Abstract
Esophageal cancer is one of the fatal cancers around the world. Dexmedetomidine (DEX) is widely used during anesthesia of esophageal cancer surgery. Nevertheless, the role of DEX in the progression of esophageal cancer remains barely known. The proliferation, apoptosis and metastasis of esophageal cancer cells were detected by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, flow cytometry, transwell migration and invasion assays and Western blot assay. The expression of miR-143-3p was measured by quantitative real-time PCR in esophageal cancer tissues and cells. The binding sites between miR-143-3p and epidermal growth factor receptor pathway substrate 8 (EPS8) were predicted by Starbase online software, and the combination was verified by dual-luciferase reporter assay. The murine xenograft model was established using KYSE150 cells to verify the function of DEX in vivo. DEX inhibited the proliferation and metastasis while accelerated the apoptosis of esophageal cancer cells. The abundance of miR-143-3p was lower in esophageal cancer tissues and cells than that in paring normal tissues and normal esophageal mucosal cells Het-1A. MiR-143-3p could be induced by DEX treatment in esophageal cancer cells, and miR-143-3p also suppressed the development of esophageal cancer. EPS8 was a functional target of miR-143-3p, and it played an oncogenic role in esophageal cancer. DEX inhibited the growth of tumor via miR-143-3p/EPS8 in vivo. DEX suppressed the growth and metastasis while facilitated the apoptosis of esophageal cancer cells through upregulating the abundance of miR-143-3p and reducing the level of EPS8 in vivo and in vitro, providing promising target for the treatment of esophageal cancer.
Keywords: apoptosis, dexmedetomidine, epidermal growth factor receptor pathway substrate 8, esophageal cancer, miR-143-3p, metastasis, proliferation
Introduction
Esophageal cancer is one of the most commonly diagnosed cancers globally. There were 572 034 new cases and 508 585 deaths of esophageal cancer patients around the world in 2018 [1]. Although the therapeutic methods have been improved, the prognosis of esophageal cancer patients remains dismal. Therefore, finding novel diagnostic markers and uncovering the underlying mechanism are pivotal for esophageal cancer treatment.
Dexmedetomidine (DEX) is widely used in esophageal cancer surgery. DEX can reduce the cardiovascular response of endotracheal intubation and extubation in esophageal cancer surgery and the dosage of anesthetics [2–4]. However, the mechanism by which DEX regulating the proliferation, metastasis and apoptosis of esophageal cancer cells remains poorly understood.
MicroRNAs (miRNAs) could reduce the abundance of target messenger RNAs (mRNAs) or suppress their translation by directly binding to their 3′ untranslated region (UTR) [5,6]. They are involved in many physiological and pathological processes, including the initiation and progression of many cancers [7,8]. The aberrant downregulation of miRNAs and the upregulation of their paring proteins have been observed in multiple cancers. MiR-143-3p suppressed the progression of many cancers, including triple-negative breast cancer (TNBC), ovarian cancer, osteosarcoma and esophageal squamous cell carcinoma (ESCC). Li et al. [9] claimed that the enhancement of miR-143-3p suppressed TNBC progression through reducing the abundance of LIM domain kinase 1. Shi et al. [10] demonstrated that the growth and metastasis of ovarian cancer cells were restrained with the overexpression of miR-143-3p. Sun et al. [11] demonstrated that miR-143-3p suppressed the development of osteosarcoma partly through FOSL2. He et al. [12] proved that miR-143-3p inhibited the proliferation, metastasis and epithelial–mesenchymal transition of ESCC cells through reducing the level of QKI-5. Nevertheless, the biological significance of miR-143-3p and the underlying molecular mechanism remain largely unknown.
Epidermal growth factor receptor pathway substrate 8 (EPS8) has been reported to be an underlying therapeutic target of multiple cancers [13,14]. EPS8 accelerated the initiation and development of many cancers [15–21]. Chen et al. [22] reported that the enrichment of EPS8 was higher in breast cancer tissues than that in matching nontumor breast tissues, and the high expression of EPS8 was related to the growth and metastasis of breast cancer cells. He et al. [23] claimed that EPS8 was overexpressed in acute lymphoblastic leukemia (AML), and the high level of EPS8 was associated with the dismal prognosis of AML patients. However, the role and precise mechanism of EPS8 in esophageal cancer are not fully addressed.
In this article, we investigated the function of DEX on the growth, apoptosis and metastasis of esophageal cancer cells, and then we explored the signal network by which DEX inhibiting the proliferation and metastasis while promoting the apoptosis of esophageal cancer cells.
Materials and methods
Cell culture
The normal esophageal mucosal cell line Het-1A and esophageal cancer cell lines KYSE150 and ECA-109 were purchased from BeNa Culture Collection (Beijing, China). All cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, California, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 10% penicillin (100 units/mL)-streptomycin (100 μg/mL) solution and incubated at 37°C with 5% CO2.
