Abstract
Targeting metabolic reprogramming is an emerging strategy in cancer therapy. However, clinical attempts to target metabolic reprogramming have been proved to be challenging, with metabolic heterogeneity of cancer being one of many reasons that causes treatment failure. Here, we stratified non-small cell lung cancer (NSCLC) cells, mainly lung adenocarcinoma, based on their metabolic phenotypes and demonstrated that the aerobic glycolysis-preference NSCLC cell subtype was resistant to the OXPHOS-targeting inhibitors. We identified that monocarboxylate transporter 4 (MCT4), a lactate transporter, was highly expressed in the aerobic glycolysis-preference subtype with function supporting the proliferation of these cells. Glucose could induce the expression of MCT4 in these cells through a ΔNp63α and Sp1-dependent pathway. Next, we showed that knockdown of MCT4 increased intracellular lactate concentration and induced a reactive oxygen species (ROS)-dependent cellular apoptosis in the aerobic glycolysis-preference NSCLC cell subtype. By scanning a panel of monoclonal antibodies with MCT4 neutralizing activity, we further identified a MCT4 immunoglobulin M (IgM) monoclonal antibody showing capable anti-proliferation efficacy on the aerobic glycolysis-preference NSCLC cell subtype. Our findings indicate that the metabolic heterogeneity is a critical factor for NSCLC therapy and manipulating the expression or function of MCT4 can be an effective strategy in targeting the aerobic glycolysis-preference NSCLC cell subtype.
Keywords: NSCLC, cancer metabolism, metabolic heterogeneity, aerobic glycolysis, MCT4, Sp1, ΔNp63α, MCT4-neutralizing antibody
Graphical Abstract
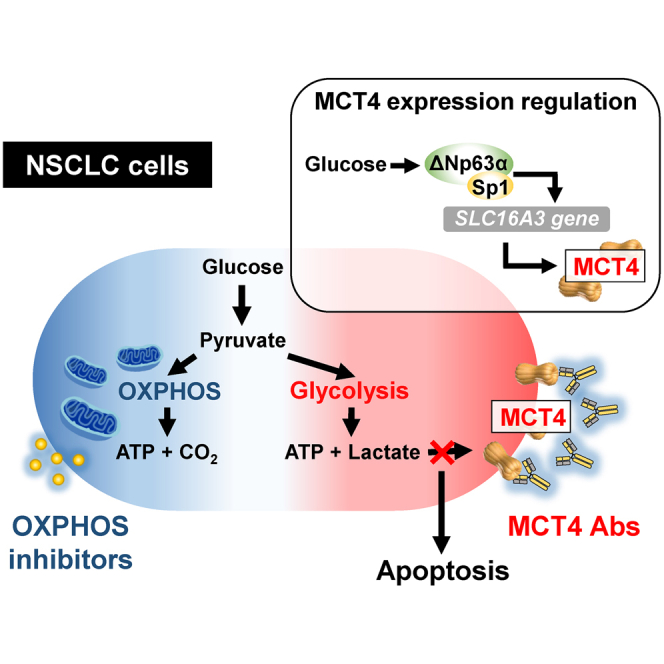
Yang and colleagues stratified NSCLC cells based on their metabolic phenotypes and identified that monocarboxylate transporter 4 (MCT4) was highly expressed in the aerobic glycolysis-preference subtype with function supporting the proliferation of these cells. Additionally, they discovered a MCT4-neutralizing antibody with cell-inhibitory activity on this aerobic glycolysis-preference NSCLC cell subtype.
Introduction
Molecularly targeted therapy has recently become mainstream in cancer treatment. However, cancer cells frequently become resistant to targeted therapy after a period of effective treatments.1,2 To improve the therapeutic efficacy in cancer treatment, targeting fundamental cellular changes in cancer cells may provide promising solutions. Among the ten cancer hallmarks proposed by Weinberg and Hanahan,3 metabolic reprogramming is highlighted as an important aspect in cancer treatment.4,5 Various small molecules that target different pathways involved in cancer metabolism have now been identified,6,7 with some already undergoing clinical trials.8 Unexpectedly, the efficacy of many metabolism-targeting drugs is inadequate. Some of these drugs even require combinations with other pharmacological agents to achieve their therapeutic efficacy.9 One possible reason for this unsuccessful outcome is that the metabolic alterations in cancer cells are dynamically adaptive to their environmental influences.4,5 Many studies indicate that cancer cells in the same population could further evolve into different metabolic subtypes with distinct sensitivities to metabolism-targeting drugs.10, 11, 12
Under normoxic conditions, the majority of ATP in cells are produced in the mitochondria through aerobic respiration. However, Otto Warburg first observed that highly proliferative cancer cells rely primarily on glycolysis for energy production even when oxygen is abundant, creating the term aerobic glycolysis.13 Because aerobic glycolysis provides a faster rate of ATP production compared with that in oxidative phosphorylation (OXPHOS), cancer cells tend to use aerobic glycolysis to meet their fast-paced energy needs, with pyruvate breaking down into lactate for an extra NAD+ instead of entering the citric acid cycle as acetyl coenzyme A (CoA).14 But simultaneously, the intracellular lactate concentration can be 40-fold higher in the highly proliferative cancer cells compared to that in normal cells.15 Thus, lactate has to be exported outside of cancer cells and into the tumor microenvironment. Because lactate is hydrophilic and a weak acid, it requires specific transporters to move across membranes. Monocarboxylate transporters (MCTs) are proton-linked membrane transporters that can carry lactate across the cellular membranes.16 Among the fourteen members of the MCT family, MCT1 and MCT4 are studied intensively in lactate transport. MCT1 can transport lactate bi-directionally, while MCT4 primarily facilitates lactate efflux from cells.17,18 MCT4 can manipulate the concentration of lactate in the tumor microenvironment and further regulate cancer cell proliferation, migration, and angiogenesis.19 High expression of MCT4 has been observed in many types of cancers, while most cases are under hypoxic conditions.20,21 As for the regulation of MCT4 expression, hypoxia-inducible factor-1 alpha (HIF-1α) is among the first transcription factor studied thoroughly. HIF-1α controls the transcriptional activation of SLC16A3 that encodes MCT4 via a hypoxia response element in the promoter region of SLC16A3.22 In contrast to the regulation of MCT4 expression under hypoxia conditions, there is much less description for MCT4 regulation under normoxic conditions. For instance, the PI3K/AKT and KRAS/MEK signaling pathways were recently reported to be associated with MCT4 expression.23,24
Here we report that high MCT4 expression is strongly associated with the aerobic glycolysis-preference NSCLC cell subtype. We show that the expression of MCT4 can be induced by glucose through a ΔNp63α and Sp1-dependent pathway, which supports the proliferation of such subtype of NSCLC cells. Knockdown of MCT4 increases the intracellular lactate concentration and results in hindered aerobic glycolysis, thereby inducing a reactive oxygen species (ROS)-dependent cellular apoptosis. Furthermore, we develop an MCT4 immunoglobulin M (IgM) monoclonal antibody showing capable anti-proliferation efficacy on the aerobic glycolysis-preference NSCLC cell subtype. Our findings therefore reveal a novel therapeutic strategy for targeting MCT4 in the aerobic glycolysis-preference NSCLC cell subtype.
