Abstract

Paracoccus sp. MKU1, a metabolically versatile bacterium that encompasses diverse metabolic pathways in its genome for the degradation of aromatic compounds, was investigated for catechol bioremediation here for the first time to our knowledge. Paracoccus sp. MKU1 degraded catechol at an optimal pH of 7.5 and a temperature of 37 °C, wherein 100 mg/L catechol was completely mineralized in 96 h but required 192 h for complete mineralization of 500 mg/L catechol. While investigating the molecular mechanisms of its degradation potential, it was unveiled that Paracoccus sp. MKU1 employed both the ortho and meta pathways by inducing the expression of catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O), respectively. C23O expression at transcriptional levels was significantly more abundant than C12O, which indicated that catechol degradation was primarily mediated by extradiol cleavage by MKU1. Furthermore, poly(MAA-co-BMA)-GO (PGO) microcomposites containing Paracoccus sp. MKU1 were synthesized, which degraded catechol (100 mg/L) completely within 48 h with excellent recycling performance for three cycles. Thus, PGO@Paracoccus microcomposites proved to be efficient in catechol degradation at not only faster rates but also with excellent recycling performances than free cells. These findings accomplish that Paracoccus sp. MKU1 could serve as a potential tool for bioremediation of catechol-polluted industrial wastewater and soil.
1. Introduction
Biodegradation of aromatic compounds and their derivatives in both soil and water is of major concern because of their toxicity and carcinogenic nature.1 Many aromatic compounds like benzene, toluene, xylene, naphthalene, benzoate, and phenol and its derivatives are utilized by microorganisms as carbon and energy sources exclusively and degraded aerobically into more reactive dihydroxylated intermediates such as catechol, protocatechuate, gentisate, and benzoquinol.2 Catechol (1,2-dihydroxybenzene) is one of the inevitable substances present in various industries such as petroleum, cosmetic, chemical, rubber, medicine, pesticide, and textile industries.3,4 Annually, 25 000 metric tons of catechol is manufactured worldwide due to its extensive usage in various industries and is unduly released into the environment in large quantities.5 Catechol is also said to be genotoxic in nature; it causes mutations, DNA breakage, and chromosomal aberrations, such as aneuploidy.6 Since catechol is highly miscible in water, it infiltrates and migrates easily into both soil and aquatic environments. Therefore, its existence is widespread in the environment and thus becomes a serious threat to human health.7
Biological degradation of xenobiotics is evidenced to be superior to physical and chemical means in terms of its efficiency, cost effectiveness, complete mineralization with innocuous end products, and simplicity in operation.8 Catechol is aerobically degraded through two pathways, such as ortho- and meta cleavage pathways, in which the intradiol and extradiol ring cleavages are catalyzed by catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O) enzymes, respectively.9,10 Both the degradative pathways result in the products that end with the tricarboxylic acid (TCA) cycle (Figure S1).11 The existence of catechol dioxygenases (CDOs) in a microbe is renowned as a common biomarker to assess its bioremediation potential of aromatic compounds.12 To date, several aerobic catechol degraders have been isolated and characterized from classes of bacteria, yeast, and fungi, such as Pseudomonas cepacia,13Arthrobacter sp., Acinetobacter sp., Sphingomonas subarctica, and Ralstonia pickettii; Comamonas testosterone,14Achromobacter xylosoxidans,15 and Halomonas campisalis(16) and Aspergillus awamori, respectively.17
The genus Paracoccus is Gram-negative, denitrifying bacteria that consists of 17 species currently,18 which exhibit a diverse range of metabolic flexibility in utilizing a wide array of hazardous pollutants and thus gained renowned attention by researchers all over the world for bioremediation applications.19 Several species of Paracoccus are known to bioremediate aromatic compounds like pyridine, halobenzoate, and pyrene.20 Furthermore, diverse catabolic clusters have also been detected in the genome of Paracoccus sp. that includes N-methylpyrrolidone,21 chloroacetamide herbicides,22 piperazine,23 and dimethylformamide,24 which evidently signifies the metabolic versatility of Paracoccus and justifies the suitability for its exploitation in bioremediation of recalcitrant pollutants. To the best of our knowledge, although there have been many reports concerning catechol degradation by other bacteria, a metabolically versatile, denitrifying bacterium Paracoccus has not yet been investigated for its catechol bioremediation potential. Therefore, it is of great importance to explore the efficacy of catechol biodegradation by Paracoccus sp. MKU1, a dimethylformamide (DMF) degrader isolated at our laboratory from the textile industry effluent.25 Subsequent genome sequencing of Paracoccus sp. MKU1 evidenced the presence of genes corresponding to diverse xenobiotics degradation such as aromatic amines, biphenyls, chromate reductase, mercuric ion reductase, toluene, benzoate, catechol, and chloroaromatic compounds.26 Furthermore, metabolic pathways corresponding to aromatic hydrocarbon degradation like salicylate and gentisate catabolism, protocatechuate and catechol branch from β-ketoadipate pathways, and homogentisate pathways were found in this novel bacterium.
In this study, we have evaluated the catechol degradation potential of metabolically versatile Paracoccus sp. MKU1 under different growth conditions including pH, incubation temperature, and additional supplementation of different carbon sources. We have also provided insights into both ortho and meta cleavage pathways and their regulation during catechol biodegradation by Paracoccus sp. Further, immobilization of Paracoccus sp. MKU1 on poly(MAA-co-BMA)-GO (PGO) microcomposites resulted in enhanced biodegradation of catechol with excellent recycling performance for three cycles. To our knowledge, this is the first study describing the efficiency of a metabolically versatile Paracoccus sp. in catechol bioremediation.
2. Results and Discussion
Paracoccus sp. MKU1 was originally isolated in our laboratory from an industrial effluent as a DMF degrader,25 which was later shown to contain catechol ring-cleaving dioxygenases by its genome sequencing.26 To our surprise, no study has characterized catechol degradation by the genera Paracoccus, which are metabolically versatile with greater flexibility in metabolizing a wide range of organic and inorganic compounds as substrates for their growth. Therefore, it is necessary to prove the existence of catechol degradative nature and the properties of catechol dioxygenases in the strain Paracoccus sp. MKU1.
