Abstract
Disruption of physiological aging of the placenta can lead to pregnancy complications and increased risk for cardiometabolic diseases during childhood and adulthood. Maternal metabolic and genetic factors need to operate in concert with placental development for optimal pregnancy outcome. However, it is unknown whether maternal cardiometabolic status and genetic ancestry contribute to differences in placental epigenetic age acceleration (PAA). We investigated whether maternal pre-pregnancy obesity, gestational weight gain (GWG), blood pressure, and genetic ancestry influence PAA. Among 301 pregnant women from four race/ethnic groups who provided placenta samples at delivery as part of the NICHD Fetal Growth Studies, placental DNA methylation age was estimated using 62 CpGs known to predict placental aging. PAA was defined to be the difference between placental DNA methylation age and gestational age at birth. Percentage of genetic ancestries was estimated using genotype data. We found that a 1 kg/week increase in GWG was associated with up to 1.71 (95% CI: −3.11, −0.32) week lower PAA. Offspring Native American ancestry and African ancestry were associated, respectively, with higher and lower PAA among Hispanics, and maternal East Asian ancestry was associated with lower PAA among Asians (p<0.05). Among mothers with a male offspring, blood pressure was associated with lower PAA across all three trimesters (p<0.05), pre-pregnancy obesity compared to normal weight was associated with 1.24 (95% CI: −2.24, −0.25) week lower PAA. In summary, we observed that maternal cardiometabolic factors and genetic ancestry influence placental epigenetic aging and some of these influences may be male offspring-specific.
Keywords: obesity, gestational weight gain, blood pressure, ancestry, placental aging, epigenetic aging
Introduction
The placenta, a tissue with endocrine and substrate transport function during pregnancy undergoes a physiologic aging process. Disruption of the physiologic aging of the placenta can deter its hormonal and transport function and may lead to several adverse pregnancy outcomes, including preeclampsia and impaired fetal growth1–5 and later-life cardiometabolic complications.6,7 Investigation of maternal influences on placental development will be key to our understanding of placental markers of future cardiovascular illnesses. Pathological studies suggest that some placentas may show signs of accelerated aging than others.8, 9 Premature placental senescence, the most studied trigger of accelerated tissue aging induced by oxidative stress, DNA damage and epigenomic disruption,10–12 is associated with fetal growth restriction, preeclampsia, spontaneous preterm birth, and intrauterine fetal death.1–5
The epigenetic clock, estimated using genome-wide DNA methylation levels of CpGs, is demonstrated to be the most promising molecular estimator of biological age.13–17 The epigenetic clock method developed by Horvath identified 353 CpG sites from multiple tissues that predicted chronological age with high accuracy.12 Age acceleration of several tissues, the difference between epigenetic age and chronological age, predicted age-related phenotypes including cancer, cardiovascular diseases and mortality in adults.13, 18, 19 Accelerated epigenetic age in neonatal cord blood was associated with lower offspring birthweight and Apgar score.20–22 In a recent study, an epigenetic tool has been developed for predicting DNA methylation-based placental age using 62 CpGs, and placental age acceleration (PAA) is associated with early onset of preeclampsia.23 DNA methylation-based prediction of fetal developmental maturity using gestational age has the potential to improve epidemiological and clinical research.24
Understanding the factors that influence DNA methylation-based placental aging may provide intervention opportunities to improve pregnancy outcomes by establishing the clinical predictors of placental aging. Maternal obesity and offspring sex influenced DNA methylation of the placenta, where methylation percentages were highest in placentas from male offspring born to obese mothers compared with placentas from female offspring born to lean mothers.25 Race/ethnicity and sex are associated with DNA methylation age acceleration in blood26. Variations in genetic ancestry influence DNA methylation27 and gene expression,28 underscoring the importance of considering genetic ancestry in epigenetic aging studies among populations from diverse race/ethnic groups.28,29 Histopathological study also suggests male fetuses have higher placental production of endotoxin-induced tumor necrosis factor-response than female fetuses.30 Epigenetic clock studies in neonatal cord blood have shown that maternal age, preeclampsia, gestational diabetes mellitus (GDM), neonatal sex, neonatal birth size and maternal smoking are associated with epigenetic age acceleration.20, 22, 31 However, to what extent maternal cardiometabolic factors and genetic ancestry relate to molecular markers of placental aging is unknown. We investigated the overall and offspring sex-specific associations of maternal cardiometabolic traits (pre-pregnancy obesity, gestational weight gain (GWG), and blood pressure) and ancestry (self-identified race/ethnicity and percentage of genetic ancestry per race/ethnic group) with PAA.
Methods
Study setting and study population
The current study included 312 women who provided placenta samples at delivery as part of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Fetal Growth Studies - Singletons. Women without major pre-existing medical conditions from each of four self-identified race/ethnic groups (i.e, non-Hispanic White [White], non-Hispanic Black [Black], Hispanic, Asian/Pacific Islander [Asian]) were recruited from 12 participating clinical sites in the United States from 2009 to 2013.32 Gestational age was determined using the date of the last menstrual period and confirmed by ultrasound between 8 weeks to 13 weeks and 6 days of gestation.33 The study was approved by institutional review boards at NICHD and each of the participating clinical sites.
