Abstract
A pathogenic GGGCCC hexanucleotide expansion in the first intron/promoter region of the C9orf72 gene is the most common mutation associated with amyotrophic lateral sclerosis (ALS). The C9orf72 gene product forms a complex with SMCR8 (Smith-Magenis Syndrome Chromosome Region, Candidate 8) and WDR41 (WD Repeat domain 41) proteins. Recent studies have indicated roles for the complex in autophagy regulation, vesicle trafficking, and immune response in transgenic mice, however a direct connection with ALS etiology remains unclear. With the aim of increasing understanding of the multi-functional C9orf72-SMCR8-WDR41 complex, we determined by mass spectrometry analysis the proteins that directly associate with SMCR8. SMCR8 protein binds many components of the ubiquitin-proteasome system, and we demonstrate its poly-ubiquitination without obvious degradation. Evidence is also presented for localization of endogenous SMCR8 protein to cytoplasmic stress granules. However, in several cell lines we failed to reproduce previous observations that C9orf72 protein enters these granules. SMCR8 protein associates with many products of genes associated with various Mendelian neurological disorders in addition to ALS, implicating SMCR8-containing complexes in a range of neuropathologies. We reinforce previous observations that SMCR8 and C9orf72 protein levels are positively linked, and now show in vivo that SMCR8 protein levels are greatly reduced in brain tissues of C9orf72 gene expansion carrier individuals. While further study is required, these data suggest that SMCR8 protein level might prove a useful biomarker for the C9orf72 expansion in ALS.
Keywords: Amyotrophic lateral sclerosis, Autophagy, Biomarker, Mass spectrometry, Proteasome, Stress granules, Ubiquitin
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that afflicts about 1 in 50,000 people each year and involves loss of upper and lower motor neurons [1]. Death typically follows 2 to 3 years after first onset. About 95% of cases are sporadic, while the rest have a family history of the disease. ALS also has overlapping clinical presentations with frontotemporal lobar degeneration (FTLD) and its most common subtype frontotemporal dementia (FTD), a neurological condition affecting the frontal and temporal lobes and marked by cognitive and behavioral impairment [2]. About 20% of ALS patients also exhibit FTLD, and ALS and FTLD have been considered to be part of a continuous disease spectrum [3].
A pathogenic GGGCCC hexanucleotide expansion in the first intron/promoter region of the C9orf72 gene is the most common mutation associated with both ALS (∼11% of all cases) and FTLD/FTD (∼13%) [3–6]. Three possible non-exclusive mechanisms have been proposed by which the C9orf72 repeat expansion may cause ALS-FTD: 1) haploinsufficiency and loss of C9orf72 protein function, 2) repeat-associated non-AUG (RAN) translation of the hexanucleotide repeats generating dipeptide repeats that aggregate in toxic neuronal cytoplasmic and nuclear aggregates, and 3) toxic gain-of-function from repeat-containing RNA which forms nuclear foci that sequester hexanucleotode repeat-binding proteins (reviewed in [7–10]). While most studies have focused on increasedtoxicity, accumulating evidence argues that a loss-of-function mechanism may also contribute to neurodegeneration. Consistently, various studies have reported a reduction in C9orf72 mRNA and/or protein expression in brain and induced pluripotent stem cell (iPSC)-derived neuronal lines of some C9orf72 ALS (C9ALS) and FTD patients [4–6, 11–25].
A series of studies have shown that the long isoform of human C9orf72 protein forms a complex with SMCR8 (Smith-Magenis Syndrome Chromosome Region, Candidate 8) and WDR41 (WD Repeat domain 41) proteins [22, 26–35]. The SMCR8 gene is within the deleted region of chromosome 17 associated with Smith-Magenis Syndrome (SMS), a developmental disorder of children involving intellectual disability, distinctive facial features, and behavioral problems, but no reported motor defects [36, 37]. WDR41 is a member of the WD-repeat family of proteins that act as protein-protein or protein-DNA interaction scaffolds for a variety of cellular functions [38]. SNPs within the WDR41 gene region have been associated with human caudate volume [39].
Bioinformatic analyses first identified both C9orf72 and SMCR8 proteins as having DENN (Differentially Expressed in Normal and Neoplastic cells) domains that are present in guanine nucleotide exchange factors (GEFs) for Rabs, multi-functional small GTPases involved in intracellular membrane trafficking and fusion, vesicle formation and transport, and autophagy [40–42]. The autolysomal-autophagy pathway involves generation of the autophagosome, an organelle surrounded by a double lipid bilayer. Autophagosomes engulf cytoplasmic components, such as protein aggregates, damaged organelles, and foreign pathogens, and fuse with lysosomes to generate autolysosomes that mediate degradation of the cargo. Autophagosomes also fuse with endosomes, forming an intermediate organelle called the amphisome, before fusion with lysosomes. Various studies have linked wild-type C9orf72 protein with proteostasis, showing that, in complex with SMCR8 and WDR41, it bind Rabs and plays roles in autophagy and initiation of autophagosome formation, as well as being linked by function and colocalization to endocytosis and lysosomal and endosomal trafficking ([9, 21–23, 26, 28, 29, 31, 32, 34, 43–48], and Discussion for review). A role in the endo-lysosome pathway has also been shown for the C. elegans C9orf72 ortholog alfa-1 [49]. Aoki et al. [46] linked the interaction of C9orf72 and RAB7L1 with regulation of vesicle trafficking, and WDR41 is necessary for recruitment of the C9orf72 complex to lysosomes [35, 50]. Thus, C9orf72 is a regulator of cellular proteostasis.
Additional roles for the C9orf72 complex have also been reported. C9orf72 alters phosporylation of cofilin and activates the small GTPase ADP-ribosylation factor-1/2 (ARF1/2) involved in actin dynamics [51]. Altered C9orf72 protein levels also causes changes in glutamatergic receptor levels, glutamate cycling and endothelin signaling, and excitotoxicity in response to glutamate, as well as widespread transcriptional changes [21, 52–54]. However, the consequences of loss of C9orf72 protein for motor neuron function remain unclear. In vivo, diminished motor function and axonal degeneration of motor neurons have been reported in zebrafish and C. elegans depleted of C9orf72 [55, 56]. However, subsequent studies detected no or only mild motor function defects in mice deficient for murine C9orf72 ortholog 3110043O21Rik [45]. On the other hand, in a gain-of-function C9ALS/FTD mouse model, Shao et al. [57] found that 3110043O21Rik haploinsufficiency or loss was associated with increased motor behavior deficits in a dose-dependent manner, while Liang et al. [25] reported that Smcr8 knockout (KO) mice displayed motor behavior defects and axonal swelling. While effects on motor function are uncertain, immune system pathology, spleen and lymph node enlargement, defects in macrophage, myeloid and microglial cell function, altered lysosomal trafficking, and decreased body weight and survival have all been reported for C9orf72 or SMCR8 knockout mice [21, 28, 32, 45, 58–65]. Despite these findings, so far no pathogenic loss-of-function coding mutation in C9orf72, SMCR8 or WDR41 genes has been found [66].
To increase understanding of the diverse functions of the C9orf72-SMCR8-WDR41 complex, we sought to determine by mass spectroscopy (MS) analyses the interactome composition of the SMCR8 component. Notably, we found that the SMCR8 complex includes numerous ubiquitin-related proteins and products of genes associated with numerous Mendelian neurological disorders. MS analyses, co-IP experiments, and association of SMCR8 with cytoplasmic stress granules (SGs) in cultured cells support a link between SMCR8 and the ubiquitin pathway. Furthermore, we reinforce previous observations that SMCR8 and C9orf72 protein levels are positively linked, now showing in vivo that SMCR8 might prove to be a useful biomarker for the C9orf72 expansion mutation in ALS patients.
Materials and methods
Plasmid constructs
Ultimate ORF cDNA clones (Invitrogen), with V5-epitope tags and tobacco etch virus (TEV) protease cleavage sites on their N-termini, were recloned by shuttling them from pENTR221 vector into pcDNA3.1/nV5-DEST using Gateway Technology (Invitrogen). Ultimate ORF Clones included AIFM1 (clone identifier IOH61019), BAG5 (IOH26366), C9orf72 (IOH45695), DARS (IOH4209), DCTN1 (IOH42830), DDB1 (IOH13816), DNAJC7 (IOH14566), FKBP5 (IOH13816), G3BP1 (IOH7337), GTF2I (IOH62625), HSPB1 (IOH10530), MAGED2 (IOH3451), PARK7 (IOH3149), PPP2R1A (IOH13670), PSMA7 (IOH5011), PSMC5 (IOH3508), PSMD14 (IOH40112), QARS (IOH3529), RAB1A (IOH2921), RAB7A (IOH40569), RAB11A (IOH13764), RANGAP1 (IOH13287), RO60 (IOH22411), RNF40 (IOH6599), RUVBL2 (IOH3426), SLC25A6 (IOH5815), SQSTM1 (IOH5103), STIP1 (IOH5061), STUB1 (IOH56981), SUGT1 (IOH2955), and TBK1 (IOH21006).
Clones purchased or obtained as gifts included FLAG-UBR5 (pCMV-Tag2B EDD; Addgene plasmid #37188 [67]), pcDNA5 FRT/TO FLAG-SMCR8 (FL-SMCR8), pcDNA5 FRT/TO HA-SMCR8, and pcDNA5 FRT/TO GFP SMCR8 (MRC PPU Reagents and Services, University of Dundee), GFP-(GA)50 (L. Petrucelli, Mayo Clinic, Florida, [68]) pRK5-myc-TDP-43 (J. Wang, Johns Hopkins University, [69]), mRFP-UBB (Addgene plasmid #11935, [70]), HA-UBB (V. Dawson, Johns Hopkins University, [71]), and pDest51-USP9X-V5 (R. Hughes, Buck Institute for Research on Aging, [72]). FL-UBB was generated using Ultimate ORF Clone IOH56688 and modified Gateway vector pEZYflag (Addgene plasmid #18700, Y.-Z. Zhang). C9orf72-FL, RO60-FL, SMCR8-V5, and WDR41-FL constructs were generated by PCR-amplification of Ultimate ORF cDNA clones, using a primer with AAG linker and C-terminal FLAG- or V5-tag, and cloning of the products in pcDNA6/myc-His B (pcDNA6, Invitrogen).
Cell line and tissue samples
Human embryonal carcinoma 2102Ep (a gift from P.K. Andrews, University of Sheffield, UK) and nTERA2D1 cells, human cervical cancer HeLa-JVM cells [73], human embryonic kidney (HEK) 293T cells (ATCC), human neuroblastoma SH-SY-5Y cells (ATCC CRL-2266), and mouse neuroblastoma Neuro2A cells (a gift from D. Hackam, JHU) were grown in Dulbecco’s modified Eagle’s Medium. Mouse hybrid spinal cord neuron/neuroblastoma NSC-34 cells (a gift from D. Griffen, JHU) and human neuroblastoma SK-N-SH cells (a parental line of SH-SY-5Y cells; gifts of D. Valle, JHU) were grown in Eagle’s Minimum Essential Medium. Medium was supplemented with 10% FBS (Hyclone), GlutaMax, and Pen-Strep (Invitrogen). Plasmid transfections used FuGENE HD (Promega) reagent. As necessary, cells were treated with 20 μM MG-132 proteasome inhibitor (Cell Signaling) for about 20 h to inhibit protein degradation, or with 0.25 mM sodium arsenite (NaAsO2) or 3 mM dithiothreitol (DTT, Sigma) for 80 min and 2 h, respectively, to induce cytoplasmic SGs.
Post-mortem ALS spinal cord and unaffected control brain motor cortex tissues were obtained from Drs. J. Ravits and R. Batra of the Department of Neurosciences, University of California San Diego School of Medicine [74], and C9ALS and unaffected control samples were from the Target ALS Multicenter Postmortem Tissue Core (Table S4). C9ALS samples had been confirmed for the C9orf72 expansion using repeat-primed PCR (RP-PCR) and Illumina Expansion Hunter (M. Harms, Columbia University, pers. comm.). Frozen spinal cord tissues were obtained from the University of Maryland Brain and Tissue Bank of the NIH NeuroBioBank. Frozen C9ALS and unaffected control cerebrospinal fluid (CSF) samples were from the Northeast ALS Consortium (NEALS). CSF was resuspended directly in 3X SDS loading buffer or first concentrated by tricholoracetic acid precipitation, and then analyzed by Western blotting using α-SMCR8 antibodies. Up to 20 μg of CSF total protein were loaded per well.
Protein isolation and immunoprecipitation
For MS sequence determination, HEK 293T cells in T75 flasks were transfected using FuGENE HD (Promega) with 15 μg of FL-SMCR8, C9orf72-FL, or pcDNA6/myc-His B empty vector and expanded for approximately 45 h, followed by whole cell lysate preparation by sonication using a Diagenode Bioruptor. IP and sample recovery were as previously described [75, 76]. Treatment of samples with 25 μg/ml DNase-free RNase (Roche) and 25 μg/ml RNaseA (Qiagen) was conducted in the absence of RNase inhibitors.
For other protein extracts, tissues or cells were lysed in RIPA buffer (Sigma) with Mammalian Protease Inhibitor Cocktail and phenylmethanesulfonyl fluoride (Sigma) and homogenized by Diagenode Bioruptor. For tissues, 2 mm ziconium silicate beads (Next Advance, Inc.) were added to the tubes. Supernatants were recovered by centrifugation at 11K rpm at 4 °C for 15 min and resuspended in 3X SDS loading buffer.
For each interaction co-IP (Fig. 2), extracts from approximately 5 × 106 293T cells in T75 flasks transfected with tagged SMCR8 and test protein constructs were prepared in 700 μl of lysis buffer (160 mM NaCl, 50 mM Tris, 1 mM EDTA, 0.25% NP40) containing protease and phosphatase inhibitors (Sigma), and RNasin (Qiagen) and vanadyl ribonucleoside complexes (Sigma) as required, and immunoprecipitated as previously described [75, 76].
