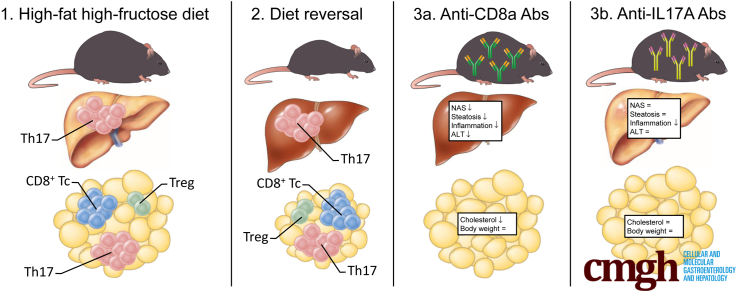
Abstract
Background & Aims
Nonalcoholic steatohepatitis (NASH) is a multisystem condition, implicating liver and adipose tissue. Although the general involvement of the innate and adaptive immune system has been established, we aimed to define the exact role of the functionally diverse T-cell subsets in NASH pathogenesis through diet reversal and immunologic modulation.
Methods
Multiple experimental set-ups were used in 8-week-old C57BL/6J mice, including prolonged high-fat high-fructose diet (HFHFD) feeding, diet reversal from HFHFD to control diet, and administration of anti-CD8a and anti–interleukin 17A antibodies. Plasma alanine aminotransferase, glucose, and lipid levels were determined. Liver and adipose tissue were assessed histologically. Cytotoxic T (Tc), regulatory T, T helper (Th) 1, and Th17 cells were characterized in liver and visceral adipose tissue (VAT) via flow cytometry and RNA analysis.
Results
HFHFD feeding induced the metabolic syndrome and NASH, which coincided with an increase in hepatic Th17, VAT Tc, and VAT Th17 cells, and a decrease in VAT regulatory T cells. Although diet reversal induced a phenotypical metabolic and hepatic normalization, the observed T-cell disruptions persisted. Treatment with anti-CD8a antibodies decreased Tc cell numbers in all investigated tissues and induced a biochemical and histologic attenuation of the HFHFD-induced NASH. Conversely, anti–interleukin 17A antibodies decreased hepatic inflammation without affecting other features of NASH or the metabolic syndrome.
Conclusions
HFHFD feeding induces important immune disruptions in multiple hepatic and VAT T-cell subsets, refractory to diet reversal. In particular, VAT Tc cells are critically involved in NASH pathogenesis, linking adipose tissue inflammation to liver disease.
Keywords: Adipose Tissue Inflammation, Cytotoxic T Cells, Regulatory T Cells, T Helper 1 Cells, T Helper 17 Cells
Abbreviations used in this paper: ALT, alanine aminotransferase; CD, control diet; DR, diet reversal; GTT, glucose tolerance testing; HFHFD, high-fat, high-fructose diet; IL, interleukin; ITT, insulin tolerance testing; MT, Masson’s trichrome; NAFLD, nonalcoholic fatty liver disease; NAS, nonalcoholic fatty liver disease activity score; NASH, nonalcoholic steatohepatitis; PPAR, peroxisome proliferator-activated receptor; SAT, subcutaneous adipose tissue; Tc, cytotoxic T; Th, T helper; Treg, regulatory T; VAT, visceral adipose tissue
Graphical abstract
Summary.
In this article, we show a critical role for differentially active T-cell subsets in visceral adipose tissue and liver in the pathogenesis of nonalcoholic steatohepatitis using multiple interventional experimental approaches in a high-fat high-fructose diet mouse model.
