Abstract
Polyploids are species in which three or more sets of chromosomes coexist. Polyploidy frequently occurs in plants and plays a major role in their evolution. Based on their origin, polyploid species can be divided into two groups: autopolyploids and allopolyploids. The autopolyploids arise by multiplication of the chromosome sets from a single species, whereas allopolyploids emerge from the hybridization between distinct species followed or preceded by whole genome duplication, leading to the combination of divergent genomes. Having a polyploid constitution offers some fitness advantages, which could become evolutionarily successful. Nevertheless, polyploid species must develop mechanism(s) that control proper segregation of genetic material during meiosis, and hence, genome stability. Otherwise, the coexistence of more than two copies of the same or similar chromosome sets may lead to multivalent formation during the first meiotic division and subsequent production of aneuploid gametes. In this review, we aim to discuss the pathways leading to the formation of polyploids, the occurrence of polyploidy in the grass family (Poaceae), and mechanisms controlling chromosome associations during meiosis, with special emphasis on wheat.
Keywords: chromosome pairing, homoeologous pairing, meiosis, Poaceae, polyploidy
Introduction
Poaceae (grasses) is a large family of monocotyledonous flowering plants that includes ~10,000 diverse species divided into 12 subfamilies, 51 tribes, and 80 subtribes (Soreng et al., 2015). This family includes the cereals, bamboos, as well as natural and cultivated grasses, and its members are found worldwide except in ice-covered areas. Their economic importance derives mainly from their utilization for food and feed production, but they also have ecological and aesthetic roles in ecosystems and for humanity. For example, maize (Zea mays), rice (Oryza sativa), and wheat (Triticum aestivum) together provide >50% of the calories consumed by all humans. Sugarcane (Saccharum officinarum) remains the major source of human-consumed sugar and is increasingly used for biofuel production. Ryegrasses (Lolium spp.), fescues (Festuca spp.), and bluegrasses (Poa spp.) are cultivated as fodder crops and for amenity purposes (i.e. sports, private and industrial lawns). Bamboos (Bambuseae) are used to construct elaborate scaffolds and the straws of cereals can serve as insulation in buildings or as raw material for paper production. All these uses make the Poaceae species a priority choice for enhancing both their quality (i.e., protein, lipid or sugar contents; cooking-quality, and digestibility, among others) and quantity (yield of grain and straw, biomass production).
Besides their great economic importance, species of the Poaceae family also serve as excellent model organisms for evolutionary studies (Kellogg, 2001). According to the pollen fossil record, grasses arose 55–70 million years ago (MYA; Jacobs et al., 1999). With ever more sequenced genomes (for details see https://bioinformatics.psb.ugent.be/plaza/), a detailed investigation of the evolutionary fate of duplicated chromosomal blocks led to the proposition of an ancestral karyotype for grasses, one structured in seven protochromosomes that contained 16,464 protogenes (Murat et al., 2014). This ancestral genome then further evolved, through the fusion and fission of chromosomes, gene duplication events as well as deletions, and chromosomal inversions and translocations. Moreover, interspecific hybridization and polyploidization (whole genome duplication; WGD) are two other key mechanisms of speciation in the Poaceae. All these phenomena have contributed to the extensive genome diversity extant within the family, including its variability in basic chromosome numbers and a wide range of polyploidy levels (Keeler, 1998). In this review, we highlight the nature of polyploidy in grasses, using wheat as a model, with a special focus on chromosome pairing during meiosis.
Polyploidy
Polyploidy plays a significant role in the evolution of higher plants, in that all angiosperms apparently underwent at least one round of WGD in their evolutionary history (Jiao et al., 2011). Polyploids can be categorized based on their origin. Autopolyploids possess three or more copies of the same chromosome set; by contrast, the multiple chromosome sets in allopolyploids are of different origin, due to the involvement of interspecific hybridization. Yet a strict boundary between these two categories is not always evident, such that a third (intermediate) group called segmental allopolyploidy is sometimes recognized in plants (Winterfeld et al., 2012). In general, autopolyploids often exhibit the formation of multivalents during meiosis and polysomic inheritance in their progeny. By contrast, allopolyploids with distant parental genomes usually exhibit formations of bivalents from homologous chromosomes (i.e., diploid-like pairing behavior), leading to disomic inheritance (Ramsey and Schemske, 1998). Nevertheless, allopolyploids sometimes carry chromosome sets that are not identical, but divergence of their sequence is insufficient to avoid the pairing of homoeologs (i.e., chromosomes originating from two related parental genomes with substantial homology); hence, they must employ an additional mechanism to ensure diploid-like behavior. Jauhar (2003) suggested that stable meiotic behavior and genome stability in allopolyploid species is achievable only after establishing a mechanism to ensure homologous chromosome recombination and segregation.
Autopolyploids
For a long time, autopolyploids were believed to suffer from various evolutionary disadvantages, leading to the conviction that autopolyploidy is rare in nature and often represents an evolutionary dead end (Clausen et al., 1945; Stebbins, 1971). This view, however, contrasts with their widespread utilization in crop production, for which many autopolyploids including potato, banana, watermelon, and sugarcane are of high economic importance. The proportion of autopolyploidy among plant species can only be debated so far, given that many autopolyploids have escaped recognition, being morphologically similar to their progenitors and concealed among common diploid taxa (Soltis et al., 2007). Recently, Barker et al. (2016) inferred that autopolyploids might be as frequent as allopolypoids among vascular plants. The Poaceae family contains many known autopolyploid species, such as Andropogon gerardii, a dominant grass of the tallgrass prairie (Keeler and Davis, 1999), several Brachiaria species (Gallo et al., 2007), the forage crop Hordeum bulbosum (Eilam et al., 2009), the sugarcane plant S. spontaneum (Wang et al., 2010), in addition to several Avena species (Ladizinsky, 1973).
Allopolyploids
Allopolyploids result from the hybridization of two more or less related species, such as Psidium guineense (Marques et al., 2016), wheat (T. aestivum) or the common oat (Avena sativa). Genomes inherited by allopolyploids vary in chromosomal homology, based on congeniality of parental species. In the case of hybridization between distantly related species, chromosomal homology can be low enough to not pair up during meiosis, frequently having different basic number of chromosomes. Conversely, allopolyploids that originated from the cross between closely related species carry chromosomes with much higher degree of homology. Accordingly, their homoeologous chromosomes have the potential to pair and recombine during meiosis (Ramsey and Schemske, 1998; Sun et al., 2017). Bread wheat is a typical example of an allopolyploid; it originated from two distinct interspecific hybridizations among three related diploid species that diverged 5–7 MYA (Marcussen et al., 2014). The first hybridization event occurred <0.82 MYA, between T. urartu and an as of yet unknown species from the Sitopsis section, closely related to Aegilops speltoides, which resulted in the development of a tetraploid species that further evolved into cultivated tetraploid wheat (T. turgidum ssp. durum; BBAA; Marcussen et al., 2014). The second hybridization took place more recently, between this newly developed tetraploid and Ae. tauschii (DD), resulting in hexaploid T. aestivum (2n = 6x = 42; BBAADD; Huang et al., 2002; Petersen et al., 2006; Marcussen et al., 2014). Similarly, oats (Avena spp.) also comprise diploid, tetraploid, and hexaploid species, either as auto- or allopolyploids. The allopolyploid oats behave diploid-like during meiosis despite having partial homology between their parental genomes (Thomas, 1992). Besides evolutionarily old allopolyploids, relativey recent allopolyploidazion events are evident in nature. For example, about 150 years ago, the two natural hybrids Spartina × neyrautii and S. × townsendii emerged through crosses between European S. maritima and S. alternifolia, the latter introduced from America. While the homoploid hybrid S. × townsendii is mostly sterile, chromosome doubling gave rise to the fertile allotetraploid species S. anglica (Hubbard, 1968) which spread rapidly throughout salt marshes in Western Europe (Gray et al., 1990; Thompson et al., 1991; Baumel et al., 2001; Salmon et al., 2005). As such, the polyploidization found in S. anglica may represent a way by which interspecific hybridization can foster evolutionary success.
Pathways Leading to Polyploidy
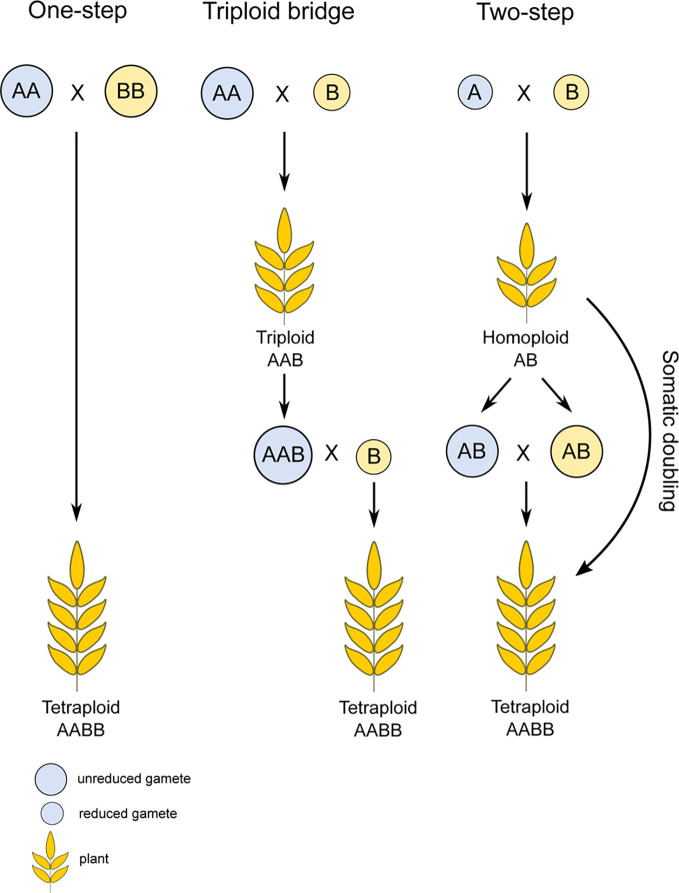
There are several routes leading to the formation of a polyploid individual. The first way is via chromosome doubling because of non-disjunction during mitosis. However, this way is rarely observed under natural conditions and is usually achieved only by exposure to chemical agents (Ramsey and Schemske, 1998; Tamayo-Ordóñez et al., 2016; Pelé et al., 2018). The more likely mechanism operating is that through the generation of unreduced gametes. The frequency of their production usually varies from 0.1% to 2% (Kreiner et al., 2017; Pelé et al., 2018) but this increases in response to stress, such as drought, low or high temperatures, and physical damage (Mason et al., 2011; Pécrix et al., 2011; De Storme et al., 2012; Vanneste et al., 2014; Kreiner et al., 2017; Van de Peer et al., 2017). This fact indicates polyploid formation could accelerate in periods of intensive environmental disturbances and rapid changes (Soltis et al., 2007). Polyploidy can be achieved in a single step process by fusing two unreduced gametes, through a so-called triploid bridge, or via a pathway involving two steps ( Figure 1 ). The triploid bridge is expected to more commonly occur than the one-step pathway, due to the low probability of fusion of two unreduced gametes in natural populations (Husband, 2004). The two-step pathway of allopolyploid formation first involves generation of a homoploid hybrid. Such an individual would either require a somatic doubling event, fusion of its two unreduced gametes, or involvement of the triploid bridge to restore its fertility (Mason and Pires, 2015). Alternatively, when the progenitors are autopolyploids, an allopolyploid can emerge immediately through the fusion of their standard (i.e., reduced) gametes (Pelé et al., 2018).
Figure 1.
Possible pathways of allopolyploid formation. Polyploidy can be achieved via multiple ways, most often through unreduced gamete formation and subsequent fertilization. In the case of the one-step pathway, two unreduced gametes merge, resulting directly in a polyploid species. Arguably, however, more steps are usually needed, where the reduced gamete merges with an unreduced gamete, forming a triploid bridge that requires an additional reduced gamete in subsequent generations. The final depicted option is the two-step pathway, through a homoploid hybrid, which needs a somatic doubling event or unreduced gamete formation to attain a polyploid state.
Polyploid species usually revert to a diploid state during evolution. The first part of this process, called cytogenetic diploidization, results in the formation of species, whose polyploid origin might be hidden by disomic inheritance and diploid-like meiosis. This step occurs rather rapidly after polyploid formation either by establishment of genetic control mechanism similar to Ph system in wheat (see below) or extensive chromosomal rearrangements. Over millions of years genomic diploidization continues. The content of the genes, which has doubled by polyloidization, is gradually returned towards one copy for each gene. For example, maize underwent an ancient WGD ~10 MYA. Since then, it has not only become cytogenetically diploid but also undergone extensive gene loss causing many genes to revert to a single-copy status in the genome (Renny-Byfield et al., 2017).
Advantages and Risks of Polyploidization
The question still stands: what is the main evolutionary advantage of polyploid formation in plants? While it may appear to have little impact on particular species (Meyers and Levin, 2006), it can also represent a significant evolutionary tool for improving possibilities of adaptation (Otto and Whitton, 2000). For example, gene redundancy offers an opportunity to better resist deleterious mutations and to diversify the extra copies of genes in subsequent evolution; in this way, new traits may be acquired without the adverse effects of losing the original genes’ function (Ha et al., 2009). From comparative analysis of collinear genes in syntenic regions of wheat and its diploid relatives Akhunov et al. (2013) confirmed the increased gene diversification conferred by polyploidy. Besides gene redundancy, allopolyploids can also benefit from the advantages of heterosis immediately upon their formation (Osborn et al., 2003; Comai, 2005), which can foster a greater biomass and accelerated development. Similarly, autopolyploidy might result in higher biomass of plants (Stebbins, 1971) and seed size, the latter enabling a more rapid rate of early development, such as in Triticum and Aegilops species (Villar et al., 1998; von Well and Fossey, 1998). All these effects of polyploidization could contribute to faster colonization of new niches, including extreme habitats (Ehrendorfer, 1980). At the chromosomal level, the existence of extra chromosomal set(s) represents a significant fitness advantage for tolerating large rearrangements in the genome that would normally lead to fatal consequences in diploid progenitors.