Drug treatment
KYSE150 and ECA-109 cells were seeded into six-well plates, and were treated with DEX (1 μM; Santa Cruz Biotechnology, Dallas, Texas, USA), Control or negative control for 48 h.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
Cells in 96-well plates were treated with 10 µL 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Invitrogen, Carlsbad, California, USA) for 4 h. After removing the cell supernatant, dimethyl sulfoxide (Sigma, St. Louis, Missouri, USA) was added to the wells. The optical density was measured by a microplate reader at 490 nm.
Cell apoptosis analysis
Flow cytometry was applied to determine the apoptosis rate of esophageal cancer cells. Esophageal cancer cells were cultured in six-well plate overnight at 37°C. After treatment with negative control or DEX, esophageal cancer cells were harvested using cold PBS buffer for twice. Then, the cells were incubated with 5 µL Annexin V combined fluorescein isothiocyanate and propidine iodide (Solarbio, Beijing, China) for 10 min at room temperature. Samples were detected by the flow cytometer (BD Biosciences, San Jose, California, USA).
Transwell migration and invasion assays
KYSE150 and ECA-109 cells were incubated in DMEM medium without serum in the upper chambers. The lower chambers were filled with medium added with 10% FBS. Cells migrated from upper chambers to lower chambers were stained and counted after 48-h incubation.
To explore the invasion ability of KYSE150 and ECA-109 cells, the BD matrigel matrix (BD Biosciences) was used to precoat the polycarbonate membrane. Then KYSE150 and ECA-109 cells were seeded into upper chambers and cultured in the serum-free medium. The following steps were the same as described above.
Western blot assay
KYSE150 and ECA-109 cells were lysed using RIPA lysis solution (Beyotime, Shanghai, China) after harvesting with cold PBS buffer. Proteins were quantified before separating by SDS-PAGE. The proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, Massachusetts, USA). After being blocked for 1 h with 5% milk, the membranes were immunoblotted with antibody against Cyclin D1 (ab134175, Abcam, Cambridge, Massachusetts USA), Bcl-2 (ab185002, Abcam), Bax (ab32503, Abcam), Vimentin (ab193555, Abcam), E-cadherin (ab1416, Abcam), EPS8 (ab96144, Abcam) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab181602, Abcam) at 4°C for 12 h. The PVDF membranes were then probed with secondary antibody (ab205718, Abcam). The enhanced chemiluminescent system (Beyotime) was used to visualize the protein signal.
Tissue specimens
In order to detect the abundance of EPS8 and miR-143-3p in patients, 25 esophageal cancer tissues and their matching control tissues were gathered from sufferers who had undergone resection in The Second Affiliated Hospital of Fujian Medical University. The esophageal cancer tissues were stored in −80°C after surgical resection. This experiment was carried out with the permission of the Ethic Committee of The Second Affiliated Hospital of Fujian Medical University, and all patients had submitted written informed consents.
Quantitative real-time PCR
TRIzol reagent (Invitrogen) was utilized to extract total RNA. M-MLV reverse transcriptase kit (Invitrogen) and All-in-OneTM miRNA First strand cDNA Synthesis Kit (GeneCopoeia, Rockville, Maryland, USA) were used to conduct the reverse transcription of EPS8 and miR-143-3p, respectively. GAPDH or U6 small RNA was treated as the internal reference. The abundance of EPS8 and miR-143-3p was detected by 2-ΔΔCt method [24]. The primer sequences involved in this study were shown as below: EPS8 (Forward, 5′-GGCAAAGTGTGGACTCAAGA-3′; Reverse, 5′-ACTAATTAGGTTTGCCTTAA-3′), miR-143-3p (Forward, 5′-GGGGTGAGATGAAGCACTG-3′; Reverse, 5′-CAGTGCGTGTCGTGGAGT-3′), U6 (Forward, 5′-CCTGCGCAAGGATGAC-3′; Reverse, 5′-GTGCAGGGTCCGAGGT-3′), GAPDH (Forward, 5′-CTGGGCTACACTGAGCACC-3′; Reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′).
Cell transfection
Lipofectamine 2000 (Invitrogen) was used to conduct transfection. Small interfering RNA negative control (si-negative control) and small interfering RNA against EPS8 (si-EPS8) were synthesized from Genepharma (Shanghai, China). MiRNA-negative control (miR-negative control), miR-143-3p, anti-miRNA-negative control (anti-miR-negative control) and anti-miR-143-3p were obtained from Ribobio (Guangzhou, China).
Dual-luciferase reporter assay
The binding between EPS8 and miR-143-3p was verified by dual-luciferase reporter assay. The 3′ UTR of EPS8 was amplified, including wild-type or mutant-type binding sites, and embedded to the luciferase reporter vector, generating WT-EPS8 or MUT-EPS8. The luciferase reporter vectors were cotransfected with miR-143-3p or miR-negative control in KYSE150 and ECA-109 cells. After 48-h transfection, the luciferase activity was examined after lysing cells.
Murine xenograft assay
Animal experiments were conducted with the permission of the Animal Research Committee of The Second Affiliated Hospital of Fujian Medical University. The tumor xenograft model was built using KYSE150 cells treated with negative control or DEX. KYSE150 cells were subcutaneously injected into the right side of the mouse back, and tumor volume was measured weekly. The mice were sacrificed after four weeks following the inoculation, and the weight of tumors was recorded. The levels of miR-143-3p and EPS8 were detected in tumor tissues.
