To the Editor – One limitation of microbial community marker-gene sequencing is that it does not provide information about the functional composition of sampled communities. PICRUSt1 was developed in 2013 to predict the functional potential of a bacterial community based on marker gene sequencing profiles, and now we present PICRUSt2 (https://github.com/picrust/picrust2), which improves upon the original method. Specifically, PICRUSt2 contains an updated and larger database of gene families and reference genomes, provides interoperability with any operational taxonomic unit (OTU)-picking or denoising algorithm, and enables phenotype predictions. Benchmarking shows that PICRUSt2 is more accurate than PICRUSt and other competing methods overall. PICRUSt2 also allows the addition of custom reference databases. We highlight these improvements and also important caveats regarding the use of predicted metagenomes.
The most common method for profiling bacterial communities is to sequence the conserved 16S rRNA gene. Functional profiles cannot be directly identified using 16S rRNA gene sequence data owing to strain variation so several methods have been developed to predict microbial community functions from taxonomic profiles (amplicon sequences) alone1–5. Shotgun metagenomic sequencing (MGS) which sequences entire genomes rather than marker genes can also be used to characterize the functions of a community, but does not work well if there is host contamination e.g. in a biopsy, or if there is very little community biomass.
PICRUSt1 (hereafter “PICRUSt1”) was the first tool developed for prediction of functions from 16S marker sequences, and is widely used but has some limitations. Standard PICRUSt1 workflows require input sequences to be OTUs generated from closed-reference OTU-picking against a compatible version of the Greengenes database6. Due to this restriction to reference OTUs, the default PICRUSt1 workflow is incompatible with sequence denoising methods, which produce amplicon sequence variants (ASVs) rather than OTUs. ASVs have finer resolution, allowing closely related organisms to be more readily distinguished. Plus, the bacterial reference databases used by PICRUSt1 have not been updated since 2013 and lack thousands of recently added gene families.