Results
Aerobic Glycolysis-Preference NSCLC Cell Subtype Demonstrates Its Resistance to OXPHOS-Targeting Inhibitors
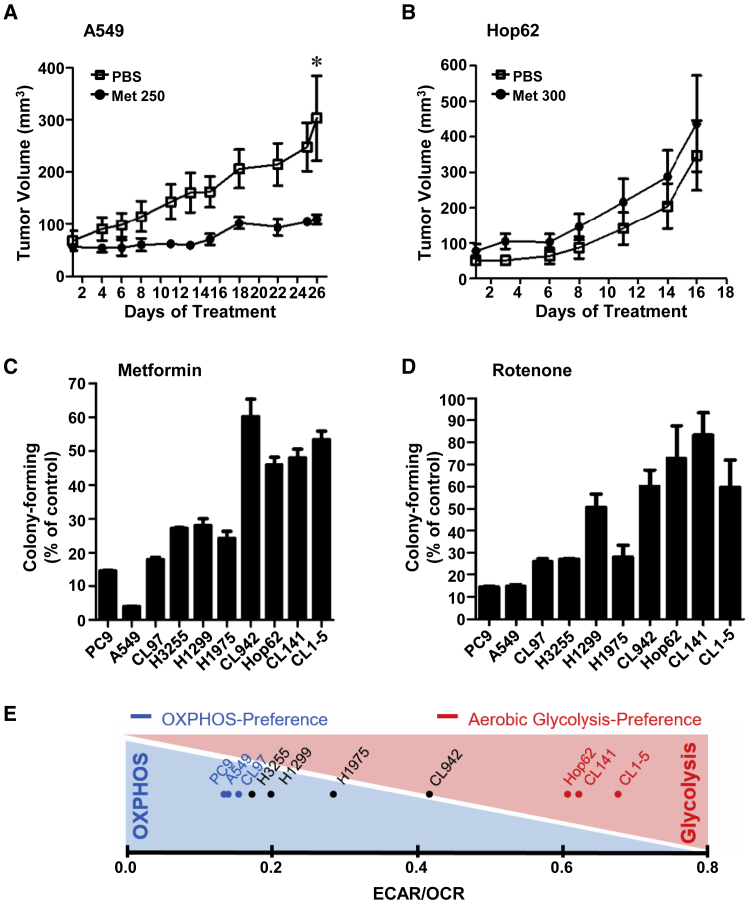
Acting as a single agent or part of a combination treatment in in vivo mouse models, the mitochondrial oxidative phosphorylation (OXPHOS)-targeting inhibitor metformin has been shown to efficiently suppress tumor growth in lung cancer.25 To investigate whether metformin could exert a broad anti-cancer efficacy for NSCLC treatment, we inoculated nude mice with two NSCLC cell lines, A549 or Hop62, and treated the subcutaneous tumors with metformin at doses of either 250 mg/kg/day or 300 mg/kg/day. We found that the growth of A549 tumors was effectively suppressed by metformin treatment (Figure 1A), whereas the growth of Hop62 tumors was not inhibited, but instead were slightly promoted by metformin (Figure 1B). To investigate whether the heterogeneity in NSCLC causes the discrepancy in response to metformin treatment, we tested the drug susceptibility of ten different NSCLC cell lines, mainly lung adenocarcinoma, to metformin and rotenone, another OXPHOS-targeting inhibitor. We found that not all of the NSCLC cell lines tested were sensitive to the treatment of OXPHOS-targeting inhibitors (Figures 1C and 1D). Notably, the NSCLC cell lines with resistance to one OXPHOS-targeting inhibitor tend to be resistant to another inhibitor as well. Since OXPHOS is a major metabolic pathway used for adenosine triphosphate (ATP) production, we used the Seahorse XF Analyzer to determine the rate of glycolytic and oxidative ATP production in these NSCLC cell lines next. The ratios of extracellular acidification rate (ECAR) to oxygen consumption rate (OCR) were measured and used to assign whether the given cell lines adopt to OXPHOS or aerobic glycolysis for the majority of their energy demands. As shown in Figure 1E, PC9, A549, and CL97 NSCLC cells with lower ECAR/OCR ratio (≤0.15) preferred to use OXPHOS, while Hop62, CL141, and CL1-5 NSCLC cells with higher ECAR/OCR ratio (≥0.50) relied on aerobic glycolysis for ATP production. Among the NSCLC cell lines, CL1-5 is a highly invasive subpopulation of cells derived from the parental CL1-0 lung cancer cells.26 The ratio of ECAR/OCR in CL1-0 cells were much lower than that in CL1-5 cells. This observation is consistent with previous studies,27 showing that aerobic glycolysis is the predominant bioenergetic pathway in cancer cells with higher migration and invasion abilities. Since the OXPHOS-targeting inhibitors had no significant growth inhibitory effects on NSCLC cells heavily relying on aerobic glycolysis for ATP production, we speculate that targeting the aerobic glycolysis pathway could be an ideal way to treat NSCLC cells showing resistance to the treatment of OXPHOS-targeting inhibitors.
Figure 1.
Aerobic Glycolysis-Preference NSCLC Cell Subtype Demonstrates Its Resistance to OXPHOS-Targeting Inhibitors
(A and B) The growth curve of A549 (A) and Hop62 (B) subcutaneous tumors in nude mice with or without metformin treatment (250 mg/kg metformin in A549 group; 300 mg/kg metformin in Hop62 group). The treatment duration was chosen as the tumor volume in the control group reached 300 mm3. Data represent mean and SD. n = 4. (C and D) The anti-proliferative effect of 10 mM metformin (C) and 100 nM rotenone (D) on the ten NSCLC cell lines. The colonies were fixed, stained, and dissolved as described in the Materials and Methods. The graph indicated the total absorbance values at 490 nm in metformin or rotenone treatment groups relative to that in the solvent control group, which were set to 100%. Data represent mean and SD. n = 3. (E) The average basal ECAR and OCR levels of the ten NSCLC cells were measured using a Seahorse XF Analyzer. The two metabolic-preference subtypes were assigned according to the ratio of cellular ECAR to OCR. Cell lines with higher ratio (larger than 0.50) were classified as the aerobic glycolysis-preference subtype; cell lines with lower ratio (smaller than 0.15) were classified as the OXPHOS-preference subtype.
Glucose Upregulates MCT4 Expression in the Aerobic Glycolysis-Preference NSCLC Cell Subtype
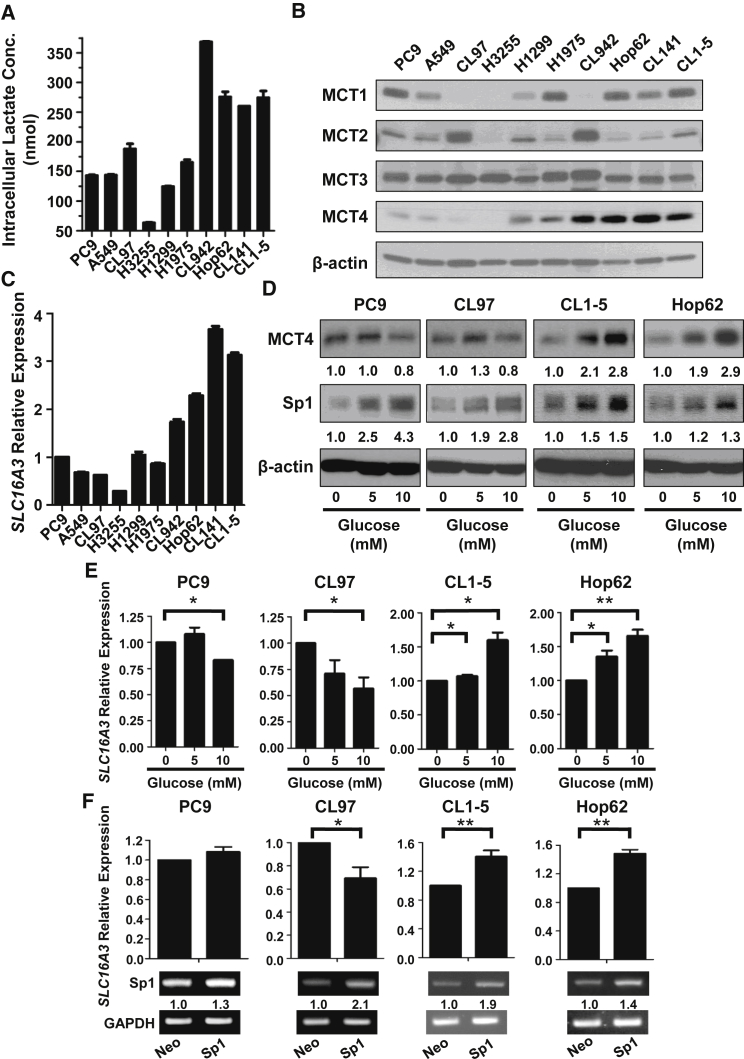
Cancer cells using aerobic glycolysis as a preferred energy source usually produce lactate along with energy productions and result in a higher intracellular lactate concentration. To avoid damages caused by the gradual accumulation of lactate, cancer cells tend to overexpress lactate transporters, such as the MCT family members, for lactate efflux. Among the MCT transporters, MCT1, MCT2, MCT3, and MCT4 are well-known monocarboxylate transporters that control the concentration of lactate within the cell and in the tumor microenvironment. As expected, we found that the intracellular lactate concentration was much higher in the aerobic glycolysis-preference NSCLC cell subtype compared with the OXPHOS-preference NSCLC cell subtype (Figure 2A). Interestingly, only the expression of MCT4 in the testing NSCLC cell lines showed a strong correlation with the ratios of ECAR to OCR and the lactate concentrations (Figure 2B). Consistent with its protein levels, the mRNA level of SLC16A3 gene, which encodes the MCT4 protein, was higher in the aerobic glycolysis-preference NSCLC cell subtype (Figure 2C). These data suggest that MCT4 plays a critical role in exporting lactate in the aerobic glycolysis-preference NSCLC cell subtype.
Figure 2.
Glucose Upregulates MCT4 Expression in the Aerobic Glycolysis-Preference NSCLC Cell Subtype
(A) Intracellular lactate concentrations of the ten NSCLC cell lines were measured by a lactate assay kit. 3 × 105 cells were collected for each cell line and the concentration was measured as described in the Materials and Methods. Data represent mean and SD. n = 3. (B) The protein levels of MCT1 to 4 in the ten NSCLC cell lines were detected in whole-cell lysates by immunoblotting. (C) The mRNA level of MCT4 (SLC16A3) in the ten NSCLC cell lines was detected by qRT-PCR and the graph indicated the levels in each cell line relative to that in PC9. Data represent mean and SD. n = 3. (D) PC9, CL97, CL1-5, and Hop62 cells were treated with 0, 5, or 10 mM of glucose for 24 h. The protein levels of MCT4 and Sp1 were detected in whole-cell lysates by immunoblotting. (E) PC9, CL97, CL1-5, and Hop62 cells were treated with 0, 5, or 10 mM of glucose for 24 h. The mRNA level of MCT4 (SLC16A3) was detected by qRT-PCR. Data represent mean and SD. n = 3. (F) The mRNA level of MCT4 (SLC16A3) was detected by qRT-PCR in PC9, CL97, CL1-5, and Hop62 cells overexpressing Neo or Sp1. Data represent mean and SD. n = 3.