2.1. Bacterial Growth and Catechol Degradation Studies
Paracoccus sp. MKU1 was cultured initially in minimal salts medium (MSM) supplemented with catechol (100 mg/L) as the exclusive carbon source at pH 7.2 and 37 °C, wherein no lag phase was observed for the acclimatization of this strain at this concentration. The maximum growth was noticed at 72 h of incubation, after which the growth started to decline. Thus, an increase in bacterial cell density in MSM containing catechol alone evidenced the utilization of such aromatic compounds. This study provides evidence, for the first time to our knowledge, for the biodegradation of catechol by a Paracoccus sp. Further, the degradation rate of catechol was determined by the 4-aminoantipyrine assay, where 81% of catechol in MSM was degraded within 48 h and almost complete degradation was observed in the stationary phase at 96 h with more than 98% of catechol degradation (Figure 1a). It is evident from the results that most of the catechol (100 mg/L) was being consumed at a faster rate at the initial concentration during the exponential phase of bacterial growth. Interestingly, the strain MKU1 could achieve complete degradation of catechol in the ranges of 200 and 500 mg/L within 120 and 192 h, respectively (Figure 1b). There are several bacteria that have been characterized to metabolize catechol including Pseudomonas sp.,3Rhodococcus sp.,32Stenotrophomonas sp., Acinetobacter sp.,14 and Achromobacter sp.15 The genus Paracoccus is ubiquitous in nature, which has been previously isolated from wastewater, contaminated soil, and activated sludge. They are characterized to utilize a wide array of organic compounds and aromatic hydrocarbons as nutrient and energy sources.20 These biodegradability results of the preliminary experiment authenticated the potential of Paracoccus sp. MKU1 in assimilating catechol as a source of carbon to support its growth.
Figure 1.
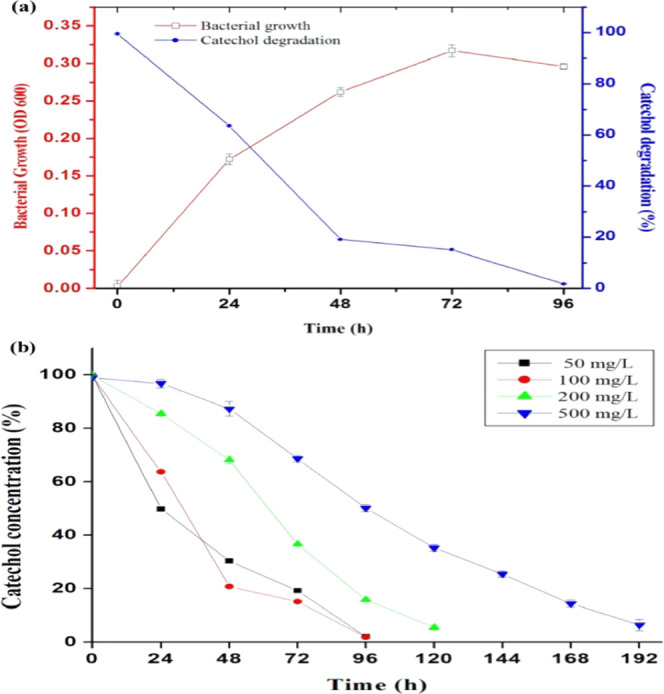
Growth of Paracoccus sp. MKU1 in minimal salt media containing catechol. (a) Batch culture of Paracoccus sp. MKU1 in MSM supplemented with catechol (100 mg/L) was raised at 37 °C and the samples were assessed for growth (□) at OD600 and residual catechol (●) using the 4-aminoantipyrine assay at different time intervals. (b) Time course of catechol degradation at different initial concentrations of catechol by Paracoccus sp. MKU1. Data are means ± SE of three separate determinations performed on three separate occasions.
2.2. Influence of pH, Temperature, and Secondary Carbon Sources on Catechol Degradation
The optimal growth conditions and catechol degradation of Paracoccus sp. MKU1 was investigated by culturing the bacteria in MSM containing catechol at various growth conditions such as pH, temperature, and secondary carbon sources. All of the biodegradation experiments were performed in batch mode at a volume of 100 mL in 250 mL Erlenmeyer flasks. Paracoccus sp. MKU1 had shown growth in MSM containing catechol having different pH values ranging from 5.0 to 8.5 (Figure 2a) with maximum growth at 72 h for all of the tested pH conditions. The percentage of catechol degradation was observed to be largely varied with different pH values and showed almost complete degradation at pH 7.5 and 96 h of incubation (Figure 2b).
Figure 2.
Effect of pH, temperature, and secondary carbon sources on catechol degradation by Paracoccus sp. MKU1. (a) Bacterial growth at different pH values. (b) Catechol degradation at different pH values. (c) Bacterial growth at different temperatures. (d) Catechol degradation at different temperatures. (e) Bacterial growth in different secondary carbon sources. (f) Catechol degradation in different secondary carbon sources. All of the assays were performed in triplicate. Values are mean ± SEM.
Catechol was degraded by Paracoccus sp. MKU1 effectively at different incubation temperatures with an optimum of 37 °C. Although the bacterial growth was slightly decreased at both 25 and 42 °C compared to that at 37 °C (Figure 2c), the temperature did not influence the efficiency of catechol degradation much (Figure 2d). The degradation rates of the cultures incubated at 25, 37, and 42 °C reached 97, 98, and 93%, respectively, within 96 h of incubation. Thus, it appears that the maximum biodegradation of catechol could occur at 37 °C as an optimum temperature for Paracoccus sp. MKU1 and that it could tolerate temperature variations without affecting the potential of catechol degradation. In contrast, the temperature has been evidenced as the crucial factor to affect the pyridine biodegradation process by Paracoccus sp. NJUST30.33
The response of Paracoccus sp. MKU1 in the presence of various secondary carbon sources (0.2%) in terms of bacterial growth and catechol degrading efficiency varied significantly. Notably, maximum growth was attained at 48 h of incubation with the addition of all of the tested secondary carbon sources, whereas it required 72 h for the cells grown in MSM containing catechol alone (Figure 2e). The addition of succinate had negatively influenced the bacterial growth with a considerable decrease than control, while the other carbon sources including glucose, sucrose, fructose, and acetate had enhanced the bacterial growth considerably (Figure 2e). Meantime, there was no increase in the catechol degradation rate in the cultures with additional carbon sources in comparison to control (98% at 96 h), while sucrose and fructose resulted only in ∼92 and ∼89% of degradation, respectively. Even though the bacterial growth was comparatively less in the succinate-containing cultures, the catechol degradation efficiency by its utilization was not affected and almost identical when compared to that of control. Catechol degradation by Paracoccus sp. MKU1 was not repressed significantly (P < 0.01) by any of the carbon sources supplied, except fructose. Similarly, fructose has been shown to reduce the degradation rate of phenol by Ralstonia eutropha(34) and chlorobenzoic acid by Enterobacter cloacae,35 even the biomass yield got improved substantially. Similar repression by glucose enrichment was observed with catechol degradation by Pseudomonas sp. CF600.36 Previously, it has been demonstrated that aromatic compounds are the preferential substrate for Pseudomonas putida CSV86 over glucose and the cometabolized aromatic compounds are preferred as substrates over the presence of glucose37 and succinate.38 Moreover, the utilization of aromatics by the strain CSV86 was not repressed either by glucose or succinate.38
