Abstract
Context.—
The rapid evolution of optical imaging modalities in recent years has opened the opportunity for ex vivo tissue imaging, which has significant implications for surgical pathology practice. These modalities have promising potential to be used as next-generation digital microscopy tools for examination of fresh tissue, with or without labeling with contrast agents.
Objective.—
To review the literature regarding various types of ex vivo optical imaging platforms that can generate digital images for tissue recognition with potential for utilization in anatomic pathology clinical practices.
Data Sources.—
Literature relevant to ex vivo tissue imaging obtained from the PubMed database.
Conclusions.—
Ex vivo imaging of tissues can be performed by using various types of optical imaging techniques. These next-generation digital microscopy tools have a promising potential for utilization in surgical pathology practice.
Anatomic pathology is essentially a field of image-based diagnostics in which light microscopic examination of stained tissues has been the mainstay for evaluating cytomorphologic and architectural features of individual cells or tissue fragments. In recent years, optical imaging modalities have rapidly evolved and can now be used for examination of tissues similar to traditional light microscopic examination. These modalities use light in the visible and adjacent spectra and harness the special properties of interaction of photons with labeled or unlabeled tissues to obtain detailed images of tissues. The rapid advancements in the field of biophotonics together with those in computer science and instrumentation technology have enabled the development of several optical imaging platforms. These modalities of tissue imaging enable the acquisition of inherently digital images, thereby allowing the visualization of tissues similar to how they are recognized under the light microscope. However, in contrast to traditional bright-field pathologic examination using a light microscope, which entails utilization of stained tissue sections cut from fresh or fixed tissue or stained smears of cytology preparations, optical imaging techniques allow in vivo or ex vivo evaluation of tissues requiring very minimal or no tissue preparation. The different types of optical imaging modalities are essentially optical sectioning microscopy techniques capable of imaging a thin layer of the intact specimen, greatly simplifying specimen preparation by eliminating the need for physical sectioning of the specimen. An added advantage of any of these optical sectioning microscopy methods is that the images acquired through these techniques are entirely digital. As a result, the digital images can be interpreted at the site of procurement in real time or remotely at any time, and they can be readily stored, retrieved, and integrated into electronic health records. Furthermore, digital images acquired through any of the currently developed platforms are amenable to machine learning, which is increasingly relevant in today’s era of the emergence of artificial intelligence in many aspects of life.
The emerging field of biomedical optical imaging has tremendous implications for the practice of anatomic pathology. Potential applications can bring revolutionary changes to the field by allowing rapid examination of tissues without the need for extensive tissue preparation and can generate digital images comparable to hematoxylin-eosin (H&E)–stained frozen sections (FSs) or formalin-fixed, paraffin-embedded (FFPE) H&E-stained tissue sections. Real-time tissue evaluation is a significant advantage that can be a major practice changer, allowing bedside tissue evaluation of the biopsied tissue at the time of procurement or permitting intraoperative tissue evaluation to guide surgeons at the time of surgery. The traditional practice for rapid evaluation of fragments of tissue for intraoperative decision-making is FS. Frozen section analysis entails freezing, cutting, and staining for conventional histopathologic examination of H&E-stained tissue sections and is the most commonly used technique for intraoperative examination of tissues. Even in practices where the pathology FS suite is adjacent to the operating rooms, turnaround times for FS analysis require 10 to 20 minutes or more. It is difficult to obtain FS diagnosis in a timely fashion for intraoperative decision-making in practices when the pathology laboratories performing FS analysis are located far away from the operating rooms. Advanced optical imaging modalities can enable acquisition of digital images of fresh unprocessed tissues with or without the use of labeling agents in the operating suites that can be interpreted by the pathologists either at the site of acquisition or remotely to support surgeons intraoperatively. Essentially these modalities can be excellent alternatives to FS analysis. Tissue imaging modalities can also facilitate procurement and quantitative assessment of high-quality tissue for biobanking. In addition, rapid bedside evaluation of core needle biopsies and other types of endoscopic and image-guided biopsies can improve the acquisition and triaging of high-quality biospecimens and hence avoid nondiagnostic results and significantly reduce the repeat biopsy rates.
The optical principles that underlie the different types of ex vivo tissue imaging modalities vary considerably. Techniques based on fluorescence confocal microscopy (FCM), optical coherence tomography (OCT), full-field optical coherence tomography (FF-OCT), structured illumination microscopy (SIM), light sheet microscopy (LSM), stimulated Raman spectroscopy (SRS), nonlinear microscopy (NLM), and microscopy using ultraviolet surface excitation (MUSE) have all been used to build different types of optical imaging platforms that can be used for ex vivo imaging of tissues. Most of the ex vivo tissue imaging platforms have been developed by individual research groups and used in the reported studies. Very few of these platforms are available commercially for imaging tissues in surgical pathology practice, and all of them are currently investigational. We describe below the different types of ex vivo optical imaging platforms that can generate digital images for ex vivo evaluation of tissues with a potential for utilization in anatomic pathology clinical practice.
CONFOCAL MICROSCOPY
Confocal microscopy allows noninvasive high-resolution imaging of fresh tissue with or without using fluorescent dyes as an external contrast agent. A confocal microscope uses a point source of light, typically generated by a laser, to illuminate a small focal spot in the tissue.1 By spatially filtering the backscattered or fluorescent emitted light from the tissue through a pinhole conjugate to the focal plane, an image of the focal plane of interest is created. Light reflected from the tissue superficially or deeply relative to the focal plane is rejected, resulting in thin optical sections of the tissue at a resolution similar to that of conventional light microscopy. Confocal microscopy can be performed in reflectance mode or in fluorescence mode using laser light sources in a variety of wavelengths combined with appropriate optical filter combinations.2 Reflectance confocal microscopy (RCM) exploits backscattered light from cellular structures to create tissue images without the need to label the tissue with exogenous contrast agents. The intensity of signals is dependent on the variable refractive index of the cellular components including nuclei, different organelles, cellular membranes, and the presence of other materials such as melanin or keratin. Conversely, FCM permits the characterization of cellular details relying on the excitation of exogenous fluorophores, which label the nuclei to generate the image contrast, thereby allowing the recognition of tissue architecture. A variety of fluorescent dyes can be used for FCM, such as acridine orange, proflavine, cresyl violet, indocyanine green, methylene blue, toluidine blue, fluorescein, and acriflavine hydrochloride. These contrast agents essentially result in at least a 1000-fold improvement in nucleus-to-cytoplasm contrast that aids in the recognition of the tissue. Acridine orange, which labels nuclear DNA and cytoplasmic RNA, has been used most frequently in FCM studies for ex vivo tissue imaging.
There are 2 commercially available confocal scanning microscopes (VivaScope 2500, Caliber Imaging and Diagnostics Inc, Rochester, New York; and Histology Scanner, SamanTree Medical SA, Switzerland) that are designed specifically for ex vivo imaging of fresh biologic tissue specimens. The system that has been used most frequently for ex vivo tissue imaging (Caliber Imaging and Diagnostics Inc) includes diode lasers that operate at wavelengths of 785 nm (used for RCM) and 488 nm (used for FCM), a 550-nm bandpass filter, maximum illumination power of 5 mM, and a 30× water immersion objective lens with a numerical aperture of 0.9. At these settings the lateral resolution is less than 1 μm and optical sectioning is less than 3.0 μm. Images can be acquired at 9 frames per second. Mosaics are captured in an automatic fashion by scanning the surface of the specimen from one end to the other, and the images are stitched to create a field of view as large as 2.0 cm × 2.0 cm in maximum dimension. The specimens can be scanned in reflectance and fluorescent modes simultaneously and can be viewed as grayscale images or as digitally colorized purple and pink images to mimic images of H&E-stain appearance. The FCM images are digitally colorized purple to mimic staining with hematoxylin and RCM images pink to mimic staining with eosin, and the 2 are overlaid such that the FCM and RCM images appear in purple and pink color contrast. This platform can image small fragments of fresh tissue measuring up to 2.0 cm × 2.0 cm. The other more recently developed and commercially available confocal laser scanning microscope (Histology Scanner, SamanTree Medical SA) is a digital microscopy scanner for ultrafast confocal imaging of fresh tissues measuring up to 8 cm in maximum dimension. This confocal microscope is an inverted microscope that provides a field of view of 16 mm × 16 mm. The platform provides a quick preview of the specimen at low resolution (10 μm/pixel) within 10 seconds following which an image of higher resolution (2 μm/pixel) can be acquired in 2.5 minutes.
Confocal microscopy has been the most commonly used ex vivo optical imaging modality for evaluation of tissues encountered in surgical pathology practice. Many studies have evaluated its utility for examining skin specimens obtained from Mohs microscopic surgery for diagnosis as well as for margin assessment of basal cell carcinoma (BCC).3–13 Diagnostic criteria have been established for evaluating grayscale images for making a diagnosis based on the FCM images. A recent study of ex vivo FCM by Longo et al12 included 753 skin sections obtained from 127 patients who underwent Mohs surgery for BCC. In comparison to FS, the sensitivity and specificity of diagnosis rendered on FCM images was 80% and 96%, respectively.12 A few preliminary reports have described the feasibility of FCM for diagnosis and margin assessment of squamous cell carcinoma in skin specimens.14 The encouraging results of the studies using FCM for evaluation of skin specimens show the potential utility of this noninvasive imaging modality for bedside diagnosis and margin assessment of skin specimens, particularly those obtained from Mohs procedure.
