Abstract
Background
Lung cancer is the leading cause of cancer-related mortality worldwide, and non-small cell lung cancer (NSCLC) accounts for over 80% of all lung cancers. Serum microRNAs (miRNAs), due to their high stability, have the potential to become valuable noninvasive biomarkers. This present study was aimed to identify the serum miRNAs expression signatures for the diagnosis and prognosis of NSCLC using bioinformatics analysis.
Methods
A total of 12 miRNAs profiling studies have been identified in Pubmed, Gene Expression Omnibus (GEO), and ArreyExpress databases. Differentially expressed miRNAs (DEmiRNAs) were analyzed according to GEO2R online tool and RRA method from R. Then, prediction of DEmiRNAs' target genes from TargetScan, PicTar, miRDB, Tarbase, and miRanda database. Furthermore, we using reverse transcription– quantitative polymerase chain reaction (RT-qPCR) to evaluate the expression levels of DEmiRNAs in serum samples obtained from NSCLC patients and healthy controls. Subsequently, the clinical significance of the tested miRNAs was determined using receiver operating characteristic (ROC) analysis and Cox regression analysis.
Results
A total of 27 DEmiRNAs were identified and 5 of them (miR-1228-3p, miR-1228-5p, miR-133a-3p, miR-1273f, miR-545-3p) were significantly up-regulated and 4 of them (miR-181a-5p, miR-266-5p, miR-361-5p, miR-130a-3p) were significantly down-regulated in NSCLC patients compared with healthy controls. RT-qPCR validated that miR-1228-3p (P =0.006) and miR-181a-5p (P =0.030) were significantly differentially expressed in the serum of NSCLC patients and healthy controls. ROC analysis on miR-1228-3p and miR-181a-5p revealed the area under the curve (AUC) of 0.685 (95% confidence interval [CI], 0.563–0.806; P =0.006) and 0.647 (95% CI, 0.506–0.758; P =0.049). ROC analysis on miR-1228-3p combined miR-181a-5p revealed the AUC of 0.711 (95% CI, 0.593–0.828; P =0.002). Multivariate Cox regression analysis demonstrated that the high serum miR-1228-3p level was an independent factor for the poor prognosis of NSCLC patients.
Conclusions
Serum miR-1228-3p and miR-181a-5p are potential noninvasive biomarkers for the diagnosis and prognosis of NSCLC patients.
1. Background
Lung cancer remains the leading cause of cancer-associated mortality worldwide, of which NSCLC accounts for over 80% of lung cancer-related deaths [1, 2]. Despite improvements in the chemotherapeutic drugs used over time, the 5-year survival rate of NSCLC patients is only 18% [3]. Besides, surgical resection is the most effective treatment for NSCLC, but most newly diagnosed patients are at the onset of advanced or metastatic stages and usually lost the chance for operation. Low-dose computed tomography (LDCT) provides excellent anatomic information in the diagnosis of early NSCLC patients. However, LDCT still have a few limitations including high false-positive rates, potential over-diagnosis, excessive cost and the potential harm related to radiation exposure. Furthermore, the response rates in subsets of NSCLC with tyrosine kinase receptors (mutant EGFR, ALK, and ROS1) were high, drug resistance has been a major challenge [4–6]. Therefore, it is vital to find an early and accurate way to diagnosis and enhance patient's chances to receive proper treatments.
Currently, considerable studies revealed miRNAs as a new opportunity in the field of noninvasive diagnosis. MiRNAs are endogenous 20–25 nucleotides long, have been found to have a profound impact on several biological and pathological processes like cell development, differentiation, proliferation and apoptosis, which play important roles in the carcinogenesis and progression of lung cancer [7, 8]. DEmiRNAs in NSCLC tissue and adjacent nontumor tissues have been reported in a previous study [9]. Circulating miRNAs also could be potential and promising biomarkers for the diagnosis and prognosis of NSCLC. However, the data from different studies are quite variable. Therefore, identification of specific circulating miRNAs reflecting investigated pathological conditions may help to develop novel noninvasive biomarkers and shed a new light on molecular processes involved in cancer and a systematical analysis of miRNA expression signature from multiple platforms and multicenter NSCLC studies is urgently needed.
In this study, due to the presence and stability of cell-free miRNAs have been clearly demonstrated in all body fluid [10, 11], we identified serum and plasma miRNAs related to NSCLC, and then screened and validated miR-1228-3p and miR-181a-5p expression level in the serum of NSCLC patients in comparison to serum of healthy volunteers.
2. Methods
2.1. Data Collection
Up to January 1, 2018, a total of 3 databases including Pubmed (http://www.ncbi.nlm.nih.gov/), GEO (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) were used for literature retrieval, and the search terms were (miR-∗ OR miRNA OR microRNA) AND (lung AND (tumor OR cancer OR carcinoma)). The selection criteria for the literature were: miRNAs detection was microarray or miRNAs sequencing; studies were published in English; patients had pathologically confirmed NSCLC; patients had no history of other cancers; none of the patients received preoperative treatment, such as radiotherapy or chemotherapy; control group was healthy normal controls; the experimental samples were derived from serum or plasma.
