Abstract
Rationale: Infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease (COVID-19), a predominantly respiratory illness. The first step in SARS-CoV-2 infection is binding of the virus to ACE2 (angiotensin-converting enzyme 2) on the airway epithelium.
Objectives: The objective was to gain insight into the expression of ACE2 in the human airway epithelium.
Methods: Airway epithelia sampled by fiberoptic bronchoscopy of trachea, large airway epithelia (LAE), and small airway epithelia (SAE) of nonsmokers and smokers were analyzed for expression of ACE2 and other coronavirus infection–related genes using microarray, RNA sequencing, and 10x single-cell transcriptome analysis, with associated examination of ACE2-related microRNA.
Measurements and Main Results: 1) ACE2 is expressed similarly in the trachea and LAE, with lower expression in the SAE; 2) in the SAE, ACE2 is expressed in basal, intermediate, club, mucus, and ciliated cells; 3) ACE2 is upregulated in the SAE by smoking, significantly in men; 4) levels of miR-1246 expression could play a role in ACE2 upregulation in the SAE of smokers; and 5) ACE2 is expressed in airway epithelium differentiated in vitro on air–liquid interface cultures from primary airway basal stem/progenitor cells; this can be replicated using LAE and SAE immortalized basal cell lines derived from healthy nonsmokers.
Conclusions: ACE2, the gene encoding the receptor for SARS-CoV-2, is expressed in the human airway epithelium, with variations in expression relevant to the biology of initial steps in SARS-CoV-2 infection.
Keywords: COVID-19, ACE2 transcriptome, coronavirus
At a Glance Commentary
Scientific Knowledge on the Subject
Coronavirus disease (COVID-19), a viral disease with severe respiratory morbidity, is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The major tropism determinant for SARS-CoV-2 is the availability of ACE2 (angiotensin-converting enzyme 2), the primary viral receptor expressed on the surface of cells. The high amount of contagion argues that inhalation of airborne virus-containing droplets is a major route of exposure, so an understanding of COVID-19 may benefit from the characterization of ACE2 expression in the airway.
What This Study Adds to the Field
We report ACE2 gene expression in the small airway, large airway, and trachea, using microarray, bulk RNA sequencing, and single-cell RNA sequencing data sets. Broad expression of ACE2 was found throughout the airway, with higher expression in proximal segments. In addition, all major epithelial cell types expressed ACE2. Smoking was associated with higher ACE2 mRNA expression in the small airway. Male smokers had the highest ACE2 expression levels, potentially providing a partial explanation for elevated COVID-19 incidence among men compared with women. ACE2 expression might be influenced by low miR-1246 expression in smokers. These data may provide insight into the pathogenesis of COVID-19 and risk factors in the population.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for coronavirus disease (COVID-19), a global pandemic characterized by fever, dry cough, dyspnea, lymphopenia, and a significant mortality rate, which is primarily due to respiratory complications (1–6). The disease is spread primarily through person‐to‐person transmission via respiratory droplets and by contact with contaminated surfaces (4, 5, 7, 8). Approximately 50% of hospitalized patients with COVID-19 have preexisting medical conditions, including diabetes, cardiovascular disease, chronic obstructive pulmonary disease, and malignancy (1, 2, 5, 9, 10), and men have increased susceptibility to infection, more severe disease, and higher mortality (1–3, 5, 9).
The SARS-CoV-2 virus is a novel coronavirus distinct from the coronaviruses causing human severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (7, 11–13). Like other closely related coronaviruses, SARS-CoV-2 interacts with cells through the virus spike protein, an envelope glycoprotein that binds to the host cell receptor, ACE2 (angiotensin-converting enzyme 2), and mediates viral entry (14–17). After binding, lineage B coronavirus spike proteins are modified by one or more cellular proteases, either at the cell surface or after endocytosis. At the cell surface, the transmembrane serine protease TMPRSS2 (transmembrane serine protease 2) can cleave the SARS-CoV-2 spike protein, leading to the exposure of a fusion peptide that can guide direct fusion of the coronavirus envelope with the plasma membrane of the target cell, as observed in other coronaviruses (14, 16). Alternatively, either cathepsin L or furin, which are intracellular proteases, can activate spike-mediated coronavirus envelope fusion with internal cellular membranes (14, 18). Secondary activation of the SARS-CoV-2 by either cathepsin L or furin, both widely expressed in airway epithelium, has not yet been demonstrated, but the sequence of the SARS-CoV-2 spike protein contains two furin cleavage sites (15). Coronavirus fusion is also affected by the activity of an enzyme, PI4KB (phosphatidylinositol 4-kinase IIIβ), a gene expressed in the airway epithelium (19). PI4KB phosphorylates phosphatidylinositol in a pathway leading to generation of inositol triphosphate, an intracellular signaling molecule. Both pharmacological inhibition of PI4KB activity and siRNA-mediated knockdown of PI4KB inhibited the infection of cells in vitro with a SARS-CoV spike protein–pseudotyped virus (19).
On the basis of the knowledge that SARS-CoV-2 infection is primarily a respiratory illness, that SARS-CoV-2 has been isolated from respiratory epithelial lining fluid, and that SARS-CoV-2 infects human airway epithelium (4, 7, 12), it is highly likely that the cells mediating the entry of SARS-CoV-2 in the majority of cases can be found in the respiratory epithelium. In this context, we searched our extensive airway epithelial transcriptome data of healthy nonsmokers and smokers for evidence of the expression of ACE2, with a focus on the extent of expression, which cell types express the receptor, whether sex and/or cigarette smoking influence ACE2 expression in airway epithelium, and biologic processes in the human airway epithelium that may be linked to ACE2 expression. Finally, we observed that the immortalized BCi-NS1.1 cell line (an immortalized airway basal cell [BC] line derived from BCs collected from the large airway epithelium [LAE] of a healthy nonsmoker) (20) and the hSABCi-NS1.1 cell line (an immortalized airway BC line derived from BCs collected from the small airway epithelium [SAE] of a healthy nonsmoker) (21) both express ACE2 and, when cultured on an air–liquid interface (ALI), the differentiated progeny express ACE2. In the context that the airway epithelium is a likely entry site for SARS-CoV-2, these cell lines should be useful investigative tools for studying SARS-CoV-2 interactions with the human airway epithelium and assessing therapeutic agents to treat the infection.
