Abstract
Numerous genes, and alterations in their expression, have been identified as risk factors for developing levodopa-induced dyskinesia (LID). However, our understanding of the complexities of molecular changes remains insufficient for development of clinical treatment. In the current study we used gene array, in situ hybridization, immunohistochemistry, and microdialysis to provide a unique compare and contrast assessment of the relationship of four candidate genes to LID, employing three genetically distinct rat strains (Sprague-Dawley (SD), Fischer-344 (F344) and Lewis-RT.1) showing differences in dyskinesia susceptibility and ‘first-ever LID’ versus ‘chronic LID’ expression in subjects displaying equal dyskinesia severity. In these studies, rat strains were easily distinguishable for their LID propensity with: 1) a majority of SD rats expressing LID (LID+) and a subset being resistant (LID−); 2) all F344 rats readily developing (LID+); and 3) all Lewis rats being LID-resistant (LID−). Following chronic levodopa, LID+ SD rats showed significant increases in candidate gene expression: Nr4a2/(Nurr1) >> Trh > Inhba = Fosb. However, SD rats with long-standing striatal dopamine (DA) depletion treated with first-ever versus chronic high-dose levodopa revealed that despite identical levels of LID severity: 1) Fosb and Nurr1 transcripts but not protein were elevated with acute LID expression; 2) FOSB/ΔFOSB and NURR1 proteins were elevated only with chronic LID; and 3) Trh transcript and protein were elevated only with chronic LID. Strikingly, despite similar levodopa-induced striatal dopamine (DA) release in both LID-expressing F344 and LID-resistant Lewis rats, Fosb, Trh, Inhba transcripts were significantly elevated in both strains; however, Nurr1 mRNA was significantly increased only in LID+ F344 rats. These findings suggest a need to reevaluate currently accepted genotype-to-phenotype relationships in the expression of LID, specifically that of Fosb, a transcription factor generally assumed to play a causal role, and Nurr1, a transcription factor that has received significant attention in PD research linked to its critical role in the survival and function of midbrain DA neurons but who’s striatal expression, generally below levels of detection, has remained largely unexplored as a regulator of LID. Finally these studies introduce a novel ‘model’ (inbred F344 vs inbred Lewis) that may provide a powerful tool for investigating the role for ‘dyskinesia-resistance’ genes downstream of ‘dyskinesia-susceptibility’ genes in modulating LID expression, a concept that has received considerably less attention and offers a new ways of thinking about antidyskinetic therapies.
Keywords: Levodopa-induced dyskinesias, Parkinson’s disease, Genes, Striatum, FosB, Nurr1
Introduction
Levodopa-induced dyskinesia (LID), motor complications of long-term dopamine (DA) replacement therapy in Parkinson’s disease (PD), remain an unmet medical need. While accounts of the occurrence of LID vary, they are estimated to occur in 50% of patients after approximately 3–5 years of treatment [1, 2], a time roughly corresponding to virtually a complete loss of striatal DA terminals [3], with the incidence of LID increasing to approximately 90% after 10 years [1, 2]. Further, while some patients develop LID early in their disease, others develop it later and a small percent never do. Similar findings have been observed in preclinical rodent models of LID [4–6]. Inarguably two key parameters necessary for the induction of LID, regardless of the species, are the extent of DA depletion and DA replacement therapy. It is notable however that severe striatal DA depletion and non-physiological DA replacement alone are insufficient to induce LID.
An abundance of genomics, transcriptomics and proteomics studies have revealed alterations of distinct genes or proteins within the striatum of dyskinetic versus non-dyskinetic parkinsonian subjects (e.g.: Table 2), suggesting that expression of one or more specific molecular triggers may induce or permit pathological changes underlying long-term adaptations of striatal circuits that result in LID. Despite a plethora of data, a void remains in successfully deciphering complex gene-related changes to allow identification of successful therapeutic targets. To further understanding of this complex issue, we report here a unique approach of comparing and contrasting gene-related changes using three genetically distinct rat strains with variable characteristics of LID susceptibility, and to the best of our knowledge, the first report of gene expression changes in rats with ‘first-ever LID’ versus ‘chronic LID’ expression in subjects displaying equal dyskinesia severity.
Table 2.
Highlights of LID-related ‘Omic Data
| # of DE Genes | Analysis Method | Citation | Highlighted Gene(s)/Modifications | Pathological Focus | Brain Region | Species | Levodopa |
|---|---|---|---|---|---|---|---|
| 709 | Transcriptome | Charbonnier-Beaupel et al., 2015 [68] | Nptx2 (Narp) | Clustering of glutamate receptors | Striatum | Mouse (C57BL/6J, M&F) | 5, 10, 20 mg/kg; 1, 2, or 15 days; sac’d 1–6 hrs post LD; with acute LD, no dyskinesias reported |
| 143 (LID+ vs LID−) | RNA-seq | Smith et al., 2016 [69] | Dynein complex genes, Integrin (Itgb4), actin (Acta1), Arc | Alterations in synapse architecture (number) and function (potentiation) | Striatum | Rat (Wistar, M) | 4 mg/kg; 2×/day for 3 wks; sac’d 60 mins post LD |
| 497 | Whole Genome Microarray | Wang et al., 2014 [71] | CYP4A1, APOA5, GABRA2, GRIA1, GRM5, NAPEPLD | Retrograde endocannabinoid signaling and related lipid metabolism processes | Striatum | Rat (SD, M) | 6 mg/kg; 1× daily; 21 days; sac’d 12 hrs post LD |
| 3100L 4603H |
Cell-type–Specific mRNA Profiling (TRAP) | Heiman et al., 2014 [13] | *** Gpr39, Fndc9, Cstb, Trh, Srxn1, Ier3, Tinf2, Cdk11b, Nr4a2 (Nurr1), Itch, Scp2, Fosl1 (Fra-1) | Dysregulation in direct pathway SPNs | Striatum | Mouse (BAC tg C57bl/6J, M) | LLow dose: 1–2 mg/kg daily, 9 days; HHigh dose: 3–6 mg/kg daily, 9 days; sac’d 2 hrs post LD |
|
19,389† 53A 71C |
ChIP-seq & RNA-seq | Sodersten et al., 2014 [14] | H3K27me3S28p positive genes | Reduced PcG repression of genes including NrA2/Nurr1 and Trh | Striatum | Mouse (C57bl/6J, F) | 10 mg/kg 1× daily; A1, or C3, C9 days; with acute LD, no dyskinesias reported |
| 55A 137C |
Transcriptome | El Atifi-Borel et al., 2009 [72] |
Apthrs, Kroz-20, CaMKIIb Cneuroserpin, Irp94, calcineurin A,CSNAP-25, VDAC3, MAP1LC3, CLDN5 |
Transcriptomic differences between acute and chronic L-DOPA treated rats | Striatum | Rat (Wistar, M) | A50 mg/kg 1× or C50 mg/kg 2×/day for 10 days; all sac’d 2 hrs post LD; with acute LD, no dyskinesias reported |
| 46 | Transcriptome | Konradi et al 2004 [5] | CA2+ homeostasis and signaling, GABAergic signaling, ATP production, ribosomal proteins | Transcriptomic differences between LID+ versus LID− rats | Striatum | Rat (SD, F) | 6mg mg/kg, 1× daily for 22 days |
| ∼20,000DMRs | DNA Methylation | Figge et al 2016 [73] | DNA demethylase expression (Tet3, Gad45b), LID-associated demethylated genes/regions (Ntrk2, Dab1, Esr1, Grin2a, Nedd4l, Rasgrp1, Arc, FosB, Rasip) | Genome-wide differential DNA methylation in LID | Striatum | Rat (SD, M) | 6 mg/kg 1× daily for 7 days |
| 67 | Proteomic | Valastro et al 2007 [74] | CRYAB, gamma-enolase, vinculin, GAMT, proteasome α-2 subunit | Differential protein expression in L-DOPA and DA agonist treated rats | Striatum | Rat (SD, M) | 6 mg/kg 1× daily for 21 days |
| NA | Histone modifications | Nicholas et al 2008 [75] | ↓H3 trimethylation, ↑H4 deacetylation | LID-associated changes in histone modifications across species | Striatum | Mouse (C57bl/6J, M) NHP (macaque) |
Mice: 200 mg/kg 2×/day for 3 days NHP: 25 mg/kg 2×/day for 120 days |
KEY:
Genes with highest correlations between expression and LD dose in dSPNs
H3K27me3 & H3K27me3 S28p positive genes
L= low dose levodopa
H= high dose levodopa
A= acute levodopa without LID
C= chronic levodopa with LID
DMRs= differentially methylated regions
To begin, we used an Affymetrix gene array and the outbred Sprague-Dawley (SD) rat strain to first identify, in our hands, the most abundantly altered LID-associated genes following chronic treatment with high-dose levodopa. SD rats are a commonly used strain for LID research because they allow comparison of presumed LID-specific gene changes between those rats that readily develop LID (LID+) and those that do not express LID (LID−) despite equivalent levels of severe striatal DA depletion and chronic levodopa administration [5, 6]. Based on gene array data from our chronically treated SD rats, together with previous reports, we chose to focus subsequent analyses on four LID-associated candidate genes: Fosb (e.g.: [7–12]), nuclear receptor related 1 protein (Nurr1/Nr4a2) [13, 14], thyrotropin releasing hormone (Trh) [15] and inhibin βA (Inhba) [13].
We next examined these candidate transcripts and protein expression levels between SD rats treated with ‘first-ever’ (i.e.: acute) versus chronic high-dose levodopa, using a paradigm that, based on the timing and degree of DA depletion, critically produced equivalent levels of LID severity in both groups. In this second study we tested the hypothesis that animals expressing equivalent LID severity in response to either ‘first-ever’ or chronic levodopa would display equivalent expression of critical molecular triggers underlying this behavior.
For the third study in this investigation, we capitalized on the fact that inbred rat strains have been useful preclinical models in understanding genetic factors that may underlie differences in behavioral phenotypes based on their homogenous genotype [16]. Lewis and Fisher-344 (F344) rats have been abundantly studied for their differences in response to psychostimulant addiction liability [16–19]. Important to the current investigation, both psychostimulant addiction and LID are thought to result from aberrant associative or motor learning, respectively, and rewiring of basal ganglia neural circuits following modulation and ‘priming’ of midbrain DA systems. As F344 rats are generally considered “addiction-resistant” and Lewis rats “addiction-prone” (e.g.: [19]), we hypothesized that these inbred strains would also exhibit similar divergent behavioral and transcript responses to levodopa.
Materials and Methods
Experimental Subjects
Adult male Sprague Dawley, F344 and Lewis RT.1 rats (Harlan, Indianapolis, IN; 225–275 g at time of lesion surgery) were employed in three parallel but distinct studies (i.e.: Experiments #1, 2, 3a and 3b) as detailed in Table 1. Rats were kept on a 12h light/dark cycle, and given food and water ad libitum. All studies were approved by the Institutional Animal Care and Use Committee at Michigan State University (MSU), the Van Andel Institute (VAI), and State University of New York (SUNY) Binghamton where these studies took place. All work was performed in accordance with the ethical standards established by the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Table 1.
Three parallel but distinct studies (i.e.: Experiments 1, 2, 3a and 3b) used to examine the relationship of genotype-to-phenotype expression in LID.
| Experiment ID | Experiment Name | Rat Strain | Principal Experimental Endpoint |
|---|---|---|---|
| 1 | Gene Array Exploration | Sprague Dawley | Differential Gene Expression in LID+ vs LID− Subjects |
| 2 | Chronic LID+ vs Acute LID+ | Sprague Dawley | Differential Transcript and Protein Expression in Chronically vs Acutely Dyskinogenic Subjects |
| 3a | F344 LID+ vs Lewis LID− | F344 & Lewis | Differential LID and Transcript Expression and DA & 5-HT content in F344 LID+ vs Lewis LID− Subjects |
| 3b | F344 LID+ vs Lewis LID− | F344 & Lewis | Differential Extracellular Striatal DA, 5-HT, & NE in F344 LID+ vs Lewis LID− Subjects |
6-OHDA Lesions
All rats were rendered unilaterally parkinsonian with a stereotaxic injection of the neurotoxin 6-hydroxydopamine (6-OHDA). Dosage of 6-OHDA was defined for each rat strain as that necessary to obtain a comparable final degree of nigrostriatal DA loss (≥ 90%) for all animals. Lewis rats have been reported to have lower dopamine transporter (DAT) levels in striatum (Gulley et al., 2007; Flores 1998) and as such we empirically determined that a slightly higher dose of 6-OHDA was necessary in this rat strain compared to the F344 and SD rats to meet the criteria of ≥ 90% nigrostriatal DA depletion. To produce equal levels of striatal DA loss, F344 and SD rats received a dose of 5μg 6-OHDA hydrobromide/μl and Lewis rats 7μg 6-OHDA hydrobromide/μl in 0.02% ascorbic acid in sterile saline. Rats were anesthetized with inhalant isoflurane (2–3%; Sigma, St. Louis, MO) and secured in a stereotaxic frame (David Kopf Instruments, Tujunga, CA, USA). Each rat received 2μl of 6-OHDA at a flow rate of 0.5μl/minute (min) into the substantia nigra pars compacta (SNpc; 4.8mm posterior, 1.7mm lateral, and 8mm ventral to bregma) and the median forebrain bundle (MFB; 4.3mm posterior, 1.6mm lateral, and 8.4mm ventral to bregma), an approach routinely used in our labs to reliably produce the severe (≥95%) nigrostriatal dopamine depletion needed to model LID (e.g.: [5, 6, 20, 21]). At completion of the surgery rats received carprofen (as Rimadyl®; 5mg/kg, 0.1cc/100g body weight) as analgesic treatment. For consistency, all lesion surgeries and levodopa priming treatments were performed at MSU by the same team of investigators.
