Abstract
Pulp regeneration is to replace the inflamed/necrotic pulp tissue with regenerated pulp-like tissue to rejuvenate the teeth. Self-assembling peptide hydrogels RADA16-I (Ac-(RADA16-I)4-CONH2) can provide a three-dimensional environment for cells. The stem cell factor (SCF) plays a crucial role in homing stem cells. Combining these advantages, our study investigated the effects of SCF-RADA16-I on adhesion, proliferation, and migration of human dental pulp stem cells (DPSCs) and the angiogenesis of human umbilical vein endothelial cells (HUVECs). The β-sheet and grid structure were observed by circular dichroism (CD), scanning electron microscopy (SEM), and atomic force microscopy (AFM). Cytoskeleton staining, living cell staining, cell viability, cell migration, angiogenesis, and western blot assays were performed, and the results indicated that all the SCF groups were superior to the corresponding non-SCF groups in cell adhesion, proliferation, migration, and angiogenesis. RADA16-I provided a three-dimensional environment for DPSCs. Besides, the SCF promoted HUVECs to form more vascular-like structures and release more vascular endothelial growth factor A. In summary, the SCF-loaded RADA16-I scaffold improved adhesion, proliferation, and migration of DPSCs and the formation of more vascular-like structures of HUVECs. SCF-RADA16-I holds promise for guided pulp regeneration, and it can potentially be applied widely in tissue engineering and translational medicine in the future.
Introduction
Dental pulp tissue, rich in nerves and blood vessels, is an important component of teeth, which plays an important role in forming dentin, nutrition, sensation, and defense. Pulp is prone to inflammation due to caries or trauma, and the continuous development of inflammation leads to necrosis of pulp and tissue around the apex. Root canal therapy is a common treatment option for dentistry. During this process, the inflamed or necrotic pulp is removed and filled with synthetic material. Although root canal therapy has proven to be quite useful, the remaining tooth structure is inactive and brittleness increases due to the loss of the natural pulp.1,2 Young permanent teeth have a certain ability to regenerate dental pulp, but mature dental pulp is difficult to regenerate. Therefore, many researchers have devoted themselves to dental pulp regeneration.
Dental pulp regeneration is designed to replace inflamed/necrotic pulp with regenerated dental pulp-like tissue, which maximizes tooth vitality and continues to develop immature teeth.3,4 Hence, most researchers pay attention to dental pulp regeneration, and the most common method is tissue engineering.
The commonly used scaffolds for pulp regeneration are synthetic polymers (such as polylactic acid and polyglycolic acid) and natural materials (such as type I collagen) in previous studies.5,6 Although those polymers are biocompatible, biodegradable, and inexpensive, they fail to mimic the complex physiological functions of natural tissues. Collagen, difficult to customize, has a fast degradation rate and issues of purity and antigenicity.7 A series of the self-assembled peptide hydrogels, represented by RADA16-I, have been synthesized since the self-assembled peptide hydrogel was first discovered in 1993.8 RADA16-I consists of positively charged arginine (R), hydrophobic alanine (A), and negatively charged aspartic acid (D). The above amino acids are periodically repeated in composition, which makes the gel formation of RADA16-I controllable.8 RADA16-I is spontaneously assembled into fibers by a neutral pH solution to form a three-dimensional hydrogel with humidity greater than 99%.8 Compared with traditional biomaterials, RADA16-I has the following advantages: (1) injectability,9,10 (2) high biocompatibility and low cytotoxicity,10,11 (3) ability to provide a true 3D nanofibrous structure for cell growth,11−16 (4) capability of being further modified by various functional amino acid fragments to obtain better biological properties,15,17 such as PRG, the functional fragment, which could promote cell adhesion and proliferation.18 Hence, RADA16-I has substantial potential for 3D cell culture and tissue engineering and as a delivery system.19
Endodontic angiogenesis is crucial to the long-term survival of regenerated pulp. In response to this point, the stem cell factor (SCF) was selected in this study. The SCF, the potent chemokine, is a glycoprotein with a molecular weight of approximately 30 kDa. As a homing agent capable of recruiting progenitor cells, the SCF has shown great potential in application prospects.18,19 In recent years, in addition to stem cells derived from dental tissue, human periapical cyst mesenchymal stem cells (hPCy-MSCs), exhibiting characteristics similar to those of other dental-derived MSCs, have received more and more attention.20 MSCs were described as promoters, enhancers, and playmakers of translational regenerative medicine in Ballini’s study,21 but the application of MSCs in dental pulp regeneration is few. Hence, dental pulp stem cells are still the most widely used in dental pulp regeneration due to their abundant sources, mature extraction technology, and thorough research. DPSCs play an important role in tissue engineering and regenerative medicine, showing great potential in becoming an ideal source of seed cells for pulp regeneration.
In the current study, the β-folded and grid structures were detected by CD, AFM, and SEM, which verified that a wrapped SCF did not affect the self-assembly process of RADA16-I. Living cell staining, proliferation, cytoskeleton staining, migration, and angiogenesis were carried out to evaluate that the SCF not only promoted angiogenesis but also promoted adhesion, proliferation, and migration of DPSCs. Furthermore, RADA16-I provided a three-dimensional growth microenvironment for DPSCs. In brief, SCF-RADA16-I has the potential to guide dental pulp regeneration.