Statistical analysis
GraphPad Prism 7 software was used to analyze all data from three independent experiments and values were displayed as mean ± SD. Differences between two groups or among multiple groups were analyzed by Student’s t-test and one-way analysis of variance. The liner relationship between the expression of EPS8 and the level of miR-143-3p in esophageal cancer tissues was evaluated by Spearman’s correlation coefficient. P less than 0.05 was considered to be significant.
Results
DEX inhibits the proliferation and metastasis while promotes the apoptosis of esophageal cancer cells
To illustrate the role of DEX in the proliferation, metastasis and apoptosis of esophageal cancer cells, KYSE150 and ECA-109 cells were treated with control, negative control or DEX. As mentioned in Fig. 1a and b, the proliferation of esophageal cancer cells was suppressed with DEX treatment compared with that in the control group. Flow cytometry was carried out to measure the apoptosis of esophageal cancer cells treated with control, negative control or DEX. DEX treatment significantly promoted the apoptosis of esophageal cancer cells (Fig. 1c). Transwell migration and invasion assays showed that the metastasis of esophageal cancer cells was prominently inhibited with DEX treatment (Fig. 1d and e). Moreover, the abundance of proliferation-associated protein Cyclin D1, antiapoptotic protein Bcl-2 and mesenchymal marker Vimentin was declined, while the enrichment of proapoptotic protein Bax and epithelial marker E-cadherin was upregulated, indicating that the DEX treatment inhibited the proliferation and metastasis while accelerated the apoptosis of esophageal cancer cells (Fig. 1f and g).
Fig. 1.
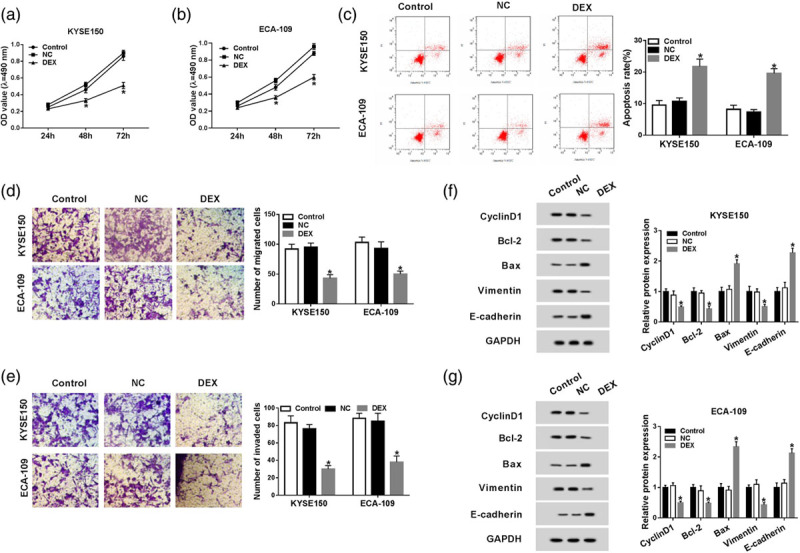
DEX inhibits the proliferation and metastasis while promotes the apoptosis of esophageal cancer cells. (a and b) The proliferation of esophageal cancer cells treated with control, negative control or DEX was determined by MTT assay. (c) Flow cytometry was carried out to measure the apoptosis of esophageal cancer cells treated with control, negative control or DEX. (d and e) The invasion and migration of KYSE150 and ECA-109 cells treated with control, negative control or DEX were determined by transwell migration and invasion assays. (f and g) Western blot assay was applied to detect the abundance of proliferation-associated protein (Cyclin D1), apoptosis-related proteins (Bcl-2 and Bax) and metastasis-associated proteins (Vimentin and E-cadherin) in KYSE150 and ECA-109 cells treated with control, negative control or DEX. *P < 0.05. DEX, dexmedetomidine; MTT assay, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
The enrichment of miR-143-3p is downregulated in esophageal cancer tissues and cells
To investigate the potential mechanism by which DEX inhibiting the proliferation and metastasis of esophageal cancer cells, we aimed to find the downstream component of DEX. MiR-143-3p was downregulated in esophageal cancer tissues compared with that in paired normal tissues (Fig. 2a). Meanwhile, the abundance of miR-143-3p was decreased in esophageal cancer cells compared with that in normal esophageal mucosal cells Het-1A (Fig. 2b). As showed in Fig. 2c, the level of miR-143-3p was positively modulated by DEX in KYSE150 and ECA-109 cells.
Fig. 2.
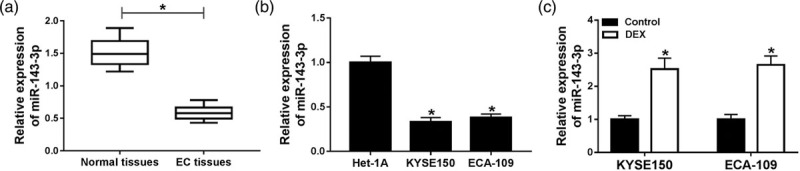
The enrichment of miR-143-3p is down-regulated in esophageal cancer tissues and cells. (a) The abundance of miR-143-3p was measured in esophageal cancer tissues (n = 25) and corresponding normal tissues (n = 25) by qRT-PCR. (b) The level of miR-143-3p was detected in esophageal cancer cells and normal esophageal mucosal cells Het-1A by qRT-PCR. (c) The expression of miR-143-3p was examined in KYSE150 and ECA-109 cells treated with control or DEX. *P < 0.05. DEX, dexmedetomidine; qRT-PCR, quantitative real time PCR.
