvious cardiac abnormalities. Together, we provide evidence that Foxc1 and Foxc2 in cNCCs are cooperatively required for proper cNCC migration, the formation of the OFT septation, and the development of the ventricles. Our data also suggests that Foxc1 expression may play a larger role in ventricular development compared to Foxc2.
Keywords: cardiac neural crest, Foxc, heart development, truncus arteriosus, ventricular septal defect
1 |. RESULTS AND DISCUSSION
NCCs are derived from the dorsal neural tube during early embryonic development and exhibit a transient cell type with multi-potency. They subsequently migrate to diverse locations in the embryo and differentiate into numerous derivatives. In particular, cardiac NCCs (cNCCs) give rise to the outflow tract (OFT) septum during embryonic heart development. In humans, defects in the cNCC lineage are associated with multiple congenital heart anoma-lies related to the impairment in rotation and septation of the cardiac OFT, including DiGeorge syndrome [Online Mendelian Inheritance in Man (OMIM) Entry #188400] and Tetralogy of Fallot (OMIM Entry #187500). However, the complex nature of cardiac abnormalities and diseases associated with cNCCs remains poorly understood (Yamagishi, 2020).
Foxc1 and Foxc2 are closely related members of the Fox transcription factor family and have numerous essential roles in cardiovascular development, including the formation of the cardiac OFT (Kume, Jiang, Topczewska, & Hogan, 2001; Seo et al., 2006; Seo & Kume, 2006). Mutations in human FOXC1 and FOXC2 are associated with cardiac OFT malformations such as Tetralogy of Fallot (Majumdar, Yasuhara, & Garg, 2019; Nees & Chung, 2019; Topf et al., 2014). Our previous studies reveal that the expression patterns of Foxc1 and Foxc2 in mice are overlapped in the developing heart, including cNCCs as well as their derivatives in the OFT cushion that contribute to the OFT septum (Iida et al., 1997; Inman et al., 2018; Seo & Kume, 2006; Winnier et al., 1999). However, the specific roles of Foxc1 and Foxc2 in cNCCs have yet to be investigated.
1.1 |. Absence of Foxc1 and Foxc2 in cardiac neural crest cells leads to heart abnormalities
We first examined compound, NC-specific Foxc1 and Foxc2 knockout (NC-Foxc1;Foxc2-DKO), mice crossed with Wnt1-Cre mice at both embryonic day (E)11.5 and E14.5 for gross morphological cardiac abnormalities. At E11.5, OFT septation into the aorta and pulmonary trunk is still progressing with the formation of the aorticopulmonary septum (Savolainen, Foley, & Elmore, 2009). We found that control (Foxc1F/F;Foxc2F/F) embryos displayed proper OFT septation with a visible aorticopulmonary septum at E11.5 (Figure 1a). In contrast, 1 of 3 NC-Foxc1;Foxc2-DKO (Wnt1-Cre;Foxc1F/F;Foxc2F/F) embryos occasionally (33%) exhibited a lack of an obvious aorticopulmonary septum, deemed partial persistent truncus arteriosus (PTA) (Figure 1b and Table 1).
FIGURE 1.
Persistent truncus arteriosus (PTA) and ventricular septal defect (VSD) in NC-specific Foxc1;Foxc2 mutants at E11.5 and E14.5. (a, c, and e) Control heart sections show normal outflow tract (OFT) and ventricular development at the respective time points in development. (b) Compound NC-Foxc1;Foxc2-DKO section at E11.5 that exhibits partial PTA. (d and f) NC-Foxc1;Foxc2-DKO sections at E14.5 that exhibit both PTA, displayed by a lack of septation between the pulmonary trunk (Pt) and aorta (Ao), and VSD, displayed by an indistinct interventricular septum. E14.5 sections cut in the transverse plane. E11.5 sections cut in the frontal plane. PTA, persistent truncus arteriosus; RV, right ventricle; LV, left ventricle; VSD, ventricular septal defect; Sep, aorticopulmonary septum; P.PTA, partial truncus arteriosus. Scale bars, 100 μm in A–B; 150 μm in C–F
TABLE 1.
