Abstract
Rationale: The management of indeterminate pulmonary nodules (IPNs) remains challenging, resulting in invasive procedures and delays in diagnosis and treatment. Strategies to decrease the rate of unnecessary invasive procedures and optimize surveillance regimens are needed.
Objectives: To develop and validate a deep learning method to improve the management of IPNs.
Methods: A Lung Cancer Prediction Convolutional Neural Network model was trained using computed tomography images of IPNs from the National Lung Screening Trial, internally validated, and externally tested on cohorts from two academic institutions.
Measurements and Main Results: The areas under the receiver operating characteristic curve in the external validation cohorts were 83.5% (95% confidence interval [CI], 75.4–90.7%) and 91.9% (95% CI, 88.7–94.7%), compared with 78.1% (95% CI, 68.7–86.4%) and 81.9 (95% CI, 76.1–87.1%), respectively, for a commonly used clinical risk model for incidental nodules. Using 5% and 65% malignancy thresholds defining low- and high-risk categories, the overall net reclassifications in the validation cohorts for cancers and benign nodules compared with the Mayo model were 0.34 (Vanderbilt) and 0.30 (Oxford) as a rule-in test, and 0.33 (Vanderbilt) and 0.58 (Oxford) as a rule-out test. Compared with traditional risk prediction models, the Lung Cancer Prediction Convolutional Neural Network was associated with improved accuracy in predicting the likelihood of disease at each threshold of management and in our external validation cohorts.
Conclusions: This study demonstrates that this deep learning algorithm can correctly reclassify IPNs into low- or high-risk categories in more than a third of cancers and benign nodules when compared with conventional risk models, potentially reducing the number of unnecessary invasive procedures and delays in diagnosis.
Keywords: early detection, risk stratification, neural networks, lung cancer, computer-aided image analysis
At a Glance Commentary
Scientific Knowledge on the Subject
It is unknown whether a deep learning algorithm applied to chest computed tomography scans of individuals presenting with indeterminate pulmonary nodules allows their reclassification into lower- or higher-risk groups.
What This Study Adds to the Field
These results suggest the potential utility of the Lung Cancer Prediction Convolutional Neural Network algorithm to revise the probability of disease for indeterminate pulmonary nodules, with the goal of decreasing invasive procedures and shortening the time to diagnosis.
Lung cancer remains the leading cause of cancer-related deaths in the United States and worldwide. In the United States alone, an estimated 228,820 adults will receive a diagnosis of lung cancer in 2020 (1). Despite recent progress in immunotherapy and other treatment modalities, the 5-year survival rate is 21.7% (2), mainly because most lung cancers are diagnosed at an advanced stage. Early diagnosis can markedly improve outcomes—the survival for patients with stage IA1 non–small cell cancer is 92% (3).
There are two principal routes to an early lung cancer diagnosis. The first is screening using low-dose computed tomography (LDCT), which has been shown to reduce lung cancer deaths by 20% in the U.S. National Lung Screening Trial (NLST) (4), and by 26% in the European NELSON (Nederlands–Leuvens Longkanker Screenings Onderzoek) trial (5). The second route is the detection of cancer as an incidental finding in patients undergoing imaging for an unrelated reason. Indeterminate pulmonary nodules (IPNs) are reported as incidental findings in ∼30% of chest CTs, and it has been estimated that 1.57 million patients with pulmonary nodules are identified in this way every year in the United States (6).
Regardless of the route to detection, the management of screen-detected and incidentally detected IPNs is a challenging clinical problem. One issue is the high false-positive rate of LDCT. The rate of positive LDCT screening tests in the NLST was reported to be ∼27% in the first two rounds and 17% in the third year of screening (4). More than 96% of all positive screens were false positives and 72% had some form of diagnostic follow-up. Variability in image interpretation among radiologists is known to be high, and this may lead to variability in management (7, 8). Moreover, CT scans on which incidental nodules occur are frequently read by generalist radiologists with limited thoracic experience.
Guidelines published by the American College of Radiology for screen-detected nodules (Lung-RADS) (9, 10), and by the Fleischner Society (11) and the British Thoracic Society (12) for incidentally detected nodules recommend management strategies based on qualitative or quantitative estimates of malignancy risk. Such estimates may incorporate clinical parameters such as patient age, smoking history, and cancer history, and radiological parameters such as nodule diameter, appearance, and location (11). Standard-of-care guidelines for incidental IPNs suggest thresholds for patient stratification (12–15); for example, nodule risks below 5% indicate interval surveillance imaging, whereas those above 65% indicate active intervention (biopsy/surgery). Such guidelines aim to optimize patient benefit given the performance of currently available tests.
Despite their availability, adherence to these guidelines can be variable and patient stratification can be subjective (16). For example, intermediate-category nodules (i.e., 5–65% for American College of Chest Physicians and American College of Radiology Lung-RADS category 4) present challenges in the clinic because the guidelines do not provide specific recommendations for what is a very broad range of risk profiles. Patients with intermediate-risk nodules are typically associated with a large number of expensive and invasive tests (17), with an unacceptable rate of surgeries on benign nodules (13, 16). Such poor stratification may result in delayed diagnosis and treatment and potential upstaging.
The current preferred option for smaller IPNs is growth assessment over time, which has been shown to contribute significantly to risk assessment (18, 19), although waiting may be difficult for patients and delay diagnosis and potential treatment. Growth identified over a short interval is less reliable for diagnosing malignancy than growth identified over longer periods (20, 21). Logistic regression–based methods, such as the Mayo and Brock risk models (22, 23), are recommended by some guidelines but are limited at least partly by their reliance on qualitative—and hence inconsistent—human interpretation of variables such as nodule size and morphology, and patients’ estimates of factors such as smoking history.
Computer-aided risk stratification using machine learning (ML) classification of benign and malignant nodules could potentially address some of these limitations, and the availability of large datasets and increasingly powerful computational resources has made the development of such techniques feasible. Such techniques work directly with the image and patient clinical data, negating the need to first describe the morphology or measure the size of the nodule. Prior ML work on previous datasets has shown that such tools have the potential to outperform conventional risk models (24–30), but their performance has not been evaluated on multiple independent datasets, including incidentally detected nodules in smokers and nonsmokers. Moreover, the published literature lacks external validation, including data acquired using heterogeneous CT technology and protocols from a variety of clinical practices. Our study offers such a level of clinical validation, which is required for future clinical trials and ultimately for clinical practice.
