Abstract
Autoreactive T cells may contribute to post-viral myocarditis induced with Coxsackievirus B3 (CVB3), but the underlying mechanisms of their generation are unclear. Here, we have comprehensively analyzed the generation of antigen-specific, autoreactive T cells in the mouse model of CVB3 infection for antigens implicated in patients with myocarditis/dilated cardiomyopathy. First, comparative analysis of CVB3 proteome with five autoantigens led us to identify three mimicry epitopes, one each from adenine nucleotide translocator 1 (ANT), sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) and cardiac troponin I. None of these induced cross-reactive T cell responses. Next, we generated major histocompatibility complex (MHC) class II dextramers to enumerate the frequencies of antigen-specific T cells to determine whether T cells with multiple antigen specificities are generated by CVB3 infection. These analyses revealed appearance of CD4 T cells positive for SERCA2a 971-990, and cardiac myosin heavy chain-α (Myhc) 334-352 dextramers, both in the periphery and also in the hearts of CVB3-infected animals. While ANT 21-40 dextramer+ T cells were inconsistently detected, the β1-adrenergic receptor 181-200/211-230 or branched chain α-ketoacid dehydrogenase kinase 111-130 dextramer+ cells were absent. Interestingly, SERCA2a 971-990, Myhc 334-352 and ANT 21-40 dextramer+ cells were also detected in the liver indicating that they may have a pathogenic role. Finally, we demonstrate that the SERCA2a 971-990-reactive T cells generated in CVB3 infection could transfer disease to naïve mice. The data suggest that CVB3 infection can lead to the generation of autoreactive T cells for multiple antigens indicating a possibility that the autoreactive T cells localized in the liver can potentially circulate and contribute to the development of viral myocarditis.
Keywords: Viral myocarditis, autoreactive T cells, CVB3, mouse model, MHC dextramers/tetramers
1. Introduction
Myocarditis can occur in association with a wide spectrum of infectious agents, systemic diseases, and hypersensitivity to drugs and toxins (Fung et al., 2016; Pollack et al., 2015). The virus-induced myocarditis in neonates and children may lead to fulminant myocarditis (N. R. Rose, 2016), and affected individuals can develop chronic myocarditis leading to dilated cardiomyopathy (DCM) and congestive heart failure. Approximately, half of these patients undergo heart transplantation due to the lack of effective treatment options (Alida LP Caforio et al., 2013; Chi et al., 2014). Enteroviruses like Coxsackievirus B (CVB)3 and adenoviruses are generally suspected as causes of viral myocarditis in North America and Europe, respectively (Blauwet and Cooper, 2010; Lasrado et al., 2020), and virus-reactive antibodies, and viral RNA have been detected in up to 50 to 70% of those affected, suggesting that CVB3 may be an important trigger in the DCM pathogenesis (Baboonian and McKenna, 2003; Cihakova and Rose, 2008; Fujinami et al., 2006; Kuhl et al., 2005). Autoimmunity is one possible mechanism as DCM patients can carry autoantibodies for cardiac myosin heavy chain-α (Myhc), adenine nucleotide translocator 1 (ANT), β1-adrenergic receptor (β1AR), branched chain α-ketoacid dehydrogenase (BCKD), sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) and cardiac troponin I (TNI) (A. L. Caforio et al., 2001; A. L. Caforio et al., 2002; Kaya et al., 2012). But, the role of autoreactive T cells, if any remains to be investigated.
Mouse models are commonly employed to determine the mechanisms of immune pathogenesis of CVB infection. CVB3 infections in susceptible mouse strains such as A/J and Balb/c are characterized by two phases that occur in continuum namely, acute myocarditis/viral phase and chronic myocarditis/non-viral phase (Cihakova and Rose, 2008; Rabausch-Starz et al., 1994; Noel R Rose et al., 1986). Affected animals show antibodies for various cardiac and non-cardiac proteins such as cardiac myosin, ANT, β1AR, BCKD, SERCA2a, TNI, laminin and muscarinic receptor suggesting a role for autoreactive T cells in the generation of autoantibodies (A. L. Caforio et al., 2005; Fairweather et al., 2012; Kaya et al., 2012). We recently demonstrated that A/J mice infected with CVB3 show Myhc 334-352-specific CD4 T cells that are pathogenic in nature (Gangaplara et al., 2012), indicating that autoreactive T cells with multiple antigen-specificities could be generated as a secondary event in infected animals. By addressing the molecular mimicry hypothesis, we found no evidence for appearance of cross-reactive T cells to ANT, SERCA2a and TNI that had sequences mimicking CVB3 proteins. However, by analyzing the frequencies of antigen-specific T cells using major histocompatibility complex (MHC) class II dextramers for five different autoantigens, we detected the presence of T cells positive for SERCA2a 971-990 and Myhc 334-352 dextramers both in the periphery and also in the infected hearts. Interestingly, dextramer (dext+) T cells for select antigens were also detected in the livers suggesting a possibility that these cells can potentially circulate and contribute to viral myocarditis in infected animals.
2. Materials and Methods
2.1. Mice
A/J (H-2a, male and female, 6 to 8 weeks old) and SJL/J (H-2s, female, 4 to 6 weeks old) mice were procured from the Jackson Laboratory (Bar Harbor, ME), and maintained by the institutional guidelines at the University of Nebraska-Lincoln. Euthanasia was performed using a carbon dioxide chamber according to the Panel on Euthanasia, the American Veterinary Medical Association.
2.2. Virus propagation, titration, and disease-induction
Vero cells (ATCC, Manassas, VA) were used to grow CVB3 (Nancy strain, ATCC) as described previously (Gangaplara et al., 2012). After confirming the cytopathic effect (CPE), tissue culture infective dose (TCID)50 values were determined according to the Spearman-Karber method (Reed and Muench, 1938), and the viral stocks were stored at −80°C. To infect mice, virus diluted in 1× phosphate-buffered saline (PBS) to a final concentration of 50 to 2000 TCID50/200 μl was administered intraperitoneally. Animals were housed in filter-top cages that were changed once in 3 days until termination. Clinically, animals were inspected daily for ruffled fur, isolation, sluggish activity or mortalities. At termination (days, 18-21 post-infection), lymphoid (spleen and lymph nodes) and non-lymphoid organs (hearts, pancreata and livers) were collected for histology and in vitro experimentation.
2.3. Peptide synthesis
Myhc 334-352 (DSAFDVLSFTAEEKAGVYK), ANT 21-40 (VSKTAVAPffiRV KLLLQVQH), β1AR 181-200 (TVWAISALVSFLPILMHWWR)/211-230 (NDPKC CDFVTNRAYAIASSV), BCKDk 111-130 (PIKDQADEAQYCQLVRQLLD), SERCA2a 971-990 (KISLPVILMDETLKFVARNY), bovine ribonuclease (RNase 43-56) (VNTFVHESLADVQA), biotinylated hen egg lysozyme (HEL) 46-61 (YNTDGSTDYGILQINSR), proteolipid protein (PLP) 139-151 (HSLGKWLGHPDKF), human immunodeficiency virus glycoprotein 120 (HIV) P18-I10 (Neopeptide, Cambridge, MA), and moth cytochrome C (MCC) 82-103 (FAGLKKANERADLIAYLKQATK) (GenScript, Piscataway, NJ), including the peptides listed in the Table S1 were synthesized by 9-fluorenylmethyloxycarbonyl chemistry (Neopeptide). The purity of peptides was confirmed to be more than 90% by high-performance liquid chromatography, and their identities were confirmed by mass spectroscopy. Peptides were dissolved in ultra-pure water or PBS and stored at −20°C.
2.4. Immunization procedures
Peptide emulsions were prepared in complete Freund’s adjuvant (CFA) containing Mycobacterium tuberculosis H37RA extract (Difco Laboratories, Detroit, MI) to a final concentration of 5 mg/ml, except that PLP 139-151 was prepared at 1 mg/ml (Miller and Karpus, 2007; Reddy et al., 2003). A/J mice were immunized twice subcutaneously in inguinal and sternal regions on days 0 and 7 (Rakesh H Basavalingappa et al., 2016; Donermeyer et al., 1995; Chandirasegaran Massilamany et al., 2011), whereas SJL/J mice received a single dose of PLP 139-151 emulsion (Miller and Karpus, 2007; Reddy et al., 2003).
2.5. Proliferation assay
Lymphocyte suspensions were prepared from a pool of spleens and lymph nodes harvested from CVB3-infected mice after lysing the erythrocytes using 1× ammonium chloride potassium buffer (Lonza, Walkersville, MD). Likewise, lymph node cells (LNCs) were prepared from immunized animals. After washing, cell pellets were suspended in RPMI containing 10% FBS, 1 mM sodium pyruvate, 4 mM L-glutamine, 1× each of non-essential amino acids and vitamin mixture, and 100 U/ml penicillin-streptomycin (Lonza; hereafter called growth medium). Cells were stimulated with the indicated peptides (0-100 μg/ml) for 2 days in growth medium at a density of 5 × 106 cells/ml in triplicates, whereas cells cultured with no peptides were used as medium controls. After pulsing with tritiated 3[H] thymidine (1 μCi per well; MP Biomedicals, Santa Ana, CA) for 16 hours, proliferative responses were measured as counts per minute (cpm) using a Wallac liquid scintillation counter (Perkin Elmer, Waltham, MA) (Rakesh H Basavalingappa et al., 2016; Chandirasegaran Massilamany et al., 2016). In some experiments, for easy depiction, stimulation indices were calculated by dividing the cpm values obtained in peptide-stimulated cultures by the values obtained in medium controls.
2.6. Creation of MHC dextramer and tetramer reagents
Our studies involved the use of MHC class II dextramers for both IAk and/or IEk alleles, and also MHC class I tetramers for H-2Dd allele (Table 1). Essentially, dextramers and tetramers were created as we have reported previously (Chandirasegaran Massilamany et al., 2016; C. Massilamany et al., 2011a; C. Massilamany et al., 2011c; Reddy et al., 2003). We had described the creation of all dextramers and tetramers (R. H. Basavalingappa et al., 2017; Bharathi Krishnan et al., 2017; B. Krishnan et al., 2018; Chandirasegaran Massilamany et al., 2016; C. Massilamany et al., 2011c) except, IAk dextramers for ANT 21-40. To generate ANT 21-40 dextramers, the nucleotide sequence of ANT 21-40 (gtctccaagacggcggtcgccccgat cgagagggtcaaactgctgctgcaggtccagcat) was assembled to IAk-β construct, and after expressing the IAk-α and IAk-β constructs in the Baculovirus using sf9 insect cells, soluble IAk/ANT 21-40 MHC monomers were purified using the antibody affinity column (clone, 10-2.16) and the proteins were biotinylated. Dextramerization was performed by mixing biotinylated protein and streptavidin-fluorophore-conjugated dextran molecules at a ratio of 20:1 to obtain fluorophore-assembled dextramers as we have described earlier (C. Massilamany et al., 2011c; Reddy et al., 2003). In experiments involving SJL mice, we used IAs dextramers for PLP 139-151 and Theiler's murine encephalomyelitis virus (TMEV) 70-86 as reported previously (C. Massilamany et al., 2014a; C. Massilamany et al., 2011c).
