Abstract
Introduction
Epidemiological studies that investigate alterations in the gut microbial composition associated with smoking are lacking. This study examined the composition of the gut microbiome in smokers compared with nonsmokers.
Aims and Methods
Stool samples were collected in a cross-sectional study of 249 participants selected from the Health Effects of Arsenic Longitudinal Study in Bangladesh. Microbial DNA was extracted from the fecal samples and sequenced by 16S rRNA gene sequencing. The associations of smoking status and intensity of smoking with the relative abundance or the absence and presence of individual bacterial taxon from phylum to genus levels were examined.
Results
The relative abundance of bacterial taxa along the Erysipelotrichi-to-Catenibacterium lineage was significantly higher in current smokers compared to never-smokers. The odds ratio comparing the mean relative abundance in current smokers with that in never-smokers was 1.91 (95% confidence interval = 1.36–2.69) for the genus Catenibacterium and 1.89 (95% confidence interval = 1.39–2.56) for the family Erysipelotrichaceae, the order Erysipelotrichale, and the class Erysipelotrichi (false discovery rate-adjusted p values = .0008–.01). A dose–response association was observed for each of these bacterial taxa. The presence of Alphaproteobacteria was significantly greater comparing current with never-smokers (odds ratio = 4.85, false discovery rate-adjusted p values = .04).
Conclusions
Our data in a Bangladeshi population are consistent with evidence of an association between smoking status and dosage with change in the gut bacterial composition.
Implications
This study for the first time examined the relationship between smoking and the gut microbiome composition. The data suggest that smoking status may play an important role in the composition of the gut microbiome, especially among individuals with higher levels of tobacco exposure.
Introduction
Despite the known and deadly risks of cigarette smoking,1 the prevalence of tobacco smoking has continued to rise in developing countries such as Bangladesh and other South Asian countries.2 Tobacco use is a risk factor for six of the eight leading causes of death in the world, according to the World Health Organization.3 The majority of the world’s current smokers now live in low to middle-income countries, with South Asia being one of the largest areas on the globe for tobacco production and consumption,4 making the Bangladeshi population of interest to study the effects of tobacco use.
The microbiota has emerged as having a potential role in smoking-related pathogenesis. The gut microbiota is composed of 1013–1014 microorganisms involved in physiological function such as digestion and metabolism, of which the majority are bacteria colonized from birth.5 Modifications of the gut microbiota have been associated with chronic conditions such as obesity and inflammatory bowel disease.6 Even with a neutralized diet, the intestinal microbial composition of smokers and nonsmokers is different.7 Several studies in humans have reported that smoking is related to differences in the microbiome measured in saliva,8 sputum,9 subgingival,10 upper gastrointestinal,11 throat,12 middle meatus,13 and bronchial wash14 samples. Mouse and rat models in the gut microbiota have shown significant changes in response to nicotine and smoke exposure.15 Although a few studies have investigated whether the gut microbiome differs by smoking status,16–18 these studies included mostly Crohn’s disease patients or had a small sample size. Epidemiological studies in the general population exploring the effect of smoking on the composition of the gut microbiome are lacking.
In this study, we examined the relationship between cigarette and/or bidi smoking and the gut microbiome in 250 participants, recruited from a large prospective cohort study, who provided fecal samples for 16S rRNA gene sequencing and completed lifestyle questionnaires.
Materials and Methods
Study Population
Data were collected for this study as part of the Health Effects of Arsenic Longitudinal Study, which is an ongoing, prospective cohort study located in Araihazar, Bangladesh. Details of the study were previously published and will be summarized briefly here.19 Between October 2000 and May 2002, an original cohort consisting of 11 746 married adults (to reduce loss to follow-up) between the ages of 18 and 75 years old were recruited from a well-defined 25 km2 geographical area. An expansion cohort of an additional 8287 participants was recruited between 2006 and 2008. Trained physicians performed in-person visits biennially to collect demographic and lifestyle data using a standardized questionnaire.19 The overall participation rate, estimated by the proportion of subjects who agreed to participate among the potential participants invited to the study, was 97%. Cohort participants received medical treatment at a field clinic that was set up for this purpose and to assist in follow-up and ancillary studies. The Ethical Committee of the Bangladesh Medical Research Council and the Institutional Review Boards of Columbia University and the University of Chicago approved all study procedures. Informed consent was obtained from all of the study participants.
The study presented here was conducted to assess the relationship between smoking and the gut microbiome. Between February 2015 and November 2016, Health Effects of Arsenic Longitudinal Study participants were recruited from the six villages surrounding the clinic to obtain fecal sample collection. Four hundred participants between the ages of 25 and 50 years old free from any major illness were randomly selected. The distribution of demographics or lifestyle variables was not significantly different from the overall Health Effects of Arsenic Longitudinal Study participants (data not shown). A total of 328 of the 400 fulfilled the eligibility requirements including absence of antibiotic use in the previous month and willingness to come to the clinic to provide stool samples and complete lifestyle questionnaires. Of the 328 eligible participants, 300 were recruited and 250 of these participants provided fecal samples.
Fecal Specimen Collection
Stool from a single bowel movement was collected in an empty ThermoFisher Scientific vial (Waltham, MA) given to participants by a senior research officer at the field clinic for fecal sample collection. Immediately stored in a −20°C freezer, samples were kept frozen until processing at the field laboratory for DNA extraction.