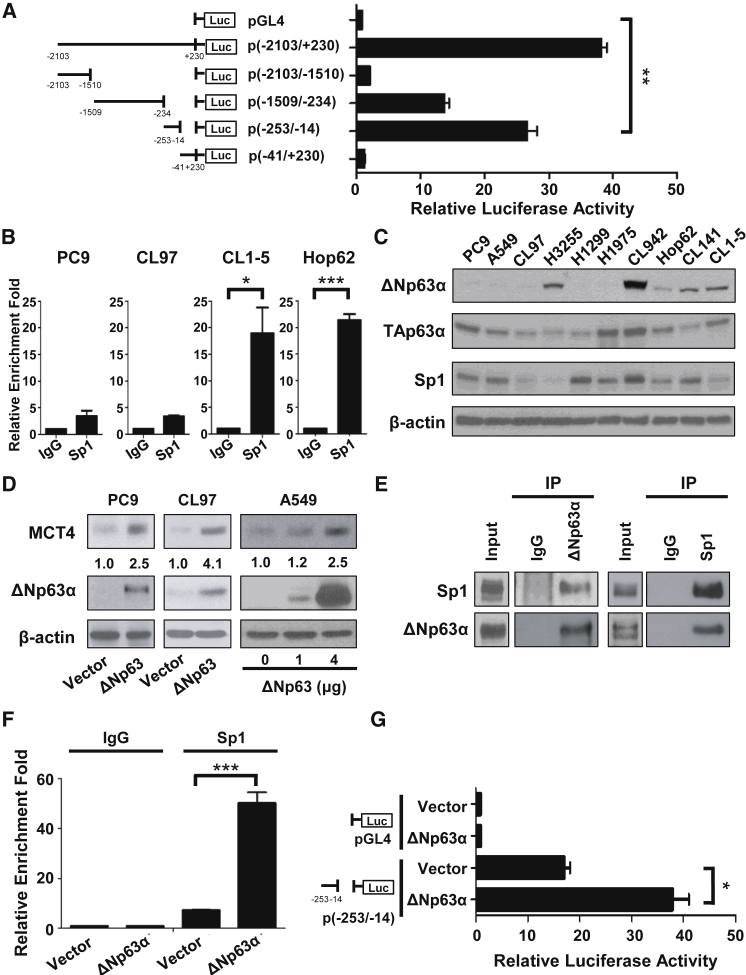
Since anaerobic glycolysis begins with the transformation of glucose to lactate, we sought to determine whether glucose could stimulate the expression of MCT4 in NSCLC cells. We chose two representative NSCLC cell lines in the aerobic glycolysis-preference NSCLC cell subtype (CL1-5 and Hop62) and two in the OXPHOS-preference NSCLC cell subtype (PC9 and CL97), respectively. We found that glucose induced MCT4 upregulation at both the mRNA and protein levels in CL1-5 and Hop62 cells, but not in PC9 and CL97 cells (Figures 2D and 2E). These data imply that glucose may stimulate the transcriptional activation of SLC16A3 gene and result in higher levels of MCT4 expression in the aerobic glycolysis-preference NSCLC cell subtype. To identify the potential transcription factors (TFs) involved in this regulation, we used the chromatin immunoprecipitation on chip (ChIP-on-chip) data from TRANSFAC database, and Sp1 appeared as one of the candidates (data not shown). To verify this result, we checked the expression of Sp1 in all the testing NSCLC cell lines and the representative cell lines under differential glucose stimulations (Figures 2D and 3C). We found that glucose increased MCT4 expression in the aerobic glycolysis preference NSCLC cell subtype, but higher concentration of glucose (10 mM) reduced MCT4 expression in the OXPHOS-preference NSCLC cell subtype (Figure 2D). Consistently, ectopic expression of Sp1 increased SLC16A3 mRNA level in the aerobic glycolysis-preference NSCLC cell subtype cell lines, CL1-5 and Hop62, but had no or inhibitory effects in the OXPHOS-preference NSCLC cell subtype cell lines, PC9 and CL97 (Figure 2F). To further identify the specific binding sites of Sp1 on the promoter region of SLC16A3 gene, we constructed a series of SLC16A3 promoter sequences and used for the luciferase reporter assays in CL1-5 cells. As shown in Figure 3A, we narrowed down the SLC16A3 promoter region to within −253 to −14, which was further confirmed to be a critical region for the transcription of SLC16A3 gene by Sp1. ChIP assays showed that Sp1 bound to the essential promoter region (−253 to −14) of the SLC16A3 gene in the aerobic glycolysis-preference NSCLC cell subtype, but not in the OXPHOS-preference NSCLC cell subtype (Figure 3B). Taken together, these results indicate that Sp1 may be involved in the glucose-dependent MCT4 upregulation while the Sp1-mediated regulatory mechanisms on the SLC16A3 gene may be different in these two metabolic subtypes of NSCLC cells.
Figure 3.
N-Terminally Truncated (ΔN) Isoform of p63α and Sp1 Coordinately Regulate the Expression of MCT4 in NSCLC Cells
(A) CL1-5 cells were co-transfected with indicated luciferase reporter constructs, together with pRL-SV40 as a transfection control. Luciferase activity was measured 48 h post-transfection. The promoter activity of different regions of SLC16A3 promoter was expressed as a ratio of firefly/Renilla luciferase activities. Data represent mean and SD. n = 3. (B) The interaction of Sp1 with the specific region between −253 and −14 on the SLC16A3 promoter was determined by ChIP assay in PC9, CL97, CL1-5, and Hop62. The ChIP assays were performed using anti-Sp1 antibody and the immunoprecipitated chromatin was analyzed using specific TaqMan probe with primers specific to the MCT4 (SLC16A3) promoter, between −253 and −14. Data represent mean and SD. n = 3. (C) The protein levels of ΔNp63α, TAp63α, and Sp1 in the ten NSCLC cell lines were detected in whole-cell lysates by immunoblotting. (D) The protein level of MCT4 was detected by immunoblotting in the three OXPHOS-preference NSCLC cell lines (PC9, CL97, and A549) with or without ΔNp63α overexpression. (E) The interaction between ΔNp63α and Sp1 was detected by the immunoprecipitation method in A549 cells overexpressing ΔNp63α. A549 cell lysates were immunoprecipitated with anti-ΔNp63α, anti-rabbit IgG (left panel), anti-Sp1, or anti-rabbit IgG (right panel) antibodies, and immunoblotted with the indicated antibodies. (F) The interaction of Sp1 with the specific region between −253 and −14 on the SLC16A3 promoter was determined by the ChIP assay in A549 cells with or without ΔNp63α overexpression. A549 cells were transfected with vector or ΔNp63α plasmids for 30 h, and ChIP assays were performed using anti-Sp1 antibody. Immunoprecipitated chromatin was analyzed using specific TaqMan probe with primers specific to the SLC16A3 promoter, between −253 and −14. Data represent mean and SD. n = 4. (G) A549 cells were co-transfected with vector or ΔNp63α plasmids, indicated luciferase reporter constructs, and pRL-SV40 as a transfection control. Luciferase activity was measured 48 h post-transfection. The promoter activity of the specific region between −253 and −14 on the SLC16A3 promoter was expressed as a ratio of firefly/Renilla luciferase activities in A549 NSCLC cells with or without ΔNp63α overexpression. Data represent mean and SD. n = 3.
The N-Terminally Truncated (ΔN) Isoform of p63α and Sp1 Coordinately Regulate the Expression of MCT4 in NSCLC Cells
We next attempted to identify the potential TFs that could promote the binding of Sp1 on the promoter region of SLC16A3 gene in the aerobic glycolysis-preference NSCLC cell subtype. P63α was identified as a candidate to interact with the promoter region of SLC16A3 gene from the TRANSFAC database (data not shown). P63 is a member of the p53 gene family that encodes for up to ten different isoforms through alternative promoter usage and alternative splicing. The TA isoforms contain the N-terminal transactivation domain (TA), whereas the ΔN isoforms are transcribed from an alternative promoter and lack the entire TA domain.28 We focused on two isoforms of p63α, TAp63α and ΔNp63α, whose expressions and functions are frequently associated with tumorigenesis. We found that the expression of ΔNp63α, but not TAp63α or Sp1, was strongly associated with higher MCT4 expression, ECAR/OCR ratio, and intracellular lactate concentration in the aerobic glycolysis-preference NSCLC cell subtype (Figures 1E, 2A, 2B, and 3C). Since ΔNp63α and TAp63α have been shown to exhibit different transactivation activities,29 we speculate that ΔNp63α and TAp63α may exert distinct functions in regulating MCT4 expression in cells. Indeed, the ectopic expression of ΔNp63α promoted MCT4 levels whereas ectopically expressed TAp63α decreased MCT4 expression in HEK293T cells (Figure S1A). Luciferase reporter experiments using the minimal SLC16A3 promoter sequence demonstrated that ΔNp63α has the potential to activate the transcription of SLC16A3 gene while TAp63α may repress the promoter activity of SLC16A3 (Figure S1B). To confirm that high levels of ΔNp63α contributes to high expression of MCT4 in NSCLC cells, we overexpressed ΔNp63α in the OXPHOS-preference NSCLC cell subtype cell lines (e.g., PC9, CL97, and A549) whose ΔNp63α levels were relatively low in comparison to that in the aerobic glycolysis-preference NSCLC cell subtype. The results showed that ΔNp63α overexpression promoted the upregulation of MCT4 in these NSCLC cell lines (Figure 3D). The ability of ΔNp63α to interact with Sp1 in vivo (Figures 3E and S2A) suggests that ΔNp63α may facilitate the binding of Sp1 to the promoter region of SLC16A3 gene. We therefore examined whether ΔNp63α facilitates the recruitment of Sp1 to the minimal SLC16A3 promoter region by ChIP experiments. Certainly, the overexpression of ΔNp63α significantly increased the binding of Sp1 to the promoter region of SLC16A3 gene and subsequently stimulated the promoter activity of SLC16A3 (Figures 3F, 3G, S2B, and S2C). Our results indicate that ΔNp63α acts synergistically with Sp1 to activate SLC16A3 gene transcription that results in high MCT4 expression in NSCLC cells.