Table 1:
The list of dextramers/tetramers used to analyze antigen-specific T cells in mice infected with CVB3
| MHC class II dextramers | |
|---|---|
| IAk | IEk |
| Myhc 334-352 | β1AR 181-200 |
| ANT 21-40 | SERCA2a 971-990 |
| β1AR 211-230 | MCC 82-103 (control) |
| BCKDk 111-130 | |
| SERCA2a 971-990 | |
| RNase 43-56 (control) | |
| MHC class I tetramers (H-2Dd) | |
| Myhc 338-348 | |
| HIV PI8-110 (control) | |
Myhc, cardiac myosin heavy chain-α
ANT, adenine nucleotide translocator 1
β1AR, β1-adrenergic receptor
BCKDk, branched chain α-ketoacid dehydrogenase kinase
SERCA2a, sarcoplasmic/endoplasmic reticulum
Ca2+ adenosine triphosphatase
RNase, bovine ribonuclease
MCC, moth cytochrome C
HIV, human immunodeficiency virus
2.7. Isolation of mononuclear cells from liver and heart tissues
After euthanasia, animals were perfused by injecting 10 ml of ice-cold PBS into the left ventricles, and hearts and livers were collected to isolate mononuclear cells (MNCs) as reported previously (Gangaplara et al., 2012; Bharathi Krishnan et al., 2017). In brief, livers were minced, and the dissociated tissues were treated with Collagenase IV (400 U/ml; Worthington, Lakewood, NJ) and DNase I (150 U/ml; Worthington) at 37°C in shaker incubator for 30 minutes. After washing, MNCs were harvested using 40%/75% percoll gradient centrifugation procedure as described previously (Bharathi Krishnan et al., 2017). After lysing erythrocytes, cell pellets were dissolved for further experimentation. For isolation of MNCs from hearts, the minced tissues were treated with Collagenase II (600 U/ml; Worthington Biochemical Corporation, Lakewood, NJ, USA) and DNase I (60 U/ml; AppliChem GmbH, Darmstadt, Germany) for 30 minutes at 37°C using tube rotator (Miltenyi Biotec Inc., Auburn, CA). Cell suspensions were filtered through 70 μM cell strainer, and after removing the residual debris using debris removal kit (Miltenyi Biotec) and lysing erythrocytes, MNC suspensions were obtained.
2.8. Dextramer and tetramer staining
Lymphocytes harvested from mice infected with or without CVB3 were stimulated with peptides for two days, and growth medium supplemented with interleukin (IL)-2 (IL-2 medium) was then added. Cells were stained with IAk/IEk dextramers during days 8 to 10 post-stimulation, followed by staining with anti-CD4 (GK1.5, Biolegend, San Diego, CA) and 7-aminoactinomycin-D (7-AAD) as we have described previously (Bharathi Krishnan et al., 2017; B. Krishnan et al., 2018; C. Massilamany et al., 2011c). Finally, cells were washed and acquired by flow cytometry and the frequencies of dext+ CD4+ T cells were enumerated within the live cells (7-AAD◌) using FlowJo software (Tree Star, Ashland, OR). This protocol was also used to stain LNC cultures prepared from SJL mice immunized with PLP 139-151 as described previously (C. Massilamany et al., 2011c). The MNCs harvested from hearts and livers from CVB3 infected and liver MNCs from PLP 139-151 immunized animals were directly stained ex vivo as above (Bharathi Krishnan et al., 2017; C. Massilamany et al., 2011c).
For staining with H-2Dd/MHC class I tetramers, cells were initially treated with 10% mouse serum and Fc blocker (Biolegend), followed by staining with anti-CD4, anti-CD8a (KT15 and CT-CD8a, Invitrogen, Carlsbad, CA) and 7-AAD on ice for 15 minutes. Staining was continued for an additional 15 minutes on ice after adding H-2Dd/MHC class I tetramers, where tetramers prepared at a ratio of 8:1 between biotinylated H-2Dd/Myhc 338-348 and streptavidin at a concentration of 30 μg/ml were previously optimized in pilot studies. After series of washes, cells were acquired by flow cytometry and the percentage of tetramer (tet)+ CD8+ cells were analyzed using FlowJo software (Tree Star).
2.9. Adoptive transfer experiments
Lymphocytes were harvested from A/J mice infected with CVB3 and cells were stimulated with SERCA2a 971-990 (20 μg/ml) for two days and IL-2 medium was then supplemented. On day 7 or 8, cells were expanded by treating with Concanavalin A (Con A, 2.5 μg/ml, 2 days). Viable cells harvested on day 9 or 10 post stimulation were administered to naïve A/J mice (3.0 to 4.0 x 107 cells/animal) intravenously that were primed with LPS on day −4 and day 0 (25 μg/animal) (Rakesh H Basavalingappa et al., 2016; Hamada et al., 2004; Chandirasegaran Massilamany et al., 2016). Each animal also received pertussis toxin (PT, 100 ng; List Biologicals, Campbell, CA) on day 0 and 2 post-transfer (Agarwal et al., 2002; Fujimoto et al., 2006; Chandirasegaran Massilamany et al., 2011). Animals were euthanized on day 14, post-transfer, and hearts and pancreata were examined by a Board-Certified, pathologist blinded to treatment groups for inflammatory changes by hematoxylin and eosin (H & E) staining as described previously (Rakesh H Basavalingappa et al., 2016; Bharathi Krishnan et al., 2017; Chandirasegaran Massilamany et al., 2011). In some experiments, heart and pancreatic sections obtained from mice infected with or without CVB3 were also examined by H & E staining as above.
2.10. Detection of CVB3 RNA
Total RNA was extracted from hearts and pancreata tissues harvested from naïve A/J mice using the Trizol method (Invitrogen) and to remove genomic DNA, 1 μg of total RNA was treated with amplification grade DNase I (Invitrogen). cDNAs were synthesized using iScript™ reverse transcription supermix kit, as recommended (Bio-Rad, Hercules, CA). PCR amplifications were performed using CVB3 viral protein (VP)1-specific primers (forward, 5’-TTGCATATGGCCCAGTGGAAG-3’; reverse, 5’-TGTGGATCCTTATT GCCTAGTAGTGGTAACTC-3’) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (forward, 5’-CGGCAAATTCAACGGCACAGTCAA-3’; reverse, 5’-CTTTCCAGAGGGGCCATCCACAG-3’) (Rakesh H Basavalingappa et al., 2016; Gangaplara et al., 2012). The PCR products were stained with ethidium bromide and resolved on 1.5% agarose gel electrophoresis.
2.11. Statistics
Right-tailed paired Wilcoxon signed rank test was used to compare frequencies of dext+ cells in splenocytes derived from CVB3 infected animals (Wilcoxon, 1945). The corresponding controls used for comparison include: RNase 43-56 for IAk dextramers (Myhc 334-352, ANT 21-40 and BCKDk 111-130), combined RNase 43-56 and MCC 82-103 for IAk and IEk dextramers (SERCA2a 971-990 and β1AR 211-230/ 181-200), and MCC 82-103 for β1AR 181-200. For comparing frequencies of dext+ cells in liver and heart MNCs, right-tailed non-parametric Wilcoxon rank sum test was used (Mann and Whitney, 1947). Student’s t-test was used to determine differences in the frequencies of CD4+ dext+ T cells between infected and non-infected (naïve) mice, ANT 21-40 dext+ cells in immunized animals, and also to analyze Myhc 338-348 tet+CD8+ T cells.
3. Results
3.1. Identification of mimicry epitopes from CVB3 proteome that have a potential to induce cross-reactive immune responses
By using LALIGN tool (Madeira et al., 2019), we sought to analyze CVB3 proteome to identify myocarditogenic mimicry epitopes by aligning CVB3 sequence with six self-antigens that are commonly implicated as autoantigens in patients with myocarditis/DCM. These include, Myhc, ANT, β1AR, BCKDk, SERCA2a and TNI (A. L. Caforio et al., 2001; A. L. Caforio et al., 2002; Kaya et al., 2012). The expectation was that the mimicry peptides can result in the generation of cross-reactive T cells due to T cell degeneracy (Gautam et al., 1998; Wooldridge et al., 2012). These analyses led us to identify a panel of 29 mimics representing ANT, β1AR, BCKDk, SERCA2a and TNI, and their percent similarities were, 35 to 43%, 30 to 47%, 33 to 40%, 32 to 46% and 28 to 47% respectively (Table S1). However, such a degree of sequence similarity was lacking for Myhc (data not shown). We also noted that the mimicry epitopes were found localized in both structural (VP1 to VP4) and non-structural regions (2B, 2C, 3C protease and 3D polymerase) of CVB3 genome. The structural proteins of viruses are generally considered as immunogenic, and VP1, is one such protein identified in many enteroviruses, including CVB3, TMEV, and poliovirus (Huber et al., 1993; Minor et al., 1986; Yauch and Kim, 1994). Similarly, several examples of both self-[ribonuclease (RNase), glucose-6-phosphate isomerase, and glutamate decarboxylase] (Atkinson et al., 1994; Basu et al., 2000; Kaufman et al., 1992; Lorenz et al., 1988) and foreign-antigens (neuraminidase and rhodanese-related sulfur transferase) (C. Massilamany et al., 2010; C. Massilamany et al., 2011b; Schaeffer et al., 1989) also exist to predict that the non-structural proteins can be immunogenic.
3.2. Analysis of T cell responses to self-peptides that mimic CVB3 proteins in A/J mice infected with CVB3
From a panel of 29 mimicry peptides, we tested a hypothesis that the CVB3 infection can lead to the induction of cross-reactive T cell responses for self-antigens listed in the Table S1. To test this hypothesis, we used 24 peptides representing the ANT, β1AR, BCKDk, SERCA2a and TNI, and the remaining five peptides could not be synthesized. After infecting A/J mice with CVB3, lymphocytes prepared on day 21 post-infection were used in 3[H]-thymidine-incorporation assay to measure proliferative responses. By using RNase 43-56 as control, T cell responses were noted for three peptides namely, ANT 33-49, SERCA2a 799-813 and TNI 138-159 to be ~1.4-fold difference (Figure S1). Although their magnitude was marginal, occurrence of T cell responses selectively to 3 out of 24 peptides may mean that the CVB3 mimicry epitopes might be immunogenic in the infected animals. But, this proposition required additional validation.
3.3. Evaluation of cross-reactive T cell responses induced by CVB3 mimicry epitopes in immunized animals
To further evaluate if mimicry epitopes can induce cross-reactive T cell responses, we utilized an immunization protocol to determine the immunogenicity of three peptide pairs in crisscross experiments. These include, ANT 33-49/CVB 1153-1169, SERCA2a 799-813/CVB 1839-1853 and TNI 138-159/CVB 1527-1548. Of note, the CVB sequences listed above are conserved in all the six serotypes of CVB (Table S2), leading to a prediction that exposure to any CVB serotype can potentially induce cross-reactive T cell responses. In the first setting, A/J mice were immunized with self-peptides (ANT 33-49, SERCA2a 799-813 and TNI 138-159), and T cell reactivity was tested to both immunizing peptides and also their mimicry epitopes, CVB 1153-1169, CVB 1839-1853 and CVB 1527-1548. The analysis revealed T cell responses occurring only to one pair (ANT 33-49/CVB 1153-1169), and responses were lacking for two other pairs (Figure S2a). Similar analysis in the second setting that involved immunization with CVB3 mimics revealed marginal responses to two CVB3 epitopes namely, CVB 1153-1169, and CVB 1839-1853 (Figure S2b). However, cross-reactive responses for their corresponding mimics from self-antigens were lacking (ANT 33-49, and SERCA2a 799-813). We had expected that the T cells sensitized with CVB 1153-1169 respond to its mimic, ANT 33-49, since ANT 33-49-sensitized T cells did respond to CVB 1153-1169 in a crisscross experiment as described above (Figure S2a, left panel). Since the CVB mimics did not result in cross-reactive T cell responses in the immunization settings that involved the use of CFA as a potent, immunestimulating adjuvant, we concluded that CVB3 infection is unlikely to lead to the induction of significant cross-reactive T cell responses to be biologically significant. However, a possibility exists that cross-reactivity can occur for other autoantigens. For example, by searching the structural databases using the crystal structure of ANT (PDB ID: 1OKC) (Pebay-Peyroula et al., 2003) , we found a peptide fragment within the LOTUS domain of the meiosis regulator and mRNA stability factor 1 (MARF1) (PDB ID: 5YAD), an orthologue of human Limkain B (LMKB) (Yao et al., 2018). As shown in the Figure S3, top panel, comparison of sequences between ANT 33-49 and CVB 1153-1169 revealed an identity of 35.3%, and in a similar comparison between MARF1 and CVB 1153-1169, the identity was found to be 41.2%. Furthermore, using the structures of ANT and MARF1, we predicted the helical structural model for CVB 1153-1169 to be similar as well (Figure S3, bottom panel). Importantly, LMKB/MARF1 appears to act as an autoantigen that may have a role in the regulation of inflammatory response (Bloch et al., 2014; Dunster et al., 2005), but we have not investigated these possibilities. However, it is also to be noted that ANT contains multiple immunodominant epitopes, and we had demonstrated that one epitope, ANT 21-40 can induce myocarditis in A/J mice by generating autoreactive T cells (Rakesh H Basavalingappa et al., 2016). We reasoned that detection of ANT 33-49-reactive T cells in CVB3-infected animals might have represented a de novo population as a result of cardiac damage induced by the virus, since 47% of residues (8/17) are similar between the two epitopes (ANT 33-49, KLLLQVQHASKQISAEK; and ANT 21-40 VSKTAVAPIERVKLLLQVQH, similar residues are underlined).
