Abstract
Purpose:
Monoclonal Gammopathy of Undetermined Significance (MGUS) is a prevalent yet largely asymptomatic precursor to multiple myeloma. Patients with MGUS must undergo regular surveillance and testing, with few known predictors of progression. We developed an algorithm to identify MGUS patients in electronic health data to facilitate large-scale, population-based studies of this premalignant condition.
Methods:
We developed a four-step algorithm using electronic health record and health claims data from men and women aged ≥50 years receiving care from a large, multispecialty medical group between 2007 and 2015. The case definition required patients to have at least two MGUS ICD-9 diagnosis codes within 12 months, at least one serum and/or urine protein electrophoresis and one immunofixation test, and at least one in-office hematology/oncology visit. Medical charts for selected cases were abstracted then adjudicated independently by two physicians. We assessed algorithm validity by positive predictive value (PPV).
Results:
We identified 833 people with at least two MGUS diagnosis codes; 429 (52%) met all four algorithm criteria. We randomly selected 252 charts for review, including 206 from patients meeting all four algorithm criteria. The PPV for the 206 algorithm-identified charts was 76% (95% CI: 70%–82%). Among the 49 cases deemed to be false positives (24%), 33 were judged to have multiple myeloma or another lymphoproliferative condition, such as lymphoma.
Conclusions:
We developed a simple algorithm that identified MGUS cases in electronic health data with reasonable accuracy. Inclusion of additional steps to eliminate cases with malignant disease may improve algorithm performance.
Keywords: algorithms; electronic health records; administrative claims, healthcare; monoclonal gammopathy of undetermined significance
Introduction
Monoclonal gammopathy of undetermined significance (MGUS) is a largely asymptomatic precursor (1, 2) to multiple myeloma, an incurable malignancy of clonal plasma cells with a 5-year survival rate of 50.7%.(3) MGUS is clinically defined by the presence of detectable serum monoclonal protein (M-protein) at concentrations less than 3 g/dL and <10% clonal plasma cells in the bone marrow, in the absence of the end-organ damage characteristic of multiple myeloma.(4) MGUS is often diagnosed incidentally; however, retrospective studies of stored blood specimens estimate that 3% of all US adults aged ≥50 years have laboratory evidence of MGUS, with higher rates among the elderly.(5) Although individuals with MGUS progress to MM at an estimated rate of 1% per year,(6, 7) they are also at an increased risk for other serious health outcomes, including lymphoproliferative disorders, and may experience a shorter lifespan compared to people without MGUS.(8) However, few biological or clinical markers, including M-protein level, immunoglobulin subtype, and serum free light chain ratio, may predict progression from MGUS to MM,(8–16) resulting in the MGUS patient experiencing heightened anxiety, repeated clinical assessment and testing, and associated medical financial costs.(1, 9, 17)
Most population-based studies of MGUS have relied on testing stored blood specimens or chart review to ascertain diagnoses,(1, 2, 18, 19) an expensive and inefficient process that makes large-scale studies of MGUS difficult to initiate. A study of the Danish National Patient Registry reported a relatively high positive predictive value (PPV; 82.3% (95% Confidence Interval [CI]: 78.1%–86.4%)) associated with the presence of an ICD-10 diagnosis code for MGUS, suggesting electronic health data may reliably identify MGUS cases.(20) However, this has not yet been tested in electronic health data from US healthcare systems. In the current study, we aimed to develop and evaluate an algorithm to accurately identify patients diagnosed with MGUS in a community-based healthcare setting using longitudinally collected automated healthcare claims and electronic health record data from 2007–2015. For purposes of clarity, and the years of the study period, the present study incorporates ICD-9 codes used prior to the transition to ICD-10. A valid and reproducible algorithm would allow for the efficient identification of true MGUS patients to facilitate large, longitudinal, population-based studies of this prevalent, premalignant condition for the study of patient outcomes. To align with this goal, we calculated the positive predictive value (PPV) for the MGUS algorithm.(21)
Methods
Data Sources
This study was conducted in a large multispecialty medical group located in Massachusetts, using healthcare data organized according to a common data model (the Health Care Systems Research Network [HCSRN] Virtual Data Warehouse [VDW]) with additions from state and national registries. The VDW follows standards, definitions, and specifications set by the HCSRN and the Cancer Research Network (www.crn.cancer.gov), and uses a common data dictionary allowing for efficient extraction of standardized information and pooling of data across health systems participating in the HCSRN.(22–24) Cancer diagnoses in this population were primarily derived from the Massachusetts Cancer Registry (1999–2015) and supplemented by an internal tumor registry maintained by the medical group (2012–15). All EHR data were de-identified, and records anonymized, before chart adjudication.
