Abstract
Diverse natural product small molecules have allowed critical insights into processes that govern eukaryotic cells’ ability to secrete cytosolically synthesized secretory proteins into their surroundings or to insert newly synthesized integral membranes into the lipid bilayer of the endoplasmic reticulum. In addition, many components of the endoplasmic reticulum, required for protein homeostasis or other processes such as lipid metabolism or maintenance of calcium homeostasis, are being investigated for their potential in modulating human disease conditions such as cancer, neurodegenerative conditions and diabetes. In this review, we cover recent findings on natural products that influence protein secretion or impact ER protein homeostasis, and serve as powerful chemical tools to understand protein flux through the mammalian secretory pathway and as leads for the discovery of new therapeutics.
Graphical Abstract
This highlight reviews functions and therapeutic potential of diverse natural products that target different components of the mammalian protein secretory pathway.
1. Introduction
Natural products (NPs) are a rich source of diverse bioactive molecules for discovery of therapeutic drugs1–3. In addition, NPs serve as valuable probes to study the inner workings of the cell and have revealed a wealth of information about dynamic cellular processes such as mechanisms of protein synthesis, secretion, and regulated turnover4. However, to gain relevant insights into biological systems and disease mechanisms, high quality probe molecules are required and many small molecules that are currently used do not fulfill the required criteria5. Current technological development, including CRISPR-based chemogenomic screening in human cells, improved methods for identifying resistance-conferring point mutations in NP target genes, proteome-wide small molecule engagement approaches and new synthetic routes to affinity-based probe molecules such as photoaffinity probes have permitted new advances in identifying cellular targets of many natural product small molecules and to examine the degree to which the observed phenotypic changes can be attributed to on-target modulation6–9. In this review, we will cover the discovery and recent literature of NPs that influence protein secretion by direct targeting of key factors influencing protein secretion or important homeostatic processes of the secretory pathway (Figure 1), including different forms of ER stress, maintenance of calcium homeostasis and ER associated protein degradation.
Figure 1.
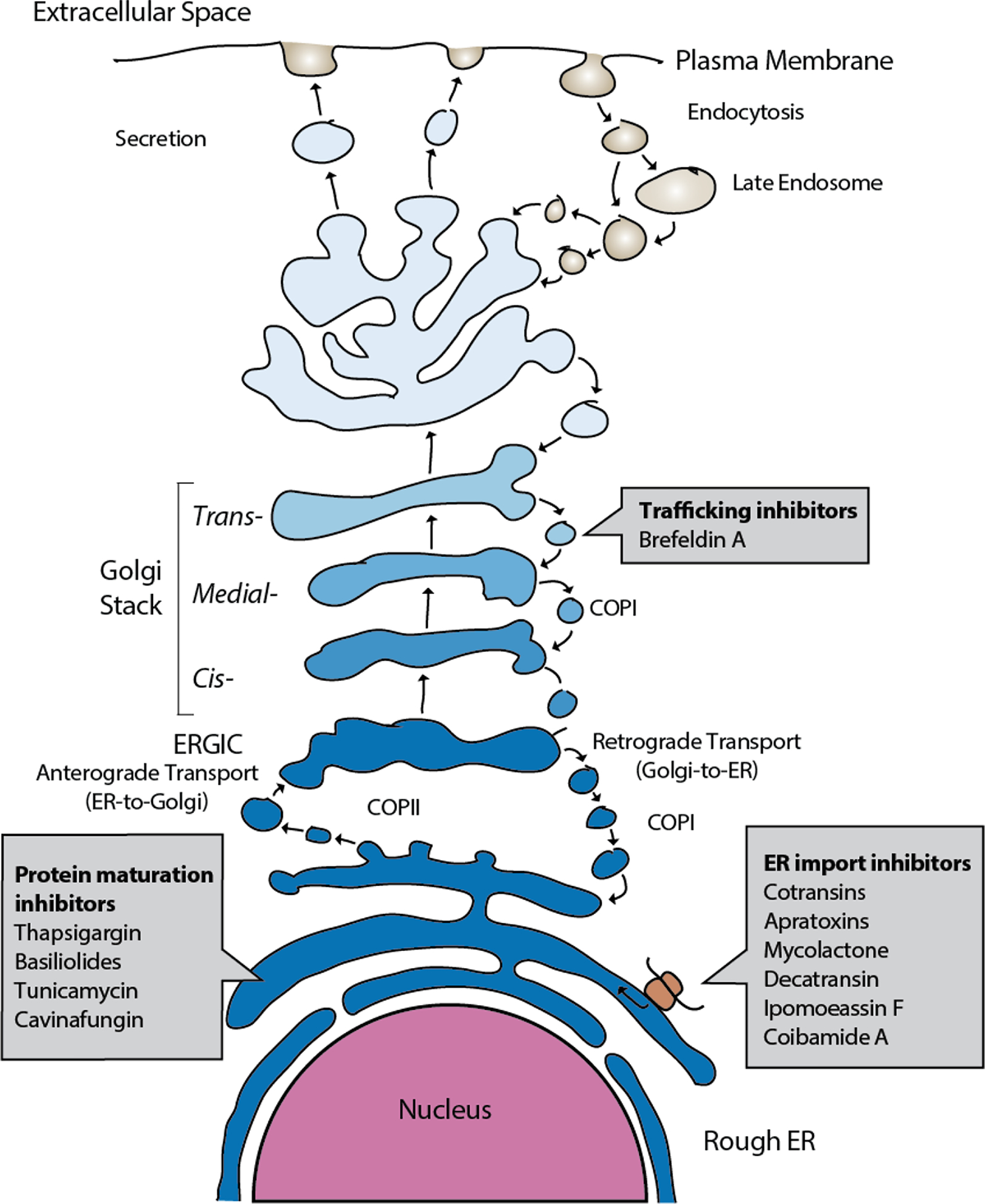
Outline of the mammalian protein secretory pathway and points of activity of different proteostasis modulating NPs.
2. Inhibitors of ER protein insertion
Protein insertion into the lumen of the ER or insertion into the ER membrane for integral membrane proteins is the first step after protein translation on the protein secretion pathway. Recent work has now revealed a dizzying array of structurally diverse, potent natural products that act by preventing protein ER translocation (reviewed in10). These compounds are produced by a host of microorganisms ranging from endosymbiotic fungi to human pathogenic bacteria, marine cyanobacteria and even medicinal plants. Unexpectedly, it now appears that nearly all of these NPs act by directly targeting the central Sec61 protein translocation channel that is responsible for ER translocation of newly synthesized secretory and membrane proteins (Figure 2). Here, we will review the discovery and current progress on understanding the mechanism of this diverse group of Sec61 modulating compounds.
Figure 2. Mechanism of inhibition of protein ER import and maturation, and location of common resistance mutations within the structure of the Sec61 protein translocon.
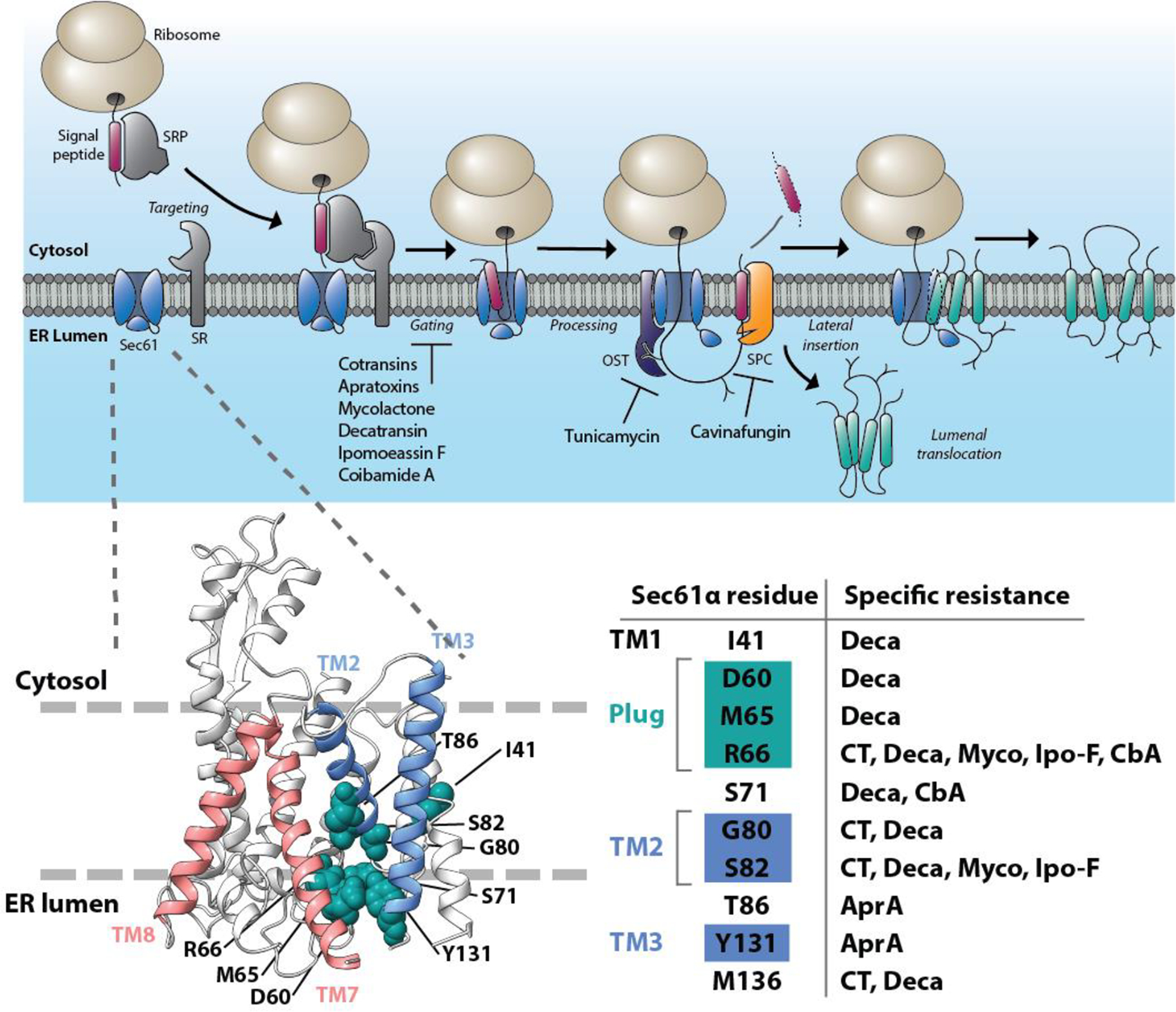
The pathway of cotranslational mammalian protein import into the ER (top). Inhibitors of Sec61 protein translocon prevent channel gating and insertion of secreted proteins into the ER lumen or diffusion of integral membrane proteins into the ER lipid bilayer. Tunicamycin is an inhibitor of cotranslational protein N-glycosylation, whereas cavinafungin prevents ER targeting signal peptide cleavage. In the structural model of mammalian Sec61 (bottom), the lateral gate helices of Sec61 are indicated in blue (TM2 and TM3) and red (TM7 and TM8) and the resistance mutations identified to confer strong resistance to most structurally diverse Sec61 inhibitors are shown in green and outline the putative NP-binding site on the lumenal end of the Sec61 lateral gate. SRP, Signal Recognition Particle. OST, Oligosaccharyl transferase complex. SPC, signal peptidase complex. CT, cotransin; Deca, decatransin; Myco, mycolactone; Ipo-F, ipomoeassin F; AprA, apratoxin A.
2.1. Cotransins
A fungal natural product HUN-7293 (Figure 3) was discovered as a potent suppressor of expression of the human cell adhesion molecule vascular endothelial cell adhesion molecule I (VCAM-1)11 and a total synthesis of HUN-7293 was established enabling detailed SAR studies12–14. Later, two independent studies revealed that HUN-7293 (later named cotransin) inhibits cell surface receptor expression by preventing ER translocation or membrane insertion of newly synthesized secreted or membrane proteins15,16. Intriguingly, cotransin only inhibits biogenesis of a subset of the thousands of Sec61 substrate proteins in a manner dependent on the N-terminal ER targeting signal peptide or transmembrane segment15,16. Photocrosslinking experiments with a photoactivatable analog of cotransin demonstrated that cotransin inhibits ER translocation by directly binding to the central pore-forming Sec61α subunit of the Sec61 translocon17. Later, an unbiased screen to identify specific resistance-conferring point mutations, suggested that cotransin allosterically blocks Sec61 gating facilitated by nascent secretory polypeptides18. Intriguingly, synthesis and testing of new cotransin analogs indicated that changes in the cotransin structure can alter the range of Sec61 substrate proteins it inhibits19. A consensus sequence for cotransin-sensitive signal peptides or N-terminal transmembrane segments has not been identified, although specific point mutations that do not interfere with protein ER targeting or insertion, yet specifically modulate cotransin sensitivity of specific membrane proteins such as TNFα and AQP2, have been described18,20. A quantitative proteomics study in human hepatocellular liver carcinoma cells using saturating (30 μM) concentration of cotransin, suggested that signal peptide-containing secreted proteins would be generally more sensitive to cotransin modulation than integral membrane proteins20.
Figure 3. Structures of inhibitors of ER protein insertion.
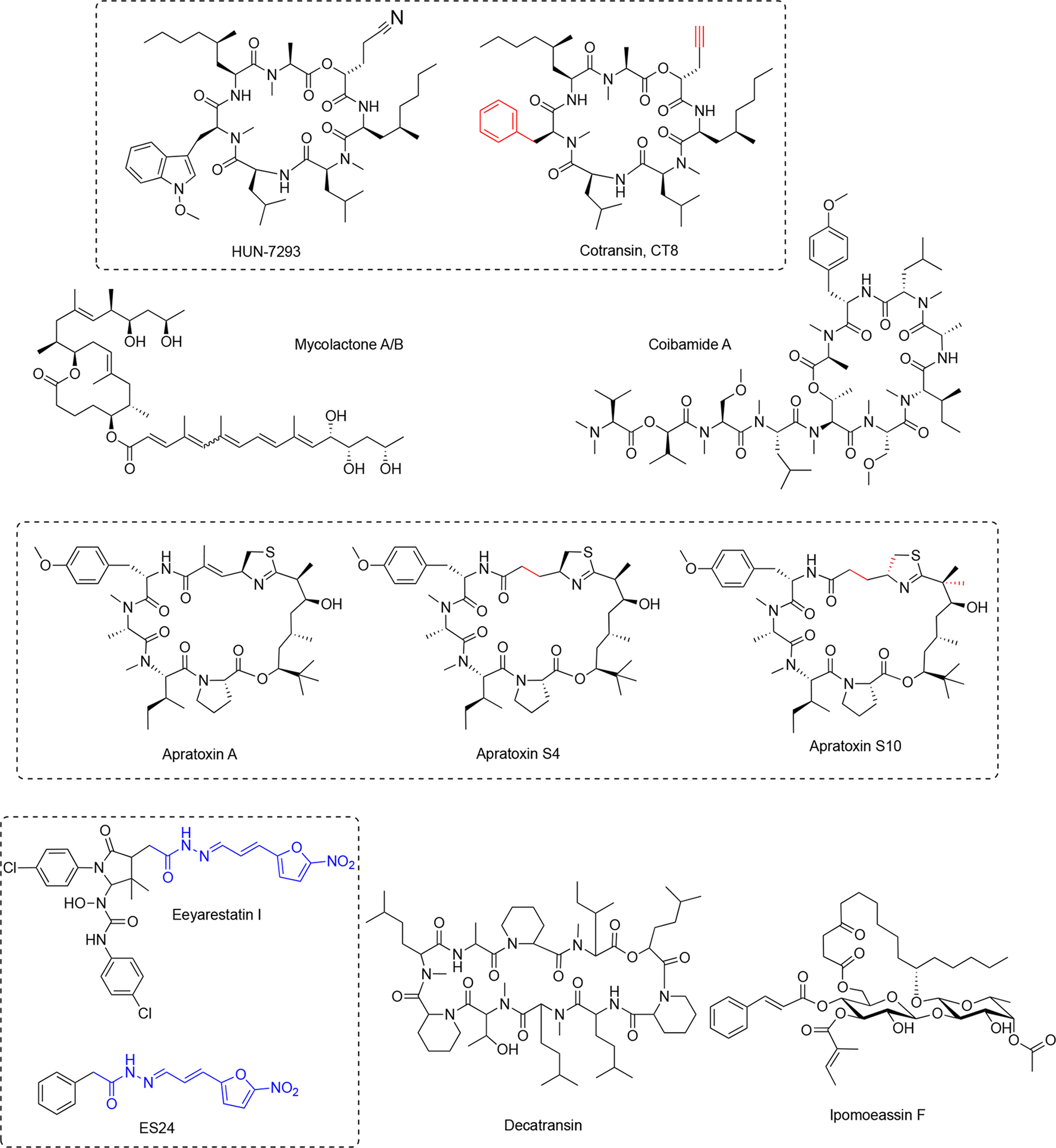
Their structural diversity translates into distinct interactions with Sec61 and differential pharmacological profiles. Red color indicates changes in synthetic compounds compared with the parent natural product. Blue indicates key fragment critical for compound activity.
