Abstract
Currently, there exists no accurate, non-invasive clinical imaging method to detect living bacteria in vivo. Our goal is to provide a positron emission tomography (PET) method to image infection by targeting bacteria-specific metabolism. Standard of care methodologies either detect morphologic changes, image immunologic response to infection, or employ invasive tissue sampling with associated patient morbidity. These strategies, however, are not specific for living bacteria and are often inadequate to detect bacterial infection during fever workup. As such, there is an unmet clinical need to identify and validate new imaging tools suitable for non-invasive, in vivo (PET) imaging of living bacteria. We have shown that D-[methyl-11C]methionine (D-[11C]Met) can distinguish active bacterial infection from sterile inflammation in a murine infection model, and is sensitive to both gram-positive and gram-negative bacteria. Here we report an automated and >99% enantiomeric excess (ee) synthesis of D-[11C]Met from a linear D-homocysteine precursor, a significant improvement over the previously reported synthesis utilizing a D-homocysteine thiolactone hydrochloride precursor with approximately 75–85% ee. Furthermore, we took additional steps towards applying D-[11C]Met to infected patients. D-[11C]Met was subject to a panel of clinically relevant bacterial strains and demonstrated promising sensitivity to these pathogens. Finally, we performed radiation dosimetry in a normal murine cohort to set the stage for translation to humans in the near future.
Keywords: Positron emission tomography, radiochemistry, PET, infection, D-amino acid
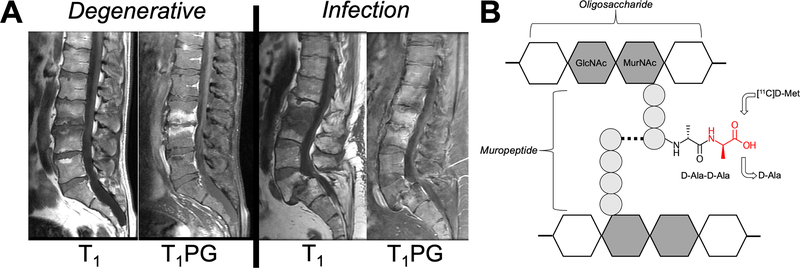
Current standard of care practice and imaging technologies fall short in their ability to accurately diagnose and monitor bacterial infection. Therefore, new tools are needed to establish the presence and location of infection, as well as monitor response to antimicrobial therapy. Our goal is to develop infection-specific positron emission tomography (PET) methods, using D-amino acid analogues that are incorporated into bacterial peptidoglycan, to image living bacteria in vivo. These methods will allow physicians to distinguish bacterial infection from sterile inflammation and other processes, and facilitate improved management of a variety of devastating clinical conditions, such as vertebral discitis-osteomyelitis (VDO) a disease for which our approach is targeted (Figure 1A).
Figure 1.
Approach to image vertebral discitis-osteomyelitis (VDO) via microorganism-specific incorporation of PET-labeled D-amino acids. (A) Variable imaging appearance of VDO via MRI. The MRIs of two patients are shown. T1-weighted imaging with and without contrast show abnormalities of the vertebral end-plates and discs that are similar for VDO and osteoarthritis (degenerative disease). (B) Structure of bacterial peptidoglycan (a high quantity present in S. aureus, the most common pathogen in VDO). The C-terminal D-Ala-D-Ala sequence of peptidoglycan muripeptide is highlighted, as well as the putative site of D-[11C]Met incorporation (red).
VDO is a challenging clinical problem, particularly for patients who are immunocompromised or abuse intravenous drugs.1 VDO symptoms are nonspecific, including neck or back pain, and clinical work-up involves leukocyte count, C-reactive protein (CRP) levels, and erythrocyte sedimentation rate (ESR).2,3 To that end, imaging studies are frequently used to support the clinical diagnosis of infection but are limited in accuracy. Present methods used in clinical practice either detect structural abnormalities (CT/ MR), metabolic changes in activated immune cells ([18F]FDG PET)4, or their recruitment to areas of infection (111In SPECT white blood cell scan)5. Moreover, invasive tissue sampling is often needed for adequate diagnosis, and poses potential complications. Current diagnostic imaging strategies are not specific for living bacteria and are often inadequate to detect bacterial infection during fever workup. Moreover, bone infection may look similar on MRI to other such conditions especially rheumatologic disorders or highly prevalent osteoarthritis.6,7 For this reason, there has been a growing interest in developing probes for imaging living bacteria in vivo. In particular, emerging PET approaches have targeted the folate biosynthesis pathway, bacterial metabolism of maltose-derived sugars, and other potential mechanisms.8–14 However, no imaging agent for the differential identification of bacterial infection from sterile inflammation has been translated for widespread use in a clinical setting. In addition, very promising metabolic tracers such as 2-[18F]-fluorodeoxysorbitol ([18F]FDS)15 lack sensitivity for gram-positive pathogens such as S. aureus, the most common causative agent of VDO. The prevalence of S. aureus in VDO and other infections has motivated us to pursue peptidoglycan-targeted PET tracers; peptidoglycan constitutes 90% of gram-positive organisms by dry weight.
