To the Editor:
In recent years, the need for enhanced rigour, reproducibility and transparency in biomedical research has been the topic of much discussion. The National Institutes of Health (NIH) led the way in this conversation with issuance of the NIH guidelines for rigour and reproducibility, which have been mandated for all grant applications since 2016 (1, 2). In support of this initiative, a recent review article by Hart and Powers, published in Diabetologia (3), with an accompanying editorial co-published in Diabetes and Diabetologia by Poitout et al (4, 5) highlighted the need for improved standardisation of human islet data and its reporting in scientific publications. Diabetologia and Diabetes have initiated a checklist that is now required at the time of manuscript submission. At a minimum, the source and isolation centre, a unique identifier number for each human islet preparation, and key demographic/clinical data must be included. Although not mandatory, the editorial encourages the inclusion of additional data, such as the cause of donor death, measurements of islet purity and viability, functional measures, ischaemia duration and culture time.
Since 2004, the Integrated Islet Distribution Program (IIDP) has served as the main source of human islets for research in the USA (6–8). Directed by Joyce C. Niland at the City of Hope National Medical Center in Duarte, CA, the IIDP is funded by the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) and the JDRF. To date, the IIDP has distributed over 250 million islets, serving more than 400 islet studies across nine different countries, with the resulting research yielding more than 640 publications. Thus, the IIDP is a major contributor to the ‘human islet research ecosystem’ as described by Hart and Powers (3), as well as an avid supporter of improved standards for reporting of human islet data.
In recognition of the growing need for improved assessment of human islet function and quality, the NIH and IIDP initiated the Human Islet Phenotyping Program (HIPP) in 2016. The HIPP programme is located at Vanderbilt University in Nashville, TN, and is directed by Marcela Brissova, with Carmella Evans-Molina, Co-Principal Investigator of the IIDP, serving as the HIPP liaison. The HIPP receives a sample of human islets from each IIDP-supported islet isolation. As depicted in the analysis workflow (Fig. 1a), the HIPP conducts post-shipment assessment of islet morphology, viability, and software-guided measurement of islet purity and number. These data are then integrated with in vitro islet functional analysis by perifusion, which simultaneously captures insulin and glucagon secretory profiles with high temporal resolution. Because human islet preparations vary in terms of their composition, the islet functional data can be further correlated with composition of three major islet cell types, alpha, beta, and delta cells, measured independently by immunofluorescence. When possible, the HIPP also collects and stores samples for future investigation, including frozen samples of handpicked islets (for RNA and protein analysis) and frozen samples of islet preparations (for DNA analysis). The sample resource is under development and will become available in the near future. In December 2018, the HIPP data were released to IIDP-affiliated investigators. To date, the HIPP has analysed nearly 200 islet preparations from a demographically diverse pool of donors (Fig. 1b). Synthesis of the rich data set generated by this unique resource can be leveraged by investigators to provide context for their own experiments and analyses, and their resulting data could potentially be integrated with IIDP–HIPP data in the future. Most importantly, the HIPP workflow yields standardised data that will enhance transparency and quality in human islet research.
Fig. 1.
