Abstract
Regular exercise has a central role in human health by reducing the risk of type 2 diabetes, obesity, stroke and cancer. How exercise is able to promote such systemic benefits has remained somewhat of a mystery but has been thought to be in part mediated by the release of myokines, skeletal muscle-specific cytokines, in response to exercise. Recent studies have revealed skeletal muscle can also release extracellular vesicles (EVs) into circulation following a bout of exercise. EVs are small membrane-bound vesicles capable of delivering biomolecules to recipient cells and subsequently altering their metabolism. The notion that EVs may have a role in both skeletal muscle and systemic adaptation to exercise has generated a great deal of excitement within a number of different fields including exercise physiology, neuroscience and metabolism. The purpose of this review is to provide an introduction to EV biology and what is currently known about skeletal muscle EVs and their potential role in the response of muscle and other tissues to exercise.
Keywords: exercise, extracellular vesicles, microRNAs, skeletal muscle
Graphical Abstract
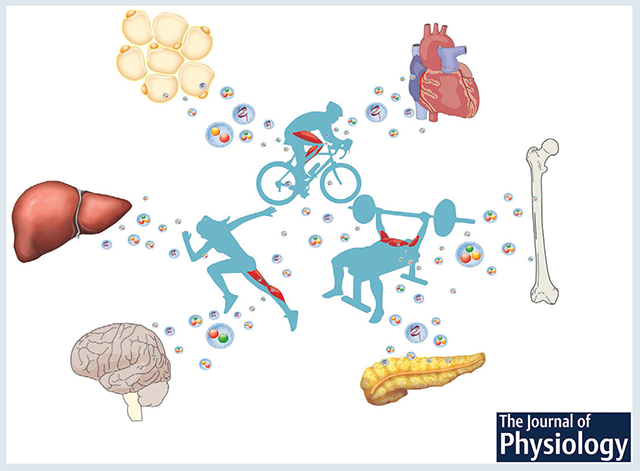
Physical activity promotes the release of extracellular vesicles from skeletal muscle into circulation which are subsequently taken up by other tissues. Extracellular vesicle uptake results in the delivery of biomolecules, such as microRNAs, which alters the metabolism of recipient cells. This mechanism is proposed to provide a basis for how regular exercise is able to improve overall health.
Introduction
Regular exercise is known to improve health by reducing the risk of type 2 diabetes, obesity, stroke and some forms of cancer (Garber et al. 2011). How exercise is able to bring about such remarkable changes in different tissues and organs of the body remains poorly understood (Neufer et al. 2015). To address this gap in knowledge, NIH, through the Common Fund, has provided $170M to establish the Molecular Transducers of Physical Activity Consortium (MoTrPAC). The goal of MoTrPAC is ‘… to catalogue the biological molecules affected by exercise in people, to assemble a comprehensive map of the molecular changes that occur in response to movement and, when possible, relate these changes to the benefits of physical activity.’ To complement the human studies, MoTrPAC has also established preclinical rodent sites which will allow for a more in-depth analyses of the biological changes that occur with exercise. The creation of MoTrPAC brings into sharp focus the recognition that exercise has a central role in human health and well-being.
One mechanism thought to mediate the systemic benefits of regular exercise is through the release of cytokines from contracting skeletal muscle known as myokines (Neufer et al. 2015). Proposed by Goldstein almost 60 years ago, muscle was viewed as an endocrine-like organ, releasing an unidentified humoral factor involved in regulating blood glucose in response to exercise (Goldstein, 1961). Years later, Pederson and colleagues identified the first myokine, interleukin-6 (IL-6), which was shown to be secreted into the bloodstream in response to muscle contractions (Pedersen et al. 2001, 2003; Pedersen & Fischer, 2007a,b ). Many other myokines have been identified since then such as myostatin, IL-15, BDNF (brain-derived neurotrophic factor) and irisin (Pedersen, 2011). Hartwigand colleagues catalogued the secretome of human primary muscle cells, identifying 548 non-redundant proteins in conditioned media, with 305 proteins classified as potential myokines (Hartwig et al. 2014).
In addition to myokines, extracellular vesicles (EVs) have emerged as a potential mechanism through which the beneficial effects of regular exercise are transmitted to other tissues. EVs are membrane-bound vesicles with the capacity to transfer functional biomolecules such as lipids, proteins, nucleic acids and sugars, thereby altering recipient cell function (Ratajczak et al. 2006; Valadi et al. 2007; Skog et al. 2008). EVs are currently divided into three subtypes: exosomes (50–150 nm in diameter), microvesicles (100–1000 nm) and apoptotic bodies (50–5000 nm) (see Fig. 1). Given the lack of specific markers for each EV subtype, the International Society for Extracellular Vesicles has suggested the use of the generic term ‘EVs’ for vesicles (delimited by a lipid bilayer) naturally released from the cell (Thery et al. 2018). Regardless of the current discussion about subtypes, EVs have been shown to mediate cell-cell communication through the transfer of functional cargo to recipient cells, expanding our understanding of how skeletal muscle (and other tissues) communicates to other organs.
Figure 1. Schematic representation of extracellular vesicle subtypes: exosomes, microvesicles and apoptotic bodies.
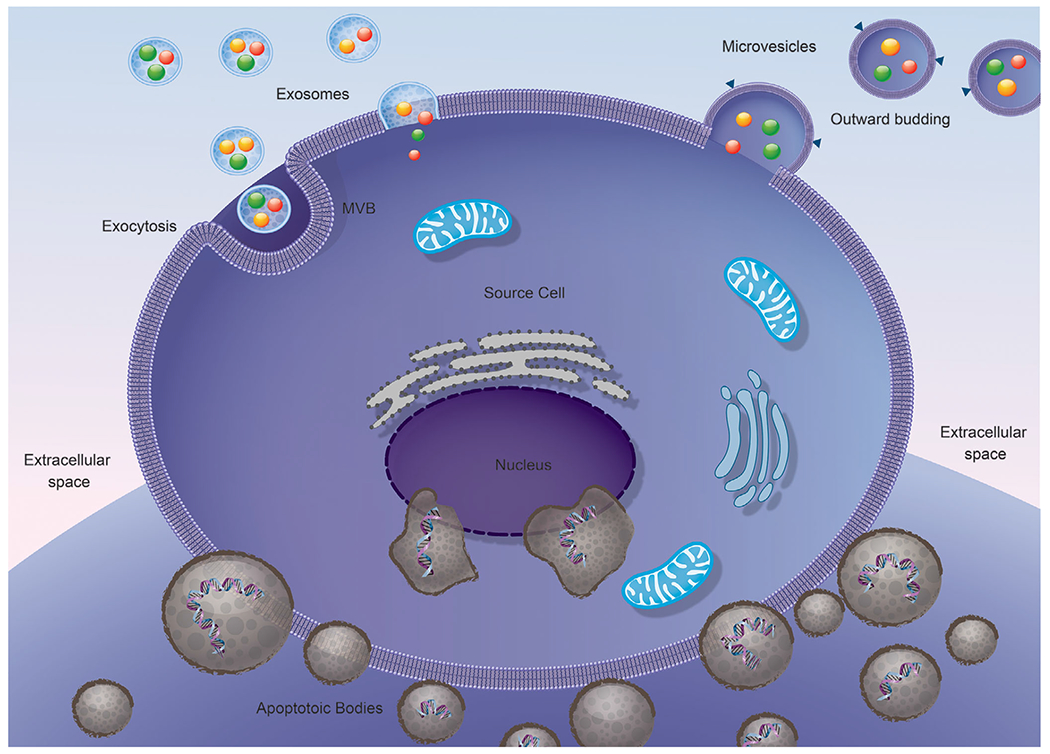
Illustration showing the release of extracellular vesicle subtypes, namely exosomes, microvesicles and apoptotic bodies through various mechanisms mainly outward budding and exocytosis.
Brief history
The intense interest in EVs belies the half-century that has passed since the original description of membrane-bound vesicles being released from cells into the extracellular environment. In the late 1960s, the first studies describing the release of EVs from either platelets or chondrocytes were published (Wolf, 1967; Anderson, 1969). A fewyears later, Trams and colleagues (Trams et al. 1981), coined the term exosome in their study showing that both normal and neoplastic mammalian cells were capable of secreting vesicles containing 5’-nucleotidase enzymatic activity that ranged in size from 40 to 1000 nm and originated directly from the plasma membrane. In contrast to this release mechanism, two independent studies (Harding et al. 1983; Pan & Johnstone, 1983) in the early 1980s provided the first evidence showing a more complex vesicle secretion pathway. These studies found the transferrin receptor (TfR) was recycled from the plasma membrane of maturing reticulocytes by selective endocytosis through internalization into a so-called multivesicular endosome (currently known as multivesicular bodies, MVBs) and then subsequently secreted from the cell. In a follow-up study, Johnstone and colleagues (Johnstone et al. 1987) proposed the term exosome for vesicles pelleted at 100,000 g of endosomal origin and released upon fusion of MVBs with the plasma membrane. As a result of these early studies, exosome biogenesis was thought to occur primarily through the endosomal pathway, though more recent work has shown that exosomes can be also secreted directly from the plasma membrane (Booth et al. 2006; Fang et al. 2007; Shen et al. 2011).
