Bashyam et al. present a deep learning network for brain MRI that provides easily accessible, robust and clinically informative estimates of ‘brain age’ from a diverse worldwide population of 14 468 people. They then leverage the network to obtain imaging signatures of Alzheimer’s disease and schizophrenia.
Keywords: brain age, deep learning, transfer learning
Abstract
Deep learning has emerged as a powerful approach to constructing imaging signatures of normal brain ageing as well as of various neuropathological processes associated with brain diseases. In particular, MRI-derived brain age has been used as a comprehensive biomarker of brain health that can identify both advanced and resilient ageing individuals via deviations from typical brain ageing. Imaging signatures of various brain diseases, including schizophrenia and Alzheimer’s disease, have also been identified using machine learning. Prior efforts to derive these indices have been hampered by the need for sophisticated and not easily reproducible processing steps, by insufficiently powered or diversified samples from which typical brain ageing trajectories were derived, and by limited reproducibility across populations and MRI scanners. Herein, we develop and test a sophisticated deep brain network (DeepBrainNet) using a large (n = 11 729) set of MRI scans from a highly diversified cohort spanning different studies, scanners, ages and geographic locations around the world. Tests using both cross-validation and a separate replication cohort of 2739 individuals indicate that DeepBrainNet obtains robust brain-age estimates from these diverse datasets without the need for specialized image data preparation and processing. Furthermore, we show evidence that moderately fit brain ageing models may provide brain age estimates that are most discriminant of individuals with pathologies. This is not unexpected as tightly-fitting brain age models naturally produce brain-age estimates that offer little information beyond age, and loosely fitting models may contain a lot of noise. Our results offer some experimental evidence against commonly pursued tightly-fitting models. We show that the moderately fitting brain age models obtain significantly higher differentiation compared to tightly-fitting models in two of the four disease groups tested. Critically, we demonstrate that leveraging DeepBrainNet, along with transfer learning, allows us to construct more accurate classifiers of several brain diseases, compared to directly training classifiers on patient versus healthy control datasets or using common imaging databases such as ImageNet. We, therefore, derive a domain-specific deep network likely to reduce the need for application-specific adaptation and tuning of generic deep learning networks. We made the DeepBrainNet model freely available to the community for MRI-based evaluation of brain health in the general population and over the lifespan.
Introduction
Normal brain development and ageing are accompanied by patterns of neuroanatomical change that can be captured by machine learning methods applied to imaging data. The construct of MRI-derived brain age has been widely adopted by the neuroscience community as an informative biomarker of brain health at the individual level (Franke et al., 2010; Cole and Franke, 2017; Cole et al., 2017a, 2019). Individuals displaying pathological or atypical brain development and ageing patterns can be identified through positive or negative deviations from typical brain age trajectories. For example, schizophrenia, mild cognitive impairment (MCI), Alzheimer’s disease, type 2 diabetes and mortality have all been linked to accelerated brain ageing at respective age ranges (Franke et al., 2013; Gaser et al., 2013; Habes et al., 2016; Cole et al., 2017b; Hajek et al., 2019).
Machine learning has offered numerous other MRI-based biomarkers of neuroanatomical change in various pathologies, including Alzheimer’s disease, MCI, schizophrenia, major depression, and autism (Arbabshirani et al., 2017; Mateos-Pérez et al., 2018). By virtue of their high sensitivity, these indices capture brain changes at very early preclinical stages (Davatzikos et al., 2009). These machine learning-based biomarkers are therefore poised to transform precision and early diagnostics by offering individualized indices of brain health.
Prior efforts to apply machine learning methods to neuroimaging have been successful in the laboratory; however, they are not generally applicable or easily adopted in practice. Traditionally, these methods require several specialized and often sophisticated preprocessing steps, such as careful bias correction, segmentation, deformable registration, and harmonization across scanners, among others. These preprocessing steps require expertise, time, effort, and are not easily reproducible especially across different scanners, populations and MRI acquisition protocols. Such complexity renders these methods impractical for use broadly in clinical settings and thus they have not been widely adopted by clinicians.
The emergence of deep learning as a powerful machine learning method offers great promise for transcending these limitations (Vieira et al., 2017). Convolutional neural networks have rapidly become the state-of-the-art in most image recognition tasks and are gaining acceptance in neuroimaging (Kamnitsas et al., 2017; Akkus et al., 2017; Anwar et al., 2018). These methods allow for complex non-linear relationships to be modelled without need for the manual feature engineering traditionally required. These models are often limited by the need to carefully adapt and fine-tune the network’s architecture to a specific problem, e.g. MRI-based classification of a specific disease.
An important requirement for deep learning applications is the availability of large and diverse samples for training the complex deep network. Although MRI data availability has rapidly increased with expanded data sharing and meta-analyses efforts (Toga et al., 2012; Thompson et al., 2014; Van Horn and Toga, 2014), sample sizes for disease-specific datasets are still relatively small, limiting the direct application of deep learning to characterize pathological neuroanatomical patterns. To address this challenge, we use a transfer learning approach leveraging a large and diverse sample of MRI scans, and demonstrate that robust brain age estimates can be obtained across scanners and populations. Our deep-learning-based brain age prediction approach is motivated by the pioneering work of Cole et al. (2017a), who demonstrated that a convolutional neural network model trained on MRI scans of n = 2001 healthy adults can obtain high predictive accuracy, with comparable performance using either preprocessed or raw T1-weighted scans. Nonetheless, we use a significantly larger and more heterogeneous dataset derived from 18 studies for training the brain age model. In a recent paper, Jonsson et al. (2019) used a dataset comparable to ours in sample size for training a deep-learning-based brain age model. However, their training set predominantly comprised data from a single study, UKBIOBANK, and application on other smaller datasets required retraining of the model. Also, the main and only focus of both studies was brain age prediction, while in our case we used brain age prediction as a tool for constructing a brain-specific deep network model using the largest sample available.