Confocal microscopy has also been investigated for recognition of tissue specimens other than skin, including surgical excision and biopsy specimens from a variety of sites such as brain, breast, thyroid, parathyroid gland, oral mucosa, lung, esophagus, stomach, colon, liver, and kidney. The early studies used confocal microscopy in the reflectance mode (RCM) that takes advantage of inherent differences in refractive indices of different subcellular structures within the tissues. Some studies used acetic acid as an exogenous contrast agent to enhance the nuclear contrast in epithelial tissue. More recent studies have used FCM with fluorescent dyes such as acridine orange, proflavine, methylene blue, fluorescein, and acriflavine hydrochloride as external contrast agents that can be applied during very brief staining steps. The imaging platform used in these studies has varied and has included custom-built instruments or more commonly the commercially available VivaScope 2500 confocal microscopy platform. Although typical sample sizes were small in most studies, the potential of FCM to facilitate histologic scale assessment of imaged tissue was evident. A few notable studies have included larger sample sizes. Dobbs et al15 used FCM and digital colorizing without, however, using the reflectance contrast to identify neoplasia in breast tissue specimens obtained from surgical excisions, stained with proflavine before imaging. Seventy specimens from 31 patients were studied, including FCM images from 235 regions of interest; these were compared in detail with corresponding light microscopic images of H&E-stained tissue sections.15 Neoplasia was identified in histologic images with a sensitivity of 93% and specificity of 97%, compared with 93% for both measures for FCM images. The authors concluded that FCM can produce images of breast tissue with recognizable architectural features comparable to conventional histology. A recent study by Ragazzi et al16 tested FCM on 35 fresh tissue samples obtained from surgical specimens of different organs including breast, lymph node, thyroid, and colon; they found that FCM images could distinguish neoplastic tissues from normal tissue. Wirth et al17 used FCM and RCM for evaluating 119 brain specimens that included tumors such as gliomas, meningiomas, and metastatic tumors as well as normal brain tissue. These authors also showed good correlation of FCM images with conventional histology. The same group also studied 37 brain biopsy and surgical specimens and confirmed the potential application of FCM in intraoperative diagnosis during neurosurgical procedures.18 Abeytunge et al19 used a confocal strip-mosaicking microscope that could image 2 cm × 2 cm tissue with cellular level resolution in 10 minutes. They imaged 34 large breast specimens obtained from 18 patients. The FCM images were interpreted by a single pathologist and the diagnosis correlated with the corresponding H&E sections of the imaged tissue in 30 specimens. Krishnamurthy et al20 recently evaluated the feasibility of FCM for ex vivo examination of 55 tissues obtained from surgical resections of breast, lung, kidney, and liver. They found good quality FCM grayscale images acquired in 5 to 10 minutes that allowed recognition of the cytomorphologic features of the tissue sufficient for accurate categorization of the specimens. They concluded that FCM has excellent potential for incorporation into surgical pathology practice, based on the ease and speed of acquisition of FCM images together with their resolution and similarity to conventional H&E-stained sections. Recently, they also reported the results of a study of 118 small (1.0 × 0.3 cm) tissue fragments obtained from surgical resections of breast, lung, kidney, and liver specimens that were sized so as to resemble core needle biopsy specimens.21 The specimens were imaged by using an FCM imaging platform optimized for evaluating small fragments in this study. The FCM images were acquired as grayscale as well as digitally colorized images in 2 to 3 minutes and could be viewed either at the site or remotely. Comparison of the diagnoses of the tissue fragments made when using FCM images with those made on the corresponding H&E-stained tissue sections indicated a sensitivity of 96.0% and specificity 97.0% in this single pathologist study.
On the basis of findings in the existing literature and emerging studies, it appears that FCM can be used for real-time tissue evaluation of small fragments of tissues encountered in surgical pathology practice; potential applications would include rapid assessment of core biopsies, endoscopic biopsies, complementary to FS and margin assessment of Mohs surgery skin specimens. Figure 1, A through C, shows grayscale and digitally colorized combined FCM and RCM images and an image of corresponding H&E tissue section of a small fragment of breast tumor tissue. The tissue was stained with acridine orange and fluorescence and reflectance imaging was performed in the fresh state by using an imaging platform (VivaScope 2500).
Figure 1.
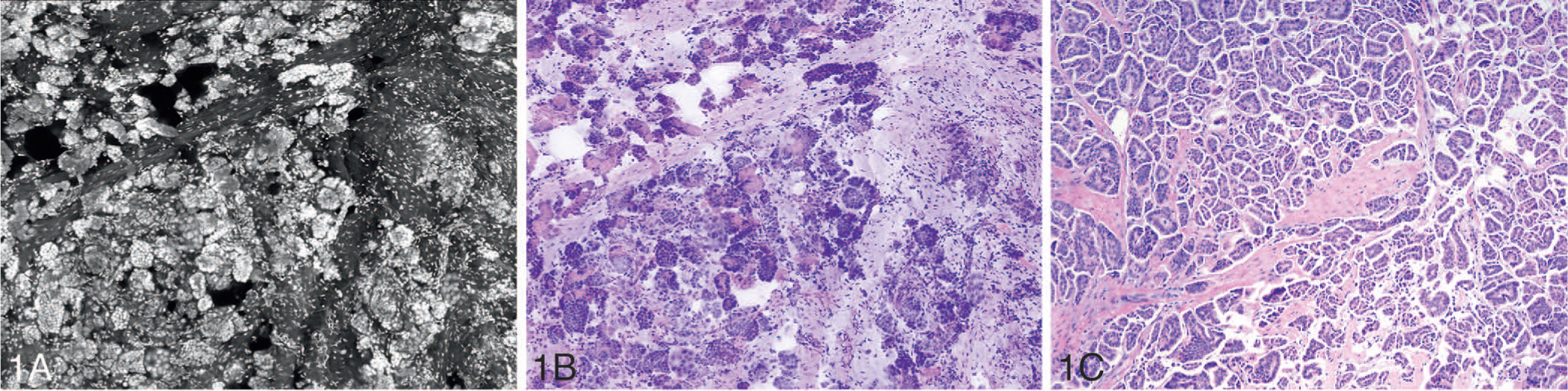
Grayscale (A) and digitally colorized (B) fluorescence confocal microscopy and reflectance confocal microscopy images and light microscopic image of the corresponding hematoxylin-eosin–stained tissue section (C) of invasive micropapillary carcinoma of the breast (original magnification ×100 [C]).
OPTICAL COHERENCE TOMOGRAPHY
Optical coherence tomography accomplishes high-resolution, cross-sectional tomographic imaging by detecting coherently gated backscattered light arising from illuminated tissue.22 The image resolution can span the range of 1 to 15 μm and imaging depth can be as much as 2 to 3 mm below the imaged surface; depth is limited by optical attenuation from tissue scattering and absorption of light. Essentially, OCT performs high-resolution interferometry imaging by measuring the echo time delay and intensity of light that is backscattered from the structures in the tissue. This imaging modality uses short coherence length light that can be obtained from short pulse lasers, compact super-luminescent diodes, or semiconductor-based light sources. Light from the source is directed onto a beam splitter, and one of the beams is incident onto the sample while the second beam travels a reference path with a variable path length and time delay. Low coherence interferometry measures the echo time delay and intensity of backscattered light by comparing it to the light that has traveled a known reference path length and time delay. The optical backscattering through a cross-section of the tissue is typically displayed as a logarithmic grayscale or digitally colorized image. The abnormalities of tissue architecture can be visualized but cellular details cannot be appreciated in conventional OCT images. Several studies have demonstrated the utility of OCT for recognition of alterations in tissue architecture.23,24 Although the potential of OCT for real-time in vivo as well as ex vivo evaluation of tissue is clearly established, most of the applications of this imaging modality in clinical practice are related to in vivo rather than ex vivo tissue imaging. This modality is routinely used currently in ophthalmology to image the retina and in cardiology for evaluation of atherosclerotic plaque.25 In addition, several OCT systems including handheld, needle, catheter-based, and balloon-based OCT probes have been commercialized by different companies for in vivo imaging for early detection of abnormalities and for obtaining targeted biopsies in organs such as skin and gastrointestinal tract. In contrast to the advancements and incorporation of OCT for in vivo imaging in clinical practice, it is only very recently that the ex vivo applications of OCT for margin assessment of surgical resections have been seriously explored.26–28 Currently, an ex vivo OCT imaging system is commercially available for margin assessment of breast mastectomy specimens (Perimeter Medical Imaging, Toronto, Ontario, Canada). In a recent study, this system was used to generate a 90-image OCT atlas that was used to train and test radiologists, pathologists, and surgeons.29 The sensitivity, specificity, and accuracy for detecting suspicious findings with histologic confirmation of cancer at the surgical margin for the 8 readers were 80%, 87%, and 87%, respectively. In addition, handheld OCT devices for breast margin assessment are also being developed.