The inhibition of miR-143-3p alleviates the suppressive effects of dexmedetomidine treatment on the proliferation and metastasis and the promoting impact on the apoptosis of esophageal cancer cells
To illustrate whether miR-143-3p was involved in DEX-mediated proliferation, apoptosis and metastasis of esophageal cancer cells, we conducted the following experiments. The knockdown efficiency of anti-miR-143-3p was measured in KYSE150 and ECA-109 cells, and the abundance of miR-143-3p was notably decreased by the transfection of anti-miR-143-3p in KYSE150 and ECA-109 cells (Fig. 3a). As indicated in Fig. 3b and c, the inhibition of miR-143-3p alleviated the inhibitory effect of DEX treatment on the proliferation of esophageal cancer cells. Flow cytometry results revealed that miR-143-3p depletion reversed the promoting impact of DEX treatment on the apoptosis of esophageal cancer cells (Fig. 3d). As showed in Fig. 3e and f, the metastasis of esophageal cancer cells was inhibited with DEX treatment, while it was recovered with the addition of anti-miR-143-3p in esophageal cancer cells. Besides, Western blot assay also revealed that miR-143-3p depletion counteracted the suppressive effects of DEX treatment on the proliferation and metastasis of esophageal cancer cells and the promoting impact on the apoptosis of esophageal cancer cells (Fig. 3g and h). Taken together, DEX facilitated the apoptosis while restrained the proliferation and metastasis of esophageal cancer cells partly through up-regulating the abundance of miR-143-3p.
Fig. 3.
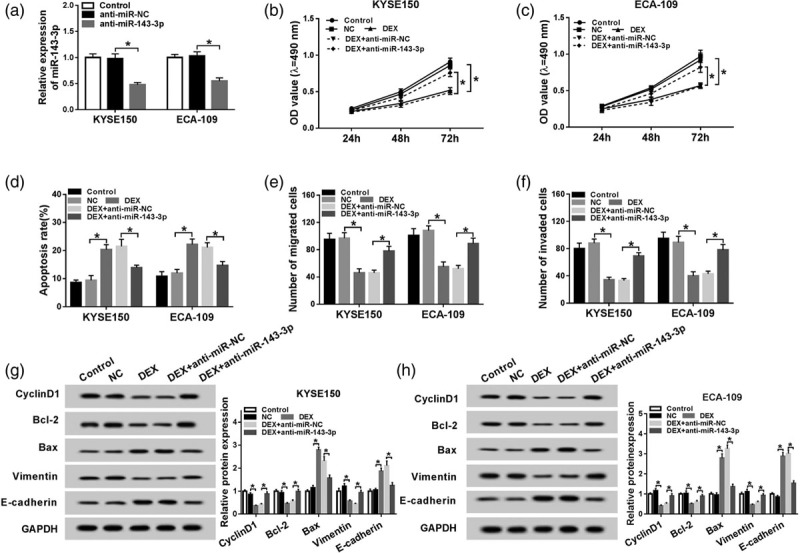
The inhibition of miR-143-3p alleviates the suppressive effects of DEX treatment on the proliferation and metastasis and the promoting impact on the apoptosis of esophageal cancer cells. (a) The level of miR-143-3p was examined in KYSE150 and ECA-109 cells transfected with control, anti-miR-negative control or anti-miR-143-3p by qRT-PCR. KYSE150 and ECA-109 cells treated with control, negative control, DEX, DEX + anti-miR-negative control or DEX + anti-miR-143-3p were used for the detection of the proliferation, metastasis and apoptosis. (b and c) The proliferation of the above esophageal cancer cells was determined by MTT assay. (d) The apoptosis of esophageal cancer cells was measured by flow cytometry. (e and f) Transwell migration and invasion assays were conducted to measure the metastasis of esophageal cancer cells. (g and h) Western blot assay was performed to detect the abundance of Cyclin D1, Bcl-2, Bax, Vimentin and E-cadherin in the above esophageal cancer cells. *P < 0.05. DEX, dexmedetomidine.
Epidermal growth factor receptor pathway substrate 8 is a target of miR-143-3p
EPS8 was predicted as a target of miR-143-3p by Starbase online software (Fig. 4a), and the combination between EPS8 and miR-143-3p was then confirmed by dual-luciferase reporter assay. The wild-type or mutant-type binding sites of EPS8 3′ UTR were inserted into luciferase reporter vector, generating WT-EPS8 or MUT-EPS8 plasmid, respectively. KYSE150 and ECA-109 cells were cotransfected with WT-EPS8 or MUT-EPS8 and miR-negative control or miR-143-3p. The luciferase activity was significantly decreased in KYSE150 and ECA-109 cells cotransfected with miR-143-3p and WT-EPS8 plasmid, whereas it remained unchanged in cells cotransfected with miR-143-3p and MUT-EPS8, demonstrating that miR-143-3p directly bound to EPS8 in KYSE150 and ECA-109 cells (Fig. 4b and c).