Placental DNA extraction, measurement and quality control of DNA methylation
Placental samples were obtained within one hour of delivery. Placental parenchymal biopsies measuring 0.5 cm × 0.5 cm × 0.5 cm were taken from the fetal side of the trophoblast, directly below the fetal surface of the placenta. Samples were placed in RNALater and frozen for molecular analysis. Genomic DNA were extracted from the placental biopsies and 500 ng was used to profile methylation data using HumanMethylation450 BeadChip as per the manufacturer’s protocols (Illumina Inc., San Diego, CA) as previously described.34 DNA methylation data were processed using Genome Studio, which calculates the fractional methylation (average beta values calculated by taking the ratio of two fluorescent signals [methylated and unmethylated signals]) at each queried CpG, after background correction, normalization to internal control probes, and quantile normalization. Normalization was performed using the Beta MIxture Quantile dilation (BMIQ) method described by Teschendorf et al.,35 and modified by Horvath.12 The method corrected for the probe design bias in the Illumina Infinium HumanMethylation450 BeadChip and achieved between-sample normalization.12, 35 After BMIQ normalization, missing CpGs were imputed by the k-nearest neighbors method setting k=10 using the impute.knn function in R as part of the impute library.36 We excluded a total of 11 samples, which included four samples showing discrepancies between phenotypic sex and genotypic sex, six samples that were outliers from the distribution of the samples’ genetic clusters based on multi-dimensional scaling plots, and one sample with a mismatching sample identifier. In addition, we removed CpG probes with mean detection p value ≥0.05 (n=36), cross-reactive (n=24,491), non-autosomal (n=14,589) and CpG sites located within 20 base pair from known SNPs (n=37,360). After quality control, 301 mothers with placenta sample (with n= 409,101 CpGs) were included in subsequent analyses. Our samples had high probability of prediction for placental tissue when tested using Horvath’s method12 implemented in glmnet R function.37
Placental DNA methylation age prediction
We predicted placental DNA methylation age using 62 CpG sites that have previously been found to predict placental epigenetic age with high accuracy.23 The data can be directly accessed from the supplementary material of Mayne et al. study.23 DNA methylation age of each of our study samples was determined by conducting a penalized regression model, known as elastic net regression, by regressing the gestational age using mean beta-values of the 62 CpG sites in glmnet R function.12, 37 We estimated the correlation between gestational age and DNA methylation age of the placenta samples in our cohort. Participants in our study, on average, had a term delivery (mean gestational age=39 weeks), and none had first and second trimester gestational age. Therefore, to assess the correlations using samples with a wider range of gestational ages, we assembled publicly available placental tissue DNA methylation data from NCBI Gene Expression Omnibus (GEO; accession numbers: GSE73375, GSE36829, GSE36829, GSE59274, GSE44667, and GSE74738). We estimated DNA methylation age of the GEO placental samples and additionally assessed the correlation between gestational age and DNA methylation age of a pooled dataset consisting the GEO samples and our samples. The GSE73375, GSE44667 and GSE74738 data were similarly assayed and normalized as in our data. The GSE36829, GSE36829, and GSE59274 data were assayed using Illumina HumanMethylation27 BeadChip. We observed similar positive correlations between gestational age and DNA methylation age by each GEO data when we evaluated the potential batch and assay differences introduced by each dataset in our assessment of the correlation (data not shown). PAA was defined to be the difference between placental DNA methylation age and gestational age at birth.
Genetic ancestry estimation
Placental DNA samples were genotyped on Illumina HumanOmni2.5 Beadchip, followed by initial data processing using Illumina’s Genome Studio. Mothers’ DNA samples extracted from stored buffy coat specimens were genotyped using the Illumina Infinium Multiethnic Global BeadChip microarray. Single nucleotide polymorphism (SNP) quality control included removing SNPs with minor allele frequency <0.05, failing Hardy-Weinberg Equilibrium test (P-value>10e-3) and those with <5% missing data. A total of 41,115 LD-pruned common SNPs were taken forward for admixture analysis. The genetic ancestry proportions of the mothers and their offspring were estimated based on previous data on composition of continental genetic ancestries of the U.S. population.29 Reference samples for European, African, East Asian, and Native American ancestries were obtained by combining genotype data on samples from the 1000 Genomes project,38 and Human Genome Diversity Project.39 In order to correct for population stratification, the percentage of genetic ancestries were estimated separately for mothers and their offspring using unsupervised clustering analysis implemented in ADMIXTURE version 1.3.40 ADMIXTURE estimated percent European ancestry in Whites, African ancestry in Blacks, African, European and Native American ancestries in Hispanics, and East Asian ancestry in Asians were subsequently used as predictors of PAA.
Maternal and offspring cardiometabolic factors
Maternal age and weight at antenatal clinical visits were abstracted from the prenatal records. Maternal age was defined by groups who were <30, between 30–35 and ≥35 to evaluate the associations particularly by advanced maternal age status, rather than a 1-year increment in age. Recalled pre-pregnancy weight and measured height were used to calculate pre-pregnancy BMI in kg/m2. Pre-pregnancy BMI was defined as a continuous variable in kilo grams per meter-squared and as a categorical variable (normal weight: <25 kg/m2; overweight: 25–30 kg/m2; and obese: >30 kg/m2). GWG was defined as rate of weight gain (kg/week) in each trimester of pregnancy, 1st (13 weeks and 6 days), 2nd (27 weeks and 6 days), and 3rd (40 weeks) as previously described.41 Systolic blood pressure (SBP) and diastolic blood pressure (DBP) repeated measurements in millimeters of mercury (mmHg) were abstracted from prenatal records. Each repeated measurement was averaged to get the blood pressure measurements by trimester. Maternal age was additionally categorized as age<30 years, 30–35 years and ≥35 years.