Nonalcoholic fatty liver disease (NAFLD) comprises a spectrum of gradually progressive disease states, characterized by hepatic steatosis. Nonalcoholic steatohepatitis (NASH) constitutes the more active form and is defined histologically by the presence of both hepatocellular ballooning, a sign of cell damage, and lobular inflammation. NASH may progress to increasing degrees of liver fibrosis, eventually leading to cirrhosis with all of its associated complications, underlining the unmet need for an efficacious medical treatment. A strong association exists between NAFLD and the metabolic syndrome, clustering visceral overweight, dyslipidemia, insulin resistance, and arterial hypertension, which has led to the assumption that the former is actually the hepatic manifestation of the latter. The pathogenesis of NASH involves multiple organ systems, including a reciprocal interaction between the liver and the adipose tissue, and entails multiple parallel hits, implicating both the innate and adaptive immune system.1
T helper (Th)1, Th17, regulatory T (Treg), and cytotoxic T (Tc) cells are differentially active T-cell subsets that exert complementary roles in physiologic conditions, most importantly in the defense against, mostly intracellular, pathogens and neoplasms. In addition, these cells are involved in the pathogenesis of various immune-mediated diseases. Concerning NAFLD, evidence has accumulated for an overall tempering effect for Treg cells, whereas Th17 and Tc cells seem to induce more liver damage and fibrosis progression. However, several studies have presented conflicting evidence and the exact role of the interplay between the adipose tissue and the liver concerning these cells still is unclear, which prompted further exploration.1 In this study, we aimed to characterize the involvement of Th1, Th17, Treg, and Tc cells in both liver and adipose tissue in a high-fat high-fructose diet (HFHFD) mouse model of NAFLD, thereby exploring the involved mechanisms using longitudinal analyses, phenotypical manipulation by means of a diet reversal (DR), and the administration of immune-altering antibodies.