Fig. 4.
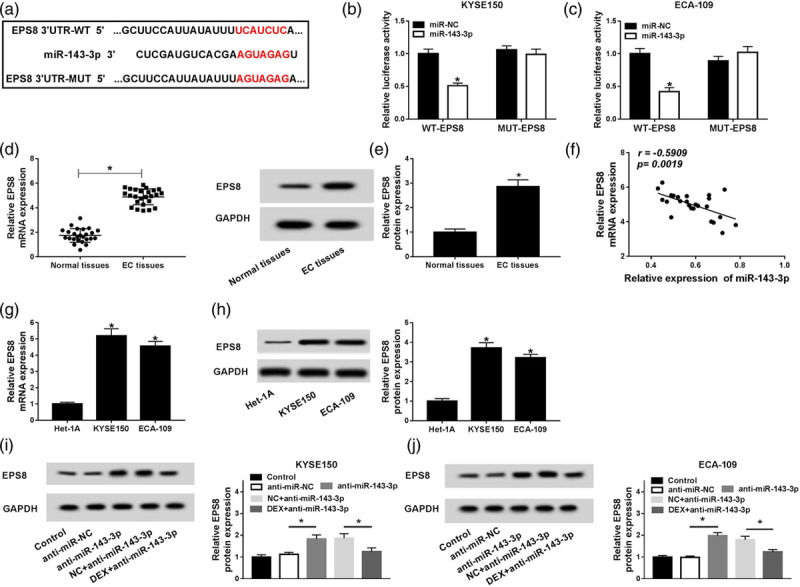
EPS8 is a target of miR-143-3p. (a) The binding sites between EPS8 and miR-143-3p were predicted by Starbase online software. (b and c) Luciferase activity was detected in KYSE150 and ECA-109 cells co-transfected with miR-negative control or miR-143-3p and WT-EPS8 or MUT-EPS8. (d and e) The abundance of EPS8 mRNA and protein was measured in esophageal cancer tissues (n = 25) and matching normal tissues (n = 25) by qRT-PCR and Western blot. (f) The correlation between the expression of EPS8 and the level of miR-143-3p was analyzed in esophageal cancer tissues (n = 25). (g and h) The abundance of EPS8 mRNA and protein was measured in esophageal cancer cells and normal esophageal mucosal cells Het-1A by qRT-PCR and Western blot. (i and j) The abundance of EPS8 was measured in KYSE150 and ECA-109 cells treated with control, anti-miR-negative control, anti-miR-143-3p, negative control + anti-miR-143-3p or DEX + anti-miR-143-3p by Western blot. *P < 0.05. DEX, dexmedetomidine EPS8, epidermal growth factor receptor pathway substrate 8.
As shown in Fig. 4d and e, the mRNA and protein expression of EPS8 were higher in esophageal cancer tissues than that in adjacent normal tissues. The expression of EPS8 was inversely correlated with the level of miR-143-3p in esophageal cancer tissues (Fig. 4f). Furthermore, the abundance of EPS8 mRNA and protein was also upregulated in esophageal cancer cells compared with that in normal esophageal mucosal cells Het-1A (Fig. 4g and h). To explore the regulatory network among EPS8, miR-143-3p and DEX, KYSE150 and ECA-109 cells were treated with control, anti-miR-negative control, anti-miR-143-3p, negative control + anti-miR-143-3p or DEX + anti-miR-143-3p. As showed in Fig. 4i and j, the abundance of EPS8 was negatively regulated by miR-143-3p and DEX in esophageal cancer cells, and the treatment of DEX reversed the effect of miR-143-3p depletion on the abundance of EPS8 in esophageal cancer cells. Collectively, EPS8 was a direct target of miR-143-3p and was inversely modulated by miR-143-3p and DEX.
The interference of EPS8 attenuates the promoting effects of miR-143-3p inhibition on the proliferation and metastasis of esophageal cancer cells and the inhibitory impact on the apoptosis of esophageal cancer cells treated with DEX
The mRNA and protein expression of EPS8 were prominently reduced by the transfection of si-EPS8 in KYSE150 and ECA-109 cells (Fig. 5a and b). MTT assay was performed to measure the proliferation of esophageal cancer cells treated with Control, DEX + anti-miR-negative control, DEX + anti-miR-143-3p, DEX + anti-miR-143-3p + si-negative control or DEX + anti-miR-143-3p + si-EPS8. The intervention of EPS8 reversed the promoting effect of miR-143-3p inhibition on the proliferation of esophageal cancer cells (Fig. 5c and d). Meanwhile, flow cytometry was conducted to measure the apoptosis of above esophageal cancer cells. The apoptosis of esophageal cancer cells was inhibited with miR-143-3p inhibition and was recovered with the addition of si-EPS8 (Fig. 5e). As indicated in Fig. 5f and g, the knockdown of EPS8 counteracted the promoting effects of miR-143-3p inhibition on the metastasis of esophageal cancer cells. Western blot assay was conducted to measure the abundance of proliferation-associated protein (Cyclin D1), apoptosis-related proteins (Bcl-2 and Bax) and metastasis-associated proteins (Vimentin and E-cadherin) in the above two cells. The knockdown of EPS8 attenuated the promoting effects of miR-143-5p inhibition on the proliferation and metastasis and the inhibitory impact on the apoptosis of esophageal cancer cells (Fig. 5h and i). These findings suggested that DEX accelerated the apoptosis while restrained the proliferation and metastasis of esophageal cancer cells via miR-143-3p/EPS8 axis in vitro.