Materials and Methods
Isolation and Culture of Cells
To isolate the DPSCs, the pulp was removed from complete wisdom teeth, placed in a humidified incubator at 37 °C with 5% CO2, digested with collagenase and neutral protease for 40 min, torn into pieces, and then inoculated on a culture dish containing DMEM (15% FBS and 1% antibiotic). All reagents were purchased from Corning. HUVECs were purchased in Sciencecell Research Laboratories.
Preparation of SCF Solution and Screening of the Optimal Concentration
SCF powder (Pepro Tech) was dissolved in ultrapure water to make a 10 mg/mL stock solution for different working solutions as needed. The cell suspension was prepared, and 8 × 103 DPSCs were seeded into a 96-well plate with three replicates for each group with different concentrations of the SCF. The samples were tested with CCK8 assay (Dojindo) on days 1, 4, and 7. The absorbance of DPSCs at 460 nm was measured, and the optimal concentration of SCF was screened.
Preparation of RADA16-I and SCF-RADA16-I Gels
The RADA16-I synthetic peptide powder (provided by School of Materials Science and Engineering Tsinghua University) was dissolved in ultrapure water to make the peptide solution at a mass concentration of 1% (10 mg/mL), and the peptide solution was sonicated with a portable ultrasonic probe (Sonics) for complete dissolution and filtered with a 0.22 μm filter. The insert chamber (Biocoat) was placed in a 24-well plate (Corning) and incubated with DMEM. A volume of 100 μL of RADA16-I polypeptide solution was added to the chamber. DMEM was repeatedly added to dilute the acid, which was released during the RADA16-I gelation process. Finally, 200 μL of DMEM was re-added and placed in an incubator overnight. The RADA16-I hydrogel formed after 24 h. The adequate amount of SCF stock solution prepared was added to the RADA16-I solution after ultrasonic dissociation to a final concentration of 100 ng/mL and re-suspended to make the SCF distribution uniform to obtain SCF-RADA16-I solution. The above gelation process was repeated by taking 100 μL of the solution therein so that the SCF-RADA16-I hydrogel was obtained.
Structure and Properties of RADA16-I and SCF-RADA16-I
A 1% concentration of RADA16-I and SCF-RADA16-I peptide solutions were diluted to 25 μmoL/L, 200 μL of them was put into the quartz tank for testing (path length of 0. 05 cm and wavelength range of 195–250 nm) by CD (Chirascan, Applied Photographysics Ltd), and 150 μL was put on the surface of a mica plate and observed by AFM. The scanning area is 1 μm × 1 μm, and the scanning frequency is 1.00 Hz. Images were recorded with a silicon scanning probe (FESP, Vecco Probe Inc., CA, U.S.A.) with a tip curvature radius of 10 nm and 225 μm length. After the two solutions (RADA16-I and SCF-RADA16-I) were separately gelatinized, they were fixed with 2.5% glutaraldehyde solution, gradually dehydrated through different volume fractions of ethanol solution, and then dried by critical-point drying (Tousimis, U.S.). After completely dried, they were sprayed with gold and placed under SEM (Jeol, Japan) for observation.
Experimental Design
The following six groups (Figure 2a) were suitable for adhesion, activity, and proliferation experiments: DPSC group: DPSCs were directly inoculated on 24-well plates; SCF + DPSC group: DPSCs were inoculated on the plates, and we added SCF to the culture medium; RADA16-I-2D group: RADA16-I was injected into the insert chamber and completed the self-assembly process, and then we inoculated DPSCs on the surface of RADA16-I; SCF-RADA16-I-2D group: we added SCF to the culture medium on the base of group RADA16-I-2D; RADA16-I-3D group: RADA16-I in the solution state was mixed with DPSCs, which were resuspended in 10% sucrose solution, and they were put into the insert chamber; and SCF-RADA16-I-3D group: RADA16-I in the solution state was mixed with the SCF and DPSCs, which were resuspended in sucrose solutions, and they were injected into the insert chamber.
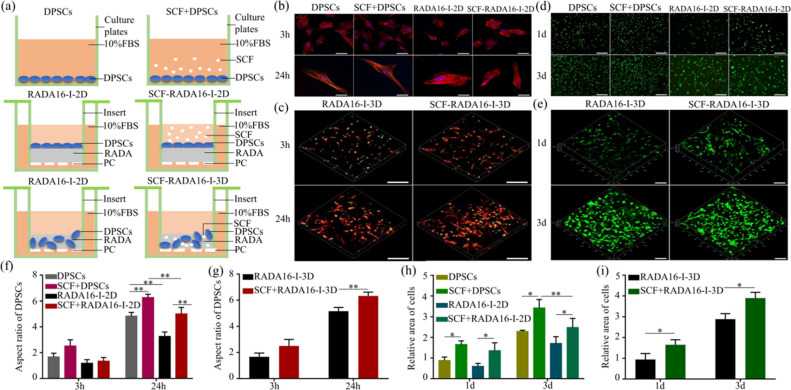
Figure 2.