High MCT4 Expression Supports Cell Growth of the Aerobic Glycolysis-Preference NSCLC Cell Subtype
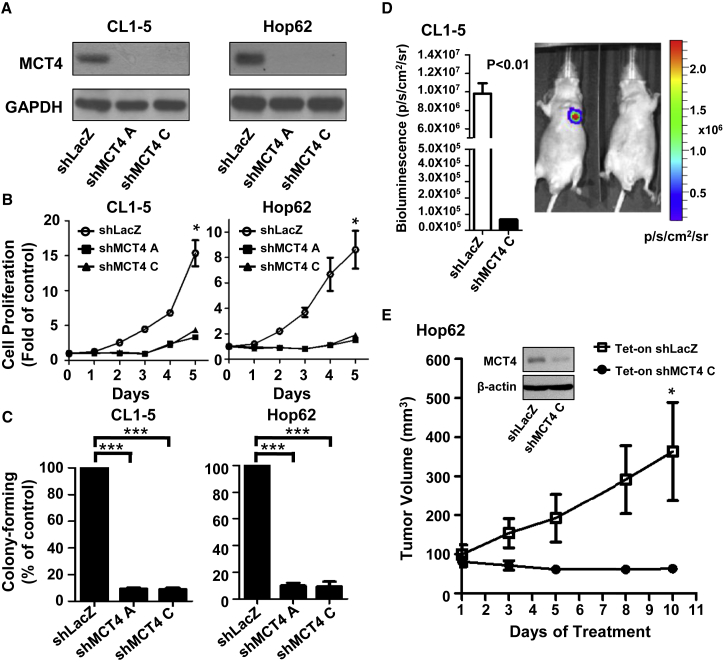
High MCT4 expression in the aerobic glycolysis-preference NSCLC cell subtype contributes to the transport of excessive lactate produced by aerobic glycolysis. This prompts us to further elucidate whether MCT4 functions in supporting cell growth of this subtype of NSCLC cells in vitro and in vivo. We knocked down the expression of MCT4 by two individual lentiviral short hairpin RNAs (shRNAs) in the representative the aerobic glycolysis-preference subtype, CL1-5 and Hop62, and the OXPHOS-preference subtype, PC9 and CL97 (Figures 4A and S3A). As shown in Figures 4B and 4C, MCT4 knockdown significantly suppressed cell growth and the colony-forming ability in the aerobic glycolysis-preference NSCLC cell subtype, while showing very mild anti-proliferation effects in the OXPHOS-preference NSCLC cell subtype (Figures S3B and S3C). In an orthotopic lung tumor mouse model, we found that CL1-5 cells could not develop into a tumor nodule in the injected lung in the absence of MCT4 expression (Figure 4D). Similarly, a doxycycline-inducible lentiviral shRNA vector significantly decreased MCT4 expression in Hop62 cells, as well as the volume of Hop62 subcutaneous tumors (Figure 4E). These data indicate that high MCT4 expression greatly supports cancer cell growth and colonization. Interestingly, although MCT4 knockdown in the OXPHOS-preference subtype PC9 and CL97 cells had little effects on cancer cell growth and colonization (Figures S3A–S3C), we observed that MCT4 knockdown could significantly inhibit the growth of PC9 subcutaneous tumors in nude mice (Figure S3D). This unexpected result might be due to the fact that the in vivo growth condition is more complicated and harsher when compared to the in vitro culture condition, and therefore, cancer cells may require more energy to support their growth in the in vivo environment.
Figure 4.
HighMCT4 Expression Supports Cell Growth of the Aerobic Glycolysis-Preference NSCLC Cell Subtype
(A) The protein level of MCT4 after transduction with shRNA virus targeting SLC16A3 gene or LacZ control was detected by immunoblotting in CL1-5 and Hop62 cells. (B) The cell proliferation rate of CL1-5 and Hop62 cells after transduction with shRNA virus targeting SLC16A3 gene or LacZ control was detected by SRB assays. The cells were fixed at the indicated day and detected as described in the Materials and Methods. The graph indicated the fold change between each day versus day 0. Data represent mean and SD. n = 3. (C) The colony formation ability of CL1-5 and Hop62 cells was detected by colony-formation assays after transduction with shRNA virus targeting SLC16A3 gene or LacZ control. The colonies were fixed, stained, and dissolved as described in the Materials and Methods. The graph indicated the total absorbance values at 490 nm in shMCT4 groups relative to that in shLacZ groups, which were set to 100%. Data represent mean and SD. n = 3. (D) Representative in vivo imaging system (IVIS) images of male nude mice captured 14 days after orthotopic implantation of CL1-5-Luc cells with or without MCT4 knockdown by shRNA virus. The luminescence intensity of photons emitted from each tumor was quantified as shown in the figure. Data represent mean and SD. n = 3. (E) Hop62 cells were transduced with inducible-shRNA virus targeting SLC16A3 gene or LacZ control and treated with doxycycline hyclate at 3 μg/mL concentration for 5 days. The MCT4 expression in these cells was shown in the western blotting plot. Following the validation, these cells were implanted subcutaneously in nude mice. Mice were treated with doxycycline hyclate (25 mg/kg) daily by oral gavage and the growth curve of Hop62 subcutaneous tumors was measured for 10 days. Data represent mean and SD. n = 6.
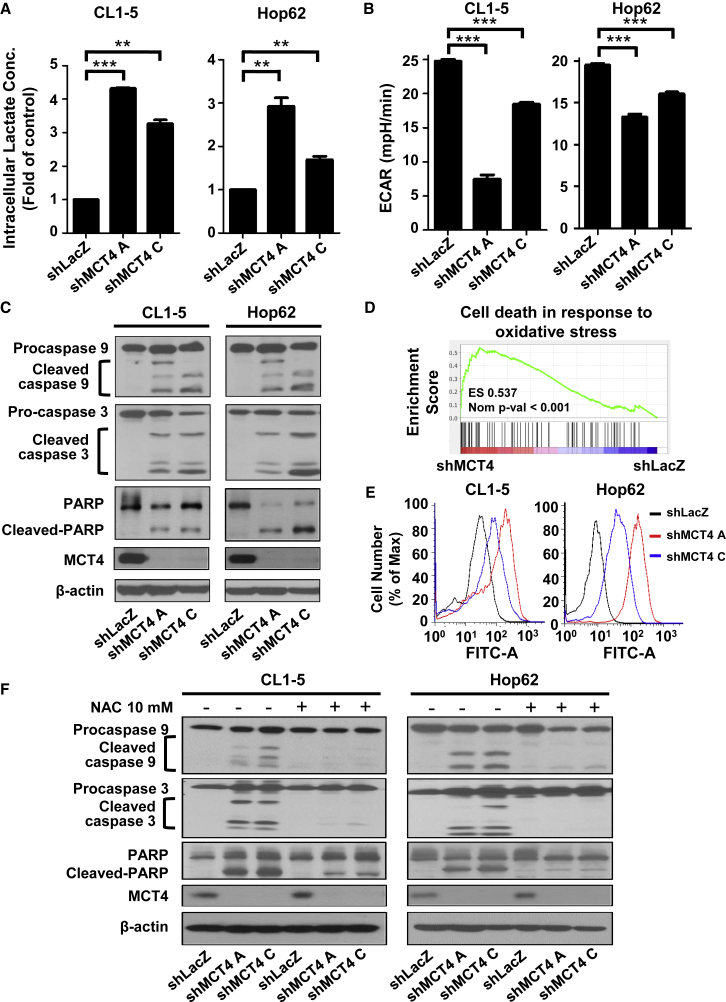
Depletion of MCT4 Hinders Aerobic Glycolysis and Causes ROS-Induced Cellular Apoptosis in the Aerobic Glycolysis-Preference NSCLC Cell Subtype
To identify the underlying mechanism that MCT4 regulates cancer cell growth, we knocked down the expression of MCT4 in the aerobic glycolysis-preference NSCLC cell subtype and measured the intracellular lactate concentration and pH levels. MCT4 knockdown significantly promoted the elevation of intracellular lactate concentration, and in turn, decreased intracellular pH level (Figure 5A; Table 1). The changes in intracellular environment would then block aerobic glycolysis and cause a significant decrease in the ECAR level (Figure 5B). The long-term inhibition of aerobic glycolysis would further trigger the proteolytic cleavage of critical intrinsic apoptosis markers, caspase-9, caspase-3, and Poly (ADP-ribose) polymerase (PARP), and induce cellular apoptosis in these cells (Figure 5C). Gene set enrichment analyses (GSEA) showed that MCT4 depletion upregulates the gene-expression programs involved in the intrinsic apoptosis pathway, with two of the top five upregulated gene sets relating to the oxidative stress-induced cell death (Table 2; Figure 5D). To verify this observation, we checked whether MCT4 depletion increases the concentration of ROS in the aerobic glycolysis-preference NSCLC cell subtype. As expected, an elevated level of intracellular ROS was detected when knocking down MCT4 in the aerobic glycolysis-preference NSCLC cell subtype (Figure 5E). Consistently, the MCT4 depletion-induced intrinsic apoptosis pathway was blocked in the aerobic glycolysis-preference NSCLC cell subtype when the intracellular ROS was effectively reduced by the antioxidant, N-acetylcysteine (NAC; Figures 5F and S4). Altogether, our results indicate that MCT4 plays a critical role in the transport of excessive lactate produced by aerobic glycolysis in NSCLC cells and loss of MCT4 may hinder aerobic glycolysis and cause a ROS-induced cellular apoptosis.
