Abstract
Temporal patterning of neural progenitors leads to the sequential production of diverse neurons. To understand how extrinsic cues influence intrinsic temporal programs, we studied Drosophila mushroom body progenitors (neuroblasts) that sequentially produce only three neuronal types: γ, then α’β’, followed by αβ. Opposing gradients of two RNA-binding proteins Imp and Syp comprise the intrinsic temporal program. Extrinsic activin signaling regulates the production of α’β’ neurons but whether it affects the intrinsic temporal program was not known. We show that the activin ligand Myoglianin from glia regulates the temporal factor Imp in mushroom body neuroblasts. Neuroblasts missing the activin receptor Baboon have a delayed intrinsic program as Imp is higher than normal during the α’β’ temporal window, causing the loss of α’β’ neurons, a decrease in αβ neurons, and a likely increase in γ neurons, without affecting the overall number of neurons produced. Our results illustrate that an extrinsic cue modifies an intrinsic temporal program to increase neuronal diversity.
Research organism: D. melanogaster
Introduction
The building of intricate neural networks during development is controlled by highly coordinated patterning programs that regulate the generation of different neuronal types in the correct number, place and time. The sequential production of different neuronal types from individual progenitors, i.e. temporal patterning, is a conserved feature of neurogenesis (Cepko, 2014; Holguera and Desplan, 2018; Kohwi and Doe, 2013; Lodato and Arlotta, 2015). For instance, individual radial glia progenitors in the vertebrate cortex sequentially give rise to neurons that occupy the different cortical layers in an inside-out manner (Gao et al., 2014; Llorca et al., 2019). In Drosophila, neural progenitors (called neuroblasts) also give rise to different neuronal types sequentially. For example, projection neurons in the antennal lobe are born in a stereotyped temporal order and innervate specific glomeruli (Jefferis et al., 2001; Kao et al., 2012; Yu et al., 2010). In both of these examples, individual progenitors age concomitantly with the developing animal (e.g., from embryonic stages 11–17 in mouse and from the first larval stage (L1) to the end of the final larva stage (L3) in Drosophila). Thus, these progenitors are exposed to changing environments that could alter their neuronal output. Indeed, classic heterochronic transplantation experiments demonstrated that young cortical progenitors placed in an old host environment alter their output to match the host environment and produce upper-layer neurons (Desai and McConnell, 2000; McConnell, 1988; McConnell and Kaznowski, 1991).
The adult Drosophila central brain is built from ~100 neuroblasts (Lee et al., 2020; Urbach and Technau, 2004; Wong et al., 2013; Yu et al., 2013a) that divide continuously from L1 to L3 (Homem et al., 2014; Sousa-Nunes et al., 2010; Yang et al., 2017). Each asymmetric division regenerates the neuroblast and produces an intermediate progenitor called ganglion mother cell (GMC) that divides only once, typically producing two different cell types (Lin et al., 2010; Spana and Doe, 1996; Truman et al., 2010). Thus, during larval life central brain neuroblasts divide 50–60 times, sequentially producing many different neuronal types. All central brain neuroblasts progress through opposing temporal gradients of two RNA-binding proteins as they age: IGF-II mRNA binding protein (Imp) when they are young and Syncrip (Syp) when they are old (Liu et al., 2015; Syed et al., 2017b; Syed et al., 2017a; Yang et al., 2016). Loss of Imp or Syp in antennal lobe or Type II neuroblasts affects the ratio of young to old neuronal types (Liu et al., 2015; Ren et al., 2017). Imp and Syp also affect neuroblast lifespan (Yang et al., 2017). Thus, a single temporal program can affect both the diversity of neuronal types produced and their numbers.
Since central brain neuroblasts produce different neuronal types through developmental time, roles for extrinsic cues have recently garnered attention. Ecdysone triggers all the major developmental transitions including progression into the different larval stages and entry in pupation (Yamanaka et al., 2013). The majority of central brain neuroblasts are not responsive to ecdysone until mid-larval life when they begin to express the Ecdysone Receptor (EcR) (Syed et al., 2017a). Expressing a dominant-negative version of EcR (EcR-DN) in Type II neuroblasts delays the Imp to Syp transition that normally occurs ~60 hr after larval hatching (ALH). This leads to many more cells that express the early-born marker gene Repo and fewer cells that express the late-born marker gene Bsh.
To further understand how extrinsic signals contribute to temporal patterning, we studied Drosophila mushroom body neuroblasts because of the deep understanding of their development. The mushroom body is comprised of ~2000 neurons (Kenyon cells) that belong to only three main neuronal types that have unique morphologies and play distinct roles in learning and memory (Cognigni et al., 2018; Ito et al., 1997; Lee et al., 1999). They receive input mainly from ~200 projection neurons that each relays odor information from olfactory receptor neurons (Vosshall and Stocker, 2007). Each projection neuron connects to a random subset of Kenyon cells and each Kenyon cell receives input from ~7 different projection neurons (Jefferis et al., 2007; Murthy et al., 2008; Turner et al., 2008). This connectivity pattern requires a large number of mushroom body neurons (~2,000) to represent complex odors (Hige, 2018). To produce this very large number of neurons, mushroom body development is unique in many respects. Mushroom body neurons are born from four identical neuroblasts that divide continuously (unlike any other neuroblast) from the late embryonic stages until the end of pupation (~9 days for ~250 divisions each) (Figure 1A; Ito et al., 1997; Kraft et al., 2016; Kunz et al., 2012; Kurusu et al., 2009; Lee et al., 1999; Pahl et al., 2019; Siegrist et al., 2010; Sipe and Siegrist, 2017). Furthermore, the two neurons born from each mushroom body GMC are identical. The neuronal simplicity of the adult mushroom body makes it ideal to study how extrinsic cues might affect diversity since the loss of any single neuronal type is obvious given that each is represented hundreds of times.
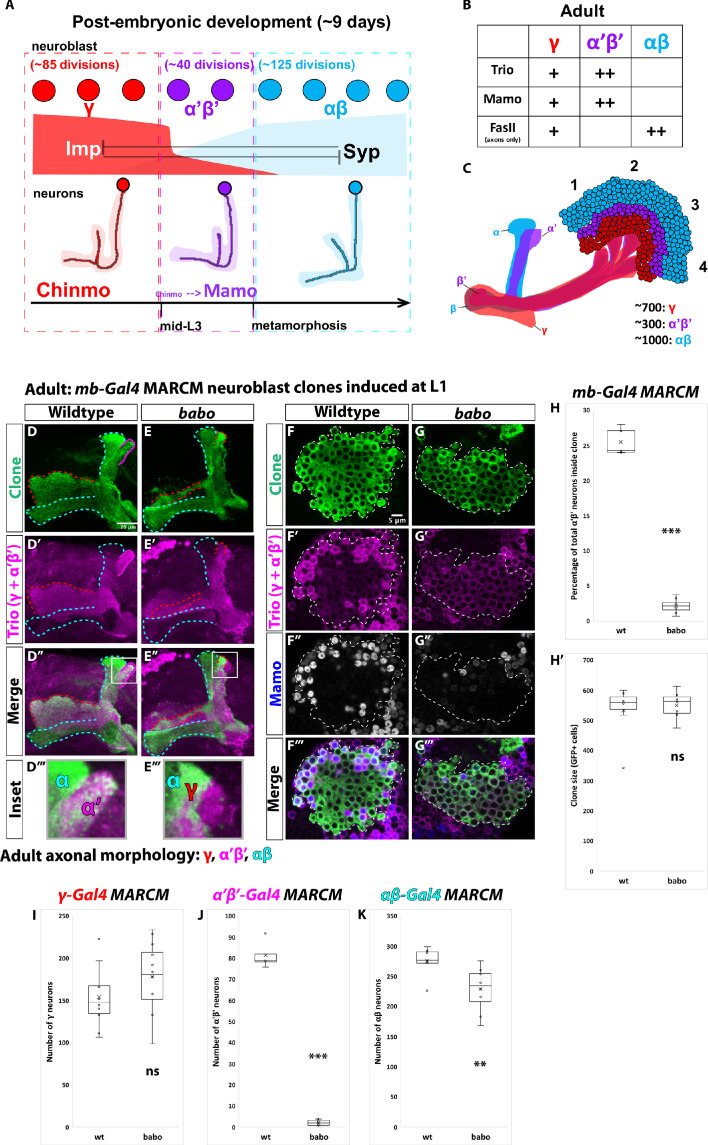
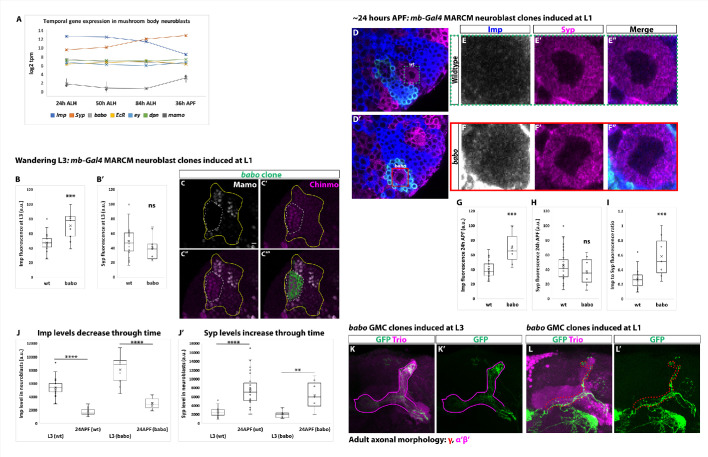
Figure 1. α’β’ neurons are not generated from babo mutant neuroblasts.