Lesion Assessment Behavioral Evaluation
Estimation of nigrostriatal DA lesion success prior to chronic levodopa treatment was done in all rats using a combination of drug-free behavioral tests including vibrissae test [22], cylinder test [23], tail hang [24], and/or forepaw bracing [22, 25] tests. In the F344 and Lewis rats amphetamine (5.0mg/kg, i.p.; Sigma-Aldrich, St. Louis, MO) rotational response was used as an initial behavioral indicator of potential differential striatal DA release between strains, which was used also as an additional indicator of lesion success. These behaviors were assessed 2–3 weeks after 6-OHDA in all rats.
Levodopa Priming and Dyskinesia Assessment
Approximately 4 to 5 weeks after 6-OHDA, all rats were single housed in cages with enrichment materials at the initiation of daily injections (M-Fr). All rats received high dose levodopa (levodopa methyl ester 12mg/kg plus 12mg/kg benserazide, Sigma-Aldrich, St. Louis MO) as detailed below. Abnormal involuntary movements (AIMs) were scored according to a LID severity rating scale for rats developed in our laboratories based on specific criteria reflective of the nature and occurrence of multiple behavioral attributes of dyskinesia as previously detailed [10, 26, 27]. Briefly, as detailed previously [10, 26, 27] both the intensity (0 = absent, 1 = mild, 2 = moderate, 3 = severe) and frequency (0= absent, 1 = 50% of rating period, 2 = 50% of the rating period, 3=constant/uninterruptable) were assessed for specific attributes of LID behaviors including: forelimb hyperkinesia, forelimb dystonia, hindlimb dystonia, trunk dystonia, neck dystonia, and orolingual dyskinesia (e.g.: https://www.youtube.com/watch?v=qvENE02Kiwo; [28]). A dyskinesia severity score for a given individual component of LID was obtained by multiplying frequency times intensity. A total daily severity score for each animal was computed by adding the severity scores of all individual components of LID behavior. Experimental animals were randomized at the beginning of the experiment and rated in the same order and by the same blinded investigator throughout each experiment.
For Experiment (Exp.) 1 (‘Gene Array Exploration’) the experimental timeline and groups are shown in Figure 1A, B. Unilaterally parkinsonian SD rats received daily injections with 12mg/kg levodopa (plus 12mg/kg benserazide) and LID behaviors were rated for a total of 7 sessions 50 mins after levodopa to differentiate LID-positive (LID+) and LID-negative (LID−) subjects (Fig. 1). For Exp. 2 (‘Chronic LID vs Acute LID’) expression patterns were compared in SD rats with: 1) long-term striatal DA depletion, naïve to levodopa 2) long-term striatal DA depletion plus a single dose levodopa and LID expression, and 3) long-term striatal DA depletion plus long-term levodopa treatment and LID expression. Rats receiving chronic levodopa were evaluated for their dyskinetic response once during weekly during the 3 weeks of levodopa priming (Fig. 2) in order to identify LID+ rats for comparison to rats exhibiting similar levels of LID in response to a single acute levodopa dose. For Exp. 3a (‘F344 LID+ vs Lewis LID−’) unilaterally lesioned F344 and Lewis rats were evaluated for LID behaviors on days 1, 7 and 15 during the 3 weeks of levodopa priming with 12mg/kg levodopa (plus 12mg/kg benserazide). In the second cohort, Exp. 3b, designated for in vivo microdialysis, F344 and Lewis rats were rendered unilaterally parkinsonian, amphetamine tested and levodopa primed in an identical manner as in Experiment 3a however, they were then transported from MSU to SUNY Binghamton one day after the final analyses. The SUNY investigators were blinded to LID priming scores and species identification until after Experiment 3b rats were sacrificed.
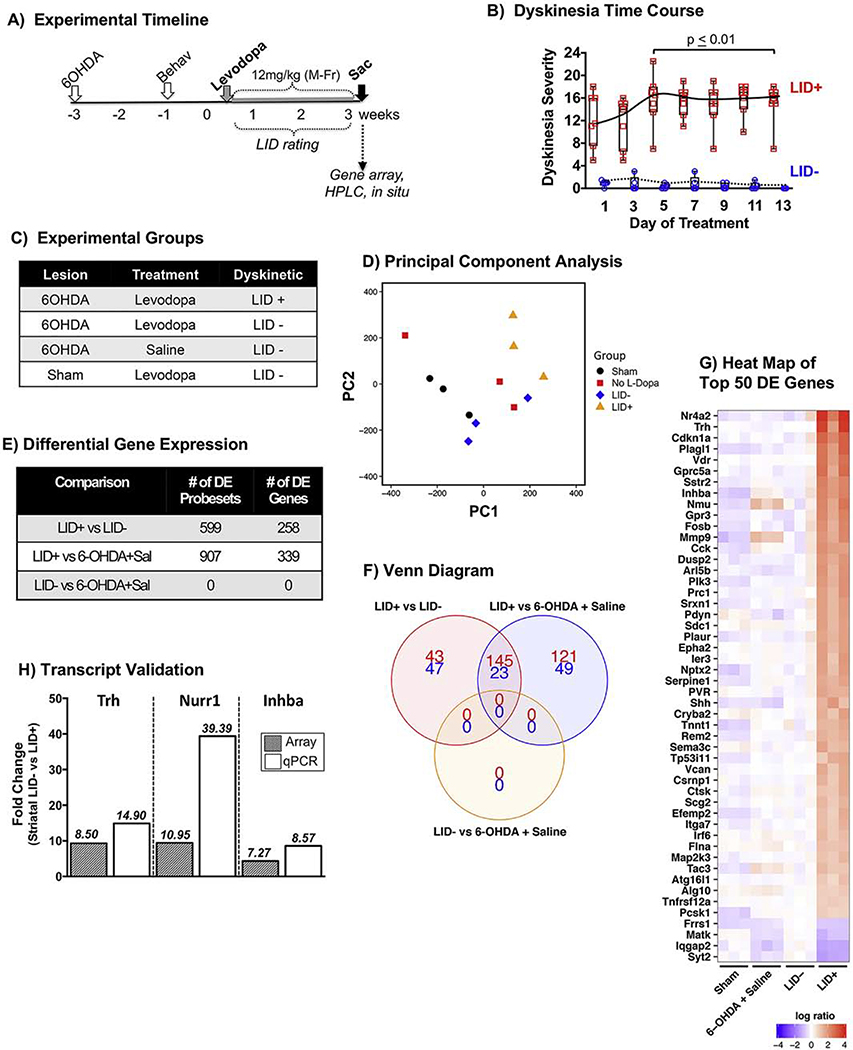
Figure 1. Differential gene expression in response to chronic levodopa in parkinsonian Sprague Dawley (SD) rats expressing dyskinesia (LID+) vs non-responders (LID−).
A) Experimental timeline. Adult male SD rats were rendered unilaterally parkinsonian with 6-OHDA delivered into the SN and MFB. Lesion status was confirmed 2 weeks post-toxin with drug-free tests as described in the methods. Rats began daily (Mon-Fri) treatment with levodopa (12mg/kg + 12mg/kg benserazide) for 2.5 weeks. B) Time course of LID in ‘responders’ and ‘non-responders’ (LID+ N=7, LID− N=5; Kruskal-Wallis, Dunn’s multiple comparison post-hoc test). C) Experimental groups. Gene expression levels were assessed from the striatum of parkinsonian rats that were: treated with levodopa and developed dyskinesias (N=3, LID+), treated with levodopa and remained resistant to dyskinesias (N=3, LID−), treated with saline (N=3, 6-OHDA + Saline); and in intact rats treated with levodopa (Sham lesion + Saline). D) Principal component analysis showing the overall relationship between groups. E) Differential expression was performed for all pairwise comparisons at the probeset level. Significant probesets (FDR adjusted p-value < 0.05) were subsequently collapsed by gene prior to visualization of the results. There were 258 genes that showed differential expression between LID+ and LID− striatum. F) Venn diagram of the three comparisons showing the total number of differentially expressed genes and their direction of change (red/top # = up-regulated; blue/bottom # = down-regulated). G) Heatmap of the top 50 differentially expressed genes between LID+ and LID− rats. Color indicates the magnitude (per ‘log ratio’ legend) and direction of change between the expression of each gene for the N=3 subjects in each treatment group as compared to the LID− rats (red=increased expression; blue= decreased expression; white= no significant change). H) qPCR validation of the top 3 transcripts showing differential expression (DE) between LID+ and LID− subjects. There is a high degree of agreement between the microarray and qPCR DE for Trh and Inhba. However, DE of Nr4a2 in the striatum of LID+ subjects was much greater when quantified by qPCR (39.39 fold) versus that observed with the microarray (9.42 fold) likely due to saturation of the Nr4a2 signal on the microarray chip, which would result in an underestimation of the differential upregulation of this transcript.
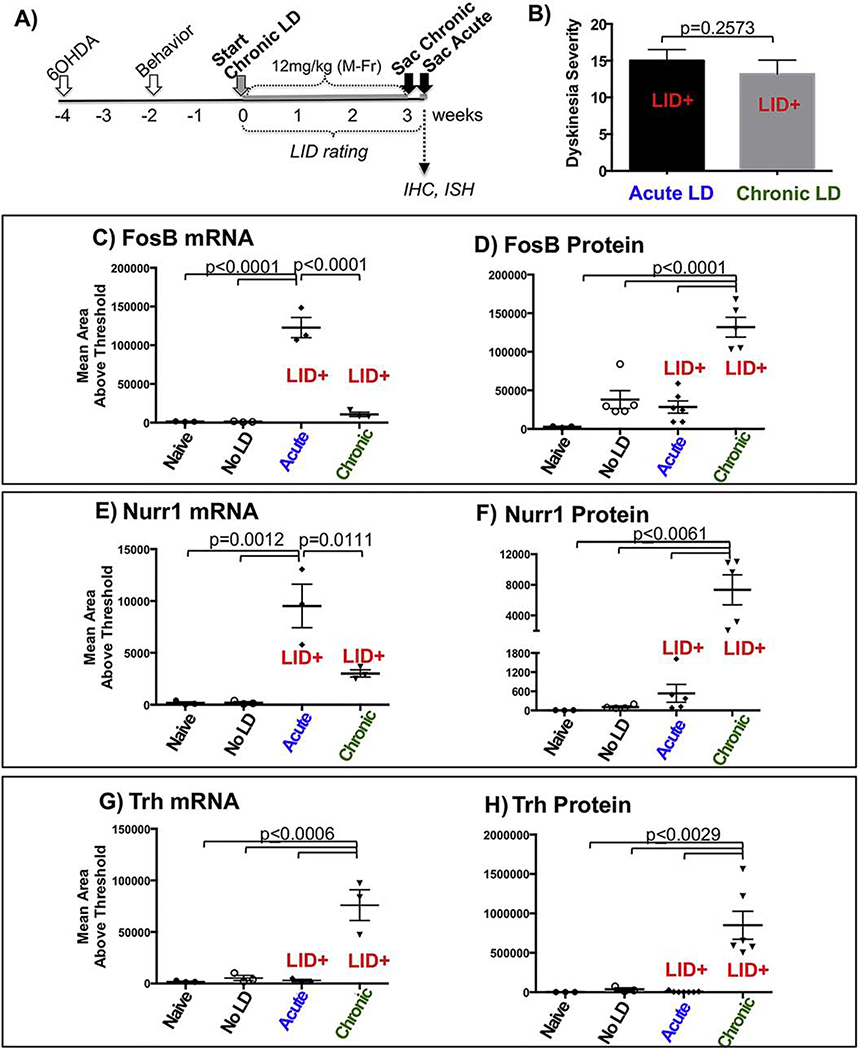
Figure 2. Comparable dyskinesia severity and differential candidate gene profiles in SD rats exposed to first-ever versus chronic levodopa.
A) Experimental timeline. Adult male SD rats were rendered unilaterally parkinsonian with 6-OHDA delivered into the SN and MFB and lesion confirmed with drug-free behavioral testing 2 weeks later. The chronically treated rats received daily (Mon-Fri) levodopa (12mg/kg + 12mg/kg benserazide) for 3 weeks. Approximately 7 weeks post-lesion, rats receiving a single acute injection or those treated with chronic levodopa were given a test dose of levodopa (12m/kg) and sacrificed approximately 90 mins post-levodopa. B) LID behavioral data showing equivalent levels of dyskinesia severity (p=0.257, Mann-Whitney U test; chronic (N=6), acute (N=12)). Transcript and protein levels of Fosb (C,D), Nurr1 (E,F), and Trh (G,H) were quantified in subsets of first-ever LID+ and chronic LID+ rats as well as parkinsonian naïve rats which served as baseline data (N=3–6 as depicted in graphs C-H; p-values indicated in these graphs derived from one-way ANOVA with multiple comparison).