Assessment of Tobacco Smoking Variables
Detailed information on status of smoking was collected around the time of stool sample collection (never, former, and current). Information on number of sticks per day was asked together regarding smoking cigarettes and bidis. Bidis are filterless locally produced thin cigarettes filled with tobacco and wrapped in leaves. Cigarettes and bidis are often sold individually in Bangladesh, however to facilitate comparison with other studies, we calculated packs per day as the number of sticks smoked per day divided by 20. Time since quitting smoking was calculated as the difference between the age at smoking cessation and the age at the time of the study visit. Current smokers of cigarettes and bidis were categorized as current smokers and past smokers of any tobacco products were classified as former smokers.
DNA Extraction and 16S rRNA Gene Sequencing
Total DNA was extracted from fecal samples at the field laboratory using the MOBIO PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA). The 16S rRNA gene was amplified targeting the hypervariable V3–V4 region using universal primer set 347F 5′-GGAGGCAGCAGTRRGGAAT and 803R 5′-GACTACHVGGGTATCTAATCC. Agencourt AMPure XP (Beckman Coulter Life Sciences, IN) was used to purify polymerase chain reaction products and Agilent 4200 TapeStation (Agilent Technologies, CA) was used to quantify polymerase chain reaction products. Performed polymerase chain reaction amplification and sequencing methods are described in further detail previously.20 The Illumina MiSeq 300-cycles (2 × 300 bp) reagent kit (Illumina, Inc, San Diego, CA) was used to pool and sequence Amplicon libraries at equimolar concentrations. One sample was not sequenced further and was excluded for failing polymerase chain reaction amplification.
Bioinformatics and Quality Control
QIIME (Quantitative Insights into Microbial Ecology) 1.8.021 was used to process sequencing data. Once demultiplexed, sequencing reads with low quality scores of less than 25 were removed. ChimeraSlayer22 was used to take out chimeric sequences.
Preprocessed sequences were clustered into operational taxonomic units (OTUs) at 97% identity using UCLUST23 against the Greengenes database 13.8 with the most abundant sequence in each OTU selected as a representative. Greengenes database 13.8 and Ribosomal Database Project Classifier 2.224 were used to assign taxonomy for each OTU from phylum to genus levels. QIIME was used to estimate several alpha diversity measures at the OTU level based on rarefied sequence count (10 000 sequences per sample). By calculating the number of observed OTUs and Chao1 richness estimator, microbial richness, a measure of the number of taxa in each sample (or the abundance of microbes) was examined.25 The Shannon diversity index was used to assess evenness, a measure of the relative number of different taxa in each sample (or the distribution of the different microbes).26
Statistical Analyses
Descriptive analysis was conducted to compare demographic, lifestyle, and alpha diversity measures by smoking status using chi-square tests for categorical variables and t tests for continuous variables.
The number of reads for each taxon was divided by the total number of reads from all taxa within each individual sample to calculate relative abundance at the levels of phylum, class, order, family, and genus.
The association between smoking status and beta diversity of the gut microbiome measured by unweighted Unifrac distance,27 weighted Unifrac distance,28 and Bray–Curtis distances matrices29 was evaluated using the microbiome regression-based kernel association test MiRKAT30 adjusting for sex, age, body mass index (BMI), betel quid use, and education. The omnibus test (optimum MiRKAT) which simultaneously considers the above listed distances matrices was conducted to report the overall result, since each individual distance matrix measurement just captures a specific association pattern.
To evaluate the association between smoking status and the relative abundance of gut bacterial taxa, we used beta regression adjusting for sex, age, BMI, betel quid use, and education, using the R package “betareg.” 31 Beta regression was considered because this model is useful in microbiome studies where the continuous nonzero mean relative microbial abundance is the variable of interest and is restricted to the interval (0, 1).32 Bacterial taxa with zero abundances were imputed as 10−6 because the logarithm of zero is undefined. The regression coefficient in the beta regression is the log odds of the mean relative abundance of the specific taxa in smokers compared to nonsmokers. The exponent of the coefficient is the odds ratio of mean relative abundance comparing the two groups. We also used logistic regression models to assess the association between smoking status and the presence/absence of the gut bacterial taxa, adjusting for sex, age, BMI, betel quid use, and education. To test this association, the abundance was recoded as 1 if it was greater than 0, that is, the taxon was present in the sample. Logistic regression model adjusting for sex, age, BMI, betel quid use, and education was then used to fit the data. Only taxa with a prevalence of greater than 10% and less than 90% were evaluated. The Benjamini–Hochberg procedure was performed for all p values at each taxonomic rank to control the false discovery rate.33 For bacterial taxa that were nominally significantly associated with smoking status, for either relative abundance or presence/absence, we further assessed their association with packs of cigarettes smoked. Because smoking was more prevalent in men (76.5%) than in women (13.5%), sensitivity analyses were also conducted in men only. However, the results were similar and therefore sex-specific results were not presented to preserve the power of the analyses. Bacterial taxa that were differentially abundant in current smokers, former smokers, and with smoking dosage, as compared with never-smokers, at either nominal or false discovery rate (FDR)-adjusted level, were plotted on a taxonomic tree (cladogram) using GraPhlAn.34 Since the study was originally designed to collect arsenic data, we performed a sensitivity analysis adding arsenic levels as an additional potential confounder in the beta regression model for the taxa that were significant in current compared to never-smokers.