The first contribution of the current study is in demonstrating that deep learning yields robust biomarkers of brain age when applied with minimal data preprocessing to a large and diverse cohort of 14 468 brain MRI scans across the lifespan and including multiple scanners, acquisition protocols, and geographic locations around the world. Moreover, we demonstrate that the common approach of prioritizing brain age models based on their fit (Franke et al., 2013;Cole et al., 2017a) may not produce the most informative brain age delta (in the remainder of the text we use the term ‘Brain age delta’ or ‘delta’ to denote the difference between the predicted brain age and the chronological age), i.e. the important deviations from typical brain development and ageing that indicate the presence of underlying pathology. Instead, we demonstrate that moderately-fit models provide the most clinically informative brain age estimates in that respective deviations from typical brain development and ageing provide the best separation of individuals with Alzheimer’s disease, MCI, schizophrenia, and major depression.
The second contribution of our study is that, by leveraging this large and diverse brain MRI dataset, it constructs a structural brain imaging network that is domain-specific, i.e. specific to brain structure, as opposed to being informed by generic databases of natural images. As a result, the DeepBrainNet was found to produce disease-specific classifiers that achieve accuracy and convergence significantly outperforming networks trained directly from patient and control datasets, or initialized with commonly used weights derived from the ImageNet natural scene database (Deng et al., 2009). We further demonstrate the advantage of using DeepBrainNet weights for classification of patients with Alzheimer’s disease, MCI, and schizophrenia, by showing robust classification accuracy as sample size decreases across these conditions.
The DeepBrainNet model is made publicly available via Github (https://github.com/vishnubashyam/DeepBrainNet). The deep learning models can also be applied on new scans using the CBICA Internet Processing Portal (IPP) (https://ipp.cbica.upenn.edu/), which allows users to apply our methods and models without the need for installing any software packages.
Materials and methods
Datasets
We used a large multisite collection of T1-weighted brain MRI scans from normal control subjects (n = 11 729) covering individuals of ages 3 to 95 for training the DeepBrainNet model and for calculating cross-validated brain age predictions. This dataset, which we refer to as LifespanCN, represented a diverse range of geographic locations, scanners, acquisition protocols, and studies. Additionally, we tested the model’s performance on an unseen site by training a model excluding the SHIP cohort and then testing it on SHIP (n = 2739). We also used three different disease-specific cohorts in order to investigate brain age deltas of the DeepBrainNet model in case of disease. These datasets included normal control, Alzheimer’s disease and MCI subjects from ADNI 1 and 2 (n = 1699, normal control = 513, MCI = 833, Alzheimer’s disease = 353), normal control and schizophrenia subjects from a multisite schizophrenia consortium (n = 835, normal control = 448, schizophrenia = 387) (Rozycki et al., 2018) and matched normal control and major depression subjects from UK Biobank (n = 408, normal control = 204, major depression = 204) (Sudlow et al., 2015). Disease-specific cohorts were also used to build and validate disease classification models through transfer learning. An overview of all datasets that are used in different models is given in Table 1. A more detailed description of the demographics of these datasets is given in the Supplementary material, section S.1.
Table 1.
Dataset description
| Study | n (male/female) | n Controls | n Disease | Mean age | Age range | Experiments |
|---|---|---|---|---|---|---|
| ADC | 76 (29/47) | 76 CN | 0 | 72.59 | 37 | LifespanCN |
| AIBL | 446 (197/249) | 446 CN | 0 | 72.77 | 32 | LifespanCN |
| BLSA 1.5 T | 90 (58/32) | 90 CN | 0 | 72.69 | 29.9 | LifespanCN |
| BLSA 3 T | 952 (436/516) | 952 CN | 0 | 67.04 | 72.7 | LifespanCN |
| CARDIA | 719 (342/377) | 719 CN | 0 | 50.29 | 14 | LifespanCN |
| PAC-WASH | 247 (95/152) | 247 CN | 0 | 61.19 | 24 | LifespanCN |
| PAC-WISC | 127 (39/88) | 127 CN | 0 | 61.47 | 34.2 | LifespanCN |
| PAC-JHU | 95 (36/59) | 95 CN | 0 | 67.75 | 44.9 | LifespanCN |
| PING | 398 (200/198) | 398 CN | 0 | 12.69 | 17.83 | LifespanCN |
| PNC | 1396 (665/731) | 1396 CN | 0 | 14.97 | 15 | LifespanCN |
| PENN PMC | 41 (19/22) | 41 CN | 0 | 72.37 | 35 | LifespanCN |
| SHIP | 2739 (1248/1491) | 2739 CN | 0 | 52.55 | 69.21 | LifespanCN |
| UK BioBank | 4402 (2067/2335) | 4403 CN | 0 | 63.2 | 34.4 | Residual Analysis |
| ADNI-1 | 747 (437/310) | 189 CN | 366 MCI, 192 AD | 75.29 | 36.5 | Residual analysis and transfer learning |
| ADNI-2 | 952 (500/452) | 324 CN | 467 MCI, 161 AD | 73.23 | 39.6 | Residual analysis and transfer learning |
| PHENOM | 835 (472/363) | 448 CN | 387 SCZ | 34.55 | 70 | Residual analysis and transfer learning |
AD = Alzheimer’s disease; CN = normal control; MD = major depression; SCZ = schizophrenia.
Data preprocessing
Raw T1-weighted scans were input to DeepBrainNet model after minimal and fully-automated preprocessing. In particular, the scans were first skull-stripped by applying an automated method based on multi-atlas label fusion (Doshi et al., 2013) consistently on each scan. A systematic quality control procedure was applied by using an automatic outlier detection followed by manual verification of flagged cases. Skull-stripped images were affinely registered to a common atlas space using FMRIB’s Linear Image Registration Tool FLIRT (Jenkinson and Smith, 2001;Jenkinson et al., 2002).
DeepBrainNet network architecture
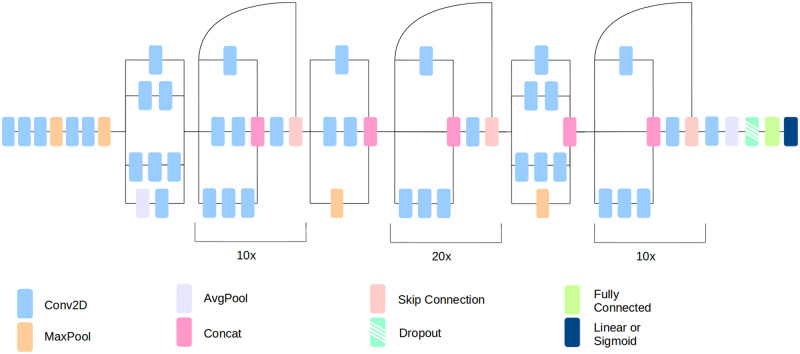
The DeepBrainNet model was built using the inception-resnet-v2 framework, which combines skip connections and inception modules (Szegedy et al., 2017). This framework is commonly used in computer vision and has been shown to perform very well on many complex imaging tasks. Confirming these findings, in our validation experiments this model obtained the highest prediction accuracy against other common architecture, although differences were not statistically significant.
