The complete genome sequence of a U.S. isolate of a Tomato brown rugose fruit virus (ToBRFV) (CA18-01) was obtained through Illumina and MinION sequencing. The U.S. ToBRFV isolate shared a high nucleic acid sequence identity (>99%) with known ToBRFV isolates. Phylogenetic analysis revealed a tight cluster for ToBRFV isolates throughout the world, suggesting a short evolutionary history.
ABSTRACT
The complete genome sequence of a U.S. isolate of a Tomato brown rugose fruit virus (ToBRFV) (CA18-01) was obtained through Illumina and MinION sequencing. The U.S. ToBRFV isolate shared a high nucleic acid sequence identity (>99%) with known ToBRFV isolates. Phylogenetic analysis revealed a tight cluster for ToBRFV isolates throughout the world, suggesting a short evolutionary history.
ANNOUNCEMENT
Tomato (Solanum lycopersicum L.), an important vegetable crop throughout the world, is susceptible to many viral diseases (1). Tomato brown rugose fruit virus (ToBRFV), an emerging tobamovirus, was capable of breaking the Tm-22 gene in tomato that is resistant to other tobamoviruses (2, 3). ToBRFV, in genus Tobamovirus and family Virgaviridae, was first identified in the Middle East (2, 4). ToBRFV has since been reported in a number of countries in Asia (5, 6), Europe (7–11), and North America (12–14) and is likely to spread to other countries. To curb virus spread, the U.S. Department of Agriculture issued a federal order in 2019 for ToBRFV (15). Here, we report the complete genome sequence of a U.S. isolate of ToBRFV using Illumina and MinION sequencing technologies. The U.S. isolate of ToBRFV, designated CA18-01, was originally collected in the fall of 2018 in southern California (14). For Illumina sequencing, libraries were prepared using the TruSeq stranded total RNA (tomato leaves) with a Ribo-Zero plant kit and sequenced in a MiSeq instrument (150 cycles, paired-end reads). A total of 1,982,141 read pairs were generated. Adapter trimming and quality filtering of reads were performed with Trimmomatic v. 0.39 (16). Clean reads were de novo assembled using the SPAdes v. 3.10.0 assembler (17) with default parameters. The MinION sequencing library was prepared using the SQK-PCS108 cDNA PCR kit (Oxford Nanopore Technologies) and sequenced in a MinION Mk1B device (MIN-101B). A total of 3,001,250 reads were generated. Adapters were removed and chimeric reads were discarded using Porechop v. 0.2.3 (https://github.com/rrwick/Porechop). Clean reads were de novo assembled using Canu v. 1.8 (18) with default Nanopore assembly parameters with readSamplingCoverage = 1000 based on a genome size of 10 kb. The resulting contigs were searched against an in-house database generated from GenBank virus and viroid sequences through BLASTn. The depth of coverage was estimated for all positively identified viruses by mapping reads to contigs using BBMap (19). A single contig with high-depth coverage (mean depth, 7,576) was mapped to the reference ToBRFV-Jordan (GenBank accession no. KT383474). To determine the exact 5′-terminal sequence of ToBRFV CA18-01, rapid amplification of cDNA ends (RACE) was used with a First Choice RLM-RACE kit (Invitrogen), and PCR products were sequenced using Sanger sequencing.