2.3. Gene Expression in Relation to Catechol Degradation
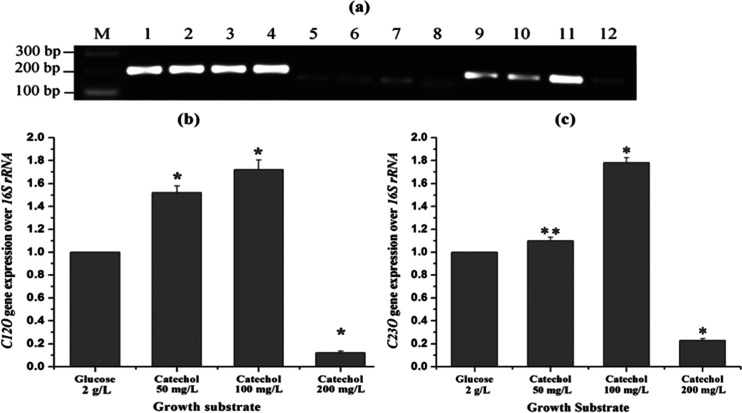
To understand the mechanism and regulation of catechol biodegradation by Paracoccus sp. MKU1, we analyzed the expression pattern of C12O and C23O genes at the transcript level in the cells grown on glucose and different concentrations of catechol (50, 100, and 200 mg/L). From the results, some interesting insights into the transcriptional regulation of genes corresponding to CDO were observed. First, the detection of C12O and C23O transcripts in the cultures grown in MSM containing catechol ensured that Paracoccus sp. MKU1 employs both the ortho and meta cleavage pathways for catechol degradation (Figure 3a). Previously, several species of Pseudomonas(13,39,40) and Sphingomonas yanoikuyae B141 have been evidenced to recruit both ortho and meta cleavage pathways simultaneously for the cleavage of the catechol ring. In contrast, Rhizobiales F11 predominantly degraded fluorobenzene via catechol with subsequent ortho cleavage only,42 while Acinetobacter sp. YAA degraded aniline to catechol, which was further fissioned through meta cleavage only.43 Second, there was a basal expression of both the CDOs (C12O and C23O) at mRNA levels when Paracoccus sp. MKU1 was cultured in the MSM containing glucose as the sole energy and carbon source (Figure 3a). These observations corroborate the findings of Li et al.,44 who evidenced the low-level expression of C12O-encoding gene catA and C23O-encoding gene nahH in the glucose-containing cultures of P. putida ND6. Likewise, the constitutive expression of C12O at low levels was observed in Acinetobacter radioresistans when grown in a nonaromatic growth substrate, acetate.45 Third, the expression of C23O at mRNA levels in Paracoccus sp. MKU1 was higher than that of C12O (Figure 3a). C23O expression at mRNA levels in the glucose-containing cultures was ∼1.9-fold higher than C12O, whereas ∼2.1-fold higher in the catechol-induced cultures of Paracoccus sp. MKU1. It has been already observed that bacteria employs the ortho pathway efficiently for converting carbon into cell biomass than the meta pathway, while meta pathway is employed for the utilization of phenolic compounds at a higher rate.46 Fourth, the expressions of both the CDO genes in the MKU1 strain were induced by catechol. Once the MKU1 cultures were grown in MSM supplemented with catechol, the expression of C12O and C23O genes at transcript levels was found to higher significantly (P < 0.01) in a concentration-dependent manner. In particular, the mRNA expression levels of C12O were increased by 1.5 and 1.7 folds with the addition of catechol at the concentrations of 50 and 100 mg/L, respectively, while catechol at a concentration of 200 mg/L strongly inhibited the C12O expression (Figure 3b). Likewise, C23O mRNA transcript levels were also found to increase with the concentrations of catechol up to 100 mg/L and decreased prominently with the catechol concentration of about 200 mg/L (Figure 3c). These observations stress that catechol could serve as a potent inducer for the expression of both the CDOs in Paracoccus sp. MKU1. This result was not consistent with previous observations with Planococcus sp. S5, in which increasing concentrations of phenol induced the expression of C23O activity alone but not C12O.47 While metabolizing phenol and benzoate by P. cepacia and P. putida, catechol has not been found to induce CDOs and both of them failed to grow on catechol when used as a sole carbon source.13,39,40 These interpretations, although conflicting, suggested that different metabolites produced during metabolism could serve as an inducer for the particular enzyme in various bacterial species.
Figure 3.
Transcript analysis for C12O and C23O expression by the semiquantitative reverse transcription polymerase chain reaction (RT-PCR). The strain MKU1 was grown in MSM supplemented with either glucose or catechol and the total RNA was prepared from the respective cultures for RT-PCR. (a) RT-PCR amplification of 16S rRNA (lanes 1–4), C12O (lanes 5–8), and C23O (lanes 9–12). Lanes 1, 5, and 9: RT-PCR products from the total RNA of 2 mg/L glucose-grown cells. Lanes 2, 6, and 10: RT-PCR products from the total RNA of 50 mg/L catechol-grown cells. Lanes 3, 7, and 11: RT-PCR products from the total RNA of 100 mg/L catechol-grown cells. Lanes 4, 8, and 12: RT-PCR products from the total RNA of 200 mg/L catechol-grown cells. (b) Relative quantification of C12O expression over 16S rRNA. (c) Relative quantification of C23O expression over 16S rRNA. Experiments were performed in triplicate. Results of a representative experiment are shown. Values are mean ± SEM. *P < 0.01; **P < 0.05.
2.4. Characterization of GO and PGO
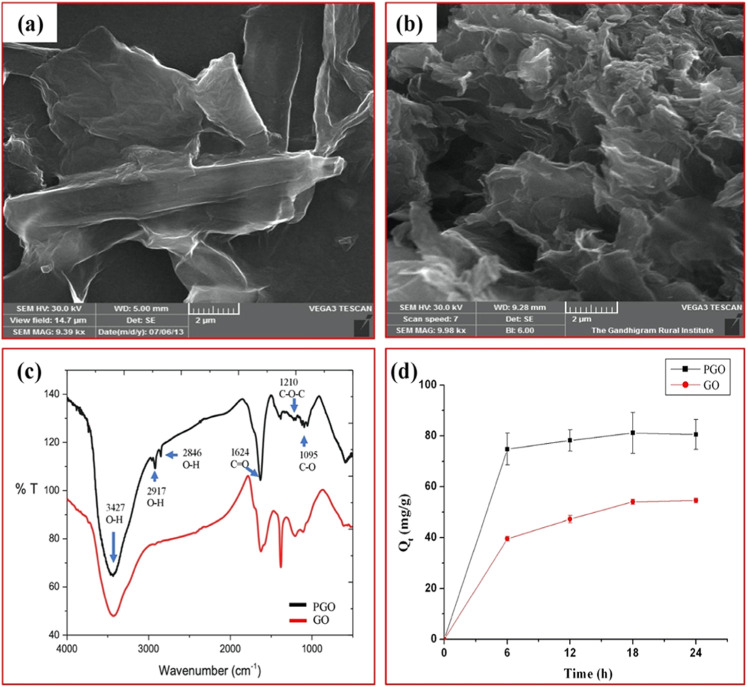
Atomic force microscopy (AFM) images of GO show irregular morphology of the initial GO nanosheets with 1–2 layers (Figure S2). Further, the morphologies of both GO and PGO were characterized by scanning electron microscopy (SEM) (Figure 4a,b). The SEM image of GO shows a sheetlike structure with a smooth surface, whereas that of PGO shows a more aggregated rough surface. GO appeared flat and smooth, whereas PGO appeared rough and rugged in nature. The conversion of the overlay surface to the rocky surface evidenced the successful polymerization with methacrylic acid (MAA) and butyl methacrylate (BMA). A similar kind of morphological conversion has been previously observed when GO undergoes polymerization with MAA and BMA.30
Figure 4.