2.2. Identification of DEmiRNAs
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is a web tool for screening DEmiRNAs by comparing two groups of samples. The procedure of GEO2R is the following: firstly, enter a series accession number in the box. Then, click “Define groups” and enter names (NSCLC and healthy control) for the groups of samples you plan to compare. After samples have been assigned to groups, click “Top 250” to run the test with default parameters. To see more than the top 250 results, or if you want to save the results, the complete results table may be downloaded using the “Save all results” button. The cut-off criterion was set as the P <0.05 and absolute fold change (FC) >1.5. In addition, the R package ggplot2 package (version 2.2.1, https://cran.r-project.org/web/packages/ggplot2) was used to perform the volcano plots of all the miRNAs among 12 miRNAs profiling. Moreover, heat maps for the DEmiRNAs was generated using the pheatmap package (version 1.0.8, https://cran.r-project.org/web/packages/pheatmap). For some literatures that did not find original data, we used the miRNAs data listed in the paper or miRNAs information in supplementary data for analysis. All miRNAs names are standardized through miRBase.
2.3. Target Gene Prediction and Functional Enrichment Analysis
Target genes of DEmiRNAs were predicted by 4 different online databases including TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), miRanda (http://www.miranda-im.org/) and miRDB (http://mirdb.org/). The target genes were screened by the intersection of TargetScan, PicTar, miRanda and miRDB. Then TarBase (http://www.microrna.gr/tarbase/) was used to validate the target genes. Then all of the target genes were sorted from the union of the front genes and the validation genes. Venn Diagram package (version 1.6.17, https://cran.r-project.org/web/packages/VennDiagram/) were applied to identify the overlapping target genes of DEmiRNAs among 12 miRNAs profiling. Furthermore, GeneCodis web tool (http://genecodis.cnb.csic.es) was used to function enrichment analysis [12–14]. The resulting gene list was submitted to GeneCodis in order to identify the targeted pathway, threshold of FDR was 0.05 and considering enrichment in Panther and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
2.4. Patients and Samples
For the current study, we recruited 50 patients with first diagnosis of NSCLC and previously untreated from the Qilu Hospital of Shandong University, from July 2017 to December, 2017. Moreover, control group consist of thirty healthy volunteers (well matched to the patients according to age and gender) was screened from the Qilu Hospital of Shandong University. This study was approved by the Ethics Committee of Qilu Hospital of Shandong University (KYLL-2013-097; 25 February 2014), and written informed consent was obtained from all patients or their guardians. Once the patient was diagnose with NSCLC, about 5 ml of peripheral blood was collected in a sterile tube without anticoagulant before any treatment was performed, and allowed to stand at room temperature for 30 – 60 min to clot, then samples were centrifuged at 4000 rpm for 15 min at the room temperature and for another 10 min at 12000 rpm at 4°C to completely remove the cell debris. Finally, the resultant serum was stored at -80°C, samples with visual evidence of hemolysis were excluded from further analysis.
2.5. Total miRNA Isolation and miRNAs Expression Analysis by RT-qPCR
Total miRNA was extracted from 200 μL of serum samples using miRcute serum/plasma miRNA isolation kit (Tiangen Biotech) according to the manufacturer's protocol. The concentration of miRNA was measured using the NanoDLite. According to the manufacturer's instructions, miRNA profiling was performed with RT-qPCR instrument StepOnePlus™ Real—Time PCR System (Thermo Fisher Scientific) using miDETECT A TrackTM miRNART- qPCR Starter Kit (RiboBio). The primers of these miRNAs and cel-miR-39 were obtained from RiboBio Corporation (Guangzhou, China). After the reactions, the ΔCt values were determined. The fold change of each miRNA expression was calculated using the 2-ΔΔCT method [15].
2.6. Statistical Analysis
The serum miRNA level was expressed as 2-ΔΔCT to maintain the normal distribution of the parameter and assure a positive correlation with the miRNA level of expression and student's t test was used to analyze miRNA expression level. Mann–Whitney tests were used to check associations between miRNA expression levels and clinicopathological features of the patients. The survival rates were estimated by the Kaplan-Meier analysis and the significance of differences was examined by log-rank test. We also performed overall survival (OS) to investigate survival outcome. OS was defined as the time between the date of surgery and the date of death or last followup. The diagnostic performance of miRNAs was assessed by the ROC curve analysis and calculated the AUC to evaluate the predictive power of candidate miRNAs for NSCLC. Multivariate analysis of the prognostic factors was performed with Cox regression model. Data was presented as mean ± standard deviation (SD) and P <0.05 were considered statistically significant. All statistical analysis was performed using the SPSS version 20.0 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
3. Results
3.1. Identification of DEmiRNAs
The general overview of the study design is shown in Figure 1. According to the selection criteria, 12 full-text studies were retrieved from July 2009 to January 2018 (Table 1) [11–18]. All these 12 studies including 413 NSCLC patients and 513 healthy controls were used to screen miRNA signature and a total of 2381 significantly up-expressed and 513 significantly down-expressed miRNAs were extracted. Then 5 GEO datasets (GSE16512, GSE70080, GSE93300, GSE94536 and GSE46729) were used to perform volcano maps and to get the DEmiRNAs (P <0.05 and absolute FC >1.5, Figure 2). According to RRA method 5 up-regulated miRNAs and 27 down-regulated miRNAs were screened (FDR<0.05). Finally we selected 9 DEmiRNAs including the top 4 down-regulated miRNAs (hsa-miR-181a-5p, hsa-miR-26b-5p, hsa-miR-361-5p and hsa-miR-130a-3p) and all 5 up-regulated miRNAs (hsa-miR-1228-3p hsa-miR-1228-5p hsa-miR-133a-3p hsa-miR-1273f hsa-miR-545-3p) for the further study based on the FDR value.
Figure 1.

The general overview of study design.
Table 1.