A significant amount of D-amino acids (DAAs) are incorporated into most bacteria, principally D-Glu and D-Ala, which are used in peptidoglycan synthesis. Peptidoglycan is a strong component of the bacterial cell wall that helps to maintain cell shape, among other processes.16 D-amino acids appear to serve several roles within the peptidoglycan, including cell signaling, protection against enzymatic activity, and are incorporated rapidly and selectively in E. coli and other microorganisms.16–21
Our group previously demonstrated uptake and incorporation of D-[methyl-11C]methionine (D-[11C]Met) in S. aureus and E. coli, with selectivity for infection over sterile inflammation (induced using heat-killed bacteria).22 The presumed mechanism of incorporation is shown in Figure 1B, where the radiotracer is “swapped” for D-alanine on peptidoglycan muripeptides. D-Met has been shown to be released by a diverse array of stationary phase bacteria, which use it as a specific signal to alter the growth of neighboring cells (Lam reference). Importantly, D-Met released into the media is avidly taken up by bacteria and incorporated into peptidoglycan. Neumann et al. reported a radiosynthesis of D-[11C]Met utilizing a cyclic precursor with 85% enantiomeric excess, 20±1% non-decay corrected yield in 21 min (EOB).22 Neumann et al. went on to show promising selectivity for D-[11C]Met in differentiating active bacterial infection from sterile inflammation in vivo in a murine myositis (infection) model. These results demand further optimization and evaluation prior to attempting in-patient studies. This includes an improved enantiomeric excess synthesis, automation, sensitivity determination across a panel of key human pathogens, and radiation dosimetry in normal mice.
RESULTS AND DISCUSSION
Clinically feasible and automated radiosynthesis of D-[11C]Met
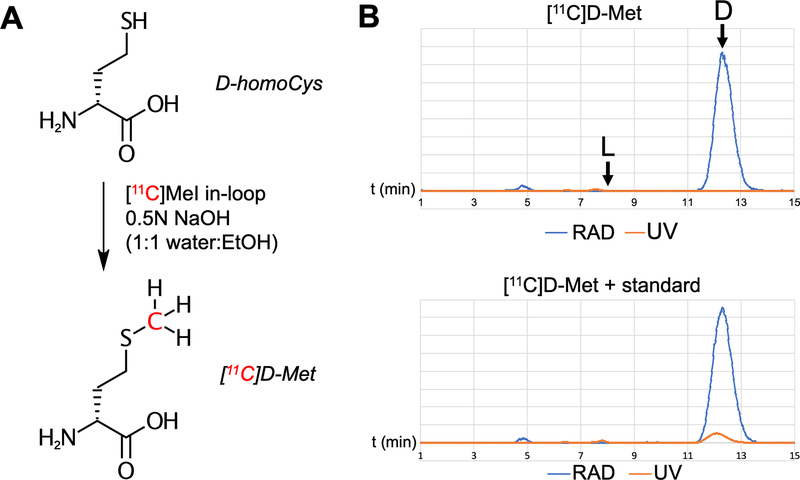
The synthesis of high enantiomeric excess D-[11C]Met is shown in Figure 2. Adapting a previously reported method23, we used the following radiosynthesis utilizing a GE TRACERlab FXc-Pro synthesis module: [11C]CO2 was produced via the 14N(p,α)11C nuclear reaction with proton bombardment of nitrogen-14 spiked with oxygen in the UCSF radiopharmaceutical facility. [11C]CO2 was trapped on a molecular sieve with nickel at room temperature. This was sealed and heated to 350oC with H2 to reduce [11C]CO2 to [11C]CH4, which was trapped on a carbosphere methane trap previously cooled to −80oC with liquid N2. The carbosphere was heated to release the [11C]CH4 which entered a recirculation loop containing an iodine column at 90oC. The recirculation tube reactor was maintained at 750oC and [11C]CH4 was converted to [11C]CH3I over a period of 5 min. The converted [11C]CH3I was trapped on a porapak column at room temperature. A stainless steel HPLC loop was previously coated with 100 μl of a solution containing 1.25 mg of D-homocysteine in 0.5M NaOH in 50/50 H2O/EtOH, followed by 300–500 μl of air to coat the Teflon loop before being placed in the synthesis module. The porapak column was heated to 180oC and the [11C]CH3I was flowed through the loop at 15 mL/min for 70–90 seconds and allowed to sit for 1 min at room temperature. The loop was rinsed with two additions of 3 mL saline to an intermediate vial containing 300 μl of NaH2PO4 (0.2 g/mL). This solution was then passed through a C18 plus Sep Pak and 0.2 micron Millex filter into the final product vial, to give D-[11C]Met. D-[11C]Met was synthesized in >99% ee, (n=9) 22±13% decay corrected radiochemical yield, and >90% radiochemical purity in all cases. A representative analytic HPLC trace is shown in Figure 2B.