Fig. 5.
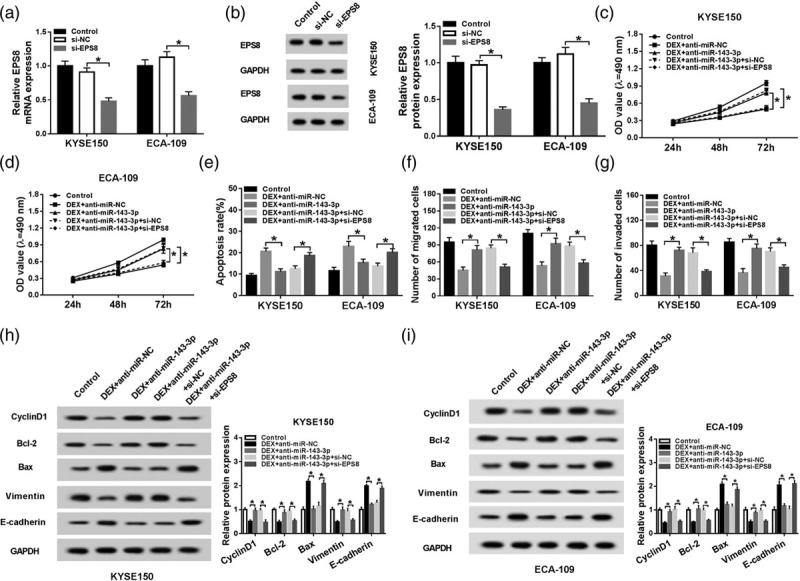
The interference of EPS8 attenuates the promoting effects of miR-143-3p inhibition on the proliferation and metastasis of esophageal cancer cells and the inhibitory impact on the apoptosis of esophageal cancer cells treated with DEX. (a and b) The enrichment of EPS8 mRNA and protein was examined in esophageal cancer cells transfected with control, si-negative control or si-EPS8 by qRT-PCR and Western blot. KYSE150 and ECA-109 cells treated with control, DEX + anti-miR-negative control, DEX + anti-miR-143-3p, DEX + anti-miR-143-3p + si-negative control or DEX + anti-miR-143-3p + si-EPS8 were used for the following experiments. (c and d) MTT assay was conducted to measure the proliferation of esophageal cancer cells. (e) Cell apoptosis was evaluated in the above esophageal cancer cells by flow cytometry. (f and g) The abilities of the migration and invasion of esophageal cancer cells were assessed by transwell migration and invasion assays. (h and i) Western blot assay was applied to detect the abundance of proliferation-associated protein (Cyclin D1), apoptosis-related proteins (Bcl-2 and Bax) and metastasis-associated proteins (Vimentin and E-cadherin) in the above esophageal cancer cells. *P < 0.05. DEX, dexmedetomidine; EPS8, epidermal growth factor receptor pathway substrate 8; MTT assay, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay; qRT-PCR, quantitative real time PCR.
dexmedetomidine inhibits esophageal cancer tumor growth in vivo
To confirm the function of DEX in vivo, we established the murine xenograft model using KYSE150 cells treated with negative control or DEX. Tumor volume was recorded once a week. The tumor was resected after four-week inoculation and was weighed. As showed in Fig. 6a and b, tumor volume and weight were less in the DEX group compared with that in the negative control group. The abundance of miR-143-3p was elevated with DEX treatment, while the level of EPS8 was declined in resected tumor tissues (Fig. 6c). The abundance of EPS8 protein was also decreased in resected tumor tissues (Fig. 6d). Taken together, DEX inhibited the growth of esophageal cancer tumor by miR-143-3p/EPS8 axis in vivo.
Fig. 6.