Another study identified CT8 (Figure 3), a highly substrate-selective cotransin as a potent inhibitor of expression of the cancer-associated cell surface pseudokinase HER3, which was found to contain a highly cotransin-sensitive signal peptide21. The reduced HER3 expression upon treatment of BT474 breast cancer cells with cotransin results from increased proteasomal turnover induced by the cytosolic displacement of the HER3 membrane protein. It is interesting that HER3 among the four signal peptide-containing HER family members is uniquely sensitive to CT8, and this opens up new therapeutic strategies to control HER activity in breast cancer. The authors further showed that CT8 efficiently enhances the efficiency of existing HER2 therapies in BT474 breast cancer cells21. Further, many studies have identified importance of the role of the ER secretory pathway for replication of different flaviviruses and in one of these studies Sec61 was identified as a host factor required for growth and infectivity of several viruses including Influenza, HIV and dengue22. Treatment of cells with noncytotoxic concentrations of CT8 potently inhibited three different strains of HIV. The ability of CT8 to prevent ER trafficking of the HIV gp120 protein and therefore biogenesis and replication of the HIV virus, suggests the possibility of targeting ER insertion with substrate-selective inhibitors as a viable antiviral strategy22.
2.2. Apratoxins
The natural secondary metabolite apratoxin A (Figure 3) was originally discovered from a marine cyanobacterium Lyngbya majuscula (later identified/reclassified as Moorea bouillonii)23 and observed to potently inhibit cancer cell proliferation by inducing cell cycle arrest and apopotosis24. Further, it was shown that treatment of cells with apratoxin A and other apratoxin family members caused downregulation of expression of many cell surface receptors, particularly receptor tyrosine kinases and components of the endoplasmic reticulum25,26. Further, it was demonstrated that apratoxin A inhibited protein translocation to the ER in biochemical experiments, suggesting that the direct cellular target may be an integral component of the ER machinery25.
More recently, photocrosslinking competition, mutagenesis27 and co-immunoprecipitation experiments28 have directly identified Sec61 as the direct cellular binding target of apratoxin A. Biochemically, apratoxin A prevents ER translocation at an earlier stage than cotransin and appears to impact ER import of Sec61 substrate proteins in a substrate-nonselective fashion27 with the exception of so-called N-tail translocating integral membrane proteins that are initially inserted by the action of the EMC complex instead of Sec6129. Intriguingly, the mutagenesis and competitive binding experiments indicate that similarly to cotransin, also apratoxin A binds Sec61α near the lumenal plug domain, suggesting that binding of different ligands to an at least partially overlapping binding site can result in inhibition of ER protein translocation with either a substrate-selective or a substrate-nonselective mechanism.
Several natural apratoxins and synthetic analogs have been synthesized, which provided clear structure-activity relationship information in cells and enabled the modulation of anticancer activity in vitro and in vivo through alteration of the ability to inhibit biogenesis of cell surface proteins and secreted factors30–33, highlighting the potential for improving the currently narrow therapeutic window of apratoxins tested in in vivo cancer models28,31,32. Specifically, removal of the Michael acceptor led to improved activity as apratoxin S4 (Figure 3) demonstrated exquisite activity in a HCT116 colon cancer xenograft model30. The next-generation apratoxin S10 (Figure 3), possessing an inverted configuration of the thiazoline ring to improve potency and a gem-dimethyl group to prevent dehydration-induced deactivation of the compound, inhibited pancreatic cancer in an orthotopic PDX mouse model31. Apratoxin S10 was able to modulate many but not all growth factors and cytokines in primary pancreatic cancer cells and tumor-associated stromal cells comprising the tumor microenvironment, suggesting the potential of apratoxin S10 to exert activity through dual inhibition of cancer growth and associated growth factor signaling but also through inhibition of growth-stimulating factors by the tumor microenvironment that are contributing to resistance. Furthermore, since VEGF and other proangiogenic factors appeared to be particularly sensitive to inhibition by apratoxins S4 and S10 in a cellular context and in vivo, these agents also have potent antiangiogenic activity30,34, which is not only relevant for the inhibition of vascularized tumors but also for treating retinal angiogenic disorders. Consequently, apratoxin S4 was recently shown to inhibit retinal vascular cell activation by suppressing several angiogenic pathways and attenuated pathological ocular neovascularization in several animal models of ocular angiogenesis35.
2.3. Mycolactone
Mycolactone (Figure 3) is produced by Mycobacterium ulcerans, the causative bacteria of a necrotising skin disease Buruli ulcers36. For a comprehensive review on mycolactone mechanisms, see37. This bacteria harbors a megaplasmid that contains the polyketide synthetase genes required for mycolactone biosynthesis38. Mycolactone is a macrolide that exerts an immunosuppressive effect at the site of bacterial infection, but also systemically prevents blood lymphocyte homing to draining lymph nodes39. Because of the central role of mycolactone in mediating bacterial immune-evasion, its mechanism of action has attracted much interest.
In immune cells, mycolactone potently inhibits cellular capacity to produce cytokines, chemokines and homing receptors, particularly inhibiting systemic production of IFN-γ40. In monocytes and macrophages, mycolactone inhibited activation-induced production of cytokines and chemokines40,41. Cellular and biochemical assays indicated that mycolactone inhibits a specific stage of ER insertion and differentially inhibits cotranslational and posttranslational protein translocation of secretory proteins40,42.
Unbiased resistance mutation mapping and competitive photocrosslinking experiments with photo-cotransin indicated that mycolactone targets Sec61α near its lumenal lateral gate, similarly as apratoxin and cotransin43. Proteomic studies in mycolactone-treated CD4+ T lymphocytes, dendritic cells and dorsal root ganglion neurons support the notion that mycolactone broadly affects a broad range of Sec61 substrates including secreted proteins and diverse integral membrane proteins excluding tail anchored proteins and Type III membrane proteins that are initially inserted through activity of the EMC complex44. Further animal work demonstrated that Sec61 is the host receptor mediating mycolactone’s diverse immunomodulatory effects43, although interactions with other factors may also contribute to mycolactone’s effects on the actin cytoskeleton45 and its analgesic properties46. Displacement of secretory proteins into the cytosol by mycolactone triggers a cytosolic integrated stress response independently of the unfolded protein response in the ER47,48. Because of its immunosuppressive properties, analogs of mycolactone are being pursued as potential anti-inflammatory agents49
2.4. Eeyarestatin I
Eeyarestatin I (ESI, Figure 3) was originally identified as a stabilizer of the known ERAD substrate MHC class I heavy chain50. In this study, ESI was shown to stabilize two known ERAD substrates without general effects for proteasomal degradation of ubiquitinated substrates. Later treatment of cells with micromolar concentrations of ESI was shown to result in inhibition of ER insertion of a number of cotranslationally inserted substrate proteins, which was also recapitulated in biochemical ER insertion assays, however 250 μM ESI was required to detect effects in these in vitro experiments51. A recent study demonstrated that ESI induces Ca2+ leakage from the lumen of the ER, which is apparently caused by ESI binding and inhibition of Sec6152, suggesting that ESI binding to Sec61 might stabilize the channel in a partially open ion-conductive configuration and that Ca2+ leakage may be an important contributor to the cytoxtoxic effects of ESI. However, it should be noted that the direct binding partner of ESI has not been demonstrated by for example direct crosslinking or affinity chromatography approaches and therefore the exact mechanism of ER import inhibition by ESI remains incompletely understood. Furthermore, it has been suggested that ESI has dual targets (Sec61 and p97) and modulate ER insertion (via Sec61) and the ERAD pathway (via p97, see below)53. It appears that a truncated version, ES24 (Figure 3) has Sec61 selectivity (over p97)52, although it remains currently unclear to what extent the observed phenotypic effects of ESI can be attributed to specific inhibition of Sec61.
2.5. Decatransin
Decatransin (Figure 3) is a novel decadepsipeptide isolated from the fungus Chaetosphaeria tulasneorum and found to inhibit growth of mammalian cells with nanomolar potency and also the yeast S. cerevisiae with an IC50 of approximately 2 μM54. Chemogenetic screens in yeast suggested that decatransin would act by targeting the Sec61 complex in a similar manner as cotransin. Further, a genome-wide mutagenesis screen was carried out in yeast cells and identified several heterozygous point mutations in the SEC61 gene, encoding the Sec61α subunit of the Sec61 translocon. Even further, a similar mutagenesis screen in human cells identified yet more decatransin resistance mutations, establishing the Sec61 translocon as the direct decatransin target in fungal and mammalian cells with high certainty. It was shown that decatransin also inhibits co- and posttranslational ER translocation in both yeast and mammalian cells. This activity was shown to be independent of the sequence of the ER targeting signal peptide, which suggests that unlike cotransin, decatransin would act as a substrate-nonselective inhibitor of Sec6154. Based on the ample mutagenesis data, it appears that binding of decatransin to Sec61 is similar, but distinct compared to cotransin.
2.6. Ipomoeassin
Ipomoeassin F (Figure 3) is one of the recent NPs described to specifically interfere with ER protein import. Ipomoeassins A-E are related amphiphilic glycolipids produced by the morning glory family plant Ipomoea squamosa, which were identified to inhibit proliferation of A2780 human ovarian cancer cells with a range of potencies55. Later, a new member of the ipomoeassin family, ipomoeassin F was discovered56 and shown to be the most potent cytotoxin of the family with single-digit nanomolar IC50 inhibition against many cancer cell lines57. Ipomoeassins contain a unique glycoconjugate structure, which prompted many groups to pursue its total synthesis57,58. Having a facile synthesis method available allowed design of several ipomoeassin F probe molecules and attempts to directly identify its cellular target. Despite containing a potent Michael acceptor electrophile, it appears that ipomoeassin F does not bind its target covalently59. However, immobilizing ipomoeassin F allowed attempts to identify its interaction partner by affinity chromatography, which revealed an approximately 40 kDa protein species which was identified as Sec61α59. This finding was supported by competitive photo-cotransin binding experiments and biochemical experiments demonstrating that also ipomoeassin F inhibited protein ER import apparently in a substrate-nonselective manner. In cells, ipomoeassin F inhibits the production of secreted and glycosylated proteins without affecting production of cytosolic proteins. Heterozygous expression of earlier characterized Sec61α point mutations conferring resistance to cotransins, apratoxin and mycolactone18,27,43 revealed broad resistance to ipomoeassin F cytotoxicity demonstrating the connection between ER import inhibition and cytotoxic effect for ipomoeassin F. The observed difference in patterns of resistance mutations against ipomoeassin F and other Sec61 inhibitors suggests it binds a similar site on Sec61α, but in a distinct manner.
2.7. Coibamide A
The latest NP to join the assorted collection of potent and specific inhibitors of Sec61 is coibamide A. Coibamide A is an N-methyl-stabilized lariat depsipeptide that was originally isolated from a Caldora species marine cyanobacteria from Panama60. Coibamide A was observed to inhibit cell cycle progression of human glioblastoma cell lines and decrease their migratory and invasive capacity. Further, in xenograft models of glioblastoma, coibamide A was shown to inhibit tumor growth, although within a narrow therapeutic window61. In addition, coibamide A induces mTOR-independent macroautophagy in mammalian cells61,62. Development of several total synthesis methods have recently resulted in refinement of the absolute configuration of the natural product63–65 and allowed pursuing its cellular target.
Coibamide A potently inhibits biogenesis of the integral membrane protein vascular endothelial growth factor receptor 2 (VEGFR-2) and its secreted ligand VEGF-A61. Very recent work explored coibamide A structure-activity relationships to develop a potent coibamide A photoaffinity probe66. Photocrosslinking experiments with this probe in isolated ER microsomes demonstrated specific crosslinking to a 40 kDa ER protein, which can be competed by known Sec61 ligands apratoxin A and mycolactone. Pulse-chase metabolic labeling experiments demonstrate that coibamide A potently inhibits cellular production of secreted and glycosylated proteins without impacting production of cytosolic proteins, consistent with substrate-nonselective inhibition of Sec61-mediated ER protein import. This notion was further supported by ER import inhibition of multiple classes of Sec61 substrate proteins in biochemical experiments using purified ER microsomes and in vitro translation66.
However, despite having apparently identical substrate-nonselective mechanisms for inhibiting Sec61 ER import, the three structurally diverse Sec61 inhibitors coibamide A, apratoxin A and mycolactone exhibit differential resistance to many Sec61 resistance mutations. Finally, a comparative analysis of the cytotoxic effects of these three Sec61 inhibitors towards cancer cell lines in the NCI-60 cancer cell line panel revealed a surprising difference in the cell types that were potently sensitive to the different Sec61 inhibitors66. This is highly surprising considering that all of the compounds target Sec61 at the same site with a seemingly identical biochemical mechanism, and further suggests that modifications of Sec61 inhibitors may allow a surprising degree of cell type selectivity against different histological cancer cell types. In support of this notion, modifications to the natural structures of apratoxin A and coibamide A have already yielded new inhibitors with reduced general cytotoxic effects while retaining their original anti-tumor efficacy in tumor xenograft models30,32,67.
3. Inhibitors of secretory protein modification and maturation
3.1. Tunicamycin
The tunicamycin antibiotics (Figure 4) were identified from Streptomyces lysosuperficus and exhibit potent antibacterial activity against various Gram-positive bacteria68. Tunicamycins are nucleoside antibiotics that contain a uracil base attached to a C11 tunicamine sugar, glycosylated at C11 by a GlcNAc sugar and N-acylated at C10 by a C12-C15 fatty acid. Tunicamycins exhibit broad antimicrobial activity against a range of Gram-positive bacterium particularly Bacillus genus members (MIC 0.1–20 μg/ml), but also exhibit significant cytotoxicity towards mammalian cells as a result of their ability to inhibit biogenesis of N-linked glycoproteins69,70. Interest in tunicamycins later led to the discovery of their biosynthetic gene cluster71. In bacteria, the direct target of tunicamycin, MraY, is a translocacase, which catalyzes the first step during the biosynthesis cycle of bacterial peptidoglycan. Tunicamycin reversibly inhibits MraY with a Ki of 0.6 μM72. Structure of tunicamycin bound to E.coli MrayY73 and its eukaryotic target enzyme, GlcNAc-1-phosphate transferase74,75 required for N-linked glycoprotein biosynthesis were reported. These structures revealed a clear difference in tunicamycin interactions between MraY and GPT and this allowed rapid structure-guided design of a modified tunicamyin analog with high selectivity for MraY over GPT (IC50 640 nM versus 15 μM, respectively)74, suggesting the possibility to develop tunicamycin as a selective antimicrobial agent.
Figure 4. Inhibitors of secretory protein modification and maturation.