Characterization of GO and PGO. (a) SEM image of GO. (b) SEM image of PGO. (c) Fourier transform infrared (FT-IR) spectra of GO and PGO. (d) Adsorption of GO and PGO with catechol.
The characteristic FT-IR spectrum of GO and PGO composites evidenced the existence of oxygen-containing functional groups, in which the major absorption band at 3427 cm–1 was designated to the O–H group stretching vibrations (Figure 4c). The absorption peak at 1624 cm–1 can be considered to C=O stretching of the carboxylic functional groups. The absorption peaks at about 1210 and 1095 cm–1 were designated to the C–O–C bond and C–O stretching vibrations.
2.5. Efficiency of GO and PGO Adsorption with Catechol
Adsorption efficiency of GO/PGO (50 mg) with 100 mg/L catechol was determined at regular 6 h intervals by the 4-aminoantipyrine assay. As shown in Figure 4d, both GO and PGO adsorbed catechol efficiently with a maximal adsorption of 54.4 mg/g for GO and 81.2 mg/g for PGO at 18 h. There was a 42% increase in the maximal adsorption of catechol with PGO than GO. Similar observations have been already reported in aniline adsorption with GO-mATP48 and DMF adsorption with GO and PGO.30 The greater adsorption efficiency of GO and PGO with catechol might be due to the presence of two hydroxyl groups in catechol, which have higher affinity toward carboxyl groups of GO and PGO. It has been already mentioned that the presence of two hydroxyl groups in catechol significantly increases its adsorption on all organic and inorganic compounds. These interactions were robust and thereby allow the catechol to adsorb even in wet circumstances.49
2.6. Immobilization of Paracoccus sp. MKU1
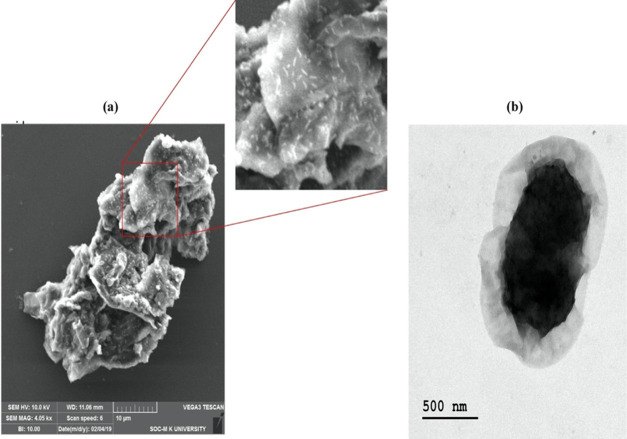
The PGO microcomposites of Paracoccus sp. MKU1 were successfully derived using N-hydroxysuccinimide as a cross-linker. The immobilization process was achieved through the amine group of the bacterial cell wall, which was chemically conjugated with the carboxyl group of PGO in the presence of N-hydroxysuccinimide. Moreover, the existence of abundant various functional groups in PGO facilitates the creation of a strong covalent bond with the bacterial cell wall surface. Finally, SEM (Figure 5a) and transmission electron microscopy (TEM) (Figure 5b) analyses confirmed the adhesion of bacteria on the surface of PGO, and the observation of clear rod-shaped cells on composites indicated the bacterial adsorption with PGO and the absence of antimicrobial activity of PGO against Paracoccus sp. MKU1.
Figure 5.
Characterization of PGO-immobilized Paracoccus sp. MKU1. (a) SEM images and (b) TEM images of immobilized Paracoccus sp. MKU1.
2.7. Biodegradation of Catechol by Immobilized Paracoccus sp. MKU1
The efficiency of catechol degradation at the initial concentration of 100 mg/L by free and PGO-immobilized Paracoccus sp. MKU1 was investigated after 24 h of treatment by estimating catechol concentrations using high-performance liquid chromatography (HPLC) analysis (Figure S3). Paracoccus sp. MKU1 degraded 39.02 mg/L in 24 h, whereas PGO-immobilized Paracoccus sp. MKU1 degraded 84.4 mg/L in 24 h, which was 2.2-fold higher compared than the free cells of Paracoccus sp. MKU1 (Figure 6a). Moreover, PGO-immobilized Paracoccus sp. MKU1 degraded catechol completely at 48 h. Previously, biodegradation of catechol using immobilized bacteria with various adsorbents has been performed (Table 1). From the observations, it is evident that the simultaneous adsorption and degradation process of PGO-coupled Paracoccus sp. MKU1 appeared advantageous in terms of effective degradation of catechol in a shorter period due to the increased adsorption of catechol to the relatively large surface area of PGO.
Figure 6.
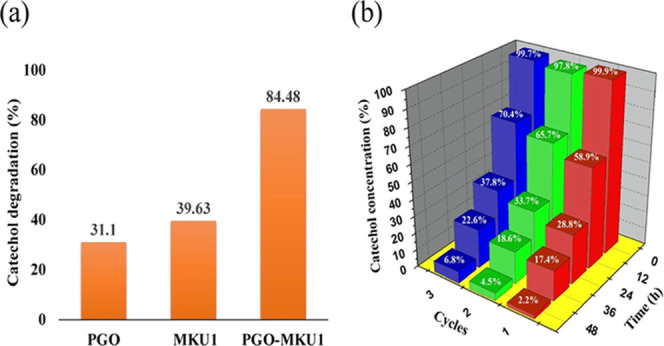
(a) Comparative analysis of catechol degradation by Paracoccus sp. MKU1 and PGO-immobilized Paracoccus sp. MKU1 at 24 h. (b) Recycling performance of PGO-immobilized Paracoccus sp. MKU1 in the catechol biodegradation process.