Statistical analyses
The Pearson correlation coefficient was used to test for correlations between PAA and maternal and fetal predictors evaluated as continuous variables. Multivariable-adjusted linear regression models were fitted incorporating PAA as the response variable, determinants of PAA as the predictor variables (maternal age, early pregnancy blood pressure, pre-pregnancy BMI, genetic ancestry, and fetal sex), and potential confounders (parity, health insurance, mode of onset of labor, marital status, educational status, preeclampsia status, and offspring sex). Offspring sex-stratified models were also fitted using linear regression models. We considered each of the tests we conducted independent tests based on a priori observations of the associations between each of the predictor variables and tissue epigenetic aging.20, 22, 31, 42, 43 In non-hypertensive pregnant women, blood pressure, most notably DBP, falls steadily until the middle of gestation and then rises again until delivery.44 In addition, longitudinal changes in GWG and its association with intrauterine fetal growth was shown in a previous study.41 Thus, we used P-value<0.05 as evidence for statistical significance. Analyses were conducted using SAS 9.4 (SAS Institute, Cary NC).
Results
Characteristics of study participants and placental DNA methylation age calibration
Overall, the mean (s.d.) of maternal age and gestational age at birth were 27.5 (5.3) years and 39.5 (1.1) weeks, respectively. Characteristics of the study participants are shown in Table 1 and Supplementary Table 1. Approximately, 64% of mothers were normal weight, 26% were overweight, and 11% were obese. The mean (s.d.) of rates of GWG were 0.22 (0.18), 0.30 (0.16), and 0.37 (0.16) kg/week in the first, second and third trimester, respectively. The mean (s.d.) of systolic and diastolic blood pressure at each trimester were 109.3 (10.7), 108.7 (9.0) and 111.1 (9.0), and 66.6 (7.5), 64.7 (6.0), 67.1 (6.3) mmHG, respectively. The mean fetal ancestry proportions within the self-reported race/ethnicity groups were 78% East Asian ancestry among Asians, 94% European ancestry among Whites, 79% African ancestry among Blacks, and 48% European, 19% African, and 30% Native-American ancestries among Hispanics, respectively. The corresponding mean maternal ancestry proportions were comparable with the fetal ones. The gestational age at delivery ranged from 36–41 weeks, and its correlation with the estimated DNA methylation age was r=0.19 (p-value=0.001). However, the correlation between gestational age and estimated DNA methylation age improved (r=0.76; p-value<0.001) after combining our data with the NCBI Gene Expression Omnibus, (GEO) data samples that had wider range of gestational ages (Supplementary Fig 1). Mean (s.d.) placental DNA methylation age and PAA were 35.7 (1.5) and −3.8 (1.7) weeks, respectively. The maternal and offspring characteristics were similar between male and female offspring.
Table 1.
Maternal and fetal characteristics of the study population
| Mean ± SD or n (%) | |||
|---|---|---|---|
| Characteristics of Study Participants | Female offspring (n=149) | Male offspring (n=152) | Combined (n=301) |
| Maternal age, y | 27.7 ± 5.4 | 27.7 ± 5.2 | 27.7 ± 5.3 |
| Maternal age, n (%) | |||
| <30 years | 89 (59.7) | 93 (61.2) | 182 (60.5) |
| 30–35 years | 44 (29.5) | 45 (29.6) | 89 (29.6) |
| ≥35 years | 16 (10.7) | 14 (9.2) | 30 (10.0) |
| Gestational age at delivery, wk | 39.6 ± 1.1 | 39.4 ± 1.2 | 39.5 ± 1.1 |
| Self-reported race/ethnicity, n (%) | |||
| White | 38 (25.5) | 39 (25.7) | 77 (25.6) |
| Black | 39 (26.2) | 33 (21.7) | 72 (23.9) |
| Hispanic | 53 (35.6) | 49 (32.2) | 102 (33.9) |
| Asian/Pacific Islander | 19 (12.8) | 31 (20.4) | 50 (16.6) |
| Pre-pregnancy body mass index in kg/m2, n (%) | |||
| <25.0 | 92 (63.0) | 96 (64.4) | 188 (63.7) |
| 25.0–29.9 | 41 (28.1) | 35 (23.5) | 76 (25.8) |
| ≥30.0 | 13 (8.9) | 18 (12.1) | 31 (10.5) |
| Trimester specific rate of GWG, kg/wk | |||
| 1st Trimester | 0.21 ± 0.17 | 0.23 ± 0.19 | 0.22 ± 0.18 |
| 2nd Trimester | 0.29 ± 0.17 | 0.30 ± 0.16 | 0.30 ± 0.16 |
| 3rd Trimester | 0.37 ± 0.16 | 0.37 ± 0.15 | 0.37 ± 0.16 |
| Trimester specific SBP, mmHG | - | - | - |
| 1st Trimester | 108.2 ± 10.4 | 110.4 ± 10.9 | 109.3 ± 10.7 |
| 2nd Trimester | 108.1 ± 9.7 | 109.3 ± 8.3 | 108.7 ± 9.0 |
| 3rd Trimester | 110.9 ± 9.3 | 111.3 ± 8.7 | 111.1 ± 9.0 |
| Trimester specific DBP, mmHG | - | - | - |
| 1st Trimester | 67.5 ± 7.4 | 67.5 ± 7.4 | 66.6 ± 7.5 |
| 2nd Trimester | 64.7 ± 5.8 | 64.7 ± 5.8 | 64.7 ± 6.0 |
| 3rd Trimester | 67.6 ± 6.1 | 67.6 ± 6.1 | 67.1 ± 6.3 |
Abbreviation: GWG, gestational weight gain; SBP, Systolic blood pressure; DBP, Diastolic blood pressure.