The basic information of the 12 studies.
| Study | References | Region | MiRNA numbers | Tumor type | Sample resource | Sample numbers | Time | Database |
|---|---|---|---|---|---|---|---|---|
| 1 | Lodes [12] | North America | 547 | Various | Serum | 2 NSCLC 14 normal |
2009.07 | Pubmed ArrayExpress GSE16512 |
| 2 | Wang [13] | China | 427 | Various | Serum | 88 NSCLC 17 normal |
2011.06 | Pubmed |
| 3 | Foss [14] | Italy | 880 | Various | Serum | 11 NSCLC 11 normal |
2011.03 | Pubmed |
| 4 | Roth [16] | Germany | 1158 | Various | Serum | 21 NSCLC 11 normal |
2012.06 | Pubmed |
| 5 | Rani [17] | Ireland | 667 | ADC | Serum | 40 NSCLC 40 normal |
2013.12 | Pubmed |
| 6 | Hu [18] | China | 723 | Various | Plasma | 73 NSCLC 34 normal |
2014.02 | Pubmed |
| 7 | Wang [19] | China | 754 | Various | Serum | 31 NSCLC 31 normal |
2015.08 | Pubmed |
| 8 | Nadal [20] | North America | 334 | Various | Serum | 70 NSCLC 22 normal |
2015.06 | Pubmed |
| 9 | Halvorsen AR | Norway | 272 | Various | Serum | 38 NSCLC 16 normal |
2016.10 | GSE70080 |
| 10 | Qu LL | China | 5915 | ADC | Plasma | 9 ADC 4 normal |
2017.01 | GSE93300 |
| 11 | Liu X | China | 5915 | Various | Plasma | 6 NSCLC 3 normal |
2017.02 | GSE94536 |
| 12 | Xu ZL | USA | 7815 | Various | NSCLC serum Normal plasma |
24 NSCLC 24 normal |
2017.05 | GSE46729 |
Figure 2.
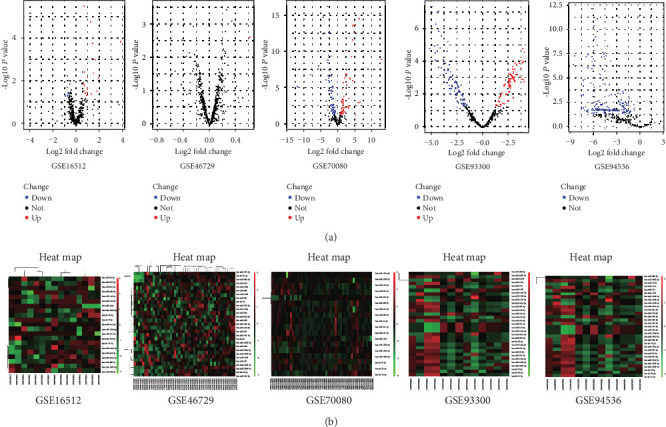
Data analysis of 5 GEO chips. (a) Volcano plots of the different miRNA expression analysis. x-axis: log 2 fold change; y-axis: -log10 p-value for each probes. (b) Two-dimensional hierarchical clustering of 27 DEmiRNA s in all samples. MiRNAs are in rows; samples are in columns.
3.2. Target Gene Prediction and Functional Enrichment Analysis
TargetScan, miRanda, miRDB and PicTar were used to predict target genes of 9 DEmiRNAs, overlap predicated target genes from 4 databases (Figure 3) and Tarbase was used to validate target genes. Then, the overlapped genes plus validated genes was defined as target genes. Furthermore, removing 108 repeated target genes, we gained a total of 8002 target genes of DEmiRNAs (Table 2). 8002 target genes were used to perform functional enrichment analysis. The gene ontology (GO) analysis showed that these target genes were mainly involved in the regulation of transcription, DNA-dependent (GO:0006355), Nucleus (GO:0005634) and Protein binding (GO:0005515) (Table 3). Further Panther and KEGG pathway analysis were performed to investigate the significance of target genes in the development of NSCLC, the results showed that these genes were significantly enriched in Pathways in cancer and Wnt signaling pathway (Figure 4).
Figure 3.

Overlap of 9 DEmiRNAs target gene prediction using 4 miRNA prediction databases.
Table 2.
Target gene prediction of 9 DEmiRNAs.
| 4 target gene prediction databases | 1 verified database | Target gene numbers | |||||
|---|---|---|---|---|---|---|---|
| miRNA | TargetScan | Pic tar | mirDB | miRanda | Overlap | Tarbase | Union |
| miR-1228-3p | 4529 | 224 | 214 | 7103 | 51 | 3 | 54 |
| miR-1228-5p | 1360 | 139 | 34 | 3218 | 5 | 15 | 20 |
| miR-133a-3p | 571 | 339 | 310 | 5314 | 85 | 298 | 383 |
| miR-1273f | 5400 | 274 | 401 | 1445 | 3 | 0 | 3 |
| miR-545-3p | 5435 | 472 | 794 | 4678 | 48 | 499 | 547 |
| miR-181a-5p | 1365 | 508 | 887 | 7846 | 186 | 1884 | 1884 |
| miR-26b-5p | 1042 | 543 | 508 | 7465 | 145 | 2949 | 2949 |
| miR-361-5p | 295 | 218 | 340 | 5615 | 41 | 685 | 726 |
| miR-130a-3p | 1026 | 578 | 652 | 7751 | 173 | 1543 | 1544 |
| Total | 8110 | ||||||
Table 3.