As is common in other deep-learning-based neuroimaging applications, we used a 2D convolutional architecture. The preference for a 2D rather than a 3D architecture was motivated by two main reasons: first, for the initialization of our networks we used a model pretrained on ImageNet, a natural scene database consisting of over 14 million hand annotated images belonging to over 1000 categories. Using a 2D architecture allowed us to use ImageNet for initialization, which has been shown to lead to more consistent and accurate models (Tajbakhsh et al., 2016). Second, the increase in the parameter space resulting from 3D kernels may make them impractical for use on MRI data, as the sample sizes are typically too small compared to the dimensionality of the data, even with >10 000 scans.
We represented each scan as a collection of 80 slices in the axial plane. During training, each slice is considered as an independent sample, resulting in a training set of ∼1 million images for the LifespanCN dataset.
We performed online data augmentation, with random vertical and horizontal flips and intensity and contrast variations obtained by randomly scaling intensities within 95% to 105% of their initial values, to make the network further invariant to imaging variations and site effects.
Inception-resnet-v2 convolutional layers were connected to a global max pooling layer, followed by a fully connected layer of size 1024 with 80% dropout and RELU (rectified linear units) activation. We used dropout after the fully connected layer during model training to prevent overfitting (Srivastava et al., 2014). Dropout rates were chosen a priori. We preferred a large dropout value because of the large number of fully connected nodes in the final layers (n = 1024 nodes). Dropout randomly removes some percentage of the inputs to a layer with the intention of reducing the network’s reliance on any single node. During the testing, the dropout function is inactive and all nodes are used. A single node with a linear activation is added as the output layer. The outline of the inception-resnet-v2 architecture is shown in Fig. 1.
Figure 1.
DeepBrainNet architecture.
The network is trained from a random weight initialization using the Adam optimizer (Kingma and Ba, 2014) with mean squared error as the loss function. The learning rate for training is set to 1 × 10−4 and decreased by a factor of 10 if the training loss remains constant for five epochs. The network is trained until the training loss remains constant for 10 consecutive epochs or until the validation loss increases for five consecutive epochs.
To obtain the final age prediction for a test sample, each of 80 slices of the test scan was input to the trained model independently and the median prediction is calculated as the predicted brain age.
We implemented our model in Tensorflow and Keras (Abadi et al., 2016). This model was trained using a NVIDIA P6000 Quadro graphics processing unit with 24 GB video RAM. Cross-validated experiments were conducted using 5-fold validations. Computational time for training in each fold was around 10 h.
Transfer learning models for disease classification
Deep learning models typically leverage large pretrained networks for initialization. For example, in computer vision it is standard practice to use ImageNet weights to initialize a network. A network that is trained on a large and varied dataset can learn a feature representation that has been shown to be highly generalizable to many other tasks (Donahue et al., 2014). The weights of the pretrained networks can be refined in specific classification problems using the available training data specific for that task. This process of transfer learning is critically important for successful training in problems that do not offer such large training sets. Medical imaging belongs to this category of problems, as the overwhelming majority of disease-specific classification studies rarely have access to more than 1000 patient scans, and often much less.
We used a similar inception-resnet-v2 based network model with transfer learning for disease classification tasks. Importantly, for the initialization of transfer learning models we used the weights from the best performing fold of the age prediction task on LifespanCN dataset. Note that the initialization preserved only the weights from the convolutional layers. The final fully connected layers from the age prediction model are removed and replaced with a fully connected layer of 1024 nodes with 85% dropout and RELU activation. A slightly larger value (85%) was chosen for the classification task since the smaller amount of training data available for these tasks may have resulted in more overfitting. This is followed by an output layer with one node with a sigmoid activation function. The fully connected layers are then trained for one epoch with the convolutional layers frozen. This is to ensure that the weights in the convolutional layers are not excessively disturbed by the large gradient caused by the random initialization of the final fully connected layers. Finally, all layers are unfrozen, and the network is trained until convergence or until the validation loss increases. The network is trained using the Adam optimizer with a learning rate of 5 × 10−5 with binary cross-entropy as the loss function.
In testing, similar to the brain age model, the final classification label is decided by calculating the median of the output probabilities for individual slices of the test scan.
Statistical testing
While comparing the effect of varying levels of regularization on brain age deltas we conduct appropriate testing to examine whether the brain age gap values differentiate disease (e.g. Alzheimer’s disease, MCI, schizophrenia or depression) and controls subjects, and whether such discrimination differ by the chose models (loose, middle and tight) (Supplementary material, section S.14). Hence, we are testing the difference (by models) of the difference (by diagnosis) in brain age gaps. We use a mixed effects model for this task because the model-specific brain age deltas are generated from the same subject’s data. Hence for any pairwise comparison such as middle fit versus tight fit, the data might be correlated within subject. Mixed effects models with subject-specific random intercepts are known to provide valid inference for correlated outcome data.