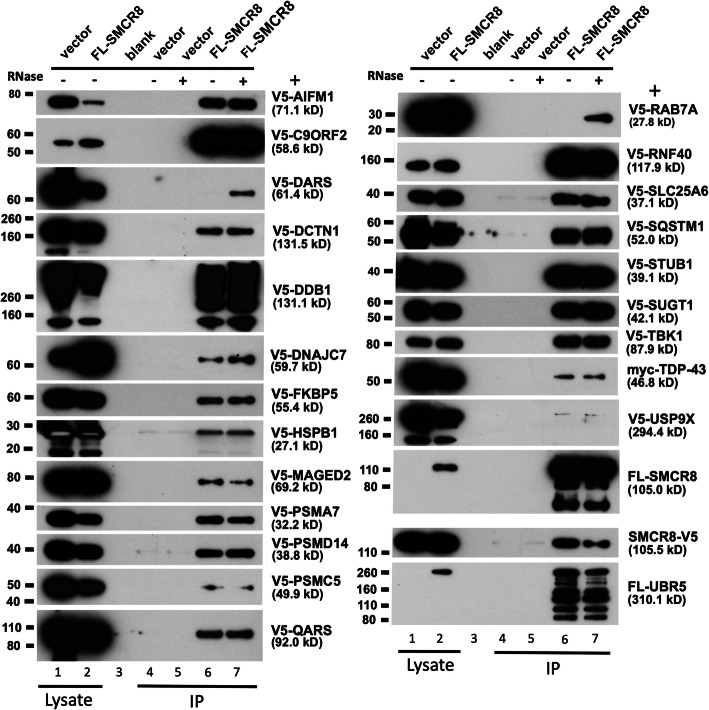
Fig. 2.
Confirmation of proteins in the SMCR8 complex. Selected proteins detected in the SMCR8 interactome by MS sequencing were tagged and coexpressed with SMCR8 in HEK 293T cells. Most were found to specifically co-IP on α-FLAG agarose with FL-SMCR8 but not empty vector. Approximately 1% of the input lysate (lanes 1, 2) and 30% of the immunoprecipitate (lanes 4–7) were loaded on gels. IP reactions were in the presence or absence of 50 μg/ml RNases. Also included is a panel representative of tagged FL-SMCR8 protein present in the input and IP fractions (detected by α-FLAG antibody) and showing that RNase treatment did not affect SMCR8 immunoprecipitation (lower right). Test proteins were detected by α-V5 antibody, except FL-UBR5, which was detected by α-FLAG antibody (bottom right). The molecular weight of each test protein, including its epitope tag, is shown in brackets. Protein molecular weight markers are those of Novex Sharp Pre-stained Protein Standard (Thermo Fisher)
Antibody analyses
Commercial antibodies included mouse (ms) α-V5-tag (Invitrogen), rabbit (rb) α-DYKDDDDK-tag (α-FLAG) (D6W5B), ms α-HA-tag (6E2), rb α-Myc-tag (71D10), rb α-HSP90, and ms α-Ubiquitin (UBB) (P4D1) (Cell Signaling Technology), rb α-4E-T, rb α-C9orf72 (S-14), goat (gt) α-eIF3η (N-20), ms α-p70 S6 kinase (which recognizes HEDLS/EDC4 [77]), gt α-TIA1 (C-20), and rb α-WDR41 (S-12) (Santa Cruz Biotechnology), rb α-SMCR8 (ab186504, epitope region 841–890) and rb α-SMCR8 (ab202283, epitope region 600–650) (Abcam), rb α-C9orf72 (22637–1-AP), rb α-SMCR8 (21125–1-AP, epitope region 588–937), and rb α-WDR41 (26817–1-AP) (Proteintech), and rb α-SMCR8 (A304-694A, epitope region 600–650) (Bethyl). Monoclonal ms α-4H1-ORF1 was kindly provided by Dr. K. Burns (JHU, USA, also Millipore MABC1152, [78]), rb α-C9-L and rb α-C9-S antibodies by Dr. J. Robertson (U. Toronto, Canada, [19, 79]), and α-GW182 human serum (IC-6) by Dr. M. Fritzler (U. Calgary, [80]). Donkey Cy3-, DyLight 488-, or DyLight 549-conjugated, and HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories.
Western blotting, immunofluorescence (IF) microscopy, and fluorescent in situ hybridization (FISH) were performed as described [81, 82]. All Western blotting used NuPAGE 4–12% Bis-Tris gels and MOPS buffer, except brain tissue analyses of Fig. 5b,c which used NuPAGE 3–8% Tris-acetate gels (Thermo Fisher). Detection used Supersignal West Pico or West Pico Plus Chemiluminescent Substrate (Thermo Fisher) with ECL Hyperfilm (MilliporeSigma).
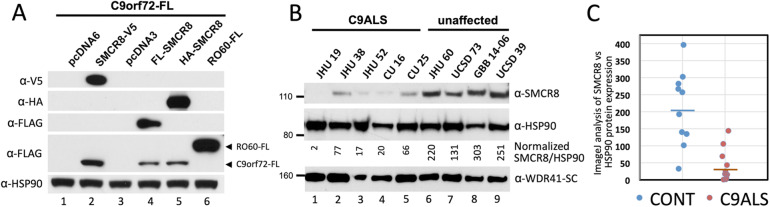
Fig. 5.
Expression of C9orf72 and SMCR8 proteins are positively correlated in cell lines and human brain tissues. a C9orf72-FL was coexpressed in HEK 293T cells with 3 different epitope-tagged SMCR8 constructs, FLAG-tagged RO60 protein, or empty vectors (pcDNA3 and pcDNA6 myc/his B). A Western blot of whole cell lysates was probed sequentially with rb α-FLAG, ms α-HA, ms α-V5, and rb α-HSP90 antibodies, the latter as a loading control. At the exposure time for the film shown, expression of C9orf72-FL was not seen in the presence of empty vector or RO60-FL, but signal was robust in the presence of SMCR8. b Western blot of brain motor cortex tissue lysates of C9ALS patients (lanes 1–5) and unaffected control individuals (lanes 6–9) probed with α-SMCR8 and α-HSP90 antibodies. Sample names are shown above the panels (see Table S4). Numbers below the middle panel are normalized ratios of SMCR8 to HSP90 expression determined by ImageJ analysis of band intensities and calculated as described in the text. The lower panel shows the approximately 150-kD unspecified band detected by α-WDR41-SC antibody in human brain tissue lysates (see Fig. S1F): this panel is included only as an additional loading control and is not intended to show expression of canonical WDR41 protein. Approximtely 50 μg of protein was loaded in each lane. c Dot plot of ratios of SMCR8 to HSP90 protein band intensities determined by ImageJ analyses of brain tissues lysates from 11 C9ALS and 10 control individuals. Each sample point is the average of 2 to 4 independent Western blot analyses. A short horizontal line indicates mean values. The presence of a C9orf72 hexanucleotide expansion in each C9ALS carrier individual was confirmed by Columbia University and Target ALS using RP-PCR and Illumina Expansion Hunter, but expansion copy numbers are not known
Immunostained cells were examined using a Nikon Eclipse Ti-A1 confocal microscope with NIS-Elements AR software.
MS sequencing and data analyses
MS sequencing and database analyses was performed by the Johns Hopkins Mass Spectrometry and Proteomics Facility as previously described [75, 76]. Peptide sequences were identified using Proteome Discoverer and Mascot software (Matrix Science) to search the NCBInr 167 database, including gly-gly modifiation on lysine as a variable protein modification. False discovery rate (FDR) was set at 1.0. Mascot search result *.dat files were processed in Scaffold (Proteome Software, Inc.) to validate protein and peptide identifications. Exclusion criteria for proteins are described in the Results section.
Functional analyses of gene lists obtained by MS sequencing utilized DAVID v6.7 (Database for Annotation, Visualization and Integrated Discovery [83]). Protein multiple alignments used Clustal Omega 1.2.1 (EMBL-EBI) and the ESPript 3.0 web sever (http://espript.ibcp.fr/ESPript/ESPript/). RNA-Seq datasets from public databases (GEO GSE67196 and NeuroLINCS dbGaP Study phs001231 SRP098831) were analyzed for gene expression using the software package TETranscripts as previously described [84, 85]. Ubiquitination prediction algorithms included UbPred (http://www.ubpred.org, [86]), BDM-PUB (http://bdmpub.biocuckoo.org), and UbiSite (http://csb.cse.yzu.edu.tw/UbiSite/).
Results
Confirmation of the SMCR8-C9orf72-WDR41 complex and detection using commercial antibodies
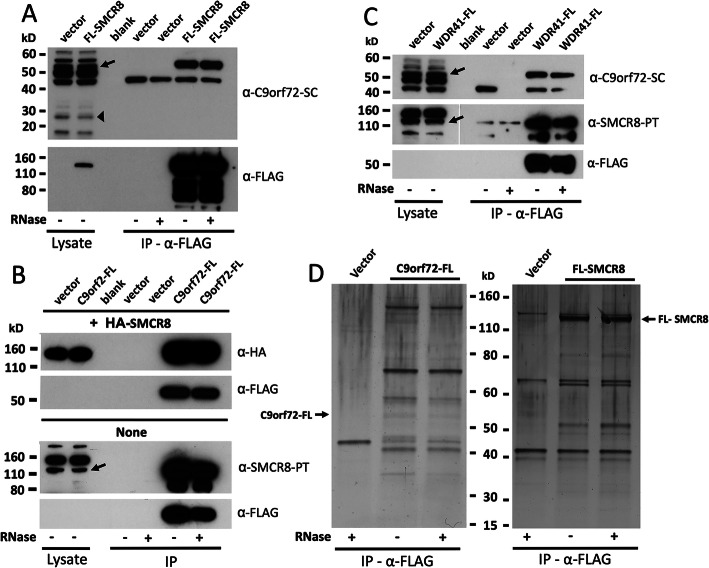
Previous studies characterizing the C9orf72 proteome identified a complex consisting of the proteins C9orf72, SMCR8 and WDR41 [22, 26–35]. By means of co-immunoprecipitation (co-IP) and MS sequencing, here we sought to identify proteins that associate with SMCR8. First, we confirmed that C9orf72, SMCR8 and WDR41 proteins formed a stable complex in our overexpression experimental system using human embryonic kidney (HEK) 293T cells. Consistent with previous reports, even in the presence of RNase, epitope-tagged SMCR8 and C9orf72 co-IPed C9orf72 and SMCR8 proteins, respectively (Fig. 1a, b). Furthermore, we confirmed that FLAG-tagged WDR41 interacts with endogenous C9orf72 and SMCR8 (Fig. 1c).
Fig. 1.
Protein interaction analyses by Western blotting and co-IP of the SMCR8 complex in HEK 293T cells (see Fig. S1 for antibody analyses). a Endogenous C9-L (arrow) co-IPs with FLAG-tagged SMCR8. The thick arrowhead marks a band consistent in size with C9-S. b FLAG-tagged C9orf72 co-IPs both endogenous and co-transfected HA-tagged SMCR8. c FLAG-tagged WDR41 protein co-IPs both endogenous C9orf72 and SMCR8 proteins (indicated by arrows). Tagged C9orf72 and WDR41 proteins of (b) and (c) are not visible in whole cell lysates at the Western blot film exposure times shown. d C9orf72-FL, FL-SMCR8, and empty vector were immunoprecipitated on α-FLAG agarose from transfected 293T whole cell lysates, resolved on a polyacrylamide gel, and silver-stained. IP reactions were in the presence or absence of 50 μg/ml RNases. Complex immunoprecipitate samples were analyzed by MS sequencing. Arrows indicate full-length protein bands. Protein molecular weight markers are those of Novex Sharp Pre-stained Protein Standard (Thermo Fisher Scientific)
We next assessed the efficacy of several commercial antibodies against these proteins. Human C9orf72 expresses a 54-kilodalton (kD) long protein isoform (C9-L) and a 25-kD short isoform (C9-S). It has been noted that commercial C9orf72 antibodies often detect additional bands other than C9-L or fail to detect C9-S [17, 19, 23]. Consistent with this observation, the Santa Cruz S-14 antibody (α-C9orf72-SC) detected multiple bands in whole cell lysates as well as products of size consistent with both endogenous C9-L and C9-S; only C9-L co-IPed with tagged SMCR8 or WDR41 (Fig. 1a,c). Similarly, in both cultured cells and brain and spinal cord tissue lysates, the Proteintech 22,637–1-AP antibody (α-C9orf72-PT) marked a major band consistent with C9-L (arrow), plus additional products (Fig. S1A). A small number of non-commercial C9orf72-specific antibodies have also been described [17, 23, 30, 79].
Both mouse and human SMCR8 have two predicted isoforms, the full length 105-kD protein and a C-terminal truncated 87.4-kD isoform generated by alternate splicing [36]. Our search of GenBank revealed additional human SMRC8 mRNA isoforms potentially encoding 35.9-kD (accession numbers BC001018, BC005067), 75.2-kD (AK296847.1), and 93-kD (BC101116, BC101117) protein products. Tissue-specific transcripts of various sizes have also been experimentally observed for human SMCR8 [36]. The Proteintech, Bethyl (A304-694A), and Abcam (ab186504 and ab202283) α-SMCR8 antibodies all marked a band consistent in size with full-length SMCR8 (i.e. 105 kD, arrows in Fig. S1B-E), plus additional bands of unknown specificicy, but which could in part relate to the above desribed SMCR8 protein isoforms. The Bethyl and Abcam ab202283 α-SMCR8 antibodies have been used in other studies, and our observations are similar [34, 50, 65, 79]. Interestingly, although expression of full length SMCR8 protein was detected in human brain tissues, none was seen in spinal cord tissue lysates of multiple samples (Fig. S1B-E).
WDR41 has two predicted isoforms of 51.7 and 45.5 kD (Swiss Prot. Q9HAD4–1, Q9HAD4–2). For selected cancer cell lines, both the Santa Cruz (S-12) and Proteintech (26817–1-AP) polyclonal antibodies detected doublet bands consistent in size with these isoforms (Fig. S1F, G). These bands were very faint (Proteintech) or absent (Santa Cruz) from human brain and spinal cord tissue lysates, although bands of larger and smaller sizes were visible by Western blotting of cultured cells.