GO analysis of target genes.
| Function enrichment | FDR | Target |
|---|---|---|
| Go biological process (BP) | ||
| GO:0006355: Regulation of transcription, DNA-dependent | 4.51E-119 | 621 |
| GO:0045893: Positive regulation of transcription, DNA-dependent | 7.41E-46 | 204 |
| GO:0045944: Positive regulation of transcription from RNA polymerase II promoter | 3.83E-45 | 232 |
| GO:0006915: Apoptotic process | 3.61E-41 | 229 |
| GO:0045892: Negative regulation of transcription, DNA-dependent | 9.24E-39 | 174 |
| GO:0000122: Negative regulation of transcription from RNA polymerase II promoter | 1.65E-37 | 176 |
| GO:0007165: Signal transduction | 2.73E-36 | 353 |
| GO:0010467: Gene expression | 5.03E-34 | 168 |
| GO:0006468: Protein phosphorylation | 2.91E-33 | 164 |
| GO:0044419: Interspecies interaction between organisms | 1.12E-32 | 144 |
| Go cellular component(CC) | ||
| GO:0005634: Nucleus | 0 | 2024 |
| GO:0005737: Cytoplasm | 0 | 1859 |
| GO:0005634: GO:0005737: Nucleus, cytoplasm | 5.851E-210 | 975 |
| GO:0005829: Cytosol | 5.38E-155 | 813 |
| GO:0005730: Nucleolus | 5.43E-134 | 606 |
| GO:0005634: GO:0005730: Nucleus, nucleolus | 2.29E-129 | 584 |
| GO:0005737: GO:0005829: Cytoplasm, cytosol | 9.11E-123 | 606 |
| GO:0005622: Intracellular | 1.24E-108 | 685 |
| GO:0016020: Membrane | 9.19E-97 | 1104 |
| GO:0005654: Nucleoplasm | 1.86E-95 | 394 |
| Go molecular function(MF) | ||
| GO:0005515: Protein binding | 0 | 1770 |
| GO:0046872: Metal ion binding | 9.92E-150 | 975 |
| GO:0000166: Nucleotide binding | 1.10E-134 | 771 |
| GO:0003677: DNA binding | 7.10E-134 | 688 |
| GO:0008270: Zinc ion binding | 1.24E-115 | 693 |
| GO:0046872: GO:0008270: Metal ion binding, zinc ion binding | 3.22E-108 | 648 |
| GO:0005524: ATP binding | 8.41E-97 | 550 |
| GO:0005515: GO:0000166: Protein binding, nucleotide binding | 6.55E-95 | 363 |
| GO:0005524: GO:0000166: ATP binding, nucleotide binding | 4.73E-89 | 506 |
| GO:0003677: GO:0005515: DNA binding, protein binding | 8.64E-80 | 278 |
Figure 4.
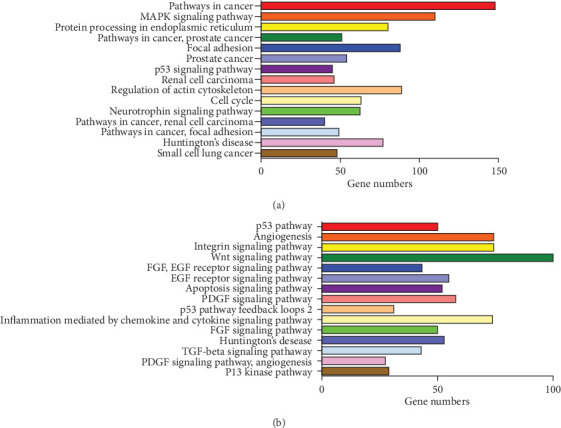
Target gene networks identified through KEGG (a) and pather (b) pathway analysis.
3.3. Association between Expression Levels of Serum miRNAs and Clinicopathological Characteristics
Among the 50 NSCLC patients, there were 34 males and 16 females, 37 adenocarcinomas (ADCs) and 13 squamous cell carcinomas (SCCs), 14 patients are at stage I, 7 at stage II, 10 at stage III and 19 at stage IV. The clinicopathological characteristics of patients with NSCLC and healthy volunteers are presented in Table 4.
Table 4.
The demographic and clinical features of the study.
| Characteristic | Healthy control (n =30) | NSCLC (n =50) | P value |
|---|---|---|---|
| Age (years) | 62 ± 7 | 62 ± 9 | 0.292 |
| Gender | 0.481 | ||
| Male | 20 | 34 | |
| Female | 10 | 16 | |
| Smoking status | 0.146 | ||
| Nonsmokers | 20 | 25 | |
| Smokers | 10 | 25 | |
| Type of NSCLC tissue | — | ||
| SCC | — | 13 | |
| ADC | — | 37 | |
| Size of NSCLC | — | ||
| Diameter < =3 cm | — | 28 | |
| Diameter>3 cm | — | 22 | |
| Lymph node metastasis | — | ||
| Yes | — | 22 | |
| No | — | 28 | |
| TNM stage | — | ||
| I | — | 14 | |
| II | — | 7 | |
| III | — | 10 | |
| IV | — | 19 |
The current study revealed that miR-1228-3p, miR-133a-3p and miR-545-3p were significantly up-regulated (P =0.006, P =0.043 and P =0.047, respectively), while miR-181a-5p and miR-361-5p were significantly down-regulated (P =0.029 and P =0.006) in NSCLC patients compared with healthy controls (Figure 5(a)-5(e)). Among NSCLC patients, miR-1228-3p expression level (P =0.009) in ADC patients was higher compared with healthy controls, while the expression levels of miR-181a-5p (P =0.031) and miR-361-5p (P =0.006) were lower than healthy controls (Figure 5(f)-5(h)). In SCC patients, miR-545-3p expression level (P =0.034) was higher compared with healthy controls (Figure 5(i)).
