Abstract
Usher syndrome is a genetic disorder causing neurosensory hearing loss and blindness from retinitis pigmentosa (RP). Adaptive techniques such as braille, digital and optical magnifiers, mobility training, cochlear implants, or other assistive listening devices are indispensable for reducing disability. However, there is currently no treatment to reduce or arrest sensory cell degeneration. There are several classes of treatments for Usher syndrome being investigated. The present article reviews the progress this research has made towards delivering commercial options for patients with Usher syndrome.
Keywords: usher syndrome, gene therapy, cell therapy, ipscs, gene editing, adeno-associated virus, antisense oligonucleotides
Introduction
Usher syndrome is a group of autosomal recessive disorders characterized by congenital neurosensory hearing loss, progressive night vision impairment, and constriction of the visual field due to retinitis pigmentosa (RP). Some forms of Usher syndrome may also have varying levels of vestibular dysfunction resulting in loss of balance. It is the most common form of inherited deaf-blindness (El-Amraoui and Petit, 2014) affecting an estimated 1 in 6,000 people worldwide (Kimberling et al., 2010). Usher subtypes (1, 2, and 3; Table 1) are graded according to the severity of symptoms and age of onset. Type 1 patients are born profoundly deaf and experience pre-pubertal onset of progressive vision loss caused by RP. The majority of type 1 patients also have developmental motor delays caused by vestibular dysfunction. Type 2 patients have mild to moderate congenital hearing loss with RP diagnosed during puberty. Hearing loss in type 3 patients is progressive and post-lingual, while RP onset may be delayed until mid-adulthood (Reiners et al., 2006). The clinical presentation of RP begins with night blindness caused by the degeneration of rod photoreceptor cells. Subsequent constriction of the visual field results in a “tunnel vision” effect caused by the centripetal progression of cone photoreceptor cell loss. In classical RP, the death of cones may be secondary to rod degeneration and this may ultimately lead to complete loss of vision in advanced age (Hartong et al., 2006). Many other inherited retinal diseases are associated with deafness (Table 2) such as cone-rod dystrophy and hearing loss-1 (CRDHL1, OMIM #617236), diabetes and deafness, maternally inherited (MIDD, OMIM #520000) and Leber congenital amaurosis (LCA) with early-onset deafness (LCAEOD, OMIM #617879). This review is limited to the combination of RP and deafness, the classical presentation of Usher syndrome.
Table 1.
Genes and proteins associated with various Usher syndrome subtypes.
| Type | Subtype (OMIM REF.) | Gene | Protein | % Cases* | Transcripts | Major transcript | Exons |
|---|---|---|---|---|---|---|---|
| 1 | 1B (#276900) | MYO7A | MYOSIN 7A | 21 | 14 | 7,483 bp; NM_000260.4 | 56 |
| 1C (#276904) | USH1C | HARMONIN | 2 | 11 | 2,232 bp; NM_005709.4 | 29 | |
| 1D (#601067) | CDH23 | CADHERIN 23 | 6 | 19 | 11,138 bp; NM_022124.6 | 71 | |
| 1F (#602083) | PCDH15 | PROTOCADHERIN 15 | 3 | 36 | 6,983 bp; NM_001142763.2 | 48 | |
| 1G (#606943) | USH1G | SANS | 1 | 2 | 3,558 bp; NM_173477.5 | 4 | |
| 2 | 2A (#276901) | USH2A | USHERIN | 50 | 5 | 6,372 bp; NM_007123.6 | 72 |
| 2C (#605472) | ADGRV1 | ADHESION G-PROTEIN COUPLED RECEPTOR-V1 | 5 | 37 | 19,557 bp; NM_032119.4 | 91 | |
| 2D (#611383) | WHRN | WHIRLIN | 0.4 | 9 | 3,989 bp; XM_011518485.1 | 21 | |
| 3 | 3A (#276902) | CLRN1 | CLARIN-1 | 2 | 8 | 2,087 bp; NM_174878.3 | 6 |
| 3B (#614504) | HARS | HISTIDYL-TRNA SYNTHETASE | - | 16 | 1,948 bp; NM_002109.6 | 13 | |
| Modifier | - | PDZD7** |
*Frequencies were calculated in a 2019 study (Jouret et al., 2019) of 684 patients with dual vision and hearing loss. A proportion of these patients did not have mutations in any of the Usher genes tested. Candidate genes are not shown. **Denotes the modifier gene PDZD7, which contributes to the phenotype of Usher 2A patients through interactions with USHERIN, but is not independently causal of Usher syndrome (Ebermann et al., 2010).
Table 2.
List of non-Usher syndromes that cause hearing loss and inherited retinal disease.
| Disease | Omim reference | Gene | Retinal phenotype | Systemic phenotype/s |
|---|---|---|---|---|
| Cone-rod dystrophy and hearing loss 1 (CRDHL1) | #617236 | CEP78 | Cone-rod dystrophy | Hearing loss |
| Cone-rod dystrophy and hearing loss 2 (CRDHL2) | #618358 | CEP250 | Cone-rod dystrophy | Early-onset sensorineural hearing loss |
| Leber congenital amaurosis with early-onset deafness (LCAEOD) | #617879 | TUBB4B | Leber congenital amaurosis | Early-onset deafness |
| Polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract | #612674 | ABHD12 | Retinitis pigmentosa | Hearing loss, polyneuropathy, ataxia |
| Diabetes and deafness, maternally inherited (MIDD) | #520000 | MTTL1 | Macular dystrophy | Adult-onset sensorineural hearing loss and diabetes, ptosis, cardiomyopathy, myopathy, renal failure, neuropsychiatric symptoms |
See www.omim.org for further information.
The Genetics and Biology of Usher Syndrome
Usher syndrome is caused by autosomal recessive inheritance of mutations in Usher genes known to encode proteins involved in transmembrane adhesion, scaffolding and motor transport. Ten causative genes have so far been identified. Inheritance of hypomorphic alleles with missense mutations often causes non-syndromic deafness, while nonsense and cryptic splice-site mutations result in Usher syndrome (Ahmed et al., 2008; Bademci et al., 2016). Certain types of mutations in Usher genes may also cause non-syndromic RP (Seyedahmadi et al., 2004). Digenic inheritance of mutations in separate Usher genes has also been proposed to be causative of Usher syndrome (Zheng et al., 2005; Bonnet et al., 2011), but remains controversial (Jouret et al., 2019). Additionally, the disparity in phenotypes and progression rates between monozygotic twins is suggestive of environmental influence (Liu et al., 1999). A summary of identified genes and their proteins can be seen in Table 1.
Usher proteins are localized both to the inner ear and retina. In the inner ear, Usher proteins are present in the cochlea and vestibular organs, accounting for the balance and deafness phenotypes. Usher genes instruct the differentiation of mechano-sensitive hair cells and affect hair bundle organization during development. Functional Usher proteins are also essential for the transduction of electrical signals in mature stereocilia (Grati and Kachar, 2011; Cosgrove and Zallocchi, 2014; El-Amraoui and Petit, 2014; Pepermans et al., 2014; Mathur and Yang, 2015, 2019; Han et al., 2018). In the retina, Usher proteins are found in the light-sensitive photoreceptor neurons. They have been implicated in intracellular trafficking at the connecting cilium, which links the photoreceptor’s inner and outer segments, and facilitates the movement of phototransduction proteins and lipids to the outer segment (Liu et al., 1997; Mathur and Yang, 2015). Knowledge of the function of Usher proteins in the retina is limited by a lack of effective animal models (El-Amraoui and Petit, 2014), perhaps due to their association with calyceal processes, the microvilli which protrude from the apical region of the inner segment and surround the connecting cilium of human, but not murine, photoreceptors (Sahly et al., 2012; Schietroma et al., 2017).
Current Trials and Pre-clinical Studies for Usher Syndrome Treatment
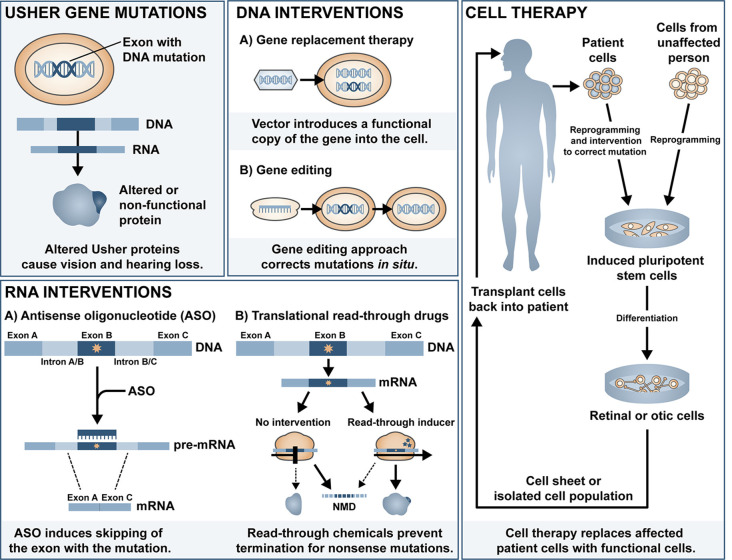
Cochlear implants are often provided to patients with type 1 Usher syndrome due to the profound deafness at birth. Those with type 2 or 3 Usher syndrome may benefit from hearing aids or cochlear implant later in life if the mild congenital deafness progresses. However, there is currently no treatment available to prevent or reverse the inevitable retinal degeneration associated with RP. There is some evidence for use of dietary supplements to delay the progression of RP, including vitamin A (Berson et al., 1993, 2018), omega-3 fish oils (Berson et al., 2004, 2012), N-acetylcysteine (Campochiaro et al., 2020)1 and the antioxidant taurine (Trouillet et al., 2018). Although some of these have been disputed (Rayapudi et al., 2013; Hoffman et al., 2014), the lack of strong evidence for efficacy may be related to the heterogeneity of study cohort, small sample size, and short follow-up study design. Additionally, vector-induced expression of ciliary neurotrophic factor (CNTF) was shown to be neuroprotective in a mouse model of RP (Lipinski et al., 2015), yet long-term follow up of three RP patients treated with sustained-release CNTF delivered intravitreally via encapsulated cell technology implant (NT-501, Neurotech Pharmaceuticals Inc.), one of which had Usher-associated RP, showed no significant difference in visual acuity (Talcott et al., 2011). In this review article, we will summarise the major classes of ongoing investigations for gene and mutation-specific treatment of hearing and visual loss and tissue regeneration and the progress and challenges in delivering therapeutic outcomes to patients with Usher syndrome. The main approaches discussed can be divided into DNA intervention, RNA intervention and cell replacement and an overview of the different approaches is shown in Figure 1.
Figure 1.
Overview of the different therapeutic approaches for Usher syndrome.
DNA Interventions
Non-viral Systems
DNA interventions are possible due to engineered or recombinant vectors (broadly classified as viral and non-viral) that are synthesized to carry genetic material (Sengillo et al., 2016). Non-viral vectors (nanoparticles) are non-immunogenic, readily customizable, and may package up to 20 kb of material, allowing the delivery of large therapeutic genes (Moore et al., 2018).
Lipid-based nanoparticles, which form a protective bilayer around transported genetic components, have been explored as potential treatments for hearing loss. Zou et al. (2017) investigated the functional, inflammatory, and apoptotic response to liposome nanoparticle delivery to the murine inner ear and found no adverse reactions. Additionally, Gao et al. (2018) reported that cationic lipid-mediated delivery of Cas9:guideRNA complexes to the Beethoven (Bth) mouse model of deafness selectively disrupted the dominant mutant Tmc1 allele, reducing hearing loss. Inner hair cells (IHCs) and outer hair cells (OHCs) had significantly improved survival rates and auditory brainstem response (ABR) thresholds compared to uninjected controls (Gao et al., 2018). Lipid-based nanoparticles have also been used to deliver base editing machinery in a proof-of-concept study to ameliorate hearing loss (discussed further in section Base Editing; Yeh et al., 2018).