Our objective in this study was to derive and validate a computer-aided tool to classify benign and malignant nodules—a “digital biomarker” for use in patient stratification and management of IPNs. We aimed to investigate the performance of a deep learning risk stratification tool developed using the LDCT arm of the NLST (i.e., current/former smokers, 55–75 yr old, ≥30 pack-years) and internally and externally validated on multiple cohorts, including never-smokers. The eventual goal of this tool is to accelerate the diagnosis and treatment of malignant nodules, and to avoid unnecessary imaging and invasive procedures in patients with benign disease. Some of the results of these studies were reported in 2018 in the form of abstracts (31–33).
Methods
Study Design
In this study we used a prospective-specimen collection, retrospective-blinded-evaluation design (34). The Lung Cancer Prediction Convolutional Neural Network (LCP-CNN) developed by Optellum was derived and internally validated using the NLST dataset with cross-validation. Two independent external validation datasets were obtained from Vanderbilt University Medical Center (VUMC) and Oxford University Hospitals National Health Service Foundation Trust (OUH). These datasets included incidentally detected IPNs that had been brought to the attention of pulmonary physicians. The LCP-CNN was applied without modification to nodules from these populations (characterized in Table E1 in the online supplement).
Datasets
Deep learning methods require large representative datasets for training. Therefore, the derivation and internal validation dataset contained CT images of all solid and semisolid nodules of at least 6 mm in diameter from the NLST dataset. Ground-glass opacities were then excluded because there were too few malignant examples to train the system reliably. Working under the supervision of expert thoracic radiologists from OUH, a team of doctors and medical students performed an extensive data curation process (summarized in Figure E1). The final dataset contained 14,761 benign nodules from 5,972 patients and 932 malignant nodules from 575 patients. Note that each patient had up to three annual images, hence a nodule could be present on up to three CTs.
The VUMC external validation dataset contains prospectively collected data from patients with incidental pulmonary nodules who were referred to a lung nodule clinic (Table E1). Patients of either sex, ≥18 years of age, with a CT scan reporting a solid pulmonary nodule 5–30 mm in diameter were included, provided that the patient had no history of a cancer diagnosis within 2 years before the nodule was detected. Nodules were only included if they had a diagnosis provided by histology or 2-year stability based on diameter. When multiple images of a nodule were available, the earliest study for which a thin-slice CT section (≤ 1.25 mm thick) was available was selected. The VUMC external validation dataset contained 116 nodules (52 benign [including at least 3 histoplasmosis] and 64 malignant) from 116 patients.
The OUH external validation dataset contained retrospectively collected data from patients with incidental IPNs (Table E1). The same inclusion criteria as described above were used, except for a size range of 5–15 mm, a 5-year cancer cutoff, and no more than five nodules per patient. Although the criteria specified a diameter of 5–15 mm, all longitudinal studies were collected, and the earliest study for which a noncontrast CT was available was selected; therefore, the dataset included nodules up to 18.8 mm in diameter. The dataset contained 463 nodules from 427 patients. These included 63 cancer nodules from 62 different patients. Deidentified NLST datasets were obtained through the National Cancer Institute’s Cancer Data Access System (35). This research was approved by the OUH (Health Research Authority Integrated Research Application System ID: 214451) and VUMC institutional review boards (000616 and 030763).
Derivation of the LCP-CNN Model
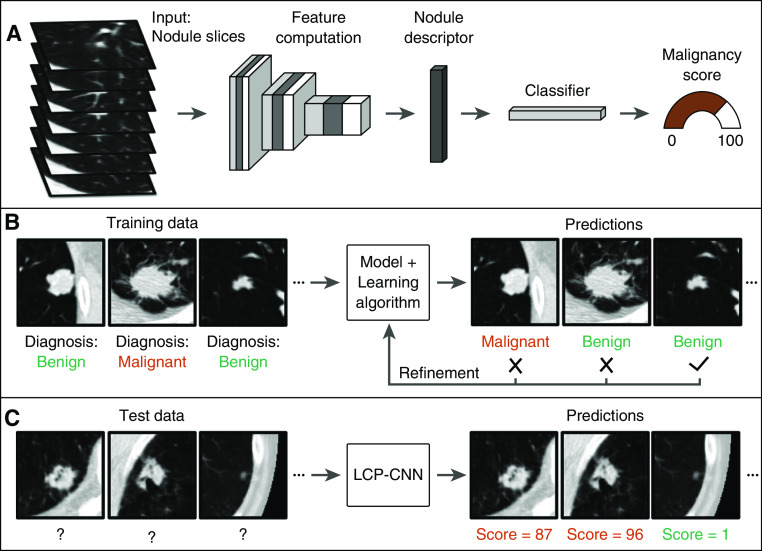
The LCP-CNN system is based on the Dense Convolutional Network (36), a widely used type of deep learning CNN architecture (37) that was designed for computer vision tasks (Figure 1). An eightfold cross-validation strategy was used for training and validation on the NLST data, and the datasets were split into eight approximately equal subsets (see Figure E1). This approach allowed us to report results that were not used for training. In all reported results, the output of the LCP-CNN model is a score between 0% and 100% to represent a likelihood of malignancy. During development, it was found that clinical variables (e.g., age, sex, and smoking history) did not contribute significantly to the performance of the model, and hence they were excluded. Further details are provided in the online supplement.
Figure 1.
Schematics showing the (A) Lung Cancer Prediction Convolutional Neural Network (LCP-CNN) architecture, (B) the training procedure, and (C) application of the trained model to novel data. The input to the network is a three-dimensional anisotropically resampled box ∼56 mm in width.
Performance Metrics and Statistical Analysis
We measured the performance of the LCP-CNN model in three different ways. First, we examined the area under the curve (AUC) for the LCP-CNN classifier over all testing data and compared the results with those obtained for relevant risk models. Second, we examined its impact on patient stratification by conducting a reclassification analysis, using a rule-in threshold of >65% and a rule-out threshold of <5% according to the American College of Chest Physicians guidelines (11). We reported net reclassification indices (NRIs) for cases and controls separately (38, 39) for both thresholds to measure the LCP-CNN’s potential to change management. A two-way reclassification analysis was performed. For example, at 65%, we calculated the fraction of cancers that were correctly classified compared with the Mayo model (“net cancer”) by counting the cancers that scored >65% using LCP-CNN but scored ≤ 65% using Mayo (“cancer up”), and subtracting the number of cancers that scored >65% with Mayo and ≤ 65% with LCP-CNN (“cancer down”).