Results
Sequencing Results
From the 249 samples, approximately 8.6 million quality-filtered sequencing reads were obtained with an average of 34 520 reads per sample that were clustered into 108 987 OTUs. There was a mean of 3297 (standard deviation = 1295) OTUs and a range of 38–8909 OTUs per sample and the OTUs were classified into 19 phyla, 39 classes, 78 orders, 142 families, and 273 genera. Taxa with a relative abundance of less than 0.01% or without a name at each taxonomic level were removed, which left a total of 8 phyla, 16 classes, 24 orders, 40 families, and 54 genera in the final analyses.
Characteristics of the Study Participants
Characteristics of the study population are shown in Table 1. Mean age of the study participants was 48.6 ± 7.9 (mean ± standard deviation). Average BMI for this lean population was 21.5 ± 4.1 kg/m2. Participants had an average of 2.4 years of formal education. Twenty-five percent of study participants were current smokers (n = 62) and 14% were former smokers (n = 36), while 61% never smoked (n = 151). An average of 0.50 ± 0.31 packs of cigarettes/bidis per day were smoked by current smokers. Former smokers similarly consumed 0.47 ± 0.49 packs per day. Current and former smokers were predominantly male (88.7% and 63.9%, respectively), slightly older and leaner than those who never smoked and were more likely to have ever used betel quid. Current smokers were less educated compared to never-smokers. Blood pressure was not associated with smoking status, nor were alpha diversity measurements.
Table 1.
Distribution of Population Characteristics and Alpha Diversity Metrics by Smoking Status
| Smoking status | ||||||
|---|---|---|---|---|---|---|
| Overall (n = 249) | Never (n = 151) | Former (n = 36) | Current (n = 62) | p* Former | p † Current | |
| BMI, kg/m2 | 21.5 ± 4.1 | 22.5 ± 4.2 | 20.8 ± 3.4 | 19.6 ± 3.3 | .03 | <.001 |
| Missing | — | 1 | — | — | ||
| Male, % | 41.0 | 15.9 | 63.9 | 88.7 | <.001 | <.001 |
| Female, % | 59.0 | 84.1 | 36.1 | 11.3 | <.001 | <.001 |
| Age, years | 48.6 ± 7.9 | 46.4 ± 7.5 | 54.5 ± 5.8 | 50.5 ± 7.8 | <.01 | <.01 |
| Education, yearsa | 2.4 ± 3.3 | 2.7 ± 3.6 | 1.8 ± 3.0 | 1.8 ± 1.1 | .15 | .03 |
| Ever betel quid use, % | 50.6 | 43.1 | 69.4 | 58.1 | <.01 | .046 |
| Packs per day | 0.19 ± 0.34 | 0 | 0.47 ± 0.49 | 0.50 ± .31 | <.001 | <.001 |
| Systolic blood pressure, mm Hg | 112.0 ± 17.4 | 112.1 ± 17.8 | 116.9 ± 20.1 | 108.8 ± 14.2 | .15 | .15 |
| Diastolic blood pressure, mm Hg | 71.1 ± 11.5 | 71.3 ± 11.3 | 72.3 ± 12.4 | 69.8 ± 11.5 | .63 | .40 |
| Observed OTUs | 3297 ± 1295 | 3209 ± 1331 | 3497 ± 1045 | 3394 ± 1336 | .23 | .36 |
| Chao1 richness estimator | 9943 ± 3587 | 9693 ± 3695 | 10 623 ± 3068 | 10 156 ± 3589 | .16 | .40 |
| Shannon diversity index | 4.5 ± 0.5 | 4.5 ± 0.6 | 4.5 ± 0.5 | 4.5 ± 0.5 | .62 | .83 |
| Simpson diversity index | 0.92 ± 0.04 | 0.92 ± 0.04 | 0.92 ± 0.04 | 0.92 ± 0.04 | .72 | .87 |
BMI = body mass index; OTUs = operational taxonomic units.
Data are presented as mean ± standard deviation.
aAssessed at baseline.
*p values were computed with the t test or chi-square in comparison to never-smokers.
† n = 248 (missing data for one person).
Alpha diversity measurements compared by smoking status also did not exhibit significant differences (Table 1). The association between smoking status and microbiome beta diversity measured by Unifrac distance, weighted Unifrac distance, and Bray–Curtis was not significant (Table 2).
Table 2.
p Values of the Association Tests Between Microbiome Diversity and Smoking Status Using MiRKATa
| Smoking status | Unweighted Unifrac | Weighted Unifrac | Bray–Curtis | Omnibusb |
|---|---|---|---|---|
| Former vs. never | 0.6869 | 0.6586 | 0.7917 | 0.8480 |
| Current vs. never | 0.0941 | 0.5342 | 0.2413 | 0.2210 |
| Smoking dosage (pack) | 0.2241 | 0.8524 | 0.6630 | 0.3630 |
aThe model has been adjusted for sex, age, body mass index, betel quid use, and education.
bThe omnibus test simultaneously considers unweighted Unifrac, weighted Unifrac and Bray–Curtis distances.
