Abstract
Leishmaniasis constitutes the 9th largest disease burden among all infectious diseases. Control of this disease is based on a short list of chemotherapeutic agents headed by pentavalent antimonials, followed by miltefosine and amphotericin B; drugs that are far from ideal due to host toxicity, elevated cost, limited access, and high rates of drug resistance. Knowing that the composition of extracellular vesicles (EVs) can vary according to the state of their parental cell, we hypothesized that EVs released by drug-resistant Leishmania infantum parasites could contain unique and differently enriched proteins depending on the drug-resistance mechanisms involved in the survival of their parental cell line. To assess this possibility, we studied EV production, size, morphology, and protein content of three well-characterized drug-resistant L. infantum cell lines and a wild-type strain. Our results are the first to demonstrate that drug-resistance mechanisms can induce changes in the morphology, size, and distribution of L. infantum EVs. In addition, we identified L. infantum’s core EV proteome. This proteome is highly conserved among strains, with the exception of a handful of proteins that are enriched differently depending on the drug responsible for induction of antimicrobial resistance. Furthermore, we obtained the first snapshot of proteins enriched in EVs released by antimony-, miltefosine- and amphotericin-resistant parasites. These include several virulence factors, transcription factors, as well as proteins encoded by drug-resistance genes. This detailed study of L. infantum EVs sheds new light on the potential roles of EVs in Leishmania biology, particularly with respect to the parasite’s survival in stressful conditions. This work outlines a crucial first step towards the discovery of EV-based profiles capable of predicting response to antileishmanial agents.
Author summary
Visceral leishmaniasis is a life-threatening disease caused by Leishmania infantum parasites, which are transmitted by sand flies. In the absence of vaccines, current control of this disease is based on chemotherapy, which is comprised of a very limited arsenal threatened by the emergence and spread of drug-resistant strains. In the shadow of growing concern and treatment failure due to resistance, the characterization of extracellular vesicles (EVs) released by drug-resistant L. infantum parasites could shed some light on the complex nature of drug resistance in Leishmania and increase our understanding of the biology of the parasite. EVs are vesicles secreted by all eukaryotic cells whose contents (proteins, DNA/RNAs, lipids) vary as a function of their cellular origin. Our results demonstrate for the first time that EVs released by drug-resistant parasites are enriched in unique protein markers that reflect the drug-resistance mechanisms involved in the survival of parental cells. These unique proteins included several virulence and transcription factors, as well as drug-resistance genes; this offers a potential benefit for drug-resistant parasites in terms of parasite-to-parasite communication and host-parasite interactions. Collectively, our initial results could serve as a jumping-off point for the future development of novel EV-based diagnostic tools for the detection and appraisal of antimicrobial-resistant Leishmania populations.
Introduction
The vector-borne protozoan parasite Leishmania affects 15 million people worldwide, and in the absence of effective preventive and therapeutic treatments, is spreading with ≈1.5M new cases/year [1, 2]. Clinical disease ranges from the self-healing cutaneous manifestation to life-threatening visceral leishmaniasis (VL), a systemic disease with a fatality rate as high as 100% within 2 years, especially if left untreated [3–5]. L. infantum is the main causative agent of VL in both humans and dogs, the latter of which serve as the main reservoir for infection [6]. While many efforts have been employed to tackle leishmaniasis, the incidence of VL is increasing in the Americas [7], as well as re-emerging in the form of epidemic outbreaks in Europe, India, and Eastern Africa [8–11]. Control of the disease is based on a very short list of chemotherapeutic agents headed by pentavalent antimonials (Sb), followed by miltefosine (MF) and amphotericin B (AmB). These drugs are far from ideal due to host toxicity, elevated cost, and limited access, but in particular due to the high rates of drug resistance [2]. The latter should be not neglected, especially when both humans and dogs are treated with the same molecules [1, 12].
Extracellular vesicles (EVs) are particles formed by a lipid bilayer containing proteins and nucleic acids; EVs originate from and are released by many types of cells, including eukaryotic cells [13–16]. Although EVs can be separated into numerous subclasses, these particles are often categorized as exosomes, microvesicles or apoptotic bodies, according to their size, origin, and components [13, 15, 17]. During EVs biogenesis, different cellular components are loaded into the vesicles, while specific proteins are integrated into their lipid bilayer. Consequently, EVs’ content varies as a function of their cellular origin [18]. EVs have been the focus of numerous studies due to their implication in intercellular communication [19]. There is growing evidence that EVs play a major role not only in parasite-to-parasite communication, but also in host-parasite interactions [20–28]. Of note, it has been determined that Leishmania exosomes (a subclass of EVs formed from late endosomes) isolated from in vitro cultures are as effective in modulating some early macrophage and host inflammatory responses as whole Leishmania parasites [29–31]. Additionally, Leishmania exosomes have recently been shown to serve as an envelope and shuttle for complete LRV1 viruses, facilitating virus transmission and increasing infectivity in the mammalian host [32]. Exosomes and other EVs are reported to be involved in cell stress responses and resistance to chemotherapeutic agents in eukaryotic cells [33]. Drug-resistance in Leishmania relies on a number of mechanisms, and different key players have been identified in the last decade. For example, Leishmania resists Sb by altering its detoxifying pathways, drug uptake, and efflux, and sequestrating Sb-thiol complexes into intracellular vesicles using ABC transporter MRPA [34–36]. Both MF and AmB affect the integrity of different membranes (cell, mitochondria, etc.), and resistance to these drugs induces important changes in specific subsets of the proteome of the parasite [35, 37].
While several studies have demonstrated the utility of whole-parasite proteomics in differentiating sensitive and resistant isolates and in in vitro-generated strains [35, 37], the composition of EVs released by drug-resistant Leishmania parasites remains unexplored. Knowing that the composition of EVs can vary according to the parental cell [38–40], we hypothesized that EVs released by drug-resistant L. infantum parasites contain differently enriched proteins that could serve as proxy for the different drug-resistance and compensatory mechanisms deployed by the releasing cell line. To assess this possibility, we used quantitative label-free proteomics to study EVs production, size, morphology, and protein content of three drug-resistant L. infantum cell lines and a wild-type strain. These experiments lead to the identification of key proteins within EVs issued from drug-resistant parasites, which are essential to understanding how drug-resistant EVs may interact and modulate not only other parasites, but also host cells and the sand fly vector. Moreover, our powerful insights open the door for further exploration of Leishmania EV-profiles as potential biomarkers for the identification and evaluation of drug-resistant parasites.
Methods
Leishmania cultures
The Leishmania infantum (MHOM/MA/67/ITMAP-263) wild-type strain (WT) and the in vitro generated resistant mutants Sb2000.1, AmB1000.1 and MF200.5 [41–46], which are resistant to 2000 μM of Sb, 1000 nM of AmB and 200 μM MF, respectively, were grown in M199 medium at 25°C supplemented with 10% fetal bovine serum, 5 μg/mL of haemin at pH 7.0 and 2000 μM Sb (Potassium antimonyl tartrate, Sigma-Aldrich), 200 μM of MF (Miltefosine, Cayman Chem.) or 1 μM AmB (Amphotericin B solution, Sigma). Antileishmanial values in promastigotes were determined by monitoring the growth of parasites after 72 h of incubation at 25°C in the presence of increasing antimony concentrations, by measuring A600 using a Cytation 5 machine (BioTek, USA). EC50 values were calculated based on dose-response curves analyzed by non-linear regression with GraphPad Prism 8.0 software (GraphPad Software, La Jolla California, USA). An average of at least three independent biological replicates was performed for each determination.
Purification of Leishmania extracellular vesicles
The purification of Leishmania EVs was performed as we previously described [29, 32]. Briefly, 1 L of Leishmania parasites (2.5–5.0 x 107 parasites/mL) was grown in drug-free M199 at 25°C (supplemented with 10% FBS, 5 μg/mL of haemin at pH 7.0), and left to divide in 10 non-ventilated 75 cm2 culture flasks (Corning, USA) until they reached late-log phase. Next, parasites were washed twice in PBS and resuspended in 5 mL RPMI-1640 medium without FBS and phenol red (Life Technologies) at a final concentration of 2.5–5.0 x 108 parasites/mL, in non-ventilated 25 cm2 culture flasks (Corning, USA) at 37°C. Parasites were then incubated for 4 h at 37°C to stimulate the release of EVs in the medium [21]. The viability of parasites was evaluated by propidium-iodide staining before and after incubation at 37°C. Only cultures with a viability > 95.0% were submitted to subsequent EV-purification steps. After the 4-h incubation, samples were centrifuged twice to eliminate parasites (10 min at 3 000 g) and clear out debris (10 min at 8 500 g), followed by two subsequent filtrations using 0.45 μm and 0.20 μm syringe filters. EVs were then recovered by a 1-h centrifugation at 100 000 g and resuspended in the EVs buffer (137 mM NaCl, 20 mM Hepes, pH 7.5). EVs’ proteins were then dosed using the Micro BCA Protein Assay Kit (Pierce Biotechnology, USA). EVs were conserved in aliquots in EVs buffer at -80°C for subsequent analyses. Aliquots were slowly thawed on ice before being used. Once thawed, EVs were never refrozen. Three independent purifications were performed for each strain.
Transmission Electron Microscopy (TEM)
EVs purified from the different L. infantum WT and drug-resistant strains were directly coated on formvar carbon grids, fixed with 1% glutaraldehyde in 0.1 M sodium cacodylate buffer for 1 min, and stained with 1% uranyl acetate for 1 min. Formvar grids coated with isolated EVs were recorded using a FEI Tecnai 12 120 kV transmission electron microscope. The resulting images were captured with the AMT XR-80C CCD Camera System (Facility for Electron Microscopy Research, McGill University).
Nanoparticle Tracking Analysis (NTA)
EVs’ concentration and size distribution were characterized by NTA using a NanoSight LM 10 Instrument (Malvern Panalytical, Worcestershire, UK) available in the laboratory of Dr Janusz Rak (RI-MUCH McGill University). To determine the size and number of particles, five sequential 30-s videos were collected using the default parameter settings of the machine. EVs buffer was used as the negative control. The average size and concentration of the particles were calculated by integrating the averages of the modes from three independent records, obtained for each of three independent biological replicates.
Protein digestion and liquid chromatography–MS/MS (LC-MS/MS)
LC-MS/MS was performed, as previously described [21, 32], at the proteomic platform of the Institut de Recherches Cliniques de Montréal (Montréal, Canada). Briefly, proteins derived from purified Leishmania EVs were precipitated with 15% TCA/acetone and processed for LC–MS/MS analysis. Protein extracts were then solubilized using a 6M urea buffer, reduced by the addition of reduction buffer (45 mM DTT, 100 mM ammonium bicarbonate) for 30 minutes at 37°C, with a final concentration of 25%, then alkylated by the addition of alkylation buffer (100 mM iodoacetamide, 100 mM ammonium bicarbonate) for 20 minutes at 24°C in the dark, with a final concentration of 25%. Subsequently, an in-solution digestion was performed by the addition of trypsin at a ratio of 1:25 protease/protein, followed by an overnight incubation at 37°C. Finally, formic acid was added to a 0.2% final concentration to quench the reaction. Samples were cleaned with C18 Zip Tip pipette tips (Millipore, USA) before MS analysis. Extracted peptides were injected into a Zorbax Extended-C18 desalting column (Agilent) and separated by chromatography on a Biobasic 18 Integrafrit capillary column (Thermo Scientific, USA) on a Nano high-performance LC system (1100 series unit; Agilent). Eluted peptides were electrosprayed as they exited the capillary column and were analyzed on a QTRAP 4000 linear ion trap mass spectrometer (SCIEX/ABI).
Protein identification
The peak list files were generated with Distiller version 2.1.0.0 software (www.matrixscience.com/distiller) using the following parameters: minimum mass set to 500 Da, maximum mass set to 6,000 Da, no grouping of MS/MS spectra, precursor charge set to auto, and minimum number of fragment ions set to 5. Protein database searching was performed with Mascot 2.6 (Matrix Science) against the L. infantum (txid5671, 16,895 proteins) protein database (RefSeq assembly accession: GCF_000002875.2; last access November 2019). The mass tolerances for precursor and fragment ions were set to 10 ppm and 0.6 Da, respectively. Trypsin was used as the enzyme, allowing for up to 1 missed cleavage. Cysteine carbamidomethylation was specified as a fixed modification, and methionine oxidation as variable modifications. Data analysis was performed using Scaffold (version 4.11.0). Peptide identifications were accepted if they could be established with a probability greater than 80.0%. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [47]. Proteins that contained similar peptides and could not be differentiated using MS/MS analysis alone were grouped to satisfy the principles of parsimony. The final number of peptides per protein was represented by the average of the three biological replicates after normalization to the total number of peptides. Normalization of total spectrum counts (TSC) for each protein was performed automatically using Scaffold. Briefly, Scaffold calculates the total number of spectra in each replicate, as well as the average number of spectra across all replicates. The software then multiplies each spectrum count in each sample by the average count over the replicate’s total spectral count, leading to normalized TSC values. Normalized TSC were subsequently used to establish the different comparisons depicted in Tables 1–5.