Methods
The assessment of expression of ACE2 and associated genes in the airway epithelium of nonsmokers and smokers was derived from multiple databases of our laboratory’s assessment of the transcriptome of nonsmokers and smokers, representing 744 independent samples of airway epithelium derived from 267 subjects. We obtained airway epithelium by fiberoptic bronchoscopy and brushing of the trachea, large airway (1–5 generations, brushing typically at 3–4 generations), and small airway (6–23 generations, brushing typically at 10–12 generations) (22–24) from phenotypically normal nonsmokers and smokers. Details regarding inclusion/exclusion criteria for nonsmokers and smokers are presented in the online supplement, as are the details regarding sample processing and analysis. Expression was assessed by microarray, RNA sequencing (RNA-seq) or 10x single-cell analysis (23–27), and comparisons were made between nonsmokers and smokers; P < 0.05 was considered significant. The majority of the ACE2 transcriptome data are from our previously published datasets. The study population, samples, and transcriptome quantification methodology for each table and figure are detailed in Table E1 in the online supplement, together with references and Gene Expression Omnibus accession numbers in the public repository of data in the Gene Expression Omnibus website (https://www.ncbi.nlm.nih.gov/geo/) if relevant. If the dataset is new, it is listed in Table E1 as “unpublished results.” The quantification of SAE microRNAs (miRNAs) that could bind to ACE2 mRNA is based on our publication by Wang and colleagues (27). In addition to the published databases, we used the following: 1) microarrays (HG-U133 Plus 2.0; Affymetrix) to assess the expression of ACE2 on a well-differentiated airway epithelium cultured at the ALI using primary trachea BC from healthy nonsmokers; 2) RNA-seq (HiSeq 2500; Illumina) to assess ACE2 expression in two immortalized BCi-NS1.1 (LAE) and hSABCi-NS1.1 (SAE) cell lines (20, 21); and 3) single-cell RNA-seq (10x Genomics) to analyze the expression of ACE2 in SAE from five nonsmokers and five smokers. In addition to the expression of ACE2, we assessed the expression of genes that have been identified as participating in the initial steps of other similar coronaviruses, with the likelihood that some of these genes participate in the early events of SARS-CoV-2 infection. Finally, we assessed the data of O’Beirne and colleagues (25) for the effects, if any, of New York City pollution levels over time on SAE ACE2 levels.
Results
Expression of ACE2 in Normal Airway Epithelium
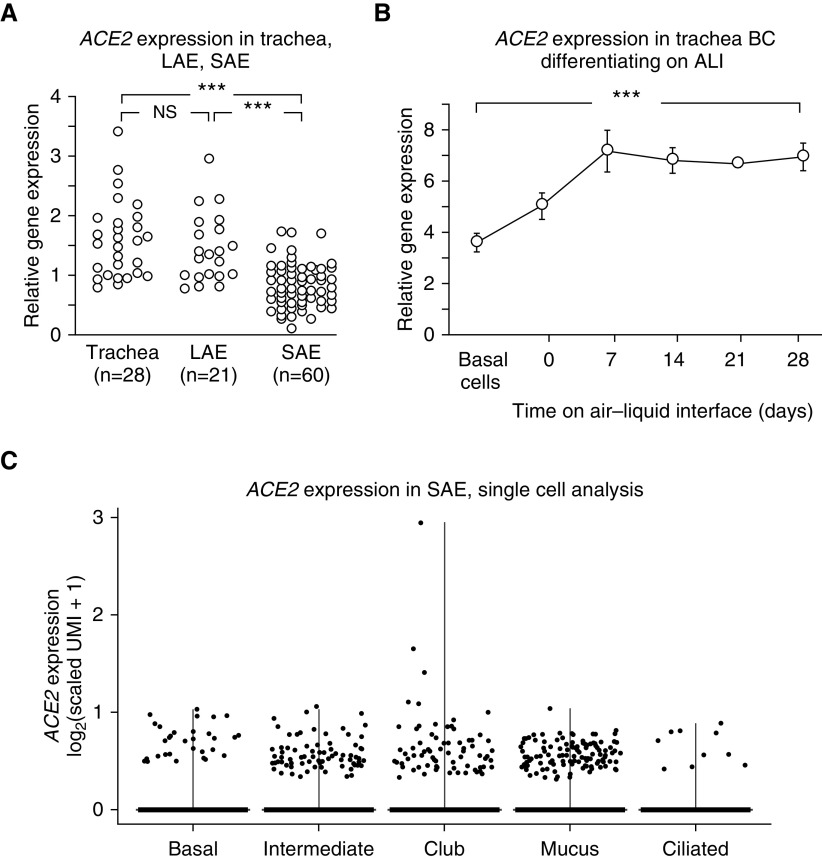
Analysis of trachea, LAE, and SAE demonstrated that ACE2 is expressed in all regions of the tracheobronchial tree of healthy nonsmokers, with higher expression in the trachea and LAE than in the SAE (Affymetrix HG-U133 Plus 2.0 microarray; Figure 1A). The assessment of primary trachea basal stem/progenitor cells of healthy nonsmokers differentiating on ALI showed that BCs express ACE2, as do, to a greater extent, the differentiated progeny of the BCs (Affymetrix HG-U133 Plus 2.0 microarray; Figure 1B). Single-cell transcriptome analysis identified all the major cell types of the SAE of healthy nonsmokers, including basal, intermediate, club, mucus, and ciliated cells, as well as ionocytes, macrophages, T cells, and mast cells (10x, Figure E1). ACE2 expression was noted mainly in epithelial cells, including basal, intermediate, club, mucus, and ciliated cells (Figure 1C). The low percentage of positive cells is partially a consequence of the technology that samples a fraction of the transcripts in a given cell (28). As a result, low abundance transcripts are not detected in every cell that expresses the gene. Of interest, the single-cell data of Reyfman and colleagues (29) derived from the lung parenchyma of individuals without lung disease demonstrated that alveolar type 2 cells and other epithelial cells express ACE2 (Figure E2A). The similar percentages of epithelial cells that were observed to express ACE2 in the Reyfman study (29) compared with those in this study support the observation that ACE2 is expressed in a broad array of epithelial cells. Similarly, an analysis of data from Duclos and colleagues (30), who obtained bronchial epithelium via bronchoscopic brushing in a similar manner to this study, reveals the presence of ACE2 in cells in a variety of epithelial cells (Figure E2B). Of note, ACE2 was detected in a higher percentage of mucus cells than other epithelial cells in the Duclos study (30), possibly indicating a difference between the large airway bronchial epithelia analyzed in that study compared with the SAE analyzed in this study. From this data, we conclude that the ACE2 receptor for SARS-CoV-2 is distributed throughout the lung epithelial surface and that the site of infection would be dictated by the size of the inhaled droplet and respiration parameters (31).
Figure 1.