Associations of Smoking With Microbial Composition
The relative abundances of 14 taxa were nominally significantly associated with smoking status when comparing current smokers and never-smokers (Table 3). Bacterial taxa along the Erysipelotrichi-to-Catenibacterium lineage were significantly enriched among current smokers compared with never-smokers after correction for multiple comparisons. The odds ratios comparing the mean relative abundance in current smokers with that in never-smokers were 1.91 for genus Catenibacterium (FDR-adjusted p = .01), 1.89 for the family Erysipelotrichaceae (FDR-adjusted p = .002), 1.89 for order Erysipelotrichales (FDR-adjusted p = .001), and 1.89 for class Erysipelotrich (FDR-adjusted p = .0008). Bacterial taxa along the Coriobacteriia-to-Collinsella lineage were also enriched among current smokers in comparison to never-smokers; however, the FDR-adjusted p values were not significant.
Table 3.
Odds Ratio of Mean Relative Abundances Comparing Current Smokers With Never Smokers
| Taxonomy | Odds ratioa (95% CI) | p value | FDR-adjusted p value† |
|---|---|---|---|
| Class Erysipelotrichi | 1.89 (1.39, 2.56) | .0001 | .0008 |
| Class Coriobacteriia | 1.46 (1.03, 2.06) | .03 | .25 |
| Order Erysipelotrichales | 1.89 (1.39, 2.56) | .0001 | .0012 |
| Order Coriobacteriales | 1.46 (1.03, 2.06) | .03 | .35 |
| Order Turicibacterales | 1.48 (1.01, 2.17) | .04 | .35 |
| Family Erysipelotrichaceae | 1.89 (1.39, 2.56) | .0001 | .002 |
| Family Mogibacteriaceae | 1.52 (1.05, 2.19) | .02 | .34 |
| Family Coriobacteriaceae | 1.46 (1.03, 2.06) | .03 | .34 |
| Family Peptostreptococcaceae | 1.44 (1.02, 2.03) | .04 | .34 |
| Family Turicibacteraceae | 1.48 (1.01, 2.17) | .04 | .34 |
| Genus Catenibacterium | 1.91 (1.36, 2.69) | .0002 | .01 |
| Genus Collinsella | 1.61 (1.13, 2.30) | .01 | .21 |
| Genus Turicibacter | 1.48 (1.01, 2.17) | .04 | .50 |
| Genus Rothia | 1.43 (1.01, 2.05) | .047 | .50 |
CI = confidence interval.
aOdds ratio of the mean relative abundance of taxa comparing current smokers to never-smokers adjusting for age, sex, body mass index, education, and betel quid use using beta regression with logit transformation based on continuous variable of relative abundance of taxa, replacing zeros with 10−6. A total of 142 bacterial taxa were tested. Only the ones with nominal significance were listed.
† p values were corrected for multiple testing using the Benjamini–Hochberg procedure to control the false discovery rate (FDR) ≤5%.
The presence/absence of five taxa was nominally significant in current smokers compared to never-smokers (Table 4), with the class Alphaproteobacteria remaining significant after multiple comparisons correction (FDR-adjusted p = .04). Current smokers were 4.85 times more likely to have this class present in their microbiome than never-smokers.
Table 4.
Odds ratio of the Presence/Absence of Bacterial Taxa Comparing Current Smokers With Never Smokers
| Taxonomy | Odds ratioa (95% CI) | p value | FDR-adjusted p value† |
|---|---|---|---|
| Class Alphaproteobacteria | 4.85 (1.53, 15.39) | .01 | .04 |
| Order GMD14H09 | 0.30 (0.11, 0.82) | .02 | .19 |
| Family Christensenellaceae | 3.62 (1.00, 13.09) | .0499 | .63 |
| Genus Slackia | 2.89 (1.12, 7.41) | .03 | .27 |
| Genus Mitsuokella | 3.84 (1.08, 13.67) | .04 | .27 |
CI = confidence interval.
aOdds ratio of the presence probability of taxa comparing current smokers to never-smokers adjusting for age, sex, body mass index, education, and betel quid use using logistic regression based on the dichotomous variable for presence/absence of taxa with a prevalence of greater than 10% and less than 90%. A total of 142 bacterial taxa were tested. Only the ones with nominal significance were listed.
† p values were corrected for multiple testing using the Benjamini–Hochberg procedure to control the false discovery rate (FDR) ≤5%.
For bacterial taxa that were nominally significantly associated with smoking status for either relative abundance or presence/absence, we further assessed their association with packs of cigarettes smoked. All of these four taxa in the Erysipelotrichi-to-Catenibacterium lineage exhibited a dose–response relationship (FDR-adjusted p = .01 for Catenibacterium, FDR-adjusted p = .004 for Erysipelotrichi) with increasing mean relative abundance as packs per day of cigarettes/bidis increased (Table 5). An increase in 1 pack per day was associated with an odds ratio of 1.88–1.90 for mean relative abundance for these specific taxa. The family Peptostreptococcaceae, the genera Slackia, and Collinsella also had increasing mean relative abundance with increasing smoking dosage with odds ratios of 1.67, 2.75, and 1.73 associated with an increase of 1 pack per day (FDR-adjusted p = .04, .0004, and .03), respectively. The presence of the genera Slackia and Mitsuokella was also related to increased smoking dosage and was 28.5 and 20.7 more likely to be present with an increase of 1 pack per day, respectively (FDR-adjusted p = .001, .048) (Table 5).
