Summary
Induced pluripotent stem cells (iPSCs) from diverse humans offer the potential to study human functional variation in controlled culture environments. A portion of this variation originates from an ancient admixture between modern humans and Neandertals, which introduced alleles that left a phenotypic legacy on individual humans today. Here, we show that a large iPSC repository harbors extensive Neandertal DNA, including alleles that contribute to human phenotypes and diseases, encode hundreds of amino acid changes, and alter gene expression in specific tissues. We provide a database of the inferred introgressed Neandertal alleles for each individual iPSC line, together with the annotation of the predicted functional variants. We also show that transcriptomic data from organoids generated from iPSCs can be used to track Neandertal-derived RNA over developmental processes. Human iPSC resources provide an opportunity to experimentally explore Neandertal DNA function and its contribution to present-day phenotypes, and potentially study Neandertal traits.
Keywords: induced pluripotent stem cells, Neandertal genomics, cerebral organoids, archaic introgression, single-cell transcriptomics
Highlights
-
•
Large human induced pluripotent stem cell resources harbor extensive Neandertal DNA
-
•
A database of Neandertal DNA content and functionally relevant alleles in each cell line
-
•
Organoid systems allow exploration into the developmental effects of Neandertal DNA
-
•
Single-cell transcriptomics tracks Neandertal DNA activity over cell differentiation
In this article, Camp and colleagues show that large stem cell resources carry extensive Neandertal DNA, including many functionally relevant Neandertal alleles. The authors demonstrate that organoid systems can be used to explore Neandertal DNA activity during development.
Introduction
Protocols exist to differentiate human embryonic and induced pluripotent stem cells (iPSCs) into many different cell types of the human body (Williams et al., 2012). In addition, stem cells can self-organize into complex three-dimensional structures containing multiple cell types that resemble human tissues (such as the brain, liver, stomach, intestine, skin, and kidney) (Clevers, 2016). These stem cell-derived systems can be used to explore how natural variation between human individuals impacts development, cell biology, and susceptibility to disease (Banovich et al., 2018, Bonder et al., 2019, Carcamo-Orive et al., 2017, Kilpinen et al., 2017, Lancaster and Knoblich, 2014). Some of the variation in present-day humans has been shown to derive from admixture between modern and archaic hominins. Analyses of Neandertal genomes revealed that Neandertals and modern humans interbred approximately 55,000 years ago as the latter migrated out of Africa. As a consequence, around 2% of the genomes of all present-day non-Africans derive from Neandertal ancestors (Green et al., 2010, Prüfer et al., 2014, Prüfer et al., 2017). Because the segments of DNA inherited from Neandertals varies between individuals, it has been estimated that at least 40% of the Neandertal genome survives in people today (Vernot and Akey, 2014). Recent genome-wide association studies suggest that the DNA relics from this admixture left a phenotypic legacy, influencing, for example, skin and hair color, immune response, lipid metabolism, skull shape, bone morphology, blood coagulation, sleep patterns, and mood disorders (Dannemann and Kelso, 2017, Dannemann et al., 2016, Gunz et al., 2019, Khrameeva et al., 2014, Quach et al., 2016, Sams et al., 2016, Sankararaman et al., 2014, SIGMA Type 2 Diabetes Consortium et al., 2014, Simonti et al., 2016, Vernot and Akey, 2014). In addition, it has been reported that Neandertal-introgressed DNA has a significant effect on gene expression in adult human tissues possibly as a result of selection acting on Neandertal variants in regulatory regions (Dannemann et al., 2017, McCoy et al., 2017, Petr et al., 2019, Silvert et al., 2019, Telis et al., 2019). However, these associations have been observed in living people or in tissues, where there is limited opportunity for controlled experimentation. Furthermore, there are few opportunities to study the impact of Neandertal-introgressed DNA on developmental processes in modern humans. The Human Induced Pluripotent Stem Cell Initiative (HipSci) has generated and characterized a large resource of human iPSCs with genome-wide genotype data (Kilpinen et al., 2017). Repositories, such as HipSci present an unprecedented opportunity to identify carriers of Neandertal alleles of interest, which could be used for controlled experiments in vitro to explore the genetic mechanisms underlying Neandertal and modern human phenotypes. However, there has been no detailed evaluation of Neandertal DNA composition within stem cell resources, and it is unknown which Neandertal-introgressed alleles are available for functional testing using such experiments.
Results
The Neandertal DNA Content in HipSci Cell Lines
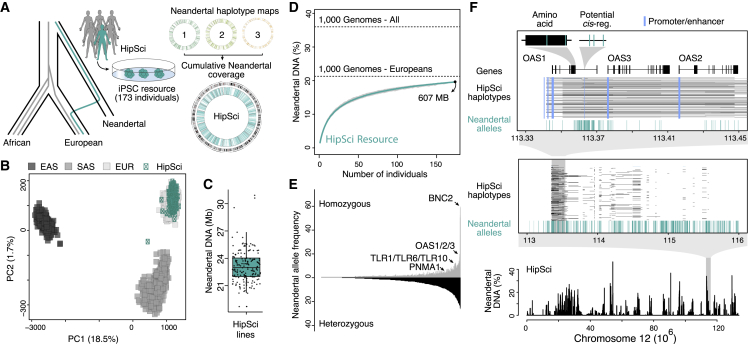
We have analyzed the genome sequences from 173 individuals (mostly Europeans) within the HipSci resource and identified the modern human and Neandertal component of each individual's ancestry (Figures 1A and 1B). We used alleles in present-day humans that are shared with the Vindija Neandertal and absent in Yoruba individuals, along with a linkage disequilibrium-based test for incomplete lineage sorting (ILS), to identify haplotypes that are likely of Neandertal origin. We used the Vindija Neandertal genome to identify Neandertal haplotypes because it is more similar to the introgressing Neandertals than the Altai Neandertal genome, thus providing additional power to detect haplotypes (Prüfer et al., 2014, Prüfer et al., 2017). Based on these inferred haplotypes, we find that cumulatively 19.6% (661 Mb) of the Neandertal genome is represented in these cell lines, with between 18.7 and 30.9 Mb Neandertal DNA per individual (Figure 1C; Tables 1 and S1). We found that 98% of inferred haplotypes overlap previously identified introgressed sequence and that the cumulative amount of Neandertal DNA present in this resource approaches the total amount that has been identified in Europeans (1000 Genomes Project Consortium et al., 2015, Sankararaman et al., 2014; Vernot et al., 2016) (21.3%, Figure 1D). Of the detected archaic variants, 22.1% are found in a homozygous state in at least one cell line (Figure 1E). Some of these homozygotic variants tag high-frequency Neandertal haplotypes close to genes with phenotype associations, including BNC2 (associated with skin color) (Dannemann and Kelso, 2017, Vernot and Akey, 2014), TLR1/TLR6/TLR10 and OAS1/OAS2/OAS3 (Figure 1F, both associate with innate immune response) (Dannemann et al., 2016, Mendez et al., 2013, Sams et al., 2016). We note that the addition of samples from non-European populations would extend the amount of Neandertal DNA that can be tested for its phenotypic effects even further. For example, an additional 16.4% of the Neandertal genome has been identified in east and south Asians (1000 Genomes Project Consortium et al., 2015), but is absent from the HipSci resource as individuals are largely of European ancestry.