Figure 5.
Depletion of MCT4 Hinders Aerobic Glycolysis and Causes ROS-Induced Cellular Apoptosis in the Aerobic Glycolysis-Preference NSCLC Cell Subtype
(A) Intracellular lactate concentration of CL1-5 and Hop62 cells after transduction with shRNA virus targeting SLC16A3 gene or LacZ control was measured by a lactate assay kit. Data represent mean and SD. n = 3. (B) The basal ECAR level of CL1-5 and Hop62 cells after transduction with shRNA virus targeting SLC16A3 gene or LacZ control was measured using a Seahorse XF analyzer. Data represent mean and SD. n = 5. (C) The protein levels of procaspase-9, cleaved caspase-9, procaspase-3, cleaved caspase-3, PARP, cleaved PARP, and MCT4 were detected in CL1-5 and Hop62 NSCLC cells at 72 h after transduction with shRNA virus targeting SLC16A3 gene or LacZ control by immunoblotting. (D) GSEA revealed that cell_death_in_response_to_oxidative_stress was one of the top five gene sets positively associated with MCT4 knockdown in CL1-5 cells. (E) The intracellular ROS level in CL1-5 or Hop62 cells at 72 h after transduction with shRNA virus targeting SLC16A3 gene or LacZ control was detected by flow cytometry with DCFH-DA dye. (F) The protein levels of procaspase-9, cleaved caspase-9, procaspase-3, cleaved caspase-3, PARP, cleaved PARP, and MCT4 were detected in CL1-5 and Hop62 NSCLC cells with or without 10 mM NAC treatment at 72 h after transduction with shRNA virus targeting SLC16A3 gene or LacZ control by immunoblotting.
Table 1.
Intracellular pH of CL1-5 and Hop62 NSCLC Cells after Transduction with shRNA Virus Targeted SLC16A3 Gene or LacZ Control
| Cell Line | Intracellular pH |
||
|---|---|---|---|
| shLacZ | shMCT4 A | shMCT4 C | |
| CL1-5 | 7.49 ± 0.01 | 6.98 ± 0.11 | 6.83 ± 0.07 |
| Hop62 | 7.39 ± 0.09 | 6.99 ± 0.10 | 7.00 ± 0.05 |
Table 2.
The Top Five Most Positively Enriched Gene Sets in MCT4 Knockdown Group
| GO Terms | Normalized Enrichment Score | Nominal p Value |
|---|---|---|
| Cellular response to glucose starvation | 2.38 | <0.001 |
| Perk mediated unfolded protein response | 2.20 | <0.001 |
| Cell death in response to oxidative stress | 2.08 | <0.001 |
| Regulation of oxidative stress induced cell death | 2.04 | <0.001 |
| Spindle midzone | 2.04 | <0.001 |
MCT4 Neutralizing Antibodies Can Effectively Target MCT4 and Inhibit Cell Growth of the Aerobic Glycolysis-Preference NSCLC Cell Subtype
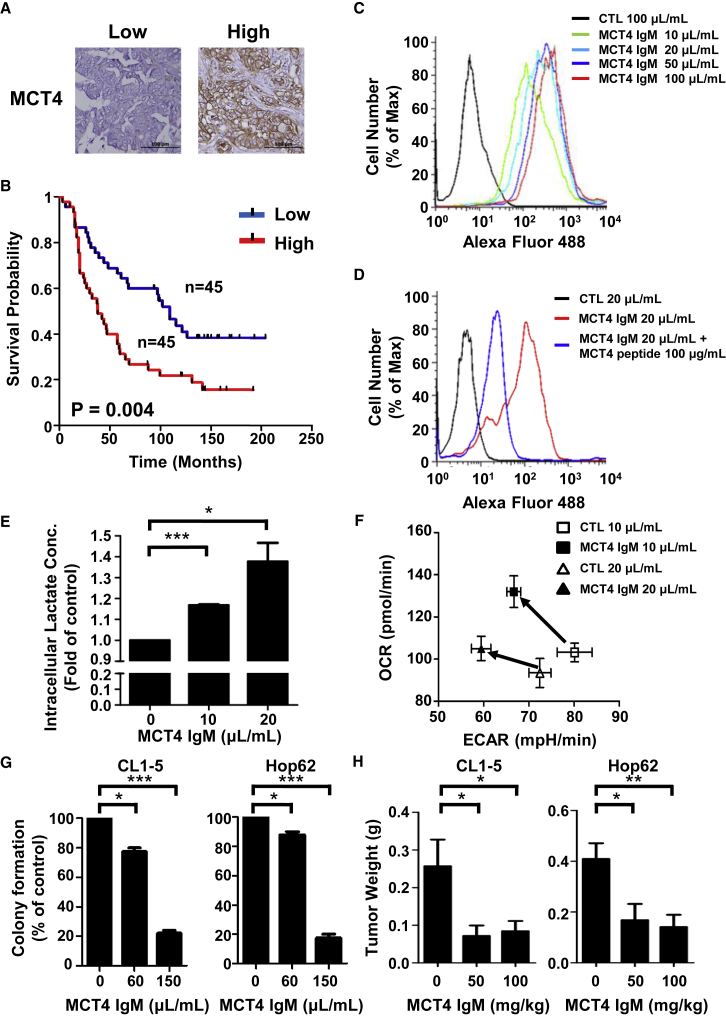
Clinically, high MCT4 expression acts as a poor prognostic factor for the survival of NSCLC patients. This association could be observed in NSCLC patients from the Shedden cohort and the KM-plotter database (Figures S5A and S5B). The hazard ratios of high MCT4 expression in these databases were 1.72 and 1.21, respectively (Table S1). This similar trend could also be observed in our own NSCLC patient dataset from the National Taiwan University Hospital (hazard ratio of high MCT4 expression, 1.81; Figures 6A and 6B; Table S1). Additionally, high MCT4 expression could be found in numerous cancers (e.g., kidney, colon, bladder, uterus, and cervix) implying the therapeutic potential of targeting MCT4 in other cancers (Figure S5C). To the best of our knowledge, only virally delivered shRNA techniques have been used to block the function of MCT4 in cancer cells. However, viral vectors are limitedly applied in the clinical setting due to safety concerns. Hence, we attempted to develop a neutralizing antibody to target MCT4 and inhibit the function of MCT4 for clinical applications.
Figure 6.
MCT4 Neutralizing Antibodies Can Effectively Inhibit the Proliferation of the Aerobic Glycolysis-Preference NSCLC Cell Subtype
(A) Representative images of immunohistochemical staining of MCT4 in NSCLC patients with high or low MCT4 expression levels. (B) The correlation between MCT4 expression level and overall survival probability of NSCLC patients. The correlation was measured using Kaplan-Meier survival curve analysis in a cohort of 90 NSCLC patients from the National Taiwan University Hospital. (C) The binding of our MCT4 IgM antibody with cell-surface MCT4 in CL1-5 cells was detected using Alexa Fluor 488 conjugated anti-mouse IgM antibody by flow cytometry. (D) The competition effect of the original immunogenic peptide to the interaction between our MCT4 IgM antibody and cell-surface MCT4 was detected using Alexa Fluor 488 conjugated anti-mouse IgM antibody by flow cytometry. (E) Intracellular lactate concentration of CL1-5 and Hop62 cells was measured by a lactate assay kit after incubating with MCT4 IgM antibody-containing ascites or control ascites for 6 h. Data represent mean and SD. n = 4. (F) The basal ECAR and OCR levels of CL1-5 and Hop62 cells was measured using a Seahorse XF analyzer after incubating with MCT4 IgM antibody-containing ascites or control ascites for 48 h. Data represent mean and SD. n = 3. (G) The anti-proliferative effect of MCT4 antibody in CL1-5 and Hop62 cells. The colonies were fixed, stained, and dissolved as described in the Materials and Methods after incubating with MCT4 IgM antibody-containing ascites or control ascites for 7 days. The graph indicated the total absorbance values at 490 nm in MCT4 IgM antibody-containing ascites group relative to that in control ascites group, which were set to 100%. Data represent mean and SD. n = 3. (H) The weight of CL1-5 and Hop62 subcutaneous tumors treated with different concentrations of MCT4 IgM antibody for 23 days. Data represent mean and SD. n = 6.