We hypothesized that optimizing genome prediction would improve accuracy of functional predictions. Therefore, the PICRUSt2 algorithm (Fig 1a) includes steps that optimize genome prediction, including placing sequences into a reference phylogeny rather than relying on predictions limited to reference OTUs (Fig 1b); basing predictions on a larger database of reference genomes and gene families (Fig 1c); more stringent prediction of pathway abundance (Supp Fig 1); enabling predictions of complex phenotypes and integration of custom databases.
Figure 1: PICRUSt2 algorithm.
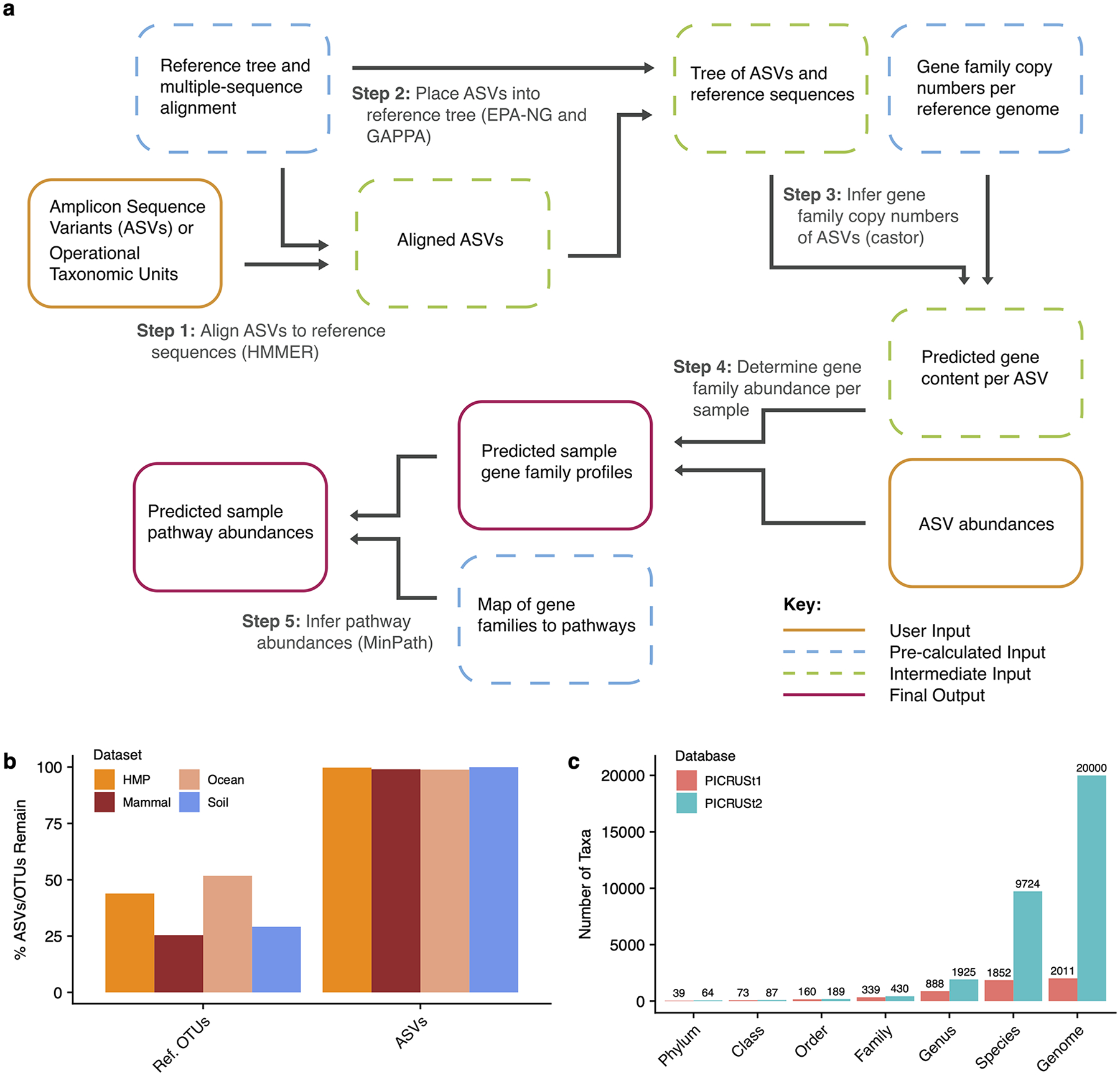
(a) The PICRUSt2 method consists of phylogenetic placement, hidden-state-prediction and sample-wise gene and pathway abundance tabulation. ASV sequences and abundances are taken as input, and gene family and pathway abundances are output. All necessary reference tree and trait databases for the default workflow are included in the PICRUSt2 implementation. (b) The default PICRUSt1 pipeline restricted predictions to reference operational taxonomic units (Ref. OTUs) in the Greengenes database. This requirement resulted in the exclusion of many study sequences across four representative 16S rRNA gene sequencing datasets. PICRUSt2 relaxes this requirement and is agnostic to whether the input sequences are within a reference or not, which results in almost all of the input amplicon sequence variants (ASVs) being retained in the final output. (c) An increase in the taxonomic diversity in the default PICRUSt2 database is observed compared to PICRUSt1.
PICRUSt2 integrates existing open-source tools to predict genomes of environmentally sampled 16S rRNA gene sequences. ASVs are placed into a reference tree, which is used as the basis of functional predictions. This reference tree contains 20,000 full 16S rRNA genes from bacterial and archaeal genomes in the Integrated Microbial Genomes (IMG) database7. Phylogenetic placement in PICRUSt2 is based on running three tools: HMMER (www.hmmer.org) to place ASVs, EPA-ng8 to determine the optimal position of these placed ASVs in a reference phylogeny, and GAPPA9 to output a new tree incorporating the ASV placements. This results in a phylogenetic tree containing both reference genomes and environmentally sampled organisms, which is used to predict individual gene family copy numbers for each ASV. This procedure is re-run for each input dataset, allowing users to utilize custom reference databases as needed, including those that may be optimized for the study of specific microbial niches.
