Abstract
Rationale: Obesity-related asthma disproportionately affects minority children and is associated with nonatopic T-helper type 1 (Th1) cell polarized inflammation that correlates with pulmonary function deficits. Its underlying mechanisms are poorly understood.
Objectives: To use functional genomics to identify cellular mechanisms associated with nonatopic inflammation in obese minority children with asthma.
Methods: CD4+ (cluster of differentiation 4–positive) Th cells from 59 obese Hispanic and African American children with asthma and 61 normal-weight Hispanic and African American children with asthma underwent quantification of the transcriptome and DNA methylome and genotyping. Expression and methylation quantitative trait loci revealed the contribution of genetic variation to transcription and DNA methylation. Adjusting for Th-cell subtype proportions discriminated loci where transcription or methylation differences were driven by differences in subtype proportions from loci that were independently associated with obesity-related asthma.
Measurements and Main Results: Obese children with asthma had more memory and fewer naive Th cells than normal-weight children with asthma. Differentially expressed and methylated genes and methylation quantitative trait loci in obese children with asthma, independent of Th-cell subtype proportions, were enriched in Rho-GTPase pathways. Inhibition of CDC42 (cell division cycle 42), one of the Rho-GTPases associated with Th-cell differentiation, was associated with downregulation of the IFNγ, but not the IL-4, gene. Differential expression of the RPS27L (40S ribosomal protein S27-like) gene, part of the p53/mammalian target of rapamycin pathway, was due to nonrandom distribution of expression quantitative trait loci variants between groups. Differentially expressed and/or methylated genes, including RPS27L, were associated with pulmonary function deficits in obese children with asthma.
Conclusions: We found enrichment of Rho-GTPase pathways in obese asthmatic Th cells, identifying them as a novel therapeutic target for obesity-related asthma, a disease that is suboptimally responsive to current therapies.
Keywords: obesity-related asthma, children, gene expression, DNA methylation, expression and methylation quantitative trait loci
At a Glance Commentary
Scientific Knowledge on the Subject
Obesity-related asthma is distinct from normal-weight asthma. Obese children with asthma have evidence of systemic nonatopic T-helper type 1 (Th1) cell polarization that correlates with pulmonary function deficits. However, the mechanistic pathways underlying nonatopic Th-cell immune responses in childhood asthma, including obesity-related asthma, are not well understood.
What This Study Adds to the Field
Analysis of the transcriptome and DNA methylome of the CD4+ (cluster of differentiation 4–positive) Th cells collected from Hispanic and African American children, adjusted for genetic variants and Th-cell subtype proportions, revealed upregulation and differential methylation of genes in the Rho-GTPase pathways in those with obesity-related asthma. We also found that genetic variants associated with differential DNA methylation (methylation quantitative trait loci) were enriched for Rho-GTPase pathways. Differentially expressed and methylated genes in obese asthma Th cells were enriched in their association with FEV1/FVC ratio and expiratory reserve volume, two pulmonary function indices classically associated with obesity-related asthma. Silencing of CDC42, a Rho-GTPase associated with differentiation of naive Th cells, was associated with lower IFN-γ expression but no effect on IL-4 expression, supporting its selective contribution to Th1 differentiation. Together, these findings suggest that Rho-GTPases and their downstream signaling pathways may be novel targets for therapeutic investigation for nonatopic immune patterns found in pediatric obesity–related asthma.
Pediatric obesity-related asthma is a distinct asthma endotype that is more severe, with worse pulmonary function and decreased responsiveness to medications, than normal-weight asthma (1–4). Epidemiologic studies have consistently reported that obesity-related asthma is nonatopic (5, 6). Obese children with asthma have T-helper type 1 cell (Th1) polarization in peripheral blood that correlates with pulmonary function deficits (2, 3). However, the cellular mechanisms that underlie systemic Th1 polarization are not known. Although the disease burden of obesity-related asthma is higher among minority populations (1), the contribution of genotype to the cellular mechanisms is not known. An analysis of the Th-cell transcriptome revealed upregulation of genes in the CDC42 (cell division cycle 42) pathway in obese children with asthma relative to normal-weight children with asthma (7), suggesting a role for Rho-GTPase pathways. However, the contributions of genotype and DNA methylation, a measure of the environmental modulation of genes, to obesity-related asthma are not known.
To address these knowledge gaps, we used a multiomics approach to identify cellular mechanisms that underlie obesity-related asthma (8, 9). We investigated the association of the obese asthma phenotype with the Th-cell transcriptome and DNA methylome, as well as the overlap between them, because both influence each other (10). Given the Th1 polarization in obese individuals with asthma (3) and the cell subtype specificity of the transcriptome and DNA methylome (11–13), we examined the influence of Th-cell subtype proportions (14) on the obese asthma transcriptome and DNA methylome. To identify the contribution of genetic variants (15, 16), we investigated the overlap between expression quantitative trait loci (eQTLs) and methylation quantitative trait loci (meQTLs) and their target genes (e-genes and me-genes) and the obese asthma transcriptome and methylome. Taking this integrated omics analytic approach, we tested the hypothesis that the Th-cell transcriptome and DNA methylome are distinct in obese individuals with asthma, are independent of Th-cell subtype proportions, are influenced by genetic variation, and are associated with the obese asthma phenotype (3).
Methods
Additional details on the study population and the methods are included in the online supplement.
Study Population
One hundred twenty African American and Hispanic children aged 7–11 years with obesity-related asthma (n = 59) or normal-weight asthma (n = 61) were recruited between July 2013 and August 2016 and comprised the primary cohort. A validation cohort, including 20 obese and 20 normal-weight children with asthma and 15 obese and 15 normal-weight children without asthma, was recruited between January 2017 and August 2018. Obesity was defined as a body mass index above the 95th percentile for age and sex (17). Asthma was based on physician diagnosis confirmed in medical records. All participants underwent a blood draw and pulmonary function testing according to the American Thoracic Society guidelines (18). Percent predicted values for spirometric indices were calculated using the National Health and Nutrition Examination Survey prediction equations, and percent predicted values for lung volume indices were calculated using equations developed at a American Thoracic Society workshop (19, 20). The institutional review board at Albert Einstein College of Medicine approved the study.
Study Measures
Isolation of Th cells and quantification of metabolic measures
From fasting blood, CD4+ (cluster of differentiation 4–positive) Th cells were isolated by magnetic bead–based negative selection (STEMCELL Technologies), achieving 95–98% purity (7). Insulin and lipids were quantified in serum.
Quantification of Th-cell transcriptome
RNA from 2 × 106 unstimulated Th cells underwent directional RNA-sequencing (RNA-seq) library preparation and sequencing (7). Of the 120 samples, RNA and RNA-seq libraries from 48 obese asthma and 55 normal-weight asthma samples passed quality control criteria and were included in the analysis.
Quantification of Th-cell subtype proportions
Using the deconvolution approach in the Seurat toolkit (21), we analyzed 10x Genomics reference RNA-seq datasets on naive (CD4+CD45RA+CD25−), regulatory (CD4+CD25+), and memory (CD4+CD45RO+) Th cells and identified seven cell clusters (Figures E1A and E1B in the online supplement). The top 30 genes for each cluster are summarized in Table E1. Cluster 0 overlapped with naive Th cells; clusters 1, 2, and 3 overlapped with memory Th cells; and cluster 4 overlapped with memory and regulatory Th cells (Figures E1A and E1B). Cluster 5, overlapping with natural killer cells, was absent from our samples, and cluster 6 corresponded to B cells and their precursors; these were excluded from downstream analysis. Using CIBERSORT (22), Th-cell subtype proportions were estimated for each study sample (Figure E1C).
Quantification of Th-cell DNA methylome
The Th-cell DNA methylome was quantified using the enzyme digestion–based HELP (HpaII Tiny Fragment Enrichment by Ligation-mediated PCR)-tagging assay (23). Libraries from 45 obese and 54 normal-weight asthma samples passed quality control criteria and were included in the analysis. Of the 1.4 × 106 CGs with quantifiable methylation, 135,947 CGs mapped to a gene promoter, and 685,377 CGs mapped to a gene body, defined by Ensembl release 83, and 55,924 CGs mapped to an enhancer or open chromatin region (24).
Identification of eQTLs and meQTLs in Th cells
Genomic DNA was genotyped using the Infinium Multi-Ethnic Genotyping Array (Illumina). For the 91 individuals included for eQTL and the 76 individuals included for meQTL analyses, genetic variants were converted to the Watson(+) strand, using the Genome Build and Allele Definition Conversion Tool (http://genomics.w3.uvm.edu/software/gact/), to correspond to the RNA-seq and HELP-tagging datasets. For eQTLs, variants within 1 Mb of a gene’s transcription start site, and for meQTLs, variants within 1 Mb of a CG, were tested for their association with gene expression and CG methylation, respectively. Analyses were limited to autosomal variants, removing those with a minor allele frequency <0.1 or those that did not follow the Hardy-Weinberg equilibrium (P < 0.000001). Adjusted P values were calculated by running 10,000 permutations using QTLtools cis, and the final eQTL and meQTL sets were defined by a conditional pass. Results are summarized as Manhattan plots).
Validation Analyses
To verify RNA-seq findings, expression of PAK3 (p21 protein-activated kinase 3) as a representative biologically relevant gene, and expression of RPS27L (40S ribosomal protein S27-like) as the e-gene, was measured using quantitative PCR (qPCR) and analyzed using the comparative cycle threshold method. Differential expression of PAK3 was validated in the validation cohort and also compared with obese and normal-weight nonasthma samples.
To confirm the biological relevance of Rho-GTPase pathways in the nonatopic obese asthma phenotype, we transfected cells with siRNA to silence CDC42, one of the three Rho-GTPases, and quantified gene expression of IFNγ and TNF (tumor necrosis factor) as pertinent downstream Th1 cytokines and of IL-4 as a representative Th2 cytokine. The experiment was done in triplicate.
Statistical Analysis
In this multiomics analysis conducted using R version 3.2.2 software (Figure 1), we quantified differences between the obese and normal-weight asthma Th-cell transcriptome and Th-cell methylome, including the influence of Th-cell subtype proportions on these differences. We then investigated the overlap between the obese asthma Th-cell transcriptome and methylome. To quantify the contribution of genetic variants, we identified the overlap between the obese asthma Th-cell transcriptome and methylome with eQTLs and meQTLs (and their target e-genes and me-genes respectively).