SMCR8 interactome contains many central nervous system (CNS) disease proteins
Because of the possible non-specific protein interactions described above, we considered commercial antibodies unsuitable for co-IP interactome studies. Therefore, we exploited a co-IP/MS protocol that we have successfully used in previous studies [75, 76]. We transfected C9-L with C-terminal FLAG (FL)-tag, full-length SMCR8 with N-terminal FLAG-tag, or empty vector control in HEK 293T cells and performed α-FLAG IP from whole cell extracts in the presence or absence of RNase (Fig. 1d). Complex immunoprecipitated samples were analyzed by liquid chromatography tandem MS. After excluding ribosomal proteins and likely contaminants (such as keratins), 340 and 201 proteins having three or more spectra and not detected in vector only control cell lysates were associated with FL-SMCR8 and C9-L-FL, respectively (Tables 1, S1, S2). Furthermore, 71 proteins were found in both proteomes, although it should be noted that C9-L-FL was expressed at significantly lower levels than FL-SMCR8 (Fig. 1d), as previously reported [29]. Tables S1 and S2 also note interacting partners of C9orf72 or SMCR8 proteins reported in previous studies [19, 28, 29, 31, 32, 34, 51, 87, 88]. During the course of our investigations, another MS experiment was published that listed 1532 proteins that co-IPed with HA-tagged SMCR8 from 293T cells [34], and a total of 272 of these (80%) were also present in our dataset (Table S2).
Table 1.
Summary of the C9orf72 and SMCR8 protein interactomes and selected functional categories
| Gene Name | Protein Name | Total # Spectra | |
|---|---|---|---|
| RNase | |||
| - | + | ||
| A. C9orf72-FL | |||
| C9orf72 | chromosome 9 open reading frame 72 | 108 | 103 |
| TOP 20 C9orf72-INTERACTING PROTEINS | |||
| SMCR8 | Smith-Magenis syndrome chromosome region, candidate 8 | 165 | 217 |
| WDR41 | WD repeat domain 41 | 69 | 85 |
| FLG2 | filaggrin family member 2 | 33 | 6 |
| LRPPRC | leucine rich pentatricopeptide repeat containing | 23 | 25 |
| IARS | isoleucyl-tRNA synthetase | 21 | 22 |
| SERPINB3 | serpin family B member 3 | 21 | 0 |
| CALML5 | calmodulin like 5 | 20 | 0 |
| HADHA | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, alpha subunit | 18 | 14 |
| CDSN | corneodesmosin | 17 | 8 |
| EEF2 | eukaryotic translation elongation factor 2 | 15 | 18 |
| FABP5 | fatty acid binding protein 5 | 15 | 0 |
| COPB1 | coatomer protein complex subunit beta 1 | 14 | 14 |
| DSC1 | desmocollin 1 | 14 | 15 |
| U2AF2 | U2 small nuclear RNA auxiliary factor 2 | 14 | 6 |
| LUC7L3 | LUC7 like 3 pre-mRNA splicing factor | 13 | 4 |
| EIF2S3 | eukaryotic translation initiation factor 2 subunit gamma | 12 | 10 |
| HNRNPH2 | heterogeneous nuclear ribonucleoprotein H2 | 12 | 9 |
| KHDRBS1 | KH RNA binding domain containing, signal transduction associated 1 | 12 | 6 |
| PSMC2 | proteasome 26S subunit, ATPase 2 | 12 | 25 |
| B. FL-SMCR8 | |||
| SMCR8 | Smith-Magenis syndrome chromosome region, candidate 8 | 1135 | 1182 |
| TOP 25 SMCR8-INTERACTING PROTEINS | |||
| DNAJA1 | DnaJ heat shock protein family member A1 | 70 | 76 |
| GTF2I | general transcription factor IIi | 55 | 61 |
| CAD | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | 62 | 48 |
| ATP1A1 | ATPase Na+/K+ transporting subunit alpha 1 | 38 | 46 |
| EPRS | glutamyl-prolyl-tRNA synthetase | 36 | 40 |
| MAGED1 | MAGE family member D1 | 26 | 33 |
| WDR41 | WD repeat domain 41 | 32 | 33 |
| DNAJC7 | DnaJ heat shock protein family member C7 | 30 | 32 |
| SLC25A3 | solute carrier family 25 member 3 | 22 | 32 |
| STUB1 | STIP1 homology and U-box containing protein 1 | 30 | 31 |
| GCN1 | GCN1, eIF2 alpha kinase activator homolog | 37 | 30 |
| DDB1 | damage specific DNA binding protein 1 | 31 | 29 |
| DYNC1H1 | dynein cytoplasmic 1 heavy chain 1 | 38 | 29 |
| CHD4 | chromodomain helicase DNA binding protein 4 | 13 | 28 |
| SLC25A11 | solute carrier family 25 member 11 | 14 | 28 |
| RANBP2 | RAN binding protein 2 | 29 | 25 |
| RUVBL2 | RuvB like AAA ATPase 2 | 30 | 25 |
| ATP2A2 | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 | 19 | 22 |
| DNAJA3 | DnaJ heat shock protein family member A3 | 19 | 22 |
| IARS | isoleucyl-tRNA synthetase | 18 | 22 |
| STIP1 | stress induced phosphoprotein 1 | 37 | 22 |
| COPA | coatomer protein complex subunit alpha | 23 | 20 |
| HUWE1 | HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase | 29 | 20 |
| AIFM1 | apoptosis inducing factor, mitochondria associated 1 | 18 | 18 |
| C9orf72 | chromosome 9 open reading frame 72 | 15 | 18 |
| AMINOACYL-tRNA SYNTHESIS | |||
| EPRS | glutamyl-prolyl-tRNA synthetase | 40 | 13 |
| IARS | isoleucyl-tRNA synthetase | 18 | 22 |
| DARS | aspartyl-tRNA synthetase | 11 | 8 |
| MARS | methionyl-tRNA synthetase | 10 | 6 |
| RARS | arginyl-tRNA synthetase | 9 | 8 |
| LARS | leucyl-tRNA synthetase | 9 | 14 |
| QARS | glutaminyl-tRNA synthetase | 8 | 7 |
| TARS2 | threonyl-tRNA synthetase 2, mitochondrial (putative) | 6 | 4 |
| TARS | threonyl-tRNA synthetase | 4 | 0 |
| FARSA | phenylalanyl-tRNA synthetase alpha subunit | 0 | 6 |
| CELL CYCLE | |||
| SMC3 | structural maintenance of chromosomes 3 | 12 | 8 |
| CDK4 | cyclin dependent kinase 4 | 5 | 0 |
| PCNA | proliferating cell nuclear antigen | 5 | 4 |
| MCM7 | minichromosome maintenance complex component 7 | 4 | 6 |
| YWHAZ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 4 | 9 |
| RAD21 | RAD21 cohesin complex component | 3 | 0 |
| CDC7 | cell division cycle 7 | 3 | 0 |
| PLK1 | polo like kinase 1 | 3 | 9 |
| PPP6C | protein phosphatase 6 catalytic subunit | 3 | 0 |
| CDKN2A | cyclin dependent kinase inhibitor 2A | 0 | 3 |
| MCM2 | minichromosome maintenance complex component 2 | 0 | 4 |
| MCM5 | minichromosome maintenance complex component 5 | 0 | 4 |
| SMC1A | structural maintenance of chromosomes 1A | 0 | 4 |
| HEAT SHOCK PROTEINS / CHAPERONES | |||
| DNAJA1 | DnaJ heat shock protein family member A1 | 70 | 76 |
| DNAJC7 | DnaJ heat shock protein family member C7 | 30 | 32 |
| DNAJA3 | DnaJ heat shock protein family member A3 | 19 | 22 |
| DNAJA3 | DnaJ heat shock protein family member A3 | 19 | 22 |
| HSPH1 | heat shock protein family H member 1 | 19 | 8 |
| BAG5 | BCL2 associated athanogene 5 | 18 | 11 |
| DNAJA2 | DnaJ heat shock protein family member A2 | 15 | 16 |
| BAG2 | BCL2 associated athanogene 2 | 11 | 16 |
| BAG6 | BCL2 associated athanogene 6 | 11 | 8 |
| SERPINH1 | serpin family H member 1 | 11 | 0 |
| HSPB1 | heat shock protein family B member 1 | 10 | 9 |
| MDN1 | midasin AAA ATPase 1 | 10 | 14 |
| FKBP8 | FK506 binding protein 8 | 8 | 0 |
| DNAJB6 | DnaJ heat shock protein family member B6 | 6 | 4 |
| PFDN2 | prefoldin subunit 2 | 5 | 4 |
| DNAJB1 | DnaJ heat shock protein family member B1 | 4 | 0 |
| HSPA4 | heat shock protein family A member 4 | 4 | 3 |
| HSPA4 | heat shock protein family A member 4 | 4 | 3 |
| RBBP7 | RB binding protein 7, chromatin remodeling factor | 3 | 4 |
| SERPINB3 | serpin family B member 3 | 3 | 0 |
| BAG3 | BCL2 associated athanogene 3 | 2 | 3 |
| UNC45A | unc-45 myosin chaperone A | 2 | 5 |
| DNAJB11 | DnaJ heat shock protein family member B11 | 0 | 6 |
| DNAJC10 | DnaJ heat shock protein family member C10 | 0 | 5 |
| PARK7 | Parkinsonism associated deglycase | 0 | 4 |
| MISMATCH REPAIR | |||
| RFC3 | replication factor C subunit 3 | 5 | 0 |
| SSBP1 | single stranded DNA binding protein 1 | 5 | 0 |
| MSH6 | mutS homolog 6(MSH6) | 5 | 5 |
| POLD3 | DNA polymerase delta 3, accessory subunit | 2 | 3 |
| PCNA | proliferating cell nuclear antigen | 0 | 4 |
| POLD1 | DNA polymerase delta 1, catalytic subunit | 0 | 3 |
| PHAGOSOME/AUTOPHAGY | |||
| DYNC1H1 | dynein cytoplasmic 1 heavy chain 1 | 38 | 29 |
| SEC16A | SEC16 homolog A, endoplasmic reticulum export factor | 14 | 12 |
| TUBB4A | tubulin beta 4A class IVa | 11 | 10 |
| IRS4 | insulin receptor substrate 4 | 10 | 13 |
| TUBB3 | tubulin beta 3 class III | 9 | 13 |
| TUBB6 | tubulin beta 6 class V | 9 | 18 |
| PPP2R1A | protein phosphatase 2 scaffold subunit Aalpha | 7 | 5 |
| CDK5 | cyclin dependent kinase 5 | 7 | 0 |
| SEC61B | Sec61 translocon beta subunit | 6 | 0 |
| HLA-A | major histocompatibility complex, class I, A | 5 | 5 |
| ANKRD28 | aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | 5 | 5 |
| RAB1B | RAB1B, member RAS oncogene family | 5 | 0 |
| SLC3A2 | solute carrier family 3 member 2 | 4 | 8 |
| SEC24C | SEC24 homolog C, COPII coat complex component | 4 | 3 |
| TFRC | transferrin receptor | 4 | 0 |
| AP3M1 | adaptor related proteincomplex 3 Mu 1 subunit | 3 | 0 |
| PPP6C | protein phosphatase 6 catalytic subunit | 3 | 0 |
| BAG3 | BCL2 associated athanogene 3 | 2 | 3 |
| WDR5 | WD repeat domain 5 | 0 | 5 |
| YME1L1 | YME1 like 1 ATPase | 0 | 4 |
| KEAP1 | kelch like ECH associated protein 1 | 0 | 4 |
| DYNC1I2 | dynein cytoplasmic 1 intermediate chain 2 | 0 | 4 |
| CDKN2A | cyclin dependent kinase inhibitor 2A | 0 | 3 |
| HLA-B | major histocompatibility complex, class I, B | 0 | 3 |
| PROTEIN PROCESSING IN THE ENDOPLASMIC RETICULUM | |||
| DNAJA1 | DnaJ heat shock protein family (Hsp40) member A1 | 70 | 76 |
| STUB1 | STIP1 homology and U-box containing protein 1 | 30 | 31 |
| HSPH1 | heat shock protein family H (Hsp110) member 1 | 19 | 8 |
| DNAJA2 | DnaJ heat shock protein family (Hsp40) member A2 | 15 | 16 |
| BAG2 | BCL2 associated athanogene 2 | 11 | 16 |
| SEC61B | Sec61 translocon beta subunit | 6 | 0 |
| RPN1 | ribophorin I | 5 | 3 |
| TXNDC5 | thioredoxin domain containing 5 | 5 | 2 |
| DNAJB1 | DnaJ heat shock protein family (Hsp40) member B1 | 4 | 0 |
| SEC24C | SEC24 homolog C, COPII coat complex component | 4 | 3 |
| DDOST | dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kD subunit | 3 | 4 |
| SSR4 | signal sequence receptor subunit 4 | 2 | 3 |
| DNAJB11 | DnaJ heat shock protein family (Hsp40) member B11 | 0 | 6 |
| DNAJC10 | DnaJ heat shock protein family (Hsp40) member C10 | 0 | 5 |
| RRBP1 | ribosome binding protein 1 | 0 | 3 |
| PROTEOSOME | |||
| PSMC2 | proteasome 26S subunit, ATPase 2 | 22 | 18 |
| PSMD2 | proteasome 26S subunit, non-ATPase 2 | 18 | 10 |
| PSMA7 | proteasome subunit alpha 7 | 17 | 16 |
| PSMD11 | proteasome 26S subunit, non-ATPase 11 | 14 | 9 |
| PSMC3 | proteasome 26S subunit, ATPase 3 | 12 | 9 |
| PSMD3 | proteasome 26S subunit, non-ATPase 3 | 12 | 8 |
| PSMA5 | proteasome subunit alpha 5 | 11 | 6 |
| PSMC5 | proteasome 26S Subunit, ATPase 5 | 10 | 10 |
| PSME3 | proteasome activator subunit 3 | 10 | 11 |
| PSMA6 | proteasome subunit alpha 6 | 9 | 0 |
| PSMD6 | proteasome 26S subunit, non-ATPase 6 | 9 | 7 |
| PSMA3 | proteasome subunit alpha 3 | 8 | 3 |
| PSMD1 | proteasome 26S subunit, non-ATPase 1 | 8 | 8 |
| PSMA1 | proteasome subunit alpha 1 | 7 | 7 |
| PSMB1 | proteasome subunit beta 1 | 7 | 7 |
| PSMB5 | proteasome subunit beta 5 | 6 | 6 |
| PSMC1 | proteasome 26S subunit, ATPase 1 | 6 | 6 |
| PSMB2 | proteasome subunit beta 2 | 5 | 5 |
| PSMB6 | proteasome subunit beta 6 | 5 | 5 |
| PSMC4 | proteasome 26S subunit, ATPase 4 | 5 | 0 |
| PSMC6 | proteasome 26S subunit, ATPase 6 | 5 | 6 |
| PSMD12 | proteasome 26S subunit, non-ATPase 12 | 4 | 2 |
| PSMD7 | proteasome 26S subunit, non-ATPase 7 | 3 | 0 |
| PSMD13 | proteasome 26S subunit, non-ATPase 13 | 0 | 3 |