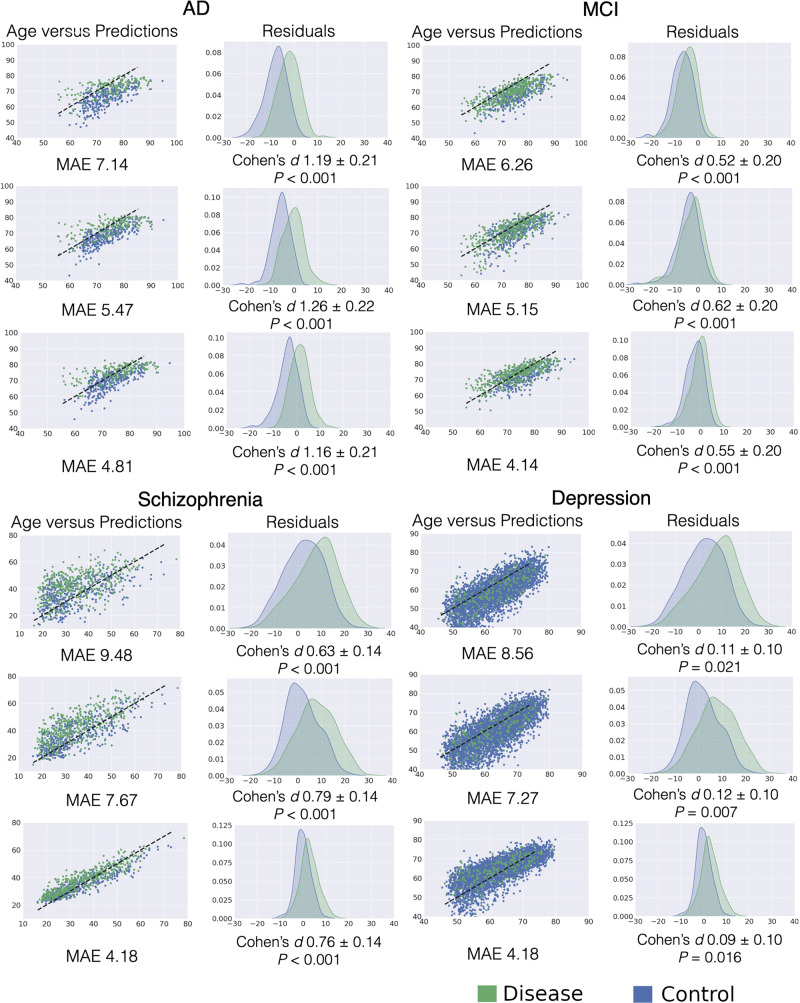
The P-values shown in Fig. 3 are generated from t-tests performed on the respective controls versus disease groups for each level of model fit. The significance of the differential discrimination from the mixed effects models were determined based on likelihood ratio (LRT) tests of the fixed effects.
Figure 3.
Distribution of brain age residuals for disease versus normal control groups for different regularizations of the brain age model. The rows in each subplot show the results for loose, moderate and tight-fit models, respectively. The left columns show predicted versus actual ages. The right columns show histograms of brain age residuals for normal control and diseased subject groups and the significance of group differences. The Cohen’s d effect size between the two groups are reported with 95% confidence intervals. AD = Alzheimer’s disease; MAE = mean absolute error.
Data availability
The data that support the findings of this study are available, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data may be available from the authors upon reasonable request and with permission.
Results
Brain age prediction
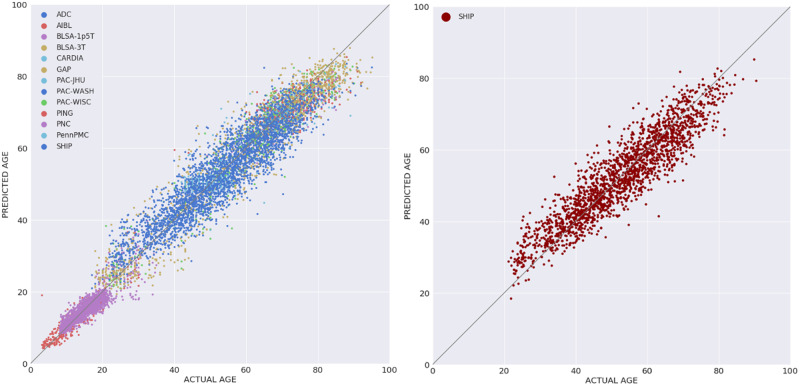
The DeepBrainNet model using the inception-resnet-v2 framework was trained on LifespanCN dataset (n = 11 729). The model was applied for predicting the brain age with 5-fold cross validation, i.e. it was trained and optimized on 80% of the data and tested on the remaining 20%, repeating this procedure for each five folds. The model obtained a mean absolute error (MAE) = 3.702 in the prediction of brain age for the complete LifespanCN dataset. Alternative network architectures, i.e. DenseNet169 (Huang et al., 2017), VGG16 (Simonyan and Zisserman, 2014) and Resnet50 (He et al., 2015), obtained lower predictive accuracy, although differences between architectures were not statistically significant (Supplementary material, section S.2). The correlation between the chronological and predicted brain ages of the subjects was r = 0.978 (Fig. 2A). The prediction accuracies in each fold were consistent (Supplementary material, section S.3). The distribution of the brain age deltas per site is shown in Supplementary material, section S.4.
Figure 2.
Brain age predictions using DeepBrainNet. Left: Predictions for the complete LifespanCN dataset. DeepBrainNet was trained and tested on LifespanCN dataset with 5-fold cross-validation. Right: Performance on previously unseen site. DeepBrainNet was trained using LifespanCN data excluding SHIP and was applied on the SHIP data.
Brain age deltas for male and female subjects were similar (MAE = 3.68 and MAE = 3.72, respectively; Supplementary material, section S.5). We further investigated gender differences by training separate male and female models with 5-fold cross validation on the LifespanCN dataset. Brain age obtained by mixed-gender and gender-specific models were highly correlated (98% and 97% for males and females respectively), suggesting that the gender bias does not significantly affect the results of the age prediction models (Supplementary material, section S.6).
To evaluate out of sample performance of the DeepBrainNet model we trained the model using the LifespanCN dataset excluding the SHIP cohort, and then applied it on the SHIP data (n = 2739). We obtained an MAE of 4.12 for the SHIP subjects (Fig. 2B). This result is comparable to intra-site predictions of similarly aged individuals, thus showing that the age prediction is highly generalizable across sites.
We repeated all experiments on LifespanCN and SHIP datasets using input images processed with additional preprocessing steps, specifically bias correction (Tustison et al., 2010) and histogram equalization. The results of these experiments indicated that the performance was comparable with or without additional preprocessing (Supplementary material, section S.7).
We evaluated the effect of the data sample age range on prediction, particularly considering that our sample included both paediatric and adult subjects. We trained and applied models separately for paediatric datasets (PING and PNC, age range 3–22) and all other datasets together (age range 18–95). Brain age obtained by mixed-age and age-specific models were highly correlated (97% for the paediatric and 95% for the adult subjects, respectively; Supplementary material, section S.8). These results indicate that the DeepBrainNet model was capable to capture complex imaging signatures associated with significantly different processes of brain development and brain ageing within a single network.