3.4. Derivation of MHC dextramers to analyze antigen-specific, autoreactive T cells in CVB3-infected mice
By using MHC dextramers (Fig S4a), we addressed a hypothesis that CVB3 infection can lead to the generation of autoreactive T cells with multiple antigen-specificities, and our previous work with the demonstration of Myhc-reactive T cells in CVB3-infected animals also supported this proposition (Gangaplara et al., 2012). We targeted a total of five autoantigens namely, Myhc, ANT, β1AR, BCKDk and SERCA2a, and we and others had previously characterized the myocarditis-inducing, immunodominant epitopes in all of these antigens (Rakesh H Basavalingappa et al., 2016; R. H. Basavalingappa et al., 2017; Daniels et al., 2008; Bharathi Krishnan et al., 2017; B. Krishnan et al., 2018; Chandirasegaran Massilamany et al., 2011). We also had reported creation and validation of MHC class II dextramers for Myhc, β1AR, BCKDk and SERCA2a (Table 1). These include IAk dextramers for Myhc 334-352 (C. Massilamany et al., 2011c), BCKDk 111-130 (Bharathi Krishnan et al., 2017), SERCA2a 971*990 (B. Krishnan et al., 2018) and β1AR 211-230 (R. H. Basavalingappa et al., 2017), and also IEk dextramers for SERCA2a 971-990 (B. Krishnan et al., 2018) and β1AR 181-200 (R. H. Basavalingappa et al., 2017). The reason to create IAk and IEk dextramers is that SERCA2a 971-990 was found to bind both IAk and IEk MHC class II alleles (B. Krishnan et al., 2018), whereas β1AR has two epitopes, with one epitope each, binding to IAk(β1AR 211-230) or IEk molecule (β1AR 181-200) (R. H. Basavalingappa et al., 2017). To define specificity, we used two control dextramers with one each, for IAk (RNase 43-56), and IEk allele (MCC 82-103). In this report, we have described the generation of dextramers for ANT 21-40 and validated using LNCs obtained from ANT 21-40-immunized animals by flow cytometry (Figure S4b). The analysis revealed that ANT 21-40 dextramers bind CD4 T cells antigen-specifically, since staining for control dextramers was negligible (ANT 21-40: 2.02 ± 0.66 vs 0.33 ± 0.03%, P=0.043; Fig S4b). We thus, generated a panel of seven dextramers to evaluate the detection of antigen-specific, CD4 T cells in CVB3 infected animals (Table 1).
3.5. CVB3 infection leads to the generation of autoreactive T cells with multiple antigen-specificities
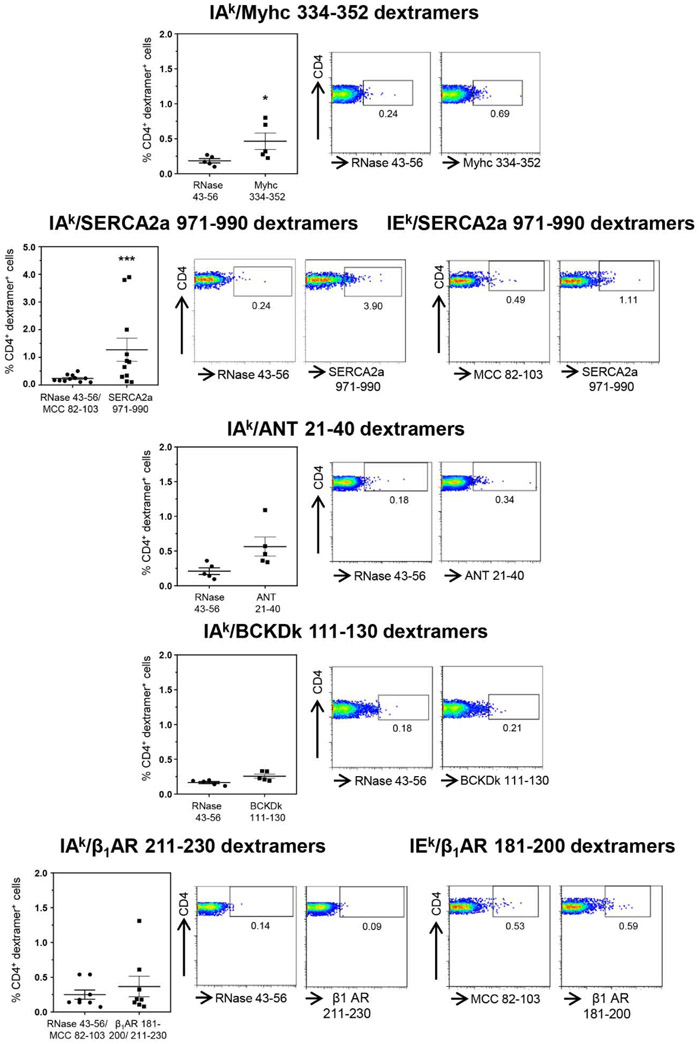
Groups of A/J mice were infected with CVB3, and lymphocytes harvested approximately 20 days post-infection were stimulated with or without Myhc 334-352, ANT 21-40, BCKDk 111-130, SERCA2a 971-990 and β1AR 181-200, β1AR 211-230, and dextramer staining was performed during 8 to 10 days post-stimulations as illustrated in the Figure S5. The flow cytometric analysis revealed detection of Myhc 334-352 dext+ CD4+ T cells to be higher than the control (RNase 43-56) as expected (0.41 ± 0.11 vs 0.18 ± 0.03%, p=0.031) (Figure 1). Similarly, by analyzing the SERCA2a 971-990-reactive T cells using IAkand IEk dextramers, we noted significant increase (five-fold) in their frequencies as compared to RNase 43-56/MCC 82-103 controls (1.28 ± 0.42 vs 0.24 ± 0.04%, p=0.00098) (Figure 1). Although, similar trends were noted for ANT 21-40 dextramers relative to control (0.56 ± 0.14 vs 0.21 ± 0.05%), no significant differences were noted for other dextramers namely, BCKDk 111-130 (0.26 ± 0.03 vs 0.17 ± 0.02%), and β1AR. 211-230 or β1AR. 181-200 (0.37 ± 0.15 vs 0.25 ± 0.07%). We also analyzed the frequencies of cells positive for Myhc 334-352 (0.26 ± 0.03 vs 0.41 ±0.11, p=0.03) and SERCA2a 971-990 (0.31 ± 0.05 vs 1.28 ± 0.42, p=0.05) dextramers to be higher in infected mice as compared to naïve animals (Fig S6). Thus, detection of dext+ T cells only for two from a panel of five antigens suggest that they may have a role in the CVB3 pathogenesis.
Figure 1: Detection of autoreactive T cells with multiple antigen-specificities in animals infected with CVB3.
Lymphocytes obtained from animals infected with CVB3 were stimulated with Myhc 334-352, SERCA2a 971-990, ANT 21-40, BCKDk 111-130, and β1AR 181-200 or β1AR 211-230 for three days, and cells were rested in IL-2 medium. Cells harvested on day 8 to 10 post-stimulation were stained with the indicated IAk and/or IEk dextramers, anti-CD4 and 7-AAD. After washing, and acquiring by flow cytometry, dext+ CD4+ cells were enumerated. Mean ± SEM values obtained from three to six individual experiments, each involving three to six mice are indicated in the left panel for each dextramer reagent. Representative flow cytometric plots for control and specific dextramers are shown on the right. RNase 43-56, control for IAk dextramers; and MCC 82-103, control for IEk dextramers. *P < 0.05 and ***P < 0.001.
3.6. Autoreactive T cells can infiltrate into the livers of CVB3-infected mice
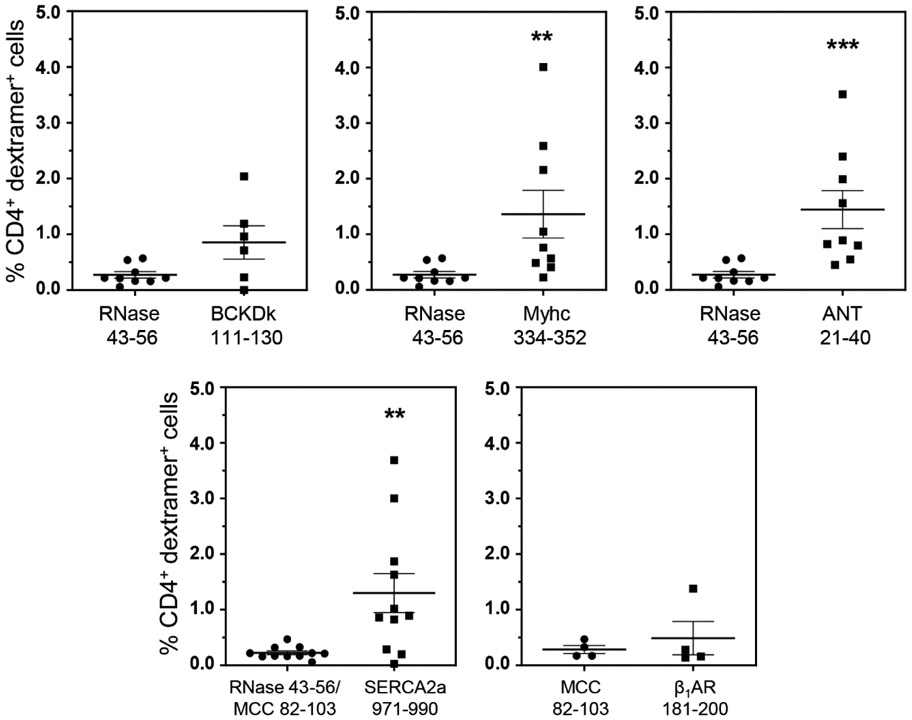
In addressing a hypothesis that the T cells specific to promiscuously-expressed antigens can infiltrate various organs, we had previously demonstrated the detection of BCKDk 111-130-specific T cells in the livers from animals immunized with BCKDk 111-130 (Bharathi Krishnan et al., 2017). Thus, we expected that the BCKDk-reactive T cells may infiltrate livers in CVB3-infected animals. We isolated the liver MNCs from CVB3-infected animals and performed dextramer staining using BCKDk 111-130 dextramers as shown in the Figure S7. The analysis revealed no significant differences in the number of BCKDk 111-130 dext+ T cells as compared to control (RNase 43-56) (0.85% vs 0.27%, p=0.083) (Figure 2). We then extended this observation to determine whether autoreactive T cells specific to other autoantigens can be detected in the livers of CVB3 infected animals by taking advantage of the availability of other dextramers (IAk dextramers: Myhc 334-352 and ANT 21-40; IAk and IEk dextramers: SERCA2a 971-990; and IEk dextramers: β1AR 181-200) (Table 1). These analyses revealed detection of significant number of CD4 T cells positive for Myhc 334-352 (1.36% vs 0.27%, p=0.003), ANT 21-40 (1.44% vs 0.27%, p=0.0003) and SERCA2a 971-990 (1.30% vs 0.22%, p=0.009), but not β1AR 181-200 dextramers (0.49% vs 0.29%) as compared to their corresponding controls (Figure 2). The data suggest that detection of autoreactive T cells in the liver might not have occurred as a random event. We made this proposition since liver MNCs obtained from animals immunized with an irrelevant antigen, PLP 139-151 in SJL mice did not reveal T cells positive for PLP 139151 dextramers (Figure S8, top panel), in spite of their presence in the LNCs ((Figure S8, bottom panel). However, we needed to address whether the autoreactive T cells generated in CVB3-infected animals can infiltrate the target organ, heart.