We compared the internal validation dataset, which contained screening data, with the Brock model (23), as that model is appropriate for screening patients (i.e., older patients with a significant smoking history). The external validation dataset contained only incidentally detected nodules, including those detected in smokers and nonsmokers; therefore, we compared it with the more generally applicable Mayo model (22). Data regarding a family history of cancer or emphysema, which are necessary for the Brock model, were missing from many of the external datasets. To enable a cross-comparison, we also included Mayo results on the NLST.
Third, we calculated the diagnostic likelihood ratio (DLR) to evaluate the clinical value added. Nonparametric bootstrapping with 10,000 samples was used for all confidence intervals and P values (40, 41).
Results
AUC Performance
The model was first internally validated using cross-validation on the NLST dataset (Figure E1). The AUC over all the testing data for the LCP-CNN classifier was 92.1% (95% confidence interval [CI], 91.2–92.9%), compared with 85.6% (95% CI, 84.3–86.8%) for the Brock model (Figure 2A) (P < 0.001) and 85.2% (95% CI, 84.1–86.4%) for the Mayo model (P < 0.001). The performances of the Brock and Mayo models were not statistically different on the NLST (P = 0.126).
Figure 2.
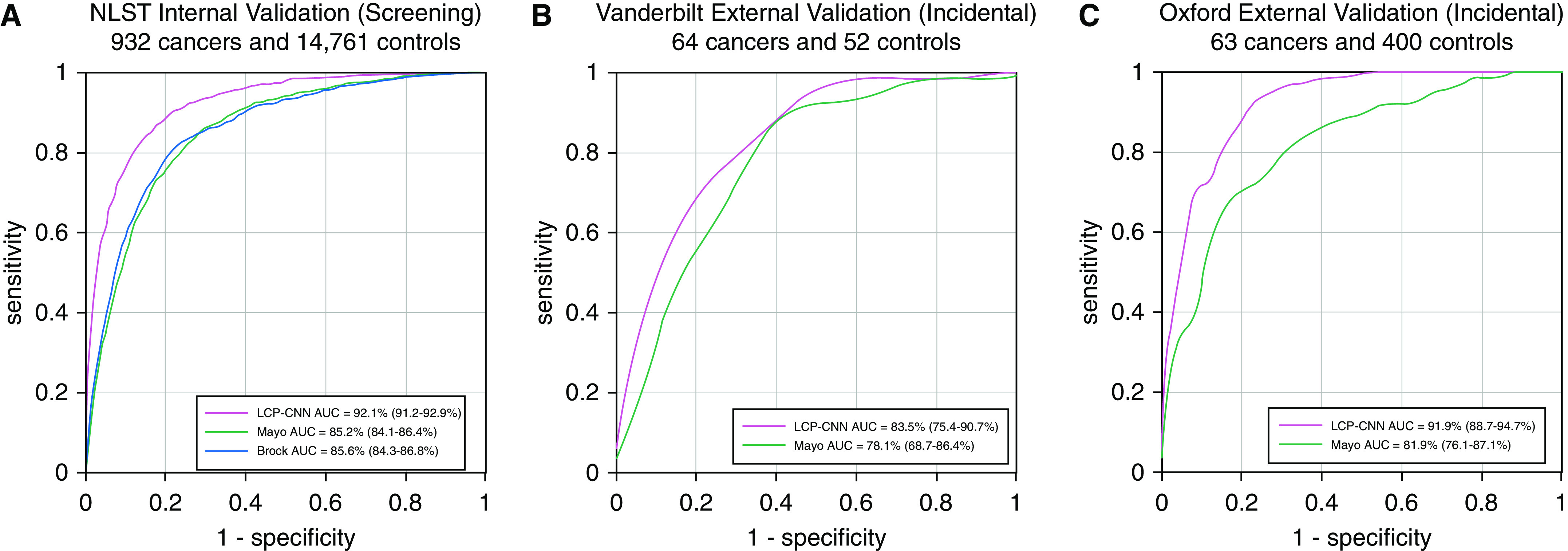
Receiver operating characteristic curves and area under the curve (AUC) analysis of the (A) internal National Lung Screening Trial (NLST) dataset using eight-way cross-validation, (B) external Vanderbilt dataset, and (C) external Oxford dataset. The Brock model was used as a comparator for the screening population, and the Mayo model was used for the incidental nodule populations for the two independent validation datasets. LCP-CNN = Lung Cancer Prediction Convolutional Neural Network.
To demonstrate generalizability beyond the NLST data, we tested the LCP-CNN on the two independent, nonscreening external cohorts. The AUCs on these represented an improvement of 5–10 percentage points of AUC compared with existing clinical prediction tools. On the OUH data, the AUC for the LCP-CNN classifier was 91.9% (95% CI, 88.7–94.7%) versus 81.9% (95% CI, 76.1–87.1%) for Mayo (P = 0.018). On the VUMC data, the AUC for the LCP-CNN classifier was 83.5% (95% CI, 75.4–90.7%) versus 78.1% (95% CI, 68.7–86.4%) for Mayo (P = 0.082). Figures 2B and 2C show the corresponding receiver operating characteristic curves.
Reclassification Performance
We analyzed the model by comparing its ability to reclassify benign and malignant nodules with that of conventional risk models using >65% (rule-in) and <5% (rule-out) thresholds. Figure 3 illustrates the benefit of the LCP-CNN in reclassifying nodules compared with the Brock and Mayo models selected for the clinical setting (screening or incidental). Table E2 provides a numerical annotation of Figure 3. The reclassification indices (42) for <5% and >65% risk thresholds were calculated separately, defining low- and high-risk categories, and are shown in Table 1. NRI results for Mayo applied to the NLST are shown in Figure E6, and reclassifications against other guideline-relevant thresholds are included in Table E3.
Figure 3.