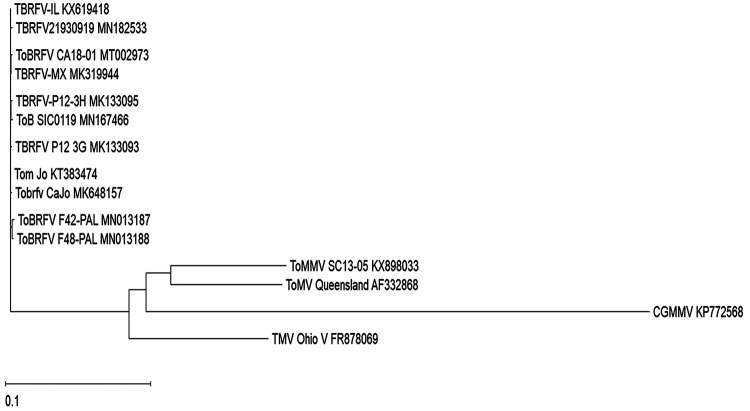
The completed genome of ToBRFV CA18-01 (GenBank accession no. MT002973) had 6,389 nucleotides (GC content, 41.51%) comprising four open reading frames (ORFs), with ORF1 encoding a 126-kDa replicase protein, ORF 2 encoding an RdRP protein of 183 kDa, ORF3 encoding a movement protein of 30 kDa, and ORF4 encoding a coat protein of 17.5 kDa. The 5′ and 3′ untranslated regions spanned 72 and 199 nucleotides, respectively. A multiple sequence alignment (Lasergene 16) using 10 ToBRFV isolates revealed 99.6 to 99.9% nucleotide sequence identities among each other and less than 82.2% identities to other tobamoviruses. Such high sequence identities among ToBRFV isolates suggest a short evolutionary history of this novel virus. Phylogenetic analysis showed a tight cluster of ToBRFV isolates separating from other tobamoviruses (Fig. 1). The complete genome sequence of the ToBRFV isolate from the United States will enhance our understanding of the genetic diversity of this emerging virus as it spreads further across the world.
FIG 1.
Phylogenetic relationship of the complete genome sequence of Tomato brown rugose fruit virus (ToBRFV) isolate CA18-01 (GenBank accession no. MT002973) with those of 10 other ToBRFV isolates and four related tobamoviruses, including Tomato mosaic virus (ToMV), Tomato mottle mosaic virus (ToMMV), Tobacco mosaic virus (TMV), and Cucumber green mottle mosaic virus (CGMMV). A multiple sequence alignment was created using ClustalOmega in the MegAlign Pro program of Lasergene 16 (DNAStar, USA). The phylogenetic tree was generated using BIONJ with default parameters.
Data availability.
The high-throughput sequencing data sets were submitted to the SRA database with the accession no. SRR11794481 (Illumina reads) and SRR11794480 (MinION reads) and the BioProject no. PRJNA632574. The genome sequence of ToBRFV CA18-01 was deposited in GenBank with accession no. MT002973.
ACKNOWLEDGMENT
This work was supported in part by the USDA-ARS, National Plant Disease Recovery System Program (to K.-S.L.).
REFERENCES
- 1.Hanssen MI, Lapidot M, Thomma PHJ. 2010. Emerging viral diseases of tomato crops. Mol Plant Microbe Interact 23:539–548. doi: 10.1094/MPMI-23-5-0539. [DOI] [PubMed] [Google Scholar]
- 2.Luria N, Smith E, Reingold V, Bekelman I, Lapidot M, Levin I, Elad N, Tam Y, Sela N, Abu-Ras A, Ezra N, Haberman A, Yitzhak L, Lachman O, Dombrovsky A. 2017. A new Israeli tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS One 12:e0170429. doi: 10.1371/journal.pone.0170429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maayan Y, Pandaranayaka E, Srivastava DA, Lapidot M, Levin I, Dombrovsky A, Harel A. 2018. Using genomic analysis to identify tomato Tm-2 resistance-breaking mutations and their underlying evolutionary path in a new and emerging tobamovirus. Arch Virol 163:1863–1875. doi: 10.1007/s00705-018-3819-5. [DOI] [PubMed] [Google Scholar]
- 4.Salem N, Mansour A, Ciuffo M, Falk BW, Turina M. 2016. A new tobamovirus infecting tomato crops in Jordan. Arch Virol 161:503–506. doi: 10.1007/s00705-015-2677-7. [DOI] [PubMed] [Google Scholar]
