Summary
Pluripotency is tightly regulated and is crucial for stem cells and their implementation for regenerative medicine. Non-coding RNAs, especially long non-coding RNAs (lncRNAs) emerged as orchestrators of versatile (patho)-physiological processes on the transcriptional and post-transcriptional level. Cyrano, a well-conserved lncRNA, is highly expressed in stem cells suggesting an important role in pluripotency, which we aimed to investigate in loss-off-function (LOF) experiments. Cyrano was described previously to be essential for the maintenance of mouse embryonic stem cell (ESC) pluripotency. In contrast, using different genetic models, we here found Cyrano to be dispensable in murine and human iPSCs and in human ESCs. RNA sequencing revealed only a moderate influence of Cyrano on the global transcriptome. In line, Cyrano-depleted iPSCs retained the potential to differentiate into the three germ layers. In conclusion, different methods were applied for LOF studies to rule out potential off-target effects. These approaches revealed that Cyrano does not impact pluripotency.
Keywords: lncRNA, Cyrano, iPSCs, pluripotency, CRISPR/Cas, non-coding RNA, OIP5-AS1, stem cells
Highlights
-
•
lncRNA Cyrano does not impact pluripotency and differentiation capacity of PSCs
-
•
Genetic deletion of Cyrano has no pluripotency phenotype in mouse iPSCs
-
•
CRISPRi- and siRNA-mediated knockdown revealed same effects in human PSCs
LncRNA Cyrano is species conserved and highly expressed in stem cells, suggesting an important role that was previously assigned to pluripotency of mouse ESCs. Here, Bär and colleagues show in contrast that Cyrano is dispensable in different genetic models in murine, human iPSCs, and human ESCs. Neither pluripotency nor the differentiation capacity of PSCs is disrupted by silencing Cyrano.
Introduction
The use of embryonic stem cells (ESCs) has been an invaluable tool for basic cell and biomedical research for more than two decades. The discovery of cellular reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006) revolutionized the field of regenerative medicine. These iPSCs can be generated from patient material and subsequently differentiated into a desired cell type for allogenic or ideally autologous cell therapy. In addition, iPSC technology enabled disease-in-a-dish modeling, for example, to better understand monogenetic disorders or to perform drug screenings in a personalized manner.
In stem cells the maintenance of pluripotency and self-renewal are crucial processes necessary to retain the differentiation capacity into various cell types of the three germ layers. In addition to the core pluripotency factors Oct4, Sox2, and Nanog, chromatin- and RNA-mediated mechanisms are involved in the regulation of pluripotency, which are not yet fully understood (Li and Belmonte, 2017). During differentiation, pluripotency and developmental genes undergo various changes, including altered chromatin interactions, histone modifications, and subnuclear localization. This leads to a change in expression patterns of genes connected to pluripotency, features of stem cells, and differentiated cell types.
Another level of complexity in the regulation of pluripotency is added by non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNA. lncRNAs constitute the largest and most heterogeneous subclass of ncRNAs classified by a transcript length of over 200 nucleotides. They play crucial roles in many physiological and pathophysiological processes (Beermann et al., 2016) and regulate gene expression transcriptionally and post-transcriptionally by manifold mechanisms (Bär et al., 2016, Nelson et al., 2016). However, due to their large number (lncipedia.org currently annotates 56,946 human lncRNA genes) and diversity, lncRNA research is still in its infancy and many lncRNA-controlled processes remain elusive. Additional challenges for studying lncRNAs are their frequently low level of expression and relatively poor sequence conservation, even between closely related species.
In contrast, lncRNA Cyrano (OIP5-AS1) is both locus- and sequence-conserved with a high complementary miRNA-7 binding site in an ultra-conserved region (Ulitsky et al., 2011). Cyrano was first described in zebrafish embryogenesis, necessary for the development of eyes and brain (Ulitsky et al., 2011). In HeLa cells Cyrano acts as a sponge for the RNA-binding protein HuR which influences proliferation. If HuR is not sponged by Cyrano, it stabilizes its target mRNAs, which code for pro-proliferative proteins, such as cyclin A2 and D1 (Kim et al., 2016). Apoptosis is regulated by Cyrano in vascular endothelial cells upon treatment with oxidative low-density lipoprotein (Wang et al., 2019). Moreover, Cyrano is part of a regulatory network of ncRNAs, including miRNA-7 in murine brains (Kleaveland et al., 2018), and a regulatory role in pluripotency and self-renewal via miRNA-7 was proposed in murine ESCs (Smith et al., 2017).
The high conservation of Cyrano between vertebrates and its stem cell-enriched expression suggested an important regulatory role of this lncRNA, which prompted us to generate Cyrano knockout (KO) iPSCs. Surprisingly, Cyrano was concurrently described as an essential lncRNA for the maintenance of pluripotency and self-renewal in ESCs (Smith et al., 2017). Since this was in marked contrast to our results in Cyrano KO iPSCs, we decided to further investigate the specific role of Cyrano in pluripotency and self-renewal using different CRISPR/Cas9 and small interfering RNA (siRNA) approaches in murine and human PSCs.