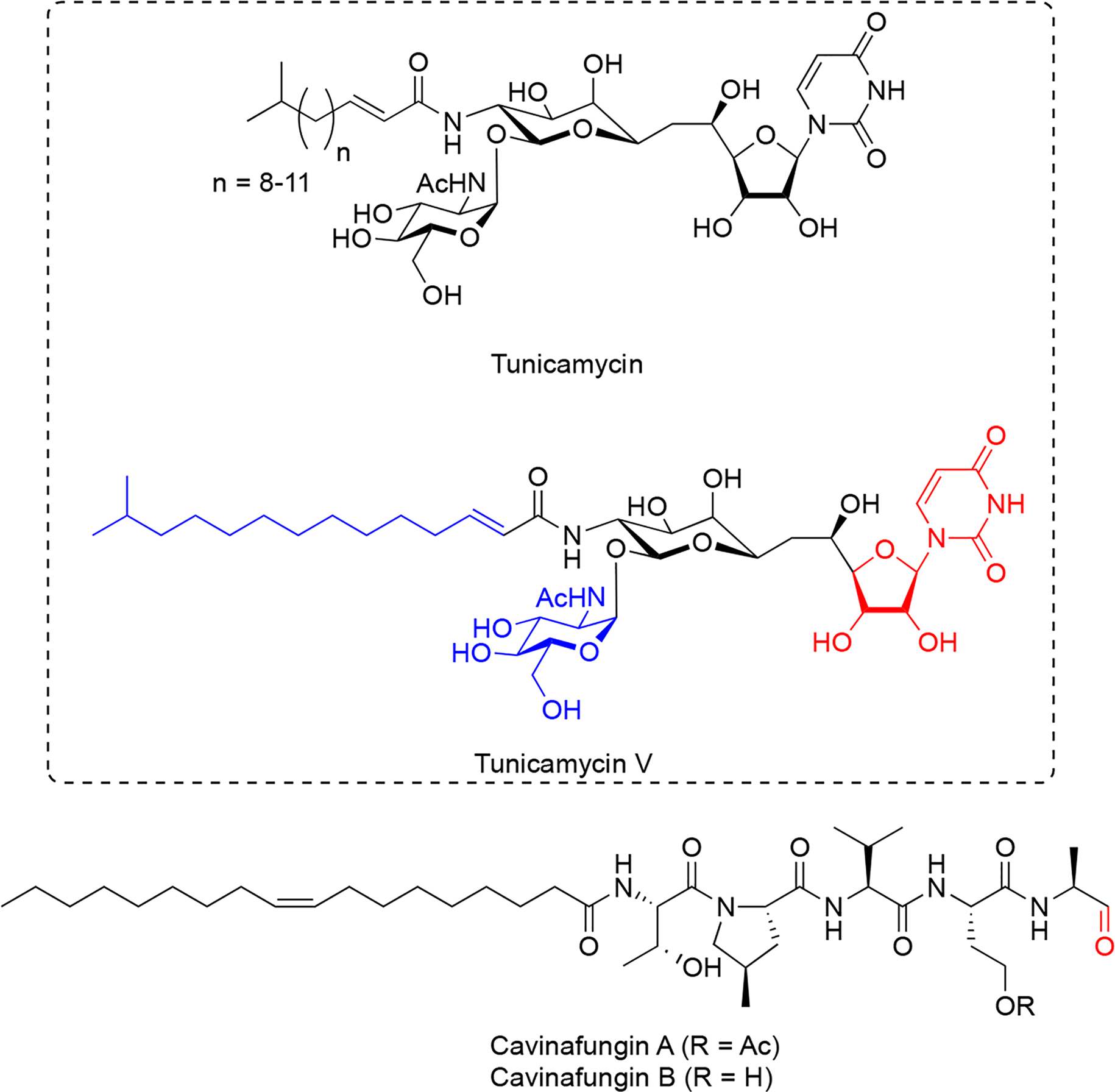
Structures of N-glycosylation inhibitors, tunicamycins, and signal peptide processing inhibitors, cavinafungins. Red color indicates key moieties for activity. Structural features in blue are contributory to activity.
Very recently, the total synthesis of tunicamycin V (Figure 4) has been described76, which led to synthesis of a series of tunicamycin analogs and generation of a structure-activity dataset for tunicamycin’s antibacterial activity77. Inhibition of N-glycosylation by tunicamycin treatment has been shown to overcome chemoresistance in a multidrug-resistant gastric cancer cell line78. Because of its ability to robustly induce ER stress resulting from rapid misfolding of newly synthesized glycoproteins within the ER lumen, tunicamycin has become a powerful probe for studying ER protein misfolding and ER stress pathways79.
3.2. Cavinafungin
Cavinafungins (Figure 4) were initially extracted from cultures of fungus Colispora cavincola isolated from plant matter from Argentina and demonstrated to exhibit antifungal activity against many fungal species.80. Determination of the absolute configuration demonstrated that cavinafungins are linear lipopeptides that contain a terminal aldehyde residue80 and exhibit broad antifungal activity against Candida species (MIC 0.4–4 μg/mL) and a reduced potency against the filamentous fungi Aspergillus fumigatus (MIC 8 μg/ml). Cavinafungins A and B are nearly identical lipopentapeptides, which only differ in acetylation of a serine sidechain (Figure 4). A one-pot solid-phase total synthesis of cavinafungin B has recently been reported81.
Cavinafungin A was discovered to inhibit replication of dengue and Zika viruses with a potency ranging from 4 to 400 nM IC50, and strongly depending on presence of the terminal aldehyde, suggesting a covalent mechanism of inhibition82. A genome-wide siRNA screen in mammalian cells suggested the ER signal peptidase complex as a possible target of cavinafungin A and this was further supported by chemogenetic screens carried out in yeast cells. A mutagenesis approach focused on ER signal peptidase subunit genes revealed a range of mutations conferring cavinafungin A resistance exclusively in the catalytic signal peptidase subunit SEC11, whereas mutations in the other three subunits were not observed, suggesting that cavinafungin A directly targets the SEC11 subunit in the signal peptide binding cleft, possibly directly competing with substrate binding82. Cavinafungin A prevents dengue virus replication at considerably lower (IC50 1–5 nM) concentrations compared to what is required to induce host cell cytotoxicity (CC50 800–18000 nM)82. Further, because most flaviviruses appear to depend on the ER signal peptidase activity for their replication, cavinafungins could be attractive lead compounds for development of broadly acting therapeutics against flavivirus infections.
4. Inhibitors of protein trafficking
4.1. Brefeldins
Brefeldin A (BFA, Figure 5) is a fungal natural product lactone that was originally isolated in the 1950s from Penicillium decumbens83. More recently, several analogues have been identified from fungi of the genus Penicillium derived from soil and marine sediments, including the marine Penicillium spp. PSU-F4484 and DT-F2985, and terrestrial Penicillium janthinellum86. The recent synthesis and discovery of new brefeldin A analogs has been reviewed in detail87. Numerous analogues were synthesized to improve drug-like properties and various prodrug strategies were considered as well (Figure 5)87.
Figure 5. Structures of brefeldin A, and natural and synthetic prodrug analogues that retained potent activity.
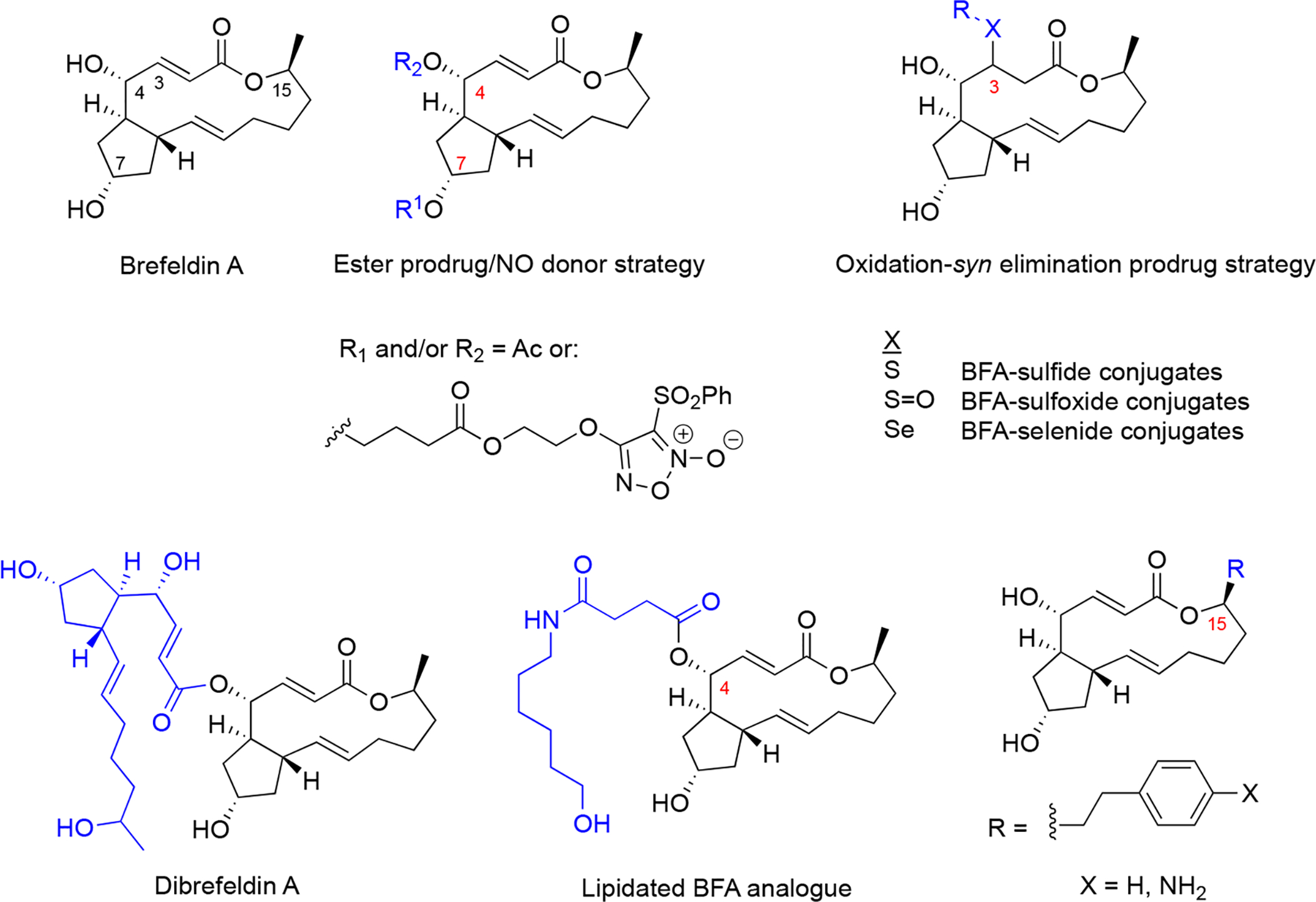
Sites of modification of the core skeleton to develop prodrugs are numbered and modifications indicated in blue.
BFA is a classical tool compound used in cell biology studies as a secretion inhibitor because of its action on the Golgi apparatus and endosomes88,89. The elucidation of its mechanism of action led to the concept of interfacial inhibition as a way to stabilize protein-protein interactions (PPI) instead of inhibiting PPI90 (Figure 6). BFA displays specific effects on membrane trafficking by affecting small G-proteins of the ADR-ribosylation factor (ARF) family, which regulate cellular traffic by GDP/GTP cycling and consequent capacity to interact with GTP-bound effectors. ARF proteins are N-myristoylated to promote membrane association when GTP bound89. The activation of ARF is facilitated by Arf-guanine nucleotide exchange factors (ArfGEFs) which are characterized by a conserved catalytic domain consisting of ~200 amino acids, termed Sec7 domain, involved in Arf binding91. BFA is an uncompetitive inhibitor of ARF at the Golgi, blocking the ArfGEF mediated Arf-GDP/GTP guanine nucleotide exchange reaction92.
Figure 6. ARF GTPase activation by GDP/GTP exchange factors to regulate vesicle formation and unusual uncompetitive mechanism of action of BFA, which led to the concept of interfacial inhibition, where protein complexes are stabilized near interacting interfaces.
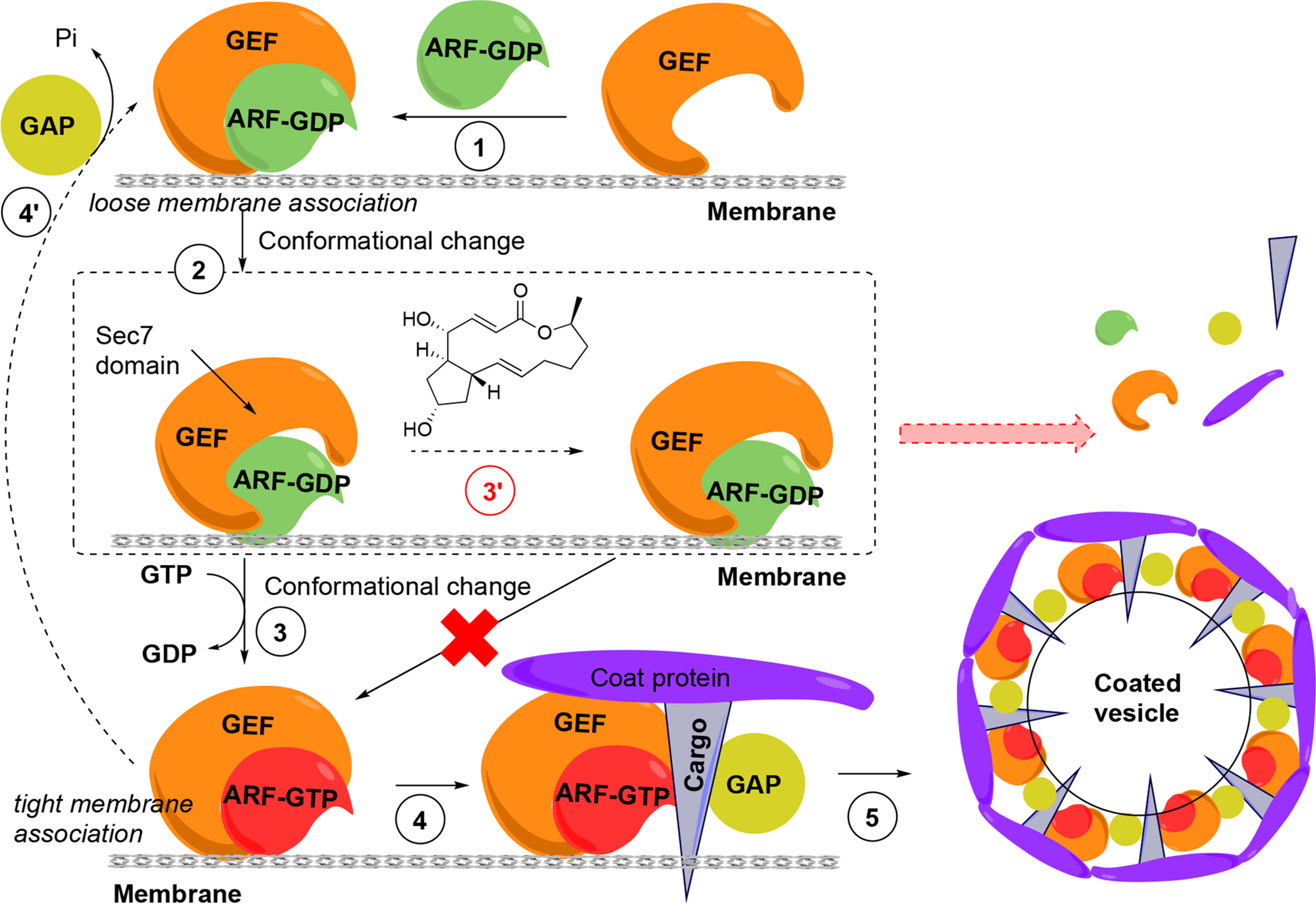
(1) GDP-bound ARF associates with GEF. (2) Conformational change of ARF is required to stimulate GDP dissociation. (3) GEF catalyzes the activation of ARF through GDP/GTP exchange and the complex now tightly associates with the donor membrane. (4) ARF-GTP recruits effectors, including coat components (coat proteins, cargo) from the cytosol, and also GTPase activating protein (GAP), which usually catalyzes the inactivation of ARF-GTP, closing the GDP-GTP cycle (4’). (5) GAP appears to be inactive when bound to the coat-containing complex, presumably driving coat polymerization, budding and vesicle formation. GAP becomes active in this conformational complex state, leading to GTP hydrolysis coat and GAP release and the uncoated vesicle is able to fuse with an acceptor membrane (not shown). (3’) In the presence of BFA the low-affinity complex is trapped through binding of BFA at the interface of ARF-GDP and the Sec7 domain of GEF. Binding of BFA to the Arf-GDP-Sec7 domain complex blocks GDP/GTP exchange on ARFs and consequently vesicle coat recruitment and vesicle formation. The ultimate result is the release of coat components into the cytosol and prevention of membrane trafficking.