Figure 5.

(a-e) The relative expression of 5 DEmiRNAs in NSCLC group and normal control group. (f-h) The expression levels of 3 DEmiRNAs in ADC group and normal control group. (i) The expression level of 1 DEmiRNA in SCC group and normal control group. ∗P<0.05, ∗∗P<0.01.
As for TNM stage, the expression level of miR-181a-5p and were significantly lower than healthy controls in TNM stage I (P =0.028), as well as miR-361-5p in both TNM stage III (P =0.016) and IV (P =0.048). On the contaray, the expression level of miR-1228-3p in TNM stage III (P =0.007) and IV (P =0.026) were significantly higher compared with healthy controls, as well as miR-545-3p in TNM stage IV (P =0.013) (Figure 6(a)). When the tumor diameter < =3 cm, the expression levels of miR-1228-3p, miR-133a-3p and miR-545-3p were significantly higher compared with healthy controls (P =0.020, P =0.005 and P =0.037) and miR-1228-3p expression level in tumor diameter>3 cm group was significantly higher compared with healthy controls (P = 0.017). In contrast, the expression level of miR-361-5p in tumor diameter>3 cm group was significantly lower compared with healthy controls (P =0.006) (Figure 6(b)). The high expression level of miR-1228-3p and low expression level of miR-181a-5p were related to lymph node metastasis (P =0.005, P =0.003, respectively), while the high expression level of miR-545-3p was related to no lymph node metastasis (P =0.045). As for miR-361-5p, its expression level in no lymph node metastasis group was lower compared with healthy controls (P =0.027), as well as lymph node metastasis group compared with healthy controls (P =0.015) (Figure 6(C)).
Figure 6.

(a) The relative expression level of miRNAs between different TNM stages of NSCLC group and normal control group; (b) The relative expression level of miRNAs between normal control group and NSCLC with different tumor size; (c) The relative expression level of miRNA between normal control group and NSCLC group with or without lymph node metastasis. ∗P<0.05, ∗∗P<0.01.
3.4. Diagnostic Value of Serum miRNAs NSCLC Patients
ROC curve analysis was used to investigate the diagnostic value of miR-1228-3p, miR-133a-3p, miR-545-3p, miR-181a-5p and miR-361-5p in distinguishing NSCLC patients from normal controls. Expression levels of 5 serum miRNAs were measured from NSCLC patients and healthy controls. The diagnostic relevance of each miRNA, both single and combination, were analyzed (Table 5). The results showed that ROC analysis revealed the AUC for miR-1228-3p was 0.685 (95% confidence interval [CI], 0.563-0.806; P =0.006), for miR-133a -3p was 0.636 (95% CI, 0.512-0.760; P =0.043), for miR-545-3p was 0.635 (95% CI, 0.514-0.756; P =0.045), for miR-181a-5p was 0.647 (95% CI, 0.506-0.758; P =0.049) and the AUC for miR-361-5p was 0.635 (95% CI, 0.508-0.761; P =0.045). ROC analysis indicated that the combination of miR-1228-3p and miR-181a-5p provided best diagnostic discriminant with an AUC of 0.711(95% CI 0.593-0.828; P =0.002).
Table 5.
ROC curve analysis of differential miRNAs.
| miRNAs | AUC | P value | 95% CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| miR-1228-3p | 0.685∗ | 0.006 | 0.563 | 0.806 |
| miR-133a-3p | 0.636∗ | 0.043 | 0.512 | 0.760 |
| miR-545-3p | 0.635∗ | 0.045 | 0.514 | 0.756 |
| miR-181a-5p | 0.647∗ | 0.049 | 0.506 | 0.758 |
| miR-361-5p | 0.635∗ | 0.045 | 0.508 | 0.761 |
| miR-1228-3p + miR-133a-3p | 0.615 | 0.087 | 0.490 | 0.739 |
| miR-1228-3p + miR-545-3p | 0.622 | 0.069 | 0.500 | 0.744 |
| miR-1228-3p + miR-181a-5p | 0.711∗ | 0.002 | 0.593 | 0.828 |
| miR-1228-3p + miR-361-5p | 0.651∗ | 0.025 | 0.520 | 0.781 |
| miR-133a-3p + miR-545-3p | 0.705∗ | 0.002 | 0.592 | 0.818 |
| miR-133a-3p + miR-181a-5p | 0.679∗ | 0.008 | 0.554 | 0.804 |
| miR-133a-3p + miR-361-5p | 0.637∗ | 0.041 | 0.510 | 0.764 |
| miR-545-3p + miR-181a-5p | 0.585 | 0.207 | 0.460 | 0.710 |
| miR-545-3p + miR-361-5p | 0.611 | 0.097 | 0.485 | 0.738 |
| miR-181a-5p + miR-361-5p | 0.646∗ | 0.030 | 0.520 | 0.772 |
| miR-1228-3p + miR-133a-3p + miR-545-3p | 0.698∗ | 0.003 | 0.585 | 0.811 |
| miR-1228-3p + miR-133a-3p + miR-181a-5p | 0.661∗ | 0.017 | 0.535 | 0.787 |
| miR-1228-3p + miR-133a-3p + miR-361-5p | 0.616 | 0.084 | 0.489 | 0.743 |
| miR-1228-3p + miR-545-3p + miR-181a-5p | 0.588 | 0.19 | 0.463 | 0.713 |
| miR-1228-3p + miR-545-3p + miR-361-5p | 0.609 | 0.105 | 0.483 | 0.735 |
| miR-1228-3p + miR-181a-5p + miR-361-5p | 0.647∗ | 0.029 | 0.517 | 0.776 |
| miR-133a-3p + miR-545-3p + miR-181a-5p | 0.609 | 0.103 | 0.486 | 0.733 |
| miR-133a-3p + miR-545-3p + miR-361-5p | 0.643∗ | 0.033 | 0.519 | 0.768 |
| miR-133a-3p + miR-181a-5p + miR-361-5p | 0.663∗ | 0.015 | 0.538 | 0.789 |
| miR-545-3p + miR-181a-5p + miR-361-5p | 0.605 | 0.116 | 0.475 | 0.736 |
| miR-1228-3p + miR-133a-3p + miR-545-3p + miR-181a-5p |
0.614 | 0.089 | 0.491 | 0.737 |
| miR-1228-3p + miR-133a-3p + miR-545-3p + miR-361-5p |
0.640∗ | 0.037 | 0.516 | 0.764 |
| miR-1228-3p + miR-133a-3p + miR-181a-5p + miR-361-5p |
0.663∗ | 0.015 | 0.538 | 0.789 |
| miR-1228-3p + miR-545-3p + miR-181a-5p + miR-361-5p |
0.607 | 0.112 | 0.477 | 0.737 |
| miR-133a-3p + miR-545-3p + miR-181a-5p + miR-361-5p |
0.623 | 0.067 | 0.493 | 0.753 |
| miR-1228-3p + miR-133a-3p + miR-545-3p + miR-181a-5p + miR-361-5p |
0.623 | 0.067 | 0.493 | 0.753 |
Note: ∗P <0.05, ∗∗P <0.01.