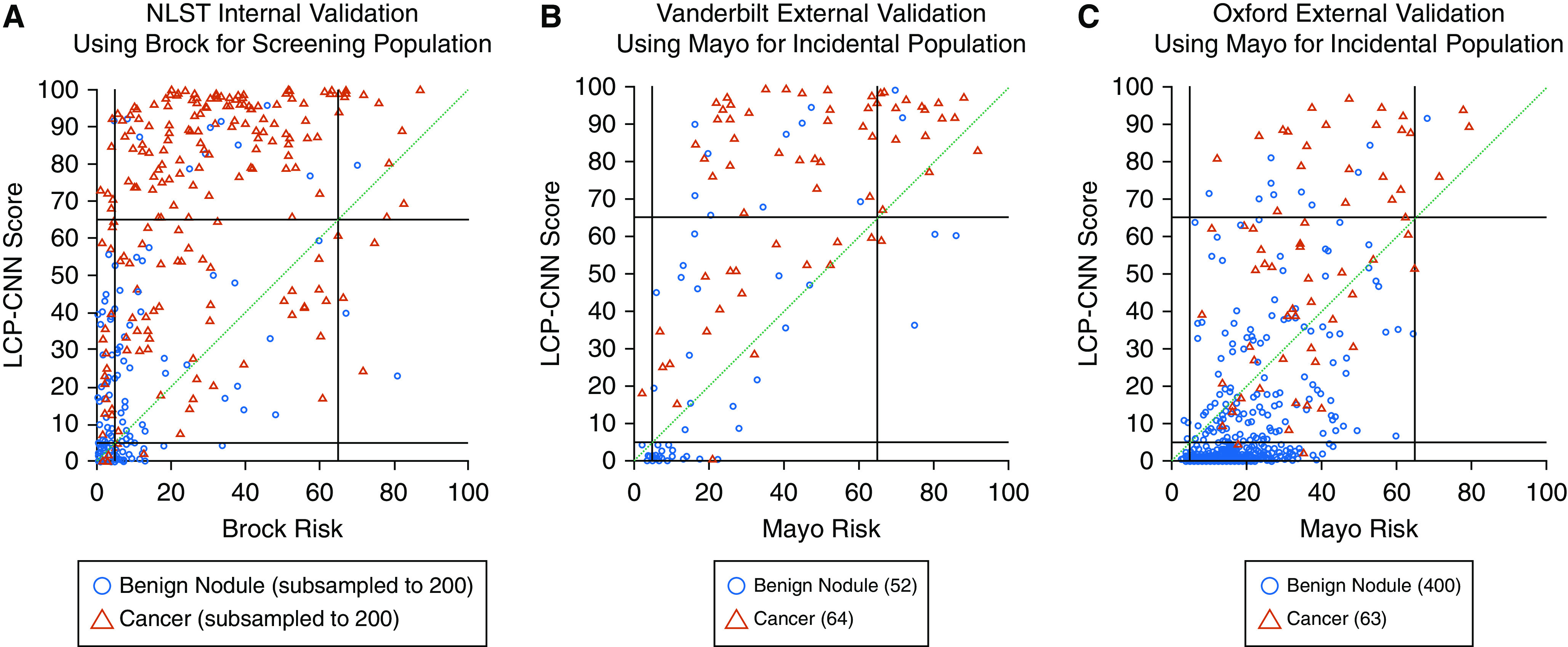
Reclassification diagrams. (A) National Lung Screening Trial (NLST) dataset for 200 cases and 200 benign nodules (randomly selected; numbers were limited for readability of the figure). (B) Vanderbilt University Medical Center dataset. (C) Oxford University Hospitals dataset. Reclassification diagrams are a useful way to visualize the impact of a new biomarker compared with a reference at predefined thresholds. Here we use rule-out and rule-in thresholds at 5% and 65%, respectively, as shown by the black lines. Red triangles indicate cancers, and blue circles indicate controls. If a new biomarker improves classification of cancers compared with the reference, then one would expect, for example, cases (red triangles) that were below 65% on the horizontal axis to move above 65% to the vertical axis, that is, from the central rectangular region to the region immediately above it. For example, on the Vanderbilt and Oxford datasets, 45% and 32% of the cancers, respectively, are reclassified up compared with the Mayo model. Similarly, a new biomarker improves benign classification compared with the reference if it moves controls (blue circles) that were above the 5% threshold on the horizontal axis to below 5% on the vertical axis. For nodules that stay within the three square regions intersected by the green diagonal, the Lung Cancer Prediction Convolutional Neural Network (LCP-CNN) does not add value because none of the nodules are correctly reclassified compared with the Brock or Mayo model. On the Vanderbilt and Oxford datasets, 33% and 61% of the benign nodules, respectively, are reclassified down compared with the Mayo model.
Table 1.
Reclassification of Indeterminate Pulmonary Nodules with the Lung Cancer Prediction Convolutional Neural Network
| National Lung Screening Trial Reclassification: Compared with Brock (Screening Population) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target (%) | Cancer Up (95% CI) | Cancer Down (95% CI) | Net Cancer (95% CI) | Net Cancer P Value | Benign Up (95% CI) | Benign Down (95% CI) | Net Benign (95% CI) | Net Benign P Value | Overall (95% CI) | Overall P Value |
| 5 | 0.11 (0.09 to 0.13) | 0.02 (0.01 to 0.02) | 0.09 (0.07 to 0.11) | <0.0001 | 0.16 (0.15 to 0.16) | 0.12 (0.12 to 0.13) | −0.04 (−0.04 to 0.03) | <0.0001 | 0.06 (0.03 to 0.08) | <0.0001 |
| 65 | 0.54 (0.51 to 0.57) | 0.02 (0.01 to 0.03) | 0.52 (0.49 to 0.56) | <0.0001 | 0.05 (0.04 to 0.05) | 0.00 (0.00 to 0.01) | −0.04 (−0.05 to 0.04) | <0.0001 | 0.48 (0.45 to 0.51) | <0.0001 |
| Vanderbilt Reclassification: Compared with Mayo (Incidental Population) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target (%) | Cancer Up (95% CI) | Cancer Down (95% CI) | Net Cancer (95% CI) | Net Cancer P Value | Benign Up (95% CI) | Benign Down (95% CI) | Net Benign (95% CI) | Net Benign P Value | Overall (95% CI) | Overall P Value |
| 5 | 0.02 (0.00 to 0.05) | 0.02 (0.00 to 0.05) | 0.00 (−0.05 to 0.05) | 0.34 | 0.00 (0.00 to 0.00) | 0.33 (0.19 to 0.46) | 0.33 (0.19 to 0.46) | <0.0001 | 0.33 (0.20 to 0.47) | <0.0001 |
| 65 | 0.47 (0.34 to 0.59) | 0.02 (0.00 to 0.05) | 0.45 (0.33 to 0.58) | <0.0001 | 0.17 (0.08 to 0.29) | 0.06 (0.00 to 0.13) | −0.12 (−0.25 to 0.00) | 0.0439 | 0.34 (0.15 to 0.52) | 0.0004 |
| Oxford Reclassification: Compared with Mayo (Incidental Population) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target (%) | Cancer Up (95% CI) | Cancer Down (95% CI) | Net Cancer (95% CI) | Net Cancer P Value | Benign Up (95% CI) | Benign Down (95% CI) | Net Benign (95% CI) | Net Benign P Value | Overall (95% CI) | Overall P Value |
| 5 | 0.00 (0.00 to 0.00) | 0.03 (0.00 to 0.08) | −0.03 (−0.08 to 0.00) | 0.1364 | 0.01 (0.00 to 0.02) | 0.62 (0.57 to 0.67) | 0.61 (0.56 to 0.66) | <0.0001 | 0.58 (0.51 to 0.64) | <0.0001 |
| 65 | 0.32 (0.21 to 0.43) | 0.00 (0.00 to 0.00) | 0.32 (0.21 to 0.43) | <0.0001 | 0.02 (0.01 to 0.04) | 0.00 (0.00 to 0.00) | −0.02 (−0.04 to 0.01) | <0.0001 | 0.29 (0.18 to 0.41) | <0.0001 |
Definition of abbreviation: CI = confidence interval.