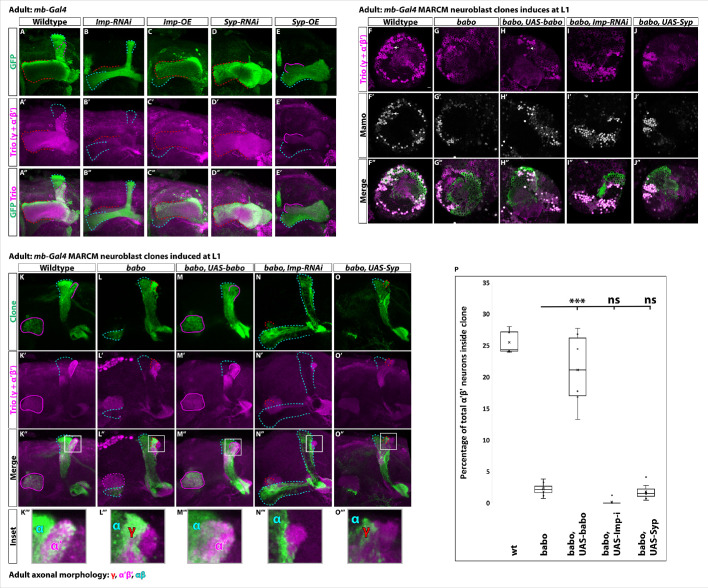
(A) Summary of intrinsic temporal patterning mechanism operating during mushroom body development. During early larval stages, mushroom body neuroblasts express high levels of Imp (red) and Chinmo (red) in neurons to specify γ identity for ~85 neuroblast divisions (red-dashed box). From mid-L3 to metamorphosis, when Imp and Syp (cyan) are both at low levels, the same neuroblast divides ~40 times to produce α’β’ neurons (magenta-dashed box). Low Chinmo regulates the expression Mamo, a terminal selector of α’β’ identity. From the beginning of metamorphosis throughout pupal development, high Syp leads to αβ neurons (cyan-dashed outline). (B) Known molecular markers can distinguish between the three mushroom body neuronal types in the adult. (C) Mushroom body projections originating from neurons born from four neuroblasts (numbered 1 to 4) per hemisphere fasciculate into a single bundle (peduncle) before branching into the five mushroom body lobes. The first-born γ neurons (red) remodel during development to project into a single, medial lobe in the adult. This lobe is the most anterior of the medial lobes. Axons from α’β’ neurons (magenta) bifurcate to project into the vertical and medial α’ and β’ lobes. The β’ lobe is posterior to the γ lobe. The last-born αβ neurons (cyan) also bifurcate their axons into the vertical projecting α lobe and medial projecting β lobe. The α lobe is positioned adjacent and medial to the α’ lobe. The β lobe is the most posterior medial lobe. (D-E) Representative max projections showing adult axons of clonally related neurons born from L1 stage in wildtype and babo conditions. UAS-CD8::GFP is driven by mb-Gal4 (OK107-Gal4). Outlines mark GFP+ axons, where γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. A white box outlines the Inset panel. Trio (magenta) is used to label all γ and α’β’ axons for comparison to GFP+ axons. (D) In wildtype, GFP+ axons (green, outlined in red, magenta and cyan) are visible in all observable mushroom body lobes. (E) In babo mutant clones, γ neurons (red outline) remain unpruned. GFP+ axons are missing inside the Trio+ α’ lobe, indicating the absence of α’β’ neurons. (F-G) Representative, single z-slices from the adult cell body region of clones induced at L1 in wildtype and babo conditions. UAS-CD8::GFP is driven by mb-Gal4. (F) Wildtype clones show the presence of strongly expressing Trio (magenta) and Mamo (blue, gray in single channel) neurons, indicative of α’β’ identity. (G) In babo mutant clones, cells strongly expressing Trio and Mamo are not present. (H) Quantification of MARCM clones marked by mb-Gal4, which labels all mushroom body neuronal types. The number of α’β’ neurons are quantified in wildtype (n = 7) and babo (n = 8) conditions. Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within clones, consistent with our expectation since each mushroom body is made from four neuroblasts. In babo clones, only 2.2 ± 0.4% of α’β’ neurons are within clones. (H’) There are no significant differences between the average clone sizes (wildtype:533.6 ± 33.3; babo:551.3 ± 17.6). (I) Quantification of γ neurons marked by γ-Gal4 (R71G10-Gal4) in MARCM clones. Plotted is the total number of γ neurons marked by GFP and Trio in wildtype (n = 10) and babo mutant (n = 12) clones. In wildtype, the average number of γ neurons is 154.3 ± 11.4. In babo mutants, the average is 178.4 ± 11.9. (J) Quantification of α’β’ neurons marked by α’β’-Gal4 (R41C07-Gal4) in MARCM clones. Plotted is the total number of α’β’ neurons marked by GFP and strong Trio in wildtype (n = 4) and babo mutant (n = 8) clones. In wildtype, the average number of α’β’ neurons is 81.5 ± 3.4. In babo mutants, the average is 2.1 ± 0.5. (K) Quantification of αβ neurons marked by αβ-Gal4 (R44E04-Gal4) in MARCM clones. Plotted is the total number of GFP+ cells in wildtype (n = 7) and babo mutant (n = 8) clones. In wildtype, the average number is 276 ± 9.1. In babo mutants, the average number is 228.9 ± 13.2. A two-sample, two-tailed t-test was performed. ***p<0.001, **p<0.01, ns: not significant. Scale bars: D, 20 µm; F, 5 µm.
Figure 1—figure supplement 1. α’β’ neurons are lost from the adult neuropil in activin signaling mutant clones.
Figure 1—figure supplement 2. γ neuron numbers likely increase, while αβ numbers decrease, in babo mutant clones.
The three main neuronal types that make up the adult mushroom body are produced sequentially during neurogenesis: first γ, followed by α’β’, and then αβ neurons (Lee et al., 1999; Figure 1A), representing the simplest lineage in the central brain. The γ temporal window extends from L1 (the first larval stage) until mid-L3 (the final larval stage) when animals attain critical weight and are committed to metamorphosis; the α’β’ window from mid-L3 to the beginning of pupation, and the αβ window from pupation until eclosion (the end of development). Like all other central brain neuroblasts Imp and Syp are expressed by mushroom body neuroblasts, but in much shallower gradients through time, which accounts for their extended lifespan (Liu et al., 2015; Yang et al., 2017). Imp and Syp are inherited by newborn neurons where they instruct temporal identity. Imp positively and Syp negatively regulate the translation of chronologically inappropriate morphogenesis (chinmo), a gene encoding a transcription factor that acts as a temporal morphogen in neurons (Kao et al., 2012; Ren et al., 2017; Zhu et al., 2006). The first-born γ neurons are produced for the first ~85 cell divisions, when Imp levels in neuroblasts, and thus Chinmo in neurons, are high. α’β’ neurons are produced for the next ~40 divisions, when Imp and Syp are at similar low levels that translate into lower Chinmo levels in neurons. Low Chinmo then regulates the expression in neurons of maternal gene required for meiosis (mamo), which encodes a transcription factor that specifies the α’β’ fate and whose mRNA is stabilized by Syp (Liu et al., 2019). αβ neurons are generated for the final ~125 neuroblast divisions, when Syp levels are high, Imp is absent in neuroblasts, and thus Chinmo and Mamo are no longer expressed in neurons.
Extrinsic cues are known to have important roles in regulating neuronal differentiation during mushroom body neurogenesis. The ecdysone peak that controls entry into pupation regulates γ neuron axonal remodeling (Lee et al., 2000). Ecdysone was also proposed to be required for the final differentiation of α’β’ neurons (Marchetti and Tavosanis, 2017). EcR expression in γ neurons is timed by activin signaling, a member of the TGFβ family, from local glia (Awasaki et al., 2011; Zheng et al., 2003). Activin signaling from glia is also required for the α’β’ fate (Marchetti and Tavosanis, 2019): Knocking-down the activin pathway receptor Baboon (Babo) leads to the loss of α’β’ neurons. It was proposed that activin signaling in mushroom body neuroblasts regulates the expression of EcR in prospective α’β’ neurons and that when the activin pathway is inhibited, it leads to the transformation of α’β’ neurons into later-born pioneer-αβ neurons (a subclass of the αβ class) (Marchetti and Tavosanis, 2019).
Although there is strong evidence that extrinsic cues have important functions in neuronal patterning in the Drosophila central brain, it remains unknown how extrinsic temporal cues interface with the Imp and Syp intrinsic temporal program to regulate neuronal specification. Here we address this question using the developing mushroom bodies. We independently discovered that activin signaling from glia is required for α’β’ specification. However, we show that activin signaling lowers the levels of the intrinsic factor Imp in mushroom body neuroblasts to define the mid-α’β’ temporal identity window. Removing the activin receptor Babo in mutant clones leads to the loss of α’β’ neurons, to fewer last-born αβ neurons, and to the likely generation of additional first-born γ neurons without affecting overall clone size. This appears to be caused by a delayed decrease in Imp levels, although the intrinsic temporal clock still progresses even in the absence of activin signaling. We also demonstrate that ecdysone signaling is not necessary for the specification of α’β’ neurons, although it might still be involved in later α’β’ differentiation. Our results provide a model for how intrinsic and extrinsic temporal programs operate within individual progenitors to regulate neuronal specification.
Results
α’β’ neurons are not generated from babo mutant neuroblasts
The production of the three different mushroom body neuronal types occurs within specific developmental stages of larval and pupal development. That is, the γ window extends from L1 to mid-L3, the α’β’ window from mid-L3 to pupation, and the αβ window from pupation to eclosion (Figure 1A; Lee et al., 1999). This means that extrinsic cues could play a role in controlling or fine-tuning transitions between these temporal windows. Additionally, the specification of neuronal identity within each temporal window could be aided by extrinsic cues. To test these hypotheses, we used Mosaic Analysis with a Repressible Cell Marker (MARCM) (Lee and Luo, 1999) to test the function of receptors for inter-cellular signaling pathways with known roles either in mushroom body neurogenesis (Activin and Ecdysone) (Lee et al., 2000; Marchetti and Tavosanis, 2017; Marchetti and Tavosanis, 2019; Zheng et al., 2003) or more broadly during nervous system development (Hedgehog and juvenile hormone) (Figure 1—figure supplement 1A–H; Baumann et al., 2017; Chai et al., 2013). We induced mushroom body neuroblast clones at L1 and compared the axonal morphologies of adult neurons born from mutant neuroblasts to neurons born from surrounding wildtype neuroblasts. To identify mushroom body axonal lobes (both mutant and wildtype), we used antibodies to the Rho guanine exchange factor Trio (a weak γ and strong α’β’ cytoplasmic marker) and to the cell adhesion molecule Fasciclin II (FasII) (an axonal γ and αβ marker) (Figure 1B; Awasaki et al., 2000; Crittenden et al., 1998). To visualize mushroom body neurons within clones we expressed UAS-CD8::GFP under the control of OK107-Gal4 (referred to as mb-Gal4 hereafter), a Gal4 enhancer trap in eyeless and a common mushroom body Gal4 driver that strongly labels all mushroom body neuronal types during development and in the adult, and weakly mushroom body neuroblasts and young neurons throughout development (Connolly et al., 1996; Liu et al., 2015; Zhu et al., 2006).
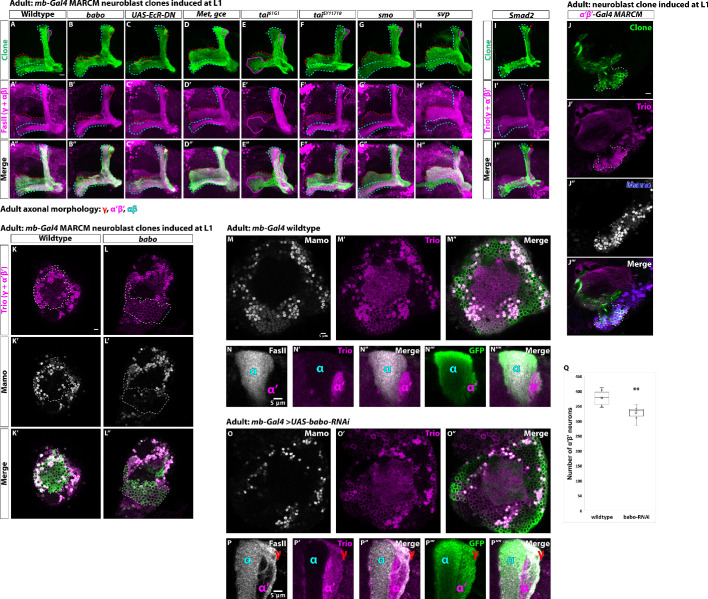
In wildtype clones induced at L1, GFP+ axons projected to all five mushroom body lobes: α, α΄, β, β’ (hidden behind the γ lobe in max projections), and γ (Figure 1D, Figure 1—figure supplement 1A). In clones mutant for babo, we did not detect GFP+ axons within the α’β’ lobes, which remained visible by Trio staining due to the presence of wildtype α’β’ neurons (Figure 1E, Figure 1—figure supplement 1B). In addition, and as previously described, γ neurons within babo mutant clones remained unpruned (visualized by vertical GFP+ axons that were Trio+ and FasII+), providing a positive control since γ remodeling is known to require activin signaling (Figure 1E, Figure 1—figure supplement 1B; Awasaki et al., 2011; Yu et al., 2013b; Zheng et al., 2003).
Babo is the sole Type I receptor in the activin pathway (a member of the TGFβ family of signaling molecules). Babo with its Type II co-receptors binds four different activin ligands and acts through the transcription factor Smad on X (Smad2) (Brummel et al., 1999; Upadhyay et al., 2017). We induced Smad2 mutant clones at L1 and characterized adult axonal morphologies. Similar to babo mutant clones, Smad2 clones were missing α’β’ neurons and also contained unpruned γ neurons (Figure 1—figure supplement 1I).