Clearly then, polyploid species are evolutionarily successful. In many cases (e.g., T. aestivum) they can grow in broad geographical areas and occupy a range of habitats (Feldman and Levy, 2005; Dubcovsky and Dvorak, 2007) as well as colonize extreme environments, like S. anglica has done (Hubbard, 1968; Gray et al., 1990; Thompson et al., 1991; Baumel et al., 2001; Salmon et al., 2005). Van de Peer et al. (2009) argued the higher competitiveness of polyploids could be explained by an ability to produce more diverse phenotypes than diploid species. Finally, it is worth noting that many staple crops are in fact polyploid species, and humankind has been using artificial polyploidization techniques and wide hybridization as a tool for their breeding and crop improvement. The use of wild relatives to enhance crops dates back to the early 1940s but gained prominence during the 1970s and 1980s (Hajjar and Hodgkin, 2007). Specifically, allopolyploidization is implemented to widen the target species’ genetic diversity or to introgress beneficial alleles from relatives into cultivated crops. For example, while the natural genetic diversity of elite sown material is significantly lower than that observed in its landraces, breeding programs have introduced new sources of diversity into wheat’s cultivars. To date, novel alleles have been introgressed from more than 50 related species representing 13 genera, highlighting the importance of these alien introgressions for improved wheat breeding (Wulff and Moscou, 2014). Perhaps the most well-known case is the rye (Secale cereale) 1RS translocation that harbors genes involved in a plant’s resistance to multiple diseases (Pm8/Sr31/Lr26/Yr9) and its yield enhancement. Other examples of introgressions include that of Sr36/Pm6 from T. timopheevii, Lr28 from Ae. speltoides, and Pch1 and Sr38/Lr37/Yr17 from Ae. ventricosa, which provided resistance to severe diseases such as stem and leaf rust and powdery mildew. Some of these introgressions were implemented gobally in commercial lines; for example, the 1RS.1BL translocation now found in 10% of the world’s genetic wheat diversity (Balfourier et al., 2019).
Nontheless, in addition to its positive impacts, polyploidy may have negative aspects. Perhaps the most obvious issue is the presence of more than one pairing partner in meiosis. Unless it is properly processed, it could result in multivalent formation and the production of aneuploid gametes, and thus, lower fertility or complete sterility (Ramsey and Schemske, 2002). Among the adaptive mechanisms described for autopolyploids, there is one based on a reduction in the number of cross-overs to one per chromosome pair, thereby ensuring only bivalents form from any two random homologs (Lloyd and Bomblies, 2016). This mechanism was observed in natural accessions of autotetraploid Arabidopsis arenosa (Carvalho et al., 2010; Pecinka et al., 2011; Yant et al., 2013; Pelé et al., 2018). By contrast, recognition of homologous chromosomes is critical for diploid-like pairing in allopolyploids. In allopolyploids containing distinct genomes, it is usually maintained by sequence variation between homoeologous chromosomes. In allopolyploids containing closely-related genomes, homolog recognition seems to be genetically controlled (Jenczewski and Alix, 2004). However, some allopolyploid and homoploid hybrids do not necessarily display significantly reduced fecundity, despite the pairing of homoeologous chromosomes. In such case, aneuploidy, chromosome rearrangements, and the predominance of one of the parental genomes could be observed, as described for ×Festulolium hybrids (Kopecký et al., 2006). Hereon, we focus on mechanisms controlling chromosome pairing in some crops belonging to the grass family (Poaceae).
Control of Chromosome Pairing in Polyploid Grasses
Meiosis is a crucial process for sexual reproduction and gamete formation. It ensures reduction of genetic material to half resulting in restoration of normal chromosomal constitution in progeny. As noted above, some allopolyploids have evolved molecular mechanisms that govern homologous chromosome pairing. Such regulators were observed and identified in several species, including those of Triticum, Avena, and Festuca. The origin of the genes responsible for regulating chromosome pairing is not known yet, however. Nonetheles, several hypotheses explaining the possible emergence of such mechanisms have been proposed.
The first hypothesis works by presuming the presence of these pairing regulators in diploid progenitors (Waines, 1976; Jenczewski and Alix, 2004). In this model, a stable allopolyploid would emerge after a rare event, in which the appropriate combination of such genes is achieved (Waines, 1976). Indeed, several regulators acting as suppressors of homoeologous chromosome pairing were believed to exist in diploid relatives of allopolyploids, such as Lolium spp., Hordeum vulgare (Gupta and Fedak, 1985), Hirschfeldia incana (Eber et al., 1994), Secale cereale (Riley and Law, 1965), Elytrigia elongata (Dvorak, 1987), Triticum monococcum (Shang et al., 1989), and Ae. tauschii (Attia et al., 1979). In Lolium, the pairing suppressors were found present in some accessions of L. multiflorum and L. perenne, where they influenced the number of chiasmata during the first meiotic division of their homoploid hybrid. This chiasma reduction was accounted for exclusively by homoeologous pairing, as revealed by artificially tetraploidized hybrids (Evans and Aung, 1985; Jenczewski and Alix, 2004). Another example of how chromosome-pairing control is induced through a combination of genotypes or genes was found in rice. Generally, rice intersubspecific autotetraploid hybrids display meiotic instability such as chromosome lagging and the formation of univalents and trivalents (Cai et al., 2007). Yet two lines PMeS-1 and PMeS-2 were distinguished as being stable, presumably due to the presence of one or more active meiotic regulator PMeS (polyploid meiosis stability) genes (Cai et al., 2007). These two lines display regular meiotic behavior, with bivalents and quadrivalents. The existence of genetic chromosome pairing PMeS control was confirmed by the persistent meiotic stability of the two lines even after several generations (Xiong et al., 2019).
The second hypothesis posits that the regulators of chromosome pairing emerge during or immediately after the formation of polyploids, by a mutation or multiple, successive mutations (Riley and Law, 1965; McGuire and Dvořák, 1982). This can happen via conversion of a gene that promotes chromosome pairing in the diploid progenitor into a repressor in the polyploidy individual (Riley and Kempanna, 1963; Feldman, 1966b). This phenomenon was described in hexaploid wheat, where a mutation in a pairing promoter gene on the long arm of its chromosome 5D caused a reduction of homoeologous chromosome paring in several interspecific hybrids. Such mutations provide a more pronounced effect than does being 5D nullisomic, which suggests the mutation is antimorphic, changing the gene’s function from pairing-promotion to suppression (Viegas et al., 1980). Those authors argued that this allele more likely arose from spontaneous mutation of a pairing-promoter known to be located on 5DL than from the transfer of Ph1 from chromosome 5B.
The third hypothesis proposes that such regulators of chromosome pairing could be transferred via accessory B chromosomes (Riley et al., 1973; Sears, 1976). Early allopolyploid species would have depended on the presence of a B chromosome(s), until the gene was transferred to an A chromosome by translocation, with the subsequent loss of the B chromosome from the karyotype (Jenczewski and Alix, 2004). Many studies have investigated the role of B chromosomes in the repression of homoeologous pairing (Evans and Macefield, 1972; Evans and Macefield, 1973; Aung and Evans, 1985). It seems that one or more B chromosomes from a specific source could complement one copy of the aforementioned homoeologous-pairing suppressor into a functional complex. Evans and Aung (1986) found homoeologous pairing dramatically reduced in the hybrids of F. arundinacea × L. perenne carrying B chromosomes. Also, the average number of chromosome arms joined by chiasmata is reduced in the presence of B chromosomes in a diploid meadow fescue when compared to the control plants lacking B chromosomes (Kopecký et al., 2009). In the hybrids of Ae. mutica and Ae. speltoides, the B chromosomes can also complement a missing Ph1 locus (Dover and Riley, 1972). Mechanisms controling chromosome pairing in allopolyploids seems to be specific among individual taxa, with very little known of the molecular pathways contributing to this phenomenon. In this respect, the best-elucidated molecular mechanism concerning the Ph genes is that of hexaploidy wheat (T. aestivum), which we describe in greater detail later on.
Apart from specific genetic systems to ensure proper chromosome pairing in particular species, various other (more general) genes are involved during process of meiosis that could increase the frequency of cross-overs between homologous chromosomes while suppressing them between homoeologs. Recently, Gonzalo et al. (2019) studied the effect of MSH4 upon homo- and homoeologous cross-overs, by using the EMS (ethylmethanesulphonate) mutant population in Brassica napus. They discovered that, when the MSH4 gene returns to a single copy status, the frequency of homologous cross-overs remained at the same frequency, whereas that of homoeologous cross-overs decreased drastically compared with the presence of two functional copies of the gene. Gonzalo et al. (2019) also studied the copy numbers of other genes of the synapsis-initiation complex (SIC, or alternatively ZMM-pathway) vis-à-vis diploid relatives, deducing that the acquisition of additional copies of such genes through small-scale duplications is a rare event; an example its occurrence is ZIP4 in wheat (Rey et al., 2017). Furthermore, the rapid reduction in the number of copies for ZMM genes in many species after whole genome duplication—namely for MSH4, MSH5, MER3, and ZIP4—supports the hypothesis that ensuring fewer copies of such genes could be a general process of meiotic stabilization (Lloyd et al., 2014; Gonzalo et al., 2019). Another study found no evidence for an increased loss of those genes after polyploidization in hexaploid wheat (including MSH4), in that most meiotic genes were retained in three homoeologous variants at similar expression levels (Lloyd et al., 2014). However, because wheat underwent its two hybridization events rather recently (Marcussen et al., 2014), the potential ZMM pathway gene reduction cannot be ruled out. Alternatively, the machinery established via Ph genes might have weakened the selective pressure for fewer copies of these genes.
Chromosome Pairing in Wheat
Allohexaploid bread wheat (T. aestivum L.; 2n = 6x = 42; BBAADD) can serve as a model plant for meiotic behavior analyses of allopolyploids. Despite the coexistence of three highly similar genomes, its meiotic behavior is strictly diploid-like, with 21 bivalents between homologous chromosomes forming in metaphase I of meiotic division. It has been known for more than 60 years that bread wheat developed genetic control of precise formation of homologous chiasmata, which is enforced by Ph (pairing homoeologous) genes (Sears and Okamoto, 1958; Riley and Chapman, 1958). The hexaploid nature of wheat allowed for the development of various aneuploid stocks, permitting the identification of several key genes involved in the regulation of meiosis (Sears and Okamoto, 1958; Sears, 1976; Sears, 1977; Sears, 1982; Sears, 1984).
It was proposed that premeiotic chromosome associations in interphase nucleus also play role in homolog recognition (Brown and Stack, 1968; Comings, 1968; Loidl, 1990; Aragón-Alcaide et al., 1997; Schwarzacher, 1997; Mikhailova et al., 1998; Martínez-Pérez et al., 1999). Nevertheless, different studies disagree in the extent and role of premeiotic chromosome associations, where they start and how long they last (Schwarzacher, 1997; Mikhailova et al., 1998; Martínez-Pérez et al., 1999). However, all these studies partially agree with Feldman (1966a), who suggested that Ph1 controls spatial organization of chromosomes in premeiotic interphase nuclei. In wheat, the arrangement of chromosomes in interphase nuclei is done through distribution of centromeres and telomeres in opposite sides of nuclei into Rabl configuration (Fussell, 1987), whereas this configuration is being maintained in premeiotic cells (Naranjo, 2015). This organization plays a role in the recognition of homologs, as it reduces the homolog search and simplifies the subsequent alignment (Pernickova et al., 2019). The telomeres are then recruited to the nuclear envelope and form a telomere bouquet (Dawe, 1998; Harper et al., 2004), which is believed to be essential for homolog identification and initiation of synapsis (Bass et al., 2000; Scherthan, 2001; Bass, 2003; Harper et al., 2004; Scherthan, 2007). The molecular mechanisms driving these changes are, however, mostly unknown.
Formation of chiasmata in wheat is driven by both suppressors and promoters, of which several have already been identified. The most important gene regulating homologous chiasmata is Ph1 (Pairing homoeologous 1), located on the long arm of chromosome 5B (Sears and Okamoto, 1958; Riley and Chapman, 1958). Another gene affecting chromosome behavior during meiosis, called Ph2, is located on the short arm of chromosome 3D but it exerts a weaker effect than does Ph1 (Mello-Sampayo, 1971). The least effective regulator, Ph3, is located on the short arm of chromosome 3A (Driscoll, 1972; Mello-Sampayo and Canas, 1973). Similar effects of Ph2 and Ph3 genes and their location on the same chromosomes of different parental genomes suggest these two genes are probably paralogs. During metaphase I of meiosis, ph mutants typically display fewer ring bivalents (with two or more chiasmata) and more univalents, rod bivalents and multivalents when compared to the wild type ( Table 1 ).
Table 1.