Results
HFHFD Feeding Induced Metabolic Disruption, Adipose Tissue Inflammation, and NAFLD
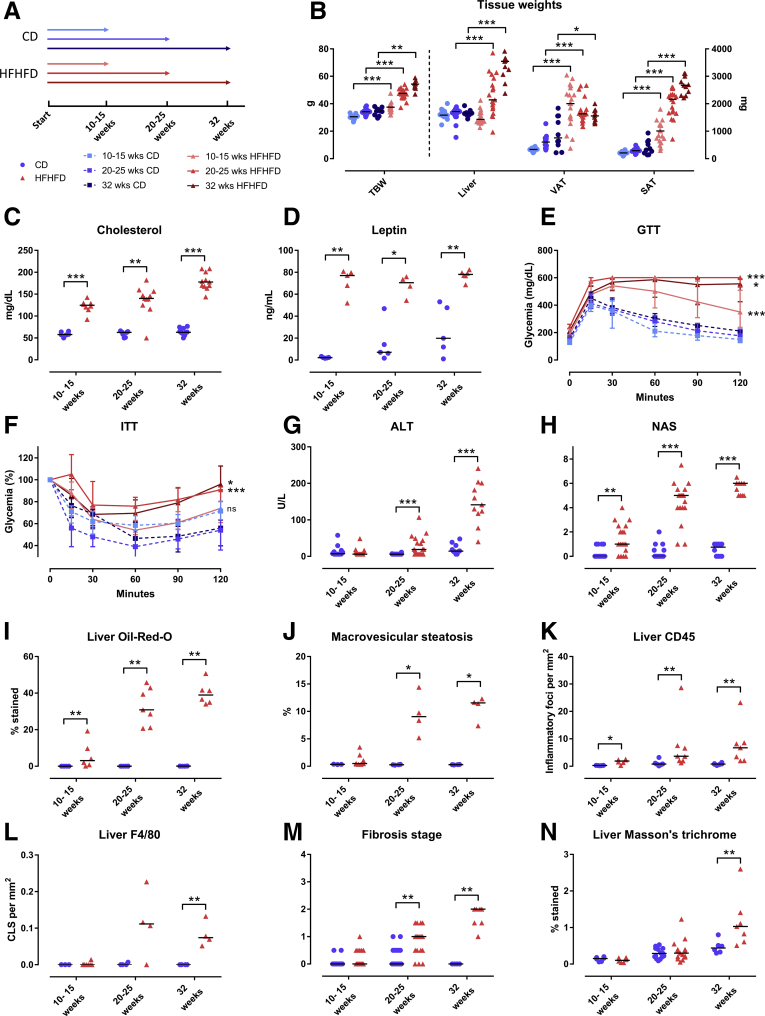
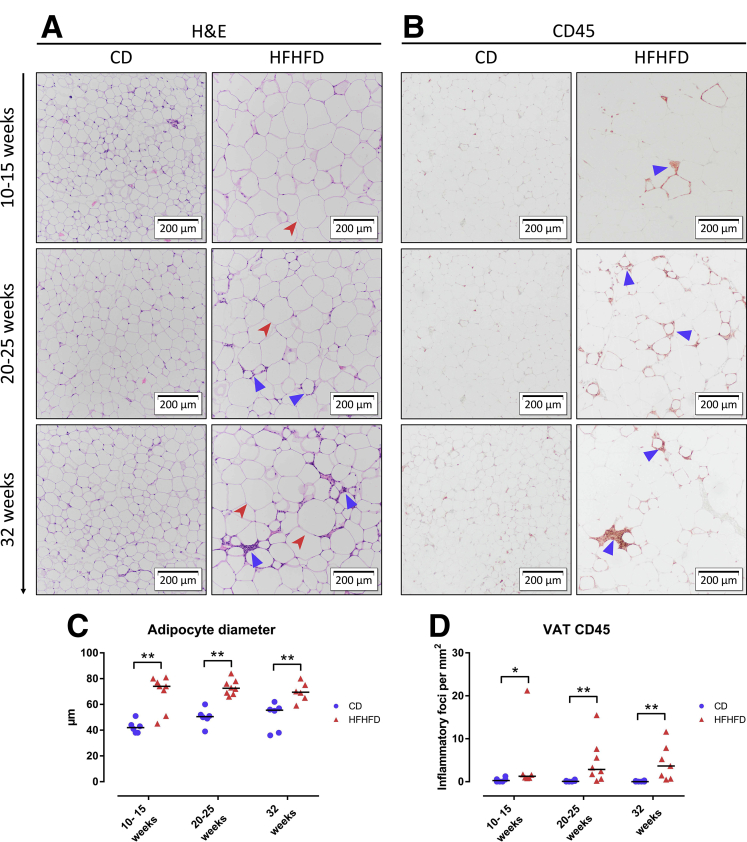
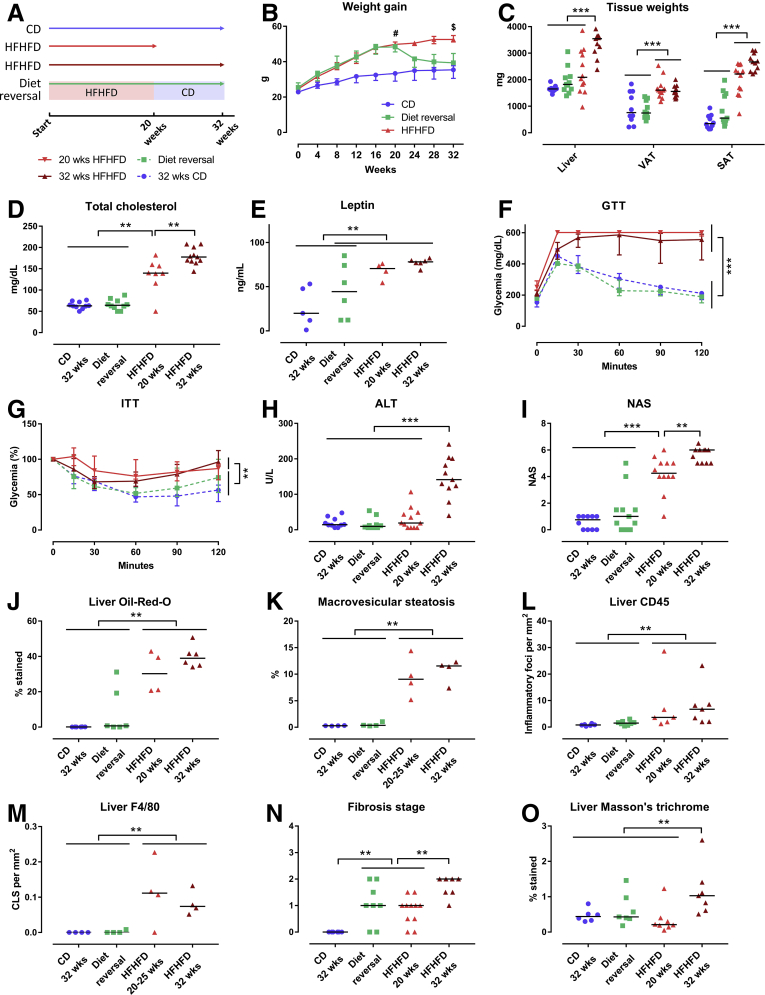
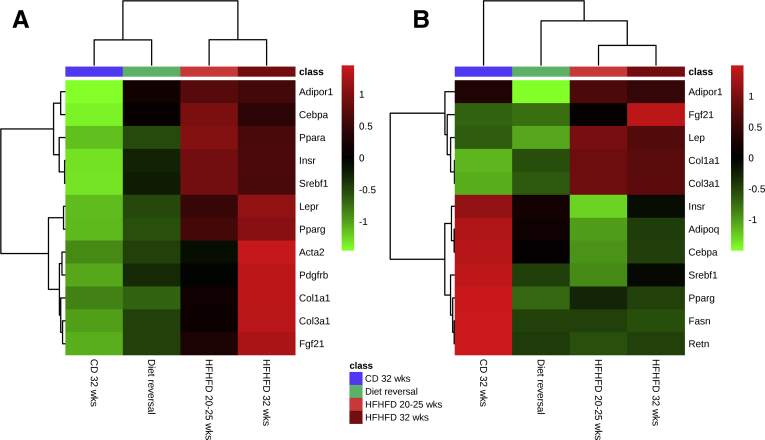
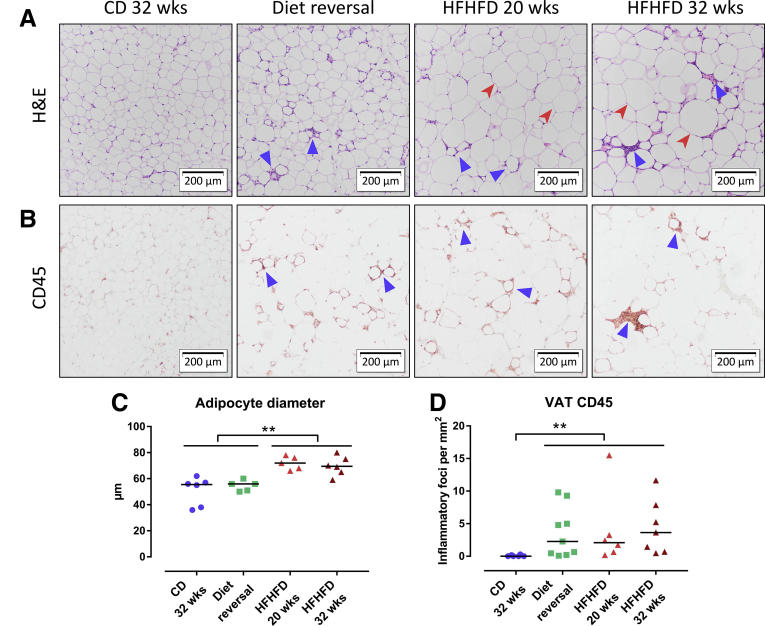
HFHFD feeding for 10–32 weeks (Figure 1A) resulted in progressively increasing total body weight (Figure 1B), adipose tissue mass (Figure 1B), plasma cholesterol (Figure 1C), leptin (Figure 1D), adiponectin, fasting glucose, low-density lipoprotein cholesterol, and high-density lipoprotein cholesterol levels (Table 1). Furthermore, glucose tolerance testing (GTT) and insulin tolerance testing (ITT) showed decreased glucose tolerance from 10 to 15 weeks and decreased insulin sensitivity from 20 to 25 weeks based on the area under the curve (Figure 1E and F). Plasma alanine aminotransferase (ALT) levels (Figure 1G) and liver weight (Figure 1B) were significantly higher in HFHFD-fed mice compared with control diet (CD)-fed mice from 20 to 25 weeks onward. Histologic assessment of the livers of HFHFD-fed mice showed progressive stages of NAFLD, including features of NASH, with increasing NAFLD activity score (NAS) the longer the HFHFD was administered (Figures 1H–L and 2A–C). Moreover, from 20 to 25 weeks significantly more fibrosis was observed (Figures 1M and N and 2D). Hepatic RNA analysis showed the up-regulation of regulatory metabolic genes from 20 to 25 weeks in HFHFD-fed mice (Figure 3A), including Ppara, Pparg, Fgf21, Cebpa, and Srebf1, respectively, coding for peroxisome proliferator-activated receptor (PPAR) α, PPARγ, fibroblast growth factor 21, CCAAT/enhancer-binding protein α (C/EBPα), and sterol regulatory element-binding transcription factor 1 (SREBF1). Moreover, Acta2 