Abstract
Genetic variations such as single nucleotide polymorphisms (SNPs) can cause susceptibility to cancer. Although thousands of genetic variants have been identified to be associated with different cancers, the molecular mechanisms of cancer remain unknown. There is not a particular dataset of relationships between cancer and SNPs, as a bipartite network, for computational analysis and prediction. Link prediction as a computational graph analysis method can help us to gain new insight into the network. In this article, after creating a network between cancer and SNPs using SNPedia and Cancer Research UK databases, we evaluated the computational link prediction methods to foresee new SNP-Cancer relationships. Results show that among the popular scoring methods based on network topology, for relation prediction, the preferential attachment (PA) algorithm is the most robust method according to computational and experimental evidence, and some of its computational predictions are corroborated in recent publications. According to the PA predictions, rs1801394-Non-small cell lung cancer, rs4880-Non-small cell lung cancer, and rs1805794-Colorectal cancer are some of the best probable SNP-Cancer associations that have not yet been mentioned in any published article, and they are the most probable candidates for additional laboratory and validation studies. Also, it is feasible to improve the predicting algorithms to produce new predictions in the future.
Keywords: Cancer, SNP, link prediction, bipartite network
Introduction
Cancer has a significant impact on human mortality. It stands among the leading causes of death worldwide. The number of cancer cases is increasing at an alarming rate annually. It is believed that some behavioral and environmental triggers can lead to cancer, including diet, lifestyle, chronic, and viral infection. Increasing life span is another leading cause of cancer, and the researchers estimate that about two-thirds of the increase is due to aging. Although thousands of genetic variants (including single nucleotide polymorphisms [SNPs]) have identified to associate with different cancers, the molecular mechanisms of cancers have remained unknown. Therefore, researchers are continuing to explore this field.1-4
Genetic variations like SNPs can cause susceptibility to cancer. For example, the SNPs in a promoter site can affect the gene expression, and even in some tumors, it can affect the patient’s overall state of health and mortality risks.5,6 Most recent, genome-wide association studies (GWASs), show relations between some of the known cancers with the specific SNPs.5,6 For example, Guo et al7 reviewed 45 SNPs involved in prostate cancer. Also, Zhang et al8 studied the effect of rs920778 in the HOTAIR gene on esophageal cancer, and Li et al’s9 study of the effect of rs13252298 SNP in the PRNCR1 gene showed a relation to gastric cancer. A recent case10 identifies the link between SNP rs10800708 and breast cancer. There are also numerous examples of the relationships between different SNPs and cancers.
However, GWAS studies have several limitations. First, at least one-third of the known variants are in non-coding regulating regions, which affect the transcription factor binding. Second, many GWAS studies show heterogeneity in allele frequencies in different populations.11,12 So, more studies are needed to identify the relationships between cancers and SNPs. Computational methods can facilitate finding and predicting cancer-SNP relationships. There are some algorithmic studies for predicting cancer-SNP relationships. Those researches are mostly based on machine learning algorithms such as classification that need SNP profile data of case and control groups to predict the relationships.13-15 The challenges of these studies are simultaneous need to case and control data, limitation to one or few numbers of the cancers in each study, computational complexity, and so on. So, there is a tangible need for a general low complexity computational solution with few pre-requirements to predict cancer-SNP relationships. To the best of our knowledge, there is no study based on link prediction forecasting cancer-SNP relationships.
Link prediction and its importance
Link prediction, as a technique to analyze the graphs, dates back to the emergence of social networks.16 Later, it was applied to other networks such as biological ones.17 The primary purpose of link prediction in its basic definition is to find connections in the network that are missing or may be formed in the future, although it is possible to use link prediction to remove weak or spurious relations from the network.18 Even in some recent articles, it is practicable to predict the addition and removal of links in the network at the same time.19
| Link Prediction Algorithm |
| Input: matrix N with n*n dimensions, represent the investigating network |
| Output: best N(i, j) link to be established |
| 1. imax = 1, jmax = 2 |
| 2. Max = 0 |
| 3. for i = 1 to n |
| 4. for j = i + 1 to n |
| 5. if N[i, j] = 0 |
| 6. Rank = score (i, j) |
| 7. if Rank > Max |
| 8. Max = score (i, j) |
| 9. imax = i, jmax = j |
| 10. return i, j, Score (i, j) |
To use the link prediction algorithms, it is first necessary to present the problem network with graph structure. In a graph, entities or elements are considered as vertices or nodes and their relationships as links or edges. The modeling graph may be one of the common types, such as simple, bipartite, weighted, or directed graphs, and the link prediction algorithms have been customized and utilized for all of these graph types. For the computation, the graph can be stored in the computer memory as an adjacency matrix. Link prediction ranks the zero entries of the associated adjacency matrix to find the best promising relationships in the graph to establish.18 The link prediction algorithm for proposing the most probable edge in a simple graph is as follows:
Self-loop for the nodes is not allowed in the simple graph used by the link prediction algorithm. This means that N(i, i) should always be zero. In addition, because of the symmetry of the corresponding adjacency matrix with respect to the main diagonal, it is enough to calculate the score for only half of it, because the calculated score for (i, j) is equal to the calculated score for (j, i). Also, the score (i, j) function in the above algorithm can be calculated in a variety of ways; one of the simplest methods is based on network topological properties such as the properties of neighbor nodes.20 Table 1 consists of the most popular topological scoring methods.
Table 1.
Link prediction score functions for topology-based node neighborhood metrics.
| Score metric | Formula |
|---|---|
| CN | |
| JC | |
| PA | |
| AA |
Abbreviations: AA, Adamic and Adar; CN, common neighbors; JC, Jaccard; PA, preferential attachment.
refers to the set of neighbors of the x vertex, and represents the number of members of the set or the number of neighbors of the x vertex. For example, for the common neighbors (CN) criterion, the corresponding formula indicates that the score of the candidate edge is calculated by counting the number of CN of the 2 vertices x and y.