Comparison of chromosome associations in hexaploid and tetraploid wheat plants and particular ph mutants during metaphase I (Martínez et al., 2001a; Martínez et al., 2001b).
| Genotype | Chromosome number | Univalents | Rod bivalents | Ring bivalents | Multivalents | Chiasmata per cell |
|---|---|---|---|---|---|---|
| Hexaploid WT | 42 | 0.02 | 1.48 | 19.50 | 0.00 | 40.49 |
| ph1b | 42 | 2.76 | 4.76 | 14.5 | 0.77 | 38.57 |
| ph2b | 42 | 0.48 | 2.95 | 17.78 | 0.00 | 34.22 |
| Tetraploid WT | 28 | 0.04 | 0.34 | 13.64 | 0.00 | 27.62 |
| ph1c | 28 | 0.94 | 3.69 | 9.46 | 0.19 | 23.16 |
Pairing Homoeologous 1 (Ph1)
Among those genes controlling chiasmata formation during meiosis in wheat, Ph1 has the strongest effect on ensuring the correct recognition of homologous chromosomes. Although the presence of this control element was discovered over 60 years ago, its molecular effect was uncovered in part only recently. Its existence was first proposed by Sears and Okamoto (1958) and Riley and Chapman (1958) in haploid lines of wheat lacking chromosome 5B, in which the formation of both bivalents and trivalents had been observed. This contrasted with the meiotic behavior of lines carrying a copy of 5B. Subsequent gene mapping was carried out using the Ph1 mutant called ph1b (Sears, 1977), which helped to delimit the gene’s location. Later mapping, by Gill et al. (1993), used deletion lines to narrow down the genome region harboring the gene, which was cytogenetically estimated to be ~70 Mb. A more recent estimate of this deletion’s length put its at 54.6 Mb (Gyawali et al., 2019). Countless studies have shown that when Ph1 is missing, the chiasmata formation is no longer strictly diploid-like and chromosomes will form multivalents in more than 50% of pollen mother cells (Riley and Chapman, 1958; Riley, 1960). Work by Sánchez-Morán et al. (2001) confirmed that stark irregularities, such as aneuploidy and genomic rearrangements, are observable in lines lacking Ph1.
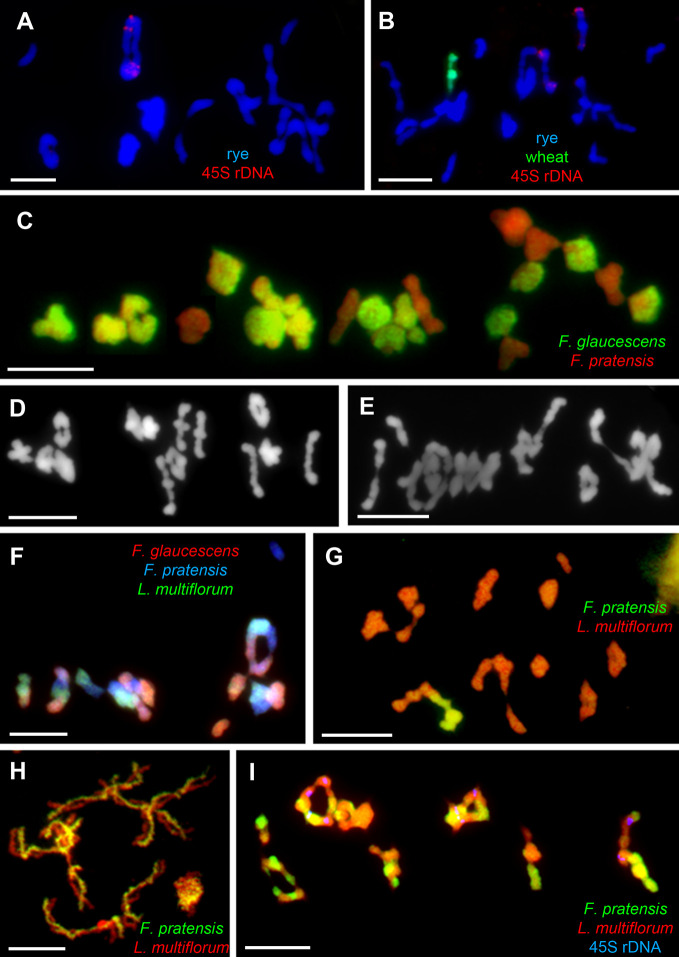
The Ph1 locus is present in tetraploid wheat plants as well, such as T. turgidum subsp. durum (Dvorak et al., 1984) and T. timopheevi subsp. timopheevi (Feldman, 1966b). In the latter, a mutant for this particular gene was developed, called ph1c, having a similar phenotype as the hexaploid mutant ph1b, i.e., increased homoeologous chromosome chiasmata in metaphase I (Jauhar et al., 1999). In a comparative study assessing the effectiveness of Ph1 gene in tetraploid and hexaploid wheat, Ozkan and Feldman (2001) crossed Ae. peregrina with hexaploid wheat and derivative lines, wherein chromosome 5B was replaced by its variant from tetraploid wheat (either from T. turgidum subsp. dicoccoides or T. timopheevi subsp. Timopheevi). With 5B from tetraploid wheat present, a higher frequency of homoeologous chromosome associations was observed in hybrids relative to the presence of endogenous 5B, indicating the tetraploid variant of Ph1 gene might operate with lower effectiveness. Interestingly, once Ph1 is introgressed from wheat into related species, its ability to modify chromosome bahavior is also preserved in the host genome ( Figures 2A, B ; Lukaszewski and Kopecký, 2010).
Figure 2.
Chromosome assocaitions in allo- and autopolyploids from the Poaceae family. Chromosome pairing in autotetraploid rye (2n = 4x = 28, RRRR) differs depending on the presence or absence of Ph1 located on the introgressed 5BL chromosome arm of wheat. In (A), trivalents and quadrivalents are commonly observed in the control line (2I+4II+2III+3IV), in (B), multivalent chromosome formation is reduced in the line (6I+7II+2IV), where 5B and 5BL are introgressed. In both (A, B), genomic DNA of Triticum aestivum was labeled with digoxigenin (green coloring), 45S rDNA was labeled with biotin (red), and genomic DNA of Secale cereale served as blocking DNA; all chromosomes counterstained with DAPI (blue). In (C), the chromosome-pairing control system similar to that of Ph1 found in allohexaploid Festuca arundinacea (2n = 6x = 42) hampers the associations of homeologous chromosomes and multivalent formation (21II). Genomic DNA of F. glaucescens was labeled with digoxigenin (green), while genomic DNA of F. pratensis was used as blocking DNA; all chromosomes were counterstained with DAPI (red pseudocolor). In (D), the homoeolog suppressor was probably inherited from one of the progenitors, F. glaucescens, as this species also forms only bivalents during meiosis (14II). Conversely, in (E), multivalent formation was detected in the autotetraploid form of the other progenitor, F. pratensis (2I+7II+3IV). The system is hemizygous-ineffective, thus allowing for promiscuous homeologous chromosome associations in tetraploid hybrids of F. arundinacea × Lolium multiflorum, where only one copy of the gene(s) is present (F). Here, genomic DNA of F. glaucescens was labeled with biotin (red coloring) and that of L. multiflorum labeled with digoxigenin (green), while that of F. pratensis was used as blocking DNA; all chromosomes were counterstained with DAPI (blue). In (G), homeologous chromosomes of F. pratensis and L. multiflorum pair freely in the substitution lines (1I+8II+1III+2IV) as well as in diploid Festuca × Lolium hybrids (7II), as seen in diplotene shown in (H), due to the absence of any chromosome pairing system and the phylogenetic relationship of both genomes. Note many chiasmata between homeologous chromosomes. This results in frequent homeologous recombinations and massive chromosome rearrangements in successive generations (I), as can be seen in the tetraploid L. multiflorum × F. pratensis cv. ‘Sulino’ (7IV). In panels (G–I), genomic DNA of F. pratensis was labeled with digoxigenin (green coloring), while genomic DNA of L. multiflorum served as blocking DNA and all chromosomes were counterstained with DAPI (red pseudocolor).
The Ph1 regulator probably acts in multiple ways during meiosis. In early prophase I, it promotes the formation and subsequent correction of synapses (Holm, 1986; Martínez et al., 2001a), but later on, it affects the frequency of cross-over formation (Martín et al., 2014). Originally, the Ph1 gene was thought to function as a suppressor of homoeologous synapses (Holm and Wang, 1988), but the current view is that it works primarily by promoting and stabilizing homologous synapses (Martín et al., 2017). During metaphase I in hexaploid wheat, ring bivalents are predominantly formed between homologous chromosomes, with some rod bivalents occurring in all meiocytes (Martín et al., 2014). In the ph1b mutant, only ~50% of meiocytes wil display similar meiotic behavior with increased frequency of rod bivalents; in the other half, variable numbers of multivalents and univalents were instead detected. This means that roughly half of the meiocytes display chiasmata only between homologous chromosomes (Martín et al., 2014). Similarly, other studies could not find homoeologous chiasmata in significant fractions of meiocytes in other Ph1 mutants (Roberts et al., 1999; Al-Kaff et al., 2008; King et al., 2016). This suggests the promotion of homologous synapses is the main function of the Ph1 gene, rather than suppression of homoeologous ones (Martín et al., 2017). This hypothesis is further supported by the higher occurrence of univalents in ph1b mutants than in the wild type or ph2b mutant ( Table 1 ).
Griffiths et al. (2006) performed a screen for a ph1-like phenotype in the population of EMS mutants. Yet they failed to find an individual showing the full ph1b-like phenotype. This indicates the Ph1 phenotype might not be under the control of a single gene. The Ph1 locus was further narrowed down to a 2.5-Mb region on the long arm of the 5B chromosome (Griffiths et al., 2006), which contains a duplicated segment from chromosome 3B composed of a cluster of Cdk2-like kinases and methyl-transferase genes (Griffiths et al., 2006; Al-Kaff et al., 2008; Martín et al., 2017). The Cdk-like kinases in the locus show close homology to the mammalian Cdk2, which is essential for homologous chromosome recognition and recombination (Ortega et al., 2003; Viera et al., 2009). Two groups of researchers disagree on which of the genes located in this particular region is the one responsible for promotion of homologous chiasmata. Bhullar et al. (2014) proposed C-Ph1 (RAFTIN1-like protein containing BURP domain) to be a putative Ph1 gene, but deletion lines for C-Ph1 locus failed to produce the same phenotype as the ph1b mutant (Al-Kaff et al., 2008). Moreover, the rice homolog and wheat paralog of this gene were already shown to be specific to tapetal cells (Jeon et al., 1999; Wang et al., 2003). The other group proposed a different candidate, a paralog of ZIP4. The encoded protein affects the homologous cross-overs in Arabidopsis and rice, supporting the assumption that this gene could be responsible for the Ph1 phenotype (Chelysheva et al., 2007; Shen et al., 2012; Rey et al., 2017). Both EMS and CRISPR mutations for this gene (named TaZIP4-B2) promoted homoeologous cross-overs in hybrids between wheat and Ae. variabilis (Rey et al., 2017; Rey et al., 2018). But these hybrids did not show the same extent of multivalent formation or an increase in univalents as typically observed in hybrids between the ph1b mutant and Ae. variabilis. Nevertheless, these results do suggest the TaZIP4-B2 plays an important role in the control of homoeologous pairing in wheat (Rey et al., 2017; Rey et al., 2018; Naranjo, 2019). The putative additional effector in this region has yet to be identified.
Pairing Homoeologous 2 (Ph2)
Another gene, called Ph2, has a weaker effect (than Ph1) on homologous chromosome pairing in wheat. That gene was assigned to chromosome 3D by Mello-Sampayo (1968; 1971) who observed multivalent formation in metaphase I in the absence of chromosome 3D in pentaploid hybrids between T. aestivum and T. durum, as well as in hybrids between T. aestivum and Aegilops. Two Ph2 mutants were since developed; the X-ray-induced mutant ph2a carrying a large deletion (Sears, 1982), and the chemically-induced (EMS) mutant ph2b (Wall et al., 1971). Using both mutants, the Ph2 phenotype was studied and the locus narrowed down, using synteny with rice, to a terminal 80 Mb of the short arm of chromosome 3D (Sutton et al., 2003). More recently, however, Svačina et al. (2020) showed that this deletion in the ph2a mutant is actually larger than expected, comprising about 125 Mb terminal part of the short arm of chromosome 3D.
The Ph2 gene operates in a different way than does Ph1 (Benavente et al., 1998; Martinez et al., 2001a). Both Martinez et al., (2001a) and Sánchez-Morán et al. (2001) evaluated the effect of its mutations in hexaploid wheat, finding no visible influence upon homoeologous chiasmata when Ph1 is present and Ph2 absent, apart from a slight increase in univalent formations. Earlier, Sears (1977; 1982) had shown that in hybrids of wheat and closely related species, moderate frequency of homoeologous chiasmata happened in the absence of Ph2 but in the presence of Ph1. In the case of wheat-rye hybrids lacking the Ph2 locus, Prieto et al. (2005) also observed an intermediate number of homoeologous chiasmata; however, according to their GISH analysis, the chromosome associations only occur between wheat chromosomes, whereas wheat–rye associations were rare similarly to the wild-type hybrid. This contrasts with the ph1b mutant, for which some frequency of wheat–rye associations was detectable (refer to Table 2 ; Prieto et al., 2005). These findings suggest to us that Ph2 plays a diminished functional role when homologous chromosomes are present ( Table 1 ). Yet, in the absence of homologs, it may well suppress associations among homoeologous chromosomes. Furthermore, researchers discovered that Ph2 has a different function to that of Ph1 as it is not involved in recognition of homologous chromosomes but instead affects the progression of synapsis (Martinez et al., 2001a; Prieto et al., 2005). We should also not overlook possible cooperation between Ph1 and Ph2 in their modes of action, as suggested by the work of Boden et al. (2009).
Table 2.