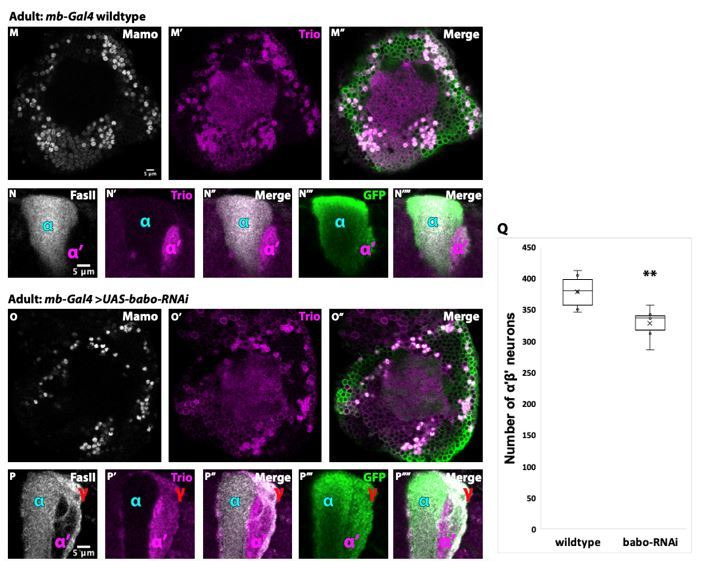
The absence of GFP+ axons within the α’β’ lobes in babo mutant clones could be due to the loss of axonal projections, or to the loss of neuronal identity. Using antibodies against Trio and Mamo that strongly label α’β’ neuron cell bodies in the adult (Figure 1B, Figure 1—figure supplement 1J; Alyagor et al., 2018; Awasaki et al., 2000; Croset et al., 2018; Liu et al., 2019), we detected strong Trio+ and Mamo+ cells within adult GFP+ clones induced at L1 (Figure 1F, Figure 1—figure supplement 1K). In babo mutant clones however, the vast majority of strong Trio+ and Mamo+ cells inside clones were missing compared to surrounding wildtype neurons (Figure 1G, Figure 1—figure supplement 1L), suggesting that α’β’ neurons were not specified. We quantified the number of α’β’ neurons in wildtype and babo clones by counting the number of strong Mamo+ cells within a clone versus the total number of strong Mamo+ cells outside the clone. In wildtype MARCM clones affecting a single mushroom body neuroblast (n = 7), the percentage of all α’β’ neurons that were present within the clones was 25.5 ± 0.7%, the expected ratio since each mushroom body is built from four identical neuroblasts (Figure 1H; Ito et al., 1997; Lee et al., 1999). In comparison, in babo mutant clones (n = 8) the percentage of α’β’ neurons within clones was 2.2 ± 0.4% (Figure 1H). Interestingly, although there was a decrease in the number of adult α’β’ neurons upon expression of UAS-babo-RNAi with mb-Gal4, the majority of α’β’ neurons were not lost (Figure 1—figure supplement 1M–Q). Importantly, γ neurons in these brains did not remodel (Figure 1—figure supplement 1P–P’’’), indicating that the babo-RNAi worked efficiently. This difference with babo clones is likely due to the weak expression of mb-Gal4 in neuroblasts and newborn neurons and suggested to us that activin signaling is necessary for α’β’ specification by acting in neuroblasts (see below).
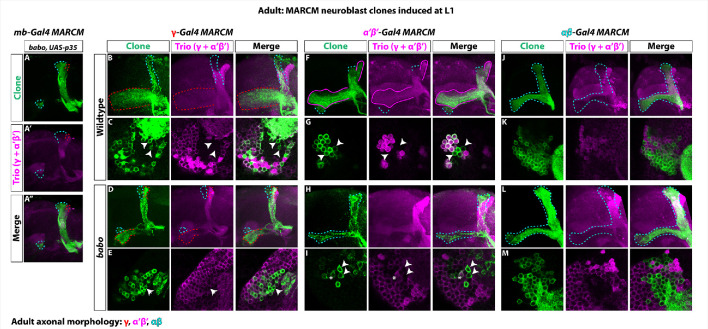
We next sought to determine the fate of the missing α’β’ neurons in babo clones, particularly since there was no significant difference in average clone sizes between mutant and control clones labeled with mb-Gal4 (wildtype: clone size = 533.6 ± 33.3, n = 7; babo: clone size = 551.3 ± 17.6, n = 7) (Figure 1H’), which suggests that there is no defect in neuroblast proliferation, and that α’β’ neurons are not lost by cell death in babo clones. However, to directly test whether cell death played a role, we expressed the caspase inhibitor P35 in babo mutant clones (Figure 1—figure supplement 2A). However, α’β’ neurons were still missing in the adult (Figure 1—figure supplement 2A), indicating that α’β’ neurons are not generated and then die. We thus tested whether the γ or αβ temporal windows were extended in babo mutant clones. We made MARCM clones in which the γ, α’β’ or αβ neurons were specifically marked with different Gal4 lines, and then quantified the total number of GFP+ neurons in wildtype versus babo mutant clones (Figure 1I–K, Figure 1—figure supplement 2B–M). Using R71G10-Gal4 (Issman-Zecharya and Schuldiner, 2014) (referred to as γ-Gal4), the average number of γ neurons trended higher in babo mutant clones, although not significantly (wildtype: 154.3 ± 11.4, n = 10; babo: 178.4 ± 11.9, n = 12) (Figure 1I, Figure 1—figure supplement 2B–E), likely because the number of γ neurons directly depends on the time of clone induction. α’β’ neurons, marked by R41C07-Gal4 (referred to as α’β’-Gal4), were mostly missing in babo mutant clones compared to wildtype clones (wildtype: 81.5 ± 3.6, n = 4; babo: 2.1 ± 0.5, n = 8,) consistent with our previous results when counting strong Mamo+ cells in babo clones marked by mb-Gal4 (Figure 1H, Figure 1J, Figure 1—figure supplement 2F–I). The average number of αβ neurons, marked by R44E04-Gal4 (referred to as αβ-Gal4), was significantly reduced in babo versus wildtype clones (wildtype: 276 ± 9.1, n = 7,; babo: 228.9 ± 13.2, n = 8) (Figure 1K, Figure 1—figure supplement 2J–M). Together, these results suggest that additional γ neurons are likely produced, and that fewer αβ neurons are generated, in babo mutant clones. Although this might appear to contradict a recent report that showed that a subclass of αβ neurons (pioneer-αβ) increases (Marchetti and Tavosanis, 2019), it is likely that the earliest born pioneer-αβ are increased because of the delayed decrease in the Imp gradient, but that the later-born αβ neurons do not have time to form (see below and Discussion). We note that the total number of neurons labeled by our neuron type specific Gal4 drivers did not add up to the expected number of ~500 neurons in babo mutant clones, which is likely explained by the large variability in the number of γ neurons labeled by γ-Gal4. Next, we focused on understanding whether and how activin signaling interacts with the intrinsic Imp and Syp temporal program.
Activin signaling acts in neuroblasts to lower Imp levels and specify α’β’ neurons
Given that α’β’ neuronal specification is intrinsically controlled by Imp and Syp (Liu et al., 2015), we asked whether activin signaling acts through or in parallel to this intrinsic temporal system, specifically at L3 when α’β’ neurons are being produced. We first asked whether babo is expressed at L3 in mushroom body neuroblasts. Based on published transcriptome data collected from mushroom body neuroblasts at different developmental stages (Liu et al., 2015; Yang et al., 2016), babo is expressed evenly through time in mushroom body neuroblasts, unlike the two RNA binding proteins Imp and Syp (Figure 2—figure supplement 1A). Although this measure does not take into account the possibility of post-transcriptional regulation, it is likely that the activin signaling pathway is temporally controlled by ligand interaction and not by differential expression of babo.
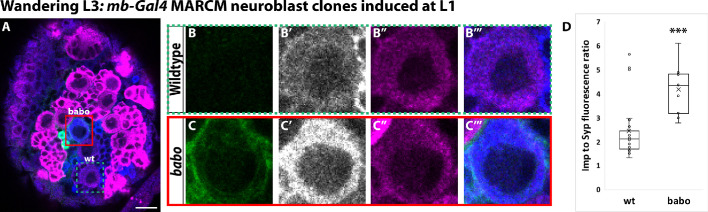
To directly test whether activin signaling acts on Imp and Syp to affect α’β’ specification, we induced MARCM clones for babo at L1 and compared the Imp to Syp protein ratio in mutant mushroom body neuroblasts to surrounding wildtype neuroblasts at wandering L3 (Figure 2). The average Imp to Syp ratio was significantly higher in babo neuroblasts (ratio: 4.2 ± 0.4; n = 9 from 4 different brains) compared to wildtype neuroblasts (ratio: 2.4 ± 0.2; n = 23 from the same 4 brains as babo) at L3, driven by a significantly higher Imp level in mutant neuroblasts (Figure 2, Figure 2—figure supplement 1B) while Syp was not significantly different (Figure 2—figure supplement 1B’). In addition, the α’β’ neuronal marker Mamo (Liu et al., 2019) was lost in babo mutant clones at L3 while the level of Chinmo was higher in these neurons (Figure 2—figure supplement 1C–C’’’), consistent with the notion that high Imp levels block α’β’ specification through maintained higher levels of Chinmo that likely lead to the increased production of γ neurons. The significantly higher Imp to Syp ratio in babo mutant neuroblasts persisted even ~24 hr After Pupal Formation (APF) (babo ratio: 0.58 ± 0.11; n = 7 from 6 different brains; wildtype ratio: 0.27 ±. 02; n = 27 from the same 6 brains as babo), once again driven by higher Imp levels (Figure 2—figure supplement 1D–I). Together, these results indicate that activin signaling lowers Imp levels at late larval and early pupal stages. Importantly, although Imp was higher in babo mutant neuroblasts and persisted longer, the absolute level of Imp still decreased significantly albeit with prolonged kinetics, while the absolute level of Syp was higher in babo mutant neuroblasts at ~24 hr APF vs. L3 (Figure 2—figure supplement 1J–J’): Thus, these changes are either intrinsically regulated or are affected by additional extrinsic factors. Our finding that Imp and Chinmo were higher in babo mutant neuroblasts and neurons at L3 is also consistent with our suggestion that additional γ neurons are produced during the α’β’ time window. The lack of α’β’ neurons in babo mutant clones even though Imp levels were finally low at ~24 hr APF suggests that α’β’ specification may only occur from L3 to the start of pupation.
Figure 2. Activin signaling acts in neuroblasts to lower Imp levels.
(A) Representative image of a babo mushroom body neuroblast marked by UAS-CD8::GFP driven by mb-Gal4 (red box) adjacent to a wildtype neuroblast (green-dashed box) in the same focal plane from a wandering L3 stage brain, immunostained for Imp (blue, gray in single channel) and Syp (magenta). (B) Close-up view of wildtype neuroblast (green-dashed box in A). (C) Close up view of babo mutant neuroblast (red box in A). (D) Quantification of the Imp to Syp ratio in babo neuroblasts (4.2 ± 0.4, n = 9 from 4 different brains) compared to wildtype (2.4 ± 0.3, n = 23 from the same 4 brains as babo neuroblasts). A two-sample, two-tailed t-test was performed. ***p<0.001, ns: not significant. Scale bar: 10 µm.
Figure 2—figure supplement 1. Activin signaling lowers Imp levels but the Imp to Syp transition does not depend on activin signaling.
We have shown that activin signaling functions in mushroom body neuroblasts to decrease Imp during L3. However, previous studies have shown that Babo also acts post-mitotically in mushroom body γ neurons where it times the expression of EcR for their remodeling, indicating that Babo can act independently in neuroblasts and in neurons (Zheng et al., 2003). To test if activin signaling functions post-mitotically in prospective α’β’ neurons, we characterized the morphology of babo mutant neurons born from ganglion mother cell (GMC) clones induced during mid-late L3, the time at which α’β’ neurons are born. GMCs are intermediate progenitors that divide only once to produce two neurons. In this way, the role of Babo in prospective α’β’ neurons can be tested without affecting mushroom body neuroblasts: α’β’ neurons were present in babo GMC clones (n = 34/34), observable by axonal projections into the Trio labeled α’β’ lobes (Figure 2—figure supplement 1K–K’). As a positive control for the efficiency of babo GMC clones, we also made babo GMC clones at L1 to target γ neurons. In the majority of cases, γ axons remained unpruned (n = 8/10, Figure 2—figure supplement 1L–L’; Zheng et al., 2003). These results show that activin signaling acts in mushroom body neuroblasts, and not in neurons, to specify the α’β’ fate.