Guide Cannulae Surgeries and In Vivo Microdialysis (Exp. 3b)
As previously detailed by [29], 1 week after arrival, rats that were previously DA-lesioned and primed with levodopa for 3 weeks were anesthetized with inhalant isofluorane (2–3%; Sigma) in oxygen and placed in a stereotaxic apparatus (David Kopf Instruments, Tujunga, CA, USA). Rats were fitted unilaterally with plastic microdialysis guide cannulae (CMA 12 Elite; Stockholm, Sweden) targeting the dorsal striatum ipsilateral to lesion (AP, 1.2 mm; ML, 2.8 mm; DV, −3.7 mm; relative to bregma; Paxinos & Watson, 1998). Cannulae were positioned and affixed to the skull with screws and liquid and dental acrylic (Lang Dental, Wheeling, IL). At the completion of surgery, animals were single housed in enriched cages and allowed to recover with ad libitum food and water. Pre-surgery and 1 day post-surgery, rats received Buprenex (buprenorphine HCl; 0.03 mg/kg, i.p.; Reckitt Benckiser Pharmaceuticals Inc., Richmond, VA) as analgesic treatment. Soft chow was also provided and rats were monitored for 7 days post-surgery to ensure full recovery. Three rats did not survive post-surgical care and final groups were carried on through testing (F344, n = 9; Lewis, n = 6).
Approximately 10 days after cannulation surgery, rats were injected with levodopa (levodopa methyl ester 6 mg/kg s.c. + benserazide 15 mg/kg s.c.; Sigma) and LID behaviors quantified; specifically axial, limb, and orolingual abnormal involuntary movements (ALO AIMs) every 10 min for the following 3 h. Microdialysis tested commenced approximately 3 days later. On the night before testing, striatal probes (CMA 12 Elite; membrane length = 3 mm; 20000 Dalton; Stockholm, Sweden) were inserted into the rats’ guide cannulae and locked into place so that the dialysis membrane extended −3.7 to −6.7 mm ventral to bregma. The next day, rats were habituated for 1 h with artificial cerebral spinal fluid (aCSF; 2.0 μL/min; in mM: 128 NaCl, 2.5 KCl, 1.3 CaCl2, 2.1 MgCl2, 0.9 NaH2PO4, 2.0 Na2HPO4, and 1.0 glucose, brought to a pH of 7.4). At this point, rats received a systemic treatment injection of vehicle (0.9% NaCl + 0.1% ascorbic acid, s.c.) to determine injection-induced changes and striatal dialysate samples were collected every 20 min for 3 h to determine baseline levels of norepinephrine (NE), DA, and 5-HT. After baseline, rats received systemic injections of levodopa (6 mg/kg) followed by sample collection, LID ratings and rotations counting every 20 min for 3 h (Figs. 1 and 7G, H). This lower dose of levodopa (compared to the 12mg/kg priming dose) was used for microdialysis to allow detection of potential differences in DA release between strains free from possible saturation effects that might have occurred with higher doses.
Euthanasia and Tissue Collection
In all studies rats were sacrificed to allow examination of experimental endpoints during peak LID expression (i.e.: 90–120 minutes after a final levodopa injection). In experiments involving gene array and/or high performance liquid chromatography (HPLC) (Exp. 1 & 3), rats were anesthetized with Beuthanasia (100mg/kg; Merck Animal Health, Madison, NJ) 120 minutes after a final levodopa injection and decapitated. Brains were rapidly removed (<1min) and submerged for 30 seconds (s) in a 250 ml beaker of isopentane super-cooled in powdered dry ice. The brains were stored at −80°C until time of dissection. At time of dissection, frozen brains were warmed to approximately −20°C by placing them into a −20°C freezer for 1–2 hours (h) prior to ‘punch dissection’. Brains were blocked caudally to remove the cerebellum, affixed to a cryostat chuck and placed into a −15°C cryostat (Microm HM 560, Thermo-Scientific, Waltham, MA). To obtain striatal tissue, brain tissue rostral to the striatum was removed by sectioning the brain until anterior aspect of the striatum (~1.8mm anterior to bregma) was visible.
Using specifically designed tissue punches, frozen striatal tissue was extracted for gene array or HPLC. For HPLC, a punch of striatal tissue 1mm wide x 2mm deep from the centromedial striatum was extracted with a slightly lateral trajectory from rostral to caudal using a15G tissue punch (Fine Science Tools, Inc., I.D. 1mm). Tissue from both the lesioned and unlesioned striata were placed into empty 1ml plastic eppendorf tubes (Eppendorf, Hamburg, Germany), frozen on dry ice and stored at −80°C until time of assay. For microarray analysis, a 2mm wide x 2mm deep cylinder of striatal tissue was extracted, placed in 1ml of trizol, (Life Technologies, Carlsbad, CA), homogenized by hand with a disposable plastic pestle, frozen on dry ice and stored at −80°C in preparation for RNA isolation.
Following punch extraction of striatal tissue from the Exp. 3a subjects, the remaining brain was immersion fixed in 4% buffered paraformaldehyde for 24 hours at 4°C followed by immersion in 30% sucrose. Sections were cut at 40 μm using a sliding microtome thru the extent of the striatum to collect tissue for RNAScope® in situ hybridization (ISH).
Rats in Exp. 2 (Chronic LID+ vs Acute LID+) were were anesthetized with Beuthanasia (100mg/kg; Merck Animal Health, Madison, NJ) 90 minutes after a final levodopa injection and perfused with 0.1 M phosphate-buffered saline (0.1 M PBS) followed by 4% paraformaldehyde (PF) per our usual protocol [6, 30] to allow for examination of transcript using ISH and protein using immunohistochemistry (IHC).
Microarray Gene Analysis
RNA extraction was performed using the RNA Clean and Concentrator kit (Zymo Research, Irvine, CA) and eluted into 15μl H2O. RNA quality was assessed using the RNA Nano 6000 Assay on an Agilent Bioanalyzer, (Santa Clara, CA). RNA quality was measured using the 10-point scale associated with the RNA Integrity Number (RIN). Only samples with RIN values ≥ 7 qualified for inclusion in microarray analyses.
Isolated RNA from tissue samples were processed for microarray hybridization on the Rat Gene 1.0 ST Array at the Gene Expression Microarray Core of Cincinnati Children’s Hospital Medical Center, Cincinnati, OH. Fifty-120ng of total RNA was converted to biotin–labeled sense-strand cDNA for hybridization using the Ambion WT Expression Kit (Life Technologies, Carlsbad, CA) combined with the GeneChip WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA). Chips were incubated at 45°C for 17 hours in the GeneChip Hybridization Oven 640, washed and stained in the Fluidics Station 450 (Affymetrix, Santa Clara, CA), and scanned using an Affymetrix Gene Chip Scanner 3000 7G (Affymetrix, Santa Clara, CA).
Only arrays meeting all of the quality control measures defined by the Affymetrix Expression Control Program were included in this study. Specific quality control metrics included signal histogram, relative log expression signal, Pearson’s correlation, perfect match (PM) mean (average signal intensity of probes), and positive and negative area under the curve (AUC). Also measured were the expression values of spiked-in poly-A RNA controls, and values of spiked-in hybridization controls. Raw data are available on the NCBI Gene Expression Omnibus Repository (http://www.ncbi.nlm.nih.gov/geo; GSE88726).
qPCR Validation of Striatal Microarray Gene Expression Changes
To validate expression differences observed from microarray analysis, quantitative polymerase chain reaction (qPCR) was performed to quantify levels of Nr4a2, Trh and Inhba. These three particular transcripts were chosen for validation due both to their high levels exclusively in the LID+ striatum; and for Inhba because it, together with Nurr1 is critically associated with synaptic plasticity, dendritic spine morphology, and learning and memory processes that are considered to be central to LID [6, 31, 32]. The mRNA levels in frozen striatal micropunches were derived from the same subjects. Total RNA was converted to cDNA using Superscript VILO Mastermix (Invitrogen/Life Technologies, Carlsbad, CA). PCR reactions were run in 30 μl using target specific, FAM™ labeled Taqman hydrolysis probes (Applied Biosystems/Life Technologies, Carlsbad, CA) multiplexed with a VIC® labeled primer-limited probe set to the glyceraldehyde 3-phosphate dehydrogenase (Gapdh) reference gene, which was confirmed to not vary with respect to the different treatment conditions (data not shown). Normalized gene expression was determined by differences in the cycle thresholds (Ct) between genes of interest and Gapdh (ΔCt) on a Viia7 qPCR System (Life Technologies). Differences between these normalized values across treatment groups (ΔΔCt) were then linearized to determine fold change differences (2−ΔΔCt) and compared to fold changes observed from samples subjected to microarray hybridization and Quantitation (Fig. 1H).
In Situ Hybridization (ISH), Immunohistochemistry (IHC) and Image Analysis
The relative abundance of LID-associated transcripts of interest (i.e.: Nr4a2, Trh, Inhba, FosB) were examined in the dorsolateral striatum of SD (Exp. 2) and F344 and Lewis (Exp. 3a) rats using RNAscope (Advanced Cell Diagnostics, Newark, CA) ISH. RNAscope ISH technology utilizes a branched/“tree” in situ method to allow ultrasensitive, single transcript detection using standard microscopy as detailed previously [33]. Custom RNAscope target probes were generated against Nr4a2 (54278, NM_019328.3, target region 1004–1924), Trh (25569,NM_013046.3, target region 240–1382), Inhba (N/A, NM_017128.2, target region 2–1083). A commercially available RNAscope target probe for FosB (455821, NM_013046.3, target region 84–1218) was used. The ISH signal was detected using 3, 3’-diaminobenzidine to produce brown punctate staining.
To estimate degree of respective protein increase, individual series (1-in-6) of sections (40um thickness) were processed for Nurr1 (R&D Systems, goat anti-Nurr1, 1.5ug/ml), FosB (Santa Cruz, rabbit anti-delta FosB, 1:2,000) or TRH protein (generous gift from E. Nillni, rabbit anti-TRH 1:2,000) IHC using free-floating sections as previously described [10, 34].
We found suboptimal tissue staining with several commercially available InhbA antibodies, and based on its close protein structure and potential cross-reactivity with InhbB and activinA [35] chose not to include analyses of its transcript and protein in this study.
The ISH and immunostaining analyses were performed using ImageJ® software (NIH) using the threshold function. Three separate images in the dorsolateral intact and DA-lesioned striatum were obtained for each animal (as illustrated in Fig. 5a) using an OlympusBX51 light microscope at 20x by a blinded investigator. All microscope and camera settings were identical for all images within a given transcript or antibody. Data are represented as the mean area above threshold.
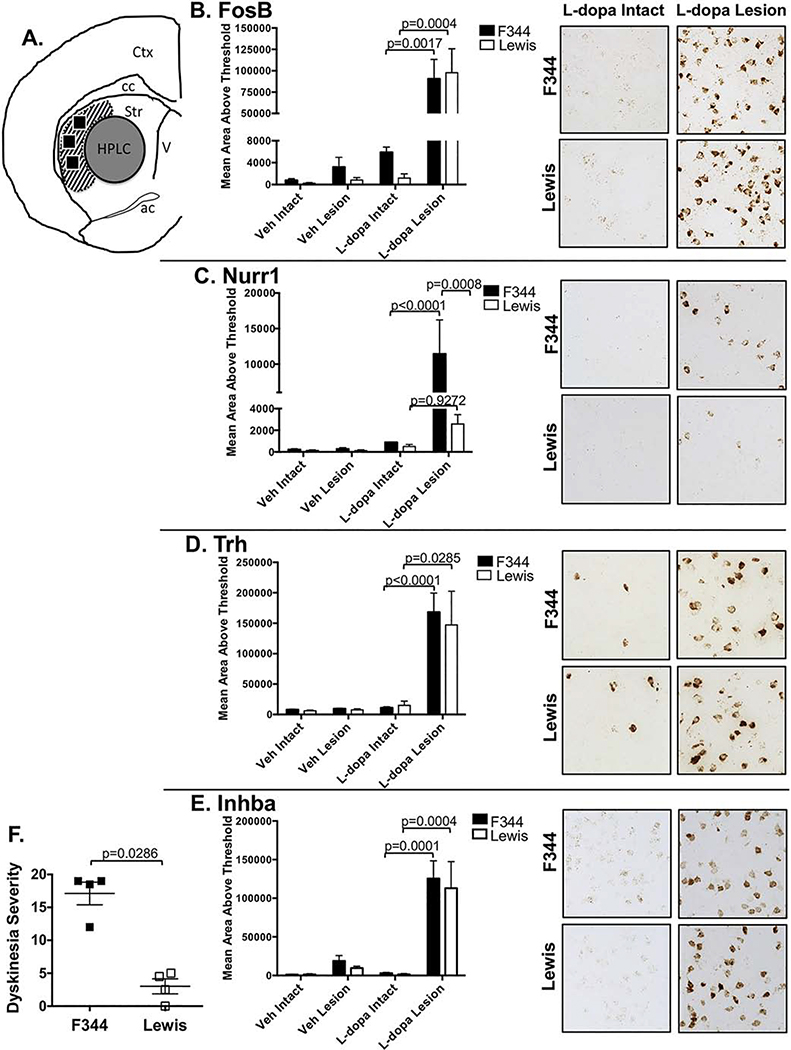
Figure 5. Effect of strain on mRNA induction of LID candidate genes.