As in PICRUSt1, hidden state prediction approaches are used in PICRUSt2 to infer the genomic content of sampled sequences. The castor R package10, which is substantially faster than the approach used in PICRUSt1, is used for core hidden state prediction functions. As in PICRUSt1, ASVs are corrected by their 16S rRNA gene copy number and then multiplied by their functional predictions to produce a predicted metagenome. PICRUSt2 also provides the ASV contribution of each predicted function allowing for taxonomy-informed statistical analyses to be conducted. Lastly, pathway abundances are inferred based on structured pathway mappings, which are more conservative than the ‘bag-of-genes’ approach used in PICRUSt1.
The PICRUSt2 default genome database is based on 41,926 bacterial and archaeal genomes from the IMG database7 (November 8, 2017) which is a >20-fold increase over the 2,011 IMG genomes used by PICRUSt1. Many of the additional genomes are from strains of the same species and have identical 16S rRNA genes. We de-replicated the identical 16S rRNA genes across these genomes, which resulted in 20,000 final 16S rRNA gene clusters. The taxonomic diversity of the PICRUSt2 reference database is increased compared with PICRUSt1 (Fig. 1c). The clearest increases in diversity is at the species and genus levels (5.3-fold and 2.2-fold increases respectively) but all taxonomic levels are more diverse including the phylum level where the coverage increased from 39 to 64 phyla (1.6-fold increase).
PICRUSt2 predictions based on several gene family databases are supported by default, including the Kyoto Encyclopedia of Genes and Genomes11 (KEGG) orthologs (KO) and Enzyme Commission numbers (EC numbers) (Supp Table 1). PICRUSt2 distinctly improves on PICRUSt1 by including gene families more recently added to the KEGG database. Specifically, the total number of KOs is 10,543 in PICRUSt2 compared to 6,909 in PICRUSt1, a 1.5-fold increase.
We validated PICRUSt2 metagenome predictions using samples from seven published datasets generated using both 16S rRNA marker-gene and MGS. We used three human-associated microbiome datasets: 57 stool samples from Cameroonian individuals, 91 stool samples from Indian individuals, and 137 samples spanning the human body (from the Human Microbiome Project [HMP]). We used four non-human-associated datasets including 77 non-human primate stool samples, eight mammalian stool samples, six ocean samples, and 22 bulk soil and blueberry rhizosphere samples. These datasets present a good variation of types of sequences and environments. (Supp Table 2).
PICRUSt2 KO predictions from 16S rRNA marker gene data were produced for each dataset. We compared these predictions to KO relative abundances profiled from the corresponding MGS metagenomes, which served as a gold-standard to evaluate prediction performance. We performed the same analyses with four alternative prediction pipelines: PICRUSt1, Piphillin2, PanFP3 and Tax4Fun24,5. We calculated Spearman correlation coefficients (hereafter “correlations”) for matching samples between the predicted KO abundance and MGS KO abundance tables after filtering all tables to the 6,220 KOs that could be output by all tested databases (Fig 2). The correlation metric represents the similarity in rank ordering of KO abundances between the predicted and observed data. The correlations based on PICRUSt2 KO predictions ranged from a mean of 0.79 (standard deviation [sd] = 0.028; primate stool) to 0.88 (sd = 0.019; Cameroonian stool dataset). For all seven datasets, PICRUSt2 predictions were either better than or comparable with the best prediction method (paired-sample, two-tailed Wilcoxon tests [PTW] P < 0.05). Correlations based on PICRUSt2 predictions were substantially better for non-human associated datasets. This result could indicate an advantage of phylogenetic-based methods over non-phylogenetic-based methods, such as Piphillin, for environments poorly represented by reference genomes.
Figure 2: PICRUSt2 performance characteristics.