Activin signaling is sufficient to expand production of α’β’ neurons
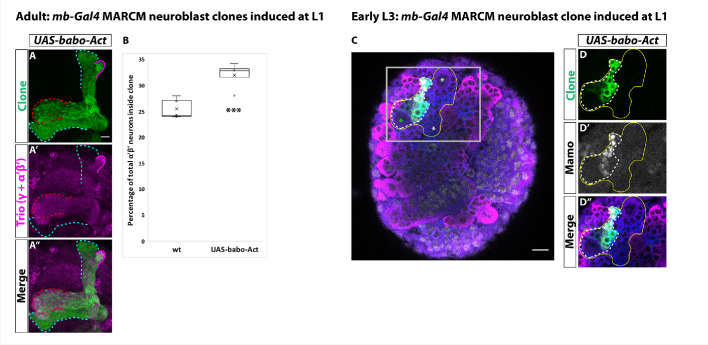
Since activin signaling functions in mushroom body neuroblasts and is necessary for α’β’ specification, we next investigated whether it is sufficient for the α’β’ fate. We expressed a constitutively active form of the Babo receptor (UAS-Babo-Act) throughout development in MARCM clones with mb-Gal4 and assessed the total number of α’β’ neurons in the adult by strong Mamo expression. While in wildtype clones the percentage of α’β’ neurons was 25.5 ± 0.7% (n = 7), the number of α’β’ neurons present within UAS-Babo-Act clones significantly increased to 32 ± 1.4% (n = 4) (Figure 3A–B). To ask when these additional α’β’ neurons were produced, we characterized the expression of the α’β’ marker Mamo in young neurons at early L3, when γ neurons are being produced. In comparison to adjacent wildtype neurons, Mamo was expressed in neurons in UAS-Babo-Act clones at this stage (Figure 3C–D). These results confirm the precocious specification of α’β’ neurons, likely at the expense of γ neurons (which would normally be produced at this stage). Importantly, constitutively expressing an activated version of Babo did not result in adult clones consisting entirely of α’β’ neurons, highlighting that activin signaling alone is not master regulator of the α’β’ fate and that other components are necessary to specify this neuronal type.
Figure 3. Activin signaling is sufficient to expand production of α’β’ neurons.
(A) Expression of UAS-Babo-Act by mb-Gal4 leads to additional α’β’ neurons but does not convert all mushroom body neurons into this fate. (B) Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. The number of α’β’ neurons is quantified in wildtype (n = 7, replotted from data in Figure 1H) and UAS-babo-Act (n = 4). In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within a clone while precociously activating the activin pathway increased the percentage to 32 ± 1.4%. (C) A representative image of an early L3 brain in which a single mushroom body neuroblast is expressing UAS-babo-Act driven by mb-Gal4 (white-dashed line). Imp (blue) and Syp (magenta), along with GFP (green), are used to identify mushroom body neuroblasts (asterisks) and neurons. (D) Inset (gray box in C) showing that Mamo (gray) is expressed inside GFP+ cells that express UAS-babo-Act but not outside in adjacent wildtype mushroom body neurons (yellow line) (n = 3/3). A two-sample, two-tailed t-test was performed. ***p<0.001.
Glia are the source of the activin ligand Myoglianin to specify α’β’ neurons
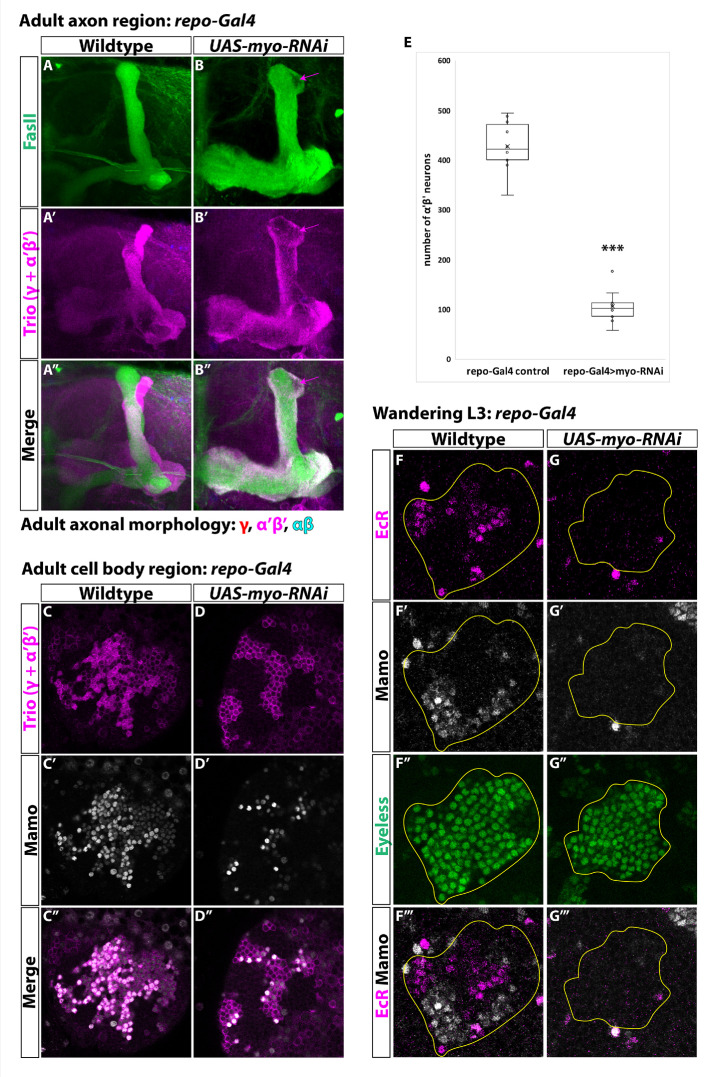
Our finding that activin signaling plays an important role in specifying α’β’ identity during mushroom body development led us to question from where the activin ligand originates. Glia secrete the activin ligand Myoglianin (Myo) to initiate γ neuron remodeling by activating EcR at L3 (Awasaki et al., 2011; Yu et al., 2013b). Therefore, we hypothesized that Myo from glia may also regulate α’β’ specification. To test this, we knocked-down myo by expressing UAS-myo-RNAi with repo-Gal4, a driver expressed in all glia, and quantified the total number of α’β’ neurons based on strong Mamo expression in the adult (Figure 4A–E). In comparison to control (428.9 ± 16.2, n = 10), the number of α’β’ neurons was dramatically reduced (106.6 ± 11.4; n = 10) (Figure 4E). Mamo was also not expressed in mushroom body neurons at L3 (Figure 4F–G). Importantly, EcR was not expressed in γ neurons at this stage, providing a positive control for the efficiency of UAS-myo-RNAi (Awasaki et al., 2011). We note that even though the number of α’β’ neurons was reduced in this experiment, myo knockdown was weaker than babo mutant clones, possibly due to incomplete knockdown of myo or because more than one ligand (or more than one source) contribute to α’β’ specification (see Discussion). Our results are consistent with a recent report that also showed that glia are the source of Myo for α’β’ specification (Marchetti and Tavosanis, 2019).
Figure 4. Glia are the source of the activin ligand Myo to specify α’β’ neurons.
(A-B) Representative images of adult mushroom body lobes labeled by FasII (green) and Trio (magenta). (A) In wildtype controls (428.9 ± 16.2, n = 10) (repo-Gal4 only) all three neuronal types are present based on axonal projections. (B) Expressing UAS-myo-RNAi (106.6 ± 11.4, n = 10) causes γ neurons not to remodel and to the loss of the majority of α’β’ neurons, however some still remain (purple arrow, FasII- region). (C-D) Representative images of adult mushroom body cell body region. Trio (magenta) and Mamo (gray) are used to distinguish between the three neuronal types. Expressing UAS-myo-RNAi leads to loss of the majority of strong Mamo+ and Trio+ cells, indicating the loss of α’β’ neurons. (E) Quantification of phenotypes presented in A-D. (F) At L3, EcR (magenta) and Mamo (gray) are expressed in mushroom body neurons labeled by Eyeless (green, yellow outline). Mamo+ cells are newborn α’β’ neurons. G. Expressing UAS-myo-RNAi with repo-Gal4 leads to loss of both Mamo and EcR in mushroom body neurons. A two-sample, two-tailed t-test was performed. ***p<0.001.
α’β’ neurons are specified by low Imp levels at L3
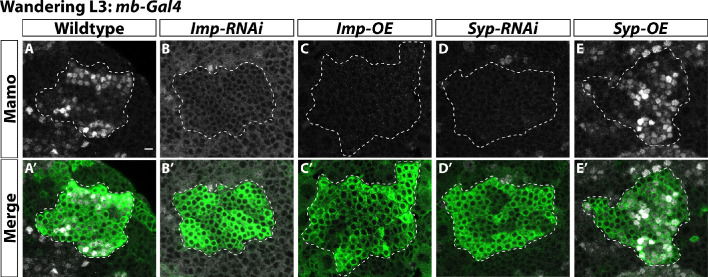
We and others have shown that activin signaling is necessary for α’β’ specification (Marchetti and Tavosanis, 2019). We have shown that activin signaling acts by lowering Imp levels at L3. Although Imp is required for α’β’ specification (Figure 5—figure supplement 1A–E; Liu et al., 2015), we wanted to determine whether low Imp levels are required at L3. We therefore characterized Mamo expression in young neurons at L3 following knockdown (UAS-Imp-RNAi) or overexpression (UAS-Imp-OE) of Imp with mb-Gal4. (Figure 5A–C; Liu et al., 2015). Consistent with our model, Mamo was not expressed in either condition. In comparison, although knocking-down Syp by expressing UAS-Syp-RNAi led to the loss of Mamo, its early overexpression (UAS-Syp-OE) did not (Figure 5D–E). The loss of Mamo in Syp knockdown is consistent with its role in stabilizing mamo transcripts at L3 (Liu et al., 2019). We conclude that low Imp and low or high Syp levels are required for α’β’ specification. Consistent with this, we were unable to rescue the loss of α’β’ neurons in babo mutant clones by constitutively repressing Imp with UAS-Imp-RNAi (0.2 ± 0.2%, n = 7) (Figure 5—figure supplement 1I, Figure 5—figure supplement 1N, Figure 5—figure supplement 1P), likely due Imp reduction below the threshold required for α’β’ specification (see Discussion). However, we could rescue babo mutant clones by expressing UAS-babo (21.1 ± 2.4%, n = 6) (Figure 5—figure supplement 1F–H, Figure 5—figure supplement 1K–M, Figure 5—figure supplement 1P). Overexpressing Syp (UAS-Syp-OE) to reduce the altered Imp:Syp ratio in babo mutant clones also did not rescue α’β’ neurons (1.8 ± 0.5%, n = 7) (Figure 5—figure supplement 1J, Figure 5—figure supplement 1O–P), further highlighting that Imp but not Syp levels are important for α’β’ specification.
Figure 5. α’β’ neurons are specified by low Imp levels at L3.
(A-A’) Representative image of wildtype mushroom body neurons labeled by mb-Gal4 driving UAS-CD8::GFP (green, white-dashed outline) during the wandering L3 stage. Mamo (gray) is used as a marker for α’β’ neurons. (B-B’) When mb-Gal4 is used to drive UAS-Imp-RNAi, Mamo is not expressed. (C-C’) Similarly, Mamo expression is lost when overexpressing Imp (UAS-Imp-overexpression (OE)). (D-D’). Expressing UAS-Syp-RNAi also leads to the loss of Mamo. (E) Expressing UAS-Syp-overexpression (OE) does not affect Mamo. Scale bar: 5 µm.
Figure 5—figure supplement 1. Low Imp levels are required for α’β’ specification.