Method
Network construction
To study and predict the SNPs associated with various cancers in human, a list of SNPs should be provided at first. For this purpose, the SNPedia database (www.snpedia.com) was used. In this database, information of each SNP, including its effects on cancers, has been gathered from the valid journals. The total number of SNPs in SNPedia reaches 109 530. Also, to determine the relationship between SNPs and cancers, we need a complete list of all cancer names. We extract this list from the Cancer Research UK online database (www.cancerresearchuk.org). It has been active since 2002 in the field of cancer research and information. Each cancer usually has some subgroups that have not always been explicitly mentioned in the articles, so there are many challenges to create an SNP-Cancer network such as the following:
Sometimes the articles refer only to the main category and general name of cancer.
Occasionally, in the articles on the relationship between cancer and the SNPs, associated SNPs are found for all subtypes of a cancer.
Sometimes in the papers, the SNPs associated with cancer are present only for some of the subtypes of that cancer, and no evidence is found for other subtypes.
In some cases, in various articles, specific cancer is mentioned with several names.
In some cases, cancerous tissue is mentioned without the determination of the exact cancer name.
Every so often, there are no data about one cancer on the SNPedia website and no articles showing associated SNP.
For this reason, the authors of the article were forced to extract and check the data manually for each cancer. First, the information on the Cancer Research UK website was studied for all cancers. Next, for each cancer, a variety of different names and different types were examined and categorized. Then each cancer was searched manually on the website (www.snpedia.com), with both the main name and its subtypes. The output was the SNPs, whose association with cancer has been reported. So the list only included the cancers, primary cancer name, or subcategory, which their SNP was found in the search.
In the second step, a Java code was implemented to connect the website (www.snpedia.com) and automatically detect the SNPs associated with the cancers that were categorized in the previous step. In cases in which an SNP was found for the primary cancer name, and there were no data about its associativity for the cancer subtypes, we considered it linked to all subtypes and not for primary cancer name. In cases where SNP was found only for a few subtypes of the main type, the SNPs associated with subtypes were generalized to the main types, and only the name of the main type was entered into the final list of cancers, and the name of the subtypes was not included, for uniformity of the cancer names. In cases where there were several names for a cancer, a name was chosen, and the SNPs found for other alias cancer names made united to only the selected name, and just the selected name entered to the final list. For instances of cancerous tissues that there were some associated SNPs, we neglected them because of the ambiguity of the cancer name, and we avoided to incorporate these data into the final list.
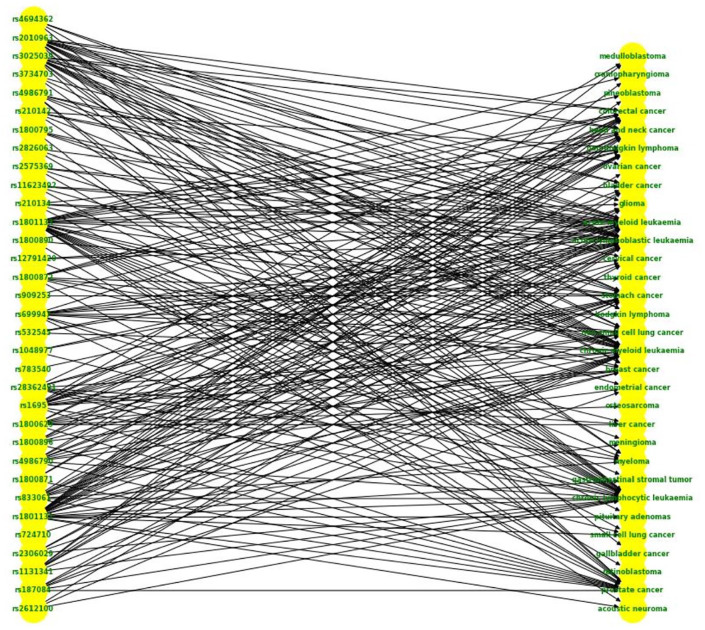
Thereupon the SNP-Cancer network was constructed. Steps performed to prepare the data are shown in Figure 1. The created network is a bipartite one, with 7599 edges or relations, 50 cancers, and 4723 SNP vertices (Figure 2). As the representation of the whole network cannot be informative due to existence of a large number of non-labeled overlapping nodes, we presented only a sub-network regarding a selected subset of cancers in Figure 3 to make the representation more understandable—a bipartite graph demonstration with nodes labeled that would be a complement representation here. In case the network is still enormous in this case, the number of SNPs has also been limited (31 cancers, 33 SNPs, and 222 relationships).
Figure 1.
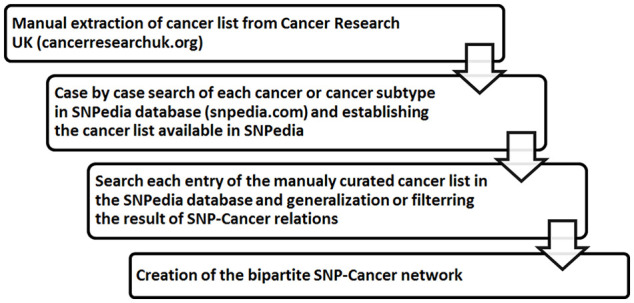
SNP-Cancer network construction steps. SNP indicates single nucleotide polymorphism.
Figure 2.