Table 1. Differential enrichment of the common proteins identified in EVs released by L. infantum WT and the three mutant strains.
| Gene ID | Protein | WT | Sb2000.1 | MF200.5 | AmB1000.1 |
|---|---|---|---|---|---|
| LINJ_26_0150 | 60S ribosomal protein L7 putative | 93.3 | 77.0 (0.83) | 46.7 (0.50) | 5.7 (0.06) |
| LINJ_36_4100 | Adenosyl homocysteinase | 13.7 | 7.7 (0.56) | 12.7 (0.93) | 7.0 (0.51) |
| LINJ_32_0410 | ATP-dependent RNA helicase putative | 95.7 | 49.0 (0.51) | 89.7 (0.94) | 9.7 (0.10) |
| LINJ_17_0180 | Elongation factor 1-alpha | 75.3 | 65.7 (0.87) | 115.7 (1.54) | 47.0 (0.62) |
| LINJ_17_0190 | Elongation factor 1-alpha | 112.3 | 92.0 (0.82) | 148.3 (1.32) | 66.3 (0.59) |
| LINJ_36_0190 | Elongation factor 2 | 21.3 | 5.0 (0.23) | 16.7 (0.78) | 6.7 (0.31) |
| LINJ_14_1240 | Enolase | 37.0 | 43.3 (1.17) | 45.0 (1.22) | 21.7 (0.59) |
| LINJ_10_0520 | GP63-leishmanolysin | 24.7 | 14.3 (0.58)* | 53.0 (2.15) | 9.7 (0.39)* |
| LINJ_10_0490 | GP63-leishmanolysin | 113.3 | 65.7 (0.58)** | 243.3 (2.15) | 66.7 (0.59)* |
| LINJ_10_0800 | GP63-leishmanolysin | 58.7 | 34.0 (0.58)* | 163.3 (2.78) | 42.3 (0.72) |
| LINJ_10_0530 | GP63-leishmanolysin | 74.7 | 45.0 (0.60)* | 123.3 (1.65) | 49.0 (0.66) |
| LINJ_33_0350 | Heat shock protein 83–1 | 36.0 | 37.7 (1.05) | 49.7 (1.38) | 21.3 (0.59) |
| LINJ_28_3000 | Heat shock protein (Fragment) | 73.7 | 61.3 (0.83) | 85.0 (1.15) | 41.0 (0.56) |
| LINJ_26_1220 | Heatshock70-related protein 1 mitochondrial precursor putative | 14.7 | 18.7 (1.27) | 18.7 (1.27) | 12.0 (0.82) |
| LINJ_18_1350 | Heatshock protein 110 putative | 18.7 | 15.3 (0.82) | 31.3 (1.68) | 5.0 (0.27) |
| LINJ_15_0010 | Histone H4 | 13.0 | 15.3 (1.18) | 24.0 (1.85) | 5.3 (0.41) |
| LINJ_32_3100 | Nucleoside diphosphate kinase | 14.7 | 8.3 (0.57)* | 19.0 (1.30) | 7.3 (0.50)** |
| LINJ_09_0950 | Polyubiquitin | 13.3 | 8.7 (0.65) | 7.7 (0.58) | 6.3 (0.48) |
| LINJ_04_0750 | Putative 60S ribosomal protein L10 | 86.7 | 70.7 (0.82) | 46.3 (0.53) | 14.3 (0.17) |
| LINJ_16_0470 | Putative 60S ribosomal protein L21 | 42.3 | 35.7 (0.84) | 44.0 (1.04) | 6.7 (0.16) |
| LINJ_34_3440 | Putative 60S ribosomal protein L21 | 42.0 | 36.7 (0.87) | 44.3 (1.06) | 6.7 (0.16) |
| LINJ_27_2350 | Putative heat shock protein DNAJ | 9.3 | 4.7 (0.50) | 10.3 (1.11) | 4.0 (0.43) |
| LINJ_28_3060 | Putative heat shock protein hsp70 | 90.3 | 72.7 (0.80) | 92.3 (1.02) | 48.7 (0.54) |
| LINJ_27_0620 | Putative small GTP-binding protein Rab1 | 9.3 | 3.7 (0.39) | 9.3 (1.00) | 4.3 (0.46) |
| LINJ_31_1240 | Pyrophosphate-energized vacuolar membrane proton pump1 putative | 36.3 | 9.3 (0.26) | 47.0 (1.24) | 10.7 (0.29) |
| LINJ_35_5450 | Pyruvate kinase | 10.7 | 6.0 (0.56) | 10.0 (0.94) | 11.0 (1.03) |
| LINJ_36_1420 | Transitional endoplasmic reticulum ATPase putative | 12.7 | 5.7 (0.45) | 11.3 (0.89) | 4.3 (0.34) |
| LINJ_13_1450 | Tubulin alpha chain | 52.7 | 44.7 (0.85) | 91.3 (1.73)* | 25.7 (0.49) |
| LINJ_08_1290 | Tubulin beta chain | 46.3 | 39.0 (0.84) | 130.3 (2.81)* | 28.3 (0.61) |
| LINJ_08_1290 | Tubulin beta chain | 45.0 | 37.0 (0.82) | 129.7 (2.88)* | 29.3 (0.65) |
| LINJ_26_1960 | Uncharacterized protein | 13.7 | 14.0 (1.02) | 18.3 (1.34) | 12.7 (0.93) |
Numbers correspond to average Total Spectrum Count (n = 3). Numbers between parentheses depict the average fold change of the mutant compared with the WT.
* p < 0.05
** p < 0.01 (unpaired t-test).
Table 5. Enriched and unique proteins identified in EVs isolated from the AmB1000.1 L. infantum strain.
| Gene ID | Protein | Total Spectrum Count1 | (Trans)membrane protein | Unconventional secretion prediction2 | Signal peptide3 | |
|---|---|---|---|---|---|---|
| 1 | LINJ_27_1630* | Hypothetical protein conserved | 37.7 | no | no (0.522) | no |
| 2 | LINJ_04_1250* | Actin | 15.3 | no | no (0.513) | no |
| 3 | LINJ_29_0110 | C2 domain in Dock180 and Zizimin proteins putative | 14.0 | no | no (0.272) | no |
| 4 | LINJ_31_0520* | Cytoskeleton-associated protein | 9.3 | no | no (0.465) | no |
| 5 | LINJ_26_2510 | Hypothetical protein conserved | 9.0 | no | no (0.107) | no |
| 6 | LINJ_05_0580* | Uncharacterized protein | 8.0 | yes | no (0.196) | no |
| 7 | LINJ_13_0090* | Putative carboxypeptidase | 7.3 | no | no (0.384) | no |
| 8 | LINJ_23_0410* | Putative NADP dependent alcohol dehydrogenase | 7.3 | no | no (0.303) | no |
| 9 | LINJ_35_1030* | Putative casein kinase | 7.0 | no | no (0.343) | no |
| 10 | LINJ_31_2890* | ADP-ribosylation factor putative | 5.7 | no | no (0.512) | no |
| 11 | LINJ_34_1630* | p25-alpha putative | 5.7 | no | yes (0.617) | no |
| 12 | LINJ_36_0080* | Stress-inducible protein STI1 homolog | 4.7 | no | yes (0.645) | no |
| 13 | LINJ_19_0250* | Kinesin-like protein | 4.3 | no | no (0.136) | no |
| 14 | LINJ_27_1770* | Putative trypanothione synthetase | 3.7 | no | no (0.417) | no |
| 15 | LINJ_23_1200* | Hydrophilic acylated surface protein a (HASPA1) | 2.7 | no | no (0.283) | no |
| 16 | LINJ_25_2100 | Hypothetical protein conserved | 2.7 | no | no (0.487) | no |
| 17 | LINJ_36_3360 | 14-3-3 protein-like protein | 2.3 | no | no (0.240) | no |
| 18 | LINJ_26_1480 | Hypothetical protein conserved | 1.7 | no | yes (0.867) | no |
| 19 | LINJ_23_0290 | Pentamidine resistance protein 1 | 1.7 | yes | no (0.497) | no |
| Total number of proteins | 2 (10.5%) | 3 (15.8%) | 0 (0.0%) | |||
1 Mean Total Spectrum Count obtained with the LC–MS/MS analyses.
2 NM score calculated using the SecretomeP 2.0 Server. Proteins with an NM score higher than 0.600 that are not predicted to have a signal peptide are considered non-classically secreted proteins.
3Signal peptides were predicted using SignalP Server integrated in SecretomeP 2.0 Sever.
*Unique protein specific to EVs released by this strain.
Bioinformatics analysis
Shared proteins among the three replicates of each strain (WT: 152; Sb2000.1: 194; MF200.5: 264; and AmB1000.1: 70 proteins) were annotated with GO terms from goa_uniprot_all.gaf (UniProt GOA knowledge base; downloaded 12-Nov-2019) using the integrated functions of Scaffold version 4.11.0. Protein information was retrieved from UniprotKB (L. infantum reference proteome; last modified January 15, 2020) [48] and TriTrypDB v46 (last modified November 6, 2019) [49]. Presence of membrane/transmembrane domains was evaluated for retained proteins using UniprotKB (https://www.uniprot.org/). To this end, Gene IDs (Tables 2–5) were used to retrieve the summary page of each individual protein (last access November 2019), which includes the subcellular localization of the query protein predicted via InterPro [50]. Venn diagrams were constructed using the online tool http://bioinformatics.psb.ugent.be/webtools/Venn/. Heat maps of total spectral counts for the common proteins between the WT and the resistant lines were generated using the Heatmapper server [51]. Prediction of non-classical protein secretion was investigated using the SecretomeP 2.0 server (http://www.cbs.dtu.dk/services/SecretomeP/; last access May 2020) [52]. Briefly, the sequence of each protein was recovered from TriTrypDB v46 (https://tritrypdb.org/tritrypdb/) using its unique Gene ID (Tables 2–5). Sequences were assembled in a bulk FASTA file and submitted to SecretomeP 2.0 server. For each input sequence, the server predicted the possibility of non-classical secretion (NN-score). Moreover, SecretomeP 2.0 server integrates SignalP5.0 [53], which allowed us to determine the potential presence of signal peptides on the different proteins. For eukaryotic organisms, proteins with an NN score > 0.600 that are not predicted to have a signal peptide are considered non-classically secreted proteins.
Table 2. Enriched and unique proteins identified in EVs isolated from the WT L. infantum strain.
| Gene ID | Protein | Total Spectrum Count1 | (Trans)membrane protein | Unconventional secretion prediction2 | Signal peptide3 | |
|---|---|---|---|---|---|---|
| 1 | LINJ_31_0450 | Putative cytoskeleton associated protein CAP5.5 | 21.3 | no | no (0.465) | no |
| 2 | LINJ_31_0820 | C2 domain protein—putative | 8.3 | no | no (0.154) | no |
| 3 | LINJ_31_1860 | Aminoacid permease | 8.0 | yes | yes (0.662) | no |
| 4 | LINJ_13_1500 | Programmed cell death 6 protein-like protein | 7.7 | no | yes (0.937) | no |
| 5 | LINJ_35_2080 | Putative calcium motive P-type ATPase | 7.7 | yes | yes (0.612) | no |
| 6 | LINJ_12_0668* | Surface antigen protein 2-putative | 7.3 | yes | no (0.295) | yes |
| 7 | LINJ_12_0490 | Glucose-6-phosphate isomerase | 7.0 | no | no (0.452) | no |
| 8 | LINJ_31_2670 | Putative calreticulin | 7.0 | no | no (0.498) | yes |
| 9 | LINJ_19_1280 | SPFH domain / Band 7 family—putative | 4.7 | no | no (0.526) | no |
| 10 | LINJ_06_1360 | CLN3 protein putative | 4.3 | yes | no (0.280) | no |
| 11 | LINJ_28_2050 | Zinc transporter 3-putative | 3.7 | yes | no (0.514) | yes |
| 12 | LINJ_16_1450 | ADP-ribosylationfactor-like-putative | 3.3 | no | no (0.342) | no |
| 13 | LINJ_06_1330* | Coproporphyrinogen III oxidase | 3.3 | no | no (0.556) | no |
| 14 | LINJ_31_0810* | Putative c2 domain protein | 3.3 | no | no (0.336) | no |
| 15 | LINJ_29_2610 | Vacuolar protein sorting ssociated protein 4 | 3.3 | no | no (0.387) | no |
| 16 | LINJ_36_2340 | Plasma-membrane choline transporter putative | 3.0 | yes | yes (0.863) | no |
| 17 | LINJ_10_0380 | Pteridine transporter putative | 3.0 | yes | no (0.361) | no |
| 18 | LINJ_09_0960 | Putative ef-hand protein 5 | 3.0 | no | no (0.311) | no |
| 19 | LINJ_36_1680 | Putative universal minicircle sequence binding protein | 3.0 | no | yes (0.822) | no |
| 20 | LINJ_14_0490 | Amastin surface glycoprotein putative | 2.3 | yes | yes (0.838) | no |
| 21 | LINJ_31_0050* | BT1 family MFS sugar transport protein putative | 2.3 | yes | no (0.466) | no |
| 22 | LINJ_03_0190 | Delta-1-pyrroline-5-carboxylate dehydrogenase putative | 2.3 | no | no (0.559) | no |
| 23 | LINJ_27_2520 | Putative cysteine peptidase, Clan CA, family C2 | 1.3 | no | sequence too long to be analyzed | |
| Total number of proteins | 9 (39.1%) | 5 (26.0%) | 3 (13.0%) | |||
1 Mean Total Spectrum Count obtained with the LC–MS/MS analyses.
2 NM score calculated using the SecretomeP 2.0 Server. Proteins with an NM score higher than 0.600 that are not predicted to have a signal peptide are considered non-classically secreted proteins.
3Signal peptides were predicted using SignalP Server integrated in SecretomeP 2.0 Server.
*Unique protein specific to EVs released by this strain.
Statistical analyses of data
Statistical analyses were performed using the unpaired Student’s t-test (*p ≤ 0.05, **p ≤ 0.01) and Shapiro-Wilk Test for normality, followed by Kruskal-Wallis rank test (****p ≤ 0.0001) and post-hoc Nemenyi (Tukey and Kramer) test (**p ≤ 0.01, ****p ≤ 0.0001). The data were analyzed using GraphPad Prism 8.0 software (GraphPad Software, La Jolla California, USA) and R 3.6.1 software [54].
Results
Drug resistance affects Leishmania EVs’ quantity, size and distribution
Composition and quantity of EVs released by eukaryotic cells is known to vary according to cell stress conditions (pH, temperature, etc.) and metabolic state [38–40]. To establish whether the capacity to produce and release EVs (including their size and distribution) is altered in drug-resistant Leishmania parasites, we selected four well-characterized strains: L. infantum WT and drug-resistant mutants Sb2000.1 (antimony), AmB1000.1 (amphotericin B), and MF200.5 (miltefosine). Before proceeding to purification of EVs, the four strains were analyzed in terms of drug sensitivity to confirm their previously reported phenotypes [41–46]. As expected, the WT was sensitive to all three drugs, while all mutants were highly resistant to their respective selection drug (S1 Table). EVs were extracted and purified from temperature-stressed cultures of stationary WT and drug-resistant parasites through multiple centrifugation, wash, and filtration steps (as detailed in the materials and methods section). It is important to note that while drug-resistant cultures were maintained under drug pressure, this pressure was removed during EVs production and purification to avoid direct effects of the drug on the membrane of the parasites (e.g. MF and AmB).
Next, purified vesicles were submitted to three nanoparticle-tracking analyses (NTA) per biological replicate to accurately determine the distribution and concentration of the particles (Fig 1A). Following MISEV2018 recommendations [55], NTA analyses and total protein quantification (microBCA) were used to calculate the particle/μg protein ratio for the different EVs’ purifications. All ratios ranged from 109 to 1010, with mean values of 2.51 x 1010 for the WT, 7.69 x 109 for Sb2000.1, 1.57 x 1010 for MF200.5 and 2.98 x1010 for AmB1000.1. No significant differences were observed among the strains.