Table 1. Comparison of Catechol Degradation by Various Immobilized Cells.
| bacteria | immobilization material | concentration (mg/L) | time (h) | degradation (%) | reference |
|---|---|---|---|---|---|
| Rhizobium | sodium alginate | 220 | 60 | 98.0 | (50) |
| Pseudomonas sp. | calcium alginate | 44 | 72 | 83.2 | (51) |
| Pseudomonas sp. NGK1 | polyurethane foam | 4400 | 72 | 75.0 | (52) |
| A. xylosoxidans 15DKVB | sodium alginate | 500 | 42 | 98 | (15) |
| Paracoccus sp. MKU1 | graphene oxide | 100 | 48 | 97.8 | present study |
| 200 | 72 | 97.24 | present study |
2.8. Recycling Potential of PGO-Coupled Paracoccus sp. MKU1
The recycling performance of PGO-coupled Paracoccus sp. MKU1 was analyzed by continuous batch experiments, wherein the inoculum was reused for three batches under constant conditions. The degradation of catechol was measured at regular 6 h time intervals using 4-aminoantipyrine, and the results of the recycle batch experiment are shown in Figure 6b. The efficiency of simultaneous catechol adsorption and degradation in the first cycle by immobilized Paracoccus sp. MKU1 was 71.2% within 24 h, whereas the complete removal of catechol (97.8%) occurred at 48 h. Meantime, the second cycle (66.3% at 24 h and 95.5% at 48 h) and third cycle (62.2% at 24 h and 93.2% at 48 h) performances of catechol degradation were comparatively less than the first cycle, albeit it exhibited far better degradation performance in the third cycle. These observations were consistent with a prior study, wherein the third cycle performance with PGO-immobilized bacteria showed better degradation of DMF.30
3. Conclusions
Paracoccus sp. MKU1, a heterotrophic denitrifying bacterium with great metabolic flexibility toward various organic and inorganic compounds, is investigated in this study for the complete mineralization of catechol for the first time. Paracoccus sp. MKU1 could degrade catechol (100 mg/L) completely within 96 h, wherein both the ortho and meta cleavage pathways were employed. Additionally, the simultaneous adsorption and biodegradation (SAB) process was employed to immobilize Paracoccus sp. MKU1 on PGO microcomposites, which proved to be efficient in the complete removal of catechol at faster rates (48 h). Recycling performance of PGO-immobilized Paracoccus sp. MKU1 was also assessed for three cycles, which evidenced the effective removal of catechol (>93%) even in the third cycle. These results prompted that Paracoccus sp. MKU1 could serve as a valuable tool for effective bioremediation of catechol-contaminated industrial wastewater and soil. To our knowledge, this study is the first to report the processes and mechanisms of catechol degradation by Paracoccus sp. MKU1, a metabolically versatile denitrifying bacterium.
4. Materials and Methods
4.1. Bacterial Growth and Culture Conditions
Pure culture of Paracoccus sp. MKU1 (NCBI accession no. KC753233.1) maintained on LB agar was transferred into 100 mL of minimal salts medium (MSM) containing catechol as seed culture for its enrichment. The composition of MSM (pH 7.2) is (in g/L) as follows: K2HPO4, 1.8; KH2PO4, 1.2; NH4Cl, 4; MgSO4, 0.2; FeSO4, 0.01; NaCl, 0.1; and catechol, 0.01. The bacteria were cultivated at 37 °C in shaking conditions at 200 rpm for 48 h. Further, the acclimatization process was successively achieved by inoculating the bacteria into fresh MSM containing different concentrations of catechol ranging from 0.01 to 0.1 g/L. Finally, the enriched culture was used as the standard inoculum for further experiments.
4.2. Optimization of Catechol Degradation Conditions
A single-factor experiment was performed to delineate the optimal conditions to achieve maximum degradation of catechol using Paracoccus sp. MKU1 by varying the pH and temperature. For the determination of optimal pH conditions for catechol degradation, axenic cultures of Paracoccus sp. MKU1 (1% inoculum) were inoculated in MSM containing catechol (100 mg/L) with different pH values ranging from 5.0 to 8.5. To assess the influence of temperature on catechol degradation, the MS medium with pH 7.2 was incubated at different temperatures including 25, 37, and 45 °C. All of the cultures were maintained in an orbital shaker with constant agitation at about 200 rpm. The samples were collected aseptically at every 24 h interval and assayed for cell mass at OD600. The samples collected at different time points were centrifuged at 5000 rpm for 20 min and assayed for the presence of residual catechol using 4-aminoantipyrine.27 Shortly, 1 mL of culture supernatant was directly added to 3 mL of 4-aminoantipyrine reagent (100 mL of distilled water contains 100 mg of 4-aminoantipyrine, 10 mL of 20% Na2CO3, and 2 mL of 1 N NaOH). The OD was measured at 540 nm after 20 min of incubation.
The influence of different carbon sources (0.2%) such as glucose, sucrose, fructose, acetate, and succinate on catechol degradation was also studied separately by culturing Paracoccus sp. MKU1 in MS media containing catechol. Pure culture of Paracoccus sp. MKU1 was grown all through the experiment with catechol (100 mg/L) at pH 7.5 and temperature 37 °C. All of the assays were performed thrice.
4.3. Semiquantitative RT-PCR
The bacteria were grown in 100 mL of MSM containing different concentrations of catechol (50, 100, and 200 mg/L). At the exponential growth phase of bacteria, total RNA was purified using the TRIZOL reagent. With 3 μg of total RNA, cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen). Semiquantitative RT-PCR was performed with 25 PCR cycles at the logarithmic phase and an annealing temperature of 52 °C using gene-specific primers for C12O and C23O with 16S rRNA as a control. The gene-specific primers used for semiquantitative RT-PCR are as follows: C12O, forward 5′-GCGCATATCCACTTCTTCGT-3′, reverse 5′-AATGCTTTCCTCGTCAGTGC-3′; C23O, forward 5′-GGACTGGGGAT CTTCACCTT-3′, reverse 5′-AGAAGCCGTTCGTGTCGTAG-3′; and 16S rRNA, forward 5′-GCAGCAGTGGGGAATCTTAG-3′, reverse 5′-CGCTTTACGCCCAGTAATTC-3′. The PCR amplicons were electrophoresed on 2% gels and imaged, and the densitometry analysis was performed using ImageJ software as mentioned earlier.28 Relative expression of CDOs at transcript levels was quantitated with the bacterial cultures grown in catechol after normalization with house-keeping gene 16S rRNA and expressed in folds, where the mRNA level obtained with the cultures grown in glucose is set as 1.
4.4. Preparation of Graphene Oxide (GO)
Graphite powder was used for the synthesis of GO by the modified Hummer’s method.29 H2SO4 (70 mL) was added to 6 g of graphite powder and 3 g of NaNO3 in an RB flask, and the temperature was decreased to 4 °C. KMnO4 (9 g) was added slowly into the reaction mixture at 20 °C. Then, the temperature of the reaction mixture was increased to 35 °C and the mixture agitated for about 30 min. Later, 400 mL of water was added and the mixture was heated at 98 °C for 15 min. Finally, 30% of H2O2 was added to terminate the reaction. Then, the product was cooled and washed with 0.1 M HCl to detach the excessive metal ions. It was subjected to continuous wash with water until the color of the solution become brown-yellow, and the synthesized GO was dried at room temperature and stored for further studies.