Summary of cardiac abnormalities in global and NC-specific mutant mice for Foxc1 and Foxc2
| Penetrance values disorders | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | Time point | PTA | VSD | PTA;VSD | None | Genetic background | Reference |
| Foxc1 mutation | |||||||
| Global Foxc1−/− | P0 (n = 11) | 0% | 73% | 0% | 27% mixed | Winnier et al., 1999 | |
| NC-Foxc1-KO | E14.5 (n = 3) | 0% | 33% | 0% | 66% | Mixed | This paper |
| Foxc2 mutation | |||||||
| Global Foxc2−/− | P0 | + | C57BL/6 | Iida et al., 1997 | |||
| E13.5/E16.5 (n = 9) | 0% | 44% | 0% | 56% | Mixed | Winnier et al., 1999 | |
| E12.5 | + | + | 129 s6/SvEv | Inman et al., 2018 | |||
| NC-Foxc2-DKO | E11.5 (n = 4) | 0% | 0% | 0% | 100% | Mixed | This paper |
| E14.5 (n = 7) | 0% | 0% | 0% | 100% | Mixed | This paper | |
| Foxc1;Foxc2 mutation | |||||||
| Global Foxc1+/−;Foxc2−/− | E11.5 | + | Mixed | Seo et al., 2006 | |||
| NC-Foxc1;Foxc2-DKO | E11.5 (n = 3) | 33% | 0% | 0% | 67% | Mixed | This paper |
| E14.5 (n = 5) | 40% | 20% | 20% | 20% | Mixed | This paper | |
| NC-Foxc1;Foxc2-DKO;mTmG | E14.5 (n = 4) | 100% | 0% | 0% | 0% | Mixed | This paper |
At E14.5, normal heart development results in a completely divided OFT, forming the aorta and pulmonary trunk; a clear interventricular septum, separating the left and right ventricle; and normal semilunar valve formation. We found that control embryos displayed normal, clear septation of the outflow tract and a clear interventricular septum at E14.5 (Figure 1c,e). In contrast, 3 of 5 NC-Foxc1;Foxc2-DKO embryos exhibited PTA (60%), in which the OFT lacked clear separation as a single duct (Figure 1d and Table 1). This observation is consistent with our previous study showing that compound, global mutant mice for Foxc1 and Foxc2 also have PTA (Seo & Kume, 2006) (Table 1). Further, 2 of 5 NC-Foxc1;Foxc2-DKO embryos (40%) displayed an indistinct interventricular septum with ventricles composed of thin, wispy trabeculae (Figure 1f). Phenotypic variability and penetrance are shown in Table 1.
1.2 |. Abnormal cardiac neural crest cell migration in compound, NC-specific Foxc1;Foxc2 mutant embryos
Migrating cNCCs give rise to the cardiac OFT cushion that forms the aorticopulmonary septum (Waldo, Miyagawa-Tomita, Kumiski, & Kirby, 1998; Yamagishi, 2020). Thus, visualization of cNCCs was done to both further characterize migration patterns of cNNCs with and without the Foxc genes and gain insight into the cell type’s role in the pathogenesis of cardiac phenotypes stemming from their deficiency. We generated a Wnt1-Cre;Foxc1;Foxc2;mTmG mouse line to accomplish the lineage tracing of cNCCs in both the control (Foxc1F/F; Foxc2F/F;mTmG/+) and Foxc1/c2-mutant (Wnt1-Cre;Foxc1F/F;Foxc2F/F;mTmG/+) embryos at E14.5 with cNCCs labeled by AP-2alpha immunostaining in control and labeled by EGFP in Foxc1/c2-mutant embryos (Figure 2). The tdTomato fluorescent reporter is expressed in all tissue and cell types. After NC-specific Cre recombination occurs, EGFP fluorescent reporter will visualize Cre activity in NCCs (Muzumdar, Tasic, Miyamichi, Li, & Luo, 2007). Analyzing this mouse line, AP-2alpha + cNCCs in control embryos migrated into the OFT to form the aorticopulmonary septum (Figure 2a–c). Specifically, labeled cNCCs lined the complete inner border of the aorta with little cNCCs migrating and differentiating into the pulmonary trunk. In contrast, NC-Foxc1;Foxc2-DKO embryos exhibited abnormal cNCCs migration, accompanied by PTA (Figure 2d–f). Foxc1/c2-mutant cNCCs failed to migrate to the aorticopulmonary septum by only lining the truncus arteriosus duct (Figure 2d). Interestingly, these mutant cNCCs populated only the left portion of the duct lining (Figure 2d,e), which may have been the aortic portion of the OFT. This observation indicates that Foxc1/c2 expression in cNCCs is required for proper cNCC migration into the OFT to form the aorticopulmonary septum.