RNAscope® in situ hybridization (ISH) was used to examine expression levels of mRNA of LID-associated genes in representative rats from same cohort shown in Figs. 3 and 4. A) Schematic showing post-mortem utilization of brains. A central region of the striatum (gray circle, “HPLC”) was punch extracted for examining monoamine content. Following tissue punch extraction, the remaining frozen block was sectioned on the cryostat and 20 μm thick sections processed for ISH for B) Fosb, C) Nurr1, D) Trh, E) Inhba mRNA. Three regions within the dorsolateral striatum (black boxes in area with slanted stripes) were analyzed for semiquantitative densitometry with ImageJ® in representative F344, N=4 and Lewis, N=4 rats with LID severity scores depicted in ‘F’. Differences in strain (F344 vs Lewis), treatment (vehicle vs levodopa), and lesion (intact vs lesion), and strain x treatment x time (2 × 2 × 2) interactions with 3-way ANOVAs. All significant ANOVA effects were further analyzed using Tukey HSD post-hoc tests. Comparison of representative LID scores were analyzed with Mann Whitney U-test. Abbreviations: Veh = vehicle; L-dopa = levodopa; Ctx = cortex; cc = corpus callosum; str = striatum; ac = anterior commissure; V = ventricle
HPLC for Striatal Monoamines
Exp. 1 & 3a
Final assessment of lesion status and potential differences in tissue content of DA and serotonin (5-HT) and their metabolites were done using HPLC as described previously [36]. Briefly, striatal samples were sonicated into 250μl of a 0.4 N perchlorate, 1.34 mM EDTA and 0.53 mM sodium metabisulfite solution. A 20μl aliquot of the homogenate was reserved for protein determination and the remaining homogenate was centrifuged at 10,500 rpm for 10 min at 4°C. The supernatant was stored in a separate tube at −80°C. Sample separation was performed on a 250×4.6mm Microsorb MV C18 100–5 column (Agilent, Santa Clara, CA). DA and 5-hydroxytryptophan (5HT; serotonin) levels were detected and quantitated using a 12-channel CoulArray 5200 coulometric array detector (ESA, Chelmsford, MA). The mobile phase consisted of 100mM Citric Acid, 75mM Na2HPO4Na, 80μM 1-heptanesulfonate monohydrate, sodium salt, 5% MeOH, pH 4.25. Samples values were interpolated against a 6-point standard curve. The final values were standardized based on protein content (BCA Protein Assay Kit, Pierce Inc., Rockford, IL). Striatal DA depletion of ≥90% in the lesioned hemisphere as compared to the unlesioned hemisphere was used as a final criterion for inclusion in the study.
Exp. 3b
At least 3 days after last microdialysis, rats were rapidly decapitated and tissue was dissected and frozen at −80 °C for later analysis for monoamine and metabolite content via HPLC with electrochemical detection. Reverse-phase HPLC was performed on bilateral striatal tissue, according to a previous protocol [37, 38]. The limit of detection was 10−10 M for monoamines and metabolites. The final oxidation current values were plotted on a standard curve of known concentrations from 10−6 M to 10−9 M, adjusted to respective tissue weights and expressed as picograms (pg) of monoamine per milligram tissue and as percent change from the intact striatum. As previously described in [39, 40], dialysate samples were measured for monoamine levels via HPLC (Eicom USA, San Diego, CA). NE, DA, and 5-HT peaks were quantified using peak heights of standard solutions. The system detection limit is reliably <50 picomolar (pM) for NE, DA and 5-HT and well within the range for detecting striatal monoamines [41, 42]. Chromatographs were analyzed using the software program Envision (provided by Eicom, USA). Data are presented as pM concentrations and as percent change from baseline.
Statistical Analyses
All LID behavioral data were analyzed by non-parametric statistics including Kruskal-Wallis with Dunn’s multiple comparison tests (for between subjects tests) or Mann-Whitney U-tests (for between-subject contrasts) and Friedman tests with Dunn’s multiple comparison tests (for within subjects tests). In Exp. 3a, amphetamine rotations and ISH data were analyzed by unpaired two-tailed t-tests and ANOVAs with Tukey HSD post-hoc comparisons, respectively. Monoamine and metabolite tissue values (pg/mg) were analyzed by mixed 2-way lesion x strain (2 × 2) ANOVAs while percent DA turnover was examined with 2-tailed unpaired t-tests. For correlation of LID severity with DA or 5-HT in DA-lesioned striata, best-fit linear regression analyses and non-parametric Spearman correlation tests were used. In Exp. 3b, averaged baseline and treatment dialysate samples were tested for lesion and strain differences using mixed 2-way (2 × 2) ANOVAs per monoamine. Percent baseline changes across time were analyzed for differences in strain, treatment, and time with mixed 3-way (2 × 2 × 9) ANOVAs per monoamine. Though constituting <1% of the data, when outliers, defined as 2 standard deviations outside the mean, were detected in microdialysis data, they were replaced with means of surrounding time point values. Strain differences at each microdialysis time point were determined using Tukey HSD post-hocs. Finally, levodopa-induced rotations were analyzed for strain and time differences using a mixed 2-way (2 × 9) ANOVA. Exp. 3b analyses (post-priming behavior after arrival at SUNY-Binghamton and neurochemistry) were performed with Statistica software ‘98 (Statsoft Inc., Tulsa, OK, USA) and alpha was set at p < 0.05. All other analyses performed at MSU were performed with Prism (v7 for Mac OS X, GraphPad Software, Inc) and alpha was set at p < 0.05.
Results
‘Gene Array Exploration’ (Exp. 1)
Similar to previous reports [5, 6] chronic daily levodopa administered to unilaterally parkinsonian SD rats resulted in pronounced LID in a majority of mean severity score across of 15.0±1.4 (mean ± SEM, N=7) on the last day of treatment. LID− rats exhibited a mean severity score of 0.0±0.0 (N=5). For all time points examined, the LID+ group showed significantly greater dyskinesia severity than the LID− group (Kruskal-Wallis, Dunn’s multiple comparison post-hoc test; p-values provided in Fig. 1B). The sham lesion (intact) + levodopa group was characterized by last day of treatment mean severity score of 0.0±0.0 (mean ± SEM; N=4)(data not shown).
Microarray analyses were performed on a subset of rats (N=3) from each of the four treatment groups in the ‘Experimental Groups’ table (Fig. 1C), chosen to have statistically similar LID severity scores to the group mean. The final LID severity scores for the gene array rats were 16.0±0.6 (LID+), 0.0±0.0 (LID−), 0.0±0.0 (6OHDA+Saline) and 0.0±0.0 (Sham Lesion+Levodopa). HPLC verified ≥95% striatal DA depletion with no difference in the level of striatal DA depletion in the parkinsonian rats in which gene array analyses were examined (98.3±0.3% (LID+); 97.9±0.8% (LID−); 99.5±0.5% (6OHDA + saline); Kruskal-Wallis p=0.064).
Principal component analysis indicated the striata of LID+ animals are transcriptomically distinct from that of LID− and 6OHDA+saline striata (Fig. 1D). Differential expression (DE) for all pairwise comparisons from the striata of parkinsonian rats revealed 258 genes that were differentially expressed between LID+ and LID− animals (Fig. 1E). Ninety genes showed unique DE specifically between LID+ and LID− striatum (43 upregulated, 47 downregulated; Fig. 1F) suggesting that these genes are of high relevance to LID expression. The top 50 differentially expressed genes are listed in Fig. 1G, with data broken out by probe set in Supplemental Table 1. The top 20 genes with the greatest differential expression in striatum were Nr4a2, Trh, Cdkn1a, Plagl1, Vdr, Gprc5a, Sstr2, Inhba, Nmu, Gpr3, Fosb, Mmp9, Cck, Dusp2, Arl5b, Plk3, Prc1, Srxn1, Pdyn, and Sdc1.
Based on data from our gene array, and historical precedence from other (e.g.: [7–9, 11–13, 15]), for our follow-up studies (Exp. 2 & 3) we examined differential expression patterns of four candidate LID-associated genes: Trh, Nurr1, Inhba, Fosb.
‘Chronic LID vs Acute LID’ (Exp. 2)
In this experiment we asked the question, if a particular gene change underlies expression of LID, should it not be expressed to similar levels in subjects with equivalent levels of LID severity regardless of their exposure to either chronic daily or a single ‘first-ever’ acute injection? To examine this question, SD rats treated either chronically with daily high-dose (12mg/kg) levodopa over 3 weeks or levodopa naïve rats received a single ‘challenge dose’ of levodopa (12mg/kg) 7 weeks after 6-OHDA lesioning (Fig. 2A). Both groups showed equivalent levels of LID when challenged with high-dose levodopa after long-term striatal DA depletion (Fig. 2B; chronic LID=13.8±1.9 (N=6); acute LID=15.0±1.5 (N=12); p=0.257, Mann-Whitney) and equivalent nigral DA neuron loss confirmed by total enumeration of tyrosine hydroxylase (TH)-positive neurons [43] (chronic LID=98.6%±0.003; first-ever LID=96.5%±0.012; p=0.235, Mann-Whitney U). While intervening with levodopa at earlier time points after lesioning (e.g.: 2–3 weeks) and/or administering lower doses generally results in a gradual increase in LID severity over time, our experience and that of others (for review see [44]) is that given a sufficient degree of DA depletion, sufficient time post-lesion and sufficient dose, maximal LID severity develop with the first-ever levodopa challenge.
Examination of Fosb, Nurr1, and Trh transcripts and corresponding proteins, which in the case of FOSB included FOSB/ΔFOSB (hereafter referred to as FOSB) in a subset of dyskinetic rats from each group as well as in drug-naïve parkinsonian rats (N=3–6) revealed significantly different expression profiles despite nearly identical levels of LID severity. Specifically, Fosb and Nurr1 transcripts but not proteins were elevated with acute LID expression; FOSB and NURR1 proteins were elevated only with chronic LID (Fig. 2C–F). Both Trh transcript and protein were elevated only after chronic LID (Fig. 2G,H). Statistics and sample numbers are detailed in Figure 2. As discussed in the methods, Inhba/INHBA was not examined here due to inability to reliably quantify the protein.
‘F344 versus Lewis’ (Exp. 3a & 3b)
To continue to explore the relationship of gene expression changes to LID expression, we next capitalized on the fact that inbred rat strains, based on their homogenous genotype, provide useful preclinical models for elucidating genetic factors underlying differences in behavioral phenotypes [16]. We first examined LID liability in parkinsonian F344 and Lewis rats administered daily high-dose levodopa for 3 weeks (Fig. 3A). As previously reported [10, 26, 34, 45], inbred F344 rats displayed a rapid and robust expression of LID with increasing severity over time (Fig. 3C, upper graph). In contrast, inbred Lewis rats displayed distinct resistance to LID expression despite the same daily high-dose levodopa over 3 weeks (Fig. 3C,D). While the LID severity in Lewis rats remained significantly less than F344, there was a modest but significant increase from Day 1 to 15 (Fig. 3C). At each time point examined peak-dose dyskinesia severity (80 mins post-levodopa; Fig. 3D) was significantly greater in F344 compared to Lewis rats (Fig. 3C).
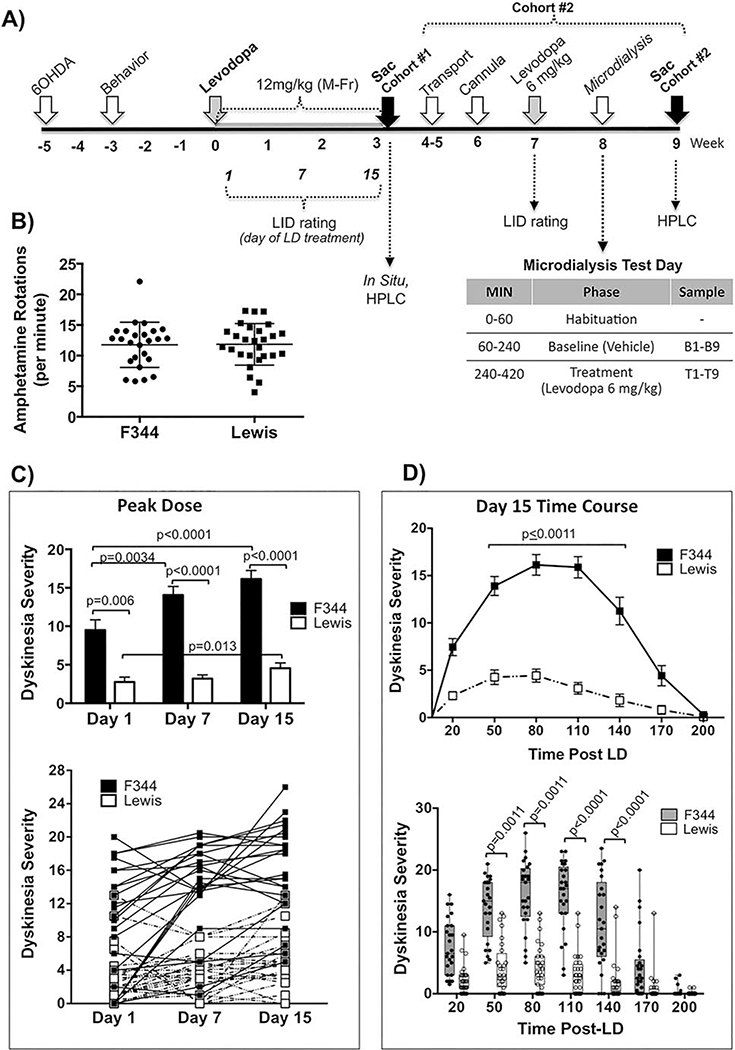
Figure 3. Effect of strain on levodopa-induced dyskinesia (LID) behavior in genetically inbred parkinsonian LID-susceptible (F344) versus LID-resistant (Lewis) rats.