Number of chromosome-arm associations in metaphase I in haploid hybrids derived from the crossing of rye with euploid wheat (CS, ‘Chinese Spring’) and ph1b and ph2b mutants (Prieto et al., 2005).
| Genotype | CS × rye | ph2b × rye | ph1b × rye |
|---|---|---|---|
| Chromosome number | 28 | 28 | 28 |
| Wheat–wheat | 0.48 | 1.68 | 7.14 |
| Wheat–rye | 0.08 | 0.08 | 0.59 |
| Rye–rye | 0.02 | 0.04 | 0.05 |
| Total | 0.58 | 1.80 | 7.78 |
The ph2a mutant has been exploited in trying to identify candidate genes underlying its phenotype. Many have been proposed, such as TaMSH7, the homolog of the MSH6 DNA mismatch repair gene in yeast (Dong et al., 2002), in addition to the WM5 (Thomas, 1997) and WM1 gene family members (Ji and Langridge, 1994; Whitford, 2002). Sutton et al. (2003) used comparative genetics to further identify the putative genes involved in the Ph2 phenotype; however, no clear candidate producing a mutant phenotype similar to the ph2a has been identified.
Meiotic Behavior in Hybrids of ph Mutants and Wild-Type Wheat With Closely Related Species
The pairing of homoeologous chromosomes is mostly studied in haploids or interspecific hybrids, that is, in the absence of homologous chromosomes, the natural partners for pairing. The exent of chromosome associations during metaphase I of meiosis, in hybrids of wild-type hexaploid wheat or ph2b and ph1b mutants with various relatives, will differ based on the degree of homology between the genomes involved. The frequency of homoeologous chromosome chiasmata increases when there is a closer phylogenetic relationship of the parents. The fewest homoeologous associations were observed in the hybrids between hexaploid wheat and rye ( Table 3 ; Naranjo et al., 1987; Naranjo et al., 1988). This can be explained by the fact that lineages towards wheat and rye split about 7 MYA while Aegilops diverged from wheat 2.5–5.0 MYA (Huang et al., 2002). Accordingly, the Aegilops chromosomes are more closely related to wheat chromosomes than those of rye. The highest frequency of homoeologous chromosome associations was observed in the hybrid of hexaploid wheat and Ae. speltoides (Maestra and Naranjo, 1998; Table 3 ); the latter is a species closely related to the donor of the B genome in wheat, and thus highly similar to one of the wheat genomes (Huang et al., 2002; Petersen et al., 2006). These observations suggest the Ph system’s recognition of homologous chromosomes begins to fail with increasing homology between genomes in the hybrid, resulting in homoeologous chromosome chiasmata. Alternatively, there may exist genes that suppress or interfere with the Ph system in certain species used for hybridization with wheat (see below).
Table 3.
Associations of homoeologous chromosomes in metaphase I in various hybrids of wild-type wheat (WT) and ph1b and ph2b mutants with closely related plant species (Naranjo et al., 1987; Naranjo et al., 1988; Naranjo and Maestra, 1995; Maestra and Naranjo, 1997; Maestra and Naranjo, 1998).
| Hybrid | Chromosome number | Univalents | Rod bivalents | Ring bivalents | Multivalents | Chiasmata percell |
|---|---|---|---|---|---|---|
| WT × rye | 28 | 26.31 | 0.80 | 0.03 | 0.01 | 0.88 |
| ph2b × rye | 28 | 19.23 | 3.4 | 0.57 | 0.51 | 5.26 |
| ph1b × rye | 28 | 11.76 | 2.33 | 2.36 | 2.16 | 12.35 |
| WT × Ae. longissima | 28 | 24.55 | 1.59 | 0.06 | 0.05 | 1.81 |
| ph2b × Ae. longissima | 28 | 14.93 | 5.8 | 0.58 | 0.55 | 7.44 |
| ph1b × Ae. longissima | 28 | 3.48 | 4.4 | 2.99 | 2.86 | 18.28 |
| WT × Ae. sharonensis | 28 | 25.21 | 1.18 | 0.03 | 0.03 | 1.29 |
| ph2b × Ae. sharonensis | 28 | 10.16 | 5.58 | 1.42 | 1.13 | 11.17 |
| ph1b × Ae. sharonensis | 28 | 4.37 | 3.74 | 3.79 | 2.39 | 17.93 |
| WT × Ae. speltoides | 28 | 3.97 | 4.9 | 3.11 | 2.61 | 17.79 |
| ph2b × Ae. speltoides | 28 | 3.25 | 3.41 | 3.28 | 3.2 | 19.41 |
| ph1b × Ae. speltoides | 28 | 2.53 | 3.36 | 4.29 | 2.68 | 20.08 |
Homoeologous Chromosome Associations in the Presence of Ph Genes
Ph genes ensure that only homologous chromosome chiasmata occur in polyploid wheat during meiosis. However, the functioning of these genes can be suppressed in some hybrids, resulting in increased homoeologous chromosome associations; e.g., in hybrids of T. aestivum with Ae. speltoides or Ae. mutica (Riley, 1960; Dover and Riley, 1972; Dvorak et al., 2006a). For the wheat × Ae. speltoides hybrid, Dvorak et al. (2006b) identified two suppressors on chromosomes 3S (Su1-Ph1) and 7S (Su2-Ph1) that affected homoeologous chromosome associations, varying from 7.0 to 16.4 chiasmata per cell. The Su1-Ph1 was introgressed into both hexaploid and tetraploid wheat, opening new possibilities in inducing homoeologous chromosome recombinations for introgression into wheat (Li et al., 2017). This phenomenon can also be observed in lines where only a single chromosome was introgressed into the wheat background. In particular, the presence in wheat of chromosome 5U from Ae. umbellulata (Riley et al., 1973), or that of chromosome 5E from Elytrigia elongata (Dvorak, 1987), promotes homoeologous chromosome chiasmata with the formation of trivalents and bivalents in the haploids (ABD + 5U; ABD + 5E). This outcome suggests that introducing some alien chromosomes can suppress the functioning of Ph genes (Koo et al., 2017). Another case of homoeologous chromosome associations in the presence of Ph genes was reported on by Liu et al. (2011), who observed frequent recombination between 5Mg and 5D chromosomes in substitution lines containing 5Mg from Ae. geniculata. Later, Koo et al. (2017) used two different 5Mg chromosomes from different accessions in the wheat background and observed differential associations between 5Mg and 5D in both lines, for which chiasmata between 5Mg and 5D were detected in 6.7% and 21.7% of ensuing meiocytes. This might have been caused by the presence of genes located on the particular alien chromosome either actively promoting homoeologous chromosome chiasmata or repressing Ph1. Additionally, homoeologous associations probably occurred only between the 5Mg and 5D chromosome, as no multivalent was detected (Koo et al., 2017). In another example, homoeologous barley chromosomes fully associated in pairs in the presence of Ph1 (Martín et al., 2017; Calderón et al., 2018). However, these homoeologous chromosomes did not cross-over, suggesting that Ph1 does not prevent chromosome pairing between homoeologs, but supresses its recombination (Calderón et al., 2018).
In a natural population of the Chinese landrace of hexaploid wheat ‘Kaixianluohanmai’ (KL), another gene promoting homoeologous chiasmata in wheat–alien hybrids (presumably in presence of Ph) was posited (Luo et al., 1992). Meiosis is regular and normal in KL wheat by itself, as in other wheat landraces (Fan et al., 2019), but a moderate frequency of homoeologous chromosome associations occurs in hybrids of KL wheat with rye and Aegilops variabilis (similar as that between ph1b × rye and ph2b × rye hybrids) ( Table 4 ; Luo et al., 1992; Liu et al., 1998; Liu et al., 2003; Xiang et al., 2005). In hybrids arising between KL wheat and Psathyrostachys huashanica, the frequency of homoeologous chromosome chiasmata even exceeded that of the ph1b × P. huashanica hybrid (Kang et al., 2008). This locus, named phKL, is most probably not allelic to either Ph1 or Ph2 (Liu et al., 2003; Hao et al., 2011). The analysis of monosomics did show that a locus on chromosome 6A in KL might be responsible for the phKL phenotype (Liu et al., 1997). However, using two mapping populations, Fan et al. (2019) recently identified a QTL locus possibly responsible for homoeologous associations on chromosome arm 3AL.
Table 4.
Chromosome associations in metaphase I in hybrids derived from crossings of rye with the wheat KL landrace, “Chinese Spring” (CS), and the Chinese Spring ph1 (CSph1b) and ph2 (CSph2a) mutants (Hao et al., 2011).
| Genotype | Number of associations per cell | |||
|---|---|---|---|---|
| Rod | Ring | Multivalent | Chiasmata | |
| KL × rye | 4.73 | 0.20 | 0.11 | 5.40 |
| CSph1b × rye | 4.85 | 1.87 | 0.47 | 9.53 |
| CSph2a × rye | 1.74 | 0.00 | 0.02 | 1.78 |
| CS × rye | 0.54 | 0.00 | 0.00 | 0.54 |
Chromosome-Pairing Regulators in Other Poaceae Taxa
Bread wheat is undoubtedly the most studied and well-understood species concerning the mechanism of homologous chromosome recognition in the Poaceae family. Nonetheless, clues to the presence of similar machinery has been observed in other grass species, namely in Avena spp. (Ladizinsky, 1973), Oryza spp. (Cai et al., 2004), Festuca spp. (Jauhar, 1993), polyploid Hordeum spp. (Gupta and Fedak, 1985), or Alopecurus spp. (Murray et al., 1984). Several examples of chromosome associations in allo- and autopolyploids from the Poaceae family are shown in Figure 2 .
The genus Festuca comprises over 500 species having a wide range of ploidy levels, from diploids to dodecaploids (Loureiro et al., 2007). Agriculturally most important are those species from the subgenus Schedonorus comprising broad-leaved fescues, the majority of which are polyploids, from tetraploids to decaploids (Kopecký et al., 2008b). Molecular and cytogenetic analyses have revealed that all these studied polyploid species arose from interspecific hybridization (Humphreys et al., 1995; Catalán and Olmstead, 2000; Hand et al., 2010; Ezquerro-López et al., 2017); hence, they are of allopolyploid origin. All these allopolyploid species—including the tetraploids F. mairei, F. apennina, and F. glaucescens, hexaploid F. arundinacea, and octoploids F. arundinacea subsp. atlantigena and decaploid F. arundinacea var. letourneuxiana—possess diploid-like pairing behavior during meiosis, with bivalent formation (reviewed in Jauhar, 1993). Jauhar (1975) had proposed the existence of a homoeologous-pairing suppressor in tall fescue (F. arundinacea, 2n = 6x = 42; FpFpFgFgFg’Fg’) ( Figure 2C ). He found frequent multivalent formations in haploid plants of tall fescue (2n = 3x = 21) and speculated on the haplo-insufficiency or hemizygous-ineffectivity of the system: meaning that two copies of such gene(s) must be present for the induction of strict homologous pairing. This differentiates the fescues’ system from Ph1 of wheat and the regulator found in oats (Jauhar, 1993). Another difference is that Ph1 can supress homeologous recombination and/or promote homologous ones, while the control system in tall fescue seems to be responsible for the formation of homologous bivalents. Colchicine-induced dodecaploid wheat was able to form quadrivalents composed of four homologous chromosomes, whereas only homologous bivalents formed in the synthetically derived dodecaploid tall fescue plant (Jauhar, 1975).
Where the gene(s) underpinning diploid-like pairing system is located on one or more particular chromosomes or even subgenomes of tall fescue plants remains unknown. In tetraploid tall fescue (FpFpFgFg’), homoeologous chromosomes form chiasmata frequently; moreover, the frequent formation of quadrivalents was recorded in colchicine-induced autotetraploids of F. pratensis ( Figure 2E ; Kopecký et al., 2009). Thus, one of the subgenomes originating from F. glaucescens must harbor the responsible gene(s) ( Figure 2D ). In early work, Jauhar (1975) analyzed a set of monosomic lines of tall fescue and found one line with disrupted diploid-like behavior, probably due to an absence of the chromosome carrying the gene(s) for diploid-like pairing behavior. Unfortunately, this line was lost over time and so it cannot be further investigated. Later, Kleijer and Morel (1984) speculated that disruption of strictly homologous associations in a single plant is more likely to be only a consequence of normal variation among plants. The system may also interfere with other systems present in the genus, or in closely related genera. A high frequency of quadrivalents was observed in the tetraploid Lolium multiflorum × F. arundinacea hybrid (LmFpFgFg’) ( Figure 2F ), which exceeded that of quadrivalents in tetraploid F. arundinacea (FpFpFgFg’) (Kopecký et al., 2009).
The origin of the system in polyploid fescues is not known, but several scenarios are plausible. It could have developed in a currently unknown diploid species, which served as a progenitor of all recent polyploid species. Alternatively, such a system arose in an early-day polyploid (presumably an allotetraploid), since involved in the evolution of other allopolyploids. Support for both scenarios lies in the fact that the system in all species has the same (rare) attribute: haplo-insufficiency. The third possible scenario involves multiple origins of the system in different species during their evolutionary history. Or, the system is the outcome of two scenarios combined. It does seem that the systems found in various species are compatible in some hybrid combinations yet dysfunctional in others. Eizenga et al. (1990) found that multivalents were rare in the hybrids of tall fescue and giant fescue (F. gigantea). Similarly, hybrids of F. mairei × F. glaucescens show preferential formation of bivalents with a very low frequency of multivalents (nine quadrivalents and one trivalent among 200 PMCs [pollen mother cells]) (Malik and Thomas, 1967). By contrast, the hybrids of Continental and Mediterranean morphotypes of tall fescue all display high levels of multivalent formation (Kopecký et al., 2019), suggesting incompatibility of the two regulatory systems, or some epistatic effects. Therefore, we cannot unambiguously clarify if the system evolved once or twice (or even more times). However, if it did develop just once, the system diverged in different species during evolution to reach a level of incompatibility, as evinced from the analyses of interspecific hybrids.