Reclassification indices for cancers and benign nodules on the National Lung Screening Trial, Vanderbilt, and Oxford University Hospitals datasets for the rule-out test with a 5% threshold and the rule-in test with a 65% threshold are shown. For each threshold, the proportion of cancers that moved above a given threshold (i.e., scored below the threshold on the comparator model and above the threshold on the Lung Cancer Prediction Convolutional Neural Network) is designated as “cancer up.” Movement of cancers and benign nodules is recorded in both the up and down directions as a proportion of the total number of cancers or benign nodules, respectively. The “net cancer” movement is positive when more cancers are reclassified above the threshold than are reclassified below the threshold, and conversely, the “net benign” movement is positive when more benign nodules are reclassified below the threshold.
Rule-in Test (>65%)
On the VUMC dataset, the NRI was 0.34 (95% CI, 0.15 to 0.52; P = 0.0004). Of the 64 cancers, 45 (70%) were classified as high-risk by the LCP-CNN, compared with 16 (25%) classified by Mayo (net cancer: 0.45 [95% CI, 0.33 to 0.58]; P < 0.0001). The LCP-CNN false-positive rate was slightly closer to that expected at this probability threshold compared with Mayo. Of 52 benign nodules, 11 (21%) were false positives with LCP-CNN, compared with 5 (10%) with Mayo (net benign: −0.12 [95% CI, −0.25 to 0.00]; P = 0.0439). On the OUH dataset, the NRI was 0.29 (95% CI 0.18 to 0.41; P < 0.0001). Of 63 cancers, 23 (36%) were classified as high-risk, compared with 3 (5%) classified by Mayo (net cancer: 0.32 [95% CI, 0.21 to 0.43]; P < 0.0001). Among the benign nodules, the LCP-CNN had 10 (2.5%) false positives and Mayo had 1 (0.25%), resulting in a false-positive rate slightly closer to that expected at this risk threshold (net benign: −0.02 [95% CI −0.04 to −0.01]; P < 0.0001).
Rule-out Test (<5%)
The VUMC NRI was 0.33 (95% CI, 0.20–0.47; P < 0.0001). Of the 52 benign nodules in VUMC, 23 (44%) were ruled out by LCP-CNN and 6 (12%) were ruled out by Mayo (net benign: 0.33 [95% CI, 0. 19–0.46]; P < 0.0001), and both had 1 (2%) false negative. The OUH NRI was 0.58 (95% CI, 0.51–0.64; P < 0.0001). Of the 400 benign nodules in OUH, 257 (64%) were ruled out by LCP-CNN and 12 (3%) were ruled out by Mayo (net benign: 0.62 [95% CI, 0.57–0.67]; P < 0.0001). There were two (3%) false negatives for LCP-CNN and none for Mayo.
DLR Performance
Table 2 presents the sensitivity and specificity of all models, with the corresponding positive and negative DLRs. The positive DLR for rule-in at >65% using the LCP-CNN was 3.32 for VUMC and 14.6 for OUH, and the negative DLR for rule-out (<5% threshold) was 0.04 for VUMC and 0.05 for OUH.
Table 2.
Sensitivity, Specificity, and Diagnostic Likelihood Ratio Testing Associated with the Lung Cancer Prediction Convolutional Neural Network at Specific Thresholds
| Threshold (%) | NLST |
|||||
|---|---|---|---|---|---|---|
|
Brock |
LCP-CNN |
|||||
| Sensitivity | Specificity | DLR− | Sensitivity | Specificity | DLR− | |
| 5 | 86.5 (84.1–88.6) | 66.5 (65.8–67.2) | 0.20 (0.17–0.24) | 95.6 (94.2–96.9) | 62.9 (62.1–63.7) | 0.07 (0.05–0.09) |
| Threshold (%) | Sensitivity | Specificity | DLR+ | Sensitivity | Specificity | DLR+ |
|---|---|---|---|---|---|---|
| 65 | 9.4 (7.5–11.4) | 99.4 (99.2–99.5) | 14.67 (10.91–19.54) | 61.8 (58.4–64.6) | 95.0 (94.6–95.3) | 12.26 (11.26–13.28) |
| Threshold (%) | VUMC |
|||||
|---|---|---|---|---|---|---|
|
Mayo |
LCP-CNN |
|||||
| Sensitivity | Specificity | DLR− | Sensitivity | Specificity | DLR− | |
| 5 | 98.4 (94.9–100.0) | 11.5 (3.8–20.8) | 0.14 (0.00–0.72) | 98.4 (94.8–100.0) | 44.2 (30.9–57.9) | 0.04 (0.00–0.13) |
| Threshold (%) | Sensitivity | Specificity | DLR+ | Sensitivity | Specificity | DLR+ |
|---|---|---|---|---|---|---|
| 65 | 25.0 (14.7–36.1) | 90.4 (81.6–98.0) | 2.60 (1.13–11.72) | 70.3 (58.6–81.2) | 78.8 (67.3–89.3) | 3.32 (2.08–6.67) |
| Threshold (%) | OUH |
|||||
|---|---|---|---|---|---|---|
|
Mayo |
LCP-CNN |
|||||
| Sensitivity | Specificity | DLR− | Sensitivity | Specificity | DLR− | |
| 5 | 100.0 (100.0–100.0) | 3.0 (1.5–4.7) | 0.00 (0.00–0.00) | 96.8 (91.7–100.0) | 64.3 (59.6–68.7) | 0.05 (0.00–0.13) |
| Threshold (%) | Sensitivity | Specificity | DLR+ | Sensitivity | Specificity | DLR+ |
|---|---|---|---|---|---|---|
| 65 | 4.8 (0.0–10.7) | 99.8 (99.2–100.0) | 19.05 (0.00–Inf) | 36.5 (24.6–47.9) | 97.5 (96.0–99.0) | 14.60 (7.96–34.93) |
Definition of abbreviations: DLR = diagnostic likelihood ratio; Inf = infinity; LCP-CNN = Lung Cancer Prediction Convolutional Neural Network; NLST = National Lung Screening Trial; OUH = Oxford University Hospitals; VUMC = Vanderbilt University Medical Center.