3.5. Associations of Serum miRNAs Expression Levels with OS
To explore whether serum miRNAs expression levels will affect the clinical outcomes, we constructed a prognostic classifier using Kaplan-Meier analysis on 50 NSCLC patients. It showed that miR-1228-3p and miR-181a-5p expression levels were significantly associated with the OS of NSCLC patients (both P =0.041) (Figure 7). As for miR-133a -3p, miR-545-3p and miR-361-5p, the expression levels of all these 3 miRNAs have no significance with OS statistically (P =0.236, P =0.709, P =0.199, respectively). The median OS in miR-133a -3p, miR-545-3p and miR-361-5p low expression group were both 8 months whereas in high expression group were all 7 months. The multivariate Cox hazard regression analysis demonstrated that expression level of serum miR-1228-3p were an independent prognostic indicator of NSCLC (hazard ratio(HR) 1.487, 95% CI 1.130-1.958; P =0.005).
Figure 7.
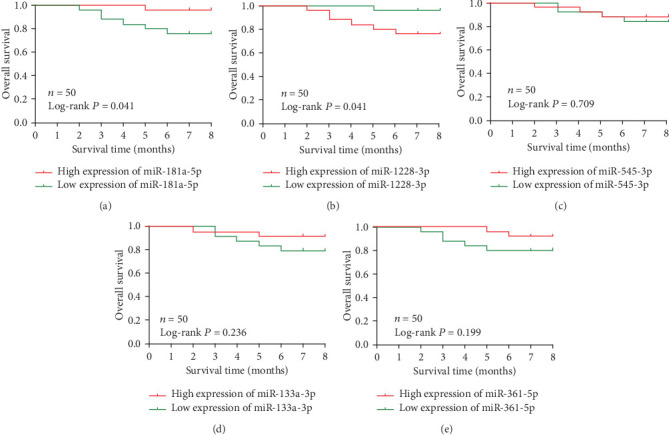
Kaplan-Meier survival curves by different miRNA expression levels of of miR-181a-5p, miR-1228-3p, miR-545-3p, miR-133a -3p and miR-361-5p in an independent NSCLC chort. (a) OS between low and high miR-181a-5p expression. (b) OS between low and high miR-1228-3p expression. (c) OS between low and high miR-545-3p expression. (d) OS between low and high miR-133a -3p Aexpression. (e) OS between low and high miR-361-5p expression.
4. Discussion
In the current study, we integrated expression profiles of 413 NSCLC patients and 513 healthy controls in 5 datasets from GEO database and identified a panel of 32 DEmiRNAs. According to FDR value, we finally identified 9 DEmiRNAs for further study. Then we used 5 online databases and screened a total of 8002 target genes of these 9 DEmiRNAs, functional enrichment analysis showed that these target genes were mainly involved in the regulation of transcription, DNA-dependent, Nucleus, Protein binding and significantly enriched in Pathways in cancer, especially in Wnt signaling pathway. The high expression levels of miR-1228-3p, miR-133a-3p and miR-545-3p and low expression levels of miR-181a-5p and miR-361-5p were also validated via an independent NSCLC cohort from Qilu Hospital of Shandong University. The result indicated that the expression level of miR-1228-3p was related to TNM stage, tumor diameter and lymph node metastasis, the expression level of miR-545-3p was related with TNM stage and tumor diameter, the expression levels of miR-181a-5p and miR-361-5p were related to TNM stage and lymph node metastasis, the expression level of miR-133a-3p was related with tumor diameter only. Furthermore, the expression levels of miR-1228-3p and miR-181a-5p were significantly associated with the OS of NSCLC patients.