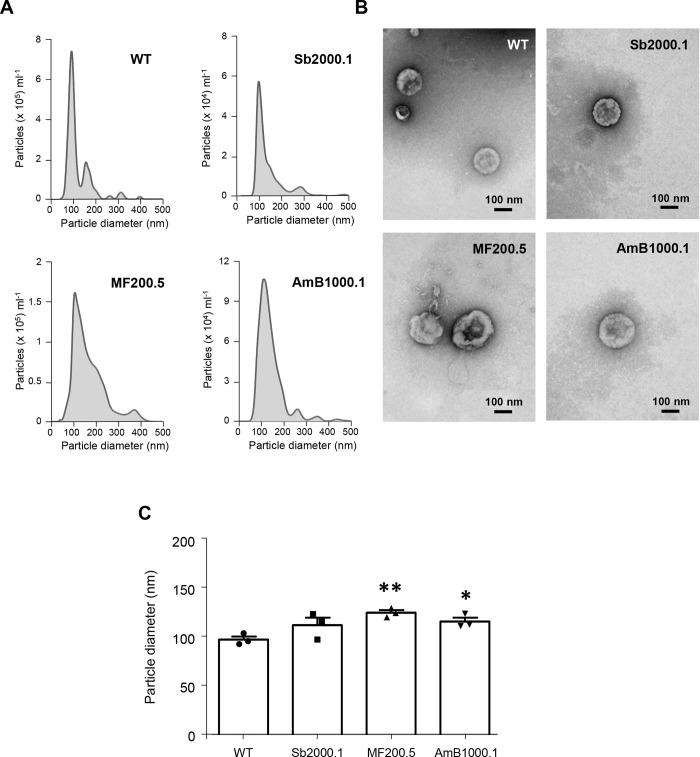
Fig 1. EVs released by L. infantum parasites differ in morphology and size depending on their drug-resistance background.
(A) Particle size distributions obtained by nanoparticle tracking analysis of L. infantum WT, and drug resistant Sb2000.1, MF200.5 and AmB1000.1 strains. Results were normalized per 106 parasites. (B) EVs derived from L. infantum WT, and drug resistant Sb2000.1, MF200.5 and AmB1000.1 promastigotes were prepared for TEM by negative staining. (C) Comparison of the modal average size of the EVs isolated from WT, Sb2000.1, MF200.5 and AmB1000.1 promastigotes. Differences were statistically evaluated by unpaired t-test (*p ≤ 0.05, (**p ≤ 0.01). In a–c, the results are representative of at least three independent experiments with similar data.
EVs were also evaluated by transmission electron microscopy (Fig 1B), confirming the predominant presence of lipid bilayer-enclosed nano-sized structures compatible with exosomes (~50 to 200 nm in diameter) and other small EVs. NTA and microscopy analyses revealed probable differences in size and morphology in EVs released by drug-resistant strains. To determine the average size of EVs released by the different strains, the modes of each distribution were determined (Sb2000.1: 112.4 nm; MF200.5: 125.0 nm and AmB1000.1: 116.2 nm) and compared with the WT (97.7 nm) (Fig 1C). While all three resistant strains showed a tendency to release larger EVs, only EVs released by MF200.5 and AmB1000.1 were significantly larger (unpaired t-test; p ≤ 0.01 and p ≤ 0.05, respectively).
The next step was to evaluate if the shift in terms of average size of EVs was affecting the global distribution, or rather specific subpopulations, of said particles. To this end, the NTA datasets corresponding to sizes ranging from 0 to 500 nm, previously identified in Fig 1A, were further analyzed. As depicted in Fig 2A (S1 Fig), the number of particles secreted per size seemed to follow a different trend depending on the strain. To verify this, the global distribution of particles released by the WT and the drug-resistant strains were compared (S2 Fig), revealing significant differences among the four strains (S2 Fig; Kruskal-Wallis rank test; p ≤ 0.01). Nemenyi post-hoc test (S2 Fig) pinpointed significant differences between WT and Sb2000.1 EV distributions (p ≤ 0.0001), as well as between Sb2000.1 and the other two resistant strains (p ≤ 0.01 for Sb2000.1 vs. MF200.5; p ≤ 0.0001 for Sb2000.1 vs. AmB1000.1).
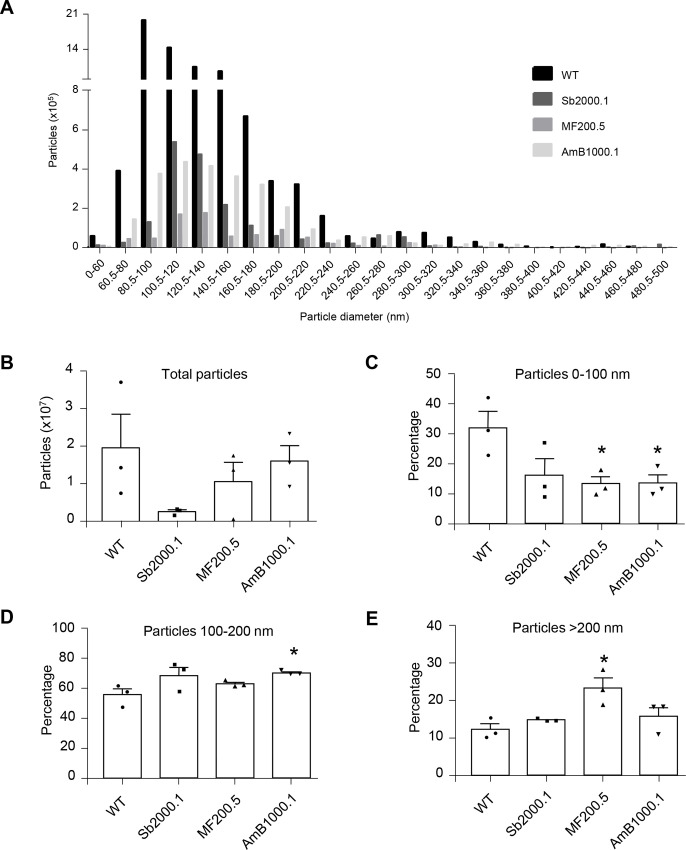
Fig 2. EVs released by drug-resistant L. infantum parasites show altered particle profiles and EV secretion capacity.
(A) L. infantum WT and drug-resistant strains were compared in terms of number of particles per category (size) between 0 nm and 500 nm using NTA. One representative image of three experiments is shown (experiments 2 and 3 are shown in S1). (B) Total number of particles, from 0 nm to 500 nm, secreted by WT, Sb2000.1, MF200.5 and AmB1000.1. (C) Percentage of particles corresponding to 0–100 nm. (D) Percentage of particles corresponding to 100–200 nm. (E) Percentage of particles corresponding to 200–500 nm. In A–E, the results are representative of at least three independent experiments with similar data. Differences were statistically evaluated by unpaired t-test (*p < 0.05).
Different quantitative analyses using NTA datasets were performed to verify the likelihood of a direct link between drug resistance and the capacity of Leishmania parasites to release different EV subpopulations. For all these comparisons, the number of particles was normalized to 106 parasites. Strain Sb2000.1 was the only cell line showing a strong tendency toward reduced EV secretion. However, this difference was not significant when compared with the WT (Fig 2B; p = 0.065). As depicted in Fig 2C, particles < 100 nm accounted for approximately 32.0% of the total particles produced by the WT strain. The percentage of particles corresponding to this range was significantly decreased (approximately 13.0% in both cases, p ≤ 0.05) in MF200.5- and AmB1000.1-resistant strains. While not statistically significant (p = 0.057), Sb2000.1 showed a tendency to produce less particles < 100 nm. Particles spanning from 100 to 200 nm represented approximately 55–70.0% of all the particles secreted by the four strains analyzed (Fig 2D). AmB1000.1 showed a significant increase in the percentage of particles within this range (p ≤ 0.05). Finally, particles > 200 nm accounted for 12–15.0% of total isolated EVs for the WT, Sb2000.1 and AmB1000.1 (Fig 2E). The percentage of particles corresponding to this range was significantly increased in MF200.5 (24.0%) when compared to the WT (p ≤ 0.05).
Protein diversity and abundance are altered in EVs released by L. infantum drug-resistant strains
To better understand the functions of EVs, including their potential role in cellular survival in the presence of stress (e.g. drug resistance), their molecular content (which specifically reflects the phenotype of parent cells) should be analyzed. Since proteins are a major component of EVs and the Leishmania proteome is specifically modified in drug-resistant strains, we decided to identify common and unique proteins present in EVs released by the WT and the three resistant strains using untargeted, label-free, shotgun LC–MS/MS proteomics. In this study, we included three independent biological replicates for each strain to minimize the any potential effect derived from EVs’ heterogeneity (S1 Data). We were therefore able to identify common and unique proteins within the same strain, as well as among the different cell lines. Proteomic data was first analyzed using the Scaffold software, which was able to identify a high number of proteins and protein clusters. Subsequent analyses were performed using independent proteins rather than clusters (S1 Data).
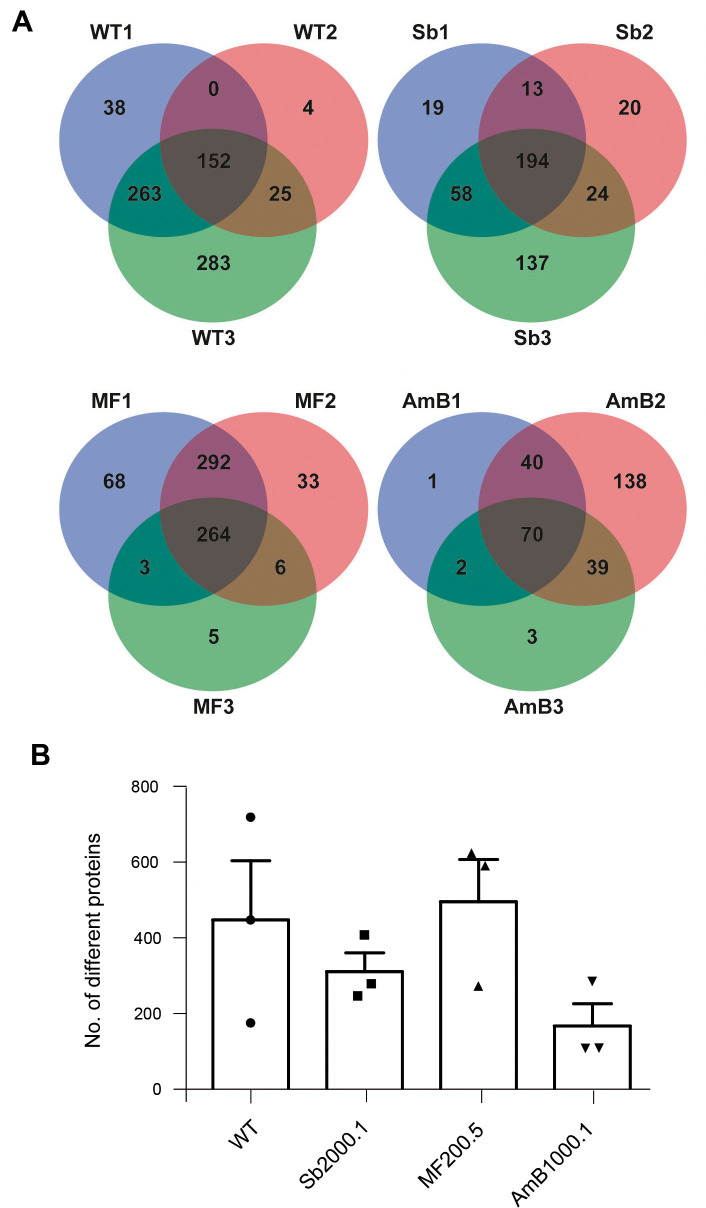
LC–MS/MS analysis identified 152, 194, 264 and 70 proteins shared through three biological replicates for the WT, Sb2000.1, MF200.5, and AmB1000.1 mutants, respectively (Fig 3A). Moreover, we evaluated the total protein diversity of EVs belonging to each strain (Fig 3B). The global proteomic profile of the EVs released by the resistant strains showed significant differences when compared with the WT (S3 Fig). The diversity was variable depending on the replicate, especially for the WT. This variation was less important in the case of the resistant strains, for which two out of three replicates were always very similar. AmB1000.1 EVs showed a tendency to harbour less different proteins than the other three strains (Fig 3B). However, no significant differences were found between the resistant strains and the WT when comparing their average amount of proteins (unpaired t-test, p > 0.05). Only proteins shared among the three replicates of each strain were retained for subsequent analyses.
Fig 3. Characterization of EVs’ protein diversity is L. infantum parasites.
(A) Venn diagrams summarizing the distribution of the identified proteins by their presence/absence in three biological replicates per strain. (B) Total number of different proteins identified in EVs isolated from each strain. Horizontal lines indicate the mean value for each strain (n = 3).
Next, using the automated features of Scaffold 4.11, we generated a mean-deviation scatterplot to evaluate the variation of protein abundance in EVs derived from the four strains. (S4 Fig). This analysis shows an estimate of the abundance (mean value of normalized spectral counts) of proteins previously retained for each strain (shared among the three replicates). Proteins recovered from WT (152) and Sb2000.1 (194) EVs were similarly distributed. However, most of the 70 conserved proteins identified for the AmB1000.1 strain (red squares in S4 Fig) were located on the right side of the scatterplot, corresponding to higher abundance levels. High-abundance proteins detected in AmB1000.1 could be masking the detection of low-abundance proteins, leading to a lower total protein diversity. In contrast, proteins identified in MF200.5 EVs were found more frequently on the left section of the scatterplot, which corresponds to lower abundance levels.