| PSMD14 | proteasome 26S subunit, non-ATPase 14 | 0 | 3 |
| PSMD8 | proteasome 26S subunit, non-ATPase 8 | 0 | 6 |
| RNA TRANSPORT | |||
| RANBP2 | RAN binding protein 2 | 29 | 25 |
| XPO1 | exportin 1 | 13 | 13 |
| NUP93 | nucleoporin 93 | 11 | 10 |
| NUP133 | nucleoporin 133 | 10 | 15 |
| THOC2 | THO complex 2 | 9 | 6 |
| RANGAP1 | Ran GTPase activating protein 1 | 8 | 11 |
| KPNB1 | karyopherin subunit beta 1 | 7 | 5 |
| KPNA2 | karyopherin subunit alpha 2 | 6 | 4 |
| EIF4A1 | eukaryotic translation initiation factor 4A1 | 5 | 4 |
| EIF4G1 | eukaryotic translation initiation factor 4 gamma 1 | 5 | 4 |
| NUP155 | nucleoporin 155 | 4 | 6 |
| NUP210 | nucleoporin 210 | 4 | 0 |
| EIF3F | eukaryotic translation initiation factor 3 subunit F | 3 | 3 |
| FXR2 | FMR1 autosomal homolog 2 | 3 | 0 |
| NUP160 | nucleoporin 160 | 3 | 4 |
| NUP205 | nucleoporin 205 | 3 | 4 |
| EIF3E | eukaryotic translation initiation factor 3 subunit E | 2 | 4 |
| FMR1 | fragile X mental retardation 1 | 2 | 0 |
| NUP98 | nucleoporin 98 | 2 | 5 |
| NUP153 | nucleoporin 153 | 0 | 5 |
| THOC3 | THO complex 3 | 0 | 4 |
| XPOT | exportin for tRNA | 0 | 4 |
| UBIQUITINATION / SUMOYLATION | |||
| STUB1 | STIP1 homology and U-box containing protein 1 | 30 | 31 |
| HUWE1 | HECT, UBA and WWE domain containing 1, E3 ubiquitin ligase | 29 | 20 |
| RANBP2 | RAN binding protein 2, E3 SUMO ligase | 29 | 25 |
| RNF40 | ring finger protein 40, E3 ubiquitin ligase | 4 | 10 |
| USP7 | ubiquitin specific peptidase 7 | 4 | 9 |
| USP9X | ubiquitin specific peptidase 9, X-linked | 4 | 5 |
| RNF2 | ring finger protein 2, , E3 ubiquitin protein ligase | 3 | 0 |
| UBR5 | ubiquitin protein ligase E3 component n-recognin 5 | 3 | 12 |
| USP15 | ubiquitin specific peptidase 15 | 3 | 0 |
| SUMO3 | small ubiquitin-like modifier 3 | 0 | 3 |
| MKRN2 | makorin ring finger protein 2, E3 ubiquitin ligase | 0 | 3 |
To further confirm the effectiveness of our MS analyses, we next analyzed some of the interactors identified. To do that, a subset of cDNAs identified from the SMCR8 proteome were cloned with an N-terminal V5-TEV (tobacco etch virus)-epitope tag or were obtained as gifts. Notably, following cotransfection in 293T cells, 73% (22/30) of proteins tested directly co-IPed with FL-SMCR8 on α-FLAG agarose, further confirming the efficiency of our protocol (Fig. 2). In almost all cases, interactions were resistant to RNase digestion. Some proteins bound non-specifically to the agarose (BAG5, PPP2R1A, RUVBL2) or failed to bind FL-SMCR8 (G3BP1, GTF2I, RAB1A, RANGAP1, STIP1). It is possible some of these latter proteins are only able to bind SMCR8 when in complex with over-expressed C9orf72 and/or WDR41.
Several studies have proposed a role for C9orf72 in the regulation of autophagy by Rab GTPases, although with disagreement concerning which of the many Rab family members binds the C9orf72/SMCR8/WDR41 complex. Farg et al. [43] first reported C9orf72 to interact with RAB1, RAB5, RAB7 and RAB11. Webster et al. [22] confirmed that C9orf72 associates with GTP-bound RAB1A and the ULK1 complex, and it has been demonstrated that C9orf72 in complex with SMCR8 and WDR41 is a GEF for RAB8A, RAB11A, and RAB39B, and that its loss perturbs autophagy in neurons [27, 29, 31, 89]. We detected only RAB1B in our SMCR8 and C9orf72 interactomes (Tables S1, S2), but failed to confirm binding of V5-tagged RAB1A, a paralog highly similar in sequence to RAB1A, with SMCR8 in direct co-IP experiments. However, we also tested and confirmed weak binding of V5-RAB7A with overexpressed SMCR8 (Fig. 2) and C9orf72 (not shown), but only in the presence of RNase.
Significantly, when we queried the OMIM (Online Mendelian Inheritence in Man) database (https://omim.org/), we found that 65 (19%) of our putative SMCR8-interacting proteins are associated with neurodegenerative and neurological genetic disorders (Table 2). These include 8 proteins linked with ALS and/or FTD, 14 with other neurodegenerative diseases (including 4 associated with spinocerebellar ataxias), 7 with Charcot-Marie Tooth disease, 5 with hypomyelinating leukodystrophy, and 13 with mental retardation. Thus, SMCR8 may recruit some of these proteins to its complex with C9orf72 and WDR41, predicting roles for the complex in central nervous system (CNS) disorders.
Table 2.
Proteins of the SMCR8 interactome associated with CNS disease according to the Online Mendelian Inheritance in Man (OMIM) database
| GENE NAME | DISEASE NAME | SYMBOL | OMIM # |
|---|---|---|---|
| ALS/FTD | |||
| ATXN2 | Amyotrophic lateral sclerosis 13 | ALS13 | 183,090 |
| C9orf72 | Frontotemporal dementia and/or amyotrophic lateral sclerosis 1 | FTDALS1 | 105,550 |
| DCTN1 | Amyotrophic lateral sclerosis | ALS | 105,400 |
| FIG4 | Amyotrophic lateral sclerosis 11 | ALS11 | 612,577 |
| FUS | Amyotrophic lateral sclerosis 6, with or without frontotemporal dementia | ALS6 | 608,030 |
| MATR3 | Amyotrophic lateral sclerosis 21 | ALS21 | 606,070 |
| SQSTM1 | Frontotemporal dementia and/or amyotrophic lateral sclerosis 3 | FTDALS3 | 616,437 |
| TARDBP | Amyotrophic lateral sclerosis 10 | ALS10 | 612,069 |
| OTHER NEURODEGENERATION DISEASES | |||
| AIFM1 | Combined oxidative phosphorylation deficiency 6 | COXPD6 | 300,816 |
| ATXN2 | Spinocerebellar ataxia 2 | SCA2 | 183,090 |
| CTSD | Ceroid lipofuscinosis, neuronal, 10 | CLN10 | 610,127 |
| DYNC1H1 | Spinal muscular atrophy, lower extremity-predominant 1, autosomal dominant | SMALED1 | 158,600 |
| EIF4G1 | Parkinson disease 18 | PARK18 | 614,251 |
| NOP56 | Spinocerebellar ataxia 36 | SCA36 | 614,153 |
| PARK7 | Parkinson disease 7 | PARK7 | 606,324 |
| PCNA | Ataxia-telangiectasia-like disorder 2 | ATLD2 | 615,919 |
| PUM1 | Spinocerebellar ataxia 47 | SCA47 | 617,931 |
| QARS | Microcephaly, progressive, with seizures and cerebral and cerebellar atrophy | MSCCA | 615,760 |
| RARS | Leukodystrophy, hypomyelinating, 9 | HLD9 | 616,140 |
| SPG20 | Spastic paraplegia 20, autosomal recessive | SPG20 | 275,900 |
| STUB1 | Spinocerebellar ataxia, autosomal recessive, 16 | SCAR16 | 615,768 |
| WNK1 | Neuropathy, hereditary sensory and autonomic, 2A | HSAN2A | 201,300 |
| OTHER NEUROLOGICAL CONDITIONS | |||
| ADAR | Aicardi-Goutieres syndrome 6 | AGS6 | 615,010 |
| ADNP | Helsmoortel-van der Aa syndrome | HVDAS | 615,873 |
| AIFM1 | Cowchock syndrome | COWCK | 310,490 |
| AIMP1 | Leukodystrophy, hypomyelinating, 3 | HLD3 | 260,600 |
| AIMP2 | Leukodystrophy, hypomyelinating, 17 | HLD17 | 618,006 |
| ALDH18A1 | Spastic paraplegia 9A, autosomal dominant | SPG9A | 601,162 |
| ALDH18A1 | Spastic paraplegia 9B, autosomal recessive | SPG9B | 616,586 |
| ALDH3A2 | Sjogren-Larsson syndrome | SLS | 270,200 |
| ARHGEF2 | Neurodevelopmental disorder with midbrain and hindbrain malformations | NEDMHM | 617,523 |
| ATAD3A | Harel-Yoon syndrome | ATAD3A | 612,316 |
| ATP1A1 | Charcot-Marie-Tooth disease, axonal, type 2DD | CMT2DD | 618,036 |
| CAD | Epileptic encephalopathy, early infantile, 50 | AR | 616,457 |
| CDK5 | Lissencephaly 7, with cerebellar hypoplasia | LIS7 | 616,342 |
| CLTC | Mental retardation, autosomal dominant 56 | MRD56 | 617,854 |
| COPB2 | Microcephaly 19, primary, autosomal recessive | MCPH19 | 617,800 |
| DARS | Hypomyelination with brainstem and spinal cord involvement and leg spasticity | HBSL | 615,281 |
| DCTN1 | Neuronopathy, distal hereditary motor, 7B | HMN7B | 607,641 |
| DCTN1 | Perry syndrome | PERRYS | 168,605 |
| DOCK7 | Epileptic encephalopathy, early infantile, 23 | AR | 615,859 |
| DYNC1H1 | Charcot-Marie-Tooth disease 2O | CMT2O | 614,228 |
| DYNC1H1 | Mental retardation, autosomal dominant 13 | MRD13 | 614,563 |
| EPRS | Leukodystrophy, hypomyelinating, 15 | HLD15 | 617,951 |
| FIG4 | Polymicrogyria, bilateral temporooccipital | BTOP | 612,691 |
| FIG4 | Charcot-Marie-Tooth disease 4 J | CMT4J | 611,228 |
| FKBP5 | {Major depressive disorder and accelerated response to antidepressant drugs} | MDD | 608,516 |
| FMR1 | Fragile X syndrome | FRAX | 300,624 |
| FMR1 | Fragile X tremor/ataxia syndrome | FXTAS | 300,623 |
| GATAD2B | Mental retardation, autosomal dominant 18 | MRD18 | 615,074 |
| HCFC1 | Mental retardation, X-linked 3 | MRX3 | 309,541 |
| HNRNPH2 | Mental retardation, X-linked, syndromic, Bain type | MRXSB | 300,986 |
| HPRT1 | Lesch-Nyhan syndrome | LNS | 300,322 |
| HSD17B4 | Perrault syndrome 1 | PRLTS1 | 233,400 |
| HSPB1 | Neuropathy, distal hereditary motor, type IIB | HMN2B | 608,634 |
| HSPB1 | Charcot-Marie-Tooth disease 2F | CMT2F | 606,595 |
| HUWE1 | Mental retardation, X-linked 17 | MRX17 | 300,705 |
| HUWE1 | Mental retardation, X-linked, syndromic, Turner type | MRXST | 300,706 |
| LAS1L | Wilson-Turner syndrome | WTS | 309,585 |
| LMNA | Charcot-Marie-Tooth disease 2B1 | CMT2B1 | 605,588 |
| LRPPRC | Leigh syndrome French-Canadian type | LSFC | 220,111 |
| MARS | Charcot-Marie-Tooth disease 2 U | CMT2U | 616,280 |
| MTHFD1 | Neural tube defects, folate-sensitive | NTDFS | 601,634 |
| NSUN2 | Mental retardation, autosomal recessive 5 | MRT5 | 611,091 |
| PDK3 | Charcot-Marie-Tooth disease, X-linked dominant, 6 | CMTX6 | 300,905 |
| PPP2R1A | Mental retardation, autosomal dominant 36 | MRD36 | 616,362 |
| PSMD12 | Stankiewicz-Isidor syndrome | STISS | 617,516 |
| RANBP2 | Encephalopathy, acute, infection-induced, 3 | IIAE3 | 608,033 |
| SAMHD1 | Aicardi-Goutieres syndrome 5 | AGS5 | 612,952 |
| SLC25A1 | Combined D-2- and L-2-hydroxyglutaric aciduria | D2L2AD | 615,182 |
| SLC25A22 | Epileptic encephalopathy, early infantile, 3 | EIEE3 | 609,304 |
| SPTLC1 | Neuropathy, hereditary sensory and autonomic, type IA | HSN1A | 162,400 |
| TECR | Mental retardation, autosomal recessive 14 | MRT14 | 614,020 |
| THOC2 | Mental retardation, X-linked 12 | MRX12 | 300,957 |
| TUBB3 | Cortical dysplasia, complex, with other brain malformations 1 | CDCBM1 | 614,039 |
| TUBB4A | Leukodystrophy, hypomyelinating, 6 | HLD | 612,438 |
| USP9X | Mental retardation, X-linked 99 | MRX99 | 300,919 |
| USP9X | Mental retardation, X-linked 99, syndromic, female-restricted | MRXS99F | 300,968 |
SMCR8 associates with proteins of the ubiquitination and protein decay pathways
Our MS analyses also showed that both SMCR8 and C9orf72 proteins bind the proteasome. Our lists of SMCR8- and C9orf72-interactors were examined for KEGG pathways using the DAVID Functional Annotation Tool (NIAID, NIH, [83]). In the case of SMCR8-associated proteins, proteasome (7.6% of total proteins), RNA transport (6.2%), and protein processing in the endoplasmic reticulum (4.4%) pathways predominated (Fig. S2A). For C9orf72, proteins associated with the proteasome and RNA transport pathways accounted for 4.5 and 8% of the total, respectively, while spliceosome pathway proteins (9%) were most abundant (Fig. S2B). Twenty-six proteasome subunits co-IPed with FL-SMCR8, and 9 subunits were detected within the C9orf72-FL interactome. Furthermore, based on KEGG and Autophagy Database (http://www.tanpaku.org/autophagy/) analyses, 24 proteins involved with phagosome and autophagy pathways were detected in the SMCR8 interactome, including proteins linked with neurological diseases (CDK5, DYNC1H1, HUWE1, PPP2R1A, RANBP2, STUB1, TUBB3, TUBB4A, and USP9X; Tables 1, 2, S2).
