Abstract
Resting state functional connectivity magnetic resonance imaging (rsfcMRI) has become a key component of investigations of neurocognitive and psychiatric behaviors. Over the past two decades, several methods and paradigms have been adopted to utilize and interpret data from resting-state fluctuations in the brain. These findings have increased our understanding of changes in many disease states. As the amount of resting state data available for research increases with big datasets and data-sharing projects, it is important to review the established traditional analysis methods and recognize areas where research methodology can be adapted to better accommodate the scale and complexity of rsfcMRI analysis. In this paper, we review established methods of analysis as well as areas that have been receiving increasing attention such as dynamic rsfcMRI, independent vector analysis, multiband rsfcMRI and network of networks.
Keywords: resting-state-fMRI, functional-connectivity, dynamic-connectivity, IVA, big data
Brain function in functional magnetic resonance imaging (fMRI) can utilize task-based paradigms, which require subjects to perform cognitive tasks, or resting state, in which subjects are instructed to let their minds wander in the absence of a task or stimulus. Resting state functional connectivity was first described in 1995, when Biswal et al. observed temporally correlated low-frequency signals (0.01–0.1 Hz) in spatially distinct regions of the brain in subjects at rest1. These signals were significant even after correcting for cardiac and respiratory noise, suggesting that the signals arose from spontaneous resting brain functions. Currently, resting state functional connectivity magnetic resonance imaging (rsfcMRI) is widely used to measure patterns of synchronous and spontaneous activation in the “task-negative” brain in healthy subjects and patients with different neurologic diseases2. This review provides a broad overview of conventional analytic methods used in rsfcMRI and discusses recent developments in this field together with perspectives on future research.
1. Resting State Networks
Functional connectivity has become a powerful tool for the definition of several resting state brain networks that are reliably detectable and consistently reproducible, both at individual and group levels when using a wide variety of analysis methods3–6. These include the primary sensorimotor network1, language networks7, visual networks8, the default mode network9, the salience network, the central executive network10, among others2,11,12. RsfcMRI has provided a new modality to examine these networks in healthy function and disease states including autism13,14, schizophrenia15–17, neurodegenerative diseases18,19, and brain tumor20. Figure 1 displays commonly identified resting state networks using independent component analyses.
Figure 1.
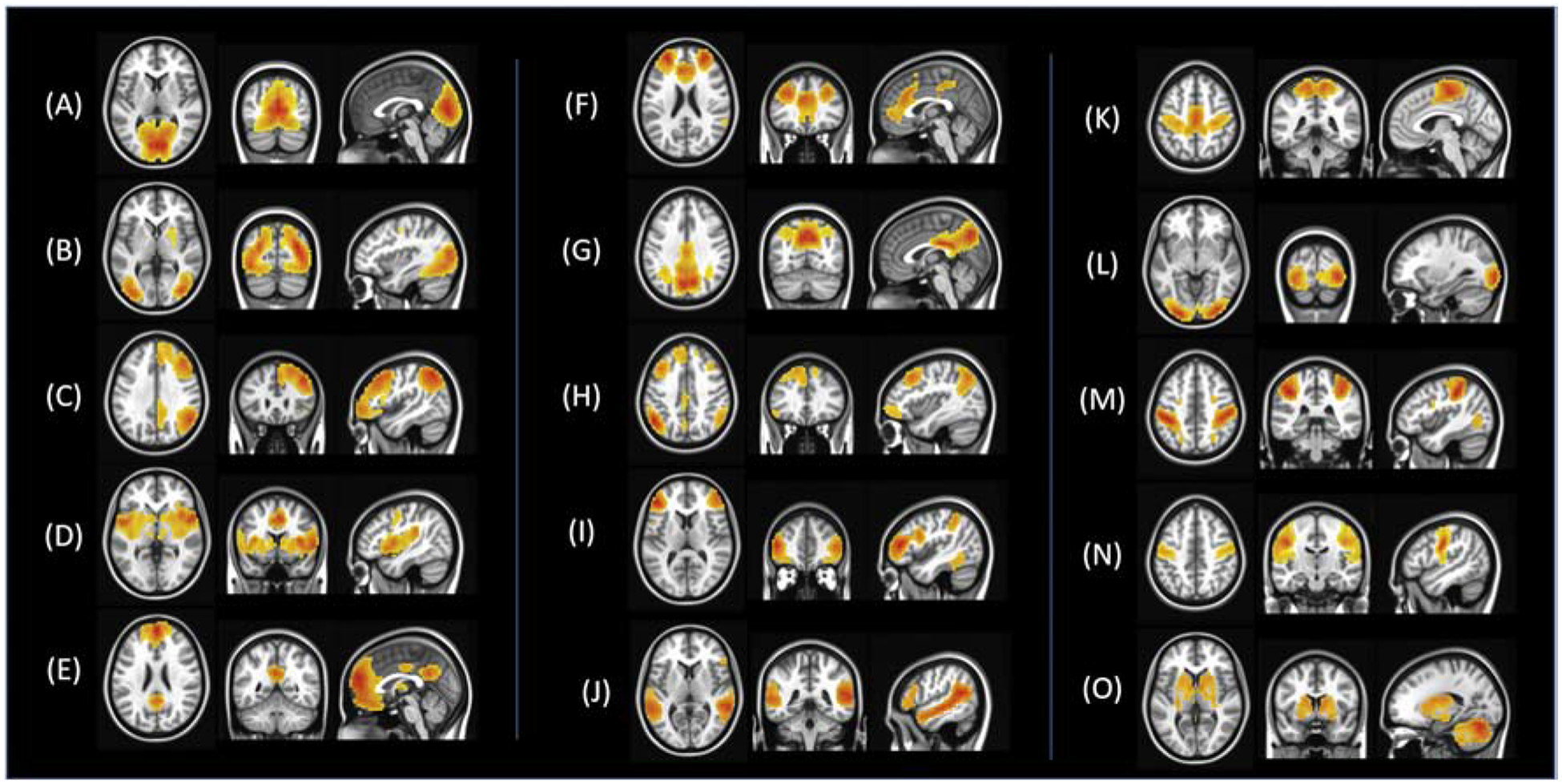
Resting state networks identified using a group-level independent component analysis (ICA) on a sample of 50 healthy participants. (A) Lingual gyrus (B) Higher visual network (C) Right central executive network (D) Bilateral insula network (E) Anterior default mode network (F) salience network (G) Posterior default mode network (H) Left central executive network (I) Bilateral middle frontal gyrus (J) Bilateral temporal gyrus network (K) Motor network (L) Visual network (M) Dorsal attention network (N) Bilateral precentral gyrus (O) Basal ganglia network
2. Conventional Methods of Analysis of rsfcMRI
Before analyzing the data, several preprocessing steps are generally performed including correction for section-dependent time shifts, regression of head motion and other nuisance regressors, spatial smoothing and band-pass filtering to retain frequencies between 0.01–0.1 Hz. Images are then either registered to individual subject structural space or registered to anatomic space to allow spatial concordance between subjects. Following preprocessing, several different approaches can be used to analyze resting state data, with some relying on a priori identification of regions of interest (ROI) and others that are data-driven and model-free as described in the sections below.