We chose one of the MCT4 extracellular domains as an immunogenic peptide to produce various monoclonal antibodies and selected the candidates by screening their ability to promote the elevation of intracellular lactate concentration in the aerobic glycolysis-preference NSCLC cell subtype. One IgM monoclonal antibody showed promising efficacy in neutralizing MCT4 activity and received further testing. Flow cytometry analyses showed that this IgM monoclonal antibody could bind to MCT4 on the plasma membrane of CL1-5 cells and the interaction between the antibody and MCT4 could be competed out by the MCT4 immunogenic peptide (Figures 6C and 6D). The incubation of this IgM monoclonal antibody with CL1-5 cells significantly increased the intracellular lactate concentration and decreased the ratio of ECAR/OCR (Figures 6E and 6F). Moreover, this MCT4 IgM monoclonal antibody not only suppressed the colony-forming abilities of CL1-5 and Hop62 cells, but also inhibited the tumor growth of CL1-5 and Hop62 subcutaneous tumors in the in vivo xenograft mouse models (Figures 6G and 6H). We observed that this MCT4 IgM monoclonal antibody is relatively safe to the mice during treatments, resulting in minimal weight loss and no obvious toxicity to the kidney or liver tissue (Figure S6).
Discussion
While under an energy-limited microenvironment, cancer cells can reprogram their metabolic pathways to meet their energy demands. The dynamic alterations in cancer cell metabolism cooperate with the tumor microenvironment, which occurs in different stages of tumor progression.30 Because cancer cells may display distinct metabolic phenotypes in the same population, several metabolic profiling methods have been developed to sub-classify cancer cell types. For instance, Daemen et al.11 divided pancreatic ductal adenocarcinomas into three subtypes—slow proliferating, glycolytic, and lipogenic—according to the levels of 256 different metabolites. Baek et al.24 also classified pancreatic cancers into different metabolic subtypes based on different expression levels of metabolism-related genes. They further found that dysregulation of the KRAS/MEK signaling was associated with the upregulation of MCT4 expression in pancreatic cancer cells, and MEK inhibitors could effectively attenuate MCT4 expression in most of the testing pancreatic cancer cell lines. Nevertheless, the classification of lung cancer according to cancer cell metabolic phenotypes has so far been poorly described. Upon the treatment of OXPHOS-targeting inhibitors, we found that not all of the testing NSCLC cell lines, mainly lung adenocarcinoma, were sensitive to the treatment. The resistant cell lines were then further characterized as an aerobic glycolysis-preference phenotype that showed higher ECAR/OCR ratio. This subtype of NSCLC cells had higher intracellular lactate concentration and tend to express a higher level of MCT4 for lactate efflux. Thus, our data indicate that MCT4 is a key regulator for lactate efflux in cancer cells heavily relying on aerobic glycolysis.
High expression of MCT4 has been reported in numerous cancers,19,20 while little is known about the regulation of MCT4 expression in response to cancer cell metabolism. In this study, we observed different expression levels of MCT4 in two different metabolic subtypes of NSCLC cells (Figure 2B), indicating the possibility of a distinct regulatory mechanism for MCT4 expression in different cellular contexts. Indeed, we found that glucose increased MCT4 expression in the aerobic glycolysis preference NSCLC cell subtype, but a higher concentration of glucose reduced MCT4 expression in the OXPHOS-preference NSCLC cell subtype (Figure 2D). We also found that glucose induced an upregulation of Sp1 in both subtypes of NSCLC cells, while this observation could not explain the distinct MCT4 regulations. Interestingly, we uncovered a novel mechanism by which two p63α isoforms, ΔNp63α and TAp63α, play critical roles in this discrepancy. We observed different expression patterns of ΔNp63α and TAp63α in these two metabolic subtypes. TAp63α was expressed equally in all the testing NSCLC cell lines, while ΔNp63α was expressed at a higher level in the aerobic glycolysis-preference NSCLC cell subtype (Figure 3C). We further demonstrated that ΔNp63α binds to Sp1 (Figure 3E) and enhances the binding of Sp1 on the promoter region of SLC16A3 (Figure 3F), which results in higher levels of MCT4 expression in the aerobic glycolysis-preference NSCLC cell subtype (Figure 3D). This suggests that ΔNp63α can upregulate the transactivation activity of Sp1 via a direct association between ΔNp63α, Sp1, and target promoters. By contrast, in NSCLC cells lacking ΔNp63α expression, TAp63α alone could not promote MCT4 expression even when the expression of Sp1 was induced by glucose in the OXPHOS-preference NSCLC cell subtype (Figures 2D and 3C). This might suggest different regulatory mechanisms from these two p63α isoforms in the regulation of MCT4 expression. Supporting this idea, we found that ΔNp63α and TAp63α exert different transcriptional activities in the regulation of SLC16A3 promoter and MCT4 expression in HEK293T cells (Figure S1). Consistent with our observations, ΔNp63α and TAp63α have been shown to exhibit different transactivation specificities that depend on distinct DNA binding abilities.29 Additionally, a TAp63 isoform has been reported to directly interact with Sp1, impairing its DNA binding activity.31 Thus, the different regulatory mechanisms of ΔNp63α and TAp63α in gene regulation may explain their distinct functions in cancer development. TAp63α usually induces senescence and suppresses tumorigenesis while ΔNp63α exhibits primarily oncogenic features.32,33
Our data reveal a novel ΔNp63-Sp1-MCT4 regulatory axis that may contribute to cell growth and survival of NSCLC cell lines heavily relying on aerobic glycolysis for their energy demands. The NSCLC cell lines tested in this study are mainly lung adenocarcinoma. Supporting our findings, high expression of MCT4 is closely associated with poor prognosis in patients with lung adenocarcinoma.34,35 High Sp1 levels have also been identified in early stages of patients with lung adenocarcinoma.36 The role of ΔNp63α in regulating cancer cell metabolism has been observed in various cancers.37,38 ΔNp63α is one of the most important markers to distinguish the two main subtypes of NSCLC, adenocarcinoma and squamous cell carcinoma. However, interestingly, the current evidence indicates that high expression of ΔNp63α could be frequently detected in lung squamous cell carcinoma, but rare in lung adenocarcinoma.39 Hence, the biological relevance of ΔNp63, Sp1, and MCT4 expression in NSCLC patients remains unclear. Since the co-expression profiling of ΔNp63, Sp1, and MCT4 in the tumor specimens of NSCLC patients is still missing, further investigations are needed to unravel the ΔNp63-Sp1-MCT4 regulatory pathway in NSCLC and to explain our observations in clinical tumor specimens.
Despite the MCT4 IgM monoclonal antibody developed in this study exhibiting an excellent cell-inhibitory effect in the aerobic glycolysis-preference NSCLC cell subtype, the pentamer structure of the IgM antibody might limit clinical applications. Based on this concern, we selected an MCT4 IgG1 monoclonal antibody via the same screening procedures. In comparison with the IgM monoclonal antibody, the IgG1 monoclonal antibody also showed high-binding affinity to MCT4 on the plasma membrane of CL1-5 and Hop62 cells (Figure S7A), and resulted in increased intracellular lactate concentration, decreased ECAR to OCR ratio, and lowered colony-forming abilities in these NSCLC cells (Figures S7B–S7D). However, the cell-inhibitory activity of the IgG1 monoclonal antibody was weaker than that of the IgM monoclonal antibody. One possible reason for this is that the IgM monoclonal antibodies with pentameric structure can display higher avidity to the target antigen compared to the IgG monoclonal antibodies.40,41 However, the pentameric structure of IgM monoclonal antibodies could also restrict its distribution and administration during treatment. The neutralizing activity of the MCT4 IgG1 monoclonal antibody warrants further improvement.
Due to the vital roles of MCTs in transporting lactate across membranes, several small molecules have been developed to evaluate the therapeutic potentials of targeting MCT in cancer treatment. The first MCT inhibitor, AR-C155858, was developed to target MCT1 and MCT2 and was shown to effectively suppress lactate export, glycolysis, and tumor growth in a RAS-transformed fibroblast model.42 A potent MCT1 inhibitor, AZD3965, was then designed with modifications on AR-C155858 and had been applied to various phase I/II clinical trials.43,44 Nevertheless, unwanted side effects were detected due to the widespread expression of MCT1 in normal tissues. Furthermore, several studies have identified that high MCT4 expression could be a potential resistance mechanism to MCT1 inhibition.45,46 In accord with this view, the MCT4 neutralizing antibody developed in this study not only can be used to suppress MCT4 function in the aerobic glycolysis-preference NSCLC cell subtype, but can also can be used as monotherapy or combination therapy with the MCT1 inhibitor for a large number of cancers. In summary, our findings provide new insights into the molecular mechanism by which ΔNp63α and Sp1 coordinately regulate MCT4 transcriptional expression in the aerobic glycolysis-preference NSCLC cell subtype. The newly developed MCT4 neutralizing antibody showing anti-proliferation efficacy may offer new therapeutic strategy for treatments to prevent cancer cell growth.
Materials and Methods
Please see the Supplemental Materials and Methods for more details.
Cell Lines
The human lung adenocarcinoma cell line PC9 was a gift from Dr. C.H. Yang (Graduate Institute of Oncology, Cancer Research Center, National Taiwan University). The human lung adenocarcinoma cell line A549, H1299, H3255, H1975, and Hop62 were purchased from American Type Culture Collection (Rockville, MD). The human lung cancer cell line CL1-5 was previously selected using an in vitro modified Boyden Chamber invasion assay in our laboratory.26 The human lung cancer cell lines, CL97, CL942, and CL141, were isolated from pleural effusion of NSCLC patients.47 In general, PC9, Hop62, CL1-5, CL97, H3255, H1975, CL942, and CL141 cells were cultured in RPMI-1640 medium with 10% FBS (v/v) and penicillin (100 units/mL)/ streptomycin (100 μg/mL). A549 and H1299 cells were cultured in DMEM with 10% FBS (v/v) and penicillin (100 units/mL)/ streptomycin (100 μg/mL). Culture dishes were maintained in a humidified incubator at 37°C in 5% CO2/95% air.