Our SMCR8 interactome also contained 9 ubiquitination pathway factors, including ubiquitin ligases and peptidases (Table 1). Therefore, we examined MS-sequenced peptides deriving from immunoprecipitated FL-SMCR8 for ubiquitin modification (72% coverage of the total protein). A total of 9 high confidence modified lysine residues were predicted by at least 5 peptides in two independent experiments, suggesting that SMCR8 is highly ubiquitinated. Eight of these lysines were also identified by at least one of three ubiquitination prediction algorithms, including UbPred [86], BDM-PUB (http://bdmpub.biocuckoo.org), and UbiSite (http://csb.cse.yzu.edu.tw/UbiSite/) (Table S3). We then considered the phylogenic conservation of these lysines by aligning SMCR8 protein sequences from 8 vertebrate (human, chimpanzee, dog, mouse, rat, chicken, zebrafish, and frog) and two mollusc (freshwater snail and sea slug) species (Fig. S3). Eight of the 9 lysines detected by MS as modified were conserved among at least 8 species, including 2 residues (K232, K479) found in both molluscs, suggesting that these post-translational modifications (PTMs) might be functionally relevant.
Immunoprecipitating FLAG-tagged SMCR8 and probing with α-ubiquitin on Western blots reveals high-molecular weight (HMW) proteins consistent with polyubiquitinated SMCR8 and/or other large ubiquitinated proteins bound in the SMCR8 complex (Fig. 3a). In whole cell lysates, SMCR8-V5, in the presence of the proteasome inhibitor MG-132 and/or coexpressed ubiquitin, showed HMW products consistent with multiple PTMs (Fig. 3b). Furthermore, FLAG-tagged ubiquitin coimmunoprecipitates on α-FLAG agarose, and so by implication is conjugated to cotransfected HA- or V5-tagged SMCR8 (Fig. 3c). Although treatment with MG132 caused accumulation of HMW SCMR8 protein species, suggesting their regulation by the ubiquitin-proteasome system (UPS), full-length SMCR8 signal was little decreased in the presence of coexpressed ubiquitin (Fig. 3b,c).
Fig. 3.
Evidence that SMCR8 protein is poly-ubiquitinated. a The FL-SMCR8 construct was transfected in 293T cells and immunoprecipitated with α-FLAG antibody-bound agarose. A Western blot of whole cell lysates probed with α-FLAG antibody shows expression of full-length FL-SMCR8 protein plus HMW products consistent with PTMs (left). Probing with α-UBB antibody marks HMW products in immunoprecipitates consistent with either poly-ubiquitinated FL-SMCR8 protein or the presence of other HMW ubiquitinated proteins that co-IP with the SMCR8 complex (right). IP reactions were in the presence or absence of 50 μg/ml RNases. b C-terminal V5-tagged SMCR8 and empty vector or HA-tagged ubiquitin were coexpressed in 293T cells and treated or not treated with the proteasome inhibitor MG132. Expression of SMCR8-V5 protein and empty vector, in the presence but not absence of MG132, produces HMW bands on Western blots that are consistent with post-translational modification of SMCR8 at multiple sites. SMCR8-V5 protein coexpressed with HA-UBB and without MG132 shows the same HMW bands, which increase in signal intensity upon incubation with MG132. c V5- or HA-epitope-tagged SMCR8 was coexpressed with empty vector or FLAG-tagged UBB in 293T cells and incubated overnight in the presence or absence of MG132. Cell lysates were subjected to immunoprecipitation with α-FLAG agarose, followed by Western blotting and probing with α-HA (top left panel), α-V5 (top right) or α-FLAG (bottom left) antibodies. A HMW smear seen in immunoprecipitates is consistent with poly-ubiquitination of tagged SMCR8 proteins. In general, overexpression of ubiquitin does not lead to a significant decrease in full-length SMCR8 protein levels
Using confocal IF microsopy, we observed that overexpression of red fluorescent protein (RFP)-tagged ubiquitin induces formation of a large aggregate consistent with the aggresome and marked by colocalization with coexpressed and therefore likely UBB-bound FL-SMCR8 (Fig. 4a). Aggresomes appear mainly within an indentation of the nucleus at the microtubule-organizing center and form when the protein-degradation machinery of the cell is overwhelmed [90]. Misfolded and ubiquitinated proteins, including perhaps SMCR8, are transported to the aggresome along the microtubule network by means of the dynein motor complex (which includes cytoplasmic dyneins DYNC1H1 and DYNC1I2, both detected in FL-SMCR8 iimmunoprecipitates, Table S2). An alternative ubiquitin-independent pathway involves interaction of STUB1 and BAG3, which transfer misfolded proteins to heat shock protein 70 (all proteins that co-IPed with FL-SMCR8, Table S2) and the dynein motor complex to promote formation of aggresomes [91, 92].
Fig. 4.
Immunofluorescence microscopy shows evidence for association of endogenous SMCR8 protein with cytoplasmic aggregates. a FLAG-tagged SMCR8 and RFP-tagged ubiquitin transfected in 2102Ep cells colocalize in a structure consistent with the aggresome. b Overexpression of V5-tagged C9orf72 does not induce stress granule formation in unstressed U2OS cells. c Exogenously expressed HA-SMCR8 protein is not observed in SGs of U2OS cells stressed with NaAsO2. d WDR41-FL protein does not colocalize with SG marker protein TIA1 in U2OS cells stressed with NaAsO2. e Endogenous C9orf72 protein detercted by the α-SMCR8-SC antibody does not colocalize with SGs in NaAsO2-stressed U2OS cells (see also Fig. S4E). f Endogenous C9orf72 protein detected by the C9-L antibody [54] does not colocalize with SGs in DTT-stressed U2OS cells. g,h Endogenous SMCR8 detected by the α-SMCR8-ab202283 antibody localizes to SGs of stressed (h), but not unstressed (g) U2OS cells (see also Fig. S4G-I). i The α-WDR41-SC antibody does not detect endogenous protein in SGs of NaAsO2-stressed 2102Ep cells. NT: no treatment. Cell nuclei were stained with Hoechst 33342 (right-most panels). Size bars are 10 μm
Thus, SMCR8 protein is bound by ubiquitin and may recruit UPS complexes to the vicinity of its other associated cellular proteins, numbers of which have been linked with neuropathologies (Table 2).
Evidence that endogenous SMCR8 accumulates in cytoplasmic stress granules
The accumulation of neuronal RNA and protein aggregates, including cytoplasmic stress granules, is a pathogenic hallmark of a number of neurodegenerative diseases, among them FTD and ALS [93–95]. SGs assemble rapidly under cellular stress and include the small, but not large, ribosomal subunits bound to translation initiation factors such as eIF2 and eIF3 (reviewed in [96]). Processing-bodies (PBs) and SGs are dynamic cytoplasmic aggregates that participate in mRNA decay, and SGs in mammalian cells are heavily ubiquitinated [97]. Because previous publications implicated C9orf72 protein expression in the metabolism of SGs [88, 98], we wished to determine if the C9orf72 binding partner SMCR8 associates with SGs in various tumor cell lines.
As reported by others [28], we observed epitope-tagged SMCR8 and C9orf72 proteins to both have a diffuse cytoplasmic distribution with protein also observed in nuclei, although nuclear localization was more evident for C9orf72 (Fig. 4b,c, S4A-C). However, although Maharjan et al. [98] reported that SGs were induced in a majority of unstressed mouse Neuro2A (N2A) neuroblastoma cells when transfected with myc-tagged C9-L, we failed to observe this phenomenon for tagged C9-L or SMCR8 proteins transfected alone or in combination (not shown) in unstressed human osterosarcoma U2OS, HEK 293T, or neuroblastoma cell lines (Fig. 4b, S4B). Furthermore, when cells were treated with 250 μM of the oxidative stressor sodium arsenite (NaAsO2) for 80 min, tagged C9orf72, SMCR8, or WDR41 protein very rarely colocalized in aggregates with endogenous canonical SG marker protein TIA1 in multiple cell lines (Figs. 4c,d, S4C).
We next examined localization of endogenous C9orf72 and SMCR8 proteins in cells. The α-C9orf72-SC and α-C9orf72-PT antibodies both detected nuclear and cytoplasmic distribution for C9orf72 protein, with fine cytoplasmic granululazion visible in unstressed cells that was more evident for the latter antibody (Fig. S4D). However, contrary to previous studies that used these antibodies to report SG localization [88, 98], we failed to detect endogenous C9orf72 in stress-induced U2OS (Figs. 4e, S4E) or 2102Ep (not shown) cells, although C9orf72 infrequently justaposed or overlapped with SGs and/or PBs in N2A cells (Fig. S4F.G). To confirm further these observations, two polyclonal antibodies developed by the Robertson lab [19, 79], and specific for C9-L (Fig. 4f) and C9-S (not shown) isoforms, were also tested but failed to show obvious C9orf72 protein presence in TIA1-marked SGs in DTT- or NaAsO2-stressed cells of multiple lines, including U2OS and 2012Ep cells. Thus, detection of C9orf72 in SGs appears to be cell line and possibly antibody dependent.
We also used the α-SMCR8-PT and α-SMCR8-ab202283 antibodies to examine endogenous SMCR8 protein localization. In unstressed U2OS cells, endogenous SMCR8 was nuclear and more prominently cytoplasmic with speckled staining (Fig. 4g). However, when cells were stressed with NaAsO2, SMCR8 redistributed to large intensely staining foci that colocalized with TIA1 (Fig. 4h). Fig. S4H shows SMCR8 protein in large cytoplasmic aggregates of NaAsO2-stressed HEK 293T cells that costain with a different endogenous SG marker, the LINE-1 retrotransposon-encoded ORF1 protein [81], while Fig. S4I shows costaining with SG-marker eIF3η in human neuroblastoma SK-N-SH cells. In N2A cells treated with the endoplasmic reticulum stressor thapsigargin, SMCR8 granules were marked by a p70 S6 kinase antibody known to recognize HEDLS/EDC4, a PB component (Fig. S4J, [77]): PBs frequently overlap or juxtapose with SGs in stressed cells [99]. Endogenous SMCR8 granules in unstressed N2A cells also partially colocalized with GW182 autoantigen, which marks PBs (Fig. S4K) [80]. However, as noted above, SMCR8 commercial antibodies detect multiple protein species (Fig. S1B-E), some possibly non-specific, and we cannot be certain that canonical full-length endogenous SMCR8 proteins are what we see in SGs. Nevertheless, our data suggest that in stressed cells a fraction of endogenous SMCR8 protein is directed to cytoplasmic SGs.
Our analyses showed that TAR DNA binding protein 43 (TDP-43, product of the TARDBP gene) binds SMCR8 (Fig. 2; Table 2). Mutations in TARDBP are involved in about 4% of familial and 1% of sporadic ALS (sALS) cases. However, even wild-type TDP-43, while mostly nuclear in healthy cells, is cleaved and hyperphosphorylated and accumulates in ubiquitinated cytoplasmic aggregates in neurons of almost all ALS and about half of FTLD patients (reviewed in [100]). We tested if endogenous or overexpressed SMCR8 protein colocalizes with TDP-43 protein in cytoplasmic granules but found this not to be the case in unstressed or stressed U2OS or 2102Ep cells (Fig. S4L).
Hexanucleotide expansions within transcripts of the C9orf72 ALS gene may undergo non-conventional repeat-associated non-ATG (RAN) translation and generate dipeptide repeats that aggregate in the cytoplasm of neuronal cells of C9ALS patients (reviewed in [101]). To see if such aggregates might colocalize with SMCR8, we coexpressed in 293T cells FL-SMCR8 and a C9orf72 RAN translation product of 50 GA-dipeptide repeats tagged with EGFP [68]. Overexpressed dipeptide proteins formed one to three large cytoplasmic aggrgates in each cell that were were ringed by, but mostly excluded SMCR8 (Fig. S4M).
Finally, the α-WDR41-SC antibody marks WDR41 protein as predominantly nuclear but also with faint cytoplasmic granules that fail to colocalize with SGs in unstressed or stressed U2OS, 2102Ep, 293T, or N2A cells (Fig. 4i and not shown). On the other hand, the α-WDR41-PT antibody colocalizes with a minor subset of granules positive for 4-ET, a marker of PBs (Fig. S4N). However, while the α-WDR41-SC antibody recognizes only bands consistent in size with WDR41 isoforms in HEK 293T, 2102Ep, and SK-N-SH cells (Fig. S1F), the α-WDR41-PT antibody detects other non-canonical protein species (Fig. S1G), and the specificity of its SG staining is thus uncertain.
Searching the Mammalian Stress Granules Proteome Database (https://msgp.pt) [102], we found that 18% of the SMCR8 proiein interactome (61/340) and 26% (35/201) of the C9orf72 interactome are known SG-associated proteins. It is thus possible that SG components bind endogenous SMCR8-C9orf72 complexes and shepherd them to SGs, although why this would not also be the case for overexpressed exogenous SMCR8 or C9orf72 proteins is unclear.