The Institutional Review Board at the University of Massachusetts Medical School approved this study. Data from the Massachusetts Cancer Registry were provided with approval from the Massachusetts Department of Public Health.
Study Population
Study participants were selected from all men and women aged 50 years or older (5) who received care from the medical group between January 1, 2007 and December 31, 2015. Eligible participants received care from the medical group for at least one year prior to the first MGUS diagnosis code in their EHR and had evidence of care for at least 6 months following the first MGUS diagnosis code to allow for follow-up. Patients were excluded if they were diagnosed with multiple myeloma prior to, or within 3 months after, the first MGUS diagnosis code, as they likely had multiple myeloma when the MGUS diagnosis code was entered into their medical record, and are unlikely to be a true MGUS case. Multiple myeloma diagnoses were identified through tumor registry data (International Classification of Diseases 9th Revision, clinical modification [ICD-9-CM] code 203.0; or the ICD for Oncology, 3rd edition [ICD-O-3] morphology code 9732).
Case Identification
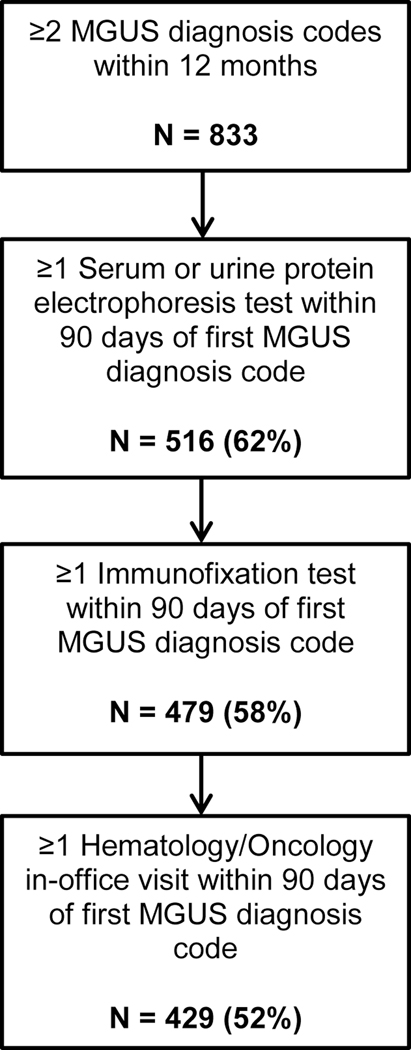
We developed a four-step algorithm to identify MGUS cases using EHR- and claims-derived datasets (Figure 1). The first appearance of the ICD-9 diagnosis code for MGUS (273.1) in a patient’s EHR was considered the index date for potential cases identified by the algorithm. We did not incorporate ICD-10 diagnosis codes in this analysis, as ICD-10 was not adopted until October 2015. Cases were first required to have at least two MGUS diagnosis codes relating to healthcare services provided on different dates within 12 months. We further required potential MGUS cases to have had two key diagnostic laboratory tests that contribute to a clinical diagnosis of MGUS. Specifically, among patients with two eligible MGUS diagnosis codes, we identified those who had a serum or urine protein electrophoresis test (defined by CPT codes 84165, 84166, 84155, and 84156) within 90 days of the index date. We then identified patients who also had a serum or urine immunofixation test within 90 days of the index date (defined by CPT codes 86334 and 86335). As a last step, we excluded potential MGUS cases who did not have an in-office (ambulatory) hematology/oncology visit within 90 days of the index date to eliminate patients for whom the tests were conducted to rule out an MGUS diagnosis.
Figure 1.
Flowchart depicting four-step algorithm to identify cases of monoclonal gammopathy of undetermined significance (MGUS) in electronic health data
Abbreviations: MGUS - monoclonal gammopathy of undetermined significance
Because protein electrophoresis and immunofixation test results are often reported as free text and are presently not incorporated in the VDW database, we did not know the actual reported levels or type of M-protein. Thus, these two algorithm steps were met if a patient had completed protein electrophoresis and immunofixation tests recorded in their EHR. We considered requiring other tests that are often used in the diagnosis of MGUS patients, including serum free light chain tests (SFLC; CPT code 83883), quantifiable immunoglobulin assays (CPT code 82784), and bone marrow biopsies (CPT codes 38220 and 38221). However, SFLC is a relatively recent test, and did not appear consistently in the database. In addition, because we did not have complete access to the results of immunofixation tests without chart review, and thus could not electronically identify specific abnormal immunoglobulins, the results of quantifiable immunoglobulin assays were not informative to the algorithm. Finally, we were unable to reliably detect bone marrow biopsies in the electronic health data through CPT codes. For these reasons, we did not include these tests in the final MGUS case-finding algorithm.