The inability to appreciate cellular details in OCT images has been overcome to a large extent with the development of FF-OCT and dynamic (D)–FFOCT techniques that can generate high-resolution, 3-dimensional real-time tomographic images from unprocessed and unlabeled tissue.30–33 Similar to conventional OCT systems, the FF-OCT imaging technique is based on the principle of white-light interference microscopy. A simple tungsten halogen lamp is used as a spatially incoherent source to illuminate the entire field of a water-immersion microscope objective. Owing to the low temporal coherence of the source, interference occurs only when the optical path lengths of the 2 interferometer arms are identical to within 1 μm of each other. When a biological object is placed in the object arm, the light reflected by the reference mirror interferes with the light reflected or backscattered by the sample structures contained in a limited volume. The reflected light is recorded by a detector array charge-coupled device or a complementary metal oxide semiconductor (CMOS) detector to generate en face tomographic images. The FF-OCT images allow recognition of tissue architecture. However, cellular details cannot be recognized in the grayscale images. Apelian et al32,33 recently developed a new approach to reveal subcellular metabolic contrast in fresh tissues, taking advantage of the time dependence of FF-OCT interferometric signals, which they referred to as dynamic FF-OCT imaging. This method captures the signals linked to the local activity of endogenous scattering elements, essentially exploiting the micrometer scale transverse resolution of FF-OCT to image intracellular dynamic metabolic status. Dynamic FFOCT is complementary to FF-OCT and allows recognition of cellular details including their metabolic activity. The development of D-FFOCT has made it possible to acquire images that exploit the ability of FF-OCT for imaging fixed and highly backscattering structures complemented by the ability of D-FFOCT for imaging low-level scattering structures; this combination thereby facilitates recognition of cellular details. Figure 2, A and B, shows a D-FFOCT image of invasive ductal carcinoma of the breast obtained by imaging unlabeled breast tumor tissue with an FF-OCT/DFFOCT platform (LLTech, Paris, France) with the corresponding H&E-stained section of the imaged tissue.
Figure 2.
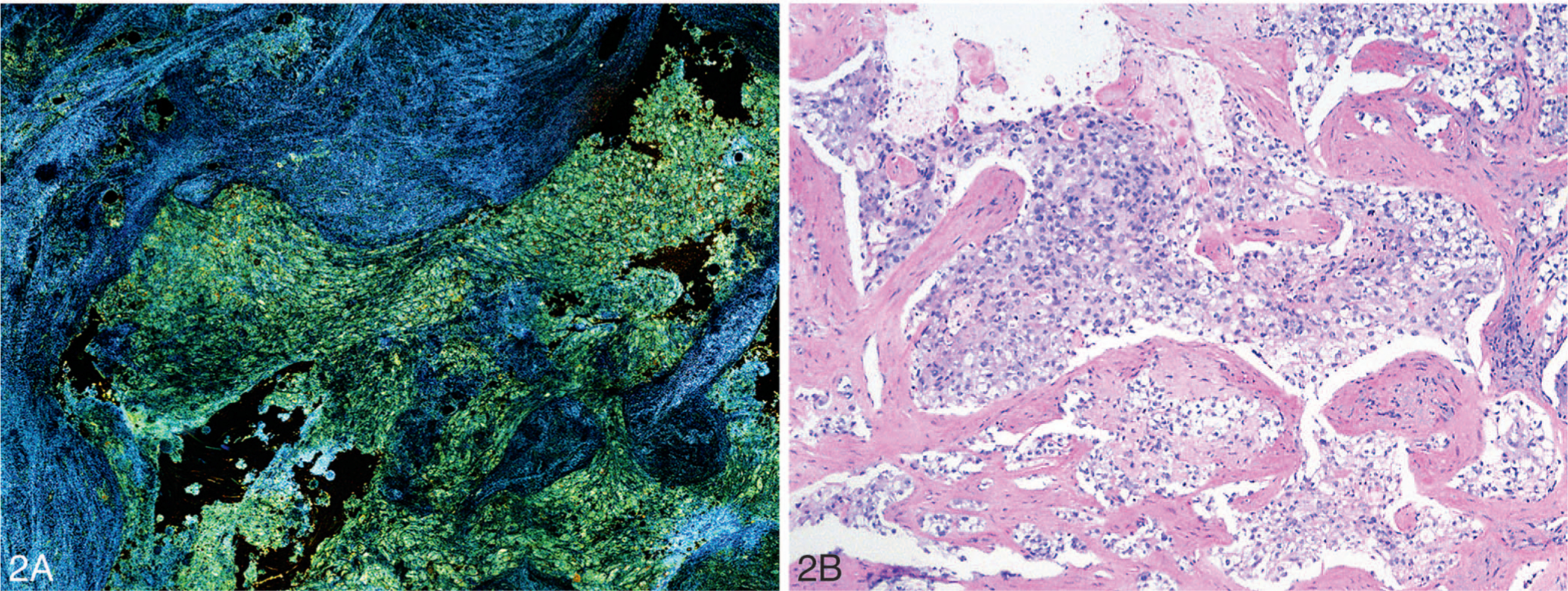
Dynamic full-field optical coherence tomography image (A) and light microscopic image of the corresponding hematoxylin-eosin–stained tissue section (B) of invasive ductal carcinoma of the breast (original magnification ×100 [B]).
A commercially available FF-OCT/D-FFOCT platform (LLTech) has been used in all the reported studies that have evaluated the utility of this platform for recognition of tissues suitable for surgical pathology practice. The instrument is compact, measuring 310 mm × 310 mm × 800 mm, and the optical power incident on the sample is less than 1 mW/mm2. There is no risk of thermal damage to tissue over the few minutes’ duration of imaging owing to the relatively low intensity of the light source used. The entire process from tissue handling to acquisition of image of a 2.0 × 2.0 × 1.0 cm3 tissue can be accomplished within 10 minutes. The field of view with this platform is about 1 mm2, resolution is 1.4 μm transversally and 1 μm axially, and pixel size on the sample is 0.8 μm. A mosaic of images that are stitched to display a larger field of view is created by using a stitching algorithm that uses a global optimization technique available with Fiji–Image J software. The imaging is rapid and noninvasive and requires neither contrast agent nor tissue preparation, which makes it suitable for medical imaging applications.
Several investigators have used the commercially available FF-OCT platform (LLTech) for imaging tissues obtained from different organs and have shown the use of the platform for rapid ex vivo imaging for recognition of the tissue. Jain et al34 imaged normal and neoplastic lung tissue obtained from 13 lobectomy specimens and compared the diagnoses rendered on the FF-OCT images with diagnoses from histology images. While all malignant tissues were correctly identified in this study, benign tissue was accurately recognized in only half the cases. The false-positive interpretation of FF-OCT images of benign lung tissue occurred in cases of lung tissue with collapse or when there were abundant alveolar macrophages. Jain et al35 also imaged renal tumor and normal tissue sections obtained from 25 nephrectomy specimens, using the same FF-OCT platform. An expert genitourinary pathologist not only could correctly distinguish the benign from malignant renal tissues but also could accurately type the renal tumor with a diagnostic accuracy of 80%. Although the cellular details were not appreciated very well, they could identify the unique cytoplasmic features in some of the kidney tumors that allowed accurate recognition of the type of kidney tumor. van Manen36 et al investigated the ability of an expert gastrointestinal pathologist and a pathology trainee to categorize 100 FF-OCT images that were generated from 29 patients who underwent pancreatic resection. They demonstrated a sensitivity of 72% and specificity of 73% for distinguishing benign from malignant pancreatic tissue. Difficulty was encountered when imaging atrophy, fibrosis, serous cystadenoma, and neuroendocrine tumors and resulted in the less than desired sensitivity and specificity in this study. Assayag et al37 conducted a prospective study using neurosurgical specimens from 18 patients to evaluate the utility of FF-OCT in brain tumor diagnosis. They studied temporal lobe chronic epileptic parenchyma, brain tumors including meningiomas and gliomas, and choroid plexus papilloma and demonstrated the feasibility of FF-OCT in real time for label-free noninvasive imaging to assess brain tissue in the intraoperative setting. The utility of FF-OCT for evaluating skin specimens was investigated by Durkin et al,38 who studied 18 tissues obtained from Mohs surgery from 11 patients; these tissues were interpreted by a dermatopathologist and the diagnoses were compared with diagnoses for H&E-stained FSs of the imaged tissue. All 9 benign and 6 malignant specimens were correctly interpreted on FF-OCT images; FF-OCT images of 3 specimens could not be categorized in this study. Recently, Lopater et al39 studied 38 consecutive patients with elevated prostate-specific antigen levels and/or suspicious results from digital rectal examination that raised concern for prostate carcinoma. From each patient, 1 to 10 cores were randomly selected and imaged with FF-OCT immediately after procuring the core biopsy. The FF-OCT images of a total of 119 cores were interpreted by 3 pathologists. The overall diagnostic accuracy of FF-OCT for prostate cancer detection was 70%, sensitivity was 63%, specificity was 74%, positive predictive value was 56%, and negative predictive value was 80% in this study. Assayag et al40 studied 75 breast specimens obtained from 22 patients by using high-resolution FF-OCT. Two breast pathologists interpreted the images, and they were able to distinguish normal from malignant tissue with individually determined sensitivities of 94% and 90% and specificities of 75% and 79%. Although FF-OCT has been evaluated for tissue recognition with reasonable success, there are no studies yet using D-FFOCT that allow better recognition of cellular details together with evaluation of cellular metabolic status of the cells.
MICROSCOPY USING SURFACE ULTRAVIOLET EXCITATION
MUSE is a recently reported optical imaging microscopy technique that can generate digital images resembling light microscopic images of H&E-stained tissue sections.41 Excitation of tissues with ultraviolet light, particularly at wavelengths shorter than 300 nm, can penetrate tissues to a depth of about 10 to 20 μm beneath the surface and can thus generate optically sectioned images that are suitable for tissue recognition. The emission from the imaged tissue can be bright if the tissue is stained with fluorescent dyes that can be excited at 280 nm and that emit photons in the visible range, which is the case for many fluorophores. The emitted photons can be captured by using simple-to-operate and inexpensive conventional glass-based microscope optics using either grayscale or color cameras. The original fluorescence MUSE images can be digitally colorized by using a number of color mapping approaches to generate colored images resembling H&E images. An important advantage of the MUSE system is that both fresh and fixed tissues of any size can be stained with fluorescent dyes to generate images at 3 to 10 frames within a few minutes.