BFA does not bind to GEF alone but to the Arf-GDP-Sec7 domain complex (Figure 6), sequestering GEFs in an abortive conformation and acting as a “dominant negative”93. In a remarkable mechanism, BFA inserts into the hydrophobic pocket transiently formed in the Arf-GDP-Sec7 complex, preventing conformational changes required for nucleotide exchange and requisite catalytic contact. The crystal structure has been solved94,95, providing the basis for the discovery of novel GEF inhibitors and possibly modulate their specificity. Because BFA contacts both ARF-GDP and ArfGEF in this complex, BFA has specificity for a subset of both ArfGEFs and ARF GTPases96. ArfGEF family members, 15 of which are encoded in the human genome, can be classified based on their BFA sensitivity.
GTP bound Arf recruits coat proteins including the COPI complex to induce budding of coated carrier vesicles (Figure 6). ArfGEFs confer spatial and temporal specificity to vesicular transport90. Phenotypically, BFA induces the release of vesicle coat proteins, including COPI and clathrin coat subunits, into the cytosol. BFA affects integrity of subcellular compartments including Golgi apparatus and endosomes, inhibiting the secretion and re-distributing Golgi membrane proteins to the endoplasmic reticulum (ER). Of note, several BFA effects are species specific, which requires caution when interpreting results in certain systems88,89.
4.2. Discovery of GEF and other secretory pathway inhibitors inspired by brefeldin A
BFA provide the inspiration for structure-based screening, which identified LM11 (Figure 7) that targets BFA-insensitive GEFs, including ARNO (CYTH2)97. Both BFA and LM11 affect activation of class I ARFs (ARF1–3) and class II ARFs (ARF4 and 5) but not ARF6. Phenotypic screens of endocytic and secretory transports yielded Secin H3 (Figure 7), which also bind to Sec7 of ArfGEFs. Secin H3 inhibits cytohesins, which leads to insulin resistance98. Golgicide A (Figure 7) was discovered to selectively inhibit GBF1, which is the ArfGEF that activates ARF1 and recruits COPI to the cis-Golgi membranes. The mechanism of action by BFA might be generally exploited to discover more selective drugs, and not only at the level of ARF and GEFs. Similarly, inhibitors of other GTPases besides ARF have been identified, including inhibitors of RhoGEFs activation. Ras GTPases might also be targeted in a similar fashion96.
Figure 7.
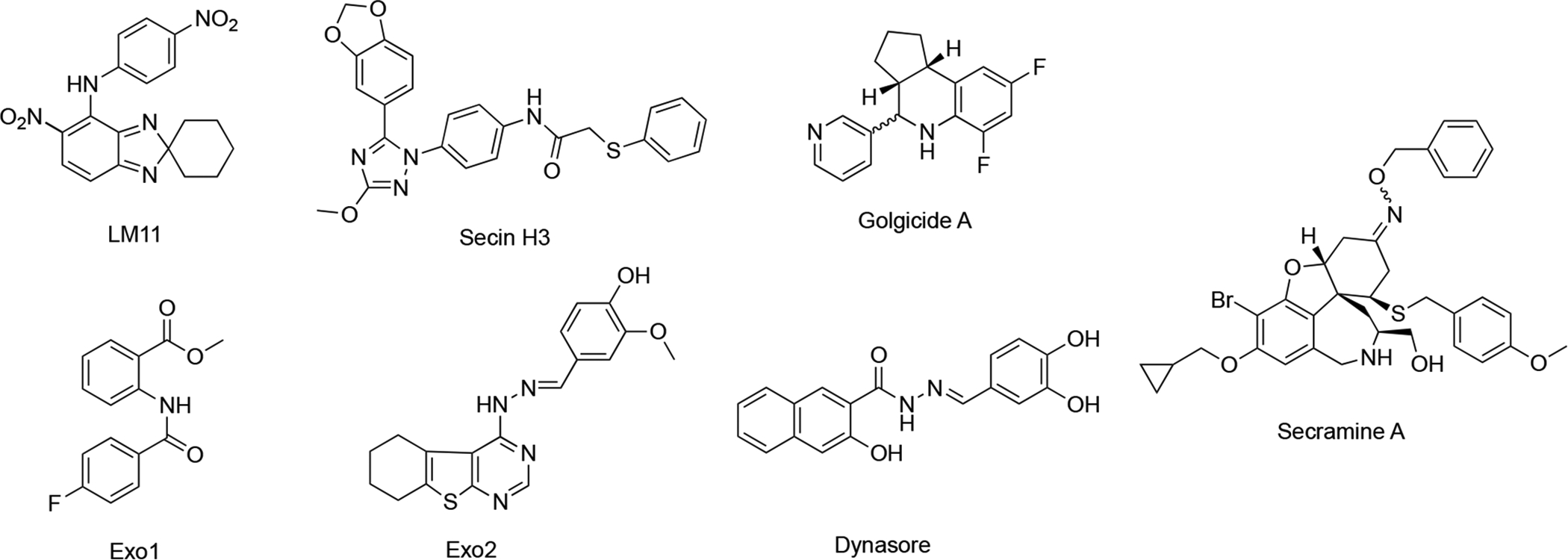
BFA-inspired secretory pathway inhibitors via structure-based or phenotypic screening.
In other phenotypic screens, Exo1 and Exo2 (Figure 7) were identified using an image-based screens99,100. Dynasore (Figure 7), a cell permeable inhibitor of dynamin as probe for dynamin-dependent clathrin coat formation was discovered to block synaptic vesicle endocytosis101,102. Secramine (Figure 7) was discovered as a potential new tool to dissect Cdc42 function from that of other Rho GTPases103.
5. Natural products that indirectly influence secretory capacity of the ER
5.1. SERCA inhibitors: thapsigargin and basiliolides
Thapsigargin (Figure 8) is a structurally complex sesquiterpene lactone that was originally isolated in 1978 from roots and fruits of the plant Thapsia garganica and observed to potently liberate histamine from mast cells of the skin104. In early 1990’s several studies demonstrated the ability of thapsigargin to induce apoptosis in several cancer cells105–108. However, later studies demonstrated that thapsigargin is similarly cytotoxic against malignant and normal cells, limiting its therapeutic potential. Thapsigargin induces cytotoxicity by elevating cytoplasmic Ca2+ levels at subnanomolar concentrations by inhibiting the sarco-endoplasmic reticulum Ca2+-ATPases (SERCAs)109,110. The X-ray crystal structure of SERCA bound to thapsigargin revealed the thapsigargin structural conformation and detailed interactions with SERCA111. In the crystal structure, a large rearrangement of the SERCA channel was observed, providing a structural explanation for how thapsigargin binding induces a passive leakage of ER lumenal Ca2+ into the cytosol. Despite many challenges, several synthetic routes for synthesis of thapsigargin and analogues have been developed by several laboratories (reviewed in112).
Figure 8. Structures of SERCA inhibitors (thapsigargin and its prodrug mipsagargin, and basiliolide A1) and the K+ ionophore valinomycin as indirect modulators of secretory capacity though cation import regulation.
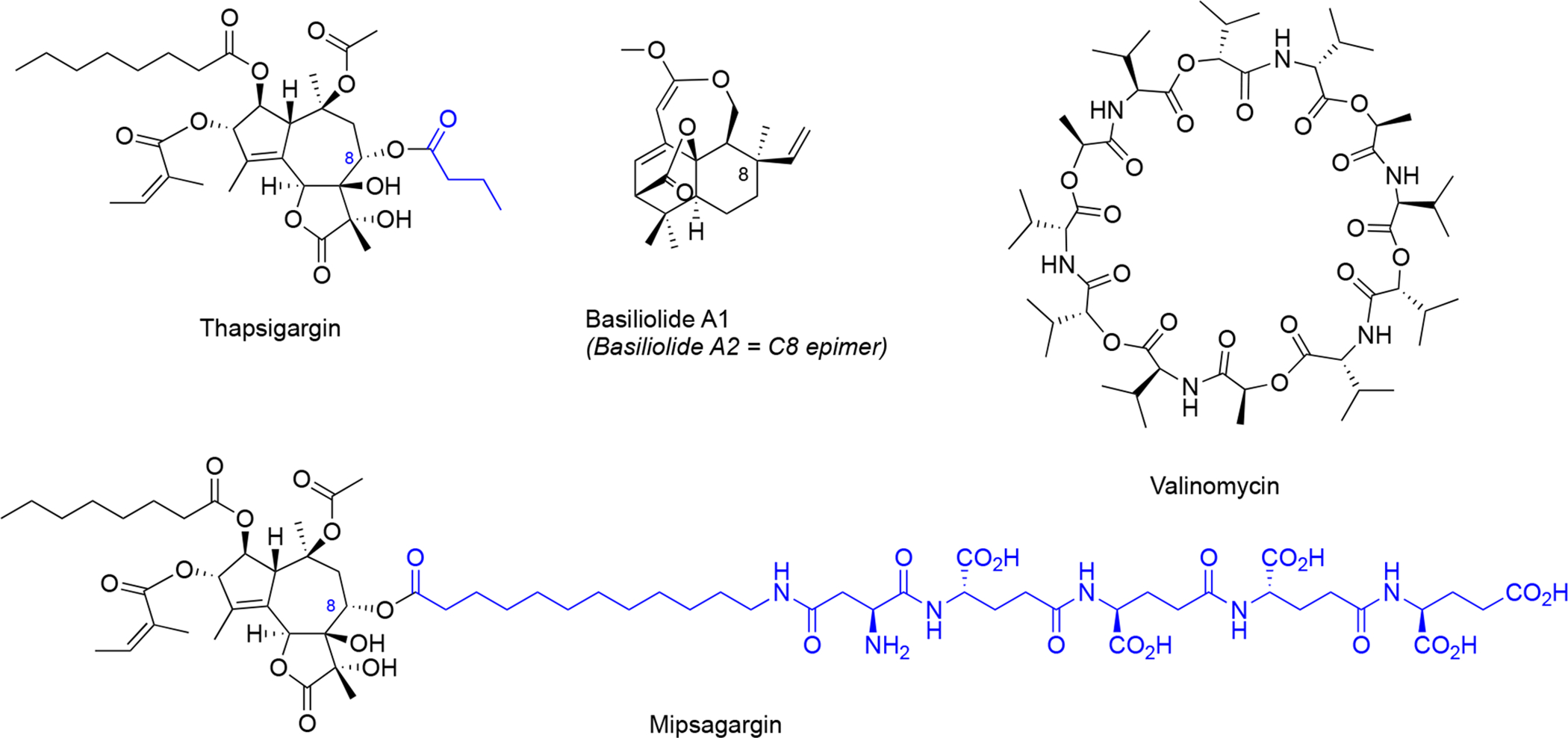
Structural differences between thapsigargin and mipsagargin are indicated in blue.
The generally cytotoxic nature of thapsigargin has prevented its development as an anticancer agent, but recently its use as a cancer-targeting prodrug has been explored. In this strategy, a polypeptide was attached to the thapsigargin C8 position (Figure 8), which targets the fusion compound to metastatic deposits of androgen independent prostate cancer113. This fusion peptide is designed to be cleaved by a prostate tissue-specific protease (PSA), which is only produced in high levels in the extracellular space of metastatic prostate cancer cells. After extracellular cleavage, the released hydrophobic thapsigargin can enter the cancer cells and promote Ca2+-mediated cell death. Importantly, the prodrug mipsagargin (Figure 8), is inactive against SERCA, which helps to minimize off-target cellular cytotoxicity. Mipsagargin is currently investigated in late stage clinical trials against prostate cancer, progressive glioblastoma and hepatocellular carcinoma114.
Later, basiliolides, a class of sesquiterpenes also isolated from T. garganica, were identified as structurally distinct SERCA inhibitors in sea urchin eggs115 and subsequently in mammalian cells116. Despite triggering ER Ca2+ release, basiliolide A1 (Figure 8) does not trigger apoptosis similarly as thapsigargin, suggesting it inhibits the SERCA pump with a distinct mechanism116. A synthetic strategy for basiliolides has been reported117.
5.2. Potassium ionophore: valinomycin
Valinomycin (Figure 8) is a cyclic dodecadepsipeptide isolated from several Streptomyces species118,119. Valinomycin acts as a highly K+-specific ionophore120 making biological membranes selectively permeable for K+ 121.Valinomycin was discovered to prevent upregulation of BIP expression as a response to ER stress122. The valinomycin biosynthetic gene cluster has been identified123,124. Interestingly, valinomycin was recently identified in a phenotypic screen for substrate-selective small molecule inhibitors of prion protein125. In this study, valinomycin surprisingly influenced biogenesis of hamster prion protein without effects on biogenesis of human prion protein and this difference was attributed to differences in the sequence of the prion protein ER targeting signal peptide125. However, how perturbation of cellular K+ homeostasis leads to signal peptide-dependent inhibition of prion protein ER insertion remains unknown.
5.3. UPR modulators
At least some of the Sec61 inhibitors discussed above in sections 2.1–2.7, which do not enter the ER, were shown to downregulate the UPR by depleting some of the key proteins (BIP/GRP78 and others). In turn, activation of the UPR upon accumulation of unfolded/misfolded proteins in the ER causes ER stress and also affects protein secretion, along with other processes, depending on the specific branch of UPR being modulated. In mammalian cells, there are three UPR signaling branches: 1) IRE1α-XBP1, 2) PERK-ATF4-CHOP, and 3) ATF6 cassette126. GRP78 regulates all three branches (Figure 9).
Figure 9. The branches of the ER stress pathways.
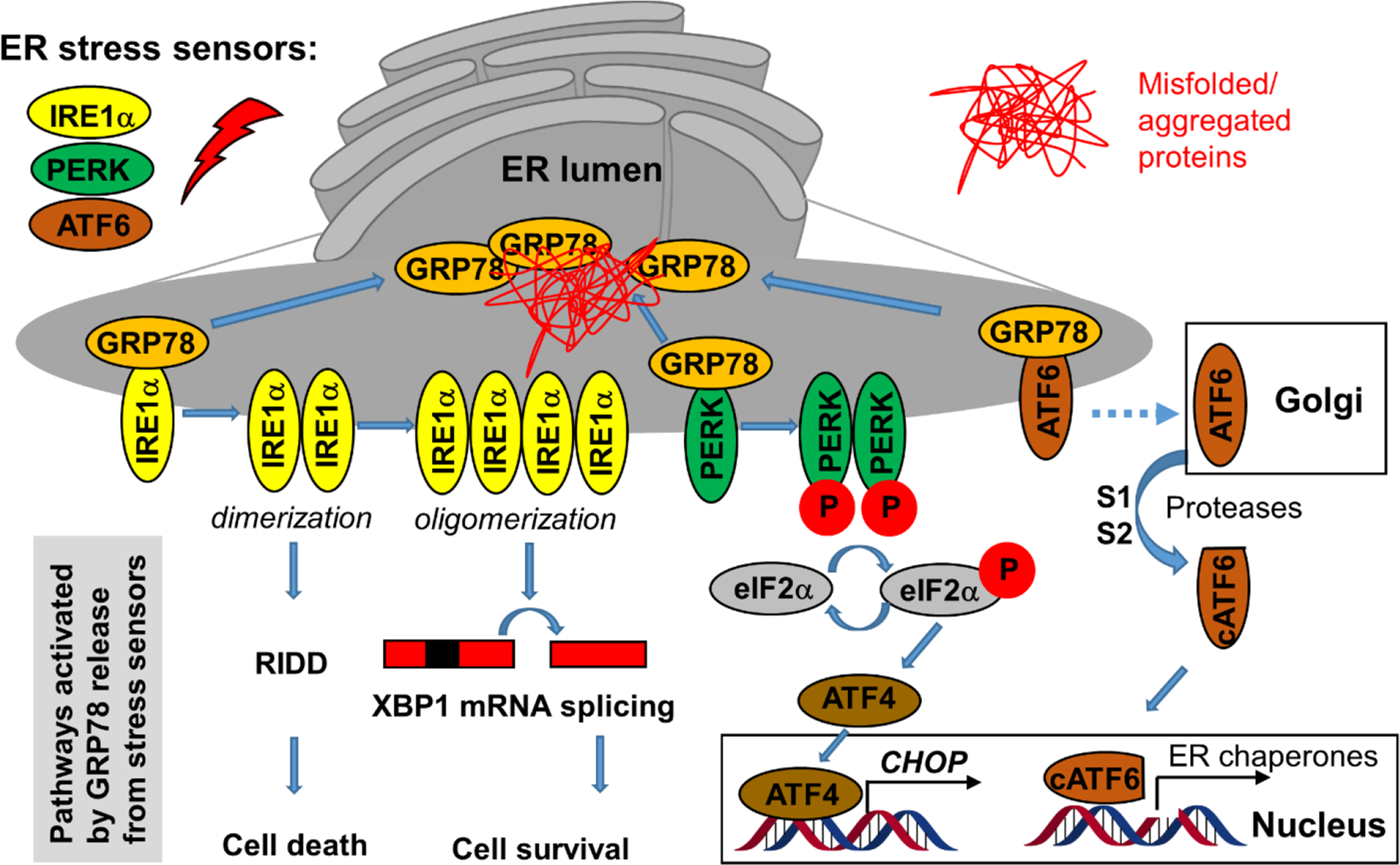
Under unstressed conditions, GRP78 associates with the three membrane-bound ER stress sensors but upon activation by unfolded proteins GRP78 dissociates from them, which triggers parallel pathways that can be pharmacologically modulated, especially the IRE1α and PERK pathways (see text).