The incidence of lung cancer is the leading factor in malignant tumors. Up to date, the gold standard in diagnosing NSCLC is pathologic evidence of malignant cells, which typically requires a surgical procedure or an invasive examination. It is mostly at advanced stage as long as lung cancer is diagnosed. The 5-year survival rate of advanced lung cancer is less than 20%, but the 5-year survival rate of stage IA lung cancer can reach 60% [21]. Early diagnosis is the key strategy to improve the outcome of lung cancer. Current methods including CEA level and CT screening cannot predict the risk of NSCLC for patients who have small lung nodules accurately. Therefore, specific and sensitive biomarkers for the detection of malignancies are urgently required to reduce the worldwide morbidity and mortality caused by NSCLC.
MiRNAs have been identified as potential biomarkers for lung cancer, it can be used to evaluate the invasion, metastasis, treatment response and prognosis of cancer. Although tumor sample miRNAs have been demonstrated to be associated with the development of tumors in many studies, it is difficult to obtain tumor samples in clinical practice. Recent studies have supported that circulating miRNAs have potential diagnostic effects for NSCLC. Studies [22, 23] have shown that there is a significant difference between serum miRNAs and blood cell miRNAs in patients with lung cancer, and blood cells can affects the detection rate of whole blood miRNAs [18, 24]. So that we choose serum miRNAs as the source of hematology of the subjects. Numerous circulating miRNA signatures have been reported for the detection of NSCLC, but the miRNAs signature identified by different groups vary from one another because of the inconsistencies platforms, it is necessary to find a better way to screen different miRNAs. The RRA approach is as good way to eliminate differences among various platforms, by which reordered miRNAs according to the FDR value.
MiR-1228 is located in the LRP1 gene on chromosome 12. This gene is mainly involved in basic metabolism and cell structure, which is a key component of maintaining cell survival [25]. There are many researches of miR-1228-3p in various diseases. It has been reported that miR-1228-3p expression level was involved in drug resistant of breast cancer, chronic heart failure, endometrial carcinoma, it can be expressed steadily in the prostate cancer, colorectal cancer and secretions of hepatocellular carcinoma [26–31]. There are another two studies about miR-1228-3p on NSCLC. One is about miR-1228-3p differentially expressed in NSCLC exocrine [32] and another suggested that miR-1228-3p can be used as an endogenous reference gene [24]. It means that miR-1228-3p can be stable in the exocrine and circulatory and further confirms that it can be released to the cell through the exocrine.
The miR-181 family includes miR-181a, miR-181b, miR-181c and miR-181d, contains the same seed sequence, which can display the functional redundancy of the gene in mRNA [33]. The role of miR-181a-5p as a tumor suppressor has been confirmed in previous studies. For example, lower expression level of miR-181a-5p was associated with a worse survival rate in colorectal cancer [34]. In gastric cancer and lung cancer, the expression of miR-181a-5p through target BCL2 increased the sensitivity of cancer cells to cisplatin and vincristine, which further induced the apoptosis of cancer cells [35]. In addition, miR-181a-5p can reduce the metastasis in breast and colon cancer cells [34]. All the results suggested that miR-181a-5p can affect the survival, invasion and metastasis of tumor cells, and even the therapeutic response to chemotherapeutic drugs, while the further role of miR-181a-5p in NSCLC remains to further explore.
5. Conclusion
In conclusion, our study indicated that miR-181a-5p play an important role in the early diagnosis of NSCLC and the combined expression levels of miR-1228-3p and miR-181a-5p have certain diagnosis efficancy for NSCLC. Furthermore, high expression level of miR-1228-3p and low expression level of miR-181a-5p have a shorter survival time, which indicated that miR-1228-3p and miR-181a-5p can be used as noninvasive diagnostic and prognostic biomarkers for NSCLC. However, it is vital to conduct more in-depth studies to explore the molecular roles of serum miR-1228-3p and miR-181a-5p in the future.
Acknowledgments
This work was supported by grants from the Major Scientific and Technological Innovation Project of Shandong Province (2018CXGC1212), the Science and Technology Foundation of Shandong Province (2014GSF118084 and 2016GSF121043), the Medical and Health Technology Innovation Plan of Ji'nan City (201805002) and the National Natural Science Foundation of China (81372333).
Abbreviations
- NSCLC:
Non-small cell lung cancer
- miRNAs:
microRNAs
- GEO:
Gene expression omnibus
- DEmiRNAs:
Differentially expressed miRNAs
- RRA:
Robust rank Aggreg
- RT-qPCR:
Reverse transcription- quantitative polymerase chain reaction
- ROC:
Receiver operating characteristic
- AUC:
Area under the curve
- CI:
Confidence interval
- LDCT:
Low-dose computed tomography
- KEGG:
Kyoto encyclopedia of genes and genomes
- OS:
Overall survival
- SD:
Standard deviation
- ADC:
Adenocarcinoma
- SCC:
Squamous cell carcinoma
- HR:
Hazard ratio.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
This study has been approved by the Ethics Committee of Qilu Hospital of Shandong University (KYLL-2013-097; 25 February 2014), and written informed consent was obtained from all patients or their guardians.
Conflicts of Interest
The authors declare that they have no conflicts of interests.