4.5. Synthesis of Poly(MAA-co-BMA)-GO Composite (PGO)
PGO was prepared from the polymerization of monomeric units methacrylic acid (MAA) and butyl methacrylate (BMA) with GO using azobisisobutyronitrile (AIBN) as the initiator.30 For this, 100 mg of dried GO and 268 μL of triethylamine (TEA) were dispersed in 40 mL of dimethylacetamide (DMAC) and stirred under constant pressure in a funnel that contained 188 μL of methylacetamide (MAC) and 20 μL of DMAC for neutralizing the HCl produced during the reaction. Then, the mixture was transferred to a conical flask dropwise under a N2 atmosphere at 0 °C, heated to room temperature, and agitated for 24 h. After centrifugation, the pellet was washed with acetone and then dried in a vacuum oven to get the solid product, which was then dissolved in 20 mL of DMF. Afterward, the monomer mixture containing 70 μL of MAA and 385 μL of BMA along with 8 mg of AIBN was added to initiate the reaction process in stirring conditions, followed by heating to 70 °C for about 8 h under N2 atmospheric conditions. After cooling to room temperature, the mixture was precipitated using ether, filtered, and then washed to remove the excessive monomers. Finally, the mixture was dried in a vacuum oven to obtain PGO.
4.6. Preparation of PGO Microcomposites of Paracoccus sp. MKU1
The PGO microcomposites of Paracoccus sp. MKU1 were successfully derived as detailed below. Initially, 100 mg of synthesized PGO and 100 mg of N-hydroxysuccinimide (NHS) were dispersed in 40 mL of DMF. Subsequently, 300 mg of N,N′-dicyclohexylcarbodiimide (DCC) and 4-dimethyl aminopyridine (DMAP) in a ratio of 1:1 was dispersed in a flask to catalyze the reaction and mixed by incubating at room temperature for 24 h. Later, the mixture was centrifuged and treated with phosphate-buffered saline (PBS) several times to remove the residual DMF. Meantime, Paracoccus sp. MKU1 grown in minimal media containing catechol was harvested by centrifugation, washed thrice, and finally dissolved in 50 mL of PBS. Further, the bacterial suspension was added to the PGO mixture under sterile conditions and maintained at 37 °C for 24 h at 150 rpm for its immobilization. The bacterial density and the amount of PGO were used as described previously,30 and the proportion of PGO and bacterial density were 120 mg of PGO and 300 mg of bacterial wet weight contains 173 × 108 CFU/mL (Figure S4). The immobilized bacterial blends on the surface of PGO were confirmed by SEM analysis.
4.7. Adsorption Capacity of GO and PGO
The adsorption range of catechol with PGO was estimated by following the equation from the literature.31
where Q is the quantity of the catechol adsorption with PGO, C0 is the starting concentration of catechol, Ct is the residual catechol concentration, V is the total volume of deionized water, and M is the amount of PGO used for the adsorption experiment.
The adsorption capacity of catechol on GO and PGO was investigated by adding the known concentration of catechol (100 mg/L) with 50 mg of GO and PGO in 50 mL of sterile deionized water separately. The residual catechol concentration was estimated by the 4-aminoantipyrine assay at regular 6 h intervals until 24 h. The catechol adsorption with GO and PGO was quantified by the standard calibration curve.
4.8. Simultaneous Adsorption and Biodegradation of Catechol
The comparative analysis of catechol degradation by Paracoccus sp. MKU1 and Paracoccus sp. MKU1 microcomposites was carried out by inoculating the bacteria in MSM containing catechol (100 mg/L) and incubating at 37 °C. Simply, 1 mL of culture supernatant was withdrawn at different time intervals for centrifugation and the supernatant was filter-sterilized through a 0.22 μm filter membrane. The residual concentration of catechol was determined by high-performance liquid chromatography using an LC-18 column (15 cm × 4.6 mm I.D., 5 μm, Shimadzu, Japan) with a photoarray detector. The mobile phase for HPLC was 20 mM phosphoric acid and acetonitrile (75:25). The sample (20 μL) was injected with 1.5 mL/min flow rate, and the compound was detected at the wavelength of 270 nm.
4.9. Recycling Performance of (SAB) Process on Catechol
The bacterial microcomposites were pelleted by centrifugation at 3000 rpm for 10 min, and the pellet was dissolved in 1 mL of PBS solution, which was further used as the standard inoculum for batch experiments. For this, MSM containing catechol (100 mg/L) was inoculated with microcomposites and maintained at 37 °C for 48 h. Next, the culture was collected by centrifugation and recycled for two more batch experiments to determine the performance of SAB in each cycle by estimating the residual catechol by the 4-aminoantipyrine assay.
Acknowledgments
The authors sincerely acknowledge the financial support from the Department of Biotechnology, Government of India, New Delhi through a research grant to B.A. and S.A.J. (BT/PR20469/BCE/8/1394/2016). The authors gratefully acknowledge DST-PURSE and UPE programs of Madurai Kamaraj University for infrastructure and other facilities.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01693.