Figure 2.
In-loop radiosynthesis of D-[11C]Met. The approach we previously employed used a D-homocysteine thiolactone precursor whose alkylation conditions rendered the compound susceptible to racemization. Here a non-cyclic chiral precursor was employed. (A) Radiosynthesis of D-[11C]Met from D-homocysteine. (B) Characterization of D-[11C]Met by HPLC. A sample of L-Met was analyzed separately (see supplementary information) and confirmed > 99% ee for the desired D-enantiomer.
D-[11C]Met shows broad sensitivity across a panel of clinically relevant gram-negative and gram-positive pathogens
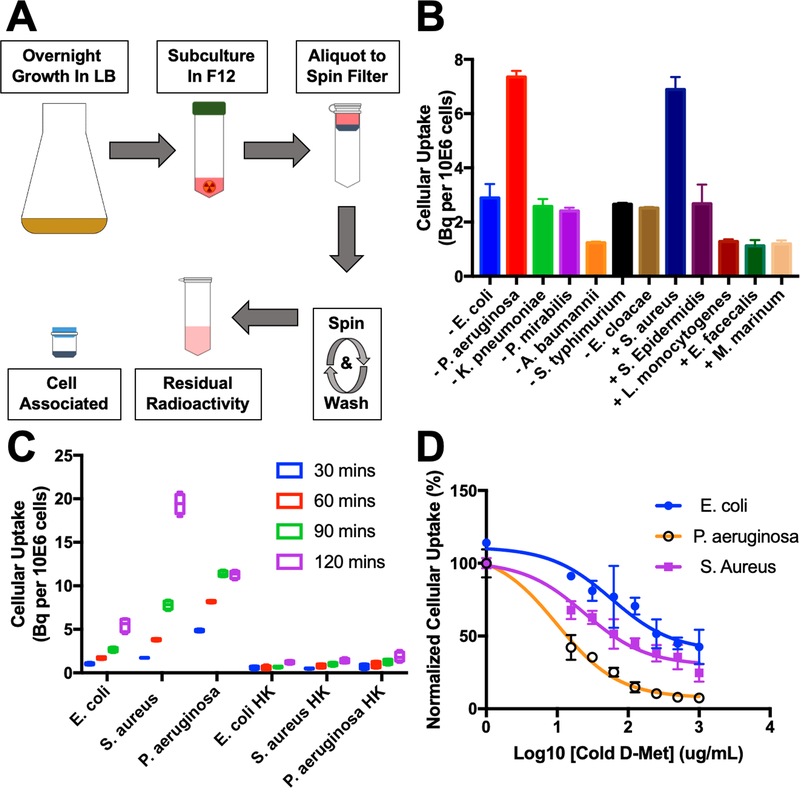
An in vitro screen was performed in small bacterial cultures to explore the versatility of D-[11C]Met. A protocol was developed to rapidly screen numerous organisms simultaneously for 11C metabolite (t½ = 20 min) uptake (Figure 3A). The following pathogens were screened (n=4, 10μCi/mL, 90 min incubation): E. coli, P. aeruginosa, K. pneumoniae, P. mirabilis, A. baumanni, S. typhimurium, E. cloacea, S. aureus, S. Epidermidis, L. monocytogenes, E. faecalis, M. marinum. The biomedical relevance of these organisms is highlighted in Table 1. Uptake was normalized to the number of colony-forming units (CFUs) by serially diluting and plating all organisms. The highest uptake was in gram-negative P. aeruginosa and gram-positive S. aureus. These data are summarized in Figure 3B. Additional experiments explored time-dependent accumulation of E. coli, S. aureus and P. aeruginosa, and demonstrated the lack of accumulation of D-[11C]Met in heat-killed organisms (Figure 3C). Finally, the uptake of D-[11C]Met was explored in the presence of varying concentrations of unlabeled D-Met demonstrating effective blocking (Figure 3D). These studies suggest specificity of D-[11C]Met for living pathogens.