Authors' Contributions
Wei-Xiao Xue and Meng-Yu Zhang designed the research, performed the experiments, analyzed the data, and wrote the paper; Rui Li provided the patient samples; Xiao Liu and Yun-Hong Yin performed the data analysis and interpreted the data; Yi-Qing Qu assisted with the study design and revised the manuscript. All authors read and approved the final manuscript. Wei-Xiao Xue and Meng-Yu Zhang contributed equally to this work.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: a Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: a Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2016. CA: a Cancer Journal for Clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Rotow J., Bivona T. G. Understanding and targeting resistance mechanisms in NSCLC. Nature Reviews. Cancer. 2017;17(11):637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 5.McCoach C. E., Le A. T., Gowan K., et al. Resistance mechanisms to targeted therapies inROS1+andALK+Non–small cell lung cancer. Clinical Cancer Research. 2018;24(14):3334–3347. doi: 10.1158/1078-0432.CCR-17-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaishnavi A., Schubert L., Rix U., et al. EGFR mediates responses to small-molecule drugs targeting oncogenic fusion kinases. Cancer Research. 2017;77(13):3551–3563. doi: 10.1158/0008-5472.CAN-17-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C., Yin Y., Liu X., Xi X., Xue W., Qu Y. Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget. 2017;8(15):24564–24578. doi: 10.18632/oncotarget.15596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Research. 2012;40(W1):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell P. S., Parkin R. K., Kroh E. M., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turchinovich A., Samatov T. R., Tonevitsky A. G., Burwinkel B. Circulating miRNAs: cell-cell communication function? Frontiers in Genetics. 2013;4:p. 119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lodes M. J., Caraballo M., Suciu D., Munro S., Kumar A., Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4(7, article e6229) doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z. X., Bian H. B., Wang J. R., Cheng Z. X., Wang K. M., de W. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. Journal of Surgical Oncology. 2011;104(7):847–851. doi: 10.1002/jso.22008. [DOI] [PubMed] [Google Scholar]
- 14.Foss K. M., Sima C., Ugolini D., Neri M., Allen K. E., Weiss G. J. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. Journal of Thoracic Oncology. 2011;6(3):482–488. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 15.Livak K. J., Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔ _C_T Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Roth C., Stückrath I., Pantel K., Izbicki J. R., Tachezy M., Schwarzenbach H. Low levels of cell-free circulating miR-361-3p and miR-625∗ as blood-based markers for discriminating malignant from benign lung tumors. PLoS One. 2012;7(6, article e38248) doi: 10.1371/journal.pone.0038248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rani S., Gately K., Crown J., O’Byrne K., O’Driscoll L. Global analysis of serum microRNAs as potential biomarkers for lung adenocarcinoma. Cancer Biology & Therapy. 2014;14(12):1104–1112. doi: 10.4161/cbt.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J., Wang Z., Liao B.-Y., et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. International Journal of Cancer. 2014;135(5):1187–1194. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]
- 19.Wang C., Ding M., Xia M., et al. A Five-miRNA Panel Identified From a Multicentric Case-control Study Serves as a Novel Diagnostic Tool for Ethnically Diverse Non-small-cell Lung Cancer Patients. eBioMedicine. 2015;2(10):1377–1385. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadal E., Truini A., Nakata A., et al. A novel serum 4-microRNA signature for lung Cancer detection. Scientific Reports. 5(1) doi: 10.1038/srep12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z., Chen X., Zhao Y., et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. Journal of Clinical Oncology. 2010;28(10):1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 22.Nogales-Cadenas R., Carmona-Saez P., Vazquez M., et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Research. 2009;37(Web Server):W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmona-Saez P., Chagoyen M., Tirado F., Carazo J. M., Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biology. 2007;8(1):p. R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q., Zeng H., Ye P., Shi Y., Guo J., Long X. Differential microRNA profiles between fulvestrant-resistant and tamoxifen-resistant human breast cancer cells. Anti-Cancer Drugs. 29(6):539–548. doi: 10.1097/CAD.0000000000000623. [DOI] [PubMed] [Google Scholar]
- 25.Cakmak H. A., Coskunpinar E., Ikitimur B., et al. The prognostic value of circulating microRNAs in heart failure. Journal of Cardiovascular Medicine (Hagerstown, Md.) 2015;16(6):431–437. doi: 10.2459/JCM.0000000000000233. [DOI] [PubMed] [Google Scholar]
- 26.Torres A., Torres K., Pesci A., et al. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. International Journal of Cancer. 2013;132(7):1633–1645. doi: 10.1002/ijc.27840. [DOI] [PubMed] [Google Scholar]
- 27.Zhao H., Ma T. F., Lin J., et al. Identification of valid reference genes for mRNA and microRNA normalisation in prostate cancer cell lines. Scientific Reports. 2018;8(1):p. 1949. doi: 10.1038/s41598-018-19458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danese E., Minicozzi A. M., Benati M., et al. Reference miRNAs for colorectal cancer: analysis and verification of current data. Scientific Reports. 2017;7(1):p. 8413. doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornari F., Ferracin M., Trerè D., et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 10(10):p. e0141448. doi: 10.1371/journal.pone.0141448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giallombardo M., Borrás J. C., Castiglia M., et al. Exosomal miRNA analysis in Non-small cell lung Cancer (NSCLC) Patients' plasma through qPCR: a feasible liquid biopsy tool. Journal of Visualized Experiments. 2016;111(111):p. 53900. doi: 10.3791/53900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor M. A., Sossey-Alaoui K., Thompson C. L., Danielpour D., Schiemann W. P. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. The Journal of Clinical Investigation. 2013;123(1):150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichler M., Winter E., Ress A. L., et al. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. Journal of Clinical Pathology. 2014;67(3):198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W., Shan X., Wang T., Shu Y., Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. International Journal of Cancer. 2010;127(11):2520–2529. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Kuscu C., Banach A., et al. miR-181a-5p inhibits Cancer cell migration and angiogenesis via Downregulation of matrix Metalloproteinase-14. Cancer Research. 2015;75(13):2674–2685. doi: 10.1158/0008-5472.CAN-14-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikh A., Lee C., Joseph P., et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nature Communications. 5(1) doi: 10.1038/ncomms3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