Identification and characterization of L. infantum EVs’ core proteome
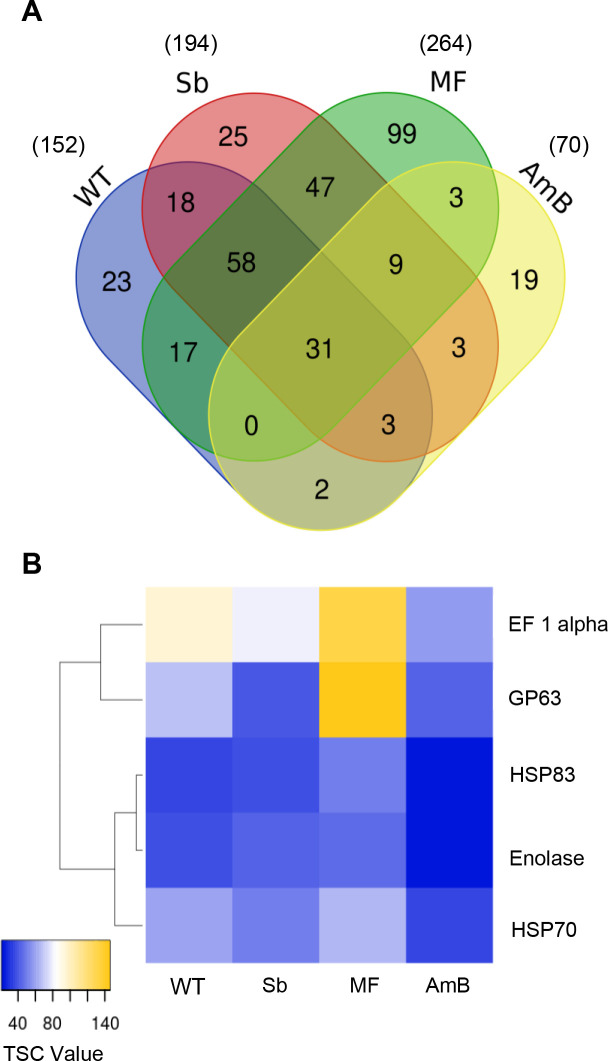
In order to identify and further characterize the core proteome of L. infantum EVs (proteins conserved among the WT and the three resistant strains), we relied only on those proteins common to all three biological replicates for each of the four cell lines (WT: 152; Sb2000.1: 194; MF200.5: 264; and AmB1000.1: 70, as depicted in Fig 3B). In this way, the Venn diagram in Fig 4A shows 31 shared proteins between the WT and the drug-resistant mutants (detailed in Table 1), as well as conserved proteins for each strain (WT: 23; Sb2000.1: 25; MF200.5: 99; and AmB1000.1: 19).
Fig 4. Identification of L. infantum EVs’ conserved proteins.
(A) Venn diagram showing all the proteins identified in EVs isolated from L. infantum WT and drug-resistant strains Sb2000.1, MF200.5 and AmB1000.1. Venn diagram shows common and exclusive proteins in the different groups. The numbers of proteins in the overlapping and non-overlapping areas are indicated (only proteins found in three replicates were included). (B) Heatmap of leishmanial EVs’ markers in particles isolated from WT, Sb2000.1, MF200.5 and AmB1000.1 L. infantum strains. Values correspond to the mean total number of spectra identified for each protein in three replicates.
Next, and in accordance with MISEV2018 recommendations [55], we proceed to identify and quantify the abundance of specific protein markers previously reported for Leishmania EVs [32, 56]. As depicted in the heatmap comparing the WT with the drug-resistant mutants (Fig 4B), vesicles isolated from the four strains contained specific Leishmania-EV markers, such as metalloprotease GP63, enolase, heat shock proteins HSP70/HSP83 and elongation factor 1 alpha. Importantly, this heatmap revealed that these markers could be enriched differently in the different strains. In order to validate this hypothesis, we evaluated the levels of enrichment of the 31-shared proteins in the drug-resistant strains, compared with the WT (Table 1). While the levels of most of the conserved proteins remained stable among the different mutants and the WT, our results revealed punctual significant, strain-dependent differences. A fold change > 1.60 was set as the up-regulation threshold, and < 0.60 for down-regulation. As depicted in Table 1, Sb2000.1 EVs showed significantly decreased levels of metalloprotease GP63 (approximately 0.60-fold change depending on the gene involved) and a nucleoside diphosphate kinase (0.57-fold change). These two proteins were also downregulated in EVs released by AmB1000.1 (0.39–0.59 and 0.50-fold, respectively). EVs released by the MF200.5 strain were significantly enriched in tubulin alpha (1.73-fold increase) and tubulin beta chains (2.80-fold increase). While not statistically significant, GP63 showed a tendency toward enrichment in EVs released by MF200.5. Of note, our analyses also revealed the existence of nine proteins common to the three resistant strains but absent in the WT (Fig 4A). These proteins included ribosomal proteins, both 40S and 60S, histone H3, along with transcription factor CBF/NF-Y (S2 Table).
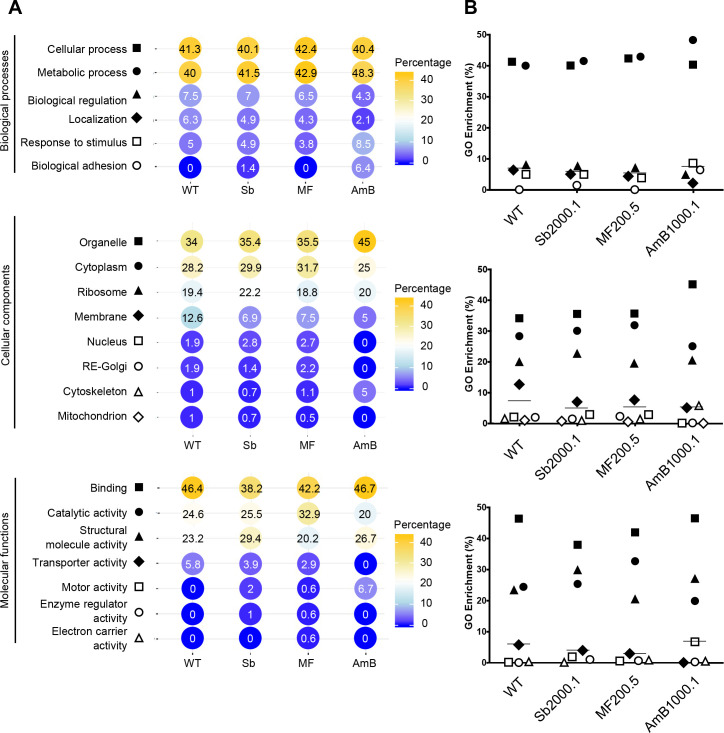
Finally, proteins (Table 1) were analyzed in terms of Gene Ontology (GO) annotation, including specific associations related to biological processes, cellular components, and molecular functions (Fig 5). An important limit of our GO analyses in Leishmania EVs is the high percentage of unknown proteins present in these particles (approximately 60.0%). Unknown proteins were not included in the GO analysis. For all four strains, EVs proteins were annotated for a wide variety of functions, including cellular and metabolic processes, cytoplasmic and ribosomal components, as well as binding functions (Fig 5A). Among the proteins harboring domains enabling functional assignment, about 40.0% of EVs proteins were functionally ascribed to the metabolism group, approximately 40.0% assigned to the cellular process group, and 4–7.0% to biological regulation processes. The other functional classes (less than 10.0% each) were represented by proteins identified as involved in protein localization, response to stimulus and biological adhesion. Our analysis did not reveal any significant difference between WT and drug-resistant EVs in terms of GO-annotations general distributions (Fig 5B). Nonetheless, we observed specific differences between WT and AmB1000.1 EVs. Proteins found in EVs released by AmB1000.1 were more frequently ascribed to “biological adhesion” (biological process), “cytoskeleton” (cellular component) and “motor activity” (molecular functions) subsets. No protein belonging to AmB1000.1 EVs was annotated within the “transporter activity” subcategory (Fig 5A). Of note, EVs released by the three resistant cell lines, but not by the WT, contained variable percentages of proteins annotated for “motor activity”. These represented approximately 7.0% of proteins identified in AmB1000.1 EVs. With regard to the cellular compartment of origin of the different proteins, most of them belonged to organelle, cytoplasmic, and ribosomal compartments. As previously reported for EVs, very few proteins were derived from the nucleus, Golgi apparatus, endoplasmic reticulum, or mitochondria [55].
Fig 5. GO analysis of proteins shared by L. infantum WT and drug-resistant strain EVs.
(A) Shared proteins among the three replicates of each strain (WT: 152; Sb2000.1: 194; MF200.5: 264; and AmB1000.1: 70 proteins) were annotated with GO terms from UniProt GOA knowledge base using the basic Scaffold version 4.11.0. GO terms were classified according to Biological Process, Cellular Component and Molecular Function, as defined by the GO consortium. (B) General distribution of the GO hits identified in (A). Horizontal lines indicate the median values.
Drug-resistance leads to differently enriched profiles and unique proteins in L. infantum EVs
Drug-resistant Leishmania parasites modify their general proteome according to the drug that has been involved in the in vitro selection or in vivo treatment process [34, 35]. Our search for enriched proteins present in EVs specific to each drug-resistance background led to the identification of 23, 25, 99 and 19 proteins for WT, Sb2000.1, MF200.5 and AmB1000.1 EVs, respectively, as shown in the Venn diagram in Fig 4A.
Tables 1–4 depict the complete list of enriched proteins identified in EVs released by the WT and the three drug-resistant strains. The total number of hypothetical conserved proteins and uncharacterized proteins varied for each strain: 0/23 for the WT, 7/25 for Sb2000.1, 11/99 for MF200.5 and 4/19 for AmB1000.1. MF200.5 was the strain showing the greatest diversity, as well as the highest enrichment in terms of total spectrum count (TSC) for the most enriched proteins, which were headed by an ATP-dependent 6-phosphofructokinase (LinJ.29.2620; TSC: 69.3) and a glycosome phosphoenolpyruvate carboxykinase (LinJ.27.1710; TEC: 66.3) (Table 3). The most enriched unique protein in WT EVs was a putative cytoskeleton associated protein CAP5.5 (LinJ.31.0450; TSC: 21.3) (Table 1). An enrichment of different ribosomal proteins (S14 and L37), a hypothetical protein (LinJ.07.0960; TEC: 17.3), a SPRY-domain ubiquitin ligase protein (LinJ.13.1510; TEC: 14.3), and an Ecotin-like protein (LinJ.15.0540; TSC: 10.0) were identified in EVs isolated from Sb2000.1 (Table 2). AmB1000.1 EVs were characterized by a high abundance of a hypothetical conserved protein (LinJ.27.1630; TSC: 37.7), followed by actin (LinJ.04.1250; TSC: 15.3), and a C2 domain protein LinJ.29.0110; TSC: 14.0).
Table 4. Enriched and unique proteins identified in EVs isolated from the MF200.5 L. infantum strain.