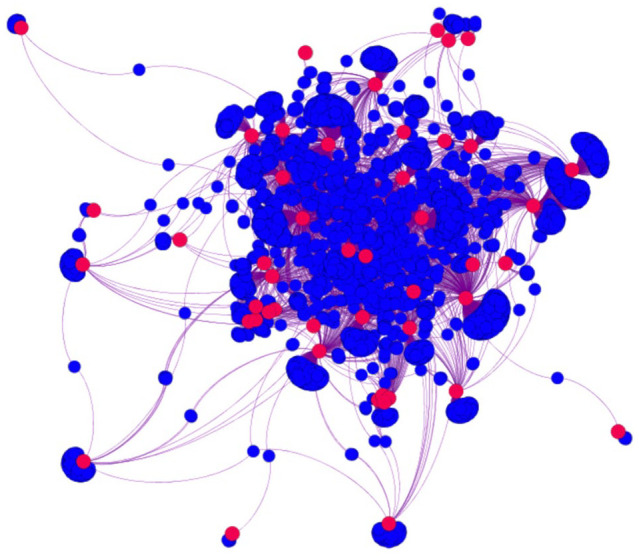
Visualization of SNP-Cancer bipartite graph. Red circles are cancers surrounded by SNPs. SNP indicates single nucleotide polymorphism.
Figure 3.
An alternative representation for the SNP-Cancer network. A sub-network with 33 SNPs, 31 cancers, and 222 relationships is presented in the bipartite form to make a better view of the real network. SNP indicates single nucleotide polymorphism.
Bipartite link prediction
After creating a bipartite graph of links between SNPs and cancers, we have a network that identifies which SNPs are associated with which cancers and vice versa. From here onward, the discovery of hidden links in the bipartite graph will be the result of calculations and link prediction algorithms. But, we only introduced the link prediction in its basic form for simple graphs earlier in the “Introduction” section. So, it is necessary to adopt the ranking scores for bipartite networks.
The bipartite network does not involve self-loop relations. Also, the scoring function, which ranks the probable links in the bipartite graph, is different. Because none of the nodes in each part has relationships with the nodes in the same part, direct relation between SNP pairs or cancer pairs is not important here in this research. In this graph type, only the links between the vertices of one part and the vertices from the other part are important. Thus, we chose the scoring functions based on reference.21 To clarify the customized formulas, it is necessary to define its elements first. If x node is in the first part and y node is in other part of the graph, refers to the set of neighbors of y node, and refers to the neighboring set of neighbors of y or simply . For example, counts the number of common neighbors of x, which is from one part of the network, and neighbors of neighbors of y, that y is from another part. In other words, neighbors of y have not any intersection with x. So, the probable relationship between x from one part of the network and y from another part will be ranked based on the neighbors of x and indirect (2 steps away) neighbors of y. This modification is not necessary for the preferential attachment (PA) scoring method, because it does not depend on vicinity intersection. It only depends on the degree of the nodes likely to connect from each graph partition (Table 2).
Table 2.
Link prediction score function for topology-based node neighborhood metrics in bipartite graphs.
| Score metric | Score formula |
|---|---|
| CN | |
| JC | |
| PA | |
| AA |
Abbreviations: AA, Adamic and Adar; CN, common neighbors; JC, Jaccard; PA, preferential attachment.
Consequently, we are willing to know which of the scoring methods has the best prediction accuracy for the SNP-Cancer bipartite network, and then we would like to know what new connections between SNPs and cancers are discovered and suggested according to the best edge scoring method. Next, we should prove these findings computationally, and finally, we should be able to validate the proposed results based on the evidence we find on the scientific databases or pass it on to new in vitro experiments.
The accuracy of predictions depends on the properties of the examined networks, such as scale-free or small-world attributes,22 and none of the scoring algorithms have complete superiority over the others in advance. Therefore, the calculations will be done for each of the scoring methods in Table 2, and after the accuracy evaluation, the best method is introduced for more practical investigation of the results.
Evaluation criteria
To compare the efficiency of the link scoring functions, it is computationally common to measure the performance based on the known network information, ie, the edges that already exist. It is recommended to apply one of the area under the receiver operating characteristic curve (AUC) or precision measures for evaluation.23 In our work, AUC is used for quantifying the accuracy of the prediction method. To do so, if the set of edges in the network is E, we divide it into 2 distinct parts, ET and EV, which their intersection is an empty set, and their union includes all the edges in the network. ET stands for a training set of edges as existing information, and EV is validation set of edges which we delete from the network randomly and provide no information on them to the scoring functions, and we are going to predict them accurately. To ensure that all validation links are tested, we will use the 10-fold cross-validation process, which does the prediction 10 times for 10 disjoint sets. Each set includes 10% of the randomly removed edges of the network, EV. After that, we report the AUC of the prediction for each score function as the average of the values of 10-fold cross-validation for each function, and larger percentage of the AUC will show the better performance of the scoring method for link prediction. Here, the AUC means the probability that a randomly chosen missing connection is given a higher score by our algorithm than a randomly chosen pair of unconnected vertices. Thus, the degree to which the AUC exceeds 0.5 indicates how much better our predictions are than chance.24 Therefore, calculating the average score is as follows
| (1) |
where n is the total number of the random edge selection, is the total number of times that randomly chosen missing link has the higher score, and has the same score.
Of course, the real data of related researches can also be used for further validation. For this purpose, we will search online scientific databases, Google Scholar and PubMed, for the predicted links that are not currently available on the SNPedia website, and if we find evidence in research papers and articles, we have another proof for the accuracy of the operation of the algorithms.
Results
Based on the AUC evaluation results among link prediction scoring functions, the PA method is more effective in predicting potential links between SNPs and cancers (Table 3). Accordingly, the most likely 15 predicted links between SNP and various cancers are as follows for PA method, first 2 columns of Table 4.
Table 3.