Ecdysone signaling is not necessary for α’β’ specification
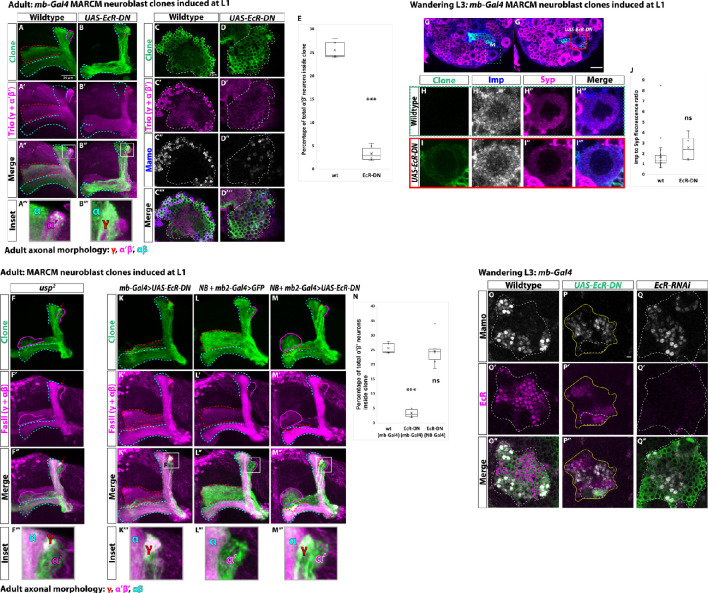
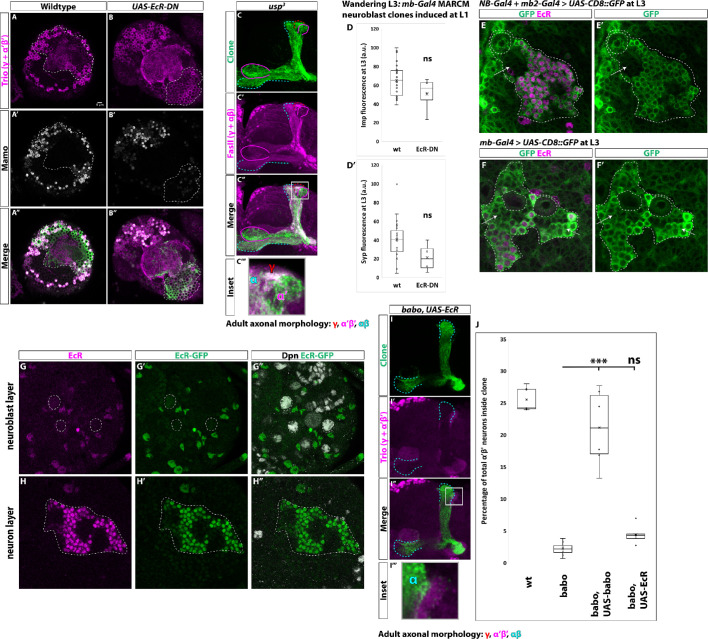
It has been proposed that activin signaling in mushroom body neuroblasts leads to EcR expression in neurons and that ecdysone signaling at late L3 induces differentiation of α’β’ neurons (Marchetti and Tavosanis, 2017; Marchetti and Tavosanis, 2019). The role of ecdysone was tested by expressing a dominant-negative ecdysone receptor (UAS-EcR-DN). We confirmed these results by also expressing UAS-EcR-DN driven by mb-Gal4 and were unable to detect GFP+ mutant axons within adult α’β’ lobes marked by Trio (Figure 6A–B). In addition, strong Trio+ and Mamo+ cells were missing inside UAS-EcR-DN mutant clones compared to wildtype clones (wildtype: 25.5 ± 0.7%, n = 7; UAS-EcR-DN: 3.4 ± 0.6%, n = 6) (Figure 6C–E, Figure 6—figure supplement 1A–B). However, we were surprised to find that α’β’ neurons were still present in mutant clones for the EcR co-receptor ultraspiracle (usp) (Figure 6F, Figure 6—figure supplement 1C). Therefore, we sought to better understand how expressing UAS-EcR-DN blocks α’β’ specification.
Figure 6. Ecdysone signaling is not necessary for α’β’ specification.
(A-B) Representative max projections showing adult axons of clonally related neurons born from L1 stage in wildtype and UAS-EcR-DN conditions. UAS-CD8::GFP is driven by mb-Gal4 (OK107-Gal4). Outlines mark GFP+ axons, where γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. A white box outlines the Inset panel. Trio (magenta) is used to label all γ and α’β’ axons for comparison to GFP+ axons. (A) In wildtype, GFP+ axons are visible in all mushroom body lobes. (B) α’β’ axons are lost, and γ neurons do not remodel, in UAS-EcR-DN expressing clones. (C-D) Representative, single z-slices from the adult cell body region of clones induced at L1 in wildtype and UAS-EcR-DN conditions. UAS-CD8::GFP is driven by mb-Gal4. (C) Wildtype clones show the presence of strongly expressing Trio (magenta) and Mamo (blue, gray in single channel) neurons, indicative of α’β’ identity. (D) In UAS-EcR-DN clones, strong Trio and Mamo cells are not present. (E) Quantification of MARCM clones marked by mb-Gal4, which labels all mushroom body neuronal types. The number of α’β’ neurons are quantified in wildtype (n = 7, replotted from data in Figure 1H) and UAS-EcR-DN (n = 6) conditions. Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within clones. In UAS-EcR-DN clones, only 3.4 ± 0.6% of α’β’ neurons are within clones. (F) usp mutant clones contain α’β’ neurons. FasII (magenta) is used to label γ and αβ lobes. Red arrow indicates unpruned γ neurons. (G) Representative image of an UAS-EcR-DN expressing neuroblast marked by UAS-CD8::GFP driven by mb-Gal4 (red box) ventral to a wildtype neuroblast (green-dashed box) from the same wandering L3 stage brain, immunostained for Imp (blue, gray in single channel) and Syp (magenta). (H) Close-up view of wildtype neuroblast (green-dashed box in G). (I) Close-up view of UAS-EcR-DN neuroblast (red box in G). (J) Quantification of the Imp to Syp ratio in UAS-EcR-DN neuroblasts (n = 4 from four different brains) compared to wildtype (n = 27 from the same four brains as UAS-EcR-DN neuroblasts). (K) A representative adult mushroom body clone (green) induced at L1 expressing UAS-EcR-DN driven by mb-Gal4. α’β’ neurons (GFP+ (green), FasII- (magenta)) are not observed and γ neurons do not remodel (GFP+, FasII+, red outline). (L) A representative adult wildtype clone induced at L1 driven by NB + mb2-Gal4. All three neuron types are present, including α’β’ neurons (GFP+, FasII-, magenta outline). (M) α’β’ neurons are also present when UAS-EcR-DN is driven by NB + mb2-Gal4 although γ neurons do not remodel. (N) Quantification of MARCM clones in which UAS-EcR-DN is driven by mb-Gal4 (n = 6, replotted from data in E) or NB + mb2-Gal4 (n = 6) compared to wildtype (n = 7, replotted from data in Figure 1H). In UAS-EcR-DN clones driven by NB + mb2-Gal4, 24.6 ± 2.1% of α’β’ neurons are within a clone, similar to wildtype. O. At L3, Mamo (gray) is expressed in young mushroom body neurons (α’β’) while EcR (magenta) can only be detected in more mature neurons (mainly γ at this stage). Note that there is no overlap between Mamo and EcR. (P) Expressing UAS-EcR-DN with mb-Gal4 (green, white outline) leads to the loss of Mamo expression (gray) inside the clone but not in surrounding wildtype mushroom body neurons. (Q) In contrast, expressing UAS-EcR-RNAi drivenE by mb-Gal4 abolishes EcR expression but does not affect Mamo. For E and J a two-sample, two-tailed t-test was performed. For N, a Tukey test was performed. ***p<0.001, ns: not significant. Scale bars: A, 20 µm; G, 10 µm; P, 5 µm.
Figure 6—figure supplement 1. Ecdysone signaling is not necessary for α’β’ specification.
First, unlike our result in babo mutant neuroblasts, we did not observe a significant difference in the average Imp to Syp protein ratio at L3 in UAS-EcR-DN expressing mushroom body neuroblasts with mb-Gal4 (UAS-EcR-DN ratio: 2.6 ± 0.7, n = 4 from four different brains; wildtype ratio: 1.7 ± 0.3, n = 27 from the same four brains as UAS-EcR-DN) (Figure 6G–J, Figure 6—figure supplement 1D–D’). Driving even stronger expression of UAS-EcR-DN in mushroom body neuroblasts with inscuteable-Gal4 (referred to as NB-Gal4) and labeling all adult neurons with R13F02-Gal4 (referred to as mb2-Gal4) (Jenett et al., 2012) also did not lead to the loss of α’β’ neurons in the adult (Figure 6K–N, Figure 6—figure supplement 1E–F). These results indicate that EcR-DN only blocks α’β’ specification when expressed in newborn mushroom body neurons, not in neuroblasts.
Given these results, we next asked whether expressing UAS-EcR-DN affects Mamo during development, which labels newborn postmitotic α’β’ neurons (Liu et al., 2019); Mamo expression was lost in UAS-EcR-DN expressing clones driven by mb-Gal4 (Figure 6P) but it was not affected by the expression of UAS-EcR-RNAi (Figure 6Q), although the RNAi was effective since we could not detect EcR protein in mushroom body neurons (Figure 6O, Figure 6Q). Given these contradictory results, we compared Mamo and EcR expression at L3. However, Mamo and EcR were mutually exclusive as EcR was not expressed in newborn α’β’ neurons (see Figure 6O’’), which precludes the possibility that EcR-DN inhibits EcR function in these neurons and might therefore act through off-target inhibition of Mamo. These results confirm the lack of EcR protein in mushroom body neuroblasts and young neurons at wandering L3 (Figure 6—figure supplement 1G–H), although EcR was clearly expressed in mature (mostly γ) neurons at this stage (Figure 6—figure supplement 1H–H’’; Lee et al., 2000; Liu et al., 2015; Marchetti and Tavosanis, 2017). Finally, we were unable to rescue the loss of α’β’ neurons in babo mutant clones by expressing UAS-EcR-B1 (Figure 6—figure supplement 1I–J), consistent with the notion that EcR does not function in α’β’ specification downstream of activin signaling (see Discussion). In summary, α’β’ neurons were only lost in adult clones when expressing UAS-EcR-DN (with mb-Gal4) and not in usp mutant clones, and α’β’ neurons were still present when expressing EcR-RNAi in L3. Most importantly, EcR protein was not detected in Mamo+ cells during development, although expressing UAS-EcR-DN blocked Mamo expression at L3. Thus, it is unlikely that ecdysone signaling is involved in α’β’ specification although it might still be used later during α’β’ differentiation (Marchetti and Tavosanis, 2017). We conclude that the loss of α’β’ neurons when expressing UAS-EcR-DN is caused by artifactual inhibition of Mamo (see Discussion).
Discussion
Establishment of mushroom body neuronal identities
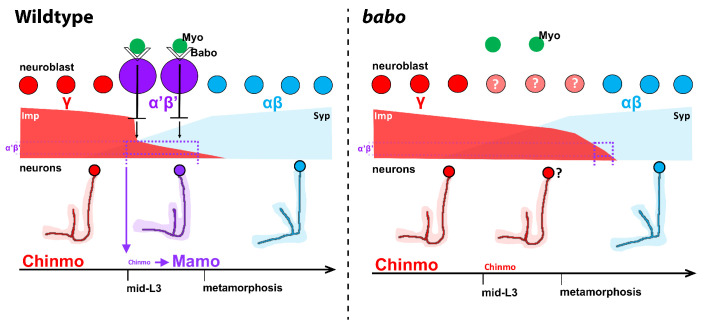
Mushroom body neurogenesis is unique and programmed to generate many copies of a few neuronal types. During the early stages of mushroom body development, high Imp levels in mushroom body neuroblasts are inherited by newborn neurons and translated into high Chinmo levels to specify γ identity. As in other central brain neuroblasts, as development proceeds, inhibitory interactions between Imp and Syp help create a slow decrease of Imp and a corresponding increase of Syp. However, at the end of the γ temporal window (mid-L3), activin signaling from glia acts to rapidly reduce Imp levels in mushroom body neuroblasts without significantly affecting Syp, establishing a period of low Imp (and thus low Chinmo in neurons) and also low Syp. This is required for activating effector genes in prospective α’β’ neurons, including Mamo, whose translation is promoted by Syp (Liu et al., 2019). The production of αβ identity begins when Imp is further decreased and Syp levels are high during pupation (modeled in Figure 7). Low Chinmo in αβ neurons is also partly regulated by ecdysone signaling through the activation of Let-7-C, which targets chinmo for degradation (Kucherenko et al., 2012; Wu et al., 2012). Based on our model, α’β’ neurons could not be rescued by knocking-down Imp in babo clones (Figure 5—figure supplement 1I,N,P), since low Imp is required for α’β’ specification while the knockdown reduces its level below this requirement. We might expect to rescue α’β’ neurons if Imp levels were specifically reduced to the appropriate levels at L3. However, reducing Imp levels might not be the only function of activin signaling, which may explain why α’β’ neurons are not simply made earlier (e.g., during L1-L2) when Imp is knocked-down.