Supraparticles are colloidal nanoparticle aggregates with a larger drug loading capacity relative to individual nanoparticles (Sperling and Gradzielski, 2017). Supraparticles have already been used to deliver the developmental neurotrophin, brain-derived neurotrophic factor (BDNF), to the inner ear of a hearing loss guinea pig model (Wang et al., 2014). BDNF is required for the maintenance of spiral ganglion neurons (SGNs; Ylikoski et al., 1993) and may serve to protect or regenerate SGNs as well as promote synaptic regeneration at the ribbon synapse (Suzuki et al., 2016). Supraparticles offer sustained longer-term release of BDNF and maintained near-wild-type numbers of SGNs in the guinea pig cochlea (Wang et al., 2014). However, delivery of supraparticles may mediate the unintentional movement of small nanoparticles from the inner ear to the cerebral spinal fuid via the cochlear aqueduct (Zhang et al., 2013).
Another significant advancement in the non-viral delivery platform was the development of the polyethylene glycol-substituted 30-mer lysine peptide (CK30-PEG) nanoparticles, which have been used successfully in a cystic fibrosis clinical trial (Konstan et al., 2004). CK30-PEG nanoparticles may also be effective for the treatment of ocular diseases, as they show a retinal targeting efficiency comparable to viral vectors up to 2 weeks post-injection (Farjo et al., 2006; Han et al., 2012a). Appreciable transgene expression mediated by CK30-PEG nanoparticle gene delivery has been reported in retinal degeneration models, including an autosomal dominant model of RP (Cai et al., 2010; Han et al., 2012b). In the autosomal RP model study, nanoparticles containing the mouse opsin promoter and the Prph2 gene were subretinally delivered to Rds mice carrying a haploinsufficiency mutation, resulting in wild-type-level recovery of cone function but a modest restoration of rod function (Cai et al., 2010). Modified CK30-PEG nanoparticles with a photoreceptor-specific promoter also led to the structural and functional rescue of Stargardt-associated pathology in the Abca4−/− mouse model of vision loss (Han et al., 2012b). Recent studies have used solid lipid nanoparticles (SLNs) as a more efficient non-viral delivery system and a study on a mouse model of x-linked juvenile retinoschisis showed transduction of retinal pigment epithelium (RPE) and photoreceptors and improved retinal phenotype (Apaolaza et al., 2016).
Though nanoparticle delivery is promising and continues to be developed, there are several roadblocks to its clinical application, including biodegradability, biocompatibility, and non-specific transfection (Yin et al., 2014; Chen et al., 2016). Topical delivery is the simplest and most patient-friendly form of administration. However, ocular barriers and the long diffusion pathway prevent therapeutic levels from reaching the retina (Bisht et al., 2018). The intraocular injection route would, therefore, be preferable, though the transient expression associated with non-viral gene delivery (Bisht et al., 2018; Huang and Chau, 2019) would necessitate repeated injections and consequently, an increase in their associated risks. Thus far, no pre-clinical studies have investigated nanoparticle delivery of Usher genes but the large packaging capacity of non-viral vectors does increase the potential of this type of platform for treatment development for Usher-causing mutations in the large genes such as CDH23, PCDH15, ADGRV1, MYO7A, and USH2A (Table 1).
Viral-Based Gene Replacement Therapy
With the advantage of increased efficiency over non-viral vectors (Nayerossadat et al., 2012), viral vectors currently represent the most promising approach to therapeutic genetic interventions. Recombinant viral vectors utilize the inherent capability of viruses for cellular transduction to deliver genetic material to donor cells in vivo. However, viral vectors come with their limitations, including limited packaging capacities. Potential immunogenicity is also an obstacle to clinical application, though advances in molecular biology have allowed for the separation of wild-type viral coding genes and cis-acting sequences. Consequently, segregation can now produce viral vectors that do not reconstitute by recombination into productive viral particles but still maintain viral infectivity capacity (Kay et al., 2001).
Gene replacement therapy involves the introduction of a non-mutant copy of the affected gene to restore expression to the host cell or tissue (Sengillo et al., 2016). One of the first described, and most frequently used vectors, adenovirus, has a large (~40 kb) packaging capacity and has been used in over 400 clinical trials (Wold and Toth, 2013). In a study by Bennett et al. (1996), an adenovirus-based vector was used to deliver the Phosphodiesterase β subunit gene to Rd1 mice, delaying photoreceptor degeneration. However, many people have circulating antibodies against adenoviruses, so their use in gene therapies is limited due to a high immunogenic response (DiCarlo et al., 2018). Furthermore, adenovirus has shown to have low tropism for photoreceptor cells in vivo (Li et al., 1994), an additional limitation that instigated the search for other types of viral vectors with improved targeting in the eye. The sections below will discuss the two most promising vectors: lentivirus and adeno-associated virus (AAV).
Lentiviral Vectors
Lentiviral vectors, derived from the human immunodeficiency virus, were also initially favored by researchers for clinical application due to their relatively large packaging capacity (reviewed in Zheng et al., 2018). However, whilst their immunogenicity is low, their tendency to integrate into the genome calls into question their clinical applicability (Zheng et al., 2018). Pioneering work by Hashimoto et al. (2007) led to the development of a lentiviral-mediated MYO7A gene delivery vector that produced wild-type protein levels in RPE cells in culture, as well as rescuing phenotypic RPE defects in vitro. In vivo, melanosome mislocalization and opsin accumulation at the photoreceptor connecting cilium was corrected in the Myo7a-deficient shaker1 mouse model of Usher 1B (Hashimoto et al., 2007). Subsequently, UshStat, a therapeutic recombinant vector expressing the human MYO7A, was developed using the equine infectious anaemia virus (Zallocchi et al., 2014), a non-pathogenic, non-primate-derived lentivirus capable of transducing human cells (Mitrophanous et al., 1999; Mazarakis et al., 2001). Subretinal delivery of UshStat to shaker1 mice was shown to protect photoreceptors from light-induced degeneration and demonstrated tolerability in non-human primates, which led to this approach progressing to a clinical trial for the prevention of RP in Usher 1B patients (NCT02065011; results pending).
Adeno-Associated Viral Vectors (AAV)
Safety is of principal concern in all viral vector technologies and as AAVs are not known to cause any human pathogenesis, they have emerged as the vector of choice for gene therapy applications worldwide. AAVs have low immunogenicity, high cellular specificity, and long-lasting gene expression. AAVs typically persist as episomes [Zheng et al., 2018; circular genomes which replicate independently of their host (Watanabe and Fukasawa, 1961)], reducing the risk of insertional mutagenesis. Vandendriessche et al. (2007) directly compared lentiviral and AAV vectors and found that AAV serotypes 8 and 9 induced higher expression of transgenic factor IX protein than lentiviruses in mouse models of hemophilia B and severe combined immunodeficiency, without interacting with proinflammatory cytokines. AAVs also hold potential for the treatment of Usher retinal degeneration as they can efficiently transduce photoreceptors and RPE (Rodrigues et al., 2018). Successful treatment of vision loss in LCA type 2 (LCA2) and choroideremia via AAV-mediated delivery of the RPE65 and CHM genes, respectively, have been demonstrated in human clinical trials (reviewed in Russell et al., 2017). The LCA2 trials have now resulted in the first-ever Food and Drug Administration (FDA)-approved and European Medicines Agency (EMA)-approved AAV-based gene therapy drug for LCA2 due to recessive RPE65 mutations (Luxturna® or Luxturna™/voretigene neparvovec-ryzl or voretigene neparvovec, Roche and Novartis).
A major disadvantage of AAV vectors is their relatively small (4.7 kb) packaging capacity (Zheng et al., 2018); however, in vivo delivery of the smaller Usher genes has been investigated using AAV vectors. Delivery of rAAV2/8 containing USH1G cDNA to the inner ear improved hearing and hair cell disorganization in the Usher 1G mice (Emptoz et al., 2017). Durable inner ear expression of Usher genes has also resulted in in vivo protein restoration of WHIRLIN (Zou et al., 2011) and CLARIN-1 in several studies (Dinculescu et al., 2016; Geng et al., 2017; Dulon et al., 2018; György et al., 2019).
Transduction efficiency is a crucial factor in the success of gene therapy and is highly cell and serotype-dependent. Several synthetically-produced AAV variants have been investigated for Usher syndrome treatments. One example is the synthetic rAAV2/Anc80L65, which may transduce close to 100% of IHCs and 90% of OHCs in mice (Landegger et al., 2017). Using rAAV2/Anc80L65, Pan et al. (2017) demonstrated gene and protein recovery of Harmonin in an Usher 1C mouse model. Deverman et al. (2016) reported a highly efficient synthetic vector, AAV9-PHP.B, which showed robust transduction efficiency in the retina of a dominant RP mouse model (Giannelli et al., 2017). Further, AAV9-PHP.B carrying a green fluorescent protein (GFP) reporter transduced the inner ear and retina of wild-type mice at a rate of 60–80% for IHCs, 30–40% for OHCs and 70–80% for photoreceptors (György et al., 2019). When used to package the Clrn-1 gene, AAV9-PHP.B mediated rescue of low-frequency hearing (4–8 kHz) in a mouse model of Usher 3A deafness (György et al., 2019). However, the tropism and potential toxicity of the AAV9-PHP.B vector in the central nervous system (CNS) of non-human primates is still under study (Hordeaux et al., 2018; reviewed in Deverman et al., 2018). The potential, therefore, for treatment of Usher syndrome using certain AAV-based vectors is high, but the large size of some Usher genes, including some of the most common forms of Usher such as USH1B (MYO7A), does present unique challenges for the field.
Oversized Adeno-Associated Viral Vectors
As mentioned above, the packaging limitation of AAVs precludes delivery of several of the larger Usher genes, including MYO7A (USH1B; Jaijo et al., 2006) and PCDH15 (USH1F; Alagramam et al., 2001). Some studies, however, have pushed the limits of AAV packaging capacity by using oversized AAV transgene constructs (fAAV). A study by Allocca et al. (2008) identified rAAV2/5 as being an efficient packager of up to 8.9 kb of genetic material, allowing, in theory, the large retinal disease genes MYO7A, ABCA4 and CEP290 to be packaged and delivered subretinally. They showed that fAAV2/5 ABCA4 delivery led to a stable improvement of morphological abnormalities and retinal dysfunction associated with a mouse model of Stargardt disease. Additionally, Colella et al. (2013) identified defects in light/dark adaptation in shaker1 mice, which was improved by fAAV2/5 delivery of MYO7A. However, other studies have shown that these vectors do not contain full-length genes, but instead tend to contain heterogeneous mixtures of gene fragments, most of which are truncated and typically less than 5 kb (Dong et al., 2010). Furthermore, Grieger and Samulski (2005) demonstrated that AAV vectors carrying genomes larger than 5 kb had less efficient transduction capacity due to a post entry preferential degradation of AAVs carrying larger genomes. Though the truncated genomes may reassemble before transcription and form full-length proteins inside the cell (Lopes et al., 2013), the lack of characterization and heterogeneity of this approach limits its clinical use.
Multi-Adeno-Associated Viral Vector Systems
As an alternative to the oversized AAV approach, different research groups have tested the use of dual and triple AAV vector systems. Multi or dual AAV systems use the inherent concatemerization of AAV genomes in vivo to form full-length cDNA which can then be transcribed into functional mRNA within the transduced cell (Trapani et al., 2014). Different methods are used to deliver dual/multivectors and are usually divided into trans-splicing, overlap, and hybrid approaches. Trans-splicing vectors, first described by Yan et al. (2000), separates the gene of interest into 5′ (left) and 3′ (right) halves, with the 3′ end of the left construct containing a splice donor site, which concatemerises with the splice acceptor site located on the 5′ end of the right construct. In the overlap approach, the 3′ end of the right half and the 5′ end of the left half contain a highly recombinogenic overlapping region which mediates homologous recombination between gene segments (Duan et al., 2001). This sequence can be from an external gene, like alkaline phosphatase, or directly from the gene of interest. Finally, hybrid vectors contain both the splice donor/acceptor sites of trans-splicing vectors and the recombinogenic properties of overlapping vectors, allowing reconstitution to occur via either method (Ghosh et al., 2008).
Several research groups have published proof-of-concept studies using multi or dual AAV systems, including for inherited retinal diseases (Colella et al., 2014; Dyka et al., 2014, 2019; McClements et al., 2019). Multi-AAV systems have been used to express MYO7A in vivo and in vitro with equal or higher efficiency than fAAV delivery (Dyka et al., 2014), and to produce full-length mRNA with 100% fidelity to the target cDNA (Dyka et al., 2014). Additionally, Maddalena et al. (2018) expanded the transfer capacity further by using a triple therapeutic vector system to incorporate large genes such as ALMS1 and the Usher 1D gene, CDH23. Truncated protein products were detected in eyes treated with CDH23 but not ALMS1 triple AAV vectors. A functional response was not recorded in CDH23-treated mice but treated Alms1−/− mice showed a non-significant increase in outer nuclear layer thickness and transient (2–6 months) improvement of electroretinogram a- and b-wave measurements (Maddalena et al., 2018).