The sensitivity, specificity, and DLRs for the NLST, VUMC, and OUH datasets at 5% and 65% probability thresholds are shown. For rule-out at 5%, a good risk model is one that can provide the greatest specificity while maintaining an adequately high sensitivity. For rule-in at 65%, a high specificity indicates few unnecessary procedures for patients with benign nodules. The LCP-CNN has a much higher sensitivity for most of these operating points, indicating that many more cancers than indicated by the Brock or Mayo model could be ruled in for fast-tracked interventions.
Discussion
The management of screen-detected and incidentally detected IPNs is a challenging and growing clinical problem. Pulmonary nodules are detected in up to 30% of chest CT studies, and the vast majority of these are benign. Importantly, establishing a definite diagnosis of IPNs can take up to 2 years and can result in many follow-up procedures, including imaging, biopsy, and surgery.
In this study, we report on the derivation and validation of the LCP-CNN, a deep learning lung cancer malignancy prediction tool, to classify and risk stratify IPNs from screening and nonscreening data. This study is the first to validate such a tool on multiple independent cohorts, including a large multicenter screening dataset (n = 15,693) and real-world clinical nodules (n = 579), and to show a reclassification performance that is significantly superior to that of existing risk models (net reclassification of at least 30% on the external validation cohorts compared with Mayo) and could potentially change patient management.
Although previous studies demonstrated the potential of radiomics/ML for predicting IPN malignancy (43–47), most of these studies used small datasets (e.g., ∼100 nodules) or did not perform external validation. Recent studies by Ardila and colleagues (30) and Huang and colleagues (48) also trained on the NLST. The former used an external validation dataset from one center that included 27 cancers, and the latter is not directly comparable because it used human-reported parameters rather than CT images directly.
In contrast, our model exhibited a robust performance on multiple independent, real-world, heterogeneous datasets (acquired with many different imaging protocols and scanners; see Table E4) across two continents, independently of differences in patient demographics. Also, for the first time, we demonstrate that a deep learning method that is appropriately trained on screening data generalizes well to the complex problem of incidentally detected nodules, including those in smokers and nonsmokers.
Performance on the internal validation NLST data is not directly comparable between our study and that by Ardila and colleagues (30). In our work, each malignant nodule was tracked to earlier CTs and considered malignant. Ardila and colleagues used only the CT nearest in time to the diagnosis; therefore, our NLST dataset had a greater number of smaller, more difficult to detect cancers. Moreover, Ardila and colleagues combined both detection and classification steps and classified at the image level, whereas the LCP-CNN performs only classification and considers each nodule separately.
Quantitative measures of IPN growth, such as the volume doubling time, were shown to provide excellent classification performance within the NELSON screening trial (19). However, such measures have not been extensively validated for incidental IPNs, and additionally require at least a second follow-up CT and accurate segmentation, which may fail in a substantial number of patients. The LCP-CNN uses only one CT image, often the earliest, and segmentation is not required.
Although logistic regression–based risk models, such as the Brock (23), Mayo (22), and Gould (14) models, may be helpful for standardizing nodule management, they require information about the patient and nodule, and their results are dominated by nodule size. They typically use radiologist-reported parameters, which are subject to variability (7, 8, 49). In contrast, the LCP-CNN is both more performant and unaffected by such subjective assessments, deriving its information directly from the image.
The LCP-CNN demonstrates superiority to the Mayo or Brock models, encouraging further exploration of its utility. The test provides excellent negative predictive value, and hence LCP-CNN scores below 5% would indicate the need for surveillance according to Fleischner Society guidelines (3). Above 65%, it would indicate the need for a tissue diagnosis. Because patients’ preferences with regard to management depend on their understanding of the risks involved, a reliable estimate of the probability of cancer would be helpful in shared decision-making. Although the NRI analysis was performed over the full range of IPNs, an inspection of Figures 3B and 3C and Table E3 shows that the LCP-CNN left fewer patients in the intermediate-risk region than the Mayo model on both validation sets, and correctly reclassified many of Mayo’s intermediate cases.
Figures E2–E5 show examples of IPNs and LCP-CNN scores. An examination of these results is useful for gaining an intuitive understanding of the tool. For example, many intrapulmonary lymph nodes are assigned very low scores (typically <0.5%), whereas more complex benign cases (infection/immune response) tend to score higher, perhaps because of their suspicious appearance, which more closely resembles a malignancy. The three lowest-scoring cancer nodules from the Vanderbilt population (Figure E5) were rather small, indolent tumors, and a diagnosis was only available 606, 1,872, and 537 days, respectively, after the CT on which the LCP-CNN score was calculated, and hence may be safely monitored with follow-up imaging.
An inspection of misclassifications also provides excellent feedback for refining our digital biomarker. For example, the fourth lowest-scoring nodule was a carcinoid. These nodules were underrepresented in the training set and typically had a benign-looking, smooth, round appearance. One histoplasmoma was misclassified as high risk; however, granulomas are known to represent a significant clinical challenge (38, 39).
Using a model trained on screening data but tested on incidental IPNs is adequate for biomarker validation studies, which use large numbers of patients from heterogeneous populations (50). Although an inability to generalize to “real-world” clinical care can cause many biomarkers to fail in validation studies, our LCP-CNN demonstrated efficacy on nodules obtained from two separate pulmonary clinics with very different populations and disease prevalence. The guideline thresholds for low and high risk are chosen as a function of the accuracy of the available tests and may shift as better tests become available. We demonstrated the advantage of LCP-CNN over traditional prediction models using multiple metrics of biomarker performance, including discrimination (51), reclassification (42), and likelihood statistic testing.