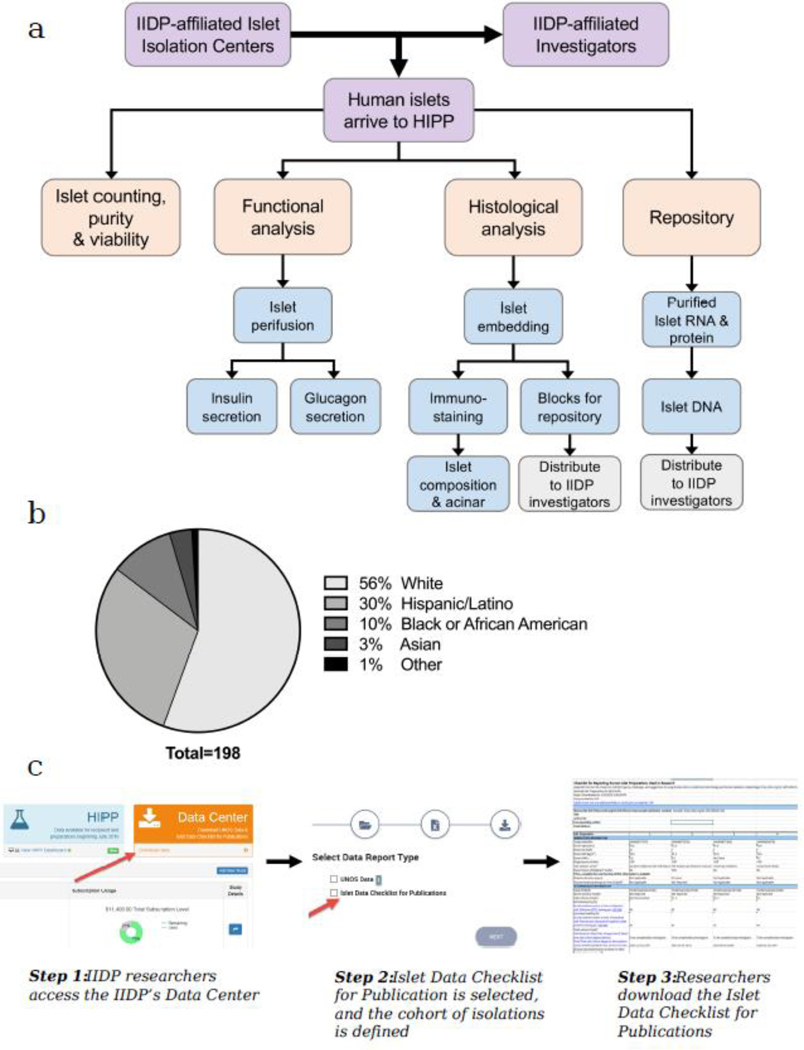
The IIDP–HIPP integrates a broad spectrum of islet phenotypic features with donor characteristics. (a) Analysis workflow performed by the HIPP on human islets from each IIDP-supported islet isolation. Human islets from the same preparation are distributed to HIPP and IIDP-affiliated investigators. (b) The IIDP–HIPP database contains data from a large and demographically diverse pool of islet donors. Shown is the racial background of islet donors for 198 preparations analysed by HIPP since 2016. (c) Steps required for automated publication checklist generation from the IIDP Data Center. The IIDP–HIPP database and the automated online tool for checklist generation can be accessed by authorised IIDP users through the dashboard of the IIDP password-protected website (https://iidp.coh.org/secure/isletavail/)
As another recent enhancement to assist investigators in meeting the new mandatory criteria for reporting of human islet data as put forth by Diabetologia and Diabetes, the IIDP has developed an automated online tool to compile and export the mandated information for manuscript submission. All of the required data have been collected by our IIDP-affiliated islet isolation centres since January 2010, and are available to investigators so that the information can be provided when submitting manuscripts to Diabetes/Diabetologia (and, possibly, additional journals in the future).
Data on the functional assessment of the islet preparations, classed as ‘recommended information’ in the reporting checklist, has been provided by the HIPP since July 2016. Using this online reporting tool, investigators will be able to directly export islet checklist data into an Excel spreadsheet that can be uploaded to the journal. Islet preparations are selected for inclusion in the checklist based on a unique Research Resource Identifier [RRID] (9) (Fig. 1c). Both the IIDP–HIPP database and the automated online tool for checklist generation can be accessed by authorised IIDP users through the dashboard of the IIDP password-protected website (https://iidp.coh.org/secure/isletavail/).