Fereidouni et al41 recently described the optical design of the MUSE platform built by their research team and the results of imaging fresh and formalin-fixed tissues using this platform. They used widely available LED sources for 280-nm UV excitation. Specimens ranging in size from core biopsies to excisions could be imaged with speeds similar to conventional whole-slide scanners (1–3 minutes per specimen). The acquired MUSE images not only resembled conventional histopathology but also provided quasi–3-dimensional surface topography that cannot be appreciated with 2-dimensional conventional histopathology. In addition, the color gamut was broader in MUSE images than in H&E-stained sections. Stromal components could be particularly distinguished more readily than in H&E-stained sections. MUSE was capable of revealing subcellular details such as nuclear chromatin texture and mitotic figures. They also demonstrated that tissue integrity was well preserved following MUSE imaging for subsequent conventional staining and molecular analysis. These authors imaged 66 fresh and fixed normal and neoplastic tissues obtained from different organs including ovary, lung, kidney, breast, and brain. The tissues were stained for 10 seconds with rhodamine and Hoechst dyes and then subjected to MUSE imaging; multiple fields of view were captured at ×10 magnification. The MUSE images were digitally colorized by using a Python language–based utility, resulting in images that resembled H&E-stained sections. Forty-two imaged tissues were read by 2 independent pathologists, and a concordance rate of 93% was achieved in this study for the interpretation made on MUSE images in comparison with conventional H&E-stained tissue sections.
Recently, Yoshitake et al42 also reported the potential of MUSE as a low-cost optical sectioning technique for evaluating tissues in surgical pathology practice. These authors designed and characterized air and water immersion MUSE systems using high-incident angle illumination delivered with illumination light-guiding optics. Propidium iodide was used as a nuclear stain and eosin yellow as a counterstain in this study. Virtual H&E color rendering with a virtual transillumination microscopy algorithm was used to generate images similar to H&E from 2-channel fluorescent detection. Images were acquired with low-cost machine vision cameras. These authors performed a preliminary evaluation of surgical specimens, using the MUSE system developed by them for 2 potential applications, namely, evaluation of surgical margins of Mohs surgery for BCC in skin and evaluation of specimens of breast lumpectomy. MUSE images of BCC showed concordance with FS histology; however, concordance in breast tissue between MUSE and paraffin-embedded H&E histology was limited owing to the thicker optical sectioning in MUSE, indicating that further development was needed for breast tissue imaging. There are few other recent reports demonstrating the utility of MUSE for tissue recognition.43 Figure 3, A through C, shows MUSE fluorescence, digitally colorized, and H&E images of pancreatic ductal adenocarcinoma.
Figure 3.
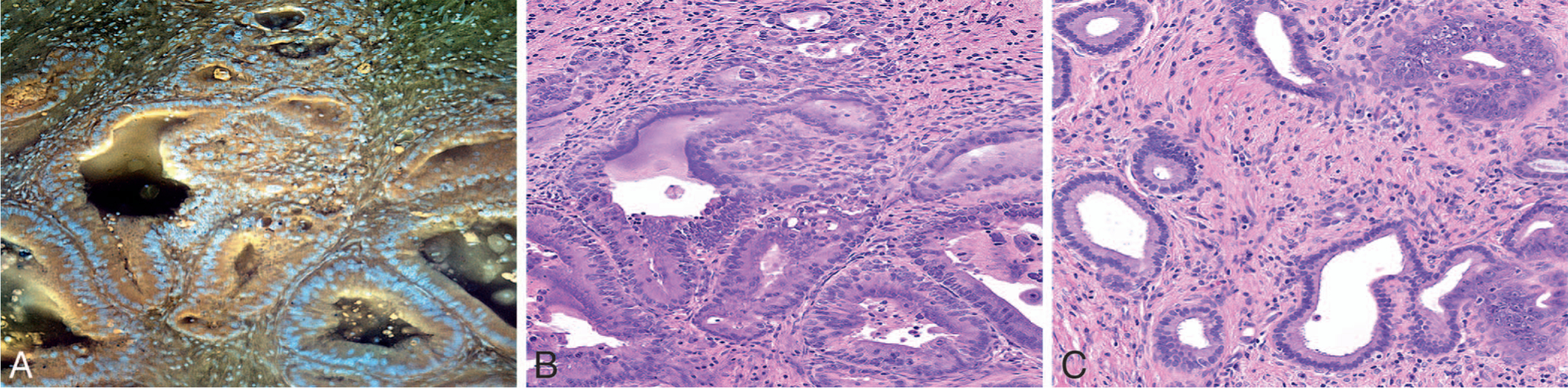
Fluorescence microscopy using ultraviolet surface excitation (MUSE) image (A), digitally colorized (MUSE) image (B), and light microscopic image of corresponding hematoxylin-eosin–stained tissue section (C) of pancreatic ductal adenocarcinoma (original magnification ×100 [C]) (This image is courtesy of Richard Levenson, MD, UC Davis, Davis, California).
STRUCTURED ILLUMINATION MICROSCOPY
Structured illumination microscopy is an optical sectioning microscopy method that uses patterned fluorescence excitation to preferentially modulate and retain the in-focus emission separately from the out-of-focus background.44–46 It has the advantage of parallel pixel acquisition such that the overall pixel scaling frequency correlates with the pixel count of the detector rather than the pixel exposure time. This distinct advantage of SIM makes it a light-efficient, wide-field technique in which the speed is decoupled from the field of view because all pixels are acquired in parallel and the pixel resolution scales with camera specifications. The disadvantage of SIM is that it has limited ability to obtain high-quality optical sections deep into the tissue. High-quality images of large tissue surfaces can be obtained, and with effective mosaicking these images can resemble light microscopic images of H&E-stained tissue sections. The utilization of the latest high-speed digital spatial light modulators and scientific CMOS camera can aid in the acquisition of high-resolution, optically sectioned images of fluorescently stained samples that can closely resemble H&E-stained sections.
Wang et al47 used a 4.2-megapixel high-speed scientific CMOS camera and fast ferroelectric spatial light modulators for rapid pattern generation and achieved video rate (VR)–SIM for optical sectioning. The VR-SIM system built by these authors is constructed around an automated epifluorescence microscope platform (RAMM, Applied Scientific Instrumentation, Eugene, Oregon) that incorporates a 7-mm/s motorized XY specimen stage and a motorized Z objective positioner. Blue excitation light at 475 nm provided by an LED (Thorlabs, Newton, New Jersey) was used for excitation. In this implementation of VR-SIM, incoherent fluorescence-structured illumination is used, and the excitation light is transmitted through a polarizing beam splitter and imaged onto a liquid crystal layer atop a silicon spatial light modulator. Wang et al47 used VR-SIM for imaging 34 unfixed and uncut 5-mm punch biopsy specimens of prostate obtained from radical prostatectomies. The specimens were stained with acridine orange before SIM imaging. High-quality grayscale mosaic images of the prostate biopsies could be obtained in seconds, using the SIM platform built by this team to achieve area-throughput rates for mosaicking microscopy of 4.4 cm2/min with 1.3-μm lateral resolution. The images had excellent contrast and detail, exhibiting similarity to H&E images. The SIM images were read by 2 independent pathologists and the diagnosis was compared with the interpretation made on FFPE tissue sections of the imaged tissue stained by the H&E method. The sensitivity ranged from 63% to 88% and specificity from 78% to 89% in this first study that used SIM for rapid evaluation of human tissue. The potential application of VR-SIM for rapid high-resolution diagnostic imaging of prostate biopsies suitable for point-of-care procedures was clearly demonstrated in this study.
The same authors subsequently studied the feasibility of using VR-SIM for margin assessment of 19 intact prostatectomy specimens.48 They could image the circumferential surfaces of 4 prostate specimens in an hour and demonstrated gigapixel panorama images of the intact prostate surface covering up to 60 cm2 (15 billion pixels) with relevant contrast and subcellular details. VR-SIM confirmed positive margins in 3 of 4 prostatectomy specimens. This study showed the potential utility of VR-SIM for margin assessment of large and fully intact surgical specimens. Recently, Elfer et al49 demonstrated the spectrally compatible combination of the nuclear stain DRAQ5 and the anionic counterstain eosin as a dual-component fluorescent staining analogue to H&E, which can be used in fresh tissues for optical sectioning fluorescence microscopy. Together with digital colorizing algorithms, DRAQ5 and eosin (D&E) staining and SIM imaging can enable very fast pseudo-H&E rendering of imaged tissues at the point of acquisition with minimal tissue handling and processing. These authors applied D&E staining to 18G renal core needle biopsy specimens and large needle core prostate biopsy specimens that were imaged with SIM. They showed the ability of this technique to obtain high-resolution, histology-like images of fresh tissues similar to those for H&E. Liu et al50 studied 65 kidney biopsy specimens (18G in size) obtained from 19 patients undergoing partial or radical nephrectomy. The biopsy specimens were stained with D&E, and the optically sectioned SIM images obtained from the specimen surfaces resembled H&E-stained sections. A single board-certified pathologist reviewed SIM images demonstrating sensitivity and specificity of 79% and 95%, respectively. Figure 4, A and B, shows an illustration of a SIM image of punch biopsy of prostate with the corresponding image of an H&E-stained section.