A) Experimental timeline. Adult male F344 and Lewis rats were rendered parkinsonian with 6-OHDA into the SN and MFB. ‘Cohort #1’ represents Exp. 3a sacrificed for ISH and HPLC and ‘Cohort #2’ represents Exp. 3b dedicated to in vivo microdialysis measurement of extracellular striatal DA, 5HT and NE. B) Amphetamine-induced Rotational Behavior. Two weeks after 6-OHDA surgery F344 and Lewis rats were exposed to a single dose of amphetamine (5mg/kg, i.p.). Both strains of rats show equal high level of rotational behavior in response to amphetamine (p=0.6248, t=0.4924 df=46, 2-tailed unpaired t-test). Data represents counter clockwise minus clockwise (CCW-CW) rotations over 90 mins. C) Effect of strain on PEAK DOSE levodopa-induced dyskinesia (LID) behavior over time. Peak dose LID (80 mins post-levodopa) in F344 rats increased significantly over time from day 1 to day 15. At all time points examined Lewis rats remained LID resistant displaying significantly less LID severity compared to F344 rats at all time points as statistically indicated within the graphs. Upper graph: Data represent mean ± SEM; Lower graph: Individual data for each subject. Data were analyzed for effect of time using non-parametric Friedman test with post-hoc Dunn’s multiple comparison. Impact of strain on Peak Dose LID was examined with a 2-tailed Mann-Whitney U-test. D) Effect of strain TIME COURSE of levodopa-induced dyskinesia (LID) behavior. Data shown here represents the time course of LID on the final day (Day 15) of levodopa treatment. Upper graph: Data as mean ± SEM. Lower graph: Data as Box & Whisker graph to show data distribution between strains. All time points between 50 and 140 mins post-levodopa (LD) Lewis rats displayed significantly less LID severity compared to F344 rats. Data were analyzed using non-parametric Kruskal-Wallis test (H=236.5, p<0.0001) with post-hoc Dunn’s multiple comparison. All graphed data (B-D) includes rats used in Exp. 3a and 3b, as well as extra rats not presented here (N=25 for F344; N=27 for Lewis).
To examine whether difference in striatal DA release could account for the differences in LID between F344 and Lewis rats we first examined amphetamine-induced rotational behavior, which was also used to confirm lesion status (Fig. 3A,B). Despite significant differences in addiction liability of F344 and Lewis rats following chronic amphetamine (e.g.: [46, 47]), a single acute injection of amphetamine (5mg/kg) resulted in virtually identical high rates of ipsiversive rotations between strains (Fig. 3B; F344=11.76±0.74 rotations/min (n=25), Lewis=11.84±0.65 rotations/min (n=27), p=0.932, two-tailed unpaired t-test).
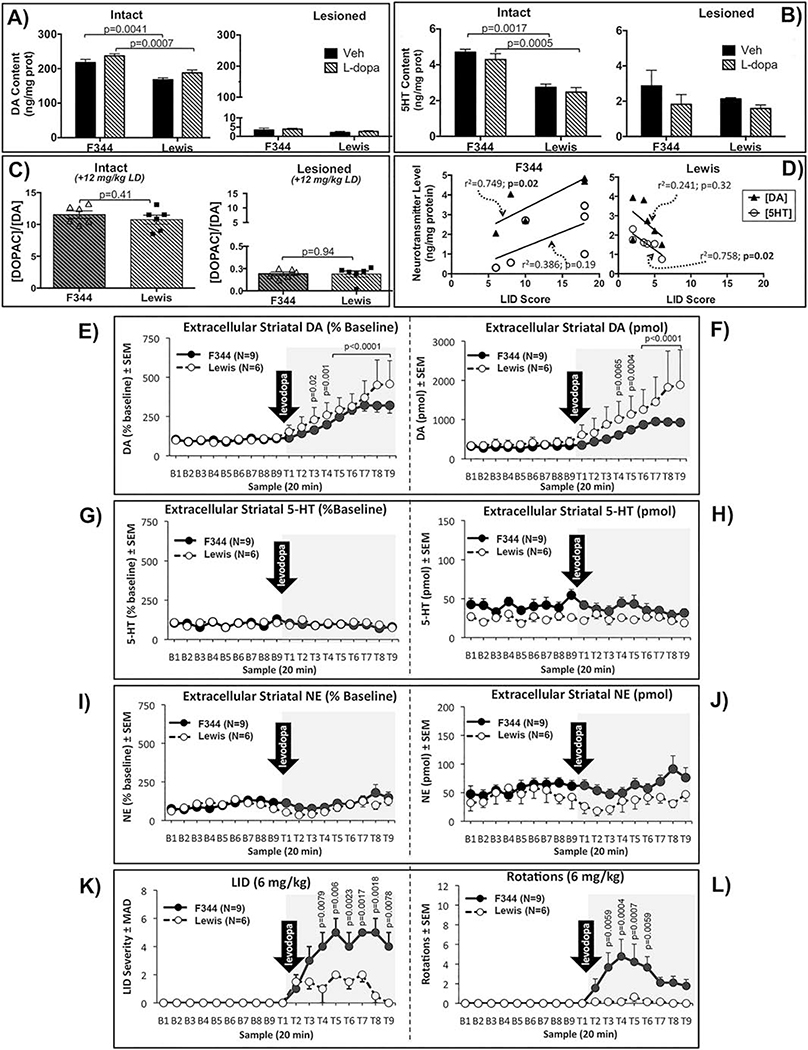
We also employed HPLC to examine the impact of levodopa and strain on striatal tissue levels/content of DA, its metabolite 3,4-dihydroxyphenylacetic acid (DOPAC) and 5-HT in a subset of parkinsonian F344 and Lewis rats primed with 3 weeks of levodopa (N=6, F344+Levodopa; N=6, Lewis+Levodopa; N=4, F344+Vehicle; N=4, Lewis+Vehicle) following an injection of levodopa (12mg/kg) or the vehicle saline. Rats were sacrificed 110–120 mins after levodopa or saline, a time when rats were still expressing peak LID behaviors (e.g.: Fig. 3D). Statistical analyses revealed equivalent levels of striatal DA depletion in the lesioned striatum (F344+Levodopa=98.4%±0.2; Lewis+Levodopa=98.46%±0.2; F344+Vehicle= 98.5%±0.5; Lewis+Vehicle=98.8%±0.4; p=0.786, Kruskal-Wallis test) and no differences in DA or 5HT content in the lesioned/DA-depleted striatum in the presence or absence of levodopa between strains (Fig. 4 A,B; DA: p=0.9068; 5HT: p=0.6277; two-way ANOVA). However, the intact Lewis striatum, in the presence and absence of levodopa, showed significantly less DA and 5HT content (Fig. 4 A,B; DA: Veh, p=0.0041, Levodopa, p=0.0007; 5HT: Veh, p=0.0017, Levodopa, p=0.0005). There was no apparent difference in turnover of striatal DA in the presence of levodopa between F344 and Lewis rats indicated by similar ratios of DOPAC/DA in both the intact (p=0.41) and lesioned striatum (p=0.94, two-tailed unpaired t-test; Fig. 4C). There was a positive correlation of increasing LID severity with increasing striatal DA content in the lesioned striatum in F344 rats (Fig. 4D, DA: r2=0.749; p=0.02, best fit linear regression; Spearman correlation p=0.05), but not in Lewis rats. In contrast there was an inverse correlation between LID severity and 5-HT content in Lewis rats (Fig. 4D, 5HT: r2=0.758; p=0.02 best fit linear regression; Spearman correlation p=0.0056), but not in F344 rats.
Figure 4. Effect of strain on baseline and treatment levels of monoamine tissue content and extracellular levels in the lesioned striatum, as well as on LID behaviors and levodopa-induced rotations in levodopa primed F344 and Lewis rats.
A-C) Tissue content of DA and 5HT in intact and lesioned striatum. As depicted in Figure 3A, F344 and Lewis rats from cohort #1 (Exp. 3a) were sacrificed for HPLC determination of tissue content of DA, DOPAC and 5HT 120 mins after levodopa (LD, 12mg/kg, N=6) or the vehicle saline (Veh, N=4). Data represent mean ± SEM and was analyzed with two-way ANOVA and post-hoc Tukey’s HSD test (A, B,) or two-tailed unpaired t-test (C). D) Relationship of DA and 5HT content in the lesioned striatum to dyskinesias severity between F344 and Lewis. There is a positive correlation of increasing LID severity with increasing striatal DA content in F344 (r2=0.749; p=0.02), but not in Lewis rats. In contrast there is an inverse correlation of LID severity with 5HT content in Lewis (r2=0.758; p=0.02), but not in F344 rats. Data were analyzed with best-fit linear regression and non-parametric Spearman correlation analyses. Best-fit linear regression: values shown in graph; Spearman correlation: Lewis (N=6) 5HT p=0.0056, DA p=0.2722; F344 (N=6) 5HT p=0.0667, DA p=0.050. E-L) Extracellular monoamine levels. As detailed in Figure 3A, rats from cohort #2 (Exp. 3b) were tested using in vivo microdialysis where, after a 1 hr habituation, striatal dialysate samples were collected every 20 min for a total of 18 samples. Rats were injected with the levodopa vehicle (saline) 10 min prior to baseline (B1 – 9) and again with levodopa (6 mg/kg; arrow) 10 mins prior to treatment time point sampling (T1 – 9). Monoamine levels (mean ± SEM) are presented as percent change from baseline (E, G, I) and pmol (F, H, J). Differences in strain, treatment (baseline vs levodopa), time, and strain x treatment x time (2 × 2 × 9) interactions with 3-way ANOVAs. All significant ANOVA effects were further analyzed using Tukey HSD post-hoc tests. K) LID behaviors and rotations were rated every 20 mins just prior to dialysate collection. LID data are presented as medians ± median absolute deviation (MAD) and were analyzed for strain differences using Mann Whitney U tests at each time point. L) Levodopa-induced rotations are presented as means ± SEM and were analyzed for differences in strain, time, and strain x time (2 × 9) interactions with 2-way ANOVAs. Statistical indications: E, F: vs corresponding Baseline time point; K: F344 vs. Lewis; L: vs. T1.
Based on the importance of elevated striatal DA in LID [48, 49], in vivo microdialysis was used to more definitively determine whether differences in LID between strains were related to differences in extracellular DA release following levodopa. This cohort of rats (‘Cohort #2’; Fig. 3A) was lesioned and primed with levodopa at MSU, and sent blinded to Binghamton University for in vivo microdialysis. Statistical analyses revealed no differences between F344 and Lewis rats following levodopa (6mg/kg) in magnitude or temporal response of increased extracellular DA in the lesioned striatum (Fig. 4E,F; Time x Strain (p=0.1228); Strain (p=0.2268); Time (p=0.0000). The 6mg/kg dose of levodopa was used to allow detection of potential differences in DA release free from possible saturation effects that might have occurred with higher doses. The main effect of time revealed significant elevations in DA in both F344 and Lewis rats at post-levodopa test-time (T) points T4-T9 compared to corresponding baseline (B) time points B4-B9 (Fig. 4 E,F; B4 v T4 p=0.0064; B5 v T5 p=0.0004; B6–9 v T6–9 p<0.0001).
There were no significant elevations of extracellular striatal 5-HT or NE following levodopa (Fig. 4 G–J). However, there was trend for less 5-HT in the Lewis compared to F344 rats (Fig. 4H; 5-HT (pmol), Strain (p=0.0541) and for change over time for NE (Fig. 4J, NE (pmol), Time (p=0.0505)).
The differential expression of LID rated by a second blinded investigator at Binghamton corroborated LID data in these same animals rated at MSU during the priming phase (Fig. 4K). Specifically, LID severity rated during microdialysis was significantly less in Lewis rats compared to F344 rats despite identical increased levels of extracellular striatal DA (T4: p=0.0079; T5: p=0.0060; T6: p=0.0023; T7: p=0.0017; T8: p=0.0018; T9: p=0.0078; Kruskal-Wallis ANOVA). Similar to dyskinetic behavior, levodopa-induced rotational behavior analyzed for differences in strain, time, and strain x time (2×9) interactions with 2-way ANOVAs showed significantly fewer rotations in the Lewis rats compared to F344 (Figure 4L; Time x Strain (p=0.1728); Time (p=0.0507); Strain (p=0.0179), Tukey’s post hoc: T1 vs T3 p=0.0058, T1 v T4 p=0.0004, T1 v T5 p=0.0007, T1 v T6 p=0.0058).