AUC of different node neighborhood similarity–based link prediction scores over bipartite SNP-Cancer network.
| Algorithm | AUC |
|---|---|
| CN | 0.90 |
| JC | 0.84 |
| PA | 0.95 |
| AA | 0.89 |
Abbreviations: AA, Adamic and Adar; AUC, area under the receiver operating characteristic curve; CN, common neighbors; JC, Jaccard; PA, preferential attachment; SNP, single nucleotide polymorphism.
Table 4.
Top 15 SNP-Cancer relationships predicted by PA, CN, JC, and AA scoring link prediction approach.
| PA | CN | JC | AA | ||||
|---|---|---|---|---|---|---|---|
| SNP | Cancer | SNP | Cancer | SNP | Cancer | SNP | Cancer |
| rs1801133 | Non-small cell lung cancer | rs1801133 | Pancreatic cancer | rs25489 | Cholangiocarcinoma | rs1801133 | Pancreatic cancer |
| rs1801131 | Non-small cell lung cancer | rs1801133 | Non-small cell lung cancer | rs20417 | Cholangiocarcinoma | rs1801133 | Non-small cell lung cancer |
| rs1048943 | Stomach cancer | rs1801133 | Gallbladder cancer | rs13181 | Laryngeal cancer | rs1801133 | Gallbladder cancer |
| rs1048943 | Prostate cancer | rs1801133 | Hodgkin lymphoma | rs17851045 | Thymoma | rs1801133 | Hodgkin lymphoma |
| rs1799793 | Ovarian cancer | rs1801133 | Thyroid cancer | rs587781525 | Thymoma | rs1801133 | Thyroid cancer |
| rs1805794 | Stomach cancer | rs1801131 | Pancreatic cancer | rs1057519984 | Thymoma | rs1801131 | Pancreatic cancer |
| rs4646903 | Prostate cancer | rs1801131 | Non-small cell lung cancer | rs764146326 | Thymoma | rs1801131 | Non-small cell lung cancer |
| rs1801394 | Non-small cell lung cancer | rs1801131 | Bladder cancer | rs1057520000 | Thymoma | rs1801131 | Bladder cancer |
| rs4880 | Non-small cell lung cancer | rs1801131 | Myeloma | rs28934874 | Thymoma | rs1801131 | Myeloma |
| rs1800566 | Stomach cancer | rs1801131 | Retinoblastoma | rs104894228 | Thymoma | rs1801131 | Retinoblastoma |
| rs3212227 | Stomach cancer | rs1801131 | Hodgkin lymphoma | rs1801131 | Retinoblastoma | rs1801131 | Hodgkin lymphoma |
| rs1805794 | Colorectal cancer | rs1801131 | Thyroid cancer | rs1799793 | Laryngeal cancer | rs1801131 | Thyroid cancer |
| rs2736100 | Breast cancer | rs1801131 | Gallbladder cancer | rs2736100 | Laryngeal cancer | rs1801131 | Gallbladder cancer |
| rs1801133 | Pancreatic cancer | rs1801133 | Skin cancer | rs1801133 | Gallbladder cancer | rs1801133 | Skin cancer |
| rs699947 | Stomach cancer | rs1801133 | Osteosarcoma | rs1801133 | Hodgkin lymphoma | rs13181 | Hodgkin lymphoma |
Abbreviations: AA, Adamic and Adar; CN, common neighbors; JC, Jaccard; PA, preferential attachment; SNP, single nucleotide polymorphism.
Because of the novelty of the idea of predicting unknown links between cancer and SNP and for more investigation and better comparison, the results of other scoring functions are also summarized in Table 4.
Discussion
Single nucleotide polymorphisms in the human genome are of the most common genetic variations and are located in different positions of genes such as exon, promoter, intron, 5′-UTR, and 3′-UTR. Due to their position in the genes, SNPs have different levels of control in various diseases, such as cancer, and the results of studies have proven the role of SNPs in cancer in terms of regulation, repair, DNA mismatch, metabolism, cell cycle, and immunity.25-27 Our understanding of the role of SNPs in cancer susceptibility depends on our molecular understanding of the pathogenicity of cancer.28 In clinical trials, people are usually identified in the advanced stages of the disease, and the main goal is to prevent the progression of disease in patients, and the SNP biomarker data are essential for predicting and screening individuals that are at hazard.
Checking the validity of the AUC results was also done through search in the popular scientific databases, Google Scholar and PubMed, to confirm the probable relationships based on reported pieces of evidence. Type of the reported relations between SNP and cancer is also noted as positive or negative associativity effects, with Yes or No in Tables 5 to 8. Results of the investigation of the evidence affirm the computational link prediction calculation.
Table 5.
Validation of the prediction results for new SNP-Cancer relationships in PA scoring method.
| Row | SNP | Cancer | SNPedia | PubMed | Google Scholar | References | Association |
|---|---|---|---|---|---|---|---|
| 1 | rs1801133 | Non-small cell lung cancer | X | ✓ | ✓ | Ding et al30 | Yes |
| 2 | rs1801131 | Non-small cell lung cancer | X | ✓ | ✓ | Li et al31 | Yes |
| 3 | rs1048943 | Stomach cancer | X | ✓ | ✓ | Hidaka et al32 | No |
| 4 | rs1048943 | Prostate cancer | X | ✓ | ✓ | Koda et al33 | Yes |
| 5 | rs1799793 | Ovarian cancer | X | ✓ | ✓ | Assis et al34 | No |
| 6 | rs1805794 | Stomach cancer | X | ✓ | ✓ | Zhou et al35 | Yes |
| 7 | rs4646903 | Prostate cancer | X | ✓ | Porchia et al36 | No | |
| 8 | rs1801394 | Non-small cell lung cancer | X | ||||
| 9 | rs4880 | Non-small cell lung cancer | X | ||||
| 10 | rs1800566 | Stomach cancer | X | ✓ | ✓ | Yadav et al37 | Yes |
| 11 | rs3212227 | Stomach cancer | X | ✓ | ✓ | Yin et al38 | Yes |
| 12 | rs1805794 | Colorectal cancer | X | ||||
| 13 | rs2736100 | Breast cancer | X | ✓ | ✓ | Aydin et al39 | No |
| 14 | rs1801133 | Pancreatic cancer | X | ✓ | ✓ | Nakao et al40 | No |
| 15 | rs699947 | Stomach cancer | X | ✓ | Ke et al41 | No |
Abbreviations: PA, preferential attachment; SNP, single nucleotide polymorphism.