Catechol degradation pathway (Figure S1); AFM and SEM images of graphene oxide (Figure S2); HPLC analysis of catechol (Figure S3); and OD vs CFU plot (Figure S4) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Costantini A. S.; Gorini G.; Consonni D.; Miligi L.; Giovannetti L.; Quinn M. Exposure to benzene and risk of breast cancer among shoe factory workers in Italy. Tumori J. 2009, 95, 8–12. 10.1177/030089160909500102. [DOI] [PubMed] [Google Scholar]
- Vaillancourt F. H.; Bolin J. T.; Eltis L. D. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 241–267. 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Kumar S.; Kumar S. Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem. Eng. J. 2005, 22, 151–159. 10.1016/j.bej.2004.09.006. [DOI] [Google Scholar]
- Wang C. L.; You S. L.; Wang S. L. Purification and characterization of a novel catechol 1, 2-dioxygenase from Pseudomonas aeruginosa with benzoic acid as a carbon source. Process Biochem. 2006, 41, 1594–1601. 10.1016/j.procbio.2006.03.008. [DOI] [Google Scholar]
- Ran N.; Zhao L.; Chen Z.; Tao J. Recent applications of biocatalysis in developing green chemistry for chemical synthesis at the industrial scale. Green Chem. 2008, 10, 361–372. 10.1039/B716045C. [DOI] [Google Scholar]
- Topping D. C.; Bernard L. G.; O’Donoghue J. L.; English J. C. Hydroquinone: acute and subchronic toxicity studies with emphasis on neurobehavioral and nephrotoxic effects. Food Chem. Toxicol. 2007, 45, 70–78. 10.1016/j.fct.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Tang L.; Zeng G.; Chen J.; Cai Y.; Zhang Y.; Yang G.; Liu Y.; Zhang C.; Tang W. Mesoporous carbon nitride based biosensor for highly sensitive and selective analysis of phenol and catechol in compost bioremediation. Biosens. Bioelectron. 2014, 61, 519–525. 10.1016/j.bios.2014.05.063. [DOI] [PubMed] [Google Scholar]
- Moussavi G.; Heidarizad M. The performance of SBR, SCR, and MSCR for simultaneous biodegradation of high concentrations of formaldehyde and ammonia. Sep. Purif. Technol. 2011, 77, 187–195. 10.1016/j.seppur.2010.11.028. [DOI] [Google Scholar]
- Bugg T. D. Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron 2003, 59, 7075–7101. 10.1016/S0040-4020(03)00944-X. [DOI] [Google Scholar]
- Palaniandavar M.; Mayilmurugan R. Mononuclear non-heme iron (III) complexes as functional models for catechol dioxygenases. C. R. Chim. 2007, 10, 366–379. 10.1016/j.crci.2007.01.001. [DOI] [Google Scholar]
- Harayama S.; Kok M.; Neidle E. L. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 1992, 46, 565–601. 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- Shen F. T.; Lin J. L.; Huang C. C.; Ho Y. N.; Arun A. B.; Young L. S.; Young C. C. Molecular detection and phylogenetic analysis of the catechol 1, 2-dioxygenase gene from Gordonia spp. Syst. Appl. Microbiol. 2009, 32, 291–300. 10.1016/j.syapm.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Hamzah R. Y.; Al-Baharna B. S. Catechol ring-cleavage in Pseudomonas cepacia: the simultaneous induction of ortho and meta pathways. Appl. Microbiol. Biotechnol. 1994, 41, 250–256. 10.1007/BF00186968. [DOI] [Google Scholar]
- Di Gioia D.; Barberio C.; Spagnesi S.; Marchetti L.; Fava F. Characterization of four olive-mill-wastewater indigenous bacterial strains capable of aerobically degrading hydroxylated and methoxylated monocyclic aromatic compounds. Arch. Microbiol. 2002, 178, 208–217. 10.1007/s00203-002-0445-z. [DOI] [PubMed] [Google Scholar]
- Bramhachari P. V.; Reddy D. R. S.; Kotresha D. Biodegradation of catechol by free and immobilized cells of Achromobacter xylosoxidans strain 15DKVB isolated from paper and pulp industrial effluents. Biocatal. Agric. Biotechnol. 2016, 7, 36–44. 10.1016/j.bcab.2016.05.003. [DOI] [Google Scholar]
- Alva V. A.; Peyton B. M. Phenol and catechol biodegradation by the haloalkaliphile Halomonas campisalis: influence of pH and salinity. Environ. Sci. Technol. 2003, 37, 4397–4402. 10.1021/es0341844. [DOI] [PubMed] [Google Scholar]
- Stoilova I.; Krastanov A.; Stanchev V.; Daniel D.; Gerginova M.; Alexieva Z. Biodegradation of high amount of phenol, catechol, 2,4 dicholophenol and 2,6 dimethyoxyphenol by Aspergillus awamori cells. Enzyme Microb. Technol. 2006, 39, 1036–1041. 10.1016/j.enzmictec.2006.02.006. [DOI] [Google Scholar]
- Bartosik D.; Szymanik M.; Baj J. Identification and distribution of insertion sequence of Paracoccus solventivorans. Appl. Environ. Microbiol. 2003, 69, 7002–7008. 10.1128/AEM.69.12.7002-7008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C.; Ferguson S. J.; Ludwig B.; Page M. D.; Richter O. M. H.; van Spanning R. J. Molecular Genetics of the Genus Paracoccus: Metabolically Versatile Bacteria with Bioenergetic Flexibility. Microbiol. Mol. Biol. Rev. 1998, 62, 1046–1078. 10.1128/MMBR.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azziz G.; Illarze G.; Irisarri P.. Heterotrophic Denitrification and Paracoccus spp. as Tools for Bioremediation. In Microbial Models: From Environmental to Industrial Sustainability; Springer: Singapore, 2016; pp 209–226. [Google Scholar]
- Cai S.; Cai T.; Liu S.; Yang Q.; He J.; Chen L.; Hu J. Biodegradation of N-methylpyrrolidone by Paracoccus sp. NMD-4 and its degradation pathway. Int. Biodeterior. Biodegrad. 2014, 93, 70–77. 10.1016/j.ibiod.2014.04.022. [DOI] [Google Scholar]
- Zhang J.; Zheng J. W.; Liang B.; Wang C. H.; Cai S.; Ni Y. Y.; He J.; Li S. P. Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro. J. Agric. Food Chem. 2011, 59, 4614–4621. 10.1021/jf104695g. [DOI] [PubMed] [Google Scholar]
- Cai S.; Li X.; Cai T.; He J. Degradation of piperazine by Paracoccus sp. TOH isolated from activated sludge. Bioresour. Technol. 2013, 130, 536–542. 10.1016/j.biortech.2012.12.095. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Jin W.; Sun C.; Gao S. H.; Chen C.; Wang Q.; Han S. F.; Tu R.; Latif M. A.; Wang Q. Microbial degradation of N, N-dimethylformamide by Paracoccus sp. strain DMF-3 from activated sludge. Chem. Eng. J. 2018, 343, 324–330. 10.1016/j.cej.2018.03.023. [DOI] [Google Scholar]
- Nisha K. N.; Devi V.; Varalakshmi P.; Ashokkumar B. Biodegradation and utilization of dimethylformamide by biofilm forming Paracoccus sp. strains MKU1 and MKU2. Bioresour. Technol. 2015, 188, 9–13. 10.1016/j.biortech.2015.02.042. [DOI] [PubMed] [Google Scholar]