Results
Cyrano Is Highly Expressed in iPSCs
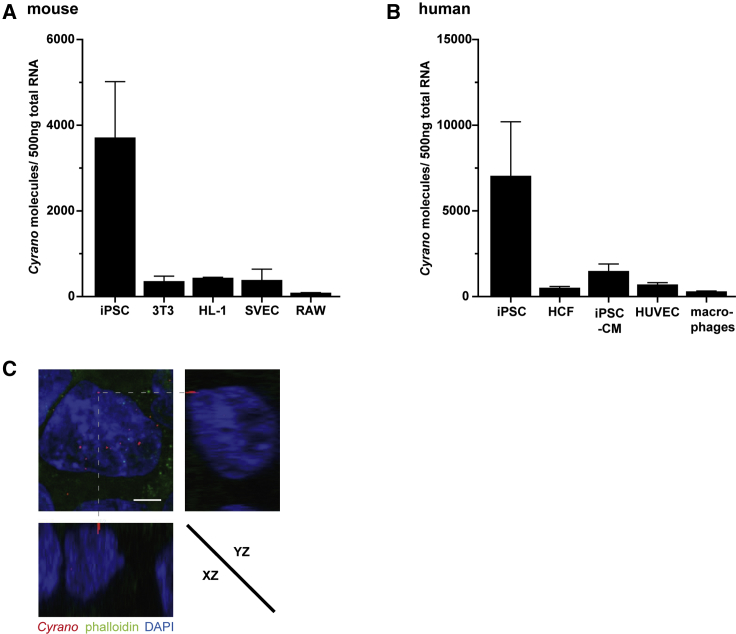
The remarkable species conservation prompted us to quantify Cyrano molecules in commonly used murine and human cell lines, as the expression of lncRNAs is often cell-type-specific (Beermann et al., 2016). A plasmid standard containing a sequence of the murine or human Cyrano transcript, respectively, was used for absolute quantification by qRT-PCR. Both human and murine Cyrano were strongly enriched in iPSCs compared with somatic cell lines (Figures 1A and 1B). RNA fluorescence in situ hybridization (FISH) with a Cyrano-specific probe followed by confocal microscopy and 3D reconstruction revealed a cytoplasmic localization in iPSCs (Figures 1C and S1).
Figure 1.
Cyrano Is Highly Enriched in iPSCs
Absolute quantification of murine (A) and human (B) Cyrano in different cell lines. qRT-PCR and a plasmid standard containing parts of murine or human Cyrano were used for quantification, n = 3 technical replicates. SVEC, seminal vesicle epithelial cells; RAW264.7 macrophages; HCF, human cardiac fibroblasts; iPSC-CM, iPSC-derived cardiomyocytes; HUVEC, human umbilical vein endothelial cells; macrophages, peripheral blood mononuclear cell-derived macrophages.
(C) Representative RNA FISH images stained with a Cyrano-specific probe and phalloidin in unmodified human iPSCs as 3D reconstruction. Scale bar, 5 μm.
Cyrano Is Dispensable for Maintenance of Pluripotency in Murine iPSCs
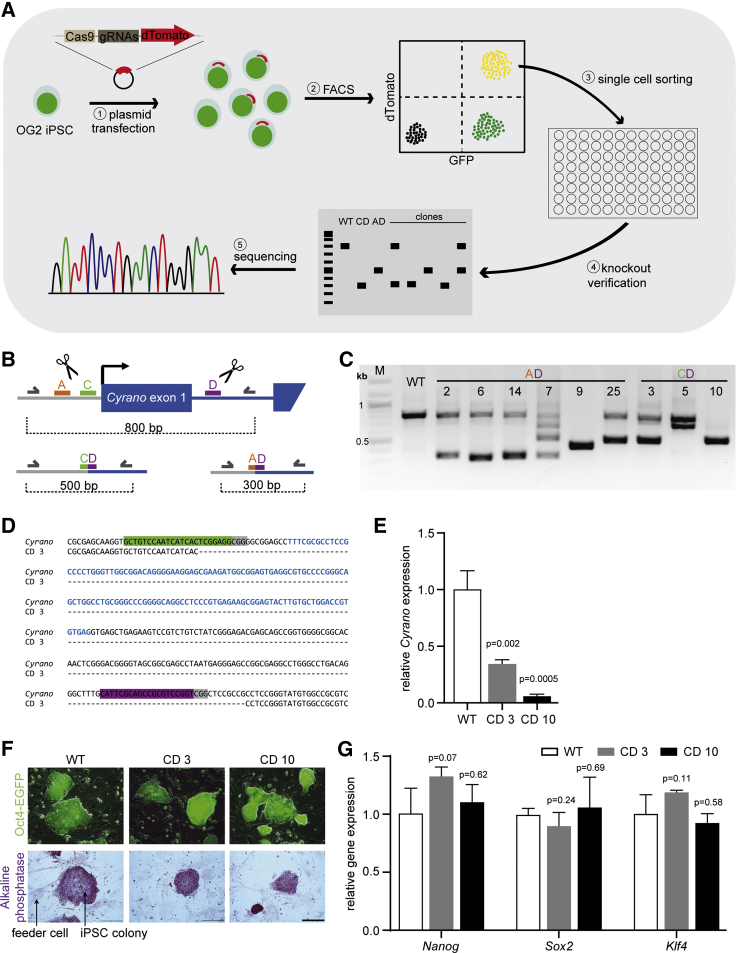
To address the role of Cyrano in pluripotency, OG2 iPSCs (expressing a pOCT4-EGFP transgene) (Kensah et al., 2013) were used to generate Cyrano KO cells by a dual guide RNA (gRNA) CRISPR/Cas9 approach (Heckl et al., 2014) (Figure 2A). The gRNAs were designed to excise the first exon of Cyrano, including parts of the promoter (Figure 2B). The dual gRNA vector, which also carries a dTomato fluorescence marker, was transiently transfected into OG2 iPSCs. Transfected cells were sorted by FACS and seeded as single cells for clonal expansion. Homo- and heterozygous KO clones for both gRNA combinations were