Figure 7. Model of how activin signaling defines the α’β’ temporal identity window.
In wildtype, as development proceeds, mushroom body neuroblasts incorporate an activin signal (Myo) from glia through Babo to lower the level of the intrinsic temporal factor Imp (magenta dashed line). The lower Imp levels inherited by newborn neurons leads to lower Chinmo levels to control the expression of the α’β’ effector Mamo, defining the mid-temporal window (magenta dashed lines). In babo mutants, Imp remains higher for longer, leading to the loss of Mamo (and likely many other targets) during mid-late L3 in neurons. In this model, γ neuron numbers increase, α’β’ neurons are lost, and fewer αβ neurons are produced. Nonetheless, the Imp to Syp transition still occurs, allowing for young (γ) and old (αβ) fates to be produced.
In babo mutant clones, we speculate that additional γ neurons are produced at the expense of α’β’ neurons since Imp levels in neuroblasts (as well as Chinmo in neurons) are higher for a longer time during development; There was also a significant decrease in the total number of αβ neurons in babo mutant clones that contrasts with a recent report by Marchetti and Tavosanis that instead concluded that additional pioneer-αβ neurons are produced (Marchetti and Tavosanis, 2019). We believe that there is both an increase in the number of γ neurons and of the pioneer-αβ neuron subclass because pioneer-αβ neurons are the first of the αβ class to be specified (when Imp is still present at very low levels) during pupation. We speculate that pioneer-αβ neurons are produced during the extended low Imp window that we detect during pupation in babo clones. However, this does not leave the time for the remaining population of αβ neurons to be formed, which explains why their number is reduced.
In this study, we have focused on the three main classes of mushroom body neurons although at least seven subtypes exist: 2 γ, 2 α’β’ and 3 αβ (Aso et al., 2014; Shih et al., 2019). The subtypes are specified sequentially (Aso et al., 2014) suggesting that each of the three broad mushroom body temporal windows can be subdivided further, either by fine-scale reading of the changing Imp and Syp gradients, by additional extrinsic cues, or perhaps by a tTF series as in other neuroblasts.
Temporal patterning of Drosophila central brain neuroblasts
Postembryonic central brain neuroblasts are long-lived and divide on average ~50 times. Unlike in other regions of the developing Drosophila brain, rapidly progressing series of tTFs have not yet been described in these neuroblasts (Doe, 2017; Holguera and Desplan, 2018; Kohwi and Doe, 2013; Rossi et al., 2017). Instead, they express Imp and Syp in opposing temporal gradients (Liu et al., 2015; Ren et al., 2017; Syed et al., 2017a). Conceptually, how Imp and Syp gradients translate into different neuronal identities through time has been compared to how morphogen gradients pattern tissues in space (Liu et al., 2019; Liu et al., 2015). During patterning of the anterior-posterior axis of the Drosophila embryo, the anterior gradient of the Bicoid morphogen and the posterior Nanos gradient are converted into discrete spatial domains that define cell fates (Briscoe and Small, 2015; Liu et al., 2019). Since gradients contain unlimited information, differences in Imp and Syp levels through time could translate into different neuronal types. Another intriguing possibility is that tTF series could act downstream of Imp and Syp, similarly to how the gap genes in the Drosophila embryo act downstream of the anterior-posterior morphogens. We have shown that another possibility is that temporal extrinsic cues can be incorporated by individual progenitors to increase neuronal diversity. In mushroom body neuroblasts activin signaling acts directly on the intrinsic program, effectively converting two broad temporal windows into three to help define an additional neuronal type. We propose that subdividing the broad Imp and Syp temporal windows by extrinsic cues may be a simple way to increase neuronal diversity in other central brain neuroblasts.
We have also shown that activin signaling times the Imp to Syp transition for mushroom body neuroblasts, similar to the function of ecdysone for other central brain neuroblasts (Syed et al., 2017a). In both cases however, the switch still occurs, indicating that a separate independent clock continues to tick. This role for extrinsic cues during Drosophila neurogenesis is reminiscent of their roles on individual vertebrate progenitors. For example, hindbrain neural stem cells progressively produce motor neurons followed by serotonergic neurons before switching to producing glia (Chleilat et al., 2018; Dias et al., 2014). The motor neuron to serotonergic neuron switch is fine-tuned by TGFβ signaling. It would be interesting to determine if hindbrain neuronal subtypes are lost in TGFβ mutants, similar to how α’β’ identity is lost in the mushroom bodies in babo mutants.
Ecdysone signaling is not necessary for α’β’ specification
The specification of α’β’ neurons begins at mid-L3 with the onset of Mamo expression (Liu et al., 2019). In contrast, high levels of EcR are detected in mature mushroom body neurons starting at late L3 (Lee et al., 2000). At this stage, both γ and α’β’ neurons already exist and new α’β’ neurons are still being generated. Thus, Mamo expression precedes EcR expression. These non-overlapping expression patterns suggest that ecdysone signaling does not regulate Mamo and therefore cannot control the specification of α’β’ neurons. Furthermore, expression of UAS-EcR-RNAi or mutants for usp do not lead to the loss of α’β’ neurons. We note that our usp results contradict the loss of α’β’ neuron reported by Marchetti and Tavosanis, 2017 in usp clones. However, we could see α’β’ neurons in these clones based on the morphology of these neurons but the remodeling defect of γ neurons makes α’β’ neurons difficult to identify. Nevertheless, ecdysone might still function later during α’β’ differentiation, particularly during pupation when all mushroom body neurons express EcR.
We and Marchetti and Tavosanis have both shown that expression of UAS-EcR-DN leads to the loss of α’β’ neurons by acting in mushroom body neurons but not in neuroblasts (Marchetti and Tavosanis, 2017). However, EcR must be first be expressed in the target cells of interest in order to make any conclusions about ecdysone function using UAS-EcR-DN. Since we cannot detect EcR protein in Mamo+ cells at L3, but expressing UAS-EcR-DN inhibits Mamo in those cells, we conclude that EcR-DN artifactually represses Mamo and leads to the loss of α’β’ neurons. This explains why expressing UAS-EcR-B1 does not rescue α’β’ neurons in babo clones. However, Marchetti and Tavosanis did rescue babo-RNAi by expressing EcR (Marchetti and Tavosanis, 2019). This is likely because our experiments were performed using babo MARCM clones in which the loss of α’β’ neurons is much more severe than with babo-RNAi used in their experiments (Figure 1—figure supplement 1Q; Marchetti and Tavosanis, 2019). Indeed, when we attempted to eliminate α’β’ neurons using a validated UAS-babo-RNAi construct (Awasaki et al., 2011), γ neurons did not remodel but there was only a minor (but significant) decrease in the number of α’β’ neurons. This indicates that knocking-down babo with mb-Gal4 that is only weakly expressed in neuroblasts and newborn neurons is not strong enough to inhibit α’β’ specification. Thus, we speculate that the LexA line used by Marchetti and Tavosanis (GMR26E01-LexA) may not be a reliable reporter for α’β’ neurons upon babo knockdown, and that it might be ecdysone sensitive later in α’β’ differentiation. Since EcR expression in all mushroom body neurons at L3 may be dependent on activin signaling directly in neurons, as it is in γ neurons for remodeling (Zheng et al., 2003), expressing UAS-EcR-B1 together with UAS-babo-RNAi using OK107-Gal4 might both reduce the effectiveness of the RNAi while also allowing for the re-expression of GMR26E01-LexA.
Glia are a source of the activin ligand myo, which is temporally expressed in brain glia starting at L3 to initiate the remodeling of mushroom body γ neurons (Awasaki et al., 2011) and α’β’ specification (Figure 4; Marchetti and Tavosanis, 2019). However, knocking-down Myo from glia is not as severe as removing Babo from mushroom body neuroblasts. This might be due to incomplete knockdown of myo or to other sources of Myo, potentially from neurons. For example, in the vertebrate cortex, old neurons signal back to young neurons to control their numbers (Parthasarathy et al., 2014; Seuntjens et al., 2009; Toma et al., 2014; Wang et al., 2016). It is also possible the Babo is activated by other activin ligands, including Activin and Dawdle (Upadhyay et al., 2017). An intriguing hypothesis is that the temporal expression of myo in glia beginning at mid-L3 is induced by the attainment of critical weight and rising ecdysone levels. It would be interesting to determine whether blocking ecdysone signaling in glia leads to the loss of α’β’ specification, similar to how blocking ecdysone reception in astrocytes prevents γ neuron remodeling (Hakim et al., 2014).
Conserved mechanisms of temporal patterning
It is well established that extrinsic cues play important roles during vertebrate neurogenesis, either by regulating temporal competence of neural stem cells or by controlling the timing of temporal identity transitions (reviewed in Kawaguchi, 2019). Competence changes mediated by extrinsic cues were demonstrated in classic heterochronic transplantation studies that showed that young donor progenitors produce old neuronal types when placed in older host brains (Desai and McConnell, 2000; Frantz and McConnell, 1996; McConnell, 1988). Recent studies show that the reverse is also true when old progenitors are placed in a young environment (Oberst et al., 2019).
Mechanisms of intrinsic temporal patterning are also conserved (Alsiö et al., 2013; Elliott et al., 2008; Holguera and Desplan, 2018; Konstantinides et al., 2015; Mattar et al., 2015; Shen et al., 2006). For example, vertebrate retinal progenitor cells use an intrinsic tTF cascade to bias young, middle, and old retinal fates (Elliott et al., 2008; Liu et al., 2020; Mattar et al., 2015). Two of the factors (Ikaros and Casz1) used for intrinsic temporal patterning are orthologs to the Drosophila tTFs Hb and Cas. tTF series might also exist in cortical radial glia progenitors and even in the spinal cord (Delile et al., 2019; Gao et al., 2014; Llorca et al., 2019; Telley et al., 2016; Telley et al., 2019). Recent results also show the importance of post-transcriptional regulation in defining either young or old cortical fates (Shu et al., 2019; Zahr et al., 2018), which can be compared to the use of post-transcriptional regulators that are a hallmark of neuronal temporal patterning in Drosophila central brain neuroblasts. These studies highlight that the mechanisms driving the diversification of neuronal types are conserved.
Materials and methods
Drosophila strains and MARCM
Flies were kept on standard cornmeal medium at 25°C. For MARCM experiments, embryos were collected every 12 hr. After 24 hr, L1 larvae were placed at 37°C for 2 hr for neuroblast clones or 15 min for GMC clones. To target GMCs at L3, larvae were aged for 84 hr and then placed at 37°C for 15 min. Brains were dissected from 1 to 5 day old adults.