Figure 1.
Identification of Neandertal Haplotypes in Human iPSC Lines
(A) The Human Pluripotent Stem Cell Initiative (HipSci) created and characterized induced pluripotent stem cells lines from 173 individuals with genome-wide genotype data, which we analyzed for Neandertal ancestry (Kilpinen et al., 2017). Circos plots show Neandertal haplotype coverage across each chromosome for three individuals (top) and the entire resource (bottom) (Kilpinen et al., 2017).
(B) Principal-component analysis on SNPs that distinguish East Asian (EAS) (dark gray), Southeast Asian (SAS) (gray), and European (light gray) individuals suggests that each HipSci individual (teal) has a major European component to their ancestry.
(C) Boxplot shows Neandertal DNA in megabases (Mb) per individual detected in 173 HipSci individuals.
(D) The cumulative percentage of the human genome covered by Neandertal haplotypes in the HipSci resource. The percentage is also shown for European and all non-African individuals from the 1000 Genome Project.
(E) Frequency distribution plot showing the HipSci resource partitioned by the contribution of variants found in heterozygous (lower, black) and homozygous state (upper, gray). Highlighted are four selected SNPs (chr9:16720122, BNC2; chr4:38760338, TLR1/TLR6/TLR10; chr12:113366899, OAS1/OAS2/OAS3; chr11:3867350, PNMA1, Table 2) tagging high-frequency Neandertal haplotypes close to genes with phenotype associations.
(F) Neandertal DNA percentage, alleles, and haplotypes present in the HipSci resource covering the OAS locus. In this example, the gene OAS1 contains Neandertal alleles in non-protein coding, potential regulatory regions (blue bars), and those that change the amino acid sequence, present at a frequency of approximately 33% in HipSci individuals. Gene, promoter, and enhancer annotations are from ENSEMBL (GRCh37, Experimental Procedures).
Table 1.
Breakdown of Neandertal DNA Coverage in the HipSci Resource
| Average across Cell Lines (Range) | Cumulative across Cell Lines (Range) | |
|---|---|---|
| Neandertal ancestry (bps) | 22,985,635 (18,725,364–30,931,585) | 607,404,493 |
| Haplotype length (bps) | 48,014 (42,613–56,968) | – |
| Neandertal alleles | 15,845 (12,829–20,060) | 205,032 |
| Missense Neandertal alleles | 59 (38–90) | 719 |
| Neandertal alleles in enhancer/promoter regions | 235 (169–340) | 2,852 |
| Neandertal PheWAS alleles | 45 (29–60) | 924 |
| Neandertal GTEx eQTLs | 45 (29–60) | 270 |
| Neandertal alleles showing allele-specific expression | 119 (75–176) | 957 |
The Presence of Putative Functional Neandertal Alleles in HipSci Cell Lines
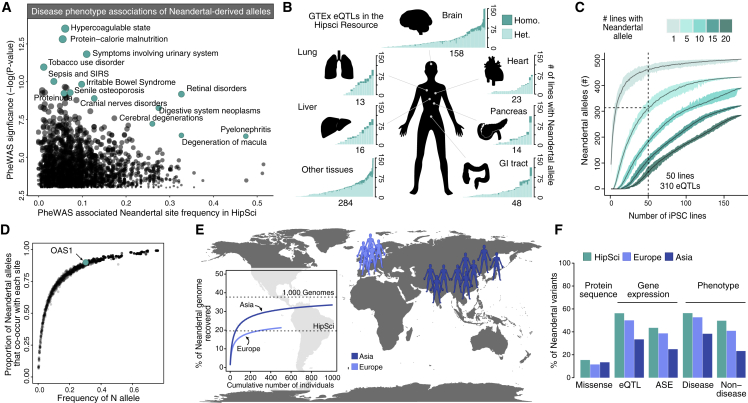
We next analyzed the prevalence of functionally relevant Neandertal DNA within the HipSci resource. We collected recently published Neandertal-derived phenotype and disease-associated alleles (Dannemann and Kelso, 2017, Dannemann et al., 2016, Quach et al., 2016, Sams et al., 2016, Sankararaman et al., 2014, Simonti et al., 2016) and found that most (22/24) of the alleles that reached genome-wide significance are present in the resource in more than 1 iPSC line (Figure 2A; Table 2). These alleles are associated with a variety of processes, including digestive function, nutrition, skin color, coagulatory protein production, and immune response. In addition, we identified hundreds of alleles that alter amino acids, are expression quantitative trait loci (eQTL) (Dannemann et al., 2017), or show allele-specific expression (McCoy et al., 2017) (Table S1). We performed a power analysis to determine how many Neandertal-associated eQTLs are present in a set of randomly sampled lines from the HipSci resource. As an example, we find that 50 HipSci lines chosen at random will allow the interrogation of approximately 310 Neandertal-associated eQTLs with each site represented in at least 5 cell lines (Figures 2B, 2C, and S1).
Figure 2.
The HipSci Resource Harbors Extensive Functionally Relevant Neandertal DNA
(A) The frequency of single nucleotide changes introgressed from Neandertals that have significant associations with human disease phenotypes (Simonti et al., 2016), with select associations highlighted in teal.