There is a delicate balance whether UPR induction causes apoptosis or cellular protection, depending on pathway/branch and target but also extent/duration of activation etc., and modulators in both directions might be of therapeutic relevance126.
5.3.1. Inhibitors of XBP1 mRNA splicing: trierixins and toyocamycin
IRE1α is an ER-resident transmembrane kinase/endoribonuclease (RNase) that associates with GRP78. Upon dissociation from GRP78, IRE1α oligomerizes and autophosphorylates, which in turn activates the RNase activity and subsequent XBP1 mRNA splicing to create an active XBP1 transcription factor. XBP1 mRNA splicing is believed to promote cell survival126, particularly of multiple myeloma cells127. However, upon dimerization (instead of oligomerization), IRE1α can also cleave ER-associated RNAs through Regulated IRE1-Dependent Decay (RIDD) which rather triggers apoptosis126, necessitating tools to direct the downstream phenotypic response to ultimately develop therapeutics.
Using an XBP1-luciferase reporter screen in HeLa cells, trierixin and quinotrierixin (Figure 10)128,129 were discovered from Streptomyces spp. as inhibitors of XBP1 activation by thapsigargin- and tunicamycin-induced ER stress128,129. This activity was further validated by monitoring endogenous XBP1 splicing using RT-qPCR at nanomolar concentrations (IC50 30 ng/mL). However, these and related compounds of the triene-ansamycin family of natural products possess pleiotropic biological activities, limiting their use as potential probes. Nevertheless, this activity might contribute to the antitumor properties and partially explain the inhibition of EGFR signaling.
Figure 10. Inhibitors of XBP1 mRNA splicing.
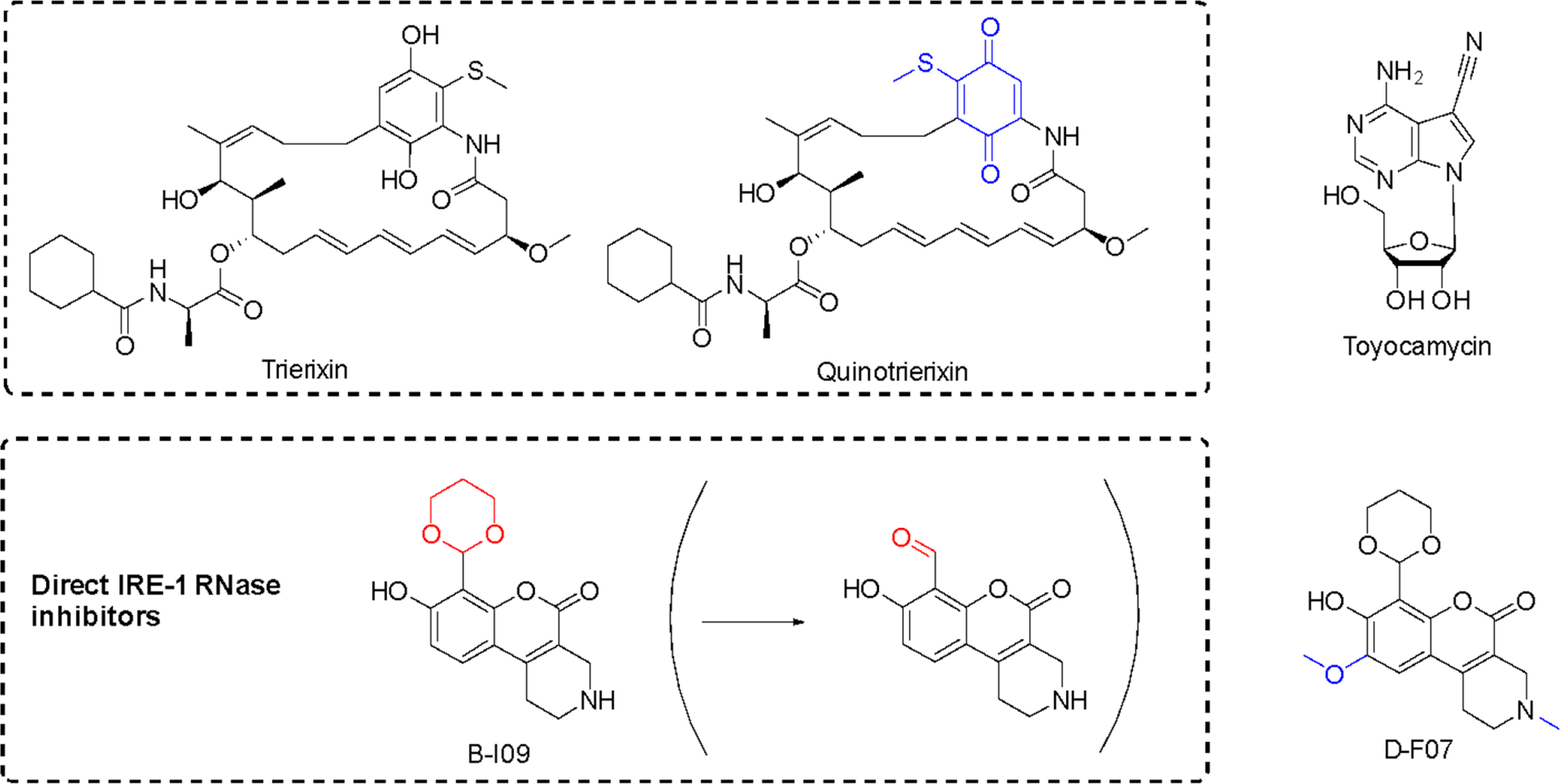
Structural changes relative to respective parent compounds are indicated in blue and the masked and reactive group for prodrug B-I09 in red.
Toyocamycin (Figure 10) is produced by an actinomycete strain and identified as an inhibitor of ER-stress induced XBP1 activation127. This natural product selectively affects the IRE1α-XBP1 branch without impinging on the activation of PERK and ATF6. It also does not affect IRE1α phosphorylation, acting more downstream at the level of XBP1127. While toyocamycin is also an RNA synthesis inhibitor, the XBP1 splicing inhibition occurs at ~150 fold lower concentrations (IC50 12 μM vs 0.08 μM in HeLa cells). SAR studies indicated the importance of the adenine moiety for the splicing inhibition. MM cells with higher levels of spliced XBP1 were particularly susceptible to toyocamycin127.
Recently, a potent covalent natural product-like IRE1α RNase inhibitor was developed based on a tricyclic chromenone scaffold, targeting the RNase domain of IRE1. This compound (B-I09) contains a masked aldehyde functionality acting as a prodrug to target lysine 907 in the RNase domain of IRE1α through specific Schiff base formation130. The efficacy of B-I09 in B-cell cancer provided evidence that the IRE-1/XBP1 pathway reduces leukemic cell survival. In addition to multiple myeloma, these inhibitors show potential to treat mature B cell leukemia and lymphoma. Its application can further be extended to other c-Myc driven cancers131. A fluorescent version of the inhibitor (D-F07) was recently reported that enables precise temporal control for the prodrug activation132.
5.3.2. Activators of PERK pathway: cephalostatin and ritterostatin GN1N
The bis-steroidal pyrazine spiroketals, ritterazines and cephalostatin (Figure 11), are natural products derived from marine tunicates and tube worms133–135. Cephalostatin 1 (Figure 11) was found to activate the ASK1-JNK pathway and induce apoptosis via an unusual caspase 4-dependent mechanism (upstream of caspase 9) linked to UPR induction. In leukemia cells it was first shown to induce GRP78 expression and transiently and rapidly increase levels of cell death modulator CHOP/GADD153 and phosphorylation levels of EIF2α by PERK136.
Figure 11. PERK pathway activators with differential selectivity.
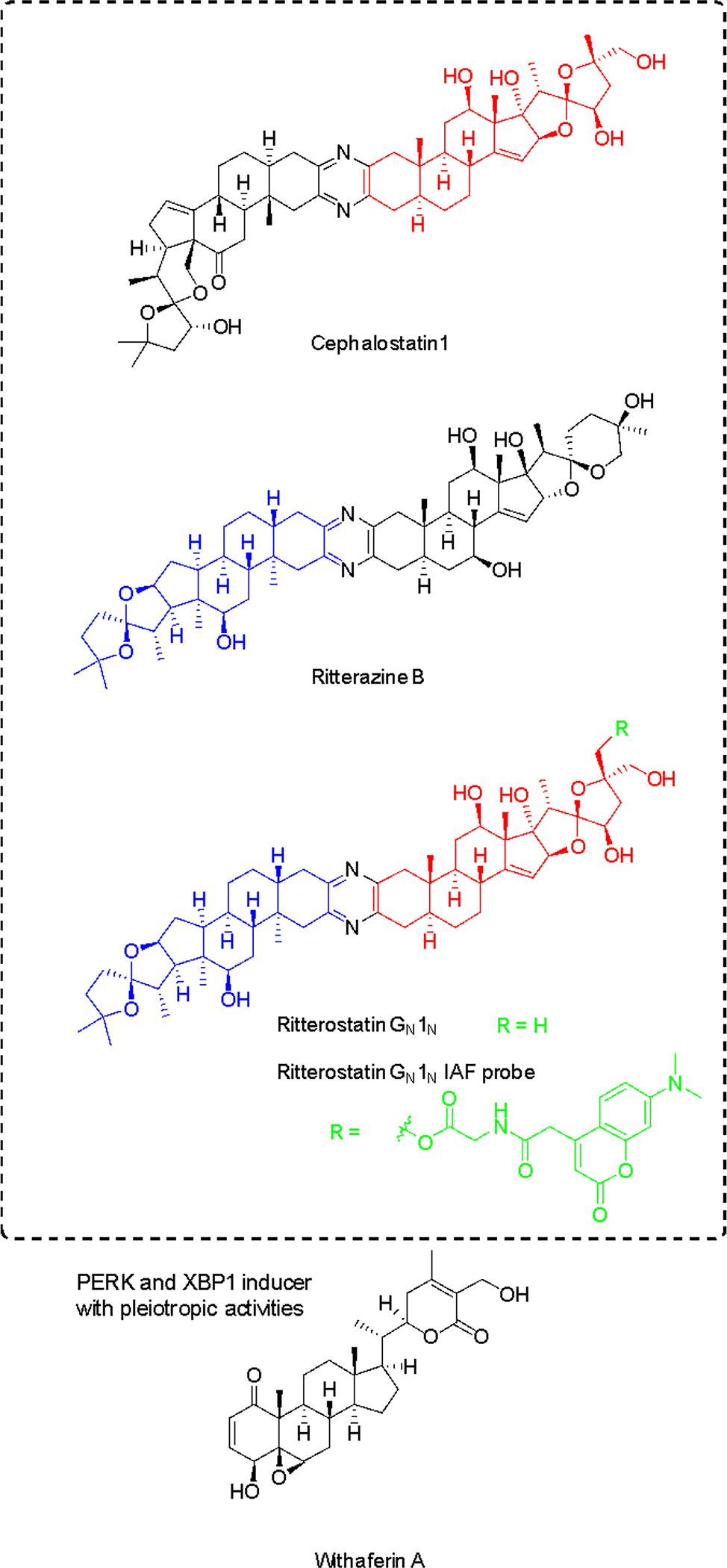
Cephalostatin-ritterazine hybrid was shown to have selectivity for the PERK branch, while withaferin A is non-selective. For ritterostatin GN1N, red and blue colors indicate cephalostatin 1 and ritterazine B fragments, respectively. Probe position and label are indicated in green.
While these compounds have been found to target oxysterol-binding protein (OSBP) and related proteins137, there is no obvious correlation to ER stress induction and apoptosis136. Using immunoaffinity chromatography with ritterostatin GN1N (ritterazine-cephalostatin hybrid, Figure 11), the ER-resident chaperone GRP78 (BIP) was identified as the dominant specific intracellular target of the compound, along with other HSP70 heat shock proteins to minor degrees138. ITC with recombinant protein and hybrid GN1N (KD 190 nM) or cephalostatin (KD 679 nM) validated the direct interactions between GRP78 and this compound class. The binding site was identified to be the C-terminal domain by testing both domains of GRP78 individually. The N-terminal ATPase site was not targeted, consistent with the lack of inhibition of the ATPase activity. However, KD values for other HSP70 proteins are similar and GRP78 selectivity confirmed to be a result of the predominant localization in the ER138. Obviously the relevant pharmacological effects are the result of a combination of affinity and subcellular localization.
GRP78 binding has been linked to UPR induction and the most affected branch was found to be the PERK arm since EIF2α phosphorylation was increased at lower concentration compared with downstream targets of the IRE1α arm (see above), including XBP1 and ATF4 (Figure 9)138. GRP78 as a marker of the UPR was also strongly increased in HCT116 cells. While not confirmed yet, it is plausible that the induction of the UPR via the PERK arm is responsible for the antitumor and apoptotic activity of this compound class138, although a net effect from multiple target interactions might also be likely.
5.3.3. PERK and XBP1 mRNA splicing inducer with pleiotropic activities: withaferin A
Withaferin A (Figure 11) isolated from the medicinal plant Withania somnifera is a steroidal lactone that exerts its anticancer activity in various cell types via multiple mechanisms. Like for cephalostatins, the apoptotic activity is due to caspase 4 activation, characteristic for several ER stressors139. However, there is no selectivity among the UPR branches as the compound induced GRP78, CHOP and ATF4 levels as well as XBP1 mRNA splicing and eIF2α phosphorylation at the same concentrations and time points in a given cell line139. Withaferin A has also been shown to affect many other signaling pathways, impairing NFκB, Akt and Notch, while inducing p35 MAPK, FOXO3a, Par-4, Bim and death receptor 5 (DR5) (see references in139). The UPR and ER stress induction appears to be a downstream event that then triggers apoptosis. Furthermore, it was shown to induce autophagy that gets impaired at a later stage and inhibits proteasomal degradation simultaneously140. The PERK arm can activate autophagy in most cancers.
5.3.4. Kinase Inhibitors of IRE1α and PERK: ATP mimics
ATP-competitive inhibitors are ATP mimics and can therefore be considered natural product inspired. Type I and type II kinase inhibitors (Figure 12) that target different conformations can be used to modulate inhibit IRE1α and modulate RNase activity in opposing fashion141. Type I inhibitors stabilize the active conformation and allosterically activate the adjacent RNase domain, stimulating XBP1 mRNA splicing in the absence of ER stress. In turn, type II inhibitors, which stabilize an inactive conformation of the ATP-binding site, and thereby suppress RNase activity and inhibit XBP1 mRNA splicing. However, there is no evidence that the effects on IRE1α by kinase inhibitors have UPR-related anticancer activity126.