Associations of maternal blood pressure and adiposity during pregnancy with placental epigenetic age acceleration
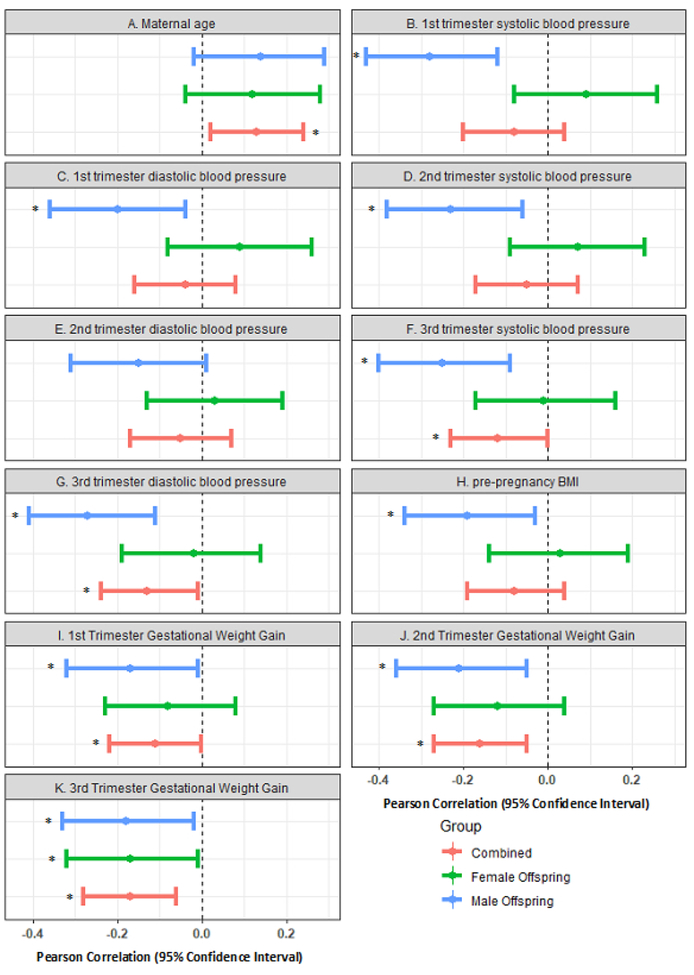
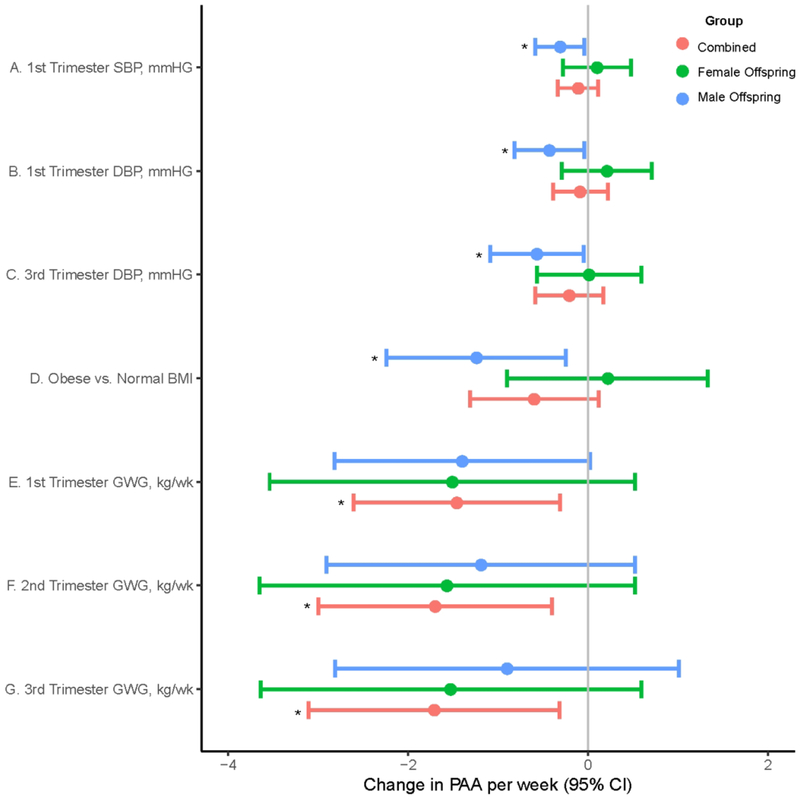
PAA had significant positive correlation with maternal age (r=0.13 in overall and r=0.12 in mothers with a female offspring), but significant inverse correlation with maternal pre-pregnancy BMI (r=−0.19 among those with a male offspring) and maternal blood pressure (range r=−0.20 to −0.28 among those with a male offspring) (Fig 1, Supplementary Table 2). In multivariable-adjusted models, each 1 kg/week increase in GWG in the first, second and third trimester were significantly associated with 1.46 (95%CI: −2.61, −0.31), 1.70 (−3.00, −0.40), and 1.71 (−3.11, −0.32) week lower PAA, respectively. Among mothers with a male offspring, pre-pregnancy obesity compared to pre-pregnancy normal weight was significantly associated with 1.24 (−2.24, −0.25) week lower PAA. Among mothers with a male offspring, each 10 mmHg increase in first trimester systolic blood pressure, first trimester and third trimester diastolic blood pressure was significantly associated with 0.31 (95%CI: −0.59, −0.04), 0.43 (−0.82, −0.04), and 0.57 (−1.09, −0.05) week lower PAA, respectively (Fig 2, Supplementary Table 3).
Fig 1.
Correlations between maternal cardiometabolic factors and placental epigenetic age acceleration. A. Maternal age, B. 1st trimester SBP, C. 1st trimester DBP, D. 2nd trimester SBP, E. 2nd trimester DBP, F. 3rd trimester SBP, G. 3rd trimester DBP, H. pre-pregnancy BMI, I. 1st trimester GWG, J. 2nd trimester GWG, and K. 3rd trimester GWG. Statistically significant correlations (P<0.05) are annotated by Asterix.
Fig 2.