Expression of ACE2 (angiotensin-converting enzyme 2) in the human airway epithelium of healthy nonsmokers. (A and B) Expression level is presented as relative gene expression compared with all other genes on the array. See the online supplement for details on normalization. (A) Comparison of ACE2 expression in trachea epithelium, large airway epithelium, and small airway epithelium (SAE). Quantification was by Affymetrix HG-U133 Plus 2.0 microarrays. The data were generated from the data sets of Gene Expression Omnibus accession numbers 13933, 10135, and 11784 (23, 24, 26) and were compared using a two-way ANOVA (sex was identified as a source of variation). (B) ACE2 expression during in vitro differentiation of airway epithelium derived from primary tracheal basal cells (BCs). RNA was collected by brushing from freshly isolated, purified tracheal BCs and from cells derived from the BCs on an air–liquid interface culture at the initiation of the culture (Day 0) and at Days 7–28 of culture. ACE2 levels (determined by Affymetrix HG-U133 Plus 2.0 microarrays) increased as BCs differentiated into airway epithelial cells (ACE2 levels at Day 28 compared with Day 0, P < 10−5). (C) Single-cell 10x analysis of ACE2 expression in the different cell populations from the normal SAE of healthy nonsmokers. All the major cell types express ACE2, including basal, intermediate, club, mucus, and ciliated cells. Each data point represents a single cell. ACE2 was detected in a minority of epithelial cells from each cluster (detected in 1.2% of BCs, 2.6% of intermediate cells, 1.7% of club cells, 2.4% of mucus cells, and 1.0% of ciliated cells). These values are useful for comparison among the epithelial cell types but underestimate the actual percentage of cells expressing the gene (28). See the online supplement for markers used to define each cell type and for details on the calculation of scaled unique molecular identifiers and transformation for data presentation. ***P < 0.001. ALI = air–liquid interface; LAE = large airway epithelium; NS = nonsignificant; UMI = unique molecular identifiers.
We also analyzed cells recovered by BAL for ACE2 expression. Consistent with our analysis of the data from Reyfman and colleagues (29) (Figure E2A), data obtained using microarray, RNA-seq, and 10x single-cell transcriptome analysis indicated that ACE2 expression was undetectable or rarely detected in alveolar macrophages, T cells, B cells, or dendritic cells (data not shown).
Smoking and Sex Influence on ACE2 Expression
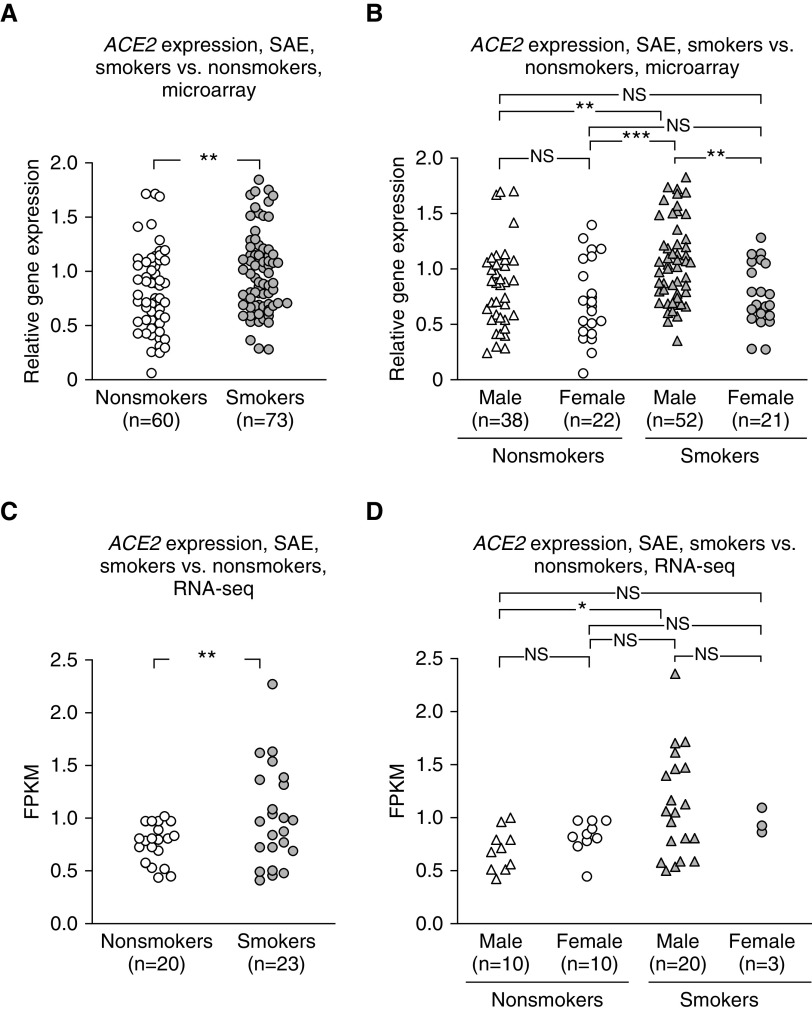
Based on the clinical data that SARS-CoV-2 lung infection is characterized in chest imaging as distal infiltrates (32), it is likely that the SAE is an important site of SARS-CoV-2 binding. ACE2 expression was higher in the SAE of smokers than nonsmokers (Affymetrix HG-U133 Plus 2.0 microarray; P < 10−5; Figure 2A). When nonsmokers and smokers were divided by sex, male smokers exhibited significantly higher ACE2 expression levels than female smokers or nonsmokers of either sex (P < 0.005, all comparisons; Figure 2B). RNA-seq analysis of a different dataset confirmed that SAE ACE2 levels were higher in smokers than nonsmokers (HiSeq 2500; Illumina) (Figure 2C), with the ACE2 levels in SAE of male smokers higher than those of male nonsmokers (Figure 2D; a low n = 3 in females obviated comparison of smoker versus nonsmoker ACE2 levels in females). Analysis of ACE2 levels in the LAE showed no differences relevant to sex or smoking, except that ACE2 levels were significantly higher in male versus female nonsmokers when assessed by Affymetrix HG-U133 Plus 2.0 microarray (Figure E3).
Figure 2.
Effect of smoking and sex on ACE2 (angiotensin-converting enzyme 2) expression in the small airway epithelium. (A and B) Expression level is presented as relative gene expression compared with all other genes on the array. See the online supplement for details on normalization. (A) Healthy smokers (gray symbols) versus nonsmokers (white symbols), with male and female sexes combined (Affymetrix HG-U133 Plus 2.0 microarrays). The data were generated from the data set of Tilley and colleagues (26), Gene Expression Omnibus accession number 11784. (B) Male sex versus female sex for smokers versus nonsmokers (Affymetrix HG-U133 Plus 2.0 microarrays), using the same data set as in A. (C) Smokers versus nonsmokers, male and female sexes combined, RNA sequencing (RNA-seq) (HiSeq 2500; Illumina). (D) Male versus female sex for smokers versus nonsmokers (RNA-seq), using the same data set as in C. A two-way ANOVA (sex was identified as a source of variation) was used for analysis. *P < 0.05, **P < 0.01, and ***P < 0.001. FPKM = fragments per kilobase of exon per million fragments sequenced; NS = nonsignificant; SAE = small airway epithelium.