Case Confirmation
We designed a standardized data abstraction form (Supplement 1) with input from study clinicians (JG, AR), and randomly selected 252 charts from MGUS cases identified by the algorithm for EHR review. One of three trained nurse abstractors reviewed all EHR data available from each patient during the study period, focusing on the period of 12 months before and after the first MGUS diagnosis code, and completed one data abstraction form per chart. Abstracted data included diagnosis dates; dates and results of relevant laboratory tests, including protein electrophoresis, immunofixation, and SFLC testing; results from bone marrow biopsies, relevant imaging studies, and genetic and molecular tests; and additional blood test results, including hemoglobin, serum calcium, and serum creatinine. Two pilot chart review sessions were conducted with 10 charts reviewed in each session, and the abstraction form was revised accordingly. Of the 252 charts reviewed, 206 charts (82%), were randomly selected from the 429 patients meeting all four algorithm criteria, and these patients represent the population in which the positive predictive value (PPV) was calculated. To investigate the utility of identifying cases by diagnosis codes alone, 31 charts were abstracted from patients meeting only the first algorithm step (two MGUS diagnosis codes) for a secondary, pilot analysis. Furthermore, 15 charts were randomly selected from patients meeting only the first 2 or 3 algorithm criteria in order to validate EHR data at all steps of the algorithm. A comprehensive coding manual was developed to assist nurse abstractors and serve as a resource for physician adjudicators.
A chart adjudication form was developed to assist three study clinicians (JG, KF, SS) in adjudicating the abstracted charts (Supplement 2). Adjudicators were provided with completed chart abstraction forms and de-identified copies of reports detailing results of bone marrow biopsies, imaging studies, and genetic and molecular tests relevant to the MGUS diagnosis. Each abstracted chart was independently adjudicated by two clinicians and assigned a status as: 1) definite MGUS; 2) probable MGUS; 3) possible MGUS; 4) no evidence of MGUS; or determined to be 5) smoldering myeloma; 6) multiple myeloma; or 7) another disorder. Adjudicators could also select “unable to determine.” MGUS status was confirmed if both adjudicators classified the case as definite or probable. For this study, we classified smoldering myeloma cases as true positives. If the two adjudicators did not agree on case status, the adjudicators convened to attempt to reconcile. Consensus was reached for all cases.
Statistical Analysis
The initial adjudicated case status was compared between adjudicators; percent agreement and Cohen’s kappa statistic were calculated to assess inter-rater reliability.
The algorithm was assessed by calculating the PPV among cases that met all four algorithm criteria. The PPV is the number of algorithm-identified MGUS cases who were confirmed through chart review divided by the total number of potential MGUS cases identified by the algorithm. In a secondary, pilot analysis, we decided a posteriori to also calculate the PPV for a small selection of cases meeting only the first step of the algorithm (two MGUS diagnosis codes within 12 months). We calculated 95% confidence intervals (CI) around all estimates. Based on the literature evaluating similar algorithms based in administrative health data,(25–28) we set a target PPV of 75% for the application of the MGUS algorithm to this patient population. Descriptive statistics were calculated for all algorithm-identified cases and adjudicated true and false positives. Data analyses were conducted using SAS version 9.4 (Cary, NC).
Results
A total of 833 patients were identified with at least two MGUS diagnosis codes on different dates within a 12-month period between 2007 and 2015 in the standardized electronic VDW database among 119,627 eligible members of the provider group (Figure 1). This suggests a prevalence of 0.70% in our population according to diagnosis codes alone. Of these, 516 (62%) had at least one serum or urine protein electrophoresis test within 90 days of their first MGUS diagnosis code (index date), and of those patients, 479 (93%) also had at least one immunofixation test during that period. Among individuals meeting the first three algorithm criteria, 429 (90%; or 52% of the original population) also had documented evidence of an ambulatory hematology/oncology visit within 90 days of index date. We observed a trend of increasing use of SFLC tests over time, ranging from 15% of MGUS cases diagnosed in 2007, to 74% of MGUS cases diagnosed in 2015.
The 429 patients satisfying all four algorithm criteria were 49% female and mostly Caucasian (85%), with a mean age at diagnosis of 74.5 years, and about six years of enrollment in the health system prior to their first MGUS diagnosis code (Table 1).