Effect of SCF-RADA16-I on adhesion and activity of DPSCs. (a) Schematic diagram of groups: DPSC group: DPSCs were directly inoculated on 24-well plates; SCF + DPSC group: DPSCs were inoculated on the plates, and we added SCF to the culture medium; RADA16-I-2D group: RADA16-I was injected into the insert chamber and completed the self-assembly process, and then we inoculated DPSCs on the surface of RADA16-I; SCF-RADA16-I-2D group: we added SCF to the culture medium on the base of group RADA16-I-2D; and RADA16-I-3D group: RADA16-I in the solution state was mixed with DPSCs, which were resuspended in sucrose solution, and they were put into the insert chamber; SCF-RADA16-I-3D group: RADA16-I in the solution state was mixed with SCF and DPSCs, which were resuspended in sucrose solutions, and they were injected into the insert chamber. (b, d) Representative images of two-dimensional cultures and (f, h) the corresponding statistical analysis . (c, e) The images of three-dimensional cultures were taken and (g, i) statistically analyzed. F-actin was dyed into red by phalloidin. The nucleus was dyed into blue by DAPI and dyed into green by SYTOX Green. (*P < 0.05, **P < 0. 01). Scale bar: 100 μm (b, d) and 200 μm (c, e).
Cytoskeleton Staining Experiment
Each sample was prepared as shown in (Figure 2a) in which the cell density was 1 × 104 DPSCs/well. Samples were collected at hours 3 and 24, fixed in 4% paraformaldehyde, permeated with 0.1% Triton® X100, blocked with 1% bovine serum albumin (BSA), incubated with rhodamine-labeled phalloidin staining solution ( Invitrogen) for 40 min, and dyed with SYTOX Green (Invitrogen ) or DAPI (Beyotime). Then, they were observed by LSCM (Zeiss 780) and analyzed with GraphPad Prism 7.
Cell Activity Assay
Each sample was prepared as shown in (Figure 2a) in which the cell density was 1 × 105 cells/well. Samples were collected separately on days 1 and 3, washed with PBS, and dyed with Calcein AM dye (Dojindo) for 30 min. Then, they were photographed by LSCM and quantified with Imagine J.
Cell Proliferation Assay
Each sample was prepared as shown in (Figure 2a) in which the cell densities of the two-dimensional and three-dimensional cultures were 4 × 104 and 1 × 104 cells/well, respectively. Furthermore, in the three-dimensional culture, the functional fragment PRG (repetitive array of PRGDS and YRGDS polypeptide sequences) was ligated at the C-terminus of RADA16-I, and group RADA16-I-PRG + DPSCs together with group SCF-RADA16-I-PRG + DPSCs was added. Samples were respectively harvested on days 1, 4, and 7, and the mixtures of cell lysate and CyQUANT NF reagent (Cy QUANTTM Cell Proliferation Assay Kit) were completely reacted with the cells according to the instructions. The fluorescence intensity was tested by a microplate reader (Biotek, U.S.) and analyzed with GraphPad Prism 7.
Cell Migration Assay
This experiment was divided into DPSC, SCF + DPSC, RADA16-I, and SCF-RADA16-I groups and HUVECs, SCF + HUVEC, RADA16-I, and SCF-RADA16-I groups. Cells (2 × 104 cells/well) were added to the Transwell chamber (nested 24-well plate), and 300 μL of medium with or without SCF and RADA16-I was added to the lower chamber. After 24 h, Transwell chambers were taken out, washed with PBS, fixed with 4% paraformaldehyde, dyed with 0.1% crystal violet dye working solution after the un-migrated cells were wiped clean, and then photographed by a microscope (Leica) and analyzed with GraphPad Prism 7.
Angiogenesis Experiment In Vitro
Two groups, HUVECs and SCF + HUVECs, were included in this assay. BD Matrigel (200 μL/well) was added into the plate precooled at 4 °C and placed at 37 °C for 30 min. HUVECs were added to the well plate (2 × 105 cells/well) with Matrigel, and the SCF was separately added into the SCF + HUVEC group. The pictures were collected with an inverted microscope in 3, 6, and 9 h. Finally, the samples were dyed with calcein AM and photographed again under a fluorescence microscope.
Western Blot Analysis
HUVECs with or without SCF were lysed with RIPA lysate (Pro Tech) at hours 3, 6, and 9. Protein concentrations were tested with a protein assay kit (Bosterbio). The total proteins were separated by SDS-PAGE, transferred to a PVDF membrane (the membrane was blocked with 5% non-fat milk), incubated with a vascular endothelial growth factor A (VEGFA) (Wanleibio, WL00009b) primary antibody at 4 °C overnight, and then incubated with a secondary antibody (ZSGBBI O, ZB2301) after washing. The signal of the immunoreactive protein was visualized using a hypersensitive ECL chemiluminescence reagent (Boster, AR1171) and then analyzed by using Image J software.