FIGURE 2.

Visualization of cNCCs and overall heart development in control and NC-specific compound Foxc1;Foxc2 mutants at E14.5. (a) Control heart section displaying normal outflow tract septation between the Aorta (Ao) and pulmonary trunk (Pt). (b and c) Control heart sections displaying outflow tract septation with AP-2alpha positive cardiac neural crest cells (cNCCs) labeled in bright pink. (d–f) NC-Foxc1;Foxc2-DKO heart sections displaying persistent truncus arteriosus (PTA) with cNCCs labeled in green. Scale bars are 150 μm
1.3 |. Absence of Foxc1 in cardiac neural crest cells leads to ventricular septal defects and normal outflow tract development
Our previous studies show that global deletion of Foxc1 in mice results in cardiac abnormalities such as VSDs (Winnier et al., 1999) (Table 1). Thus, we generated NC-specific Foxc1 mutant (NC-Foxc1-KO) mice crossed with Wnt1-Cre mice to further test the specific role of Foxc1 in cNCCs during heart development. Both control (Foxc1F/F) and NC-Foxc1-KO (Wnt1-Cre;Foxc1F/F) embryos exhibited normal OFT development, displayed by distinct aorticopulmonary septum formation (Figure 3a,b). However, 1 of 3 NC-Foxc1-KO embryos (33%) exhibited VSD, displayed by an indistinct membranous ventricular septum (Figure 3d and Table 1), compared to normal ventricular development as shown in the control (Figure 3c).
FIGURE 3.
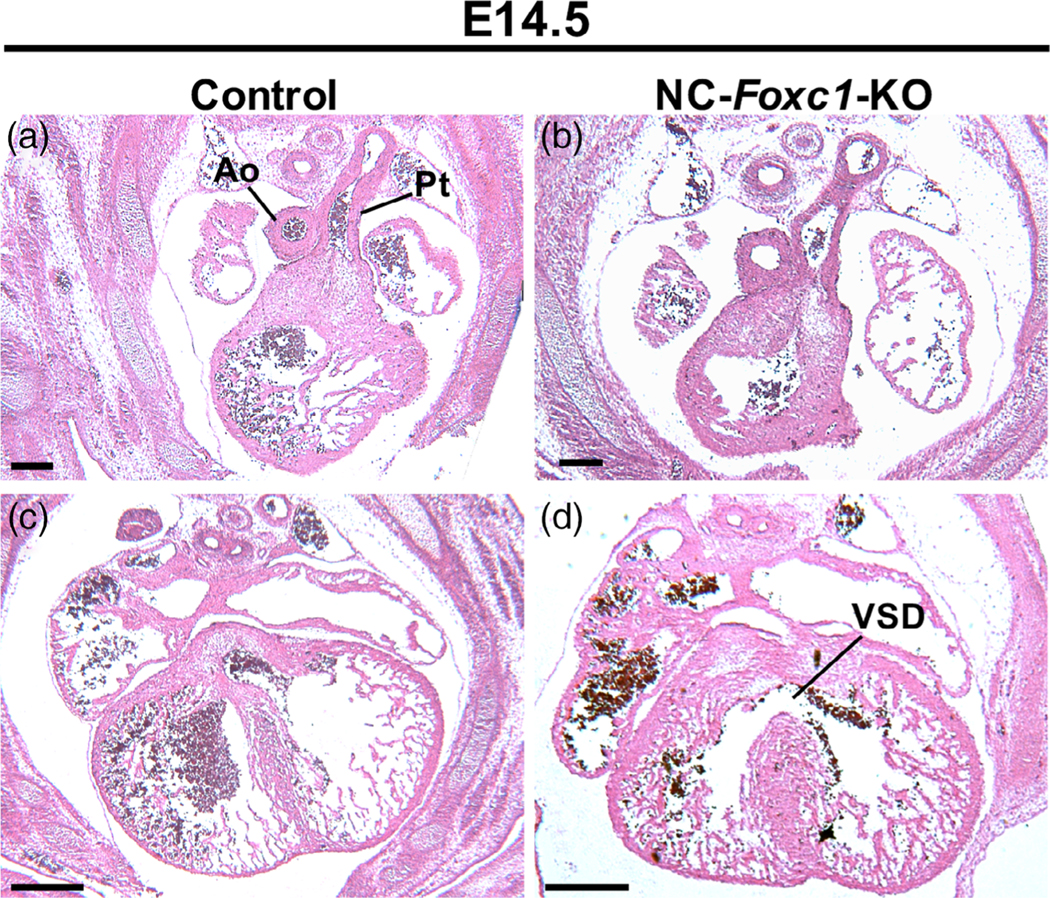
NC-specific Foxc1 mutants at E14.5 have normal outflow tract (OFT) development with ventricular septal defects. (a, c) Control heart sections show normal OFT and ventricular development at the E14.5 time point. (b) NC-Foxc1-KO section at the E14.5 time point that exhibits normal OFT development, displayed by a distinct aorticopulmonary septum. (d) NC-Foxc1 KO section at the E14.5 time point that exhibits VSD, displayed by an indistinct interventricular septum. E14.5 sections cut in the transverse plane. Ao, aorta; Pt, pulmonary trunk; VSD, ventricular septal defect; Scale bars, 150 μm in A–D
1.4 |. Absence of Foxc2 in cardiac neural crest cells leads to no obvious cardiac abnormalities
Previous studies describe global mutant mice for Foxc2 that exhibit cardiac abnormalities such as VSDs, PTA, and hypoplastic ventricles (Iida et al., 1997; Inman et al., 2018; Winnier et al., 1999) (Table 1), whereas specific deletion of Foxc2 in the mesenchyme/endothelium of the pharyngeal arches results in VSDs (Uddin et al., 2015).