Figure 4.
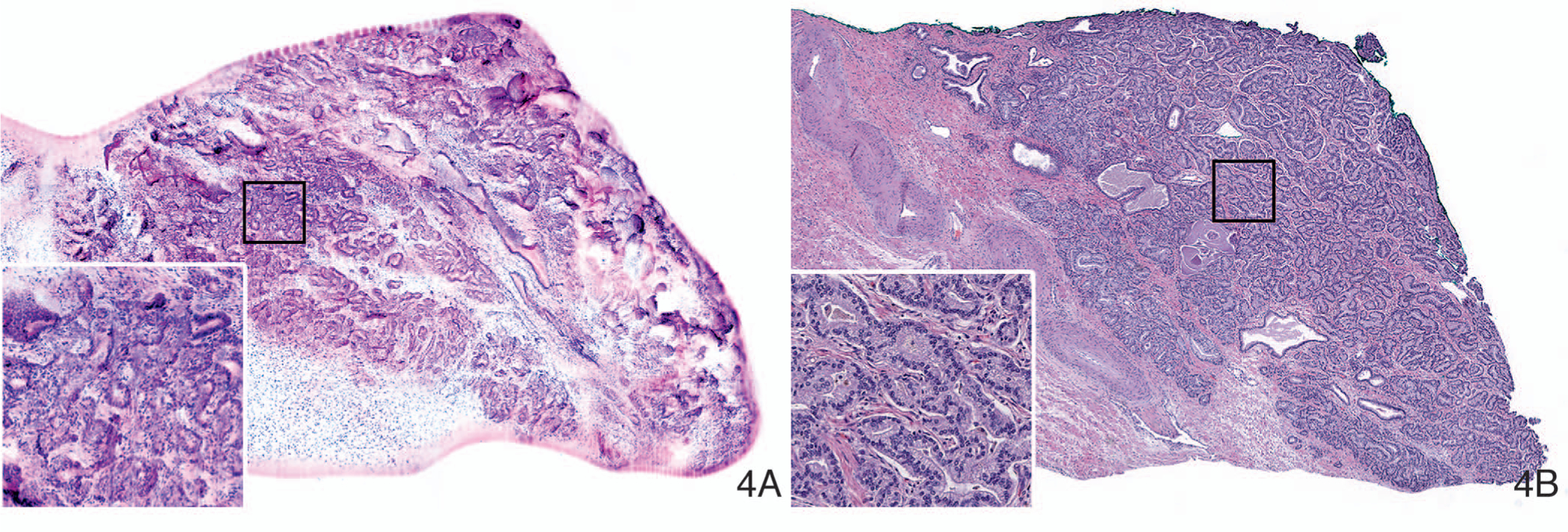
Digitally colorized structured illumination microscopy image (A) and inset of the same, and light microscopic image of corresponding hematoxylin-eosin–stained tissue section (B) of punch biopsy of prostate adenocarcinoma (original magnifications ×100 [B] and ×200 [inset B]). (This image is courtesy of David Tulman, PhD, Jonathon Quincy Brown, PhD, and Mei Wang, PhD, Tulane University, New Orleans, Louisiana.)
LIGHT SHEET MICROSCOPY
Light sheet microscopy is an imaging technique that achieves optical sectioning by rejecting out-of-focus light, using a thin selective illumination plane that generates fluorescence signals to be imaged in the orthogonal direction.51–53 The illumination and collection beam paths are decoupled, and each of these paths can be individually optimized in this technique. This is a distinct advantage in comparison with conventional single axis microscopes in which the illumination and collection beams travel along an identical path and where the depth of focus and resolution are coupled by the laws of diffraction. The depth of focus of an LSM can be extended to hundreds of micrometers by using a moderately thick illumination beam. Despite the low numerical aperture of the optics generating the moderately thick light sheet, it is still possible to achieve high spatial resolution sufficient to visualize nuclei by using a higher numerical aperture for the orthogonal collection beam path. The distinct advantage of the LSM technique in comparison with essentially camera-based planar microscopy systems such as SIM and MUSE is its effectiveness for volumetric imaging applications. However, it is to be noted that such 3D imaging typically requires a previous tissue-clearing step.
Glaser et al54 developed an open top LSM optimized for nondestructive slide-free tissue imaging of large slices of tissue as well as for core needle biopsy specimens. This imaging technique allowed rapid imaging at high resolution over large 2-dimensional and 3-dimensional fields of view, with the same level of detail as traditional pathology. They studied 25 large human prostate slices measuring approximately 3.1 cm × 3.5 cm × 0.4 cm and a large slice of breast tissue measuring 2.0 cm × 2.0 cm × 0.4 cm, which were stained with acridine orange and imaged by using the open top LSM. The average imaging time for prostate tissues was 10 minutes per slice. In the case of breast tissue, which was not as firm as the prostate tissue, with reduced tissue surface irregularities, the imaging speed was even faster: 4 cm2 area of tissue in less than 1 minute. A prostate core biopsy 2 cm in length and 1 mm in diameter was optically cleared overnight, stained with DRAQ5 and eosin, and imaged with open top LSM. Volumetric dual-channel imaging of the entire thickness of the optically cleared specimen could be achieved in 14 minutes. The 2-fluorescent color channels were digitally colorized to mimic H&E-stained sections.55,56 These authors therefore demonstrated the utility of this technique for wide area surface microscopy to triage surgical specimens using prostate tissue slices, rapid intraoperative assessment of margins using a breast tissue slice, and volumetric assessment of optically cleared core needle biopsies using a prostate core biopsy specimen. Chen at al55 developed a novel fluorescent analog of H&E including SYBR Gold and ATTO 655 N-hydroxysuccinimide ester for staining fresh tissues and optimized an open top LSM for imaging the stained tissues. They reported the feasibility of using the open top LSM for comprehensive examination of surgical margins of breast lumpectomy specimens. The open top LSM could image the specimens at a rate of 1.5 cm2/min. The image quality was better than FS and comparable to corresponding H&E sections. Reder et al56 recently presented an image atlas of open top LSM images of 9 prostate core biopsies that were obtained from radical prostatectomy specimens. The biopsy specimens were fixed in formalin, dehydrated in ethanol, stained with TO-PRO3 and eosin, optically cleared, and then imaged with an open top LSM. Three-dimensional open top LSM microscopy images were obtained that were similar to conventional H&E images providing useful 3D structural information.
Recently, Abadie et al57 performed volumetric evaluation of optically cleared FFPE specimens of skin by using LSM and showed that in-depth optical sectioning was possible, allowing 3-dimensional image analysis and reconstruction of the skin biopsy, which was useful for visualizing and quantifying histologic abnormalities. Noe et al58 used LSM to perform 3-dimensional imaging of 26 optically cleared sections of pancreas for morphologic evaluation as well as localization of immunolabeled cytokeratin 19 in pancreatic parenchyma. These authors showed the value of 3-dimensional volumetric imaging, which allowed recognition of unique 3-dimensional architectural features not possible using conventional 2-dimensional evaluation of light microscopic images. Several protocols are available for clearing fresh and fixed tissue specimens for volumetric imaging using LSM.
STIMULATED RAMAN SCATTERING MICROSCOPY
Coherent Raman imaging, including coherent anti-Stokes Raman scattering microscopy and SRS microscopy, allows imaging based on intrinsic vibrational properties of molecules such as lipids and proteins in the cells.59,60 Essentially, coherent Raman imaging relies on chemical contrast created by the vibrational properties of these molecules, allowing optical sectioning by nonlinear excitation and without the need for labeling the tissue with extrinsic agents. The more recently developed SRS microscopy techniques offer several advantages including elimination of image artifacts due to nonresonant background, excitation spectra similar to spontaneous Raman spectra, and a linear relationship between signal and concentration of the target molecules.61
Orringer et al62 recently engineered a portable SRS microscope using dual-fiber lasers, including erbium and ytterbium, that could be used in neurosurgery operating rooms. Histologic quality images of fresh unstained surgical specimens obtained from neurosurgical procedures were created by mapping 2 Raman shifts: 2845 cm−1, which corresponds to CH2 bonds that are abundant in lipids, and 2939 cm−1, which corresponds to CH3 bonds that are abundant in proteins and DNA. Assigning a subtracted CH3-CH2 image to a blue channel and a CH2 image to a green channel, these investigators could produce an image with the needed contrast that was suitable for recognition of brain tissue specimens. They also described a method for processing the SRS microscopy images to resemble light microscopic images of H&E-stained tissue sections, which they referred to as stimulated Raman histology (SRH). The images of the fields of view were acquired at a speed of 2 seconds per frame and stitched to create a mosaic in 2.5 minutes that could then be viewed either at the site of acquisition in the operating room or remotely at any connected network station. The SRS microscope was placed in the neurosurgery suite and used for imaging brain tissue obtained from 101 patients undergoing neurosurgical procedures. A simulation of intraoperative pathology consultation was tried for 30 patients. They demonstrated a remarkable concordance of the interpretations made on SRH images with conventional histology for making the diagnosis using the imaged tissue and achieved an accuracy exceeding 92%. In addition to demonstrating the ability of the SRS microscope to generate SRH images that allowed excellent recognition of the tissue, they also built and validated a supervised machine learning algorithm called a multilayer perceptron for enabling automated tissue diagnosis based on quantified SRH image attributes. This algorithm derived from SRH images predicted brain tumor subtypes with 90% accuracy.