Figure 2: Detection of autoreactive T cells in the livers from CVB3-infected mice.
Livers from CVB3 infected mice were processed to obtain MNCs as described in the methods section. MNCs were stained with the indicated IAk and/or IEk dextramers, followed by anti-CD4, and 7-AAD. After acquiring the cells by flow cytometry, dext+ cells were analyzed in the live (7-AAD−), CD4 subset. Mean ± SEM values from four to nine individual experiments, each involving three to six mice are shown. RNase 43-56 (control for IAk dextramers); and MCC 82-103 (control for IEk dextramers). **P < 0.01, and ***P < 0.001.
3.7. Heart infiltrates from CVB3-infected animals show detection of autoreactive CD4 and CD8 T cells
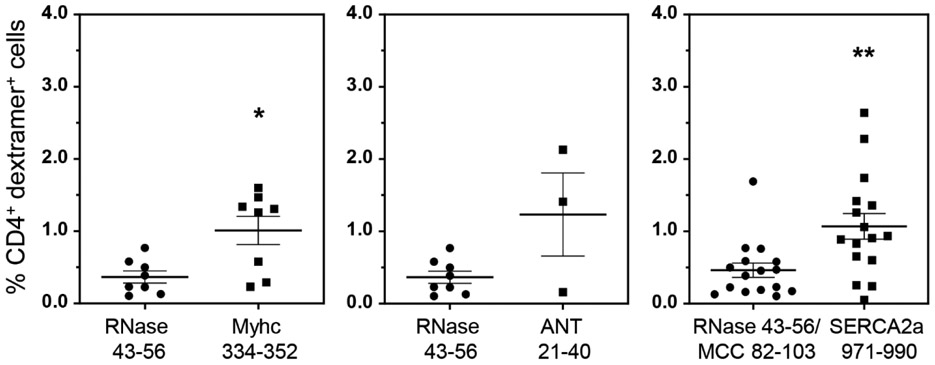
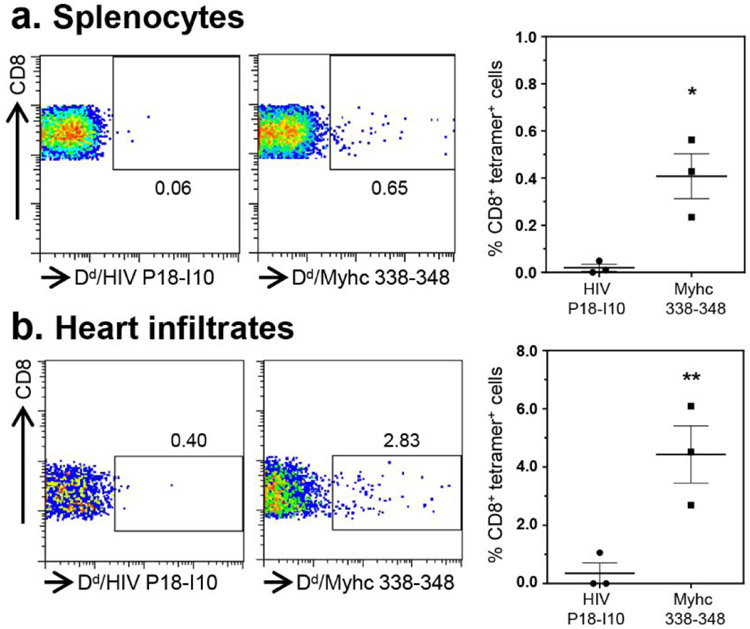
In our previous study, we had demonstrated the detection of Myhc-reactive T cells in hearts from CVB3 infected animals (Gangaplara et al., 2012). Since, the current study revealed appearance of SERCA2a 971-990 dext+ T cells, but to a lesser degree for ANT 21-40 dextramers, we asked if SERCA2a 971-990-reactive T cells can infiltrate hearts in infected animals. We performed dextramer staining on MNCs from hearts harvested from CVB3-infected animals using Myhc 334-352, ANT 21-40 and SERCA2a 971-990 dextramers as shown in the Figure S7. The analysis revealed detection of CD4+ T cells staining with Myhc 334-352 (1.01% vs 0.37%, p=0.021) and SERCA2a 971-990 (1.07% vs 0.46%, p=0.0027), but not ANT 21-40 dextramers (1.23% vs 0.37%, p=0.279) when compared with their controls (Figure 3). Furthermore, we had previously noted that the heart infiltrates from CVB3-infected animals contain both CD4 and CD8 T cells (Gangaplara et al., 2012). In addition, we also had demonstrated that the Myhc 334-352 encompasses epitopes for both CD4 and CD8 T cells (Chandirasegaran Massilamany et al., 2016). Since, we noted the detection of Myhc 334-352 dext+ CD4+ T cells in the hearts from CVB3-infected mice, we asked whether CD8 T cells specific to Myhc 338-348 could be detected in the hearts from CVB3-infected animals. To this end, we created MHC class I/H-2Dd tetramers for Myhc 338-348. By using HIV P18-I10 tetramers as controls, we detected the Myhc 338-348 tet+ CD8+ T cells in significant frequencies in splenocytes from CVB3-infected animals (0.4% vs 0.02%, p=0.015) (Figure 4a, top panel), but such a frequency was low in non-infected animals (Figure S9). We also noted significant number of CD8 T cells binding to Myhc 338-348 tetramers in the heart MNCs as compared to control tetramers (4.0% vs 0.38%, p=0.005) (Figure 4b, bottom panel). The data suggest that the Myhc-reactive CD8 T cells infiltrating the hearts may also contribute to cardiac damage.
Figure 3: Heart infiltrates from CVB3-infected mice revealed the presence of SERCA2a 971-990 dext+ CD4 T cells.
After perfusing the CVB3-infected animals at termination, hearts were collected and processed to obtain MNCs as described in the Methods section. Cells were then stained with the indicated dextramers, anti-CD4 and 7-AAD. After acquiring by flow cytometry, the frequencies of dext+ CD4+ T cells were determined. The data sets represent mean ± SEM values derived from three to eight experiments each involving three to six mice. Controls, RNase 43-56 for IAk dextramers and MCC 82-103 for IEk dextramers. *P < 0.05 and **P < 0.01.
Figure 4: CVB3-infection leads to the generation of Myhc 338-348 tet+ CD8+ T cells.
(a) Splenocytes. Between 18 to 21 days post-infection with CVB3, splenocytes were prepared and stimulated with Myhc 334-352 for three days. After resting in IL-2 medium, cells harvested on day 8 or 9 were stained with CD4 and CD8 antibodies and 7-AAD and H-2Dd/Myhc 338-348 (specific) and HIV P18-I10 (control) tetramers. Cells were washed and acquired by flow cytometry to determine the frequencies of tet+ cells in the live (7-AAD−) CD8 subset. (b) Heart infiltrates. Hearts collected from CVB3-infected animals were processed to obtain MNCs. Cells were stained as above, and after acquiring by flow cytometry, frequencies of live (7-AAD−), tet+ CD8+ cells were enumerated. Representative flow cytometric plots and mean ± SEM values derived from four experiments each involving two to three mice are shown. *P<0.05 and **P<0.01.