Whereas efforts towards standardised reporting of human islet data are a critical first step in ensuring rigour and reproducibility in diabetes research, an unintended consequence of any new mandate can be an additional burden on investigators both in time and resources. Our hope is that both the data provided by the IIDP isolation centres pre-shipment and the rich phenotyping data provided by the HIPP post-shipment can assist investigators in meeting these new requirements. Other programmes have begun similar initiatives to share phenotyping and multi-omics data from isolated islets, including the Human Pancreas Analysis Program (HPAP) of the Human Islet Research Network (HIRN) and the Islet Core at the Alberta Diabetes Institute. Making these types of centrally collected data available can both enhance rigour and reproducibility and accelerate scientific discoveries. In addition, they provide cost savings to the diabetes research community by reducing the need for individual investigators to collect the same type of information and/or perform redundant assays. We believe that the recent IIDP–HIPP initiatives described here will help to set new standards in human islet research, and we are pleased to offer solutions that improve human islet research and satisfy new journal reporting requirements, without adding undue burden to individual investigators.
Acknowledgements
The authors would like to acknowledge R. Aramandla, V. DiBacco, H. Durai, A. Coldren, S. Mei, G. Poffenberger and C. Reihsmann at the Vanderbilt University Medical Center, for their assistance with the development of HIPP workflows and the design of the IIDP–HIPP database. We acknowledge J.-N. Chuang (Database Administrator), and M. Pineda and H. Sibley (Senior Software Developers), all from the Department of Diabetes & Cancer Discovery Science, Diabetes and Metabolism Research Institute, City of Hope National Medical Center, for their work in developing the IIDP–HIPP database and the Islet Data Checklist for Publication. In addition, the authors would like to acknowledge K. Abraham (Project Scientist for the IIDP) of NIDDK, for her advice and guidance on this project.
Funding
The Integrated Islet Distribution Program is funded by National Institute of Diabetes and Digestive and Kidney Diseases (UC4DK098085) with additional support from the JDRF (1-RSC-2019–712-S-B). We also acknowledge support from the Vanderbilt Diabetes Research and Training Center (P60 DK020593) in providing resources, core infrastructure and support for HIPP-related activities.
Abbreviations
- HIPP
Human Islet Phenotyping Program
- IIDP
Integrated Islet Distribution Program
- NIH
National Institutes of Health
Footnotes
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
This Editorial is being simultaneously published in Diabetologia and Diabetes.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES:
- 1.Collins FS, Tabak LA (2014) Policy: NIH plans to enhance reproducibility. Nature 505(7485):612–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Institutes of Health. Rigor and reproducibility. Available from https://grants.nih.gov/policy/reproducibility/guidance.htm. Accessed 4 Apr 2019
- 3.Hart NJ, Powers AC (2019) Use of human islets to understand islet biology and diabetes: progress, challenges and suggestions. Diabetologia 62(2):212–22. doi: 1007/s00125-018-4772-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poitout V, Satin LS, Kahn SE, et al. (2019) A call for improved reporting of human islet characteristics in research articles. Diabetes 68(2):239–40. doi: 10.2337/dbi18-0055. [DOI] [PubMed] [Google Scholar]
- 5.Poitout V, Satin LS, Kahn SE, et al. (2019) A call for improved reporting of human islet characteristics in research articles. Diabetologia 62(2):209–11. doi: 10.1007/s00125-018-4784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaddis JS, Olack BJ, Sowinski J, Cravens J, Contreras JL, Niland JC (2009) Human pancreatic islets and diabetes research. 301(15):1580–7. doi: 10.1001/jama.2009.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaddis JS, Hanson MS, Cravens J, et al. (2013) Standardized transportation of human islets: an islet cell resource center study of more than 2,000 shipments. Cell Transplant. 22(7):1101–11. doi: 10.3727/096368912X653219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niland JC, Stiller T, Cravens J, Sowinski J, Kaddis J, Qian D. (2010) Effectiveness of a web-based automated cell distribution system. Cell Transplant 19(9):1133–42. doi: 10.3727/096368910X505486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandrowski A, Brush M, Grethe JS, et al. (2016) The resource identification initiative: a cultural shift in publishing. Neuroinformatics 14(2):169–82. doi: 10.1002/brb3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]