SMCR8 expression in ALS patient brain tissues
Despite its strong association with protein-degradation factors, SMCR8 overexpression does not stimulate degradation of C9orf72 protein with which it is in complex. Contrarily, multiple studies in cells and knockout mice have shown that protein but not RNA levels of SMCR8 and C9orf72 are positively correlated, suggesting that in complex the two proteins stabilize and protect each other from degradation [26, 28, 29, 32, 47, 54, 65, 103]. On the other hand, increased SMCR8 protein reportedly has little effect on WDR41 levels in KO mice or cells [32, 35]. We confirmed in 293T cells that overexpression of SMCR8 with various tags strongly increased levels of cotransfected FL-C9-L protein, while cotransfection of empty vectors or an unrelated protein (RO60) did not (Fig. 5a). Considering the interplay between SMCR8 and C9orf72 proteins, and the fact that C9orf72 RNA expression is reduced in some C9ALS patient cohorts, we asked if SMCR8 expression levels are altered in the brains of C9ALS patients compared with non-affected controls.
We first examined transcription levels of C9orf72, SMCR8, and WDR41 genes in RNA-Seq datasets from several sequence read archives that contain C9ALS sample data. GEO dataset GSE67196 includes cerebellum and frontal cortex samples of 9 healthy, 8 C9ALS, and 10 sALS individuals. Using TEtranscripts [84] to analyze C9orf72 gene expression levels, we found a significant log2 0.96-fold decrease (padj 4.6E-5) in the frontal cortex of C9ALS vs sALS individuals and a 1.1-fold decrease (padj 1.6-E4) in the cerebellum of C9ALS vs control individuals; however, in neither case was decrease in SMCR8 expression significant. The NeuroLINCS dbGaP Study phs001231 (SRP098831) consists of poly(A) + non-stranded mRNA of iPSC-derived motor neurons from 4 C9ALS, 3 spinal muscular atrophy (SMA), and 3 unaffected individuals (2 or 3 replicates each). No significant changes in C9orf72 or SMCR8 transcript levels were seen in this dataset, although WDR41 sequence read numbers were reduced about 0.35-fold in both C9ALS vs control and SMA vs control samples (padj< 0.01). Finally, a recent RNA-Seq study comparing C9 FTLD and FTLD/motor neuron disease patients with unaffected control individuals reported a highly significant decrease in C9orf72 RNA levels in C9 FTLD samples; however, this data showed no significant change in SMCR8 or WDR41 RNA expression [24].
We next assayed endogenous SMCR8 protein expression levels in the context of the C9orf72 hexanucleotide expansion. Motor cortex brain tissue lysate samples of 11 C9ALS and 10 unaffected control individuals were analyzed by Western blotting with α-SMCR8 antibodies (Fig. 5b, Table S4). Multiple film exposures were made to optimize signal to noise. Individual band intensities were quantitated with ImageJ software [104] and normalized against the summed exposures of all equivalent bands on the same gel. SMCR8 signal was then normalized to endogenous HSP90 protein signal detected on the same gel after reprobing with α-HSP90 antibody. Remarkably, an average 5-fold reduction in SMCR8 protein signal was seen in C9ALS vs control tissues (Fig. 5b). We also tested by Western blotting cerebrospinal fluid samples from 5 C9ALS patients and 5 unaffected controls, but were unable to detect full-length SMCR8 protein signal with either the α-SMCR8-PT or α-SMCR8-ab202283 antibodies (not shown). We also plotted normalized SMCR8 protein signal against ALS disease duration in months (Table S4), finding a weak negative but non-significant correlation (r = 0.34). Nevertheless, altogether our data recommend further investigation of SMCR8 protein level as a potential biomarker of the C9orf72 expansion disease mutation.
Discussion
In this study we characterized the SMCR8 protein interactome and found it to include numerous components of the ubiquitin-proteasome system, including ubiquitin ligases and peptidases. Of note, the IP method used here exploited FLAG-tagged proteins and so overcame limitations imposed by differences in isoform expression and non-specific protein species recognized by C9orf72 and SMCR8 antibodies. Despite evidence that SMCR8 itself is ubiquitinated at multiple residues, its degradation is not significantly induced in the presence of overexpressed ubiquitin suggesting other roles linking it with the UPS. Recruitment of UPS components to autophagy complexes could be one such role, and our SMCR8 interactome contains 24 autophagy pathway-associated proteins (Table 1). Ubiquitin plays a fundamental role not only in proteasome-mediated protein degradation but also in the targeting of proteins for degradation by autophagic complexes. Protein ubiquitination also regulates multiple steps of the autophagy pathway (reviewed in [105, 106]). For example, E3 ubiquitin ligase STUB1, a protein that co-IPs with SMCR8 (Table 1, Fig. 2), regulates autophagy by targeting TFEB for degradation by the UPS [107]. Also, E3 ligase HUWE1 (Table 1) mediates the ubiquitination and proteasomal degradation of WIPI2, a protein involved in autophagosome formation [108].
Recruitment of ubiquitination factors would be consistent with reported roles of the C9orf72-SMCR8 complex in autophagy and endosomal-lysosomal metabolism. The C9orf72/SMCR8/WDR41 complex associates with the ATG8 autophagy receptor network and influences activity of the ULK1-RB1CC1-ATG13-ATG101 autophagy initiation complex (RB1CC1 was detected in both our SMCR8 and C9orf72 protein interactomes, Tables S1, S2) [22, 28, 29, 31, 34]. C9orf72 and SMCR8 reportedly also play roles in regulating mTORC1 and TFEB autophagy and lysosomal gene transcription factors upstream of autophagy [26, 31, 32, 34, 57, 103, 109, 110]. Ugolino et al. [32], however, indicated a negative effect of C9orf72 on autophagy, and Yang et al. [31] reported increased autophagic flux in C9orf72 knockdown MEF cells, opposite to the reduction they saw in SMCR8-deficient cells. Unlike some C9orf72-deficient mice [45], Smcr8 KO mice showed motor behavior defects, including axonal swelling due to impaired autophagy and motor neuron axonal transport of autophagosomes [25]. Apparently, although in complex, some biological roles of C9orf72 and SMCR8 proteins differ.
Association of the SMCR8-C9orf72 complex with the UPS and autophagy would also be consistent with stress granule localization, since protein ubiquitination regulates SG dynamics. Components of the UPS, including ubiquitin, co-localize with SGs, while proteasome inhibition, and consequent increase in ubiquitinated proteins, induces SG formation [111–113]. Recent evidence also suggests SGs are regulated by autophagy [114, 115], and it has been proposed that improper metabolism of SGs could be involved in ALS pathology [93, 94]. Interestingly, Chitiproulu et al. [88] proposed that C9orf72 protein associates with autophagy cargo receptor p62 (encoded by the SQSTM1 gene) to control SG elimination rather than assembly by forming a complex that eliminates by autophagy SG proteins dimethylated on arginines (of note, we found p62 in the SMCR8 but not C9orf72 interactomes; Fig. 2, Table 2S).
However, our data disagree in some aspects with previously published results concerning C9orf72 colocalization with SGs. While, Maharjan et al. [98] reported that overexpression of myc-tagged C9-L led to the spontaneous appearance of SGs in a majority of N2A cells and cortical neurons in the absence of cellular stress, we failed to reproduce these observations in either U2OS or N2A cells for tagged C9orf72 or SMCR8 proteins, overexpressed together or separately. Furthermore, using the α-C9orf72-PT antibody (Fig. S1A), Maharjan et al. [98] noted that endogenous C9orf72 protein colocalized with a fraction of SGs in neuronal cell lines and cortical neurons in response to DTT and heat shock-induced cell stress, and that C9orf72 depletion inhibited SG assembly, impaired expression of proteins required for their formation, and increased cell sensitivity to stress. However, despite testing several antibodies, cell lines and conditions, we could not detect endogenous C9orf72 in SGs of selected non-neuronal cancer-derived cell lines, and we saw only minor colocalization of C9orf72 with SGs and PBs of N2A cells. Thus, association of C9orf72 protein to SGs appears to be cell line-dependent.
On the other hand, we observed endogenous, but not exogenously expressed, SMCR8 protein localization to SGs of all chemically stressed cell lines tested. Interestingly, about one-fifth of the putative interacting proteins we identified as members of our C9orf72 and SMCR8 interactomes are known SG proteins, which themselves might play a role in targeting of SMCR8 complexes to granules. It is conceivable that SMCR8-C9orf72 SG association is sensitive to cell type, cellular conditions, and levels of interacting proteins as determinants of entry into SGs, and perhaps these factors explain discrepancies between our data and previously published observations.
As reported in other studies, we also presented supporting evidence that C9orf72 protein levels are positively correlated with those of SMCR8 in cultured cells [26, 28, 29, 32, 47, 54, 65, 103]. Furthermore, we now show that SMCR8 protein expression is reduced in the brains of C9ALS patients compared with unaffected controls (and as also recently noted by [25]. To date, it has been reported that a small number of proteins, including neurofilament proteins, are differentially expressed in the CSF of ALS and FTD proteins and have been proposed as candidate biomarkers for the C9orf72 mutation [116, 117]. Whether or not SMCR8 protein can also be an effective CSF or plasma biomarker for C9 expansion patients remains to be determined and is likely contingent upon the development of better α-SMCR8 antibodies.
Conclusions
In this study we characterized the protein interactome of SMCR8, which binds the protein product of C9orf72, the major susceptibility gene for ALS. Using a robust and highly specific protocol, we demonstrated ubiquitination without significant degradation of SMCR8 protein and its association with many components of the ubiquitin-proteasome system. Evidence was presented for localization of endogenous SMCR8 protein to cytoplasmic stress granules, although in several cell lines we failed to reproduce previous observations that C9orf72 protein enters these granules. SMCR8 protein levels were downregulated in whole tissue brain lysates of C9ALS patients compared with unaffected controls, suggesting the potential usefulness for SMCR8 as a biomarker of the disease state.
In addition to ALS and FTD, the C9orf72 gene expansion mutation has been linked with other neurodegenerative and psychiatric disorders, although etiological roles remain unknown [118–123]. We have shown that SMCR8, whose cellular levels positively correlate with C9orf72 protein expression, associates not only with many factors of protein metabolism and stress granule dynamics, but also with numerous products of genes linked with a range CNS disorders (65/340 in total, Table 2). It is therefore reasonable in future studies to consider a role for SMCR8 in these diverse neuropathologies, perhaps relating to recruitment of the UPS with consequent effects on protein homeostasis.
Supplementary information
Additional file 1 Fig. S1. Western blotting of cell line and brain tissue lysates using commercial antibodies against C9orf72, SMCR8, and WDR41 proteins. Antibodies shown are (A) α-C9ORF72-PT (Proteintech Group 22637–1), (B) α-SMCR8-PT (Proteintech 21125–1-AP, (C) α-SMCR8-B (Bethyl Laboratories A304-694A), (D) α-SMCR8-ab186504 (Abcam ab186504), (E) α-SMCR8-ab202283 (Abcam ab202283), (F) α-WDR41-SC (Santa Cruz sc-137923), and (G) α-WDR41-PT (Proteintech 26817–1-AP). Large arrows mark full-length protein products. The small arrows in (F) and (G) mark the presumed minor 45.5 kD WDR41 isoform. Samples were run on NuPAGE 4–12% Bis-Tris Protein gels (Novex) and Western blotting was performed according to [81, 82]. Molecular weight markers are Novex Sharp Pre-stained Protein Standard. Motor: motor cortex, Occ: occipital cortex, S.C.: spinal cord; sample identifier numbers are also shown. Fig. S2. Pie chart of results of DAVID (Database for Annotation, Visualization and Integrated Discovery, [83]) analyses of KEGG pathways showing selected functional categories for candidate member proteins of the (A) FL-SMCR8 and (B) C9orf72-FL protein interactomes. Percentages of the total number of proteins identified (Tables S1 and S2) for each category are shown within the slices. Fig. S3. Phylogenetic multi-sequence alignment of SMCR8 protein sequences for ten species. Alignments were made with Clustal Omega 1.2.1 (EMBL-EBI) followed by BoxShade 3.2 (http://sourceforge.net/projects/boxshade). Pink shading marks amino acid residues identical in at least 8 species, while green includes conservative replacements. Lysine residues predicted by MS sequencing to be ubiquitinated are boxed in blue (see Table S3). Species shown are: Homo sapiens, human; Pan troglodytes, chimpanzee; Canus lupus familiaris, dog; Mus musculus, house mouse; Rattus norvegicus, brown rat; Gallus gallus domesticus, chicken; Danio rerio, zebrafish; Xenopus tropicalis, western clawed frog; Biomphalaria glabrata, freshwater snail; Aplysia californica, California sea hare. Fig. S4. Immunofluorescence microscopy evidence for association of SMCR8 protein with cytoplasmic aggregates. (A) V5-tagged SMCR8 does not significantly enter SGs of NaAsO2-stressed N2A cells. (B) Overexpression of FLAG-tagged C9orf72 (left) or SMCR8 (right) protein does not induce formation of cytoplasmic granules in neuroblastoma cells. (C) V5-tagged exogenously expressed C9orf72 fails to enter SGs in NaAsO2-treated U2OS cells. (D) Endogenous C9orf72 protein speckles observed in unstressed N2A cells are not marked by TIA1, a SG protein. (E) C9orf72 protein detected by the α-C9orf72-PT antibody fails to enter SGs of NaAsO2-stressed U2OS cells. (F,G), Endogenous C9orf72 protein detected by the α-C9orf72-SC antibody colocalizes or justaposes with only a minor subset of SGs (marked by TIA1) or PBs (marked by 4-ET) in stressed mouse neuroblastoma N2A cells (see arrows). (H) In NaAsO2-stressed 293T cells, endogenous SMCR8 detected by the α-SMCR8-PT antibody colocalizes with granules marked by ORF1p, a protein encoded by LINE-1 retrotransposons [81]. (I) In NaAsO2-stressed human neuroblastoma SK-N-SH cells, endogenous SMCR8 localizes to SGs marked by α-eIF3η antibody. (J) In N2A cells treated with the endoplasmic reticulum stressor thapsigarin, endogenous SMCR8 protein colocalizes to granules marked by HEDLS/EDC4 (detected by α-p70 S6 kinase antibody, [77]), a component of PBs. (K) In unstressed N2A cells, endogenous SMCR8 localizes in granules with GW142, a PB marker. (L) Exogenously expressed TDP-43 and endogenous SMCR8 proteins do not colocalize in cytoplasmic granules of stressed U2OS cells. (M) SMCR8 rings but is generally excluded from overexpressed GFP-(GA)50 dipeptide aggregates. (M) In 293T cells, the α-WDR41-PT antibody detects endogenous protein in or beside a subset granules marked by 4-ET (see arrows). NT: no treatment. Cell nuclei were stained with Hoechst 33342 (right-most panels). Size bars are 10 μm. Table S1. The C9orf72 protein interactome determined by mass spectrometry. Table S2. The SMCR8 protein interactome determined by mass spectrometry. Table S3. Ubiquitinated SMCR8 lysine residues determined by MS sequencing, prediction algorithms, and the comparative phylogenetic analyses of Fig. S3.Table S4. Post-mortem brain motor cortex tissue samples used for the analyses of Fig. 5b,c.