Multivariable adjusted significant associations of maternal blood pressure and adiposity with placental epigenetic age acceleration. A. 1st trimester SBP, B. 1st trimester DBP, and C. 3rd trimester DBP) (D. pre-pregnancy BMI, E. 1st trimester GWG, F. 2nd trimester GWG, and G. 3rd trimester GWG). Models included parity, health insurance, mode of onset of labor, marital status, educational status, preeclampsia status, and offspring sex as adjustment variables. Estimates for maternal blood pressure were change in PAA per 10mmHG increase in DBP and SBP. Statistically significant estimates (P<0.05) are annotated by Asterix. Abbreviation: GWG, gestational weight gain; SBP, Systolic blood pressure; DBP, Diastolic blood pressure; PAA, placental epigenetic age acceleration.
Associations of genetic ancestry with placental epigenetic age acceleration
Among Hispanics, each 10 percent increase in maternal and offspring Native-American ancestry was associated with 0.20 (95%CI: 0.02, 0.40) and 0.30 (95%CI: 0.20, 0.50) week higher PAA, respectively; each 10 percent increase in offspring African ancestry was associated with 0.40 (−0.60, −0.20) week lower PAA, respectively. Among Asians, each 10 percent increase in maternal Asian ancestry was associated with 0.20 (95%CI: −0.40, −0.04) week lower PAA (Table 2).
Table 2.
Change in placental epigenetic age acceleration per 10% increase in maternal and offspring genetic ancestry component
| Female offspring | Male offspring | Combined | ||||
|---|---|---|---|---|---|---|
| Self-reported race/ethnicity and genetic ancestry | Mean PAA change (95% CI) in weeks1 | P-value | Mean PAA change (95% CI) in weeks1 | P-value | Mean PAA change (95% CI) in weeks1 | P-value |
| Maternal genetic ancestry2 | ||||||
| White | ||||||
| European | 0.30 (−0.10, 0.70) | 0.17 | 0.00 (−0.80, 0.80) | 0.98 | 0.20 (−0.20, 0.60) | 0.33 |
| Blacks | ||||||
| African | −0.10 (−0.60, 0.40) | 0.65 | −0.10 (−0.10, 0.20) | 0.68 | −0.10 (−0.40, 0.20) | 0.67 |
| Hispanic | ||||||
| European | −0.10 (−0.50, 0.30) | 0.54 | −0.10 (−0.40, 0.20) | 0.63 | −0.10 (−0.30, 0.10) | 0.38 |
| African | −0.30 (−0.70, 0.03) | 0.07 | −0.10 (−0.40, 0.20) | 0.39 | −0.20 (−0.50, 0.00) | 0.05 |
| Native American | 0.30 (0.00, 0.50) | 0.06 | 0.10 (−0.10, 0.30) | 0.34 | 0.20 (0.02, 0.40) | 0.03 |
| Asian | ||||||
| East Asian | −0.30 (−0.80, 0.30) | 0.27 | −0.10 (−0.40, 0.10) | 0.29 | −0.20 (−0.40, −0.04) | 0.02 |
| Offspring genetic ancestry2 | ||||||
| White | ||||||
| European | 0.10 (−0.30, 0.50) | 0.60 | −0.10 (−0.70, 0.40) | 0.59 | 0.10 (−0.20, 0.40) | 0.42 |
| Blacks | ||||||
| African | 0.20 (−0.30, 0.70) | 0.35 | −0.10 (−0.30, 0.20) | 0.47 | 0.05 (−0.20, 0.30) | 0.72 |
| Hispanic | ||||||
| European | −0.20 (−0.60, 0.10) | 0.24 | −0.10 (−0.40, 0.20) | 0.57 | −0.20 (−0.40, 0.10) | 0.15 |
| African | −0.40 (−0.80, −0.10) | 0.01 | −0.20 (−0.50, 0.20) | 0.36 | −0.40 (−0.60, −0.20) | 0.001 |
| Native American | 0.50 (0.20, 0.70) | <0.001 | 0.10 (−0.10, 0.30) | 0.38 | 0.30 (0.20, 0.50) | <0.001 |
| Asian | ||||||
| East Asian | −0.08 (−0.60, 0.40) | 0.73 | −0.10 (−0.30, 0.20) | 0.56 | −0.10 (−0.30, 0.10) | 0.20 |
Models adjusted for maternal age, pre-pregnancy BMI, race/ethnicity, parity, health insurance, mode of onset of labor, marital status, educational status, and preeclampsia status; variables were taken out of the model when entered as a predictor; offspring sex was taken out of the model for stratified analyses; Statistically significant estimates are highlighted in bold
per 10% increase in percent ancestry
Abbreviation: PAA, placental epigenetic age acceleration
Discussion
In this first study of determinants of PAA, we found that maternal cardiometabolic factors and genetic ancestry are associated with epigenetic aging of the placenta. We also found that some of these associations were offspring sex-specific. Specifically, maternal weight gain during pregnancy was associated with lower PAA. Maternal blood pressure during pregnancy, and pre-pregnancy obesity were associated with lower PAA among mothers with a male offspring. Offspring Native American ancestry and African ancestry percent were associated, respectively, with higher and lower PAA among Hispanics, and maternal East Asian ancestry percent was associated with lower PAA among Asians.