Pollution Effects on ACE2 Expression
Correlations between ambient pollution levels and COVID-19 cases have been reported (33). To assess whether low levels of air pollution might affect SAE ACE2 expression, we analyzed the dataset of O’Beirne and colleagues (25), a study performed to evaluate the relationship between gene expression in the SAE of healthy nonsmokers and smokers in New York City with average monthly pollution levels (PM2.5) as reported by the U.S. Environmental Protection Agency. Although no differences were observed between SAE ACE2 expression and PM2.5 levels (nonsmokers and smokers combined or separately; Figure E4), the peak 30-day mean PM2.5 concentration in New York City during this study was 18 μg/m3, a level considered safe by the U.S. Environmental Protection Agency (25).
Possible Influences of miRNA Expression on ACE2
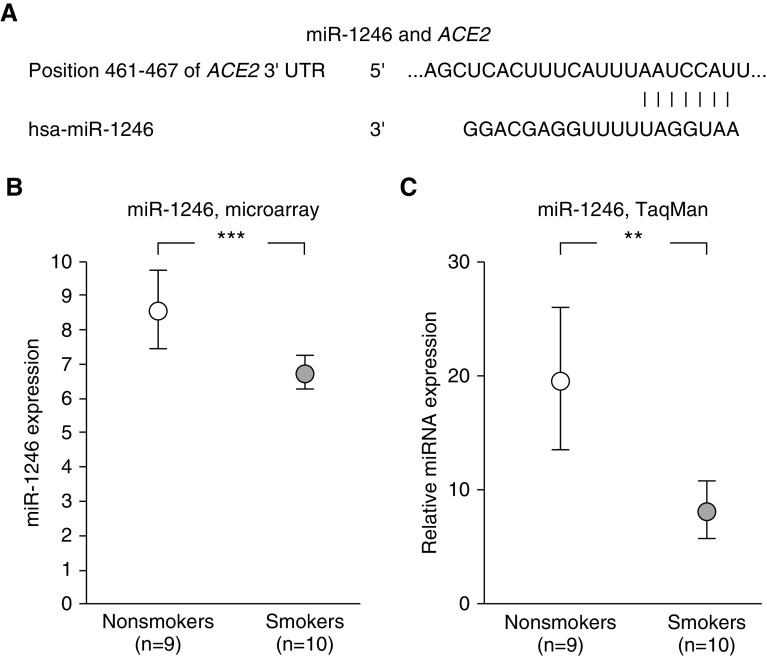
Our database was assessed for miRNAs homologous to the 3′ end of the ACE2 gene that are significantly modulated by smoking. Of interest, miR-1246 has homology to ACE2, and miR-1246 is downregulated in SAE of smokers compared with nonsmokers (27) (Affymetrix miRNA 2.0 arrays, Figure 3). We have insufficient data to determine if this smoking-related decrease in miR-1246 plays a role in ACE2 expression in the SAE, but this is a possible mechanism to be assessed in future studies.
Figure 3.
Possible relationship of miR-1246 levels to modulate the levels of ACE2 (angiotensin-converting enzyme 2) in the small airway epithelium (SAE). Assessment of the data set of Wang and colleagues (27) (Gene Expression Omnibus accession number 53519) of healthy nonsmokers (n = 9; white circles) and healthy smokers (n = 10; gray circles) for smoking-related significant changes in levels of microRNA (miRNA) in the SAE for miR-1246 with sequences that complement the sequence of the 3′ untranslated region (3′UTR) of ACE2 mRNA. (A) Predicted pairing of target region in ACE2 3′UTR (top) and human miR-1246 (hsa-miR-1246, bottom) analyzed by TargetScanHuman 7.2 (http://www.targetscan.org/vert_72/). (B and C) miR-1246 levels are decreased in the SAE of smokers compared with those of nonsmokers. A two-way ANOVA (age was identified as a source of variation) was used for analysis. (B) Assessment by Affymetirx miRNA 2.0 arrays. Expression of miRNA is presented as relative miRNA expression compared with all other human mature miRNA. See online supplement for details. (C) Assessment by TaqMan PCR. Data are from Wang and colleagues (27). **P < 0.01 and ***P < 0.001.
Expression of Genes Related to the Initial Steps of Coronavirus Infection
In addition to evaluating the expression of ACE2, the gene encoding the primary SARS-CoV-2 receptor, we assessed the SAE mRNA expression levels of other cellular proteins reported to be related to the early steps in infection pathway based on lineage B coronaviruses and, thus, possibly relevant to airway epithelial infection by SARS-CoV-2.