To test the precise role of Foxc2 in cNCCs, we generated NC-specific Foxc2 mutant (NC-Foxc2-KO) mice crossed with Wnt1-Cre mice. While control (Foxc2F/F) embryos displayed normal heart formation at E11.5 and E14.5 (Figure 4a,C), NC-Foxc2-KO embryos exhibited no obvious cardiac abnormalities at both time points (Figure 4b,D and Table 1). In this study, we generated and analyzed NC-Foxc2-KO mice on a mixed genetic background, containing those of an inbred 129/SvEv mouse strain and an outbred Black Swiss mouse strain. Global deletion of Foxc2 in mice on the 129 s6/SvEv inbred genetic background results in disruption of cNCC migration, accompanied by PTA (Inman et al., 2018); however, PTA is not observed in global Foxc2 mutant mice on other inbred (C57BL/6) or mixed genetic backgrounds (Iida et al., 1997; Winnier et al., 1999) (Table 1). This indicates the presence of genetic modifiers that are associated with PTA.
FIGURE 4.

NC-specific Foxc2 mutants at E11.5 and E14.5 have normal outflow tract (OFT) and ventricular development. (a and c) Control heart sections show normal OFT development at E11.5 and E14.5. (b and d) NC-Foxc2-KO embryos at both E11.5 and E14.5 show no obvious OFT abnormalities. (e and f) Control and NC-Foxc2-KO embryos at E14.5 show normal ventricular development. E14.5 sections cut in the transverse plane. E11.5 sections cut in the frontal plane. Ao, aorta; Pt, pulmonary trunk; Sep, aorticopulmonary septum. Scale bars, 100 μm in A–B; 150 μm in C–F
In this study, we show that Foxc1 and Foxc2 cooperatively function in the cNCC lineage during heart development. Specific deletion of both Foxc1 and Foxc2 in NCCs leads to persistent truncus arteriosus (PTA), ventricular septal defects (VSDs), and hypoplasia of the ventricular myocardium, whereas NC-specific Foxc2 deletion alone does not result in cardiac abnormalities. However, NC-specific Foxc1 deletion leads to VSDs. Together, our data indicate an overlapping role of Foxc1 and Foxc2 in cNCCs during cardiac development. In summary, our studies suggest that Foxc1 and Foxc2 have overlapping functions in cNCCs during proper formation of the OFT septum and that Foxc1 expression in cNCCs may play a larger role in ventricular development compared to Foxc2.