Despite the promising potential of dual AAV approaches, most studies show very low levels of protein expression using this system. Dyka et al. (2014) quantified the relative expression of full-length cDNA mediated by dual AAV reconstitution and found hybridizing vectors to be 2–3 fold more efficient than overlap and trans-splicing counterparts in MYO7A-transfected HEK293T cells. Data published by Colella et al. (2014) found that hybrid and trans-splice dual AAV reconstitution achieved approximately one-quarter of the photoreceptor expression levels induced by single AAV vectors in the large white pig retina. In their 2018 article, Maddalena et al. (2018) reported that the co-transduction rate for triple AAV vectors was <6% of single vector expression in HEK293T cells. Interestingly, this efficiency increased when vectors were delivered subretinally to mice and pigs (photoreceptor expression = 27 ± 6% and 39 ± 17% of that induced by single AAV, respectively). In a recent study, Carvalho et al. (2017) tested the in vitro and in vivo expression of the different dual AAV approaches and found hybrid vectors to have superior rates of reconstitution. They showed that reconstitution efficiency in HEK293T cells for trans-splicing, hybrid, and overlapping vectors was 10.3%, 15.3%, and 17.4%, respectively. The efficiency of overlapping vectors was found to be gene-specific as it was dependent on the length of the recombinogenic region and showed no detectable levels in vivo. In vivo subretinal delivery to the mouse retina resulted in full-length protein expression in 9.07% and 1.78% of cells transduced with hybrid and trans-splicing vectors, respectively (Carvalho et al., 2017). Interestingly, they were the first to show a discrepancy in reconstituted mRNA and protein levels after dual AAV delivery which may be explained by transcript instability of longer mRNA sequences (Feng and Niu, 2007).
An alternative to dual AAV systems may be dual-intein splicing, which is based on protein rather than mRNA reconstitution, but is still delivered by AAV vectors. Inteins are segments of proteins that have been described as “protein introns,” as they can excise themselves from the sequence of peptides and join the flanking portions (the exteins) together. Recently, Tornabene et al. (2019) demonstrated reporter protein levels in C57BL/6J mice retinae induced by dual-intein splicing to be comparable to single AAV transduction and significantly higher than dual AAV transduction. This approach was also shown to alleviate retinal degeneration in animal models of Leber Congenital Amaurosis (Rd16 mice) and Stargardt disease (Abca4−/− mice). Physiological symptoms of retinal degeneration, including RPE lipofuscin accumulation in Abca4−/− mice, and outer nuclear layer thinning in Rd16 mice, were significantly reduced in the dual-intein treated mice. Additionally, pupillary light responses increased by ~20% in treated Rd16 mice compared to untreated Rd16 controls (Tornabene et al., 2019). However, the viability of this approach for the delivery of large genes has not yet been tested in models of Usher syndrome.
DNA Editing
While gene delivery aims to replace a defective gene with a functional copy, DNA editing attempts to directly correct the mutation in vivo, which also allows the repaired gene to be expressed under endogenous regulators. Furthermore, the size of the gene is not a limiting factor. Repair of single base transitions or transversions at the DNA level can be achieved through the induction of a DNA break to facilitate incorporation of the correct DNA base (Kim et al., 1996; Christian et al., 2010; Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013) while targeted conversion of a single DNA base can reverse a transitional mutation (Komor et al., 2016; Gaudelli et al., 2017).
Gene Editing
The first attempt using gene editing for Usher syndrome was a study by Overlack et al. (2012) that used zinc finger nucleases (ZFNs) to target the p.R31X mutation in the human Ush1c gene. Their in vitro results show partial repair of the Ush1c gene and recovery of harmonin protein in a p.R31X cell line after transfection with ZFNs. In recent years, however, the discovery of the clustered, regularly interspaced, palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system have changed the field of gene editing profoundly. Their higher efficiency, simplicity, and targeting capacity have made them the preferred technology of choice for gene editing (Khan, 2019).
The CRISPR/Cas9 system uses an adapted microbial immune technique for precise editing of the genome (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013). A guide RNA is engineered to target a specific locus, which is then digested by the Cas9 endonuclease (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013). Either of two endogenous mechanisms repairs double-stranded breaks in the genome. The clinically-favorable homology-directed repair relies on an intact template strand whilst error-prone non-homologous end-joining is independent of a template sequence, and often results in insertions or deletions (Adli, 2018).
CRISPR/Cas9 has been used to study potential treatments for inherited retinal diseases. Moreno et al. (2018) used the viral delivery of CRISPR components to downregulate Nrl in mouse models of non-Usher RP. Nrl is a transcriptional regulator that indirectly determines whether photoreceptors develop into rods or cones (Cheng et al., 2004; Moreno et al., 2018). The knockdown of Nrl expression transformed rods into cone-like cells that did not experience rod-specific degeneration. This leads to a decrease in rods and therefore night blindness, but an increase in daylight vision (Moreno et al., 2018). However, mouse models of Usher 1C and 1G have cones that are sensitive to oxidative stress, indicating that increasing the number of cone-like cells may not be a viable option for Usher RP treatment (Trouillet et al., 2018).
Viral delivery of CRISPR/Cas9 to fibroblasts derived from an Usher 2A patient showed some rescue of Usherin expression, however, the efficiency of restoration was not significant enough to proceed to clinical trials (Fuster-García et al., 2017). Additionally, induced pluripotent stem cells (iPSCs) derived from a patient with a mutation in MYO7A were investigated by Tang et al. (2016). Stereocilia-like protrusions from patient iPSCs developed disorganized morphology which was rescued by CRISPR/Cas9 editing (Tang et al., 2016), indicating a potential treatment avenue for Usher 1B patients that deserves further investigation.
Recently, the CRISPR/Cas9-based tool EDIT-101 received FDA approval for a clinical trial in LCA10 patients (NCT03872479), leading to the first-ever direct human administration of CRISPR/Cas92. EDIT-101 is a gene editing therapeutic which utilizes a Staphylococcus aureus Cas9 guide RNA which has high specificity for the c.2991 + 1655A >G transversion in intron 26 (IVS26) of the CEP290 gene, limiting off-target effects (Maeder et al., 2019). Recently, the company behind EDIT-101, Editas3, has publically announced the utilization of a new therapeutic, EDIT-102, to target Usher type 2A4. Another recent update is the introduction of a new gene editing tool, “prime editing,” which utilizes a synthetic fusion of an altered Cas9 endonuclease and reverse transcriptase, directed by a gene editing guide RNA (Anzalone et al., 2019). This technology shows promising specificity and broad applicability to a large number of human pathogenic mutations, although further studies will be needed to confirm its viability for treating Usher syndrome.
Though the potential of CRISPR/Cas9 to make precise and patient-specific gene corrections with high targeting capacity is attractive, the efficacy of in vivo delivery is still in doubt due to potential off-target effects (Fu et al., 2013; Kuscu et al., 2014). Advancements in the field since its original application in gene editing, including the development of anti-CRISPR proteins to limit non-specific activity (Pawluk et al., 2016; Rauch et al., 2017; Nakamura et al., 2019), have potential to address some limitations of this technology and further studies could help validate CRISPR/Cas9-based approaches for the treatment of Usher syndrome.
Base Editing
The advent of CRISPR/Cas9 gene editing systems has also allowed for the development of base editing, which utilizes inactivated Cas nucleases and single-stranded editing enzymes to replace one nucleobase with another without creating double-stranded breaks (Rees and Liu, 2018). Two classes of base editing technology have so far been reported; cytosine base editors (CBEs; Komor et al., 2016) which convert C-G base pairs to T-A, and adenine base editors (ABEs), which convert A-T to G-C (Gaudelli et al., 2017). Both CBEs and ABEs introduce transition mutations between chemically similar base pairs, which can theoretically correct over 60% of human pathogenic point mutations (Rees and Liu, 2018). However, transversion base editing (e.g., G-C to C-G) remains an elusive target.
Base editing has already been used in proof-of-concept studies in mouse models of Duchenne muscular dystrophy (Ryu et al., 2018), phenylketonuria (Villiger et al., 2018), hereditary tyrosinemia type I (Song et al., 2020) and hypercholesterolemia through targeting the PCSK9 gene (Chadwick et al., 2017). Importantly, base editing has also been used to investigate enhanced cellular reprogramming of supporting cells to cochlea hair cells to mediate hearing loss (Yeh et al., 2018). C-T conversion of a single base in the CTNNB1 gene prevented phosphorylation and degradation of β-catenin protein, leading to a 7-fold increase of β-catenin levels in HEK293T cultures. Consequently, there was an increase in signaling to the Wnt pathway, which is crucial to the development of sensory hair cells. When delivered to mice via intracochlear injection, base editing resulted in the differentiation of cochlea supporting cells to MYO7A-expressing hair cells (Yeh et al., 2018).
RNA Intervention
RNA splicing can be altered to either restore exons lost due to aberrant splicing induced by the mutation, or induce exon skipping to remove nonsense or missense mutations in coding regions. RNA intervention resulting in altered splicing is achieved through the binding of an antisense oligonucleotide to RNA strands (reviewed by Havens et al., 2013). Base editing of RNA is also possible (Bass, 2002). Finally, translational readthrough techniques aim to suppress protein truncation by overriding mutations that cause premature termination of the translation machinery. The reader is referred to a review outlining the therapeutic progress of translational readthrough-inducing compounds in the treatment of inherited diseases (Nagel-Wolfrum et al., 2016).
RNA-Based Drug Interventions
RNA molecules designed to silence or interfere with toxic gain-of-function mutations have been investigated in the treatment of several non-Usher models of genetic hearing loss. Notably, Maeda et al. (2005) identified a short interfering RNA (siRNA) that targets the R75W allele variant in GJB2, a common cause of autosomal recessive hearing loss. The synthetic siRNA suppressed GJB2 in HEK293T cells by >80% for more than 120 h. When administered to the GJB2 mouse model, siRNA treatment suppressed GJB2 expression by >70% and prevented hearing loss (Maeda et al., 2005, 2007). Another study by Shibata et al. (2016) developed a microRNA targeting non-syndromic deafness caused by gain-of-function mutations in TMC1. The microRNA was packaged via AAV2/9 and delivered to Bth mice, resulting in significant preservation of hearing for up to 21 weeks relative to untreated controls. The animals who responded best to treatment maintained ABR thresholds 40 dB greater than untreated counterparts (Shibata et al., 2016).
Antisense Oligonucleotides
ASOs are short nucleic acid sequences that modulate gene expression via complementary binding to mRNA. ASOs are often synthesized to activate ribonuclease H (RNAse-H, which degrades mRNA) or target splicing defects (Goyal and Narayanaswami, 2018). ASOs can then be designed to target a pathogenic mutation; hence, the size of the gene is not an obstacle as with gene delivery techniques. However, the half-life of the drug means re-occurring invasive administration to the eye rather than one-off treatments, as is the case with a gene therapy approach.
ASO treatment has been widely investigated for retinal disease therapies (Huang et al., 2017). Recently, up to nine antisense-oligonucleotide variants were identified for the treatment of Stargardt disease caused by the intronic c.4539 + 2001G >A mutation in the large ABCA4 gene (Garanto et al., 2019). In a separate study, screening of ASOs led to the discovery of QR-110 (Dulla et al., 2018), a therapeutic for LCA type 10 (LCA10), caused by the c.2991 + 1655A >G mutation, in the CEP290 gene (Den Hollander et al., 2006). Like Usher syndrome, the pathogenesis of LCA10 is caused by defects in the photoreceptor connecting cilium. QR-110 was able to restore wild-type CEP290 transcript and protein expression in mutant fibroblasts and decrease ciliopathy in 3-dimensional (3D) retinal organoids (explained further in section Cell Therapy for Retinal Degeneration). The drug was also well-tolerated in mice, rabbits, and non-human primates (Dulla et al., 2018), leading to the approval of phase I/IIa clinical trial (NCT03140969) by ProQR Therapeutics5.