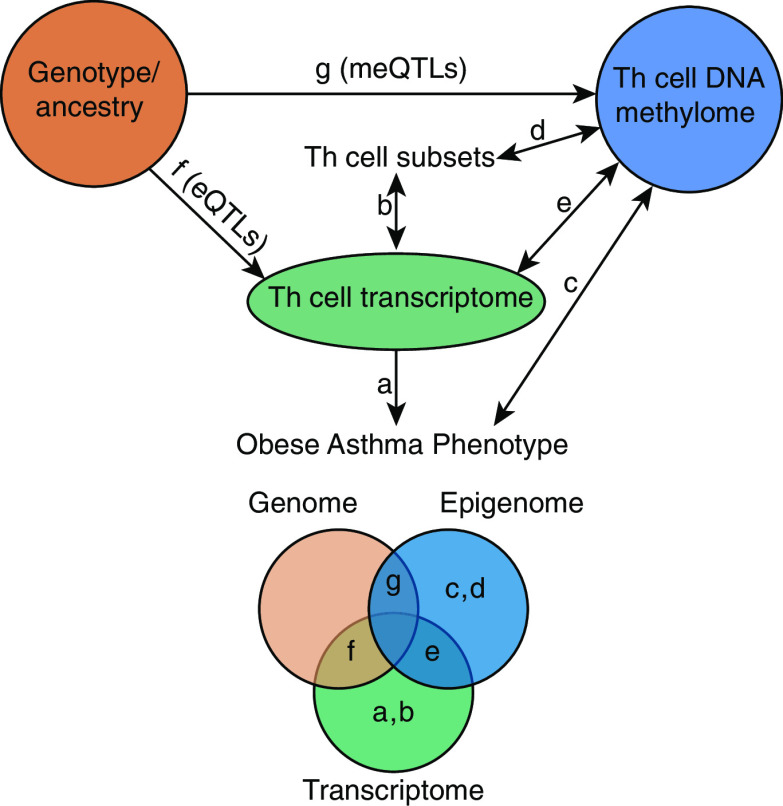
Figure 1.
Summary of the analytic approach. In this multiomics analysis, we compared the T-helper (Th)-cell transcriptome and DNA methylome between obese asthma and normal-weight asthma and quantified the contribution of the genome (genotype) to the transcriptome and DNA methylome. We quantified (a) differences between obese and normal-weight asthma Th-cell transcriptome and (b) the contribution of the Th-cell subtype proportions to differential gene expression. We similarly quantified (c) differences between obese and normal-weight asthma Th-cell DNA methylome and (d) the contribution of Th-cell subtype proportions to differential methylation. (e) We then investigated the overlap between differentially methylated and expressed genes in obese asthma Th cells. To quantify the contribution of genetic variants to the transcriptome and DNA methylome, we identified (f) expression quantitative trait loci (eQTLs) and (g) methylation quantitative trait loci (meQTLs), their target genes (e-genes and me-genes, respectively), and their overlap with differentially expressed and methylated genes. The Venn diagram illustrates these relationships as areas of overlap.
Using principal component analysis, we identified the technical and biological covariates, including the Th-cell subtype proportions, associated with variance of normalized gene expression counts (Figure 2A) and percentage DNA methylation (Figure 3A) and adjusted for these covariates in linear regression modeling, including age, sex, and ethnicity as pertinent demographic variables. To identify the influence of Th-cell proportions on differentially expressed or methylated genes, we conducted linear regression modeling first without and then with inclusion of the Th-cell subtype proportions as covariates. To identify the Gene Ontology pathways enriched in obese asthma samples, genes differentially expressed among obese children with asthma, controlling for false discovery rate at 0.05, as well as CGs with a between-group methylation difference of 10%, a between-group P value < 0.05, and false discovery rate P value < 0.05 in multivariable analysis, were analyzed using NetworkAnalyst software (25).
Figure 2.
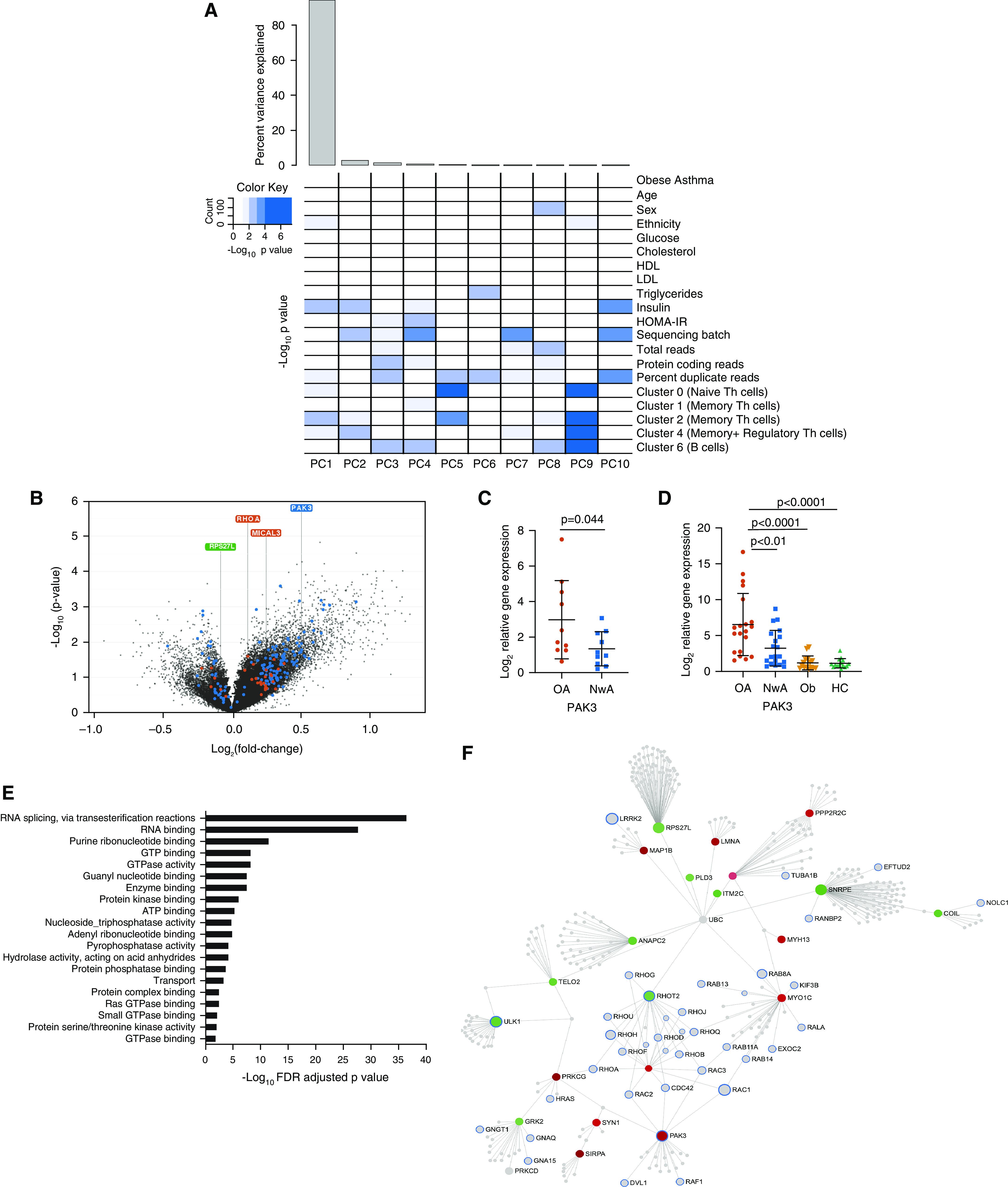
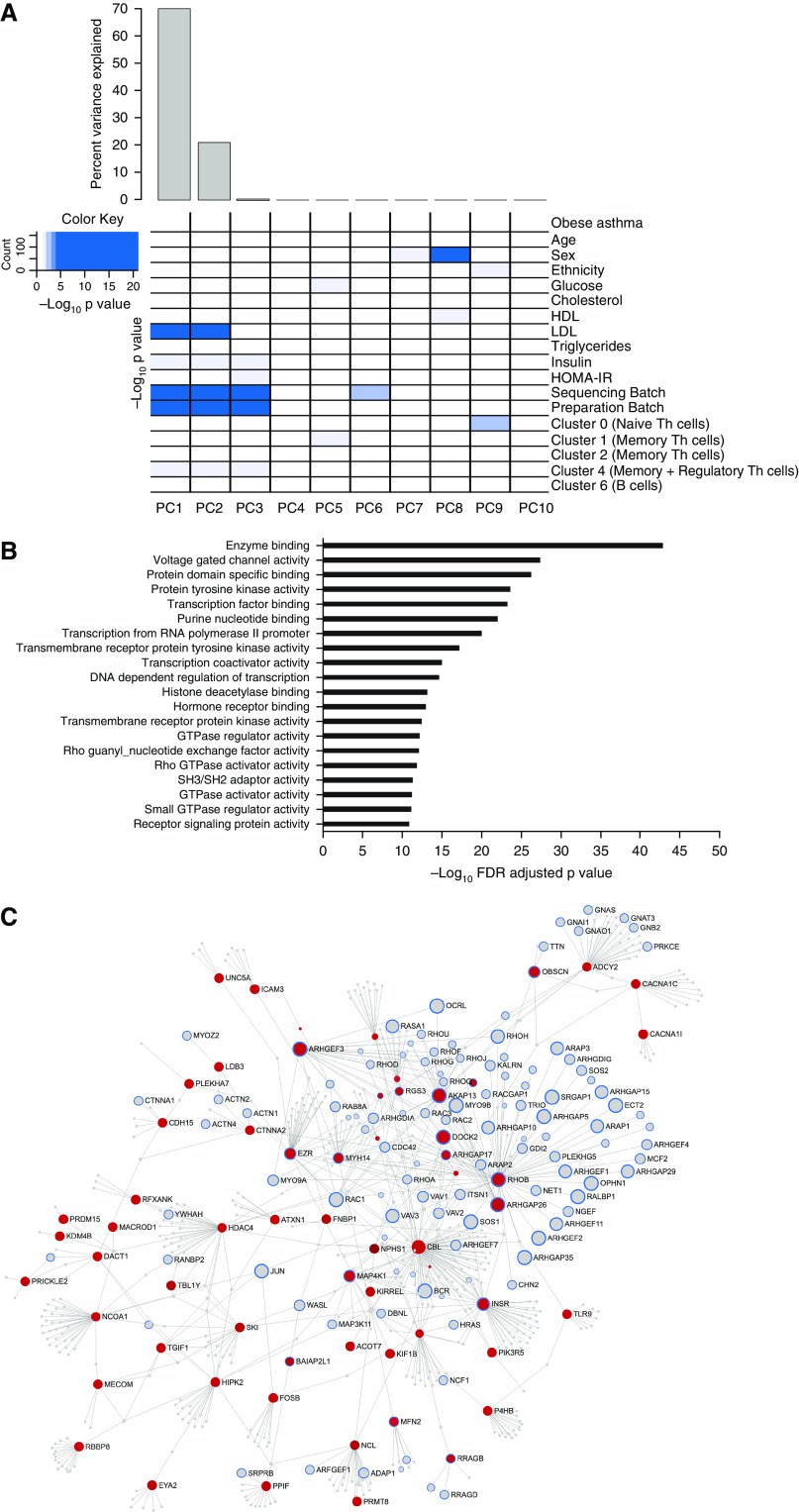
Differential gene expression in obese asthma T-helper (Th) cells. (A) Gene expression variance was influenced by both technical and biological factors. The heat map summarizes the principal components (PCs) for gene expression variance, with >85% variance explained by PC1. Shades of blue denote the significance of the contribution of each covariate to the PC. Apart from technical factors, proportions of naive (cluster 0), memory (clusters 1, 2, and 4), and regulatory Th cells (cluster 4) and serum insulin concentrations were significant contributors to the variance of the Th-cell transcriptome. (B) The volcano plot summarizes the differential obese asthma transcriptome. More genes were upregulated than downregulated in obese asthma Th cells relative to normal-weight asthma Th cells. Genes marked in blue are those that were retained or gained after Th-cell subtype adjustment, whereas those marked in orange were those that were rendered nonsignificant when adjusted for Th-cell subtype proportions. Although RHOA (Ras homolog family member A) and MICAL3 (microtubule-associated monooxygenase, calponin, and LIM domain containing 3) are two representative genes that were rendered nonsignificant after adjusting for cell subtype proportions, PAK3 (p21 protein-activated kinase 3) was not influenced by cell subtype proportions, and RPS27L (40S ribosomal protein S27-like) was identified after adjustment for cell subtype proportions. (C) Higher PAK3 expression identified on RNA-sequencing analysis was confirmed by quantitative PCR in a subset of 10 samples in each group (obese asthma [OA] and normal-weight asthma [NwA]). (D) Differential PAK3 expression was further validated in a separate cohort of 20 obese and 20 normal-weight asthma samples; the potential role of obesity alone (Ob) contributing to PAK3 upregulation was addressed by quantifying it in 15 obese and 15 normal-weight nonasthma or healthy control (HC) samples. (E) The 157 genes differentially expressed in obese asthma, independent of Th-cell subtype proportions, were enriched for Rho-GTPase/small GTPase pathways in Gene Ontology pathway analysis; the top 20 Gene Ontology pathways are shown (false discovery rate [FDR]-adjusted P < 0.05). (F) Network analysis of the differentially expressed genes is shown with genes in the GTPase pathways marked with a blue outline. Those marked in red were upregulated, and those in green were downregulated, in obese asthma Th cells relative to normal-weight asthma Th cells. Genes marked in gray were not significantly differentially expressed in obese relative to normal-weight asthma Th cells. GTP = guanosine triphosphate; HDL = high-density lipoprotein; HOMA-IR = homeostatic model assessment of insulin resistance; LDL = low-density lipoprotein.