2 |. METHODS
2.1 |. Animal husbandry
Conditional floxed Foxc1F/F, Foxc2F/F, and Foxc1F/F;Foxc2F/F mice were described previously (Sasman et al., 2012). Wnt1-Cre;Foxc1F/F(NC-Foxc1-KO), Wnt1-Cre;Foxc2F/F(NC-Foxc2-KO) and Wnt1-Cre;Foxc1F/F; Foxc2F/F (NC-Foxc1;Foxc2-DKO) mice were generated on a mixed background, containing 129/SvEv and Black Swiss, by crossing Foxc-floxed mice (Foxc1F/F, Foxc2F/F, and Foxc1F/F;Foxc2F/F) with mice expressing Wnt1-Cre (Danielian, Muccino, Rowitch, Michael, & McMahon, 1998), respectively, as described previously (Seo et al., 2012; Seo et al., 2017). For cell fate mapping, Wnt1-Cre; Foxc1F/F;Foxc2F/F;mTmG/+ (NC-Foxc1;Foxc2-DKO;mTmG) mice were generated by crossing Foxc1F/F;Foxc2F/F;mTmG/mTmG mice with Wnt1-Cre; Foxc1F/F;Foxc2F/F (NC-Foxc1;Foxc2-DKO) mice. The embryonic age was defined as embryonic day 0.5 (E0.5) at noon on the day of the vaginal plug. Genotyping of NC-specific Foxc conditional mutants was performed by Transnetyx Inc. All experimental protocols and procedures used in this study were approved by the Institutional Animal Care and Use Committee at Northwestern University.
2.2 |. Tissue processing
Whole mouse embryos were fixed in 4% paraformaldehyde (PFA) overnight in 4°C, dehydrated with ethanol the following day, and then cut into 8-μm sections. For frozen sections (mTmG reporter line), whole embryos were fixed in 4% PFA overnight, and then immersed in a fresh-made 30% sucrose solution in PBS overnight the following day. Samples were then embedded in OCT compound (Tissue-Tek) and frozen in an ethanol/dry ice bath. Frozen samples were then cut into 8-μm sections.
2.3 |. Histological and immunohistochemical analyses
All paraffin sections were stained with hematoxylin and eosin (H&E), using non-acidified hematoxylin. Samples were then imaged using an Olympus Vanox AHBT3 microscope.
For cell-fate mapping of neural crest cells, frozen sections were prepared for immunohistochemical analysis. All frozen sections were blocked with a 10% Donkey serum (Sigma-Aldrich Corp.) and 0.2% Triton X-100 (Sigma-Aldrich Corp.) in PBS solution for 30 min at room temperature. Control frozen sections were then incubated in a blocking buffer with a NC-marker with primary AP-2alpha monoclonal antibody 3b5–S (Developmental Studies Hybridoma Bank, Catalog # 3b5), diluted 1:100, overnight in 4°C and then incubated with secondary Alexa Fluor™ 594 goat anti-mouse IgG2b (y2b) (Thermo Fisher Scientific, Catalog # A-21145), diluted 1:250 with blocking solution for 1 hr. Then, control sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature, and then mounted with aqueous PermaFluor mounting medium (Thermo Scientific). Similarly, mutant sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature, and then mounted with aqueous PermaFluor mounting medium (Thermo Scientific). All fluorescent sections were then imaged using a Zeiss Observer D1 microscope.
ACKNOWLEDGMENTS
We thank Andrew McMahon for providing the Wnt1-Cre transgenic mice. This work was supported by the NIH (R01HL126920 and R01HL144129 to T. Kume).