- 5.Alkowni R, Alabdallah O, Fadda Z. 2019. Molecular identification of Tomato brown rugose fruit virus in tomato in Palestine. J Plant Pathol 101:719–723. doi: 10.1007/s42161-019-00240-7. [DOI] [Google Scholar]
- 6.Yan Z, Ma H, Han S, Geng C, Tian Y, Li X. 2019. First report of Tomato brown rugose fruit virus infecting tomato in China. Plant Dis 103:2973–2973. doi: 10.1094/PDIS-05-19-1045-PDN. [DOI] [Google Scholar]
- 7.Beris D, Malandraki I, Kektsidou O, Theologidis I, Vassilakos N, Varveri C. 2020. First report of Tomato brown rugose fruit virus infecting tomato in Greece. Plant Dis 104:PDIS-01-20-0212-PDN. doi: 10.1094/PDIS-01-20-0212-PDN. [DOI] [Google Scholar]
- 8.Fidan H, Sarikaya P, Calis O. 2019. First report of Tomato brown rugose fruit virus on tomato in Turkey. New Dis Rep 39:18. doi: 10.5197/j.2044-0588.2019.039.018. [DOI] [Google Scholar]
- 9.Menzel W, Knierim D, Winter S, Hamacher J, Heupel M. 2019. First report of Tomato brown rugose fruit virus infecting tomato in Germany. New Dis Rep 39:1. doi: 10.5197/j.2044-0588.2019.039.001. [DOI] [Google Scholar]
- 10.Panno S, Caruso AG, Davino S. 2019. First report of Tomato brown rugose fruit virus on tomato crops in Italy. Plant Dis 103:1443–1443. doi: 10.1094/PDIS-12-18-2254-PDN. [DOI] [Google Scholar]
- 11.Skelton A, Buxton-Kirk A, Ward R, Harju V, Frew L, Fowkes A, Long M, Negus A, Forde S, Adams IP, Pufal H, McGreig S, Weekes R, Fox A. 2019. First report of Tomato brown rugose fruit virus in tomato in the United Kingdom. New Dis Rep 40:12. doi: 10.5197/j.2044-0588.2019.040.012. [DOI] [Google Scholar]
- 12.Camacho-Beltran E, Perez-Villarreal A, Rodríguez-Negrete EA, Ceniceros-Ojeda E, Leyva-López NE, Mendez-Lozano J. 2019. Occurrence of Tomato brown rugose fruit virus infecting tomato crops in Mexico. Plant Dis 103:1440–1440. doi: 10.1094/PDIS-11-18-1974-PDN. [DOI] [Google Scholar]
- 13.Cambrón-Crisantos JM, Rodríguez-Mendoza J, Valencia-Luna JB, Alcasio-Rangel S, García-Ávila CJ, López-Buenfil JA, Ochoa-Martínez DL. 2018. First report of Tomato brown rugose fruit virus (ToBRFV) in Michoacan, Mexico. Rev Mex Fitopatol 37:185–192. doi: 10.18781/R.MEX.FIT.1810-5. [DOI] [Google Scholar]
- 14.Ling K-S, Tian T, Gurung S, Salati R, Gilliard AC. 2019. First report of Tomato brown rugose fruit virus infecting greenhouse tomato in the United States. Plant Dis 103:1439. doi: 10.1094/PDIS-11-18-1959-PDN. [DOI] [Google Scholar]
- 15.USDA. 2019. Tomato brown rugose fruit virus. USDA Federal Order https://www.aphis.usda.gov/aphis/ourfocus/planthealth/import-information/federal-import-orders/tobrfv/tomato-brown-rugose-fruit-virus.
- 16.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bushnell B. 2017. BBMap: short read aligner, and other bioinformatic tools. https://sourceforge.net/projects/bbmap/files/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The high-throughput sequencing data sets were submitted to the SRA database with the accession no. SRR11794481 (Illumina reads) and SRR11794480 (MinION reads) and the BioProject no. PRJNA632574. The genome sequence of ToBRFV CA18-01 was deposited in GenBank with accession no. MT002973.