We next examined whether the candidate striatal genes exhibited strain-dependent differences between F344 and Lewis rats using N=4 subjects from each strain, with LID scores of these subjects being representative of each strain (Fig. 5F). Results of 2×2×2 ANOVA with multiple comparison for effect of strain, levodopa treatment, and lesion revealed that there was a main effect of levodopa treatment on all 4 mRNA (Fig. 5B–E) (Treatment, Fosb: p<0.0001, Nurr1: p=0.0003, Trh: p<0.0001, Inhba: p<0.0001). Curiously of the mRNA examined in this study, striatal expression levels for 3 of the 4 candidate transcripts were robustly elevated, with no statistical difference between the LID+ F344 and LID− Lewis subjects (Fig. 5B,D,E; Strain, Fosb: p=0.9753, Trh: p=0.1084, Inhba: p=0.5757). The one exception was Nurr1 which showed significant elevation only in F344 LID+ rats with no significant elevation in the LID− Lewis (Strain, Nurr1: p=0.0122) (Fig. 5C).
Discussion
Parsimony suggests that genes critical to development and expression of LID, should show similar patterns and/or magnitudes of change across the abundant ‘omic’ data currently available (e.g.: Table 2). Despite significant variability in genes highlighted as important between studies, some consensus LID targets have emerged including those investigated in the current study [7, 9, 11–15]. Yet, clinical treatment of dyskinesias based on such knowledge remains elusive. In the current study, we first used gene array methodology to determine, in our hands, which genes showed the most prominent change. We then selected four representative genes based principally on their magnitude of change in our chronically levodopa treated LID+ compared to LID− SD rats and prevalent association with LID [7, 9, 11–15], and examined their relationship to LID expression under two distinct scenarios. Our approach of comparing and contrasting expression changes across diverse models sheds novel insight into the complex relationship of genotype-to-phenotype expression, challenging assumptions about the causative role of single genes, particularly Fosb and Nurr1. Below, we focus discussion on how the unique findings of our ‘compare and contrast’ approach might guide future investigations.
Acute versus Chronic LID: Same Phenotype, Different Genotype
While there are different concepts surrounding the term ‘priming’ [44], the common view for LID is that repeated exposure of the DA depleted striatum to pulsatile DA fluctuations during levodopa treatment modifies and sensitizes the brain such that over time the chance of eliciting dyskinetic behavior increases. However, Nadjar and colleagues [44] posit the interesting proposition that “priming does not exist per se but is the direct and intrinsic consequence of the loss of dopamine innervation of the striatum… meaning that the first injections of dopaminergic drugs only exacerbate those mechanisms (sensitization) but do not induce them”. In support of this concept, a significant portion of parkinsonian rats and monkeys with severe DA depletion develop LID during the first-ever administration of levodopa as opposed to the gradual development generally espoused (reviewed in [44]); a phenomenon that is arguably dependent on dose as well as the degree and length of time of DA depletion. In idiopathic PD patients there is a more insidious loss of striatal DA that results in a more gradual development of LID, however whether the first molecular events responsible for the expression of LID (i.e.: priming) are a feature of the disease or the treatment remains debatable, and unfortunately impossible to explore in patients [44].
Our preclinical data, similar to abundant other data [7, 44, 50–54], clearly demonstrate that a priming event is not required for LID expression, but that the lesion itself, of established severity and appropriate duration, is the primary prerequisite. This is supported by the fact that LID is not seen with comparable levodopa in intact subjects. However, as introduced previously, while severe striatal DA depletion and non-physiological DA replacement are necessary, they are not always sufficient to induce LID. It remains unclear, why some individuals (rodent, non-human primate or human) do not develop LID, or show remarkable resistance. In understanding the importance of gene alterations in the induction or suppression of LID, there remains a void in understanding causality-specific alterations, harkening back to the lack of understanding whether levodopa priming is a feature inherent to the treatment (i.e.: levodopa) or the disease/individual.
In trying to better understand the complex relationship of gene changes to LID, in our first experiment (Exp. 1), we, like many before us asked the question of what genes are upregulated to (presumably) “cause” LID. We reasoned that if a given gene alteration is causative/permissive/necessary for LID expression, a first exposure of levodopa that resulted in an equivalent level of dyskinesias as did chronic levodopa (Exp. 2) should reveal an equivalent level of expression in important genes. Just like turning on a lamp produces light, turning on a causative gene should produce LID. While we only examined four candidate genes in this proof-of-principle investigation, this supposition is clearly not supported by our data. Specifically, Nurr1 and Fosb transcript and protein showed significantly different expression levels in rats with first-ever LID compared to those with chronic LID despite nearly identical levels of LID expression. Indeed even in the presence of identical LID severity there is a complex and dynamic regulation of transcript and protein. For example, we show that both Nurr1 and FosB mRNA are higher than their respective protein levels following acute dyskinogenic levodopa; and the inverse occurs with chronic levodopa. We postulate that these effects are the results of altered regulation of protein and mRNA following the two different treatment paradigms. Indeed, it is well known that FosB proteins are highly stable and that the protein accumulates over time with repeated levodopa (e.g.: [55, 56]). In contrast, we observed FosB mRNA increases most significantly after acute, but not chronic, stimulation; in keeping with previously reported dynamic regulation of FosB/ΔFosB transcript over a 24 hour time course post-levodopa [57]. Much less is known about Nurr1 outside of midbrain dopamine neurons, but it is conceivable that similar regulatory pathways contribute to the discrepant findings.
Further, the neuromodulatory hormone TRH, which has a literature dating back 25 years or more, suggesting a modulatory relationship between TRH and DA (e.g.: [58–61]) has previously been suggested to underlie pathologic neuroplasticity driving LID based on its dramatic and selective up-regulation in the sensorimotor striatum of dyskinetic rats [15]. However, our data distinctly demonstrate that it is only upregulated after chronic levodopa but not in severely LID+ rats following a single acute administration. Similarly Inhba, which has yet to be explored specifically in LID, is 1 of 9 genes that exhibit increased expression associated with synaptic activity-dependent acquired neuroprotection, termed Activity-regulated Inhibitor of Death (AID) genes that are induced via a cascade of glutamatergic NMDA receptor activation and nuclear calcium signaling [62, 63]. In the context of LID, Inhba upregulation may represents an adaptive neuroprotective response (termed adaptogenomics [62]) that has been shown to be associated with altered NMDA receptor expression and toxic calcium influx [62], both associated with LID [28, 64–66]. However, Inhba is equally induced to very high levels in the LID+ (SD, F344) and LID− (Lewis) striata. These data emphasize that elevation/alteration of a gene does not necessitate causation of a phenotype as is often attributed to such changes.
With regard to the LID− outbred SD rats, it could be suggested that these are individuals that never will be “primed” due to DA-depletion or levodopa treatment because they are genetically different from the LID+ rats; a supposition confirmed with our gene array data demonstrating no increased expression of any of the four candidate (or top 50 DE) genes in this study, and similarly per [5]. This could be taken to suggest that in the absence of elevated expression of Fosb, Nurr1, Trh and/or Inhba there would be a lack of LID expression. Accordingly, therapies aimed at preventing the induction of one or more of these molecular targets should prevent LID in parkinsonian subjects, which has been demonstrated with infusion of a Fosb antisense oligonucleotide in the striatum [7]. However, by introducing data from a new model system (i.e.: inbred rats with distinct LID phenotypes) this assumption is challenged.
F344 versus Lewis: Different Phenotype, Similar Alterations in Genotype
F344 and Lewis rats have been used for nearly three decades to model genetic vulnerability to drug addiction based on their divergent responses to drugs of abuse (e.g.: [16, 17, 67]). We provide here the first data demonstrating that these inbred rat strains, when rendered hemiparkinsonian, also show differential liability to LID expression. In our study we demonstrate that Lewis rats are dyskinesia-resistant and F344 are dyskinesia-prone, the apparent opposite of that seen with addiction where Lewis rats are considered addiction-prone and F344 addiction-resistant. Given that LID and addiction are both thought to involve DA priming and sensitization of forebrain DA systems, this finding was surprising. However, addiction studies are considerably more complex involving numerous abusive substances including heroin, morphine, nicotine, cocaine and amphetamine, are dependent on motivational aspects of drug seeking behavior, and outcomes vary depending on doses of the drugs employed (for review [67]). That said, there are some observations in addiction research involving the mesolimbic DA system that suggest that our findings involving the nigrostriatal DA system may not actually be disparate, at least related to psychostimulant drugs (e.g.: cocaine, amphetamine). Specifically, Lewis rats are said to be addiction prone based in part on the fact that “Lewis rats escalate both cocaine and heroin intake to a greater extent than Fischer rats” [19]. It could be reasoned that the ‘need to escalate’ cocaine intake in Lewis rats, compared to the non-escalating Fischer rats could be related to resistance of Lewis rats to rewarding quality (“reward-resistance”) of these drugs similar to their resistance to levodopa in developing LID reported here. In addition, while some conditioned place preference (CPP) studies in F344 and Lewis rats have demonstrated that morphine, heroin, and nicotine are more rewarding in Lewis than in F344 rats, the one exception is with amphetamine where F344 rats display preference for the drug paired compartment but Lewis rats do not [67]. Finally, there numerous differences in gene regulation that have been observed in the mesolimbic DA systems between these rats strains that have not been seen in the nigrostriatal DA system [68, 69], thus direct comparison of addiction phenotype characteristics with dyskinesia phenotype in these strains is not possible.
Dysregulated DA release is a significant contributor to LID (for review [49]), and strain differences in response to some drugs of abuse have been ascribed to differences in DA neurotransmission [16, 46, 47]. However, we demonstrate here that despite significant differential LID expression, F344 and Lewis rats displayed similar levels of striatal DA depletion, ipsiversive rotation in response to acute amphetamine, and importantly levodopa-induced increases in extracellular DA in the lesioned striatum. Indeed despite evidence of decreased basal striatal DA content in Lewis compared F344 rats, which is similar to that previously reported [70], indications of DA release using in vivo microdialysis, showed a modest albeit non-significant trend for elevated extracellular DA following levodopa in the striatum of LID-resistant Lewis rats compared to F344 rats. The physiological significance, or a lack of, for a decrease in lower striatal DA tissue content in Lewis rats is uncertain. In fact, despite a great deal of work reporting differences in DA function in the mesolimbic DA system, comparatively little has been performed characterizing the nigrostriatal system (see [67] for review). In striatal tissue we found that Lewis rats display 20% less striatal tissue DA content. The origins of this difference are not clear, but noteworthy as there are no differences in tyrosine hydroxlase (TH) levels in substantia nigra or striatum between strains [68]. Interestingly F344 rats do express higher striatal DA transporter (DAT) levels and in vivo clearance compared to Lewis rats [46], which could indicate the severe 6-OHDA-induced DAT loss could differentially influence striatal DA dynamics and thus LID. This certainly requires further investigation.
Similar to a decrease in striatal DA content, we found a decrease in 5HT content in Lewis compared to F344 rats, which again is similar to that previously reported [70]. While there is an apparent decrease in tissue content of these transmitters in Lewis compared to F344 rats, the level of DA or 5HT turnover is either not different between species (for DA; Fig. 4C & [70]), or increased in Lewis rats (for 5HT; [70]). Further, we found no difference in extracellular 5HT release between strains under basal conditions or in response to levodopa. We did however observe an apparent differential relationship of DA and 5HT content with LID behavior between rat strains. Specifically, while severe LID in the F344 rats showed a positive correlation with DA content, the mild degree of LID expressed in Lewis rats showed an inverse correlation with 5HT content. In the parkinsonian striatum where there is a significant degree of striatal DA terminal loss, a leading hypothesis is that levodopa is taken up and converted to DA into serotonergic neurons that sprout into the denervated striatum from the dorsal raphe nucleus [49, 71]. Indeed 5HT neurons contain aromatic amino acid decarboxylase (AADC) allowing conversion of levodopa to DA. However, these neurons do not express the regulatory mechanisms (i.e.: DA autoreceptors) to regulate DA synthesis and release, which is hypothesized to result in dysregulated release of DA leading to LID. While our data demonstrate no differences in 5HT or DA content in the lesioned striatum between strains, the significance of these correlational differences is uncertain. However, it is clear that in response to levodopa there are equal elevations in extracellular striatal DA in F344 and Lewis rats suggesting that mechanisms downstream of synaptic DA are most likely responsible for their divergent LID phenotypes.