Table 1.
Characteristics of the study population, potential cases of monoclonal gammopathy of undetermined significance identified by an algorithm using electronic health data, 2007–15
| Characteristic | Algorithm positive (N=429)† | Algorithm negative (N=119,198)‡ | True Positive (N=157) § | False Positive (N=49)¶ |
|---|---|---|---|---|
| Male, N (%) | 217 (51%) | 54,163 (45) | 79 (50%) | 27 (55%) |
| Female | 212 (49) | 65,035 (55) | 78 (50) | 22 (45) |
| Caucasian | 365 (85) | 74,727 (63) | 132 (84) | 37 (76) |
| Age at first MGUS diagnosis, years (mean ± SD, range) | 74.5 ± 10.4 (50–97) | -- | 75.0 ± 10.3 (51–95) | 74.0 ± 10.0 (52–95) |
| Number of SPEP codes in complete record | 12.9 ± 10.3 (2–56) | 0.07 ± 1.39 (0–134) | 12.7 ± 9.3 (2–42) | 10.0 ± 8.3 (2–37) |
| Years of enrollment prior to MGUS diagnosis | 6.1 ± 4.8 (1–35) | -- | 5.9 ± 4.6 (1–30) | 6.2 ± 4.2 (1–20) |
Abbreviations: MGUS - monoclonal gammopathy of undetermined significance; SD standard deviation; SPEP - serum protein electrophoresis
Individuals meeting all four algorithm criteria
Eligible individuals from the source population who did not meet algorithm criteria
Cases judged to be true positives following adjudication of 206 charts from participants meeting all four algorithm criteria
Cases judged to be false positives following adjudication of 206 charts from participants meeting all four algorithm criteria
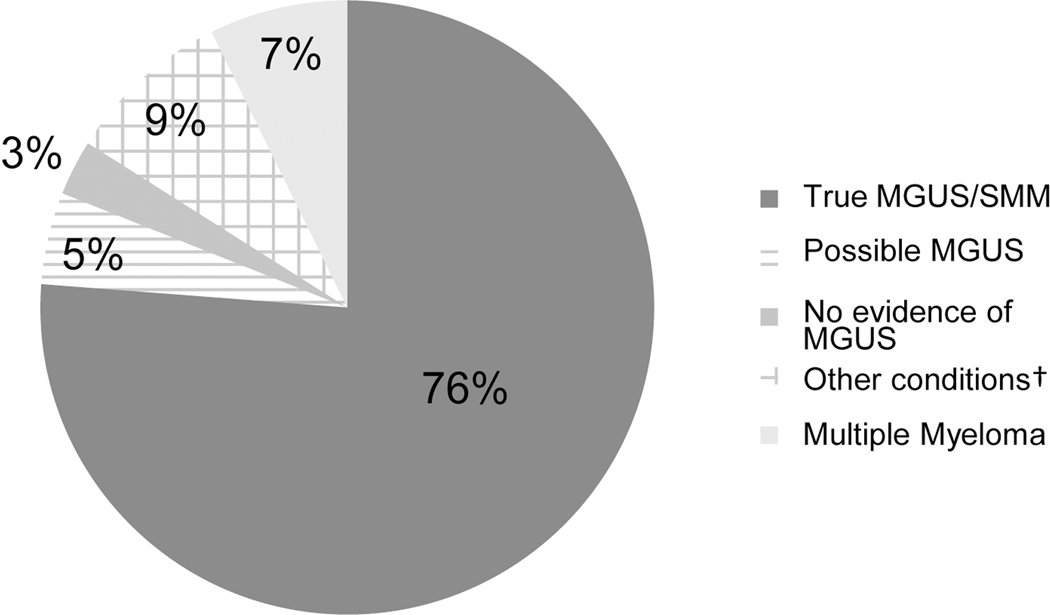
Of the 206 patients randomly selected for chart review who met all four algorithm criteria, 157 (76%) were adjudicated to be definite or probable MGUS cases, or determined to have smoldering multiple myeloma, and were classified as true cases. Forty-nine cases (24%) were judged to be false positives (Figure 2). Ten of the false positive cases were classified as possible cases or lacking sufficient data, and an additional six cases had no evidence of MGUS in their EHR. The adjudicators determined that 15 cases had clinical evidence of multiple myeloma and another 18 cases had clinical evidence of a different condition, including B-cell lymphoma (N=7) or Waldenstrom macroglobulinemia (N=6).