2.1. Frequency-Domain Analyses
The amplitude of low frequency fluctuations (ALFF) for a voxel’s time series is the total power in the low frequency range (0.01 – 0.1 Hz)21. Specifically, the time series for each voxel is transformed to the frequency domain and the power spectrum is obtained. The square root is calculated at each frequency of the power spectrum and the average square root is obtained across the 0.01–0.1 Hz frequency range for each voxel. This ALFF of each voxel is then divided by the individual global mean of ALFF with a brain mask. The resultant ALFF values are believed to reflect spontaneous regional neural activity21 but can be contaminated by non-neural physiologic fluctuations from respiration, cardiac activity and motion22. To improve on the original ALFF approach, a modified measure called fractional ALFF (fALFF) was introduced, examining the ratio of the power of each frequency at the low-frequency range to that of the entire frequency range23. Both ALFF and fALFF are used to study regional activation changes in sensorimotor tasks24, ADHD21, Alzheimer’s Disease25, OCD26, bipolar disorder27, schizophrenia28 and psychosis29. Although both measures are related, they are not entirely the same: the reliability of ALFF in gray matter regions is better than fALFF and it is more sensitive to differences between groups and individuals23,30. However, ALFF is more likely to be affected by noise from physiological sources5. Therefore, it is recommended that both measures be evaluated and reported in papers that examine these frequency domain parameters5.
2.2. Regional Homogeneity Analysis
Regional homogeneity (ReHo) is a voxel-based measure of brain activity that evaluates the synchronization between the time series of a given voxel and its nearest neighbors using Kendall’s coefficient of concordance31. ReHo requires no apriori definition of ROIs and has a high test-retest reliability32. ReHo is usually calculated within the frequency range between 0.01 to 0.1 Hz and can be subdivided into different frequency bands33. Moreover, although several studies demonstrated the frequency dependence of ReHo changes in different neurologic disorders34–36, the exact biologic meaning of ReHo within these different frequency bands remains elusive limiting its use in the research and clinical realm. However, like ALFF, ReHo methods are used to identify local neural activity of the brain and are sometimes implemented to define a region of interest (ROI) for seed-based connectivity analysis37.
2.3. Seed-Based Connectivity Analysis
The earliest form of rsfcMRI analysis was a seed-based approach used by Biswal et al.1 to identify the sensorimotor network. Seed-based analysis is a modelbased approach that relies on defining a particular ROI or set of ROIs and correlating the BOLD fMRI time series of this region against the time series of all other regions, resulting in a functional connectivity map. The seed can be chosen based on a priori knowledge or could be isolated based on task-based activation2. Seed-based connectivity analysis is used in numerous studies due to the easy interpretability of the method and because of its test-retest reliability3. However, although seed-based analysis produces more precise measurements, it can only capture coactivations with the defined ROIs. Thus, it can provide finer detail but is very much user/definition dependent and cannot be used to analyze a large number of nodes38. Figure 2 demonstrates seed-based functional connectivity analysis performed with the left Brodmann Area 44 (BA44) for the language network and the left precentral gyrus for the motor network as seed regions.
Figure 2.
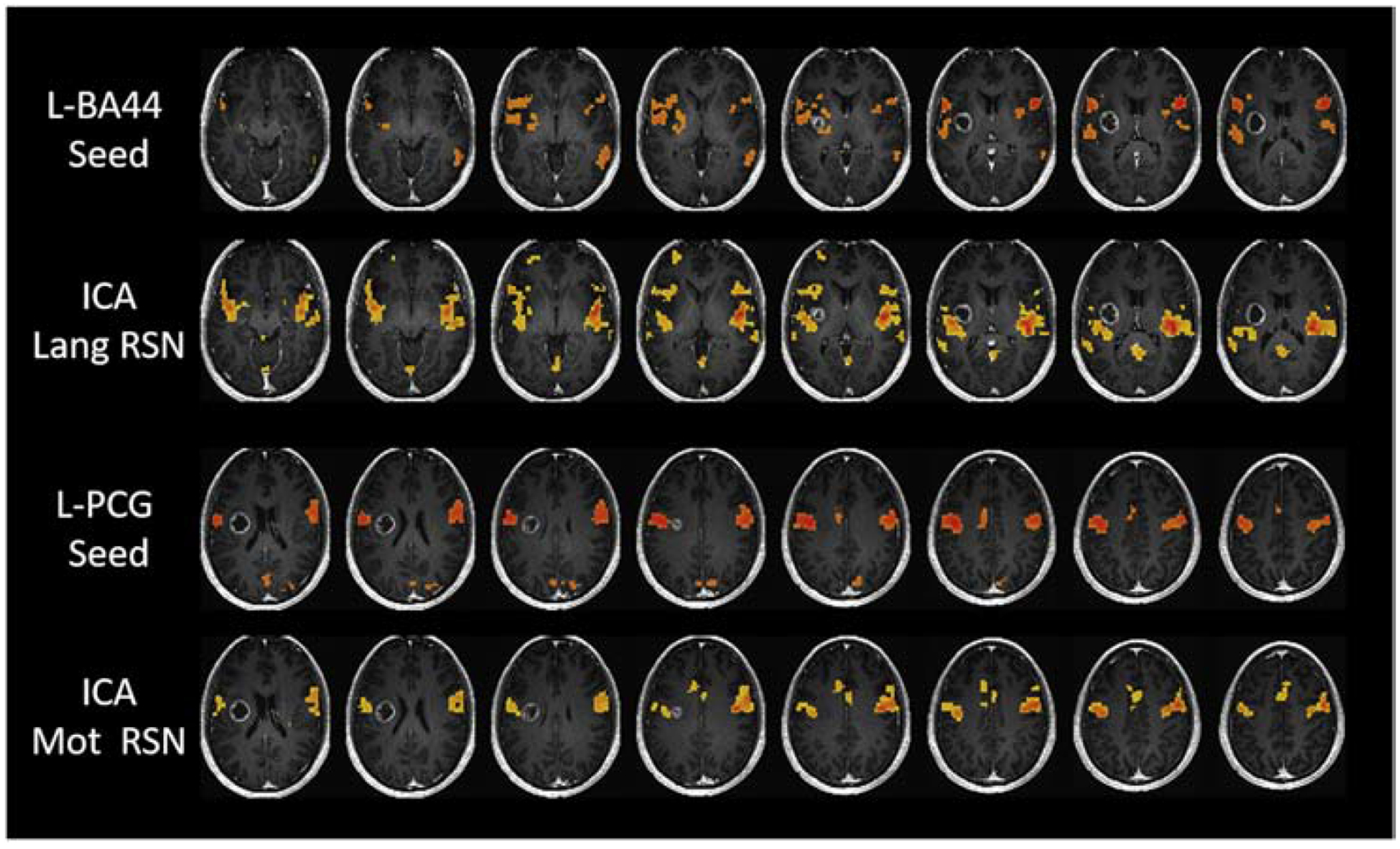
Seed-based correlation and ICA maps for a representative subject with a right-sided glioblastoma. L-BA44 seed and L-PCG seed maps represent seed-based correlation maps while ICA-Lang RSN and ICA-Motor RSN represent the independent component maps identified for the same subject. L-BA44: Left Brodmann Area 44. L-PCG: Left Precentral Gyrus
2.4. Independent Component Analysis
One of the most popular model-free methods applied to rsfcMRI is independent component analysis (ICA). ICA, like other model-free methods, analyzes signals from all voxels of the brain. This is distinct from the more limited seed-based approach, in which all voxel correlations were only calculated against one seed ROI. The central assumption of ICA is that each voxel’s signal output is composed of many different sources of activation and noise and the different sources of signal (whether they are neuronal, or artifact based) can be parsed apart by looking at similarities in BOLD signal across brain regions. ICA programs group areas of the brain based on the degree of similarity between their voxel activation time series, into a user-specified number of groups, or “components”8,39. ICA is useful because it is data-driven and does not depend on user-selected a priori ROI. An additional advantage of ICA is that unlike seed-based analysis which extracts only networks specific to the ROI, ICA extracts all networks within the subject simultaneously. However, ICA programs can be computationally demanding and produce results that may be hard to interpret, as users must discern which components represent noise signals and which represent true neuronal activation based on a priori understanding40. Figure 2 illustrates the language and motor networks obtained by ICA analysis and seed-based analysis, demonstrating that both seed-based and ICA methods produce similar results in a patient with brain tumor.