Figure 3.
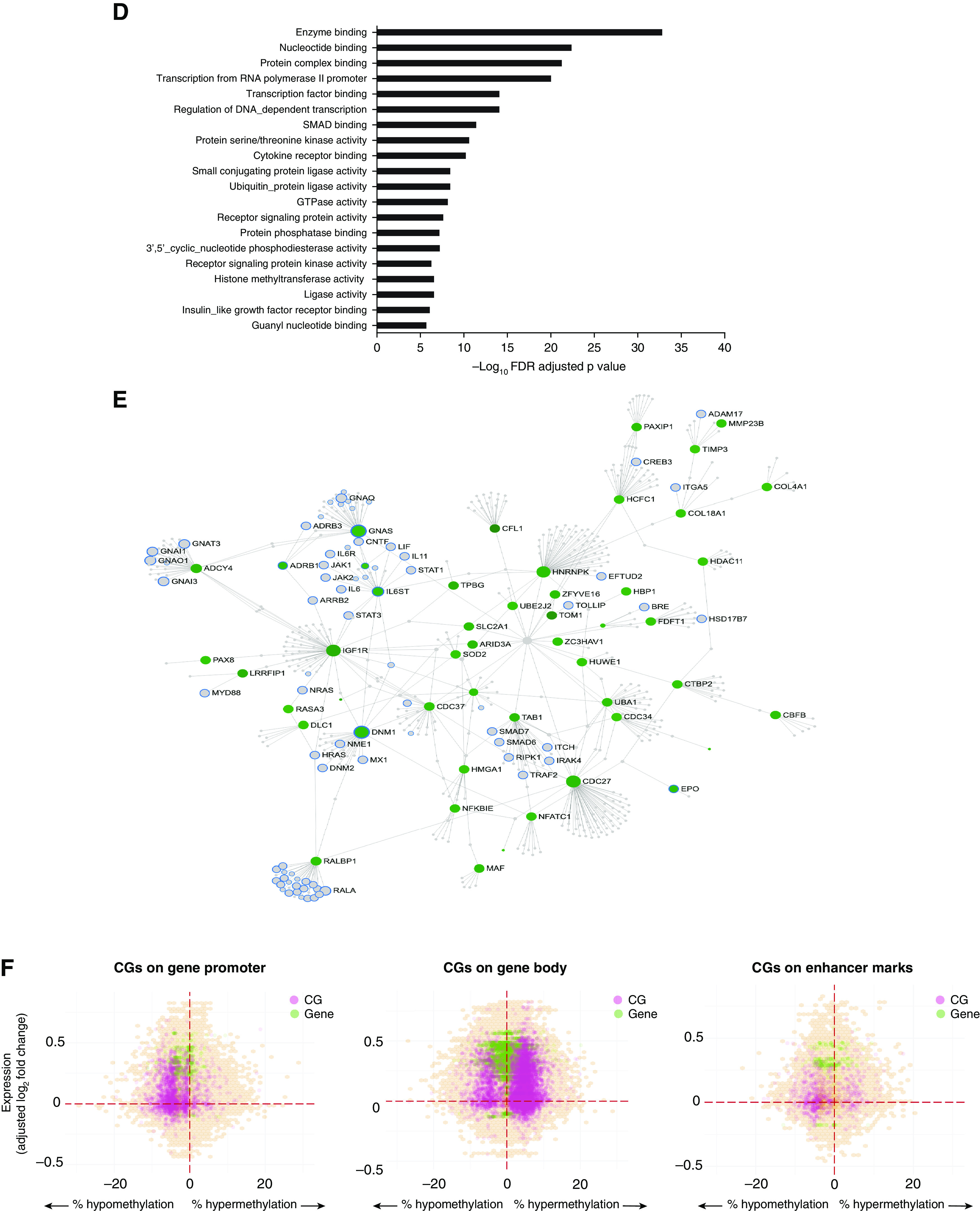
Differential DNA methylation in obese asthma T-helper (Th) cells. (A) DNA methylation variance was also influenced by both technical and biological factors. The heat map summarizes the principal components (PCs) for gene expression variance, with greater than 80% of the variance explained by the first PC. Shades of blue denote the significance of the contribution of each covariate to the PC. Apart from technical factors, proportions of memory and regulatory Th cells (cluster 4) and serum insulin and low-density lipoprotein (LDL) concentrations were significant contributors to variance of the Th-cell methylome. Gene Ontology (GO) pathway analysis revealed that both (B) hypermethylated CGs, marked in red in C, and (D) hypomethylated CGs, marked in green in E, in obese asthma Th cells relative to normal-weight asthma Th cells were enriched for Rho-GTPase/small GTPase pathways; the top 20 GO pathways are included (false discovery rate [FDR]-adjusted P < 0.05). In the networks (C and E), genes in GTPase pathways are marked with blue outline; genes marked in gray were not significantly differentially methylated in obese asthma relative to normal-weight asthma Th cells. (F) The hexagonal plots summarize the association between gene expression and DNA methylation. Statistically significant differentially expressed genes are marked in green, and differentially methylated CGs are marked in pink. Overlapping genes and CGs that did not reach statistical significance are shown in orange, with higher density denoted with darker shades. Both hypo-and hypermethylated CGs were associated with gene upregulation; few CGs were associated with gene downregulation. In keeping with existing literature, more CGs in promoters and enhancers were hypomethylated, and in gene body were hypermethylated, in association with gene upregulation in obese asthma Th cells relative to normal-weight asthma Th cells. GTP = guanosine triphosphate; HDL = high-density lipoprotein; HOMA-IR = homeostatic model assessment of insulin resistance.
To investigate the clinical relevance of differentially expressed genes, with/without overlap with differential methylation, Pearson correlation coefficients were calculated for the association of these genes with FEV1/FVC ratio and expiratory reserve volume (ERV). To determine if differentially expressed genes were not randomly enriched for their association with obese asthma phenotype, we ran a 1,000-iteration permutation analysis to determine the mean number of genes with a nonzero expression value randomly associated with FEV1/FVC ratio and/or ERV. The mean number of randomly associated genes was compared with the number of genes differentially expressed in obese children with asthma that were associated with the pulmonary function variables (Figures E2A–E2C). Gene expression, DNA methylation and genotyping data, and patient characteristics are available at dbGaP (database of Genotypes and Phenotypes) study identifier 33254.
Results
Study Population
Demographic and clinical characteristics of the primary study cohort are summarized in Table 1. Demographics and medication use did not differ between obese and normal-weight children with asthma. Obese children with asthma had lower FEV1/FVC ratio, ERV, FRC, and residual volume/TLC ratio and higher systemic insulin concentrations. Compared with normal-weight children with asthma, obese children with asthma had fewer naive (cluster 0) and more memory (cluster 1) Th cells (Figure E1C). Characteristics of the validation cohort (Table E2) were similar to those of the primary cohort.
Table 1.
Characteristics of Study Participants in Primary Cohort
| Obese Asthma (n = 59) | Normal-Weight Asthma (n = 61) | P Value | |
|---|---|---|---|
| Demographic characteristics | |||
| Age, yr | 9.1 ± 1.7 | 8.9 ± 1.3 | 0.84 |
| Sex, M | 33 (55.9) | 35 (57.4) | 1 |
| Hispanic | 40 (67.8) | 39 (63.9) | 0.70 |
| BMI, z-score | 1.83 ± 0.53 | 0.12 ± 0.87 | <0.001 |
| Insulin | 16.7 ± 11.4 | 12.6 ± 7.3 | 0.03 |
| Atopic sensitization* | 32 (54.2) | 44 (72.1) | 0.04 |
| | |||
| Pulmonary function indices† | |||
| FVC | 102.7 ± 17.2 | 97.7 ± 14.9 | 0.23 |
| FEV1 | 93.3 ± 19.5 | 94.3 ± 17.3 | 0.50 |
| FEV1/FVC | 79.5 ± 12 | 84.4 ± 7.6 | <0.001 |
| TLC | 94.3 ± 17 | 94.4 ± 12.4 | 0.63 |
| RV | 95.7 ± 38.6 | 111.2 ± 30.4 | 0.01 |
| RV/TLC | 24.0 ± 7.5 | 28.1 ± 6.9 | 0.001 |
| ERV | 71.5 ± 20.1 | 90.7 ± 29.6 | <0.001 |
| FRC | 87.5 ± 23.2 | 104.3 ± 25.3 | <0.001 |
| | |||
| Medication use | |||
| Inhaled steroids | 28 (47.4) | 27 (44.3) | 0.85 |
| Montelukast | 41 (69.5) | 37 (60.7) | 0.34 |
| Combination therapy | 16 (27.1) | 15 (24.6) | 0.83 |
Definition of abbreviations: BMI = body mass index; ERV = expiratory reserve volume; RV = residual volume.