We used the following transgenic and mutant flies in combination or recombined in this study. {} enclose individual genotypes, separated by commas. Stock numbers refers to BDSC unless otherwise stated:
{y, w, UAS-mCD8::GFP, hsFlp; FRTG13, tub-Gal80/CYO;; OK107-Gal4 (gift from Oren Schuldiner)}, {hsFLP, y, w; FRTG13, UAS-mCD8::GFP (#5131)}, {hsFLP, tubP-GAL80, w, FRT19A; UAS-mCD8::GFP/CyO; OK107-Gal4 (#44407)}, {hsFLP, y, w, UAS-mCD8::GFP; FRT82B, tubP-GAL80/TM3, Sb1; OK107-Gal4 (#44408)}, {hsFLP, y1, w*, UAS-mCD8::GFP; tubP-GAL80, FRT40A; OK107-Gal4 (#44406)}, {UAS-EcR.B1-DeltaC655.W650A (#6872)}, {y, w, FRT19A (#1744)}, {FRTG13, babo52 (gift from Dr. Michael B. O’Connor)}, {w; FRTG13 (#1956)}, {w1118; repo-Gal4/TM3, Sb1 (#7415)}, {w; GMR71G10-GAL4 (#39604)}, {w; GMR41C07-GAL4/TM3, Sb1 (#48145)}, {w; GMR13F02-GAL4 (#48571)}, {w; GMR44E04-GAL4 (#50210)}, {w*; insc-Gal4Mz1407 (#8751)}, {usp2/FM7a (#31414)}, {Met27, gce2.5K/FM7c, 2xTb1-RFP, sn+ (gift from Dr. Lynn Riddiford)}, {yd2, w1118, ey-FLP; taiEY11718 FRT40A/CyO, y+ (DGRC #114680)}, {dpyov1, tai61G1, FRT40A/CyO (#6379)}, {w*; smo119B6, al1, dpyov1, b1, FRT40A/CyO (#24772)}, {FRT82B, svp1/TM3 (gift from Tzumin Lee)}, {y1, w*, UAS-mCD8::GFP, SmoxMB388, FRT19A/FM7c (#44384)}, {w*;; UAS-p35 (#5073)}, {y1, w; Mi{PT-GFSTF.1}EcR[MI05320-GFSTF.1]/SM6a, (#59823)}, {y1, w*; PinYt/CyO; UAS-mCD8::GFP (#5130)}, {w*;; UAS-EcR.B1 (#6469)}, {y, w;; UAS-babo-a/TM6 (gift from Dr. Michael O’Connor)}, {UAS-Imp-RNAi (#34977)}, {UAS-Imp-RM-Flag (gift from Dr. Tzumin Lee)}, {UAS-Syp-RNAi (VDRC 33012, gift from Dr. Tzumin Lee)}, {UAS-Syp-RB-HA (gift from Dr. Tzumin Lee)}, {y1, v1; UAS-myoglianin-RNAi (#31200)}, {w*; OK107-Gal4/In4, ciD (#854)}; {w, UAS-EcR-RNAi (#9326)}; {w, UAS-EcR-RNAi (#9327)}; {yw, UAS-babo.Q302D (#64293)}; {UAS-babo-RNAi (#44400)}.
Immunohistochemistry and microscopy
Fly brains were dissected in ice-cold PBS and fixed for 15–20 min in 4% Formaldehyde (v/w) in 1XPBS. Following a 2 hr wash in PBST (1XPBS + 0.3% Triton X-100), brains were incubated for 1–2 days in primary antibodies diluted in PBST, followed by overnight with secondary antibodies diluted in PBST. After washes, brains were mounted in Slowfade (Life Technologies) and imaged on either a Leica SP5 or SP8 confocal. Images were processed in Fiji and Adobe Illustrator (CC18).
We used the following antibodies in this study: sheep anti-GFP (1:500, Bio-Rad #4745–1051; RRID:AB_619712), mouse anti-Trio (1:50, DSHB #9.4A anti-Trio; RRID:AB_528494), guinea pig anti-Mamo (1:200, this study, Genscript), mouse anti-FasII (1:50, DSHB #1D4 anti-Fasciclin II; RRID:AB_528235), rat anti-Imp (1:200, this study, Genscript), rabbit anti-Syp (1:200, this study, Genscript), guinea pig anti-Dpn (1:1000, Genscript), rabbit anti-FasII (1:50, this study, Genscript), mouse anti-EcR-B1 (1:20, DSHB #AD4.4(EcR-B1); RRID:AB_528215), mouse anti-Dac2-3 (1:20, DSHB #mAbdac2-3; RRID:AB_528190), guinea pig anti-Chinmo (1:200, this study, Genscript), rat anti-Chinmo (1:200, gift from Dr. Cedric Maurange), rat anti-DNcad (1:20, DSHB #DN-Ex #8; RRID:AB_528121), donkey anti-sheep Alexa 488 (1:500, Jackson ImmunoResearch #713-545-147; RRID:AB_2340745), donkey anti-mouse Alexa 555 (1:400, Thermo Scientific #A-31570; RRID:AB_2536180), donkey anti-rabbit Alexa 555 (1:400, Thermo Scientific #A-31572; RRID:AB_162543), donkey anti-rat Alexa 647 (1:400, Jackson Immunochemicals #712-605-153; RRID:AB_2340694), donkey anti-guinea pig Alexa 647 (1:400, Jackson Immunochemicals #706-605-148; RRID:AB_2340476), donkey anti-rabbit 405 (1:100, Jackson Immunochemicals #711-475-152; RRID:AB_2340616), donkey anti-rat Cy3 (1:400, Jackson Immunochemicals #712-165-153; RRID:AB_2340667), donkey anti-mouse 405 (1:100, Jackson Immunochemicals #715-475-150; RRID:AB_2340839).
Polyclonal antibodies were generated by Genscript (https://www.genscript.com/). The epitopes used for each immunization are listed below.
Mamo: amino acids 467–636 of the full length protein:
MDDRLEQDVDEEDLDDDVVVVGPATAMARGIAQRLAHQNLQRLHHTHHHAQHQHSQHHHPHSQHHHTPHHQQHHTHSDDEDAMPVIAKSEILDDDYDDEMDLEDDDEADNSSNDLGLNMKMGSGGAGGGGGVDLSTGSTLIPSPLITLPSSSAAAAAAAAAAMESQRSTPHHHHHH.
Imp: amino acids 76–455 (of isoform PB) of the full length protein:
ADFPLRILVQSEMVGAIIGRQGSTIRTITQQSRARVDVHRKENVGSLEKSITIYGNPENCTNACKRILEVMQQEAISTNKGEICLKILAHNNLIGRIIGKSGNTIKRIMQDTDTKITVSSINDINSFNLERIITVKGLIENMSRAENQISTKLRQSYENDLQAMAPQSLMFPGLHPMAMMSTPGNGMVFNTSMPFPSCQSFAMSKTPASVVPPVFPNDLQETTYLYIPNNAVGAIIGTRGSHIRSIMRFSNASLKIAPLDADKPLDQQTERKVTIVGTPEGQWKAQYMIFEKMREEGFMCGTDDVRLTVELLVASSQVGRIIGKGGQNVRELQRVTGSVIKLPEHALAPPSGGDEETPVHIIGLFYSVQSAQRRIRAMML.
Syp: amino acids 35-231(of isoform PA) of the full length protein:
MAEGNGELLDDINQKADDRGDGERTEDYPKLLEYGLDKKVAGKLDEIYKTGKLAHAELDERALDALKEFPVDGALNVLGQFLESNLEHVSNKSAYLCGVMKTYRQKSRASQQGVAAPATVKGPDEDKIKKILERTGYTLDVTTGQRKYGGPPPHWEGNVPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMM.
FasII: amino acids 770–873 (of isoform PA) of the full length protein:
MHHHHHHDLLCCITVHMGVMATMCRKAKRSPSEIDDEAKLGSGQLVKEPPPSPLPLPPPVKLGGSPMSTPLDEKEPLRTPTGSIKQNSTIEFDGRFVHSRSGEIIGKNSAV.
Chinmo: amino acids 494–604 (of isoform PF) of the full length protein:
MLNVWNATKMNNKNSVNTADGKKLKCLYCDRLYGYETNLRAHIRQRHQGIRVPCPFCERTFTRNNTVRRHIAREHKQEIGLAAGATIAPAHLAAAAAASAAATAAAS NHSPHHHHHH.
Cell counts quantification
All confocal images were taken with a step size of three microns. Using Fiji, each image was cropped to limit the area to a region containing mostly mushroom body cell bodies. In all cases, GFP+ cells were manually counted. To count α’β’ neurons, images were split into their individual channels and the channel containing Mamo staining was automatically binarized to account for weak and strong Mamo expression using either Default or RenyiEntropy thresholding. Binarized images were processed further using the Watershed method to differentiate between contacting cells. The number of particles (i.e., strong Mamo cells) measuring between 50-infinity squared pixels were automatically counted using the Analyze Particles function and a separate channel containing bare outlines of the counts was produced and inverted. This method automatically produced the total number of strong Mamo+ cells. Individual channels were then remerged. Outlines drawn from the Analyze Particles function that overlapped with GFP+ cells were defined as α’β’ neurons within a clone. In the eight cases where two mushroom body neuroblasts were labeled in a single hemisphere (wildtype:1; babo, UAS-EcR: 3; babo, UAS-Syp: 2), the total number of α’β’ neurons within clones was divided by 2.
Imp and Syp fluorescence quantification
All brains used for quantifying Imp and Syp fluorescence values in babo or UAS-EcR-DN mutants were prepared together. Additionally, all images used for quantification were imaged using the same confocal settings for each channel. Fluorescence measurements were made in Fiji. Values for Imp and Syp were measured within the same hand-drawn area encompassing the entire neuroblast from a single z-slice.
Statistics
Statistical tests were performed in Excel or R. The exact tests used are reported in the figure legends. In all cases, whisker plots represent the minimum value (bottom whisker), first quartile (bottom of box to middle line), inclusive median (middle line), third quartile (middle line to top of box) and maximum value (top whisker). The ‘x’ represents the average value. Outliers are 1.5 times the distance from the first and third quartile. Reported are averages ± standard error of mean (SEM).
Acknowledgements
We would like to thank the fly community, the Bloomington and the DGRC stock centers for flies and reagents; Nikos Konstantinides for discussion throughout the project and comments on the manuscript, and the three reviewers for constructive feedback on our paper; Tzumin Lee for sharing data prior to publication and for discussions; Tzumin Lee, Lynn Riddiford, Oren Schuldine, Cedric Maurange and Michael B O’Connor for fly stocks and antibodies; And all the Desplan lab members for their discussion and comments on the manuscript. Funding: This work was supported by grants from NIH (R01 EY017916 and R21 NS095288) and from NYSTEM (DOH01-C32604GG) to CD. AMR was partly supported by funding from NIH (T32 HD007520), and by NYU’s GSAS MacCracken Program and a Dean’s Dissertation Fellowship.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Anthony M Rossi, Email: amr808@nyu.edu.
Claude Desplan, Email: cd38@nyu.edu.
Chris Q Doe, Howard Hughes Medical Institute, University of Oregon, United States.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
Funding Information
This paper was supported by the following grants:
National Eye Institute R01 EY017916 to Claude Desplan.
National Institute of Neurological Disorders and Stroke R21 NS095288 to Claude Desplan.
National Institutes of Health T32 HD007520 to Anthony M Rossi.
New York University GSAS MacCracken Program to Anthony M Rossi.
New York State Stem Cell Science DOH01-C32604GG to Claude Desplan.
Additional information
Competing interests
Reviewing editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Software, Formal analysis, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing - original draft, Project administration, Writing - review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Project administration, Writing - review and editing.