3.8. Autoreactive T cells generated in CVB3-infected mice are pathogenic in nature
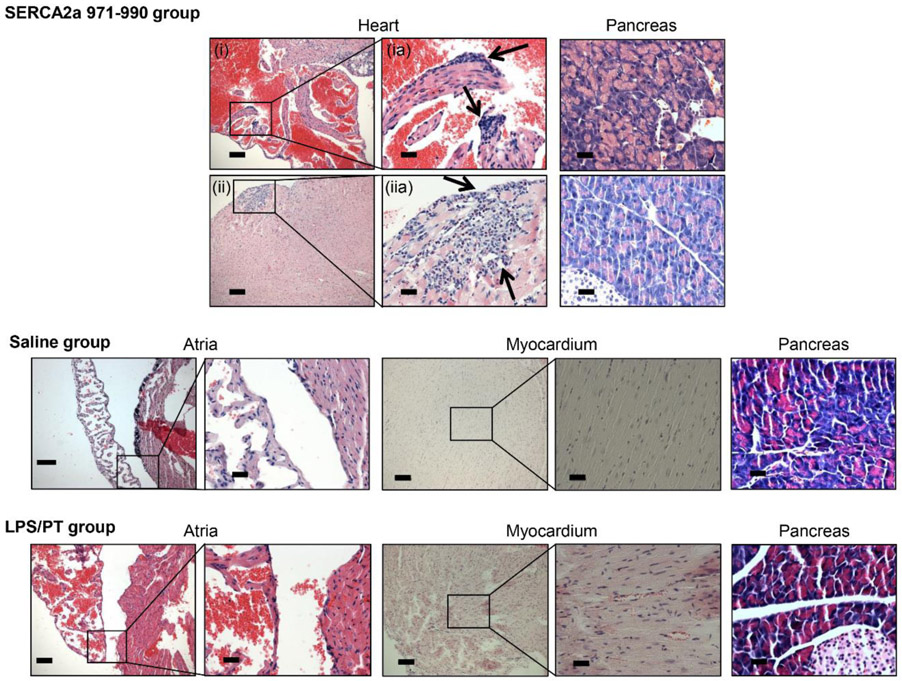
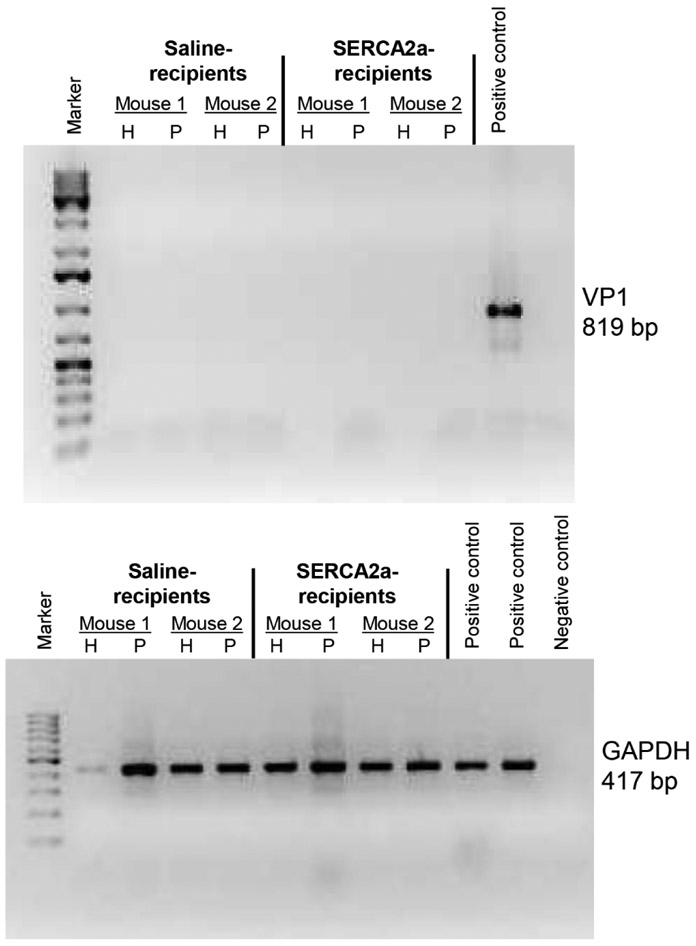
To determine the pathogenicity of autoreactive T cell responses generated in CVB3-infected mice, we used adoptive transfer protocol with an expectation that the autoreactive T cells can transfer disease to naïve animals as we had previously demonstrated for Myhc 334-352 (Gangaplara et al., 2012). In this study, we tested the pathogenic potential of SERCA2a 971-990-reactive T cells generated in CVB3-infected mice. As shown in the Figure 5, top left [atrium, (i), and myocardium of ventricle, (ii)] and their corresponding insets in the middle panels (ia, and iia), we noted that the naïve mice receiving T cells sensitized with SERCA2a 971-990 had infiltrations in the epicardium and myocardium with the lesions being apparent in the atria as compared to ventricles. However, pancreatic sections from these animals did not reveal any inflammatory changes (Figure 5, top right panel), indicating that the disease-inducing ability of SERCA2a 971-990-sensitized T cells was limited to heart. Similarly, heart, and pancreatic sections from control groups (saline and LPS/PT) (Figure 5, bottom panel) and also those from uninfused naïve mice (Figure S10, top panel) had no pathological changes as expected. Furthermore, by comparing the heart and pancreatic inflammatory changes in CVB3-infected mice with the recipients of SERCA2a 971-990-reactive T cells, we noted that sections from CVB3-infected mice had a widespread inflammatory foci accompanied with necrosis and mineralization in both hearts and pancreas (Figure S10, bottom panel). As reported previously, such a diffused nature is expected in infected animals that reflects damage resulting from both cytolytic properties of virus and immune response (Gangaplara et al., 2012; Garmaroudi et al., 2015; C. Massilamany et al., 2014b). We then sought to rule out a possibility of residual viral nucleic acid, if any in the cell suspensions used for adoptive transfer experiments. We performed RT-PCR analysis on the RNA samples prepared from hearts and pancreata harvested from the recipients of saline or cells to detect the presence of viral RNA using primers specific to CVB3 VP1 (Figure 6). The PCR analysis revealed no amplifications for VP1 suggesting that the inflammatory changes noted in the hearts from animals that received SERCA2a 971-990-reactive T cells are not due to residual virus. Taken together, our data provide evidence that CVB3-infection can result in the generation of pathogenic autoreactive T cells with specificities for multiple antigens supporting the notion that pathogens primarily affecting the target organs can secondarily result in the generation of pathogenic autoimmune responses.
Figure 5: SERCA2a-reactive T cells from CVB3-infected mice can induce myocarditis in naïve A/J mice.
Splenocytes were obtained from animals infected with CVB3 between 10 to 21 days post-infection, and the cells were stimulated with SERCA2a 971-990 for two days. After resting in IL-2 medium, SERCA2a-specific cells were expanded with Con-A and the viable cells were administered into naïve A/J mice that were primed with LPS and PT as described in the methods section, where, saline-recipients and LPS/PT-primed animals uninfused with cells were used as controls. At termination on day 14 post-transfer, hearts and pancreata were collected for histological evaluation by H and E staining. Sections in the top panel represent animals that received SERCA2a 971-990-reactive T cells, where infiltrations were noted in the atria (i, and inset, ia) and myocardium (ii, and inset, iia) are shown with arrows. The bottom panel represents sections from saline and LPS/PT-groups. Normal pancreatic sections are shown in the right panels in all the groups. Representative sections from groups of four animals are shown.
Figure 6: Evaluation of CVB3 RNA in mice that received SERCA2a 971-990-reactive T cells.
Total RNA was extracted from hearts and pancreata representing the indicated groups with two animals in each, and the cDNAs derived from these samples were evaluated for the presence of CVB3 VP1 (top panel) and GAPDH (housekeeping gene) by PCR using the sequence-specific primers. The PCR products analyzed by ethidium bromide (1.5%) in the agarose gel electrophoresis are shown. VP1 PCR product, positive control; water without template, negative control.
4. Discussion
In this report, we have described the characterization of autoreactive T cells, including cross-reactive T cell responses in viral myocarditis induced with CVB3. Previous reports indicate that antibodies and T cells specific to streptococcal M protein could cross-react with cardiac myosin, suggesting that cross-reactive T cells can contribute to the pathogenesis of rheumatic heart disease (Dale and Beachey, 1985; Pruksakorn et al., 1994). Additionally, while antibodies for ANT and CVB3 could cross-react with each other (Schwimmbeck et al., 1993), antibodies generated against one self-antigen, namely cardiac myosin could cross-react with another self-antigen β1AR, potentially leading to apoptosis of cardiomyocytes, thus supporting a role for antibodies in the CVB pathogenesis (Li et al., 2006). However, it was unknown whether pathogenic cross-reactive T cells can be generated in CVB3 infection for antigens that are commonly implicated as autoimmune targets in DCM patients. These include cardiac (Myhc, TNI, β1AR and SERCA2a) and/or non-cardiac (ANT and BCKDk) antigens (A. L. Caforio et al., 2001; A. L. Caforio et al., 2002; Kaya et al., 2012; D. A. Neumann et al., 1991). We identified three mimicry epitopes, CVB 1153-1169, CVB 1839-1853 and CVB 1527-1548, respectively for ANT 33-49, SERCA2a 799-813 and TNI 138-159. However, by analyzing their cross-reactive T cell responses in the immunization settings, none of them induced significant responses except that T cell reactivity was more apparent for ANT 33-49 than others, raising questions as to the relevance of molecular mimicry hypothesis in the causation of CVB3 myocarditis. First, the finding that lack of detection of cross-reactive T cells for antigens tested in our studies suggests that they do not contribute to the initiation and progression of myocarditis induced with CVB3. However, a possibility exists that the antibodies generated against these antigens may still be relevant to CVB3 pathogenesis. In support of this notion, immune complexes have been demonstrated for ANT and BCKDk in the serum from CVB3-infected animals (D. Neumann et al., 1994), but their pathogenic role remains unclear. Likewise, our data also do not rule out a possibility for appearance of cross-reactive T cells for other antigens that were not investigated in our studies. One such potential antigen that we have identified is LMKB/MARF1 (Dunster et al., 2005). In these scenarios however, pathogenicity of cross-reactive T cells needs to be demonstrated in adoptive transfer experiments to provide a proof-of concept that they are biologically relevant to CVB3 pathogenesis. Second, antibody cross-reactivity has been noted between ANT, actin, β1AR and myosin with CVB3 (Cunningham et al., 1992; Maisch et al., 1993; Mascaro-Blanco et al., 2008; Schulze and Schultheiss, 1995) that can also potentially result from formation of anti-idiotypic antibodies (Root-Bernstein and Fairweather, 2015). However, direct evidence is lacking as to the pathogenic role of such cross-reactive antibodies in CVB3 myocarditis. Nevertheless, it is possible that cross-reactive antibodies may indirectly contribute to viral pathogenesis as demonstrated with cardiac myosin and β1AR as described above (Li et al., 2006; Mascaro-Blanco et al., 2008).
We next investigated whether CVB3-infection can lead to the generation of autoreactive T cells with multiple antigen-specificities, since we had previously reported that CVB3 infection can result in the induction of pathogenic Myhc-reactive CD4 T cells (Gangaplara et al., 2012). We analyzed the frequencies of antigen-specific, CD4 T cells using a panel of seven MHC class II dextramers, and the analyses revealed detection of Myhc 334-352 dext+ CD4+ T cells as expected (Gangaplara et al., 2012). In addition, CD4 T cells positive for SERCA2a 971-990 dextramers also were detected in the splenocytes from CVB3 infected animals. These data reinforce the notion that the cardio tropic pathogens like CVB3 can cause damage to the myocardium leading to release of intracellular antigens such as Myhc and SERCA2a that can later become immunogenic in infected animals. Lack of detection of significant proportions of T cells for ANT 21-40, β1AR 181-200/211-230 and BCKDk 111-130 dextramers may imply that these antigens might not be released in CVB3 infection or these antigenic determinants might not have been naturally presented in the infected animals.
We had previously made an observation that CD4 T cells sensitized with BCKDk 111-130 could induce both autoimmune hepatitis and myocarditis in A/J mice immunized with BCKDk 111-130 (Bharathi Krishnan et al., 2017). We had proposed that BCKDk may be an autoimmune target for induction of hepatitis because its expression can occur in the liver (Muller and Danner, 2004). Additionally, CVB3 can also infect liver (Liu et al., 2013). Therefore, we performed dextramer analysis using the liver infiltrates in CVB3 infected animals leading to the detection of T cells positive for SERCA2a 971-990, Myhc 334-352 and ANT 21-40 dextramers, but not for other antigens. Detection of ANT 21-40-reactive T cells in the liver was not expected as T cells positive for ANT 21-40 dextramers were not significantly elevated in the splenocytes. Previous reports suggest that antigen-sensitized CD8 T cells, including autoreactive T cells, can migrate to liver and die by apoptosis (Crispe et al., 2000; Park et al., 2002). Although, we do not discount the possibility that the autoreactive T cells being detected in our studies may represent those that are destined to die, additional studies are needed to determine as to why only SERCA2a, Myhc and ANT-reactive CD4 T cells were selectively detected in the liver. It is also possible that SERCA2a 971-990 and ANT 21-40-sensitized T cells can cross-react with their isoforms, SERCA2b and ANT-2, respectively, whose expression can also occur in the liver (Arai et al., 1992; Levy et al., 2000). Importantly, whether autoreactive T cells localized in the liver can recirculate in infected animals also need detailed analysis. Likewise, although CD4 T cell exhaustion is poorly understood as compared to CD8 T cells (Saeidi et al., 2018), whether the autoreactive CD4+ T cells that we have observed in our studies possess exhaustion phenotype may be worth investigating in the future. However, since liver can be affected in CVB3 infection (Liu et al., 2013), autoreactive T cells localized in the liver may have a pathogenic role in CVB3 infection.