Figure 12. Representative ATP-competitive IRE1α and an advanced PERK inhibitor but with relevant RIPK1-inhibitory activity.
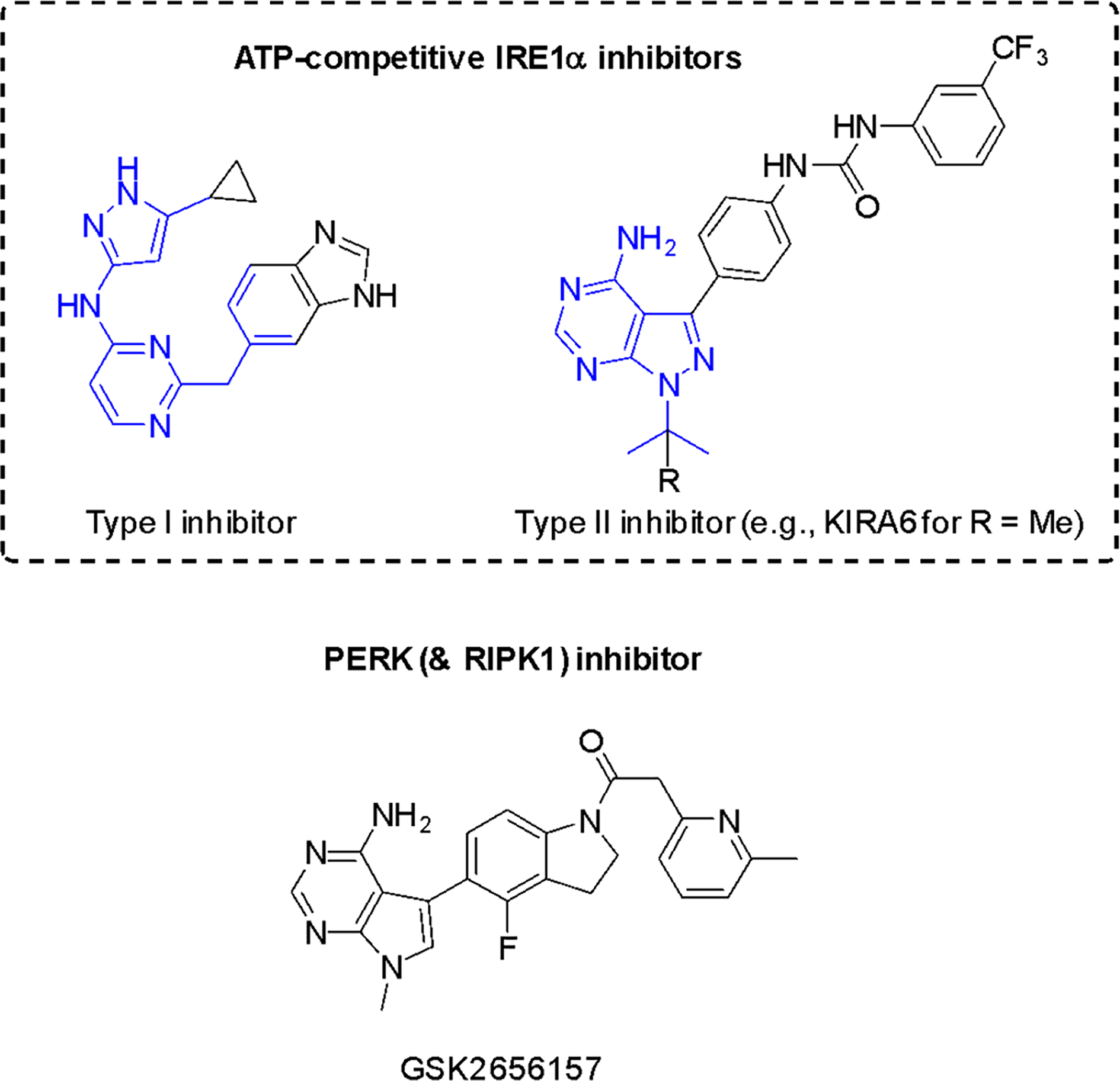
Adenine-pocket interacting units (mimics) are indicated in blue.
The PERK branch can be selectively inhibited by GSK2656157 and related compounds142. These compounds prevent ER-stress induced autophosphorylation of PERK and consequently inhibit EIF2α substrate phosphorylation in vitro143. However, in vivo data did not provide evidence of PERK inhibition; specificity is an issue with the current lead compound and RIPK1 inhibition is probably more relevant144.
In addition to natural products and natural product-like compounds, there are now numerous synthetic pharmacological agents that target the various branches of the UPR as positive and negative regulators, and their efficacy is being assessed in various disease states where unfolded protein accumulation plays a critical role145.
5.4. ERAD
ER-associated Degradation (ERAD) alleviates ER stress and maintains ER homeostasis. It is a complementary mechanism to autophagy which can also clear protein aggregates generated by improper protein folding in the ER. Proteins commonly undergo N-glycosylation upon ER entry and this multi-step process requires the action by several enzymes. The resulting N-glycan structures promote glycoprotein folding, while mannose-trimmed structures are directed towards ERAD126. In mammalian cells, ER Degradation-Enhancing α-Mannosidase-like proteins (EDEMs), ER Mannosidase I (ER ManI) and Osteosarcoma amplified-9 (OS-9) ensure the routing of misfolded proteins, while p97 ATPase is required for functional ERAD146.
The proteasome in the cytosol takes part in ERAD at a later stage by ubiquitin-dependent degradation. Several proteasome inhibitors are FDA approved, including carfilzomib based on epoxomicin produced by actinomycetes147. In this brief section, however, we focus on the upstream ER events.
5.4.1. Mannosidase I inhibitors: kifunensine and 1-deoxymannojirimycin
Kifunensine (Figure 13), isolated from the actinomycete Kitatosporia kifunense 9482, is an unusual oxalamide-fused iminosugar that inhibits α-mannosidases of glycoside hydrolase (GH) family 47 with high specificity at low-nanomolar concentrations148. Kifunensine-sensitive α-mannosidases include ER mannosidase I, EDEMs 1, 2 and 3 as well as Golgi mannosidase I. This compound has become a powerful tool compound for the targeted manipulation of the N-glycan structure. The co-crystal structure with Homo sapiens GH47 α-mannosidase has been solved at 1.75 Å, providing the structural basis and essential role of Ca2+ by stabilizing the 1C4 conformation, which is the same “ring-flipped” conformation as for 1-deoxymannojirimycin (Figure 13)149. The catalytic mechanism appeared to be novel and more recent conformational analyses with kifunensine based on quantum mechanics and structural studies highlighted the unique ability of this compound to access conformational free-energy landscapes (FELs) not accessed by other GH inhibitors150. Kifunensine mimics the product state of GH47 enzymes without being able to mimic relevant conformational states for other glycosidase families, guaranteeing the observed specificity. The mechanism highlights the critical importance of shape, achieved here through fusion of the bridge and sp2 hybridization of the endocyclic amide nitrogen. The natural product structure as well as mechanism can provide the inspiration for the design of other selective GH inhibitors.
Figure 13. Structures of GH47 family α-mannosidase inhibitors as ERAD inhibitors.
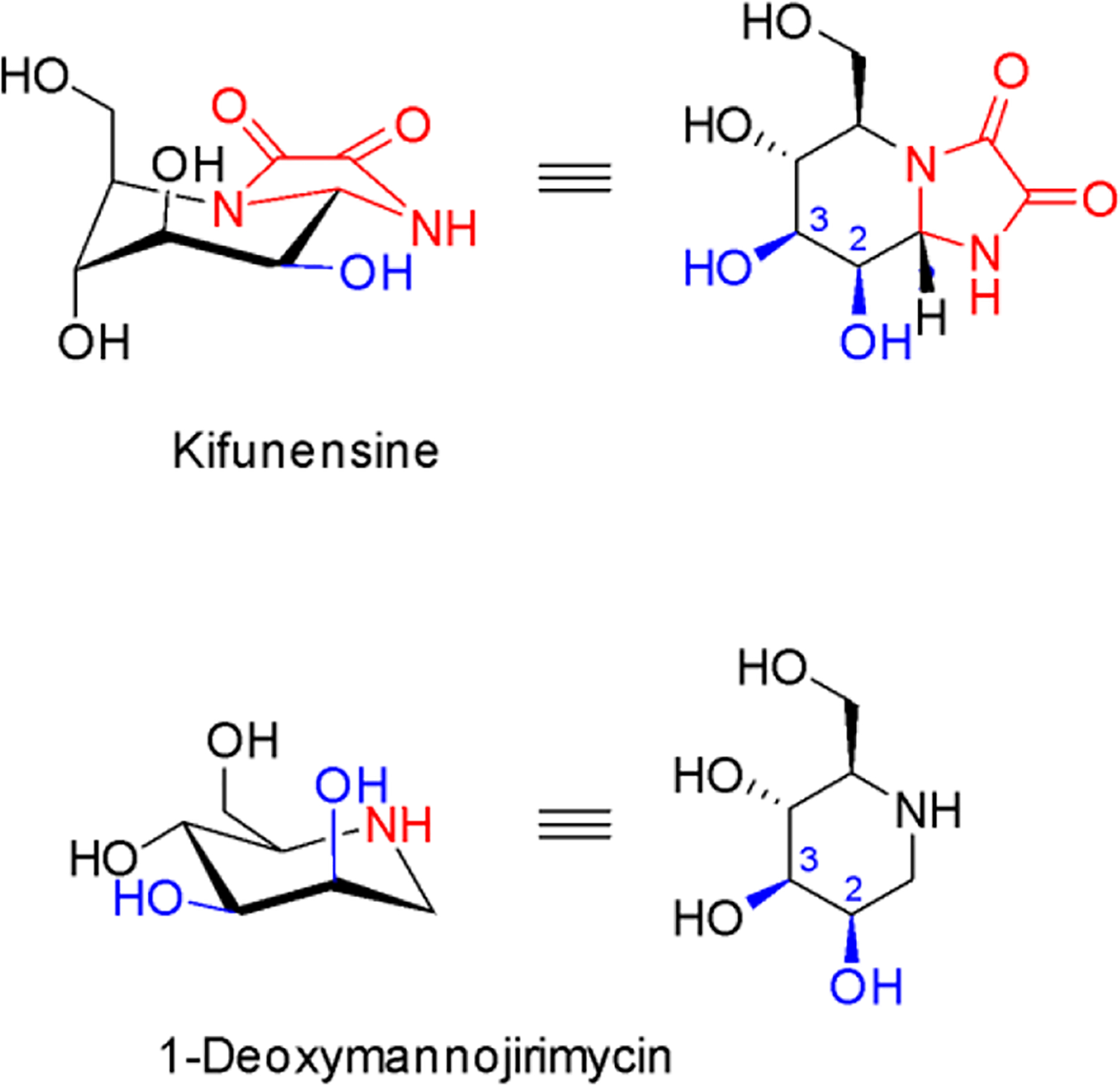
Kifunensine is 100-fold more potent than 1-deoxymannojirimycin. Structural differences are indicated in red. Hydroxy groups that coordinate the essential Ca2+ in the active site are indicated in blue.
5.4.2. Eeyarestatin I
In addition to its capability to inhibit protein import into the ER (discussed in section 2.4), Eeyarestatin I (ESI, Figure 3) also has distinct effects on turnover of proteins that are retrotranslocated by the ERAD pathway for cytosolic destruction50. ESI was demonstrated to interact with the cytosolic ERAD factor p97 required for extraction of polyubiquitinated ERAD substrate proteins from the ER membrane, which could account for the ability of ESI to stabilize ERAD substrate proteins in cells151. Another study demonstrated that ESI interacts with p97 in biochemical experiments with an estimated KD of 5–10 μM53. Further support for targeting of p97 was provided by the finding that p97 knockdown by siRNA abolished ESI’s ability to inhibit ERAD substrate degradation. The ability of ESI to trigger an ER stress response may contribute to its cytotoxicity against mammalian cells152. In another study, ESI was demonstrated to interfere with vesicular trafficking of a Shiga-like toxin153. This suggests that mechanisms by which ESI modulates different aspects of protein trafficking may be more complex than previously appreciated and it is currently difficult to correlate the diverse cellular phenotypes induced by ESI to modulation of a specific target factor.
6. Therapeutic potential and opportunities for protein secretion modulation
The secretory pathway has emerged as an attractive yet complex target for therapeutic intervention, due the multitude of secretory molecules and their disease involvement145,154–156. Since protein secretion is an essential pathway for mammalian cell physiology, selective modulation of pathway components is required for ultimate translational success and for development of selective pharmacological tools to further understand the biology which is by no means fully illuminated yet. As we have described, in many cases natural products identified through phenotypic screening campaigns have provided the motivation and starting points for chemical biology approaches to identify and characterize the direct biological targets or target complexes. As one example, inhibitors of protein maturation and trafficking have historically been identified and have served as important chemical tools to provide insight into the pathways they inhibit, once their mechanisms had been successfully established. BFA is a prime example for the fact that studying the mechanism of action of natural products can yield unique mechanisms of action. Subsequently these natural products provide the inspiration for other screening and synthetic campaigns. Until recently the therapeutic development of secretion pathway modulators has not been perceived feasible, which has prevented their rigorous pursuit in part due to on-target or pleiotropic off-target toxic effects. However, in recent years increased efforts have been directed to more seriously interrogate the therapeutic potential of protein secretion modulators through analogue synthesis and prodrug strategies, while developing an increased understanding of the underlying biology. The availability of structural information at the protein level and refinement of screening technologies have enabled increasing compound specificity and/or identification of new chemical scaffolds. There is renewed interest in targeting components of the UPR network since we now are able to selectively target branches of the UPR that ultimately affect the pharmacological and toxicological profile of compounds, with promising XBP1 mRNA splicing inhibitors such as B-I09 as candidates for further development for multiple myeloma. The greatest level of activity in recent years has been arguably at the stage of ER insertion, with the availability of the first ER insertion inhibitors targeting Sec61, many of which appear to have differential pharmacological profiles due to binding to different sites and potentially conformations of Sec61 in a cell type-dependent manner. While cancer has been the predominant disease indication for all subsets of secretory pathway inhibitors, particularly the selective modulation of Sec61 gating opens up opportunities for application to other diseases where secretory substrates play a role, including ocular angiogenic diseases, as has been already demonstrated for specific synthetic apratoxins, whereas cotransins showed initial promise as antivirals.
We predict that secretory pathway modulating compounds hold great therapeutic potential and that natural products will continue to deliver structurally diverse starting points to directly or indirectly modulate secretion. Natural products possess unique chemical and consequently biological properties as a result of evolutionary refinement, including unusual mechanisms of action and ability to stabilize or inhibit protein-protein interactions. The examples showcased in this review alone highlight the structural diversity of natural products targeting the secretory pathway at different stages, but even seemingly structurally distinct compounds were shown to interact with the same target in a different manner, such as Sec61. The continuous search for natural products and their rigorous pharmacological characterization will undoubtedly allow us to extend the druggable genome and validate previously unknown or undruggable targets or target complexes.
8. Acknowledgements
This work was supported by a National Institutes of Health grant R01CA172310 (HL), National Institute of General Medical Sciences grant R01GM132649 (VOP), Academy of Finland grants 289737, 314672 and 330255 (VOP) and a grant from the Sigrid Juselius Foundation (VOP). We thank Dr. Dale Tranter for helpful discussions and comments on the manuscript and the figures.
Biographies
Author Biographies
Dr. Hendrik Luesch

Hendrik Luesch received his Diplom in Chemistry at the University of Siegen (Germany) in 1997. He studied marine natural products chemistry at the University of Hawaii at Manoa and obtained his Ph.D. with Professor Richard E. Moore in 2002. He undertook three years of postdoctoral studies as an Irving S. Sigal Fellow at The Scripps Research Institute with Professor Peter G. Schultz in functional genomics and chemical biology. Since 2005 he has been faculty at the University of Florida (UF) and is currently Professor and Chair of the Department of Medicinal Chemistry. He holds the endowed Debbie and Sylvia DeSantis Chair in Natural Products Drug Discovery and Development and leads a multidisciplinary marine natural products program that integrates isolation, synthesis, pharmacology, mechanism of action and early development studies. He is UF’s Director of the Center for Natural Products, Drug Discovery and Development (CNPD3). Since 2018, he is Visiting Professor at the Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore.