Figure 2.
Results of adjudication of 206 electronic health records from potential cases of monoclonal gammopathy of undetermined significance identified by a four-step algorithm
Abbreviations: MGUS - monoclonal gammopathy of undetermined significance; SMM - smoldering multiple myeloma
†Other conditions include B-cell lymphoma (N=7), Waldenstrom macroglobulinemia (N=6), other lymphoma or leukemia (N=5)
The percent agreement among clinician adjudicators following chart review was 79% for all adjudicated cases, and for the subset meeting all four algorithm criteria. Cohen’s kappa suggests fair inter-rater reliability across all 252 charts reviewed (κ=0.37) with three experienced physician adjudicators. Patients confirmed to be true MGUS cases were slightly more likely to be female (50% vs. 45%; p=0.56) and had a similar age at diagnosis (75 vs. 74 years; p=0.56) compared to those incorrectly identified by the algorithm (Table 1). True positive cases also had slightly more codes for serum protein electrophoresis tests in their EHR than false positives (12.7 vs. 10; p=0.06).
The PPV for the four-step algorithm was 76% (95% CI: 70%–82%), indicating that among all potential MGUS cases identified by the algorithm, 76% were true cases. In the pilot analysis of the one-step algorithm, we examined the ability of the algorithm to correctly identify MGUS cases meeting only the first algorithm step (two diagnosis codes within a 12-month period). Of the 31 patients selected for chart review, 25 were adjudicated to be true MGUS cases, resulting in a similar, yet imprecise PPV of 80.6% (95% CI: 67%–95%).
Discussion
We used a large, standardized electronic database of claims- and EHR-derived datasets to develop a four-step algorithm that can identify cases of MGUS in a community-based setting with reasonable accuracy. The calculated PPV slightly exceeded our initial target of 75%, and suggests that about three-quarters of all MGUS cases identified by the algorithm will be true cases. The four steps of the algorithm were defined by the presence of MGUS diagnosis codes and relevant procedure codes in patients’ electronic health data collected over a nine-year period. We were unable to include test result data in this algorithm as the results did not appear in an informative format in the electronic database. However, this may be considered a strength of this algorithm, as it may be applicable to a broader range of datasets that contain only claims-type data.
The algorithm required procedure codes for two essential diagnostic tests: protein electrophoresis and immunofixation. In our database, 93% of the patients who had an electrophoresis test also had an immunofixation test, as the health system serving as the setting for this study often orders an immunofixation test as a reflex test following abnormal protein electrophoresis; this practice may differ in other clinical settings. Also, we found that both true and false positive MGUS cases had multiple codes for protein electrophoresis tests in their EHR, representing frequent use of this diagnostic test. Future iterations of the algorithm using more contemporary data may include the SFLC test to increase accuracy. The SFLC test has increased in utilization in recent years, although it was uncommon early in our study period.(29) Because the long-term goal of this algorithm is to identify true MGUS cases for inclusion in follow-up studies of patient outcomes, we acknowledge that we may have erroneously excluded some true MGUS cases by requiring cases to meet all four criteria. For example, we may have excluded MGUS cases who were referred to a hematologist-oncologist but did not attend the appointment. However, we believe that we also minimized the number of false positives through the inclusion of these criteria, since the majority of true MGUS cases would be diagnosed through these laboratory tests and through consultation with a hematologist-oncologist.
The kappa statistic for this study, 0.37, suggests fair inter-rater reliability among three experienced physician adjudicators. Disagreement between adjudicators was more likely for algorithm-identified cases that were ultimately judged to have another condition, including multiple myeloma and other cancers. Despite the relatively low kappa value, the agreement rate between clinician adjudicators was high (79%), illustrating a paradox where agreement may be high, and kappa low, due to the low prevalence of the condition (MGUS) in the population.(30) Thus, both measures of inter-rater reliability should be considered when interpreting the results of this study.(31)
We excluded patients diagnosed with multiple myeloma prior to, or within 3 months of MGUS diagnosis based on tumor registry data. We were unable to determine whether some of those cases truly had smoldering multiple myeloma, an intermediate precursor condition between MGUS and multiple myeloma that lacks its own diagnosis code, and may be coded as multiple myeloma or as MGUS. Future versions of the algorithm could be improved by including further steps to rule out malignant disease and reduce misclassification. In addition, we could not access results of key diagnostic tests, namely protein electrophoresis and immunofixation, in our electronic health database. Actual test result data may improve the ability of the algorithm to distinguish true MGUS cases from patients receiving a rule-out MGUS diagnosis code. However, due to the text-based nature of these test results, more sophisticated methods, such as natural language processing, may be required to extract essential information.