A major challenge of deep learning algorithms is the interpretation of the imaging patterns that are learned by the network. A direct visualization of these patterns is not possible because of the complexity of the network (Zeiler and Fergus, 2014). We used the technique suggested in Kotikalapudi and contributors (2017) to create saliency maps that show the voxels with the highest activation in different axial image slices at different age ranges (Supplementary material, section S.11).
Effect of regularization on brain age deltas for diseased subjects
The clinical significance of brain age is obviously not in determining someone’s age, but in identifying individuals who deviate from typical brain development and ageing, i.e. individuals who have positive or negative brain age deltas. In that respect, accurate age predictions don’t necessarily yield the most clinically informative brain age deltas, since the deep learning model might focus on imaging features and patterns that are not affected by pathologies, in an effort to match brain age and chronological age in individuals with such pathologies. To address this issue, we developed three different models with varying levels of fitness to the data. Specifically, the model was saved after each epoch of training, and for comparative evaluations we selected three models with smaller to larger number of epochs, such that the average MAE on the training set was 7.651, 5.922 and 3.701 for each model, respectively, for the loose, moderate, and tight fits. We then evaluated the resultant deltas on the groups with pathology for each model. These deltas were corrected for age using a linear model, as has been suggested in the literature in order to remove age-related bias (Le et al., 2018).
In line with our hypothesis, all cohorts with pathologies displayed positive brain age deltas on average, i.e. their brain age was older than their chronological age. However, the model with highest age prediction accuracy was not the best one in terms of yielding brain age deltas with the highest discrimination between patients and controls (Fig. 3). Indeed, the moderately-fit model, with the mid-range MAE, had brain age deltas with more significant group differences and the largest effect sizes between disease and normal control groups across all brain pathologies tested. Further statistical testing with a mixed effects model confirmed that the moderate fit was most discriminative across Alzheimer’s disease, MCI, and schizophrenia, with significant differences between the models for Alzheimer’s disease and MCI, but not for schizophrenia and depression (Supplementary material, section S.14).
Transfer learning for pathology-specific classification
We tested the hypothesis that the DeepBrainNet network, which was trained on the LifespanCN data for the brain age prediction task, would provide a better platform for transfer learning for disease-specific classification in the Alzheimer’s disease, MCI, schizophrenia and major depression groups, compared to alternative initializations. Our hypothesis was that while many of the lower level features captured by the ImageNet weights are useful for neuroimaging tasks, the higher level abstractions might not be. This approach presents an opportunity for more specialized network weights, i.e. model weights that will better capture high-level abstract neuroimaging features. In particular, we constructed four independent classifiers, one for each of these four patient cohorts, using transfer learning. These models were initialized with network weights of the DeepBrainNet model, and further trained and tested for the specific task with 5-fold cross-validation. For the comparative evaluations, we also constructed models by training from scratch (random initialization), and by initializing the model with pretrained ImageNet weights. Both these models were retrained with scans from the disease groups during the cross-validation analysis.
It should be noted that the major depression classifier did not converge with any initialization.
BrainNet-based classification models outperformed models initialized using ImageNet consistently for Alzheimer’s disease, MCI, schizophrenia and major depression classification tasks, with a significant increase in both accuracy and area under the curve (AUC) values for all tasks (Table 2). Additionally, using DeepBrainNet weights for initialization allowed the model to consistently converge faster in all tasks. The network with random initialization failed to converge for at least 1-fold for all diseases tested.
Table 2.
Transfer learning performance comparison
| Task | Model | Accuracy | AUC | Epochs to converge (average) |
|---|---|---|---|---|
| AD versus CN | DeepBrainNet | 0.86 | 0.91 | 3.4 |
| ImageNet | 0.849 | 0.893 | 4.6 | |
| Random Init.a | No convergence | No convergence | No convergence | |
| MCI versus CN | DeepBrainNet | 0.702 | 0.743 | 4.2 |
| ImageNet | 0.628 | 0.645 | 5.6 | |
| Random Init. | No convergence | No convergence | No convergence | |
| SCZ versus CN | DeepBrainNet | 0.735 | 0.791 | 3.4 |
| ImageNet | 0.702 | 0.774 | 5 | |
| Random Init. | No convergence | No convergence | No convergence |
aThe model did not converge in all folds.
AD = Alzheimer’s disease; SCZ = schizophrenia.
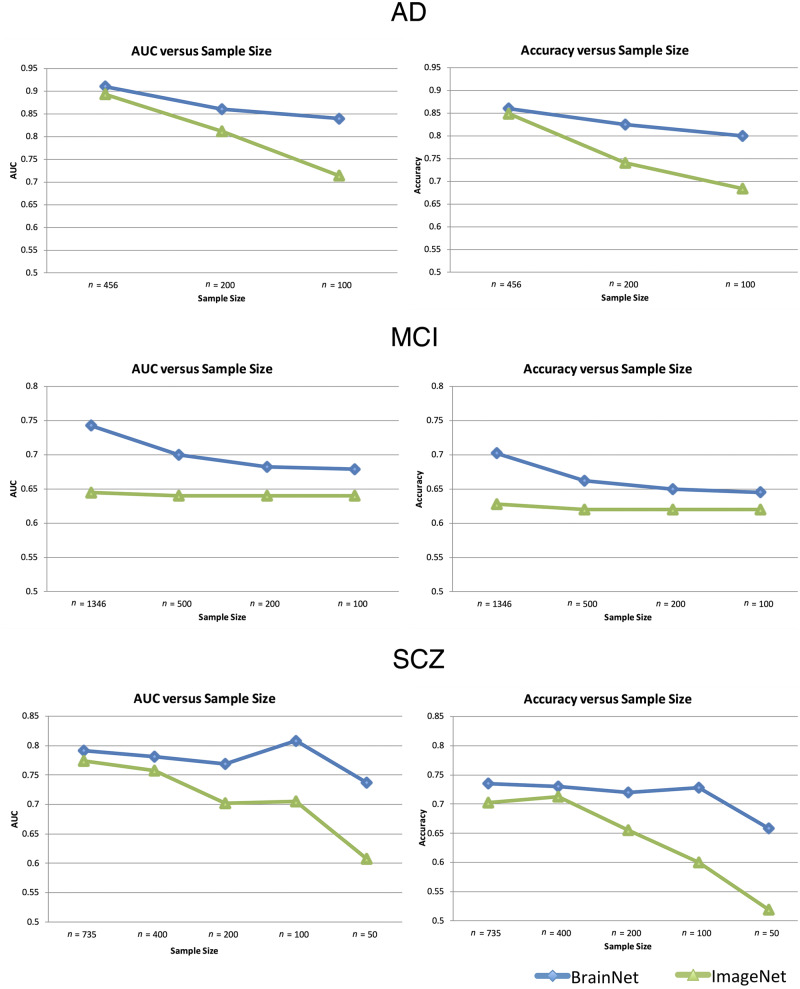
We performed a series of additional experiments to evaluate the classification performance of network models using BrainNet and ImageNet initializations on problems where smaller sample sizes are available for training. For this purpose, we subsampled each disease-specific subset and created new datasets with decreasing sample sizes. A stratified subsampling technique was used to preserve the initial normal control versus diseased subjects’ ratio in the new datasets. The two deep learning models, initialized using DeepBrainNet or ImageNet weights, were applied on each new subsampled dataset with cross-validation, similar to experiments that were performed on the complete samples. We repeated each small sample experiment two additional times with different stratified randomizations to obtain robust estimates of the performance with gradually decreasing sample size. These experiments show similar results to Fig. 4. It should be noted that at n = 50, the schizophrenia classifier failed to converge in one sampling. This may indicate that the convergence at this sample size is not reliable (Supplementary material, section S.12).