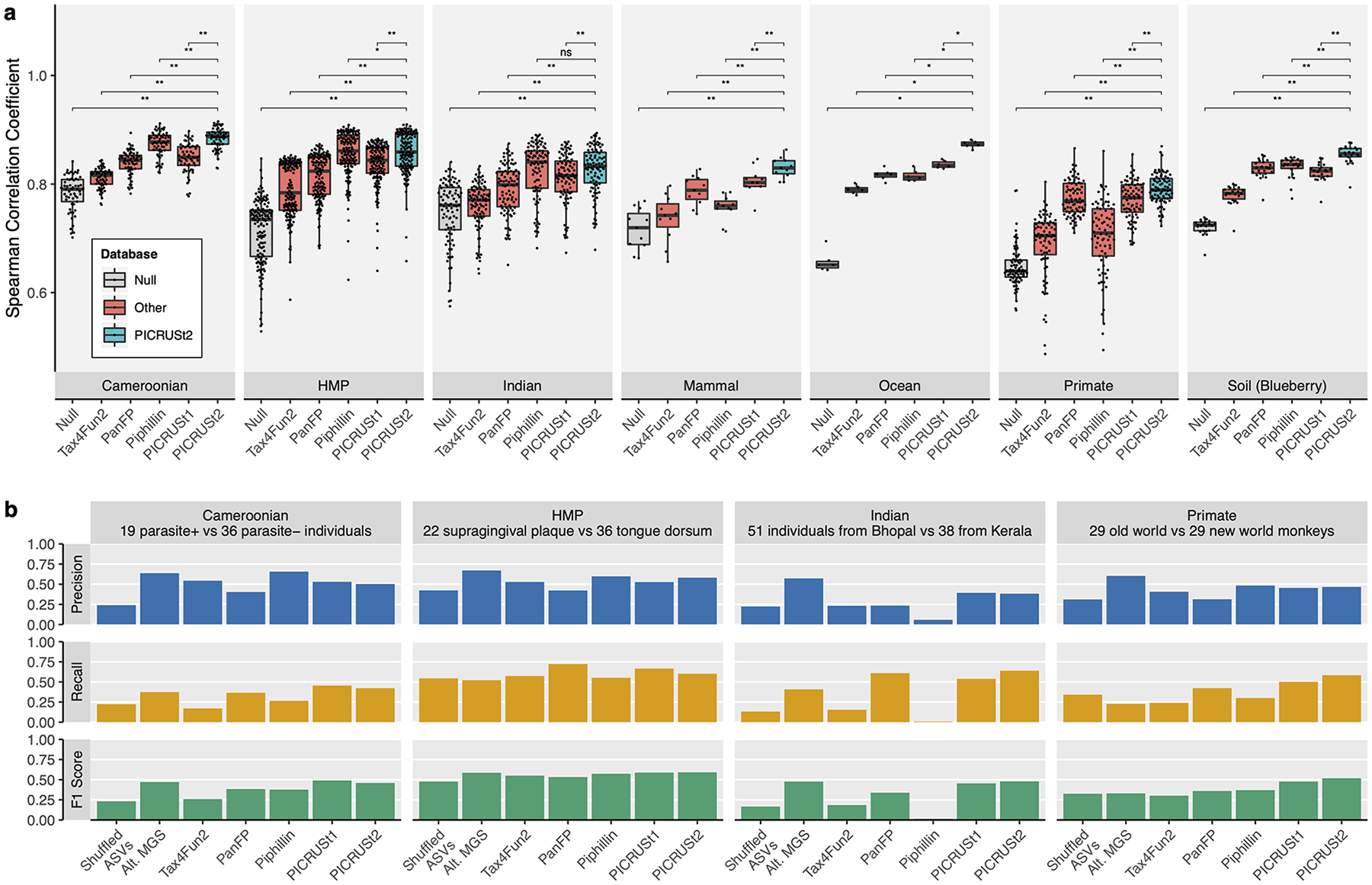
Validation of PICRUSt2 KEGG ortholog (KO) predictions comparing metagenome prediction performance against gold-standard shotgun metagenomic sequencing (MGS). (a) Boxplots of Spearman correlation coefficients observed in stool samples from Cameroonian individuals (n=57), the human microbiome project (HMP, n=137), stool samples from Indian individuals (n=91), non-human primate stool samples (n=77), mammalian stool (n=8), ocean water (n=6), and blueberry soil (n=22) datasets. The significance of paired-sample, two-tailed Wilcoxon tests is indicated above each tested grouping (*, **, and ns correspond to P < 0.05, P < 0.001, and not significant respectively). (b) Comparison of significantly differentially abundant KOs between predicted metagenomes and MGS. Precision, recall, and F1 score are reported for each category compared to the MGS data. Precision corresponds to the proportion of significant KOs for that category also significant in the MGS data. Recall corresponds to the proportion of significant KOs in the MGS data also significant for that category. The F1 score is the harmonic mean of these metrics. The subsets of the four datasets compared are indicated above each panel (the Cameroonian parasite is Entamoeba). Wilcoxon tests were performed on the KO relative abundances after normalizing by the median number of universal single-copy genes per sample. Significance was defined at a false discovery rate < 0.05. The “Shuffled ASVs” category corresponds to PICRUSt2 predictions with ASV labels shuffled per dataset. The “Alt. MGS” category corresponds to an alternative MGS processing pipeline with reads aligned to the KEGG database rather than the default HUMAnN2 pipeline.