All continuous variables are reported as mean ± SD, and categorical variables are reported as n (%).
Atopic sensitization was defined as a positive skin test reaction to two or more allergens.
All pulmonary function indices, other than FEV1/FVC ratio, are reported as percent predicted values.
Differential Gene Expression in Obese Asthma Th Cells and the Contribution of Th-Cell Subtypes
In addition to technical factors and insulin concentrations, proportions of naive (cluster 0), memory (clusters 1, 2 and 4), and regulatory Th cells (cluster 4) were associated with gene expression variance (Figure 2A). Regression analysis without adjusting for Th-cell subtype proportions identified 129 differentially expressed genes in obese asthma Th cells, which were enriched for Rho-GTPase function in Gene Ontology pathway analysis. After adjusting for cell subtype proportions, differential expression of 29 of the 129 genes was rendered nonsignificant (Table E3 and Figure 2B). These 29 genes included RHOARas homolog family member A), a GTPase, and MICAL3 (microtubule-associated monooxygenase, calponin, and LIM domain containing 3), associated with actin polymerization, both of which were upregulated in memory Th cells (cluster 1) and downregulated in naive Th cells (cluster 0). However, the differential expression of PAK3, verified by qPCR (Figure 2C) and validated in a separate cohort of children, including control subjects without asthma (Figure 2D), downstream in the Rho-GTPase pathway, was not influenced by Th-cell subtype proportions, suggesting that downstream genes in the Rho-GTPase pathway may be more robust indicators of primary cellular reprogramming.
An additional 57 genes gained significance after adjusting for cell subtype proportions (Table E4), demonstrating that cell subtype adjustment can reveal genuinely differentially expressed genes that would otherwise be missed. The 100 genes that retained significance and the 57 genes that gained significance after adjustment for cell subtype proportions remained enriched for Rho-GTPase pathways (Figures 2E and 2F) and were retained for further analysis as differentially expressed genes in obese asthma Th cells, independent of Th-cell subtype proportions.
Differential DNA Methylation in Obese Asthma Th Cells and the Contribution of Th-Cell Subtypes
Next, we investigated differences in DNA methylation between obese and normal-weight asthma Th cells and the overlap between differential DNA methylation and gene expression. Similarly to the transcriptome, we identified an association of technical and biological factors (insulin and low-density lipoprotein concentrations) and proportions of regulatory/memory Th cells (cluster 4) with DNA methylation variance (Figure 3A). However, unlike the transcriptome, only 92 CGs were rendered nonsignificant after adjusting for Th-cell subtype proportions; thus, the majority of CGs were not influenced by Th-cell subtype proportions. Among these CGs not influenced by cluster 4 proportions, 836 CGs were ≥10% differentially methylated between obese and normal-weight Th cells. Of these, 452 CGs mapped to a gene promoter, gene body, or cis-regulatory region, of which 237 were hypomethylated and 215 were hypermethylated in obese asthma Th cells (Table E5). Both hyper- and hypomethylated CGs were enriched for Rho-GTPase signaling (Figures 3B–3E). Hypomethylated CGs were also enriched for regulation of cellular metabolic processes and protein modification processes.
Interaction between Differential DNA Methylation and Gene Expression in Obese Asthma Th Cells
Having identified enrichment for Rho-GTPase pathways in both transcriptome and DNA methylome studies, we investigated the potential contribution of DNA methylation to expression of genes enriched in these pathways (Figure 3F). Four genes (DUT[deoxyuridine 5′-triphosphate nucleotidohydrolase], ARHGAP19-SLIT1 [naturally occurring read-through transcription between the neighboring Rho-GTPase activating protein 19 and slit homolog 1], ADAMTS2 [a disintegrin and metalloproteinase with thrombospondin type 1, motif 2], and SLC13A3 [solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3]) were differentially expressed and contained CGs >10% differentially methylated between obese and normal-weight samples (Table E6). ARHGAP19-SLIT1 is a paralog of Rho-GTPase activating protein 19; ADAMTS2 encodes the ADAM metallopeptidase with thrombospondin type 1 motif 2 that modifies the actin cytoskeleton (26); and DUT encodes deoxyuridine 5′-triphosphatase, which plays a role in Th-cell differentiation (27). This suggests that these differentially regulated genes in obese asthma Th cells influence cellular mechanisms with properties in common with those influenced by Rho-GTPase pathways.
Contribution of Genotype to Differential Gene Expression and DNA Methylation in Obese Asthma Th Cells
We then investigated the contribution of genotype to the obese asthma Th-cell transcriptome and DNA methylome. We found 441 e-genes (influenced by eQTLs) in an analysis inclusive of obese and normal-weight samples, as shown in the Manhattan plot in Figure 4A (see also Table E7). Although the e-genes were not enriched for Rho-GTPase pathways, RPS27L was the one e-gene that was among the 157 differentially expressed genes and was downregulated in obese asthma Th cells (Figure 4A), as verified by qPCR (Figure 4B). RPS27L expression did not differ between groups among the major allele homozygous (T/T), major/minor heterozygous (T/C), or minor allele homozygous (C/C) individuals (Figure 4C), although all children homozygous for the minor allele were in the obese asthma group, suggesting that genotype rather than study group was associated with gene expression. The minor allele, more common in the Latino and Afro-Caribbean populations than in white populations (Figure 4D), was an independent predictor of lower gene expression (Table 2). Furthermore, RPS27L inversely correlated with PAK3 concentrations, an association that did not differ by study group (Figure 4E).
Figure 4.
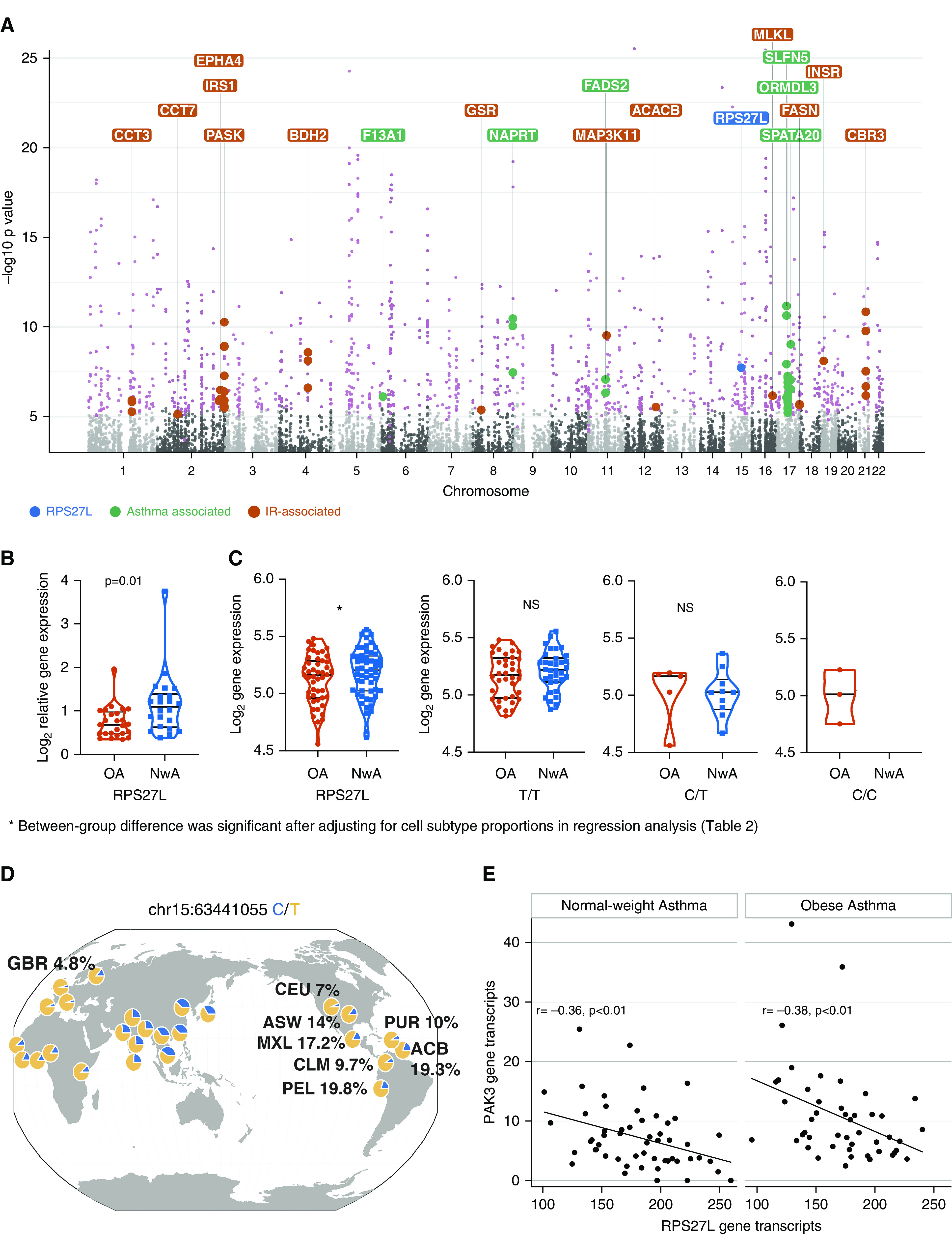
Contribution of genotype to differential gene expression. (A) Manhattan plot of expression quantitative trait loci analysis, inclusive of obese and normal-weight samples, summarizes the 441 expression quantitative trait loci target genes (e-genes; labeled in pink). Of these, several e-genes (labeled in green) overlapped with genes whose variants have previously been associated with childhood asthma, supporting a role of genetic variance in differential expression in childhood asthma. We also found several e-genes in the insulin metabolism pathway (labeled in orange) that suggest a genetic propensity for altered insulin-mediated T-helper cell metabolism. RPS27L (40S ribosomal protein S27-like; labeled in blue) was the one e-gene that was differentially expressed (downregulated) in obese children with asthma and (B) was verified by quantitative PCR. (C) Gene expression differences in RPS27L were driven by nonuniform distribution of genetic variants between obese asthma (OA) and normal-weight asthma (NwA) samples, with only obese children with asthma and no normal-weight children with asthma being homozygous for the minor allele. (D) The minor allele is enriched in Americans of African ancestry in the southwestern United States (ASW), Afro-Caribbeans in Barbados (ACB), and Latino populations (Mexican ancestry from Los Angeles [MXL], Colombians [CLM], Puerto Ricans [PUR], and Peruvians [PEL]) as compared with Utah residents of northern and western European ancestry (CEU) and British (GBR) (mapped on the Genomics of Genetic Variants Browser; popgen.uchicago.edu/ggv). (E) The number of RPS27L gene transcripts inversely correlated with number of PAK3 (p21 protein-activated kinase 3) gene transcripts. IR = insulin resistance; NS = nonsignificant.