ASOs have also been developed for autosomal dominant RP (Naessens et al., 2019), as well as Usher 2A-associated RP (Slijkerman et al., 2016). An engineered ASO targeting a pseudo-exon-causing mutation in USH2A (c.7595–2144A >G) displayed splice-correcting properties in patient-derived fibroblasts (Slijkerman et al., 2016). The same company that funded this study, ProQR Therapeutics, has also investigated an ASO, QR-421a, for the treatment of Usher 2A-associated RP caused by a common exon 13 mutation (c.2299delG; Van Diepen et al., 2019). The efficacy of this treatment was initially demonstrated in patient-derived retinal organoids and animal models (Table 3), with exon-skipping capability maintained in cynomolgus monkeys for more than 100 days post-treatment. They also showed that Usherin protein was present in wild-type zebrafish larvae at the photoreceptor connecting cilium but absent in untreated c.2299delG zebrafish. Partial restoration of correctly localized usherin expression was observed in treated larvae as well as improved ERG recordings compared to untreated zebrafish (Van Diepen et al., 2019; conference abstract). The results of this study have allowed for advancement to a clinical trial (NCT03780257), which has already led to the first treatment to the eyes of an Usher 2A patient using QR-421a6 (preliminary findings reported on the 20th of April 2020 here: https://ir.proqr.com/news-releases/news-release-details/proqr-announces-positive-findings-interim-analysis-phase-12).
Table 3.
Summary of transgenic animal disease models discussed in this review.
| Human disease (equivalent) | Model | Animal | Gene | Mutation | Therapy tested | Reference |
|---|---|---|---|---|---|---|
| Usher 1B | Shaker1 (Sh1−/–) | Mouse | Myo7a | G720X induced by ENU | AAV2/2-mediated gene delivery | Colella et al. (2013) |
| Usher 1C caused by c.216G >A mutation | Ush1c c.216G >A | Mouse | Ush1C | c.216G >A | AAV2/Anc80L65-mediated gene delivery | Pan et al. (2017) |
| ASO-mediated suppression of exon 3 cryptic splice site (ASO-29) | Lentz et al. (2013), Vijayakumar et al. (2017), Donaldson et al. (2018) and Ponnath et al. (2018) | |||||
| Usher 1G | Ush1g−/– | Mouse | Ush1g | Ush1gfl/fl mice targeting exon 2, crossed with PGK-cre mice | rAAV2/8-mediated gene delivery | Emptoz et al. (2017) |
| Usher 2A | Ush2amcm1 | Zebrafish | Ush2a | Homozygous premature stop mutations in exon 13 | ASO-mediated exon 13 skipping (QR-421a) | Van Diepen et al. (2019) |
| Usher 2D caused by compound heterozygous Q103 ×/c.837 + 1G >A mutation | Whirlin−/– | Mouse | Whrn | Targeted deletion of exon 1 | AAV2/8-mediated gene delivery | Zou et al. (2011) |
| Usher 3A | Clrn1 KO | Mouse | Clrn1 | Targeted deletion of Clrn1 upstream promoter, first exon, 269 bp of the first intron | AAV-mediated gene delivery | Dinculescu et al. (2016), Geng et al. (2017) and György et al. (2019) |
| Clrn1ex4−/−−/– | Mouse | Clrn1 | Early ubiquitous deletion of Clrn1 exon 4 | AAV-mediated gene delivery | Dulon et al. (2018) | |
| Clrn1ex4fl/flMyo15-Cre+/− | Mouse | Clrn1 | Hair cell-specific postnatal deletion of Clrn1 exon 4 | AAV-mediated gene delivery | Dulon et al. (2018) | |
| Non-syndromic hearing loss (DFNA36) | Beethoven (Bth) | Mouse | Tmc1 | c.1235T>A | Lipid-mediated delivery of cas9:gRNA complex | Gao et al. (2018) |
| Non-syndromic hearing loss (SLC26A4) | Slc26a4-null | Mouse | Slc26a4 | Mutations not reported | iPSC otic progenitors | Chen et al. (2018) |
| Non-syndromic hearing loss (GJB2) | GJB2_R75W-eGFP | Mouse | Gjb2 | R75W | siRNA | Maeda et al. (2005) |
Abbreviations: ENU, N-ethyl-N-nitrosourea; AAV, adeno-associated virus; ASO, Antisense oligonucleotides; iPSC, induced pluripotent stem cells; siRNA, small interfering RNA.
Several studies have investigated ASO-29 treatment of the Usher 1C, c.216G >A mouse model, which exhibits hearing loss as well as disrupted exploratory movements, indicating vestibular dysfunction. ASO-29 administration to the inner ear of c.216G >A mouse pups has been shown to improve both the auditory and vestibular response (Lentz et al., 2013; Vijayakumar et al., 2017; Donaldson et al., 2018). However, mounting evidence has demonstrated an age threshold for effective delivery, with early-treated mice responding better to treatment. Ponnath et al. (2018) demonstrated that both outer and inner hair cell function was effectively restored by ASO-29 treatment, but the threshold for effective outer hair cell treatment (post-natal day 5) was lower than that of inner hair cell treatment (post-natal day 7). Considering these promising results in the auditory and vestibular response of USH1C mice, it would be interesting to assess the efficacy of ASO-29 towards the vision loss phenotype. However, the limited visual phenotype of the USH1C mice model means alternative models will need to be used for testing ASO-29.
Translational Readthrough Inducing Drugs
Nonsense mutations cause premature termination codons (PTCs), leading to the production of a truncated polypeptide or targeting of the transcript by nonsense-mediated decay (NMD). When a translation termination codon (UAA, UGA, UAG) enters the ribosomal A site, it is recognized by eukaryotic release factor 1 (eRF1), which changes conformation when bound to eRF3, initiating a signal cascade that results in hydrolytic cleavage of the polypeptide by eRF1. Translation termination is a competition between recognition by eRF1 leading to termination and continuation of the translation process, leading to readthrough or natural suppression. Natural suppression occurs >0.1% of the time in non-mutant transcripts, but PTCs increase the rate of natural suppression to up to 1% (Keeling et al., 2012). Nonsense suppression therapies aim to increase translational readthrough, avoiding the production of a truncated protein. The two main obstacles with this type of therapy are avoiding mRNA degradation by NMD, and avoiding the promotion of non-specific readthrough (Frischmeyer and Dietz, 1999; Keeling and Bedwell, 2011; Wang and Gregory-Evans, 2015; Dabrowski et al., 2018; Campofelice et al., 2019).
Nonsense suppression has clear advantages over gene-based therapies; they are not gene- or mutation-specific, the size of the gene is not an issue, and they allow the cell to maintain expression regulation. However, they are associated with nephrotoxic (Mingeot-Leclercq and Tulkens, 1999) and ototoxic (Selimoglu, 2007) effects and often result in the incorporation of a non-cognate amino acid, which may alter protein function. A recent advancement in the field was the development of anticodon engineered transfer RNAs (ACE-tRNAs), which recognize and suppress stop codons while encoding the cognate amino acid of the non-mutant polypeptide (Lueck et al., 2019). This method induced a specific readthrough of CFTR mutations in vitro. Low levels of off-target suppression were detected, depending on the genetic environment of the mutation, but the authors suggest that endogenous cellular pathways to degrade incorrectly transcribed proteins would be sufficient to offset these effects (Lueck et al., 2019).
Several studies have investigated nonsense-mediated therapies for the treatment of Usher syndrome. Initial studies focussed on nb30, a synthetic derivative developed from the commercial aminoglycoside, paromomycin (Nudelman et al., 2006). Nb30 was shown to induce significant nonsense suppression of a common USH1C mutation (p.R31X) with reduced toxicity and increased biocompatibility compared to commercial aminoglycosides (Goldmann et al., 2010). Similarly, favorable toxicity profiles were observed in nb30-mediated suppression of PCDH15 nonsense mutations relative to traditional antibiotics (Rebibo-Sabbah et al., 2007). Subsequently, a second paromomycin derivative, nb54, was identified as a promising drug candidate for nonsense suppression which demonstrated readthrough ability in several prominent disease genes, including Usher 1F syndrome (Nudelman et al., 2009). In this study, Nudelman et al. (2009) show that nb54 is capable of inducing in vitro stop codon suppression 3–5-fold times more efficient for PCDH15 (Usher 1F) mutations p.R3X and p.R245X compared to three other aminoglycoside compounds.
PTC-124 (Ataluren) is another translational readthrough inducer which has been studied for application in cystic fibrosis and Duchenne muscular dystrophy cases (Welch et al., 2007), though success has thus far been limited in clinical trials (Wilschanski et al., 2011; Kerem et al., 2014; McDonald et al., 2017). PTC-124-induced readthrough in vitro has been demonstrated in the common p.Trp3955* mutation, which accounts for 13% of USH2A mutations (Neuhaus et al., 2017). Additionally, PTC-124 treatment induced in vitro and in vivo readthrough of USH1C mutations, leading to the recovery of protein function with superior biocompatibility to gentamicin and nb30 (Goldmann et al., 2011). Finally, Goldmann et al. (2012) compared nb30, nb54, and PTC-124 for translational readthrough of USH1C mutations and identified both PTC-124 and nb54 as ideal drug candidates. Though promising, functional rescue of Usher phenotypes in vivo will nonetheless be necessary to evaluate the efficacy of both PTC-124 and nb54.
Cell Replacement Therapy
Cellular therapies are therapeutic approaches that aim to regenerate damaged tissue by introducing replacement donor cells (Zakrzewski et al., 2019). This approach relies on the survival of connecting interneurons when the stimulus-receiving neuron has already degenerated in the cochlea or retina. Typically, progenitor cells derived from stem cells are used as an unlimited source of donor cells. Stem cells are pluripotent progenitors that can differentiate into cell lineages from each of the three germ layers. Early stem cell studies relied on the controversial use of human embryonic stem cells (hESCs; Omole and Fakoya, 2018) until the discovery in 2007 by Shinya Yamanaka and colleagues that adult human somatic cells, such as adult fibroblasts, could be reprogrammed back into a pluripotent state (Takahashi et al., 2007). They achieved this through the expression of characteristic pluripotent markers SOX2, OCT3/4, KLF4, and c-MYC, delivered to the cell via retroviral vectors (Takahashi et al., 2007). As the stem cells carry the same genetic information as the somatic cells from which they were made, this breakthrough allowed human diseases to be replicated and studied non-invasively in the laboratory for the first time (Omole and Fakoya, 2018).
Cell Therapy for Retinal Degeneration
iPSCs can aggregate to form “embryoid bodies” (aggregates of cells thought to mimic the early embryo), which can be prompted to differentiate into specific lineages (Omole and Fakoya, 2018), including 3D retinal organoids (Nakano et al., 2012; Phillips et al., 2012; Zhong et al., 2014; Mellough et al., 2015, 2019). These organoids closely resemble in vivo human eye development, forming eye field-like domains that form 3D retinal neuroepithelium and RPE. The neural retina and RPE domains transition into a pseudo optic cup-like structure and retinal progenitor cells differentiate into neurons including horizontal cells, amacrine cells, and retinal ganglion cells (Zhong et al., 2014). Organoids develop a defined outer nuclear layer containing radially-aligned photoreceptors with detectable inner and outer segments connected by a connecting cilium (Zhong et al., 2014; Mellough et al., 2015, 2019; Lowe et al., 2016; Parfitt et al., 2016). These photoreceptors express phototransduction proteins, including opsins, and can respond to a light stimulus (Zhong et al., 2014; Hallam et al., 2018; Mellough et al., 2019).