The genus oat (Avena spp.) consists of diploid, tetraploid, and hexaploid species, including the important crop A. sativa. Polyploid oats include both auto- and allopolyploid forms, whose diploid-like behavior in meiosis is preserved despite partial homology between their genomes, suggesting the existence of a Ph-like system (Thomas, 1992). Oats comprise four cytologically distinct genomes (A, B, C, and D), however the genomes B and D occur only in polyploid taxa (Leggett and Thomas, 1995). Similar to wheat, the system found in tetraploid and hexaploid oats is hemizygous effective and haplo-sufficient, and susceptible to dosage effects and genetic repressibility. The locus that contains the gene(s) for meiotic regulation is likely localized to the A genome (Jauhar, 1977). Unfortunately, surpisingly little is known about the genes whose activity maintains homologous chromosome pairing in oats, apart from their existence being proven by increased associations among homoeologous chromosomes in some nulli-haploid A. sativa lines (Gauthier and McGuinnis, 1968).
Polyploidy and Homoeologous Chromosome Pairing in Plant Breeding
Besides its key role in plant speciation, polyploidization and hybridization are popular tools in plant breeding. The most straightforward agronomical effect of polyploidy is an increased cell size, potentially resulting in larger organs, including fruits, roots, flowers, leaves, and seeds (Stebbins, 1950). Another frequent consequence of polyploidy is sterility, which generally has an agronomically negative effect; however, for seedless fruit production it can be a desirable trait, as in triploid seedless watermelon (Crow, 1994). The fixation of heterozygosity in allopolyploid species often leads to heterosis, resulting in higher vigor of the hybrids compared with their diploid progenitors, such as in hexaploid wheat T. aestivum (Sattler et al., 2016). Wide hybridization coupled to whole genome duplication is commonly used to merge beneficial inheritable traits from both parents, namely in the introgression of a chromosome segment carrying genes for a desirable trait from the wild relative to elite crop cultivars, or for simply widening the gene pool. One of the most promising artificially developed hybrids is Triticale, which originated from the crossing of wheat and rye with a subsequent chromosome doubling (Meister and Tjumjakoff, 1928).
One of the key components for the successful utilization of wide hybridization in plant breeding is the control of homoeologous chromosome associations. In countless studies, the ph1b mutant of wheat has been used to induce homoeologous chromosome recombinations between chromosomes of wheat and related species, for transferring desirable traits into the wheat genome (Marais et al., 2010; Niu et al., 2011; Ayala-Navarrete et al., 2013; Rey et al., 2015a; Rey et al., 2015b; Han et al., 2016; King et al., 2019). After the introgression of the chromosomal segment from a related species, it is necessary to immediately re-activate the Ph1 gene to avoid risking the rapid elimination of the segment. Nevertheless, some hybrids without meiotic regulation but with homoeologous chromosome pairing can be valuable also and remain relatively stable. Complementary attributes of ryegrasses (i.e., high yield and nutrition) and fescues (i.e., abiotic stress tolerance) can be combined in their hybrids called Festulolium. In last 50 years, many agriculturally successful cultivars have been released via several breeding programs (Ghesquière et al., 2010). To do this, the breeders often used tetraploid parents for the initial mating. Such F1 Festulolium hybrids are all allotetraploids and possess two sets of chromosomes from both parental species. One would presume that homologous chromosome associations would be the predominant mode of action due to variation in the DNA sequence. The repetitive elements from these two genera diverged sufficiently that it is now possible to distinguish chromosomes of Festuca from those of Lolium by genomic in situ hybridization (GISH) (Thomas et al., 1994). Yet, frequent formation of homoeologous chromosome chiasmata has been detected in F1 hybrids, as well as in monosomic and disomic substitution lines of L. multiflorum × F. pratensis ( Figures 2G, H ; Kopecký et al., 2008a). Such massive homoeologous associations and recombination leads to highly variable karyotypes differing from plant to plant ( Figure 2I ). An outcrossing mode of reproduction augments this variability within each population of hybrids over subsequent generations. Consequently, both high variability and heterosis ensue within the bred plant material. It is nevertheless possible to uniform the breeding material at a phenotypic level to the extent that it passes DUS tests for registration as a commercial cultivar. While the proportion of parental genomes was relatively stable in subsequent generations of three commercial hybrids (Kopecký et al., 2008a), substantial variability was found within populations of each generation of those cultivars.
Besides those amphiploid (or allotetraploid) cultivars, introgression breeding may also be used to develop Festulolium cultivars. Doing this involves at least one round of backcrossing of F1 hybrids with one of the parental species (usually Lolium), giving rise to plants similar to the parental species but with improved characteristics, such as frost tolerance or higher survivorship (reviewed in Kopecký et al., 2008b). Karyologically, these plants usually carry only one or few chromosome segments of Festuca. Such introgression lines are usually highly unstable and the introgressed segment(s) is/are often lost in subsequent generations (Kopecký et al., 2019). Accordingly, implementing any system capable of preventing associations of homoeologous chromosomes is arguably desirable to stabilize the genomic composition of hybrids. In amphiploids, immediate introgression of the system would be required to keep both parental subgenomes intact. To date, most cultivars have originated from the cross of L. multiflorum × F. pratensis, though none of the parents carry a homoeologous suppressor. Instead, tetraploid wild relatives, such as F. glaucescens, F. apennina and F. mairei, which possess a meiotic regulator hampering homoeologous pairing, should be considered for future crosses as they are known for their tolerance to biotic and abiotic stress, which might complement the high yield and nutrition traits of ryegrasses. In this respects, first attempts have been made and the cultivar of L. multiflorum × F. glaucescens ‘Lueur’ was registered in France (Ghesquière et al., 2010) and other similar cross combinations are used in breeding programs in both the UK and Czech Republic. Considering the haplo-insufficiency of the system found in polyploid fescues, evidently the F1 hybrids will possess some level of homoeologous associations. Still, it should be possible to select F2 plants that have two copies of the gene(s) of the system and then intercross them. Doing this should facilitate the stabilization of the hybrid genome in successive generations. For the corresponding introgression lines, the segment carrying the gene(s) of the system must be present among the introgressions. Thereafter, haploidization, followed by either spontaneous or induced chromosome doubling, should result in the establishment of plants having two copies of such gene(s) required for its/their functionality as the homoeologous pairing suppressor(s). Clearly, though, further investigation of chromosome behavior in fescues is necessary if we hope to foster genetically stable grass hybrids.
We envisage that with more knowledge of the mechanisms responsible for correct chromosome associations, the efficient employment of targeted interspecific hybridization techniques will become available in the near future. Perhaps the most challenging task is the developing and operating of an “OFF” and “ON” switch to control recombination of homoeologous chromosomes. It would be immensely helpful for breeders to switch “OFF” the system in wheat and other allopolyploids with an established and functional regulatory system for introgressing the specific segment from a wild relative. Once the segment is transferred, the switch to “ON” would then stabilize the segment and permit its proper transmission into successive generations. Similarly, introgression of the system into a hybrid (originally lacking the regulator) with desirable combinations of parental chromatin would assist in further stabilizing the hybrid genome composition. To conclude, additional research broadening our knowledge of the mechanisms governing meiotic chromosome behavior in allopolyploids is necessary to ensure further success in future breeding of grass plants.
Author Contributions
RS, PS, DK, and JB wrote the manuscript.
Funding
This work was supported by the Czech Science Foundation (grant award 17-05341S) and the ERDF project “Plants as a tool for sustainable global development” (CZ.02.1.01/0.0/0.0/16_019/0000827).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Akhunov E. D., Sehgal S., Liang H., Wang S., Akhunova A. R., Kaur G., et al. (2013). Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 161, 252–265. 10.1104/pp.112.205161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kaff N., Knight E., Bertin I., Foote T., Hart N., Griffiths S., et al. (2008). Detailed dissection of the chromosomal region containing the ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling. Ann. Bot. 101, 863–872. 10.1093/aob/mcm252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón-Alcaide L., Reader S., Beven A., Shaw P., Miller T., Moore G. (1997). Association of homologous chromosomes during floral development. Curr. Biol. 7, 905–908. 10.1016/S0960-9822(06)00383-6 [DOI] [PubMed] [Google Scholar]
- Attia T., Ekingen H., Röbbelen G. (1979). Origin of 3D-suppressor of homoeologous pairing in hexaploid wheat. Z. Pflanzenzüchtg 83, 121–126. [Google Scholar]
- Aung T., Evans G. M. (1985). The potential for diploidizing Lolium multiflorum × L. perenne tetraploids. Can. J. Genet. Cytol. 27, 506–509. 10.1139/g85-075 [DOI] [Google Scholar]
- Ayala-Navarrete L., II, Mechanicos A. A., Gibson J. M., Singh D., Bariana H. S., Fletcher J., et al. (2013). The Pontin series of recombinant alien translocations in bread wheat: single translocations integrating combinations of Bdv2, Lr19 and Sr25 disease-resistance genes from Thinopyrum intermedium and Th. ponticum . Theor. Appl. Genet. 126, 2467–2475. 10.1007/s00122-013-2147-0 [DOI] [PubMed] [Google Scholar]
- Balfourier F., Bouchet S., Robert S., DeOliveira R., Rimbert H., Kitt J., et al. (2019). Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 5 (5), eaav0536. 10.1126/sciadv.aav0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. S., Arrigo N., Baniaga A. E., Li Z., Levin D. A. (2016). On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210, 391–398. 10.1111/nph.13698 [DOI] [PubMed] [Google Scholar]
- Bass H. W., Riera-Lizarazu O., Ananiev E. V., Bordolo S. J., Rines H. W., Phillips R. L., et al. (2000). Evidence for the coincident initiation of homologue pairing and synapsis during the telomere clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Bass H. W. (2003). Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell. Mol. Life Sci. 60, 2319–2324. 10.1007/s00018-003-3312-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumel A., Ainouche M. L., Levasseur J. E. (2001). Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France). Mol. Ecol. 10, 1689–1701. 10.1046/j.1365-294X.2001.01299.x [DOI] [PubMed] [Google Scholar]
- Benavente E., Orellana J., Fernández-Calvín B. (1998). Comparative analysis of the meiotic effects of wheat ph1b and ph2b mutations in wheat×rye hybrids. Theor. Appl. Genet. 96, 1200–1204. 10.1007/s001220050857 [DOI] [Google Scholar]
- Bhullar R., Nagarajan R., Bennypaul H., Sidhu G. K., Sidhu G., Rustgi S., et al. (2014). Silencing of a metaphase I-specific gene results in a phenotype similar to that of the pairing homeologous 1 (Ph1) gene mutations. Proc. Natl. Acad. Sci. U.S.A. 111, 14187–14192. 10.1073/pnas.1416241111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden S. A., Langridge P., Spangenberg G., Able J. A. (2009). TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 57, 487–497. 10.1111/j.1365-313X.2008.03701.x [DOI] [PubMed] [Google Scholar]
- Brown W. V., Stack S. M. (1968). Somatic pairing as a regular preliminary to meiosis. Bull. Torrey Bot. Club 95, 369–378. 10.2307/2483872 [DOI] [Google Scholar]
- Cai D. T., Chen D. L., Chen J. G., Liu Y. Q. (2004). A method of induction polyploidy rice with high frequency through tissue culture together with chemical agent induction. China Patent: ZL01133529.7. [Google Scholar]
- Cai D. T., Chen J., Chen D., Dai B. C., Song Z. J., Yang Z. F., et al. (2007). The breeding of two polyploid rice lines with the characteristic of polyploid meiosis stability. Sci. China Ser. C. 50, 356–366. 10.1007/s11427-007-0049-6 [DOI] [PubMed] [Google Scholar]