Table 5.
Odds Ratio of Mean Relative Abundances or Presence/Absence of Bacterial Taxa Associated With One Increase in Packs Smoked
| Taxonomy | Odds ratioa (95% CI) | p value |
|---|---|---|
| Abundance association testb | ||
| Class Erysipelotrichi | 1.88 (1.22, 2.89) | .004 |
| Order Erysipelotrichales | 1.88 (1.22, 2.89) | .004 |
| Family Erysipelotrichaceae | 1.88 (1.22, 2.89) | .004 |
| Family Peptostreptococcaceae | 1.67 (1.02, 2.69) | .04 |
| Genus Slackia | 2.75 (1.57, 4.76) | .0004 |
| Genus Catenibacterium | 1.90 (1.17, 3.03) | .01 |
| Genus Collinsella | 1.73 (1.05, 2.89) | .03 |
| Presence/absence association testc | ||
| Genus Slackia | 28.5 (3.9, 208.51) | .001 |
| Genus Mitsuokella | 20.7 (1.03, 411.58) | .048 |
CI = confidence interval.
aOdds ratio of the mean relative abundance of taxa (abundance association test) or the mean presence probability of taxa (presence/absence association test) comparing smokers to never-smokers adjusting for age, sex, body mass index, education, and betel quid use.
bBeta regression with logit transformation based on the continuous variable of relative abundance of taxa, replacing zeros with 10−6.
cLogistic regression based on the dichotomous variable for presence/absence of taxa with a prevalence of greater than 10% and less than 90%.
In former smokers compared to never-smokers, the relative abundances of four taxa, including three that remained significant after adjusting the p values for multiple comparisons testing in current smokers (the family Erysipelotrichaceae, order Erysipelotrichales, and class Erysipelotrichi) were nominally associated with smoking status (Supplemental Table 1, available at Nicotine and Tobacco Research online). A total of 12 taxa had higher prevalence among former smokers relative to never-smokers at the nominal level (Supplemental Table 2, available at Nicotine and Tobacco Research online), although none of the associations remained significant after correcting for multiple comparisons.
For the four taxa that were significantly associated with current smoking (class Erysipelotrichi, order Erysipelotrichale, family Erysipelotrichaceae, and genus Catenibacterium) after adjusting for multiple comparisons (three of which were nominally significant in the former smokers), there was also a dose–response relationship between their relative abundance and the intensity of smoking. However, there was no correlation between the time since quitting and the relative abundance of these four taxa using linear regression adjusting for sex, age, BMI, education, and betel quid use. However, our sample size was limited for this analysis. Future studies with a larger sample size are needed to confirm the findings.
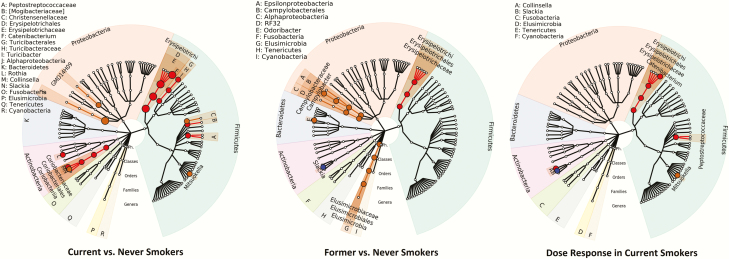
Figure 1 shows an association map of the taxa that were significantly related to smoking status and smoking dosage (packs per day). The nodes on the tree first branch into each phylum, which are shown by differently colored sections, then class, order, family, and genera. The red highlighted taxa are those whose abundance was associated with smoking. The orange highlighted taxa represent those for which their presence/absence was associated with smoking. The taxa for which both abundance and presence/absence were associated with smoking are highlighted in blue.
Figure 1.
The association mapping results of current smokers versus never-smokers, former smokers versus never-smokers, and smoking dosage (packs per day) on the taxonomic tree (generated using GraPhlAn34). The nodes on the tree from inner to outer circles are the phylum, class, order, family and genus rank. The corresponding annotations are written in the reverse order. Taxa whose abundance is associated with smoking status are highlighted in red; taxa whose presence/absence is associated with smoking status are highlighted in orange; taxa where both presence/absence and abundance is associated with smoking status are highlighted in blue. Small circles represent nominal p value <0.05; larger circles represent adjusted p value <0.05 (FDR = 5%). FDR = false discovery rate.
Sensitivity Analysis
There were no significant differences in arsenic levels between the current smokers, former smokers and control group. Adding arsenic as an additional potential confounder into the beta regression model for the relative abundance of the four taxa along the Catenibacterium-to-Erysipelotrichi lineage that were significantly different between current and never-smokers, after adjusting for multiple comparisons, did not change the results. In this model, the odds ratio comparing the mean relative abundance in current smokers with that in never-smokers was 1.89 (95% confidence interval = 1.34–2.66) for the genus Catenibacterium (p value = .0002) and 1.89 (95% confidence interval = 1.39–2.56) for the family Erysipelotrichaceae, the order Erysipelotrichale, and the class Erysipelotrichi (p values <.0001).