identified by PCR in genomic DNA (Figure 2C) and successful excision confirmed by Sanger sequencing (Figure 2D). One heterozygous and one homozygous KO clone were selected for further analysis which, as expected, showed a significant reduction or complete elimination of Cyrano RNA expression as determined by qRT-PCR (Figure 2E).
Figure 2.
Cyrano Is Dispensable for Self-Renewal and Maintenance of Pluripotency in Cyrano KO Murine iPSCs
(A) Schematic overview of the generation of murine Cyrano KO iPSCs by CRISPR/Cas9.
(B) Graphical representation of three gRNAs (A, C, D) up- and downstream of the first exon of Cyrano. Primers used for PCR and sequencing are depicted as gray arrows, which amplify fragments of 800 bp in wild-type (WT) and 500 bp (using gRNA C and D) or 300 bp (using gRNA A and D) after CRISPR/Cas9 excision.
(C) Verification of Cyrano KO via PCR. Unmodified cells were loaded as control (WT).
(D) Example of Sanger sequencing results of clone CD 3 compared with WT Cyrano sequence. gRNA sequences are highlighted in green and purple with PAM sequences in gray, the sequence of the first exon is depicted in blue letters.
(E and G) Relative expression levels in KO and unmodified cells (WT). Expression levels were measured by qPCR, mean ± SD of three independent experiments are shown, unpaired t test was performed for statistical analysis.
(F) Representative images Oct4-EGFP expression and alkaline phosphatase staining of KO and unmodified cells (WT). iPSCs were cultured on feeder cells. Scale bar, 100 μm.
Based on a previous report linking Cyrano to pluripotency maintenance (Smith et al., 2017), Cyrano KO iPSCs were analyzed for their pluripotency. The morphology of the cells appeared regular for iPSCs and the expression of pOCT4-EGFP showed no differences. In addition, the expression of alkaline phosphatase was not altered in Cyrano KO iPSCs compared with wild-type iPSCs (Figure 2F). The mRNA expression levels of Nanog, Sox2, and Klf4 did not change in response to the loss of Cyrano (Figure 2G). Thus, our data demonstrate that Cyrano is not required for pluripotency in murine iPSCs.