Table 8.
Validation of the prediction results for new SNP-Cancer relationships in AA scoring method.
| Row | SNP | Cancer | SNPedia | PubMed | Google Scholar | References | Association |
|---|---|---|---|---|---|---|---|
| 1 | rs1801133 | Pancreatic cancer | X | ✓ | ✓ | Nakao et al40 | No |
| 2 | rs1801133 | Non-small cell lung cancer | X | ✓ | Ding et al30 | Yes | |
| 3 | rs1801133 | Gallbladder cancer | X | ✓ | ✓ | Dixit et al42 | No |
| 4 | rs1801133 | Hodgkin lymphoma | X | ✓ | ✓ | Sud et al43 | Yes |
| 5 | rs1801133 | Thyroid cancer | X | ✓ | ✓ | Zara-Lopes et al44 | Yes |
| 6 | rs1801131 | Pancreatic cancer | X | ✓ | ✓ | Nakao et al40 | No |
| 7 | rs1801131 | Non-small cell lung cancer | X | ✓ | ✓ | Li et al31 | Yes |
| 8 | rs1801131 | Bladder cancer | X | ✓ | ✓ | De Maturana et al45 | No |
| 9 | rs1801131 | Myeloma | X | ✓ | ✓ | Ma et al46 | Yes |
| 10 | rs1801131 | Retinoblastoma | X | ✓ | ✓ | Soleimani et al47 | No |
| 11 | rs1801131 | Hodgkin lymphoma | X | ||||
| 12 | rs1801131 | Thyroid cancer | X | ✓ | ✓ | Yang et al48 | No |
| 13 | rs1801131 | Gallbladder cancer | X | ✓ | ✓ | De Maturana et al45 | No |
| 14 | rs1801133 | Skin cancer | X | ✓ | ✓ | Xie et al49 | No |
| 15 | rs13181 | Hodgkin lymphoma | X |
Abbreviations: AA, Adamic and Adar; SNP, single nucleotide polymorphism.
Of the 15 not included links in SNPedia, that has been predicted by the PA link prediction algorithm, 12 cases have been addressed in the papers; 6 were confirmed by the experiments, 6 were rejected, and the other 3 were not yet declared. While other link prediction methods, which have fewer points than PA in terms of AUC (CN, Jaccard [JC], and Adamic and Adar [AA] methods), have fewer predicted positive associations than PA in literature survey. In particular, JC, which has the weakest predictability power in link prediction researches,29 has also least positive findings and returned more results that have not been verified at all in the literature (Tables 5-8).
Table 6.
Validation of the prediction results for new SNP-Cancer relationships in CN scoring method.
| Row | SNP | Cancer | SNPedia | PubMed | Google Scholar | References | Association |
|---|---|---|---|---|---|---|---|
| 1 | rs1801133 | Pancreatic cancer | X | ✓ | ✓ | Nakao et al40 | No |
| 2 | rs1801133 | Non-small cell lung cancer | X | ✓ | Ding et al30 | Yes | |
| 3 | rs1801133 | Gallbladder cancer | X | ✓ | ✓ | Dixit et al42 | No |
| 4 | rs1801133 | Hodgkin lymphoma | X | ✓ | ✓ | Sud et al43 | No |
| 5 | rs1801133 | Thyroid cancer | X | ✓ | ✓ | Zara-Lopes et al44 | Yes |
| 6 | rs1801131 | Pancreatic cancer | X | ✓ | ✓ | Nakao et al40 | No |
| 7 | rs1801131 | Non-small cell lung cancer | X | ✓ | ✓ | Li et al31 | Yes |
| 8 | rs1801131 | Bladder cancer | X | ✓ | ✓ | De Maturana et al45 | No |
| 9 | rs1801131 | Myeloma | X | ✓ | ✓ | Ma et al46 | Yes |
| 10 | rs1801131 | Retinoblastoma | X | ✓ | ✓ | Soleimani et al47 | No |
| 11 | rs1801131 | Hodgkin lymphoma | X | ||||
| 12 | rs1801131 | Thyroid cancer | X | ✓ | ✓ | Yang et al48 | No |
| 13 | rs1801131 | Gallbladder cancer | X | ✓ | ✓ | De Maturana et al45 | Yes |
| 14 | rs1801133 | Skin cancer | X | ✓ | ✓ | Xie et al49 | No |
| 15 | rs1801133 | Osteosarcoma | X |
Abbreviations: CN, common neighbors; SNP, single nucleotide polymorphism.
Table 7.