The early studies on EVs viewed them as a mechanism for the cell to dispose of unwanted cellular components. It was not until the late 1990s that this view of EVs began to change, when it was shown that EVs were able to transfer protein, RNA and even organelles from one to cell to another thereby acting as mediators of cell-cell communication (Raposo et al. 1996; Mesri & Altieri, 1998; Zitvogel et al. 1998; Hess et al. 1999). While the cancer field had described in significant detail tumor-derived EVs and their ability to transfer membrane receptors, protein and mRNA to recipient cells, it was the study by Valadi and coworkers that is often credited with kick-starting the current excitement about EVs and their potential role in intercellular communication (Ratajczak et al. 2006; Valadi et al. 2007). It is not clear why this study in particular generated so much interest in EVs but it might be the fact that it was the first study to show that EVs contained functional microRNAs (miRNA) capable of regulating target cell gene expression (Valadi et al. 2007). Since then, the interest in EVs as a vehicle for intercellularcommunication has increased exponentially. The purpose of this review is to discuss our current understanding of EV biogenesis and cellular uptake, and the emerging field of skeletal muscle EVs and their potential role skeletal muscle and systemic adaptation to exercise.
Extracellular vesicle biogenesis
It is well established that virtually all cells of the body can secrete distinct types of EVs (Deatherage & Cookson, 2012). The different types of EVs are generated at specific subcellular locations and share common as well as distinct features. As the methods used to isolate EVs has advanced, it has become clear the EV population is a heterogeneous mixture of small vesicles. It is also important to note that the current methods used to isolate EVs are not capable of isolating pure populations of each EV subtype, especially if only a single method is used (Kowal et al. 2016). Given this limitation, it is important to keep in mind that most of the biological findings regarding cell-cell communication may not necessarily be induced by exosomes alone. Besides isolation, the biogenesis process is also complex and is not completely understood. For example, most of the current literature describes the biogenesis of exosome and microvesicles as being completely different; microvesicles are made through budding off of the plasma membrane, while exosomes are formed by an endocytic process, followed by exocytosis. While several lines of evidence support this distinction, there is also evidence suggesting that exosome biogenesis could occur directly at the plasma membrane (Anderson et al. 2005; Shen et al. 2011) (see Fig. 2).
Figure 2. Pathways involved in extracellular vesicle biogenesis.
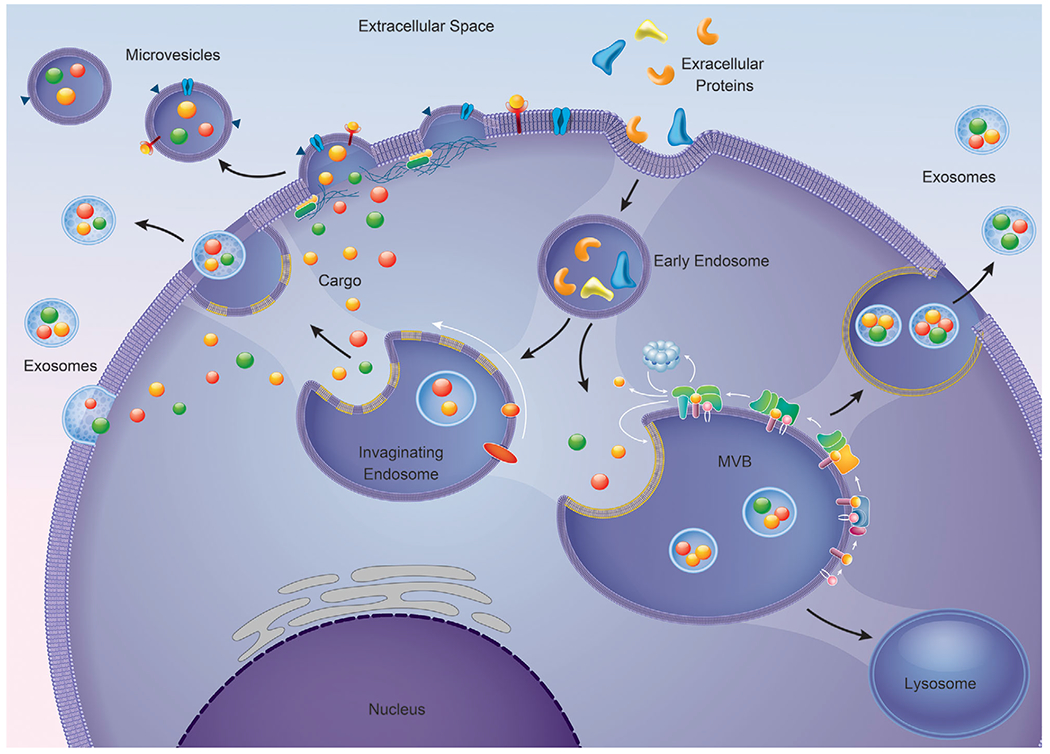
In the classical pathway, extracellular proteins can be internalized to form early endosomes. Invagination of the early endosome are responsible for the origin of the intraluminal vesicles (ILV)/multivesicular body (MVB). The MVB can fuse directly to the plasma membrane releasing its content, exosomes, or fuse with the lysosomes and have its content degraded. Another mechanism of exosomes biogenesis is direct outward budding of the plasma membrane. Microvesicle biogenesis occurs by the generation of small cytoplasmic protrusions that cause the cellular membrane to pinch off giving rise to microvesicles.
Microvesicle biogenesis
Microvesicles, also known as shedding vesicles, ectosomes and oncosomes, are important vehicles for intercellular communication (Cantaluppi et al. 2012; Kalra et al. 2016). Microvesicles have been studied for more than three decades but only recently have gained considerable attention (Tkach et al. 2018). Usually reported in the range of 100–1000 nm in size, microvesicles are membrane-bound vesicles that are shed from the plasma membrane (Kalra et al. 2016). Although the biogenesis of microvesicles remains to be described in complete detail, prior to shedding, small cytoplasmic protrusions are generated which cause a pinching of the cell membrane by a fission event, releasing the nascent microvesicle into the extracellular space (Cocucci et al. 2007; Kalra et al. 2016). The cytoplasmic protrusions which initiate the release of microvesicles are usually distinct and thought to be caused by localized changes in the protein and lipid composition of the plasma membrane such that it promotes membrane curvature and rigidity (McMahon & Boucrot, 2015). While microvesicle biogenesis is not associated with the endosomal pathway, the budding process requires endosomal machinery for its formation (Tricarico et al. 2017). Nabhan and colleagues reported that components of the endosomal sorting complexes required for transport (ESCRT) machinery, particularly TSG101, were able to promote microvesicle formation (Nabhan et al. 2012). They found that membrane budding was driven by a specific interaction of TSG101 with a tetrapeptide PSAP motif of an accessory protein, arrestin domain-containing protein 1 (ARRDC1). This interaction caused the relocation of TSG101 from endosomes to the plasma membrane, facilitating the release of microvesicles that contained TSG101, ARRDC1 and other cellular proteins. Another protein associated with the endosomal pathway that appears to be involved in microvesicle formation is the Ras-related GTPase ADP-ribosylation factor 6 (ARF6) (Muralidharan-Chari et al. 2009). In this study, the authors showed that ARF6 activated phospholipase D, leading to the activation of extracellular-signal-regulated kinase (ERK). Once activated, ERK phosphorylated myosin light-chain kinase (MLCK), which in turn activated myosin light chain via phosphorylation, leading to microvesicle release subsequent to actomyosin contraction of the microvesicle ‘neck’.
In addition to proteins known to have a role in promoting membrane curvature, there is evidence to suggest that protein crowding at the plasma membrane itself may be sufficient to induce microvesicle formation via membrane curvature. For instance, green fluorescent protein (GFP) has been shown to be capable of membrane bending when concentrated within the membrane (Stachowiak et al. 2012; Snead et al. 2017). While more studies are necessary to determine if protein crowding alone is sufficient to generate microvesicles, the enrichment of protein cargo at sites of microvesicle budding may be adequate to drive de novo microvesicle formation (Tricarico et al. 2017).