Results
Screening the Optimal Concentration of the SCF
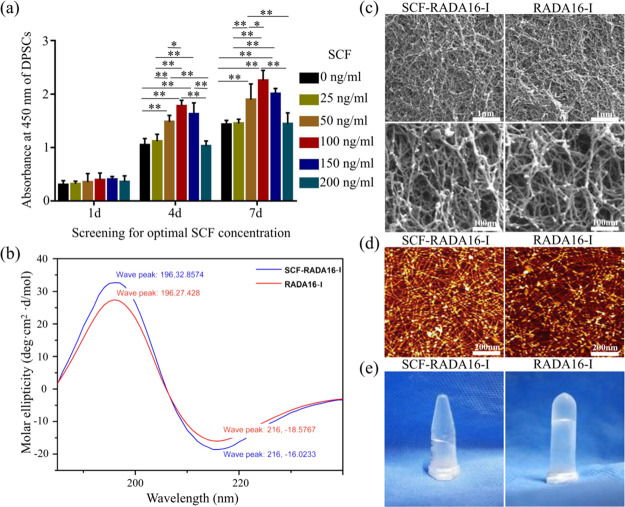
In order to screen the optimal concentration of the SCF, the proliferation of DPSCs was evaluated. As shown in Figure 1a, the proliferation of DPSCs with the SCF was better than that without the SCF, which indicated that the SCF promoted the proliferation of DPSCs. On the fourth day, under a concentration of 100 ng/mL, it was found that the higher the concentration, the faster the proliferation. When the SCF concentration increased to 150 ng/mL up to 200 ng/mL, the number of cells showed a downward trend. There was a statistical difference (P < 0.01) between group 100 ng/mL and groups 0, 25, and 200 ng/mL, and there was no statistical difference between group 100 ng/mL and groups 50, 100, and 150 ng/mL (P > 0.05). On the seventh day, although there was no statistical difference between group 100 ng/mL and group 150 ng/mL (P > 0.05), there was a statistical difference between groups 100 and 50 ng/mL (P < 0.05). Hence, 100 ng/mL was used to complete the following experiment because the number of cells at 100 ng/mL was always the largest.
Figure 1.
Screening optimal concentration of SCF and schematic illustrations of RADA16-I and SCF-RADA16-I. (a) Screening optimal concentration of SCF (*P < 0.05, **P < 0. 01). (b) Typical CD spectrum of SCF-RADA16-I and RADA16-I. (c) Typical SEM morphologies of SCF-RADA16-I and RADA16-I. (d) Representative AFM images of SCF-RADA16-I and RADA16-I. (e) SCF-RADA16-I and RADA16-I form hydrogel scaffolds under physiological conditions.
Structure and Properties of RADA16-I and SCF-RADA16-I
A stable β-sheet secondary structure of SAPs is required for peptide self-assembly and nanofiber formation.22 As shown in Figure 1b, a typical spectrum of β-sheet structures with a positive maximum molar ellipticity at 196 nm and a negative maximum molar ellipticity at 216 nm was observed in group SCF-RADA16-I, which was similar to that in group RADA16-I. The result of SEM (Figure 1c) revealed interweaving nanofibers and porous structures with diameters of 10–30 nm and pores between 5 and 200 nm, which provide a true 3D microenvironment for cells by mimicking the extracellular matrix (ECM).11,14,20 The results of AFM (Figure 1d) showed that SCF-RADA16-I formed long uniform and interwoven nanofibers similar to those of RADA16-I, which was consistent with SEM results. SCF-RADA16-I and RADA16-I form hydrogel scaffolds under physiological conditions (Figure 1e). Those results indicated that loaded SCF did not affect the β-sheet structures and self-assembly process of RADA16-I.
SCF-RADA16-I Promoted the Adhesion of DPSCs
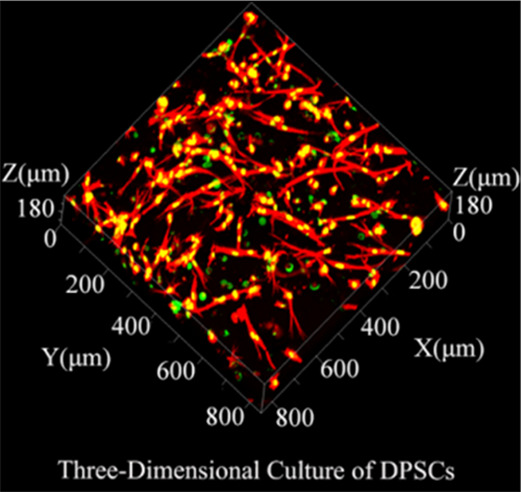
To evaluate the adhesion of DPSCs, cytoskeletal staining was performed (Figure 2(b, c)). By calculating and analyzing the aspect ratio of DPSCs (Figure 2f,g), the aspect ratio of groups with the SCF was better than that of corresponding groups without the SCF at 24 h (P < 0.01). In addition, the state of DPSCs was irregularly polygonal, and the 3D cultured cells have an aspect ratio superior to that of the 2D cultured. In summary, SCF-RADA16-I promoted the adhesion of DPSCs and provided a three-dimensional environment for cells simultaneously.
SCF-RADA16-I Promoted the Activity of DPSCs
To evaluate the activity of DPSCs, living-cell staining was carried out. The activity of 3D cultured DPSCs (Figure 2e) was close to that of DPSCs cultured on plates (Figure 2d) and superior to that of 2D cultured DPSCs (Figure 2d). In 3D culture, the cells exhibited agglomerated growth. The statistical results (Figure 2h,i) showed that the activity of cells in groups with the SCF was significantly more than that in the corresponding SCF-free groups whether in 2D or 3D culture (P < 0.05). In short, SCF-RADA16-I promoted the activity of DPSCs and provided simultaneously a three-dimensional growth environment for cells.