The work presented here has limitations. Although we compared the performance of LCP-CNN with that of relevant clinical risk models, we did not report its potential to change clinical decision-making. Because some clinical parameters were missing, not all risk models could be run on all datasets. In the future, comparisons with multiple models would be desirable (52, 53). Because of the smaller size of the VUMC dataset (n = 116), the difference in AUC was not significant (P = 0.082), although all VUMC reclassification results were significant. As discussed above, despite the differences in disease prevalence and patient populations across the three validation datasets, the same linear calibration between the LCP-CNN and risk was used for all the results shown in Figure 3; however, the results may be further optimized by a population-specific calibration. For example, although the reclassification of VUMC and OUH datasets was very good, on the NLST, 3.5% of controls were incorrectly classified as intermediate risk compared with Brock, because of the low prevalence of disease. The OUH dataset did not capture the patients’ history of cancer, which is necessary to calculate the Mayo risk scores, although patients who had received a cancer diagnosis in the last 5 years were excluded. Therefore, in calculating the Mayo scores, it was assumed that the OUH patients had no history of cancer. Although the results are at the nodule level rather than the patient level, the VUMC dataset only had one nodule per patient, and the mean number of nodules per patient in the OUH dataset was 1.08.
In summary, using an ML method as a diagnostic algorithm, our LCP-CNN model provided a significant improvement in AUC over the clinically validated risk models (Brock and Mayo). Furthermore, it achieved a strong improvement in DLRs in both clinical validation sets, which included different patient populations. Our model is intended to be improved over time as data collections are added and structured curation efforts continue. Although more stringent clinical validations on additional (external and independent) datasets are needed, our results suggest that it may be possible to address a major problem in the management of individuals presenting with IPNs by using an ML-derived prediction model.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Zoe Sandford for her contribution to dataset curation.
Footnotes
Supported by National Cancer Institute grants CA186145 and CA152662 (P.P.M.) and by Optellum.
Author Contributions: P.P.M., C.A., T.K., L.C.P., and F.G. initiated and conceived the study. P.P.M. and T.K. supervised the study. S. Antic, S. Ather, C.A., J.B., J.D., D.D., W.H., J.K., B.A.L., R.F.M., P.N., H.P., L.C.P., C.S., G.T.S., and A.T. provided clinical and imaging review of the data. C.A., H.C., and L.C.P. performed statistical analyses. P.P.M, T.K., and L.C.P. wrote the manuscript. P.P.M., T.K., and F.G. proofread the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201903-0505OC on April 24, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.American Lung Association. State of lung cancer. 2019. [accessed 2019 Feb 1]. Available from: https://www.lung.org/research/state-of-lung-cancer.
- 3.American Cancer Society. Cancer facts & figures. 2018. [accessed 2019 Feb 1]. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2020/incidence-and-mortality-rates-race-and-ethnicity-2012-2017.pdf.
- 4.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Koning HJ.NELSON study shows CT screening for nodule volume management reduces lung cancer mortality by 26 percent in men. Presented at the IASLC 19th World Conference on Lung Cancer. Sept 23–26, 2018; Toronto, Canada [Google Scholar]
- 6.Gould MK, Tang T, Liu IL, Lee J, Zheng C, Danforth KN, et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med. 2015;192:1208–1214. doi: 10.1164/rccm.201505-0990OC. [DOI] [PubMed] [Google Scholar]
- 7.Nair A, Bartlett EC, Walsh SLF, Wells AU, Navani N, Hardavella G, et al. Lung Nodule Evaluation Group. Variable radiological lung nodule evaluation leads to divergent management recommendations. Eur Respir J. 2018;52:1801359. doi: 10.1183/13993003.01359-2018. [DOI] [PubMed] [Google Scholar]
- 8.Penn A, Ma M, Chou BB, Tseng JR, Phan P. Inter-reader variability when applying the 2013 Fleischner guidelines for potential solitary subsolid lung nodules. Acta Radiol. 2015;56:1180–1186. doi: 10.1177/0284185114551975. [DOI] [PubMed] [Google Scholar]
- 9.American College of Radiology. Lung CT screening reporting and data system (Lung-RADSTM) [accessed 2019 Feb 1]. Available from: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads.
- 10.McKee BJ, Regis SM, McKee AB, Flacke S, Wald C. Performance of ACR lung-RADS in a clinical CT lung screening program. J Am Coll Radiol. 2015;12:273–276. doi: 10.1016/j.jacr.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 11.MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 2017;284:228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 12.Baldwin DR, Callister ME Guideline Development Group. The British Thoracic Society guidelines on the investigation and management of pulmonary nodules. Thorax. 2015;70:794–798. doi: 10.1136/thoraxjnl-2015-207221. [DOI] [PubMed] [Google Scholar]
- 13.Gould MK, Donington J, Lynch WR, Mazzone PJ, Midthun DE, Naidich DP, et al. Evaluation of individuals with pulmonary nodules: when is it lung cancer: diagnosis and management of lung cancer, 3rd ed. American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e93S–e120S. doi: 10.1378/chest.12-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould MK, Ananth L, Barnett PG Veterans Affairs SNAP Cooperative Study Group. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest. 2007;131:383–388. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiga AW, Deppen SA, Massion PP, Callaway-Lane C, Pinkerman R, Dittus RS, et al. Communication about the probability of cancer in indeterminate pulmonary nodules. JAMA Surg. 2018;153:353–357. doi: 10.1001/jamasurg.2017.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanner NT, Aggarwal J, Gould MK, Kearney P, Diette G, Vachani A, et al. Management of pulmonary nodules by community pulmonologists: a multicenter observational study. Chest. 2015;148:1405–1414. doi: 10.1378/chest.15-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair A, Baldwin DR, Field JK, Hansell DM, Devaraj A. Measurement methods and algorithms for the management of solid nodules. J Thorac Imaging. 2012;27:230–239. doi: 10.1097/RTI.0b013e31824f83e1. [DOI] [PubMed] [Google Scholar]