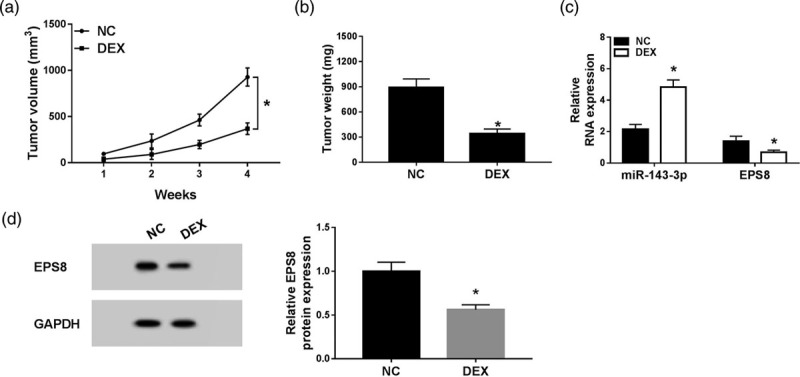
DEX inhibits esophageal cancer tumor growth in vivo. KYSE150 cells treated with negative control or DEX were subcutaneously injected into the murine. (a) Tumor volume was recorded once a week. (b) Murine xenograft tumor was weighed following four-week inoculation. (c) The expression of miR-143-3p and EPS8 was analyzed in resected tumor tissues by qRT-PCR. (d) The protein level of EPS8 was determined in resected tumor tissues by Western blot. *P < 0.05. DEX, dexmedetomidine; EPS8, epidermal growth factor receptor pathway substrate 8; qRT-PCR, quantitative real time PCR.
Discussion
DEX has a significant application value in the anesthesia of esophageal cancer surgery. Nevertheless, the regulatory role of DEX in the progression of esophageal cancer remains to be determined. In this study, we found miR-143-3p was upregulated in esophageal cancer cells treated with DEX, and DEX restrained the growth and motility while facilitated the apoptosis of esophageal cancer cells through miR-143-3p/EPS8 axis.
We first explored the role of DEX in the proliferation, metastasis and apoptosis of esophageal cancer cells. The proliferation and metastasis of esophageal cancer cells treated with DEX were inhibited, while the apoptosis was accelerated. MiR-143-3p was a tumor suppressor in multiple cancers [9–12]. He et al. [12] reported that the enrichment of miR-143-3p was downregulated in ESCC tissues, and the low expression of miR-143-3p was related to the poor prognosis of ESCC patients. Consistent with the above results, the abundance of miR-143-3p was decreased in esophageal cancer tissues and cells compared with that in normal tissues and normal esophageal mucosal cells Het-1A. To illustrate the modulatory relationship between miR-143-3p and DEX, KYSE150 and ECA-109 cells were treated with control or DEX. The abundance of miR-143-3p was increased in the two cells treated with DEX. We wondered whether miR-143-3p was involved in DEX-mediated proliferation, metastasis and apoptosis of esophageal cancer cells, and the following experiments were conducted. esophageal cancer cells were treated with control, negative control, DEX, DEX + anti-miR-negative control or DEX + anti-miR-143-3p, and MTT assay, flow cytometry, transwell migration and invasion assays and Western blot assay were performed to detect the proliferation, apoptosis and metastasis of the above esophageal cancer cells. The depletion of miR-143-3p alleviated the inhibitory effects of DEX treatment on the proliferation and metastasis of esophageal cancer cells and the promoting impact on the apoptosis of esophageal cancer cells.
EPS8 was predicted as a direct target of miR-143-3p by Starbase bioinformatic software, and the combination between EPS8 and miR-143-3p was validated by dual-luciferase reporter assay. To clarify the role of EPS8 in esophageal cancer, we first measured the abundance of EPS8 in esophageal cancer tissues and cells. The abundance of EPS8 mRNA and protein was higher in esophageal cancer tissues and cells than that in corresponding normal tissues and normal esophageal mucosal cells Het-1A. As mentioned above, the expression of miR-143-3p was declined in esophageal cancer tissues and cells, showing an inverse trend with the expression of EPS8. The statistic analysis revealed the same result. There was a negative correlation between the expression of EPS8 and the enrichment of miR-143-3p. Subsequently, we found EPS8 was inversely modulated by DEX and miR-143-3p, and DEX treatment attenuated the impact of miR-143-3p depletion on the expression of EPS8 in esophageal cancer cells.
EPS8 played an oncogenic role in many cancers. Huang et al. [25] found that EPS8 accelerated the growth of BCR-ABL positive cells through BCR-ABL/PI3K/AKT/mTOR axis. Shin et al. [26] demonstrated that PTK6 facilitated cell growth and migration by phosphorylating EPS8. To clarify the role of EPS8 in esophageal cancer cells, KYSE150 and ECA-109 cells treated with control, DEX + anti-miR-negative control, DEX + anti-miR-143-3p, DEX + anti-miR-143-3p + si-negative control or DEX + anti-miR-143-3p + si-EPS8 were used to perform MTT assay, flow cytometry, transwell migration and invasion assays and Western blot assay. The knockdown of EPS8 attenuated the promoting effects of miR-143-3p inhibition on the proliferation and metastasis of esophageal cancer cells and the suppressive impact on the apoptosis of esophageal cancer cells. These findings indicated that DEX restrained the proliferation and metastasis while promoted the apoptosis of esophageal cancer cells through miR-143-3p/EPS8 axis in vitro.
The murine xenograft model was built using KYSE150 cells treated with negative control or DEX to evaluate the function of DEX in vivo. DEX treatment suppressed the growth of tumor via miR-143-3p/EPS8 axis. More efforts are needed to investigate the mechanism by which EPS8 promoting the progression of esophageal cancer.