A preliminary secondary analysis suggests that a simple one-step algorithm requiring two MGUS diagnosis codes within 12 months may also perform well, with a PPV of 80.6%, although wide confidence intervals (67%–95%) reflect the imprecision of this analysis. This observation is in line with a recent study of the Danish National Patient Register, which observed a PPV of 82.3% (95% CI: 78.1%–86.4%) for patients registered with an ICD-10 diagnosis code for MGUS.(20) However, since this pilot analysis was not adequately powered to draw definitive conclusions, a larger study population is required to validate this simpler algorithm, which could potentially have greater generalizability to settings without EHR data.
Because an MGUS diagnosis must occur in a clinical setting, and thus to someone with access to care, the results of this study are generalizable to other healthcare systems with similar data structure. The algorithm does not detect undiagnosed MGUS, a large percentage of total MGUS cases, as undiagnosed patients would lack the requisite diagnosis codes. In the future, specific test result data may allow us to detect clinically relevant, undiagnosed MGUS. We may have also underestimated the prevalence of MGUS in our population by requiring two diagnosis codes; the estimated prevalence of 0.70% is slightly less than published estimates from comparable populations.(32, 33)
We restricted our study population to adults aged 50 and older since this age is clinically relevant to the development and increasing prevalence of MGUS. Since the algorithm PPV is influenced by disease prevalence, we chose to limit our study to adults aged 50 and older as a balance between the lower prevalence of MGUS experienced by younger adults, and the higher prevalence experienced by older adults.(5, 34) If the analysis was restricted to adults aged 70 and older, the PPV would likely increase, reflecting a higher prevalence of disease in this age group.
In addition, our study population was largely white. Future studies should apply the algorithm to more diverse populations to assess generalizability across race/ethnic groups since the risk of MGUS varies by race, including a two to three-fold increased prevalence in people of African descent compared to Caucasians.(18, 35–37) Lastly, our study, conducted using data from 2007 to 2015, utilized ICD-9 diagnosis codes to identify MGUS patients. Future iterations of the algorithm that encompass time periods after October 2015 should use ICD-10 codes to identify all potential MGUS cases (ICD-10-CM code D47.2).
MGUS is a prevalent, yet understudied, premalignant condition that requires repeated clinical follow-up for diagnosed patients. Current guidelines from the International Myeloma Working Group recommend initial follow-up with serum protein electrophoresis testing at six months post-diagnosis, and then annually for life, although low-risk patients may be seen less often.(4) As few reliable markers of progression to multiple myeloma or other diseases are known,(6, 7) patients live with uncertainty regarding their risk of a malignant diagnosis. Larger population-based datasets are needed to expand research into this important condition and identify patterns of care and potential markers to distinguish patients at risk for progression, and of equal importance, patients who may not require frequent monitoring. Because MGUS is not reportable to cancer registries, applying the algorithm developed in this study to large electronic health databases with similar data structure could identify large series of MGUS cases for prospective study. A strength of the current study is the use of healthcare data organized according to a common data model via the HCSRN VDW. Successful cancer algorithms have previously been developed in the setting of the VDW,(38–40) which lends support for validating this MGUS case-finding algorithm at other HCSRN sites.
In summary, we developed a simple four-step algorithm that can identify patients with MGUS with reasonable accuracy in electronic health data sources, including health claims and EHR data. Since MGUS is an understudied condition, it is essential to develop tools to identify large numbers of MGUS patients for population-based research. The current algorithm is a first step towards identifying a cohort of MGUS patients for future longitudinal studies that can lead to insights about MGUS etiology, progression, and health service utilization.
Five key points:
-
1)
This study developed an algorithm to identify monoclonal gammopathy of undetermined significance (MGUS) patients in electronic health data to facilitate large-scale, population-based studies of this premalignant condition;
-
2)
The algorithm incorporated diagnosis and procedure codes, as well as provider type data, to identify MGUS cases;
-
3)
The positive predictive value of the algorithm was 76%, suggesting more than three-quarters of MGUS cases identified by the algorithm were true cases.
Acknowledgements:
We would like to acknowledge Brenda Valenti, BSN; Marcia Kirkpatrick, RN, BSN, CCRC; Sarah Cutrona, MD, MPH; Tejaswini Dhawale, MD; Jessica Chubak, PhD; and Christopher Delude, BA for their assistance in data collection, interpretation, analysis, and presentation. This work was funded in part by the National Cancer Institute grant R03 CA199383 (to MME). MME is supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2TR001454.