| Gene ID | Protein | Total Spectrum Count1 | (Trans)membrane protein | Unconventional secretion prediction2 | Signal peptide3 | |
|---|---|---|---|---|---|---|
| 1 | LINJ_29_2620 | ATP-dependent 6-phosphofructokinase | 69.3 | no | no (0.295) | no |
| 2 | LINJ_27_1710 | Glycosomal phosphoenol pyruvate carboxykinase, putative | 66.3 | no | no (0.279) | no |
| 3 | LINJ_21_0310 | Phosphotransferase | 52.0 | no | no (0.454) | no |
| 4 | LINJ_11_1000 | Pyruvate, phosphate dikinase | 34.3 | no | no (0.442) | no |
| 5 | LINJ_36_1320 | Fructose-bisphosphate aldolase | 32.0 | no | no (0.547) | no |
| 6 | LINJ_35_1190 | Putative NADH-dependent fumarate reductase | 30.3 | no | no (0.285) | no |
| 7 | LINJ_02_0430 | Voltage-dependent anion-selective channel—putative | 30.0 | no | no (0.550) | no |
| 8 | LINJ_30_2480 | Heatshock70-related protein1 -mitochondrial precursor putative | 28.0 | no | no (0.234) | no |
| 9 | LINJ_36_7320 | Putative eukaryotic translation initiation factor 3 subunit 8 | 26.0 | no | no (0.129) | no |
| 10 | LINJ_16_0560 | Orotidine 5 phosphate decarboxylase/orotate phosphoribosyl transferase putative | 19.0 | no | yes (0.850) | no |
| 11 | LINJ_35_3150 | ATP-dependent RNA helicase putative | 18.0 | no | no (0.171) | no |
| 12 | LINJ_17_0870 | GMP reductase | 17.7 | no | yes (0.655) | no |
| 13 | LINJ_29_2310 | Dynamin-1-like protein | 14.7 | no | no (0.454) | no |
| 14 | LINJ_36_0270 | Eukaryotic translation initiation factor 3 subunit L | 14.3 | no | no (0.259) | no |
| 15 | LINJ_17_0010 | Hypothetical protein conserved | 14.3 | no | no (0.309) | no |
| 16 | LINJ_36_7070 | ATP synthase delta (OSCP) subunit putative | 12.0 | no | yes (0.807) | no |
| 17 | LINJ_32_3470 | Chaperonin alpha subunit-putative | 11.3 | no | no (0.414) | no |
| 18 | LINJ_17_1390 | Translation initiation factor putative | 11.3 | no | no (0.266) | no |
| 19 | LINJ_35_3750 | Putative Gim5A protein | 10.3 | no | yes (0.705) | no |
| 20 | LINJ_18_0510 | Aconitate hydratase | 10.0 | no | no (0.493) | no |
| 21 | LINJ_30_3430 | Phosphoglycerate kinase | 10.0 | no | no (0.501) | no |
| 22 | LINJ_30_0120 | Alkyl dihydroxy acetone phosphate synthase O | 9.3 | no | no (0.400) | no |
| 23 | LINJ_32_3200 | Leucine-rich repeat protein-putative | 9.3 | no | no (0.568) | no |
| 24 | LINJ_22_1390 | Alanine—tRNA ligase | 9.0 | no | no (0.324) | no |
| 25 | LINJ_36_1000 | 40S ribosomal protein S18-putative | 8.3 | no | no (0.667) | no |
| 26 | LINJ_10_0070 | Dehydrogenase like protein | 8.3 | yes | 0.820, but prediction of a peptide signal | yes |
| 27 | LINJ_23_0050 | Peroxidoxin | 8.3 | no | yes (0.854) | no |
| 28 | LINJ_24_2150 | Transketolase | 8.3 | no | no (0.569) | no |
| 29 | LINJ_35_1490 | Arginase | 8.0 | no | no (0.376) | no |
| 30 | LINJ_29_0120 | Proteasome regulatory non-ATPase subunit putative | 7.7 | no | no (0.519) | no |
| 31 | LINJ_07_0710 | Hypothetical protein conserved | 7.3 | no | yes (0.609) | no |
| 32 | LINJ_05_0280 | Protein tyrosine phosphatase putative | 7.3 | yes | no (0.100) | no |
| 33 | LINJ_32_2280 | Hypothetical protein conserved | 7.0 | no | yes (0.628) | no |
| 34 | LINJ_11_0100 | Seryl-tRNA synthetase putative | 7.0 | no | no (0.330) | no |
| 35 | LINJ_24_0870 | Triosephosphate isomerase | 7.0 | no | no (0.413) | no |
| 36 | LINJ_23_0810 | Uncharacterized protein | 7.0 | yes | no (0.254) | yes |
| 37 | LINJ_30_3080 | Eukaryotic translation initiation factor 3 subunit 7-like protein | 6.7 | no | no (0.196) | no |
| 38 | LINJ_19_1590 | Inosine-5'-monophosphate dehydrogenase | 6.7 | no | no (0.331) | no |
| 39 | LINJ_25_2520 | Uncharacterized protein | 6.7 | no | no (0.281) | no |
| 40 | LINJ_35_1000 | Aldose 1 epimerase putative | 6.3 | no | no (0.598) | no |
| 41 | LINJ_32_3510 | Dihydrolipoyl dehydrogenase | 6.3 | no | no (0.463) | no |
| 42 | LINJ_36_3220 | Fibrillarin | 6.0 | no | no (0.334) | no |
| 43 | LINJ_28_1830 | Hypothetical protein conserved | 6.0 | no | no (0.129) | no |
| 44 | LINJ_29_2300 | Hypothetical protein conserved | 6.0 | yes | 0.675, but prediction of a peptide signal | yes |
| 45 | LINJ_19_1560 | Peptidylprolyl isomerase | 6.0 | no | yes (0.665) | no |
| 46 | LINJ.20.0120 | Phosphoglycerate kinase | 6.0 | no | no (0.226) | no |
| 47 | LINJ_11_1100 | Sterol14-alpha-demethylase putative | 6.0 | no | 0.829, but prediction of a peptide signal | yes |
| 48 | LINJ_36_3940 | 40S ribosomal protein S27-1 putative | 5.7 | no | yes (0.681) | no |
| 49 | LINJ_27_1770 | Trypanothione synthetase putative | 5.7 | no | no (0.415) | no |
| 50 | LINJ_19_1100 | Proteasome regulatory non-ATPase subunit9 putative | 5.3 | no | no (0.532) | no |
| 51 | LINJ_36_4030 | Putative glycyl tRNA synthetase | 5.3 | no | no (0.436) | no |
| 52 | LINJ_34_0010 | Uncharacterized protein | 5.3 | no |
yes (0.738) |
no |
| 53 | LINJ_32_3460 | Glucosamine-6-phosphate isomerase | 5.0 | no | yes (0.835) | no |
| 54 | LINJ_36_6080 | Nitroreductase family putative | 5.0 | no | no (0.511) | no |
| 55 | LINJ_36_7280 | Protein disulfide-isomerase | 5.0 | no | 0.649, but prediction of a peptide signal | yes |
| 56 | LINJ_35_0370 | ATP-dependent DEAD-box RNA helicase putative | 4.7 | no | no (0.407) | no |
| 57 | LINJ_32_1060 | Chaperon incontaining t-complex protein putative | 4.7 | no | no (0.407) | no |
| 58 | LINJ_15_0270 | Lysine-tRNA ligase | 4.7 | no | no (0.337) | no |
| 59 | LINJ_22_0310 | 40S ribosomal protein S15 putative | 4.3 | no | no (0.453) | no |
| 60 | LINJ_10_0560* | Glycerol-3-phosphate dehydrogenase [NAD(+)] | 4.3 | no | yes (0.633) | no |
| 61 | LINJ_27_1150 | Putative T-complex protein 1, beta subunit | 4.3 | no | no (0.393) | no |
| 62 | LINJ_30_3480* | Protein_mkt1_-_putative | 4.3 | no | no (0.505) | no |
| 63 | LINJ_36_4380 | Zeta-crystallin/NADPH-oxidoreductase-like protein | 4.3 | no | yes (0.670) | no |
| 64 | LINJ_27_1230 | Arginyl-tRNA synthetase-putative | 4.0 | no | no (0.393) | no |
| 65 | LINJ_27_2020 | RNA-binding protein putative | 4.0 | no | no (0.378) | no |
| 66 | LINJ_36_3750 | Cysteine synthase | 3.7 | no | no (0.411) | no |
| 67 | LINJ_36_4070 | Eukaryotic translation initiation factor 3 subunit I | 3.7 | no | no (0.405) | no |
| 68 | LINJ_02_0330 | Putative casein kinase II, alpha chain | 3.7 | no | no (0.598) | no |
| 69 | LINJ_28_2480 | Eukaryotic translation initiation factor 3 subunit E | 3.3 | no | no (0.557) | no |
| 70 | LINJ_25_1670 | Hypothetical protein conserved | 3.3 | no | no (0.451) | no |
| 71 | LINJ_36_2510 | Methyltransferase | 3.3 | no | no (0.339) | no |
| 72 | LINJ_29_2270 | Rab GDP dissociation inhibitor | 3.3 | no | no (0.541) | no |
| 73 | LINJ_36_5330 | Hypothetical protein conserved | 3.3 | no | no (0.344) | no |
| 74 | LINJ_06_0120 | Peptidyl-prolyl cis-trans isomerase | 3.0 | no | 0.842, but prediction of a peptide signal | yes |
| 75 | LINJ_03_0670 | DEAD/DEAH box helicase /Type III restriction enzyme res subunit—putative | 3.0 | no | no (0.324) | no |
| 76 | LINJ_19_0090 | Fibrillarin putative | 2.7 | no | yes (0.659) | no |
| 77 | LINJ_18_0270 | Glycogen synthase kinase 3 putative | 2.7 | no | no (0.499) | no |
| 78 | LINJ_11_0640 | Putative aminopeptidase | 2.7 | no | no (0.499) | no |
| 79 | LINJ_10_0210 | Putative nucleolar protein | 2.7 | no | no (0.396) | no |
| 80 | LINJ_29_2680 | Putative serine/threonine-protein kinase | 2.7 | no | yes (0.613) | no |
| 81 | LINJ_32_2330 | SAC3/GANP/Nin1/mts3/eIF-3 p25 family /COP9 signalosome—subunit CSN8—putative | 2.7 | no | yes (0.850) | no |
| 82 | LINJ_29_2350 | Aminopeptidase | 2.3 | no | yes (0.606) | no |
| 83 | LINJ_36_6170 | Halo acid dehalogenase-like hydrolase putative | 2.3 | no | 0.714, but prediction of a peptide signal | yes |
| 84 | LINJ_18_0670 | HEAT repeats/HEAT repeat putative | 2.3 | no | no (0.439) | no |
| 85 | LINJ_09_1130 | Translation initiation factor EIF-2B gamma subunit putative | 2.3 | no | yes (0.655) | no |
| 86 | LINJ_35_2400 | Aminopeptidase P putative | 2.0 | no | yes (0.680) | no |
| 87 | LINJ_08_0960 | Cysteine peptidase B (CPB) | 2.0 | no | 0.790, but prediction of a peptide signal | yes |
| 88 | LINJ_34_3160* | Glucose-6-phosphate 1-epimerase | 2.0 | no | yes (0.606) | no |
| 89 | LINJ_28_3050 | Hypothetical protein conserved | 2.0 | no | no (0.183) | no |
| 90 | LINJ_35_2090 | Kinesin putative | 2.0 | no | no (0.548) | no |
| 91 | LINJ_03_0240 | Ribosomal protein L38 putative | 2.0 | no | yes (0.723) | no |
| 92 | LINJ_15_1170* | Cyclic nucleotide-binding domain containing protein—putative | 2.0 | no | no (0.367) | no |
| 93 | LINJ_27_1300 | 60S acidic ribosomal protein P0 | 1.7 | no | no (0.208) | no |
| 94 | LINJ_21_0980 | Hypoxanthine phosphoribosyl transferase | 1.7 | no | no (0.587) | no |
| 95 | LINJ_36_1390 | N-acetyltransferase subunit Nat1 putative | 1.7 | no | no (0.346) | no |
| 96 | LINJ_31_3080 | Acetyl-CoA carboxylase putative | 1.3 | no | no (0.316) | no |
| 97 | LINJ_35_0840 | Aspartate aminotransferase putative | 1.3 | no | no (0.587) | no |
| 98 | LINJ_31_0890 | Ras-like small GTPases putative | 1.3 | no | no (0.208) | no |
| 99 | LINJ_27_1920 | GMP-PDE-delta subunit-putative/Pfam:PF05351 | 1.0 | no | yes (0.664) | no |
| Total number of proteins | 4 (4.0%) | 22 (21.8%) | 8 (7.9%) | |||
1 Mean Total Spectrum Count obtained with the LC–MS/MS analyses.
2 NM score calculated using the SecretomeP 2.0 Server. Proteins with an NM score higher than 0.600 that are not predicted to have a signal peptide are considered non-classically secreted proteins.
3Signal peptides were predicted using SignalP Server integrated in SecretomeP 2.0 Sever.
*Unique protein specific to EVs released by this strain.
Table 3. Enriched proteins identified in EVs isolated from the Sb2000.1 L. infantum strain.
| Gene ID | Protein | Total Spectrum Count1 | (Trans)membrane protein | Unconventional secretion prediction2 | Signal peptide3 | |
|---|---|---|---|---|---|---|
| 1 | LINJ_07_0960 | Hypothetical protein conserved | 17.3 | yes | 0.693, but prediction of a peptide signal | yes |
| 2 | LINJ_30_3650 | 40S ribosomal protein S14 | 15.3 | no | yes (0.693) | no |
| 3 | LINJ_28_1050 | 40S ribosomal protein S14 | 15.3 | no | yes (0.695) | no |
| 4 | LINJ_13_1510 | SPRYdomain/HECT-domain (Ubiquitin-transferase) putative | 14.3 | no | sequence too long to be analyzed | |
| 5 | LinJ_15_0540 | Ecotin-like protein 3 | 10.0 | no | no (0.343) | no |
| 6 | LINJ_33_2070 | Ribosomal protein L37 | 6.7 | no | yes (0.783) | no |
| 7 | LINJ_34_3420 | Uncharacterized protein | 5.7 | yes | 0.825, but prediction of a peptide signal | yes |
| 8 | LINJ_16_0510 | Hypothetical protein conserved | 5.3 | yes | no (0.126) | yes |
| 9 | LINJ_02_0340 | Putative proteasome regulatory non-ATPase subunit 6 | 4.7 | no | no (0.097) | no |
| 10 | LINJ_27_0500 | Putative calpain-like cysteine peptidase | 4.0 | no | sequence too long to be analyzed |
|
| 11 | LINJ_19_0560 | Hypothetical protein conserved | 3.7 | yes | 0.653, but prediction of a peptide signal | yes |
| 12 | LINJ_22_0440 | Putative proteasome regulatory ATPase subunit 1 | 3.7 | no | yes (0.659) | no |
| 13 | LINJ_28_0090 | Adenylate cyclase-like protein | 3.3 | no | no (0.358) | no |
| 14 | LINJ_20_1310 | Domain of unknown function (DUF1935)—putative | 3.3 | no | no (0.599) | no |
| 15 | LINJ_21_0830 | Hypothetical protein conserved | 3.0 | no | no (0.402) | no |
| 16 | LINJ_28_3090 | Malate dehydrogenase | 3.0 | no | no (0.447) | no |
| 17 | LINJ_16_1550 | Putative kinesin | 2.7 | no | no (0.394) | no |
| 18 | LINJ_32_4040 | Cysteine peptidase—putative | 2.3 | no | yes (0.620) | no |
| 19 | LINJ_30_1710 | SmallGTP-binding protein Rab28 putative | 2.3 | no | no (0.450) | no |
| 20 | LINJ_01_0010 | Protein of unknown function (DUF2946) | 2.0 | yes | no (0.085) | yes |
| 21 | LINJ_29_2550 | Phosphodiesterase putative | 1.7 | no | no (0.354) | no |
| 22 | LINJ_25_1640 | Putative casein kinase I | 1.3 | no | no (0.559) | no |
| 23 | LINJ_31_1130 | Putative N-acyl-L-amino acid amidohydrolase | 1.3 | no | no (0.530) | no |
| 24 | LINJ_32_2610 | Hypothetical protein conserved | 1.3 | no | no (0.408) | no |
| 25 | LINJ_20_0160 | Hypothetical protein conserved | 1.0 | no | no (0.080) | no |
| Total number of proteins | 5 (20.0%) | 5 (20.0%) | 5 (20.0%) | |||
1 Mean Total Spectrum Count obtained with the LC–MS/MS analyses.
2 NM score calculated using the SecretomeP 2.0 Server. Proteins with an NM score higher than 0.600 that are not predicted to have a signal peptide are considered non-classically secreted proteins.
3Signal peptides were predicted using SignalP Server integrated in SecretomeP 2.0 Sever.
Proteins summarized in Tables 2–5 constitute the conserved proteomic profile of EVs released by the different strains studied. These proteins were shared by the three replicates of one specific strain, but were not shared by the three replicates of any of the other strains. While these profiles point to drug-dependent enrichments in the different strains, this data does not pinpoint unique proteins that could be proposed as biomarkers. To determine if some of the enriched proteins could be considered strain-specific (absent in all the replicates of the rest of the strains), we performed a cross-correlation comparison (S5 Fig) between excluded and common pools of proteins previously retained to generate the EV profiles (reported in Fig 3A). This analysis revealed 4 unique enriched proteins in EVs released by WT (Table 2) and MF200.5 (Table 4) strains, and 13 unique proteins in the AmB1000.1 strain (Table 5).
Previous studies have reported that unconventional secretion is a key feature of EVs’ proteins in Leishmania [57]. To validate this among the unique, enriched proteins identified in our four strains, we used SecretomeP 2.0 proteomic tool [52] for identification of non-classical protein secretion. Of note, this server integrates SignalP5.0 for further confirmation of the presence of signal peptides [53]. Our analyses showed that approximately 26.0% of WT-unique proteins are unconventionally secreted (Table 2; NN-score > 0.600 and absence of signal peptide). This value was slightly lower for the three drug-resistant strains, with values of 20.0%, 21.8%, and 15.8% for Sb2000.1, MF200.5, and AmB1000.1 EVs, respectively (Tables 3–5). On the other hand, according to SignalP prediction, 13.0% of the unique proteins identified in WT EVs have signal peptides (Table 2). Regarding drug-resistant strains, 20.0% of Sb2000.1 and 7.9% of MF200.5 EVs unique proteins were predicted with signal peptides (Tables 3 and 4). No signal peptide was identified among of the proteins identified in AmB1000.1 EVs (Table 5). As EVs usually include various membrane proteins, we manually assessed the presence of membrane/transmembrane proteins using UniprotKB [48]. As summarized in Tables 2–5, the number of unique, enriched membrane/transmembrane proteins for each strain was very variable: 9 (39.1%) for the WT, 5 (20.0%) for Sb2000.1, 4 (4.0%) for MF200.5 and 2 (10.5%) for AmB1000.1.