Table 2.
Differential Expression of RPS27L in Obese Children with Asthma Is Associated with T-Helper Cell Subtypes and an Expression and Quantitative Trait Locus
| Variables | Model 1 |
Model 2 |
Model 3* |
|||
|---|---|---|---|---|---|---|
| β-Value (95% CI) | P Value | β-Value (95% CI) | P Value | β-Value (95% CI) | P Value | |
| Study group | −0.08 (−0.15 to 0.001) | 0.053 | −0.07 (−0.14 to −0.003) | 0.049 | −0.06 (−0.13 to 0.01) | 0.1 |
| Cluster 0 | — | — | 0.49 (−0.30 to 1.27) | 0.22 | 0.33 (−0.50 to 1.16) | 0.43 |
| Cluster 1 | — | — | 1.01 (−0.04 to 0.01) | 0.06 | 0.75 (−0.39 to 1.89) | 0.19 |
| Cluster 2 | — | — | −0.54 (−0.97 to −0.11) | 0.01 | −0.59 (−1.06 to −0.12) | 0.01 |
| Cluster 4 | — | — | −0.59 (−1.31 to 0.12) | 0.10 | −0.64 (−1.38 to 0.09) | 0.08 |
| Genotype (T/C)† | — | — | — | — | −0.17 (−0.26 to −0.08) | <0.001 |
| Genotype (C/C)† | — | — | — | — | −0.16 (−0.34 to 0.03) | 0.09 |
Definition of abbreviations: CI = confidence interval; RPS27L = 40S ribosomal protein S27-like.
The proportion of memory T-helper cells (cluster 2) and the minor allele were independent predictors of gene expression.
Major allele homozygous (T/T) was the reference group.
Moreover, several e-genes (ORMDL3 [ORMDL sphingolipid biosynthesis regulator 3], SLFN5 [Schlafen family member 5], NAPRT [nicotinate phosphoribosyltransferase], SPATA20 [spermatogenesis-associated protein 20], FADS2 [fatty acid desaturase 2], and F13A1 [coagulation factor XIII A chain]) overlapped with genes previously identified in genome-wide association studies (GWAS) of asthma (Figure 4A) (28–30), and six e-genes (SCGB3A1 [secretoglobin family 3A member 1], CHURC1 [Churchill domain containing 1], ERAP2 [endoplasmic reticulum aminopeptidase 2], DDX11 [DEAD/H-box helicase 11], RPS26 [ribosomal protein S26], and PEX6 [peroxisomal biogenesis factor 6]) overlapped with an eQTL analysis of Puerto Rican pediatric asthma samples (31), supporting that our study population is similar to those included in larger studies. Furthermore, several e-genes, including ACACB (acetyl-coenzyme A carboxylase-β), FASN (fatty acid synthase), PASK (PAS domain-containing serine/threonine-protein kinase), MLKL (mixed lineage kinase domain like pseudokinase), MAP3K11 (mitogen-activated protein kinase kinase kinase 11), CBR3 (carbonyl reductase 3), GSR (glutathione-disulfide reductase), BDH2 (3-hydroxybutyrate dehydrogenase 2), CCT3 (chaperonin containing T-complex protein 1, subunit 3), CCT (chaperonin containing T-complex protein 1), EPHA4 (Ephrin type-A receptor 4), INSR (insulin receptor), and IRS1 (insulin receptor substrate 1), were enriched for the insulin metabolism pathway (Figure 4A). Because insulin is associated with gene expression variance (Figure 2A) and insulin resistance is associated with Th1 polarization in obese individuals with asthma (3), these findings indicate a genetic predisposition to altered insulin-mediated Th-cell metabolism in obese individuals with asthma.
The 1,263 genes proximal to the cytosine targets for meQTLs (me-genes) (Figure 5A and Table E8) were enriched for Rho-GTPase pathways (Figure 5B). Of these, 12 me-genes (PRKCG [protein kinase Cγ], PPP2R2C [serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit Bγ], ADAMTS2, MAP1B [microtubule-associated protein 1B], ALDH1L1 [aldehyde dehydrogenase 1 family member L1], PACSIN2 [protein kinase C and casein kinase substrate in neurons 2], ITM2C [integral membrane protein 2C], CA10[carbonic anhydrase-related protein 10], KLHDC2 [Kelch domain-containing 2], LARGE2[LARGE xylosyl- and glucuronyltransferase 2], ANKRD33 [ankyrin repeat domain 33], and MYO1C [myosin IC]) were among the list of 157 differentially expressed genes. In particular, PRKCG and PPP2R2C are in Rho-GTPase pathways, and ADAMTS2 and MYO1C, which modify the actin cytoskeleton, have molecular functions that overlap with Rho-GTPase activities in the cell.
Figure 5.
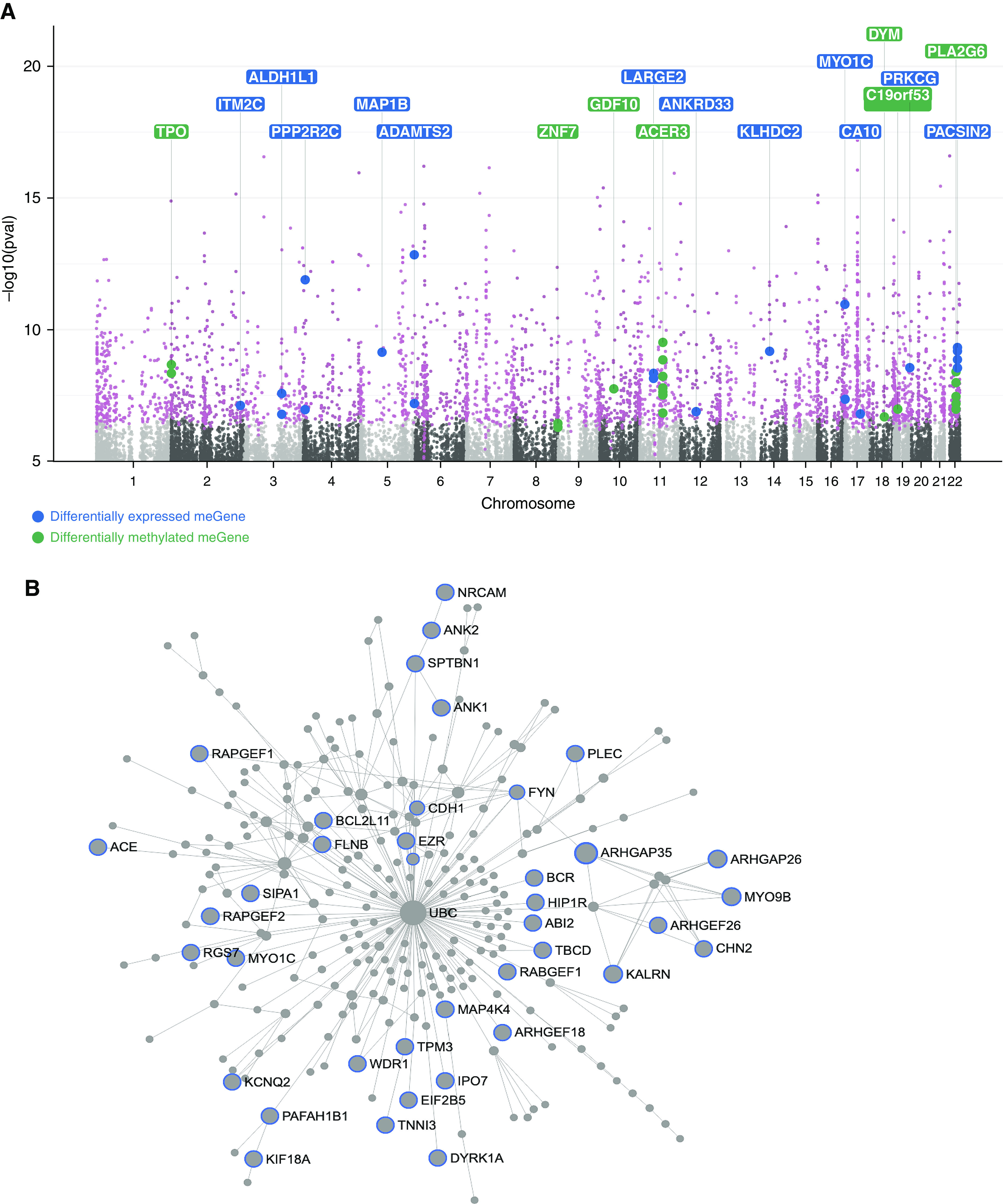
Contribution of genotype to differential methylation. (A) Manhattan plot of methylation quantitative trait loci analysis, inclusive of obese and normal-weight samples, summarizes the 1,263 methylation quantitative trait loci target genes (me-genes; labeled in pink). Of these, 12 me-genes (labeled in blue) were among the 157 differentially expressed genes in obese asthma T-helper (Th) cells, of which PPP2R2C (serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit Bγ) and PRKCG (protein kinase Cγ) are in the Rho-GTPase pathway. These findings suggest that genetic polymorphisms influence gene expression through DNA methylation. We also found that several me-genes (labeled in green) were differentially methylated in the obese asthma relative to normal-weight asthma Th cells, supporting a role of genetic variance in differential methylation in childhood obesity-related asthma. (B) Overall, me-genes were enriched for Rho-GTPase/small GTPase pathways. Genes in GTPase pathways are marked with blue outline.