- Calderón M. C., Rey M. D., Martín A., Prieto P. (2018). Homoeologous Chromosomes From Two Hordeum Species Can Recognize and Associate During Meiosis in Wheat in the Presence of the Ph1 Locus. Front. Plant Sci. 9, 585. 10.3389/fpls.2018.00585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Delgado M., Barão A., Frescatada M., Ribeiro E., Pikaard C. S., et al. (2010). Chromosome and DNA methylation dynamics during meiosis in the autotetraploid Arabidopsis arenosa . Sex Plant Reprod. 23, 29–37. 10.1007/s00497-009-0115-2 [DOI] [PubMed] [Google Scholar]
- Catalán P., Olmstead R. G. (2000). Phylogenetic reconstruction of the genus Brachypodium P. Beauv. (Poaceae) from combined sequences of chloroplast ndhF gene and nuclear ITS. Pl. Syst. Evol. 220, 1–19. 10.1007/BF00985367 [DOI] [Google Scholar]
- Chelysheva L., Gendrot G., Vezon D., Doutriaux M. P., Mercier R., Grelon M. (2007). Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana . PloS Genet. 3, e83. 10.1371/journal.pgen.0030083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen J., Keck D. D., Hiesey W. M. (1945). Experimental studies on the nature of species, II. Plant evolution through amphiploidy and autopolyploidy, with examples from the Madiinae (Washington, DC.: Carnegie Institute of Washington; ). [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. 10.1038/nrg1711 [DOI] [PubMed] [Google Scholar]
- Comings D. E. (1968). The rational for an ordered arrangement of chromatin in the prophase nucleus. Am. J. Hum. Genet. 20, 440–460. [PMC free article] [PubMed] [Google Scholar]
- Crow J. F. (1994). Hitoshi Kihara, Japan’s pioneer geneticist. Genetics 137, 891–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe R. K. (1998). Meiotic chromosome organization and segregation in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 49, 371–395. 10.1146/annurev.arplant.49.1.371 [DOI] [PubMed] [Google Scholar]
- De Storme N., Copenhaver G. P., Geelen D. (2012). Production of diploid male gametes in Arabidopsis by cold-induced destabilization of postmeiotic radial microtubule arrays. Plant Physiol. 160, 1808–1826. 10.1104/pp.112.208611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Whitford R., Langridge P. (2002). A DNA mismatch repair gene links to the Ph2 locus in wheat. Genome 45, 116–124. 10.1139/g01-126 [DOI] [PubMed] [Google Scholar]
- Dover G. A., Riley R. (1972). Prevention of pairing of homoeologous meiotic chromosomes of wheat by an activity of supernumerary chromosomes of Aegilops . Nature 240, 159–161. 10.1038/240159a0 [DOI] [Google Scholar]
- Driscoll C. J. (1972). Genetic suppression of homoeologous chromosome pairing in hexaploid wheat. Can. J. Genet. Cytol. 14 (1), 39–42. 10.1139/g72-004 [DOI] [Google Scholar]
- Dubcovsky J., Dvorak J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866. 10.1126/science.1143986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J., Chen K. C., Giorgi B. (1984). The C-banding pattern of a Ph-mutant of durum wheat. Can. J. Genet. Cytol. 26, 360–363. 10.1139/g84-056 [DOI] [Google Scholar]
- Dvorak J., Akhunov E. D., Akhunov A. R., Deal K. R., Luo M. C. (2006. a). Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23, 1386–1396. 10.1093/molbev/msl004 [DOI] [PubMed] [Google Scholar]
- Dvorak J., Deal K. R., Luo M. C. (2006. b). Discovery and mapping of wheat Ph1 suppressors. Genetics 174, 17–27. 10.1534/genetics.106.058115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J. (1987). Chromosomal distribution of genes in diploid Elytrigia elongata that promote or suppress pairing of wheat homoeologous chromosomes. Genome 29, 34–40. 10.1139/g87-006 [DOI] [Google Scholar]
- Eber F., Chèvre A. M., Baranger A., Vallée P., Tanguy X., Renard M. (1994). Spontaneous hybridization between a male-sterile oilseed rape and two weeds. Theor. Appl. Genet. 88, 362–368. 10.1007/BF00223646 [DOI] [PubMed] [Google Scholar]
- Ehrendorfer F. (1980). ““Polyploidy and Distribution,”,” in Polyploidy. Basic Life Sciences, vol. 13 . Ed. Lewis W. H. (Boston, MA: Springer; ), 45–60. 10.1007/978-1-4613-3069-1_3 [DOI] [PubMed] [Google Scholar]
- Eilam T., Anikster Y., Millet E., Manisterski J., Feldman M. (2009). Genome size in natural and synthetic autopolyploids and in a natural segmental allopolyploid of several Triticeae species. Genome 52, 275–285. 10.1139/G09-004 [DOI] [PubMed] [Google Scholar]
- Eizenga G. C., Burner D. M., Buckner R. C. (1990). Meiotic and isozymic analyses of tall fescue × giant fescue hybrids and amphiploids. Plant Breed. 104, 202–211. 10.1111/j.1439-0523.1990.tb00424.x [DOI] [Google Scholar]
- Evans G. M., Aung T. (1985). Identification of a diploidizing genotype of Lolium multiflorum . Can. J. Genet. Cytol. 27, 498–505. 10.1139/g85-074 [DOI] [Google Scholar]
- Evans G. M., Aung T. (1986). The influence of the genotype of Lolium perenne on homoeologous chromosome association in hexaploid Festuca arundinacea . Heredity 56, 97–103. 10.1038/hdy.1986.13 [DOI] [Google Scholar]
- Evans G. M., Macefield A. J. (1972). Suppression of homoeologous pairing by B chromosomes in a Lolium species hybrid. Nat. New Biol. 236, 110–111. 10.1038/newbio236110a0 [DOI] [PubMed] [Google Scholar]
- Evans G. M., Macefield A. J. (1973). The effect of B chromosomes on homoeologous pairing in species hybrids. Chromosoma 41, 63–73. 10.1007/BF00284074 [DOI] [Google Scholar]
- Ezquerro-López D., Kopecký D., Aramendía L. (2017). Cytogenetic relationships within the Maghrebian clade of Festuca subgen. Schedonorus (Poaceae), using flow cytometry and FISH. Anales del Jardín Botánico Madrid 74, 1. 10.3989/ajbm.2455 [DOI] [Google Scholar]
- Fan C., Luo J., Zhang S., Liu M., Li Q., Li Y., et al. (2019). Genetic mapping of a major QTL promoting homoeologous chromosome pairing in a wheat landrace. Theor. Appl. Genet. 132, 2155–2166. 10.1007/s00122-019-03344-x [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A. (2005). Allopolyploidy – a shaping force in the evolution of wheat genomes. Cytogenet. Genome. Res. 109, 250–258. 10.1159/000082407 [DOI] [PubMed] [Google Scholar]
- Feldman M. (1966. a). The effect of chromosomes 5B, 5D and 5A on chromosomal pairing in Triticum aestivum . Proc. Natl. Acad. Sci. U.S.A. 55, 1447–1453. 10.1073/pnas.55.6.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. (1966. b). The mechanism regulating pairing in Triticum timopheevii . Wheat Inf. Serv. 21, 1–2. [Google Scholar]
- Fussell C. P. (1987). “The Rabl orientation: a prelude to synapsis,” in Meiosis. Ed. Moens P. B. (Orlando: Academic Press; ), 275–299. [Google Scholar]
- Gallo P. H., Micheletti P. L., Boldrini K. R., Risso-Pascotto C., Pagliarini M. S., do Valle C. B. (2007). 2n Gamete formation in the genus Brachiaria (Poaceae: Paniceae). Euphytica 154, 255–260. 10.1007/s10681-006-9294-1 [DOI] [Google Scholar]
- Gauthier F. M., McGuinnis R. C. (1968). The meiotic behaviour of a nulli-haploid plant in Avena sativa L. Can. J. Genet. Cytol. 10, 186–189. 10.1139/g68-025 [DOI] [Google Scholar]
- Ghesquière M., Humphreys M. W., Zwierzykowski Z. (2010). ““Festulolium,” in Fodder Crops and Amenity Grasses,” in Handbook of Plant Breeding, vol. 5 . Eds. Boller B., Posselt U., Veronesi F. (New York, NY: Springer; ), 288–311. 10.1007/978-1-4419-0760-8_12 [DOI] [Google Scholar]
- Gill K. S., Gill B. S., Endo T. R., Mukai Y. (1993). Fine physical mapping of Ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics 134, 1231–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo A., Lucas M., Charpentier C., Sandmann G., Lloyd A., Jenczewski E. (2019). Reducing MSH4 copy number prevents meiotic crossovers between non-homologous chromosomes in Brassica napus . Nat. Commun. 10, 2354. 10.1038/s41467-019-10010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray A. J., Benham P. E. M., Raybould A. F. (1990). ““Spartina anglica — the evolutionary and ecological background,”,” in Spartina anglica — A Research Review. Eds. Gray A. J., Benham P. E. M. (London, UK: Institute of Terrestrial Ecology, Natural Environment Research Council; ), 5–10. [Google Scholar]
- Griffiths S., Sharp R., Foote T. N., Bertin I., Wanous M., Reader S., et al. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752. 10.1038/nature04434 [DOI] [PubMed] [Google Scholar]
- Gupta P. K., Fedak G. (1985). Genetic control of meiotic chromosome pairing in polyploids in the genus Hordeum . Can. J. Genet. Cytol. 27, 515– 530. 10.1139/g85-077 [DOI] [Google Scholar]
- Gyawali Y., Zhang W., Chao S., Xu S., Cai X. (2019). Delimitation of wheat ph1b deletion and development of ph1b-specific DNA markers. Theor. Appl. Genet. 132, 195–204. 10.1007/s00122-018-3207-2 [DOI] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K. D., Chapman E. J. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. U.S.A. 106, 17835–17840. 10.1073/pnas.0907003106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar R., Hodgkin T. (2007). The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156, 1–13. 10.1007/s10681-007-9363-0 [DOI] [Google Scholar]
- Han C., Zhang P., Ryan P. R., Rathjen T. M., Yan Z., Delhaize E. (2016). Introgression of genes from bread wheat enhances the aluminium tolerance of durum wheat. Theor. Appl. Genet. 129, 729–739. 10.1007/s00122-015-2661-3 [DOI] [PubMed] [Google Scholar]
- Hand M. L., Cogan N. O., Stewart A. V., Forster J. W. (2010). Evolutionary history of tall fescue morphotypes inferred from molecular phylogenetics of the Lolium-Festuca species complex. BMC Evol. Biol. 10, 303. 10.1186/1471-2148-10-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao M., Luo J. T., Yang M., Zhang L. Q., Yan Z. H., Yuan Z. W., et al. (2011). Comparison of homoeologous chromosome pairing between hybrids of wheat genotypes Chinese Spring ph1b and Kaixian-luohanmai with rye. Genome 54, 959–964. 10.1139/g11-062 [DOI] [PubMed] [Google Scholar]
- Harper L., Golubovskaya I., Cande W. Z. (2004). A bouquet of chromosomes. J. Cell Sci. 117, 4025–4032. 10.1242/jcs.01363 [DOI] [PubMed] [Google Scholar]
- Holm P. B., Wang X. (1988). The effect of chromosome 5B on synapsis and chiasma formation in wheat, Triticum aestivum cv. Chin. spring. Carls. Res. Communs. 53, 191–208. 10.1007/BF02907179 [DOI] [Google Scholar]
- Holm P. B. (1986). Chromosome pairing and chiasma formation in allohexaploid wheat, Triticum aestivum analyzed by spreading of meiotic nuclei. Carlsberg. Res. Commun. 51, 239. 10.1007/BF02906837 [DOI] [Google Scholar]
- Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., Haselkorn R., et al. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. U.S.A. 99, 8133–8138. 10.1073/pnas.072223799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J. C. E. (1968). Grasses. 2nd edn (London: Penguin Books; ). [Google Scholar]
- Humphreys M. W., Thomas H. M., Morgan W. G., Meredith M. R., Harper J. A., Thomas A., et al. (1995). Discriminating the ancestral progenitors of hexaploid Festuca arundinacea using genomic in situ hybridization. Heredity 75, 171–174. 10.1038/hdy.1995.120 [DOI] [Google Scholar]
- Husband B. C. (2004). The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biol. J. Linn. Soc 82, 537–546. 10.1111/j.1095-8312.2004.00339.x [DOI] [Google Scholar]
- Jacobs B. F., Kingston J. D., Jacobs L. L. (1999). The origin of grass-dominated ecosystems. Ann. Mo. Bot. Gard. 86, 590–643. 10.2307/2666186 [DOI] [Google Scholar]
- Jauhar P. P., Almouslem A. B., Peterson T. S., Joppa L. R. (1999). Inter- and intra-genomic chromosome pairing in haploids of durum wheat. J. Hered. 90, 437–445. 10.1093/jhered/90.4.437 [DOI] [Google Scholar]
- Jauhar P. P. (1975). Genetic regulation of diploid-like chromosome pairing in the hexaploid species, Festuca arundinacea Schreb. and F. rubra L. (Gramineae). Chromosoma 52, 363–382. 10.1007/BF00364020 [DOI] [Google Scholar]
- Jauhar P. P. (1977). Genetic regulation of diploid-like chromosome pairing in Avena . Theor. Appl. Genet. 49, 287–295. 10.1007/BF00275135 [DOI] [PubMed] [Google Scholar]
- Jauhar P. P. (1993). Cytogenetics of the Festuca-Lolium complex: relevance to breeding (Berlin: Springer; ). [Google Scholar]
- Jauhar P. P. (2003). Formation of 2n gametes in durum wheat haploids: sexual polyploidization. Euphytica 133, 81–94. 10.1023/A:1025692422665 [DOI] [Google Scholar]
- Jenczewski E., Alix K. (2004). From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23, 21–45. 10.1080/07352680490273239 [DOI] [Google Scholar]
- Jeon J., Chung Y., Lee S., Yi G., Oh B., An G. (1999). Isolation and characterization of an anther-specific gene, RA8, from rice (Oryza sativa L.). Plant Mol. Biol. 39, 35–44. 10.1023/A:1006157603096 [DOI] [PubMed] [Google Scholar]
- Ji L., Langridge P. (1994). An early meiosis cDNA clone from wheat. Molec. Gen. Genet. 243, 17–23. 10.1007/BF00283871 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N., Ayyampalayam S., Chanderbali A. S., Landherr L., Ralph P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. 10.1038/nature09916 [DOI] [PubMed] [Google Scholar]