Discussion
Different parasite molecular modifications, such as ABC transporters, membrane composition changes or oxidative stress, among others, can contribute both directly and indirectly to the phenomena of drug resistance [35, 46, 58]. Moreover, it has been shown that Leishmania is able to actively alter macrophage and neutrophil environments to resist current antileishmanial agents [59]. Recently, different drug-resistant field isolates have shown either efficient survival inside the insect vector or increased infectivity in the mammalian host [60–64]. This highlights the importance of not only parasite-to-parasite communication, but also a more complex host-parasite and vector-parasite environment. EVs have become known as an integral part of the parasite’s infectious life cycle as powerful cell-cell communication mediators. Leishmania infection, parasite survival (both inside the vector and the host), as well as the resulting clinical manifestations, are highly influenced by EVs [21–23, 29, 30, 65]. While traditional studies have focused on the survival of drug-resistant parasites in the host during treatment (amastigote form), the impact of drug resistance on the survival of the parasites inside the sand-fly vector (promastigote form) and the first moments of infection has recently begun to be explored. The latter could have a major impact on the spread and perpetuation of drug-resistant Leishmania populations [60–62, 66]. To better understand the potential roles of EVs in both the development of Leishmania parasites and the spread of drug-resistant strains, we have isolated and characterized the EVs released by the promastigote form of three drug-resistant L. infantum strains, as well as their WT counterpart. Our results showed, for the first time, that EV quantities, morphology, distribution, and protein cargo are altered in drug-resistant Leishmania parasites. Moreover, these changes appear to be specific to the antileishmanial drug involved in the resistant phenotype.
First, we investigated the impact of drug resistance on the production and distribution of EVs, which could lead to these extracellular elements serving different biological functions. While all four strains were able to produce particles compatible with EVs, parasites resistant to both miltefosine (MF200.5) and amphotericin B (AmB1000.1) produced larger EV subpopulations with altered morphology, especially in MF200.5, as shown by TEM and nanoparticle-tracking analysis. It is known that resistance to miltefosine and amphotericin B in Leishmania can lead to drastic changes in different lipids species, predominantly in sterols and phospholipids, some of which are enriched with cyclopropanated fatty acids, as well as to an increase in inositolphosphoceramide species [46, 67]. Consequently, MF- and AmB-resistant parasites tend to show a significant increase in membrane rigidity [67, 68]. Similarly, AmB1000.1 parasites are greatly depleted in ergosterol and 5-dehydroepisterol, but highly enriched in 4-methyl-8,24-cholestadienol. Likewise, MF200.5 cells contain strikingly low levels of ergosterol, with episterol as predominant sterol [46]. Due to the generation of exosomes from late endosomes [38], these vesicles are highly enriched in sterols (up to 43%) in eukaryotic cells [69]. As such, alterations in sterols, and thus membrane fluidity, could be responsible for the marked morphological changes observed in EVs from the MF200.5 strain. Specific EV-targeting lipid analysis should be conducted to further support this hypothesis. Nanoparticle-tracking analyses revealed a significant increase in the modal particle size of MF200.5 and AmB.1000.1 EVs. This is in correlation with previous TEM studies in which L. infantum parasites were stimulated with G418 (geneticin), leading to an increased release of larger vesicles in response to death-inducing stimuli [70]. Likewise, gentamicin-resistant pathogenic E. coli increased the modal size of their outer membrane vesicles (OMVs) when exposed to high concentrations of gentamicin [71]. Similarly, a recent study showed that β-lactam antibiotic-resistant E. coli produced significantly larger OMVs than its sensitive counterpart [72]. Particle-size distribution analyses revealed significant differences between the four strains, as well as important levels of heterogeneity, which is in accordance with previous reports involving different eukaryotic-cell lines [73]. MF200.5 and AmB1000.1 showed significantly increased amounts of particles within the ranges 100–200 and >200 nm, respectively. Among biomolecules, sterols, especially cholesterol, play a major role in the formation of EVs. As such, the above-mentioned modifications in sterol species in the MF200.5 and AmB1000.1 mutants [46] could potentially be inducing changes in the formation and release of small EVs (normally associated with exosomes). Moreover, while there is a growing body of knowledge suggesting a pathway analogous to that of the mammalian ESCRT-dependent pathway previously reported, the precise mechanism in Leishmania has yet to be fully elucidated (reviewed in [65]). It is therefore possible that this pathway, as well as different molecular sorting mechanisms, have been altered in drug-resistant mutants. For example, EVs released by Sb2000.1 and AmB1000.1 showed significantly reduced metalloprotease GP63 content, which has been shown to participate in the sorting of protein content of Leishmania exosomes [30].
During their biogenesis, EVs are hypothesized to selectively capture cell-specific proteins that may then become a part of the EVs “molecular signature”. However, the mechanism of such selective packaging remains unknown [74]. To investigate the proteomic “molecular signature” of Leishmania EVs, we first evaluated the protein diversity found in the EVs released by the WT and three drug-resistant strains. No significant differences between strains were observed in terms of number of different proteins. This could be explained by the fact that proteins in EVs account for more than half of the total secretion of parasite proteins [75], and EVs, due to their size, have a limited space to enclose large quantities. Moreover, Leishmania genomic organization and control leads to great individual parasite-to-parasite variations resulting in mosaic aneuploidy [76, 77], which ultimately modulates protein expression [35]. Due to the specialization of drug-resistant parasites required for their survival (compared with the WT), a lower parasite-to-parasite variability in these strains could be expected, which, coupled with the larger size of drug-resistant EVs, could lead to lower variability within the same strain when compared with the WT. MF200.5 EVs showed a tendency to contain a more diverse EV protein population, which could be facilitated by their larger size (when compared to the other strains). On the other hand, despite its EVs being significantly larger than those of the WT, the AmB1000.1 strain showed the lowest EV protein diversity. This lower diversity was accompanied by an increase in protein abundance. That said, high-abundance proteins could be masking the detection of low-abundance proteins in AmB1000.1, leading to a lower total protein diversity [78]. We also identified the presence of several conserved markers in the EVs isolated from the three replicates of the four strains. These corresponded to EV-specific markers previously reported for Leishmania and other eukaryotic cells [32, 55, 56]. The zinc-metalloprotease GP63, in addition to the protein sorting functions discussed above [30], is a virulence factor that greatly influences host cell signaling mechanisms and related functions [79]. This analysis allowed us to identify a number of correlations between EVs’ composition and the whole-cell proteome of the drug-resistant strains, demonstrating that EVs could reflect the drug-resistance background of their parental cell line. In this way, previous proteomic studies targeting drug-resistant promastigotes reported a downregulation of GP63 in both Sb2000.1- and AmB1000.1-resistant parasites (Sb/WT ratio: -1.91 and AmB/WT ratio: -4.14, respectively) [42, 44]. In our experiments, GP63 was significantly underrepresented in Sb2000.1 and AmB1000.1 EVs. Likewise, proteins enriched in drug-resistant EVs, such as 14-3-3 Protein-like protein (in AmB1000.1) or Tryparedoxin peroxidase (in Sb2000.1), were also enriched in whole-parasite preparations (AmB/WT ratio: +1.98 and Sb/WT ratio: +1.38, respectively) [42, 44]. Another example is a kinesin coded by the gene LinJ.16.1550, which was enriched 2.75-fold in Sb-resistant parasites compared with their WT counterpart in SILAC experiments [42]. Similarly, all three replicates of EVs released by Sb2000.1, but none of the other investigated strains, showed the presence of this specific protein. This is the first time it has been demonstrated that the proteomic signature profile of EVs–and thus protein sorting–of Leishmania parasites could vary according to their drug-resistance profile, as previously observed for cancer cells and bacteria [38–40].
The mechanisms of drug-resistance in Leishmania parasites are far from completely elucidated, and some may be non-specific adaptations that provide a general fitness gain allowing the parasite to survive in stressful conditions. In the search for non-specific mechanisms of drug resistance, we identified an interesting subset of nine EVs’ proteins enriched in the three replicates of the three drug-resistant strains, but absent in the WT. These proteins included histone H3, core histone-like transcription factor (CBF/NF-Y), and different ribosomal proteins. Histones play an important role in DNA packaging, transcription and gene regulation in the parasite, which also includes drug-resistant genes. This is in accordance with previous studies in L. donovani, in which Sb-resistant strains isolated from patients showed an overexpression of histones, highlighting the potential role of these proteins as traits of drug resistance [80]. While it is well documented that Leishmania EVs are effective modulators of early macrophage and host inflammatory responses [29–31], the exact mechanisms behind this modulation are yet to be characterized. Strikingly, a recent study discovered a new virulence mechanism in L. major, by which Leishmania H3, one of the histones only present in our drug-resistant EVs is secreted by the parasite and forms a nucleosome with the human histones in the host chromatin during infection. This leads to a relaxed conformation of chromatin that impacts gene expression pattern, facilitating survival of the parasite [81]. Transcription factor CBF/NF-Y has been shown to be regulated in different cell types when submitted to different types of stress, such as mechanical stress, ER stress, and DNA damage-related stress, among others [82]. Its presence in EVs derived from drug-resistant parasites would reflect general adaptation mechanisms against drug-generated stress. Moreover, drug-resistant EVs, but not naïve EVs, would have the potential to modulate transcription in recipient Leishmania parasites with the potential to mediate different downstream pathways. Similarly, it has recently been shown that cancer cells may exploit exosomes to confer transcriptome reprogramming that leads to cancer-associated pathologies [83]. Drug-resistant EVs were also enriched in ribosomal proteins. Similarly, several ribosomal proteins were upregulated in Sb-resistant L. donovani field isolates, decreasing the sensitivity of the parasites to Sb, MF, and paromomycin by increasing the proliferation of parasites [84].
As previously discussed, EVs capture cell-specific proteins during their formation and release. For this reason, we were interested in what EVs could potentially tell us about the biology, immunopathology, and drug-resistance mechanisms deployed by each strain. Recent evidence points to a possible correlation between Sb-resistance and virulence in Leishmania field isolates [66]. Interestingly, among the 25 enriched proteins present in Sb2000.1 EVs, one of the most upregulated proteins was a SPRY domain-containing ubiquitin ligase. The SPRY domain-containing SOCS box protein 2 was shown to recruit an E3 ubiquitin ligase complex to polyubiquitinate iNOS, targeting it for proteasomal degradation and therefore reducing macrophage killing of Leishmania parasites [85]. Moreover, Sb2000.1 EVs contained two proteasome regulatory subunits (an ATPase and a non-ATPase). This suggests that Sb2000.1-derived EVs could be involved in the modulation of redox control and protein degradation of recipient parasites, leading to increased tolerance to stress-inducing drugs as well as to the intracellular environment. Another important protein highly enriched in Sb2000.1 EVs was an ecotin-like protein, which has an important role in the inhibition of peptidases and promotion of Leishmania survival inside the sand fly vector. In addition, this protein modulates parasite differentiation and increases macrophage internalization due to the upregulation of phagocytosis by a mechanism dependent on serine peptidase activity [86, 87]. With respect to the MF200.5 strain, released EVs were enriched in glycolytic (e.g. ATP-dependent 6-phosphofructokinase and glycosomal phosphoenol pyruvate carboxykinase) and lipid-related (e.g. inosine 5’-monophosphate dehydrogenase and sterol 14α-demethylase) pathways, reflecting MF’s mode of action and mechanisms of drug resistance previously described in whole-parasite studies [46, 59]. MF200.5 EVs carried two ATP-dependent RNA helicases. One of them was a member of the DEAD-box family, which has been shown to play a central role in preventing ROS-mediated damage and in maintaining mitochondrial protein quality control in Leishmania [88]. This would facilitate the survival of parasites against mitochondrial MF-induced ROS. In addition, several partners of the DEAD-box helicase [88] were also found in MF200.5 EVs. For example, the HS70-related protein-1 mitochondrial precursor (HSPA9B) has been recently linked to miltefosine resistance and response to oxidative stress in L. donovani [89]. In addition to GP63, several other virulence factors, such as disulfide-isomerase and cysteine peptidase B (CPB) proteins, were found in MF200.5 EVs, further confirming that Leishmania EVs play a major role as virulence factors [21, 65]. This also raises the concern about a potentially increased virulence induced by EVs secreted by drug-resistant parasites. Last, analysis of AmB1000.1 EVs led to the identification of 19 enriched proteins. Among these, we identified a trypanothione synthetase (TryS) and an ATP-binding cassette protein (MRPA/P-gpA), the latter of which was also present in 2/3 replicates of Sb2000.1. Resistance to AmB in Leishmania has been associated with increased expression of enzymes involved in thiol metabolism. Equbal and collaborators showed that TryS mRNA is upregulated in AmB-resistant L. donovani field isolates when compared with sensitive strains [90]. Increased levels in the thiol pathway could also explain the presence of MRPA, which is implicated in the sequestration of thiol conjugates near the flagellar pocket (a specific site for exosome accumulation and secretion in Leishmania [56, 57]), and subsequent exocytosis outside the cell [91]. Moreover, in relation to MRPA, there is considerable evidence that P-gp and other MDR transporters can be transmitted from drug-resistant to drug-sensitive tumor cells by exosomes in vivo and in vitro [92]. Ongoing research will shed light on the potential occurrence of similar mechanisms in Leishmania parasites.
The final question we broached was the potential use of unique proteins present in EVs as biomarkers of drug resistance. Since Leishmania EVs are highly enriched within the sand fly and are co-egested with the parasite during the insect’s bite [56], drug-specific biomarkers would represent a major epidemiological tool for the evaluation of drug-resistant Leishmania populations, as well as increase our knowledge on how drug-resistant parasites adapt to mammalian and insect hosts in order to guarantee survival. Our results demonstrate that different proteomic-signature profiles can be obtained depending on the drug-resistance background of the parental cell. Moreover, we were able to identify two sets of four unique proteins present only in EVs released by the WT and MF200.5, as well as 13 in AmB1000.1 particles. While our study represents the first step towards the discovery of EV-based drug-resistance biomarkers, our work presents two limitations that should be addressed in future studies: future analyses should target larger sets of strains and replicates in order to reduce the observed variability and improve the robustness of potential biomarkers. Furthermore, evaluation and comparison of EV production and composition directly from resistant field strains is required.