Clinical Relevance of Rho-GTPase Pathway Enrichment in Obese Asthma Th Cells
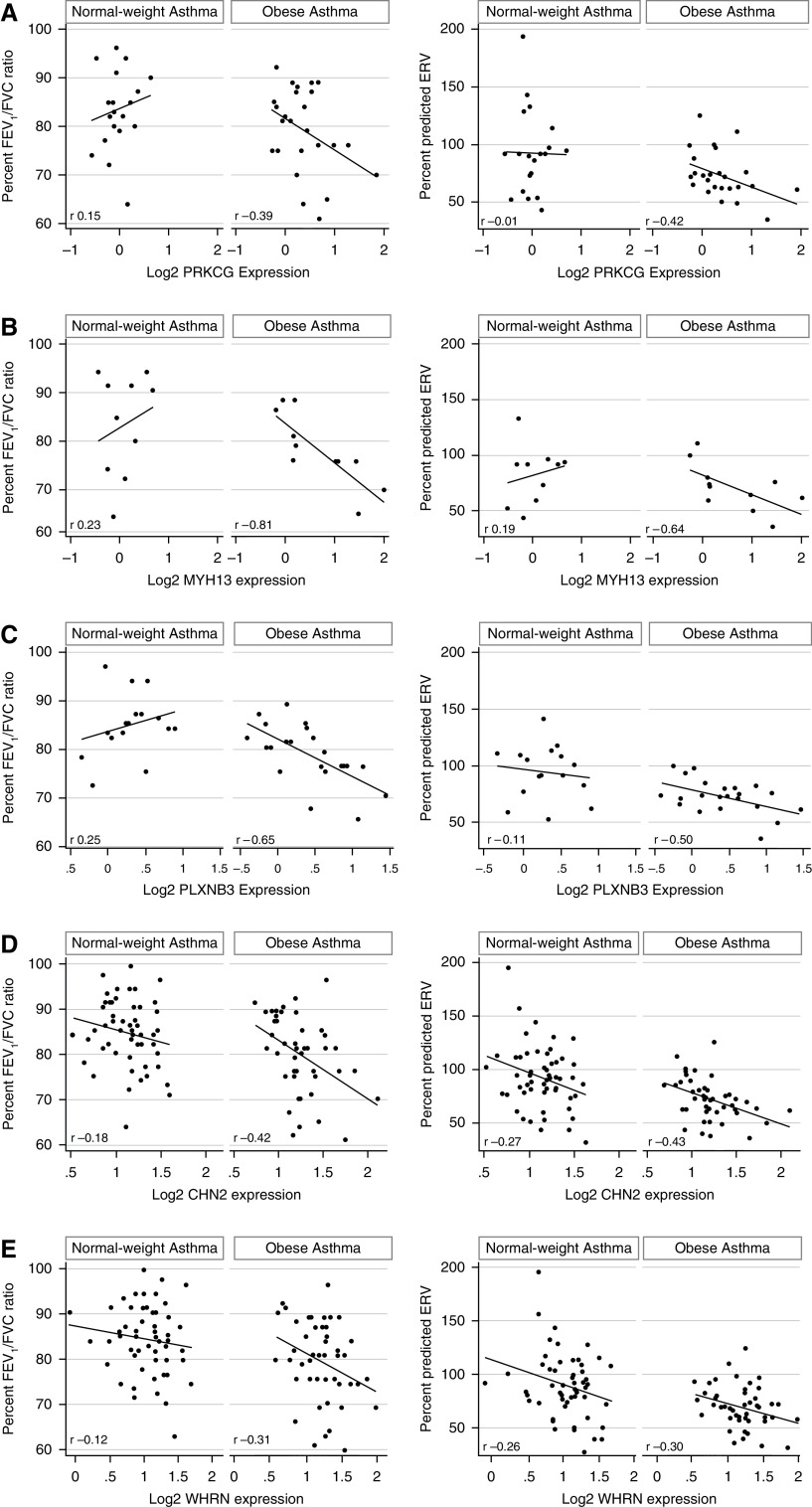
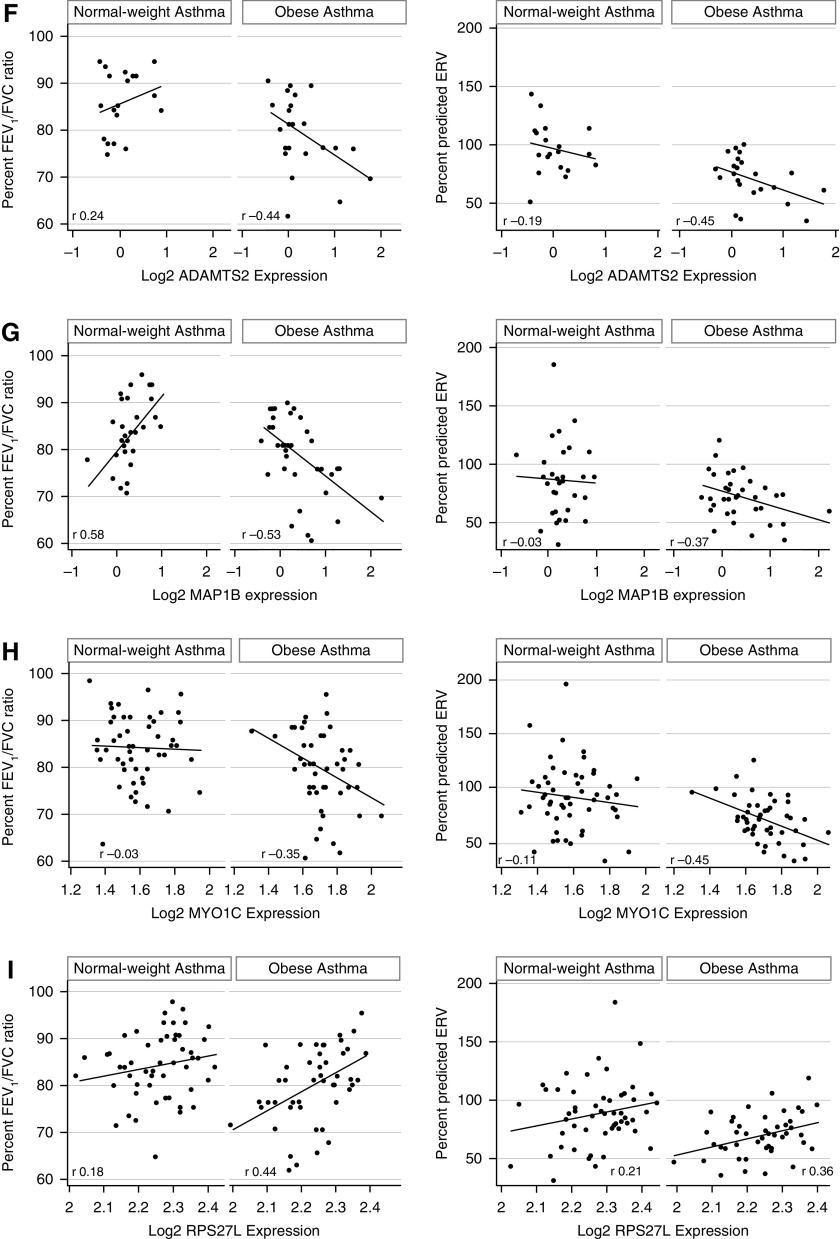
We then tested the clinical relevance of Rho-GTPase pathway enrichment in obese asthma Th cells. We found that the 157 differentially expressed genes were enriched in their association with pulmonary function deficits found in obese individuals with asthma (3). Of the 157 genes, 55 correlated with percentage FEV1/FVC ratio, and 39 correlated with ERV percent predicted, of which 24 genes correlated with both FEV1/FVC ratio and ERV (Figures E2A–E2C). Representative correlations of FEV1/FVC and ERV are shown for genes enriched in Rho-GTPase pathways (PRKCG, MYH13 [myosin heavy chain 13], and PLXNB3 [plexin B3]); genes that influence Rho-GTPases (CHN2 [chimerin 2] and WHRN [whirlin]); and genes associated with cellular mechanisms overlapping with those of Rho-GTPases, including actin polymerization (MYO1C, MAP1B, and ADAMTS2) (Figures 6A–6H). Because RPS27L, the one differentially expressed e-gene, correlated with both FEV1/FVC ratio and ERV (Figure 6I), in multivariable analysis, we found that gene expression and genetic variant were independent predictors of FEV1/FVC (Table 3), whereas gene expression, but not the genetic variant, was an independent predictor of ERV (Table 4).
Figure 6.
Correlation between gene expression and pulmonary function indices associated with obesity-related asthma. Expression of several genes correlated with percentage FEV1/FVC ratio and percent predicted expiratory reserve volume (ERV) in obese children with asthma but not in normal-weight children with asthma. Representative correlations of FEV1/FVC and ERV with differentially expressed genes enriched in Rho-GTPase pathways (A) PRKCG (protein kinase Cγ), (B) MYH13 (myosin heavy chain 13), and (C) PLXNB3 (plexin B3), genes that influence Rho-GTPases (D) CHN2 (chimerin 2) and (E) WHRN (whirlin) and genes associated with cellular mechanisms related to Rho-GTPases, including actin polymerization, (F) MYO1C (myosin IC), (G) MAP1B (microtubule-associated protein 1B), and (H) ADAMTS2 (a disintegrin and metalloproteinase with thrombospondin type 1, motif 2) are shown. (I) RPS27L (40S ribosomal protein S27-like) was the one expression quantitative trait loci target gene that was differentially expressed in obese asthma T-helper cells and was also associated with FEV1/FVC ratio and ERV.
Table 3.
Association of RPS27L Gene Expression with FEV1/FVC Ratio
| Variables | Model 1 |
Model 2 |
Model 3* |
|||
|---|---|---|---|---|---|---|
| β-Value (95% CI) | P Value | β-Value (95% CI) | P Value | β-Value (95% CI) | P Value | |
| Gene expression | 12.56 (5.41 to 19.71) | 0.001 | 11.38 (4.40 to 18.36) | 0.002 | 14.07 (5.59 to 22.55) | 0.001 |
| Obese asthma | — | — | −4.10 (−7.07 to −1.12) | 0.007 | −3.72 (−6.95 to −0.48) | 0.03 |
| Genotype (T/C)† | — | — | — | — | 0.67(−3.81 to 5.14) | 0.77 |
| Genotype (C/C)† | — | — | — | — | −10.04(−18.95 to −1.13) | 0.03 |
Definition of abbreviations: CI = confidence interval; RPS27L = 40S ribosomal protein S27-like.
Both gene expression and genetic variant were associated with percentage FEV1/FVC ratio.
Major allele homozygous (T/T) was the reference group.
Table 4.
Association of RPS27L Gene Expression with Percent Predicted ERV
| Variables | Model 1 |
Model 2 |
Model 3* |
|||
|---|---|---|---|---|---|---|
| β-Value (95% CI) | P Value | β-Value (95% CI) | P Value | β-Value (95% CI) | P Value | |
| Gene expression | 35.88 (11.76 to 60.00) | 0.004 | 30.62 (7.74 to 53.49) | 0.009 | 30.00 (1.49 to 58.51) | 0.04 |
| Obese asthma | — | — | −18.25 (−28.00 to −8.50) | 0.007 | −18.37 (−29.26 to −7.49) | 0.03 |
| Genotype (T/C)† | — | — | — | — | −3.40 (−18.46 to 11.66) | 0.65 |
| Genotype (C/C)† | — | — | — | — | −11.95 (−41.91 to 18.01) | 0.43 |
Definition of abbreviations: CI = confidence interval; ERV = expiratory reserve volume; RPS27L = 40S ribosomal protein S27-like.
Gene expression, but not the genetic variant, was associated with ERV percent predicted.
Major allele homozygous (T/T) was the reference group.