Gene families regularly co-occur within genomes, so the use of correlations to assess gene-table similarity may be limited by the lack of independence of gene families within a sample (Supp Fig 2). To address this dependency, we compared the observed correlations between paired MGS and predicted metagenomes to correlations between MGS functions and a null reference genome, comprised of the mean gene family abundance across all reference genomes. For all datasets, PICRUSt2 metagenome tables were more similar to MGS values than the null (Fig 2a). However, this increase over the null expectation is predominately driven by each dataset’s predicted genome content (rather than that of individual samples). This is demonstrated by the fact that these correlations are actually only slightly significantly higher than those observed when ASV labels are shuffled within a dataset (Supp Fig 3). The observed correlations for the shuffled ASVs ranged from a mean of 0.77 (sd = 0.196; primate stool) to 0.84 (sd = 0.178; blueberry rhizosphere). Biologically these results are consistent with several patterns. First, gene families are correlated in copy number across diverse taxa (as captured by the ‘Null’ dataset). Second, these correlations are stronger within than between environments (as shown by the difference between the ‘Null’ and ‘Shuffled ASV’ results). Lastly, environment-to-environment differences tend to be larger than sample-to-sample differences within an environment (as shown by the differences between PICRUSt2 predictions and the ‘Shuffled ASV’ results).
A complementary approach for validating metagenome predictions is to compare the results of differential abundance tests on 16S-predicted metagenomes to MGS data. A recent analysis of Piphillin suggested that this tool out-performs PICRUSt2 based on this approach12. We similarly performed this evaluation on the KO predictions for four validation datasets (Fig 2b; see Supplementary Text). Overall, PICRUSt2 displayed the highest F1 score, the harmonic mean of precision and recall, compared to other prediction methods (ranging from 0.46–0.59; mean=0.51; sd=0.06). However, all prediction tools displayed relatively low precision, the proportion of significant KOs that were also significant in the MGS data. In particular, precision ranged from 0.38–0.58 (mean=0.48; sd=0.08) for PICRUSt2 and 0.06–0.66 (mean=0.45; sd=0.27) for Piphillin. In all cases, PICRUSt2 predictions out-performed ASV-shuffled predictions, which ranged in precision from 0.22–0.42 (mean=0.30; sd=0.09). In addition, differential abundance tests performed on MGS-derived KOs from an alternative MGS-processing workflow resulted in only marginally higher precision (ranging from 0.57–0.67; mean=0.62; sd=0.04). Taken together, these results highlight the difficulty of reproducing microbial functional biomarkers with both predicted and actual metagenomics data.
MetaCyc pathway abundances are now the main high-level predictions output by PICRUSt2 by default. The MetaCyc database13 is an open-source alternative to KEGG and is also a major focus of the widely-used metagenomics functional profiler, HUMAnN214. MetaCyc pathway abundances are calculated in PICRUSt2 through structured mappings of EC gene families to pathways. These pathway predictions performed better than the null distribution for all metrics overall (PTW P < 0.05; Fig 3a and Supp Fig 4–5) compared to MGS-derived pathways. Similar to our previous analysis, shuffled ASV predictions representing overall functional structure within each dataset accounted for the majority of this signal (Supp Fig 4). In addition, differential abundance tests on these pathways showed high variability in F1 scores across datasets and statistical methods with the ASV shuffled predictions contributing the majority of this signal (Supp Fig 6; F1 scores ranged from 0.23–0.62 [mean=0.41; sd=0.17] and 0.22–0.60 [mean=0.34; sd=0.18] for the observed and ASV shuffled PICRUSt2 predictions, respectively). Again, these results suggest that identifying robust differentially abundant metagenome-wide pathways is difficult and highlights the challenge of analyzing microbial pathways in general.