Finally, to determine the pathogenic significance of autoreactive T cells in the causation of myocarditis, we performed dextramer staining analysis on heart infiltrates and the analysis revealed the presence of CD4 T cells positive for SERCA2a 971-990 and Myhc 334-352 dextramers. Since, Myhc 334-352 was found to harbor a CD8 T cell determinant (Myhc 338-348) (Chandirasegaran Massilamany et al., 2016), we used MHC class I tetramers (H-2Dd) for Myhc 338-348. We demonstrate that the heart infiltrates from CVB3-infected animals contain Myhc 338-348 tet+ CD8+ T cells suggesting that Myhc-reactive CD8 T cells may contribute to the disease pathogenesis. In support of this proposition, we had previously demonstrated that the CD8 T cells generated in response to Myhc 334-352-immunization, induced myocarditis in naïve mice in adoptive transfer experiments (Chandirasegaran Massilamany et al., 2016). In this study, we demonstrate that the SERCA2a 971-990-reactive T cells generated in CVB3 infected animals are pathogenic as indicated by their ability to transfer myocarditis to naïve mice with the inflammatory lesions being frequently detected in the atria. This was an expected phenotype because A/J mice immunized with SERCA2a 971-990 develop mainly atrial myocarditis (B. Krishnan et al., 2018) that can be attributed to abundant expression of SERCA2a in the atria as compared to ventricles (Lüss et al., 1999).
In summary, by testing the molecular mimicry hypothesis, we did not observe cross-reactive T cells for autoantigens tested in our studies. Whether CVB3 infection can lead to the generation of cross-reactive T cells for other antigens needs further investigations. Such a possibility exists for LMKB/MARF1 as described above (Dunster et al., 2005). Conversely, our data support a possibility that CVB3 infection can lead to the generation of pathogenic autoreactive T cells with multiple antigen specificities. We envision that a combination of virus-induced damage to cardiomyocytes due to cytolytic properties of CVB3, and inflammatory damage induced by the host response may lead to the release of various intracellular and surface antigens including cryptic epitopes, and trigger the generation of autoreactive T cells, which in turn can infiltrate hearts and aggravate inflammation. Such a repertoire of cells can also be generated consequent to engulfment of dying cardiomyocytes resulting from virus-induced apoptosis (Esfandiarei and McManus, 2008) and autophagy (Garmaroudi et al., 2015) processes. A proportion of these autoreactive T cells may remain dormant with a possibility that their activation may occur through bystander activation up on exposure to non-specific infections, and they may contribute to the development of chronic myocarditis/DCM in CVB3-infected animals. Since T cells occupy a central role in the induction of autoimmunity, and their help is critical for B cells to produce antibodies, T cells may be the main mediators of cardiac autoimmunity in CVB3 infection.
Supplementary Material
Figure S1: Determination of T cell responses to self-antigens that possess sequences similar to CVB3 proteins. Splenocytes prepared from CVB3-infected animals were stimulated with the indicated peptides (100 μg/ml) for two days. Proliferative responses were measured as cpm after pulsing with3[H]-thymidine for 16 hours and stimulation indices were then calculated. Data are mean ± SEM of triplicate samples and are representative of two individual experiments with each involving 15 mice. RNase 43-56, control.
Figure S2: Analysis of T cell responses induced by mimicry epitopes in immunized animals, a. Immunization with self-peptides and testing for reactivity to self and CVB3 mimicry epitope. LNCs obtained from animals immunized with self-peptides were stimulated with both immunizing (self-peptides) and non-immunizing peptides (CVB3) for two days, and after pulsing with 3[H]-thymidine for 16 hours, proliferative responses were measured as cpm. b. Immunization with CVB3 mimicry epitopes and testing for reactivity to CVB3 mimicry and self-epitopes. LNCs obtained from CVB3 peptides were stimulated with the immunizing (CVB3 peptides) and self-peptides and the proliferative responses were measured as above. Mean ± SEM values representing two individual experiments each with five to ten mice are shown. RNase 43-56, control.
Figure S3: Structural characterization of CVB3 1153-1169 in relation to epitopes from ANT and MARF1. The top panel denotes sequence comparisons between mimicry peptides from CVB3 1153-1169 and ANT 33-49, and MARF1 874-890, where the identical residues are highlighted in green. The bottom left and middle panels respectively represent the helical structures of ANT and MARF1 (yellow), whereas the bottom right panel denotes the predicted helical structural model of CVB3 epitope.
Figure S4: Creation and validation of ANT 21-40 dextramers. a. Creation of MHC dextramers. The IAk monomers bearing the ANT 21-40 sequence were expressed in the Baculovirus system, and the monomers were then purified through an affinity column followed by biotinylation. Dextramers were then derived by mixing the optimized ratios of biotinylated IAk/ANT 21-40 protein and dextran molecules containing streptavidin and fluorophore at a ratio of 20:1 which were then used to stain CD4 T cells. b. Staining with ANT 21-40 dextramers. LNCs obtained from animals immunized with ANT 21-40 were restimulated with ANT 21-40 for two days and cells were rested in IL-2 medium. Cells were stained with the indicated dextramers between days 7 to 9 post-stimulation, followed by staining anti-CD4 and 7-AAD. After acquiring by flow cytometry, cells positive for dextramers were enumerated. Mean ± SEM values obtained from two individual experiments (left panel) with each involving three mice, and representative flow cytometric plots (right panel) are shown. RNase 43-56, control dextramer.
Figure S5: Schematic representation of the analysis of dextramer staining of lymphocytes harvested from CVB3-infected mice. Lymphocytes harvested from CVB3-infected mice were stimulated with antigens and rested in IL-2 medium. Cells were stained with dextramers, anti-CD4 and 7-AAD and the dext+CD4+ T cells were then analyzed as described in the methods section. An example of flow cytometric plots showing the gating strategies are shown.
Figure S6: Analysis of autoreactive CD4+ T cells in naïve mice by dextramer staining. Lymphocytes obtained from naïve animals were stimulated with Myhc 334-352, and SERCA2a 971-990 for three days, and cells were rested in IL-2 medium. Staining was performed using the indicated dextramers during 8 to 10 post-stimulation, anti-CD4 and 7-AAD and the dext+ CD4+ cells were then enumerated flow cytometrically. Mean ± SEM values are shown on the left panel (n=4 to 5) for each dextramer, whereas the right panels indicate the representative flow cytometric plots. RNase 43-56, control for IAk dextramers; and MCC 82-103, control for IEk dextramers.
Figure S7: Schematic representation of the analysis of dextramer staining of heart and liver MNCs harvested from CVB3-infected mice. Hearts and livers harvested from CVB3-infected mice were processed by enzymatic digestions to obtain MNC suspensions. Cells were stained with dextramers, anti-CD4 and 7-AAD and the dext+CD4+ T cells were then analyzed as described in the methods section. An example of flow cytometric plots showing the gating strategies for liver MNCs are shown.
Figure S8: Evaluation of PLP 139-151 dext+CD4+ T cells in SJL mice immunized with PLP 139-151. SJL mice were immunized with PLP 139-151, and after 10 days, livers and lymph nodes were harvested. The MNCs obtained from livers were stained ex vivo with IAs/PLP 139151 or TMEV 70-86 dextramers, anti-CD4 and 7-AAD to analyze the dext+ CD4+ T cells in the live subset (7-AAD−) (top panel). LNC were stimulated with PLP 139-151 and the cells rested in IL-2 medium were stained as above to analyze the frequencies of CD4+dext+ cells (bottom panel). Flow cytometric plots from three mice are shown. TMEV 70-86, control.
Figure S9: Analysis of Myhc 338-348 tet+ CD8+ T cells in naïve mice. Lymphocytes harvested from naïve mice (n=3) were stimulated with Myhc 334-352 for three days. After resting in IL-2 medium, cells harvested at two time points (day 8 and day 10) were stained with anti-CD8, indicated tetramers [H-2Dd/Myhc 338-348 (specific) and HIV P18-I10 (control)] and 7-AAD and the tet+ CD8+ T cells were then analyzed. The left panels denote the representative flow cytometric plots, whereas the mean ± SEM values are shown in the right panel.
Figure S10: Histological analysis of heart and pancreatic sections obtained from mice infected with or without CVB3. Heart and pancreatic sections were prepared from animals infected with or without CVB3. After H & E staining, sections were analyzed for inflammatory changes. While the top panel indicates representative sections from non-infected mice (n=5), the bottom panel denotes staining of sections from CVB3-infected animals (n=6), where the enlarged images are shown with insets in each. Arrows indicate inflammatory changes (atrium, infiltrates; myocardium, infiltrates with necrosis and mineralization; and pancreas, infiltrates, atrophy, and necrosis/mineralization).
Highlights.
CVB3 contains mimicry epitopes for ANT, SERCA2a and TNI, but none induced cross-reactive T cells
CVB3 infection can lead to the generation of T cells with specificities for multiple antigens
SERCA2a-reactive T cells were pathogenic
Autoreactive T cells were found localized in the livers of CVB3-infected animals
Acknowledgements
This research was supported by the National Institutes of Health [HL114669], the Transformational Grant by the American Heart Association [18TPA34170206], and the Biomedical Research Seed Grant by the University of Nebraska-Lincoln.
Abbreviations
- DCM
Dilated cardiomyopathy
- CVB3
Coxsackievirus B3
- Myhc
cardiac myosin heavy chain-α
- ANT
adenine nucleotide translocator 1
- β1AR
β1-adrenergic receptor
- BCKD
branched chain α-ketoacid dehydrogenase
- SERCA2a
sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a
- TNI
cardiac troponin I
- MHC
major histocompatibility complex
- CPE
cytopathic effect
- RNase
bovine ribonuclease
- HEL
hen egg lysozyme
- PLP
proteolipid protein
- HIV
human immunodeficiency virus
- MCC
moth cytochrome c
- CFA
complete Freund’s adjuvant
- LNCs
lymph node cells
- RPMI
Roswell Park Memorial Institute
- FBS
fetal bovine serum
- 3[H]
tritiated-thymidine
- cpm
counts per minute
- sf
Spodoptera frugiperda
- MNCs
mononuclear cells
- IL
Interleukin
- 7-AAD
7-aminoactinomycin-D
- DNasel
deoxyribonuclease I
- VP
viral protein
- TMEV
Theiler’s murine encephalomyelitis virus
- MARF1
meiosis regulator and mRNA stability factor 1
- LMKB
human Limkain B
- H & E
Hematoxylin and eosin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare no competing interests.