Figure 4.
Classification performance of transfer learning based networks. Classification performance of transfer learning based networks using two different initializations (DeepBrainNet and ImageNet) on three classification tasks [Alzheimer’s disease (AD) versus normal control, MCI versus normal control and schizophrenia (SCZ) versus normal control] trained and tested on datasets with different sample sizes. The sample sizes used in different experiments are shown in the x-axis of each plot, with larger (initial) to smaller (subsampled) sample sizes. Each model was run using 5-fold cross-validation.
Classification accuracy and AUC values for the two models on datasets with decreasing sample sizes are shown in Fig. 4. DeepBrainNet obtained superior performance in all classification tasks. Importantly, DeepBrainNet based models have maintained performance relatively well with smaller sample sizes, compared to ImageNet-based models that showed a consistently lower accuracy. In Alzheimer’s disease versus normal control and schizophrenia versus normal control classification tasks, DeepBrainNet’s performance on small samples was particularly stable, while ImageNet based models showed a significant decrease in accuracy with smaller sample sizes. Both networks performed well for these classification tasks with large sample sizes. For the MCI versus normal control classification, DeepBrainNet had a larger decrease in performance with decreasing samples while ImageNet had a stable but lower accuracy.
Discussion
We developed a deep brain network, DeepBrainNet, derived from and tested on collectively 14 468 diverse structural brain MRI scans, which generates estimates of an individual’s brain age. We showed that minimal preparation and preprocessing of the brain MRI scans is sufficient for DeepBrainNet to produce informative estimates of brain age. Most importantly, we found that using DeepBrainNet as a foundation for further deriving disease-specific networks via transfer learning resulted in better accuracy and convergence across all tested diseases, especially for relatively smaller sample sizes, when compared to deep learning models without prior training with brain MRIs. This result underlines the importance of domain-specific deep learning networks that don’t require specialized adaptation and fine-tuning to specific problems. Finally, we found that moderately fitted brain age models are optimal, in terms of providing brain age deltas that correlate with four different clinical categories: MCI, Alzheimer’s disease, schizophrenia, and major depression, compared to tightly or loosely-fitted brain age models. This finding challenges current trends to achieve the tightest possible brain age estimates, and provides guidelines as to how this increasingly popular biomarker should be used.
Deep learning-based age prediction from minimally processed scans achieves high accuracy
It is well established that brain structure shows consistent patterns of developmental and ageing related changes through the lifespan. Yet, the degree of change is highly heterogeneous across different brain structures and different phases of life, resulting in complex non-linear age trajectories of regional brain changes (Fjell and Anders, 2013). The concept of estimating brain age from MRI scans has been previously explored (Dosenbach, 2010; Franke et al., 2010; Brown et al., 2012; Habes et al., 2016; Madan and Kensinger, 2018), showing that it is possible to accurately predict the chronological age of subjects from volumetric or voxelwise imaging features using machine learning or multivariable regression techniques. However, most prior studies have been restricted to relatively homogeneous sets of data and small samples. Perhaps most importantly, prior attempts to provide robust brain age estimates have relied on carefully preprocessed datasets using sophisticated and often delicate segmentation and deformable registration tools. These tools are not easily accessible to clinicians who want to readily obtain a brain health index, such as brain age. Critically, these preprocessing steps often need to be carefully adjusted to the particular characteristics of a study’s, scanner’s, or centre’s images, and typically require human supervision for quality checks, which limits their broad applicability.
Deep learning methods provided a valuable opportunity for overcoming these limitations. As demonstrated in Cole et al. (2017a), a deep-learning-based model using convolutional neural networks obtained high predictive accuracy when the model was trained on MRI scans of n = 2001 healthy adults, with performance comparable using either preprocessed or raw T1-weighted scans. Motivated by these results, we derived a unique brain age index from a large and diverse set of brain MRI scans using minimal preprocessing and with fully automated procedures. The size and diversity of our dataset, as well as our results, bolster confidence that DeepBrainNet can provide an index of brain age that can be useful in initial screening for the presence of many pathologies that cause deviation from typical brain development and ageing. Further, as our training set is highly diverse, the network is robust to confounding site effects, as evidenced by the strong performance on the out of sample validation, which may allow it to succeed with diverse acquisition and clinical scenarios.
Deep learning revealed conserved patterns of brain change throughout the lifespan, which allowed DeepBrainNet to achieve a quite accurate estimation of brain age. Although this success might be somewhat expected, especially in typical brain development that involves well-coordinated brain growth and maturation (Erus et al., 2015), it is still quite surprising in several ways. Our ability to estimate someone’s age from their brain MRI scan within an average ∼4 years implies that the brain changes constantly throughout the lifespan, in subtle but well-coordinated ways that allowed us to determine a highly predictive brain age network. Notably, such brain changes are also present in ages 25–55, an age range previously considered to be mostly stable in terms of brain structural morphology.