Retinal organoids are a good resource of photoreceptor progenitors for transplantation. These progenitors can be enriched before transplantation for a homogenous cell population via cell sorting for dissociated cell transplants (Lakowski et al., 2015, 2018). MacLaren et al. (2006) transplanted developing mouse photoreceptors into the retinae of degenerative mouse models with successful integration and differentiation, noting an optimal therapeutic window corresponding to the specification of rod cell fate. Subsequently, photoreceptor transplantation was reported by several research groups (Pearson et al., 2012; Barber et al., 2013; Gonzalez-Cordero et al., 2013; Singh et al., 2013; Santos-Ferreira et al., 2016), including Barber et al. (2013) who looked into six mouse models of inherited retinal degenerations (IRDs) with environmental, disease course, and genetic factors influencing the integration outcome (Barber et al., 2013). Though these initial photoreceptor transplantation experiments seemed promising, the transfer of cytoplasmic material between labeled donors and host photoreceptors has been attributed to the seemingly high rates of integration (Santos-Ferreira et al., 2016; Singh et al., 2016). Alternatively, a stem cell-derived retinal sheet may be transplanted into the recipient’s eye (Assawachananont et al., 2014). Though clinical trials have focussed on RPE transfer in patients with age-related macular degeneration (AMD) and Stargardt Disease (NCT03102138, NCT02941991, NCT01469832), none thus far have reported results on photoreceptor transplants. However, a first-in-human phase I/II dose-escalation study is currently examining the safety and tolerability of human retinal progenitor cells in patients with RP (NCT02464436, estimated completion date: July 2021). Furthermore, proof-of-principle studies in mouse and non-human primates showed survival of retinal sheets [containing hESC-derived retinal cells (Shirai et al., 2016), hiPSC-derived RPE cells (Mandai et al., 2017) or hiPSC-derived retinal progenitors (Tu et al., 2019)] post-transplantation, and improvement of light sensitivity (Shirai et al., 2016; Mandai et al., 2017; Tu et al., 2019).
Cell Therapy for Neurosensory Deafness
Stem cell therapy also has the potential to treat hearing loss by replacing cochlear hair cells, although this is a difficult environment for this approach. The hostile high potassium luminal fluid (endolymph) environment of the cochlear duct poses significant challenges for cell survival. Further, the robust tight junctions in the auditory epithelium in the organ of Corti make donor cellular integration difficult. A recent article reported the ability of iPSC-derived otic progenitors to form connections with co-cultured spiral ganglion-like cells (Chen et al., 2018). Subsequently, these cells were detached and administered to the inner ear via round window injection in hearing loss Slc26a4 mice. Cells positive for the hair cell marker, MYO7A (also causative of Usher 1B), were detected in the organ of Corti, indicating the potential of this method to deliver progenitor cells capable of migrating, differentiating and forming appropriate connections in the inner ear (Chen et al., 2018). However, the progenitors did not form an organized stereocilial arrangement, which is critical to hearing, and disrupted in Usher cases (Mathur and Yang, 2015).
Limitations of Cell Therapies
The development of cellular therapies presents a promising and broad approach to treat a host of sensory diseases regardless of genetic etiology. However, several safety concerns remain to be solved. Of principal concern is the potential for donor cells to cause neoplastic changes in the tissue, particularly those that are derived from stem cells. Mouse embryonic stem cells injected into the subretinal space have been shown to induce tumor formation (Arnhold et al., 2004), though human iPSC-derived photoreceptor progenitor delivery to rd1 mice found no evidence of abnormal cell growth (Barnea-Cramer et al., 2016). Additionally, Chen et al. (2018) injected mouse models with hiPSC-derived otic progenitors and hiPSC controls to examine the tumorigenicity of hiPSC-derived cells. They reported tumor formation in hiPSC-injected tissue, but not in tissue injected with hiPSC-derived progenitors (Chen et al., 2018). Exclusion of undifferentiated stem cells before transplantation could further decrease the risk of neoplastic changes in the host. In a recent study, Gagliardi et al. (2018) demonstrated the potential of this strategy by safely transplanting CD73+ photoreceptor precursors separated from a cell population using magnetic-activated cell sorting into the eyes of P23H-transgenic rats.
Despite the multitude of challenges in delivering stem cell therapeutics safely and effectively, regenerative medicine remains attractive to patients, industry, and commercial clinics. Several stem cell-based clinical trials are listed on clinicaltrials.gov for AMD (NCT01736059, NCT01920867) and other ocular diseases (NCT01920867), including RP (NCT02320812). Recently, the stem cell ophthalmology treatment study (SCOTS) published results from a phase I/IIa study of five Usher syndrome patients with bilaterally-treated eyes (Weiss and Levy, 2019). Autologous bone marrow-derived stem cells were separated from bone marrow aspirate and administered into patients’ eyes via a combination of retrobulbar, sub-Tenon, intravitreal, subretinal, and intra-optic injections, as chosen by the patient and depending on the extent of visual loss and relative risks. Increases in visual acuity were noted in 80% of treated eyes with a statistical significance of p < 0.01. There was no reported visual loss nor any complications up to a year post-treatment (Weiss and Levy, 2019). However, a continual follow up is needed to confirm the long-term visual acuity effects and determine whether the patients remain free of adverse events.
Conclusions
Significant advancements have been made in the last several years towards the treatment and prevention of sensory loss in Usher patients. Particularly, gene and cell therapies pose attractive, potentially one-off solutions that would reduce the burden of invasive re-administration of medications to the eye. Though the majority of studies are currently proof-of-principle treatment strategies using animal models of disease (summarized in Table 3), it is highly possible that the results from current ongoing clinical trials may translate into effective new treatments (see Table 4 for a summary of ongoing clinical trials). However, more temporary therapeutics such as ASOs and translational readthrough inhibitors may also offer a significant reduction in disability. Continuing to improve the safety and efficacy of these treatment approaches is of utmost importance to provide commercially available options for Usher syndrome patients.
Table 4.
Active clinical trials for Usher patients.
| Identifier | Status | Intervention | Usher subtypes | Country |
|---|---|---|---|---|
| NCT02065011 | NR | UshStat—lentiviral delivery of MYO7A | Usher 1B | USA, France |
| NCT01530659 | NR | NT-501—ciliary neurotrophic factor intraocular implant | Usher types 2 and 3 | USA |
| NCT03780257 | R | QR-421a—antisense oligonucleotide to induce skipping of exon 13 | Usher 2A due to mutations in exon 13 | USA, Belgium, Canada, France |
| NCT03011541 | R | Autologous bone marrow-derived stem cells | All | USA |
All trials listed are for the treatment or prevention of Usher-associated retinal degeneration. R, currently recruiting; NR, not recruiting. See www.clinicaltrials.gov for further information.
Author Contributions
LF wrote the first draft of the manuscript. FC, CM, and LC contributed to manuscript revision, reading, and approval of the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by funding from Retina Australia (LC, FC, and CM), Usher 1 Collaborative (LC), Genetics Cures Australia (LC), Lions Eye Institute/Save Sight Foundation Brian King Fellowship (CM), Australian National Health and Medical Research Council (MRF1142962, GNT1188694, and GNT1116360; FC).
References
- Adli M. (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9:1911. 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z. M., Riazuddin S., Aye S., Ali R. A., Venselaar H., Anwar S., et al. (2008). Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum. Genet. 124, 215–223. 10.1007/s00439-008-0543-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam K. N., Murcia C. L., Kwon H. Y., Pawlowski K. S., Wright C. G., Woychik R. P. (2001). The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet. 27, 99–102. 10.1038/83837 [DOI] [PubMed] [Google Scholar]
- Allocca M., Doria M., Petrillo M., Colella P., Garcia-Hoyos M., Gibbs D., et al. (2008). Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J. Clin. Invest. 118, 1955–1964. 10.1172/JCI34316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apaolaza P. S., del Pozo-Rodríguez A., Solinís M. A., Rodríguez J. M., Friedrich U., Torrecilla J., et al. (2016). Structural recovery of the retina in a retinoschisin-deficient mouse after gene replacement therapy by solid lipid nanoparticles. Biomaterials 90, 40–49. 10.1016/j.biomaterials.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Arnhold S., Klein H., Semkova I., Addicks K., Schraermeyer U. (2004). Neurally selected embryonic stem cells induce tumor formation after long-term survival following engraftment into the subretinal space. Invest. Ophthalmol. Vis. Sci. 45, 4251–4255. 10.1167/iovs.03-1108 [DOI] [PubMed] [Google Scholar]
- Assawachananont J., Mandai M., Okamoto S., Yamada C., Eiraku M., Yonemura S., et al. (2014). Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Reports 2, 662–674. 10.1016/j.stemcr.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G., Foster J., II., Mahdieh N., Bonyadi M., Duman D., Cengiz F. B., et al. (2016). Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet. Med. 18, 364–371. 10.1038/gim.2015.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber A. C., Hippert C., Duran Y., West E. L., Bainbridge J. W. B., Warre-Cornish K., et al. (2013). Repair of the degenerate retina by photoreceptor transplantation. Proc. Natl. Acad. Sci. U S A 110, 354–359. 10.1073/pnas.1212677110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea-Cramer A. O., Wang W., Lu S.-J., Singh M. S., Luo C., Huo H., et al. (2016). Function of human pluripotent stem cell-derived photoreceptor progenitors in blind mice. Sci. Rep. 6:29784. 10.1038/srep29784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846. 10.1146/annurev.biochem.71.110601.135501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J., Tanabe T., Sun D., Zeng Y., Kjeldbye H., Gouras P., et al. (1996). Photoreceptor cell rescue in retinal degeneration (Rd) mice by in vivo gene therapy. Nat. Med. 2, 649–654. 10.1038/nm0696-649 [DOI] [PubMed] [Google Scholar]
- Berson E. L., Rosner B., Sandberg M. A., Hayes K. C., Nicholson B. W., Weigel-DiFranco C., et al. (1993). A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch. Ophthalmol. 111, 761–772. 10.1001/archopht.1993.01090060049022 [DOI] [PubMed] [Google Scholar]
- Berson E. L., Rosner B., Sandberg M. A., Weigel-DiFranco C., Willett W. C. (2012). Ω-3 intake and visual acuity in patients with retinitis pigmentosa receiving vitamin A. Arch. Ophthalmol. 130, 707–711. 10.1001/archophthalmol.2011.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson E. L., Rosner B., Sandberg M. A., Weigel-DiFranco C., Moser A., Brockhurst R. J., et al. (2004). Further evaluation of docosahexaenoic acid in patients with retinitis pigmentosa receiving vitamin A treatment: subgroup analyses. Arch. Ophthalmol. 122, 1306–1314. 10.1001/archopht.122.9.1306 [DOI] [PubMed] [Google Scholar]
- Berson E. L., Weigel-DiFranco C., Rosner B., Gaudio A. R., Sandberg M. A. (2018). Association of vitamin a supplementation with disease course in children with retinitis pigmentosa. JAMA Ophthalmol. 136, 490–495. 10.1001/jamaophthalmol.2018.0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht R., Mandal A., Jaiswal J. K., Rupenthal I. D. (2018). Nanocarrier mediated retinal drug delivery: overcoming ocular barriers to treat posterior eye diseases. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 10:e1473. 10.1002/wnan.1473 [DOI] [PubMed] [Google Scholar]