(B) Barplots illustrating the number of iPSC lines that contain each Neandertal-associated eQTL (y axis sorted by frequency of alleles in HipSci), from the GTEx dataset present in the HipSci resource, colored by tissue and by homozygous (dark) or heterozygous (light). Neandertal-associated eQTLs in the brain represent the pool of Neandertal-associated eQTLs that are found in at least one of the 13 GTEx brain tissues.
(C) A power analysis shows the average of how many Neandertal eQTLs are present in at least a certain number of cell lines (color) out of a random sample of X lines (x axis) from the HipSci resource (grey). The range across the random samplings is shown in respective colors for each number of lines.
(D) Plot showing the co-occurrence of Neandertal alleles within individuals. The frequency of each Neandertal allele is plotted against the proportion of all other Neandertal alleles with which it co-occurs in the HipSci resource. For example, an OAS1 Neandertal-derived allele is found at relatively high frequency (0.33, and present in 53% of all individuals) in the HipSci resource and is therefore paired with the majority of other Neandertal alleles.
(E) Inferred Neandertal haplotypes in the 5 European (light blue, total of 505 individual) and 10 Asian (dark blue, total of 1,009 individuals) populations of the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2015) recovered 21.3% and 33.5% of the Neandertal genome in Europeans and Asians, respectively.
(F) Barplots show the presence of putative functional Neandertal variants at frequencies greater than 5% in the HipSci cohort as well as 1000 Genomes European and Asian populations (allele-specific expression [ASE]). We found that amino acid-modifying variants were slightly more prevalent in Asian populations, consistent with the higher amount of recovered Neandertal DNA in that population. However, other functional variants derived from association studies with gene expression and phenotype data were more often found in Europeans and the mostly European HipSci individuals, consistent with a detection bias inherited from the association analyses that were conducted in cohorts with a mostly European ancestry.
See also Figure S1.
Table 2.
Prevalence of Phenotype-Associated Neandertal Alleles in the HipSci Resource
| Publication | Phenotype | Neandertal Allele Frequency in HipSci Resource (%) | tag SNP |
|---|---|---|---|
| Sankararaman et al., 2014 | smoking behavior | 11.3 | chr9:136478355 |
| Crohn disease | 26.6 | chr10:64415184 | |
| optic disc size | 4.9 | chr10:70019371 | |
| Crohn disease | 1.2 | chr12:40601940 | |
| Dannemann et al., 2016 | allergy susceptibility H. pylori status |
17.9 | chr4:38760338 |
| Simonti et al., 2016 | hypercoagulable state | 0.6 | chr1:169593113 |
| protein-calorie malnutrition | 5.5 | chr1:234099819 | |
| tobacco use disorder | 0.3 | chr3:10962315 | |
| symptoms involving urinary system | 10.7 | chr11:3867350 | |
| Quach et al., 2016 | pathogen response | 16.2 | chr14:74152316 |
| Sams et al., 2016 | pathogen response | 32.9 | chr12:113366899 |
| Dannemann and Kelso, 2017 | chronotype | 11.3 | chr2:239316043 |
| hair color | 0 | chr6:503851 | |
| skin color | 9 | chr6:45553288 | |
| pulse rate | 1.7 | chr6:121947984 | |
| skin tanning | 65.6 | chr9:16720122 | |
| skin color | 7.2 | chr9:16904635 | |
| sitting height | 4.9 | chr10:70019371 | |
| narcolepsy | 0.6 | chr10:94574048 | |
| skin color | 2.9 | chr11:89996325 | |
| hair color | 5.2 | chr14:92793206 | |
| impedance of leg | 22.3 | chr15:85114447 | |
| hair color | 0 | chr16:89947203 | |
| height at age 10 | 20.2 | chr19:31033240 |
We note that each Neandertal allele present in the HipSci resource exists in a primarily modern human background. However, in each individual many Neandertal alleles co-occur (Figure 2D). For example, in the HipSci resource, one of the Neandertal alleles at the OAS1 locus (chr12:113425154, Figure 1F), a locus with the highest Neandertal frequencies in present-day humans (Mendez et al., 2013, Sams et al., 2016), is paired with 90% of the other introgressed Neandertal alleles in at least one cell line. It may thus be possible to leverage such co-occurrences to study epistatic interactions among Neandertal alleles. Given a large sample size, one could, for example, test for differences in gene expression or immune phenotype associations of OAS1 in the presence of other immune-related Neandertal alleles. Restricting to such interactions may reduce the search space enough to make “NxN” epistasis a more tractable problem than the identification of more general epistatic interactions (Huang et al., 2013).
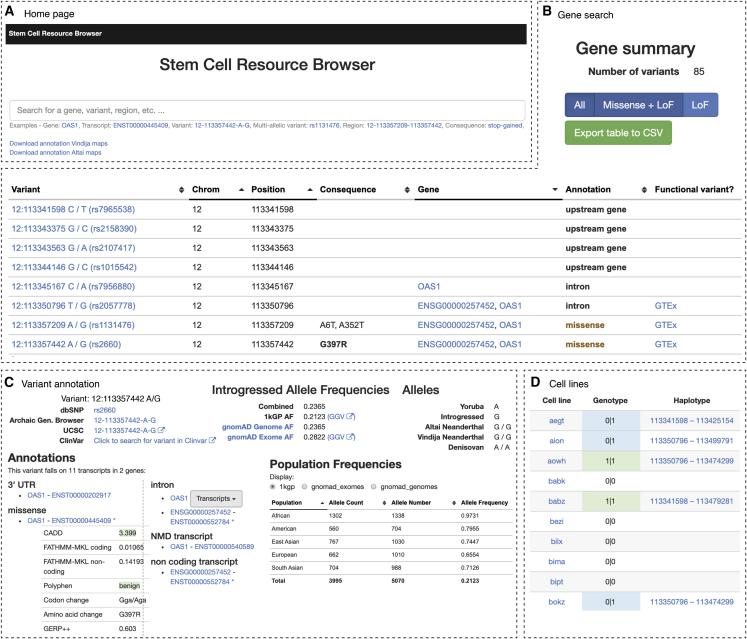
Association of Neandertal variants with gene expression and phenotype variation were conducted in cohorts mostly of European ancestry, a bias present in many genome-wide association analyses (Mills and Rahal, 2019, Sirugo et al., 2019); therefore, our knowledge of the functional potential of Neandertal DNA is limited to variants that are on average at increased frequencies in European populations (Figures 2E and 2F). In addition, as iPSC resources continue to expand to include individuals from other non-European populations, it will not only become possible to explore the phenotypic contribution of Neandertal DNA enriched in those populations, but also study alleles derived from other archaic hominins, such as Denisovans (Meyer et al., 2012), a distant Asian relative of Neandertals that made even larger genetic contributions to present-day people in parts of Oceania (Qin and Stoneking, 2015, Sankararaman et al., 2016, Vernot et al., 2016). We provide a database of the inferred introgressed Neandertal alleles for each of the 173 HipSci resource individuals together with the annotation of the predicted functional variants, which can be accessed at https://bioinf.eva.mpg.de/stemcellbrowser (Figure 3; for more information see description in the Supplemental Information). The website contains information about the presence of Neandertal variants for each individual with a link to those variants that modify the protein sequence or have previously been associated with effects on gene expression and disease and non-disease phenotypes. Furthermore, for each variant the information of which cell line carries it is displayed and is designed to provide a practical resource to query the HipSci for available cell lines to study the effects of individual variants.