SCF-RADA16-I Promoted the Proliferation and Migration of DPSCs
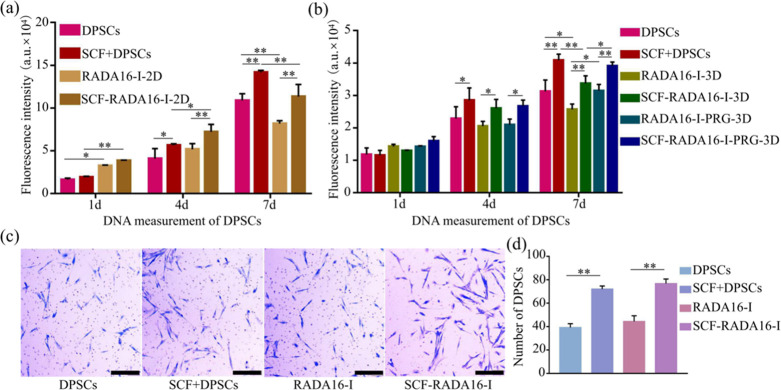
The proliferation of DPSCs was detected by a DNA content determination method, and fluorescence intensity reflected the number of cells. The results showed that the cells showed good growth and proliferation whether in 2D culture (Figure 3a) or 3D culture (Figure 3b), and the trend was not different from TCP, which further proved the good compatibility of the hydrogel. The addition of the SCF increased proliferation of DPSCs (P < 0.05), which further verified the previous screening of the SCF concentration. Moreover, the number of 3D cultured cells was still inferior to that of DPSCs cultured on plates, and surprisingly, this gap was compensated by connecting the amino acid sequence (PRG) at the C-end of RADA16-I (Figure 3b) (P < 0.05).
Figure 3.
Effect of SCF-RADA16-I on proliferation and migration of DPSCs. The proliferation of DPSCs cultured in (a) 2D and (b) 3D. (c) Migration images of DPSCs and (d) the corresponding statistical results. (n = 5, *P < 0.05, **P < 0. 01).
The migration results of DPSCs are shown in Figure 3c. According to statistical analysis (d), the number of DPSCs that passed through the polycarbonate membrane of in group SCF + DPSCs was more than that of group DPSCs (P < 0.01), and the SCF-RADA16-I group was more than RADA16-I. No significant difference was found between group RADA16-I and group DPSCs (P > 0.05).
SCF Promoted the Angiogenesis of HUVECs
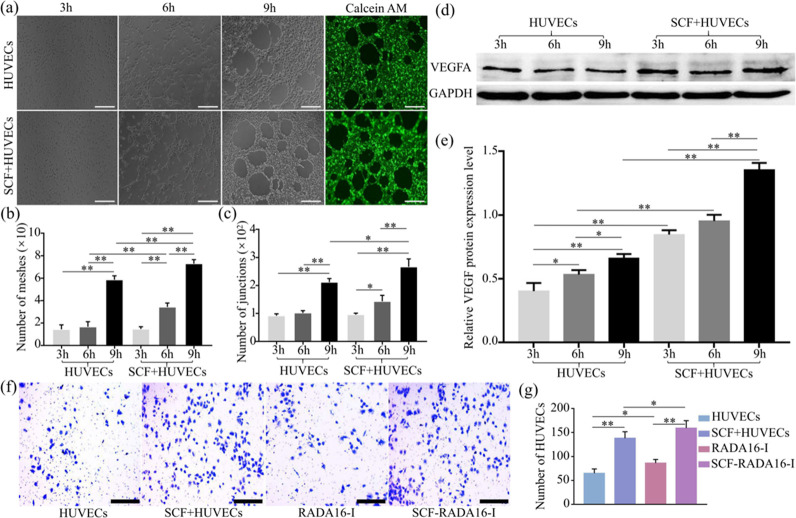
The vascular-like structures formed in group HUVECs and group SCF + HUVECs at hours 3, 6, and 9 are exhibited in Figure 4a. After analysis, the number of meshes (Figure 4b) in the SCF + HUVEC group was more than that in group HUVECs both at hours 6 and 9 (P < 0.01), and the number of junctions (Figure 4c) in the SCF + HUVEC group was more than that in the HUVEC group at hour 9 (P < 0.05), which indicated that the SCF promoted HUVECs to form more vascular-like structures.
Figure 4.
(a–c) Angiogenesis experiments in vitro. (a) Vascular-like structures formed in 3, 6, and 9 h, and the vascular-like structures formed in the ninth hour were dyed with Calcein AM for easy observation. Statistical results include (b) meshes and (c) junctions. (d, e) Level of VEGFA released by HUVECs. (d) Western blotting strips and (e) statistical results. (f) Representative migration images of migration and (g) the statistical results of migration. (*P < 0.05, **P < 0. 01). Scale bar: 200 μm.
The level of VEGFA was detected by western blotting (Figure 4d). After analysis (Figure 4e), the expression of VEGFA increased gradually from hours 3 to 9 whether in group SCF + HUVECs or group HUVECs. Most importantly, the VEGFA protein in group SCF + HUVECs was more than that in group HUVECs whether at hours 3, 6, or 9 (P < 0.01), which indicated that the SCF promoted the expression of VEGFA.