- Bonnet C., Grati M. H., Marlin S., Levilliers J., Hardelin J.-P., Parodi M., et al. (2011). Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J. Rare Dis. 6:21. 10.1186/1750-1172-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Conley S. M., Nash Z., Fliesler S. J., Cooper M. J., Naash M. I. (2010). Gene delivery to mitotic and postmitotic photoreceptors via compacted DNA nanoparticles results in improved phenotype in a mouse model of retinitis pigmentosa. FASEB J. 24, 1178–1191. 10.1096/fj.09-139147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campochiaro P. A., Iftikhar M., Hafiz G., Akhlaq A., Tsai G., Wehling D., et al. (2020). Oral N-acetylcysteine improves cone function in retinitis pigmentosa patients in phase I trial. J. Clin. Invest. 130, 1527–1541. 10.1172/JCI132990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campofelice A., Lentini L., Di Leonardo A., Melfi R., Tutone M., Pace A., et al. (2019). Strategies against nonsense: oxadiazoles as translational readthrough-inducing drugs (TRIDs). Int. J. Mol. Sci. 20:3329. 10.3390/ijms20133329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho L. S., Turunen H. T., Wassmer S. J., Luna-Velez M. V., Xiao R., Bennett J., et al. (2017). Evaluating efficiencies of dual AAV approaches for retinal targeting. Front. Neurosci. 11:503. 10.3389/fnins.2017.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick A. C., Wang X., Musunuru K. (2017). In vivo base editing of pcsk9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscler. Thromb. Vasc. Biol. 37, 1741–1747. 10.1161/atvbaha.117.309881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo Z., Tian H., Chen X. (2016). Production and clinical development of nanoparticles for gene delivery. Mol. Ther. Methods Clin. Dev. 3:16023. 10.1038/mtm.2016.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Hong F., Zhang C., Li L., Wang C., Shi H., et al. (2018). Differentiation and transplantation of human induced pluripotent stem cell-derived otic epithelial progenitors in mouse cochlea. Stem Cell Res. Ther. 9:230. 10.1186/s13287-018-0967-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Khanna H., Oh E. C., Hicks D., Mitton K. P., Swaroop A. (2004). Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 13, 1563–1575. 10.1093/hmg/ddh173 [DOI] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., Hummel A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella P., Sommella A., Marrocco E., Di Vicino U., Polishchuk E., Garcia Garrido M., et al. (2013). Myosin7a deficiency results in reduced retinal activity which is improved by gene therapy. PLoS One 8:e72027. 10.1371/journal.pone.0072027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella P., Trapani I., Cesi G., Sommella A., Manfredi A., Puppo A., et al. (2014). Efficient gene delivery to the cone-enriched pig retina by dual AAV vectors. Gene Ther. 21, 450–456. 10.1038/gt.2014.8 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D., Zallocchi M. (2014). Usher protein functions in hair cells and photoreceptors. Int. J. Biochem. Cell Biol. 46, 80–89. 10.1016/j.biocel.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E. (2018). Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons. Mol. Med. 24, 25–25. 10.1186/s10020-018-0024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hollander A. I., Koenekoop R. K., Yzer S., Lopez I., Arends M. L., Voesenek K. E. J., et al. (2006). Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 79, 556–561. 10.1086/507318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman B. E., Pravdo P. L., Simpson B. P., Kumar S. R., Chan K. Y., Banerjee A., et al. (2016). Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209. 10.1038/nbt.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman B. E., Ravina B. M., Bankiewicz K. S., Paul S. M., Sah D. W. (2018). Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov. 17, 641–659. 10.1038/nrd.2018.110 [DOI] [PubMed] [Google Scholar]
- DiCarlo J. E., Mahajan V. B., Tsang S. H. (2018). Gene therapy and genome surgery in the retina. J. Clin. Invest. 128, 2177–2188. 10.1172/jci120429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinculescu A., Stupay R. M., Deng W. T., Dyka F. M., Min S. H., Boye S. L., et al. (2016). AAV-mediated clarin-1 expression in the mouse retina: implications for USH3A gene therapy. PLoS One 11:e0148874. 10.1371/journal.pone.0148874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson T. N., Jennings K. T., Cherep L. A., McNeela A. M., Depreux F. F., Jodelka F. M., et al. (2018). Antisense oligonucleotide therapy rescues disruptions in organization of exploratory movements associated with Usher syndrome type 1C in mice. Behav. Brain Res. 338, 76–87. 10.1016/j.bbr.2017.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Nakai H., Xiao W. (2010). Characterization of genome integrity for oversized recombinant AAV vector. Mol. Ther. 18, 87–92. 10.1038/mt.2009.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D., Yue Y., Engelhardt J. F. (2001). Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 4, 383–391. 10.1006/mthe.2001.0456 [DOI] [PubMed] [Google Scholar]
- Dulla K., Aguila M., Lane A., Jovanovic K., Parfitt D. A., Schulkens I., et al. (2018). Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c.2991+1655A>G LCA10 models. Mol. Ther. Nucleic Acids 12, 730–740. 10.1016/j.omtn.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulon D., Papal S., Patni P., Cortese M., Vincent P. F., Tertrais M., et al. (2018). Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J. Clin. Invest. 128, 3382–3401. 10.1172/jci94351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyka F. M., Boye S. L., Chiodo V. A., Hauswirth W. W., Boye S. E. (2014). Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum. Gene Ther. Methods 25, 166–177. 10.1089/hgtb.2013.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyka F. M., Molday L. L., Chiodo V. A., Molday R. S., Hauswirth W. W. (2019). Dual ABCA4-AAV vector treatment reduces pathogenic retinal A2E accumulation in a mouse model of autosomal recessive stargardt disease. Hum. Gene Ther. 30, 1361–1370. 10.1089/hum.2019.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebermann I., Phillips J. B., Liebau M. C., Koenekoop R. K., Schermer B., Lopez I., et al. (2010). PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J. Clin. Invest. 120, 1812–1823. 10.1172/JCI39715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Amraoui A., Petit C. (2014). The retinal phenotype of Usher syndrome: pathophysiological insights from animal models. C. R. Biol. 337, 167–177. 10.1016/j.crvi.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Emptoz A., Michel V., Lelli A., Akil O., Boutet de Monvel J., Lahlou G., et al. (2017). Local gene therapy durably restores vestibular function in a mouse model of Usher syndrome type 1G. Proc. Natl. Acad. Sci. U S A 114, 9695–9700. 10.1073/pnas.1708894114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo R., Skaggs J., Quiambao A. B., Cooper M. J., Naash M. I. (2006). Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS One 1:e38. 10.1371/journal.pone.0000038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Niu D.-K. (2007). Relationship between mRNA stability and length: an old question with a new twist. Biochem. Genet. 45, 131–137. 10.1007/s10528-006-9059-5 [DOI] [PubMed] [Google Scholar]
- Frischmeyer P. A., Dietz H. C. (1999). Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8, 1893–1900. 10.1093/hmg/8.10.1893 [DOI] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster-García C., García-García G., González-Romero E., Jaijo T., Sequedo M. D., Ayuso C., et al. (2017). USH2A gene editing using the CRISPR system. Mol. Ther. Nucleic Acids 8, 529–541. 10.1016/j.omtn.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi G., Ben M’Barek K., Chaffiol A., Slembrouck-Brec A., Conart J.-B., Nanteau C., et al. (2018). Characterization and transplantation of CD73-positive photoreceptors isolated from human iPSC-derived retinal organoids. Stem Cell Reports 11, 665–680. 10.1016/j.stemcr.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Tao Y., Lamas V., Huang M., Yeh W.-H., Pan B., et al. (2018). Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221. 10.1038/nature25164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garanto A., Duijkers L., Tomkiewicz T. Z., Collin R. W. J. (2019). Antisense oligonucleotide screening to optimize the rescue of the splicing defect caused by the recurrent deep-intronic ABCA4 variant c.4539+2001G >A in stargardt disease. Genes 10:452. 10.3390/genes10060452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N. M., Komor A. C., Rees H. A., Packer M. S., Badran A. H., Bryson D. I., et al. (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R., Omar A., Gopal S. R., Chen D. H., Stepanyan R., Basch M. L., et al. (2017). Modeling and preventing progressive hearing loss in usher syndrome III. Sci. Rep. 7:13480. 10.1038/s41598-017-13620-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Yue Y., Lai Y., Duan D. (2008). A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol. Ther. 16, 124–130. 10.1038/sj.mt.6300322 [DOI] [PubMed] [Google Scholar]
- Giannelli S. G., Luoni M., Castoldi V., Massimino L., Cabassi T., Angeloni D., et al. (2017). Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum. Mol. Genet. 27, 761–779. 10.1093/hmg/ddx438 [DOI] [PubMed] [Google Scholar]
- Goldmann T., Overlack N., Möller F., Belakhov V., Van Wyk M., Baasov T., et al. (2012). A comparative evaluation of NB30, NB54 and PTC124 in translational read-through efficacy for treatment of an USH1C nonsense mutation. EMBO Mol. Med. 4, 1186–1199. 10.1002/emmm.201201438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T., Overlack N., Wolfrum U., Nagel-Wolfrum K. (2011). PTC124-mediated translational readthrough of a nonsense mutation causing Usher syndrome type 1C. Hum. Gene Ther. 22, 537–547. 10.1089/hum.2010.067 [DOI] [PubMed] [Google Scholar]
- Goldmann T., Rebibo-Sabbah A., Overlack N., Nudelman I., Belakhov V., Baasov T., et al. (2010). Beneficial read-through of a USH1C nonsense mutation by designed aminoglycoside NB30 in the retina. Invest. Ophthalmol. Vis. Sci. 51, 6671–6680. 10.1167/iovs.10-5741 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cordero A., West E. L., Pearson R. A., Duran Y., Carvalho L. S., Chu C. J., et al. (2013). Photoreceptor precursors derived from three-dimensional embryonic stem cell cultures integrate and mature within adult degenerate retina. Nat. Biotechnol. 31, 741–747. 10.1038/nbt.2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal N., Narayanaswami P. (2018). Making sense of antisense oligonucleotides: a narrative review. Muscle Nerve 57, 356–370. 10.1002/mus.26001 [DOI] [PubMed] [Google Scholar]
- Grati M. H., Kachar B. (2011). Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl. Acad. Sci. U S A 108, 11476–11481. 10.1073/pnas.1104161108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger J. C., Samulski R. J. (2005). Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 15, 9933–9944. 10.1128/jvi.79.15.9933-9944.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B., Meijer E. J., Ivanchenko M. V., Tenneson K., Emond F., Hanlon K. S., et al. (2019). Gene transfer with AAV9-PHP.B rescues hearing in a mouse model of usher syndrome 3A and transduces hair cells in a non-human primate. Mol. Ther. Methods Clin. Dev. 13, 1–13. 10.1016/j.omtm.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam D., Hilgen G., Dorgau B., Zhu L., Yu M., Bojic S., et al. (2018). Human-induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient-dependent efficiency. Stem Cells 36, 1535–1551. 10.1002/stem.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Conley S. M., Makkia R. S., Cooper M. J., Naash M. I. (2012a). DNA nanoparticle-mediated ABCA4 delivery rescues Stargardt dystrophy in mice. J. Clin. Invest. 122, 3221–3226. 10.1172/jci64833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Conley S. M., Makkia R., Guo J., Cooper M. J., Naash M. I. (2012b). Comparative analysis of DNA nanoparticles and AAVs for ocular gene delivery. PLoS One 7:e52189. 10.1371/journal.pone.0052189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Liu X., Xie S., Gao M., Liu F., Yu S., et al. (2018). Knockout of ush2a gene in zebrafish causes hearing impairment and late onset rod-cone dystrophy. Hum. Genet. 137, 779–794. 10.1007/s00439-018-1936-6 [DOI] [PubMed] [Google Scholar]
- Hartong D. T., Berson E. L., Dryja T. P. (2006). Retinitis pigmentosa. Lancet 368, 1795–1809. 10.1016/S0140-6736(06)69740-7 [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Gibbs D., Lillo C., Azarian S. M., Legacki E., Zhang X. M., et al. (2007). Lentiviral gene replacement therapy of retinas in a mouse model for Usher syndrome type 1B. Gene Ther. 14, 584–594. 10.1038/sj.gt.3302897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens M. A., Duelli D. M., Hastings M. L. (2013). Targeting RNA splicing for disease therapy. Wiley Interdiscip. Rev. RNA 4, 247–266. 10.1002/wrna.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D. R., Hughbanks-Wheaton D. K., Pearson N. S., Fish G. E., Spencer R., Takacs A., et al. (2014). Four-year placebo-controlled trial of docosahexaenoic acid in X-linked retinitis pigmentosa (DHAX trial): a randomized clinical trial. JAMA Ophthalmol. 132, 866–873. 10.1001/jamaophthalmol.2014.1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J., Wang Q., Katz N., Buza E. L., Bell P., Wilson J. M. (2018). The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 26, 664–668. 10.1016/j.ymthe.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chau Y. (2019). Intravitreal nanoparticles for retinal delivery. Drug Discov. Today 24, 1510–1523. 10.1016/j.drudis.2019.05.005 [DOI] [PubMed] [Google Scholar]