Discussion
In a Bangladeshi population, we found that the relative abundance of bacterial taxa along the Erysipelotrichi-to-Catenibacterium lineage, including the genus Catenibacterium, the family Erysipelotrichaceae, order Erysipelotrichales, and class Erysipelotrich, was significantly higher in current smokers compared to never-smokers, with each taxon exhibiting a dose–response relationship with packs of cigarettes smoked per day. The class Alphaproteobacteria was significantly more prevalent in current compared to never-smokers (FDR-adjusted p = .04). Genus Slackia, genus Collinsella, and other bacterial taxa along the same linages, including family Coriobacteriaceae, order Coriobacteriales, and class Coriobacteriia were also enriched among current smokers in comparison to never-smokers; however, the FDR-adjusted p values were not significant. The data suggest a critical role of smoking in gut microbiome composition.
Data from this study suggest effect of cigarette smoking on the relative abundance of certain bacterial taxa in the gut remained even after smoking cessation. Of all the bacterial taxa in the gut that had significantly higher levels or had a significantly higher proportion of presence at the nominal level in current smokers compared with never-smokers, the levels or presence of five bacterial taxa were also nominally significantly higher in former smokers, compared with never-smokers. These taxa included class Alphaproteobacteria, class Erysipelotrichi, order Erysipelotrichale, family Erysipelotrichaceae, and genus Slackia (comparing results in Table 3 vs. Supplemental Table 1, available at Nicotine and Tobacco Research online, in Table 4 vs. Supplemental Table 2, available at Nicotine and Tobacco Research online). The association comparing former smokers and never-smokers was weaker than that comparing current smokers and never-smokers, suggesting that the effect, although lasting after quitting smoking, may diminish overtime. All other taxa that were nominally significant in the current versus never-smokers are not seen in former smokers.
Several explanations have been proposed for the dysbiosis created by smoking, the simplest being cigarette exposure directly causes specific bacteria to enter the gut.35 Other hypotheses include immunomodulation,15,36 oxidative stress,36–38 or molecular changes to the cellular tight junctions and mucin composition of the gut.15,36
Epidemiological studies on the association between cigarette smoking and gut microbiome measured in stool samples is limited. Several studies in humans have reported that smoking is related to differences in the microbiome measured in saliva,8 sputum,9 subgingival,10 upper gastrointestinal, throat,12 middle meatus,13 and bronchial wash11,14 samples. Given variations in the gut microbiome between communities, it is difficult to directly compare across studies. Nonetheless, we found some similarities to other studies. Gram-positive class Coriobacteriia was nominally significant in our study in current versus never-smokers, and was identified as enriched in current smokers in the oral microbiome.8,39 This class has been implicated in oral diseases such as periodontitis, halitosis, and endodontic infections6 and these diseases have also been associated with smoking.40,41
Several studies have investigated cigarette smoking in relation to the gut microbiome in patients with Crohn’s disease. One study showed a decrease in richness, genus and species diversity and reduced relative abundance of Collinsella, Enterohabdus, and Gordonibacter.17 Our study found a significant increase in Collinsella among current smokers, in contrast to the findings from this study, however we also found a dose–response with Collinsella rendering the increase likely to be associated with smoking. Collinsella is well suited to colonize mucosal surfaces, metabolizes amino acids, and may directly interact with the host.6 Higher abundance of Collinsella has been related to type 2 diabetes42,43 and symptomatic atherosclerosis.44 A study evaluating the effect of smoking cessation showed a decrease in Bacteroidetes (Prevotella spp. and Bacteroides spp.) and the classes Alphaproteobacteria and Betaproteobacteria after smoking cessation.18 Interestingly, we found that the class Alphaproteobacteria, a Gram-negative class in the phylum Proteobacteria, was 4.85 times more prevalent in current smokers than those who never smoked in our study. Alphaproteobacteria has been directly detected in cigarette samples.35
Our finding of the higher abundance of the Erysipelotrichi-to-Catenibacterium lineage and the higher prevalence/abundance of the genus Slackia in current smokers are highly significant and novel. The genus Catenibacterium is a Gram-positive, nonspore-forming and anaerobic genus from the family Erysipelotrichidae, which produces short chain fatty acids as products of glucose fermentation. Catenibacterium has been associated with higher fat intake in a study of fecal samples in Seoul & Jeju Island in S. Korea.45 The class Erysipelotrich was found to be increased in murine gut in high-fat diets46 and related to systemic inflammation.47 The family Erysipelotrichaceae has been linked to dyslipidemic phenotypes and systemic inflammation.48 The genus Slackia has been suggested to be involved in lipid and xenobiotic metabolism,49 proinflammatory, and enriched in prediabetic individuals. Xenobiotic degradation pathways are crucial in bacterial upregulation to detoxify cigarette smoke. A metagenomics study found an increase in genes involved in xenobiotic metabolism in smokers.50 It is likely that an increase in tobacco exposure would lead to an increase in bacteria involved in the xenobiotic degradation of the toxins associated with smoking, and our study found a positive correlation between the genus Slackia and the amount of cigarettes/bidis smoked per day and also an increased likelihood for this genus to be present with increasing tobacco exposure (28.5 times more likely per 1 pack of cigarettes/bidis), which may explain the increase found in current smokers in our study. Given the many toxicants found in cigarette smoke, it is not surprising to find the higher prevalence and/or abundance of these inflammatory bacteria in smokers. However, whether their enrichment could contribute to smoking-related diseases warrants future studies.