In exploring the four candidate ‘downstream’ genes that we chose, it is puzzling that within the DA-depleted striatum of LID-expressing F344 and LID-resistant Lewis rats there were significant increases in three of these candidate LID-associated transcripts (i.e.: Fosb, Trh, Inhba). This was particularly unexpected for Fosb in the LID− Lewis rats since it has been demonstrated that Fosb and FOSB/ΔFOSB expression in striatum is significantly correlated with LID severity, striatal overexpression can induce LID, and interference with its expression reduces LID expression [7–12, 57, 72, 73]. FosB proteins, which dimerize with Jun proteins, form AP-1 complexes that impact a multitude of molecular pathways thought to be critically involved in aberrant plasticity underlying DA sensitization phenomenon including LID (e.g.: [8, 74, 75]). Thus, Fosb is generally considered a surrogate marker of LID [73]. However, we found that in the DA-depleted striatum of dyskinesia-resistant Lewis rats treated with chronic high dose levodopa there was a 407-fold increase in Fosb mRNA compared to a 116-fold increase in the dyskinesia-prone F344 rats compared to basal striatal expression levels. These massive increases in Fosb transcript expression in both strains would appear to suggest that upstream regulators of Fosb do not appear to underlie the significantly disparate expression of LID behaviors. While protein levels were not measured in the current investigation, the curious and excessive elevation of Fosb in the absence of notable LID behaviors in Lewis rats serves as a springboard for future investigations, suggesting reassessment of the role of this transcription factor and/or its molecular partners in mediating LID. These findings further suggest that additional investigations into potential differences between F344 and Lewis rats in expression and/or targets of the FOSB proteins, which act as transcription factors altering expression of other genes [57, 74] are warranted. Interestingly, these findings could also be taken to suggest that there may be differential induction of LID-resistance genes downstream of Fosb in Lewis rats.
The only transcript investigated in the current study demonstrating differential upregulation between strains and whose upregulation was specifically associated with elevated LID severity was Nurr1. This transcription factor has received a great deal of attention for its role in the survival and maintenance of SN DA neurons that degenerate in PD [76–80], but has only recently begun to be considered for its potential role in LID. Interestingly, Nurr1 has been reported to play a role in remodeling of mesolimbic basal ganglia circuits during drug addiction [81] and within the hippocampus is critically associated learning and memory processes [82–85]. Based on these known biological functions associated with synaptic plasticity together with the high degree of association of striatal Nurr1 with LID demonstrated by others [13, 14] and in our studies in three diverse rat strains, it is reasonable to suggest that induction of striatal Nurr1 may play an integral role in LID. While normally not expressed (or expressed at very low levels) in the striatum [14], recent evidence suggests that, in the parkinsonian striatum levodopa activation of DA D1 receptors (D1r) promotes the expression of genes, including Nurr1 that are normally repressed in the adult striatum [14]. Indeed, the induction of the Nurr1 gene in striatum of dyskinetic parkinsonian mice shows strong correlation with dose and duration of levodopa/LID [13, 14]. While we did not observe a difference in LID severity between the acute and chronic L-DOPA treatment paradigms, we hypothesize that this is the result of a “ceiling effect” related to Nurr1. Indeed, as we recently reported in a follow up set of studies (Sellnow et al. J. Neurosci in press [86]) prompted by the current studies, AAV-mediated overexpression of Nurr1 resulting in significantly higher levels than that observed herein, was not capable of further driving the already high levels of dyskinesia severity in LID-susceptible SD and Fisher rats. In these follow-up studies [86] we did however corroborate the importance of striatal Nurr1 in LID by demonstrating: 1) the Nurr1 agonist amodiaquine (AQ) when given chronically with either levodopa or the D2/D3 receptor agonist ropinirole significantly enhance dyskinesia severity; 2) that viral vector mediated over overexpression of Nurr1 in the parkinsonian striatum of Lewis LID-resistant rats promotes development of severe LID behavior; and 3) viral vector mediated over overexpression of Nurr1 in the absence of levodopa increases firing activity of direct pathway neurons in the DA-depleted striata and alters dendritic spines on striatal medium spiny neurons, mimicking changes seen in dyskinetic rats.
Conclusions
One important point that can be taken from the collection of studies presented here, which is underscored by previous findings (e.g.: [57] [87]), is that moving forward studies aimed at identifying gene targets for therapy development need to appreciate that there is a complex relationship of genotype alteration to phenotype expression. Specifically, as we show here, identical LID behavior can be associated with different gene/protein profiles (‘acute vs chronic LID’), and disparate LID behavior can also be associated with comparable gene/protein profiles (‘F344 vs Lewis’). This challenge, faced in all ‘omic studies, is attributable, at least in part, to the fact that there appears to be desensitization of genes with chronic drug treatment [57] and levels of gene or protein expression vary over a time course following drug administration, with behavioral correlation dependent upon the timing of sacrifice [57]. For example, striatal FosB/ΔFosb transcript levels have been found to correlate with LID behavior only one hour after sacrifice but not at three or 24 hours post sacrifice [57]. Further, a given time course of effect also can vary depending upon whether administration of the drug is acute versus chronic. For example, activator protein-1 (AP-l), a transcription factor composed of proteins including FOSB, and is induced by cocaine. In the case of a single acute cocaine injection AP-1 expression is maximal at 2 – 4 hours and disappears within 12 – 18 hours; however, following chronic cocaine, the AP-l-binding complex is expressed at maximal levels 18 hours following the last cocaine injection [87].
Another point of value from the current studies is the discovery the inbred rat strains employed here may provide a novel model system that could provide new insight into the complex relationships of gene alterations that contribute to LID. Specifically, while Nurr1 and FosB may both be critical triggers of LID, the 407-fold increase in Fosb mRNA that we observed in the LID-resistant Lewis rats compared to the more modest 116-fold increase in the severely dyskinetic F344 rats suggests that future studies comparing changes in Lewis rats to those in F344 rats may provide a useful tool for exploring the role of ‘dyskinesia-resistance’ genes downstream of ‘dyskinesia-susceptibility’ genes and/or identifying difference in downstream genes activated by a gene of interest (i.e.: Fosb). The idea that there may be dyskinesia resistance genes or differential genes modulators impacting the onset and/or severity LID in individuals with PD is a concept that has received considerably less attention and offers new ways of thinking about development of antidyskinetic therapies. Overall, our intention is that the current studies provide a springboard for novel ways of thinking about and continuing investigations into resilience versus risk modifier genes/proteins that will lead to therapeutic advancement for this unmet medical need.
Supplementary Material
Highlights.
Identical dyskinesia severity during first-ever vs chronic levodopa induces differential gene expression.
Inbred F344 and Lewis rats show divergent dyskinesia expression and offer a novel model for understanding genetic modulators of dyskinesias.
The currently accepted genotype-to-phenotype relationships of Fosb is brought into question.
Striatal Nurr1 may represent a novel anti-dyskinesia target.
Acknowledgments
This study was supported by the National Institute of Neurological Disorders and Stroke P50 NS058830 (TJC, KSC, CES, JWL) and the MSU CHM Dean’s DFI Fund (KSC). We would like to acknowledge the excellent technical assistance of Dr. Nicole Polinski, Dr. Luke Fischer, Rory Kruithoff and Paul Francoeur. We would like to acknowledge the services of the Van Andel Institute Bioinformatics and Biostatistics and Vivarium and Transgenics Cores.
Footnotes
Financial Disclosures/Conflicts: KSC, FPM, TJC, JWL are co-inventors listed on a patent entitled “Nurr1 as a genetic target for treating levodopa-induced dyskinesias in Parkinson’s disease” (U.S. Patent Application No.: 15/316,726). The authors have no other conflicts to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hauser RA, et al. , ADS-5102 (Amantadine) Extended-Release Capsules for Levodopa-Induced Dyskinesia in Parkinson’s Disease (EASE LID 2 Study): Interim Results of an Open-Label Safety Study. J Parkinsons Dis, 2017. 7(3): p. 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huot P, et al. , The pharmacology of L-DOPA-induced dyskinesia in Parkinson’s disease. Pharmacol Rev, 2013. 65(1): p. 171–222. [DOI] [PubMed] [Google Scholar]
- 3.Kordower JH, et al. , Disease duration and the integrity of the nigrostriatal system in Parkinson’s disease. Brain, 2013. 136(Pt 8): p. 2419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lundblad M, et al. , A model of L-DOPA-induced dyskinesia in 6-hydroxydopamine lesioned mice: relation to motor and cellular parameters of nigrostriatal function. Neurobiol Dis, 2004. 16(1): p. 110–23. [DOI] [PubMed] [Google Scholar]
- 5.Konradi C, et al. , Transcriptome analysis in a rat model of L-DOPA-induced dyskinesia. Neurobiol Dis, 2004. 17(2): p. 219–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. , Aberrant restoration of spines and their synapses in L-DOPA-induced dyskinesia: involvement of corticostriatal but not thalamostriatal synapses. J Neurosci, 2013. 33(28): p. 11655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson M, Hilbertson A, and Cenci MA, Striatal fosB expression is causally linked with l-DOPA-induced abnormal involuntary movements and the associated upregulation of striatal prodynorphin mRNA in a rat model of Parkinson’s disease. Neurobiol Dis, 1999. 6(6): p. 461–74. [DOI] [PubMed] [Google Scholar]
- 8.Cenci MA, Transcription factors involved in the pathogenesis of L-DOPA-induced dyskinesia in a rat model of Parkinson’s disease. Amino Acids, 2002. 23(1–3): p. 105–9. [DOI] [PubMed] [Google Scholar]
- 9.Lindgren HS, et al. , Putaminal upregulation of FosB/DeltaFosB-like immunoreactivity in Parkinson’s disease patients with dyskinesia. J Parkinsons Dis, 2011. 1(4): p. 347–57. [DOI] [PubMed] [Google Scholar]
- 10.Maries E, et al. , Focal not widespread grafts induce novel dyskinetic behavior in parkinsonian rats. Neurobiol Dis, 2006. 21(1): p. 165–80. [DOI] [PubMed] [Google Scholar]
- 11.Padovan-Neto FE, et al. , Anti-dyskinetic effect of the neuronal nitric oxide synthase inhibitor is linked to decrease of FosB/deltaFosB expression. Neurosci Lett, 2013. 541: p. 126–31. [DOI] [PubMed] [Google Scholar]
- 12.Pavon N, et al. , ERK phosphorylation and FosB expression are associated with L-DOPA-induced dyskinesia in hemiparkinsonian mice. Biol Psychiatry, 2006. 59(1): p. 64–74. [DOI] [PubMed] [Google Scholar]
- 13.Heiman M, et al. , Molecular adaptations of striatal spiny projection neurons during levodopa-induced dyskinesia. Proc Natl Acad Sci U S A, 2014. 111(12): p. 4578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sodersten E, et al. , Dopamine signaling leads to loss of Polycomb repression and aberrant gene activation in experimental parkinsonism. PLoS Genet, 2014. 10(9): p. e1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantuti-Castelvetri I, et al. , Levodopa-induced dyskinesia is associated with increased thyrotropin releasing hormone in the dorsal striatum of hemi-parkinsonian rats. PLoS One, 2010. 5(11): p. e13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brodkin ES, et al. , Genetic analysis of behavioral, neuroendocrine, and biochemical parameters in inbred rodents: initial studies in Lewis and Fischer 344 rats and in A/J and C57BL/6J mice. Brain Res, 1998. 805(1–2): p. 55–68. [DOI] [PubMed] [Google Scholar]
- 17.Fole A, et al. , Lewis and Fischer 344 rats as a model for genetic differences in spatial learning and memory: Cocaine effects. Prog Neuropsychopharmacol Biol Psychiatry, 2017. 76: p. 49–57. [DOI] [PubMed] [Google Scholar]
- 18.Miguens M, et al. , Depotentiation of hippocampal long-term potentiation depends on genetic background and is modulated by cocaine self-administration. Neuroscience, 2011. 187: p. 36–42. [DOI] [PubMed] [Google Scholar]
- 19.Valenza M, et al. , Strain and cocaine-induced differential opioid gene expression may predispose Lewis but not Fischer rats to escalate cocaine self-administration. Neuropharmacology, 2016. 105: p. 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cenci MA and Crossman AR, Animal models of l-dopa-induced dyskinesia in Parkinson’s disease. Mov Disord, 2018. 33(6): p. 889–899. [DOI] [PubMed] [Google Scholar]
- 21.Morin N, Jourdain VA, and Di Paolo T, Modeling dyskinesia in animal models of Parkinson disease. Exp Neurol, 2014. 256: p. 105–16. [DOI] [PubMed] [Google Scholar]
- 22.Schallert T, Behavioral tests for preclinical intervention assessment. NeuroRx, 2006. 3(4): p. 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schallert T, et al. , CNS plasticity and assessment of forelimb sensorimotor outcome in unilateral rat models of stroke, cortical ablation, parkinsonism and spinal cord injury. Neuropharmacology, 2000. 39(5): p. 777–87. [DOI] [PubMed] [Google Scholar]