- Huang D., Fletcher S., Wilton S. D., Palmer N., McLenachan S., Mackey D. A., et al. (2017). Inherited retinal disease therapies targeting precursor messenger ribonucleic acid. Vision 1:22. 10.3390/vision1030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaijo T., Aller E., Oltra S., Beneyto M., Najera C., Ayuso C., et al. (2006). Mutation profile of the MYO7A gene in Spanish patients with Usher syndrome type I. Hum. Mutat. 27, 290–291. 10.1002/humu.9404 [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouret G., Poirsier C., Spodenkiewicz M., Jaquin C., Gouy E., Arndt C., et al. (2019). Genetics of Usher syndrome: new insights from a meta-analysis. Ontol. Neurotol. 40, 121–129. 10.1097/mao.0000000000002054 [DOI] [PubMed] [Google Scholar]
- Kay M. A., Glorioso J. C., Naldini L. (2001). Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 7, 33–40. 10.1038/83324 [DOI] [PubMed] [Google Scholar]
- Keeling K. M., Bedwell D. M. (2011). Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip. Rev. RNA 2, 837–852. 10.1002/wrna.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K. M., Wang D., Conard S. E., Bedwell D. M. (2012). Suppression of premature termination codons as a therapeutic approach. Crit. Rev. Biochem. Mol. Biol. 47, 444–463. 10.3109/10409238.2012.694846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem E., Konstan M. W., De Boeck K., Accurso F. J., Sermet-Gaudelus I., Wilschanski M., et al. (2014). Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir. Med. 2, 539–547. 10.1016/S2213-2600(14)70100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. H. (2019). Genome-editing technologies: concept, pros and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol Ther. Nucleic Acids 16, 326–334. 10.1016/j.omtn.2019.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. G., Cha J., Chandrasegaran S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U S A 93, 1156–1160. 10.1073/pnas.93.3.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberling W. J., Hildebrand M. S., Shearer A. E., Jensen M. L., Halder J. A., Trzupek K., et al. (2010). Frequency of Usher syndrome in two pediatric populations: implications for genetic screening of deaf and hard of hearing children. Genet Med. 12, 512–516. 10.1097/gim.0b013e3181e5afb8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstan M. W., Davis P. B., Wagener J. S., Hilliard K. A., Stern R. C., Milgram L. J. H., et al. (2004). Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum. Gene Ther. 15, 1255–1269. 10.1089/hum.2004.15.1255 [DOI] [PubMed] [Google Scholar]
- Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32, 677–683. 10.1038/nbt.2916 [DOI] [PubMed] [Google Scholar]
- Lakowski J., Gonzalez-Cordero A., West E. L., Han Y.-T., Welby E., Naeem A., et al. (2015). Transplantation of photoreceptor precursors isolated via a cell surface biomarker panel from embryonic stem cell-derived self-forming retina. Stem Cells 33, 2469–2482. 10.1002/stem.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski J., Welby E., Budinger D., Di Marco F., Di Foggia V., Bainbridge J. W. B., et al. (2018). Isolation of human photoreceptor precursors via a cell surface marker panel from stem cell-derived retinal organoids and fetal retinae. Stem Cells 36, 709–722. 10.1002/stem.2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegger L. D., Pan B., Askew C., Wassmer S. J., Gluck S. D., Galvin A., et al. (2017). A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 35, 280–284. 10.1038/nbt.3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz J. J., Jodelka F. M., Hinrich A. J., McCaffrey K. E., Farris H. E., Spalitta M. J., et al. (2013). Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 19, 345–350. 10.1038/nm.3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Adamian M., Roof D. J., Berson E. L., Dryja T. P., Roessler B. J., et al. (1994). In vivo transfer of a reporter gene to the retina mediated by an adenoviral vector. Invest. Ophthalmol. Vis. Sci. 35, 2543–2549. [PubMed] [Google Scholar]
- Lipinski D. M., Barnard A. R., Singh M. S., Martin C., Lee E. J., Davies W. I. L., et al. (2015). CNTF gene therapy confers lifelong neuroprotection in a mouse model of human retinitis pigmentosa. Mol. Ther. 23, 1308–1319. 10.1038/mt.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Z., Hope C., Liang C. Y., Zou J. M., Xu L. R., Cole T., et al. (1999). A mutation (2314delG) in the Usher syndrome type IIA gene: high prevalence and phenotypic variation. Am. J. Hum. Genet. 64, 1221–1225. 10.1086/302332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Vansant G., Udovichenko I. P., Wolfrum U., Williams D. S. (1997). Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil. Cytoskeleton 37, 240–252. [DOI] [PubMed] [Google Scholar]
- Lopes V. S., Boye S. E., Louie C. M., Boye S., Dyka F., Chiodo V., et al. (2013). Retinal gene therapy with a large MYO7A cDNA using adeno-associated virus. Gene Ther. 20, 824–833. 10.1038/gt.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe A., Harris R., Bhansali P., Cvekl A., Liu W. (2016). Intercellular adhesion-dependent cell survival and ROCK-regulated actomyosin-driven forces mediate self-formation of a retinal organoid. Stem Cell Reports 6, 743–756. 10.1016/j.stemcr.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueck J. D., Yoon J. S., Perales-Puchalt A., Mackey A. L., Infield D. T., Behlke M. A., et al. (2019). Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 10:822. 10.1038/s41467-019-08329-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren R. E., Pearson R. A., MacNeil A., Douglas R. H., Salt T. E., Akimoto M., et al. (2006). Retinal repair by transplantation of photoreceptor precursors. Nature 444, 203–207. 10.1038/nature05161 [DOI] [PubMed] [Google Scholar]
- Maddalena A., Tornabene P., Tiberi P., Minopoli R., Manfredi A., Mutarelli M., et al. (2018). Triple vectors expand AAV transfer capacity in the retina. Mol. Ther. 26, 524–541. 10.1016/j.ymthe.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Fukushima K., Kawasaki A., Nishizaki K., Smith R. J. (2007). Cochlear expression of a dominant-negative GJB2 R75W construct delivered through the round window membrane in mice. Neurosci. Res. 58, 250–254. 10.1016/j.neures.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Maeda Y., Fukushima K., Nishizaki K., Smith R. J. H. (2005). In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum. Mol. Genet. 14, 1641–1650. 10.1093/hmg/ddi172 [DOI] [PubMed] [Google Scholar]
- Maeder M. L., Stefanidakis M., Wilson C. J., Baral R., Barrera L. A., Bounoutas G. S., et al. (2019). Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25, 229–233. 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., Dicarlo J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M., Fujii M., Hashiguchi T., Sunagawa G. A., Ito S.-I., Sun J., et al. (2017). iPSC-derived retina transplants improve vision in rd1 end-stage retinal-degeneration mice. Stem Cell Reports 8, 69–83. 10.1016/j.stemcr.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur P., Yang J. (2015). Usher syndrome: hearing loss, retinal degeneration and associated abnormalities. Biochim. Biophys. Acta 1852, 406–420. 10.1016/j.bbadis.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur P. D., Yang J. (2019). Usher syndrome and non-syndromic deafness: functions of different whirlin isoforms in the cochlea, vestibular organs and retina. Hear. Res. 375, 14–24. 10.1016/j.heares.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarakis N. D., Azzouz M., Rohll J. B., Ellard F. M., Wilkes F. J., Olsen A. L., et al. (2001). Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum. Mol. Genet. 10, 2109–2121. 10.1093/hmg/10.19.2109 [DOI] [PubMed] [Google Scholar]
- McClements M. E., Barnard A. R., Singh M. S., Issa P. C., Jiang Z., Radu R. A., et al. (2019). An AAV dual vector strategy ameliorates the stargardt phenotype in adult Abca4−/− mice. Hum. Gene Ther. 30, 590–600. 10.1089/hum.2018.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C. M., Campbell C., Torricelli R. E., Finkel R. S., Flanigan K. M., Goemans N., et al. (2017). Ataluren in patients with nonsense mutation Duchenne muscular dystrophy (ACT DMD): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 1489–1498. 10.1016/S0140-6736(17)31611-2 [DOI] [PubMed] [Google Scholar]
- Mellough C. B., Collin J., Khazim M., White K., Sernagor E., Steel D. H. W., et al. (2015). IGF-1 signaling plays an important role in the formation of three-dimensional laminated neural retina and other ocular structures from human embryonic stem cells. Stem Cells 33, 2416–2430. 10.1002/stem.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellough C. B., Collin J., Queen R., Hilgen G., Dorgau B., Zerti D., et al. (2019). Systematic comparison of retinal organoid differentiation from human pluripotent stem cells reveals stage specific, cell line and methodological differences. Stem Cells Transl. Med. 8, 694–706. 10.1002/sctm.18-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingeot-Leclercq M.-P., Tulkens P. M. (1999). Aminoglycosides: nephrotoxicity. Antimicrob. Agents Chemother. 43, 1003–1012. 10.1128/AAC.43.5.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanous K. A., Yoon S., Rohll J. B., Patil D., Wilkes F. J., Kim V. N., et al. (1999). Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 6, 1808–1818. 10.1038/sj.gt.3301023 [DOI] [PubMed] [Google Scholar]
- Moore N. A., Morral N., Ciulla T. A., Bracha P. (2018). Gene therapy for inherited retinal and optic nerve degenerations. Expert Opin. Biol. Ther. 18, 37–49. 10.1080/14712598.2018.1389886 [DOI] [PubMed] [Google Scholar]
- Moreno A. M., Fu X., Zhu J., Katrekar D., Shih Y. V., Marlett J., et al. (2018). in situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation. Mol. Ther. 26, 1818–1827. 10.1016/j.ymthe.2018.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naessens S., Ruysschaert L., Lefever S., Coppieters F., De Baere E. (2019). Antisense oligonucleotide-based downregulation of the g56r pathogenic variant causing NR2E3-associated autosomal dominant retinitis pigmentosa. Genes 10:363. 10.3390/genes10050363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel-Wolfrum K., Moller F., Penner I., Baasov T., Wolfrum U. (2016). Targeting nonsense mutations in diseases with translational read-through-inducing drugs (TRIDs). BioDrugs 30, 49–74. 10.1007/s40259-016-0157-6 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Srinivasan P., Chavez M., Carter M. A., Dominguez A. A., La Russa M., et al. (2019). Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 10:194. 10.1038/s41467-018-08158-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., et al. (2012). Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785. 10.1016/j.stem.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Nayerossadat N., Maedeh T., Ali P. A. (2012). Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 1:27. 10.4103/2277-9175.98152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus C., Eisenberger T., Decker C., Nagl S., Blank C., Pfister M., et al. (2017). Next-generation sequencing reveals the mutational landscape of clinically diagnosed Usher syndrome: copy number variations, phenocopies, a predominant target for translational read-through and PEX26 mutated in Heimler syndrome. Mol. Genet. Genomic Med. 5, 531–552. 10.1002/mgg3.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman I., Rebibo-Sabbah A., Cherniavsky M., Belakhov V., Hainrichson M., Chen F., et al. (2009). Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations. J. Med. Chem. 52, 2836–2845. 10.1021/jm801640k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman I., Rebibo-Sabbah A., Shallom-Shezifi D., Hainrichson M., Stahl I., Ben-Yosef T., et al. (2006). Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg. Med. Chem. Lett. 16, 6310–6315. 10.1016/j.bmcl.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Omole A. E., Fakoya A. O. J. (2018). Ten years of progress and promise of induced pluripotent stem cells: historical origins, characteristics, mechanisms, limitations and potential applications. PeerJ 6:e4370. 10.7717/peerj.4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overlack N., Goldmann T., Wolfrum U., Nagel-Wolfrum K. (2012). Gene repair of an Usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Invest. Ophthalmol. Vis. Sci. 53, 4140–4146. 10.1167/iovs.12-9812 [DOI] [PubMed] [Google Scholar]