The migration results of HUVECs are shown in Figure 3f. According to statistical analysis in Figure 3e, the number of HUVECs passed through the polycarbonate membrane in groups with the SCF was more than that in corresponding SCF-free groups (P < 0.01). Furthermore, there was a statistical difference between group RADA16-I and group HUVECs (P < 0.05) and group SCF-RADA16-I and group SCF + HUVECs (P < 0.05). The above results indicated that SCF-RADA16-I promoted the migration of HUVECs.
Discussion
Pulp regeneration aims at replacing inflamed or infected pulp with regenerated pulp-like tissue, maximizing the vitality of teeth and continuing to develop immature teeth.3 The key to pulp regeneration is to provide a regenerative microenvironment in the root canal to promote the growth of dental pulp stem cells. In addition, accelerating vascularization is equally important for regenerative pulp tissues.
In this study, the optimal concentration of the SCF was first screened. In previous studies, the SCF was found to promote the migration of dental progenitor cells and neovascularization.23 In the existing literature reports, the additive amount of SCF differs by more than 10 times.19,24 Therefore, we studied the relationship between the SCF concentration and proliferation of DPSCs, and 100 ng/mL was chosen as the optimum concentration, which differed from the study of Zheng et al. where the optimum concentration of the SCF for in vitro expansion of hematopoietic stem cells was 50 ng/mL.23,25 We hypothesized that the optimal working concentration of SCF varies with the type of cells.
The results of CD, SEM, and AFM showed that a wrapped SCF had no effect on the self-assembly process of RADA16-I. Then, RADA16-I composed of nanosized fibers is an ideal microenvironment-simulating extracellular matrix by evaluating the bio-behaviors of DPSCs. In addition, RADA16-I could load the SCF and maintain the activity. The results of this study also proved that RADA16-I wrapping the SCF promoted the bio-behaviors of DPSCs and provided a 3D environment for DPSCs. Interestingly, DPSCs exhibited irregular polygons and aggregate growth during 3D culture in living-cell staining and cytoskeleton staining, which may be related to the grid structure of RADA16-I15 and chemotaxis of the SCF,23,25−28,30 and although it no longer presented the typical long spindle growth, the three-dimensional culture is closer to the real environment in vivo.16,31 It is worth noting that RADA16-I inhibited the proliferation of DPSCs to a certain extent, which may be related to the average diameter of cells being larger than the pore diameter of RADA16-I. In order to make up for the gap between the RADA16-I group and the control group, the functional amino acid sequence was linked at the C-terminal of RADA16-I (RADA16-I-PRG and SCF- RADA16-I-PRG in Figure 3b), which is supported by the relevant literature.14,29
We found that the SCF promoted the migration of DPSCs and HUVECs, while RADA16-I promoted the migration of HUVECs but has no significant effect on DPSCs, which is similar to that in the study of Pan et al. where the SCF promoted the migration of dental progenitor cells23 and the study of Wang et al. where RADA16-I promoted migration of endothelial cells.32 Finally, experiments related to in vitro angiogenesis were carried out, and the result showed that the SCF obviously promoted HUVECs to form more vascular-like structures, and the SCF promoted HUVECs to secrete more VEGFA, providing a possibility for revascularization in the root canal in vivo.
Whether it is the SCF solution or DPSC cell suspension, most of them would be lost when they are inoculated into the root canal. The outstanding advantage of RADA16-I is the injectability;9,10 the root canal has an irregular geometry, and the hydrogel with humidity greater than 99% could well fill the root canal of various shapes, so RADA16-I could be used as a carrier to encapsulate the SCF and DPSCs into the root canal and provide a 3D environment, which is closer to the internal environment for DPSCs. During the culture process, the cells were fully embedded in the three-dimensional environment of the RADA16-I in order to better simulate the internal microenvironment. The 3D culture is between the monolayer cell culture and the animal experiment, allowing the cells to grow in all directions, which could not only simulate the internal environment to the greatest extent but also have the advantages of demonstrating the intuitiveness and conditional controllability of a cell culture, providing a good platform for further study of cell characteristics. RADA16-I could also be further modified by various functional amino acid fragments or growth factors required to obtain better biological properties.15,18 If RADA16-I wrapping DPSCs and the SCF is injected into the root canal sterilized thoroughly, DPSCs will adhere and proliferate in a three-dimensional environment and the SCF will recruit periapical endothelial cells into the root canal to form blood vessels, thus realizing the regeneration of pulp tissue rich in vascular-like structures in vivo.
In addition, SCF-RADA16-I has great potential to become a carrier or study model for therapeutic delivery. The SCF will gradually be released with the degradation of RADA16-I. In Barry et al.’s study, using nanomaterials for the delivery of protein-based therapeutics has shown promise for the treatment of osteoporosis.33 Therefore, the novel approaches with nanotechnologies are expected to treat more diseases.
Meanwhile, there is a limitation in this study, the pH value of the RADA16-I solution is 3 before gel formation, which may affect cell growth if it is not diluted timely and adequately. Therefore, the above results are required to be further verified by in vivo experiments.