Figure 3.
Overview of the Stem Cell Resource Browser
Screenshots showing pages of the Neandertal Stem Cell Resource Browser (A). The website contains information about the presence of Neandertal variants for each individual in the HipSci resource (B,D) with a link to those variants that modify the protein sequence or have previously been associated with effects on gene expression and disease and non-disease phenotypes (B,C). The information of which cell lines carries the variant is displayed, and is designed to provide a practical resource to query the HipSci for available cell lines to study the effects of individual variants.
Using HipSci Cell Lines to Model Neandertal DNA Activity in the Developing Brain
We next wanted to indicate the potential of resources, such as the HipSci to explore Neandertal DNA activity. Previous comparative RNA sequencing (RNA-seq) expression analysis across the HipSci lines in a pluripotent state identified 76 significant eQTLs (false discovery rate [FDR] < 0.05) (Kilpinen et al., 2017) that are tagged by an SNP that is defining a Neandertal haplotype. We find that between 0% and 29.6% of these iPSC eQTLS are significant eQTLs in 48 GTeX adult tissues (p < 0.01, Figure S2) (Dannemann et al., 2017). The brain is of particular interest since previous studies have shown that Neandertal alleles are downregulated in the brain (McCoy et al., 2017) and there is significant contribution of Neandertal variants to neurological and behavioral phenotypes (Dannemann and Kelso, 2017, Simonti et al., 2016). We find that the overlap of iPSC eQTLs with the 13 brain tissues was particularly low (0%–15.4%, comparison of overlap brain versus other, p = 0.001, Mann-Whitney U test). Among the 105 Neandertal eQTLs in cortex tissues and 79 in HipSci only 3 are shared. These data highlight that relevant brain cell states need to be differentiated from iPSCs in order to study brain-related Neandertal alleles.
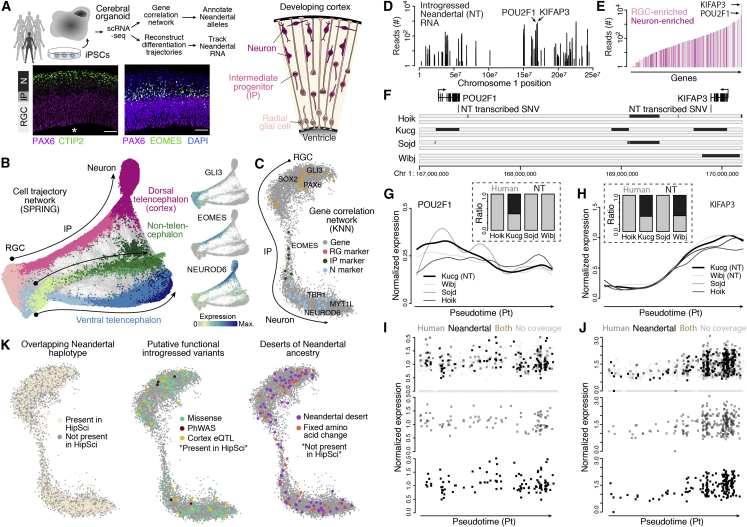
Recently, 2-month-old cerebral organoids from seven pluripotent stem cell lines (including four HipSci lines) were analyzed using single-cell RNA-seq (Kanton et al., 2019). These organoids were composed of progenitors and neurons from the dorsal (cortex) and ventral forebrain, midbrain, and hindbrain, and the single-cell data could be used to reconstruct progenitor-to-neuron differentiation trajectories with the forebrain regions (Figures 4A–4C and S3). Intriguingly, we find that Neandertal RNA molecules transcribed from within Neandertal DNA haplotypes can be tracked over developmental processes. For the HipSci lines, 763 genes have transcribed sequences that overlap a Neandertal haplotype and have a Neandertal-informative variant in their transcript in at least one of the individuals (Figures 4D and 4E). Of these genes, we detect Neandertal-introgressed RNA from 535 genes, and many of these genes have a dynamic expression pattern as progenitor cells differentiate into neurons in the developing human cortex (Figures 4F–4J). Most of these Neandertal-like RNAs are specific to one of the four HipSci individuals. Together, these data suggest that iPSC-derived organoids from diverse individuals could be used to track Neandertal-derived RNA molecules during cellular differentiation and other developmental processes.
Figure 4.
Tracking RNA from Neandertal-Introgressed Haplotypes during Cortex Development
(A) Overview of analysis using single-cell RNA sequencing (scRNA-seq) data from cerebral organoids from multiple individuals with whole-genome genotype data. Schematic and immunohistochemistry show the structure of cortical-like regions within cerebral organoids. Radial glial (RG), intermediate progenitor (IP), and neuronal (N) (Mora-Bermúdez et al., 2016) cells were extracted from the organoid scRNA-seq data, and used to identify correlated gene expression patterns during cortical neuron differentiation. scRNA-seq data is from Kanton et al. (2019), and stainings are from Mora-Bermúdez et al. (2016).
(B) SPRING reconstruction based on organoid scRNA-seq data from seven individuals (including four HipSci iPSC lines), with clusters colored by cell types. All subsequent analysis was based on RGCs, IPs, and neurons in the cortical trajectory.
(C) A correlation network (knn, k = 70) using 7,349 genes (gray) that highly correlate in expression (r > 0.6) with transcription factors that have been shown to regulate progenitor proliferation/self-renewal and neuron differentiation (Camp et al., 2015) is shown with transcription factors that represent individual cell-type-specific expression colored (orange, RG; green, IP; blue, N).