Funding: This work was funded in part by the National Cancer Institute grant R03 CA199383 (to MME). MME is supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2TR001454.
References
- 1.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood 2009;113:5418–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood 2009;113:5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975–2015, National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/csr/1975_2015/, based on November 2017 SEER data submission, posted to the SEER web site, April 2018. [Google Scholar]
- 4.Kyle RA, Durie BG, Rajkumar SV, Landgren O, Blade J, Merlini G, et al. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia 2010;24:1121–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyle RA, Therneau TM, Rajkumar SV, Larson DR, Plevak MF, Offord JR, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med 2006;354:1362–1369. [DOI] [PubMed] [Google Scholar]
- 6.Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med 2002;346:564–569. [DOI] [PubMed] [Google Scholar]
- 7.Kyle RA, Therneau TM, Rajkumar SV, Remstein ED, Offord JR, Larson DR, et al. Long-term follow-up of IgM monoclonal gammopathy of undetermined significance. Blood 2003;102:3759–3764. [DOI] [PubMed] [Google Scholar]
- 8.Kyle RA, Larson DR, Therneau TM, Dispenzieri A, Kumar S, Cerhan JR, et al. Long-Term Follow-up of Monoclonal Gammopathy of Undetermined Significance. N Engl J Med 2018;378:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landgren O Monoclonal gammopathy of undetermined significance and smoldering myeloma: new insights into pathophysiology and epidemiology. Hematology Am Soc Hematol Educ Program 2010;2010:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landgren O, Kyle RA, Rajkumar SV. From myeloma precursor disease to multiple myeloma: new diagnostic concepts and opportunities for early intervention. Clin Cancer Res 2011;17:1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyle RA, Remstein ED, Therneau TM, Dispenzieri A, Kurtin PJ, Hodnefield JM, et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N Engl J Med 2007;356:2582–2590. [DOI] [PubMed] [Google Scholar]
- 12.Rajkumar SV, Lacy MQ, Kyle RA. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. Blood Reviews 2007;21:255–265. [DOI] [PubMed] [Google Scholar]
- 13.Bida JP, Kyle RA, Therneau TM, Melton LJI, Plevak MF, Larson DR, et al. Disease Associations With Monoclonal Gammopathy of Undetermined Significance: A Population-Based Study of 17,398 Patients. Mayo Clin Proc 2009;84:685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristinsson SY, Tang M, Pfeiffer RM, Bjorkholm M, Goldin LR, Blimark C, et al. Monoclonal gammopathy of undetermined significance and risk of infections: a population-based study. Haematologica 2012;97:854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Donk NW, Palumbo A, Johnsen HE, Engelhardt M, Gay F, Gregersen H, et al. The clinical relevance and management of monoclonal gammopathy of undetermined significance and related disorders: recommendations from the European Myeloma Network. Haematologica 2014;99:984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roeker LE, Larson DR, Kyle RA, Kumar S, Dispenzieri A, Rajkumar SV. Risk of acute leukemia and myelodysplastic syndromes in patients with monoclonal gammopathy of undetermined significance (MGUS): a population-based study of 17 315 patients. Leukemia 2013;27:1391–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren DF, Alexanian R, Goeken JA, Gorevic PD, Kyle RA, Tomar RH. Guidelines for clinical and laboratory evaluation patients with monoclonal gammopathies. Arch Pathol Lab Med 1999;123:106–107. [DOI] [PubMed] [Google Scholar]
- 18.Landgren O, Katzmann JA, Hsing AW, Pfeiffer RM, Kyle RA, Yeboah ED, et al. Prevalence of monoclonal gammopathy of undetermined significance among men in Ghana. Mayo Clin Proc 2007;82:1468–1473. [DOI] [PubMed] [Google Scholar]
- 19.Wu SP, Minter A, Costello R, Zingone A, Lee CK, Au WY, et al. MGUS prevalence in an ethnically Chinese population in Hong Kong. Blood 2013;121:2363–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregersen H, Larsen CB, Haglund A, Mortensen R, Andersen NF, Norgaard M. Data quality of the monoclonal gammopathy of undetermined significance diagnosis in a hospital registry. Clin Epidemiol 2013;5:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chubak J, Pocobelli G, Weiss NS. Tradeoffs between accuracy measures for electronic health care data algorithms. J Clin Epidemiol 2012;65:343–349 e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross TR, Ng D, Brown JS, Pardee R, Hornbrook MC, Hart G, et al. The HMO Research Network Virtual Data Warehouse: A Public Data Model to Support Collaboration. EGEMS (Wash DC) 2014;2:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner EH, Greene SM, Hart G, Field TS, Fletcher S, Geiger AM, et al. Building a research consortium of large health systems: the Cancer Research Network. J Natl Cancer Inst Monogr 2005:3–11. [DOI] [PubMed] [Google Scholar]