Collectively, DEX facilitated the apoptosis while impeded the proliferation and metastasis of esophageal cancer cells through miR-143-3p/EPS8 axis. MiR-143-3p/EPS8 axis might be a potential therapeutic target for esophageal cancer therapy.
Acknowledgements
This work was supported by Fujian Medical Innovation Project: Effects of different doses of dexmedetomidine combined with morphine on postoperative analgesic effect and immune stress in patients with esophageal cancer (No. 2015-cxb-19).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 2.Scott-Warren VL, Sebastian J. Dexmedetomidine: its use in intensive care medicine and anaesthesia. BJA Education. 2016; 16:242–246 [Google Scholar]
- 3.Liu HL, Zhang Y, Zheng GL. Clinical observation of injection of dexmedetomidine in anaesthesia for esophageal cancer operation. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2011; 27:495–497 [PubMed] [Google Scholar]
- 4.Sottas CE, Anderson BJ. Dexmedetomidine: the new all-in-one drug in paediatric anaesthesia? Curr Opin Anaesthesiol. 2017; 30:441–451 [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal micrornas. Nature. 2004; 431:350–355 [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. Microrna signatures in human cancers. Nat Rev Cancer. 2006; 6:857–866 [DOI] [PubMed] [Google Scholar]
- 7.Croce CM, Calin GA. Mirnas, cancer, and stem cell division. Cell. 2005; 122:6–7 [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. Micrornas: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297 [DOI] [PubMed] [Google Scholar]
- 9.Li D, Hu J, Song H, Xu H, Wu C, Zhao B, et al. Mir-143-3p targeting LIM domain kinase 1 suppresses the progression of triple-negative breast cancer cells. Am J Transl Res. 2017; 9:2276–2285 [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, Shen H, Xu J, Zhao S, Yao S, Jiang N. Mir-143-3p suppresses the progression of ovarian cancer. Am J Transl Res. 2018; 10:866–874 [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X, Dai G, Yu L, Hu Q, Chen J, Guo W. Mir-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci Rep. 2018; 8:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Z, Yi J, Liu X, Chen J, Han S, Jin L, et al. Mir-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016; 15:51. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang Q, Yong L. Eps8, a therapeutic potential target for cancer. Hum Pathol. 2018; 82:300–301 [DOI] [PubMed] [Google Scholar]
- 14.Li YH, Xue TY, He YZ, Du JW. Novel oncoprotein EPS8: a new target for anticancer therapy. Future Oncol. 2013; 9:1587–1594 [DOI] [PubMed] [Google Scholar]
- 15.Cappellini E, Vanetti C, Vicentini LM, Cattaneo MG. Silencing of eps8 inhibits in vitro angiogenesis. Life Sci. 2015; 131:30–36 [DOI] [PubMed] [Google Scholar]
- 16.Wen Q, Jiao X, Kuang F, Hou B, Zhu Y, Guo W, et al. Foxo3a inhibiting expression of EPS8 to prevent progression of NSCLC: a new negative loop of EGFR signaling. Ebiomedicine. 2019; 40:198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoenherr C, Serrels B, Proby C, Cunningham DL, Findlay JE, Baillie GS, et al. Eps8 controls src- and FAK-dependent phenotypes in squamous carcinoma cells. J Cell Sci. 2014; 127:5303–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorsic LK, Stark AL, Wheeler HE, Wong SS, Im HK, Dolan ME. EPS8 inhibition increases cisplatin sensitivity in lung cancer cells. PLoS One. 2013; 8:e82220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding X, Zhou F, Wang F, Yang Z, Zhou C, Zhou J, et al. Eps8 promotes cellular growth of human malignant gliomas. Oncol Rep. 2013; 29:697–703 [DOI] [PubMed] [Google Scholar]
- 20.Yeudall A, Patel V. EPS8 signaling as a therapeutic target in oral cancer. Oral Dis. 2018; 24:128–131 [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Teh MT, Ji Y, Patel V, Firouzabadian S, Patel AA, et al. EPS8 upregulates FOXM1 expression, enhancing cell growth and motility. Carcinogenesis. 2010; 31:1132–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Liang Z, Huang W, Li X, Zhou F, Hu X, et al. Eps8 regulates cellular proliferation and migration of breast cancer. Int J Oncol. 2015; 46:205–214 [DOI] [PubMed] [Google Scholar]
- 23.He YZ, Liang Z, Wu MR, Wen Q, Deng L, Song CY, et al. Overexpression of EPS8 is associated with poor prognosis in patients with acute lymphoblastic leukemia. Leuk Res. 2015; 39:575–581 [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001; 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25.Huang R, Liu H, Chen Y, He Y, Kang Q, Tu S, et al. EPS8 regulates proliferation, apoptosis and chemosensitivity in BCR-ABL positive cells via the BCR-ABL/PI3K/AKT/mtor pathway. Oncol Rep. 2018; 39:119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin WS, Shim HJ, Lee YH, Pyo M, Park JS, Ahn SY, Lee ST. PTK6 localized at the plasma membrane promotes cell proliferation and migration through phosphorylation of eps8. J Cell Biochem. 2017; 118:2887–2895 [DOI] [PubMed] [Google Scholar]