2.5. Clustering Analysis
Clustering analysis is an additional method that groups voxels together by similarities in time series11,41. Although also data-driven, it differs from ICA by directly grouping voxels together by their similarities without requiring user-dependent filtering of components42. ICA, seed-based, and clustering methods have been shown to produce concurrent results2.
2.6. Graph Theory
The transdisciplinary approach of graph theory, used in network science, has become germane to the study of functional connectivity41,43–45. Using a graph theory approach, the brain’s networks are modeled with nodes (regions of interest) and edges (connections between those regions of interest). By examining measures of this graph, such as average distance between nodes, number of edges and nodes, and how they are arranged in space, we can calculate network parameters that characterize these networks, such as global and local efficiency, node degree, centrality, and modularity46,47. Graph theory approaches allow for the examination of not only connections within specialized networks (segregation) but also how those networks and nodes interact or overlap with each other (integration). Graph theory applications to rsfcMRI are centered on the idea of constructing a functional connectome, a matrix of all possible paired connections between brain regions. The concept of a connectome was first introduced in reference to the anatomical connections of the brain48 but has since been applied more broadly to functional connectivity49.
Several network characteristics of resting state functional networks are understood. First, that brain networks have small world architecture, characterized by short path lengths between nodes and a high clustering coefficient between nodes12,43,50,52. Second, that brain networks can also be described as scale-free, meaning that although the average number of connections at each node are low, there is still a high level of global connectivity due to a few hub nodes in the network with very high number of connections44,46,53,54. Scale-free networks tend to be resilient against random attacks, due to the robustness of the many non-critical nodes, but are vulnerable against targeted attacks on the hub nodes, such as connectivity diseases47. Figure 3 illustrates a representative flow chart for graph theoretical analysis of resting state fMRI data.
Figure 3.
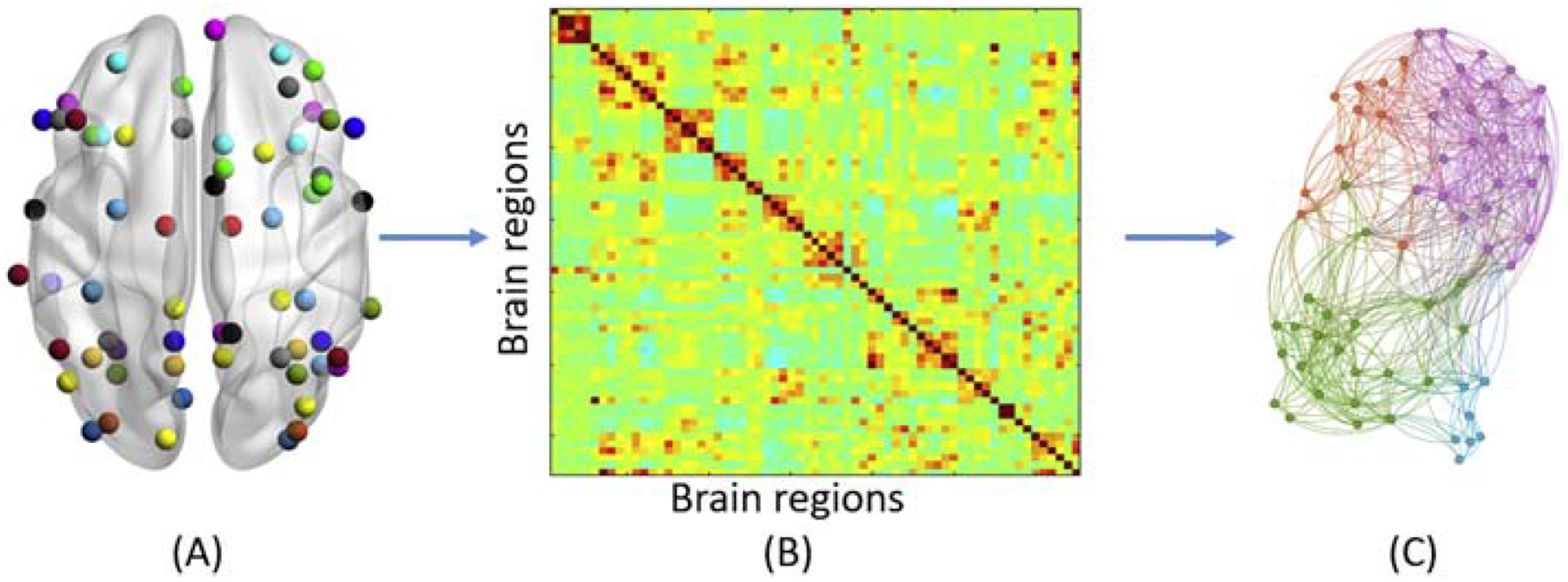
Representative flow chart for graph theoretical analysis of resting state fMRI data. (A) brain regions overlaid on a Glass brain surface (B) functional connectivity matrix representing Pearson’s correlation between BOLD fMRI time series of brain regions (C) Network representation of brain network derived from connectivity matrix where circles represent nodes and straight lines represent edges demonstrating presence of significant functional connectivity between brain regions
Challenges of graph theory analyses can arise if there is poor node definition and nodes derived from a graph theory model do not fit well with subjects’ true brain regions38. This can lead to limited interpretation of the data’s biological significance. In addition, because graph theory summarizes networks by global network measures, such as efficiency and small worldness, changes in these summary metrics may not actually reflect changes in nodes but rather confounding factors2.