In conclusion, we have conducted the first comparative characterization of Leishmania EVs in the context of drug resistance. We have analyzed the EVs of three different L. infantum strains resistant to antimony, miltefosine and amphotericin B, respectively, as well as their WT counterpart. Our results showed for the first time that drug-resistance mechanisms can induce changes in the morphology, size, and distribution of EVs in Leishmania. Next, we identified the core proteome of EVs conserved in both sensitive and drug-resistant backgrounds. Moreover, we obtained the first snapshot of the enriched and unique proteins in EVs released by each drug-resistant strain. Additionally, among these enriched EVs proteins, we identified several virulence factors, transcription factors, as well as proteins coded by drug-resistance genes. This selective sorting of cargo could facilitate the survival of drug-resistant parasites, and potentially sensitive parasites in contact with drug-resistant strains, to a plethora of stressful situations including drug pressure, initial moments of infection, adaptation to the sand-fly vector, etc. Future studies may explore all of these aspects and shed light on how EVs, especially those released by drug-resistant parasites, contribute to the survival of Leishmania during its life cycle.
Supporting information
L. infantum WT and drug-resistant strains were compared in terms of number of particles per category (size) between 0 nm and 500 nm using NTA. (A-B) Two representative images of three experiments is shown (experiment 1 is shown in Fig 2A).
(PDF)
Particle distribution analysis corresponding to the range of 0 to 500 nm. Horizontal lines indicate the median value for each particle size (n = 3). Whiskers correspond to minimum and maximum values. Differences were statistically evaluated using Shapiro-Wilk Test for normality, followed by Kruskal-Wallis rank sum test (****p ≤ 0.0001). Nemenyi post-hoc test pinpointed significant differences between WT and Sb2000.1 EVs distributions (p ≤ 0.0001), as well as between Sb2000.1 and the other two resistant strains (**p ≤ 0.01 for Sb2000.1 vs. MF200.5; ****p ≤ 0.0001 for Sb2000.1 vs. AmB1000.1).
(PDF)
The volcano plot shows the intensity of protein expression between EVs from Sb2000.1 (A), MF200.5 (B) and AmB1000.1 (C) cells and EVs from L. infantum WT cells. The horizontal axis represents the log2 fold change and the vertical axis represents −log10 (Fisher exact test, P value).
(PDF)
Standard deviation scatterplot depicting the mean and standard deviation values of the abundance (normalized total spectra) of each EVs protein transformed into base-10 logarithms for WT, Sb2000.1, MF200.5 and AmB1000.1 (n = 3). X-axis: Log10 of the mean value of the estimated protein abundance across those proteins retained for each strain (shared among the three replicates).Y-axis: Log10 of standard deviation of the estimated protein abundance computed across those proteins retained for each strain (shared among the three replicates).
(PDF)
Enriched proteins common to the three replicates of EVs isolated from WT (A), Sb2000.1 (B), MF200.5 (C) and AmB1000.1 (D) and their relationship with the pool of proteins excluded because they were not conserved through the replicates of the other strains.
(TIF)
EC50 values were calculated from the dose-response curves after performing a nonlinear fitting with the GraphPad 8.0 software program. An average of three independent biological replicates is shown ± standard error of the mean.
(PDF)
(PDF)
Total Spectra of Leishmania EVs using Leishmania infantum database.
(XLSX)
Acknowledgments
Authors want to thank Prof. Marc Ouellette for the kind gift of the Sb2000.1, MF200.5 and AmB1000.1 drug-resistant strains, and Dr Aida Mínguez-Menéndez for her help with the conception of the figures.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by a Natural Sciences and Engineering Research Council (NSERC; www.nserc-crsng.gc.ca) of Canada Discovery Grant RGPIN-2017-04480 and by the Canada foundation for Innovation (www.innovation.ca), grant number 37324; both awarded to CFP. Work in the MO lab is supported by the Canadian Institutes of Health Research (CIHR; www.cihr-irsc.gc.ca) Grant PJT-159765 and the NSERC of Canada Discovery Grant RGPIN-2018-03849. ND and GD are respectively supported by the Fonds de recherche du Québec – Nature et technologies (FRQNT) and the Canadian Institutes of Health Research (CIHR) studentship programs. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fernandez-Prada C, Minguez-Menendez A, Pena J, Tunes LG, Pires DEV, Monte-Neto R. Repurposed molecules: A New Hope in Tackling Neglected Infectious Diseases. In: Roy K, editor. In Silico Drug Design 1st Edition: Repurposing Techniques and Methodologies. Netherlands: Elsevier; 2019. [Google Scholar]
- 2.Sundar S, Singh B. Emerging therapeutic targets for treatment of leishmaniasis. Expert Opin Ther Targets. 2018:1–20. Epub 2018/05/03. 10.1080/14728222.2018.1472241 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCall LI, Zhang WW, Matlashewski G. Determinants for the development of visceral leishmaniasis disease. PLoS Pathog. 2013;9(1):e1003053 10.1371/journal.ppat.1003053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7(9):581–96. 10.1016/S1473-3099(07)70209-8 . [DOI] [PubMed] [Google Scholar]
- 5.Okwor I, Uzonna J. Social and Economic Burden of Human Leishmaniasis. Am J Trop Med Hyg. 2016;94(3):489–93. 10.4269/ajtmh.15-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abranches P, Silva-Pereira MC, Conceicao-Silva FM, Santos-Gomes GM, Janz JG. Canine leishmaniasis: pathological and ecological factors influencing transmission of infection. J Parasitol. 1991;77(4):557–61. Epub 1991/08/01. . [PubMed] [Google Scholar]
- 7.PAHO. Epidemiological Report of the Americas. Leishmaniases, Pan American Health Organization. 2019.
- 8.Arce A, Estirado A, Ordobas M, Sevilla S, Garcia N, Moratilla L, et al. Re-emergence of leishmaniasis in Spain: community outbreak in Madrid, Spain, 2009 to 2012. Euro Surveill. 2013;18(30):20546 Epub 2013/08/10. 10.2807/1560-7917.es2013.18.30.20546 . [DOI] [PubMed] [Google Scholar]
- 9.Uranw S, Hasker E, Roy L, Meheus F, Das ML, Bhattarai NR, et al. An outbreak investigation of visceral leishmaniasis among residents of Dharan town, eastern Nepal, evidence for urban transmission of Leishmania donovani. BMC Infect Dis. 2013;13:21 Epub 2013/01/19. 10.1186/1471-2334-13-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abubakar A, Ruiz-Postigo JA, Pita J, Lado M, Ben-Ismail R, Argaw D, et al. Visceral leishmaniasis outbreak in South Sudan 2009–2012: epidemiological assessment and impact of a multisectoral response. PLoS Negl Trop Dis. 2014;8(3):e2720 Epub 2014/03/29. 10.1371/journal.pntd.0002720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babuadze G, Alvar J, Argaw D, de Koning HP, Iosava M, Kekelidze M, et al. Epidemiology of visceral leishmaniasis in Georgia. PLoS Negl Trop Dis. 2014;8(3):e2725 Epub 2014/03/08. 10.1371/journal.pntd.0002725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solano-Gallego L, Miro G, Koutinas A, Cardoso L, Pennisi MG, Ferrer L, et al. LeishVet guidelines for the practical management of canine leishmaniosis. Parasit Vectors. 2011;4:86 Epub 2011/05/24. 10.1186/1756-3305-4-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066 Epub 2015/05/14. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devhare PB, Ray RB. Extracellular vesicles: Novel mediator for cell to cell communications in liver pathogenesis. Mol Aspects Med. 2018;60:115–22. Epub 2017/11/11. 10.1016/j.mam.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750 Epub 2018/11/23. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao CY, Papoutsakis ET. Extracellular vesicles: exosomes, microparticles, their parts, and their targets to enable their biomanufacturing and clinical applications. Curr Opin Biotechnol. 2019;60:89–98. Epub 2019/03/07. 10.1016/j.copbio.2019.01.005 . [DOI] [PubMed] [Google Scholar]
- 17.Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11(1):R3 Epub 2010/01/12. 10.1186/gb-2010-11-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066 Epub 2015/05/17. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. Epub 2019/01/04. 10.1038/s41556-018-0250-9 . [DOI] [PubMed] [Google Scholar]
- 20.Marcilla A, Martin-Jaular L, Trelis M, de Menezes-Neto A, Osuna A, Bernal D, et al. Extracellular vesicles in parasitic diseases. J Extracell Vesicles. 2014;3:25040 Epub 2014/12/30. 10.3402/jev.v3.25040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atayde VD, Hassani K, da Silva Lira Filho A, Borges AR, Adhikari A, Martel C, et al. Leishmania exosomes and other virulence factors: Impact on innate immune response and macrophage functions. Cell Immunol. 2016;309:7–18. Epub 2016/08/09. 10.1016/j.cellimm.2016.07.013 . [DOI] [PubMed] [Google Scholar]
- 22.Olivier M, Minguez-Menendez A, Fernandez-Prada C. Leishmania Viannia guyanensis. Trends Parasitol. 2019;35(12):1018–9. Epub 2019/07/12. 10.1016/j.pt.2019.06.008 . [DOI] [PubMed] [Google Scholar]
- 23.Olivier M, Fernandez-Prada C. Leishmania and its exosomal pathway: a novel direction for vaccine development. Future Microbiol. 2019;14:559–61. Epub 2019/06/01. 10.2217/fmb-2019-0087 . [DOI] [PubMed] [Google Scholar]
- 24.Szempruch AJ, Dennison L, Kieft R, Harrington JM, Hajduk SL. Sending a message: extracellular vesicles of pathogenic protozoan parasites. Nat Rev Microbiol. 2016;14(11):669–75. Epub 2016/09/13. 10.1038/nrmicro.2016.110 . [DOI] [PubMed] [Google Scholar]
- 25.Szempruch AJ, Sykes SE, Kieft R, Dennison L, Becker AC, Gartrell A, et al. Extracellular Vesicles from Trypanosoma brucei Mediate Virulence Factor Transfer and Cause Host Anemia. Cell. 2016;164(1–2):246–57. Epub 2016/01/16. 10.1016/j.cell.2015.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantel PY, Hjelmqvist D, Walch M, Kharoubi-Hess S, Nilsson S, Ravel D, et al. Infected erythrocyte-derived extracellular vesicles alter vascular function via regulatory Ago2-miRNA complexes in malaria. Nat Commun. 2016;7:12727 Epub 2016/10/11. 10.1038/ncomms12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantel PY, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe. 2013;13(5):521–34. Epub 2013/05/21. 10.1016/j.chom.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell. 2013;153(5):1120–33. Epub 2013/05/21. 10.1016/j.cell.2013.04.029 . [DOI] [PubMed] [Google Scholar]
- 29.Hassani K, Antoniak E, Jardim A, Olivier M. Temperature-induced protein secretion by Leishmania mexicana modulates macrophage signalling and function. PLoS One. 2011;6(5):e18724 Epub 2011/05/12. 10.1371/journal.pone.0018724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassani K, Shio MT, Martel C, Faubert D, Olivier M. Absence of metalloprotease GP63 alters the protein content of Leishmania exosomes. PLoS One. 2014;9(4):e95007 Epub 2014/04/17. 10.1371/journal.pone.0095007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silverman JM, Reiner NE. Leishmania exosomes deliver preemptive strikes to create an environment permissive for early infection. Front Cell Infect Microbiol. 2011;1:26 Epub 2011/01/01. 10.3389/fcimb.2011.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atayde VD, da Silva Lira Filho A, Chaparro V, Zimmermann A, Martel C, Jaramillo M, et al. Exploitation of the Leishmania exosomal pathway by Leishmania RNA virus 1. Nat Microbiol. 2019;4(4):714–23. Epub 2019/01/30. 10.1038/s41564-018-0352-y [DOI] [PubMed] [Google Scholar]
- 33.O'Neill CP, Gilligan KE, Dwyer RM. Role of Extracellular Vesicles (EVs) in Cell Stress Response and Resistance to Cancer Therapy. Cancers (Basel). 2019;11(2). Epub 2019/01/27. 10.3390/cancers11020136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leprohon P, Fernandez-Prada C, Gazanion E, Monte-Neto R, Ouellette M. Drug resistance analysis by next generation sequencing in Leishmania. Int J Parasitol Drugs Drug Resist. 2015;5(1):26–35. Epub 2015/05/06. 10.1016/j.ijpddr.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernández-Prada C, Vincent IM, Gazanion É, Monte-Neto RL. Chapter 6 Omics and Their Impact on the Development of Chemotherapy Against Leishmania. Drug Discovery for Leishmaniasis: The Royal Society of Chemistry; 2018. p. 101–29. [Google Scholar]
- 36.Douanne N, Wagner V, Roy G, Leprohon P, Ouellette M, Fernandez-Prada C. MRPA-independent mechanisms of antimony resistance in Leishmania infantum. International Journal for Parasitology: Drugs and Drug Resistance. 2020;13:28–37. 10.1016/j.ijpddr.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundar S, Singh B. Understanding Leishmania parasites through proteomics and implications for the clinic. Expert Rev Proteomics. 2018;15(5):371–90. Epub 2018/05/03. 10.1080/14789450.2018.1468754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19 Epub 2019/03/01. 10.1186/s13578-019-0282-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Ji Q, Yang Y, Li Q, Wang Z. Exosome: Function and Role in Cancer Metastasis and Drug Resistance. Technol Cancer Res Treat. 2018;17:1533033818763450 Epub 2018/04/24. 10.1177/1533033818763450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75(2):193–208. Epub 2017/07/25. 10.1007/s00018-017-2595-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leprohon P, Legare D, Raymond F, Madore E, Hardiman G, Corbeil J, et al. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37(5):1387–99. Epub 2009/01/09. 10.1093/nar/gkn1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brotherton MC, Bourassa S, Leprohon P, Legare D, Poirier GG, Droit A, et al. Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PLoS One. 2013;8(11):e81899 Epub 2013/12/07. 10.1371/journal.pone.0081899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Fadili K, Messier N, Leprohon P, Roy G, Guimond C, Trudel N, et al. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob Agents Chemother. 2005;49(5):1988–93. Epub 2005/04/28. 10.1128/AAC.49.5.1988-1993.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brotherton MC, Bourassa S, Legare D, Poirier GG, Droit A, Ouellette M. Quantitative proteomic analysis of amphotericin B resistance in Leishmania infantum. Int J Parasitol Drugs Drug Resist. 2014;4(2):126–32. Epub 2014/07/25. 10.1016/j.ijpddr.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira W, Leprohon P, Ouellette M. Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis. 2011;2:e201 Epub 2011/09/02. 10.1038/cddis.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Prada C, Vincent IM, Brotherton MC, Roberts M, Roy G, Rivas L, et al. Different Mutations in a P-type ATPase Transporter in Leishmania Parasites are Associated with Cross-resistance to Two Leading Drugs by Distinct Mechanisms. PLoS Negl Trop Dis. 2016;10(12):e0005171 Epub 2016/12/03. 10.1371/journal.pntd.0005171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. Epub 2003/11/25. 10.1021/ac0341261 . [DOI] [PubMed] [Google Scholar]