Validation of the prediction results for new SNP-Cancer relationships in JC scoring method.
| Row | SNP | Cancer | SNPedia | PubMed | Google Scholar | References | Association |
|---|---|---|---|---|---|---|---|
| 1 | rs25489 | Cholangiocarcinoma | X | ||||
| 2 | rs20417 | Cholangiocarcinoma | X | ||||
| 3 | rs13181 | Laryngeal cancer | X | ✓ | ✓ | Sun et al50 | No |
| 4 | rs17851045 | Thymoma | X | ||||
| 5 | rs587781525 | Thymoma | X | ||||
| 6 | rs1057519984 | Thymoma | X | ||||
| 7 | rs764146326 | Thymoma | X | ||||
| 8 | rs1057520000 | Thymoma | X | ||||
| 9 | rs28934874 | Thymoma | X | ||||
| 10 | rs104894228 | Thymoma | X | ||||
| 11 | rs1801131 | Retinoblastoma | X | ✓ | ✓ | Soleimani et al47 | No |
| 12 | rs1799793 | Laryngeal cancer | X | ✓ | ✓ | Lu et al51 | No |
| 13 | rs2736100 | Laryngeal cancer | X | ||||
| 14 | rs1801133 | Gallbladder cancer | X | ✓ | ✓ | De Maturana et al45 | No |
| 15 | rs1801133 | Hodgkin lymphoma | X | ✓ | ✓ | Sud et al43 | Yes |
Abbreviations: JC, Jaccard; SNP, single nucleotide polymorphism.
However, in the 3 methods, AA, PA, and CN, there are 2 common couples of rs1801133-Non-small cell lung cancer and rs1801131-Non-small cell lung cancer that all of them confirm but with different score and positions in their sorted, ranked list. Rs1801133 and rs1801131 are also popular and have many related studies and have links to several cancers, while the most frequent cancer in the predictions is Non-small cell lung cancer. AA and CN results are almost identical except in the last position, row 15.
Consequently, there are different confirming publications for many of the SNP-Cancer predicted relationships. We briefly mentioned the latest published paper in column 7 of Tables 5 to 8, to show the recent findings of the studies. The last published paper also integrates all the previous studies and final findings of the type of association (Yes or No) between SNP and cancer.
Several factors such as sparsity or completeness of the network can affect the evaluation results. For example, the network density, number of the existing edges divided by the total possible edges, is 0.032 here, and we have a relatively sparse network. The denser the network will be, the better the performance of link prediction algorithms will get. Also, the completeness of the investigated dataset affects the accuracy of the calculations and results. SNPedia is not fully up-to-date and ideal, as we will demonstrate in the next section and our computations show. Furthermore, the AUC criterion chosen for the evaluation is not the only criterion. It can be completed by verifying the availability of the results in the literature, which is well known as domain knowledge evaluation and will be discussed further.
Another notable point is that there are several predicted relationships here, which have been studied in the literature, but the result of the researches reports no association between SNP and cancer. This is also significant because it has attracted the researchers and approves the importance and directionality of our computational prediction methods for more in vitro investigations.
Conclusions
Based on the promising results of the PA scoring method, to predict new links between SNP and cancer, we suggest examining and verifying below relationships in vitro because, to the best of our knowledge, such links have not yet been reported in scientific publications. If one or more of the following links are verified, one can consider more of these PA predictions to find new SNP-Cancer associations:
rs1801394-Non-small cell lung cancer
rs4880-Non-small cell lung cancer
rs1805794-Colorectal cancer
Numerous unreported predictions of SNP and cancer links on the SNPedia reference website indicate that this database is incomplete and can be completed using literature reviews, in vitro tests, or other methods that can also be used to validate the result of the link prediction method. This is also true for many other biological networks, and they can be enriched with the help of link prediction algorithms, or even their hidden or incomplete relations can be discovered. Also, the precision of the link prediction depends on how the network is created, network properties, and the preprocessing of the network constructor data. The more reliable and accurate the work is, the better the results will be.
Only a small number of the basic existing algorithms for link prediction are used in this research. There were unsupervised node neighborhood-based link prediction algorithms. Other methods, such as path-based or supervised machine learning–based, can also be used to increase the accuracy of the results. In particular, machine learning–based methods can take into account different related features of the network and not just network topology.18
Link prediction is not used only to predict new or missed relations. Its newer versions can be used to remove noise or misconnections. This version of the link prediction is known as the Negative Link Prediction (NLP)17 and can be used to identify and eliminate the weak associations between SNP and cancer. The effectiveness of such a method in noise elimination has been proven on experimental data extracted from high-throughput methods for protein networks.52 Finally, link prediction can also be used to develop and predict links between SNP and other non-cancerous diseases.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: SB gathered the data and carried out the calculations. SS gave the idea of the research and designed the structure of the work and prepared the draft of the manuscript with SB. MT did the analysis and search for confirmations and filled the tables accordingly. KK participated in analysis and interpretation of the results and final edit of the manuscript’s writing. All authors read and approved the final manuscript. SS managed the teamwork and cooperation.