Several studies have also implemented graph theory approaches to white-matter tracts derived using diffusion tensor imaging55–59. Similar to resting state graph theory approaches, these results have identified a small-world architecture of the brain. However, very few studies have objectively compared the brain functional architecture derived through resting state fMRI and structural architecture derived through diffusion tensor imaging studies60,61. One of the limitations of these approaches is related to the absence of structural white matter connections between regions of interest although functionally these regions may still be correlated.
3. New Methods of Analysis of rsfcMRI
3.1. Dynamic rsfcMRI
More recently, studies have been examining dynamic or non-stationary rsfcMRI, an approach that focuses on changes in network connections over a short period of time (often in the range of 10 s-2 min). Previous literature proposes dynamic rsfcMRI as a way to detect between-group differences and neurometabolic changes not evident in traditional rsfcMRI analyses62,63. Dynamic rsfcMRI evaluates fluctuations in connectivity by calculating the variations in temporal and spatial correlations over multiple time intervals of fMRI signal rather than over the full BOLD fMRI time series62. The dynamic fluctuations seen in functional connectivity may be a physiological process to balance efficient information processing and minimize metabolic demands on the brain, and the most dynamic connections are those that are spatially distant and intermodular64.
There are several approaches to defining the time series intervals. Many of these are variations of a standard sliding window approach, the prevailing dynamic rsfcfMRI methodology65–67. The sliding window method is relatively straightforward: the correlation matrix is calculated using a subset of the resting state time series, and this is repeatedly recalculated as the starting point of the window shifts incrementally down the time series with the window length and amount of desired overlap between windows defined by the user68. Figure 4 provides a schematic representation of the sliding window method of analysis adapted and modified from Valsasina et al67. The limitations of the sliding window approach primarily involve the user-dependent decision of the shape and size of the window; too large of a time segment may result in dynamic rsfcMRI approximating traditional rsfcMRI, while too short of a window may introduce spurious fluctuations67,69. A window size ranging between 30s-60s is therefore recommended for accurately capturing dynamic rsfcMRI69 and other methods have been explored to overcome the limitations of a user-driven process, such as data-driven adaptive windowing and time-frequency analysis65. Another limitation to dynamic-rsfcMRI research is the susceptibility for noise to be interpreted as dynamic fluctuations64,67.
Figure 4.
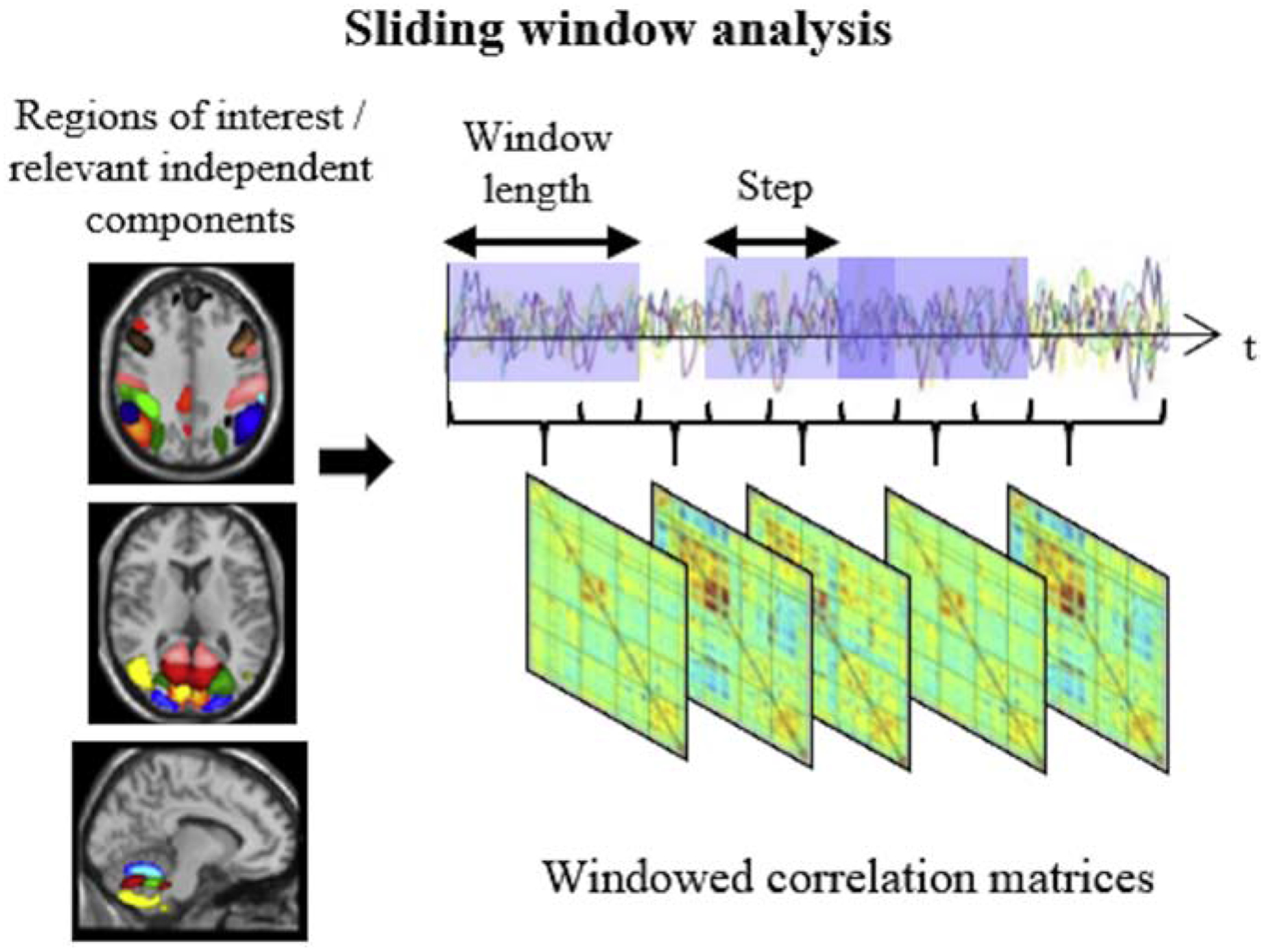
Schematic representation of sliding window analysis, the most popular approach in the assessment of time-varying functional connectivity. Adapted and reproduced with permission from Valsasina P, Hidalgo de la Cruz M, Filippi M, Rocca MA. Characterizing rapid fluctuations of resting state functional connectivity in demyelinating, neurodegenerative, and psychiatric conditions: from static to time-varying analysis. Frontiers Neurosci 2019; 13: Article 618 [doi: 10.3389/fnins.2019.00618]. (Citation #60) Creative Commons license: (https://creativecommons.org/licenses/by/4.0/).