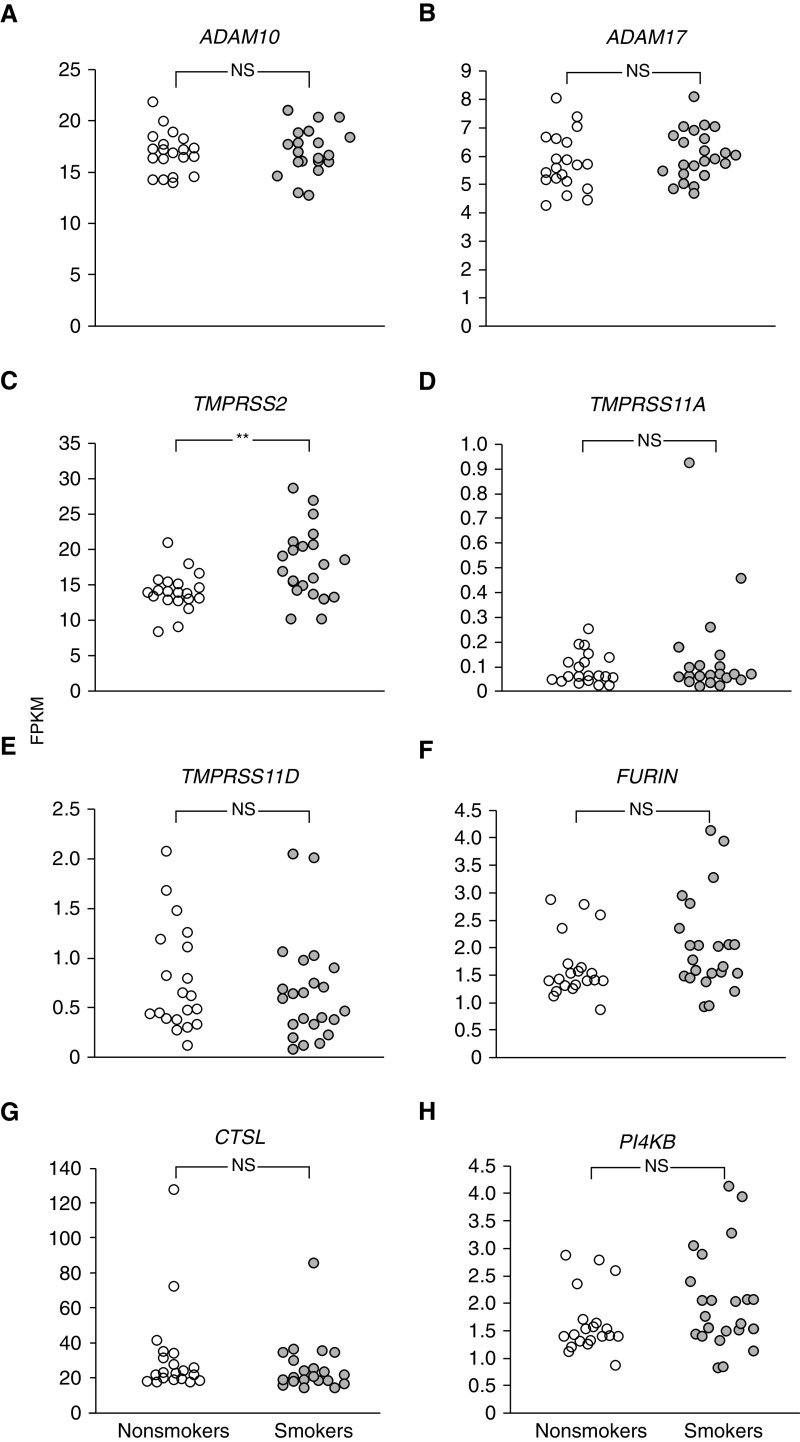
Cell-surface ACE2 levels can be regulated by ADAM10 and ADAM17, cell-surface disintegrins that mediate shedding of ACE2 from the cell surface (34). Both ADAM10 and ADAM17 are expressed in the SAE (Figures 4A and 4B). TMPRSS2, TMPRSS11A, and TMPRSS11D are proteases that cleave the SARS-CoV spike protein at the cell surface to facilitate the fusion of the coronavirus envelope with the cell membrane, a critical step in the transfer of the nucleocapsid to the cytosol (16, 35). All three enzymes are expressed in the SAE (Figures 4C–4E). Interestingly, expression of TMPRSS2 is upregulated in the SAE of smokers compared with those of nonsmokers (Figure 4C). Furin and cathepsin L, two proteases encoded by the genes FURIN and CTSL, are found in the endolysosomal pathway and can cleave the coronavirus spike protein, leading to intracellular fusion of the envelope with the organelle membrane relevant to infection by SARS and other coronaviruses (18, 36, 37). Both proteins are expressed in the SAE (Figures 4F and 4G). Finally, inhibition of PI4KB an enzyme that phosphorylates phosphatidylinositol, results in the inhibition of SARS-CoV infection (19). PI4KB is also expressed in the SAE (Figure 4H). Other than the upregulation of TMPRSS2 in smokers, there were no other smoking-related changes in SAE expression among the genes related to coronavirus infection. Separately, an analysis of single-cell RNA-seq data showed that coronavirus infection–related genes were broadly expressed in airway epithelium, including basal, intermediate, club, mucus, and ciliated cells in both healthy nonsmokers and smokers (Figure 5). As reported above, ACE2 expression was observed in all of the epithelial cell types; however, in contrast to data from RNA-seq and microarrays, ACE2 did not exhibit a smoking-dependent increase in gene expression in the single-cell transcriptome data (Figure 5A). The absence of an observed difference in ACE2 expression among nonsmokers and smokers in the single-cell RNA-seq dataset compared with the bulk RNA dataset and microarray dataset likely reflects the technical details inherent in the three types of analysis. Single-cell RNA-seq data are derived from a smaller number of individuals and a smaller number of cells per individual compared with bulk RNA-seq or microarray analysis. The preparation time and, therefore, potential changes because of RNA degradation are greater for single-cell RNA-seq than bulk RNA-seq or microarray analysis, which may lead to a disproportionate increase in variability for low abundance transcripts such as ACE2 in single-cell RNA-seq compared with bulk RNA-seq or microarray analysis. Finally, single-cell RNA-seq is also influenced by the relative size and shape of cells, with larger, fragile, and differentiated cells exhibiting higher losses during recovery from the airway epithelium. For example, ciliated cells are known to constitute 60–70% in cell differentials of epithelium taken from small airway but make up less than 20% of live single cells that survive to be included in single-cell analysis. Despite these caveats, single-cell RNA-seq provides an opportunity to gain expression data on single cell types in a mixed population, which is not possible using bulk RNA-seq or microarray analysis. Among the other host factors related to coronavirus infection, all except for TMPRSS11A were detected by single-cell RNA-seq, all were widely expressed in SAE cells, and none were significantly different in nonsmokers versus smokers (Figures 5B–5H). CLEC4M-mediated expression of CD209L, a cell-surface protein reported to have binding activity for SARS-CoV (38), was not detected in airway epithelial samples by single-cell RNA-seq. For transcriptomic analyses, including single-cell RNA-seq, microarray analysis, and bulk RNA sequencing, it is important to remember that mRNA levels do not always precisely predict protein levels in tissues and that correlative studies to assess protein levels need to be performed.
Figure 4.
Assessment of the small airway epithelia of healthy nonsmokers (n = 20; white circles) and smokers (n = 23; gray circles) for expression of genes that may be relevant to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. See text for details regarding the possible relevance of these genes to small airway epithelia infection by SARS-CoV-2. Quantification was by RNA sequencing (Illumina HiSeq 2500). A one-way ANOVA was used for analysis. (A) ADAM10. (B) ADAM17. (C) TMPRSS2. (D) TMPRSS11A. (E) TMPRSS11D. (F) FURIN. (G) CTSL. (H) PI4KB. **P < 0.01. FPKM = fragments per kilobase of exon per million fragments sequenced; NS = nonsignificant.
Figure 5.