The proposed deep learning approach has a significantly higher computational complexity compared to other machine learning or multivariable regression techniques previously suggested for brain age prediction. While these simpler methods work very well in many instances, they benefit from specialized domain-specific preprocessing and may not be generalizable to other tasks. Additionally, we should note that this complexity only involves the training phase. Once a model is trained, application of the model on new subjects is straightforward and computationally very efficient, particularly considering that the DeepBrainNet model can be directly applied on minimally preprocessed scans without requiring complex processing steps that often limit widespread usage in clinical settings.
Using DeepBrainNet with transfer learning outperforms generic ImageNet-based training for smaller sample sizes
Although the brain age delta is a simple and clinically appealing index for estimating overall brain health, it is not specific to or optimized for any particular disease. Diseases such as Alzheimer’s disease and schizophrenia display highly distinctive neuroanatomical patterns that could better be captured by specialized indices (Davatzikos et al., 2009; Rozycki et al., 2018). Our work has provided insights into how deep learning can optimize disease-specific indices. In particular, we showed that using the network weights from DeepBrainNet along with transfer learning, deep learning models are more robust across training sample size, so could be trained with dramatically fewer training examples than might be otherwise required. This saving in sample size is especially important in medical imaging where data is expensive and time consuming to collect or for under-studied diseases. Moreover, we showed that the brain MRI-specific DeepBrainNet network performs better than deep learning networks trained on orders of magnitude larger, but not brain-specific, databases like ImageNet. This finding suggests that domain-specific technologies for deep learning might perform better than generically-trained networks, which is the current practice in medical imaging, especially when relatively limited samples are available for training. Considering that there are more than a hundred pathologies that can be captured by medical scans of the brain, it is almost certain that sufficiently large databases for each pathology will not be available for many years, especially for rare diseases. The pooling and use of a large and highly diverse lifespan brain MRI database used herein played a critical role in our ability to achieve robust disease-specific indices.
For the tasks presented it is likely that the DeepBrainNet weights are closer to the global minimum of the task in the gradient decent landscape than the ImageNet weights (and certainly Random Initialization). Thus, we are more likely to converge to a minimum closer to the global minimum after disease-specific training. The reason that the ImageNet weights do not converge to the same accuracy as the DeepBrainNet weights, even given more training time, is that they get trapped in local minima during the optimization. This difference further highlights the importance of a domain-specific set of weights for transfer learning, particularly in complex non-convex optimization problems such as this (Becherer, 2017).
The final disease classifiers could likely be improved through careful hyperparameter tuning and refinements of the architectures. However, this would require a high level of expert knowledge and extensive experimental validations. On the other hand, DeepBrainNet allows for out-of-the-box convolutional neural networks architectures to be reliably applied to neuroimaging tasks.
The best brain age model is not the most sensitive in identifying pathologies
Our deep learning network was able to obtain very good estimates of brain age, with an MAE of 3.702. This raised the question of whether such accuracy was beneficial for detecting the informative discrepancies between brain age and chronological age that can be used to identify the presence of pathologies. We evaluated three levels of model tightness to the data: relatively looser, moderate, and relatively tighter. The moderately fitting brain age model yielded the most significant brain age deltas across the clinical categories examined, thereby offering evidence that this ‘middle of the road’ approach may be the best way to construct brain age indices. This finding is not unexpected, but it has been overlooked in the literature, which focuses on finding the best possible brain age fits for a given model. Tight-fitting brain age predictive models are likely to focus on brain features and patterns that are not affected by any factor other than age. However, many typically ageing individuals have various and often covert pathologies even if they are cognitively normal, such as small vessel ischaemic disease, amyloid plaques, and tau neurofibrillary tangles, amongst others. Therefore, a tightly-fitting brain age model will naturally seek to avoid the effects of such pathological processes, in its attempt to achieve the lowest MAE. The resultant brain age delta will then likely fail to capture brain-ageing effects of these pathological processes, at least to some extent. On the other hand, loosely fitting brain age models tend to miss the nuances of pathological patterns, and hence also fail to capture important features of brain-ageing. Put differently, a rough estimate of someone’s age might be obtained from the size of the ventricles or another simple feature that cannot capture subtle patterns of neuroanatomical change induced by neuropathological processes. Our experiments on four different disease groups showed that the moderate fit was more discriminative versus the others, while these differences were statistically significant for Alzheimer’s disease and MCI groups and not for schizophrenia and depression groups (Supplementary material, section S.14). Our results indicate that moderately accurate brain age models may provide the most meaningful brain age delta values and future work should investigate this further.
The trained network is available as an online resource, as well as on our online platform
Additionally, we release the weights of all models described in this paper as they will be valuable in a variety of transfer learning tasks. We have collected the DeepBrainAgeNet weights for multiple popular network architectures so that the architecture that best suits a research problem can still be selected. These architectures include VGG-16, ResNet-50 and DenseNet-169 and Inception-ResNet-v2. Files can be found at our GitHub repository (https://github.com/vishnubashyam/DeepBrainNet). The DeepBrainNet model can also be applied on any new scan to estimate the brain age of the subject using the pretrained model on the CBICA Internet Processing Portal (IPP) (https://ipp.cbica.upenn.edu/)
In summary, we present a complex and very broadly trained deep learning network, optimized on brain MRI features used to estimate brain age. In addition to providing estimates of brain age, and hence indicators of resilient versus advanced brain ageing, we found that this specialized network provides a better springboard for constructing disease-specific deep learning classifiers. Therefore, we hope to enable the development of a large family of pathology-specific deep learning networks utilizing DeepBrainNet as a foundation whose parameters are tuned and adapted to the pathology or disease of interest.
Funding
This work was supported by the National Institute on Aging (grant number 1RF1AG054409) and the National Institute of Mental Health (grant number 5R01MH112070). M.H. was supported in part by The Allen H. and Selma W. Berkman Charitable Trust (Accelerating Research on Vascular Dementia) and the National Institutes of Health (grant number R01 HL127659-04S1, R01EB022573, and HHSN271201600059C). This work was also supported in part by the Intramural Research Program, National Institute on Aging, NIH. SHIP is part of the Community Medicine Research Net of the University Medicine Greifswald, which is supported by the German Federal State of Mecklenburg- West Pomerania.