Figure 3.
In vitro analysis of D-[11C]Met in bacteria. (A) Workflow of high-throughput in vitro assay. Bacteria is grown overnight in LB broth, then subcultured in F12 media in 50 mL centrifuge tubes in the presence of a radiotracer. After the designated time, the subculture is aliquotted into 1.5 mL spin filters equipped with a 0.22 micron nylon filter. The aliquot is centrifuged and washed. The spin filter is separated and each portion analyzed on a gamma counter. (B) Sensitivity analysis of D-[11C]met using a broad panel of human pathogens indicating avid incorporation (up to approximately 7 Bq incorporation of radiolabel) D-[11C]Met by S. aureus and P. aeruginosa. (C) Dynamic cellular uptake of D-[11C]Met in E. coli, S. aureus, and P. aeruginosa in exponential-phase cultures at 30,60,90, and 120 minutes. No radioactivity was incorporated into heat-killed organisms (P < 0.05 for live versus heat-killed, all organisms at all time points). (D) Accumulation of D-[11C]Met in the presence of increasing concentrations of unlabelled D-methionine. In all cases blocked uptake suggests specific incorporation.
Table 1.
Summary of gram-negative and gram-positive organisms studied, as well as their associated clinically-relevant infections.
| Strain | Clinical Relevance |
|---|---|
| Gram-negative | |
| E. coli | Diarrhea, urinary tract infections, respiratory illness. |
| P. aeruginosa | Ear infections, skin rashes, eye infections, nosocomial (hospital acquired) infections. |
| K. pneumoniae | Sepsis, wound or surgical site infections, meningitis, and pneumonia. Nosocomial infections. |
| A. baumannii | Sepsis, wound or surgical site infections, and pneumonia. Nosocomial infections. |
| S. typhimurium | Gastrointestinal distress: diarrhea, abdominal cramps, and fever. |
| E. cloacae | Urinary tract, lower respiratory tract, skin and soft-tissue infections. |
| P. mirabilis | Catheter-associated urinary tract infections in sensitive populations. Nosocomial infections. |
| Gram-negative | |
| S. aureus | Sepsis, pneumonia, endocarditis, and osteomyelitis. Nosocomial infections. |
| E. faecalis | Urinary and intra-abdominal infections, bacteremia, and endocarditis. |
| S. epidermidis | Device-associated infections. Nosocomial infections. |
| L. monocytogenes | Gastrointestinal distress, diarrhea, and fever. |
| M. marinum | Septic arthritis, osteomyelitis, disseminated skin lesions, and bacteremia. |
| www.cdc.gov | |
Radiation Dosimetry
All animal procedures were approved by the UCSF Institutional Animal Care and Use Committee. Veterinary services for the study were provided by the UCSF Laboratory Animal Resource Center (LARC) and all studies were performed in accordance with UCSF guidelines regarding animal housing, pain management, and euthanasia. All mice used were CBA/J mice (Jackson Laboratory) aged between 8–10 weeks. Rodent PET studies were conducted using an Inveon micro PET-CT system (Siemens, Erlanen, Germany). In normal female mice (22.7±1.2 g, n = 3) and normal male mice (27.9±2 g, n = 4), anesthesia was induced and maintained with isoflurane/O2 and the animals kept warm under a heat lamp. A tail vein catheter was placed and the female and male mice injected with 633±6 μCi and 533±7 μCi of D-[11C]Met, respectively. A 90 min scan in list-mode (raw) data acquisition was performed, followed by a 10 min micro-CT scan for attenuation correction and anatomical co-registration. The 90-min list-mode data were divided to 10 sequential time frames to generate required time-activity curves for dosimetry. No adverse effects were observed during or after injection of the compound.
In normal mice, 3 female and 4 male, the effective dose estimate was 3.9 μSv/MBq for females, and 3.0 μSv/MBq for males (based on ICRP60) (Table 2, Supp. Fig. 1) Even compared to other carbon-11 tracers, the effective dose is rather low.24 The organs with the highest estimated doses are liver, kidney, and bladder, and these data are consistent with human radiation dosimetry data for L-[11C]Met.25 This and our previous biodistribution analysis suggests that D- and L-Met undergo similar metabolism by mammalian organs, a finding that will be explored in future studies.