Exosome biogenesis
The primary mechanism thought to be involved in exosome biogenesis is the inward invagination of late endosome/multivesicular bodies (MVBs) which give rise to intraluminal vesicles (ILVs); upon fusion with the plasma membrane, ILVs are released into the extracellular space as exosomes. ESCRT is the molecular machinery employed by the cell to generate exosomes (de Gassart et al. 2004; Colombo et al. 2013). The ESCRT machinery is composed of approximately 30 proteins that assemble into four complexes (ESCRT-0, -I, -II and -III) (Babst et al. 1998, 2002a,b; Katzmann et al. 2001, 2003; Thery et al. 2001). All these complexes are involved in the formation of the MVBs and ILVs, and in the sorting of proteins. ESCRT-0 recognizes phosphatidylinositol 3-phosphate and ubiquitinated proteins and sorts them to the endosomal membrane. Specifically, a component of ESCRT-0, HRS (hepatocyte growth factor-regulated tyrosine kinase substrate, official gene symbol Hgs), binds to ubiquitinated proteins and then associates with another ESCRT-0 component, STAM (signal transducing adaptor molecule) and clathrin (not-ESCRT protein). This complex recruits ESCRT-I (Tumor Susceptibility 101, TSG101) to the endosomal membrane, resulting in formation of the ESCRT-0/ESCRT-I complex. The ESCRT-0/ESCRT-I complex mobilizes ESCRT-II (Vps22), initiating reverse budding of ILVs within MVBs and cargo uptake. Finally, ESCRT-II recruits ESCRT-III (ALIX and Vps2) which promotes vesicle closure and detachment of ILVs from the membrane (Fernandez-Borja et al. 1999; Hurley, 2010; Baietti et al. 2012; Hanson & Cashikar, 2012; Nabhan et al. 2012; Bissig & Gruenberg, 2014; Chiaruttini et al. 2015; Kalra et al. 2016; Wenzel et al. 2016). Somewhat surprising was the demonstration that MVBs are still capable of forming in the absence of all four ESCRT components (Stuffers et al. 2009), thus revealing an ESCRT-independent pathway for exosome biogenesis. Confirmation of an ESCRT-independent pathway came from the work of Trajkovic and colleagues who showed exosome biogenesis was promoted by the conversion of sphingomyelin to ceramide by neutral sphingomyelinase (Trajkovic et al. 2008). The depletion of neutral sphingomyelin in cancer cells, however, did not prevent the production of exosomes, indicating that the synthesis of ceramide under particular conditions is not an absolute requirement for the production of exosomes (van Niel et al. 2011). Finally, a more recent study reported a novel role for the small integral membrane protein of the lysosome/late endosome (SIMPLE) protein in exosome biogenesis through regulating MVB formation (Zhu et al. 2013).
While the majority of studies have focused on the molecular mechanism involved in the endosomal biogenesis pathway, there is evidence suggesting that exosomes can also bud directly from the plasma membrane (Anderson et al. 2005; Booth et al. 2006; Fang et al. 2007; Shen et al. 2011; Bianchi et al. 2014; Nager et al. 2017). For example, electron microscopic evidence has shown stem cells budding small vesicles (60–100 nm in diameter) directly from the plasma membrane (Cantaluppi et al. 2012). Additionally, transmission electron microscopy of glioblastoma cells also revealed budding of small vesicles (~80 nm) from the plasma membrane. Consistent with these observations, proteins typically associated with specific regions of the plasma membrane (e.g. tetraspanin CD9 and CD81) have been shown in exosomes (Halova & Draber, 2016). Collectively, these findings clearly showthat exosome biogenesis can occur at the plasma membrane. What remains to be determined is the contribution of each of these different biogenic pathways to total exosome production within the cell and how the role of each pathway involved in exosome biogenesis may differ between cell types (i.e. myofibre and satellite cells) and in response to different stimuli.
Extracellular vesicle uptake
The discovery that EVs were capable of inducing a physiological change in recipient cells through the delivery of functional cargo, generated a great deal of interest in EVs as mediators of cell-cell communication (Valadi et al. 2007; Alvarez-Erviti et al. 2011; Wahlgren et al. 2012; Lee et al. 2018). The exact manner through which EVs are taken up by recipient cells is still under debate, though several possible mechanisms for EV uptake have been reported.
Endocytosis is a complex process that comprises several molecular pathways involved in the cellular uptake of EVs. The different endocytic pathways include clathrin-mediated endocytosis, caveolin-dependent endocytosis, macropinocytosis and phagocytosis (see Fig. 3). Clathrin-mediated endocytosis involves cellular internalization of molecules through the assembly of clathrin-coated vesicles. These vesicles deform the membrane thereby triggering an inward budding of the plasma membrane followed by fusion with the endosome (Kaksonen & Roux, 2018). Caveolin-dependent endocytosis is a clathrin-independent endocytic process involving plasma membrane invaginations called caveolae which are ultimately internalized by the cell (Kiss & Botos, 2009). Macropinocytosis involves the formation of membrane ruffles that pinch off into the extracellular space, surrounding an area of extracellular fluid which is subsequently internalized by the cell (Lim & Gleeson, 2011). Phagocytosis is the process by which the cell uses the plasma membrane to ingest large particles (>0.5 μm), creating a membrane-bound vesicle called a phagosome (Richards & Endres, 2014). All of these endocytic pathways have been shown to participate in the cellular uptake of EVs (Svensson et al. 2013; Tian et al. 2014); however, there is currently no data on whether one of these mechanisms is preferentially used for the uptake of skeletal muscle-derived EVs. While speculative, it may be that EV internalization could be cell type-specific or dependent upon the physiological status of the recipient cell. It is also likely that the heterogeneity of EVs will have an influence on the mechanism of uptake used by the recipient cell.
Figure 3. Mechanism of EV uptake by recipient cell.
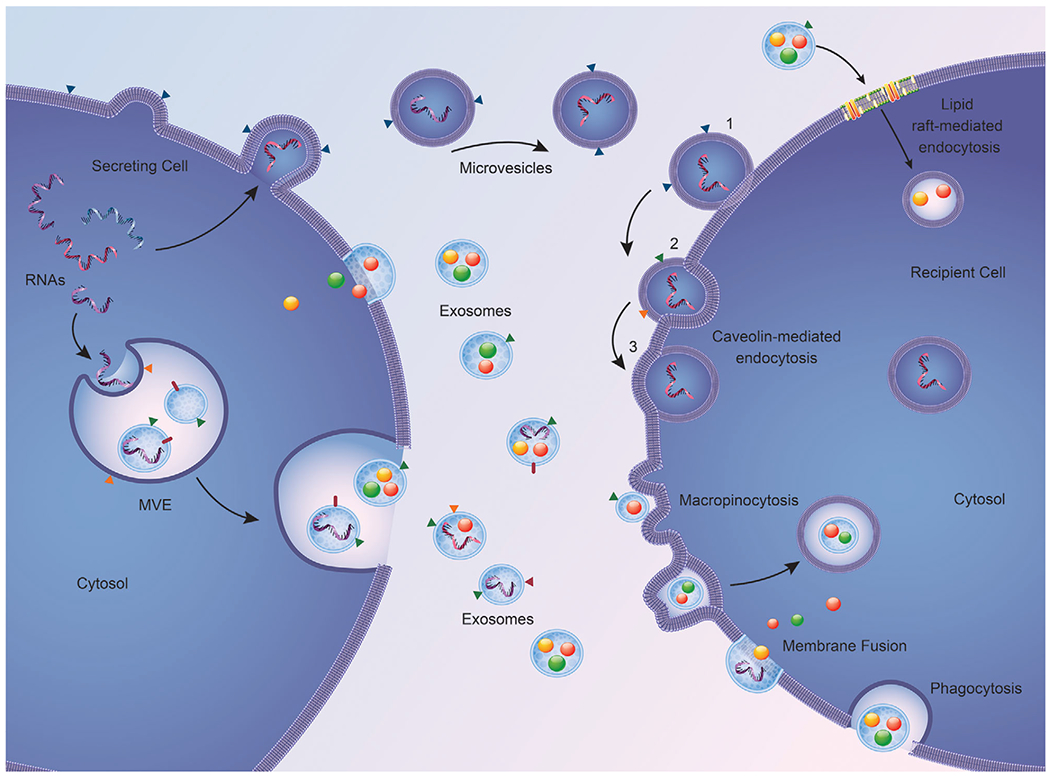
After release, EVs can be taken up by the recipient cell through: (1) lipid rafts that facilitate endocytosis, (2) caveolin-mediated endocytosis, which involves invaginations of the plasma membrane resulting in EV internalization, (3) micropinocytosis, which occurs when the membrane forms ruffles that surround the extracellular space and engulf the incoming EV, leading to entry into the cell, and (4) phagocytosis of the EV or alternatively fusion of the EV directly to the plasma membrane.
Extracellular vesicle isolation methods
Although the field has evolved considerably in the last few years, there still a lot of open questions regarding EV biology. Perhaps one of the main reasons for the limited knowledge is the challenges in isolating a single EV subtype. Most of the current methods for EV isolation are not capable of recovering a single EV population, mainly because the majority of isolation methods used are based on a single characteristic (e.g. size, buoyant density). Since many characteristics can be shared among different vesicles, the use of a method that relies on single characteristic for EV isolation will result in the recovery a mixed EV population (Van Deun et al. 2014; Kowal et al. 2016), thus preventing a clear understanding of functional properties of a specific EV subtype. Here, we discussed the most common methods used for EV isolation highlighting the approaches and respective limitations.
Differential ultracentrifugation
Ultracentrifugation (UC) is one of the most commonly used and reported methods for EV isolation. It consists of cycles of different centrifugal force to fractionate small bioparticles such as viruses, bacteria, subcellular organelles and extracellular vesicles (Li et al. 2017). For EV-based UC isolation, differential centrifugation steps with increased centrifugal force are used to produce sequential pellets of cells and cell debris (300–1000 g), microvesicles (10,000–20,000 g) and exosomes (100,000 g). To improve the purification of the samples, serial filtration through 0.22 and 0.45 μm filters before pelleting have been used (Thery et al. 2006). Although it has long been considered the gold standard technique for EV isolation due to its simplicity and minimal sample preparation, UC does not allow for a complete separation of EV population. Additionally, due to the high centrifugal forces applied to the vesicles, deformation of EV structure and aggregations are formed during the isolation process (Linares et al. 2015). The efficiency of EV isolation depends on several factors (e.g. acceleration, type of rotor, viscosity of the samples) that should be taken into consideration when using UC methods. For example, EV isolation from plasma/serum requires higher-speed UC and longer times compared with cell culture media (Thery et al. 2006).