Although there are current challenges in utilization and interpretation of dynamic rsfcMRI, this paradigm has been shown to explain more variation in behavioral measurements such as working memory, facial expression processing, and sustained attention66. Testing dynamic rsfcMRI can provide a better picture of the dynamic changes that may underly many clinical conditions that involve unstable or overly stable states69. Dynamic rsfcMRI has been used to study RSFC in several disease states67, including MS70,71 neurodegenerative diseases72–74, bipolar disease75, major depressive disorder76, schizophrenia77,78, post-traumatic stress disorder79, and stroke80. There are also indications that dynamic rsfcMRI can detect changes that happen over the course of hours or months as well, and it has been proposed that these longer-term changes may reflect learning or variable gene expression69.
3.2. Independent Vector Analysis
Another emerging data-driven rsfcMRI method is independent vector analysis (IVA), which builds upon ICA. IVA is similar to ICA in its blind source splitting approach but is proposed as a method to solve for the permutation ambiguities found in ICA output81. IVA, like ICA, assumes that elements of each source vector are independent from elements of other source vectors in the same fMRI dataset, but it differs in that it assumes increased dependence among the similar source vectors across fMRI datasets. In their paper defining IVA methodology, Kim et al. first showed that by defining a multivariate score function rather than a single-variate score function, as is used in ICA, the IVA analysis provides a well-ordered output of source vectors, compared to the scrambling of source signal elements from ICA82. IVA approaches to group-level rsfcMRI analysis can improve the isolation of true signal source elements and improve user-independence by eliminating the need for manual selection of components from each source signal required in ICA83. Additionally, IVA algorithms applied to a group level rsfcMRI analysis can result in spatially similar activation maps, which can be related to group level analysis maps as a result of general linear based modelling approaches. IVA has also been shown to be better at detecting spatial fluctuations. Recently, Ma et al. used IVA to examine group-level dynamic spatial fluctuations between pairs of resting state networks that existed in healthy controls and in schizophrenia patients84. The IVA findings resembled previous research on schizophrenia patients, finding the most spatial fluctuations in the frontoparietal, cerebellar, and temporal areas. It also found that schizophrenia patients exhibited more dynamic fluctuations in connectivity, suggesting a more disorganized way of recruiting functional areas of the brain84. A practical implementation of IVA is available through the Group ICA Toolbox (GIFT, http://mialab.mrn.org/software); however, extensive computation time and interpretation challenges have limited the application of IVA to wider clinical populations.
3.3. Multiband rsfcMRI
Until recently, most of the studies in rsfcMRI have investigated functional connectivity in clinical population in low-frequency bands of BOLD fMRI fluctuations (0.01–0.1 Hz). This has been mainly due to the significantly higher power observed by Biswal et al1 in the low-frequency ranges of 0.01–0.1Hz. Additionally, due to the limitations on temporal sampling for whole brain BOLD fMRI data, most fMRI studies have used rsfcMRI data collected at 2 second intervals, thus limiting the study of BOLD signal frequencies in the low-frequency range. Recent advancements in data acquisition techniques have enabled researchers to collect BOLD fMRI data from multiple brain slices at the same time, resulting in faster brain acquisition sequences. With the implementation of such imaging sequences referred to as multi-band imaging techniques85–87, it is now possible to acquire whole brain fMRI at sub-second temporal resolutions. This has resulted in significant improvements in temporal resolution along with enhanced capabilities to investigate resting state functional connectivity at high-frequency bands and improved characterization of cardiac and respiratory noises. Using these multiband imaging data, researchers have shown presence of resting state functional connectivity across BOLD signal frequencies higher than 0.1 Hz88,89. Studies have implemented progressively faster multiband imaging sequences, resulting in a sampling time of as little as 333 ms90. which has pushed the upper range of BOLD fMRI signal frequency that can be investigated to as high as 1.5 Hz. Recent studies have used these frequency specific resting state measures to quantify functional brain disruptions in clinical populations including epilepsy91, psychosis29,92, ADHD93, dyskinesia34, and brain tumors30. Although there are challenges associated with the effect of nuisance regression techniques94 and head motion on high-frequency resting state data95, multi-band resting state fMRI analysis represents an innovative approach to quantify functional brain disruptions.
4. Future Directions
4.1. Network of Networks
An emerging graph theoretical approach is the idea of the brain being modeled as a “network of networks” (NoN). In 2017, Morone and colleagues described a robust, modular NoN pattern that was defined by functionally specialized subnetworks within the brain. They studied a n=15 visual-auditory task paradigm and created a map of neural networks that identified critical nodes they labelled as neural collective influencers (NCI). Morone defined NCIs as the minimal set of nodes that would confer global connectivity to the network, and the identified NCIs in the visual-auditory task were the anterior cingulate cortex, the posterior parietal cortex, and the posterior occipital cortex96.
To model the network’s robustness against disease, neural influencers were removed, and global efficiency was re-evaluated by calculating the giant component G, or the largest interconnected active component of the network. In modular NoN network model, not all of the activated nodes participated in the giant component G, and their ability to activate apart from G suggests that a modular NoN is robust to cascading effects of injury (power-grid catastrophic effects). This NoN paradigm is promising in its ability to help further elucidate key areas of influence and modulation in the brain and understand the brain’s response to injury and should be extended to resting state research.
4.2. Big Data Analysis
Understanding the human connectome has been recognized as a major new frontier of research. In 2010, the NIH established the Human Connectome Project (HCP) to compile a comprehensive map of functional brain networks and improve current MRI acquisition techniques97. A group led by Washington University, the University of Minnesota, and Oxford University (WU-Minn Consortium) is collecting over 1000 subject scans, and the data is publicly available at humanconnectome.org. This has provided a large database of rsfcMRI data that can help elucidate the behavior and function of resting brain networks, and the many factors (genetics, age, environmental factors) that can affect how those networks function. Concurrently, the 1000 Functional Connectomes Project (FCP) was started in 2009 to promote resting state data sharing, publishing over 1200 rsfcMRI datasets, including many heterogenous datasets sourced from many subject groups and pathologies, and this was succeeded by the International Neuroimaging Datasharing Initiative (INDI)98.
Traditionally, fMRI analysis has involved unwieldy preprocessing pipelines to reduce noise and normalize imaging data. As big datasets become more common in research with data sharing initiatives, newer processing methods are necessary to streamline rsfcMRI analysis and standardize the outputs. The several central challenges of big data in rsfcfMRI concern high-performance computing requirements, adequate data-sharing infrastructure and standardization of processing pipelines99. Currently, no consensus exists on the ordering or utilization of optimal preprocessing steps. In addition, when examining big datasets, common noise variances can be exacerbated, leading to increased risk of interpreting spurious signal as true activation99,100. Solutions proposed have been to minimize the preprocessing steps, conduct systematic reviews of preprocessing methods and outcomes, and adopt software packages better suited for big fMRI data100. Makkie et al. also recently reviewed fMRI applications of Apache Spark and Hadoop, two open-source software suites with big data capabilities101. Standardization of fMRI processing of big data is an important area of work as big datasets continue to be an invaluable resource in neuroscience research and will allow for more efficient exploration of the underlying resting state networks that might explain common behaviors and pathologies.