- 24.Hornbrook MC, Hart G, Ellis JL, Bachman DJ, Ansell G, Greene SM, et al. Building a virtual cancer research organization. J Natl Cancer Inst Monogr 2005:12–25. [DOI] [PubMed] [Google Scholar]
- 25.Walsh KE, Cutrona SL, Foy S, Baker MA, Forrow S, Shoaibi A, et al. Validation of anaphylaxis in the Food and Drug Administration’s Mini-Sentinel. Pharmacoepidemiol Drug Saf 2013;22:1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrold LR, Yood RA, Andrade SE, Reed JI, Cernieux J, Straus W, et al. Evaluating the predictive value of osteoarthritis diagnoses in an administrative database. Arthritis Rheum 2000;43:1881–1885. [DOI] [PubMed] [Google Scholar]
- 27.Cutrona SL, Toh S, Iyer A, Foy S, Daniel GW, Nair VP, et al. Validation of acute myocardial infarction in the Food and Drug Administration’s Mini-Sentinel program. Pharmacoepidemiol Drug Saf 2013;22:40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo Re V, 3rd, Haynes K, Goldberg D, Forde KA, Carbonari DM, Leidl KB, et al. Validity of diagnostic codes to identify cases of severe acute liver injury in the US Food and Drug Administration’s Mini-Sentinel Distributed Database. Pharmacoepidemiol Drug Saf 2013;22:861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel D, Bilotti E, van Hoeven KH. Serum Free Light Chain Analysis for Diagnosis, Monitoring, and Prognosis of Monoclonal Gammopathies. Lab Med 2009;40:363–366. [Google Scholar]
- 30.Feinstein AR, Cicchetti DV. High agreement but low kappa: I. The problems of two paradoxes. J Clin Epidemiol 1990;43:543–549. [DOI] [PubMed] [Google Scholar]
- 31.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 32.Landgren O, Weiss BM. Patterns of monoclonal gammopathy of undetermined significance and multiple myeloma in various ethnic/racial groups: support for genetic factors in pathogenesis. Leukemia 2009;23:1691–1697. [DOI] [PubMed] [Google Scholar]
- 33.Therneau TM, Kyle RA, Melton LJ, 3rd, Larson DR, Benson JT, Colby CL, et al. Incidence of monoclonal gammopathy of undetermined significance and estimation of duration before first clinical recognition. Mayo Clin Proc 2012;87:1071–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wadhera RK, Rajkumar SV. Prevalence of monoclonal gammopathy of undetermined significance: a systematic review. Mayo Clin Proc 2010;85:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landgren O, Graubard BI, Katzmann JA, Kyle RA, Ahmadizadeh I, Clark R, et al. Racial disparities in the prevalence of monoclonal gammopathies: a population-based study of 12,482 persons from the National Health and Nutritional Examination Survey. Leukemia 2014;28:1537–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landgren O, Gridley G, Turesson I, Caporaso NE, Goldin LR, Baris D, et al. Risk of monoclonal gammopathy of undetermined significance (MGUS) and subsequent multiple myeloma among African American and white veterans in the United States. Blood 2006;107:904–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal A, Ghobrial IM. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: a review of the current understanding of epidemiology, biology, risk stratification, and management of myeloma precursor disease. Clin Cancer Res 2013;19:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassett MJ, Ritzwoller DP, Taback N, Carroll N, Cronin AM, Ting GV, et al. Validating billing/encounter codes as indicators of lung, colorectal, breast, and prostate cancer recurrence using 2 large contemporary cohorts. Med Care 2014;52:e65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassett MJ, Uno H, Cronin AM, Carroll NM, Hornbrook MC, Ritzwoller D. Detecting Lung and Colorectal Cancer Recurrence Using Structured Clinical/Administrative Data to Enable Outcomes Research and Population Health Management. Med Care 2017;55:e88–e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritzwoller DP, Hassett MJ, Uno H, Cronin AM, Carroll NM, Hornbrook MC, et al. Development, Validation, and Dissemination of a Breast Cancer Recurrence Detection and Timing Informatics Algorithm. J Natl Cancer Inst 2018;110:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]