A strength of this study is that it is the first study of the gut microbiome in South Asians, and includes participants who are free from frequent use of medications, supplements and antibiotics that disrupt the microbiome since it was conducted in rural community lacking basic health care services. Further, our findings align with other studies of the effect of smoking on the bacterial microbiome in other areas of the body and provide evidence of a dose–response relationship solidifying the results that certain taxa are more abundant in smokers. Limitations of the study include the cross-sectional design, instead of prospective, however it is not expected to be biased as participation in the study was not likely dependent on both the composition of the gut microbiome and smoking status. Longitudinal studies to evaluate the effect of smoking and smoking cessation on gut microbiome composition would be helpful to determine causality. The smoking dosage was based on the number of packs smoked per day since cotinine was not measured as part of the study. However, the questionnaire used to assess smoking dosage was standard and analyses using the data on all-cause mortality, prostate cancer mortality and multiple myeloma mortality, among others, generated similar data consistent with the literature.4,51–53 Lastly, 16S rRNA gene sequencing only has resolution to the genus level, and this lack prevents direct functional profiling. Metagenomic sequencing studies to evaluate the relationship of the gut microbiome and smoking are needed to better understand these mechanisms.
Our data suggest evidence of an association between cigarette/bidi smoking and the relative abundance of the genus Catenibacterium, family Erysipelotrichaceae, order Erysipelotrichales, and class Erysipelotrichi, with a dose–response indicating increased smoking exposure leads to an increase in relative abundance, and to the presence of the class Alphaproteobacteria. These changes in microbiome composition in association with smoking may be involved in the pathogenesis of several diseases. Further studies are required to investigate the underpinnings of bacterial dysbiosis related to smoking, the impact of smoking on the metagenomic content of the gut microbiome, and whether smoking-related gut microbial and/or metagenomic changes may explain smoking-related disease mechanisms.
Funding
This work was supported by National Institutes of Health grant R21 ES023421, P42 ES010349, P30 ES000260, P30 ES009089, and R01 DK110014.
Declaration of Interests
Authors have no competing interests to declare. All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. The text of this article was not reviewed by the sponsor. The manuscript is not under review by another journal.
Supplementary Material
Acknowledgments
We thank the Genome Technology Center for expert sequencing. This shared resource is partially supported by a National Cancer Institute grant, P30 CA016087, at the Laura and Isaac Perlmutter Cancer Center. ZP is staff physician at the Department of Veterans Affairs New York Harbor Healthcare System. The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs or the United States Government.
References
- 1. Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohen N. Smoking, health, and survival: prospects in Bangladesh. Lancet. 1981;1(8229):1090–1093. [DOI] [PubMed] [Google Scholar]
- 3. World-Health-Organization. WHO Report on the Global Tobacco Epidemic, 2008: The MPOWER Package Geneva, Switzerland: WHO; 2008; http://whqlibdoc.who.int/publications/2008/9789241596282_eng.pdf. 2008. Accessed July 29, 2018. [Google Scholar]
- 4. Wu F, Chen Y, Parvez F, et al. . A prospective study of tobacco smoking and mortality in Bangladesh. PLoS One. 2013;8(3):e58516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gill SR, Pop M, Deboy RT, et al. . Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clavel T, Desmarchelier C, Haller D, et al. . Intestinal microbiota in metabolic diseases: from bacterial community structure and functions to species of pathophysiological relevance. Gut Microbes. 2014;5(4):544–551. [DOI] [PubMed] [Google Scholar]
- 7. Kobayashi T, Fujiwara K. Identification of heavy smokers through their intestinal microbiota by data mining analysis. Biosci Microbiota Food Health. 2013;32(2):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu J, Peters BA, Dominianni C, et al. . Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10(10):2435–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim MY, Yoon HS, Rho M, et al. . Analysis of the association between host genetics, smoking, and sputum microbiota in healthy humans. Sci Rep. 2016;6:23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, Kumar PS. The subgingival microbiome of clinically healthy current and never smokers. ISME J. 2015;9(1):268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vogtmann E, Flores R, Yu G, et al. . Association between tobacco use and the upper gastrointestinal microbiome among Chinese men. Cancer Causes Control. 2015;26(4):581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox MJ, Hunter M, Musk AW, Cookson WO, James A, Moffatt MF. The upper airway microbiome of smokers, ex-smokers and never-smokers in busselton, Western Australia. In: American Journal of Respiratory and Critical Care Medicine. Conference: American Thoracic Society International Conference, Denver, CO: ATS. 2011;183(1 MeetingAbstracts).