Density gradient ultracentrifugation
Density gradient ultracentrifugation (DGUC) is perhaps one of the best methods for isolating EVs. There are several different adaptations for this method, but essentially DGUC exploits the buoyant density of particles in order to isolate heterogeneous EV/particle population (Thery et al. 2006). Under a specific centrifugal force, the different components of the sample will settle to their iso-density zone, thus achieving the separation of exosomes (~1.10—1.18 g ml−1) from other components in the sample. The density gradient can utilize sucrose or iodixanol, the latter preserves the size of EVs whilst in the gradient, improving on some of the limitations of the sucrose gradient (Li & Donowitz, 2014; Gupta et al. 2018; Li et al. 2018; Duong et al. 2019). The addition of the gradient to the UC method allows for a higher separation efficiency, thus improving the purity of the isolate. Additionally, if the gradient is added prior to UC, the particles are unlikely to be deformed or to form aggregates due to the high force applied during centrifugation (Duong et al. 2019). Although DGUC has several benefits in terms of isolating a purer EV population, it does present some limitations. For example, this method is time consuming (i.e. 16–90 h), requires specialized techniques, a substantial amount of sample, and can lead to co-isolation of high density lipoprotein (HDL) (Yuana et al. 2014).
Precipitation of extracellular vesicles
The isolation of exosomes by polymer-based reagents, like ExoQuick by System Biosciences, a proprietary polyethylene glycol (PEGs) precipitation reagent, have become a popular method for isolating EVs. These polymer-based reagents work by binding up water molecules and forcing the less-soluble vesicles (i.e. smaller vesicles, <200 nm) out of solution, followed by bench top centrifugation (Zeringer et al. 2015). This polymer-based solution can be optimized and used across different applications (i.e. in vivo vs. in vitro), as well as a wide range of biological fluids (i.e. plasma, serum, saliva, milk and urine). However, a major disadvantage of this application is that various sized EVs and other particles, like lipoproteins and protein aggregates, are blended together, thus making the definitive isolation and recognition of a specific size or population of particles unattainable.
Immuno-based capture of extracellular vesicles
The immuno-based capturing of EVs is made possible due to the detection of tetraspanin proteins (i.e. CD9, −63, −81), which are entwined within the membrane of EVs, using immunoaffinity-based detection with specific antibodies. The specificity and yield of EVs isolated using this method have been compared to yields reported using ultracentrifugation (Zarovni et al. 2015). One way of optimizing this method is to incorporate the use of submicron-sized magnetic beads which are coated with an antibody of choice (i.e. CD9, −63, −81) to allow for higher particle capture efficiency by expanding the detectable surface area. A potential limitation of this method is knowing whether the EV population of interest expresses the membrane protein used for immune-capture (Hong et al. 2014; Zarovni et al. 2015).
Ultrafiltration
The ultrafiltration (UF)-based EV isolation method consists of commercial membrane filters with a narrow range of pore sizes, allowing particles of a specific size to pass through or be intercepted. UF can be used independently or in conjunction with other techniques such as UC. Compared to UC technology, UF is less time consuming, recovers more EVs per sample volume and is suitable for isolating EVs in different volumes. Additionally, UF does not requires high pressure, reducing the likelihood of EV rupture aggregates (Yu et al. 2018). Although UF demonstrates many advantages in EV isolation, the inability to isolate a specific EV population remains a problem with this method, since all vesicles with similar size would be co-isolated.
Size exclusion chromatography
Size exclusion chromatography (SEC) or gel filtration uses a porous polymer typically composed of sepharose to separate molecules based on size. The polymer is typically referred to as the stationary phase and there are different sepharose matrices that can be utilized depending on the particle size of interest (Monguio-Tortajada et al. 2019). Although SEC allows for a better separation compared to precipitation methods (Gamez-Valero et al. 2016), depending on the complexity of the biological samples (e.g. serum/plasma), it may not provide the best means for isolation. For example, size-based isolation can lead to contamination with other non-vesicle particles of comparable size, which decreases the purity of the sample (Sodar et al. 2016).
Skeletal muscle as an endocrine organ
As one of the largest organs in the body, skeletal muscle represents approximately 40% of the body weight and is the largest protein reservoir in the body. In addition to its primary function in locomotion, skeletal muscle also has an important role in thermoregulation and whole-body metabolism. Over the last few decades, evidence has emerged that skeletal muscle can also function as an endocrine organ (Pedersen & Febbraio, 2012). The ability of skeletal muscle to communicate with distal tissues often occurs in response to muscle activity whereby released myokines are able to stimulate the production of glucose and lipolysis (Karsenty & Olson, 2016). Specifically, skeletal muscle has been shown to release cytokines, referred to as myokines, and/or exerkines (Pedersen et al. 2003; Pedersen & Febbraio, 2012; Whitham & Febbraio, 2016), which can act in an autocrine (e.g. interleukin-6, IL-6), paracrine (e.g. myostatin) or endocrine manner. Myokines have been reported to be taken up by various tissues such as adipose tissue (Leal et al. 2018), liver (Febbraio et al. 2004), pancreas (Plomgaard et al. 2012; Guo et al. 2017), brain (Pedersen et al. 2009) and skeletal muscle itself (Pedersen et al. 2007; Pedersen & Febbraio, 2008; Pedersen, 2009; Walsh, 2009). A large body of research has been published by the Pedersen laboratory (Febbraio & Pedersen, 2002; Febbraio et al. 2004; Pedersen & Febbraio, 2008; Broholm & Pedersen, 2010; Christiansen et al. 2010; Brandt et al. 2012) detailing how myokines such as IL-6 can induce an anti-inflammatory response both during and following a single or repeated bouts of exercise. Furthermore, it has been established that skeletal muscle possesses a secretome which consists of several hundred peptides (Bortoluzzi et al. 2006; Yoon et al. 2009; Henningsen et al. 2010) and conceptually provides a basis for how skeletal muscle may be communicating with other tissues within the body.
The modalities through which skeletal muscle can function as an endocrine organ has expanded to include EVs (Rome et al. 2019; Trovato et al. 2019). A recent study by Whitman and coworkers showed 1 h of cycling induced a significant increase in systemic EVs that were taken up by the liver. Additionally, Frtihbeis and colleagues reported systemic EV concentration was higher immediately following a single bout of exhaustive endurance exercise, with EV size and composition differing during the early recovery phase despite EV abundance remaining elevated (Fruhbeis et al. 2015). Thus, exercise serves as a strong stimulus for promoting the release of EVs into circulation, with the intensity of exercise influencing how many EVs are presumably released by skeletal muscle. A current challenge for the in vivo study of skeletal muscle-derived EVs is the inability to specifically label and track EVs released from skeletal muscle. As a proxy, researchers have used the presence of muscle-specific proteins or microRNAs to assess the change in muscle-derived EVs in response to exercise. A promising approach for isolating skeletal muscle-specific EVs was described by Guescini and colleagues, who showed that up to 5% of systemic EVs were positive for the muscle-enriched a-sarcoglycan protein and contained high levels of the skeletal muscle-specific microRNA, miR-206 – further confirmation that the α-sarcoglycan enriched EVs were derived from skeletal muscle (Guescini et al. 2015). Despite the aforementioned challenges to studying skeletal muscle-derived EVs in vivo, numerous in vitro studies have clearly demonstrated that both myoblasts and myotubes (Le Bihan et al. 2012; Romancino et al. 2013; Forterre et al. 2014a,b; Choi et al. 2016) are capable of releasing EVs. Early work in determining the cargo of EVs revealed that muscle-derived microvesicles cargo contained RNA that clustered into 6 functional groups: zinc finger C2H2-type, integral to membrane ionic channel, transcription, protein secretion, ion transport and signal transduction (Le Bihan et al. 2012). Forterre and colleagues went on to demonstrate that both myoblasts and myotubes were capable of secreting EVs with specific protein and microRNA composition that differed from the intracellular composition (Forterre et al. 2014b). Furthermore, they also showed that the cargo contained within myotube-derived EVs could be transferred to myoblasts and subsequently regulate gene expression involved in myoblast differentiation (Forterre et al. 2014a). This early work revealed that the loading of EVs does not necessarily reflect the intracellular composition of the cell but appears to be regulated by a yet-to-be described mechanism. A nice example of the selective loading of EVs was the study by Hudson and co-workers showing dexamethasone-induced myotube atrophy caused a significant decrease in myotube miR-23a expression that was paralleled by an increase in the abundance of EV miR-23a (Hudson et al. 2014). This finding demonstrated that miR-23a was being selectively exported from myotubes by EVs, which in turn led to the de-repression of atrogin-1 and MuRF-1, target genes of miR-23a known to promote muscle atrophy.