Our findings for associations between higher maternal adiposity (including BMI and weight gain during pregnancy) and lower PAA are supported by findings of histological studies. Using placentas from a group of mothers, a study demonstrated that higher maternal BMI is associated with increased placental nitrative stress but showed no signs of increase in oxidative stress,42 a marker of cellular aging.10, 11 This shift in balance between nitrative stress and oxidative stress may be a potential adaptive mechanism by the placenta to reduce the harmful effects of obesity-induced free radicals.42 Another study carried out in mice using quantitative PCR analysis of oxidative stress primers such as Gpx1, Sod1 and Sod2 and showed that there was no evidence of oxidative stress in obese placenta or embryo.45 A pathological analyses of singleton births demonstrated that the placenta was less mature in obese mothers compared with normal weight mothers.46 Similar to ours, the study found evidence for reduced placental maturity, including maternal origin vascular and villus lesions among obese mothers without obstetrical complications (e.g. chronic hypertension, GDM and preterm delivery). However, in GDM placentas, another study observed reduced apoptosis, which may contribute to increased placental tissue.47 It is possible that maternal obesity can exert adverse in-utero influence on placental pathology46 and explain the observed inverse associations with PAA in our study. The placenta’s unique ability to trigger an adaptive response if the fetus is not developing well due to hypoxia and oxidative stress was discussed previously.45, 48 Even in normal pregnancy, the placenta responds to abnormal maternal nutrient status and hypoxia by altering transporter expression, activity or epigenetic regulation of placental gene expression to maintain fetal growth.48
We also found that increase in systolic and diastolic blood pressures in the first and third trimesters were associated with lower PAA among mothers with a male offspring. Using the Grannum classification of four distinct grades of placental maturity,49 a study demonstrated that preeclampsia is associated with maturation of the placenta.43 Immune genes were found to be expressed at higher level in placentas from mothers with a female offspring compared with those with a male offspring.50 Gene expression of the placenta was shown to respond to maternal inflammatory status in an offspring sex-dependent manner.51 A male fetus is more at risk to poor outcomes such as placental insufficiency,52 which may be a result of observed higher placental TLR4 expression and a greater production of TNFα in response to lipopolysaccharide in males.51 A recent study showed higher levels of pro-inflammatory cytokines and cell death (via expressions of hypoxia and apoptotic molecules) in placentas of preeclamptic mothers with a male offspring compared with those with a female offspring.53 We have also recently showed that low HDL cholesterol in early pregnancy was associated with accelerated epigenetic ageing of the placenta in a sex-specific manner.54
Our study participants were largely composed of mothers without major obstetrical complications (e.g. n=8 preeclamptics), which may partly explain the observed negative associations between PAA and blood pressure. Early onset of preeclampsia was observed to be associated with higher PAA.23 However, we did not have enough power to detect an association between preeclampsia and PAA in our study; only one mother had high blood pressure during pregnancy and none were taking hypertensive medications. Maternal intrauterine environment could support fetal growth when blood pressure increases in the third trimester through an adaptive mechanism, as both high and low diastolic blood pressures during pregnancy are associated with small babies and high perinatal mortality.55 Previous studies have shown low blood pressure in pregnancy is associated with lower birth weights and increase in preterm deliveries.56 Third trimester concentrations of placental growth factor (P1GF) is decreased as a result of lower maternal serum concentrations of pregnancy associated plasma protein (PAPP)-A.57, 58 Lower PAPP-A was observed in pregnancies with higher placental weight, implicating reduced endocrine function.58 Future studies are needed to understand whether variations in adverse pregnancy outcomes known to be associated with obesity or higher blood pressure are mediated through accelerated aging of the placenta.
We further demonstrated the associations between genetic ancestry and PAA using estimated global ancestry among the four racial/ethnic groups. Present-day human populations have mixed genetic ancestries that vary even among people from the same self-reported race/ethnicity.59 Self-reported race/ethnicity has been found to be associated with methylation changes driven by both genes and environment,60 strengthening the potential utility of genetic ancestry as a modifying factor in epigenetic association studies in admixed populations.27, 28 The relatively stronger influence of fetal genetic ancestry than maternal genetic ancestry observed in our study is indicative of direct genetic influence on PAA. Based on this observation and given that heritability of PAA has been previously estimated to be 57.2%,23 future studies are needed to unravel the molecular genetic basis of PAA.
Our study included diverse study participants with placenta samples that are representative of the U.S. racial/ethnic groups. PAA in our study was estimated based on previously identified 62 placental CpGs that have higher accuracy in predicting epigenetic age of the placenta.23 We acknowledge that the small sample size limited the study’s power to test interactions of offspring sex and maternal factors on PAA. Absence of reference for placental cell type composition may result residual confounding by unaccounted cell type in our estimation of placental epigenetic aging.
In summary, we found that maternal adiposity, blood pressure and genetic ancestry are associated with molecular markers of placental aging. These associations could be male offspring-specific. Investigation of maternal influences on placental physiological development will be key to our understanding of placental markers of pregnancy complications and future cardiovascular illnesses. Together with future biomedical studies, these findings open intervention opportunities to improve pregnancy outcomes by establishing the pathophysiology of placental aging.
Supplementary Material
Supplementary Table 1. Additional maternal and offspring characteristics of the study population.
Supplementary Table 2. Correlation estimates between maternal/offspring factors and placental epigenetic age acceleration.
Supplementary Table 3. Change in placental epigenetic age acceleration with each unit increase in a maternal cardiometabolic factor
Supplementary Fig 1. Correlations between estimated placental DNA methylation age and gestational age at birth.
Acknowledgements:
We acknowledge the study participants of the NICHD Fetal Growth Studies. We thank research teams at all participating clinical centers (which include Christina Care Health Systems, Columbia University, Fountain Valley Hospital, California, Long Beach Memorial Medical Center, New York Hospital, Queens, Northwestern University, University of Alabama at Birmingham, University of California, Irvine, Medical University of South Carolina, Saint Peters University Hospital, Tufts University, and Women and Infants Hospital of Rhode Island). The authors also acknowledge the Wadsworth Center, C-TASC and The EMMES Corporations in providing data and imaging support. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Financial disclosure: This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health including American Recovery and Reinvestment Act funding via contract numbers HHSN275200800013C; HHSN275200800002I; HHSN27500006; HHSN275200800003IC; HHSN275200800014C; HHSN275200800012C; HHSN275200800028C; HHSN275201000009C and HHSN27500008. Additional support was obtained from the NIH Office of the Director, the National Institute on Minority Health and Health Disparities and the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Conflict of Interest: None
Ethical standards: The authors assert all procedures contributing to this work comply with the ethical standards of the NIH on human subject research and has been approved by the institutional review boards at NICHD and each of the participating clinical sites.