ORCID iD: Sadegh Sulaimany  https://orcid.org/0000-0002-4618-0428
https://orcid.org/0000-0002-4618-0428
References
- 1. Alnuaimi AD, Wiesenfeld D, O’Brien-Simpson NM, Reynolds EC, McCullough MJ. Oral Candida colonization in oral cancer patients and its relationship with traditional risk factors of oral cancer: a matched case-control study. Oral Oncol. 2015;51:139-145. doi: 10.1016/j.oraloncology.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 2. Girschik J, Heyworth J, Fritschi L. Self-reported sleep duration, sleep quality, and breast cancer risk in a population-based case-control study. Am J Epidemiol. 2013;177:316-327. doi: 10.1093/aje/kws422. [DOI] [PubMed] [Google Scholar]
- 3. Wang CC, Palefsky JM. Human papillomavirus-related oropharyngeal cancer in the HIV-infected population. Oral Dis. 2016;22:98-106. doi: 10.1111/odi.12365. [DOI] [PubMed] [Google Scholar]
- 4. Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78-85. [DOI] [PubMed] [Google Scholar]
- 5. Yuan H, Liu H, Liu Z, et al. A novel genetic variant in long non-coding RNA gene NEXN-AS1 is associated with risk of lung cancer. Sci Rep. 2016;6:34234. doi: 10.1038/srep34234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xavier-Magalhães A, Oliveira AI, de Castro JV, et al. Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J Neurooncol. 2017;132:27-34. doi: 10.1007/s11060-016-2345-0. [DOI] [PubMed] [Google Scholar]
- 7. Guo H, Ahmed M, Zhang F, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet. 2016;48:1142-1150. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 8. Zhang X, Zhou L, Fu G, et al. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062-2067. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 9. Li L, Jia F, Bai P, et al. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of gastric cancer. Tumour Biol. 2016;37:299-303. doi: 10.1007/s13277-015-3750-2. [DOI] [PubMed] [Google Scholar]
- 10. Shasttiri A, Rostamian Delavar M, Baghi M, Dehghani Ashkezari M, Ghaedi K. SNP rs10800708 within the KIF14 miRNA binding site is linked with breast cancer. Br J Biomed Sci. 2019;76:46-48. [DOI] [PubMed] [Google Scholar]
- 11. Yao L, Tak YG, Berman BP, Farnham PJ. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu MC, Kraft P, Epstein MP, et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Byun J, Han Y, Amos CI. Genetic interactions in lung cancer using machine-learning approaches in genome-wide association studies. Paper presented at: American Association for Cancer Research Annual Meeting 2019; March 29-April 3, 2019; Atlanta, GA. [Google Scholar]
- 15. Behravan H, Hartikainen JM, Tengström M, et al. Machine learning identifies interacting genetic variants contributing to breast cancer risk: a case study in Finnish cases and controls. Sci Rep. 2018;8:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liben-Nowell D, Kleinberg J. The link-prediction problem for social networks. J Am Soc Inf Sci Technol. 2007;58:1019-1031. [Google Scholar]
- 17. Sulaimany S, Khansari M, Masoudi-Nejad A. Link prediction potentials for biological networks. Int J Data Min Bioinform. 2018;20:161-184. [Google Scholar]
- 18. Wang P, Xu B, Wu Y, Zhou X. Link prediction in social networks: the state-of-the-art. Sci China Inf Sci. 2015;58:1-38. [Google Scholar]
- 19. Sulaimany S, Khansari M, Zarrineh P, et al. Predicting brain network changes in Alzheimer’s disease with link prediction algorithms. Mol Biosyst. 2017;13:725-735. doi: 10.1039/C6MB00815A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu H, Hu Z, Haddadi H, Tian H. Hidden link prediction based on node centrality and weak ties. Europhys Lett. 2013;101:18004. doi: 10.1209/0295-5075/101/18004. [DOI] [Google Scholar]
- 21. Huang Z, Li X, Chen H. Link prediction approach to collaborative filtering. Paper presented at: 5th ACM/IEEE-CS joint conference on digital libraries (JCDL ’05); June 7-11, 2005:141; Denver, CO New York, NY: ACM Press. doi: 10.1145/1065385.1065415. [DOI] [Google Scholar]
- 22. Martínez V, Berzal F, Cubero JC. A survey of link prediction in complex networks. ACM Comput Surv. 2016;49:1-33. doi: 10.1145/3012704. [DOI] [Google Scholar]
- 23. Valverde-Rebaza JC, de Andrade Lopes A. Link prediction in online social networks using group information. In: Murgante B, Misra S, Rocha AMAC, et al. , eds. Computational Science and Its Applications. Cham, Switzerland: Springer; 2014:31-45. doi: 10.1007/978-3-319-09153-2_3. [DOI] [Google Scholar]
- 24. Clauset A, Moore C, Newman MEJ. Hierarchical structure and the prediction of missing links in networks. Nature. 2008;453:98-101. [DOI] [PubMed] [Google Scholar]
- 25. Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525-530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oldridge DA, Wood AC, Weichert-Leahey N, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418-421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Von Mutius E, Drazen JM. Choosing asthma step-up care. N Engl J Med. 2010;362:1042-1043. doi: 10.1056/NEJMe1002058. [DOI] [PubMed] [Google Scholar]
- 28. Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635-110649. doi: 10.18632/oncotarget.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasan M, Al Zaki MJ. A survey of link prediction in social networks. In: Aggarwal C, ed. Social Network Data Analytics. Boston, MA: Springer; 2011:243-275. doi: 10.1007/978-1-4419-8462-3_9. [DOI] [Google Scholar]
- 30. Ding H, Wang Y, Chen Y, et al. Methylenetetrahydrofolate reductase tagging polymorphisms are associated with risk of non-small cell lung cancer in eastern Chinese Han population. Oncotarget. 2017;8:110326-110336. doi: 10.18632/oncotarget.22887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X, Shao M, Wang S, et al. Heterozygote advantage of methylenetetrahydrofolate reductase polymorphisms on clinical outcomes in advanced non-small cell lung cancer (NSCLC) patients treated with platinum-based chemotherapy. Tumour Biol. 2014;35:11159-11170. doi: 10.1007/s13277-014-2427-6. [DOI] [PubMed] [Google Scholar]