Regeneration of pulp tissue rich in vascular-like structures is essential for restoring tooth vitality. In this study, we engineered an injectable peptide scaffold wrapping the SCF and demonstrate the injectable properties crucial for clinical application. The next step in SCF-RADA16-I would be aimed at regeneration of pulp tissue rich in vascular-like structures in vivo, which will verify the viability of such a biomimetic tissue regeneration approach.
Acknowledgments
Authors X.M., L.H., Y.N., and X.W. received funding from the National Natural Science Foundation of China (no. 81970924). Author S.P. received funding from the National Natural Science Foundation of China (no. 81570963). Author L.S. received funding from the Health Commission of Heilongjiang Province (nos. 2017–015). We would like to thank Shuo Wang and Jiaju Lu at Tsinghua University for the helpful suggestion of experiments.
Author Contributions
∥ X.M. and L.S. contributed equally.
The authors declare no competing financial interest.
References
- Xuan K.; Li B.; Guo H.; Sun W.; Kou X.; He X.; Zhang Y.; Sun J.; Liu A.; Liao L.; et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci. Transl. Med. 2018, 10, eaaf3227 10.1126/scitranslmed.aaf3227. [DOI] [PubMed] [Google Scholar]
- Wang F.; Wu Z.; Fan Z.; Wu T.; Wang J.; Zhang C.; Wang S. The cell re-association-based whole-tooth regeneration strategies in large animal; Sus scrofa. Cell Proliferation 2018, 51, e12479 10.1111/cpr.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo S.; Natali A.; Pinna R.; Milia E. Dental pulp regeneration via cell homing. Int. Endod. J. 2018, 51, 405–419. 10.1111/iej.12868. [DOI] [PubMed] [Google Scholar]
- Xu F.; Qiao L.; Zhao Y.; Chen W.; Hong S.; Pan J.; Jiang B. The potential application of concentrated growth factor in pulp regeneration: an in vitro and in vivo study. Stem Cell Res. Ther. 2019, 10, 134. 10.1186/s13287-019-1247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Chen J.; Yang B.; Li L.; Luo X.; Zhang X.; Feng L.; Jiang Z.; Yu M.; Guo W.; Tian W. Combination of aligned PLGA/Gelatin electrospun sheets, native dental pulp extracellular matrix and treated dentin matrix as substrates for tooth root regeneration. Biomaterials 2015, 52, 56–70. 10.1016/j.biomaterials.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Glowacki J.; Mizuno S. Collagen scaffolds for tissue engineering. Biopolymers 2008, 338. 10.1002/bip.20871. [DOI] [PubMed] [Google Scholar]
- Galler K. M.; Hartgerink J. D.; Cavender A. C.; Schmalz G.; D’Souza R. N. A Customized Self-Assembling Peptide Hydrogel for Dental Pulp Tissue Engineering. Tissue Eng., Part A 2012, 18, 176–184. 10.1089/ten.tea.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.; Wang Z.; Guo Y.; Li H.; Chen Z. Design of a RADA16-based self-assembling peptide nanofiber scaffold for biomedical applications. J. Biomater. Sci., Polym. Ed. 2019, 30, 713–736. 10.1080/09205063.2019.1605868. [DOI] [PubMed] [Google Scholar]
- Davis M. E.; Motion J. P. M.; Narmoneva D. A.; Takahashi T.; Hakuno D.; Kamm R. D.; Zhang S.; Lee R. T. Injectable self-assembling peptide nanofibers create intramyocardial microenvironments for endothelial cells. Circulation 2005, 111, 442–450. 10.1161/01.CIR.0000153847.47301.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis-Behnke R. G.; Liang Y.-X.; Tay D. K. C.; Kau P. W. F.; Schneider G. E.; Zhang S.; Wu W.; So K.-F. Nano hemostat solution: immediate hemostasis at the nanoscale. Nanomedicine 2006, 2, 207–215. 10.1016/j.nano.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Gelain F.; Zhao X. Designer self-assembling peptide nanofiber scaffolds for 3D tissue cell cultures. Semin. Cancer Biol. 2005, 15, 413–420. 10.1016/j.semcancer.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gelain F.; Bottai D.; Vescovi A.; Zhang S. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS One 2006, 1, e119 10.1371/journal.pone.0000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes T. C. Novel peptide-based biomaterial scaffolds for tissue engineering. Trends Biotechnol. 2002, 20, 16–21. 10.1016/S0167-7799(01)01840-6. [DOI] [PubMed] [Google Scholar]
- Horii A.; Wang X.; Gelain F.; Zhang S. Biological designer self-assembling peptide nanofiber scaffolds significantly enhance osteoblast proliferation, differentiation and 3-D migration. PLoS One 2007, 2, e190 10.1371/journal.pone.0000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos S.; Zhang S. Long-term three-dimensional neural tissue cultures in functionalized self-assembling peptide hydrogels, matrigel and collagen I. Acta Biomater. 2013, 9, 5162–5169. 10.1016/j.actbio.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Liedmann A.; Rolfs A.; Frech M. J. Cultivation of human neural progenitor cells in a 3-dimensional self-assembling peptide hydrogel. J. Visualized Exp. 2012, 5ll, e3830 10.3791/3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.; Zygourakis K.; Farach-Carson M. C.; Yaszemski M. J.; Mikos A. G. Attachment, proliferation, and migration of marrow stromal osteoblasts cultured on biomimetic hydrogels modified with an osteopontin-derived peptide. Biomaterials 2004, 25, 895–906. 10.1016/S0142-9612(03)00602-1. [DOI] [PubMed] [Google Scholar]