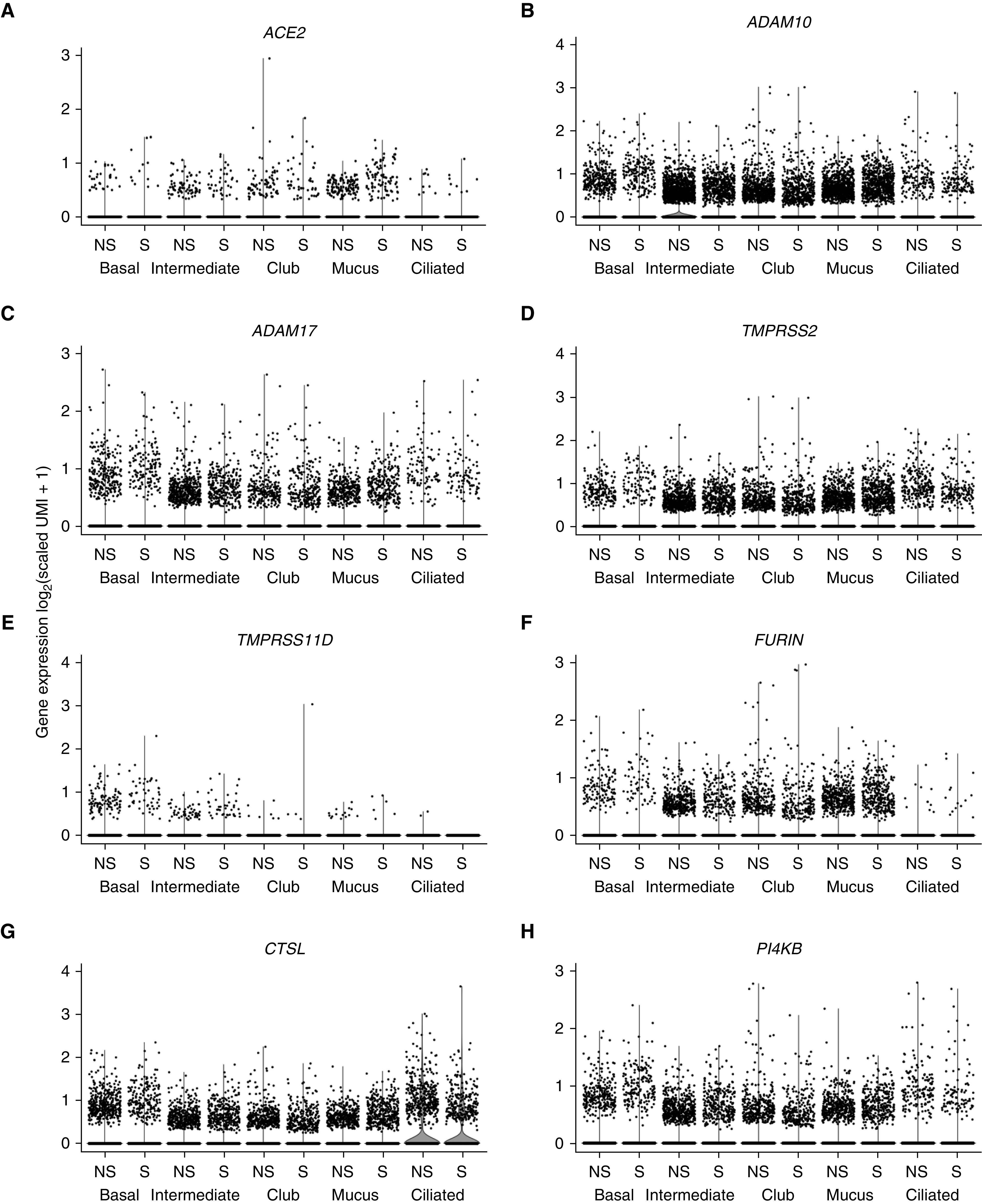
Single-cell 10x transcriptome analysis of the small airway epithelia of healthy nonsmokers (NS) (n = 5) and smokers (S) (n = 5) for expression of genes that may be relevant to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of the small airway epithelium. A total of 18,263 cells from NS and 16,678 cells from S were analyzed. ACE2 (angiotensin-converting enzyme 2) cells were detected in a minority of epithelial cells from both NS and S (percentage of cells in NS/percentage of cells in S: 1.2%/0.6% of basal cells, 2.6%/2.1% of intermediate cells, 1.7%/1.3% of club cells, 2.4%/1.9% of mucus cells, and 1.0%/1.2% of ciliated cells). These values are useful for comparison among the epithelial cell types but underestimate the actual percentage of cells expressing the gene (28). Expression of TMPRSS11A was detected by bulk RNA sequencing (HiSeq 2500; Illumina, Figure 4D) but was not detected by single-cell RNA sequencing (10x). For all comparisons of gene expression in all cell types, the differences in expression levels in NS and S were <10%; no significant differences in gene expression were observed. Statistical comparisons were performed using the Wilcoxon rank sum test, with P values adjusted using the Bonferroni correction. (A) ACE2. (B) ADAM10. (C) ADAM17. (D) TMPRSS2. (E) TMPRSS11D. (F) FURIN. (G) CTSL. (H) PI4KB. See the online supplement for markers used to define each cell type and for details on calculation of scaled unique molecular identifiers and transformation for data presentation. UMI = unique molecular identifiers.
Finally, we assessed ACE2 expression in two cell lines generated from BCs of healthy nonsmokers, including BCi-NS1.1 (derived from a single BC from the LAE of a healthy nonsmoker) (20) and hSABCi-NS1.1 (derived from a single BC from the SAE of a healthy nonsmoker) (21). With the caveat of a small n, these data were derived from RNA-seq data on a single sample of each cell line at each stage of differentiation and are subject to validation; both cell lines expressed low levels of ACE2 before differentiation. When allowed to differentiate on an ALI, both cell lines expressed elevated levels of ACE2 (RNA-seq, HiSeq 4000; Illumina, Table 1). When queried for expression levels of other genes encoding proteins important to coronavirus infection, relatively high levels of ADAM10, ADAM17, FURIN, CTSL, and PI4KB were all detected and were maintained with differentiation. TMPRSS2 expression was low in undifferentiated large airway BCs but became much more pronounced in differentiated LAE. In contrast, the level of TMPRSS2 was higher in the small airway immortalized BC line, and expression was maintained at approximately the same expression level after differentiation. Protease family members TMPRSS11A and TMPRSS11D showed low expression at both stages of differentiation. These cell lines can be grown indefinitely and should be useful for investigating the biology of SARS-CoV-2 infection, including the screening of therapies designed to inhibit the early stages of infection.
Table 1.