Guescini and co-workers were the first to describe exercise-induced release of EVs from skeletal muscle (Guescini et al. 2015). Presumptive skeletal muscle-derived EVs were immune-captured using an antibody against α-sarcoglycan, a protein highly enriched in skeletal muscle. Alpha-sarcoglycan-positive EVs represented approximately 2–5% of the total EV population and were highly enriched for the skeletal muscle-specific microRNA miR-206. These authors found that after an acute bout of endurance exercise there was a significant increase in the abundance of EV miR-181a-5p but no change in the abundance of miR-1, miR-133b, miR-206, miR-499 or miR-146a within EVs (Guescini et al. 2015). Subsequent studies confirmed the increase in serum EV abundance in response to exercise and the presence of EV miRNAs (Fry et al. 2017; Lovett et al. 2018; Oliveira et al. 2018; D’Souza et al. 2018a). Oliveira and colleagues observed a correlation between miRNA content in EVs and exercise intensity, proposing that intensity may play a role in the packaging and release of EVs (Oliveira et al. 2018). This finding was consistent with an earlier study showing that the kinetics of EV release into circulation correlated with the increase in workload (Fruhbeis et al. 2015), suggesting a sufficient exercise intensity was required to detect a significant increase in EVs within systemic circulation. This concept was supported by another study that found different modalities of exercise were able to affect the release of muscle-specific miRNAs (Uhlemann et al. 2014). For example, 4 h of cycling at 70% of the anaerobic threshold induced a significant increase in plasma miR-126 levels after 30 min, which persisted for the duration of the bout with no change in miR-133 levels. In contrast, plasma miR-133 and miR-126 levels were significantly higher following a marathon race (Uhlemann et al. 2014). The variability in the release of miRNAs into circulation was also shown when participants engaged in three different types of resistance training bouts (Cui et al. 2017). It should be noted that these studies did not specifically isolate EVs, rather they looked at miRNAs in the plasma; however, these results indicate that exercise mode and intensity are likely to be important factors to consider when investigating the release of EVs into circulation.
The identification of presumptive EV cargo was performed by Whitham and colleagues who used nano-ultra-high-performance liquid chromatography tandem mass spectrometry analysis to identify serum proteins from human participants after a bout of endurance exercise (Whitham et al. 2018). They found a significant change in the abundance of 322 proteins between rest and following exercise. Gene ontology analysis revealed enrichment of proteins with the keyword ‘glycolysis’, consistent with previous work showing that EVs can deliver glycolytic enzymes to recipient cells and alter their glycolytic rate (Garcia et al. 2016; Zhao et al. 2016; Whitham et al. 2018). While Whitham and colleagues demonstrated an exciting possible cross-talk between skeletal muscle and liver during acute exercise, the method used to isolate EVs should be taken into consideration when interpreting the results. As described in our previous section, the isolation process is still a major challenge for the field in terms of isolating a pure EV population. Based on previous studies characterizing exosomes, employing the method (2 × 1 h centrifugation at 20,000 g) described by Whitham and coworkers for human plasma EV isolation would be expected to yield exosomes as well as other vesicles such as microvesicles and apoptotic bodies (Kowal et al. 2016; Jeppesen et al. 2019).
As the preceding discussion highlights, the skeletal muscle EV field is still in its infancy, with only a handful of studies investigating how exercise affects the presumptive release of EVs from skeletal muscle, the identification of their protein and/or RNA cargo and what cells take up EVs following exercise. The study by Fry and coworkers showed activated satellite cells were required for the proper remodelling of the extracellular environment during skeletal muscle hypertrophy (Fry et al. 2017). In particular, myogenic progenitor cells were shown to release EVs which were taken up by fibroblasts, leading to down-regulation of Rrbp1 via muscle-specific miR-206; Rrbp1 is known to be a master regulator of collagen production. Through this mechanism, the remodelling of the extracellular matrix was properly regulated; however, when satellite cells were conditionally depleted, this regulatory mechanism was not operative, which caused fibrosis within the muscle as a result of collagen over-production. Most recently, Yin and co-workers demonstrated that in response to downhill running miR-1, miR-133a/b, miR-206, miR-208a and miR-499 all significantly increased in serum EVs immediately after exercise in rats (Yin et al. 2019). Interestingly, these results were not conserved after a bout of uphill running, suggesting that the EV-mediated secretion of myomiRs (muscle-specific microRNAs) into circulation are exercise-mode dependent. Furthermore, the myomiR changes after exercise are not uniform across different skeletal muscles. The downhill protocol triggered significant fluctuations of miR-1 in both the quadriceps and gastrocnemius muscles. In addition, the uphill running protocol prompted significant changes in the amount of miR-1 and miR-499 in the gastrocnemius only. However, miR-1 levels did not change in the soleus or cardiac tissue in response to either downhill or uphill running. These results suggest the release of EVs could be fibre-type or muscle dependent, owing to specific roles of EVs and their cargo in the variability of adaptation of skeletal muscle in response to exercise. Therefore, specific skeletal muscle adaptations to exercise may rely on the muscle fibre’s ability to release or uptake EV functional cargo.
To further explore a possible way in which exercise-induced EVs participate in cell-cell communication, we compiled a list of previously published miRNAs that were isolated from plasma EVs after a bout of exercise (see Table 1). We then ran a prediction analysis to determine possible targets of EV carrying miRNAs (Fig. 4). Our prediction analysis totaled 5486 possible targets of the 36 miRNAs identified in EVs. We next uploaded this gene set into gene ontology and found that the top two most significantly enriched biological processes of these genes were (1) the response to reactive oxygen species and (2) insulin secretion. The data from our analysis provide a somewhat broad idea of the possible systemic effects of exercise-induced EVs, specifically in relation to EVs carrying miRNAs. These pathways have been previously discussed in their response to exercise (He et al. 2016; Barlow & Solomon, 2018). For example, studies have shown that exercise can improve pancreatic β-cell function (Dela et al. 2004; Malin et al. 2013) and increase antioxidant enzymatic activity (Berzosa et al. 2011). These two examples provide a possible link between exercise-induced EVs and some of the systemic adaptions that have been established. While our analysis focused on the miRNA within EVs, the benefits of exercise-induced EVs could be mediated by other types of cargo.
Table 1.
Extracellular vesicle cargo identified from exercise induced EVs release during in vivo studies
| Reference | Model | Type of exercise | Protein | miRNA |
|---|---|---|---|---|
| Bei et al. 2017 | Humans and mice | Exercise stress test | CD63 | N/A |
| Bertoldi et al. 2018 | Rats | Aerobic | CD 63 Acetylcholinesterase 2′,7′-dichloroflurescein diacetate Superoxide dismutase Amyloid β 1–42 |
N/A |
| D’Souza et al. 2018b | Humans | 10 × 60 s cycling at peak power | miR-1-3p miR-16-5p miR-222-3p miR-23a-3p miR-208a-3p miR-150-5p miR-486-5p miR-378a-5p miR-126-3p miR-23b-3p miR-451a miR-186-5p |
|
| Fruhbeis et al. 2015 | Humans | Incremental cycling or running until exhaustion | Flotillin 1, Hsp70, Integrin α2b | N/A |
| Guescini et al. 2015 | Humans | Acute aerobic, 40 min of vigorous-intensity exercise on a treadmill | Alpha-sarcoglycan | miR-181a-5p miR-1 miR-133a miR-133b miR-206 miR-499 miR-136a miR-24 |
| Lovett et al. 2018 | Human | Workout: plyometric jumping and downhill running 10 sets of 10 jumps followed by 5 sets of 4 min of downhill running |
N/A | miR-1 miR-133a miR-133b miR-206 miR-208b miR-486 miR-499a |
| Oliveira et al. 2018 | Rat | 3 different exercise intensities: Low: 40 min, 14–16m/min Mod: 40m, 20–22m/min High: 40 min, 24–26m/min |
CD63 as an EV marker | miR-128-3P miR-103-3p miR-330-5p miR- 148a-3p miR-191a-5p miR-10b-5p miR-93-5p miR-25-3p miR-142-5P miR-3068-3p miR-142-3p miR-410-3p |
| Yin et al. 2019 | Rat | Downhill running: 90 min, 20m/min, −15% grade. Uphill running: 90 min, 20m/min, + 15% grade |
miR-1 miR-133a miR-133b miR-206 miR-208a miR-499 |
|
| Whitham et al. 2018 | Humans | Cycling exercise, 30 min at 55% V̇O2max, then 20 min at 70% V̇O2 max and until exhaustion at 80% V̇O2 max | 322 proteins identified as significantly different between exercise and rest. With their analysis they found 35 proteins that had significant rates of release either during or after recovery | N/A |
Figure 4. Gene pathway enrichment analysis of the predicted targets of the identified miRNAs found in EVs.