Additional files
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
The following previously published dataset was used:
Sugino K, Lee T, Liu Z, Yang C. 2015. Opposite Imp/Syp temporal gradients govern birth time-dependent neuronal fates. NCBI Gene Expression Omnibus. GSE71103
References
- Alsiö JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. PNAS. 2013;110:E716–E725. doi: 10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyagor I, Berkun V, Keren-Shaul H, Marmor-Kollet N, David E, Mayseless O, Issman-Zecharya N, Amit I, Schuldiner O. Combining developmental and Perturbation-Seq uncovers transcriptional modules orchestrating neuronal remodeling. Developmental Cell. 2018;47:38–52. doi: 10.1016/j.devcel.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26:119–131. doi: 10.1016/S0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Huang Y, O'Connor MB, Lee T. Glia instruct developmental neuronal remodeling through TGF-β signaling. Nature Neuroscience. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann AA, Texada MJ, Chen HM, Etheredge JN, Miller DL, Picard S, Warner R, Truman JW, Riddiford LM. Genetic tools to study juvenile hormone action inDrosophila sci. Rep. 2017;7:2132. doi: 10.1038/s41598-017-02264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Small S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Abdollah S, Haerry TE, Shimell MJ, Merriam J, Raftery L, Wrana JL, O'Connor MB. The Drosophila activin receptor baboon signals through dSmad2 and controls cell proliferation but not patterning during larval development. Genes & Development. 1999;13:98–111. doi: 10.1101/gad.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nature Reviews Neuroscience. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Chai PC, Liu Z, Chia W, Cai Y. Hedgehog signaling acts with the temporal cascade to promote neuroblast cell cycle exit. PLOS Biology. 2013;11:e1001494. doi: 10.1371/journal.pbio.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chleilat E, Skatulla L, Rahhal B, Hussein MT, Feuerstein M, Krieglstein K, Roussa E. TGF-β signaling regulates development of midbrain dopaminergic and hindbrain serotonergic neuron subgroups. Neuroscience. 2018;381:124–137. doi: 10.1016/j.neuroscience.2018.04.019. [DOI] [PubMed] [Google Scholar]
- Cognigni P, Felsenberg J, Waddell S. Do the right thing: neural network mechanisms of memory formation, expression and update in Drosophila. Current Opinion in Neurobiology. 2018;49:51–58. doi: 10.1016/j.conb.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learning & Memory. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- Croset V, Treiber CD, Waddell S. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife. 2018;7:e34550. doi: 10.7554/eLife.34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delile J, Rayon T, Melchionda M, Edwards A, Briscoe J, Sagner A. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development. 2019;146:dev173807. doi: 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Dias JM, Alekseenko Z, Applequist JM, Ericson J. Tgfβ signaling regulates temporal neurogenesis and potency of neural stem cells in the CNS. Neuron. 2014;84:927–939. doi: 10.1016/j.neuron.2014.10.033. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Temporal patterning in the Drosophila CNS. Annual Review of Cell and Developmental Biology. 2017;33:219–240. doi: 10.1146/annurev-cellbio-111315-125210. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/S0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, Kodish O, Huang K, Simons BD, Luo L, Hippenmeyer S, Shi SH. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim Y, Yaniv SP, Schuldiner O. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLOS ONE. 2014;9:e86178. doi: 10.1371/journal.pone.0086178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hige T. What can tiny mushrooms in fruit flies tell Us about learning and memory? Neuroscience Research. 2018;129:8–16. doi: 10.1016/j.neures.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Holguera I, Desplan C. Neuronal specification in space and time. Science. 2018;362:176–180. doi: 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CCF, Steinmann V, Burkard TR, Jais A, Esterbauer H, Knoblich JA. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014;158:874–888. doi: 10.1016/j.cell.2014.06.024. [DOI] [PubMed] [Google Scholar]
- Issman-Zecharya N, Schuldiner O. The PI3K class III complex promotes axon pruning by downregulating a Ptc-derived signal via endosome-lysosomal degradation. Developmental Cell. 2014;31:461–473. doi: 10.1016/j.devcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SC, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Yu HH, He Y, Kao JC, Lee T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 2012;73:677–684. doi: 10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A. Temporal patterning of neocortical progenitor cells: how do they know the right time? Neuroscience Research. 2019;138:3–11. doi: 10.1016/j.neures.2018.09.004. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nature Reviews Neuroscience. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N, Rossi AM, Desplan C. Common temporal identity factors regulate neuronal diversity in fly ventral nerve cord and mouse retina. Neuron. 2015;85:447–449. doi: 10.1016/j.neuron.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft KF, Massey EM, Kolb D, Walldorf U, Urbach R. Retinal homeobox promotes cell growth, proliferation and survival of mushroom body neuroblasts in the Drosophila brain. Mechanisms of Development. 2016;142:50–61. doi: 10.1016/j.mod.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. The EMBO Journal. 2012;31:4511–4523. doi: 10.1038/emboj.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz T, Kraft KF, Technau GM, Urbach R. Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development. 2012;139:2510–2522. doi: 10.1242/dev.077883. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Maruyama Y, Adachi Y, Okabe M, Suzuki E, Furukubo-Tokunaga K. A conserved nuclear receptor, tailless, is required for efficient proliferation and prolonged maintenance of mushroom body progenitors in the Drosophila brain. Developmental Biology. 2009;326:224–236. doi: 10.1016/j.ydbio.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/S0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Yang CP, Miyares RL, Huang YF, He Y, Ren Q, Chen HM, Kawase T, Ito M, Otsuna H, Sugino K, Aso Y, Ito K, Lee T. Conservation and divergence of related neuronal lineages in the Drosophila central brain. eLife. 2020;9:e53518. doi: 10.7554/eLife.53518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/S0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lin S, Lai S-L, Yu H-H, Chihara T, Luo L, Lee T. Lineage-specific effects of notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang CP, Sugino K, Fu CC, Liu LY, Yao X, Lee LP, Lee T. Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science. 2015;350:317–320. doi: 10.1126/science.aad1886. [DOI] [PubMed] [Google Scholar]
- Liu LY, Long X, Yang CP, Miyares RL, Sugino K, Singer RH, Lee T. Mamo Decodes hierarchical temporal gradients into terminal neuronal fate. eLife. 2019;8:e48056. doi: 10.7554/eLife.48056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu X, Li S, Huang X, Qian H, Jin K, Xiang M. Foxn4 is a temporal identity factor conferring mid/late-early retinal competence and involved in retinal synaptogenesis. PNAS. 2020;117:5016–5027. doi: 10.1073/pnas.1918628117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca A, Ciceri G, Beattie R, Wong FK, Diana G, Serafeimidou-Pouliou E, Fernández-Otero M, Streicher C, Arnold SJ, Meyer M, Hippenmeyer S, Maravall M, Marin O. A stochastic framework of neurogenesis underlies the assembly of neocortical cytoarchitecture. eLife. 2019;8:e51381. doi: 10.7554/eLife.51381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Arlotta P. Generating neuronal diversity in the mammalian cerebral cortex. Annual Review of Cell and Developmental Biology. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti G, Tavosanis G. Steroid hormone ecdysone signaling specifies mushroom body neuron sequential fate via chinmo. Current Biology. 2017;27:3017–3024. doi: 10.1016/j.cub.2017.08.037. [DOI] [PubMed] [Google Scholar]
- Marchetti G, Tavosanis G. Modulators of hormonal response regulate temporal fate specification in the Drosophila brain. PLOS Genetics. 2019;15:e1008491. doi: 10.1371/journal.pgen.1008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Ericson J, Blackshaw S, Cayouette M. A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron. 2015;85:497–504. doi: 10.1016/j.neuron.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK. Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. The Journal of Neuroscience. 1988;8:945–974. doi: 10.1523/JNEUROSCI.08-03-00945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell S, Kaznowski C. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.1925583. [DOI] [PubMed] [Google Scholar]
- Murthy M, Fiete I, Laurent G. Testing odor response stereotypy in the Drosophila mushroom body. Neuron. 2008;59:1009–1023. doi: 10.1016/j.neuron.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst P, Fièvre S, Baumann N, Concetti C, Bartolini G, Jabaudon D. Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature. 2019;573:370–374. doi: 10.1038/s41586-019-1515-6. [DOI] [PubMed] [Google Scholar]
- Pahl MC, Doyle SE, Siegrist SE. E93 integrates neuroblast intrinsic state with developmental time to terminate MB neurogenesis via autophagy. Current Biology. 2019;29:750–762. doi: 10.1016/j.cub.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy S, Srivatsa S, Nityanandam A, Tarabykin V. Ntf3 acts downstream of Sip1 in cortical postmitotic neurons to control progenitor cell fate through feedback signaling. Development. 2014;141:3324–3330. doi: 10.1242/dev.114173. [DOI] [PubMed] [Google Scholar]
- Ren Q, Yang C-P, Liu Z, Sugino K, Mok K, He Y, Ito M, Nern A, Otsuna H, Lee T. Stem Cell-Intrinsic, Seven-up-Triggered temporal factor gradients diversify intermediate neural progenitors. Current Biology. 2017;27:1303–1313. doi: 10.1016/j.cub.2017.03.047. [DOI] [PubMed] [Google Scholar]
- Rossi AM, Fernandes VM, Desplan C. Timing temporal transitions during brain development. Current Opinion in Neurobiology. 2017;42:84–92. doi: 10.1016/j.conb.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nature Neuroscience. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nature Neuroscience. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shih M-FM, Davis FP, Henry GL, Dubnau J. Nuclear Transcriptomes of the Seven Neuronal Cell Types That Constitute the Drosophila Mushroom Bodies. G3: Genes, Genomes, Genetics. 2019;9:81–94. doi: 10.1534/g3.118.200726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P, Wu C, Ruan X, Liu W, Hou L, Fu H, Wang M, Liu C, Zeng Y, Chen P, Yin B, Yuan J, Qiang B, Peng X, Zhong W. Opposing gradients of MicroRNA expression temporally pattern layer formation in the developing neocortex. Developmental Cell. 2019;49:764–785. doi: 10.1016/j.devcel.2019.04.017. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Haque NS, Chen C-H, Hay B, Hariharan IK. Inactivation of both foxo and reaper promotes Long-Term adult neurogenesis in Drosophila curr. Biol. 2010;20:643–648. doi: 10.1016/j.cub.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe CW, Siegrist SE. Eyeless uncouples mushroom body neuroblast proliferation from dietary amino acids in Drosophila. eLife. 2017;6:e26343. doi: 10.7554/eLife.26343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R, Cheng LY, Gould AP. Regulating neural proliferation in the Drosophila CNS. Current Opinion in Neurobiology. 2010;20:50–57. doi: 10.1016/j.conb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/S0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Syed MH, Mark B, Doe CQ. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife. 2017a;6:e26287. doi: 10.7554/eLife.26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed MH, Mark B, Doe CQ. Playing well with others: extrinsic cues regulate neural progenitor temporal identity to generate neuronal diversity. Trends in Genetics. 2017b;33:933–942. doi: 10.1016/j.tig.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley L, Govindan S, Prados J, Stevant I, Nef S, Dermitzakis E, Dayer A, Jabaudon D. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science. 2016;351:1443–1446. doi: 10.1126/science.aad8361. [DOI] [PubMed] [Google Scholar]
- Telley L, Agirman G, Prados J, Amberg N, Fièvre S, Oberst P, Bartolini G, Vitali I, Cadilhac C, Hippenmeyer S, Nguyen L, Dayer A, Jabaudon D. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019;364:eaav2522. doi: 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]
- Toma K, Kumamoto T, Hanashima C. The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. Journal of Neuroscience. 2014;34:13259–13276. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Bazhenov M, Laurent G. Olfactory representations by Drosophila mushroom body neurons. Journal of Neurophysiology. 2008;99:734–746. doi: 10.1152/jn.01283.2007. [DOI] [PubMed] [Google Scholar]
- Upadhyay A, Moss-Taylor L, Kim M-J, Ghosh AC, O’Connor MB. TGF-β family signaling inDrosophila cold spring harb. Perspect. Biol. 2017;9:a022152. doi: 10.1101/cshperspect.a022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. BioEssays. 2004;26:739–751. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annual Review of Neuroscience. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Wang W, Jossin Y, Chai G, Lien W-H, Tissir F, Goffinet AM. Feedback regulation of apical progenitor fate by immature neurons through Wnt7–Celsr3–Fzd3 signalling. Nature Communications. 2016;7:e10936. doi: 10.1038/ncomms10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V. Postembryonic lineages of the Drosophila brain: ii. identification of lineage projection patterns based on MARCM clones. Developmental Biology. 2013;384:258–289. doi: 10.1016/j.ydbio.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, Sokol NS. let-7-Complex MicroRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Developmental Cell. 2012;23:202–209. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, O'Connor MB. Ecdysone control of developmental transitions: lessons from Drosophila research. Annual Review of Entomology. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CP, Fu CC, Sugino K, Liu Z, Ren Q, Liu LY, Yao X, Lee LP, Lee T. Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development. 2016;143:411–421. doi: 10.1242/dev.129163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CP, Samuels TJ, Huang Y, Yang L, Ish-Horowicz D, Davis I, Lee T. Imp and syp RNA-binding proteins govern decommissioning of Drosophila neural stem cells. Development. 2017;144:3454–3464. doi: 10.1242/dev.149500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T. A complete developmental sequence of a Drosophila Neuronal Lineage as Revealed by Twin-Spot MARCM. PLOS Biology. 2010;8:e1000461. doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Clonal development and organization of the Adult Drosophila Central Brain. Current Biology : CB. 2013a;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XM, Gutman I, Mosca TJ, Iram T, Ozkan E, Garcia KC, Luo L, Schuldiner O. Plum, an immunoglobulin superfamily protein, regulates axon pruning by facilitating TGF-β signaling. Neuron. 2013b;78:456–468. doi: 10.1016/j.neuron.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr SK, Yang G, Kazan H, Borrett MJ, Yuzwa SA, Voronova A, Kaplan DR, Miller FD. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron. 2018;97:520–537. doi: 10.1016/j.neuron.2017.12.045. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O'Connor MB, Lee CH, Lee T. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112:303–315. doi: 10.1016/S0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]