Expression of Host Proteins Potentially Relevant to SARS-CoV-2 Infection in Undifferentiated and Differentiated Airway Basal Cell Lines*
| Gene | BCi-NS1.1† |
hSABCi-NS1.1‡ |
||
|---|---|---|---|---|
| Basal Cell Baseline | Air–Liquid Interface Day 28 | Basal Cell Baseline | Air–Liquid Interface Day 28 | |
| ACE2 | 0.4 | 1.7 | 0.4 | 2.8 |
| ADAM10 | 23.8 | 13.6 | 23.4 | 21.8 |
| ADAM17 | 9.6 | 6.9 | 14.0 | 11.8 |
| TMPRSS2 | 3.3 | 25.4 | 12.2 | 15.2 |
| TMPRSS11A | 0 | 0.5 | 0.5 | 0 |
| TMPRSS11D | 0.1 | 0.7 | 0.4 | 0.2 |
| FURIN | 14.9 | 12.5 | 46.8 | 29.5 |
| CTSL | 37.5 | 33.1 | 74.7 | 67.0 |
| PI4KB | 18.0 | 28.6 | 19.2 | 22.6 |
Definition of abbreviations: FPKM = fragments per kilobase of exon per million fragments sequenced; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Expression assessed by RNA sequencing (HiSeq 4000; Illumina); data is presented in FPKM.
The BCi-NS1.1 line was derived from large airway epithelium basal cells of a healthy nonsmoker and differentiated on air–liquid interface for 28 days (20).
The hSABCi-NS1.1 line was derived from small airway epithelium basal cells of a healthy nonsmoker and differentiated on air–liquid interface for 28 days (21).
Discussion
Biology of SARS-CoV-2 Infection
Respiratory disease is the dominant manifestation of SARS-CoV-2, which is both acquired and transmitted by the inhalation of airborne droplets and by contact routes (4, 5, 7, 8). SARS-CoV-2 expressed the spike protein that binds to ACE2 on the surface of airway epithelial cells (7, 13, 15–17). To provide additional insights into the biology of this initial interaction of SARS-CoV-2 with the airway epithelium, we assessed our published and unpublished transcriptome databases of human airway epithelium to assess the extent of expression of ACE2 in healthy nonsmokers and smokers. The data demonstrates widespread expression of ACE2 throughout the respiratory epithelial surface. In the SAE, a likely site of entry of SARS-CoV-2, all major epithelial cell types express the ACE2 gene, as indicated by analysis of our single-cell RNA-seq data as well as reanalysis of single-cell RNA-seq data from Reyfman and colleagues and Duclos and colleagues, which is presented in the online supplement (29, 30). It is important to note that although the absolute number of cells identified with ACE2 is very low in this study and in the Reyfman and Duclos studies, the percentage is necessarily an underestimate of the proportion of cells in the airway epithelium that are actually expressing ACE2 because of technical details of the single-cell RNA-seq method (28). In support of our data, Harmer and colleagues (39) found ACE2 mRNA by qRT-PCR at several levels of the airway. Using data from Duclos and colleagues (30), after supervised clustering, ACE2 was apparent in large airway basal/intermediate cells, club cells, mucus cells, and ciliated cells. Finally, several recently submitted manuscripts also indicate that ACE2 is expressed in a broad array of epithelial cells at various positions along the airway (40, 41). At the protein level, Hamming and colleagues (42) observed ACE2 staining in alveolar epithelial cells and in the basal layer of airways. A previous report by Jia and colleagues localized the majority of the ACE2 protein to ciliated cells in an in vitro differentiated large airway cell culture (43), contrasting with the broader expression in epithelial cells we identified. The level of ACE2 expression in this differentiated LAE culture was sufficient to measure ADAM10- and ADAM17-dependent ACE2 shedding (34). Data from Jia and colleagues (43) and from our analysis of undifferentiated and differentiated immortalized airway BCs suggest that in vitro cultures of airway epithelium may be useful in studying SARS-CoV-2 infection.
Of interest, the expression of the TMPRSS2 gene is upregulated in the SAE of smokers. Other members of the transmembrane serine protease family, including TMPRSS11A and TMPRSS11D (also known as HAT), share the ability to activate the fusion peptide in the spike protein via proteolysis (35). Although TMPRSS11A was not detected in the single-cell transcriptome data, both TMPRSS2 and TMPRSS11D were detected in basal, intermediate, club, mucus, and ciliated cells. TMPRSS2 exhibited strong expression through the airway epithelium, whereas TMPRSS11D was less prevalent in fully differentiated airway epithelial cells. Other than TMPRSS2, smoking did not affect the expression of other proteases of this class. Furthermore, the intracellular proteases furin and cathepsin L were also widely expressed, suggesting that SARS-CoV-2 could be induced to escape from an endosome after entry, and PI4KB was also expressed, completing the complement of cellular proteins needed to support a productive infection of most airway epithelial cell types. Additional host proteins will surely be demonstrated to play modifying roles in SARS-CoV-2 infection. For example, the IFITM family of IFN-induced transmembrane proteins has previously been shown to have anti-SARS activity, although the mechanism of this action has not yet been determined (44). Of interest, the fact that cigarette smoke blocks IFN signaling might provide yet another link between cigarette smoking and SARS-CoV-2 infection.
Control of Airway Epithelium ACE2 Gene Expression
The relative contribution of smoking to the acquisition and course of a SARS-CoV-2 infection has been a source of controversy as the COVID-19 pandemic has developed (45, 46). The finding of the smoking-specific difference in ACE2 expression in the small airway, but not in the large airway or trachea, is of interest because it suggests potential differences in airway epithelial susceptibility. Depending on droplet size, the inhalation of droplets can lead to the deposition of materials throughout the airway (31). At present, the precise avenues of infection by SARS-CoV-2 are not well understood, a point highlighted by the revelation that many contagious individuals are likely asymptomatic, suggesting that they may have active infection in the upper airway without involvement of the lower airway (47). A study of pulmonary infections caused by a closely related coronavirus in mice showed that airway infection preceded alveolar involvement and was worse in aged mice and that an ACE2-knockout model was protected from infection in the lung, all of which implicate the airway epithelium as a critical element of pathogenesis (48).
We observed that SAE ACE2 expression is higher in male smokers compared with female smokers, all nonsmokers, and male nonsmokers, separately. Combined with the observation that expression of the gene encoding the infectivity activating protease, TMPRSS2, was elevated in smokers, there are reasons that smokers, and, in particular, male smokers, would be at a greater risk of propagating a SARS-CoV-2 infection. However, whether smoking is a significant risk factor for COVID-19 infection and/or the intensity of the infection is not clearly defined at the clinical level. Cai and colleagues (49) noted a disproportionate number of men reported with COVID-19 across several epidemiological studies among Asian populations early during the pandemic and postulated that the high incidence in men was due to a higher incidence of smoking in that population. Whether the disproportionate incidence among men can be attributed to smoking-induced changes in gene expression, smoking-associated comorbidities, or some other factor remains to be determined. A mechanistic explanation for the variations in ACE2 gene expression in this study has not yet been established.