- 13. Ramakrishnan VR, Frank DN. Impact of cigarette smoking on the middle meatus microbiome in health and chronic rhinosinusitis. Int Forum Allergy Rhinol. 2015;5(11):981–989. [DOI] [PubMed] [Google Scholar]
- 14. Einarsson GG, Comer DM, McIlreavey L, et al. . Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax. 2016;71(9):795–803. [DOI] [PubMed] [Google Scholar]
- 15. Allais L, Kerckhof FM, Verschuere S, et al. . Chronic cigarette smoke exposure induces microbial and inflammatory shifts and mucin changes in the murine gut. Environ Microbiol. 2016;18(5):1352–1363. [DOI] [PubMed] [Google Scholar]
- 16. Benjamin JL, Hedin CR, Koutsoumpas A, et al. . Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18(6):1092–1100. [DOI] [PubMed] [Google Scholar]
- 17. Opstelten JL, Plassais J, van Mil SW, et al. . Gut microbial diversity is reduced in smokers with Crohn’s disease. Inflamm Bowel Dis. 2016;22(9):2070–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biedermann L, Zeitz J, Mwinyi J, et al. . Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8(3):e59260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahsan H, Chen Y, Parvez F, et al. . Health Effects of Arsenic Longitudinal Study (HEALS): description of a multidisciplinary epidemiologic investigation. J Expo Sci Environ Epidemiol. 2006;16(2):191–205. [DOI] [PubMed] [Google Scholar]
- 20. Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137(2):588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haas BJ, Gevers D, Earl AM, et al. ; Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- 24. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32(4):557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:249–325. [Google Scholar]
- 30. Zhao N, Chen J, Carroll IM, et al. . Testing in microbiome-profiling studies with MiRKAT, the Microbiome Regression-Based Kernel Association Test. Am J Hum Genet. 2015;96(5):797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeileis A, Croissant Y. Extended model formulas in R: multiple parts and multiple responses. J Stat Softw. 2010;34(1). [Google Scholar]
- 32. Peng X, Li G, Liu Z. Zero-inflated beta regression for differential abundance analysis with metagenomics data. J Comput Biol. 2016;23(2):102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 34. Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sapkota AR, Berger S, Vogel TM. Human pathogens abundant in the bacterial metagenome of cigarettes. Environ Health Perspect. 2010;118(3):351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H, Zhao JX, Hu N, Ren J, Du M, Zhu MJ. Side-stream smoking reduces intestinal inflammation and increases expression of tight junction proteins. World J Gastroenterol. 2012;18(18):2180–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Talukder MA, Johnson WM, Varadharaj S, et al. . Chronic cigarette smoking causes hypertension, increased oxidative stress, impaired NO bioavailability, endothelial dysfunction, and cardiac remodeling in mice. Am J Physiol Heart Circ Physiol. 2011;300(1):H388–H396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tharappel JC, Cholewa J, Espandiari P, Spear BT, Gairola CG, Glauert HP. Effects of cigarette smoke on the activation of oxidative stress-related transcription factors in female A/J mouse lung. J Toxicol Environ Health A. 2010;73(19):1288–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vallès Y, Inman CK, Peters BA, et al. . Types of tobacco consumption and the oral microbiome in the United Arab Emirates Healthy Future (UAEHFS) pilot study. Sci Rep. 2018;8(1):11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gautam DK, Jindal V, Gupta SC, Tuli A, Kotwal B, Thakur R. Effect of cigarette smoking on the periodontal health status: a comparative, cross sectional study. J Indian Soc Periodontol. 2011;15(4):383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rad M, Kakoie S, Niliye Brojeni F, Pourdamghan N. Effect of long-term smoking on whole-mouth salivary flow rate and oral health. J Dent Res Dent Clin Dent Prospects. 2010;4(4):110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang X, Shen D, Fang Z, et al. . Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8(8):e71108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lambeth SM, Carson T, Lowe J, et al. . Composition, diversity and abundance of gut microbiome in prediabetes and type 2 diabetes. J Diabetes Obes. 2015;2(3):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karlsson FH, Fåk F, Nookaew I, et al. . Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin JH, Sim M, Lee JY, Shin DM. Lifestyle and geographic insights into the distinct gut microbiota in elderly women from two different geographic locations. J Physiol Anthropol. 2016;35(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greiner T, Bäckhed F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab. 2011;22(4):117–123. [DOI] [PubMed] [Google Scholar]
- 47. Dinh DM, Volpe GE, Duffalo C, et al. . Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaakoush NO. Insights into the role of erysipelotrichaceae in the human host. Front Cell Infect Microbiol. 2015;5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cho GS, Ritzmann F, Eckstein M, et al. . Quantification of Slackia and Eggerthella spp. in human feces and adhesion of representatives strains to Caco-2 cells. Front Microbiol. 2016;7:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boyle JO, Gümüs ZH, Kacker A, et al. . Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev Res (Phila). 2010;3(3):266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ugai T, Ito H, Oze I, et al. . Association of BMI, smoking and alcohol with multiple myeloma mortality in Asians: a pooled analysis of more than 800,000 participants in the Asia Cohort Consortium. Cancer Epidemiol Biomarkers Prev. 2019;28(11):1861–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fowke JH, McLerran DF, Gupta PC, et al. . Associations of body mass index, smoking, and alcohol consumption with prostate cancer mortality in the Asia Cohort Consortium. Am J Epidemiol. 2015;182(5):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang JJ, Yu D, Wen W, et al. . Tobacco smoking and mortality in Asia: a pooled meta-analysis. JAMA Netw Open. 2019;2(3):e191474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.