Knockdown of Cyrano Does Not Interfere with Pluripotency in Human iPSCs
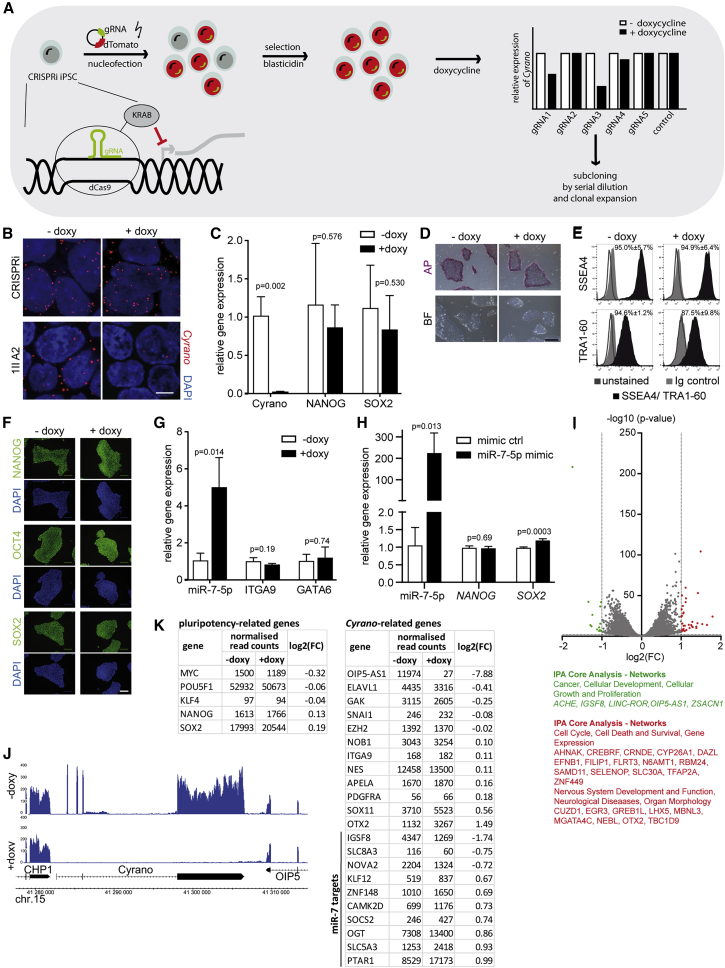
We next set out to investigate the role of Cyrano in human iPSCs. In contrast to the genetic KO approach in murine iPSCs, a CRISPRi system (Mandegar et al., 2016) was used for inducible, highly efficient Cyrano knockdown (KD) (Figure 3A). Five different gRNAs, located around 150 bp up- and downstream of the transcription start site were designed, cloned into pgRNA-CKB, and nucleofected into CRISPRi iPSCs (Figure S2A). Bulk RNA expression analysis after dCas9 induction by doxycycline showed a significant downregulation of Cyrano for gRNA3 (Figure S2B). After clonal expansion, 21 colonies were treated with doxycycline to shut down Cyrano expression, which did not result in morphological abnormalities. Four clones were selected for further analyses based on their strong repression of Cyrano (Figure S2C). The nearly complete ablation of Cyrano was verified by RNA FISH. Specific Cyrano signals were comparable in control cells (no doxycycline or no gRNA), whereas hardly any signal was detected in Cyrano KD cells after doxycycline treatment (Figure 3B).
Figure 3.
KD of Cyrano with CRISPRi Approach Has No Impact on the Pluripotency of Human iPSCs
(A) Schematic overview of the generation of inducible Cyrano KD human iPSCs using a CRISPRi approach.
(B) Representative RNA FISH images stained with a Cyrano-specific probe in unmodified cells (CRISPRi) and one clone, with and without doxycycline (doxy) treatment. Nuclei were stained with DAPI. Scale bar, 5 μm.
(C) Expression of Cyrano, NANOG, and SOX2 after doxy treatment in the monoclonal population. Expression levels were measured by qRT-PCR, mean ± SD of three independent experiments are shown, unpaired t test was performed for statistical analysis.
(D) Representative images of alkaline phosphatase staining of one KD clone with and without doxy treatment. Scale bar, 500 μm.
(E) Flow cytometry analysis of SSEA4 and TRA1-60 of one clone. Isotype controls (light gray, filled) and unstained cells (dark gray line) were used as controls. Representative plots with mean ± SD of three independent experiments are depicted.
(F) Fluorescence immunocytochemistry of NANOG, OCT4, and SOX2 was performed in Cyrano KD cells after doxy treatment. Scale bar, 100 μm.
(G) Expression levels were measured by qRT-PCR, mean ± SD of three independent experiments, unpaired t test was performed for statistical analysis.
(H) miR-7-5p was overexpressed in CRISPRi cells. Expression levels were measured by qRT-PCR, mean ± SD of three independent experiments, unpaired t test was performed for statistical analysis.
(I) Volcano plot depicting the results of the RNA sequencing of Cyrano KD clone with and without doxy treatment of three experiments. Cut off criteria −1 < log2(FC) > 1, q > 2. IPA network analysis.
(J) RNA-seq tracks of CRISPRi 1II E12 with and without doxy.
(K) Normalized read counts using DEseq2 of pluripotency-related and Cyrano-related genes.
To exclude doxycycline-specific effects, CRISPRi iPSCs without a gRNA were treated with doxycycline. No effects were observed with regard to growth dynamics, expression of pluripotency markers, or alkaline phosphatase staining (Figures S2D–S2G).
Similar to the observation in murine iPSCs, the expression levels of NANOG and SOX2 were unchanged after doxycycline-induced Cyrano KD (Figures 3C and S3A). Furthermore, neither the alkaline phosphatase staining, nor the morphology of the cells showed any changes after Cyrano KD (Figures 3D and S3B). The proliferation dynamics were also unaffected by the loss of Cyrano (Figure S3C). The pluripotency markers SSEA4 and TRA1-60 were analyzed by flow cytometry, and NANOG, OCT4, and SOX2 by immunohistochemistry. These markers did not respond to loss of Cyrano (Figures 3E, 3F, S3D, and S3E). Since Cyrano was shown to act on miR-7-5p in murine ESCs, which in turn represses Nanog and Itga9, it was therefore suggested to influence pluripotency and cell adhesion (Smith et al., 2017). We checked the expression of miR-7-5p, its proposed target ITGA9, and GATA6 as a negatively regulated target of NANOG, and saw that silencing Cyrano significantly increased only miR-7-5p levels, while ITGA9 and GATA6 remained unaffected (Figures 3G and S3F). In addition, iPSC treatment with miR-7-5p mimics had no consequences on NANOG (a suggested direct target of miR-7) or on SOX2 expression (Figure 3H). Consequently, alkaline phosphatase staining after miR-7 overexpression was indifferent from mimic controls (Figure S3G).