- Pan B., Askew C., Galvin A., Heman-Ackah S., Asai Y., Indzhykulian A. A., et al. (2017). Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat. Biotechnol. 35, 264–272. 10.1038/nbt.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt D. A., Lane A., Ramsden C., Jovanovic K., Coffey P. J., Hardcastle A. J., et al. (2016). Using induced pluripotent stem cells to understand retinal ciliopathy disease mechanisms and develop therapies. Biochem. Soc. Trans. 44, 1245–1251. 10.1042/bst20160156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., et al. (2016). Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829.e9–1838.e9. 10.1016/j.cell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R. A., Barber A. C., Rizzi M., Hippert C., Xue T., West E. L., et al. (2012). Restoration of vision after transplantation of photoreceptors. Nature 485, 99–103. 10.1038/nature10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepermans E., Michel V., Goodyear R., Bonnet C., Abdi S., Dupont T., et al. (2014). The CD2 isoform of protocadherin-15 is an essential component of the tip-link complex in mature auditory hair cells. EMBO Mol. Med. 6, 984–992. 10.15252/emmm.201403976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. J., Wallace K. A., Dickerson S. J., Miller M. J., Verhoeven A. D., Martin J. M., et al. (2012). Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest. Ophthalmol. Vis. Sci. 53, 2007–2019. 10.1167/iovs.11-9313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnath A., Depreux F. F., Jodelka F. M., Rigo F., Farris H. E., Hastings M. L., et al. (2018). Rescue of outer hair cells with antisense oligonucleotides in usher mice is dependent on age of treatment. J. Assoc. Res. Otolaryngol. 19, 1–16. 10.1007/s10162-017-0640-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch B. J., Silvis M. R., Hultquist J. F., Waters C. S., Mcgregor M. J., Krogan N. J., et al. (2017). Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150.e10–158.e10. 10.1016/j.cell.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapudi S., Schwartz S. G., Wang X., Chavis P. (2013). Vitamin A and fish oils for retinitis pigmentosa. Cochrane Database Syst. Rev. 12:CD008428. 10.1002/14651858.cd008428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebibo-Sabbah A., Nudelman I., Ahmed Z. M., Baasov T., Ben-Yosef T. (2007). In vitro and ex vivo suppression by aminoglycosides of PCDH15 nonsense mutations underlying type 1 Usher syndrome. Hum. Genet. 122, 373–381. 10.1007/s00439-007-0410-7 [DOI] [PubMed] [Google Scholar]
- Rees H. A., Liu D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. 10.1038/s41576-018-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J., Nagel-Wolfrum K., Jurgens K., Marker T., Wolfrum U. (2006). Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 83, 97–119. 10.1016/j.exer.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Rodrigues G. A., Shalaev E., Karami T. K., Cunningham J., Slater N. K. H., Rivers H. M. (2018). Pharmaceutical development of AAV-based gene therapy products for the eye. Pharm. Res. 36:29. 10.1007/s11095-018-2554-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S., Bennett J., Wellman J. A., Chung D. C., Yu Z.-F., Tillman A., et al. (2017). Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860. 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S.-M., Koo T., Kim K., Lim K., Baek G., Kim S.-T., et al. (2018). Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539. 10.1038/nbt.4148 [DOI] [PubMed] [Google Scholar]
- Sahly I., Dufour E., Schietroma C., Michel V., Bahloul A., Perfettini I., et al. (2012). Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J. Cell Biol. 199, 381–399. 10.1083/jcb.201202012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ferreira T., Llonch S., Borsch O., Postel K., Haas J., Ader M. (2016). Retinal transplantation of photoreceptors results in donor-host cytoplasmic exchange. Nat. Commun. 7:13028. 10.1038/ncomms13028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietroma C., Parain K., Estivalet A., Aghaie A., Boutet De Monvel J., Picaud S., et al. (2017). Usher syndrome type 1-associated cadherins shape the photoreceptor outer segment. J. Cell Biol. 216, 1849–1864. 10.1083/jcb.201612030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selimoglu E. (2007). Aminoglycoside-induced ototoxicity. Curr. Pharm. Des. 13, 119–126. 10.2174/138161207779313731 [DOI] [PubMed] [Google Scholar]
- Sengillo J. D., Justus S., Tsai Y. T., Cabral T., Tsang S. H. (2016). Gene and cell-based therapies for inherited retinal disorders: an update. Am. J. Med. Genet. C Semin. Med. Genet. 172, 349–366. 10.1002/ajmg.c.31534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedahmadi B. J., Rivolta C., Keene J. A., Berson E. L., Dryja T. P. (2004). Comprehensive screening of the USH2A gene in Usher syndrome type II and non-syndromic recessive retinitis pigmentosa. Exp. Eye Res. 79, 167–173. 10.1016/j.exer.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Shibata S. B., Ranum P. T., Moteki H., Pan B., Goodwin A. T., Goodman S. S., et al. (2016). RNA interference prevents autosomal-dominant hearing loss. Am. J. Hum. Genet. 98, 1101–1113. 10.1016/j.ajhg.2016.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H., Mandai M., Matsushita K., Kuwahara A., Yonemura S., Nakano T., et al. (2016). Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc. Natl. Acad. Sci. U S A 113, E81–E90. 10.1073/pnas.1512590113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. S., Balmer J., Barnard A. R., Aslam S. A., Moralli D., Green C. M., et al. (2016). Transplanted photoreceptor precursors transfer proteins to host photoreceptors by a mechanism of cytoplasmic fusion. Nat. Commun. 7:13537. 10.1038/ncomms13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. S., Charbel Issa P., Butler R., Martin C., Lipinski D. M., Sekaran S., et al. (2013). Reversal of end-stage retinal degeneration and restoration of visual function by photoreceptor transplantation. Proc. Natl. Acad. Sci. U S A 110, 1101–1106. 10.1073/pnas.1119416110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slijkerman R. W., Vaché C., Dona M., García-García G., Claustres M., Hetterschijt L., et al. (2016). Antisense oligonucleotide-based splice correction for USH2A-associated retinal degeneration caused by a frequent deep-intronic mutation. Mol. Ther. Nucleic Acids 5:e381. 10.1038/mtna.2016.89 [DOI] [PubMed] [Google Scholar]
- Song C.-Q., Jiang T., Richter M., Rhym L. H., Koblan L. W., Zafra M. P., et al. (2020). Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 4, 125–130. 10.1038/s41551-019-0357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling M., Gradzielski M. (2017). Droplets, evaporation and a superhydrophobic surface: simple tools for guiding colloidal particles into complex materials. Gels 3:15. 10.3390/gels3020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Corfas G., Liberman M. (2016). Round-window delivery of neurotrophin 3 regenerates cochlear synapses after acoustic overexposure. Sci. Rep. 6:24907. 10.1038/srep24907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Talcott K. E., Ratnam K., Sundquist S. M., Lucero A. S., Lujan B. J., Tao W., et al. (2011). Longitudinal study of cone photoreceptors during retinal degeneration and in response to ciliary neurotrophic factor treatment. Invest. Ophthalmol. Vis. Sci. 52, 2219–2226. 10.1167/iovs.10-6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.-H., Chen J.-R., Zheng J., Shi H.-S., Ding J., Qian X.-D., et al. (2016). Genetic correction of induced pluripotent stem cells from a deaf patient with MYO7A mutation results in morphologic and functional recovery of the derived hair cell-like cells. Stem Cells Transl. Med. 5, 561–571. 10.5966/sctm.2015-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornabene P., Trapani I., Minopoli R., Centrulo M., Lupo M., De Simone S., et al. (2019). Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci. Transl. Med. 11:eaav4523. 10.1126/scitranslmed.aav4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani I., Colella P., Sommella A., Iodice C., Cesi G., De Simone S., et al. (2014). Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 6, 194–211. 10.1002/emmm.201302948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillet A., Dubus E., Degardin J., Estivalet A., Ivkovic I., Godefroy D., et al. (2018). Cone degeneration is triggered by the absence of USH1 proteins but prevented by antioxidant treatments. Sci. Rep. 8:1968. 10.1038/s41598-018-20171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H.-Y., Watanabe T., Shirai H., Yamasaki S., Kinoshita M., Matsushita K., et al. (2019). Medium- to long-term survival and functional examination of human iPSC-derived retinas in rat and primate models of retinal degeneration. EBioMedicine 39, 562–574. 10.1016/j.ebiom.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Diepen H., Dulla K., Chan H., Schulkens I., Beumer W., Vorthoren L., et al. (2019). QR-421a, an antisense oligonucleotide, for the treatment of retinitis pigmentosa due to USH2A exon 13 mutations. Invest. Ophthalmol. Vis. Sci. 60:3250. [Google Scholar]
- Vandendriessche T., Thorrez L., Acosta-Sanchez A., Petrus I., Wang L., Ma L., et al. (2007). Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J. Thromb. Haemost. 5, 16–24. 10.1111/j.1538-7836.2006.02220.x [DOI] [PubMed] [Google Scholar]
- Vijayakumar S., Depreux F. F., Jodelka F. M., Lentz J. J., Rigo F., Jones T. A., et al. (2017). Rescue of peripheral vestibular function in Usher syndrome mice using a splice-switching antisense oligonucleotide. Hum. Mol. Genet. 26, 3482–3494. 10.1093/hmg/ddx234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiger L., Grisch-Chan H. M., Lindsay H., Ringnalda F., Pogliano C. B., Allegri G., et al. (2018). Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525. 10.1038/s41591-018-0209-1 [DOI] [PubMed] [Google Scholar]
- Wang X., Gregory-Evans C. Y. (2015). Nonsense suppression therapies in ocular genetic diseases. Cell. Mol. Life Sci. 72, 1931–1938. 10.1007/s00018-015-1843-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wise A. K., Tan J., Maina J. W., Shepherd R. K., Caruso F. (2014). Mesoporous silica supraparticles for sustained inner-ear drug delivery. Small 10, 4244–4248. 10.1002/smll.201401767 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Fukasawa T. (1961). Episome-mediated transfer of drug resistance in Enterobacteriaceae. I. Transfer of resistance factors by conjugation. J. Bacteriol. 81, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. N., Levy S. (2019). Stem Cell Ophthalmology Treatment Study (SCOTS): bone marrow derived stem cells in the treatment of Usher syndrome. Stem Cell Investig. 6:31. 10.21037/sci.2019.08.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch E. M., Barton E. R., Zhuo J., Tomizawa Y., Friesen W. J., Trifillis P., et al. (2007). PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91. 10.1038/nature05756 [DOI] [PubMed] [Google Scholar]
- Wilschanski M., Miller L. L., Shoseyov D., Blau H., Rivlin J., Aviram M., et al. (2011). Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis. Eur. Respir. J. 38, 59–69. 10.1183/09031936.00120910 [DOI] [PubMed] [Google Scholar]
- Wold W. S. M., Toth K. (2013). Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 13, 421–433. 10.2174/1566523213666131125095046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Zhang Y., Duan D. (2000). Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. U S A 97, 6716–6721. 10.1073/pnas.97.12.6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh W.-H., Chiang H., Rees H. A., Edge A. S. B., Liu D. R. (2018). In vivo base editing of post-mitotic sensory cells. Nat. Commun. 9:2184. 10.1038/s41467-018-04580-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Kanasty R. L., Eltoukhy A. A., Vegas A. J., Dorkin J. R., Anderson D. G. (2014). Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555. 10.1038/nrg3763 [DOI] [PubMed] [Google Scholar]
- Ylikoski J., Pirvola U., Moshnyakov M., Palgi J., Arumäe U., Saarma M. (1993). Expression patterns of neurotrophin and their receptor mRNAs in the rat inner ear. Hear. Res. 65, 69–78. 10.1016/0378-5955(93)90202-c [DOI] [PubMed] [Google Scholar]
- Zakrzewski W., Dobrzyński M., Szymonowicz M., Rybak Z. (2019). Stem cells: past, present, and future. Stem Cell Res. Ther. 10:68. 10.1186/s13287-019-1165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M., Binley K., Lad Y., Ellis S., Widdowson P., Iqball S., et al. (2014). EIAV-based retinal gene therapy in the shaker1 mouse model for usher syndrome type 1B: development of UshStat. PLoS One 9:e94272. 10.1371/journal.pone.0094272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen G., Wen L., Yang F., Shao A.-L., Li X., et al. (2013). Novel multiple agents loaded PLGA nanoparticles for brain delivery via inner ear administration: in vitro and in vivo evaluation. Eur. J. Pharm. Sci. 48, 595–603. 10.1016/j.ejps.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Zheng C. X., Wang S. M., Bai Y. H., Luo T. T., Wang J. Q., Dai C. Q., et al. (2018). Lentiviral vectors and adeno-associated virus vectors: useful tools for gene transfer in pain research. Anat. Rec. 301, 825–836. 10.1002/ar.23723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. Y., Yan D., Ouyang X. M., Du L. L., Yu H., Chang B., et al. (2005). Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum. Mol. Genet. 14, 103–111. 10.1093/hmg/ddi010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M. N., Cao L. H., et al. (2014). Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5:4047. 10.1038/ncomms5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Feng H., Sood R., Kinnunen P. K. J., Pyykko I. (2017). Biocompatibility of liposome nanocarriers in the rat inner ear after intratympanic administration. Nanoscale Res. Lett. 12:372. 10.1186/s11671-017-2142-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Luo L., Shen Z., Chiodo V. A., Ambati B. K., Hauswirth W. W., et al. (2011). Whirlin replacement restores the formation of the USH2 protein complex in whirlin knockout photoreceptors. Invest. Ophthalmol. Vis. Sci. 52, 2343–2351. 10.1167/iovs.10-6141 [DOI] [PMC free article] [PubMed] [Google Scholar]