Table 2.
Radiation dosimetry analysis of female mice (male mice in Supporting Information). Relevant organ and whole body doses are (1) low and (2) similar to those seen with L-[11C]Met.
| Absorbed Dose (mGy/MBq) | Adult female (60 kg) | ||
| Organ | |||
| Adrenals | 0.0039 | ± | 0.00005 |
| Brain | 0.0027 | ± | 0.00023 |
| Breasts | 0.0028 | ± | 0.00008 |
| Gallbladder Wall | 0.0040 | ± | 0.00007 |
| LLI Wall | 0.0037 | ± | 0.00009 |
| Small Intestine | 0.0035 | ± | 0.00009 |
| Stomach Wall | 0.0036 | ± | 0.00009 |
| ULI Wall | 0.0037 | ± | 0.00010 |
| Heart Wall | 0.0049 | ± | 0.00009 |
| Kidneys | 0.0077 | ± | 0.00152 |
| Liver | 0.0073 | ± | 0.00052 |
| Lungs | 0.0053 | ± | 0.00033 |
| Muscle | 0.0031 | ± | 0.00009 |
| Ovaries | 0.0037 | ± | 0.00009 |
| Pancreas | 0.0039 | ± | 0.00008 |
| Red Marrow | 0.0029 | ± | 0.00007 |
| Osteogenic Cells | 0.0049 | ± | 0.00015 |
| Skin | 0.0026 | ± | 0.00008 |
| Spleen | 0.0036 | ± | 0.00007 |
| Thymus | 0.0034 | ± | 0.00010 |
| Thyroid | 0.0030 | ± | 0.00010 |
| Urinary Bladder Wall | 0.0068 | ± | 0.00098 |
| Uterus | 0.0037 | ± | 0.00008 |
| Total Body | 0.0034 | ± | 0.00007 |
| Effective Dose (mSv/MBq) | 0.0039 | ± | 0.00008 |
CONCLUSION
Since mammalian cells principally use L-amino acids, our hypothesis is that PET tracers derived from D-amino acids will be highly specific to living bacteria, which incorporate DAAs into their peptidoglycans. Following our early pre-clinical evaluation of D-[11C]Met derived from a thiolactone hydrochloride precursor, we sought to improve this radiosynthesis, and explore the sensitivity of this radiotracer to a panel of clinically relevant human pathogens. The radiosynthesis reported herein, from a linear homocysteine precursor, is straightforward, automated, fast, and can be easily incorporated into a production setting. Upon applying D-[11C]Met to a panel of gram-positive and gram-negative bacteria, promising uptake was seen across all organisms suggesting the utility of D-[11C]Met as a diagnostic radiotracer for active infection (together with previously reported in vivo data).22 As a final effort to set the stage for qualification syntheses and first in-human studies with D-[11C]Met, radiation dosimetry was performed in male and female normal mice. The effective dose is low (3–3.9 μSv/MBq), and the organs (liver, kidney and bladder) with the highest dose estimated similar to that of L-[11C]Met, a radiotracer already being studied in humans. While these data indicate relatively high background accumulation in mammalian organs, the workup of patients with musculoskeletal and other infections may benefit from the use of D-[11C]Met and related tracers. Future efforts will focus on these patient studies as well as tracer sensitivity to bacteria in challenging clinical scenarios for example when surface-associated bacteria or “biofilms” are present.
METHODS
General
The D-homocysteine precursor was either prepared from D-methionine (Sigma Aldrich), using standard Birch Reduction conditions with Na/NH3 (liq) according to literature precedent26, or purchased from AChemTeck, Inc. D-methionine and L-methionine (Sigma Aldrich) were used as cold reference standards. Ethanol, sodium phosphate monobasic, sodium hydroxide, and USP grade saline were commercially available. Solid-phase exchange cartridges (Waters Sep Pak C-18) were conditioned with 5 mL ethanol and 10 mL water before use. A GE TRACERLab FXc-Pro synthesis module was modified to allow for direct collection bypassing HPLC.
D-[11C]Met Quality Control
The identity, radiochemical purity, and enantiomeric excess of D-[11C]Met was determined by chiral HPLC with a gamma and UV detector, against the cold reference standard(s). This utilized a UV detector (λ = 220nm), Chirobiotic T2 250 × 4.6 mm LC column, 1 mL/min, 75% MeCN, 25% water mobile phase. The final dose of D-[11C]Met was a clear, colorless solution, pH 7. Details regarding characterization of D-[11C]Met are presented in the Supporting Information (Supp. Fig. 2).