Using the same SRS microscope, Hollon et al63 evaluated the ability of SRH images to allow interpretation of brain tissue specimens obtained from 33 prospective pediatric brain tumor patients. Similar to the study by Orringer et al,62 they showed that accurate diagnosis of pediatric tumors was possible without the need for freezing, cutting, and staining because SRH preserved both cytologic and histoarchitectural features of fresh tumor specimens. They simulated an intraoperative consultation for 25 patients and showed that diagnosis based on SRH was concordant with conventional histology with an accuracy of 92% to 96%. A machine learning algorithm was also developed by these authors that could correctly distinguish benign from malignant and low-grade from high-grade tumors with 100% accuracy. In addition to these recent studies, a few other reports have indicated the potential utility of Raman spectroscopy for tissue recognition.
A commercial SRS imaging platform (Invenio Imaging Inc, Santa Clara, California) is currently available for intraoperative diagnosis of tissues obtained from different types of neurosurgical procedures. Figure 5, A and B, is an illustration of unlabeled liver tissue that was imaged by using the commercially available SRS imaging platform, showing poorly differentiated adenocarcinoma with the corresponding image of an H&E-stained section.
Figure 5.
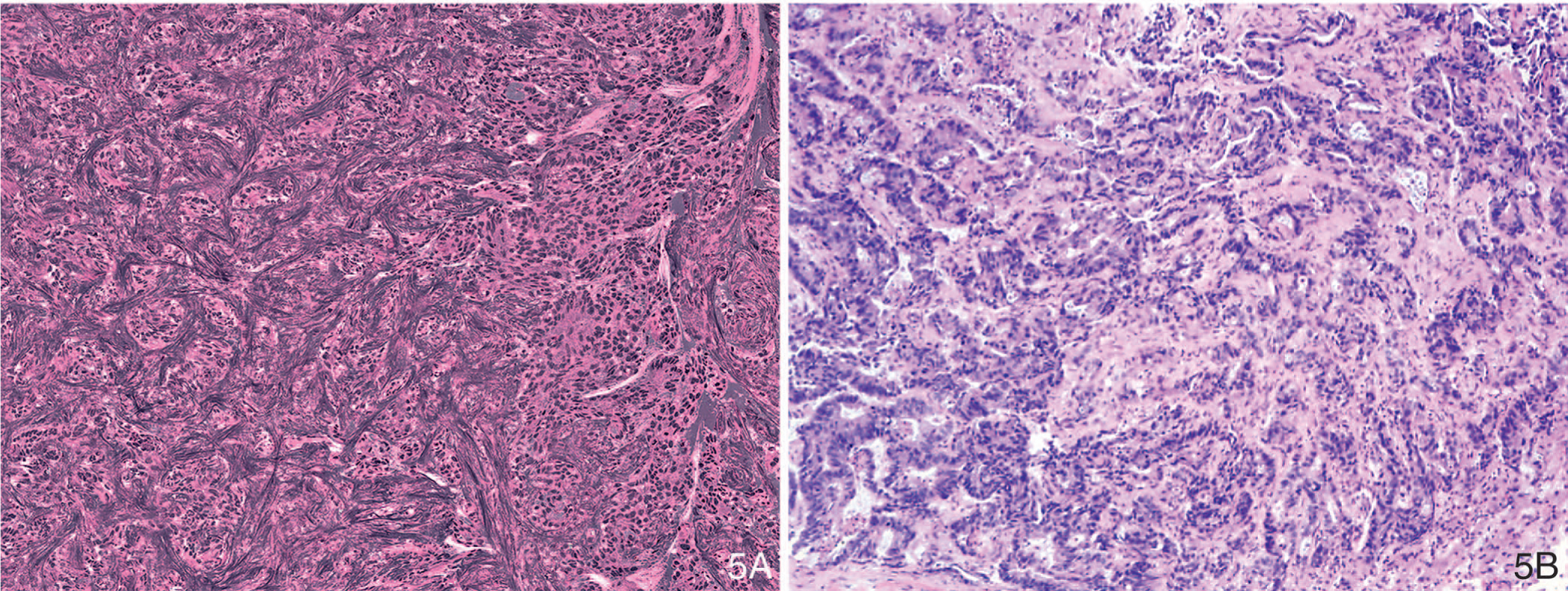
Stimulated Raman scattering microscopy image of metastatic adenocarcinoma in liver (A) and light microscopic image of corresponding hematoxylin-eosin–stained tissue section (B) (original magnification ×100 [B]).
NONLINEAR MICROSCOPY
Nonlinear microscopy including multiphoton, 2-photon, and second-harmonic generation microscopy enables high-resolution imaging in optically scattering tissues.64 Nonlinear microscopy has improved imaging depth in comparison with other optical sectioning techniques such as FCM but not as greatly as is achievable using OCT.65,66 The ability to use both intrinsic and extrinsic contrast in NLM enables visualizing nuclear size and shape as well as structural features such as reorganization of collagen. Nonlinear fluorescent excitation enables high-resolution imaging through the overlaying tissue, blood, and cellular debris into the surgical specimens at high imaging rates relative to other methods.
Tao et al67 used NLM to assess breast tissues obtained from surgical specimens. Imaging was performed by using a 100-fs tunable Ti:sapphire laser (Mira Optima 900-F, Coherent) at 740 nm with a 76-MHz repetition rate and a commercial nonlinear microscope (Thorlabs).67 Specimens were stained with acridine orange for nuclear contrast. Dual-channel photomultipliers were used to collect nuclear-bound 2-photon fluorescence generated from acridine orange bound to nuclei as well as intrinsic collagen signals arising from second-harmonic generation. The 2 channels of NLM were digitally colorized to resemble H&E images. High-magnification fields were imaged by using a 20×, 0.95 nuclear aperture objective with a 480 μm × 480 μm field with 0.44 μm × 0.44 μm × 1.21 μm resolution sampled with 1024 × 1024 pixels at 19 frames per second. The NLM images were stitched and converted to a pyramidal image format. In the reported study, 179 specimens from 50 patients were imaged with NLM. Blinded reading of the NLM images by 3 pathologists yielded an impressive sensitivity of 95.4% and specificity of 93.3% in comparison with readings based on H&E-stained sections of the imaged tissue. Tao and colleagues67 concluded that NLM can be rapidly performed on unfixed specimens with high diagnostic accuracy and that NLM is a promising method for intraoperative margin assessment not only of breast resections but also of other organs such as lung, thyroid, and head and neck. Image mosaicking was used to acquire NLM images over large fields of view for correlation with paraffin-embedded histology. Mosaicking was not optimized for speed, and a standard commercial nonlinear microscope was used. These factors contributed to the relatively low imaging speed.
Cahill et al68 recently demonstrated real-time histologic evaluation of breast cancer surgical specimens by staining specimens with acridine orange and sulforhodamine 101 and then imaging the specimens with fluorescence NLM using a compact femtosecond fiber laser. Because the femtosecond titanium-sapphire lasers used for NLM are large, fragile, expensive, and difficult to maintain, Cahill et al68 used more cost-effective, lower-power unamplified femtosecond ytterbium fiber lasers that operate at 1-μm wavelengths. A video-rate computational light absorption model was used to produce realistic virtual H&E images of tissues in real time and in 3 dimensions that were similar to H&E images. NLM imaging could be performed to depths of 100 μm below the tissue surface. Preservation of tissue integrity for subsequent fluorescence in situ hybridization testing for determination of HER2 status was also shown in this study.
Recently, Giacomelli et al69 developed a multiscale, real-time microscope with variable magnification NLM and real-time coregistered position display using a wide-field white light imaging system. They performed NLM mosaic imaging of 50 discarded breast specimens obtained from 21 patients, with the specimens stained with acridine orange and sulforhodamine 101 before imaging. Specimens were imaged by using a 1030-nm excitation wavelength generated by using a custom multiscale NLM system with a Ti:sapphire laser. Margin assessment could be performed rapidly under operator guidance to image specific regions of interest, using wide-field imaging. They showed that the margins of the breast specimens could be comprehensively imaged at cellular resolution.
Multiphoton microscopy (MPM) is another nonlinear laser-based imaging technique that can generate high-resolution images resembling images of H&E-stained sections in real time without the need for special tissue processing or extrinsic dyes.65,70 MPM has been used to image fresh and fixed human tissues for differentiating neoplastic from nonneoplastic tissues and to characterize kidney tumors.71–74 Jain et al75 used a commercially available multiphoton laser scanning microscope equipped with a Ti:sapphire femtosecond pulsed laser for imaging tissues at a wavelength of 780 nm. The platform generated images from the emission signals including (1) short-wavelength autofluorescence in the wavelength range of 420 to 490 nm that captured the emission signals derived from reduced nicotinamide adenine dinucleotide (NADH) and lipofuscin; (2) long-wavelength autofluorescence in the range of 550 to 650 nm capturing signals derived from flavin adenine dinucleotide, NADH, lipofuscin, and iron; and (3) second-harmonic generation in the range of 360 to 400 nm capturing signals from collagen. The authors demonstrated the potential of this platform to distinguish chromophobe renal cell carcinoma and oncocytic carcinoma. Jain et al76 characterized various malignant kidney tumors by using MPM and achieved a high diagnostic accuracy of 98% in subtyping the tumors.