Acknowledgements
The authors appreciate reagents provided by researchers listed in the Materials and Methods section. Human samples were received from the Target ALS Multicenter Postmortem Tissue Core (with thanks to K. Wilsbach and Dr. L. Ostrow), Drs. J. Ravits and R. Batra of the University of California San Diego, the University of Maryland Brain and Tissue Bank of the NIH NeuroBioBank, and the Northeast ALS Consortium (NEALS), as described above. Services and reagents were provided by the Johns Hopkins University School of Medicine ChemCore, Synthesis & Sequencing Facility, and Mass Spectrometry and Proteomics Core. Special thanks to Dr. Haig H. Kazazian and his laboratory (Johns Hopkins University School of Medicine) for ongoing advice and support.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- C9ALS
C9orf72 repeat expansion ALS
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- FTD
Frontotemporal dementia
- FTLD
Frontotemporal lobar degeneration
- GEF
Guanine nucleotide exchange factor
- GFP
Green fluorescent protein
- HMW
High molecular weight
- IF
Immunofluorescence
- IP
Immunoprecipitation
- iPSC
Induced pluripotent stem cell
- kD
Kilodalton
- N2A
Neuro2A
- ORF
Open reading frame
- PB
Processing (P-) body
- PCR
Polymerase chain reaction
- PTM
Post-translational modification
- RFP
Red fluorescent protein
- sALS
Sporadic ALS
- SG
Stress granule
- UPS
Ubiquitin-proteasome system
Authors’ contributions
JLG designed experiments and wrote the manuscript. AS, GCP, JLG, JLGP, and LS performed experiments. AS and JLGP assisted with manuscript editing. RNC and LRD performed MS analyses and assisted with data interpretation and manuscript review. The author (s) read and approved the final manuscript.
Funding
JLG was supported by grants from the NIH National Institute of Neurological Disorders and Stroke (1R03NS087290–01), the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development (R21HD083915-01A1), and the ALS Therapy Alliance (2013-F-067). The JLGP lab acknowledges funding from the European Research Council (ERC-Consolidator ERC-STG-2012-309433), the MINECO-FEDER (SAF2017–89745-R), and The Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund (ISFF2). RNC was partially supported by NIH/NCATS grant UL1 TR003098.
Availability of data and materials
Raw MS sequencing data used to generate Supplementary Tables S1 and S2 will be made available from the corresponding author upon request.
Ethics approval and consent to participate
All post-mortem tissues were obtained following approval of the Institutional Review Board of the JHU School of Medicine (IRB00066246 to JLG).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
John L. Goodier, Email: jgoodier@jhmi.edu
Alisha O. Soares, Email: asoares@alumni.nd.edu
Gavin C. Pereira, Email: gpereir1@jhmi.edu
Lauren R. DeVine, Email: LaurenDeVine@jhmi.edu
Laura Sanchez, Email: laura.sanchez@genyo.es.
Robert N. Cole, Email: rcole@jhmi.edu
Jose Luis García-Pérez, Email: Jose.Garcia-Perez@igmm.ed.ac.uk.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40478-020-00982-x.
References
- 1.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377(2):162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 2.Miller B, Llibre Guerra JJ. Frontotemporal dementia. Handb Clin Neurol. 2019;165:33–45. doi: 10.1016/B978-0-444-64012-3.00003-4. [DOI] [PubMed] [Google Scholar]
- 3.Abramzon YA, Fratta P, Traynor BJ, Chia R (2020) The overlapping genetics of amyotrophic lateral sclerosis and frontotemporal dementia. Front Neurosci 14:42 [DOI] [PMC free article] [PubMed]
- 4.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 7.Todd TW, Petrucelli L. Insights into the pathogenic mechanisms of chromosome 9 open reading frame 72 (C9orf72) repeat expansions. J Neurochem. 2016;138(Suppl 1):145–162. doi: 10.1111/jnc.13623. [DOI] [PubMed] [Google Scholar]
- 8.Ji AL, Zhang X, Chen WW, Huang WJ. Genetics insight into the amyotrophic lateral sclerosis/frontotemporal dementia spectrum. J Med Genet. 2017;54(3):145–154. doi: 10.1136/jmedgenet-2016-104271. [DOI] [PubMed] [Google Scholar]
- 9.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14(9):544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babic Leko M, Zupunski V, Kirincich J, Smilovic D, Hortobagyi T, Hof PR, et al. Molecular mechanisms of Neurodegeneration related to C9orf72 Hexanucleotide repeat expansion. Behav Neurol. 2019;2019:2909168. doi: 10.1155/2019/2909168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 13.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fratta P, Poulter M, Lashley T, Rohrer JD, Polke JM, Beck J, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126(3):401–409. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92(6):981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35(7):1779.e5–1779e13. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, et al. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol. 2015;130(6):863–876. doi: 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao S, MacNair L, McGoldrick P, McKeever PM, McLean JR, Zhang M, et al. Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann Neurol. 2015;78(4):568–583. doi: 10.1002/ana.24469. [DOI] [PubMed] [Google Scholar]
- 20.Rizzu P, Blauwendraat C, Heetveld S, Lynes EM, Castillo-Lizardo M, Dhingra A, et al. C9orf72 is differentially expressed in the central nervous system and myeloid cells and consistently reduced in C9orf72, MAPT and GRN mutation carriers. Acta Neuropathol Commun. 2016;4(1):37. doi: 10.1186/s40478-016-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24(3):313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35(15):1656–1676. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frick P, Sellier C, Mackenzie IRA, Cheng CY, Tahraoui-Bories J, Martinat C, et al. Novel antibodies reveal presynaptic localization of C9orf72 protein and reduced protein levels in C9orf72 mutation carriers. Acta Neuropathol Commun. 2018;6(1):72. doi: 10.1186/s40478-018-0579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson DW, Baker MC, Jackson JL, DeJesus-Hernandez M, Finch NA, Tian S, et al. Extensive transcriptomic study emphasizes importance of vesicular transport in C9orf72 expansion carriers. Acta Neuropathol Commun. 2019;7(1):150. doi: 10.1186/s40478-019-0797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang C, Shao Q, Zhang W, Yang M, Chang Q, Chen R, et al. Smcr8 deficiency disrupts axonal transport-dependent lysosomal function and promotes axonal swellings and gain of toxicity in C9ALS/FTD mouse models. Hum Mol Genet. 2019;28(23):3940–3953. doi: 10.1093/hmg/ddz230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amick J, Roczniak-Ferguson A, Ferguson SM. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell. 2016;27(20):3040–3051. doi: 10.1091/mbc.E16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciura S, Sellier C, Campanari ML, Charlet-Berguerand N, Kabashi E. The most prevalent genetic cause of ALS-FTD, C9orf72 synergizes the toxicity of ATXN2 intermediate polyglutamine repeats through the autophagy pathway. Autophagy. 2016;12(8):1406–1408. doi: 10.1080/15548627.2016.1189070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4(1):51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35(12):1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao S, MacNair L, McLean J, McGoldrick P, McKeever P, Soleimani S, et al. C9orf72 isoforms in amyotrophic lateral sclerosis and Frontotemporal lobar degeneration. Brain Res. 1647;2016:43–49. doi: 10.1016/j.brainres.2016.04.062. [DOI] [PubMed] [Google Scholar]
- 31.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016;2(9):e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A et al (2016) Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet 12(11):e1006443 [DOI] [PMC free article] [PubMed]
- 33.Corbier C, Sellier C. C9ORF72 is a GDP/GTP exchange factor for Rab8 and Rab39 and regulates autophagy. Small GTPases. 2017;8(3):181–186. doi: 10.1080/21541248.2016.1212688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung J, Nayak A, Schaeffer V, Starzetz T, Kirsch AK, Muller S, et al. Multiplex image-based autophagy RNAi screening identifies SMCR8 as ULK1 kinase activity and gene expression regulator. Elife. 2017;6:e23063. doi: 10.7554/eLife.23063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amick J, Tharkeshwar AK, Amaya C, Ferguson SM. WDR41 supports lysosomal response to changes in amino acid availability. Mol Biol Cell. 2018;29(18):2213–2227. doi: 10.1091/mbc.E17-12-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bi W, Yan J, Stankiewicz P, Park SS, Walz K, Boerkoel CF, et al. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 2002;12(5):713–728. doi: 10.1101/gr.73702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madduri N, Peters SU, Voigt RG, Llorente AM, Lupski JR, Potocki L. Cognitive and adaptive behavior profiles in Smith-Magenis syndrome. J Dev Behav Pediatr. 2006;27(3):188–192. doi: 10.1097/00004703-200606000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Jain BP, Pandey S. WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 2018;37(5):391–406. doi: 10.1007/s10930-018-9785-7. [DOI] [PubMed] [Google Scholar]
- 39.Stein JL, Hibar DP, Madsen SK, Khamis M, McMahon KL, de Zubicaray GI, et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol Psychiatry. 2011;16(9):927–937. doi: 10.1038/mp.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang D, Iyer LM, He F, Aravind L. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29(4):499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128(17):3171–3176. doi: 10.1242/jcs.166074. [DOI] [PubMed] [Google Scholar]
- 43.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23(13):3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webster CP, Smith EF, Grierson AJ, De Vos KJ. C9orf72 plays a central role in Rab GTPase-dependent regulation of autophagy. Small GTPases. 2018;9(5):399–408. doi: 10.1080/21541248.2016.1240495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351(6279):1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoki Y, Manzano R, Lee Y, Dafinca R, Aoki M, Douglas AGL, et al. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain. 2017;140(4):887–897. doi: 10.1093/brain/awx024. [DOI] [PubMed] [Google Scholar]
- 47.Leskela S, Huber N, Rostalski H, Natunen T, Remes AM, Takalo M, et al. C9orf72 proteins regulate autophagy and undergo Autophagosomal or proteasomal degradation in a cell type-dependent manner. Cells. 2019;8(10):1233. doi: 10.3390/cells8101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai JD, Ichida JK. C9ORF72 protein function and immune dysregulation in amyotrophic lateral sclerosis. Neurosci Lett. 2019;713:134523. doi: 10.1016/j.neulet.2019.134523. [DOI] [PubMed] [Google Scholar]
- 49.Corrionero A, Horvitz HR. A C9orf72 ALS/FTD ortholog acts in endolysosomal degradation and lysosomal homeostasis. Curr Biol. 2018;28(10):1522–35.e5. doi: 10.1016/j.cub.2018.03.063. [DOI] [PubMed] [Google Scholar]
- 50.Amick J, Tharkeshwar AK, Talaia G, Ferguson SM (2020) PQLC2 recruits the C9orf72 complex to lysosomes in response to cationic amino acid starvation. J Cell Biol 219(1):e201906076 [DOI] [PMC free article] [PubMed]
- 51.Sivadasan R, Hornburg D, Drepper C, Frank N, Jablonka S, Hansel A, et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci. 2016;19(12):1610–1618. doi: 10.1038/nn.4407. [DOI] [PubMed] [Google Scholar]
- 52.Selvaraj BT, Livesey MR, Zhao C, Gregory JM, James OT, Cleary EM, et al. C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca(2+)-permeable AMPA receptor-mediated excitotoxicity. Nat Commun. 2018;9(1):347. doi: 10.1038/s41467-017-02729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staats KA, Seah C, Sahimi A, Wang Y, Koutsodendris N, Lin S, et al. Small molecule inhibition of PIKFYVE kinase rescues gain- and loss-of-function C9ORF72 ALS/FTD disease processes in vivo. bioRxiv. 2019. 10.1101/685800
- 54.Xiao S, McKeever PM, Lau A, Robertson J. Synaptic localization of C9orf72 regulates post-synaptic glutamate receptor 1 levels. Acta Neuropathol Commun. 2019;7(1):161. doi: 10.1186/s40478-019-0812-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol. 2013;74(2):180–187. doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 56.Therrien M, Rouleau GA, Dion PA, Parker JA. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One. 2013;8(12):e83450. doi: 10.1371/journal.pone.0083450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao Q, Yang M, Liang C, Ma L, Zhang W, Jiang Z et al (2019) C9orf72 and Smcr8 mutant mice reveal MTORC1 activation due to impaired lysosomal degradation and exocytosis. Autophagy:1–16 [DOI] [PMC free article] [PubMed]
- 58.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li H-R, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110(47):E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA. Vieira de Sa R, et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol. 2015;78(3):426–438. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee HC, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med. 2016;8(347):347ra93. doi: 10.1126/scitranslmed.aaf6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sudria-Lopez E, Koppers M, de Wit M, van der Meer C, Westeneng HJ, Zundel CA, et al. Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathol. 2016;132(1):145–147. doi: 10.1007/s00401-016-1581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90(3):535–550. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McAlpine W, Sun L, Wang KW, Liu A, Jain R, San Miguel M, et al. Excessive endosomal TLR signaling causes inflammatory disease in mice with defective SMCR8-WDR41-C9ORF72 complex function. Proc Natl Acad Sci U S A. 2018;115(49):E11523–E11e31. doi: 10.1073/pnas.1814753115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Burberry A, Wang JY, Sandoe J, Ghosh S, Udeshi ND, et al. The C9orf72-interacting protein Smcr8 is a negative regulator of autoimmunity and lysosomal exocytosis. Genes Dev. 2018;32(13–14):929–943. doi: 10.1101/gad.313932.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harms MB, Cady J, Zaidman C, Cooper P, Bali T, Allred P, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34(9):2234.e13–2234.e19. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henderson MJ, Russell AJ, Hird S, Munoz M, Clancy JL, Lehrbach GM, et al. EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J Biol Chem. 2002;277(29):26468–26478. doi: 10.1074/jbc.M203527200. [DOI] [PubMed] [Google Scholar]