Loss of Cyrano Only Moderately Affects the Global Transcriptome
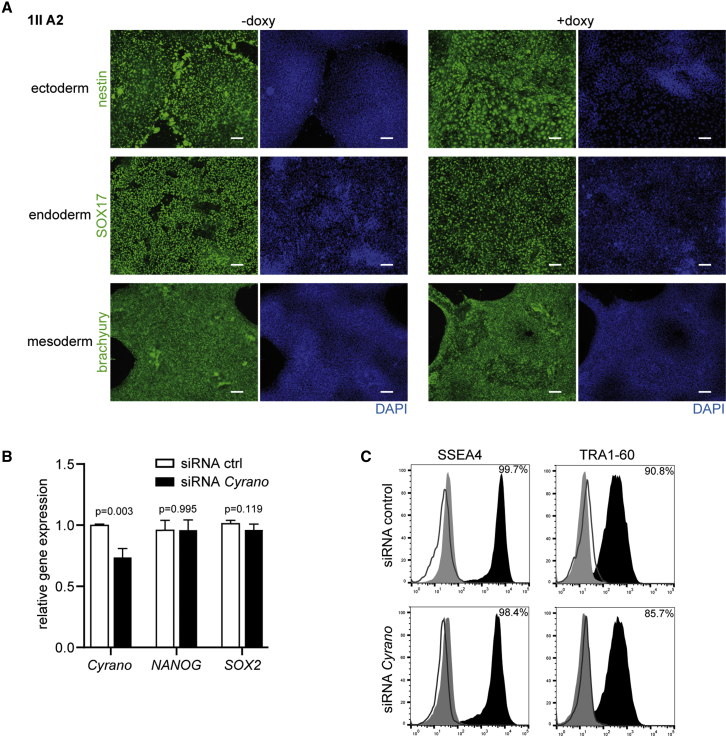
Transcriptome analysis in human iPSCs with and without doxycycline treatment, i.e., in the presence and absence of Cyrano, led to differential expression of only 50 genes: 34 genes were up- and 16 downregulated (−1<log2(FC) > 1, p < 0.01) (Figures 3I and S4A). Using Ingenuity Pathway Analysis (IPA), downregulated genes are linked to the “cancer, cellular development, cellular growth and proliferation” network while upregulated genes are part of the “cell cycle, cell death and survival, gene expression,” or “nervous system development and function, neurological disease, organ morphology” networks (Figure 3I). As shown in the RNA sequencing (RNA-seq) read mapping in a genome browser, Cyrano is nearly absent after doxycycline treatment. This had no effect on the neighboring genes CHP1 and OIP5, indicating that off-target effects in cis can be ruled out (Figures 3J and S4B). Figure 3K shows normalized RNA-seq read-count tables for genes related to pluripotency or connected to Cyrano based on literature research (more genes in Figure S4C). None of them were significantly dysregulated after Cyrano KD except for orthodenticle homeobox 2 (OTX2), which is a transcription factor in brain development (Beby and Lamonerie, 2013). In line with higher levels of miR-7 after Cyrano KD (Figure 3G), some known miR-7 targets (IGFS8, SLC8A3) (Agarwal et al., 2015) were downregulated, but none have been linked to pluripotency. The RNA-seq data were validated by qRT-PCR measurements (Figure S4D). Finally, to further demonstrate that Cyrano is dispensable for pluripotency, we performed a trilineage differentiation assay. Importantly, loss of Cyrano did not influence the potential of human iPSC to differentiate into the three different germ layers (Figures 4A and S4E).
Figure 4.
Silencing of Cyrano Does Not Influence the Differentiation Potential of Human iPSCs or Pluripotency of Human ESCs
(A) Differentiation of iPSCs in the presence and absence of Cyrano to the three germ layers with subsequent immunofluorescence staining. Scale bar, 100 μm.
(B) Gene expression of Cyrano, NANOG, and SOX2 was analyzed by qRT-PCR, mean ± SD of three independent experiments, unpaired t test was performed for statistical analysis.
(C) Flow cytometry analysis of SSEA4 and TRA1-60 after Cyrano silencing in human ESCs. Isotype controls (filled gray) and unstained cells (dark gray line) were used as controls.