References
- Agarwal RK, Sun SH, Su SB, Chan CC, Caspi RR, 2002. Pertussis toxin alters the innate and the adaptive immune responses in a pertussis-dependent model of autoimmunity. J Neuroimmunol 129, 133–140. [DOI] [PubMed] [Google Scholar]
- Arai M, Otsu K, MAcLENNAN DH, Periasamy M, 1992. Regulation of sarcoplasmic reticulum gene expression during cardiac and skeletal muscle development. American Journal of Physiology-Cell Physiology 262, C614–C620. [DOI] [PubMed] [Google Scholar]
- Atkinson MA, Bowman MA, Campbell L, Darrow BL, Kaufman DL, Maclaren NK, 1994. Cellular immunity to a determinant common to glutamate decarboxylase and coxsackie virus in insulin-dependent diabetes. J Clin Invest 94, 2125–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboonian C, McKenna W, 2003. Eradication of viral myocarditis: is there hope? J Am Coll Cardiol 42, 473–476. [DOI] [PubMed] [Google Scholar]
- Basavalingappa RH, Massilamany C, Krishnan B, Gangaplara A, Kang G, Khalilzad-Sharghi V, Han Z, Othman S, Li Q, Riethoven J-J, 2016. Identification of an Epitope from Adenine Nucleotide Translocator 1 That Induces Inflammation in Heart in A/J Mice. The American Journal of Pathology 186, 3160–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavalingappa RH, Massilamany C, Krishnan B, Gangaplara A, Rajasekaran RA, Afzal MZ, Riethoven JJ, Strande JL, Steffen D, Reddy J, 2017. beta1-Adrenergic Receptor Contains Multiple IA(k) and IE(k) Binding Epitopes That Induce T Cell Responses with Varying Degrees of Autoimmune Myocarditis in A/J Mice. Front Immunol 8, 1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Horvath S, Matsumoto I, Fremont DH, Allen PM, 2000. Molecular basis for recognition of an arthritic peptide and a foreign epitope on distinct MHC molecules by a single TCR. J Immunol 164, 5788–5796. [DOI] [PubMed] [Google Scholar]
- Blauwet LA, Cooper LT, 2010. Myocarditis. Prog Cardiovasc Dis 52, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch DB, Li P, Bloch EG, Berenson DF, Galdos RL, Arora P, Malhotra R, Wu C, Yang W, 2014. LMKB/MARF1 localizes to mRNA processing bodies, interacts with Ge-1, and regulates IFI44L gene expression. PLoS One 9, e94784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caforio AL, Daliento L, Angelini A, Bottaro S, Vinci A, Dequal G, Tona F, Iliceto S, Thiene G, McKenna WJ, 2005. Autoimmune myocarditis and dilated cardiomyopathy: focus on cardiac autoantibodies. Lupus 14, 652–655. [DOI] [PubMed] [Google Scholar]
- Caforio AL, Mahon NJ, McKenna WJ, 2001. Cardiac autoantibodies to myosin and other heart-specific autoantigens in myocarditis and dilated cardiomyopathy. Autoimmunity 34, 199–204. [DOI] [PubMed] [Google Scholar]
- Caforio AL, Mahon NJ, Tona F, McKenna WJ, 2002. Circulating cardiac autoantibodies in dilated cardiomyopathy and myocarditis: pathogenetic and clinical significance. Eur J Heart Fail 4, 411–417. [DOI] [PubMed] [Google Scholar]
- Caforio AL, Pankuweit S, Arbustini E, Basso C, Gimeno-Blanes J, Felix SB, Fu M, Helio T, Heymans S, Jahns R, 2013. Current state of knowledge on aetiology, diagnosis, management, and therapy of myocarditis: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. European heart journal 34, 2636–2648. [DOI] [PubMed] [Google Scholar]
- Chi N-H, Chou N-K, Yu Y-H, Yu H-Y, Wu I-H, Chen Y-S, Huang S-C, Ko W-J, Wang S-S, 2014. Heart Transplantation in Endstage Rheumatic Heart Disease. Circulation Journal 78, 1900–1907. [DOI] [PubMed] [Google Scholar]
- Cihakova D, Rose NR, 2008. Pathogenesis of myocarditis and dilated cardiomyopathy. Adv Immunol 99, 95–114. [DOI] [PubMed] [Google Scholar]
- Crispe IN, Dao T, Klugewitz K, Mehal WZ, Metz DP, 2000. The liver as a site of T-cell apoptosis: graveyard, or killing field? Immunol Rev 174, 47–62. [DOI] [PubMed] [Google Scholar]
- Cunningham MW, Antone SM, Gulizia JM, McManus BM, Fischetti VA, Gauntt CJ, 1992. Cytotoxic and viral neutralizing antibodies crossreact with streptococcal M protein, enteroviruses, and human cardiac myosin. Proc Natl Acad Sci U S A 89, 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JB, Beachey EH, 1985. Epitopes of streptococcal M proteins shared with cardiac myosin. J Exp Med 162, 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MD, Hyland KV, Wang K, Engman DM, 2008. Recombinant cardiac myosin fragment induces experimental autoimmune myocarditis via activation of Th1 and Th17 immunity. Autoimmunity 41, 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donermeyer DL, Beisel KW, Allen PM, Smith SC, 1995. Myocarditis-inducing epitope of myosin binds constitutively and stably to I-Ak on antigen-presenting cells in the heart. The Journal of experimental medicine 182, 1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunster K, Lai FP, Sentry JW, 2005. Limkain b1, a novel human autoantigen localized to a subset of ABCD3 and PXF marked peroxisomes. Clin Exp Immunol 140, 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiarei M, McManus BM, 2008. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol 3, 127–155. [DOI] [PubMed] [Google Scholar]
- Fairweather D, Stafford KA, Sung YK, 2012. Update on coxsackievirus B3 myocarditis. Curr Opin Rheumatol 24, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto C, Yu CR, Shi G, Vistica BP, Wawrousek EF, Klinman DM, Chan CC, Egwuagu CE, Gery I, 2006. Pertussis toxin is superior to TLR ligands in enhancing pathogenic autoimmunity, targeted at a neo-self antigen, by triggering robust expansion of Th1 cells and their cytokine production. J Immunol 177, 6896–6903. [DOI] [PubMed] [Google Scholar]
- Fujinami RS, Von Herrath MG, Christen U, Whitton JL, 2006. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clinical microbiology reviews 19, 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung G, Luo H, Qiu Y, Yang D, McManus B, 2016. Myocarditis. Circ Res 118, 496–514. [DOI] [PubMed] [Google Scholar]
- Gangaplara A, Massilamany C, Brown DM, Delhon G, Pattnaik AK, Chapman N, Rose N, Steffen D, Reddy J, 2012. Coxsackievirus B3 infection leads to the generation of cardiac myosin heavy chain-α-reactive CD4 T cells in A/J mice. Clinical immunology 144, 237–249. [DOI] [PubMed] [Google Scholar]
- Garmaroudi FS, Marchant D, Hendry R, Luo H, Yang D, Ye X, Shi J, McManus BM, 2015. Coxsackievirus B3 replication and pathogenesis. Future Microbiol 10, 629–653. [DOI] [PubMed] [Google Scholar]
- Gautam AM, Liblau R, Chelvanayagam G, Steinman L, Boston T, 1998. A viral peptide with limited homology to a self peptide can induce clinical signs of experimental autoimmune encephalomyelitis. J Immunol 161, 60–64. [PubMed] [Google Scholar]
- Hamada Y, Takata M, Kiyoku H, Enzan H, Doi Y, Fujimoto S, 2004. Monomethoxypolyethylene glycol-modified cardiac myosin treatment blocks the active and passive induction of experimental autoimmune myocarditis. Circ J 68, 149–155. [DOI] [PubMed] [Google Scholar]
- Huber S, Polgar J, Moraska A, Cunningham M, Schwimmbeck P, Schultheiss P, 1993. T lymphocyte responses in CVB3-induced murine myocarditis. Scand J Infect Dis Suppl 88, 67–78. [PubMed] [Google Scholar]
- Kaufman DL, Erlander MG, Clare-Salzler M, Atkinson MA, Maclaren NK, Tobin AJ, 1992. Autoimmunity to two forms of glutamate decarboxylase in insulin-dependent diabetes mellitus. J Clin Invest 89, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya Z, Leib C, Katus HA, 2012. Autoantibodies in heart failure and cardiac dysfunction. Circ Res 110, 145–158. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Massilamany C, Basavalingappa RH, Gangaplara A, Kang G, Li Q, Uzal FA, Strande JL, Delhon GA, Riethoven JJ, 2017. Branched chain α-ketoacid dehydrogenase kinase 111-130, a T cell epitope that induces both autoimmune myocarditis and hepatitis in A/J mice. Immunity, Inflammation and Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Massilamany C, Basavalingappa RH, Gangaplara A, Rajasekaran RA, Afzal MZ, Khalilzad-Sharghi V, Zhou Y, Riethoven JJ, Nandi SS, Mishra PK, Sobel RA, Strande JL, Steffen D, Reddy J, 2018. Epitope Mapping of SERCA2a Identifies an Antigenic Determinant That Induces Mainly Atrial Myocarditis in A/J Mice. J Immunol 200, 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, Poller W, Kandolf R, Schultheiss HP, 2005. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with "idiopathic" left ventricular dysfunction. Circulation 111, 887–893. [DOI] [PubMed] [Google Scholar]
- Lasrado N, Yalaka B, Reddy J, 2020. Triggers of Inflammatory Heart Disease. Frontiers in Cell and Developmental Biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SE, Chen YS, Graham BH, Wallace DC, 2000. Expression and sequence analysis of the mouse adenine nucleotide translocase 1 and 2 genes. Gene 254, 57–66. [DOI] [PubMed] [Google Scholar]
- Li Y, Heuser JS, Cunningham LC, Kosanke SD, Cunningham MW, 2006. Mimicry and antibody-mediated cell signaling in autoimmune myocarditis. J Immunol 177, 8234–8240. [DOI] [PubMed] [Google Scholar]
- Liu JY, Wang SM, Chen IC, Yu CK, Liu CC, 2013. Hepatic damage caused by coxsackievirus B3 is dependent on age-related tissue tropisms associated with the coxsackievirus-adenovirus receptor. Pathog Dis 68, 52–60. [DOI] [PubMed] [Google Scholar]
- Lorenz RG, Tyler AN, Allen PM, 1988. T cell recognition of bovine ribonuclease. Self/non-self discrimination at the level of binding to the I-Ak molecule. J Immunol 141, 4124–4128. [PubMed] [Google Scholar]
- Liiss T, Boknik P, Jones LR, Kirchhefer U, Knapp J, Linck B, Liiss H, Meissner A, Muller FU, Schmitz W, 1999. Expression of cardiac calcium regulatory proteins in atrium v ventricle in different species. Journal of molecular and cellular cardiology 31, 1299–1314. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R, 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch B, Bauer E, Cirsi M, Kochsiek K, 1993. Cytolytic cross-reactive antibodies directed against the cardiac membrane and viral proteins in coxsackievirus B3 and B4 myocarditis. Characterization and pathogenetic relevance. Circulation 87, IV49–65. [PubMed] [Google Scholar]
- Mann HB, Whitney DR, 1947. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Statist 18, 50–60. [Google Scholar]
- Mascaro-Blanco A, Alvarez K, Yu X, Lindenfeld J, Olansky L, Lyons T, Duvall D, Heuser JS, Gosmanova A, Rubenstein CJ, Cooper LT, Kem DC, Cunningham MW, 2008. Consequences of unlocking the cardiac myosin molecule in human myocarditis and cardiomyopathies. Autoimmunity 41, 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massilamany C, Gangaplara A, Basavalingappa RH, Rajasekaran RA, Khalilzad-Sharghi V, Han Z, Othman S, Steffen D, Reddy J, 2016. Localization of CD8 T cell epitope within cardiac myosin heavy chain-α 334-352 that induces autoimmune myocarditis in A/J mice. International journal of cardiology 202, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massilamany C, Gangaplara A, Chapman N, Rose N, Reddy J, 2011a. Detection of cardiac myosin heavy chain-alpha-specific CD4 cells by using MHC class II/IA(k) tetramers in A/J mice. J Immunol Methods 372, 107–118. [DOI] [PubMed] [Google Scholar]
- Massilamany C, Gangaplara A, Jia T, Elowsky C, Li Q, Zhou Y, Reddy J, 2014a. In situ detection of autoreactive CD4 T cells in brain and heart using major histocompatibility complex class II dextramers. J Vis Exp, e51679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massilamany C, Gangaplara A, Reddy J, 2014b. Intricacies of cardiac damage in coxsackievirus B3 infection: implications for therapy. Int J Cardiol 177, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massilamany C, Gangaplara A, Steffen D, Reddy J, 2011. Identification of novel mimicry epitopes for cardiac myosin heavy chain-α that induce autoimmune myocarditis in A/J mice. Cellular immunology 271, 438–449. [DOI] [PubMed] [Google Scholar]