References
- 1.Chen K, Chen L, Lee Y. Exploring the relationship between preterm placental calcification and adverse maternal and fetal outcome. Ultrasound in Obstetrics & Gynecology. 2011;37(3), 328–334. [DOI] [PubMed] [Google Scholar]
- 2.Chen K-H, Chen L-R, Lee Y-H. The role of preterm placental calcification in high-risk pregnancy as a predictor of poor uteroplacental blood flow and adverse pregnancy outcome. Ultrasound in medicine &biology. 2012;38(6), 1011–1018. [DOI] [PubMed] [Google Scholar]
- 3.Biron-Shental T, Sukenik-Halevy R, Sharon Y, et al. Short telomeres may play a role in placental dysfunction in preeclampsia and intrauterine growth restriction. American journal of obstetrics and gynecology. 2010;202(4), 381. e381–381. e387. [DOI] [PubMed] [Google Scholar]
- 4.Maiti K, Sultana Z, Aitken RJ, et al. Evidence that fetal death is associated with placental aging. American journal of obstetrics and gynecology. 2017;217(4), 441. e441–441. e414. [DOI] [PubMed] [Google Scholar]
- 5.Sultana Z, Maiti K, Aitken J, Morris J, Dedman L, Smith R. Oxidative stress, placental ageing-related pathologies and adverse pregnancy outcomes. American Journal of Reproductive Immunology. 2017;77(5), e12653. [DOI] [PubMed] [Google Scholar]
- 6.Thornburg K, O’tierney P, Louey S. The placenta is a programming agent for cardiovascular disease. Placenta. 2010;31, S54–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arabin B, Baschat AA. Pregnancy: An underutilized window of opportunity to improve long-term maternal and infant health—An appeal for continuous family care and interdisciplinary communication. Frontiers in pediatrics. 2017;5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sultana Z, Maiti K, Dedman L, Smith R. Is there a role for placental senescence in the genesis of obstetric complications and fetal growth restriction? American journal of obstetrics and gynecology. 2017. [DOI] [PubMed] [Google Scholar]
- 9.Polettini J, Dutta E, Behnia F, Saade G, Torloni M, Menon R. Aging of intrauterine tissues in spontaneous preterm birth and preterm premature rupture of the membranes: A systematic review of the literature. Placenta. 2015;36(9), 969–973. [DOI] [PubMed] [Google Scholar]
- 10.Rodier F, Campisi J. Four faces of cellular senescence. The Journal of cell biology. 2011, jcb. 201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501), 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14(10), 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics. 2018, 1. [DOI] [PubMed] [Google Scholar]
- 14.Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017;21, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell JT, Tsai P-C, Yang T-P, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS genetics. 2012;8(4), e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS genetics. 2009;5(8), e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd-Kirkup JD, Green CD, Wu G, Wang D, Han J- DJ. Epigenomics and the regulation of aging. Epigenomics. 2013;5(2), 205–227. [DOI] [PubMed] [Google Scholar]
- 18.Perna L, Zhang Y, Mons U, Holleczek B, Saum K-U, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clinical epigenetics. 2016;8(1), 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng SC, Widschwendter M, Teschendorff AE. Epigenetic drift, epigenetic clocks and cancer risk. Epigenomics. 2016;8(5), 705–719. [DOI] [PubMed] [Google Scholar]
- 20.Girchenko P, Lahti J, Czamara D, et al. Associations between maternal risk factors of adverse pregnancy and birth outcomes and the offspring epigenetic clock of gestational age at birth. Clinical epigenetics. 2017;9(1), 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohlin J, Håberg SE, Magnus P, et al. Prediction of gestational age based on genome-wide differentially methylated regions. Genome biology. 2016; 17(1), 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight AK, Craig JM, Theda C, et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome biology. 2016; 17(1), 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayne BT, Leemaqz SY, Smith AK, Breen J, Roberts CT, Bianco-Miotto T. Accelerated placental aging in early onset preeclampsia pregnancies identified by DNA methylation. Epigenomics. 2017;9(3), 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight AK, Conneely KN, Smith AK. Gestational age predicted by DNA methylation: potential clinical and research utility. 2017. Future Medicine. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Cartier J, Drake A, Reynolds R. DNA methylation differs between lean and obese placenta and is influenced by maternal environment and fetal sex. In Society for Endocrinology BES 2017. 2017. BioScientifica. [Google Scholar]
- 26.Horvath S, Gurven M, Levine ME, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome biology. 2016; 17(1), 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cappetta M, Berdasco M, Hochmann J, et al. Effect of genetic ancestry on leukocyte global DNA methylation in cancer patients. BMC cancer. 2015; 15(1), 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Dolan ME. Ancestry-related differences in gene expression: findings may enhance understanding of health disparities between populations. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of african americans, latinos, and european Americans across the United States. The American Journal of Human Genetics. 2015;96(1), 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeganegi M, Watson CS, Martins A, et al. Effect of Lactobacillus rhamnosus GR-1 supernatant and fetal sex on lipopolysaccharide-induced cytokine and prostaglandin-regulating enzymes in human placental trophoblast cells: implications for treatment of bacterial vaginosis and prevention of preterm labor. American journal of obstetrics and gynecology. 2009;200(5), 532. e531–532. e538. [DOI] [PubMed] [Google Scholar]