and Pdgfrb, genes associated with hepatic stellate cell activation, as well as Col1a1 and Col1a3, also were up-regulated, pointing to hepatic collagen formation. Histologic and immunohistochemical assessment of the visceral adipose tissue (VAT) showed the development of adipocyte hypertrophy and increasing amounts of inflammatory foci (Figure 4). VAT RNA analysis showed the down-regulation of regulatory metabolic genes, including Adipoq, Retn, Fasn, and Pparg, coding for adiponectin, resistin, fatty acid synthase, and PPARγ, respectively (Figure 3B). Conversely, the following genes were up-regulated: Lep, Adipor1, and Fgf21, coding for leptin, adiponectin receptor 1, and fibroblast growth factor 21, respectively. Interestingly, the expression of Col1a1 and Col3a1 also were increased, suggesting that collagen formation is activated not only in the liver, but also in the VAT.
Figure 1.
Effect of HFHFD feeding on phenotype. (A) Study design and legend. (B) Total body weight and tissue weights. (C) Plasma cholesterol. (D) Plasma leptin. (E) GTT, absolute glycemia in mg/dL, and (F) ITT, percentage of glycemia at 0 minutes. The areas under the curve were compared between the HFHFD-fed and CD-fed mice for the following groups: 10–15 weeks, 20–25 weeks, and 32 weeks. Medians with interquartile range are shown. (G) Plasma ALT. (H) NAS. (I) Quantification of hepatic steatosis through Oil-red-O staining, expressed as the percentage stained. (J) Morphometric quantification of macrovesicular steatosis on H&E-stained liver tissue, expressed as the percentage of macrovesicular steatosis. (K and L) Quantification of hepatic inflammation through (K) CD45 and (L) F4/80 immunohistochemistry, expressed as the number of inflammatory foci and crown-like structures per mm2, respectively. (M) Fibrosis stage according to NASH-clinical research network, evaluated on MT-stained liver tissue. (N) Quantification of fibrosis on MT-stained liver tissue, expressed as the percentage stained. N = 10–20 per group. Medians are depicted with a horizontal black line. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 (Mann–Whitney U). TBW, total body weight.