- Massilamany C, Steffen D, Reddy J, 2010. An epitope from Acanthamoeba castellanii that cross-react with proteolipid protein 139-151-reactive T cells induces autoimmune encephalomyelitis in SJL mice. J Neuroimmunol 219, 17–24. [DOI] [PubMed] [Google Scholar]
- Massilamany C, Thulasingam S, Steffen D, Reddy J, 2011b. Gender differences in CNS autoimmunity induced by mimicry epitope for PLP 139-151 in SJL mice. J Neuroimmunol 230, 95–104. [DOI] [PubMed] [Google Scholar]
- Massilamany C, Upadhyaya B, Gangaplara A, Kuszynski C, Reddy J, 2011c. Detection of autoreactive CD4 T cells using major histocompatibility complex class II dextramers. BMC Immunol 12, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SD, Karpus WJ, 2007. Experimental autoimmune encephalomyelitis in the mouse. Curr Protoc Immunol Chapter 15, Unit 15 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor PD, Ferguson M, Evans DM, Almond JW, Icenogle JP, 1986. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J Gen Virol 67 (Pt 7), 1283–1291. [DOI] [PubMed] [Google Scholar]
- Muller EA, Danner DJ, 2004. Tissue-specific translation of murine branched-chain α-ketoacid dehydrogenase kinase mRNA is dependent upon an upstream open reading frame in the 5’-untranslated region. Journal of Biological Chemistry 279, 44645–44655. [DOI] [PubMed] [Google Scholar]
- Neumann D, Rose N, Ansari A, Herskowitz A, 1994. Induction of multiple heart autoantibodies in mice with coxsackievirus B3-and cardiac myosin-induced autoimmune myocarditis. The Journal of Immunology 152, 343–350. [PubMed] [Google Scholar]
- Neumann DA, Lane JR, LaFond-Walker A, Allen GS, Wulff SM, Herskowitz A, Rose NR, 1991. Heart-specific autoantibodies can be eluted from the hearts of Coxsackievirus B3-infected mice. Clin Exp Immunol 86, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Murray D, John B, Crispe IN, 2002. Biology and significance of T-cell apoptosis in the liver. Immunol Cell Biol 80, 74–83. [DOI] [PubMed] [Google Scholar]
- Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G, 2003. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44. [DOI] [PubMed] [Google Scholar]
- Pollack A, Kontorovich AR, Fuster V, Dec GW, 2015. Viral myocarditis--diagnosis, treatment options, and current controversies. Nat Rev Cardiol 12, 670–680. [DOI] [PubMed] [Google Scholar]
- Pruksakorn S, Currie B, Brandt E, Phornphutkul C, Hunsakunachai S, Manmontri A, Robinson JH, Kehoe MA, Galbraith A, Good MF, 1994. Identification of T cell autoepitopes that cross-react with the C-terminal segment of the M protein of group A streptococci. Int Immunol 6, 1235–1244. [DOI] [PubMed] [Google Scholar]
- Rabausch-Starz I, Schwaiger A, Grunewald K, Muller-Hermelink HK, Neu N, 1994. Persistence of virus and viral genome in myocardium after coxsackievirus B3-induced murine myocarditis. Clin Exp Immunol 96, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy J, Bettelli E, Nicholson L, Waldner H, Jang MH, Wucherpfennig KW, Kuchroo VK, 2003. Detection of autoreactive myelin proteolipid protein 139-151-specific T cells by using MHC II (IAs) tetramers. J Immunol 170, 870–877. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H, 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27, 493–497. [Google Scholar]
- Root-Bernstein R, Fairweather D, 2015. Unresolved issues in theories of autoimmune disease using myocarditis as a framework. J Theor Biol 375, 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, 2016. Viral myocarditis. Curr Opin Rheumatol 28, 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Wolfgram LJ, Herskowitz A, Beisel KW, 1986. Postinfectious autoimmunity: Two distinct phases of coxsackievirus B3-induced myocarditis. Annals of the New York Academy of Sciences 475, 146–156. [DOI] [PubMed] [Google Scholar]
- Saeidi A, Zandi K, Cheok YY, Saeidi H, Wong WF, Lee CYQ, Cheong HC, Yong YK, Larsson M, Shankar EM, 2018. T-Cell Exhaustion in Chronic Infections: Reversing the State of Exhaustion and Reinvigorating Optimal Protective Immune Responses. Front Immunol 9, 2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer EB, Sette A, Johnson DL, Bekoff MC, Smith JA, Grey HM, Buus S, 1989. Relative contribution of "determinant selection" and "holes in the T-cell repertoire" to T-cell responses. Proc Natl Acad Sci U S A 86, 4649–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K, Schultheiss HP, 1995. The role of the ADP/ATP carrier in the pathogenesis of viral heart disease. Eur Heart J 16 Suppl O, 64–67. [DOI] [PubMed] [Google Scholar]
- Schwimmbeck PL, Schwimmbeck NK, Schultheiss HP, Strauer BE, 1993. Mapping of antigenic determinants of the adenine-nucleotide translocator and coxsackie B3 virus with synthetic peptides: use for the diagnosis of viral heart disease. Clin Immunol Immunopathol 68, 135–140. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F, 1945. Individual Comparisons by Ranking Methods. Biometrics Bulletin 1, 80–83. [Google Scholar]
- Wooldridge L, Ekeruche-Makinde J, van den Berg HA, Skowera A, Miles JJ, Tan MP, Dolton G, Clement M, Llewellyn-Lacey S, Price DA, Peakman M, Sewell AK, 2012. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem 287, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Cao G, Li M, Wu B, Zhang X, Zhang T, Guo J, Yin H, Shi L, Chen J, Yu X, Zheng L, Ma J, Su YQ, 2018. Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice. Proc Natl Acad Sci U S A 115, 11250–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Kim BS, 1994. A predominant viral epitope recognized by T cells from the periphery and demyelinating lesions of SJL/J mice infected with Theiler's virus is located within VP1(233-244). J Immunol 153, 4508–4519. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Determination of T cell responses to self-antigens that possess sequences similar to CVB3 proteins. Splenocytes prepared from CVB3-infected animals were stimulated with the indicated peptides (100 μg/ml) for two days. Proliferative responses were measured as cpm after pulsing with3[H]-thymidine for 16 hours and stimulation indices were then calculated. Data are mean ± SEM of triplicate samples and are representative of two individual experiments with each involving 15 mice. RNase 43-56, control.
Figure S2: Analysis of T cell responses induced by mimicry epitopes in immunized animals, a. Immunization with self-peptides and testing for reactivity to self and CVB3 mimicry epitope. LNCs obtained from animals immunized with self-peptides were stimulated with both immunizing (self-peptides) and non-immunizing peptides (CVB3) for two days, and after pulsing with 3[H]-thymidine for 16 hours, proliferative responses were measured as cpm. b. Immunization with CVB3 mimicry epitopes and testing for reactivity to CVB3 mimicry and self-epitopes. LNCs obtained from CVB3 peptides were stimulated with the immunizing (CVB3 peptides) and self-peptides and the proliferative responses were measured as above. Mean ± SEM values representing two individual experiments each with five to ten mice are shown. RNase 43-56, control.
Figure S3: Structural characterization of CVB3 1153-1169 in relation to epitopes from ANT and MARF1. The top panel denotes sequence comparisons between mimicry peptides from CVB3 1153-1169 and ANT 33-49, and MARF1 874-890, where the identical residues are highlighted in green. The bottom left and middle panels respectively represent the helical structures of ANT and MARF1 (yellow), whereas the bottom right panel denotes the predicted helical structural model of CVB3 epitope.
Figure S4: Creation and validation of ANT 21-40 dextramers. a. Creation of MHC dextramers. The IAk monomers bearing the ANT 21-40 sequence were expressed in the Baculovirus system, and the monomers were then purified through an affinity column followed by biotinylation. Dextramers were then derived by mixing the optimized ratios of biotinylated IAk/ANT 21-40 protein and dextran molecules containing streptavidin and fluorophore at a ratio of 20:1 which were then used to stain CD4 T cells. b. Staining with ANT 21-40 dextramers. LNCs obtained from animals immunized with ANT 21-40 were restimulated with ANT 21-40 for two days and cells were rested in IL-2 medium. Cells were stained with the indicated dextramers between days 7 to 9 post-stimulation, followed by staining anti-CD4 and 7-AAD. After acquiring by flow cytometry, cells positive for dextramers were enumerated. Mean ± SEM values obtained from two individual experiments (left panel) with each involving three mice, and representative flow cytometric plots (right panel) are shown. RNase 43-56, control dextramer.
Figure S5: Schematic representation of the analysis of dextramer staining of lymphocytes harvested from CVB3-infected mice. Lymphocytes harvested from CVB3-infected mice were stimulated with antigens and rested in IL-2 medium. Cells were stained with dextramers, anti-CD4 and 7-AAD and the dext+CD4+ T cells were then analyzed as described in the methods section. An example of flow cytometric plots showing the gating strategies are shown.
Figure S6: Analysis of autoreactive CD4+ T cells in naïve mice by dextramer staining. Lymphocytes obtained from naïve animals were stimulated with Myhc 334-352, and SERCA2a 971-990 for three days, and cells were rested in IL-2 medium. Staining was performed using the indicated dextramers during 8 to 10 post-stimulation, anti-CD4 and 7-AAD and the dext+ CD4+ cells were then enumerated flow cytometrically. Mean ± SEM values are shown on the left panel (n=4 to 5) for each dextramer, whereas the right panels indicate the representative flow cytometric plots. RNase 43-56, control for IAk dextramers; and MCC 82-103, control for IEk dextramers.
Figure S7: Schematic representation of the analysis of dextramer staining of heart and liver MNCs harvested from CVB3-infected mice. Hearts and livers harvested from CVB3-infected mice were processed by enzymatic digestions to obtain MNC suspensions. Cells were stained with dextramers, anti-CD4 and 7-AAD and the dext+CD4+ T cells were then analyzed as described in the methods section. An example of flow cytometric plots showing the gating strategies for liver MNCs are shown.
Figure S8: Evaluation of PLP 139-151 dext+CD4+ T cells in SJL mice immunized with PLP 139-151. SJL mice were immunized with PLP 139-151, and after 10 days, livers and lymph nodes were harvested. The MNCs obtained from livers were stained ex vivo with IAs/PLP 139151 or TMEV 70-86 dextramers, anti-CD4 and 7-AAD to analyze the dext+ CD4+ T cells in the live subset (7-AAD−) (top panel). LNC were stimulated with PLP 139-151 and the cells rested in IL-2 medium were stained as above to analyze the frequencies of CD4+dext+ cells (bottom panel). Flow cytometric plots from three mice are shown. TMEV 70-86, control.
Figure S9: Analysis of Myhc 338-348 tet+ CD8+ T cells in naïve mice. Lymphocytes harvested from naïve mice (n=3) were stimulated with Myhc 334-352 for three days. After resting in IL-2 medium, cells harvested at two time points (day 8 and day 10) were stained with anti-CD8, indicated tetramers [H-2Dd/Myhc 338-348 (specific) and HIV P18-I10 (control)] and 7-AAD and the tet+ CD8+ T cells were then analyzed. The left panels denote the representative flow cytometric plots, whereas the mean ± SEM values are shown in the right panel.
Figure S10: Histological analysis of heart and pancreatic sections obtained from mice infected with or without CVB3. Heart and pancreatic sections were prepared from animals infected with or without CVB3. After H & E staining, sections were analyzed for inflammatory changes. While the top panel indicates representative sections from non-infected mice (n=5), the bottom panel denotes staining of sections from CVB3-infected animals (n=6), where the enlarged images are shown with insets in each. Arrows indicate inflammatory changes (atrium, infiltrates; myocardium, infiltrates with necrosis and mineralization; and pancreas, infiltrates, atrophy, and necrosis/mineralization).