(D) Based on the reads from the scRNA-seq that overlap informative positions we are able to classify the reads as “Neandertal” or “modern human” depending on the observed allele at these sites. Plot shows the distribution of the number of reads with Neandertal informative variants across chromosome 1 from scRNA-seq data from organoids from four HipSci lines. Reads assigned to KIFAP3 and POU2F1, two genes that are part of the gene expression correlation network shown in Figure 3, are highlighted. POU2F1 and KIFAP3 show the highest expression in progenitors and neurons, respectively.
(E) Number of reads covering Neandertal-informative variants which shows differences in their genotype between the 4 HipSci individuals for 535 genes. Bars representing genes with an expression difference of at least 2-fold between progenitors and neurons are highlighted in light pink (larger expression in progenitors) and dark pink (larger expression in neurons). Genes with no expression differences are shown in gray.
(F) Gene models for KIFAP3 and POU2F1 are shown together with the locations of the Neandertal-informative SNPs, rs4519 and rs1059761, in the corresponding 3′ UTRs of these genes. The lower panel shows inferred Neandertal haplotypes in the four HipSci individuals overlapping chr1:167,000,000-170,000,000.
(G and H) Spline-interpolated gene expression trajectories across pseudotime for four HipSci cell lines for POU2F1 (G) and KIFAP3 (H). Cell lines with a chromosome carrying Neandertal haplotypes overlapping KIFAP3 and POU2F1, respectively, are labeled with “NT.” Barplots show the fraction of reads for each cell line carrying at the Neandertal-informative position the modern human (gray) and Neandertal (black) variants.
(I and J) The top panel shows the expression of all kucg cells for POU2F1 (I) and KIFAP3 (J) and whether cells have reads assigned that carry a Neandertal variant (black), a human variant (gray), both Neandertal and human variants (brown), or have no reads carrying Neandertal-informative variants (light grey). The bottom two panels highlight the cells with human-only (gray) or Neandertal-only reads.
(K) Cortical gene correlation network. Left: 1,777 genes (24% of network genes) with overlapping Neandertal haplotypes in the HipSci individuals are colored in beige. Middle: subset of genes with overlapping Neandertal haplotypes in HipSci that carry Neandertal variants that are potentially functional, including eQTLs in cortical brain regions that are linked to a Neandertal haplotype in GTEx (Dannemann et al., 2017), variants that change the amino acid sequence, and variants that have been associated with clinical phenotypes (Simonti et al., 2016). Right: among the genes without overlapping Neandertal haplotypes in the HipSci are 81 genes that are located in desert regions devoid of Neandertal ancestry (Vernot et al., 2016) and 33 genes with fixed human-derived amino acid substitutions (Prüfer et al., 2014).
See also Figures S2–S4.
We next used the cortical single-cell transcriptome data to build a gene correlation network to understand which functional Neandertal alleles could be studied in iPSC-derived cerebral organoids. The network identified genes with dynamic expression patterns across progenitor-to-neuron differentiation trajectories in the developing cortex (Figure 4C). We find that, of these 7,349 genes, 1,777 overlap a Neandertal haplotype in the HipSci resource with many of these haplotypes containing potentially functional features (e.g., amino acid changes, phenome-wide associations, cortex eQTLs) (Figure 4K). Notably, we identified homozygotic carriers in the HipSci resource for 37% (59 of 173 amino acid changes, 13 of 26 cortex eQTLs, and 4 of 6 phenome-wide association variants) of these putative functionally relevant alleles (Table S1). We note that cortical neurons within 2-month-old organoids are immature, but still have strong similarity to adult excitatory neurons (rho = 0.75, Spearman's correlation, Figures S3 and S4). We find that 21% of genes linked to previously described Neandertal-associated eQTLs in brain cortex tissues (Dannemann et al., 2017) are detected in both adult excitatory neurons and organoid-derived neurons, with 41% and 23% being detected in adult or organoid cortical neurons, respectively. Altogether, this analysis identifies the putatively functional Neandertal alleles that could be studied in brain organoids using the HipSci resource.
Perhaps even more intriguing are the expressed cortical genes that are located in genomic regions with no overlapping Neandertal haplotype in the HipSci resource. Some of these loci are completely devoid of any Neandertal introgression in any present-day individual sequenced to date (so-called deserts) or contain amino acid changes that are fixed or nearly fixed in all present-day humans and ancestral in Neandertals and the Denisovan (Figures 4D and S4). These genes could represent exciting candidates to use CRISPR/Cas gene editing to study the effect of the ancestral or Neandertal-specific alleles on cortex development.
Discussion
Our analysis suggests that the HipSci and potentially other existing stem cell resources (e.g., http://hub4organoids.eu/ or iPSCORE [Panopoulos et al., 2017]) can be used to systematically explore Neandertal allele function in diverse cell types differentiated in controlled culture environments, including the previously unexplored study of developmental processes. Improved methods to generate efficient and homogeneous-engineered cells and tissues from stem cells, together with high-throughput single-cell sequencing approaches, will make such experiments tractable in the near future. A major challenge in using iPSCs to study Neandertal alleles is that the genetic background must be considered when comparing differences between individuals, and many individuals are required to identify new eQTLs de novo. Often, several eQTLs are found to be associated with the same gene; and since Neandertal alleles are rare, they often appear as heterozygous in any given individual, limiting the power to detect associations with gene expression and other phenotypes. However, heterozygosity can provide a powerful opportunity to study the Neandertal contribution to allele-specific expression (McCoy et al., 2017), and our analysis highlights that allele-specific expression could be studied over developmental processes and in controlled environments using stem cells. To saturate Neandertal homozygote diversity, stem cell resources of thousands of individuals from multiple populations would be required. Nevertheless, stem cell resources allow for a pre-selection of cell lines based on the presence of particular variants and provide a dataset that could maximize the power for phenotype associations. Complementary approaches to create isogenic lines that have human alleles Neandertalized on both chromosomes (Pääbo, 2014) will help control for genetic background. However, eQTLs frequently are found to be in high linkage disequilibrium with other genetic variants, complicating the prediction of the causal variant. This problem is particularly important for Neandertal variants, which, due to the divergence between modern humans and Neandertals and the time of admixture, come on haplotype blocks with many genetic variants per block and it is difficult to predict which genetic change is functional. The growing number of new genomes from archaic humans are continuously providing profound new insights into the history of modern human evolution and further helps us to understand how admixture still shapes the genomes of people living today (Browning et al., 2018, Hajdinjak et al., 2018, Slon et al., 2018). In the future, as stem cell resources continue to grow, it will be exciting to search for functional hominin alleles handed down from other archaic populations, such as Denisovans. Our vision is that the merging of archaic genomics with stem cell technologies will provide a new avenue to explore the functional consequences of genetic variants contributed by archaic hominins to present-day humans.