- 32. Hidaka A, Sasazuki S, Matsuo K, et al. CYP1A1, GSTM1 and GSTT1 genetic polymorphisms and gastric cancer risk among Japanese: a nested case-control study within a large-scale population-based prospective study. Int J Cancer. 2016;139:759-768. doi: 10.1002/ijc.30130. [DOI] [PubMed] [Google Scholar]
- 33. Koda M, Iwasaki M, Yamano Y, Lu X, Katoh T. Association between NAT2, CYP1A1, and CYP1A2 genotypes, heterocyclic aromatic amines, and prostate cancer risk: a case control study in Japan. Environ Health Prev Med. 2017;22:72. doi: 10.1186/s12199-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Assis J, Pereira C, Nogueira A, Pereira D, Carreira R, Medeiros R. Genetic variants as ovarian cancer first-line treatment hallmarks: a systematic review and meta-analysis. Cancer Treat Rev. 2017;61:35-52. doi: 10.1016/j.ctrv.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 35. Zhou J, Liu Z, Li C, et al. Genetic polymorphisms of DNA repair pathways influence the response to chemotherapy and overall survival of gastric cancer. Tumour Biol. 2015;36:3017-3023. doi: 10.1007/s13277-014-2936-3. [DOI] [PubMed] [Google Scholar]
- 36. Porchia L, Meneses-Sanchez P, Ruiz-Vivanco G, Perez-Fuentes R, Gonzalez-Mejia ME. CYP1A1 MspI polymorphism and cancer susceptibility among Latinos: a meta-analysis. Meta Gene. 2017;11:197-204. [Google Scholar]
- 37. Yadav U, Kumar P, Rai V. NQO1 gene C609T polymorphism (dbSNP: rs1800566) and digestive tract cancer risk: a meta-analysis. Nutr Cancer. 2018;70:557-568. doi: 10.1080/01635581.2018.1460674. [DOI] [PubMed] [Google Scholar]
- 38. Yin J, Wang X, Wei J, et al. Interleukin 12B rs3212227 T>G polymorphism was associated with an increased risk of gastric cardiac adenocarcinoma in a Chinese population. Dis Esophagus. 2015;28:291-298. doi: 10.1111/dote.12189. [DOI] [PubMed] [Google Scholar]
- 39. Aydin M, Sümbül AT, Camuz Hilaloğullari G, Bayram S. Genetic polymorphisms in human telomerase reverse transcriptase (hTERT) gene polymorphisms do not associated with breast cancer in patients in a Turkish population: hospital-based case-control study. Cell Mol Biol (Noisy-le-Grand). 2018;64:108-115. doi: 10.14715/cmb/2018.64.1.3. [DOI] [PubMed] [Google Scholar]
- 40. Nakao H, Wakai K, Ishii N, et al. Associations between polymorphisms in folate-metabolizing genes and pancreatic cancer risk in Japanese subjects. BMC Gastroenterol. 2016;16:83. doi: 10.1186/s12876-016-0503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ke Q, Liang J, Wang L, et al. Potentially functional polymorphisms of the vascular endothelial growth factor gene and risk of gastric cancer. Mol Carcinog. 2008;47:647-651. doi: 10.1002/mc.20435. [DOI] [PubMed] [Google Scholar]
- 42. Dixit R, Singh G, Pandey M, et al. Association of methylenetetrahydrafolate reductase gene polymorphism (MTHFR) in patients with gallbladder cancer. J Gastrointest Cancer. 2016;47:55-60. doi: 10.1007/s12029-015-9794-0. [DOI] [PubMed] [Google Scholar]
- 43. Sud A, Hemminki K, Houlston RS. Candidate gene association studies and risk of Hodgkin lymphoma: a systematic review and meta-analysis. Hematol Oncol. 2017;35:34-50. doi: 10.1002/hon.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zara-Lopes T, Gimenez-Martins APA, Nascimento-Filho CHV, et al. Role of MTHFR C677T and MTR A2756G polymorphisms in thyroid and breast cancer development. Genet Mol Res. 2016;15:gmr.15028222. doi: 10.4238/gmr.15028222. [DOI] [PubMed] [Google Scholar]
- 45. De Maturana EL, Rava M, Anumudu C, Sáez O, Alonso D, Malats N. Bladder cancer genetic susceptibility. A systematic review. Bladder Cancer. 2018;4:215-226. doi: 10.3233/BLC-170159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma LM, Ruan LH, Yang HP. Meta-analysis of the association of MTHFR polymorphisms with multiple myeloma risk. Sci Rep. 2015;5:10735. doi: 10.1038/srep10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soleimani E, Saliminejad K, Akbari MT, Kamali K, Ahani A. Association study of the common polymorphisms in the folate-methionine pathway with retinoblastoma. Ophthalmic Genet. 2016;37:384-387. doi: 10.3109/13816810.2015.1107596. [DOI] [PubMed] [Google Scholar]
- 48. Yang S, Lee J, Park Y, et al. Interaction between alcohol consumption and methylenetetrahydrofolate reductase polymorphisms in thyroid cancer risk: National Cancer Center cohort in Korea. Sci Rep. 2018;8:4077. doi: 10.1038/s41598-018-22189-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie S-Z, Liu Z-Z, Yu J, et al. Association between the MTHFR C677T polymorphism and risk of cancer: evidence from 446 case-control studies. Tumour Biol. 2015;36:8953-8972. doi: 10.1007/s13277-015-3648-z. [DOI] [PubMed] [Google Scholar]
- 50. Sun Y, Tan L, Li H, Qin X, Liu J. Association of NER pathway gene polymorphisms with susceptibility to laryngeal cancer in a Chinese population. Int J Clin Exp Pathol. 2015;8:11615-11621. doi: 10.1074/jbc.M110.127233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu B, Li J, Gao Q, Yu W, Yang Q, Li X. Laryngeal cancer risk and common single nucleotide polymorphisms in nucleotide excision repair pathway genes ERCC1, ERCC2, ERCC3, ERCC4, ERCC5 and XPA. Gene. 2014;542:64-68. doi: 10.1016/j.gene.2014.02.043. [DOI] [PubMed] [Google Scholar]
- 52. Lei C, Ruan J. A novel link prediction algorithm for reconstructing protein-protein interaction networks by topological similarity. Bioinformatics. 2013;29:355-364. [DOI] [PMC free article] [PubMed] [Google Scholar]