PET Imaging and Radiation Dosimetry
Imaging data acquisition
All imaging data (n=3 for male mice; n=3 for female mice) were acquired using a small animal PET/CT scanner (Inveon, Siemens Medical Solutions, Malvern, PA) for 90 minutes in list-mode. The 90 minute list-mode data were divided to 10 sequential dynamic time frames to create time-activity curves of organs and the remainder of the body, required for dosimetry. Image reconstruction was performed using a vendor-provided ordered subsets expectation maximization algorithm. CT-based attenuation correction was performed, and quantification calibration to convert the raw reconstructed pixel value to a physical unit of Bq/ml was performed as well.
Normal tissue radiation dose estimation
Volumes of interest (VOIs) were drawn on coregistered CT images for brain, lungs, heart, liver, kidneys, and urinary bladder. All VOIs were either elliptical cylinder (5 mm long axis, 3 mm short axis, and 5 mm height for the brain) or cylinders (3 mm diameter and 3 mm height for lungs, heart, liver, kidneys, and urinary bladder), and they were placed well within the anatomical boundaries to minimize spill-over or spill-in of radioactivity. The mean values (in Bq/ml) in these VOIs were multiplied by standard mouse organ volumes (in ml) to estimate total activity (in Bq) within these organs. The total activity within the entire animal subtracted by all organ activities was used as activity in the remainder of the body. The percent of injected activity within the defined organs (%IA) in mice was extrapolated to human-equivalent values (i.e., %IA in human equivalents) by multiplying %IA in mice by ratios of standard human organ weights to mouse organ weights. Then, the adult male and female models in Organ Level INternal Dose Assessment (OLINDA) were used for human-equivalent dosimetry. The time points we used for our dosimetry calculation were from the 10 sequential time frames reconstructed over the 90 min list-mode data: 0.25, 0.75, 1.25, 1.75, 3, 7, 12.5, 27.5, 45, and 80 minutes post-injection frames. The durations of these reconstructed time frames were: 30 seconds for the 0.25, 0.75, 1.25, and 3 min data, 120 seconds for the 7 and 12.5 min data, 300 seconds for the 27.5 and 45 min data, 600 seconds for the 45 min data, and 1200 seconds for the 80 min data. The list-mode data are the most primitive data that can be divided into different frames and durations. These input %IA data for each organ and the remainder of the body were curve-fitted to derive time-integrated activity coefficients (also known as residence times) (in Bq-hr/Bq) and organ and whole-body effective doses for human equivalents were estimated using each mouse data. The data from the three animals were averaged to derive absorbed organs doses (in mGy/MBq) and whole-body effective dose (in mSv/MBq). Organ and effective dose estimations were performed using OLINDA version 1.1 using International Commission on Radiological Protection (ICRP) Publication 60 tissue weighting factors as well as OLINDA version 2.0 using ICRP Publication 103 tissue weighting factors.
In vitro tracer uptake assay
The bacterial strains, and their growth conditions are listed in Supp. Fig. 3. With the exception of M. marinum, bacteria strains were aerobically grown in their listed medias for 16 hours with agitation of 111 rpm. M. marinum was aerobically grown for 3 days with media replenishment every 24 hours. The cultures were pelleted at 3400 rpm for 5 minutes and resuspended in an equivalent amount of Ham’s F12 media (Gibco). A 1/16 dilution of the cultures were incubated with 1 μCi D-[11C]Met at 180 rpm for 90 minutes. The bacterial suspensions were transferred to filter tubes (Corning Costar Spin-X) and centrifuged at 8000 rpm for 5 minutes. Phosphate buffed saline was added to each tube and the cultures were centrifuged at 8000 rpm for 5 minutes. The pellet and supernatant were separated and counted on a γ counter (Hidex Automatic Gamma Counter). Four replicates were performed for each bacterial strain. A heat killed control for E.coli, S.aureus, and P. aeruginosa (95°C for 30 minutes) were incubated and processed the same way.
Data analysis and statistical considerations
For synthesis, radiochemical yield incorporates decay-correction for 11C (t½=20 min). In vitro data was normalized to CFU’s for sensitivity analysis to account for differential growth rates between organisms. All statistical analysis was performed using Prism 7. Data were analyzed using an unpaired two-tailed Student’s t-test. All graphs are depicted with error bars corresponding to the standard error of the mean. Other data including radiochemical yield are also reported as mean ± standard error.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to acknowledge Dr. Sanjay Jain and Alvaro Ordonez (Johns Hopkins University) for helpful discussions related to in vitro protocols.