Experimental Procedures
Principal-Component Analysis on HipSci Lines
To infer the genetic relationship between the HipSci individuals and present-day people, we performed a principal-component analysis using polymorphic sites in 1000 Genomes Eurasians (1000 Genomes Project Consortium et al., 2015) that show large population differentiation between Europeans and Asians (Fst > 0.5). Population differentiation was calculated based on Fst, using the Weir and Cockerham calculation implemented in vcftools (Danecek et al., 2011) and 100 unrelated Asians and Europeans each in the 1000 Genomes panel. While the HipSci resource contains non-European individuals, almost all of the individuals with genotypes clustered with Europeans from the 1000 Genomes panel (Figure 1B).
Detection of Neandertal Haplotypes
To define Neandertal haplotypes, we first identified a set of SNPs where one allele is likely of Neandertal origin. These Neandertal SNPs (aSNPs) have one allele that is (1) present in the genomes of the Vindija Neandertal (Prüfer et al., 2017) and (2) present in 1000 Genomes Project (phase III) Eurasian populations, but (3) absent from Yoruban, an African population with little to no Neandertal admixture (1000 Genomes Project Consortium et al., 2015). To detect putative Neandertal haplotypes we scanned for consecutive stretches of aSNPs in the genomes of the cell lines where the individual carries the Neandertal-shared alleles, with continuous SNPs located not more than 20,000 bps from one another. To define a Neandertal haplotype we required a consecutive stretch of at least three Neandertal alleles across their corresponding successive aSNPs. The sharing of such alleles can occur due to two evolutionary processes: first, ILS, a phenomenon where parts of the genome do not fit the genome-wide phylogeny, can result in allele sharing, in this case between non-Africans and Neandertals, but not Africans. These alleles are part of haplotypes that are shared with the common ancestor of Neandertals and modern humans. A second scenario that is consistent with the sharing of alleles between Neandertals and non-Africans is admixture between them. While the allele-sharing features are similar between both processes, they differ in one important aspect. Due to the rather recent admixture between Neandertals and modern humans ∼55,000 years ago, the haplotypes that these shared alleles exist on are substantially longer (Sankararaman et al., 2012). To exclude that our inferred haplotypes are a result of ILS, we computed the probability of them being compatible with ILS based on the algorithm presented by Huerta-Sánchez et al. (2014) and the age of the divergence to Neandertals of 465,000 years used in (Dannemann et al., 2016), a conservative estimate of the human mutation rate (mu = 1 × 10−8 per site per generation) and two recombination maps (Hinch et al., 2011, Kong et al., 2010). The resulting p values were corrected for multiple testing using the Benjamini-Hochberg approach. We included haplotypes with an FDR < 0.05 for ILS for at least one of the recombination maps, or if no recombination map data were available, inferred haplotypes with a length greater than 50 kb or at least 10 consecutive aSNPs with an Neandertal allele to our analyses. All inferred haplotypes for each cell line are available at https://bioinf.eva.mpg.de/stemcellbrowser. We applied the method to the genotype data for 173 individuals of the HipSci resource and all non-Africans of the 1000 Genomes project (phase III).
Detection of Neandertal Missense, Regulatory, and Phenotype-Associated Variants
We identified putatively functional Neandertal alleles that overlap confidently inferred Neandertal haplotypes (see section “Detection of Neandertal Haplotypes”) in the cell lines by detecting those that alter the protein or regulatory sequence of a gene. First, for the detection of Neandertal alleles that modify the protein sequence, we selected all Neandertal alleles within confidently inferred Neandertal haplotypes detected in any cell line and annotated them functionally using the variant effect predictor (VEP, human Ensembl version 73). We selected those alleles that were defined as “missense” by VEP. Second, we annotated Neandertal alleles likely to be involved in gene regulation by overlapping them with three datasets: (1) enhancer and promoter regions are inferred from open chromatin assays (DNase-seq), histone modification assays, and transcription factor binding assays (chromatin immunoprecipitation sequencing) from various cell types provided by the Ensembl Regulatory Build (version GRCh37, 20161117) (Zerbino et al., 2015), (2) significant eQTLs in the GTEx dataset (Dannemann et al., 2017), and (3) allele-specific expression (McCoy et al., 2017). For (1) we identified aSNPs with the Neandertal alleles directly overlapping with a regulatory motif. For (2) we selected the inferred Neandertal haplotypes with the top 20 most significant eQTLs in each of the 48 GTEx tissues with more than 50 individuals (GTEx Consortium et al., 2017) and required to have at least one aSNP to be present in a given Neandertal haplotype and iPSC individual, resulting in a total of 409 such Neandertal haplotypes. For (3) we selected all Neandertal alleles that have been identified to show allele-specific expression (FDR < 0.1). Third, we selected Neandertal alleles that have previously been associated with specific disease and non-disease phenotypes in modern humans. We selected all 925 aSNPs with significant phenotype associations detected by Simonti et al. (2016). We further selected multiple additional significant phenotypes associations for Neandertal alleles (Table 2) (Dannemann et al., 2016, Dannemann and Kelso, 2017, Quach et al., 2016, Sams et al., 2016, Sankararaman et al., 2014).
Power Analysis
The ability to study a particular Neandertal variant depends both on its effect size and its frequency within a given sample of individuals or cell lines. We cannot control the effect size, but one can—within reason—control the number of samples considered. Larger sample sizes allow more variants to be studied, but may offer diminishing returns. To determine the power of studies of certain sample sizes, we considered how many Neandertal variants would be present at particular frequency thresholds, as an effect of sample size. For each category of Neandertal allele (eQTL, amino acid change, etc.), we subsampled X cell lines, and counted the number of Neandertal variants present at least z times, for values of z = (1, 5, 10, 15, 20). This subsampling was repeated 100 times for all values of x and z. We plotted the average number of Neandertal variants present at a particular rate on the y axis. Each possible value of z is given a different color, and the range of values over 100 resamplings was shown as colored confidence intervals. For example, given a sample of 50 random cell lines, 62% of the 501 Neandertal eQTLs in the HipSci resource are present in at least 5 cell lines, and 92% are present in at least 1 cell line.