SUMMARY
The emerging plethora of optical imaging modalities has promising potential for evaluating tissues in surgical pathology practice. These ex vivo entirely digital imaging modalities can be regarded as the next-generation microscopy tools that may be potentially useful for surgical pathology practice. Each of these imaging modalities has its own advantages and limitations. Imaging modalities with the significant advantage of not requiring any extrinsic labeling agents include OCT, FF-OCT/D-FFOCT, and SRS, all of which are currently available commercially. Of these ex vivo intrinsic contrast imaging techniques, SRS microscopy and D-FFOCT alone can generate images resembling images of H&E-stained sections. The images acquired by D-FFOCT techniques have the advantage of reflecting the metabolic activity of the constituent cells in small fragments of tissues. The commercially available SRS microscopy platform can be used for intraoperative evaluation of specimens obtained from neurosurgical procedures. The FF-OCT/D-FFOCT platform can be used for evaluation of small fragments of fresh unprocessed and unlabeled tissues measuring up to 2 cm in maximum dimension. Margin assessment of breast lumpectomy specimens can be performed by using the currently available OCT-based platform (Perimeter Medical Imaging), which generates grayscale images that allow detection of tissue abnormalities without, however, the ability to visualize cellular details.
The ex vivo imaging modalities that use fluorescent dyes as contrast agents include FCM, MUSE, SIM, LSM, and NLM. A variety of fluorescent dyes such as acridine orange, proflavine, Hoechst, DRAQ5, eosin, and sulforhodamine have been used as contrast agents in the studies that evaluated these imaging modalities. All these techniques can generate images that can be digitally colorized to resemble light microscopic images of H&E-stained tissue sections. Two FCM platforms are currently available commercially. On the basis of reported studies, these platforms are suitable for rapid evaluation of small fragments of tissues such as core biopsy, endoscopic biopsies, and for margin assessment of small surgical excisions such as skin specimens obtained from Mohs surgery. The other imaging platforms with promising potential for evaluating small specimens as well as for margin evaluation of larger surgical resections include MUSE, SIM, LSM, and NLM. These platforms are not yet commercially available although activities are underway for their commercial development. Tissue integrity for conventional histopathologic examination is clearly shown in all the studies that evaluated these platforms and compared the interpretation of the digital images with the gold standard of FFPE tissue sections of the imaged tissue stained by H&E. Staining the tissues with fluorescent dyes did not interfere with subsequent genomic testing, based on the few reported studies.41,68
In essence, the recent developments in the field of ex vivo tissue imaging demonstrate the promising potential of optical imaging modalities for the different types of application in surgical pathology practice. Much of the current literature establishes the feasibility and addresses the strengths and limitations of these modalities. These next-generation digital microscopy tools have promising potential for utilization in surgical pathology practice.
Acknowledgments
The authors would like to thank Kechen Ban, PhD, and Gerald Pendleton, BS, for help with preparation of this manuscript. The authors also thank Scientific Publication, The University of Texas MD Anderson Cancer Center, for help with editing of the manuscript.
Contributor Information
Savitri Krishnamurthy, Department of Pathology and Laboratory Medicine, The University of Texas, MD Anderson Cancer Center, Houston.
Jonathan Quincy Brown, Biomedical Engineering, Tulane University, New Orleans, Louisiana.
Nicusor Iftimia, Physical Sciences Inc, Andover, Massachusetts.
Richard M. Levenson, Department of Pathology and Laboratory Medicine, University of California Davis, Davis.
Milind Rajadhyaksha, Dermatology Section, Memorial Sloan Kettering Cancer Center, New York, New York.
References
- 1.Minsky M Memoir on inventing the confocal scanning microscope. Scanning. 1988;10(4):128–138. [Google Scholar]
- 2.Ragazzi M, Longo C, Piana S. Ex vivo (fluorescence) confocal microscopy in surgical pathology: state of the art. Adv Anat Pathol. 2016;23(3):159–169. [DOI] [PubMed] [Google Scholar]
- 3.Gareau DS. Feasibility of digitally stained multimodal confocal mosaics to simulate histopathology. J Biomed Opt. 2009;14(3):034050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gareau DS, Jeon H, Nehal KS, Rajadhyaksha M. Rapid screening of cancer margins in tissue with multimodal confocal microscopy. J Surg Res. 2012;178(2): 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson B, Abeytunge S, Seltzer E, Rajadhyaksha M, Nehal K. Detection of skin cancer margins in Mohs excisions with high-speed strip mosaicing confocal microscopy: a feasibility study. Br J Dermatol. 2013;169(4):922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennassar A, Carrera C, Puig S, Vilalta A, Malvehy J. Fast evaluation of 69 basal cell carcinomas with ex vivo fluorescence confocal microscopy: criteria description, histopathological correlation, and interobserver agreement. JAMA Dermatol. 2013;149(7):839–847. [DOI] [PubMed] [Google Scholar]
- 7.Longo C, Ragazzi M, Rajadhyaksha M, et al. In vivo and ex vivo confocal microscopy for dermatologic and Mohs surgeons. Dermatol Clin. 2016;34(4): 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu EW, Lewin JM, Stevenson ML, et al. Use of digitally stained multimodal confocal mosaic images to screen for nonmelanoma skin cancer. JAMA Dermatol. 2016;152(12):1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain M, Rajadhyaksha M, Nehal K. Implementation of fluorescence confocal mosaicking microscopy by “early adopter” Mohs surgeons and dermatologists: recent progress. J Biomed Opt. 2017;22(2):24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann D, Krammer S, Bachmann MR, et al. Simple 3-criteria-based ex vivo confocal diagnosis of basal cell carcinoma. J Biophotonics. 2018;11(7): e201800062. [DOI] [PubMed] [Google Scholar]
- 11.Longo C, Borsari S, Pampena R, et al. Basal cell carcinoma: the utility of in vivo and ex vivo confocal microscopy. J Eur Acad Dermatol Venereol. 2018; 32(12):2090–2096. [DOI] [PubMed] [Google Scholar]
- 12.Longo C, Pampena R, Bombonato C, et al. Diagnostic accuracy of ex vivo fluorescence confocal microscopy in Mohs surgery of basal cell carcinomas: a prospective study on 753 margins [published online ahead of print February 20, 2019] Br J Dermatol. doi: 10.1111/bjd.17507. [DOI] [PubMed] [Google Scholar]
- 13.Peters N, Schubert M, Metzler G, Geppert JP, Moehrle M. Diagnostic accuracy of a new ex vivo confocal laser scanning microscope compared to H&E-stained paraffin slides for micrographic surgery of basal cell carcinoma. J Eur Acad Dermatol Venereol. 2019;33(2):298–304. [DOI] [PubMed] [Google Scholar]
- 14.Longo C, Ragazzi M, Gardini S, et al. Ex vivo fluorescence confocal microscopy in conjunction with Mohs micrographic surgery for cutaneous squamous cell carcinoma. J Am Acad Dermatol. 2015;73(2):321–322. [DOI] [PubMed] [Google Scholar]
- 15.Dobbs JL, Ding H, Benveniste AP, et al. Feasibility of confocal fluorescence microscopy for real-time evaluation of neoplasia in fresh human breast tissue. J Biomed Opt. 2013;18(10):106016. [DOI] [PubMed] [Google Scholar]
- 16.Ragazzi M, Piana S, Longo C, et al. Fluorescence confocal microscopy for pathologists. Mod Pathol. 2014;27(3):460–471. [DOI] [PubMed] [Google Scholar]
- 17.Wirth D, Snuderl M, Sheth S, et al. Identifying brain neoplasms using dye-enhanced multimodal confocal imaging. J Biomed Opt. 2012;17(2):026012. [DOI] [PubMed] [Google Scholar]
- 18.Snuderl M, Wirth D, Sheth SA, et al. Dye-enhanced multimodal confocal imaging as a novel approach to intraoperative diagnosis of brain tumors. Brain Pathol. 2013;23(1):73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abeytunge S, Larson B, Peterson G, et al. Evaluation of breast tissue with confocal strip-mosaicking microscopy: a test approach emulating pathology-like examination. J Biomed Opt. 2017;22(3):34002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnamurthy S, Cortes A, Lopez M, et al. Ex vivo confocal fluorescence microscopy for rapid evaluation of tissues in surgical pathology practice. Arch Pathol Lab Med. 2018;142(3):396–401. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy S, Ban K, Shaw K, et al. Confocal fluorescence microscopy platform suitable for rapid evaluation of small fragments of tissue in surgical pathology practice. Arch Pathol Lab Med. 2019;143(3):305–313. [DOI] [PubMed] [Google Scholar]
- 22.Huang D, Swanson EA, Lin CP, et al. Optical coherence tomography. Science. 1991;254(5035):1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Manen L, Dijkstra J, Boccara C, et al. The clinical usefulness of optical coherence tomography during cancer interventions. J Cancer Res Clin Oncol. 2018;144(10):1967–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Xu Y, Boppart SA. Review of optical coherence tomography in oncology. J Biomed Opt. 2017;22(12):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson EA, Fujimoto JG. The ecosystem that powered the translation of OCT from fundamental research to clinical and commercial impact [invited]. Biomed Opt Express. 2017;8(3):1638–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen FT, Zysk AM, Chaney EJ, et al. Intraoperative evaluation of breast tumor margins with optical coherence tomography. Cancer Res. 2009;69(22): 8790–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zysk AM, Chen K, Gabrielson E, et al. Intraoperative assessment of final margins with a handheld optical imaging probe during breast-conserving surgery may reduce the reoperation rate: results of a multicenter study. Ann Surg Oncol. 2015;22(10):3356–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erickson-Bhatt SJ, Nolan RM, Shemonski ND, et al. Real-time imaging of the resection bed using a handheld probe to reduce incidence of microscopic positive margins in cancer surgery. Cancer Res. 2015;75(18):3706–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha R, Friedlander LC, Hibshoosh H, et al. Optical coherence tomography: a novel imaging method for post-lumpectomy breast margin assessment-a multi-reader study. Acad Radiol. 2018;25(3):279–287. [DOI] [PubMed] [Google Scholar]