- 68.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19(5):668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang T, Baldie G, Periz G, Wang J. RNA-processing protein TDP-43 regulates FOXO-dependent protein quality control in stress response. PLoS Genet. 2014;10(10):e1004693. doi: 10.1371/journal.pgen.1004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20(10):1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ko HS, Bailey R, Smith WW, Liu Z, Shin JH, Lee YI, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106(8):2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agrawal P, Chen YT, Schilling B, Gibson BW, Hughes RE. Ubiquitin-specific peptidase 9, X-linked (USP9X) modulates activity of mammalian target of rapamycin (mTOR) J Biol Chem. 2012;287(25):21164–21175. doi: 10.1074/jbc.M111.328021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390(1):199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Batra R, Hutt K, Vu A, Rabin SJ, Baughn MW, Libby RT et al (2016) Gene expression signatures of sporadic ALS motor neuron populations. bioRxiv. 10.1101/038448
- 75.Goodier JL, Cheung LE, Kazazian HH. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013;41(15):7401–7419. doi: 10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH (2015) The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet 11(5):e1005252 [DOI] [PMC free article] [PubMed]
- 77.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7(1):72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodić N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184(5):1280–1286. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laflamme C, McKeever PM, Kumar R, Schwartz J, Kolahdouzan M, Chen CX, et al. Implementation of an antibody characterization procedure and application to the major ALS/FTD disease gene C9ORF72. eLife. 2019;8:e48363. doi: 10.7554/eLife.48363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eystathioy T, Chan EK, Takeuchi K, Mahler M, Luft LM, Zochodne DW, et al. Clinical and serological associations of autoantibodies to GW bodies and a novel cytoplasmic autoantigen GW182. J Mol Med (Berl) 2003;81(12):811–818. doi: 10.1007/s00109-003-0495-y. [DOI] [PubMed] [Google Scholar]
- 81.Goodier JL, Zhang L, Vetter MR, Kazazian HH. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27(18):6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodier JL, Mandal PK, Zhang L, Kazazian HH. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet. 2010;19(9):1712–1725. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 84.Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31(22):3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pereira GC, Sanchez L, Schaughency PM, Rubio-Roldan A, Choi JA, Planet E, et al. Properties of LINE-1 proteins and repeat element expression in the context of amyotrophic lateral sclerosis. Mob DNA. 2018;9:35. doi: 10.1186/s13100-018-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78(2):365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blokhuis AM, Koppers M, Groen EJN, van den Heuvel DMA, Dini Modigliani S, Anink JJ, et al. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016;132(2):175–196. doi: 10.1007/s00401-016-1575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G, Savard A, et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun. 2018;9(1):2794. doi: 10.1038/s41467-018-05273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang D, Sheng J, Xu L, Zhan X, Liu J, Jiang H, et al. Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a. Proc Natl Acad Sci U S A. 2020;117(18):9876–9883. doi: 10.1073/pnas.2002110117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 91.Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12(2):149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X, Qian SB. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22(18):3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bentmann E, Haass C, Dormann D. Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 2013;280(18):4348–4370. doi: 10.1111/febs.12287. [DOI] [PubMed] [Google Scholar]
- 94.Fernandes N, Eshleman N, Buchan JR. Stress granules and ALS: a case of causation or correlation? Adv Neurobiol. 2018;20:173–212. doi: 10.1007/978-3-319-89689-2_7. [DOI] [PubMed] [Google Scholar]
- 95.Baradaran-Heravi Y, Van Broeckhoven C, van der Zee J. Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol Dis. 2020;134:104639. doi: 10.1016/j.nbd.2019.104639. [DOI] [PubMed] [Google Scholar]
- 96.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172(6):803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21(24):3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maharjan N, Kunzli C, Buthey K, Saxena S. C9ORF72 regulates stress granule formation and its deficiency impairs stress granule assembly, hypersensitizing cells to stress. Mol Neurobiol. 2017;54(4):3062–3077. doi: 10.1007/s12035-016-9850-1. [DOI] [PubMed] [Google Scholar]
- 99.Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122(Pt 20):3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- 100.Kawakami I, Arai T, Hasegawa M. The basis of clinicopathological heterogeneity in TDP-43 proteinopathy. Acta Neuropathol. 2019;138(5):751–770. doi: 10.1007/s00401-019-02077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Freibaum BD, Taylor JP. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front Mol Neurosci. 2017;10:35. doi: 10.3389/fnmol.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nunes C, Mestre I, Marcelo A, Koppenol R, Matos CA, Nobrega C (2019) MSGP: the first database of the protein components of the mammalian stress granules. Database (Oxford) 2019:baz031 [DOI] [PMC free article] [PubMed]
- 103.Lan Y, Sullivan PM, Hu F. SMCR8 negatively regulates AKT and MTORC1 signaling to modulate lysosome biogenesis and tissue homeostasis. Autophagy. 2019;15(5):871–885. doi: 10.1080/15548627.2019.1569914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grumati P, Dikic I. Ubiquitin signaling and autophagy. J Biol Chem. 2018;293(15):5404–5413. doi: 10.1074/jbc.TM117.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen RH, Chen YH, Huang TY. Ubiquitin-mediated regulation of autophagy. J Biomed Sci. 2019;26(1):80. doi: 10.1186/s12929-019-0569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rao L, Sha Y, Eissa NT. The E3 ubiquitin ligase STUB1 regulates autophagy and mitochondrial biogenesis by modulating TFEB activity. Mol Cell Oncol. 2017;4(6):e1372867. doi: 10.1080/23723556.2017.1372867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wan W, You Z, Zhou L, Xu Y, Peng C, Zhou T, et al. mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Mol Cell. 2018;72(2):303–15.e6. doi: 10.1016/j.molcel.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 109.Amick J, Ferguson SM. C9orf72: at the intersection of lysosome cell biology and neurodegenerative disease. Traffic. 2017;18(5):267–276. doi: 10.1111/tra.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jung J, Behrends C. Multifaceted role of SMCR8 as autophagy regulator. Small GTPases. 2020;11(1):53–61. doi: 10.1080/21541248.2017.1346553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18(7):2603–2618. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36(12):1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie X, Matsumoto S, Endo A, Fukushima T, Kawahara H, Saeki Y et al (2018) Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J Cell Sci 131(8):jcs210856 [DOI] [PubMed]
- 114.Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, et al. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 2014;21(12):1838–1851. doi: 10.1038/cdd.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Monahan Z, Shewmaker F, Pandey UB. Stress granules at the intersection of autophagy and ALS. Brain Res. 2016;1649(Pt B):189–200. doi: 10.1016/j.brainres.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vu LT, Bowser R. Fluid-based biomarkers for amyotrophic lateral sclerosis. Neurotherapeutics. 2017;14(1):119–134. doi: 10.1007/s13311-016-0503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van der Ende EL, Meeter LH, Stingl C, van Rooij JGJ, Stoop MP, Nijholt DAT, et al. Novel CSF biomarkers in genetic frontotemporal dementia identified by proteomics. Ann Clin Transl Neurol. 2019;6(4):698–707. doi: 10.1002/acn3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. 2013;92(3):345–353. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hensman Moss DJ, Poulter M, Beck J, Hehir J, Polke JM, Campbell T, et al. C9orf72 expansions are the most common genetic cause of Huntington disease phenocopies. Neurology. 2014;82(4):292–299. doi: 10.1212/WNL.0000000000000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bourinaris T, Houlden H. C9orf72 and its relevance in parkinsonism and movement disorders: a comprehensive review of the literature. Mov Disord Clin Pract. 2018;5(6):575–585. doi: 10.1002/mdc3.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Devenney EM, Ahmed RM, Halliday G, Piguet O, Kiernan MC, Hodges JR. Psychiatric disorders in C9orf72 kindreds: study of 1,414 family members. Neurology. 2018;91(16):e1498–ee507. doi: 10.1212/WNL.0000000000006344. [DOI] [PubMed] [Google Scholar]
- 122.Silverman HE, Goldman JS, Huey ED. Links between the C9orf72 repeat expansion and psychiatric symptoms. Curr Neurol Neurosci Rep. 2019;19(12):93. doi: 10.1007/s11910-019-1017-9. [DOI] [PubMed] [Google Scholar]
- 123.Marogianni C, Rikos D, Provatas A, Dadouli K, Ntellas P, Tsitsi P, et al. The role of C9orf72 in neurodegenerative disorders: a systematic review, an updated meta-analysis, and the creation of an online database. Neurobiol Aging. 2019;84:238.e25–238.e34. doi: 10.1016/j.neurobiolaging.2019.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Fig. S1. Western blotting of cell line and brain tissue lysates using commercial antibodies against C9orf72, SMCR8, and WDR41 proteins. Antibodies shown are (A) α-C9ORF72-PT (Proteintech Group 22637–1), (B) α-SMCR8-PT (Proteintech 21125–1-AP, (C) α-SMCR8-B (Bethyl Laboratories A304-694A), (D) α-SMCR8-ab186504 (Abcam ab186504), (E) α-SMCR8-ab202283 (Abcam ab202283), (F) α-WDR41-SC (Santa Cruz sc-137923), and (G) α-WDR41-PT (Proteintech 26817–1-AP). Large arrows mark full-length protein products. The small arrows in (F) and (G) mark the presumed minor 45.5 kD WDR41 isoform. Samples were run on NuPAGE 4–12% Bis-Tris Protein gels (Novex) and Western blotting was performed according to [81, 82]. Molecular weight markers are Novex Sharp Pre-stained Protein Standard. Motor: motor cortex, Occ: occipital cortex, S.C.: spinal cord; sample identifier numbers are also shown. Fig. S2. Pie chart of results of DAVID (Database for Annotation, Visualization and Integrated Discovery, [83]) analyses of KEGG pathways showing selected functional categories for candidate member proteins of the (A) FL-SMCR8 and (B) C9orf72-FL protein interactomes. Percentages of the total number of proteins identified (Tables S1 and S2) for each category are shown within the slices. Fig. S3. Phylogenetic multi-sequence alignment of SMCR8 protein sequences for ten species. Alignments were made with Clustal Omega 1.2.1 (EMBL-EBI) followed by BoxShade 3.2 (http://sourceforge.net/projects/boxshade). Pink shading marks amino acid residues identical in at least 8 species, while green includes conservative replacements. Lysine residues predicted by MS sequencing to be ubiquitinated are boxed in blue (see Table S3). Species shown are: Homo sapiens, human; Pan troglodytes, chimpanzee; Canus lupus familiaris, dog; Mus musculus, house mouse; Rattus norvegicus, brown rat; Gallus gallus domesticus, chicken; Danio rerio, zebrafish; Xenopus tropicalis, western clawed frog; Biomphalaria glabrata, freshwater snail; Aplysia californica, California sea hare. Fig. S4. Immunofluorescence microscopy evidence for association of SMCR8 protein with cytoplasmic aggregates. (A) V5-tagged SMCR8 does not significantly enter SGs of NaAsO2-stressed N2A cells. (B) Overexpression of FLAG-tagged C9orf72 (left) or SMCR8 (right) protein does not induce formation of cytoplasmic granules in neuroblastoma cells. (C) V5-tagged exogenously expressed C9orf72 fails to enter SGs in NaAsO2-treated U2OS cells. (D) Endogenous C9orf72 protein speckles observed in unstressed N2A cells are not marked by TIA1, a SG protein. (E) C9orf72 protein detected by the α-C9orf72-PT antibody fails to enter SGs of NaAsO2-stressed U2OS cells. (F,G), Endogenous C9orf72 protein detected by the α-C9orf72-SC antibody colocalizes or justaposes with only a minor subset of SGs (marked by TIA1) or PBs (marked by 4-ET) in stressed mouse neuroblastoma N2A cells (see arrows). (H) In NaAsO2-stressed 293T cells, endogenous SMCR8 detected by the α-SMCR8-PT antibody colocalizes with granules marked by ORF1p, a protein encoded by LINE-1 retrotransposons [81]. (I) In NaAsO2-stressed human neuroblastoma SK-N-SH cells, endogenous SMCR8 localizes to SGs marked by α-eIF3η antibody. (J) In N2A cells treated with the endoplasmic reticulum stressor thapsigarin, endogenous SMCR8 protein colocalizes to granules marked by HEDLS/EDC4 (detected by α-p70 S6 kinase antibody, [77]), a component of PBs. (K) In unstressed N2A cells, endogenous SMCR8 localizes in granules with GW142, a PB marker. (L) Exogenously expressed TDP-43 and endogenous SMCR8 proteins do not colocalize in cytoplasmic granules of stressed U2OS cells. (M) SMCR8 rings but is generally excluded from overexpressed GFP-(GA)50 dipeptide aggregates. (M) In 293T cells, the α-WDR41-PT antibody detects endogenous protein in or beside a subset granules marked by 4-ET (see arrows). NT: no treatment. Cell nuclei were stained with Hoechst 33342 (right-most panels). Size bars are 10 μm. Table S1. The C9orf72 protein interactome determined by mass spectrometry. Table S2. The SMCR8 protein interactome determined by mass spectrometry. Table S3. Ubiquitinated SMCR8 lysine residues determined by MS sequencing, prediction algorithms, and the comparative phylogenetic analyses of Fig. S3.Table S4. Post-mortem brain motor cortex tissue samples used for the analyses of Fig. 5b,c.
Data Availability Statement
Raw MS sequencing data used to generate Supplementary Tables S1 and S2 will be made available from the corresponding author upon request.