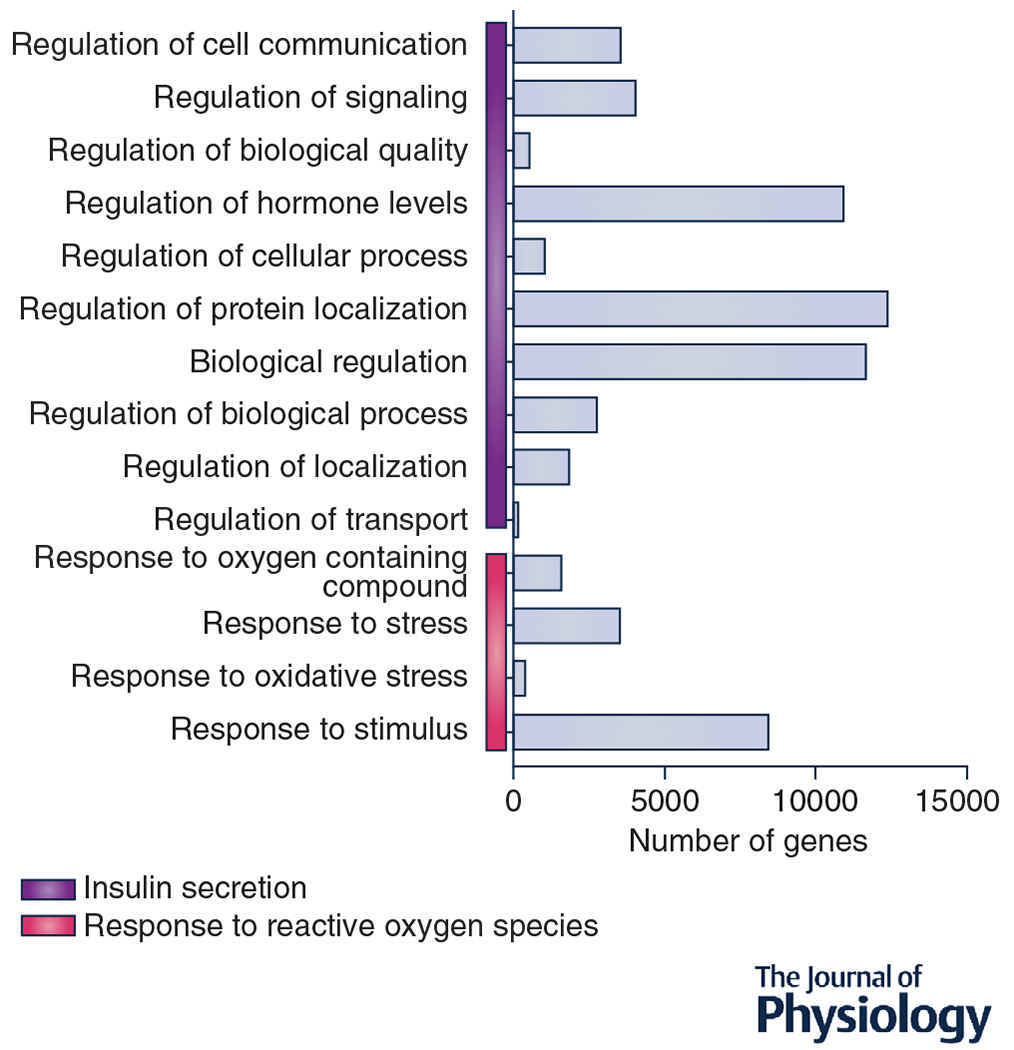
The horizontal bars represent the number of genes in each pathway. The vertical bars represent the major biological pathways. RNAhybrid and Miranda programs were used to determine the predicted target genes of the published miRNAs isolated from exercise-induced EVs.
Final thoughts
The discovery that EVs are capable of delivering their cargo to a recipient cell and altering its metabolism has opened up an exciting new field of investigation into cell-cell communication. That such intercellular communication could provide a novel mechanism for how exercise promotes systemic benefits has generated a deal of interest in the exercise physiology field, as well as other fields such neuroscience, metabolism and cancer. While there remain important challenges (i.e. the isolation and tracking of tissue-specific EVs) for the field, it is balanced by the exciting prospect of developing a deeper, more holistic, understanding of how exercise is able to promote systemic health with skeletal muscle at the centre.
Acknowledgments
Funding
The study on EVs is being supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), Grant 5R01DK119619 to John J. McCarthy.
Biography
Ivan Vechetti obtained his PhD in molecular biology from Sao Paulo State University. He studied molecular pathways associated with aerobic and resistance exercise. He is now working in Dr John McCarthy’s lab on extracellular vesicles, microRNA and skeletal muscle hypertrophy. Taylor Valentino is currently a graduate student in Dr McCarthy’s lab, where his research focus is on the interaction between the gut microbiome and skeletal muscle plasticity. C. Brooks Mobley received his PhD in exercise physiology from Auburn University studying the effects of different supplemental protein sources on skeletal muscle adaptation to resistance exercise. He is currently a postdoctoral scholar in Dr John McCarthy’s lab focusing on the transcriptome differences in myonuclei and satellite cell-derived myonuclei during skeletal muscle hypertrophy. John J. McCarthy received his PhD from the University of Oregon. The primary focus of his lab is to better understand the molecular and cellular mechanism involved in regulating skeletal muscle hypertrophy.
![]()
Footnotes
This review was presented at the 2018 ACSM “Integrative Physiology of Exercise (IPE)” conference, which took place in San Diego, California, 5–8 September 2018.
Competing interests
The authors declare no competing interests.
References
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S & Wood MJ (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29, 341–345 [DOI] [PubMed] [Google Scholar]
- Anderson HC (1969). Vesicles associated with calcification in the matrix of epiphyseal cartilage. J Cell Biol 41, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson HC, Garimella R & Tague SE (2005). The role of matrix vesicles in growth plate development and biomineralization. Front Biosci 10, 822–837. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T & Emr SD (2002a). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B & Emr SD (2002b). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3, 283–289. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ & Emr SD (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17, 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P & David G (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14, 677–685. [DOI] [PubMed] [Google Scholar]
- Barlow JP & Solomon TP (2018). Do skeletal muscle-secreted factors influence the function of pancreatic beta-cells? Am J Physiol Endocrinol Metab 314, E297–E307. [DOI] [PubMed] [Google Scholar]
- Bei Y, Chen T, Banciu DD, Cretoiu D & Xiao J (2017). Circulating exosomes in cardiovascular diseases. Adv Exp Med Biol 998, 255–269. [DOI] [PubMed] [Google Scholar]
- Bertoldi K, Cechinel LR, Schallenberger B, Corssac GB, Davies S, Guerreiro ICK, Bello-Klein A, Araujo ASR & Siqueira IR (2018). Circulating extracellular vesicles in the aging process: impact of aerobic exercise. Mol Cell Biochem 440,115–125. [DOI] [PubMed] [Google Scholar]
- Berzosa C, Cebrian I, Fuentes-Broto L, Gomez-Trullen E, Piedrafita E, Martinez-Ballarin E, Lopez-Pingarron L, Reiter RJ & Garcia JJ (2011). Acute exercise increases plasma total antioxidant status and antioxidant enzyme activities in untrained men. J Biomed Biotechnol 2011, 540458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Doe B, Goulding D & Wright GJ (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C & Gruenberg J (2014). ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24, 19–25. [DOI] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE & Gould SJ (2006). Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol 172, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi S, Scannapieco P, Cestaro A, Danieli GA & Schiaffino S (2006). Computational reconstruction of the human skeletal muscle secretome. Proteins 62, 776–792. [DOI] [PubMed] [Google Scholar]
- Brandt C, Nielsen AR, Fischer CP, Hansen J, Pedersen BK & Plomgaard P (2012). Plasma and muscle myostatin in relation to type 2 diabetes. PLoS One 7, e37236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm C & Pedersen BK (2010). Leukaemia inhibitory factor-an exercise-induced myokine. Exerc Immunol Rev 16, 77–85. [PubMed] [Google Scholar]
- Cantaluppi V, Gatti S, Medica D, Figliolini F, Bruno S, Deregibus MC, Sordi A, Biancone L, Tetta C & Camussi G (2012). Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int 82, 412–427. [DOI] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S & Roux A (2015). Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JS, Yoon HI, Lee KS, Choi YC, Yang SH, Kim IS & Cho YW (2016). Exosomes from differentiating human skeletal muscle cells trigger myogenesis of stem cells and provide biochemical cues for skeletal muscle regeneration. J Control Release 222, 107–115. [DOI] [PubMed] [Google Scholar]
- Christiansen T, Paulsen SK, Bruun JM, Pedersen SB & Richelsen B (2010). Exercise training versus diet-induced weight-loss on metabolic risk factors and inflammatory markers in obese subjects: a 12-week randomized intervention study. Am J Physiol Endocrinol Metab 298, E824–E831. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G, Podini P & Meldolesi J (2007). Enlargeosome traffic: exocytosis triggered by various signals is followed by endocytosis, membrane shedding or both. Traffic 8, 742–757. [DOI] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C & Raposo G (2013). Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 126, 5553–5565. [DOI] [PubMed] [Google Scholar]
- Cui S, Sun B, Yin X, Guo X, Chao D, Zhang C, Zhang CY, Chen X & Ma J (2017). Time-course responses of circulating microRNAs to three resistance training protocols in healthy young men. Sci Rep 7, 2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza RF, Woodhead JST, Zeng N, Blenkiron C, Merry TL, Cameron-Smith D & Mitchell CJ (2018a). Circulatory exosomal miRNA following intense exercise is unrelated to muscle and plasma miRNA abundances. Am J Physiol Endocrinol Metab 315, E723–E733. [DOI] [PubMed] [Google Scholar]
- D’Souza RF, Woodhead JST, Zeng N, Blenkiron C, Merry TL, Cameron-Smith D & Mitchell CJ (2018b). Circulatory exosomal miRNA following intense exercise is unrelated to muscle and plasma miRNA abundances. Am J Physiol Endocrinol Metab 315, E723–E733 [DOI] [PubMed] [Google Scholar]
- de Gassart A, Geminard C, Hoekstra D & Vidal M (2004). Exosome secretion: the art of reutilizing nonrecycled proteins? Traffic 5, 896–903. [DOI] [PubMed] [Google Scholar]
- Deatherage BL & Cookson BT (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun 80, 1948–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela F, von Linstow ME, Mikines KJ & Galbo H (2004). Physical training may enhance beta-cell function in type 2 diabetes. Am J Physiol Endocrinol Metab 287, E1024–E1031. [DOI] [PubMed] [Google Scholar]
- Duong P, Chung A, Bouchareychas L & Raffai RL (2019). Cushioned-Density Gradient Ultracentrifugation (C-DGUC) improves the isolation efficiency of extracellular vesicles. PLoS One 14, e0215324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Wu N, Gan X, Yan W, Morrell JC & Gould SJ (2007). Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol 5, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio MA, Hiscock N, Sacchetti M, Fischer CP & Pedersen BK (2004). Interleukin-6 is a novel factor mediating glucose homeostasis during skeletal muscle contraction. Diabetes 53, 1643–1648. [DOI] [PubMed] [Google Scholar]
- Febbraio MA & Pedersen BK (2002). Muscle-derived interleukin-6: mechanisms for activation and possible biological roles. FASEB J 16, 1335–1347. [DOI] [PubMed] [Google Scholar]
- Fernandez-Borja M, Wubbolts R, Calafat J, Janssen H, Divecha N, Dusseljee S & Neefjes J (1999). Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr Biol 9, 55–58. [DOI] [PubMed] [Google Scholar]