Browser with Neandertal Haplotypes for HipSci Resource
We provided a database of the inferred introgressed Neandertal alleles for each of the 173 HipSci resource individuals, which can be accessed at https://bioinf.eva.mpg.de/stemcellbrowser. We combined these alleles with further annotations from several external genomic databases and functional annotations of introgressed alleles. The browser layout was inspired by the browser of the Exome Aggregation Consortium.
HipSci and GTEx eQTLs
We have identified 76 of the 7,229 significant eQTLs (FDR < 0.05) in the HipSci iPSC expression data that are tagged by an SNP that is linked to a Neandertal haplotype (Kilpinen et al., 2017). We have then queried which of these haplotypes have been linked to a significant eQTL in any of 48 GTEx tissues as well (p < 0.01) (Dannemann et al., 2017). Between 0% and 29.6% of the iPSC eQTLS that are linked to a Neandertal haplotype are significant eQTLs in 48 GTeX adult tissues as well (Figure S2).
Single-Cell and Single-Nuclei Transcriptome Data Analysis
Cerebral organoid single-cell RNA-seq data were generated using the 10X Chromium Single Cell 3′ v.2 Kit following the manufacturer's instructions. The reads and expression data, together with cell type annotations and projection coordinates from 2-month-old cerebral organoids from seven individuals (four of which were lines from the HipSci resource) were acquired from Kanton et al. (2019). Extensive details on cell type annotation and data analysis can be found in the primary publication. For the four HipSci lines, we note that organoids were dissociated for all four HipSci cell lines and pooled at equal ratios to be loaded on one lane of the microfluidic device aiming for 10k cells. The data were then demultiplexed based on analysis of single nucleotide polymorphisms detected in the single-cell RNA-seq reads. Diffusion mapping (implemented in R package destiny) (Angerer et al., 2016) was applied to the cortical cells from each of the four HipSci iPSC lines using default settings, with the expression levels of the highly variable genes as the input. The ranks in diffusion component 1 were used as the pseudotimes. We have calculated gene expression correlation networks using 7,349 genes that correlate strongly (r > 0.6) with transcription factors that have previously been reported to be involved in the regulation of progenitor proliferation and neuron differentiation (Camp et al., 2015). Based on the correlation expression patterns we have generated knn networks (k = 70) using the igraph R package (Figures 4C and 4K). To classify reads as “Neandertal” or “modern human” we have selected genomic position overlapping KIFAP3 and POU2F1 for which the four HipSci cell lines differ and that are informative for the existence of a Neandertal haplotype (rs4519 and rs1059761). Using samtools (Li et al., 2009) we have extracted reads that overlap these positions and partitioned them based on whether the Neandertal allele was present or not. We have then classified cells in which KIFAP3 and POU2F1 were detected into four groups: (1) those with reads with and without the Neandertal allele, (2) only reads the Neandertal allele, (3) only reads with the modern human allele, and (4) no informative reads (Figures 4G–4J). Based on the reads from the single-cell RNA-seq that overlap informative positions we are able to classify the reads as Neandertal or modern human depending on the observed allele at these sites. We calculated relative expression differences between progenitors and neurons for genes that carry Neandertal-informative reads (Figure 4E). For this analysis we defined all cells in the lower 30% of pseudotime as progenitors and cells in the upper 30% as neurons and computed the ratios between the mean expression in each cell type. Single-nuclei RNA-seq data, including cell type annotations and projection coordinates from adult human prefrontal cortex were acquired from Kanton et al. (2019) without modification. Plots were generated using the ggplot2 (Wilkinson, 2011) and Seurat (Butler et al., 2018) R packages.
Data and Code Availability
We have provided the raw and processed data, as well as extensive metadata to the Human Cell Atlas Data Coordination Platform (HCA-DCP) as well as to ArrayExpress (E-MTAB-7552).
Author Contributions
M.D., B.V., J.G.C. have conceived and designed the study. M.D., Z.H., B.V., L.S., S.K., A.W. and J.G.C. have performed experiments and analyzed data. C.H., J.K., M.D. and J.G.C. have designed and generated the web browser. J.C.G., B.T., J.K., S.P. have supervised this study. M.D., J.G.C. have written and revised the manuscript with input from all other authors.
Acknowledgments
The GTEx data used for the analyses described in this manuscript were obtained from dbGaP accession number phs000424.v6.p1.c1 on 05/23/2016. S.K. was supported by a PhD fellowship of the Boehringer Ingelheim Fonds. We thank Rigo Schultz and the IT department of the Max Planck Institute for Evolutionary Anthropology for help with setting up the online database. This work was supported by the European Research Council (803441 ANTHROPOID to J.G.C.), the NOMIS Foundation, and the Max Planck Society.
Published: June 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.05.018.