Table 1.
Effect of HFHFD Feeding on Plasma Biochemistry
| Biochemical plasma analysis | CD | HFHFD | P |
|---|---|---|---|
| AST, U/L | |||
| 10–15 wk | 172 (124–225) | 127 (106–166) | .423 |
| 20–25 wk | 107 (88–147) | 244 (199–326) | <.001 |
| 32 wk | 110 (76-188) | 318 (266-457) | <.001 |
| ALT, U/L | |||
| 10–15 wk | 7 (6–13) | 6 (6–12) | .568 |
| 20–25 wk | 6 (6–6) | 19 (6–40) | <.001 |
| 32 wk | 15 (13–30) | 141 (121–191) | <.001 |
| Fasted glycemia, mg/dL | |||
| 10–15 wk | 128 (119–149) | 188 (159–213) | <.001 |
| 20–25 wk | 149 (130–173) | 236 (220–255) | <.001 |
| 32 wk | 155 (135–172) | 197 (179–225) | .002 |
| Total cholesterol, mg/dL | |||
| 10–15 wk | 58 (57–62) | 125 (119–129) | <.001 |
| 20–25 wk | 63 (55–64) | 140 (132–152) | .002 |
| 32 wk | 63 (61–70) | 178 (169–192) | <.001 |
| LDL, mg/dL | |||
| 10–15 wk | 7 (4–9) | 16 (13–17) | .051 |
| 20–25 wk | 7 (6–8) | 24 (19–30) | <.001 |
| 32 wk | 6 (5–8) | 26 (22–30) | <.001 |
| HDL, mg/dL | |||
| 10–15 wk | 48 (38–53) | 97 (84–106) | .014 |
| 20–25 wk | 41 (39–49) | 87 (64–133) | .035 |
| 32 wk | 59 (55–64) | 138 (116–154) | <.001 |
| Triglycerides, mg/dL | |||
| 10–15 wk | 115 (91–141) | 71 (56–91) | .073 |
| 20–25 wk | 93 (74–129) | 54 (48–74) | .035 |
| 32 wk | 107 (94–120) | 67 (64–76) | .016 |
| Leptin, ng/mL | |||
| 10–15 wk | 2 (2–2) | 77 (68–80) | .008 |
| 20–25 wk | 7 (6–14) | 70 (64–74) | .016 |
| 32 wk | 20 (12–48) | 78 (77–79) | .004 |
| Adiponectin, μg/mL | |||
| 10–15 wk | 41 (38–43) | 70 (61–74) | .004 |
| 20–25 wk | 45 (43–51) | 60 (59–61) | .030 |
| 32 wk | 64 (49–65) | 58 (55–59) | .931 |
NOTE. Data are presented as the median (interquartile range). Statistical analysis was performed with the Mann–Whitney U test. Significant P values are in bold.