4.3. RsfcMRI at Ultrahigh Fields
While earlier resting state data was acquired using 3 Tesla (3T) scanning, recently the US Food and Drug Administration (FDA) approved the next generation of ultra-high field 7 Tesla (7T) MRI magnets for clinical use. A major advantage of ultra-high field scanning using newer generations of 7T magnets over more traditional 3T imaging is higher functional contrast-to-noise ratios resulting in increased spatial resolution102. 7T scanning also reduces time-series SNR and is more sensitive to temporal correlations in BOLD signal, which can capture previously unrecognized nodes in functional networks103. Resting state connections have been seen on 7T scans that are not evident in 3T scans, particularly involving short voxel lengths between 1 and 1.5 mm102,104,105. Potential drawbacks of ultra-high field scanning include increasing sensitivity to motion and noise artifact, and longer scan times104,106. Different approaches have been suggested to correct for noise, including, for example, an autoregressive statistical approach used for 3T scans, which has been extended and proposed for ultra-high field imaging107. Although a number of academic institutions have been using ultra-high field scanning at 7T predominantly in research settings102,105,108,103,109,110 systematic studies comparing 7T and 3T functional MRI in different clinical cohorts remain scarce and are warranted.
5. Conclusion
Resting state fMRI has led to the identification of brain networks critical to affecting how humans interact, perceive, and process environmental and internal stimuli. While widely used rsfcMRI processing techniques are still the topic of discussion and refinement, interdisciplinary approaches from the realm of network science could help answer further questions about the dynamics, robustness, and interplay of these brain networks. Due to the demanding nature of fMRI data collection and analysis, it is of critical importance to engage in interdisciplinary research and implement large-scale data-sharing initiatives.
Acknowledgements
This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. Research reported in this study was also supported by the National Cancer Institute of the National Institutes of Health under Award Number R25CA020449.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. [DOI] [PubMed] [Google Scholar]
- 2.van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20(8):519–534. [DOI] [PubMed] [Google Scholar]
- 3.Cole DM, Smith SM, Beckmann CF. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front Syst Neurosci. 2010;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou YH, Panych LP, Dickey CC, Petrella JR, Chen NK. Investigation of long-term reproducibility of intrinsic connectivity network mapping: a resting-state fMRI study. AJNR Am J Neuroradiol. 2012;33(5):833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo XN, Di Martino A, Kelly C, et al. The oscillating brain: complex and reliable. Neuroimage. 2010;49(2):1432–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greicius MD, Menon V. Default-mode activity during a passive sensory task: uncoupled from deactivation but impacting activation. J Cogn Neurosci. 2004;16(9):1484–1492. [DOI] [PubMed] [Google Scholar]
- 7.Tomasi D, Volkow ND. Resting functional connectivity of language networks: characterization and reproducibility. Mol Psychiatry. 2012;17(8):841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98(2):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeley WW, Menon V, Schatzberg AF, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27(9):2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soares JM, Magalhaes R, Moreira PS, et al. A Hitchhiker’s Guide to Functional Magnetic Resonance Imaging. Front Neurosci. 2016;10:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MH, Smyser CD, Shimony JS. Resting-state fMRI: a review of methods and clinical applications. AJNR Am J Neuroradiol. 2013;34(10):1866–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hull JV, Dokovna LB, Jacokes ZJ, Torgerson CM, Irimia A, Van Horn JD. Resting-State Functional Connectivity in Autism Spectrum Disorders: A Review. Front Psychiatry. 2016;7:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Martino A, Yan CG, Li Q, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19(6):659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheffield JM, Barch DM. Cognition and resting-state functional connectivity in schizophrenia. Neurosci Biobehav Rev. 2016;61:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraldo-Chica M, Woodward ND. Review of thalamocortical resting-state fMRI studies in schizophrenia. Schizophr Res. 2017;180:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watsky RE, Gotts SJ, Berman RA, et al. Attenuated resting-state functional connectivity in patients with childhood- and adult-onset schizophrenia. Schizophr Res. 2018;197:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohenfeld C, Werner CJ, Reetz K. Resting-state connectivity in neurodegenerative disorders: Is there potential for an imaging biomarker? Neuroimage Clin. 2018;18:849–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennis EL, Thompson PM. Functional brain connectivity using fMRI in aging and Alzheimer’s disease. Neuropsychol Rev. 2014;24(1):49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox ME, King TZ. Functional Connectivity in Adult Brain Tumor Patients: A Systematic Review. Brain Connect. 2018;8(7):381–397. [DOI] [PubMed] [Google Scholar]
- 21.Zang YF, He Y, Zhu CZ, et al. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007;29(2):83–91. [DOI] [PubMed] [Google Scholar]
- 22.Kublbock M, Woletz M, Hoflich A, et al. Stability of low-frequency fluctuation amplitudes in prolonged resting-state fMRI. Neuroimage. 2014;103:249–257. [DOI] [PubMed] [Google Scholar]
- 23.Zou QH, Zhu CZ, Yang Y, et al. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods. 2008;172(1):137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Long XY, Yang Y, et al. Amplitude of low frequency fluctuation within visual areas revealed by resting-state functional MRI. Neuroimage. 2007;36(1):144–152. [DOI] [PubMed] [Google Scholar]
- 25.He Y, Wang L, Zang Y, et al. Regional coherence changes in the early stages of Alzheimer’s disease: a combined structural and resting-state functional MRI study. Neuroimage. 2007;35(2):488–500. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Xu J, Nie B, et al. Abnormal resting-state activities and functional connectivities of the anterior and the posterior cortexes in medication-naive patients with obsessivecompulsive disorder. PLoS One. 2013;8(6):e67478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas C, Lopez-Jaramillo C, Vieta E. A systematic literature review of resting state network--functional MRI in bipolar disorder. J Affect Disord. 2013;150(3):727–735. [DOI] [PubMed] [Google Scholar]
- 28.Turner JA, Damaraju E, van Erp TG, et al. A multi-site resting state fMRI study on the amplitude of low frequency fluctuations in schizophrenia. Front Neurosci. 2013;7:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gohel S, Gallego JA, Robinson DG, DeRosse P, Biswal B, Szeszko PR. Frequency specific resting state functional abnormalities in psychosis. Hum Brain Mapp. 2018;39(11):4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal S, Lu H, Pillai JJ. Value of Frequency Domain Resting-State Functional Magnetic Resonance Imaging Metrics Amplitude of Low-Frequency Fluctuation and Fractional Amplitude of Low-Frequency Fluctuation in the Assessment of Brain Tumor-Induced Neurovascular Uncoupling. Brain Connect. 2017;7(6):382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zang Y, Jiang T, Lu Y, He Y, Tian L. Regional homogeneity approach to fMRI data analysis. Neuroimage. 2004;22(1):394–400. [DOI] [PubMed] [Google Scholar]