Figure 3: PICRUSt2 accurately predicts MetaCyc pathways and phenotypes for characterizing overall environments.
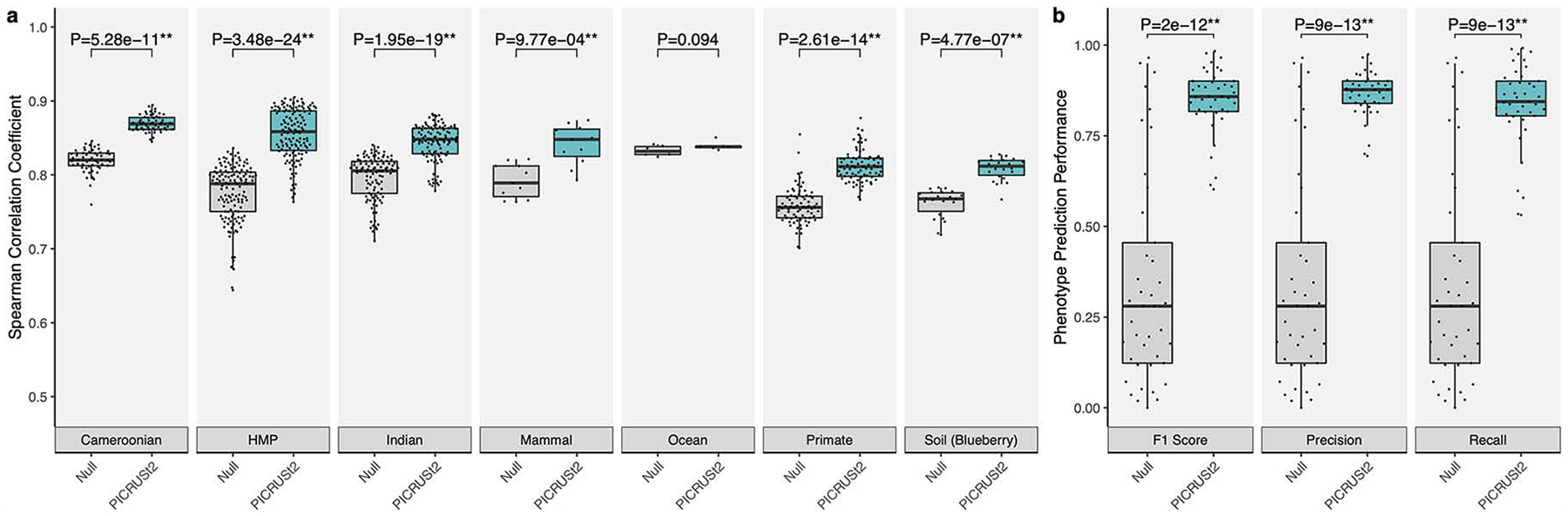
(a) Spearman correlation coefficients between PICRUSt2 predicted pathway abundances and gold-standard metagenomic sequencing (MGS). Results are shown for each validation dataset: stool from Cameroonian individuals, The Human Microbiome Project (HMP), stool from Indian individuals, mammalian stool, ocean water, non-human primate stool, and blueberry soil. These results are limited to the 575 pathways that could potentially be identified by PICRUSt2 and HUMAnN2. (b) Performance of binary phenotype predictions based on three metrics: F1 score, precision, and recall. Each point corresponds to one of the 41 phenotypes tested. Predictions assessed here are based on holding out each genome individually, predicting the phenotypes for that holdout genome, and comparing the predicted and observed values. The null distribution in this case is based on randomizing the phenotypes across the reference genomes and comparing to the actual values, which results in the same output for all three metrics. The P-values of paired-sample, two-tailed Wilcoxon tests is indicated above each tested grouping (* and ** correspond to P < 0.05 and P < 0.001, respectively). Note that in panel a the y-axis is truncated below 0.5 rather than 0 to better visualize small differences between categories. The sample sizes in panel a are 57 (Cameroonian), 137 (HMP), 91 (Indian), 8 (mammal), 6 (ocean), 77 (primate), and 22 (soil).