Funding Sources
We acknowledge NIH R01EB024014, NIH R01EB025985, DOD W81XWH1810641 and the UCSF Resource Allocation Program for support of this project.
ABBREVIATIONS
- Sv
sievert
- Bq
bequerel
- Ci
curie
Footnotes
ASSOCIATED CONTENT
Supporting Information
Detailed information regarding synthesis, in vitro, and in vivo experiments not reported in the main text.
REFERENCES
- (1).Mylona E; Samarkos M; Kakalou E; Fanourgiakis P; Skoutelis A Pyogenic Vertebral Osteomyelitis: A Systematic Review of Clinical Characteristics. Semin. Arthritis Rheum. 2009, 39 (1), 10–17. 10.1016/j.semarthrit.2008.03.002. [DOI] [PubMed] [Google Scholar]
- (2).Mazzie JP; Brooks MK; Gnerre J Imaging and Management of Postoperative Spine Infection. Neuroimaging Clin. N. Am. 2014, 24 (2), 365–374. 10.1016/j.nic.2014.01.003. [DOI] [PubMed] [Google Scholar]
- (3).Go JL; Rothman S; Prosper A; Silbergleit R; Lerner A Spine Infections. Neuroimaging Clin. N. Am. 2012, 22 (4), 755–772. 10.1016/j.nic.2012.06.002. [DOI] [PubMed] [Google Scholar]
- (4).Gafter-Gvili A; Raibman S; Grossman A; Avni T; Paul M; Leibovici L; Tadmor B; Groshar D; Bernstine H [18F]FDG-PET/CT for the Diagnosis of Patients with Fever of Unknown Origin. Qjm 2015, 108 (4), 289–298. 10.1093/qjmed/hcu193. [DOI] [PubMed] [Google Scholar]
- (5).Palestro CJ Radionuclide Imaging of Osteomyelitis. Semin. Nucl. Med. 2015, 45 (1), 32–46. 10.1053/j.semnuclmed.2014.07.005. [DOI] [PubMed] [Google Scholar]
- (6).Baker JC; Demertzis JL; Rhodes NG; Wessell DE; Rubin DA Diabetic Musculoskeletal Complications and Their Imaging Mimics. RadioGraphics 2012, 32 (7), 1959–1974. 10.1148/rg.327125054. [DOI] [PubMed] [Google Scholar]
- (7).Stacy GS; Kapur A Mimics of Bone and Soft Tissue Neoplasms. Radiol. Clin. North Am. 2011, 49 (6), 1261–1286. 10.1016/j.rcl.2011.07.009. [DOI] [PubMed] [Google Scholar]
- (8).van Oosten M; Hahn M; Crane LMA; Pleijhuis RG; Francis KP; van Dijl JM; van Dam GM Targeted Imaging of Bacterial Infections: Advances, Hurdles and Hopes. FEMS Microbiol. Rev. 2015, 39 (6), 892–916. 10.1093/femsre/fuv029. [DOI] [PubMed] [Google Scholar]
- (9).Becker W; Palestro CJ; Winship J; Feld T; Pinsky CM; Wolf F; Goldenberg DM Rapid Imaging of Infections with a Monoclonal Antibody Fragment (LeukoScan). Clin. Orthop. Relat. Res. 1996, No. 329, 263–272. [DOI] [PubMed] [Google Scholar]
- (10).Fuster D; Soriano A; Garcia S; Piera C; Suades J; Rodríguez D; Martinez JC; Mensa J; Campos F; Pons F Usefulness of 99mTc-Ciprofloxacin Scintigraphy in the Diagnosis of Prosthetic Joint Infections. Nucl. Med. Commun. 2011, 32 (1), 44–51. 10.1097/MNM.0b013e328340e6fb. [DOI] [PubMed] [Google Scholar]
- (11).Ebenhan T; Gheysens O; Kruger HG; Zeevaart JR; Sathekge MM Antimicrobial Peptides: Their Role as Infection-Selective Tracers for Molecular Imaging. Biomed Res. Int. 2014, 2014. 10.1155/2014/867381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bettegowda C; Foss C. a; Cheong I; Wang Y; Diaz L; Agrawal N; Fox J; Dick J; Dang LH; Zhou S; Kinzler KW; Vogelstein B; Pomper MG Imaging Bacterial Infections with Radiolabeled 1-(2’-Deoxy-2’-Fluoro-Beta-D-Arabinofuranosyl)-5-Iodouracil. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (4), 1145–1150. 10.1073/pnas.0408861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hernandez FJ; Huang L; Olson ME; Powers KM; Hernandez LI; Meyerholz DK; Thedens DR; Behlke MA; Horswill AR; Mcnamara JO Noninvasive Imaging of Staphylococcus Aureus Infections with a Nuclease-Activated Probe. Nat. Med. 2014, 20 (3), 301–306. 