- Liu X.; Wang X.; Wang X.; Ren H.; He J.; Qiao L.; Cui F.-Z. Functionalized self-assembling peptide nanofiber hydrogels mimic stem cell niche to control human adipose stem cell behavior in vitro. Acta Biomater. 2013, 9, 6798–6805. 10.1016/j.actbio.2013.01.027. [DOI] [PubMed] [Google Scholar]
- Kisiday J.; Jin M.; Kurz B.; Hung H.; Semino C.; Zhang S.; Grodzinsky A. J. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 9996–10001. 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatullo M.; Codispoti B.; Pacifici A.; Palmieri F.; Marrelli M.; Pacifici L.; Paduano F. Potential use of human periapical cyst-mesenchymal stem cells (hpcy-mscs) as a novel stem cell source for regenerative medicine applications. Front. Cell Dev. Biol. 2017, 5, 103. 10.3389/fcell.2017.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini A.; Scacco S.; Coletti D.; Pluchino S.; Tatullo M. Mesenchymal Stem Cells as Promoters, Enhancers, and Playmakers of the Translational Regenerative Medicine. Stem Cells Int. 2017, 2017, 1–2. 10.1155/2017/3292810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Horii A.; Zhang S. Designer functionalized self-assembling peptide nanofiber scaffolds for growth, migration, and tubulogenesis of human umbilical vein endothelial cells. Soft Matter 2008, 4, 2388–2395. 10.1039/b807155a. [DOI] [Google Scholar]
- Pan S.; Dangaria S.; Gopinathan G.; Yan X.; Lu X.; Kolokythas A.; Niu Y.; Luan X. SCF promotes dental pulp progenitor migration, neovascularization, and collagen remodeling - potential applications as a homing factor in dental pulp regeneration. Stem Cell Rev. Rep. 2013, 9, 655–667. 10.1007/s12015-013-9442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagari E.; Rand M. K.; Tayari L.; Vastardis H.; Sharma P.; Hauschka P. V.; Damoulis P. D. Expression of stem cell factor and its receptor, c-kit, in human oral mesenchymal cells. Eur. J. Oral Sci. 2006, 114, 409–415. 10.1111/j.1600-0722.2006.00388.x. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Sun A.; Han Z. C. Stem cell factor improves SCID-repopulating activity of human umbilical cord blood-derived hematopoietic stem/progenitor cells in xenotransplanted NOD/SCID mouse model. Bone Marrow Transplant. 2005, 35, 137–142. 10.1038/sj.bmt.1704751. [DOI] [PubMed] [Google Scholar]
- Reber L.; Da Silva C. A.; Frossard N. Stem cell factor and its receptor c-Kit as targets for inflammatory diseases. Eur. J. Pharmacol. 2006, 533, 327–340. 10.1016/j.ejphar.2005.12.067. [DOI] [PubMed] [Google Scholar]
- Du Z.; Cai H.; Ye Z.; Tan W.-S. Optimization of SCF feeding regimen for ex vivo expansion of cord blood hematopoietic stem cells. J. Biotechnol. 2013, 164, 211–219. 10.1016/j.jbiotec.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Du Z.; Wang Z.; Zhang W.; Cai H.; Tan W.-S. Stem cell factor is essential for preserving NOD/SCID reconstitution capacity of ex vivo expanded cord blood CD34+ cells. Cell Proliferation 2015, 48, 293–300. 10.1111/cpr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C.; Panseri S.; Villa O.; Silva D.; Gelain F. 3D culture of adult mouse neural stem cells within functionalized self-assembling peptide scaffolds. Int. J. Nanomed. 2011, 6, 943–955. 10.2147/IJN.S17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangsawasdi N.; Zehnder M.; Patcas R.; Ghayor C.; Siegenthaler B.; Gjoksi B.; Weber F. E. Effects of Stem Cell Factor on Cell Homing During Functional Pulp Regeneration in Human Immature Teeth. Tissue Eng., Part A 2017, 23, 115–123. 10.1089/ten.tea.2016.0227. [DOI] [PubMed] [Google Scholar]
- Chaudhuri O. Viscoelastic hydrogels for 3D cell culture. Biomater. Sci. 2017, 5, 1480–1490. 10.1039/C7BM00261K. [DOI] [PubMed] [Google Scholar]
- Wang X.; Qiao L. Screen and synthesis of functionalized self-assembling peptide nanofiber scaffolds for promoting vascular endothelial cells growth. Zhongguo Zuzhi Gongcheng Yanjiu yu Linchuang Kangfu. 2011, 15, 7075–7079. [Google Scholar]
- Barry M.; Pearce H.; Cross L.; Tatullo M.; Gaharwar A. K. Advances in Nanotechnology for the Treatment of Osteoporosis. Curr. Osteoporosis Rep. 2016, 14, 87–94. 10.1007/s11914-016-0306-3. [DOI] [PubMed] [Google Scholar]