- Nisha K. N.; Sridhar J.; Varalakshmi P.; Ashokkumar B. Draft genome sequence of Paracoccus sp. MKU1, a new bacterial strain isolated from an industrial effluent with potential for bioremediation. J. Genomics 2016, 4, 13. 10.7150/jgen.14847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue T. A.; Blakley E. R. Spectrophotometric determination of catechols with 4-aminoantipyrine. Anal. Chim. Acta 1964, 31, 400–403. 10.1016/S0003-2670(00)88845-5. [DOI] [Google Scholar]
- Udhayabanu T.; Karthi S.; Mahesh A.; Varalakshmi P.; Manole A.; Houlden H.; Ashokkumar B. Adaptive regulation of riboflavin transport in heart: effect of dietary riboflavin deficiency in cardiovascular pathogenesis. Mol. Cell. Biochem. 2018, 440, 147–156. 10.1007/s11010-017-3163-1. [DOI] [PubMed] [Google Scholar]
- Kovtyukhova N. I.; Ollivier P. J.; Martin B. R.; Mallouk T. E.; Chizhik S. A.; Buzaneva E. V.; Gorchinskiy A. D. Layer-by-layer assembly of ultrathin composite films from micron-sized graphite oxide sheets and polycations. Chem. Mater. 1999, 11, 771–778. 10.1021/cm981085u. [DOI] [Google Scholar]
- Zheng Y.; Chen D.; Li N.; Xu Q.; Li H.; He J.; Lu J. Efficient simultaneous adsorption-biodegradation of high-concentrated N,N-dimethylformamide from water by Paracoccus denitrificans-graphene oxide microcomposites. Sci. Rep. 2016, 6, 20003 10.1038/srep20003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W.; Rao P.; Lo I. M.; Zhang W.; Zheng W. Preparation of cross-linked magnetic chitosan with quaternary ammonium and its application for Cr (VI) and P (V) removal. J. Environ. Sci. 2014, 26, 2379–2386. 10.1016/j.jes.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Čejková A.; Masák J.; Jirků V.; Veselý M.; Pátek M.; Nešvera J. Potential of Rhodococcus erythropolis as a bioremediation organism. World J. Microbiol. Biotechnol. 2005, 21, 317–321. 10.1007/s11274-004-2152-1. [DOI] [Google Scholar]
- Wang J.; Jiang X.; Liu X.; Sun X.; Han W.; Li J.; Wang L.; Shen J. Microbial degradation mechanism of pyridine by Paracoccus sp. NJUST30 newly isolated from aerobic granules. Chem. Eng. J. 2018, 344, 86–94. 10.1016/j.cej.2018.03.059. [DOI] [Google Scholar]
- Leonard D.; Lindley N. D. Carbon and energy flux constraints in continuous cultures of Alcaligenes eutrophus grown on phenol. Microbiology 1998, 144, 241–248. 10.1099/00221287-144-1-241. [DOI] [PubMed] [Google Scholar]
- Khleifat K. M.; Sharaf E. F.; Al-limoun M. O. Biodegradation of 2-chlorobenzoic acid by Enterobacter cloacae: Growth kinetics and effect of growth conditions. Biorem. J. 2015, 19, 207–217. 10.1080/10889868.2015.1029113. [DOI] [Google Scholar]
- Mrozik A.; Piotrowska-Seget Z.; Labuzek S. FAME profiles in Pseudomonas vesicularis during catechol and phenol degradation in the presence of glucose as an additional carbon source. Pol. J. Microbiol. 2007, 56, 157. [PubMed] [Google Scholar]
- Basu A.; Apte S. K.; Phale P. S. Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl. Environ. Microbiol. 2006, 72, 2226–2230. 10.1128/AEM.72.3.2226-2230.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A.; Das D.; Bapat P.; Wangikar P. P.; Phale P. S. Sequential utilization of substrates by Pseudomonas putida CSV86: signatures of intermediate metabolites and online measurements. Microbiol. Res. 2009, 164, 429–437. 10.1016/j.micres.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Loh K. C.; Chua S. S. Ortho pathway of benzoate degradation in Pseudomonas putida: induction of meta pathway at high substrate concentrations. Enzyme Microb. Technol. 2002, 30, 620–626. 10.1016/S0141-0229(02)00016-9. [DOI] [Google Scholar]
- Cao B.; Geng A.; Loh K. C. Induction of ortho-and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl. Microbiol. Biotechnol. 2008, 81, 99. 10.1007/s00253-008-1728-3. [DOI] [PubMed] [Google Scholar]
- Song J.; Sung J.; Kim Y. M.; Zylstra G. J.; Kim E. Roles of the meta-and the ortho-cleavage pathways for the efficient utilization of aromatic hydrocarbons by Sphingomonas yanoikuyae B1. J. Microbiol. 2000, 38, 245–249. [Google Scholar]
- Carvalho M. F.; Ferreira M. I. M.; Moreira I. S.; Castro P. M.; Janssen D. B. Degradation of fluorobenzene by Rhizobiales strain F11 via ortho cleavage of 4-fluorocatechol and catechol. Appl. Environ. Microbiol. 2006, 72, 7413–7417. 10.1128/AEM.01162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T.; Takeo M.; Maeda Y. Plasmid-encoded genes specifying aniline oxidation from Acinetobacter sp. strain YAA. Microbiology 1997, 143, 93–99. 10.1099/00221287-143-1-93. [DOI] [PubMed] [Google Scholar]
- Li W.; Shi J.; Wang X.; Han Y.; Tong W.; Ma L.; Liu B.; Cai B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 2004, 336, 231–240. 10.1016/j.gene.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Pessione E.; Giuffrida M. G.; Mazzoli R.; Caposio P.; Landolfo S.; Conti A.; Giunta C.; Gribaudo G. The catechol 1, 2 dioxygenase system of Acinetobacter radioresistens: isoenzymes, inductors and gene localisation. Biol. Chem. 2001, 382, 1253–1261. 10.1515/BC.2001.156. [DOI] [PubMed] [Google Scholar]
- Kiesel B.; Müller R. H. The meta pathway as a potential energy-generating sequence and its effects on the growth rate during the utilisation of aromatics. Acta Biotechnol. 2002, 22, 221–234. . [DOI] [Google Scholar]
- Domaradzka D.; Guzik U.; Hupert-Kocurek K.; Wojcieszyńska D. Cometabolic degradation of naproxen by Planococcus sp. strain S5. Water Air Soil Pollut. 2015, 226, 297. 10.1007/s11270-015-2564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q.; Chen C.; Lei Q.; Liang J.; Zhang T.; Jiang J. Adsorption of aniline from aqueous solution using graphene oxide-modified attapulgite composites. RSC Adv. 2018, 8, 23382–23389. 10.1039/C8RA04143A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz-Poseu J.; Mancebo-Aracil J.; Nador F.; Busqué F.; Ruiz-Molina D. The Chemistry behind Catechol-Based Adhesion. Angew. Chem., Int. Ed. 2019, 58, 696–714. 10.1002/anie.201801063. [DOI] [PubMed] [Google Scholar]
- Gajendiran N.; Mahadevan A. Catechol degradation by immobilized Rhizobium sp.. Zentralbl. Mikrobiol. 1991, 146, 99–101. 10.1016/S0232-4393(11)80287-8. [DOI] [PubMed] [Google Scholar]
- Tewari L.; Malviya P. Biodegradation of catechol by fluorescent Pseudomonas for sustainable environment. J. Sci. Ind. Res. 2001, 61, 70–74. [Google Scholar]
- Patil N. K.; Sharanagouda U.; Niazi J. H.; Karegoudar T. B. Stable degradation of catechol by Pseudomonas sp. strain NGK1 encapsulated in alginate and polyurethane foam. Indian J. Biotechnol. 2004, 3, 568–572. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.