- Forterre A, Jalabert A, Berger E, Baudet M, Chikh K, Errazuriz E, De Larichaudy J, Chanon S, Weiss-Gayet M, Hesse AM, Record M, Geloen A, Lefai E, Vidal H, Coute Y & Rome S (2014a). Proteomic analysis of C2C12 myoblast and myotube exosome-like vesicles: a new paradigm for myoblast-myotube cross talk? PLoS One 9, e84153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre A, Jalabert A, Chikh K, Pesenti S, Euthine V, Granjon A, Errazuriz E, Lefai E, Vidal H & Rome S (2014b). Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts during muscle cell differentiation. Cell Cycle 13, 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruhbeis C, Helmig S, Tug S, Simon P & Kramer-Albers EM (2015). Physical exercise induces rapid release of small extracellular vesicles into the circulation. J Extracell Vesicles 4, 28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Kirby TJ, Kosmac K, McCarthy JJ & Peterson CA (2017). Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 20, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamez-Valero A, Monguio-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K & Borras FE (2016). Size-exclusion chromatography-based isolation minimally alters extracellular vesicles’ characteristics compared to precipitating agents. Sci Rep 6, 33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber CE, Blissmer B, Deschenes MR, Franklin BA, Lamonte MJ, Lee IM, Nieman DC & Swain DP (2011). American College of Sports Medicine position stand. Quantity and quality of exercise for developing and maintaining cardiorespiratory, musculoskeletal, and neuromotor fitness in apparently healthy adults: guidance for prescribing exercise. Med Sci Sports Exerc 43, 1334–1359. [DOI] [PubMed] [Google Scholar]
- Garcia NA, Moncayo-Arlandi J, Sepulveda P & Diez-Juan A (2016). Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc Res 109, 397–408. [DOI] [PubMed] [Google Scholar]
- Goldstein MS (1961). Humoral nature of the hypoglycemic factor of muscular work. Diabetes 10, 232–234. [DOI] [PubMed] [Google Scholar]
- Guescini M, Canonico B, Lucertini F, Maggio S, Annibalini G, Barbieri E, Luchetti F, Papa S & Stocchi V (2015). Muscle releases alpha-sarcoglycan positive extracellular vesicles carrying miRNAs in the bloodstream. PLoS One 10, e0125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Zhang ZK, Liang C, Li J, Liu J, Lu A, Zhang BT & Zhang G (2017). Molecular communication from skeletal muscle to bone: a review for muscle-derived myokines regulating bone metabolism. Calcif Tissue Int 100, 184–192. [DOI] [PubMed] [Google Scholar]
- Gupta S, Rawat S, Arora V, Kottarath SK, Dinda AK, Vaishnav PK, Nayak B & Mohanty S (2018). An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Curr Stem Cell Res Ther 9, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halova I & Draber P (2016). Tetraspanins and transmembrane adaptor proteins as plasma membrane organizers-mast cell case. Front Cell Dev Biol 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI & Cashikar A (2012). Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28, 337–362. [DOI] [PubMed] [Google Scholar]
- Harding C, Heuser J & Stahl P (1983). Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 97, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig S, Raschke S, Knebel B, Scheler M, Irmler M, Passlack W, Muller S, Hanisch FG, Franz T, Li X, Dicken HD, Eckardt K, Beckers J, de Angelis MH, Weigert C, Haring HU, Al-Hasani H, Ouwens DM, Eckel J, Kotzka J & Lehr S (2014). Secretome profiling of primary human skeletal muscle cells. Biochim Biophys Acta 1844,1011–1017. [DOI] [PubMed] [Google Scholar]
- He F, Li J, Liu Z, Chuang CC, Yang W & Zuo L (2016). Redox mechanism of reactive oxygen species in exercise. Front Physiol 7, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen J, Rigbolt KT, Blagoev B, Pedersen BK & Kratchmarova I (2010). Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol Cell Proteomics 9, 2482–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess C, Sadallah S, Hefti A, Landmann R & Schifferli JA (1999). Ectosomes released by human neutrophils are specialized functional units. J Immunol 163, 4564–4573. [PubMed] [Google Scholar]
- Hong CS, Muller L, Boyiadzis M & Whiteside TL (2014). Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One 9, e103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson MB, Woodworth-Hobbs ME, Zheng B, Rahnert JA, Blount MA, Gooch JL, Searles CD & Price SR (2014). miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol 306, C551–C558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH (2010). The ESCRT complexes. Crit Rev Biochem Mol Biol 45, 463–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT & Coffey RJ (2019). Reassessment of exosome composition. Cell 177, 428–445 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RM, Adam M, Hammond JR, Orr L & Turbide C (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 262, 9412–9420. [PubMed] [Google Scholar]
- Kaksonen M & Roux A (2018). Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 19, 313–326. [DOI] [PubMed] [Google Scholar]
- Kalra H, Drummen GP & Mathivanan S (2016). Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci 17,170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G & Olson EN (2016). Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell 164,1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M & Emr SD (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M & Emr SD (2003). Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol 162, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AL & Botos E (2009). Endocytosis via caveolae: alternative pathway with distinct cellular compartments to avoid lysosomal degradation? J Cell Mol Med 13,1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M & Thery C (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Nat Acad Sci U S A 113, E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal LG, Lopes MA & Batista ML Jr (2018). Physical exercise-induced myokines and muscle-adipose tissue crosstalk: a review of current knowledge and the implications for health and metabolic diseases. Front Physiol 9, 1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan MC, Bigot A, Jensen SS, Dennis JL, Rogowska-Wrzesinska A, Laine J, Gache V, Furling D, Jensen ON, Voit T, Mouly V, Coulton GR & Butler-Browne G (2012). In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J Proteomics 77, 344–356. [DOI] [PubMed] [Google Scholar]
- Lee BR, Kim JH, Choi ES, Cho JH & Kim E (2018). Effect of young exosomes injected in aged mice. Int J Nanomed 13, 5335–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wong DK, Hong KY & Raffai RL (2018). Cushioned-density gradient ultracentrifugation (C-DGUC): a refined and high performance method for the isolation, characterization, and use of exosomes. Methods Mol Biol 1740, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Kaslan M, Lee SH, Yao J & Gao Z (2017). Progress in exosome isolation techniques. Theranostics 7, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X & Donowitz M (2014). Fractionation of subcellular membrane vesicles of epithelial and non-epithelial cells by OptiPrep density gradient ultracentrifugation. Methods Mol Biol 1174, 85–99. [DOI] [PubMed] [Google Scholar]
- Lim JP & Gleeson PA (2011). Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol Cell Biol 89, 836–843. [DOI] [PubMed] [Google Scholar]
- Linares R, Tan S, Gounou C, Arraud N & Brisson AR (2015). High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles 4, 29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett JAC, Durcan PJ & Myburgh KH (2018). Investigation of circulating extracellular vesicle MicroRNA following two consecutive bouts of muscle-damaging exercise. Front Physiol 9, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT & Boucrot E (2015). Membrane curvature at a glance. J Cell Sci 128, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SK, Solomon TP, Blaszczak A, Finnegan S, Filion J & Kirwan JP (2013). Pancreatic beta-cell function increases in a linear dose-response manner following exercise training in adults with prediabetes. Am J Physiol Endocrinol Metab 305, E1248–E1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri M & Altieri DC (1998). Endothelial cell activation by leukocyte microparticles. J Immunol 161, 4382–4387. [PubMed] [Google Scholar]
- Monguio-Tortajada M, Galvez-Monton C, Bayes-Genis A, Roura S & Borras FE (2019). Extracellular vesicle isolation methods: rising impact of size-exclusion chromatography. Cell Mol Life Sci 76, 2369–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G & D’Souza-Schorey C (2009). ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol 19, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN & Lu Q (2012). Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Nat Acad Sci U S A 109, 4146–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager AR, Goldstein JS, Herranz-Perez V, Portran D, Ye F, Garcia-Verdugo JM & Nachury MV (2017). An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263 e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufer PD, Bamman MM, Muoio DM, Bouchard C, Cooper DM, Goodpaster BH, Booth FW, Kohrt WM, Gerszten RE, Mattson MP, Hepple RT, Kraus WE, Reid MB, Bodine SC, Jakicic JM, Fleg JL, Williams JP, Joseph L, Evans M, Maruvada P, Rodgers M, Roary M, Boyce AT, Drugan JK, Koenig JI, Ingraham RH, Krotoski D, Garcia-Cazarin M, McGowan JA & Laughlin MR (2015). Understanding the cellular and molecular mechanisms of physical activity-induced health benefits. Cell Metab 22, 4–11. [DOI] [PubMed] [Google Scholar]