- 31.Simpkin AJ, Hemani G, Suderman M, et al. Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Human molecular genetics. 2015;25(1), 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grewal J, Grantz KL, Zhang C, et al. Cohort Profile: NICHD Fetal Growth Studies-Singletons and Twins. International journal of epidemiology. 2017;47(1), 25–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buck Louis GM, Grewal J, Albert PS, et al. Racial/ethnic standards for fetal growth: the NICHD Fetal Growth Studies. Am J Obstet Gynecol. 2015;213(4), 449 e441–449 e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delahaye F, Do C, Kong Y, et al. Genetic variants influence on the placenta regulatory landscape. PLoS genetics. 2018;14(11), e1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2012;29(2), 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6), 520–525. [DOI] [PubMed] [Google Scholar]
- 37.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of statistical software. 2010;33(1), 1. [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium GP. A global reference for human genetic variation. Nature. 2015;526(7571), 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. science. 2008;319(5866), 1100–1104. [DOI] [PubMed] [Google Scholar]
- 40.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome research. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinkle SN, Johns AM, Albert PS, Kim S, Grantz KL. Longitudinal changes in gestational weight gain and the association with intrauterine fetal growth. European Journal of Obstetrics &Gynecology and Reproductive Biology. 2015; 190, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts VH, Smith J, McLea SA, Heizer AB, Richardson JL, Myatt L. Effect of increasing maternal body mass index on oxidative and nitrative stress in the human placenta. Placenta. 2009;30(2), 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siveska E, Jasovic V. Fetal Growth and Body Proportion during Pre-Eclamptic Pregnancy. Obstet Gynecol Int J. 2015;2(3), 00038. [Google Scholar]
- 44.Hermida RC, Ayala DE, Iglesias M. Predictable blood pressure variability in healthy and complicated pregnancies. Hypertension. 2001;38(3), 736–741. [DOI] [PubMed] [Google Scholar]
- 45.Norwood KA. Maternal Obesity Alters Fetal Development Due to Impaired Placental Function and has Lasting Effects on Adult Offspring. 2013. [Google Scholar]
- 46.Huang L, Liu J, Feng L, Chen Y, Zhang J, Wang W. Maternal prepregnancy obesity is associated with higher risk of placental pathological lesions. Placenta. 2014;35(8), 563–569. [DOI] [PubMed] [Google Scholar]
- 47.Belkacemi L, Kjos S, Nelson D, Desai M, Ross M. Reduced apoptosis in term placentas from gestational diabetic pregnancies. Journal of developmental origins of health and disease. 2013;4(3), 256–265. [DOI] [PubMed] [Google Scholar]
- 48.Myatt L. Placental adaptive responses and fetal programming. The Journal of physiology. 2006;572(1), 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker MG, Hindmarsh PC, Geary M, Kingdom JC. Sonographic maturation of the placenta at 30 to 34 weeks is not associated with second trimester markers of placental insufficiency in low-risk pregnancies. Journal of Obstetrics and Gynaecology Canada. 2010;32(12), 1134–1139. [DOI] [PubMed] [Google Scholar]
- 50.Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proceedings of the National Academy of Sciences. 2006;103(14), 5478–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott NM, Hodyl NA, Murphy VE, et al. Placental cytokine expression covaries with maternal asthma severity and fetal sex. The Journal of Immunology. 2009;182(3), 1411–1420. [DOI] [PubMed] [Google Scholar]
- 52.Clifton V. Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31, S33–S39. [DOI] [PubMed] [Google Scholar]
- 53.Muralimanoharan S, Maloyan A, Myatt L. Evidence of sexual dimorphism in the placental function with severe preeclampsia. Placenta. 2013;34(12), 1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shrestha D, Workalemahu T, Tekola-Ayele F. Maternal dyslipidemia during early pregnancy and epigenetic aging of the placenta. Epigenetics. 2019(just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steer PJ, Little MP, Kold-Jensen T, Chapple J, Elliott P. Maternal blood pressure in pregnancy, birth weight, and perinatal mortality in first births: prospective study, bmj. 2004;329(7478), 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng PH, Walters WA. The effects of chronic maternal hypotension during pregnancy. Australian and New Zealand journal of obstetrics and gynaecology. 1992;32(1), 14–16. [DOI] [PubMed] [Google Scholar]
- 57.Melo NADB, Araujo Jûnior E, Helfer TM, et al. Assessment of maternal Doppler parameters of ophthalmic artery in fetuses with growth restriction in the third trimester of pregnancy: A case-control study. Journal of Obstetrics and Gynaecology Research. 2015;41(9), 1330–1336. [DOI] [PubMed] [Google Scholar]
- 58.Lean SC, Heazell AE, Dilworth MR, Mills TA, Jones RL. Placental dysfunction underlies increased risk of fetal growth restriction and stillbirth in advanced maternal age women. Scientific reports. 2017;7(1), 9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shriner D, Tekola-Ayele F, Adeyemo A, Rotimi CN. Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Scientific reports. 2014;4, 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galanter JM, Gignoux CR, Oh SS, et al. Methylation analysis reveals fundamental differences between ethnicity and genetic ancestry. BioRxiv. 2016, 036822. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Additional maternal and offspring characteristics of the study population.
Supplementary Table 2. Correlation estimates between maternal/offspring factors and placental epigenetic age acceleration.
Supplementary Table 3. Change in placental epigenetic age acceleration with each unit increase in a maternal cardiometabolic factor
Supplementary Fig 1. Correlations between estimated placental DNA methylation age and gestational age at birth.