10.1038/nm.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bardhan NM; Ghosh D; Belcher AM Carbon Nanotubes as in Vivo Bacterial Probes. Nat. Commun. 2014, 5, 1–11. 10.1038/ncomms5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Weinstein EA; Ordonez AA; DeMarco VP; Murawski AM; Pokkali S; MacDonald EM; Klunk M; Mease RC; Pomper MG; Jain SK Imaging Enterobacteriaceae Infection in Vivo with 18F-Fluorodeoxysorbitol Positron Emission Tomography. Sci. Transl. Med. 2014, 6 (259), 259ra146 10.1126/scitranslmed.3009815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bugg TD; Walsh CT Intracellular Steps of Bacterial Cell Wall Peptidoglycan Biosynthesis: Enzymology, Antibiotics, and Antibiotic Resistance. Nat. Prod. Rep. 1992, 9 (3), 199–215. [DOI] [PubMed] [Google Scholar]
- (17).Lam H; Oh D-C; Cava F; Takacs CN; Clardy J; de Pedro M. a.; Waldor MK D-Amino Acids Govern Stationary Phase Cell Wall Remodeling in Bacteria. Science (80-. ). 2009, 325 (5947), 1552–1555. 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hammes WP; Neuhaus FC On the Mechanism of Action of Vancomycin: Inhibition of Peptidoglycan Synthesis in Gaffkya Homari. Antimicrob. Agents Chemother. 1974, 6 (6), 722–728. 10.1128/AAC.6.6.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Caparrós M; Pisabarro AG; de Pedro MA Effect of D-Amino Acids on Structure and Synthesis of Peptidoglycan in Escherichia Coli. J. Bacteriol. 1992, 174 (17), 5549–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Siegrist MS; Whiteside S; Jewett JC; Aditham A; Cava F; Bertozzi CR D-Amino Acid Chemical Reporters Reveal Peptidoglycan Dynamics of an Intracellular Pathogen. ACS Chem. Biol. 2013, 8 (3), 500–505. 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kuru E; Tekkam S; Hall E; Brun YV; Van Nieuwenhze MS Synthesis of Fluorescent D-Amino Acids and Their Use for Probing Peptidoglycan Synthesis and Bacterial Growth in Situ. Nat. Protoc. 2015, 10 (1), 33–52. 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Neumann KD; Villanueva-Meyer JE; Mutch CA; Flavell RR; Blecha JE; Kwak T; Sriram R; VanBrocklin HF; Rosenberg OS; Ohliger MA; Wilson DM Imaging Active Infection in Vivo Using D-Amino Acid Derived PET Radiotracers. Sci. Rep. 2017, 7 (1), 7903 10.1038/s41598-017-08415-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gómez V; Gispert JD; Amador V; Llop J New Method for Routine Production of L-[Methyl-11C]Methionine: In Loop Synthesis. J. Label. Compd. Radiopharm. 2008, 51 (1), 83–86. 10.1002/jlcr.1483. [DOI] [Google Scholar]
- (24).Van Der Aart J; Hallett WA; Rabiner EA; Passchier J; Comley RA Radiation Dose Estimates for Carbon-11-Labelled PET Tracers. Nucl. Med. Biol. 2012, 39 (2), 305–314. 10.1016/j.nucmedbio.2011.08.005. [DOI] [PubMed] [Google Scholar]
- (25).Deloar HM; Fujiwara T; Nakamura T; Itoh M; Imai D; Miyake M Short Communication Estimation of Internal Absorbed Dose Using Whole-Body Positron Emission Tomography. 1998, 25 (6). [DOI] [PubMed] [Google Scholar]
- (26).Langstrom B; Antoni G; Gullberg P; Halldin C; Malmborg F; Nagren K; Rimland A; Svard H Synthesis of L- and D- [ Methyl-11C ] Methionine. J Nucl Med 1987, 28 (6), 1037–1041. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.