- Kang H. Y., Zhang H. Q., Wang Y., Jiang Y., Yuan H. J., Zhou Y. H. (2008). Comparative analysis of the homoeologous pairing effects of phKL gene in common wheat × Psathyrostachys huashanica . Keng Cereal Res. Commun. 36, 429–440. 10.1556/CRC.36.2008.3.7 [DOI] [Google Scholar]
- Keeler K. H., Davis G. A. (1999). Comparison of common cytotypes of Andropogon gerardii (Andropogoneae: Poaceae). Am. J. Bot. 86, 974–979. 10.2307/2656614 [DOI] [PubMed] [Google Scholar]
- Keeler K. H. (1998). “Population biology of intraspecific polyploidy in grasses,” in Population Biology of Grasses. Ed. Cheplick G. P. (Cambridge, UK: Cambridge University Press; ), 183–206. [Google Scholar]
- Kellogg E. A. (2001). Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205. 10.1104/pp.125.3.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Grewal S., Yang C. Y., Hubbart S., Scholefield D., Ashling S., et al. (2016). A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum . Plant Biotech. J. 15, 217–226. 10.1111/pbi.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Newell C., Grewal S., Hubbart-Edwards S., Yang C. Y., Scholefield D., et al. (2019). Development of stable homozygous wheat/Amblyopyrum muticum (Aegilops mutica) introgression lines and their cytogenetic and molecular characterization. Front. Plant Sci. 10, 34. 10.3389/fpls.2019.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijer G., Morel P. (1984). Cytogenetic studies of crosses between Lolium multiflorum Lam. and Festuca arundinacea Schreb. II. The amphidiploids. Z. Pflanzenzücht 93, 23–42. [Google Scholar]
- Koo D., Liu W., Friebe B., Gill B. S. (2016). Homoeologous recombination in the presence of Ph1 gene in wheat. Chromosoma 126, 531–540. 10.1007/s00412-016-0622-5 [DOI] [PubMed] [Google Scholar]
- Kopecký D., Loureiro J., Zwierzykowski Z., Ghesquière M., Doležel J. (2006). Genome constitution and evolution in Lolium × Festuca hybrid cultivars (Festulolium). Theor. Appl. Genet. 113, 731–742. 10.1007/s00122-006-0341-z [DOI] [PubMed] [Google Scholar]
- Kopecký D., Lukaszewski A. J., Doležel J. (2008. a). Meiotic behaviour of individual chromosomes of Festuca pratensis in tetraploid Lolium multiflorum . Chromosome Res. 16, 987. 10.1007/s10577-008-1256-0 [DOI] [PubMed] [Google Scholar]
- Kopecký D., Lukaszewski A. J., Doležel J. (2008. b). Cytogenetics of Festulolium (Festuca x Lolium hybrids). Cytogenet. Genome Res. 120, 370–383. 10.1159/000121086 [DOI] [PubMed] [Google Scholar]
- Kopecký D., Bartoš J., Zwierzykowski Z., Doležel J. (2009). Chromosome pairing of individual genomes in tall fescue (Festuca arundinacea Schreb.), its progenitors, and hybrids with Italian ryegrass (Lolium multiflorum Lam.). Cytogenet. Genome Res. 124, 170–178. 10.1159/000207525 [DOI] [PubMed] [Google Scholar]
- Kopecký D., Talukder S. K., Zwyrtková J., Trammell M., Doležel J., Saha M. C. (2019). Inter-morphotype hybridization in tall fescue (Festuca arundinacea Schreb.): exploration of meiotic irregularities and potential for breeding. Euphytica 215, 97. 10.1007/s10681-019-2419-0 [DOI] [Google Scholar]
- Kreiner J. M., Kron P., Husband B. C. (2017). Evolutionary dynamics of unreduced gametes. Trends Genet. 33, 583–593. 10.1016/j.tig.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Ladizinsky G. (1973). Genetic control of bivalent pairing in the Avena strigosa polyploid complex. Chromosoma 42, 105–110. 10.1007/BF00326334 [DOI] [PubMed] [Google Scholar]
- Leggett J. M., Thomas H. (1995). “Oat evolution and cytogenetics,” in The Oat Crop. World Crop Series. Ed. Welch R. W. (Dordrecht, DE: Springer; ), 120–149. 10.1007/978-94-011-0015-1_5 [DOI] [Google Scholar]
- Li H., Deal K. R., Luo M. C., Ji W., Distelfeld A., Dvorak J. (2017). Introgression of the Aegilops speltoides Su1-Ph1 Suppressor into Wheat. Front. Plant Sci. 8, 2163. 10.3389/fpls.2017.02163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. C., Luo M. C., Yang J. L., Yen C., Lan X. J., Yang W. Y. (1997). Chromosome location of a new paring promoter in natural populations of common wheat. Xi Nan Nong Ye Xue Bao 10, 10–15. (in Chinese). [Google Scholar]
- Liu D. C., Luo M. C., Yen C., Yang J. L., Yang W. Y. (1998). “The promotion of homoeologous pairing in hybrids of common wheat cv. Kaixianluohanmai with alien species,” in Proceedings of the 9th International Wheat Genetics Symposium, vol. 4 . Ed. Slinkard A. E. (Saskatoon, CA: University Extension Press, University of Saskatchewan; ), 377–378. [Google Scholar]
- Liu D. C., Zheng Y. L., Yan Z. H., Zhou Y. H., Wei Y. M., Lan X. J. (2003). Combination of homoeologous pairing gene phKL and Ph2-deficiency in common wheat and its meiotic behaviors in hybrids with alien species. Acta Bot. Sin. 45, 1121–1128. [Google Scholar]
- Liu W., Rouse M., Friebe B., Jin Y., Gill B. S., Pumphrey M. O. (2011). Discovery and molecular mapping of a new gene conferring resistance to stem rust, Sr53, derived from Aegilops geniculata and characterization of spontaneous translocation stocks with reduced alien chromatin. Chromosom Res. 19, 669–682. 10.1007/s10577-011-9226-3 [DOI] [PubMed] [Google Scholar]
- Lloyd A., Bomblies K. (2016). Meiosis in autopolyploid and allopolyploid Arabidopsis . Curr. Opin. Plant Biol. 30, 116–122. 10.1016/j.pbi.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Lloyd A., Ranoux M., Vautrin S., Glover N. M., Fourment J., Charif D., et al. (2014). Meiotic gene evolution: can you teach a new dog new tricks? Mol. Biol. Evol. 31, 1724–1727. 10.1093/molbev/msu119 [DOI] [PubMed] [Google Scholar]
- Loidl J. (1990). The initiation of meiotic chromosome pairing: the cytological view. Genome 33, 759–778. 10.1139/g90-115 [DOI] [PubMed] [Google Scholar]
- Loureiro J., Kopecký D., Castro S., Santos C., Silveira P. (2007). Flow cytometric and cytogenetic analyses of Iberian Peninsula Festuca spp. Plant Syst. Evol. 269, 89–105. 10.1007/s00606-007-0564-8 [DOI] [Google Scholar]
- Lukaszewski A. J., Kopecký D. (2010). The ph1 locus from wheat controls meiotic chromosome pairing in autotetraploid rye (Secale cereale L.). Cytogenet. Genome Res. 129, 117–123. 10.1159/000314279 [DOI] [PubMed] [Google Scholar]
- Luo M. C., Yang Z. L., Yen C., Yang J. L. (1992). ““The cytogenetic investigation on F1 hybrid of Chinese wheat landrace,”,” in Exploration of Crop Breeding. Eds. Ren Z. L., Peng J. H. (Sichuan: Science and Technology Press; ), 169–176. [Google Scholar]
- Maestra B., Naranjo T. (1997). Homoeologous relationships of Triticum sharonense chromosomes to T. aestivum . Theor. Appl. Genet. 94, 657–663. 10.1007/s001220050463 [DOI] [Google Scholar]
- Maestra B., Naranjo T. (1998). Homoeologous relationships of Aegilops speltoides chromosomes to bread wheat. Theor. Appl. Genet. 97, 181–186. 10.1007/s001220050883 [DOI] [Google Scholar]
- Malik C. P., Thomas P. T. (1967). Cytological relationships and genome structure of some Festuca species. Caryologia 20, 1–39. 10.1080/00087114.1967.10796244 [DOI] [Google Scholar]
- Marais G. F., Marais A. S., Eksteen A., Pretorius Z. A. (2010). Modification of the Aegilops neglecta-common wheat Lr62/Yr42 translocation through allosyndetic pairing induction. Crop Sci. 49, 871–879. 10.2135/cropsci2008.06.0317 [DOI] [Google Scholar]
- Marcussen T., Sandve S. R., Heier L., Spannagl M., Pfeifer M., IWGSC (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 6194. 10.1126/science.1250092 [DOI] [PubMed] [Google Scholar]
- Marques A. M., Tuler A. C., Carvalho C. R., Carrijo T. T., Ferreira M. F., Clarindo W. R. (2016). Refinement of the karyological aspects of Psidium guineense (Swartz 1788): a comparison with Psidium guajava (Linnaeus 1753). Comp. Cytogenet. 10, 117–128. 10.3897/CompCytogen.v10i1.6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín A. C., Shaw P., Phillips D., Reader S., Moore G. (2014). Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 5, 1–5. 10.1038/ncomms5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín A. C., Rey M. D., Shaw P., Moore G. (2017). Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 126, 669–680. 10.1007/s00412-017-0630-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez M., Cuñado N., Carcelén N., Romero C. (2001. a). The Ph1 and Ph2 loci play different roles in the synaptic behaviour of hexaploid wheat Triticum aestivum . Theor. Appl. Genet. 103, 398–405. 10.1007/s00122-001-0543-3 [DOI] [Google Scholar]
- Martínez M., Naranjo T., Cuadrado C., Romero C. (2001. b). The synaptic behaviour of Triticum turgidum with variable doses of the Ph1 locus. Theor. Appl. Genet. 102, 751–758. 10.1007/s001220051706 [DOI] [Google Scholar]
- Martínez-Pérez E., Shaw P., Reader S., Aragón-Alcaide L., Miller T., Moore G., et al. (1999). Homologous chromosome pairing in wheat. J. Cell Sci. 112, 1761–1769. [DOI] [PubMed] [Google Scholar]
- Mason A. S., Pires J. C. (2015). Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 31, 5–10. 10.1016/j.tig.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Mason A. S., Nelson M. N., Yan G., Cowling W. A. (2011). Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biol. 11:, 103. 10.1186/1471-2229-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire P. E., Dvořák J. (1982). Genetic regulation of heterogenetic chromosome pairing in polyploid species of the genus Triticum sensu lato. Can. J. Genet. Cytol. 24, 57–82. 10.1139/g82-007 [DOI] [Google Scholar]
- Meister N., Tjumjakoff N. A. (1928). Rye–wheat hybrids from reciprocal crosses. J. Genet. 20, 233–245. 10.1007/BF02983142 [DOI] [Google Scholar]
- Mello-Sampayo T., Canas A. P. (1973). “Suppression of meiotic chromosome pairing in common wheat,” in Proceedings of the 4th International Wheat Genetics Symposium. Eds. Sears E. R., ER L. M. S. (Columbia, MI: Agricultural Experiment Station, College of Agriculture, University of Missouri; ), 703–713. [Google Scholar]
- Mello-Sampayo T. (1968). ““Homoeologous chromosome pairing in pentaploid hybrids of wheat,”,” in Third International Wheat Genetics Symposium. Eds. Finlay K. W., Shepherd K. W. (Canberra: Butterworth & Company; ), 179–184. [Google Scholar]
- Mello-Sampayo T. (1971). Genetic regulation of meiotic chromosome pairing by chromosome-3D of Triticum aestivum . Nat. New Biol. 230, 22. 10.1038/newbio230022a0 [DOI] [PubMed] [Google Scholar]
- Meyers L. A., Levin D. A. (2006). On the abundance of polyploids in flowering plants. Evolution 60, 1198–1206. 10.1111/j.0014-3820.2006.tb01198.x [DOI] [PubMed] [Google Scholar]
- Mikhailova E. I., Naranjo T., Shepherd K., Wennekes-van E. J., Heyting C., de Jong H. (1998). The effect of the wheat Ph1 locus on chromatin organisation and meiotic pairing analysed by genome painting. Chromosoma 107, 339–350. 10.1007/s004120050316 [DOI] [PubMed] [Google Scholar]
- Murat F., Zhang R., Guizard S., Flores R., Armero A., Pont C., et al. (2014). Shared subgenome dominance following polyploidization explains grass genome evolutionary plasticity from a seven protochromosome ancestor with 16K protogenes. Genome Biol. Evol. 6, 12–33. 10.1093/gbe/evt200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. G., Sieber V. K., Jackson R. C. (1984). Further evidence for the presence of meiotic pairing control genes in Alopecurus L. (Gramineae). Genet. 63, 13–20. 10.1007/BF00137460 [DOI] [Google Scholar]
- Naranjo T., Maestra B. (1995). The effect of ph mutations on homoeologous pairing in hybrids of wheat with Triticum longissimum . Theor. Appl. Genet. 91, 1265–1270. 10.1007/BF00220939 [DOI] [PubMed] [Google Scholar]
- Naranjo T., Roca A., Goicoechea P. G., Giráldez R. (1987). Arm homoeology of wheat and rye chromosomes. Genome 29, 873–882. 10.1139/g87-149 [DOI] [Google Scholar]
- Naranjo T., Roca A., Goicoechea P. G., Giráldez R. (1988). “Chromosome structure of common wheat: genome reassignment of chromosomes 4A and 4B,” in Proceedings of the 7th International Wheat Genetics Symposium, eds. Eds. Miller T. E., Koebner R. M. D. (Cambridge, UK: Cambridge University; ), 115–120. [Google Scholar]
- Naranjo T. (2015). Contribution of Structural Chromosome Mutants to the Study of Meiosis in Plants. Cytogenet. Genome Res. 147, 55–69. 10.1159/000442219 [DOI] [PubMed] [Google Scholar]
- Naranjo T. (2019). The effect of chromosome structure upon meiotic homologous and homoeologous recombinations in Triticeae . Agronomy 9, 552. 10.3390/agronomy9090552 [DOI] [Google Scholar]
- Niu Z., Klindworth D. L., Friesen T. L., Chao S., Jin Y., Cai X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. 10.1534/genetics.110.123588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S., Prieto I., Odajima J., Martín A., Dubus P., Sotillo R., et al. (2003). Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35, 25–31. 10.1038/ng1232 [DOI] [PubMed] [Google Scholar]
- Osborn T. C., Pires J. C., Birchler J. A., Auger D. L., Chen Z. J., Lee H. S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. 10.1016/S0168-9525(03)00015-5 [DOI] [PubMed] [Google Scholar]