- 18.Lindell RM, Hartman TE, Swensen SJ, Jett JR, Midthun DE, Mandrekar JN. 5-year lung cancer screening experience: growth curves of 18 lung cancers compared to histologic type, CT attenuation, stage, survival, and size. Chest. 2009;136:1586–1595. doi: 10.1378/chest.09-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Klaveren RJ, Oudkerk M, Prokop M, Scholten ET, Nackaerts K, Vernhout R, et al. Management of lung nodules detected by volume CT scanning. N Engl J Med. 2009;361:2221–2229. doi: 10.1056/NEJMoa0906085. [DOI] [PubMed] [Google Scholar]
- 20.Kostis WJ, Yankelevitz DF, Reeves AP, Fluture SC, Henschke CI. Small pulmonary nodules: reproducibility of three-dimensional volumetric measurement and estimation of time to follow-up CT. Radiology. 2004;231:446–452. doi: 10.1148/radiol.2312030553. [DOI] [PubMed] [Google Scholar]
- 21.Xu DM, van der Zaag-Loonen HJ, Oudkerk M, Wang Y, Vliegenthart R, Scholten ET, et al. Smooth or attached solid indeterminate nodules detected at baseline CT screening in the NELSON study: cancer risk during 1 year of follow-up. Radiology. 2009;250:264–272. doi: 10.1148/radiol.2493070847. [DOI] [PubMed] [Google Scholar]
- 22.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med. 1997;157:849–855. [PubMed] [Google Scholar]
- 23.McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med. 2013;369:910–919. doi: 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins S, Wang H, Liu Y, Garcia A, Stringfield O, Krewer H, et al. Predicting malignant nodules from screening CT scans. J Thorac Oncol. 2016;11:2120–2128. doi: 10.1016/j.jtho.2016.07.002. [Published erratum appears in J Thorac Oncol 13:280–281.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armato SG, III, Drukker K, Li F, Hadjiiski L, Tourassi GD, Engelmann RM, et al. LUNGx Challenge for computerized lung nodule classification. J Med Imaging (Bellingham) 2016;3:044506. doi: 10.1117/1.JMI.3.4.044506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul R, Hall L, Goldgof D, Schabath M, Gillies R. Predicting nodule malignancy using a CNN ensemble approach. Proc Int Jt Conf Neural Netw. 2018;2018 doi: 10.1109/IJCNN.2018.8489345. 10.1109/IJCNN.2018.8489345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang P, Park S, Yan R, Lee J, Chu LC, Lin CT, et al. Added value of computer-aided CT image features for early lung cancer diagnosis with small pulmonary nodules: a matched case-control study. Radiology. 2018;286:286–295. doi: 10.1148/radiol.2017162725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zinovev D, Feigenbaum J, Furst J, Raicu D. Probabilistic lung nodule classification with belief decision trees. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:4493–4498. doi: 10.1109/IEMBS.2011.6091114. [DOI] [PubMed] [Google Scholar]
- 29.Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One. 2017;12:e0188290. doi: 10.1371/journal.pone.0188290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 31.Kadir T, Arteta C, Pickup L, Declerck J, Massion P. Deep learning based risk stratification of patients with suspicious nodules [abstract] Am J Respir Crit Care Med. 2018;197:A4695. [Google Scholar]
- 32.Kadir T, Arteta C, Pickup L, Novotny P, Sandford Z, Brabec J, et al. Solid and part-solid lung nodule classification using deep learning on the national lung screening trial dataset [abstract] Am J Respir Crit Care Med. 2018;197:A7417. [Google Scholar]
- 33.Peschl H, Arteta C, Pickup L, Tsakok M, Ather S, Hussain S, et al. Deep learning for rule-out of unnecessary follow-up in patients with incidentally detected, indeterminate pulmonary nodules: results on an independent dataset [abstract] Radiology. 2018:SSG03–SSG06. [Google Scholar]
- 34.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Cancer Institute. Cancer data access system [accessed 2019 Feb 1]. Available from: https://cdas.cancer.gov/
- 36.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. [preprint]. arXiv; 2018 [accessed 2019 Feb]. Available from: https://arxiv.org/abs/1608.06993. [Google Scholar]
- 37.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 38.Pepe MS, Janes H, Li CI. Net risk reclassification p values: valid or misleading? J Natl Cancer Inst. 2014;106:dju041. doi: 10.1093/jnci/dju041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerr KF, Wang Z, Janes H, McClelland RL, Psaty BM, Pepe MS. Net reclassification indices for evaluating risk prediction instruments: a critical review. Epidemiology. 2014;25:114–121. doi: 10.1097/EDE.0000000000000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Efron B.An introduction to the Bootstrap. New York: Chapman and Hall; 1993 [Google Scholar]
- 41.Fu WJ, Carroll RJ, Wang S. Estimating misclassification error with small samples via bootstrap cross-validation. Bioinformatics. 2005;21:1979–1986. doi: 10.1093/bioinformatics/bti294. [DOI] [PubMed] [Google Scholar]
- 42.Paynter NP, Cook NR. Adding tests to risk based guidelines: evaluating improvements in prediction for an intermediate risk group. BMJ. 2016;354:i4450. doi: 10.1136/bmj.i4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciompi F, Chung K, van Riel SJ, Setio AAA, Gerke PK, Jacobs C, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep. 2017;7:46479. doi: 10.1038/srep46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [Published erratum appears in Nat Commun 5:4644.]24892406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul R, Hawkins SH, Schabath MB, Gillies RJ, Hall LO, Goldgof DB. Predicting malignant nodules by fusing deep features with classical radiomics features. J Med Imaging (Bellingham) 2018;5:011021. doi: 10.1117/1.JMI.5.1.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang P, Lin CT, Li Y, Tammemagi CM, Brock MV, Atkar-Khattra S, et al. Prediction of lung cancer risk at follow-up screening with low-dose CT: a training and validation study of a deep learning method. Lancet Digit Health. 2019;1:e353–e362. doi: 10.1016/S2589-7500(19)30159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Riel SJ, Sánchez CI, Bankier AA, Naidich DP, Verschakelen J, Scholten ET, et al. Observer variability for classification of pulmonary nodules on low-dose CT images and its effect on nodule management. Radiology. 2015;277:863–871. doi: 10.1148/radiol.2015142700. [DOI] [PubMed] [Google Scholar]
- 50.Fiore LD, D’Avolio LW. Detours on the road to personalized medicine: barriers to biomarker validation and implementation. JAMA. 2011;306:1914–1915. doi: 10.1001/jama.2011.1605. [DOI] [PubMed] [Google Scholar]
- 51.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Nair VS, Sundaram V, Desai M, Gould MK. Accuracy of models to identify lung nodule cancer risk in the national lung screening trial. Am J Respir Crit Care Med. 2018;197:1220–1223. doi: 10.1164/rccm.201708-1632LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Ameri A, Malhotra P, Thygesen H, Plant PK, Vaidyanathan S, Karthik S, et al. Risk of malignancy in pulmonary nodules: a validation study of four prediction models. Lung Cancer. 2015;89:27–30. doi: 10.1016/j.lungcan.2015.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.