- 30.Dubois A, Grieve K, Moneron G, et al. Ultrahigh-resolution full-field optical coherence tomography. Appl Opt. 2004;43(14):2874–2883. [DOI] [PubMed] [Google Scholar]
- 31.Dubois A, Vabre L, Boccara AC, Beaurepaire E. High-resolution full-field optical coherence tomography with a Linnik microscope. Appl Opt. 2002;41(4): 805–812. [DOI] [PubMed] [Google Scholar]
- 32.Apelian C, Harms F, Thouvenin O, Boccara AC. Dynamic full field optical coherence tomography: subcellular metabolic contrast revealed in tissues by interferometric signals temporal analysis. Biomed Opt Express. 2016;7(4):1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apelian C, Gastaud C, Boccara AC. Extracting relevant information for cancer diagnosis from dynamic full field OCT through image processing and learning. SPIE BiOS. 2017;10053:7. [Google Scholar]
- 34.Jain M, Narula N, Salamoon B, et al. Full-field optical coherence tomography for the analysis of fresh unstained human lobectomy specimens. J Pathol Inform. 2013;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain M, Robinson BD, Salamoon B, et al. Rapid evaluation of fresh ex vivo kidney tissue with full-field optical coherence tomography. J Pathol Inform. 2015; 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Manen L, Stegehuis PL, Farina-Sarasqueta A, et al. Validation of full-field optical coherence tomography in distinguishing malignant and benign tissue in resected pancreatic cancer specimens. PLoS One. 2017;12(4):e0175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assayag O, Grieve K, Devaux B, et al. Imaging of non-tumorous and tumorous human brain tissues with full-field optical coherence tomography. Neuroimage Clin. 2013;2:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durkin JR, Fine JL, Sam H, Pugliano-Mauro M, Ho J. Imaging of Mohs micrographic surgery sections using full-field optical coherence tomography: a pilot study. Dermatol Surg. 2014;40(3):266–274. [DOI] [PubMed] [Google Scholar]
- 39.Lopater J, Colin P, Beuvon F, et al. Real-time cancer diagnosis during prostate biopsy: ex vivo evaluation of full-field optical coherence tomography (FFOCT) imaging on biopsy cores. World J Urol. 2016;34(2):237–243. [DOI] [PubMed] [Google Scholar]
- 40.Assayag O, Antoine M, Sigal-Zafrani B, et al. Large field, high resolution full-field optical coherence tomography: a pre-clinical study of human breast tissue and cancer assessment. Technol Cancer Res Treat. 2014;13(5):455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fereidouni F, Harmany ZT, Tian M, et al. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat Biomed Eng. 2017;1:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshitake T, Giacomelli MG, Quintana LM, et al. Rapid histopathological imaging of skin and breast cancer surgical specimens using immersion microscopy with ultraviolet surface excitation. Sci Rep. 2018;8(1):4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho D, Fereidouni F, Levenson RM, Jagdeo J. Real-time, high-resolution, in vivo characterization of superficial skin with microscopy using ultraviolet surface excitation (MUSE). J Drugs Dermatol. 2016;15(11):1344–1346. [PMC free article] [PubMed] [Google Scholar]
- 44.Schlichenmeyer TC, Wang M, Elfer KN, Brown JQ. Video-rate structured illumination microscopy for high-throughput imaging of large tissue areas. Biomed Opt Express. 2014;5(2):366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mertz J Optical sectioning microscopy with planar or structured illumination. Nat Methods. 2011;8(10):811–819. [DOI] [PubMed] [Google Scholar]
- 46.Neil MA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt Lett. 1997;22(24):1905–1907. [DOI] [PubMed] [Google Scholar]
- 47.Wang M, Kimbrell HZ, Sholl AB, et al. High-resolution rapid diagnostic imaging of whole prostate biopsies using video-rate fluorescence structured illumination microscopy. Cancer Res. 2015;75(19):4032–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M, Tulman DB, Sholl AB, et al. Gigapixel surface imaging of radical prostatectomy specimens for comprehensive detection of cancer-positive surgical margins using structured illumination microscopy. Sci Rep. 2016;6:27419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elfer KN, Sholl AB, Wang M, et al. DRAQ5 and eosin (‘D&E’) as an analog to hematoxylin and eosin for rapid fluorescence histology of fresh tissues. PLoS One. 2016;11(10):e0165530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Wang M, Tulman D, et al. Nondestructive diagnosis of kidney cancer on 18-gauge core needle renal biopsy using dual-color fluorescence structured illumination microscopy. Urology. 2016;98:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Q, Martin BL, Matus DQ, Gao L. Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat Commun. 2016;7:11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pitrone PG, Schindelin J, Stuyvenberg L, et al. OpenSPIM: an open-access light-sheet microscopy platform. Nat Methods. 2013;10(7):598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Methods. 2017;14(4):360–373. [DOI] [PubMed] [Google Scholar]
- 54.Glaser AK, Reder NP, Chen Y, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng. 2017; 1(7):0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Xie W, Glaser AK, et al. Rapid pathology of lumpectomy margins with open-top light-sheet (OTLS) microscopy. Biomed Opt Express. 2019;10(3): 1257–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reder NP, Glaser AK, McCarty EF, et al. Open-top light-sheet microscopy image atlas of prostate core needle biopsies [published online ahead of print March 20, 2019] Arch Pathol Lab Med. doi: 10.5858/arpa.2018-0466-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abadie S, Jardet C, Colombelli J, et al. 3D imaging of cleared human skin biopsies using light-sheet microscopy: a new way to visualize in-depth skin structure. Skin Res Technol. 2018;24(2):294–303. [DOI] [PubMed] [Google Scholar]
- 58.Noe M, Rezaee N, Asrani K, et al. Immunolabeling of cleared human pancreata provides insights into three-dimensional pancreatic anatomy and pathology. Am J Pathol. 2018;188(7):1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans CL, Xie XS. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu Rev Anal Chem (Palo Alto Calif). 2008;1:883–909. [DOI] [PubMed] [Google Scholar]
- 60.Freudiger CW, Min W, Saar BG, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008; 322(5909):1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saar BG, Freudiger CW, Reichman J, et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330(6009):1368–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orringer DA, Pandian B, Niknafs YS, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng. 2017;1 pii:0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hollon T, Lewis S, Freudiger CW, Sunney Xie X, Orringer DA. Improving the accuracy of brain tumor surgery via Raman-based technology. Neurosurg Focus. 2016;40(3):E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248(4951):73–76. [DOI] [PubMed] [Google Scholar]
- 65.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21(11):1369–1377. [DOI] [PubMed] [Google Scholar]
- 66.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2(12):932–940. [DOI] [PubMed] [Google Scholar]
- 67.Tao YK, Shen D, Sheikine Y, et al. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci U S A. 2014;111(43):15304–15309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cahill LC, Giacomelli MG, Yoshitake T, et al. Rapid virtual hematoxylin and eosin histology of breast tissue specimens using a compact fluorescence nonlinear microscope. Lab Invest. 2018;98(1):150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giacomelli MG, Yoshitake T, Cahill LC, et al. Multiscale nonlinear microscopy and widefield white light imaging enables rapid histological imaging of surgical specimen margins. Biomed Opt Express. 2018;9(5):2457–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zipfel WR, Williams RM, Christie R, et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A. 2003;100(12):7075–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain M, Robinson BD, Scherr DS, et al. Multiphoton microscopy in the evaluation of human bladder biopsies. Arch Pathol Lab Med. 2012;136(5):517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain M, Narula N, Aggarwal A, et al. Multiphoton microscopy: a potential “optical biopsy” tool for real-time evaluation of lung tumors without the need for exogenous contrast agents. Arch Pathol Lab Med. 2013;138(8):1037–1047. [DOI] [PubMed] [Google Scholar]
- 73.Makino T, Jain M, Montrose DC, et al. Multiphoton tomographic imaging: a potential optical biopsy tool for detecting gastrointestinal inflammation and neoplasia. Cancer Prev Res (Phila). 2012;5(11);1280–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Najari BB, Ramasamy R, Sterling J, et al. Pilot study of the correlation of multiphoton tomography of ex vivo human testis with histology. J Urol. 2012; 188(2):538–543. [DOI] [PubMed] [Google Scholar]
- 75.Jain M, Robinson BD, Wu B, Khani F, Mukherjee S. Exploring multiphoton microscopy as a novel tool to differentiate chromophobe renal cell carcinoma from oncocytoma in fixed tissue sections. Arch Pathol Lab Med. 2018;142(3): 383–390. [DOI] [PubMed] [Google Scholar]
- 76.Jain M, Robinson BD, Aggarwal A, et al. Multiphoton microscopy for rapid histopathological evaluation of kidney tumours. BJU Int. 2016;118(1):118–126. [DOI] [PubMed] [Google Scholar]