Predictions for 41 microbial phenotypes, which are linked to IMG genomes15, can also now be generated with PICRUSt2. These represent high-level microbial metabolic activities such as “Glucose utilizing” and “Denitrifier” that are annotated as present or absent within each reference genome. We performed a hold-out validation to assess the performance of PICRUSt2 phenotype predictions, which involved comparing the binary phenotype predictions to the expected phenotypes for each reference genome. Based on F1 score (mean=84.8%; sd=9.01%), precision (mean=86.5%; sd=6.21%), and recall (mean=83.5%; sd=11.4%), these predictions performed significantly better than the null expectation (Fig 3b; Wilcoxon tests P < 0.05).
There are two main criticisms of amplicon-based functional prediction. First, the predictions are biased towards existing reference genomes, which means that rare environment-specific functions are less likely to be identified. This limitation is reducing over time as the number of high-quality available genomes continues to grow. PICRUSt2 also allows user-specified genomes to be used for generating predictions, which provides a flexible framework for studying particular environments. The second criticism is that amplicon-based predictions cannot provide resolution to distinguish strain-specific functionality. This is an important limitation of PICRUSt2 and any amplicon-based analysis, which can only differentiate taxa to the degree they differ at the amplified marker gene sequence.
PICRUSt2 provides improved accuracy and flexibility for marker gene metagenome inference. We have highlighted these improvements while also describing limitations with identifying consistent differentially abundant functions in microbiome studies. We hope that the expanded functionality of PICRUSt2 will continue to enable the identification of insights into functional microbial ecology from amplicon sequencing profiles.
Supplementary Material
Acknowledgements
We would like to thank Zhenjiang Xu and Amy Chen for providing us access to datafiles used for testing and the default reference database. We would also like to thank Heather McIntosh for her help designing the pipeline flowchart. G.M.D. is funded by an NSERC CGS-D scholarship. VJM is funded by an NIH/NIAAA Ruth L. Kirschstein National Research Service Award (F30 AA026527). JRZ is supported by NSF IOS CAREER grant 1942647. SYN is funded by an NSERC Discovery Grant. CH is funded in part by NIH NIDDK grants U54DK102557 and R24DK110499. MGIL is funded by an NSERC Discovery Grant and an NSERC Collaborative Research Development with co-funding from GlaxoSmithKline to MGIL and JRB.
Footnotes
Code and data availability
PICRUSt2 is available at: https://github.com/picrust/picrust2. The Python and R code used for the analyses and database construction described in this paper are available online at https://github.com/gavinmdouglas/picrust2_manuscript. This repository also includes the processed datafiles that can be used to re-generate the figures and findings in this paper. The accessions for all sequencing data used in this study are listed in the supplementary information.
References
- 1.Langille MGI et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwai S et al. Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One 11, e0166104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jun SR, Robeson MS, Hauser LJ, Schadt CW & Gorin AA PanFP: Pangenome-based functional profiles for microbial communities. BMC Res. Notes 8, 479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aßhauer KP, Wemheuer B, Daniel R & Meinicke P Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wemheuer F et al. Tax4Fun2: a R-based tool for the rapid prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene marker gene sequences. bioRxiv (2018). doi: 10.1101/490037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeSantis TZ et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markowitz VM et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40(D1), D115–D122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbera P et al. EPA-ng: Massively Parallel Evolutionary Placement of Genetic Sequences. Syst. Biol 68, 365–369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czech L & Stamatakis A Scalable methods for analyzing and visualizing phylogenetic placement of metagenomic samples. PLoS One 14, e0217050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louca S & Doebeli M Efficient comparative phylogenetics on large trees. Bioinformatics 34, 1053–1055 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S, Sato Y, Furumichi M & Tanabe M KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40(D1), D109–D114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayan NR et al. Piphillin predicts metagenomic composition and dynamics from DADA2-corrected 16S rDNA sequences. BMC Genomics 21, 56 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspi R et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44(D1), D471–D480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen IMA et al. Improving Microbial Genome Annotations in an Integrated Database Context. PLoS One 8, e54859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