Validation of Rho-GTPases in Nonatopic Responses in the Obese Asthma Phenotype
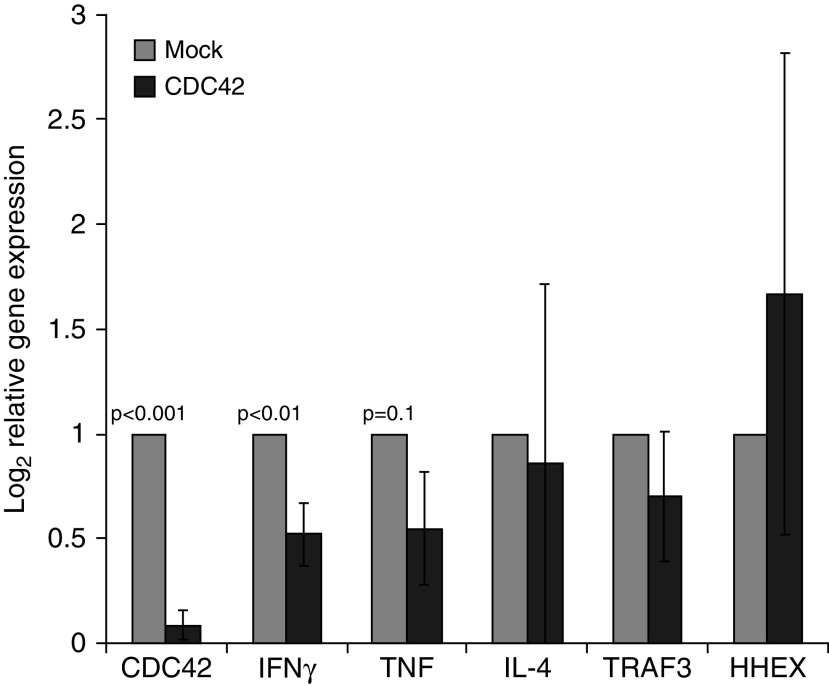
Given the clinical relevance of the Rho-GTPase pathway to the obese asthma phenotype, to investigate its role in the nonatopic immune profile, we used CDC42, one of the three Rho-GTPases, because it is upstream of and linked with PAK3 function (32). Compared with mock siRNA, CDC42 siRNA transfection led to a 95% downregulation of CDC42, which was associated with 51% downregulation of IFNγ, but no change in IL-4 expression. Furthermore, although it did not reach statistical significance, a 50% downregulation of TNF expression was observed in cells with CDC42 silencing. As expected, no effect was detected on expression of genes (TRAF3 [TNF receptor–associated factor 3] and HHEX[hematopoietically expressed homeobox protein]) not influenced by CDC42 (Figure 7).
Figure 7.
Validation of the role of Rho-GTPases in nonatopic T-helper–cell responses. siRNA-based silencing of CDC42 (cell division cycle 42; black bars), as compared with mock control (gray bars), was associated with downregulation of IFNγ, but not of IL-4, expression. Although it did not reach statistical significance, a 50% downregulation of TNF (tumor necrosis factor) expression was observed in cells with CDC42 silencing. TRAF3 (TNF receptor–associated factor 3) and HHEX (hematopoietically expressed homeobox protein), included as additional controls, supported that the findings were not due to off-target effects of gene silencing.
Discussion
In this study, using a multiomics approach that combined transcriptome and DNA methylome profiling of purified Th cells, adjusted for the influence of genetic polymorphisms and cell subtype proportions, we found evidence for enrichment of Rho-GTPase pathways in a cell repertoire enriched for memory Th cells in obese children with asthma. In addition, me-genes were also enriched in the Rho-GTPase pathways, identifying a role of the genotype in obese asthma. The differentially expressed and methylated genes in obese asthma samples were enriched in their association with two pulmonary function indices associated with the obese asthma phenotype. A gene silencing–based approach further validated a role of Rho-GTPases in nonallergic Th-cell immune responses. Together, these results provide evidence for a role of Rho-GTPases in the nonallergic immune responses associated with the obese asthma disease phenotype.
Although there is limited understanding of pathways underlying nonatopic asthma, there is substantial evidence to support the role of Rho-GTPases in Th-cell development (33). By modifying the immune synapse, which modulates Th-cell differentiation (34), Rho-GTPases influence differentiation of naive Th cells to the Th1 phenotype (35) and production of nonatopic cytokines, IFN-γ and TNF (36). We have previously reported a higher proportion of Th1 cells with higher circulating concentrations of IFN-γ and TNF in obese children with asthma (3). Our cell subtype deconvolution approach validates these findings with more memory and fewer naive Th cells in obese children with asthma. Although the transcriptome revealed an upregulation of RHOA in memory Th cells and downregulation in naive Th cells in obese children with asthma, differences in PAK3 expression, a gene downstream of CDC42, one of the Rho-GTPases, could not be attributed to Th-cell subtype proportion differences, supporting a potential key role of PAK3 in the obese asthma phenotype. Upregulation of PAK3 in obese children with asthma relative to normal-weight children with asthma, obese children without asthma, and healthy control subjects, further suggested that asthma or obesity alone does not contribute to PAK3 upregulation and confirmed its distinct role in obese children with asthma. Because PAK3 is involved in CDC42-mediated MAPK (mitogen-activated protein kinase) and downstream NF-κB (nuclear factor-κB) activation (32), and because MAPK/NF-κB activation leads to IFN-γ production (37), we tested the contribution of Rho-GTPases to the nonatopic immune responses in obese children with asthma. Selective inhibition of IFN-γ and TNF, rather than IL-4, in response to CDC42 silencing confirmed the contribution of CDC42 to Th1 responses. Our findings are also supported by enrichment of upregulated genes in the NF-κB pathway among children who developed obesity in the CAMP (Childhood Asthma Control Program) study (38) and had more severe disease (39). Furthermore, differential DNA methylation in phosphatidylinositol-3-kinase/Akt (protein kinase B), MAPK, and NF-κB signaling pathways, which are all downstream of Rho-GTPase pathways, were observed in blood from nonatopic obese adults with asthma (40). Together, our findings, in the context of prior literature, provide substantial evidence to support that the Rho-GTPase/MAPK/NF-κB pathway is central in mediating the nonatopic obese asthma phenotype.
We also identified RPS27L as the one gene that was downregulated in obese individuals with asthma, contained an eQTL that is enriched in Latino and Afro-Caribbean populations, and was associated with the obese asthma phenotype. These findings exemplify how functional genomic differences can reveal a genetic association with the studied phenotype. Disruption of RPS27L, best known for its role in p53-mediated apoptosis (41), triggers ribosomal stress and induction of p53, which modifies cellular metabolism with upregulation of the glucose cell surface receptors GLUT1 (glucose transporter 1) and GLUT4 (42), and activation of mTORC1 (mammalian target of rapamycin complex 1) pathway (43). Intriguingly, the mTORC1 pathway is also activated by insulin (44) that increases GLUT1 and GLUT4 translocation to the cell surface in the setting of Th-cell activation (45), which is dependent on increased T-cell metabolism (45). Because mTORC1 causes selective maturation of naive Th cells into Th1 and Th17 cells (46), particularly in the setting of excess nutrients (47), we speculate that RPS27L may be a key gene that links genetic susceptibility to obesity-related asthma with cellular microenvironment exposures, including hyperinsulinemia, triggering activation of the mTORC1 pathway that in turn facilitates nonatopic inflammation in obese individuals with asthma. The inverse association between RPS27L and PAK3 provides further evidence of its potential role in the obese asthma phenotype through altered immunometabolism, a hypothesis that is also supported by enrichment of several e-genes in the insulin metabolism pathway, and warrants further investigation.
Although ours is a single-site study, several eQTLs identified in our population have previously been identified in GWAS for asthma, including ORMDL3 (28), SLFN5, NAPRT, SPATA20 (29), FADS2, and F13A1 (30), suggesting that our study participants are similar to those included in large multicenter studies (29, 30). Highlighting the importance of genetic admixture in multifactorial diseases, six eQTLs in our population overlapped with those identified in a study on Puerto Rican children with asthma (31), whereas only one eQTL (rs9272346) associated with HLA-DRB1 overlapped with a meta-analysis of asthma GWAS that included individuals of European, African, Japanese, and Latino ancestry (48). The association of these genetic variants, enriched for ancestry, with gene expression further supports their role in childhood asthma.
Although our study is the first to identify meQTLs in the context of obesity-related asthma, six differentially methylated genes (ACOT7 [acyl-coenzyme A thioesterase 7], ARHGAP17 [Rho-GTPase–activating protein 17], ARID3A [AT-rich interactive domain-containing protein 3A], SHANK2 [SH3 and multiple ankyrin repeat domains protein 2], SLC2A9 [solute carrier family 2, facilitated glucose transporter member 9], and ZFPM1[zinc finger protein, FOG family member 1]) overlapped with those identified in one recent epigenome-wide meta-analysis on childhood asthma that included admixed populations; there was no overlap with the study that included only European cohorts (49, 50). Of these, ARHGAP17 is a Rho-GTPase–activating protein; SHANK2 plays a role in actin cytoskeleton formation; SLC2A9 plays a role in glucose homeostasis; and ARID3A is a direct p53 effector. Furthermore, ADAMTS2 was interesting because it correlated with pulmonary function, was upregulated in obese asthma Th cells, was targeted for increased local DNA methylation, and was proximal to a cytosine associated with an meQTL, and because it plays a role in actin polymerization, a key cellular mechanism for Th-cell activation and chemotaxis (26). Because immune synapse formation and chemotaxis, downstream of actin polymerization, require an increase in cellular metabolism (45), which specifically involves anaerobic glycolysis in Th cells, the differentially expressed genes in obese asthma Th cells may together contribute to increased Th-cell activation. Together, these findings validate prior studies that included admixed populations, highlighting the contribution of genotype to both the transcriptome and DNA methylome in urban minority obese children with asthma. These results will inform future investigation of mechanisms by which obese asthma Th cells interact with the lungs to cause disease.
We recognize that a larger sample size would have allowed greater confidence in the findings, because our sample number was further limited by exclusion of several samples due to quality control issues. A larger study would allow a more robust test of the causal link between genotype, gene expression, DNA methylation, and the obese asthma phenotype. A more comprehensive DNA methylation assay could have revealed additional informative loci at which DNA methylation was altered. Furthermore, we note that proximity, by itself, does not confirm that change in DNA methylation influences gene expression. However, by using our stringent approach, we validated our earlier observation of a role for small GTPases in Th-cell immune responses in obesity-related asthma (7), and we identified new genes influenced by genotype and DNA methylation that partly overlapped with those showing differential expression in obese individuals with asthma. Having used the deconvolution approach rather than cell subtype sorting, we do not have conclusive evidence of cell-specific DNA methylation and gene expression patterns. However, our findings identify the need for such single-cell approaches to further explore the contribution of DNA methylation to gene expression and Th-cell subtype differentiation in multifactorial diseases such as asthma.