- 32.Zuo XN, Xing XX. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci Biobehav Rev. 2014;45:100–118. [DOI] [PubMed] [Google Scholar]
- 33.Song X, Zhang Y, Liu Y. Frequency specificity of regional homogeneity in the resting-state human brain. PLoS One. 2014;9(1):e86818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZR, Miao HH, Yu Y, Ding MP, Liao W. Frequency-Specific Local Synchronization Changes in Paroxysmal Kinesigenic Dyskinesia. Medicine (Baltimore). 2016;95(13):e3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z, Wang Z, Zhang J, et al. Altered temporal variance and neural synchronization of spontaneous brain activity in anesthesia. Hum Brain Mapp. 2014;35(11):5368–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu R, Hsieh MH, Wang HL, et al. Frequency dependent alterations in regional homogeneity of baseline brain activity in schizophrenia. PLoS One. 2013;8(3):e57516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv H, Wang Z, Tong E, et al. Resting-State Functional MRI: Everything That Nonexperts Have Always Wanted to Know. AJNR Am J Neuroradiol. 2018;39(8):1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith SM, Vidaurre D, Beckmann CF, et al. Functional connectomics from resting-state fMRI. Trends Cogn Sci. 2013;17(12):666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14(3):140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8(9):700–711. [DOI] [PubMed] [Google Scholar]
- 41.Smitha KA, Akhil Raja K, Arun KM, et al. Resting state fMRI: A review on methods in resting state connectivity analysis and resting state networks. Neuroradiol J. 2017;30(4):305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee MH, Hacker CD, Snyder AZ, et al. Clustering of resting state networks. PLoS One. 2012;7(7):e40370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186–198. [DOI] [PubMed] [Google Scholar]
- 44.Reijneveld JC, Ponten SC, Berendse HW, Stam CJ. The application of graph theoretical analysis to complex networks in the brain. Clin Neurophysiol. 2007;118(11):2317–2331. [DOI] [PubMed] [Google Scholar]
- 45.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Zuo X, He Y. Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci. 2010;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Heuvel MP, Mandl RC, Kahn RS, Hulshoff Pol HE. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp. 2009;30(10):3127–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1(4):e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fornito A, Zalesky A, Bullmore ET. Fundamentals of brain network analysis. Amsterdam; Boston: Elsevier/Academic Press; 2016. [Google Scholar]
- 50.Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12(6):512–523. [DOI] [PubMed] [Google Scholar]
- 51.Huang Q, Zhang R, Hu X, et al. Disturbed small-world networks and neurocognitive function in frontal lobe low-grade glioma patients. PLoS One. 2014;9(4):e94095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bassett DS, Bullmore ET. Small-World Brain Networks Revisited. Neuroscientist. 2017;23(5):499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eguiluz VM, Chialvo DR, Cecchi GA, Baliki M, Apkarian AV. Scale-free brain functional networks. Phys Rev Lett. 2005;94(1):018102. [DOI] [PubMed] [Google Scholar]
- 54.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. [DOI] [PubMed] [Google Scholar]
- 55.Wang Q, Su TP, Zhou Y, et al. Anatomical insights into disrupted small-world networks in schizophrenia. Neuroimage. 2012;59(2):1085–1093. [DOI] [PubMed] [Google Scholar]
- 56.Pandit AS, Expert P, Lambiotte R, et al. Traumatic brain injury impairs small-world topology. Neurology. 2013;80(20):1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M, Chen H, Wang J, et al. Handedness- and hemisphere-related differences in small-world brain networks: a diffusion tensor imaging tractography study. Brain Connect. 2014;4(2):145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan H, Tian L, Wang Q, et al. Compromised small-world efficiency of structural brain networks in schizophrenic patients and their unaffected parents. Neurosci Bull. 2015;31(3):275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen W, He Y, Sachdev P. Structural brain networks and neuropsychiatric disorders. Curr Opin Psychiatry. 2011;24(3):219–225. [DOI] [PubMed] [Google Scholar]
- 60.Park C-hK SY; Kim Y-H; Kim K Comparison of the small-world topology between anatomical and functional connectivity in the human brain. Physica A: Statistical Mechanics and its Applications. 2008;387(23):5958–5962. [Google Scholar]
- 61.Hosseini SM, Kesler SR. Comparing connectivity pattern and small-world organization between structural correlation and resting-state networks in healthy adults. Neuroimage. 2013;78:402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson GJ. Neural and metabolic basis of dynamic resting state fMRI. Neuroimage. 2018;180(Pt B):448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16(3):159–172. [DOI] [PubMed] [Google Scholar]
- 64.Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. Proc Natl Acad Sci U S A. 2014;111(28):10341–10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keilholz S, Caballero-Gaudes C, Bandettini P, Deco G, Calhoun V. Time-Resolved Resting-State Functional Magnetic Resonance Imaging Analysis: Current Status, Challenges, and New Directions. Brain Connect. 2017;7(8):465–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liegeois R, Li J, Kong R, et al. Resting brain dynamics at different timescales capture distinct aspects of human behavior. Nat Commun. 2019;10(1):2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valsasina P, Hidalgo de la Cruz M, Filippi M, Rocca MA. Characterizing Rapid Fluctuations of Resting State Functional Connectivity in Demyelinating, Neurodegenerative, and Psychiatric Conditions: From Static to Time-Varying Analysis. Frontiers in Neuroscience. 2019;13(618). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2014;24(3):663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Preti MG, Bolton TA, Van De Ville D. The dynamic functional connectome: State-of-theart and perspectives. Neuroimage. 2017;160:41–54. [DOI] [PubMed] [Google Scholar]
- 70.Huang M, Zhou F, Wu L, et al. White matter lesion loads associated with dynamic functional connectivity within attention network in patients with relapsing-remitting multiple sclerosis. J Clin Neurosci. 2019;65:59–65. [DOI] [PubMed] [Google Scholar]
- 71.Cordani C, Valsasina P, Preziosa P, Meani A, Filippi M, Rocca MA. Action observation training promotes motor improvement and modulates functional network dynamic connectivity in multiple sclerosis. Mult Scler. 2019:1352458519887332. [DOI] [PubMed] [Google Scholar]
- 72.Jones DT, Vemuri P, Murphy MC, et al. Non-stationarity in the “resting brain’s” modular architecture. PLoS One. 2012;7(6):e39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quevenco FC, Preti MG, van Bergen JM, et al. Memory performance-related dynamic brain connectivity indicates pathological burden and genetic risk for Alzheimer’s disease. Alzheimers Res Ther. 2017;9(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai J, Liu A, Mi T, et al. Dynamic Graph Theoretical Analysis of Functional Connectivity in Parkinson’s Disease: The Importance of Fiedler Value. IEEE J Biomed Health Inform. 2019;23(4):1720–1729. [DOI] [PubMed] [Google Scholar]