Measurement of ECAR and OCR
The ECAR and OCR levels were measured according to the manufacturer’s instructions. Briefly, cells were seeded in XF96 cell culture microplate for 1 day before measurement. The sensor cartridges were pre-incubated with the calibration solution overnight in a non-CO2 incubator at 37°C. On the day of assay, the medium was changed to assay medium without glucose with 2% FBS. Glucose was loaded to the injection port before the measurement and automatically injected into each well according to the protocol. The OCR and ECAR were measured by the sensor probe on a Seahorse XF Extracellular Flux Analyzers (Agilent, Santa Clara, CA).
Sulforhodamine B (SRB) Assay
Cells were incubated with cell culture medium with or without indicated reagents for specific durations, and the anti-proliferation effect was assessed. Cells were first fixed using 10% (wt/vol) trichloroacetic acid for 30 min, and stained with 0.4% (wt/vol) SRB in 1% acetic acid for 30 min. The excess dye was removed by repeated washing with 1% acetic acid. The protein-bound dye was then dissolved in 10 mM Tris base solution, and the results were read by enzyme-linked immunosorbent assay (ELISA) reader (540 nm).
Determination of Intracellular pH Using pHrodo Dye
Cells were harvested by trypsinization and washed with PBS. After centrifugation, cells were suspended in PBS with 2% FBS and stained with pHrodo Red dye in power load buffer (Thermo Fisher Scientific) at 37°C for 30 min. The stained cells were then analyzed with FACScan and CellQuest software (Becton Dickinson, Mountain View, CA).
MCT4 Monoclonal Antibody Production and Purification
BALB/c mice were used to produce MCT4 monoclonal antibodies. The hybridoma supernatants and ascites fluid were obtained according to current protocols.48 The neutralizing activity of the monoclonal antibodies was tested using the lactate assay kit. The ascites fluid samples were collected and stored at −80°C until time of purification. The isotype of the antibody was determined using the Mouse Ig Isotyping Ready-SET-Go! ELISA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The anti-MCT4 IgM monoclonal antibody was further purified with 1 mL HiTrap IgM Purification HP columns (GE Healthcare Life Sciences, Chicago, IL) according to the manufacturer’s instructions.
Clinical Lung Cancer Samples and Immunohistochemistry
Lung cancer specimens from 90 NSCLC patients who underwent surgical resection at the National Taiwan University Hospital were analyzed for the expression of the MCT4 protein. The sections were fixed in formalin and embedded in paraffin. The primary antibody against MCT4 was obtained from Santa Cruz Biotechnology. The immunohistochemistry results were scored and classified into two groups according to the average staining intensity and area. The MCT4-low expression group corresponded to a positive staining of <50% of the tissue sections, and the MCT4-high expression group corresponded to a positive staining of >50% of the tissue sections. The immunostaining results were assessed and scored independently by two pathologists.
Statistics
Data were presented as mean ± SD values. The Kaplan-Meier method was used to estimate overall survival. Differences in survival between the two groups, MCT4-high and MCT4-low, were analyzed using the log-rank test. Multivariate Cox proportional hazard regression analysis with stepwise selection was used to evaluate independent prognostic factors associated with patient survival, and the expression of MCT4, gender, and tumor stage were used as covariates. Two-tailed Student’s t-test were used and p values < 0.05 were considered statistically significant. (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Ethics Approval and Consent to Participate
All animal protocols were evaluated and approved by the Institutional Animal Care and Use Committee of Academia Sinica and Council of Agriculture Guidebook for the Care and Use of Laboratory Animals. (Protocol ID: 13-11-593). The protocol of human sample collection and immunohistochemistry staining was evaluated and approved by the Institutional Review Board/Research Ethics Committee (IRB/REC) of National Taiwan University Hospital. IRB/REC was approved to waive the requirement of informed consent in this protocol (Protocol ID: 201505139RINC).
Author Contributions
Conceptualization, T.-C.K., T.-H.H., and P.-C.Y.; Methodology, T.-C.K., K.-Y.H., S.-C.Y., and Y.-L.C.; Investigation, T.-C.K., K.-Y.H., S.-C.Y., W.-C. C., and Y.-L.C.; Writing – Original Draft, T.-C.K.; Writing –Review & Editing, S.W., T.-M.H., S.-P.W., H.-Y.C., T.-H.H., and P.-C.Y.; Funding Acquisition, S.-P.W., H.-Y.C., and P.-C.Y.; Resources, H.-Y.C., T.-H.H., P.-C.Y.; Supervision, H.-Y.C., T.-H.H., P.-C.Y.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We are grateful for the technical service support from the Experimental Animal Facility and DNA Sequencing Laboratory (Institute of Biomedical Sciences, Academia Sinica) and the support of bioinformatics work and computational and storage resources from the Mathematics in Biology Group (Institute of Statistical Science, Academia Sinica) and National Center for High-Performance Computing of National Applied Research Laboratories in Taiwan. Additionally, we thank Dr. Yidong Chen (Graduate School of Biomedical Sciences, University of Texas Health Science Center at San Antonio) for using TRANSFAC database to predict the potential TFs in regulating the expression of MCT4. This research was supported by grants from Ministry of Science and Technology (MOST-106-2314-B-002-103-MY2, MOST-107-0210-01-19-01, MOST-107-2314-B-002-239, MOST-108-3114-Y-001-002, MOST-108-2314-B-002-195, MOST-108-2314-B-002-027, MOST-108-2320-B-001-007-MY2, MOST-108-2319-B-492-001, and MOST-109-0210-01-18-02) and Academia Sinica (AS-SUMMIT-109 and AS-KPQ-109-BioMed) in Taiwan.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.06.012.
Contributor Information
Hsuan-Yu Chen, Email: hychen@stat.sinica.edu.tw.
Tzu-Hung Hsiao, Email: d93921032@gmail.com.
Pan-Chyr Yang, Email: pcyang@ntu.edu.tw.
Supplemental Information
References
- 1.Zugazagoitia J., Guedes C., Ponce S., Ferrer I., Molina-Pinelo S., Paz-Ares L. Current Challenges in Cancer Treatment. Clin. Ther. 2016;38:1551–1566. doi: 10.1016/j.clinthera.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Camidge D.R., Pao W., Sequist L.V. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat. Rev. Clin. Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden M.G. Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 5.Phan L.M., Yeung S.C., Lee M.H. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014;11:1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu L., Zeng Z., Xia Q., Liu Z., Feng X., Chen J., Huang M., Chen L., Fang Z., Liu Q. Metformin attenuates hepatoma cell proliferation by decreasing glycolytic flux through the HIF-1α/PFKFB3/PFK1 pathway. Life Sci. 2019;239:116966. doi: 10.1016/j.lfs.2019.116966. [DOI] [PubMed] [Google Scholar]
- 7.Kim E.H., Jang H., Shin D., Baek S.H., Roh J.L. Targeting Nrf2 with wogonin overcomes cisplatin resistance in head and neck cancer. Apoptosis. 2016;21:1265–1278. doi: 10.1007/s10495-016-1284-8. [DOI] [PubMed] [Google Scholar]
- 8.Mullard A. Cancer metabolism pipeline breaks new ground. Nat. Rev. Drug Discov. 2016;15:735–737. doi: 10.1038/nrd.2016.223. [DOI] [PubMed] [Google Scholar]
- 9.Park J., Shim J.K., Kang J.H., Choi J., Chang J.H., Kim S.Y., Kang S.G. Regulation of bioenergetics through dual inhibition of aldehyde dehydrogenase and mitochondrial complex I suppresses glioblastoma tumorspheres. Neuro-oncol. 2018;20:954–965. doi: 10.1093/neuonc/nox243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J., Locasale J.W., Bielas J.H., O’Sullivan J., Sheahan K., Cantley L.C., Vander Heiden M.G., Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daemen A., Peterson D., Sahu N., McCord R., Du X., Liu B., Kowanetz K., Hong R., Moffat J., Gao M. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. USA. 2015;112:E4410–E4417. doi: 10.1073/pnas.1501605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haukaas T.H., Euceda L.R., Giskeødegård G.F., Lamichhane S., Krohn M., Jernström S., Aure M.R., Lingjærde O.C., Schlichting E., Garred Ø., Oslo Breast Cancer Consortium (OSBREAC) Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016;4:12. doi: 10.1186/s40170-016-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O.H. Arnold Constable; London, UK: 1930. The Metabolism of Tumours: Investigations from the Kaiser Wilhelm Institute for Biology, Berlin-Dahlem. [Google Scholar]