- Oliveira GP Jr, Porto WF, Palu CC, Pereira LM, Petriz B, Almeida JA, Viana J, Filho NNA, Franco OL & Pereira RW (2018). Effects of Acute Aerobic Exercise on Rats serum extracellular vesicles diameter, concentration and small RNAs content. Front Physiol 9, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan BT & Johnstone RM (1983). Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33, 967–978. [DOI] [PubMed] [Google Scholar]
- Pedersen BK (2009). The diseasome of physical inactivity—and the role of myokines in muscle-fat cross talk. J Physiol 587, 5559–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BK (2011). Muscles and their myokines. J Exp Biol 214, 337–346. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Akerstrom TC, Nielsen AR & Fischer CP (2007). Role of myokines in exercise and metabolism. J Appl Physiol (1985) 103, 1093–1098. [DOI] [PubMed] [Google Scholar]
- Pedersen BK & Febbraio MA (2008). Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol Rev 88, 1379–1406. [DOI] [PubMed] [Google Scholar]
- Pedersen BK & Febbraio MA (2012). Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol 8, 457–465. [DOI] [PubMed] [Google Scholar]
- Pedersen BK & Fischer CP (2007a). Beneficial health effects of exercise - the role of IL-6 as a myokine. Trends Pharmacol Sci 28, 152–156. [DOI] [PubMed] [Google Scholar]
- Pedersen BK & Fischer CP (2007b). Physiological roles of muscle-derived interleukin-6 in response to exercise. Curr Opin Clin Nutr Metab Care 10, 265–271. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Pedersen M, Krabbe KS, Bruunsgaard H, Matthews VB & Febbraio MA (2009). Role of exercise-induced brain-derived neurotrophic factor production in the regulation of energy homeostasis in mammals. Exp Physiol 94, 1153–1160. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Steensberg A, Fischer C, Keller C, Keller P, Plomgaard P, Febbraio M & Saltin B (2003). Searching for the exercise factor: is IL-6 a candidate? J Muscle Res Cell Motil 24, 113–119. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Steensberg A & Schjerling P (2001). Muscle-derived interleukin-6: possible biological effects. J Physiol 536, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomgaard P, Halban PA & Bouzakri K (2012). Bimodal impact of skeletal muscle on pancreatic beta-cell function in health and disease. Diabetes Obes Metab 14(Suppl. 3), 78–84. [DOI] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ & Geuze HJ (1996). B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P & Ratajczak MZ (2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856. [DOI] [PubMed] [Google Scholar]
- Richards DM & Endres RG (2014). The mechanism of phagocytosis: two stages of engulfment. Biophys J 107, 1542–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romancino DP, Paterniti G, Campos Y, De Luca A, Di Felice V, d’Azzo A & Bongiovanni A (2013). Identification and characterization of the nano-sized vesicles released by muscle cells. FEBS Lett 587,1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S, Forterre A, Mizgier ML & Bouzakri K (2019). Skeletal muscle-released extracellular vesicles: state of the art. Front Physiol 10, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Fang Y, Wu N & Gould SJ (2011). Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J Biol Chem 286, 44162–44176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM & Breakefield XO (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10,1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead WT, Hayden CC, Gadok AK, Zhao C, Lafer EM, Rangamani P & Stachowiak JC (2017). Membrane fission by protein crowding. Proc Nat Acad Sci U S A 114, E3258–E3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodar BW, Kittel A, Paloczi K, Vukman KV, Osteikoetxea X, Szabo-Taylor K, Nemeth A, Sperlagh B, Baranyai T, Giricz Z, Wiener Z, Turiak L, Drahos L, Pallinger E, Vekey K, Ferdinandy P, Falus A & Buzas EI (2016). Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep 6, 24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA & Hayden CC (2012). Membrane bending by protein-protein crowding. Nat Cell Biol 14, 944–949. [DOI] [PubMed] [Google Scholar]
- Stuffers S, Sem Wegner C, Stenmark H & Brech A (2009). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10, 925–937. [DOI] [PubMed] [Google Scholar]
- Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Morgelin M & Belting M (2013). Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 288, 17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G & Clayton A (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 30, 3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J & Amigorena S (2001). Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol 166, 7309–7318. [DOI] [PubMed] [Google Scholar]
- Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7,1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH & Xiao ZD (2014). Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 289, 22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, Kowal J & Thery C (2018). Why the need and how to approach the functional diversity of extracellular vesicles. Philos Trans R Soc Lond B Biol Sci 373, 20160479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B & Simons M (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247. [DOI] [PubMed] [Google Scholar]
- Trams EG, Lauter CJ, Salem N Jr & Heine U (1981). Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 645, 63–70. [DOI] [PubMed] [Google Scholar]
- Tricarico C, Clancy J & D’Souza-Schorey C (2017). Biology and biogenesis of shed microvesicles. Small GTPases 8, 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovato E, Di Felice V & Barone R (2019). Extracellular vesicles: delivery vehicles of myokines. Front Physiol 10, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann M, Mobius-Winkler S, Fikenzer S, Adam J, Redlich M, Mohlenkamp S, Hilberg T, Schuler GC & Adams V (2014). Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol 21,484–491. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ & Lotvall JO (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O & Hendrix A (2014). The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles 3, 24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E & Raposo G (2011). The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 21, 708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P & Valadi H (2012). Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res 40, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K (2009). Adipokines, myokines and cardiovascular disease. Circ J 73,13–18. [DOI] [PubMed] [Google Scholar]
- Wenzel EM, Schultz SW, Schink KO, Pedersen NM, Nahse V, Carlson A, Brech A, Stenmark H & Raiborg C (2018). Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat Commun 9, 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham M & Febbraio MA (2016). The ever-expanding myokinome: discovery challenges and therapeutic implications. Nat Rev Drug Discovery 15, 719–729. [DOI] [PubMed] [Google Scholar]
- Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE, Egan CL, Cron L, Watt KI, Kuchel RP, Jayasooriah N, Estevez E, Petzold T, Suter CM, Gregorevic P, Kiens B, Richter EA, James DE, Wojtaszewski JFP & Febbraio MA (2018). Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab 27, 237–251 e234. [DOI] [PubMed] [Google Scholar]
- Wolf P (1967). The nature and significance of platelet products in human plasma. Br J Haematol 13, 269–288. [DOI] [PubMed] [Google Scholar]
- Yin X, Zhao Y, Zheng YL, Wang JZ, Li W, Lu QJ, Huang QN, Zhang CY, Chen X & Ma JZ (2019). Time-course responses of muscle-specific microRNAs following acute uphill or downhill exercise in Sprague-Dawley rats. Front Physiol 10, 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Yea K, Kim J, Choi YS, Park S, Lee H, Lee CS, Suh PG & Ryu SH (2009). Comparative proteomic analysis of the insulin-induced L6 myotube secretome. Proteomics 9, 51–60. [DOI] [PubMed] [Google Scholar]
- Yu LL, Zhu J, Liu JX, Jiang F, Ni WK, Qu LS, Ni RZ, Lu CH & Xiao MB (2018). A comparison of traditional and novel methods for the separation of exosomes from human samples. Biomed Res Int 2018, 3634563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuana Y, Levels J, Grootemaat A, Sturk A & Nieuwland R (2014). Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles 3, 23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J & Chiesi A (2015). Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 87, 46–58. [DOI] [PubMed] [Google Scholar]
- Zeringer E, Barta T, Li M & Vlassov AV (2015). Strategies for isolation of exosomes. Cold Spring Harb Protoc 2015, 319–323. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A & Nagrath D (2016). Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5,e10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Guariglia S, Yu RY, Li W, Brancho D, Peinado H, Lyden D, Salzer J, Bennett C & Chow CW (2013). Mutation of SIMPLE in Charcot-Marie-Tooth 1C alters production of exosomes. Mol Biol Cell 24, 1619–1637, S1611–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G & Amigorena S (1998). Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med 4, 594–600. [DOI] [PubMed] [Google Scholar]