- 48.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D15. Epub 2018/11/06. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(Database issue):D457–62. Epub 2009/10/22. 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40. Epub 2014/01/24. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaak RG, Sanders MA, Bijl MA, Delwel R, Horsman S, Moorhouse MJ, et al. HeatMapper: powerful combined visualization of gene expression profile correlations, genotypes, phenotypes and sample characteristics. BMC Bioinformatics. 2006;7:337 Epub 2006/07/14. 10.1186/1471-2105-7-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17(4):349–56. Epub 2004/04/30. 10.1093/protein/gzh037 . [DOI] [PubMed] [Google Scholar]
- 53.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. Epub 2011/10/01. 10.1038/nmeth.1701 . [DOI] [PubMed] [Google Scholar]
- 54.R-Core-Team. A Language and Environment for Statistical Computing. 3.6.1 ed Vienna: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 55.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750 Epub 2019/01/15. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atayde VD, Aslan H, Townsend S, Hassani K, Kamhawi S, Olivier M. Exosome Secretion by the Parasitic Protozoan Leishmania within the Sand Fly Midgut. Cell Rep. 2015;13(5):957–67. Epub 2015/11/14. 10.1016/j.celrep.2015.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silverman JM, Chan SK, Robinson DP, Dwyer DM, Nandan D, Foster LJ, et al. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008;9(2):R35 Epub 2008/02/20. 10.1186/gb-2008-9-2-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez-Prada C, Sharma M, Plourde M, Bresson E, Roy G, Leprohon P, et al. High-throughput Cos-Seq screen with intracellular Leishmania infantum for the discovery of novel drug-resistance mechanisms. International Journal for Parasitology: Drugs and Drug Resistance. 2018;8(2):165–73. 10.1016/j.ijpddr.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hefnawy A, Berg M, Dujardin JC, De Muylder G. Exploiting Knowledge on Leishmania Drug Resistance to Support the Quest for New Drugs. Trends Parasitol. 2017;33(3):162–74. Epub 2016/12/21. 10.1016/j.pt.2016.11.003 . [DOI] [PubMed] [Google Scholar]
- 60.Van Bockstal L, Bulte D, Hendrickx S, Sadlova J, Volf P, Maes L, et al. Impact of clinically acquired miltefosine resistance by Leishmania infantum on mouse and sand fly infection. Int J Parasitol Drugs Drug Resist. 2020;13:16–21. Epub 2020/05/11. 10.1016/j.ijpddr.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hendrickx S, Van Bockstal L, Aslan H, Sadlova J, Maes L, Volf P, et al. Transmission potential of paromomycin-resistant Leishmania infantum and Leishmania donovani. J Antimicrob Chemother. 2020;75(4):951–7. Epub 2019/12/31. 10.1093/jac/dkz517 . [DOI] [PubMed] [Google Scholar]
- 62.Rai K, Cuypers B, Bhattarai NR, Uranw S, Berg M, Ostyn B, et al. Relapse after treatment with miltefosine for visceral leishmaniasis is associated with increased infectivity of the infecting Leishmania donovani strain. mBio. 2013;4(5):e00611–13. Epub 2013/10/10. 10.1128/mBio.00611-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deep DK, Singh R, Bhandari V, Verma A, Sharma V, Wajid S, et al. Increased miltefosine tolerance in clinical isolates of Leishmania donovani is associated with reduced drug accumulation, increased infectivity and resistance to oxidative stress. PLoS Negl Trop Dis. 2017;11(6):e0005641 Epub 2017/06/03. 10.1371/journal.pntd.0005641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seblova V, Oury B, Eddaikra N, Ait-Oudhia K, Pratlong F, Gazanion E, et al. Transmission potential of antimony-resistant leishmania field isolates. Antimicrob Agents Chemother. 2014;58(10):6273–6. Epub 2014/07/23. 10.1128/AAC.02406-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong G, Filho AL, Olivier M. Modulation of Host-Pathogen Communication by Extracellular Vesicles (EVs) of the Protozoan Parasite Leishmania. Front Cell Infect Microbiol. 2019;9:100 Epub 2019/04/30. 10.3389/fcimb.2019.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ouakad M, Vanaerschot M, Rijal S, Sundar S, Speybroeck N, Kestens L, et al. Increased metacyclogenesis of antimony-resistant Leishmania donovani clinical lines. Parasitology. 2011;138(11):1392–9. Epub 2011/08/09. 10.1017/S0031182011001120 . [DOI] [PubMed] [Google Scholar]
- 67.Barratt G, Saint-Pierre-Chazalet M, Loiseau PM. Cellular transport and lipid interactions of miltefosine. Curr Drug Metab. 2009;10(3):247–55. Epub 2009/05/16. 10.2174/138920009787846332 . [DOI] [PubMed] [Google Scholar]
- 68.Rakotomanga M, Saint-Pierre-Chazalet M, Loiseau PM. Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug-membrane interactions. Antimicrob Agents Chemother. 2005;49(7):2677–86. Epub 2005/06/28. 10.1128/AAC.49.7.2677-2686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60(1):9–18. Epub 2018/08/05. 10.1194/jlr.R084343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santarem N, Racine G, Silvestre R, Cordeiro-da-Silva A, Ouellette M. Exoproteome dynamics in Leishmania infantum. J Proteomics. 2013;84:106–18. Epub 2013/04/06. 10.1016/j.jprot.2013.03.012 . [DOI] [PubMed] [Google Scholar]
- 71.Chan KW, Shone C, Hesp JR. Antibiotics and iron-limiting conditions and their effect on the production and composition of outer membrane vesicles secreted from clinical isolates of extraintestinal pathogenic E. coli. Proteomics Clin Appl. 2017;11(1–2). Epub 2016/11/03. 10.1002/prca.201600091 . [DOI] [PubMed] [Google Scholar]
- 72.Kim SW, Park SB, Im SP, Lee JS, Jung JW, Gong TW, et al. Outer membrane vesicles from beta-lactam-resistant Escherichia coli enable the survival of beta-lactam-susceptible E. coli in the presence of beta-lactam antibiotics. Sci Rep. 2018;8(1):5402 Epub 2018/03/31. 10.1038/s41598-018-23656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–65. Epub 2013/10/10. 10.1242/jcs.128868 . [DOI] [PubMed] [Google Scholar]
- 74.Margolis L, Sadovsky Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019;17(7):e3000363 Epub 2019/07/19. 10.1371/journal.pbio.3000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silverman JM, Clos J, de'Oliveira CC, Shirvani O, Fang Y, Wang C, et al. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci. 2010;123(Pt 6):842–52. Epub 2010/02/18. 10.1242/jcs.056465 . [DOI] [PubMed] [Google Scholar]
- 76.Sterkers Y, Lachaud L, Crobu L, Bastien P, Pages M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol. 2011;13(2):274–83. Epub 2010/10/23. 10.1111/j.1462-5822.2010.01534.x . [DOI] [PubMed] [Google Scholar]
- 77.Ubeda JM, Legare D, Raymond F, Ouameur AA, Boisvert S, Rigault P, et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9(7):R115 Epub 2008/07/22. 10.1186/gb-2008-9-7-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev Proteomics. 2010;7(1):39–53. Epub 2010/02/04. 10.1586/epr.09.69 . [DOI] [PubMed] [Google Scholar]
- 79.Isnard A, Shio MT, Olivier M. Impact of Leishmania metalloprotease GP63 on macrophage signaling. Front Cell Infect Microbiol. 2012;2:72 Epub 2012/08/25. 10.3389/fcimb.2012.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar D, Singh R, Bhandari V, Kulshrestha A, Negi NS, Salotra P. Biomarkers of antimony resistance: need for expression analysis of multiple genes to distinguish resistance phenotype in clinical isolates of Leishmania donovani. Parasitol Res. 2012;111(1):223–30. Epub 2012/02/04. 10.1007/s00436-012-2823-z . [DOI] [PubMed] [Google Scholar]
- 81.Dacher M, Tachiwana H, Horikoshi N, Kujirai T, Taguchi H, Kimura H, et al. Incorporation and influence of Leishmania histone H3 in chromatin. Nucleic Acids Res. 2019;47(22):11637–48. Epub 2019/11/14. 10.1093/nar/gkz1040 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maity SN. NF-Y (CBF) regulation in specific cell types and mouse models. Biochim Biophys Acta Gene Regul Mech. 2017;1860(5):598–603. Epub 2016/11/07. 10.1016/j.bbagrm.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qadir F, Aziz MA, Sari CP, Ma H, Dai H, Wang X, et al. Transcriptome reprogramming by cancer exosomes: identification of novel molecular targets in matrix and immune modulation. Mol Cancer. 2018;17(1):97 Epub 2018/07/17. 10.1186/s12943-018-0846-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biyani N, Singh AK, Mandal S, Chawla B, Madhubala R. Differential expression of proteins in antimony-susceptible and -resistant isolates of Leishmania donovani. Mol Biochem Parasitol. 2011;179(2):91–9. Epub 2011/07/09. 10.1016/j.molbiopara.2011.06.004 . [DOI] [PubMed] [Google Scholar]
- 85.Kuang Z, Lewis RS, Curtis JM, Zhan Y, Saunders BM, Babon JJ, et al. The SPRY domain-containing SOCS box protein SPSB2 targets iNOS for proteasomal degradation. J Cell Biol. 2010;190(1):129–41. Epub 2010/07/07. 10.1083/jcb.200912087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verma S, Das S, Mandal A, Ansari MY, Kumari S, Mansuri R, et al. Role of inhibitors of serine peptidases in protecting Leishmania donovani against the hydrolytic peptidases of sand fly midgut. Parasit Vectors. 2017;10(1):303 Epub 2017/06/25. 10.1186/s13071-017-2239-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Eschenlauer SC, Faria MS, Morrison LS, Bland N, Ribeiro-Gomes FL, DosReis GA, et al. Influence of parasite encoded inhibitors of serine peptidases in early infection of macrophages with Leishmania major. Cell Microbiol. 2009;11(1):106–20. Epub 2008/11/20. 10.1111/j.1462-5822.2008.01243.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Padmanabhan PK, Zghidi-Abouzid O, Samant M, Dumas C, Aguiar BG, Estaquier J, et al. DDX3 DEAD-box RNA helicase plays a central role in mitochondrial protein quality control in Leishmania. Cell Death Dis. 2016;7(10):e2406 Epub 2016/10/14. 10.1038/cddis.2016.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vacchina P, Norris-Mullins B, Carlson ES, Morales MA. A mitochondrial HSP70 (HSPA9B) is linked to miltefosine resistance and stress response in Leishmania donovani. Parasit Vectors. 2016;9(1):621 Epub 2016/12/03. 10.1186/s13071-016-1904-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Equbal A, Suman SS, Anwar S, Singh KP, Zaidi A, Sardar AH, et al. Stage-dependent expression and up-regulation of trypanothione synthetase in amphotericin B resistant Leishmania donovani. PLoS One. 2014;9(6):e97600 Epub 2014/06/06. 10.1371/journal.pone.0097600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Legare D, Cayer S, Singh AK, Richard D, Papadopoulou B, Ouellette M. ABC proteins of Leishmania. J Bioenerg Biomembr. 2001;33(6):469–74. Epub 2002/01/24. 10.1023/a:1012870904097 . [DOI] [PubMed] [Google Scholar]
- 92.Sousa D, Lima RT, Vasconcelos MH. Intercellular Transfer of Cancer Drug Resistance Traits by Extracellular Vesicles. Trends Mol Med. 2015;21(10):595–608. Epub 2015/10/04. 10.1016/j.molmed.2015.08.002 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L. infantum WT and drug-resistant strains were compared in terms of number of particles per category (size) between 0 nm and 500 nm using NTA. (A-B) Two representative images of three experiments is shown (experiment 1 is shown in Fig 2A).
(PDF)
Particle distribution analysis corresponding to the range of 0 to 500 nm. Horizontal lines indicate the median value for each particle size (n = 3). Whiskers correspond to minimum and maximum values. Differences were statistically evaluated using Shapiro-Wilk Test for normality, followed by Kruskal-Wallis rank sum test (****p ≤ 0.0001). Nemenyi post-hoc test pinpointed significant differences between WT and Sb2000.1 EVs distributions (p ≤ 0.0001), as well as between Sb2000.1 and the other two resistant strains (**p ≤ 0.01 for Sb2000.1 vs. MF200.5; ****p ≤ 0.0001 for Sb2000.1 vs. AmB1000.1).
(PDF)
The volcano plot shows the intensity of protein expression between EVs from Sb2000.1 (A), MF200.5 (B) and AmB1000.1 (C) cells and EVs from L. infantum WT cells. The horizontal axis represents the log2 fold change and the vertical axis represents −log10 (Fisher exact test, P value).
(PDF)
Standard deviation scatterplot depicting the mean and standard deviation values of the abundance (normalized total spectra) of each EVs protein transformed into base-10 logarithms for WT, Sb2000.1, MF200.5 and AmB1000.1 (n = 3). X-axis: Log10 of the mean value of the estimated protein abundance across those proteins retained for each strain (shared among the three replicates).Y-axis: Log10 of standard deviation of the estimated protein abundance computed across those proteins retained for each strain (shared among the three replicates).
(PDF)
Enriched proteins common to the three replicates of EVs isolated from WT (A), Sb2000.1 (B), MF200.5 (C) and AmB1000.1 (D) and their relationship with the pool of proteins excluded because they were not conserved through the replicates of the other strains.
(TIF)
EC50 values were calculated from the dose-response curves after performing a nonlinear fitting with the GraphPad 8.0 software program. An average of three independent biological replicates is shown ± standard error of the mean.
(PDF)
(PDF)
Total Spectra of Leishmania EVs using Leishmania infantum database.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.