- 14.Liberti M.V., Locasale J.W. Correction to: ‘The Warburg Effect: How Does it Benefit Cancer Cells?’: [Trends in Biochemical Sciences, 41 (2016) 211] Trends Biochem. Sci. 2016;41:287. doi: 10.1016/j.tibs.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Walenta S., Mueller-Klieser W.F. Lactate: mirror and motor of tumor malignancy. Semin. Radiat. Oncol. 2004;14:267–274. doi: 10.1016/j.semradonc.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Doherty J.R., Cleveland J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Escuredo J., Van Hée V.F., Sboarina M., Falces J., Payen V.L., Pellerin L., Sonveaux P. Monocarboxylate transporters in the brain and in cancer. Biochim. Biophys. Acta. 2016;1863:2481–2497. doi: 10.1016/j.bbamcr.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halestrap A.P., Wilson M.C. The monocarboxylate transporter family--role and regulation. IUBMB Life. 2012;64:109–119. doi: 10.1002/iub.572. [DOI] [PubMed] [Google Scholar]
- 19.Payen V.L., Mina E., Van Hée V.F., Porporato P.E., Sonveaux P. Monocarboxylate transporters in cancer. Mol. Metab. 2020;33:48–66. doi: 10.1016/j.molmet.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyen J., Trastour C., Ettore F., Peyrottes I., Toussant N., Gal J., Ilc K., Roux D., Parks S.K., Ferrero J.M., Pouysségur J. Expression of the hypoxia-inducible monocarboxylate transporter MCT4 is increased in triple negative breast cancer and correlates independently with clinical outcome. Biochem. Biophys. Res. Commun. 2014;451:54–61. doi: 10.1016/j.bbrc.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 21.Gao H.J., Zhao M.C., Zhang Y.J., Zhou D.S., Xu L., Li G.B., Chen M.S., Liu J. Monocarboxylate transporter 4 predicts poor prognosis in hepatocellular carcinoma and is associated with cell proliferation and migration. J. Cancer Res. Clin. Oncol. 2015;141:1151–1162. doi: 10.1007/s00432-014-1888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullah M.S., Davies A.J., Halestrap A.P. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J. Biol. Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- 23.Baenke F., Dubuis S., Brault C., Weigelt B., Dankworth B., Griffiths B., Jiang M., Mackay A., Saunders B., Spencer-Dene B. Functional screening identifies MCT4 as a key regulator of breast cancer cell metabolism and survival. J. Pathol. 2015;237:152–165. doi: 10.1002/path.4562. [DOI] [PubMed] [Google Scholar]
- 24.Baek G., Tse Y.F., Hu Z., Cox D., Buboltz N., McCue P., Yeo C.J., White M.A., DeBerardinis R.J., Knudsen E.S., Witkiewicz A.K. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 2014;9:2233–2249. doi: 10.1016/j.celrep.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Gao Q., Wang D., Wang Z., Hu C. Metformin inhibits growth of lung adenocarcinoma cells by inducing apoptosis via the mitochondria-mediated pathway. Oncol. Lett. 2015;10:1343–1349. doi: 10.3892/ol.2015.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu Y.W., Yang P.C., Yang S.C., Shyu Y.C., Hendrix M.J., Wu R., Wu C.W. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell Mol. Biol. 1997;17:353–360. doi: 10.1165/ajrcmb.17.3.2837. [DOI] [PubMed] [Google Scholar]
- 27.Han T., Kang D., Ji D., Wang X., Zhan W., Fu M., Xin H.B., Wang J.B. How does cancer cell metabolism affect tumor migration and invasion? Cell Adhes. Migr. 2013;7:395–403. doi: 10.4161/cam.26345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nylander K., Vojtesek B., Nenutil R., Lindgren B., Roos G., Zhanxiang W., Sjöström B., Dahlqvist A., Coates P.J. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J. Pathol. 2002;198:417–427. doi: 10.1002/path.1231. [DOI] [PubMed] [Google Scholar]
- 29.Monti P., Ciribilli Y., Bisio A., Foggetti G., Raimondi I., Campomenosi P., Menichini P., Fronza G., Inga A. ΔN-P63α and TA-P63α exhibit intrinsic differences in transactivation specificities that depend on distinct features of DNA target sites. Oncotarget. 2014;5:2116–2130. doi: 10.18632/oncotarget.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlova N.N., Thompson C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishi H., Senoo M., Nishi K.H., Murphy B., Rikiyama T., Matsumura Y., Habu S., Johnson A.C. p53 Homologue p63 represses epidermal growth factor receptor expression. J. Biol. Chem. 2001;276:41717–41724. doi: 10.1074/jbc.M101241200. [DOI] [PubMed] [Google Scholar]
- 32.Bergholz J., Xiao Z.X. Role of p63 in Development, Tumorigenesis and Cancer Progression. Cancer Microenviron. 2012;5:311–322. doi: 10.1007/s12307-012-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X., Keyes W.M., Papazoglu C., Zuber J., Li W., Lowe S.W., Vogel H., Mills A.A. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruan Y., Zeng F., Cheng Z., Zhao X., Fu P., Chen H. High expression of monocarboxylate transporter 4 predicts poor prognosis in patients with lung adenocarcinoma. Oncol. Lett. 2017;14:5727–5734. doi: 10.3892/ol.2017.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meijer T.W., Schuurbiers O.C., Kaanders J.H., Looijen-Salamon M.G., de Geus-Oei L.F., Verhagen A.F., Lok J., van der Heijden H.F., Rademakers S.E., Span P.N., Bussink J. Differences in metabolism between adeno- and squamous cell non-small cell lung carcinomas: spatial distribution and prognostic value of GLUT1 and MCT4. Lung Cancer. 2012;76:316–323. doi: 10.1016/j.lungcan.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Hsu T.I., Wang M.C., Chen S.Y., Yeh Y.M., Su W.C., Chang W.C., Hung J.J. Sp1 expression regulates lung tumor progression. Oncogene. 2012;31:3973–3988. doi: 10.1038/onc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong L., Pan X., Lim C.B., de Polo A., Little J.B., Yuan Z.M. A functional interplay between Δ133p53 and ΔNp63 in promoting glycolytic metabolism to fuel cancer cell proliferation. Oncogene. 2018;37:2150–2164. doi: 10.1038/s41388-017-0117-8. [DOI] [PubMed] [Google Scholar]
- 38.Viticchiè G., Agostini M., Lena A.M., Mancini M., Zhou H., Zolla L., Dinsdale D., Saintigny G., Melino G., Candi E. p63 supports aerobic respiration through hexokinase II. Proc. Natl. Acad. Sci. USA. 2015;112:11577–11582. doi: 10.1073/pnas.1508871112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonaka D. A study of ΔNp63 expression in lung non-small cell carcinomas. Am. J. Surg. Pathol. 2012;36:895–899. doi: 10.1097/PAS.0b013e3182498f2b. [DOI] [PubMed] [Google Scholar]
- 40.Rudnick S.I., Adams G.P. Affinity and avidity in antibody-based tumor targeting. Cancer Biother. Radiopharm. 2009;24:155–161. doi: 10.1089/cbr.2009.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenes Z., Mulhern T.D., Hatters D.M., Ilag L.L., Power B.E., Hosking C., Hensel F., Howlett G.J., Mok Y.F. The anti-cancer IgM monoclonal antibody PAT-SM6 binds with high avidity to the unfolded protein response regulator GRP78. PLoS ONE. 2012;7:e44927. doi: 10.1371/journal.pone.0044927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ovens M.J., Davies A.J., Wilson M.C., Murray C.M., Halestrap A.P. AR-C155858 is a potent inhibitor of monocarboxylate transporters MCT1 and MCT2 that binds to an intracellular site involving transmembrane helices 7-10. Biochem. J. 2010;425:523–530. doi: 10.1042/BJ20091515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bola B.M., Chadwick A.L., Michopoulos F., Blount K.G., Telfer B.A., Williams K.J., Smith P.D., Critchlow S.E., Stratford I.J. Inhibition of monocarboxylate transporter-1 (MCT1) by AZD3965 enhances radiosensitivity by reducing lactate transport. Mol. Cancer Ther. 2014;13:2805–2816. doi: 10.1158/1535-7163.MCT-13-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan X., Rodriguez-Cruz V., Morris M.E. Cellular Uptake of MCT1 Inhibitors AR-C155858 and AZD3965 and Their Effects on MCT-Mediated Transport of L-Lactate in Murine 4T1 Breast Tumor Cancer Cells. AAPS J. 2019;21:13. doi: 10.1208/s12248-018-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polański R., Hodgkinson C.L., Fusi A., Nonaka D., Priest L., Kelly P., Trapani F., Bishop P.W., White A., Critchlow S.E. Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin. Cancer Res. 2014;20:926–937. doi: 10.1158/1078-0432.CCR-13-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curtis N.J., Mooney L., Hopcroft L., Michopoulos F., Whalley N., Zhong H., Murray C., Logie A., Revill M., Byth K.F. Pre-clinical pharmacology of AZD3965, a selective inhibitor of MCT1: DLBCL, NHL and Burkitt’s lymphoma anti-tumor activity. Oncotarget. 2017;8:69219–69236. doi: 10.18632/oncotarget.18215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh C.T., Wu A.T., Chang P.M., Chen K.Y., Yang C.N., Yang S.C., Ho C.C., Chen C.C., Kuo Y.L., Lee P.Y. Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell growth and overcomes drug resistance of lung cancer. Am. J. Respir. Crit. Care Med. 2012;186:1180–1188. doi: 10.1164/rccm.201207-1180OC. [DOI] [PubMed] [Google Scholar]
- 48.Yokoyama W.M., Christensen M., Santos G.D., Miller D. Production of monoclonal antibodies. Current Protocols in Immunology. 2006;74:2.5.1–2.5.25. doi: 10.1002/0471142735.im0205s74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.