Our data also showed that miR-1246, a miRNA with homology to ACE2, is downregulated in the SAE of smokers, providing a potential mechanism for smoking-related upregulation of ACE2. Other notable aspects of ACE2 expression imply that sex-specific gene expression would be anticipated. Despite the fact that ACE2 is located on the X chromosome and is known to escape X inactivation, the gene exhibits a variable sex- and tissue-specific bias, with lower expression observed in female lungs compared with male lungs (50). ACE2 gene expression may be one of several factors contributing to prevalence of COVID-19 in men.
BCi-NS1.1 and hSABCi-NS1.1 Cell Lines
One of the challenges in studying the early steps in virulent coronaviruses such as SARS-CoV-2 is establishing an in vitro cell culture system that reflects, as closely as possible, the interaction of the virus with the human respiratory epithelium. In the context that the primary human airway BCs express ACE2 and then when differentiated on ALI, the differentiated progeny express ACE2, BC differentiation provides an in vitro culture model to assess SARS-CoV-2 and airway epithelium interaction. Although primary normal human airway epithelium cannot be maintained for more than three to four passages in vitro, we have immortalized two cell lines, BCi-NS1.1 and hSABCi-NS1.1, each derived from a single BC of a healthy nonsmoker from LAE or SAE, respectively (20, 21). Genes encoding ACE2, TMPRSS2, and the other cellular factors that collaborate to create a successful SARS-CoV-2 infection were found to be expressed in vitro in airway epithelium differentiated from the immortalized LAE BCi-NS1.1 and SAE hSABCi-NS1.1 cell lines (20, 21); these cells lines should be useful in studying the early events of SARS-CoV-2 infection of the airway epithelium and the assessment of potential therapies to prevent the progression of COVID-19. Both lines can be genetically manipulated, and both can be passaged indefinitely. Both cell lines are available to the coronavirus community by contacting the senior author.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the Epigenomics, Genomics, and Flow Cytometry Core Facilities at Weill Cornell Medical College for assistance in generating the single-cell RNA sequencing data and N. Mohamed for editorial assistance.
Footnotes
Supported by NIH grants HL134549, HL107882, HL107882-S, HL118857, and HL118541; the Department of Genetic Medicine at Weill Cornell Medical College; Hoffmann-La Roche, Inc; and Boehringer Ingelheim Pharmaceuticals. M.R.R. was supported, in part, by NIH T32HL094284, and S.L.O’B. was supported, in part, by the Pulmonary Fibrosis Foundation and the American Thoracic Society.
Author Contributions: H.Z., M.R.R., P.L.L., Y.S.-B., and R.G.C.: data analysis and manuscript writing. J.G.M.: statistical analysis. S.L.O’B.: subject recruitment and phenotyping, bronchoscopy, and manuscript writing.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202003-0541OC on May 20, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 7.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization, Depart of Communications. Modes of transmission of virus causing COVID-19:implications for IPC precaution recommendations. 2020 [accessed 2020 Apr 30]. Available from: https://www.who.int/publications-detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations.
- 9.Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention. [The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China] Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280, e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang N, Ma P, Lang J, Zhang Y, Deng J, Ju X, et al. Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry. J Biol Chem. 2012;287:8457–8467. doi: 10.1074/jbc.M111.312561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walters MS, Gomi K, Ashbridge B, Moore MA, Arbelaez V, Heldrich J, et al. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res. 2013;14:135. doi: 10.1186/1465-9921-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Lou HH, Salit J, Leopold PL, Driscoll S, Schymeinsky J, et al. Characterization of an immortalized human small airway basal stem/progenitor cell line with airway region-specific differentiation capacity. Respir Res. 2019;20:196. doi: 10.1186/s12931-019-1140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med (Berl) 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 23.Turetz ML, O’Connor TP, Tilley AE, Strulovici-Barel Y, Salit J, Dang D, et al. Trachea epithelium as a “canary” for cigarette smoking-induced biologic phenotype of the small airway epithelium. Clin Transl Sci. 2009;2:260–272. doi: 10.1111/j.1752-8062.2009.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanni H, Kazeros A, Wang R, Harvey BG, Ferris B, De BP, et al. Cigarette smoking induces overexpression of a fat-depleting gene AZGP1 in the human. Chest. 2009;135:1197–1208. doi: 10.1378/chest.08-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Beirne SL, Shenoy SA, Salit J, Strulovici-Barel Y, Kaner RJ, Visvanathan S, et al. Ambient pollution-related reprogramming of the human small airway epithelial transcriptome. Am J Respir Crit Care Med. 2018;198:1413–1422. doi: 10.1164/rccm.201712-2526OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilley AE, O’Connor TP, Hackett NR, Strulovici-Barel Y, Salit J, Amoroso N, et al. Biologic phenotyping of the human small airway epithelial response to cigarette smoking. PLoS One. 2011;6:e22798. doi: 10.1371/journal.pone.0022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Wang R, Strulovici-Barel Y, Salit J, Staudt MR, Ahmed J, et al. Persistence of smoking-induced dysregulation of miRNA expression in the small airway epithelium despite smoking cessation. PLoS One. 2015;10:e0120824. doi: 10.1371/journal.pone.0120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duclos GE, Teixeira VH, Autissier P, Gesthalter YB, Reinders-Luinge MA, Terrano R, et al. Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution. Sci Adv. 2019;5:eaaw3413. doi: 10.1126/sciadv.aaw3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonda I. Particle deposition in the human respiratory tract. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The lung: scientific foundations. editors. Philadelphia: Lippincott-Raven; 1997. pp. 2289–2294. [Google Scholar]
- 32.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, Xie J, Huang F, Cao L. Association between short-term exposure to air pollution and COVID-19 infection: evidence from China. Sci Total Environ. 2020;727:138704. doi: 10.1016/j.scitotenv.2020.138704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia HP, Look DC, Tan P, Shi L, Hickey M, Gakhar L, et al. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch BJ, Bartelink W, Rottier PJ. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 40.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lippi G, Henry BM. Active smoking is not associated with severity of coronavirus disease 2019 (COVID-19) Eur J Intern Med. 2020;75:107–108. doi: 10.1016/j.ejim.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vardavas CI, Nikitara K. COVID-19 and smoking: a systematic review of the evidence. Tob Induc Dis. 2020;18:20. doi: 10.18332/tid/119324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimball A, Hatfield KM, Arons M, James A, Taylor J, Spicer K, et al. Public Health – Seattle & King County; CDC COVID-19 Investigation Team. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility: King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377–381. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menachery VD, Yount BL, Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. 2020;8:e20. doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, et al. GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz Landscape of X chromosome inactivation across human tissues Nature 2017550244–248.29022598 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.