Inhibition of Cyrano in Human ESCs by siRNA Has No Effect on Pluripotency
Despite large similarities of ESCs and iPSCs in expression patterns and chromatin modifications, there are also certain differences in gene expression and methylation profiles (Bilic and Belmonte, 2012). To test whether Cyrano is specifically required for the maintenance of pluripotency in human ESCs, we used an siRNA approach to silence Cyrano in these cells. NANOG and SOX2 remained unchanged after silencing (Figure 4B) and the expression of SSEA4 and TRA1-60 did not change after Cyrano modulation (Figure 4C).
Discussion
Here, we report that the highly conserved lncRNA Cyrano is enriched in PSCs which prompted us to investigate its functional role pluripotency. We used a dual gRNA CRISPR/Cas9 approach to generate a Cyrano KO in murine iPSCs, which did not result in adverse effects on pluripotency. This was in marked contrast to a simultaneously published report suggesting that Cyrano is essential for pluripotency in murine ESCs (Smith et al., 2017). To have a more clinically translatable platform and to also strengthen our own data with a different approach, we used CRISPRi in human iPSCs for a robust KD of Cyrano. As a third model, we used human ESCs in which Cyrano was silenced with specific siRNAs. Both approaches confirmed the data from murine iPSCs, showing that Cyrano has no overt effect on pluripotency in stem cells from different species. As mentioned above, this is in contrast to previously published data suggesting that Cyrano derepresses Nanog by sponging miR-7-5p. The two main differences between the studies were: firstly, Smith et al. investigated murine ESCs grown in potentially less-stringent stem cell culture conditions that may favor differentiation upon Cyrano repression. Nevertheless, when seeding human ESCs at low density, which can be considered a less-stringent condition, no effect after siRNA Cyrano treatment was observed. Secondly, Smith et al. (2017) used commercially available small hairpin RNAs (shRNAs) for Cyrano KD. Since the shRNA sequences used in this study were not accessible, we were unable to further investigate this discrepancy. Of note, Cyrano was also found as a candidate in an RNAi screen for pluripotency-related lncRNAs in murine ESCs that also used shRNA technology (Lin et al., 2015). Nevertheless, constraints on different approaches for loss-off-function studies were recently described, in particular, the comparison of screenings using CRISPR/Cas9 KO and shRNA-mediated KD showed weak correlations (Morgens et al., 2016). Moreover, the comparison of two different RNAi technologies showed poor reproducibility (Bhinder and Djaballah, 2013). CRISPR technology has gained more popularity in the last years and outperformed RNAi approaches with less noise and off-target effects (Evers et al., 2016). Hence, the readouts after modulation of transcript expression have to be validated carefully. Accordingly, we tested different approaches and cell types (mouse iPSCs, human iPSCs, and ESCs), which all showed the same effects. Thus, it is unlikely that our results are based on off-target or approach-dependent effects. It should be noted that, although unlikely given the high conservation of Cyrano, the previously described effects of shRNA-mediated KD of Cyrano may be unique to murine ESCs. But it is also unlikely that both shRNAs used by Smith et al. would show the same off-target effects, which is supported by their data showing that overexpression of miR-7-5p exhibits the same phenotype as Cyrano KD. Thus, there exists a currently unknown factor to explain the discrepancy between both studies.
We further investigated the suggested downstream mechanism via regulation of miR-7-5p. In line with previous reports, we found an upregulation of miR-7-5p after Cyrano inhibition. However, there was no evidence that miR-7-5p affects pluripotency in our systems as demonstrated in experiments with miR-7-5p mimics.
Cyrano was first described to be important for zebrafish embryogenesis and brain morphogenesis (Ulitsky et al., 2011), which goes along with the proposed role in self-renewal and maintenance of pluripotency of murine ESCs (Smith et al., 2017). In contrast to the study in zebrafish, it was recently shown that a genetic KO of Cyrano in fish did not influence embryogenesis, viability, or fertility. The lack of reproducibility of the zebrafish experiments is based on off-target effects of the two different morpholinos that were used to inhibit Cyrano (Goudarzi et al., 2019). In addition, Cyrano KO mice do not show defects in embryogenesis, viability, or fertility (Kleaveland et al., 2018), which one would expect if Cyrano was essential for the maintenance of PSCs.
Finally, global transcriptomic analysis revealed that loss of Cyrano has only very moderate effects in PSCs that are not related to know pluripotency factors. Moreover, despite the absence of Cyrano, human iPSCs can be readily differentiated into the three germ layers which require the iPSCs to be truly pluripotent.
In summary, contradictory to previous data, we did not detect any changes in the pluripotency of murine and human iPSCs and human ESCs after Cyrano inhibition. Our different approaches to silence Cyrano in PSCs are highly consistent, firmly demonstrating that Cyrano is not regulating pluripotency. Nonetheless, the high degree of conservation and the high expression in PSCs assume an important cellular function of Cyrano for whose study we generated state-of-the-art in vitro tools.