Dr. Ville Paavilainen

Ville Paavilainen is a Tenure Track Group Leader at the Institute of Biotechnology (HiLIFE) at the University of Helsinki. He received his Ph.D. degree at the University of Helsinki where he studied regulation of the cellular actin cytoskeleton using biochemistry and structural biology methods. He then moved to University of California -San Francisco to work with Prof. Jack Taunton as a Postdoctoral Fellow to develop covalent-reversible cysteine targeting kinase inhibitors and to study integral membrane protein membrane insertion using a substrate-selective inhibitor of ER protein import. His independent research group studies proteostasis modulating small molecules and specialized functions of organelle targeting polypeptides. He lives in Helsinki and is passionate about rock climbing, music and hanging out with his family.
Footnotes
Conflicts of interest
H.L. is co-founder of Oceanyx Pharmaceuticals, Inc., which has licensed patents and patent applications related to compounds discussed in this article (apratoxins).
9. References
- 1.Newman DJ and Cragg GM, J. Nat. Prod, 2016, 79, 629–661. [DOI] [PubMed] [Google Scholar]
- 2.Davison EK and Brimble MA, Curr. Opin. Chem. Biol, 2019, 52, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Liang X, Luo D and Luesch H, Pharmacol. Res, 2019, 147, 104373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson EE, ACS Chem. Biol, 2010, 5, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME, Buser-Doepner C, Campbell RM, Carter AJ, Cohen P, Copeland RA, Cravatt B, Dahlin JL, Dhanak D, Edwards AM, Frederiksen M, Frye SV, Gray N, Grimshaw CE, Hepworth D, Howe T, Huber KVM, Jin J, Knapp S, Kotz JD, Kruger RG, Lowe D, Mader MM, Marsden B, Mueller-Fahrnow A, Müller S, O’Hagan RC, Overington JP, Owen DR, Rosenberg SH, Roth B, Ross R, Schapira M, Schreiber SL, Shoichet B, Sundström M, Superti-Furga G, Taunton J, Toledo-Sherman L, Walpole C, Walters MA, Willson TM, Workman P, Young RN and Zuercher WJ, Nat. Chem. Biol, 2015, 11, 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jost M and Weissman JS, ACS Chem. Biol, 2018, 13, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schürmann M, Janning P, Ziegler S and Waldmann H, Cell Chem Biol, 2016, 23, 435–441. [DOI] [PubMed] [Google Scholar]
- 8.Park H, Ha J and Park SB, Curr. Opin. Chem. Biol, 2019, 50, 66–72. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor TM and Miller RM, Trends Pharmacol. Sci, 2017, 38, 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Puyenbroeck V and Vermeire K, Cell. Mol. Life Sci, 2018, 75, 1541–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster CA, Dreyfuss M, Mandak B, Meingassner JG, Naegeli HU, Nussbaumer A, Oberer L, Scheel G and Swoboda EM, J. Dermatol, 1994, 21, 847–854. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Bilban M, Foster CA and Boger DL, J. Am. Chem. Soc, 2002, 124, 5431–5440. [DOI] [PubMed] [Google Scholar]
- 13.Schreiner EP, Kern M, Steck A and Foster CA, Bioorg. Med. Chem. Lett, 2004, 14, 5003–5006. [DOI] [PubMed] [Google Scholar]
- 14.Coin I, Beerbaum M, Schmieder P, Bienert M and Beyermann M, Org. Lett, 2008, 10, 3857–3860. [DOI] [PubMed] [Google Scholar]
- 15.Garrison JL, Kunkel EJ, Hegde RS and Taunton J, Nature, 2005, 436, 285–289. [DOI] [PubMed] [Google Scholar]
- 16.Besemer J, Harant H, Wang S, Oberhauser B, Marquardt K, Foster CA, Schreiner EP, de Vries JE, Dascher-Nadel C and Lindley IJD, Nature, 2005, 436, 290–293. [DOI] [PubMed] [Google Scholar]
- 17.MacKinnon AL, Garrison JL, Hegde RS and Taunton J, J. Am. Chem. Soc, 2007, 129, 14560–14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackinnon AL, Paavilainen VO, Sharma A, Hegde RS and Taunton J, Elife, 2014, 3, e01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maifeld SV, MacKinnon AL, Garrison JL, Sharma A, Kunkel EJ, Hegde RS and Taunton J, Chem. Biol, 2011, 18, 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein W, Westendorf C, Schmidt A, Conill-Cortés M, Rutz C, Blohs M, Beyermann M, Protze J, Krause G, Krause E and Schülein R, PLoS One, 2015, 10, e0120886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz-Saenz A, Sandhu M, Carrasco Y, Maglathlin RL, Taunton J and Moasser MM, Oncogene, 2015, 34, 5288–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heaton NS, Moshkina N, Fenouil R, Gardner TJ, Aguirre S, Shah PS, Zhao N, Manganaro L, Hultquist JF, Noel J, Sachs D, Hamilton J, Leon PE, Chawdury A, Tripathi S, Melegari C, Campisi L, Hai R, Metreveli G, Gamarnik AV, García-Sastre A, Greenbaum B, Simon V, Fernandez-Sesma A, Krogan NJ, Mulder LCF, van Bakel H, Tortorella D, Taunton J, Palese P and Marazzi I, Immunity, 2016, 44, 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luesch H, Yoshida WY, Moore RE, Paul VJ and Corbett TH, J. Am. Chem. Soc, 2001, 123, 5418–5423. [DOI] [PubMed] [Google Scholar]
- 24.Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Izpisúa Belmonte JC and Schultz PG, Nat. Chem. Biol, 2006, 2, 158–167. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Law BK and Luesch H, Mol. Pharmacol, 2009, 76, 91–104. [DOI] [PubMed] [Google Scholar]
- 26.Gutiérrez M, Suyama TL, Engene N, Wingerd JS, Matainaho T and Gerwick WH, J. Nat. Prod, 2008, 71, 1099–1103. [DOI] [PubMed] [Google Scholar]
- 27.Paatero AO, Kellosalo J, Dunyak BM, Almaliti J, Gestwicki JE, Gerwick WH, Taunton J and Paavilainen VO, Cell Chem Biol, 2016, 23, 561–566. [DOI] [PubMed] [Google Scholar]
- 28.Huang K-C, Chen Z, Jiang Y, Akare S, Kolber-Simonds D, Condon K, Agoulnik S, Tendyke K, Shen Y, Wu K-M, Mathieu S, Choi H-W, Zhu X, Shimizu H, Kotake Y, Gerwick WH, Uenaka T, Woodall-Jappe M and Nomoto K, Mol. Cancer Ther, 2016, 15, 1208–1216. [DOI] [PubMed] [Google Scholar]
- 29.Chitwood PJ, Juszkiewicz S, Guna A, Shao S and Hegde RS, Cell, 2018, 175, 1507–1519.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Q-Y, Liu Y and Luesch H, ACS Med. Chem. Lett, 2011, 2, 861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai W, Ratnayake R, Gerber MH, Chen Q-Y, Yu Y, Derendorf H, Trevino JG and Luesch H, Invest. New Drugs, 2019, 37, 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q-Y, Liu Y, Cai W and Luesch H, J. Med. Chem, 2014, 57, 3011–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P, Cai W, Chen Q-Y, Xu S, Yin R, Li Y, Zhang W and Luesch H, Org. Lett, 2016, 18, 5400–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai W, Chen Q-Y, Dang LH and Luesch H, ACS Med. Chem. Lett, 2017, 8, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu B, Tan A, Veluchamy AB, Li Y, Murray H, Cheng W, Liu C, Busoy JM, Chen Q-Y, Sistla S, Hunziker W, Cheung CMG, Wong TY, Hong W, Luesch H and Wang X, Invest. Ophthalmol. Vis. Sci, 2019, 60, 3254–3263. [DOI] [PubMed] [Google Scholar]
- 36.Demangel C, Stinear TP and Cole ST, Nat. Rev. Microbiol, 2009, 7, 50–60. [DOI] [PubMed] [Google Scholar]
- 37.Demangel C and High S, Biol. Cell, 2018, 110, 237–248. [DOI] [PubMed] [Google Scholar]
- 38.Stinear TP, Mve-Obiang A, Small PLC, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PDR, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF and Cole ST, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guenin-Macé L, Carrette F, Asperti-Boursin F, Le Bon A, Caleechurn L, Di Bartolo V, Fontanet A, Bismuth G and Demangel C, Proc. Natl. Acad. Sci. U. S. A, 2011, 108, 12833–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall BS, Hill K, McKenna M, Ogbechi J, High S, Willis AE and Simmonds RE, PLoS Pathog, 2014, 10, e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmonds RE, Lali FV, Smallie T, Small PLC and Foxwell BM, J. Immunol, 2009, 182, 2194–2202. [DOI] [PubMed] [Google Scholar]
- 42.McKenna M, Simmonds RE and High S, J. Cell Sci, 2016, 129, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baron L, Paatero AO, Morel J-D, Impens F, Guenin-Macé L, Saint-Auret S, Blanchard N, Dillmann R, Niang F, Pellegrini S, Taunton J, Paavilainen VO and Demangel C, J. Exp. Med, 2016, 213, 2885–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guna A, Volkmar N, Christianson JC and Hegde RS, Science, 2018, 359, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guenin-Macé L, Veyron-Churlet R, Thoulouze M-I, Romet-Lemonne G, Hong H, Leadlay PF, Danckaert A, Ruf M-T, Mostowy S, Zurzolo C, Bousso P, Chrétien F, Carlier M-F and Demangel C, J. Clin. Invest, 2013, 123, 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marion E, Song O-R, Christophe T, Babonneau J, Fenistein D, Eyer J, Letournel F, Henrion D, Clere N, Paille V, Guérineau NC, Saint André J-P, Gersbach P, Altmann K-H, Stinear TP, Comoglio Y, Sandoz G, Preisser L, Delneste Y, Yeramian E, Marsollier L and Brodin P, Cell, 2014, 157, 1565–1576. [DOI] [PubMed] [Google Scholar]
- 47.Morel J-D, Paatero AO, Wei J, Yewdell JW, Guenin-Macé L, Van Haver D, Impens F, Pietrosemoli N, Paavilainen VO and Demangel C, Mol. Cell. Proteomics, 2018, 17, 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogbechi J, Hall BS, Sbarrato T, Taunton J, Willis AE, Wek RC and Simmonds RE, Cell Death & Disease, 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guenin-Macé L, Baron L, Chany A-C, Tresse C, Saint-Auret S, Jönsson F, Le Chevalier F, Bruhns P, Bismuth G, Hidalgo-Lucas S, Bisson J-F, Blanchard N and Demangel C, Science Translational Medicine, 2015, 7, 289ra85–289ra85. [DOI] [PubMed] [Google Scholar]
- 50.Fiebiger E, Hirsch C, Vyas JM, Gordon E, Ploegh HL and Tortorella D, Mol. Biol. Cell, 2004, 15, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cross BCS, McKibbin C, Callan AC, Roboti P, Piacenti M, Rabu C, Wilson CM, Whitehead R, Flitsch SL, Pool MR, High S and Swanton E, J. Cell Sci, 2009, 122, 4393–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gamayun I, O’Keefe S, Pick T, Klein M-C, Nguyen D, McKibbin C, Piacenti M, Williams HM, Flitsch SL, Whitehead RC, Swanton E, Helms V, High S, Zimmermann R and Cavalié A, Cell Chem Biol, 2019, 26, 571–583.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q, Shinkre BA, Lee J-G, Weniger MA, Liu Y, Chen W, Wiestner A, Trenkle WC and Ye Y, PLoS One, 2010, 5, e15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junne T, Wong J, Studer C, Aust T, Bauer BW, Beibel M, Bhullar B, Bruccoleri R, Eichenberger J, Estoppey D, Hartmann N, Knapp B, Krastel P, Melin N, Oakeley EJ, Oberer L, Riedl R, Roma G, Schuierer S, Petersen F, Tallarico JA, Rapoport TA, Spiess M and Hoepfner D, J. Cell Sci, 2015, 128, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao S, Guza RC, Wisse JH, Miller JS, Evans R and Kingston DGI, J. Nat. Prod, 2005, 68, 487–492. [DOI] [PubMed] [Google Scholar]
- 56.Cao S, Norris A, Wisse JH, Miller JS, Evans R and Kingston DGI, Nat. Prod. Res, 2007, 21, 872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagano T, Pospíšil J and Chollet G, A European Journal. [Google Scholar]
- 58.Fürstner A and Nagano T, J. Am. Chem. Soc, 2007, 129, 1906–1907. [DOI] [PubMed] [Google Scholar]
- 59.Zong G, Hu Z, O’Keefe S, Tranter D, Iannotti MJ, Baron L, Hall B, Corfield K, Paatero AO, Henderson MJ, Roboti P, Zhou J, Sun X, Govindarajan M, Rohde JM, Blanchard N, Simmonds R, Inglese J, Du Y, Demangel C, High S, Paavilainen VO and Shi WQ, J. Am. Chem. Soc, 2019, 141, 8450–8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medina RA, Goeger DE, Hills P, Mooberry SL, Huang N, Romero LI, Ortega-Barría E, Gerwick WH and McPhail KL, J. Am. Chem. Soc, 2008, 130, 6324–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serrill JD, Wan X, Hau AM, Jang HS, Coleman DJ, Indra AK, Alani AWG, McPhail KL and Ishmael JE, Invest. New Drugs, 2016, 34, 24–40. [DOI] [PubMed] [Google Scholar]
- 62.Wan X, Serrill JD, Humphreys IR, Tan M, McPhail KL, Ganley IG and Ishmael JE, Mar. Drugs, DOI: 10.3390/md16030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao G, Pan Z, Wu C, Wang W, Fang L and Su W, J. Am. Chem. Soc, 2015, 137, 13488–13491. [DOI] [PubMed] [Google Scholar]
- 64.Nabika R, Suyama TL, Hau AM, Misu R, Ohno H, Ishmael JE, McPhail KL, Oishi S and Fujii N, Bioorg. Med. Chem. Lett, 2015, 25, 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sable GA, Park J, Kim H, Lim S-J, Jang S and Lim D, Eur. J. Org. Chem, 2015, 2015, 7043–7052. [Google Scholar]
- 66.Tranter D, Paatero AO, Kawaguchi S, Kazemi S, Serrill JD, Kellosalo J, Vogel WK, Richter U, Thornburg CC, Oishi S, Mcphail KL, Ishmael JE and Paavilainen VO, ChemRxiv, 2019. [Google Scholar]
- 67.Yao G, Wang W, Ao L, Cheng Z, Wu C, Pan Z, Liu K, Li H, Su W and Fang L, Journal of Medicinal Chemistry, 2018, 61, 8908–8916. [DOI] [PubMed] [Google Scholar]
- 68.Takatsuki A, Arima K and Tamura G, J. Antibiot, 1971, 24, 215–223. [DOI] [PubMed] [Google Scholar]
- 69.Heifetz A, Keenan RW and Elbein AD, Biochemistry, 1979, 18, 2186–2192. [DOI] [PubMed] [Google Scholar]