- 75.Du Y, Pearlson GD, Lin D, et al. Identifying dynamic functional connectivity biomarkers using GIG-ICA: Application to schizophrenia, schizoaffective disorder, and psychotic bipolar disorder. Hum Brain Mapp. 2017;38(5):2683–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhi D, Calhoun VD, Lv L, et al. Aberrant Dynamic Functional Network Connectivity and Graph Properties in Major Depressive Disorder. Front Psychiatry. 2018;9:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yaesoubi M, Miller RL, Bustillo J, Lim KO, Vaidya J, Calhoun VD. A joint time-frequency analysis of resting-state functional connectivity reveals novel patterns of connectivity shared between or unique to schizophrenia patients and healthy controls. Neuroimage Clin. 2017;15:761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, Li S, Wang X, et al. Abnormal dynamic functional connectivity between speech and auditory areas in schizophrenia patients with auditory hallucinations. Neuroimage Clin. 2018;19:918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Zhu D, Jiang X, et al. Dynamic functional connectomics signatures for characterization and differentiation of PTSD patients. Hum Brain Mapp. 2014;35(4):1761–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J, Sun D, Shi Y, et al. Alterations of static functional connectivity and dynamic functional connectivity in motor execution regions after stroke. Neurosci Lett. 2018;686:112–121. [DOI] [PubMed] [Google Scholar]
- 81.Anderson M, Adali T, Li XL. Joint Blind Source Separation With Multivariate Gaussian Model: Algorithms and Performance Analysis. Ieee T Signal Proces. 2012;60(4):1672–1683. [Google Scholar]
- 82.Kim T, Eltoft T, Lee TW. Independent vector analysis: An extension of ICA to multivariate components. Lect Notes Comput Sc. 2006;3889:165–172. [Google Scholar]
- 83.Lee JH, Lee TW, Jolesz FA, Yoo SS. Independent vector analysis (IVA): multivariate approach for fMRI group study. Neuroimage. 2008;40(1):86–109. [DOI] [PubMed] [Google Scholar]
- 84.Ma S, Calhoun VD, Phlypo R, Adali T. Dynamic changes of spatial functional network connectivity in healthy individuals and schizophrenia patients using independent vector analysis. Neuroimage. 2014;90:196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feinberg DA, Moeller S, Smith SM, et al. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;5(12):e15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feinberg DA, Yacoub E. The rapid development of high speed, resolution and precision in fMRI. Neuroimage. 2012;62(2):720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jesmanowicz A, Nencka AS, Li SJ, Hyde JS. Two-axis acceleration of functional connectivity magnetic resonance imaging by parallel excitation of phase-tagged slices and half k-space acceleration. Brain Connect. 2011;1(1):81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boubela RN, Kalcher K, Huf W, Kronnerwetter C, Filzmoser P, Moser E. Beyond Noise: Using Temporal ICA to Extract Meaningful Information from High-Frequency fMRI Signal Fluctuations during Rest. Front Hum Neurosci. 2013;7:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gohel SR, Biswal BB. Functional integration between brain regions at rest occurs in multiple-frequency bands. Brain Connect. 2015;5(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Demetriou L, Kowalczyk OS, Tyson G, Bello T, Newbould RD, Wall MB. A comprehensive evaluation of increasing temporal resolution with multiband-accelerated protocols and effects on statistical outcome measures in fMRI. Neuroimage. 2018;176:404–416. [DOI] [PubMed] [Google Scholar]
- 91.Wang J, Zhang Z, Ji GJ, et al. Frequency-Specific Alterations of Local Synchronization in Idiopathic Generalized Epilepsy. Medicine (Baltimore). 2015;94(32):e1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Zhang Y, Long Z, et al. Frequency-specific alteration of functional connectivity density in antipsychotic-naive adolescents with early-onset schizophrenia. J Psychiatr Res. 2017;95:68–75. [DOI] [PubMed] [Google Scholar]
- 93.Yu X, Yuan B, Cao Q, et al. Frequency-specific abnormalities in regional homogeneity among children with attention deficit hyperactivity disorder: a resting-state fMRI study. Science Bulletin. 2016;61(9):682–692. [Google Scholar]
- 94.Chen JE, Jahanian H, Glover GH. Nuisance Regression of High-Frequency Functional Magnetic Resonance Imaging Data: Denoising Can Be Noisy. Brain Connect. 2017;7(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan BK, Zang YF, Liu DQ. Influences of Head Motion Regression on High-Frequency Oscillation Amplitudes of Resting-State fMRI Signals. Front Hum Neurosci. 2016;10:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morone F, Roth K, Min B, Stanley HE, Makse HA. Model of brain activation predicts the neural collective influence map of the brain. Proc Natl Acad Sci U S A. 2017;114(15):3849–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Essen DC, Smith SM, Barch DM, et al. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: the FCP/INDI experience. Neuroimage. 2013;82:683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poldrack RA, Gorgolewski KJ. Making big data open: data sharing in neuroimaging. Nat Neurosci. 2014;17(11):1510–1517. [DOI] [PubMed] [Google Scholar]
- 100.Phinyomark A, Ibáñez-Marcelo E, Petri G. Resting-State fMRI Functional Connectivity: Big Data Preprocessing Pipelines and Topological Data Analysis. IEEE Transactions on Big Data. 2017;3(4):415–428. [Google Scholar]
- 101.Makkie M, Li X, Quinn S, et al. A Distributed Computing Platform for fMRI Big Data Analytics. IEEE Trans Big Data. 2019;5(2):109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gorgolewski KJ, Mendes N, Wilfling D, et al. A high resolution 7-Tesla resting-state fMRI test-retest dataset with cognitive and physiological measures. Sci Data. 2015;2:140054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Triantafyllou C, Hoge RD, Krueger G, et al. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. Neuroimage. 2005;26(1):243–250. [DOI] [PubMed] [Google Scholar]
- 104.Hale JR, Brookes MJ, Hall EL, et al. Comparison of functional connectivity in default mode and sensorimotor networks at 3 and 7T. MAGMA. 2010;23(5–6):339–349. [DOI] [PubMed] [Google Scholar]
- 105.Agarwal S, Sair HI, Airan R, et al. Demonstration of Brain Tumor-Induced Neurovascular Uncoupling in Resting-State fMRI at Ultrahigh Field. Brain Connect. 2016;6(4):267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Martino F, Esposito F, van de Moortele PF, et al. Whole brain high-resolution functional imaging at ultra high magnetic fields: an application to the analysis of resting state networks. Neuroimage. 2011;57(3):1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang X, Holmes MJ, Newton AT, Morgan VL, Landman BA. A Comparison of Distributional Considerations with Statistical Analysis of Resting State fMRI at 3T and 7T. Proc SPIE Int Soc Opt Eng. 2012;8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lottman KK, Gawne TJ, Kraguljac NV, Killen JF, Reid MA, Lahti AC. Examining resting-state functional connectivity in first-episode schizophrenia with 7T fMRI and MEG. Neuroimage Clin. 2019;24:101959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beisteiner R, Robinson S, Wurnig M, et al. Clinical fMRI: evidence for a 7T benefit over 3T. Neuroimage. 2011;57(3):1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balchandani P, Naidich TP. Ultra-High-Field MR Neuroimaging. AJNR Am J Neuroradiol. 2015;36(7):1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]