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
- MCI
mild cognitive impairment
References
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: A system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), 2016. P. 265–83.
- Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ.. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017; 30: 449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK.. Medical image analysis using convolutional neural networks: a review. J Med Syst 2018; 42: 226. [DOI] [PubMed] [Google Scholar]
- Arbabshirani MR, Plis S, Sui J, Calhoun VD.. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. Neuroimage 2017; 145: 137–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer N. Transfer Learning in Convolutional Neural Networks for Fine-Grained Image Classification; 2017.
- Brown TT, Kuperman JM, Chung Y, Erhart M, McCabe C, Hagler DJ Jr, et al. Neuroanatomical assessment of biological maturity. Curr Biol 2012; 22: 1693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JH, Franke K.. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci 2017; 40: 681–90. [DOI] [PubMed] [Google Scholar]
- Cole JH, Marioni RE, Harris SE, Deary IJ.. Brain age and other bodily ‘ages’: implications for neuropsychiatry. Mol Psychiatry 2019; 24: 266–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JH, Poudel RP, Tsagkrasoulis D, Caan MW, Steves C, Spector TD, et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 2017; 163: 115–24. [DOI] [PubMed] [Google Scholar]
- Cole JH, Ritchie SJ, Bastin ME, Hernández MV, Maniega SM, Royle N, et al. Brain age predicts mortality. Mol Psychiatry 2018; 23: 1385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Xu F, An Y, Fan Y, Resnick SM.. Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain 2009; 132: 2026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li L, Li K, Fei-Fei L. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, 2009. P. 248–55.
- Donahue J, Jia Y, Vinyals O, Hoffman J, Zhang N, Tzeng E, et al. Decaf: a deepconvolutional activation feature for generic visual recognition. In: International Conference on Machine Learning, 2014. P. 647–55.
- Dosenbach NUF, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, et al. Prediction of individual brain maturity using fMRI. Science 2010; 329: 1358–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C.. Multi-atlas skull-stripping. Acad Radiol 2013; 20: 1566–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erus G, Battapady H, Satterthwaite TD, Hakonarson H, Gur RE, Davatzikos C.. Imaging patterns of brain development and their relationship to cognition. Cereb Cortex 2015; 25: 1676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Grydeland H, Amlien I, Espeseth T, Reinvang I, et al. Critical ages in the life course of the adult brain: nonlinear subcortical aging. Neurobiol Aging 2013; 34: 2239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Gaser C, Manor B, Novak V.. Advanced BrainAGE in older adults with type 2 diabetes mellitus. Front Aging Neurosci 2013; 5: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Ziegler G, Klöppel S, Gaser C.. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage 2010; 50: 883–92. [DOI] [PubMed] [Google Scholar]
- Gaser C, Franke K, Klöppel S, Koutsouleris N, Sauer H.. BrainAGE in mild cognitive impaired patients: predicting the conversion to Alzheimer’s Disease. PLoS ONE 2013; 8: e67346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habes M, Janowitz D, Erus G, Toledo JB, Resnick SM, Doshi J, et al. Advanced brain aging: relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Transl Psychiatry 2016; 6: e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajek T, Franke K, Kolenic M, Capkova J, Matejka M, Propper L, et al. Brain age in early stages of bipolar disorders or schizophrenia. Schizophr Bull 2019; 45: 190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR); 2016. p. 770–8.
- Huang G, Liu Z, van der Maaten L. Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017.
- Jenkinson M, Bannister PR, Brady JM, Smith SM.. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage 2002; 17: 825–41. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Smith SM.. A global optimisation method for robust affine registration of brain images. Med Image Anal 2001; 5: 143–56. [DOI] [PubMed] [Google Scholar]
- Jonsson BA, Bjornsdottir G, Thorgeirsson TE, Ellingsen LM, Walters GB, Gudbjartsson DF, et al. Brain age prediction using deep learning uncovers associated sequence variants. Nat Commun 2019; 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017; 36: 61–78. [DOI] [PubMed] [Google Scholar]
- Kingma DP, Ba J. Adam: A method for stochastic optimization. In: International conference on learning representations; 2014.
- Kotikalapudi R, & contributors. 2017. keras-vis. Available from: https://github.com/raghakot/keras-vis (October 2019, date last accessed).
- Le TT, Kuplicki RT, McKinney BA, Yeh HW, Thompson WK, Paulus MP, et al. A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Front Aging Neurosci 2018; 10: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan CR, Kensinger EA.. Predicting age from cortical structure across the lifespan. Eur J Neurosci 2018; 47: 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Pérez JM, Dadar M, Lacalle-Aurioles M, Iturria-Medina Y, Zeighami Y, Evans AC.. Structural neuroimaging as clinical predictor: a review of machine learning applications. Neuroimage Clin 2018; 20: 506–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycki M, Satterthwaite TD, Koutsouleris N, Erus G, Doshi J, Wolf DH, et al. Machine learning analysis provides a robust structural imaging signature of schizophrenia detectable across diverse patient populations and within individuals. Schizophr Bull 2018; 44: 1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. CoRR arXiv:1409.1556.
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R.. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014; 15: 1929–58. [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex disease of middle and old age. PLOS Med 2015; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI Conference on Artificial Intelligence; 2017.
- Tajbakhsh N, Shin JY, Gurudu SR, Todd Hurst R, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 2016; 35: 1299–312. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 2014; 8: 153–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Clark KA, Thompson PM, Shattuck DW, Van Horn JD.. Mapping the human connectome. Neurosurgery 2012; 71: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010; 29: 1310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Toga AW.. Human neuroimaging as a “Big Data” science. Brain Imaging Behav 2014; 8: 323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira S, Pinaya WH, Mechelli A.. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci Biobehav Rev 2017; 74: 58–75. [DOI] [PubMed] [Google Scholar]
- Zeiler MD, Fergus R. Visualizing and understanding convolutional networks In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Computer vision–ECCV 2014. ECCV 2014. Lecture notes in computer science. Cham: Springer, vol. 8689. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data may be available from the authors upon reasonable request and with permission.