- 70.Olden K, Pratt RM and Yamada KM, Cell, 1978, 13, 461–473. [DOI] [PubMed] [Google Scholar]
- 71.Pemberton JM, Vincent KM and Penfold RJ, Current Microbiology, 1991, 22, 355–358. [Google Scholar]
- 72.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT and Bugg TD, Antimicrob. Agents Chemother, 1996, 40, 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hakulinen JK, Hering J, Brändén G, Chen H, Snijder A, Ek M and Johansson P, Nat. Chem. Biol, 2017, 13, 265. [DOI] [PubMed] [Google Scholar]
- 74.Yoo J, Mashalidis EH, Kuk ACY, Yamamoto K, Kaeser B, Ichikawa S and Lee S-Y, Nat. Struct. Mol. Biol, 2018, 25, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong YY, Wang H, Pike ACW, Cochrane SA, Hamedzadeh S, Wyszyński FJ, Bushell SR, Royer SF, Widdick DA, Sajid A, Boshoff HI, Park Y, Lucas R, Liu W-M, Lee SS, Machida T, Minall L, Mehmood S, Belaya K, Liu W-W, Chu A, Shrestha L, Mukhopadhyay SMM, Strain-Damerell C, Chalk R, Burgess-Brown NA, Bibb MJ, Barry Iii CE, Robinson CV, Beeson D, Davis BG and Carpenter EP, Cell, 2018, 175, 1045–1058.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto K, Yakushiji F, Matsumaru T and Ichikawa S, Org. Lett, 2018, 20, 256–259. [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto K, Katsuyama A and Ichikawa S, Bioorg. Med. Chem, 2019, 27, 1714–1719. [DOI] [PubMed] [Google Scholar]
- 78.Wu J, Chen S, Liu H, Zhang Z, Ni Z, Chen J, Yang Z, Nie Y and Fan D, J. Exp. Clin. Cancer Res, 2018, 37, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdullahi A, Stanojcic M, Parousis A, Patsouris D and Jeschke MG, Shock, 2017, 47, 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ortiz-Lopez FJ, Monteiro MC, Gonzalez-Menendez V, Tormo JR, Genilloud O, Bills GF, Vicente F, Zhang C, Roemer T, Singh SB and Others J Nat. Prod, 2015, 78, 468–475. [DOI] [PubMed] [Google Scholar]
- 81.Zwick CR and Renata H, Tetrahedron, 2018, 74, 6469–6473. [Google Scholar]
- 82.Estoppey D, Lee CM, Janoschke M, Lee BH, Wan KF, Dong H, Mathys P, Filipuzzi I, Schuhmann T, Riedl R, Aust T, Galuba O, McAllister G, Russ C, Spiess M, Bouwmeester T, Bonamy GMC and Hoepfner D, Cell Rep, 2017, 19, 451–460. [DOI] [PubMed] [Google Scholar]
- 83.Singleton VL, Bohonos N and Ullstrup AJ, Nature, 1958, 181, 1072–1073. [DOI] [PubMed] [Google Scholar]
- 84.Trisuwan K, Rukachaisirikul V, Sukpondma Y, Phongpaichit S, Preedanon S and Sakayaroj J, Chem. Pharm. Bull, 2009, 57, 1100–1102. [DOI] [PubMed] [Google Scholar]
- 85.Hu Z-F, Qin L-L, Ding W-J, Liu Y and Ma Z-J, Nat. Prod. Res, 2016, 30, 2311–2315. [DOI] [PubMed] [Google Scholar]
- 86.Zeng F, Chen C, Al Chnani AA, Zhou Q, Tong Q, Wang W, Zang Y, Gong J, Wu Z, Liu J, Wang J, Zhu H and Zhang Y, Bioorg. Chem, 2019, 86, 176–182. [DOI] [PubMed] [Google Scholar]
- 87.Paek S-M, Mar. Drugs, DOI: 10.3390/md16040133. [DOI] [Google Scholar]
- 88.Pelham HR, Cell, 1991, 67, 449–451. [DOI] [PubMed] [Google Scholar]
- 89.Anders N and Jürgens G, Cell. Mol. Life Sci, 2008, 65, 3433–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeghouf M, Guibert B, Zeeh J-C and Cherfils J, Biochem. Soc. Trans, 2005, 33, 1265–1268. [DOI] [PubMed] [Google Scholar]
- 91.Shin H-W and Nakayama K, J. Biochem, 2004, 136, 761–767. [DOI] [PubMed] [Google Scholar]
- 92.Casanova JE, Traffic, 2007, 8, 1476–1485. [DOI] [PubMed] [Google Scholar]
- 93.Chardin P and McCormick F, Cell, 1999, 97, 153–155. [DOI] [PubMed] [Google Scholar]
- 94.Renault L, Guibert B and Cherfils J, Nature, 2003, 426, 525–530. [DOI] [PubMed] [Google Scholar]
- 95.Mossessova E, Corpina RA and Goldberg J, Mol. Cell, 2003, 12, 1403–1411. [DOI] [PubMed] [Google Scholar]
- 96.Vigil D, Cherfils J, Rossman KL and Der CJ, Nat. Rev. Cancer, 2010, 10, 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Viaud J, Zeghouf M, Barelli H, Zeeh J-C, Padilla A, Guibert B, Chardin P, Royer CA, Cherfils J and Chavanieu A, Proc. Natl. Acad. Sci. U. S. A, 2007, 104, 10370–10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hafner M, Schmitz A, Grüne I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I and Famulok M, Nature, 2006, 444, 941–944. [DOI] [PubMed] [Google Scholar]
- 99.Feng Y, Yu S, Lasell TKR, Jadhav AP, Macia E, Chardin P, Melancon P, Roth M, Mitchison T and Kirchhausen T, Proc. Natl. Acad. Sci. U. S. A, 2003, 100, 6469–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng Y, Jadhav AP, Rodighiero C, Fujinaga Y, Kirchhausen T and Lencer WI, EMBO Rep, 2004, 5, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C and Kirchhausen T, Dev. Cell, 2006, 10, 839–850. [DOI] [PubMed] [Google Scholar]
- 102.Newton AJ, Kirchhausen T and Murthy VN, Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 17955–17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pelish HE, Peterson JR, Salvarezza SB, Rodriguez-Boulan E, Chen J-L, Stamnes M, Macia E, Feng Y, Shair MD and Kirchhausen T, Nat. Chem. Biol, 2006, 2, 39–46. [DOI] [PubMed] [Google Scholar]
- 104.Rasmussen U, Brøogger Christensen S and Sandberg F, Acta Pharm. Suec, 1978, 15, 133–140. [PubMed] [Google Scholar]
- 105.Furuya Y, Lundmo P, Short AD, Gill DL and Isaacs JT, Cancer Res, 1994, 54, 6167–6175. [PubMed] [Google Scholar]
- 106.Furuya Y and Isaacs JT, Prostate, 1994, 25, 301–309. [DOI] [PubMed] [Google Scholar]
- 107.Lam M, Dubyak G and Distelhorst CW, Mol. Endocrinol, 1993, 7, 686–693. [DOI] [PubMed] [Google Scholar]
- 108.Kaneko Y and Tsukamoto A, Cancer Lett, 1994, 79, 147–155. [DOI] [PubMed] [Google Scholar]
- 109.Thastrup O, Cullen PJ, Drøbak BK, Hanley MR and Dawson AP, Proc. Natl. Acad. Sci. U. S. A, 1990, 87, 2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sagara Y and Inesi G, J. Biol. Chem, 1991, 266, 13503–13506. [PubMed] [Google Scholar]
- 111.Toyoshima C and Nomura H, Nature, 2002, 418, 605–611. [DOI] [PubMed] [Google Scholar]
- 112.Dey S and Bajaj SO, Synth. Commun, 2018, 48, 1–13. [Google Scholar]
- 113.Denmeade SR, Jakobsen CM, Janssen S, Khan SR, Garrett ES, Lilja H, Christensen SB and Isaacs JT, J. Natl. Cancer Inst, 2003, 95, 990–1000. [DOI] [PubMed] [Google Scholar]
- 114.Mahalingam D, Peguero J, Cen P, Arora SP, Sarantopoulos J, Rowe J, Allgood V, Tubb B and Campos L, Cancers, DOI: 10.3390/cancers11060833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Appendino G, Prosperini S, Valdivia C, Ballero M, Colombano G, Billington RA, Genazzani AA and Sterner O, J. Nat. Prod, 2005, 68, 1213–1217. [DOI] [PubMed] [Google Scholar]
- 116.Navarrete C, Sancho R, Caballero FJ, Pollastro F, Fiebich BL, Sterner O, Appendino G and Muñoz E, J. Pharmacol. Exp. Ther, 2006, 319, 422–430. [DOI] [PubMed] [Google Scholar]
- 117.Ma W, Liang X, Ye W, Wang Y, Min L, He S and Lee C-S, Eur. J. Org. Chem, 2018, 2018, 196–208. [Google Scholar]
- 118.Ovchinnikov YA, Eur. J. Biochem, 1979, 94, 321–336. [DOI] [PubMed] [Google Scholar]
- 119.Pressman BC, Annu. Rev. Biochem, 1976, 45, 501–530. [DOI] [PubMed] [Google Scholar]
- 120.Berezin SK, J. Membr. Biol, 2015, 248, 713–726. [DOI] [PubMed] [Google Scholar]
- 121.Rose L and Jenkins ATA, Bioelectrochemistry, 2007, 70, 387–393. [DOI] [PubMed] [Google Scholar]
- 122.Ryoo I-J, Park H-R, Choo S-J, Hwang J-H, Park Y-M, Bae K-H, Shin-Ya K and Yoo I-D, Biol. Pharm. Bull, 2006, 29, 817–820. [DOI] [PubMed] [Google Scholar]
- 123.Cheng Y-Q, Chembiochem, 2006, 7, 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Magarvey NA, Ehling-Schulz M and Walsh CT, J. Am. Chem. Soc, 2006, 128, 10698–10699. [DOI] [PubMed] [Google Scholar]
- 125.Kim J, Choi I, Park J-Y and Kang S-W, Exp. Cell Res, 2013, 319, 2049–2057. [DOI] [PubMed] [Google Scholar]
- 126.Wang M, Law ME, Castellano RK and Law BK, Crit. Rev. Oncol. Hematol, 2018, 127, 66–79. [DOI] [PubMed] [Google Scholar]
- 127.Ri M, Tashiro E, Oikawa D, Shinjo S, Tokuda M, Yokouchi Y, Narita T, Masaki A, Ito A, Ding J, Kusumoto S, Ishida T, Komatsu H, Shiotsu Y, Ueda R, Iwawaki T, Imoto M and Iida S, Blood Cancer J, 2012, 2, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tashiro E, Hironiwa N, Kitagawa M, Futamura Y, Suzuki S-I, Nishio M and Imoto M, J. Antibiot, 2007, 60, 547–553. [DOI] [PubMed] [Google Scholar]
- 129.Kawamura T, Tashiro E, Shindo K and Imoto M, J. Antibiot, 2008, 61, 312–317. [DOI] [PubMed] [Google Scholar]
- 130.Tang C-HA, Ranatunga S, Kriss CL, Cubitt CL, Tao J, Pinilla-Ibarz JA, Del Valle JR and Hu C-CA, J. Clin. Invest, 2014, 124, 2585–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xie H, Tang C-HA, Song JH, Mancuso A, Del Valle JR, Cao J, Xiang Y, Dang CV, Lan R, Sanchez DJ, Keith B, Hu C-CA and Simon MC, J. Clin. Invest, 2018, 128, 1300–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shao A, Kang CW, Tang C-HA, Cain CF, Xu Q, Phoumyvong CM, Del Valle JR and Hu C-CA, J. Med. Chem, 2019, 62, 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Menna M, Curr. Top. Med. Chem, 2014, 14, 207–223. [DOI] [PubMed] [Google Scholar]
- 134.Iglesias-Arteaga MA and Morzycki JW, Alkaloids Chem. Biol, 2013, 72, 153–279. [DOI] [PubMed] [Google Scholar]
- 135.Moser BR, J. Nat. Prod, 2008, 71, 487–491. [DOI] [PubMed] [Google Scholar]
- 136.López-Antón N, Rudy A, Barth N, Schmitz ML, Pettit GR, Schulze-Osthoff K, Dirsch VM and Vollmar AM, J. Biol. Chem, 2006, 281, 33078–33086. [DOI] [PubMed] [Google Scholar]
- 137.Burgett AWG, Poulsen TB, Wangkanont K, Anderson DR, Kikuchi C, Shimada K, Okubo S, Fortner KC, Mimaki Y, Kuroda M, Murphy JP, Schwalb DJ, Petrella EC, Cornella-Taracido I, Schirle M, Tallarico JA and Shair MD, Nat. Chem. Biol, 2011, 7, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ambrose AJ, Santos EA, Jimenez PC, Rocha DD, Wilke DV, Beuzer P, Axelrod J, Kumar Kanduluru A, Fuchs PL, Cang H, Costa-Lotufo LV, Chapman E and La Clair JJ, Chembiochem, 2017, 18, 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Choi MJ, Park EJ, Min KJ, Park J-W and Kwon TK, Toxicol. In Vitro, 2011, 25, 692–698. [DOI] [PubMed] [Google Scholar]
- 140.Ghosh K, De S, Mukherjee S, Das S, Ghosh AN and Sengupta SB, Toxicol. In Vitro, 2017, 44, 330–338. [DOI] [PubMed] [Google Scholar]
- 141.Wang L, Perera BGK, Hari SB, Bhhatarai B, Backes BJ, Seeliger MA, Schürer SC, Oakes SA, Papa FR and Maly DJ, Nat. Chem. Biol, 2012, 8, 982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Axten JM, Romeril SP, Shu A, Ralph J, Medina JR, Feng Y, Li WHH, Grant SW, Heerding DA, Minthorn E, Mencken T, Gaul N, Goetz A, Stanley T, Hassell AM, Gampe RT, Atkins C and Kumar R, ACS Med. Chem. Lett, 2013, 4, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Atkins C, Liu Q, Minthorn E, Zhang S-Y, Figueroa DJ, Moss K, Stanley TB, Sanders B, Goetz A, Gaul N, Choudhry AE, Alsaid H, Jucker BM, Axten JM and Kumar R, Cancer Res, 2013, 73, 1993–2002. [DOI] [PubMed] [Google Scholar]
- 144.Rojas-Rivera D, Delvaeye T, Roelandt R, Nerinckx W, Augustyns K, Vandenabeele P and Bertrand MJM, Cell Death Differ, 2017, 24, 1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hetz C, Axten JM and Patterson JB, Nat. Chem. Biol, 2019, 15, 764–775. [DOI] [PubMed] [Google Scholar]
- 146.van den Boom J and Meyer H, Mol. Cell, 2018, 69, 182–194. [DOI] [PubMed] [Google Scholar]
- 147.Kim KB and Crews CM, Nat. Prod. Rep, 2013, 30, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Elbein AD, Tropea JE, Mitchell M and Kaushal GP, J. Biol. Chem, 1990, 265, 15599–15605. [PubMed] [Google Scholar]
- 149.Vallee F, Karaveg K, Herscovics A, Moremen KW and Howell PL, J. Biol. Chem, 2000, 275, 41287–41298. [DOI] [PubMed] [Google Scholar]
- 150.Males A, Raich L, Williams SJ, Rovira C and Davies GJ, Chembiochem, 2017, 18, 1496–1501. [DOI] [PubMed] [Google Scholar]
- 151.Wang Q, Li L and Ye Y, J. Biol. Chem, 2008, 283, 7445–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Q, Mora-Jensen H, Weniger MA, Perez-Galan P, Wolford C, Hai T, Ron D, Chen W, Trenkle W, Wiestner A and Ye Y, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 2200–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aletrari M-O, McKibbin C, Williams H, Pawar V, Pietroni P, Lord JM, Flitsch SL, Whitehead R, Swanton E, High S and Spooner RA, PLoS One, 2011, 6, e22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang M and Kaufman RJ, Nature, 2016, 529, 326–335. [DOI] [PubMed] [Google Scholar]
- 155.Dejeans N, Manié S, Hetz C, Bard F, Hupp T, Agostinis P, Samali A and Chevet E, Trends Mol. Med, 2014, 20, 242–250. [DOI] [PubMed] [Google Scholar]
- 156.Bell SC, Mall MA, Gutierrez H, Macek M, Madge S, Davies JC, Burgel P-R, Tullis E, Castaños C, Castellani C, Byrnes CA, Cathcart F, Chotirmall SH, Cosgriff R, Eichler I, Fajac I, Goss CH, Drevinek P, Farrell PM, Gravelle AM, Havermans T, Mayer-Hamblett N, Kashirskaya N, Kerem E, Mathew JL, McKone EF, Naehrlich L, Nasr SZ, Oates GR, O’Neill C, Pypops U, Raraigh KS, Rowe SM, Southern KW, Sivam S, Stephenson AL, Zampoli M and Ratjen F, Lancet Respir Med, DOI: 10.1016/S2213-2600(19)30337-6. [DOI] [Google Scholar]