Supplemental Information
References
- 1000 Genomes Project Consortium, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer P., Haghverdi L., Büttner M., Theis F.J., Marr C., Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- Banovich N.E., Li Y.I., Raj A., Ward M.C., Greenside P., Calderon D., Tung P.Y., Burnett J.E., Myrthil M., Thomas S.M. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res. 2018;28:122–131. doi: 10.1101/gr.224436.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder M.J., Smail C., Gloudemans M.J., Frésard L., Jakubosky D., D’Antonio M., Li X., Ferraro N.M., Carcamo-Orive I., Mirauta B. Systematic assessment of regulatory effects of human disease variants in pluripotent cells. bioRxiv. 2019 doi: 10.1101/784967. [DOI] [Google Scholar]
- Browning S.R., Browning B.L., Zhou Y., Tucci S., Akey J.M. Analysis of human sequence data reveals two pulses of archaic denisovan admixture. Cell. 2018;173:53–61.e9. doi: 10.1016/j.cell.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I., Hoffman G.E., Cundiff P., Beckmann N.D., D’Souza S.L., Knowles J.W., Patel A., Papatsenko D., Abbasi F., Reaven G.M. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell. 2017;20:518–532.e9. doi: 10.1016/j.stem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M., Kelso J. The contribution of Neanderthals to phenotypic variation in modern humans. Am. J. Hum. Genet. 2017;101:578–589. doi: 10.1016/j.ajhg.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M., Andrés A.M., Kelso J. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors. Am. J. Hum. Genet. 2016;98:22–33. doi: 10.1016/j.ajhg.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M., Prüfer K., Kelso J. Functional implications of Neandertal introgression in modern humans. Genome Biol. 2017;18:61. doi: 10.1186/s13059-017-1181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H.-Y. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunz P., Tilot A.K., Wittfeld K., Teumer A., Shapland C.Y., van Erp T.G.M., Dannemann M., Vernot B., Neubauer S., Guadalupe T. Neandertal introgression sheds light on modern human endocranial globularity. Curr. Biol. 2019;29:895. doi: 10.1016/j.cub.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdinjak M., Fu Q., Hübner A., Petr M., Mafessoni F., Grote S., Skoglund P., Narasimham V., Rougier H., Crevecoeur I. Reconstructing the genetic history of late Neanderthals. Nature. 2018;555:652–656. doi: 10.1038/nature26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch A.G., Tandon A., Patterson N., Song Y., Rohland N., Palmer C.D., Chen G.K., Wang K., Buxbaum S.G., Akylbekova E.L. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wuchty S., Przytycka T.M. eQTL epistasis—challenges and computational approaches. Front. Genet. 2013;4:51. doi: 10.3389/fgene.2013.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchís-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- Khrameeva E.E., Bozek K., He L., Yan Z., Jiang X., Wei Y., Tang K., Gelfand M.S., Prufer K., Kelso J. Neanderthal ancestry drives evolution of lipid catabolism in contemporary Europeans. Nat. Commun. 2014;5:3584. doi: 10.1038/ncomms4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D.F., Masson G., Sigurdsson A., Jonasdottir A., Walters G.B., Jonasdottir A., Gylfason A., Kristinsson K.T. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy R.C., Wakefield J., Akey J.M. Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell. 2017;168:916–927.e12. doi: 10.1016/j.cell.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez F.L., Watkins J.C., Hammer M.F. Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol. Biol. Evol. 2013;30:798–801. doi: 10.1093/molbev/mst004. [DOI] [PubMed] [Google Scholar]
- Meyer M., Kircher M., Gansauge M.-T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M.C., Rahal C. A scientometric review of genome-wide association studies. Commun. Biol. 2019;2:9. doi: 10.1038/s42003-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermúdez F., Badsha F., Kanton S., Camp J.G., Vernot B., Köhler K., Voigt B., Okita K., Maricic T., He Z. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife. 2016;5:e18683. doi: 10.7554/eLife.18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo S. The human condition—a molecular approach. Cell. 2014;157:216–226. doi: 10.1016/j.cell.2013.12.036. [DOI] [PubMed] [Google Scholar]
- Panopoulos A.D., D’Antonio M., Benaglio P., Williams R., Hashem S.I., Schuldt B.M., DeBoever C., Arias A.D., Garcia M., Nelson B.C. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports. 2017;8:1086–1100. doi: 10.1016/j.stemcr.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petr M., Pääbo S., Kelso J., Vernot B. Limits of long-term selection against Neandertal introgression. Proc. Natl. Acad. Sci. U S A. 2019;116:1639–1644. doi: 10.1073/pnas.1814338116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., de Filippo C., Grote S., Mafessoni F., Korlević P., Hajdinjak M., Vernot B., Skov L., Hsieh P., Peyrégne S. A high-coverage Neandertal genome from Vindija cave in Croatia. Science. 2017;358:655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Stoneking M. Denisovan ancestry in east eurasian and native American populations. Mol. Biol. Evol. 2015;32:2665–2674. doi: 10.1093/molbev/msv141. [DOI] [PubMed] [Google Scholar]
- Quach H., Rotival M., Pothlichet J., Loh Y.-H.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams A.J., Dumaine A., Nédélec Y., Yotova V., Alfieri C., Tanner J.E., Messer P.W., Barreiro L.B. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 2016;17:246. doi: 10.1186/s13059-016-1098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Patterson N., Li H., Pääbo S., Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Mallick S., Patterson N., Reich D. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGMA Type 2 Diabetes Consortium, Williams A.L., Jacobs S.B.R., Moreno-Macías H., Huerta-Chagoya A., Churchhouse C., Márquez-Luna C., García-Ortíz H., Gómez-Vázquez M.J., Burtt N.P. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvert M., Quintana-Murci L., Rotival M. Impact and evolutionary determinants of Neanderthal introgression on transcriptional and post-transcriptional regulation. Am. J. Hum. Genet. 2019;104:1241–1250. doi: 10.1016/j.ajhg.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti C.N., Vernot B., Bastarache L., Bottinger E., Carrell D.S., Chisholm R.L., Crosslin D.R., Hebbring S.J., Jarvik G.P., Kullo I.J. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016;351:737–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G., Williams S.M., Tishkoff S.A. The missing diversity in human genetic studies. Cell. 2019;177:1080. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- Slon V., Mafessoni F., Vernot B., de Filippo C., Grote S., Viola B., Hajdinjak M., Peyrégne S., Nagel S., Brown S. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature. 2018;561:113–116. doi: 10.1038/s41586-018-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telis N., Aguilar R., Harris K. Selection against archaic DNA in human regulatory regions. bioRxiv. 2019 doi: 10.1101/708230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- Vernot B., Tucci S., Kelso J., Schraiber J.G., Wolf A.B., Gittelman R.M., Dannemann M., Grote S., McCoy R.C., Norton H. Excavating Neandertal and denisovan DNA from the genomes of melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson L. ggplot2: elegant graphics for data analysis by WICKHAM, H. Biometrics. 2011;67:678–679. [Google Scholar]
- Williams L.A., Davis-Dusenbery B.N., Eggan K.C. SnapShot: directed differentiation of pluripotent stem cells. Cell. 2012;149:1174–1174.e1. doi: 10.1016/j.cell.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Wilder S.P., Johnson N., Juettemann T., Flicek P.R. The ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have provided the raw and processed data, as well as extensive metadata to the Human Cell Atlas Data Coordination Platform (HCA-DCP) as well as to ArrayExpress (E-MTAB-7552).