Experimental Procedures
Detailed methods are provided in Supplemental Experimental Procedures.
Murine iPSC Culture and Generation
Murine iPSCs have been reprogrammed from OG2 mice expressing EGFP under the control of an Oct3/4 promoter (Kensah et al., 2013). The murine iPSCs were grown on irradiated mitotic inactive mouse embryonic fibroblasts.
To knock out the Cyrano gene in iPSCs, the dual gRNA CRISPR/Cas9 approach was used (Heckl et al., 2014). The first exon of Cyrano was excised with two gRNAs knocking out both murine transcripts.
Human iPSC Culture and Generation
CRISPRi iPSCs (Mandegar et al., 2016) were cultured on Matrigel Growth Factor Reduced (Corning) in mTesR (STEMCELL Technologies) and passaged when confluent with Accutase (STEMCELL Technologies) in mTeSR supplemented with 10 μM Y-27632 2HCI (Selleckchem).
The gRNA design, cloning, and nucleofection were performed as described previously (Mandegar et al., 2016). For KD, induction cells were treated with 2 μM doxycycline for at least 3 days.
Human ESC Culture
MIXL1-GFP human ESC (Davis et al., 2008) were cultured on Geltrex (Thermo Fisher Scientific) in homebrewed E8 medium and passaged with Accutase (Gibco), seeded in 10 μM Y-27632 (Tocris). Cells came from a conventional feeder-based culture, expanded on Geltrex for two passages before being used for experiments.
Gene Expression
Total RNA of cultured cells was isolated using peqGOLD TriFast (VWR Life Science, Radnor, PA, USA) according to the manufacturer's instructions. RNA was DNase treated using RNase-free DNase set (QIAGEN), and 100 to 1,000 ng were reverse transcribed using Biozym cDNA synthesis kit.
To analyze miRNA expression levels 100 ng RNA was reverse transcribed using a iScript select cDNA synthesis kit (Bio-Rad) and specific TaqMan probes (hsa-miR-7-5p ID 005723_mat and RNU48 ID 001006, Thermo Fisher Scientific).
Real-time quantitative PCR was performed in CFX384 Touch Real-Time PCR Detection System (Bio-Rad) or QuantStudio 7 Flex System (Applied Biosystems). For mRNA and lncRNA expression analysis, iQ SYBR Green Supermix (Bio-Rad) was used and Absolute Blue qPCR Mix (Thermo Fisher Scientific) was used for miRNA expression analysis.
Absolute Quantification
The copy number of Cyrano was quantified by qPCR and a plasmid standard curve as described previously (Feretzaki et al., 2019).
Growth Curve
iPSCs (1 × 105) were seeded on Matrigel, and the following day doxycycline treatment was started and cells were counted for 5 consecutive days with Countess II (Invitrogen).
Alkaline Phosphatase Staining
The iPSCs were stained with the Alkaline Phosphatase Detection Kit (Millipore) according to the manufacturer's guidelines. Images were acquired with an Eclipse Ti-U fluorescence microscope (Nikon) and NIS Elements BR 3.26 (Nikon) software.
Flow Cytometry
For flow cytometry, cells were harvested, stained, then washed. Measurement was performed at Guava easyCite 5 flow cytometer (Millipore) and data analyzed with FlowJo (BD).
Human ESCs were fixed and permeabilized with FIX&PERM kit 1000 (Nordic MUbio). Staining was performed as described above.
Immunocytochemistry
iPSCs were washed, permeabilized, and incubated with primary and secondary antibodies.
RNA FISH
RNA FISH was performed using a ViewRNA Cell plus Assay (Thermo Fisher Scientific) according to the manufacturer's guidelines in Nunc Lab-Tek eight-well chamber slides (Merck) with a specific Cyrano probe (ID VA1-3020478-VCP, Thermo Fisher Scientific). Images were acquired using a Leica SP8 confocal microscope and LAS X (Leica) software.
Trilineage Differentiation
Differentiation was performed using a STEMdiff Trilineage Differentiation Kit (STEMCELL Technologies) according to manufacturer's guidelines.
Transfection of siRNA
Downregulation was achieved by transfecting 200 nM siRNAs (Eurofins) with Lipofectamine 2000 (Invitrogen) according to the manufacturer's guidelines.
PCR
PCR was performed using a HotStar Mastermix Kit (QIAGEN) according to the manufacturer's guidelines in 10 μL reaction. PCR products were resolved by agarose gel electrophoresis (0.8%–1.2% agarose gel in 1× TAE).
Sanger Sequencing
To verify sequences, DNA bands were cut from an agarose gel and DNA was extracted via the QIAquick Gel Extraction Kit (QIAGEN). Five to 10 ng/μL PCR product or 50–100 ng/μL plasmid DNA were brought to 15 μL volume premixed with 2 μL primer (10 μM) and sent to Eurofins for sequencing.