Funding information
National Heart, Lung, and Blood Institute, Grant/Award Numbers: HL126920, HL144129
REFERENCES
- Danielian PS, Muccino D, Rowitch DH, Michael SK, & McMahon AP (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current Biology, 8(24), 1323–1326. [DOI] [PubMed] [Google Scholar]
- Iida K, Koseki H, Kakinuma H, Kato N, Mizutani-Koseki Y, Ohuchi H, … Miura N (1997). Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development, 124(22), 4627–4638. [DOI] [PubMed] [Google Scholar]
- Inman KE, Caiaffa CD, Melton KR, Sandell LL, Achilleos A, Kume T, & Trainor PA (2018). Foxc2 is required for proper cardiac neural crest cell migration, outflow tract septation, and ventricle expansion. Developmental Dynamics, 247(12), 1286–1296. 10.1002/dvdy.24684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, & Hogan BL (2001). The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes & Development, 15(18), 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar U, Yasuhara J, & Garg V (2019). In vivo and in vitro genetic models of congenital heart disease. Cold Spring Harbor Perspectives in Biology. 10.1101/cshperspect.a036764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, & Luo L (2007). A global double-fluorescent cre reporter mouse. Genesis, 45(9), 593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nees SN, & Chung WK (2019). Genetic basis of human congenital heart disease. Cold Spring Harbor Perspectives in Biology. 10.1101/cshperspect.a036749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasman A, Nassano-Miller C, Shim KS, Koo HY, Liu T, Schultz KM, … Kume T (2012). Generation of conditional alleles for Foxc1 and Foxc2 in mice. Genesis, 50(10), 766–774. 10.1002/dvg.22036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen SM, Foley JF, & Elmore SA (2009). Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicologic Pathology, 37(4), 395–414. 10.1177/0192623309335060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Chen L, Liu W, Zhao D, Schultz KM, Sasman A, … Kume T (2017). Foxc1 and Foxc2 in the neural crest are required for ocular anterior segment development. Investigative Ophthalmology & Visual Science, 58(3), 1368–1377. 10.1167/iovs.16-21217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Fujita H, Nakano A, Kang M, Duarte A, & Kume T (2006). The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Developmental Biology, 294(2), 458–470. [DOI] [PubMed] [Google Scholar]
- Seo S, & Kume T (2006). Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Developmental Biology, 296(2), 421–436. [DOI] [PubMed] [Google Scholar]
- Seo S, Singh HP, Lacal PM, Sasman A, Fatima A, Liu T, … Kume T (2012). Forkhead box transcription factor FoxC1 preserves corneal transparency by regulating vascular growth. Proceedings of the National Academy of Sciences of the United States of America, 109(6), 2015–2020. 10.1073/pnas.1109540109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf A, Griffin HR, Glen E, Soemedi R, Brown DL, Hall D, … Goodship JA (2014). Functionally significant, rare transcription factor variants in tetralogy of fallot. PLoS One, 9(8), e95453. 10.1371/journal.pone.0095453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin MK, Kimura W, Ishikura T, Koseki H, Yoshida N, Islam MJ, … Miura N (2015). Foxc2 in pharyngeal arch mesenchyme is important for aortic arch artery remodelling and ventricular septum formation. Biomedical Research, 36(4), 235–245. 10.2220/biomedres.36.235 [DOI] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, & Kirby ML (1998). Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: Aortic sac to ventricular septal closure. Developmental Biology, 196(2), 129–144. 10.1006/dbio.1998.8860 [DOI] [PubMed] [Google Scholar]
- Winnier GE, Kume T, Deng K, Rogers R, Bundy J, Raines C, … Conway SJ (1999). Roles for the winged helix transcription factors MF1 and MFH1 in cardiovascular development revealed by nonallelic noncomplementation of null alleles. Developmental Biology, 213(2), 418–431. [DOI] [PubMed] [Google Scholar]
- Yamagishi H (2020). Cardiac Neural Crest. Cold Spring Harbor Perspectives in Biology. 10.1101/cshperspect.a036715 [DOI] [PMC free article] [PubMed] [Google Scholar]