- 24.Borlongan CV, Hida H, and Nishino H, Early assessment of motor dysfunctions aids in successful occlusion of the middle cerebral artery. Neuroreport, 1998. 9(16): p. 3615–21. [DOI] [PubMed] [Google Scholar]
- 25.Tillerson JL, et al. , Forced nonuse in unilateral parkinsonian rats exacerbates injury. J Neurosci, 2002. 22(15): p. 6790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steece-Collier K, et al. , Embryonic mesencephalic grafts increase levodopa-induced forelimb hyperkinesia in parkinsonian rats. Mov Disord, 2003. 18(12): p. 1442–54. [DOI] [PubMed] [Google Scholar]
- 27.Bishop C, et al. , Contribution of the striatum to the effects of 5-HT1A receptor stimulation in L-DOPA-treated hemiparkinsonian rats. J Neurosci Res, 2009. 87(7): p. 1645–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steece-Collier K, et al. , Genetic silencing of striatal CaV1.3 prevents and ameliorates levodopa dyskinesia. Mov Disord, 2019. 34(5): p. 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupre KB, et al. , Local modulation of striatal glutamate efflux by serotonin 1A receptor stimulation in dyskinetic, hemiparkinsonian rats. Exp Neurol, 2011. 229(2): p. 288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine ND, et al. , Advances in thin tissue Golgi-Cox impregnation: fast, reliable methods for multi-assay analyses in rodent and non-human primate brain. J Neurosci Methods, 2013. 213(2): p. 214–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picconi B, et al. , Pathological synaptic plasticity in the striatum: implications for Parkinson’s disease. Neurotoxicology, 2005. 26(5): p. 779–83. [DOI] [PubMed] [Google Scholar]
- 32.Pisani A, et al. , Striatal synaptic plasticity: implications for motor learning and Parkinson’s disease. Mov Disord, 2005. 20(4): p. 395–402. [DOI] [PubMed] [Google Scholar]
- 33.Grabinski TM, et al. , A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS One, 2015. 10(3): p. e0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collier TJ, et al. , Interrogating the aged striatum: robust survival of grafted dopamine neurons in aging rats produces inferior behavioral recovery and evidence of impaired integration. Neurobiol Dis, 2015. 77: p. 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namwanje M and Brown CW, Activins and Inhibins: Roles in Development, Physiology, and Disease. Cold Spring Harb Perspect Biol, 2016. 8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNamara RK, et al. , Omega-3 fatty acid deficiency augments amphetamine-induced behavioral sensitization in adult DBA/2J mice: relationship with ventral striatum dopamine concentrations. Synapse, 2008. 62(10): p. 725–35. [DOI] [PubMed] [Google Scholar]
- 37.Eskow Jaunarajs KL, et al. , Potential mechanisms underlying anxiety and depression in Parkinson’s disease: consequences of l-DOPA treatment. Neurosci Biobehav Rev, 2011. 35(3): p. 556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilpatrick IC, Jones MW, and Phillipson OT, A semiautomated analysis method for catecholamines, indoleamines, and some prominent metabolites in microdissected regions of the nervous system: an isocratic HPLC technique employing coulometric detection and minimal sample preparation. J Neurochem, 1986. 46(6): p. 1865–76. [DOI] [PubMed] [Google Scholar]
- 39.Hall JM and Savage LM, Exercise leads to the re-emergence of the cholinergic/nestin neuronal phenotype within the medial septum/diagonal band and subsequent rescue of both hippocampal ACh efflux and spatial behavior. Exp Neurol, 2016. 278: p. 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano M, Hitomi E, and Mizuno T, Age-related changes in the metabolism of neurotransmitters and the effect of scavengers: an in vivo microdialysis study Arch Gerontol Geriatr, 1994. 19 Suppl 1: p. 171–6. [DOI] [PubMed] [Google Scholar]
- 41.Bhide N, et al. , Effects of the beta-adrenergic receptor antagonist Propranolol on dyskinesia and L-DOPA-induced striatal DA efflux in the hemi-parkinsonian rat. J Neurochem, 2015. 134(2): p. 222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conti MM, et al. , Monoamine transporter contributions to l-DOPA effects in hemi-parkinsonian rats. Neuropharmacology, 2016. 110(Pt A): p. 125–134. [DOI] [PubMed] [Google Scholar]
- 43.Gombash SE, et al. , Neuroprotective potential of pleiotrophin overexpression in the striatonigral pathway compared with overexpression in both the striatonigral and nigrostriatal pathways. Gene Ther, 2014. 21(7): p. 682–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadjar A, Gerfen CR, and Bezard E, Priming for l-dopa-induced dyskinesia in Parkinson’s disease: a feature inherent to the treatment or the disease? Prog Neurobiol, 2009. 87(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steece-Collier K, et al. , Effect of levodopa priming on dopamine neuron transplant efficacy and induction of abnormal involuntary movements in parkinsonian rats. J Comp Neurol, 2009. 515(1): p. 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gulley JM, Everett CV, and Zahniser NR, Inbred Lewis and Fischer 344 rat strains differ not only in novelty- and amphetamine-induced behaviors, but also in dopamine transporter activity in vivo. Brain Res, 2007. 1151: p. 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer AC and Bardo MT, Amphetamine self-administration and dopamine function: assessment of gene x environment interactions in Lewis and Fischer 344 rats. Psychopharmacology (Berl), 2015. 232(13): p. 2275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bastide MF, et al. , Pathophysiology of L-dopa-induced motor and non-motor complications in Parkinson’s disease. Prog Neurobiol, 2015. 132: p. 96–168. [DOI] [PubMed] [Google Scholar]
- 49.Sellnow RC, et al. , Regulation of dopamine neurotransmission from serotonergic neurons by ectopic expression of the dopamine D2 autoreceptor blocks levodopa-induced dyskinesia. Acta Neuropathol Commun, 2019. 7(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cenci MA, Lee CS, and Bjorklund A, L-DOPA-induced dyskinesia in the rat is associated with striatal overexpression of prodynorphin- and glutamic acid decarboxylase mRNA. Eur J Neurosci, 1998. 10(8): p. 2694–706. [PubMed] [Google Scholar]
- 51.Delfino MA, et al. , Behavioral sensitization to different dopamine agonists in a parkinsonian rodent model of drug-induced dyskinesias. Behav Brain Res, 2004. 152(2): p. 297–306. [DOI] [PubMed] [Google Scholar]
- 52.Johansson PA, et al. , Alterations in cortical and basal ganglia levels of opioid receptor binding in a rat model of l-DOPA-induced dyskinesia. Neurobiol Dis, 2001. 8(2): p. 220–39. [DOI] [PubMed] [Google Scholar]
- 53.Di Monte DA, et al. , Relationship among nigrostriatal denervation, parkinsonism, and dyskinesias in the MPTP primate model. Mov Disord, 2000. 15(3): p. 459–66. [DOI] [PubMed] [Google Scholar]
- 54.Yoo HS, et al. , Presynaptic dopamine depletion determines the timing of levodopa-induced dyskinesia onset in Parkinson’s disease. Eur J Nucl Med Mol Imaging, 2018. 45(3): p. 423–431. [DOI] [PubMed] [Google Scholar]
- 55.Alibhai IN, et al. , Regulation of fosB and DeltafosB mRNA expression: in vivo and in vitro studies. Brain Res, 2007. 1143: p. 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cenci MA, et al. , Changes in the regional and compartmental distribution of FosB- and JunB-like immunoreactivity induced in the dopamine-denervated rat striatum by acute or chronic L-dopa treatment. Neuroscience, 1999. 94(2): p. 515–27. [DOI] [PubMed] [Google Scholar]
- 57.Palafox-Sanchez V, et al. , Differential Expression of Striatal DeltaFosB mRNA and FosB mRNA After Different Levodopa Treatment Regimens in a Rat Model of Parkinson’s Disease. Neurotox Res, 2019. 35(3): p. 563–574. [DOI] [PubMed] [Google Scholar]
- 58.Przegalinski E, Jaworska L, and Budziszewska B, The role of dopamine receptors in the release of thyrotropin-releasing hormone from the rat striatum and nucleus accumbens: an in vitro study. Neuropeptides, 1993. 25(5): p. 277–82. [DOI] [PubMed] [Google Scholar]
- 59.Przegalinski E, et al. , The role of dopamine in regulation of thyrotropin-releasing hormone in the striatum and nucleus accumbens of the rat. Neuropeptides, 1991. 19(3): p. 189–95. [DOI] [PubMed] [Google Scholar]
- 60.Heal DJ, et al. , Behavioural effects of central and peripheral injection of various analogues and metabolites of thyrotropin releasing hormone (TRH). Neuropharmacology, 1981. 20(10): p. 947–57. [DOI] [PubMed] [Google Scholar]
- 61.Kreutz MR, et al. , Systemic administration of thyrotropin-releasing hormone enhances striatal dopamine release in vivo. Brain Res, 1990. 536(1–2): p. 347–52. [DOI] [PubMed] [Google Scholar]
- 62.Lau D, et al. , BDNF Reduces Toxic Extrasynaptic NMDA Receptor Signaling via Synaptic NMDA Receptors and Nuclear-Calcium-Induced Transcription of inhba/Activin A. Cell Rep, 2015. 12(8): p. 1353–66. [DOI] [PubMed] [Google Scholar]
- 63.Zhang SJ, et al. , Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet, 2009. 5(8): p. e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmed I, et al. , Glutamate NMDA receptor dysregulation in Parkinson’s disease with dyskinesias. Brain, 2011. 134(Pt 4): p. 979–86. [DOI] [PubMed] [Google Scholar]
- 65.Ghiglieri V, et al. , Corticostriatal Plastic Changes in Experimental L-DOPA-Induced Dyskinesia. Parkinsons Dis, 2012. 2012: p. 358176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Picconi B, et al. , Loss of bidirectional striatal synaptic plasticity in L-DOPA-induced dyskinesia. Nat Neurosci, 2003. 6(5): p. 501–6. [DOI] [PubMed] [Google Scholar]
- 67.Cadoni C, Fischer 344 and Lewis Rat Strains as a Model of Genetic Vulnerability to Drug Addiction. Front Neurosci, 2016. 10: p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harris HW and Nestler EJ, Immunohistochemical studies of mesolimbic dopaminergic neurons in Fischer 344 and Lewis rats. Brain Res, 1996. 706(1): p. 1–12. [DOI] [PubMed] [Google Scholar]
- 69.Ortiz J, et al. , Biochemical adaptations in the mesolimbic dopamine system in response to repeated stress. Neuropsychopharmacology, 1996. 14(6): p. 443–52. [DOI] [PubMed] [Google Scholar]
- 70.Pletnikov MV, et al. , Effects of genetic background on neonatal Borna disease virus infection-induced neurodevelopmental damage. II. Neurochemical alterations and responses to pharmacological treatments. Brain Res, 2002. 944(1–2): p. 108–23. [DOI] [PubMed] [Google Scholar]
- 71.De Deurwaerdere P, Di Giovanni G, and Millan MJ, Expanding the repertoire of L-DOPA’s actions: A comprehensive review of its functional neurochemistry. Prog Neurobiol, 2017. 151: p. 57–100. [DOI] [PubMed] [Google Scholar]
- 72.Cao X, et al. , Striatal overexpression of DeltaFosB reproduces chronic levodopa-induced involuntary movements. J Neurosci, 2010. 30(21): p. 7335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engeln M, et al. , Selective Inactivation of Striatal FosB/DeltaFosB-Expressing Neurons Alleviates L-DOPA-Induced Dyskinesia. Biol Psychiatry, 2016. 79(5): p. 354–361. [DOI] [PubMed] [Google Scholar]
- 74.Nestler EJ, Barrot M, and Self DW, DeltaFosB: a sustained molecular switch for addiction. Proc Natl Acad Sci U S A, 2001. 98(20): p. 11042–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feyder M, et al. , A Role for Mitogen- and Stress-Activated Kinase 1 in L-DOPA-Induced Dyskinesia and FosB Expression. Biol Psychiatry, 2016. 79(5): p. 362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alavian KN, et al. , The lifelong maintenance of mesencephalic dopaminergic neurons by Nurr1 and engrailed. J Biomed Sci, 2014. 21: p. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu Y, et al. , Age-related decreases in Nurr1 immunoreactivity in the human substantia nigra. J Comp Neurol, 2002. 450(3): p. 203–14. [DOI] [PubMed] [Google Scholar]
- 78.Chu Y, et al. , Nurr1 in Parkinson’s disease and related disorders. J Comp Neurol, 2006. 494(3): p. 495–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Decressac M, et al. , NURR1 in Parkinson disease--from pathogenesis to therapeutic potential. Nat Rev Neurol, 2013. 9(11): p. 629–36. [DOI] [PubMed] [Google Scholar]
- 80.Dong J, et al. , Nurr1-Based Therapies for Parkinson’s Disease. CNS Neurosci Ther, 2016. 22(5): p. 351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campos-Melo D, et al. , Nur transcription factors in stress and addiction. Front Mol Neurosci, 2013. 6: p. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colon-Cesario WI, et al. , Knockdown of Nurr1 in the rat hippocampus: implications to spatial discrimination learning and memory. Learn Mem, 2006. 13(6): p. 734–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hawk JD, et al. , NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J Clin Invest, 2012. 122(10): p. 3593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pena de Ortiz S, Maldonado-Vlaar CS, and Carrasquillo Y, Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol Learn Mem, 2000. 74(2): p. 161–78. [DOI] [PubMed] [Google Scholar]
- 85.Aldavert-Vera L, et al. , Intracranial self-stimulation facilitates active-avoidance retention and induces expression of c-Fos and Nurr1 in rat brain memory systems. Behav Brain Res, 2013. 250: p. 46–57. [DOI] [PubMed] [Google Scholar]
- 86.Sellnow RC, Steece-Coliier K., Altwal F, Sandoval IM, Kordower JH, Collier TJ, Sortwell CE, West AR, and Manfredsson FP, Striatal Nurr1 Facilitates the Dyskinetic State and Exacerbates Levodopa-Induced Dyskinesia in a Rat Model of Parkinson’s Disease. Journal of Neuroscience, 2020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hope BT, et al. , Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron, 1994. 13(5): p. 1235–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.