RNA-Seq
RNA was isolated using a miRNeasy mini kit (QIAGEN) according to the manufacturer's guidelines. Sample preparation and data processing are described in Supplemental Information.
Statistics
Statistical analyses were performed using an unpaired Student's t test. The results shown were obtained by at least three measurements (mean ± standard deviation [SD]). Statistical analysis was carried out using GraphPad Prism 8.
Data and Code Availability
RNA-seq data (GEO: GSE150421) are available in the Gene Expression Omnibus.
Author Contributions
H.J.H. performed the majority of the experiments and analyzed the data. S.C. and J.H. performed some experiments in human iPSCs. C.-K.H. contributed to RNA FISH measurements. I.G. and M.J.-A. helped with the establishment of mouse iPSC cultures. E.B. and R.Z. provided ESCs and helped with ESC experiments. C.B., T.T., and H.J.H. designed the study and prepared the manuscript. All authors approved the final version of the manuscript.
Conflicts of Interests
T.T. and C.B. filed patents in the field of ncRNAs. T.T. is founder and holds shares of Cardior Pharmaceuticals GmbH. All other authors have no conflict of interest or financial interest to declare.
Acknowledgments
We acknowledge the Core Facilities for Cell Sorting (supported in part by Braukmann-Wittenberg-Herz-Stiftung and Deutsche Forschungsgemeinschaft), for Laser Microscopy and Genomics (RCUG) at Hannover Medical School. The CRISPRi Gen1B (short CRISPRi) iPSC line was provided by the Bruce R. Conklin Laboratory at the Gladstone Institutes and UCSF. R.Z. received funding from Deutsche Forschungsgemeinschaft, Germany (DFG: ZW64/4-1 and the Cluster of Excellence REBIRTH DFG EXC62/2, EXC62/3 and KFO311 ZW64/7-1), Federal Ministry for Education and Science, Germany (grants: 13N14086, 01EK1601A, 01EK1602A and 13XP5092B) and the European Union H2020 program (TECHNOBEAT grant 66724). C.B. received funding from the Federal Ministry of Education and Research, Germany (research grant ERA-CVD JTC2018 INNOVATION, 01KL1903). T.T. received funding from European Research Council, (consolidator grant Longheart, LS4, ERC-2014-CoG).
Published: June 11, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.05.011.
Supplemental Information
References
- Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:1–38. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bär C., Chatterjee S., Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation. 2016;134:1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- Beby F., Lamonerie T. The homeobox gene Otx2 in development and disease. Exp. Eye Res. 2013;111:9–16. doi: 10.1016/j.exer.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- Bhinder B., Djaballah H. Systematic analysis of RNAi reports identifies dismal commonality at gene-level and reveals an unprecedented enrichment in pooled shRNA screens. Comb. Chem. High Trooughput Screen. 2013;16:665–681. doi: 10.2174/13862073113169990045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic J., Belmonte J.C.I. Concise review: induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- Davis R.P., Ng E.S., Costa M., Mossman A.K., Sourris K., Elefanty A.G., Stanley E.G. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Evers B., Jastrzebski K., Heijmans J.P.M., Grernrum W., Beijersbergen R.L., Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- Feretzaki M., Nunes P.R., Lingner J. Expression and differential regulation of human TERRA at several chromosome ends. RNA. 2019;25:1470–1480. doi: 10.1261/rna.072322.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M., Berg K., Pieper L.M., Schier A.F. Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility. eLife. 2019;8:1–17. doi: 10.7554/eLife.40815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensah G., Roa Lara A., Dahlmann J., Zweigerdt R., Schwanke K., Hegermann J., Skvorc D., Gawol A., Azizian A., Wagner S. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur. Heart J. 2013;34:1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- Kim J., Abdelmohsen K., Yang X., De S., Grammatikakis I., Noh J.H., Gorospe M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016;44:2378–2392. doi: 10.1093/nar/gkw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Belmonte J.C.I. Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 2017;18:180–191. doi: 10.1038/nrg.2016.156. [DOI] [PubMed] [Google Scholar]
- Lin N., Chang K., Li Z., Gates K., Rana Z.A., Zhang D., Han T., Yang C., Cunningham T.J., Head R. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2015;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar M.A., Huebsch N., Frolov E.B., Shin E., Truong A., Olvera M.P., Chan A.H., Miyaoka Y., Holmes K., Spencer C.I. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens D.W., Deans R.M., Li A., Bassik M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupers C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.N., Starmer J., Miller S.C., Sethupathy P., Magnuson T. Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Reports. 2017;9:108–121. doi: 10.1016/j.stemcr.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Liu Y., Li C., Zhang Y., Zhou X., Lu C. Long noncoding RNA OIP5-AS1 accelerates the ox-LDL mediated vascular endothelial cells apoptosis through targeting GSK-3β via recruiting EZH2. Am. J. Transl. Res. 2019;11:1827–1834. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data (GEO: GSE150421) are available in the Gene Expression Omnibus.