In conclusion, we found evidence of enrichment of Rho-GTPase pathways in Th cells enriched for memory T cells among urban minority obese children with asthma. Although it is difficult to understand how the environment influences a phenotype, we found enrichment of differential DNA methylation in the same pathways that included differentially expressed genes, identifying a role of DNA methylation for the first time in the context of childhood obesity-related asthma. Furthermore, the enrichment of meQTLs and the one e-gene, RPS27L, which was associated with PAK3, a gene downstream in the Rho-GTPase pathway, identifies a contribution of genetic variability that mediates susceptibility to environmental influences. These studies reveal a role for DNA methylation as a mechanism that may mediate, or reflect other regulatory events mediating, the effect of genetic variance on transcription, and they exemplify a gene-by-environment interaction. Together, the cellular mechanisms identified in this study provide a foundation for further exploration of the biological pathways, including those mediated by small GTPases that may underlie the nonatopic immune responses in pediatric obesity-related asthma.
Supplementary Material
Acknowledgments
Acknowledgment
The authors acknowledge the children and their families who participated in this study and Toni A. Tobias for her contribution to this study.
Footnotes
Supported by the NIH (grants K23HL118733 and HL141849 [D.R.]).
Author Contributions: D.R., F.M., and J.M.G. contributed to the study design, data analysis, and drafting of the manuscript. A.D.J. and M.S. contributed to data analysis and interpretation and critical review of the manuscript. J.N., L.N.L., and Y.J. contributed to data acquisition and critical review of the manuscript. All authors approve of the submitted manuscript and are personally accountable for their contributions. D.R. and A.D.J. contributed equally to the study.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201906-1199OC on April 7, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gross E, Lee DS, Hotz A, Ngo KC, Rastogi D. Impact of obesity on asthma morbidity during a hospitalization. Hosp Pediatr. 2018;8:538–546. doi: 10.1542/hpeds.2017-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rastogi D, Canfield SM, Andrade A, Isasi CR, Hall CB, Rubinstein A, et al. Obesity-associated asthma in children: a distinct entity. Chest. 2012;141:895–905. doi: 10.1378/chest.11-0930. [DOI] [PubMed] [Google Scholar]
- 3.Rastogi D, Fraser S, Oh J, Huber AM, Schulman Y, Bhagtani RH, et al. Inflammation, metabolic dysregulation, and pulmonary function among obese urban adolescents with asthma. Am J Respir Crit Care Med. 2015;191:149–160. doi: 10.1164/rccm.201409-1587OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGarry ME, Castellanos E, Thakur N, Oh SS, Eng C, Davis A, et al. Obesity and bronchodilator response in black and Hispanic children and adolescents with asthma. Chest. 2015;147:1591–1598. doi: 10.1378/chest.14-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinmayr G, Forastiere F, Büchele G, Jaensch A, Strachan DP, Nagel G ISAAC Phase Two Study Group. Overweight/obesity and respiratory and allergic disease in children: International Study of Asthma and Allergies in Childhood (ISAAC) Phase Two. PLoS One. 2014;9:e113996. doi: 10.1371/journal.pone.0113996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han Y-Y, Forno E, Celedón JC. Adiposity, fractional exhaled nitric oxide, and asthma in U.S. children. Am J Respir Crit Care Med. 2014;190:32–39. doi: 10.1164/rccm.201403-0565OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rastogi D, Nico J, Johnston AD, Tobias TAM, Jorge Y, Macian F, et al. CDC42-related genes are upregulated in helper T cells from obese asthmatic children. J Allergy Clin Immunol. 2018;141:539–548, e7. doi: 10.1016/j.jaci.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pecak M, Korošec P, Kunej T. Multiomics data triangulation for asthma candidate biomarkers and precision medicine. OMICS. 2018;22:392–409. doi: 10.1089/omi.2018.0036. [DOI] [PubMed] [Google Scholar]
- 9.Persson H, Kwon AT, Ramilowski JA, Silberberg G, Söderhäll C, Orsmark-Pietras C, et al. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J Allergy Clin Immunol. 2015;136:638–648. doi: 10.1016/j.jaci.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 11.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong Y, Rastogi D, Seoighe C, Greally JM, Suzuki M. Insights from deconvolution of cell subtype proportions enhance the interpretation of functional genomic data. PLoS One. 2019;14:e0215987. doi: 10.1371/journal.pone.0215987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur J Immunol. 2009;39:2076–2082. doi: 10.1002/eji.200939722. [DOI] [PubMed] [Google Scholar]
- 15.Nieuwenhuis MA, Siedlinski M, van den Berge M, Granell R, Li X, Niens M, et al. Combining genomewide association study and lung eQTL analysis provides evidence for novel genes associated with asthma. Allergy. 2016;71:1712–1720. doi: 10.1111/all.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol. 2014;15:777–788. doi: 10.1038/ni.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. Overweight & obesity. 2018 [accessed 2018 Apr 30]. Available from: https://www.cdc.gov/obesity/
- 18.Miller MR, Crapo R, Hankinson J, Brusasco V, Burgos F, Casaburi R, et al. ATS/ERS Task Force. General considerations for lung function testing. Eur Respir J. 2005;26:153–161. doi: 10.1183/09031936.05.00034505. [DOI] [PubMed] [Google Scholar]
- 19.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 20.Stocks J, Quanjer PH. Reference values for residual volume, functional residual capacity and total lung capacity: ATS Workshop and Lung Volume Measurements. Official Statement of the European Respiratory Society. Eur Respir J. 1995;8:492–506. doi: 10.1183/09031936.95.08030492. [DOI] [PubMed] [Google Scholar]
- 21.Butler A, Hoffman P, Smibert P, Paplexi E, Satija R. Integrated analysis of single cell transcriptomic data across conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rastogi D, Suzuki M, Greally JM. Differential epigenome-wide DNA methylation patterns in childhood obesity-associated asthma. Sci Rep. 2013;3:2164. doi: 10.1038/srep02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The Ensembl Regulatory Build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 26.Dubail J, Kesteloot F, Deroanne C, Motte P, Lambert V, Rakic JM, et al. ADAMTS-2 functions as anti-angiogenic and anti-tumoral molecule independently of its catalytic activity. Cell Mol Life Sci. 2010;67:4213–4232. doi: 10.1007/s00018-010-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilpo JA. Mitogen induction of deoxyuridine triphosphatase activity in human T and B lymphocytes. Med Biol. 1983;61:54–58. [PubMed] [Google Scholar]
- 28.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 29.Qiu W, Rogers AJ, Damask A, Raby BA, Klanderman BJ, Duan QL, et al. Pharmacogenomics: novel loci identification via integrating gene differential analysis and eQTL analysis. Hum Mol Genet. 2014;23:5017–5024. doi: 10.1093/hmg/ddu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma S, Zhou X, Thibault DM, Himes BE, Liu A, Szefler SJ, et al. A genome-wide survey of CD4+ lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014;134:1153–1162. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Brehm JM, Lin J, Wang T, Forno E, Acosta-Pérez E, et al. Expression quantitative trait loci (eQTL) mapping in Puerto Rican children. PLoS One. 2015;10:e0122464. doi: 10.1371/journal.pone.0122464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagrodia S, Dérijard B, Davis RJ, Cerione RA. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 33.Smits K, Iannucci V, Stove V, Van Hauwe P, Naessens E, Meuwissen PJ, et al. Rho GTPase Cdc42 is essential for human T-cell development. Haematologica. 2010;95:367–375. doi: 10.3324/haematol.2009.006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thauland TJ, Koguchi Y, Wetzel SA, Dustin ML, Parker DC. Th1 and Th2 cells form morphologically distinct immunological synapses. J Immunol. 2008;181:393–399. doi: 10.4049/jimmunol.181.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Panhuys N. TCR signal strength alters T-DC activation and interaction times and directs the outcome of differentiation. Front Immunol. 2016;7:6. doi: 10.3389/fimmu.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chemin K, Bohineust A, Dogniaux S, Tourret M, Guégan S, Miro F, et al. Cytokine secretion by CD4+ T cells at the immunological synapse requires Cdc42-dependent local actin remodeling but not microtubule organizing center polarity. J Immunol. 2012;189:2159–2168. doi: 10.4049/jimmunol.1200156. [DOI] [PubMed] [Google Scholar]
- 37.Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, et al. Interaction of NF-κB and NFAT with the interferon-γ promoter. J Biol Chem. 1997;272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 38.Croteau-Chonka DC, Chen Z, Barnes KC, Barraza-Villarreal A, Celedón JC, Gauderman WJ, et al. Gene coexpression networks in whole blood implicate multiple interrelated molecular pathways in obesity in people with asthma. Obesity (Silver Spring) 2018;26:1938–1948. doi: 10.1002/oby.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strunk RC, Colvin R, Bacharier LB, Fuhlbrigge A, Forno E, Arbelaez AM, et al. Childhood Asthma Management Program Research Group. Airway obstruction worsens in young adults with asthma who become obese. J Allergy Clin Immunol Pract. 2015;3:765–771, e2. doi: 10.1016/j.jaip.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong A, Imboden M, Ghantous A, Novoloaca A, Carsin AE, Kogevinas M, et al. DNA methylation in inflammatory pathways modifies the association between BMI and adult-onset non-atopic asthma. Int J Environ Res Public Health. 2019;16:600. doi: 10.3390/ijerph16040600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Tan J, Zhuang L, Banerjee B, Yang X, Chau JF, et al. Ribosomal protein S27-like, a p53-inducible modulator of cell fate in response to genotoxic stress. Cancer Res. 2007;67:11317–11326. doi: 10.1158/0008-5472.CAN-07-1088. [DOI] [PubMed] [Google Scholar]
- 42.Puzio-Kuter AM. The role of p53 in metabolic regulation. Genes Cancer. 2011;2:385–391. doi: 10.1177/1947601911409738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong X, Liu X, Li H, He H, Sun Y, Zhao Y. Ribosomal protein S27-like regulates autophagy via the β-TrCP-DEPTOR-mTORC1 axis. Cell Death Dis. 2018;9:1131. doi: 10.1038/s41419-018-1168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Rhodes CJ, Lawrence JC., Jr Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J Biol Chem. 2006;281:24293–24303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
- 45.Stentz FB, Kitabchi AE. Hyperglycemia-induced activation of human T-lymphocytes with de novo emergence of insulin receptors and generation of reactive oxygen species. Biochem Biophys Res Commun. 2005;335:491–495. doi: 10.1016/j.bbrc.2005.07.109. [DOI] [PubMed] [Google Scholar]
- 46.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmuller J, Ang W, et al. Australian Asthma Genetics Consortium Collaborators. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu CJ, Söderhäll C, Bustamante M, Baïz N, Gruzieva O, Gehring U, et al. BIOS Consortium. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6:379–388. doi: 10.1016/S2213-2600(18)30052-3. [DOI] [PubMed] [Google Scholar]
- 50.Reese SE, Xu CJ, den Dekker HT, Lee MK, Sikdar S, Ruiz-Arenas C, et al. BIOS Consortium. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. 2019;143:2062–2074. doi: 10.1016/j.jaci.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.