- Otto S. P., Whitton J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. 10.1146/annurev.genet.34.1.401 [DOI] [PubMed] [Google Scholar]
- Ozkan H., Feldman M. (2001). Genotypic variation in tetraploid wheat affecting homoeologous pairing in hybrids with Aegilops peregrina . Genome 44, 1000–1006. 10.1139/g01-100 [DOI] [PubMed] [Google Scholar]
- Pécrix Y., Rallo G., Folzer H., Cigna M., Gudin S., Le Bris M. (2011). Polyploidization mechanisms: temperature environment can induce diploid gamete formation in Rosa sp. J. Exp. Bot. 62, 3587–3597. 10.1093/jxb/err052 [DOI] [PubMed] [Google Scholar]
- Pecinka A., Fang W., Rehmsmeier M., Levy A. A., Scheid O. M. (2011). Polyploidization increases meiotic recombination frequency in Arabidopsis . BMC Biol. 9, 24. 10.1186/1741-7007-9-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelé A., Rousseau-Gueutin M., Chèvre A. M. (2018). Speciation success of polyploid plants closely relates to the regulation of meiotic recombination. Front. Plant Sci. 9, 907. 10.3389/fpls.2018.00907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernickova K., Linc G., Gaal E., Kopecký D., Šamajová O., Lukaszewski A. (2019). Out-of-position telomeres in meiotic leptotene appear responsible for chiasmate pairing in an inversion heterozygote in wheat (Triticum aestivum L.). Chromosoma 128, 31–39. 10.1007/s00412-018-0686-5 [DOI] [PubMed] [Google Scholar]
- Petersen G., Seberg O., Yde M., Berthelsen K. (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39, 70–82. 10.1016/j.ympev.2006.01.023 [DOI] [PubMed] [Google Scholar]
- Prieto P., Moore G., Reader S. (2005). Control of conformation changes associated with homologue recognition during meiosis. Theor. Appl. Genet. 111, 505–510. 10.1007/s00122-005-2040-6 [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. 10.1146/annurev.ecolsys.29.1.467 [DOI] [Google Scholar]
- Ramsey J., Schemske D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639. 10.1146/annurev.ecolsys.33.010802.150437 [DOI] [Google Scholar]
- Renny-Byfield S., Rodgers-Melnick E., Ross-Ibara J. (2017). Gene fractionation and function in the ancient subgenomes of maize. Mol. Biol. Evol. 34, 1825–1832. 10.1093/molbev/msx121 [DOI] [PubMed] [Google Scholar]
- Rey M. D., Calderón M. C., Prieto P. (2015. a). The use of the ph1b mutant to induce recombination between the chromosomes of wheat and barley. Front. Plant Sci. 6, 160. 10.3389/fpls.2015.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey M. D., Calderón M. C., Rodrigo M. J., Zacarías L., Alós E., Prieto P. (2015. b). Novel Bread Wheat Lines Enriched in Carotenoids Carrying Hordeum chilense Chromosome Arms in the ph1b Background. PloS One 10 (8), e0134598. 10.1371/journal.pone.0134598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey M., Martín A. C., Higgins J., Swarbreck D., Uauy C., Shaw P., et al. (2017). Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 37, 95. 10.1007/s11032-017-0700-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey M. D., Martin A. C., Smedley M., Hayta S., Harwood W., Shaw P., et al. (2018). Magnesium increases homoeologous crossover frequency during meiosis in ZIP4 (Ph1 gene) mutant wheat-wild relative hybrids. Front. Plant Sci. 9, 509. 10.3389/fpls.2018.00509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R., Chapman V. (1958). Genetic control of the cytologically diploid behavior of hexaploid wheat. Nature 182, 713–715. . 10.1038/182713a0 [DOI] [Google Scholar]
- Riley R., Kempanna C. (1963). The homoeologous nature of the non-homologous meiotic pairing in Triticum aestivum deficient for chromosome V. Heredity 18, 287–306. 10.1038/hdy.1963.31 [DOI] [Google Scholar]
- Riley R., Law C. N. (1965). Genetic variation in chromosome pairing. Adv. Genet. 13, 57–114. 10.1016/S0065-2660(08)60047-4 [DOI] [Google Scholar]
- Riley R., Chapman V., Miller T. E. (1973). “The determination of meiotic chromosome pairing,” in Proceedings of the 4th International Wheat Genetics Symposium. Eds. Sears E. R., ER L. M. S. (Columbia, MI: Agricultural Experiment Station, College of Agriculture, University of Missouri; ), 731–738. [Google Scholar]
- Riley R. (1960). The diploidization of polyploid wheat. Heredity 15, 407–429. 10.1038/hdy.1960.106 [DOI] [Google Scholar]
- Roberts M. A., Reader S. M., Dalgliesh C., Miller T. E., Foote T. N., Fish L. J., et al. (1999). Induction and characterisation of the Ph1 wheat mutants. Genetics 153, 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A., Ainouche M. L., Wendel J. F. (2005). Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14, 1163–1175. 10.1111/j.1365-294X.2005.02488.x [DOI] [PubMed] [Google Scholar]
- Sánchez-Morán E., Benavente E., Orellana J. (2001). Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma 110, 371–377 (2001). 10.1007/s004120100156 [DOI] [PubMed] [Google Scholar]
- Sattler M. C., Carvalho C. R., Clarindo W. R. (2016). The polyploidy and its key role in plant breeding. Planta 243, 281–296. 10.1007/s00425-015-2450-x [DOI] [PubMed] [Google Scholar]
- Scherthan H. (2001). A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2, 621–627. 10.1038/35085086 [DOI] [PubMed] [Google Scholar]
- Scherthan H. (2007). Telomere attachment and clustering during meiosis. Cell Mol. Life Sci. 64, 117–124. 10.1007/s00018-006-6463-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T. (1997). Three stages of meiotic homologous chromosome pairing in wheat: cognition, alignment and synapsis. Sex- Plant Reprod. 10, 324–331. 10.1007/s004970050106 [DOI] [Google Scholar]
- Sears E. R., Okamoto M. (1958). “Intergenomic chromosome relationship in hexaploid wheat,” in Proceedings of 10th International Congress of Genetics (Toronto, CA: University of Toronto Press; ), 258–259. [Google Scholar]
- Sears E. R. (1976). Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10, 31–51. 10.1146/annurev.ge.10.120176.000335 [DOI] [PubMed] [Google Scholar]
- Sears E. R. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. 10.1139/g77-063 [DOI] [Google Scholar]
- Sears E. R. (1982). A wheat mutation conditioning an intermediate level of homoeologous chromosome pairing. Can. J. Genet. Cytol. 24, 715–719. 10.1139/g82-076 [DOI] [Google Scholar]
- Sears E. R. (1984). “Mutations in wheat that raise the level of meiotic chromosome pairing,” in Gene Manipulation in Plant Improvement. Proc. 16th Stadler Genet. Symp. Ed. Gustafson J. P. (New York, NY: Plenum Press; ), 295–300. [Google Scholar]
- Shang X. M., Jackson R. C., NGuyen H. T., Huang H. T. (1989). Chromosome pairing in the Triticum monococcum complex: evidence for pairing control genes. Genome 32, 213–226. 10.1139/g89-432 [DOI] [Google Scholar]
- Shen Y., Tang D., Wang K., Wang M., Huang J., Luo W., et al. (2012). ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 125, 2581–2591. 10.1242/jcs.090993 [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., Schemske D. W., Hancock J. F., Thompson J. N., Husband B. C., et al. (2007). Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56, 13–30. 10.2307/25065732 [DOI] [Google Scholar]
- Soreng R. J., Peterson P. M., Romaschenko K., Davidse G., Zuloaga F. O., Judziewicz E. J., et al. (2015). A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 53, 117–137. 10.1111/jse.12150 [DOI] [Google Scholar]
- Stebbins G. L. (1950). Variation and Evolution in Plants (New York: Columbia University Press; ). [Google Scholar]
- Stebbins G. L. (1971). Chromosomal Evolution in Higher Plants (London: Addison-Wesley; ). [Google Scholar]
- Sun Y., Wu Y., Yang C., Sun S., Lin X., Liu L., et al. (2017). Segmental allotetraploidy generates extensive homoeologous expression rewiring and phenotypic diversity at the population level in rice. Mol. Ecol. 26, 5451–5466. 10.1111/mec.14297 [DOI] [PubMed] [Google Scholar]
- Sutton T., Whitford R., Baumann U., Dong C. M., Able J. A., Langridge P. (2003). The Ph2 pairing homoeologous locus of wheat (Triticum aestivum): identification of candidate meiotic genes using a comparative genetics approach. Plant J. 36, 443–456. 10.1046/j.1365-313X.2003.01891.x [DOI] [PubMed] [Google Scholar]
- Svačina R., Karafiátová M., Malurová M., Serra H., Vítek D., Endo T. R., et al. (2020). Development of deletion lines for chromosome 3D of bread wheat. Front. Plant Sci. 10, 1756. 10.3389/fpls.2019.01756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo-Ordóñez M. C., Espinosa-Barrera L. A., Tamayo-Ordóñez Y. J., Ayil-Gutiérrez B., Sánchez-Teyer L. F. (2016). Advances and perspectives in the generation of polyploid plant species. Euphytica 209, 1–22. 10.1007/s10681-016-1646-x [DOI] [Google Scholar]
- Thomas H. M., Morgan W. G., Meredith M. R., Humphreys M. W., Leggett J. M. (1994). Identification of parental and recombined chromosomes in hybrid derivatives of Lolium multiflorum × Festuca pratensis by genomic in situ hybridization. Theor. Appl. Genet. 88, 909–913. 10.1007/BF00220795 [DOI] [PubMed] [Google Scholar]
- Thomas H. (1992). “Cytogenetics of Avena,” in Oat Science and Technology. Monograph 33, Agronomy Series. Eds. Marshall H. G., Sorrells M. E. (Madison, WI: ASA and CSSA; ), 473–507. [Google Scholar]
- Thomas S. W. (1997). Molecular studies of homologous chromosome pairing in Triticum aestivum . [dissertation]. [Adelaide]: University of Adelaide.
- Thompson J. D., McNeilly T., Gray A. J. (1991). Population variation in Spartina anglica C.E. Hubbard. I. Evidence from a common garden experiment. New Phytol. 117, 115–128. 10.1111/j.1469-8137.1991.tb00951.x [DOI] [Google Scholar]
- Van de Peer Y., Maere S., Meyer A. (2009). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732. 10.1038/nrg2600 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Vanneste K., Baele G., Maere S., Van de Peer Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res. 24, 1334–1347. 10.1101/gr.168997.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas W. S., Mello-Sampayo T., Feldman M., Avivi L. (1980). Reduction of chromosome pairing by a spontaneous mutation on chromosomal arm 5DL of Triticum aestivum . Can. J. Genet. Cytol. 22, 569–575. 10.1139/g80-062 [DOI] [Google Scholar]
- Viera A., Rufas J. S., Martinez I., Barbero J. L., Ortega S., Suja J. (2009). CDK2 is required for proper homologous pairing, recombination and sex-body formation during male meiosis. J. Cell Sci. 122, 2149–2159. 10.1242/jcs.046706 [DOI] [PubMed] [Google Scholar]
- Villar R., Veneklaas E. J., Jordano P., Lambers H. (1998). Relative growth rate and biomass allocation in 20 Aegilops (Poaceae) species. N. Phytol. 140, 425–437. 10.1046/j.1469-8137.1998.00286.x [DOI] [PubMed] [Google Scholar]
- von Well E., Fossey A. (1998). A comparative investigation of seed germination, metabolism and seedling growth between two polyploid Triticum species. Euphytica 101, 83–89. 10.1023/A:1018320230154 [DOI] [Google Scholar]
- Waines J. G. (1976). A model for the origin of diploidizing mechanisms in poly-ploid species. Am. Nat. 110, 415– 430. 10.1086/283077 [DOI] [Google Scholar]
- Wall A. M., Riley R., Chapman V. (1971). Wheat mutants permitting homoeologous meiotic chromosomes pairing. Genet. Res. 18, 311–328. 10.1017/S0016672300012714 [DOI] [Google Scholar]
- Wang A., Xia Q., Xie W., Datla R., Selvaraj G. (2003). The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. U.S.A. 100, 14487–14492. 10.1073/pnas.2231254100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Roe B., Macmil S., Yu Q., Murray J. E., Tang H., et al. (2010). Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics 11, 261. 10.1186/1471-2164-11-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R. (2002). From intimate chromosome associations to wild sex in wheat (Triticum aestivum). [dissertation]. [Adelaide]: University of Adelaide.
- Winterfeld G., Schneider J., Perner K., Röser M. (2012). Origin of highly polyploids: different pathways of auto- and allopolyploidy in 12–18x species of Avenula (Poaceae). Int. J. Pl. Sci. 173, 1–14. 10.1086/664710 [DOI] [Google Scholar]
- Wulff B. B. H., Moscou M. J. (2014). Strategies for transferring resistance into wheat: from wide crosses to GM cassettes. Front. Plant Sci. 5, 692. 10.3389/fpls.2014.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z. G., Liu D. C., Zheng Y. L., Zhang L. Q., Yan Z. H. (2005). The effect of phKL gene on homoeologous pairing of wheat-alien hybrids is situated between gene mutants of Ph1 and Ph2 . Hereditas 27, 935–940. [PubMed] [Google Scholar]
- Xiong Y. G., Gan L., Hu Y. P., Sun W. C., Zhou X., Song Z. J., et al. (2019). OsMND1 regulates early meiosis and improves the seed set rate in polyploid rice. Plant Growth Regul. 87, 341–356. 10.1007/s10725-019-00476-4 [DOI] [Google Scholar]
- Yant L., Hollister J. D., Wright K. M., Arnold B. J., Higgins J. D., Franklin F. C. H., et al. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa . Curr. Biol. 23, 2151–2156. 10.1016/j.cub.2013.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]