AST, aspartate aminotransferase; HDL, high-density lipoprotein cholesterol; LDL low-density lipoprotein cholesterol.
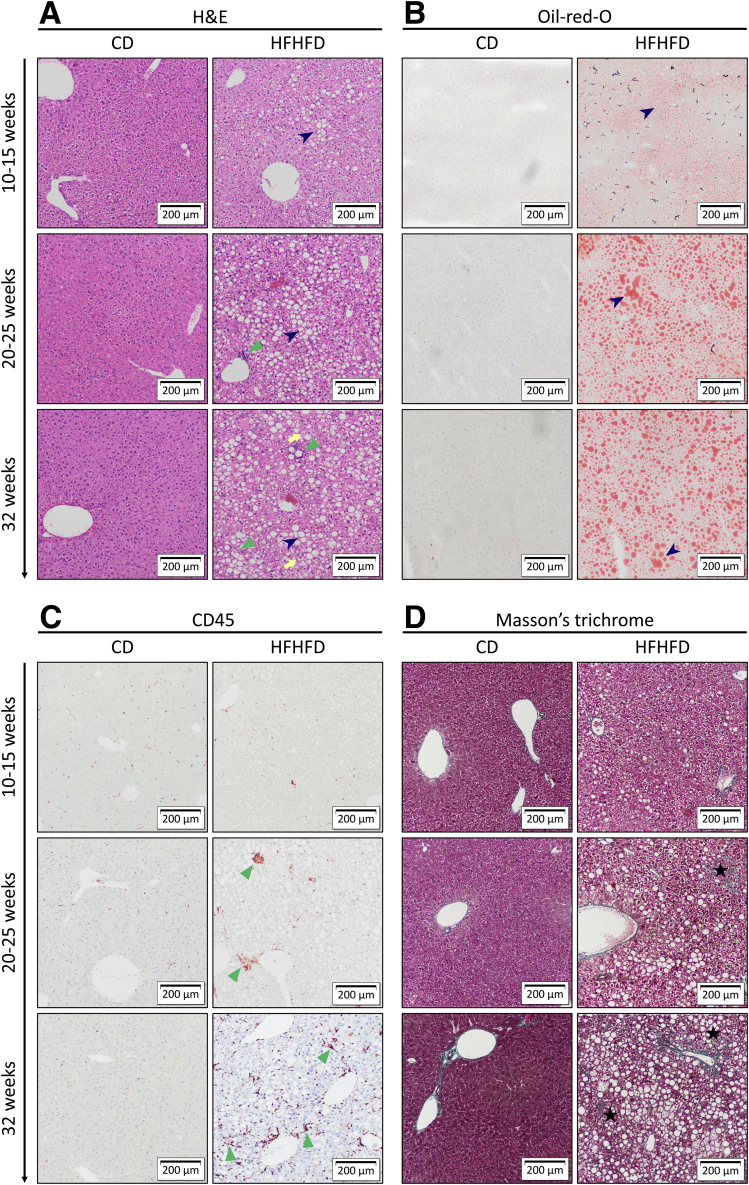
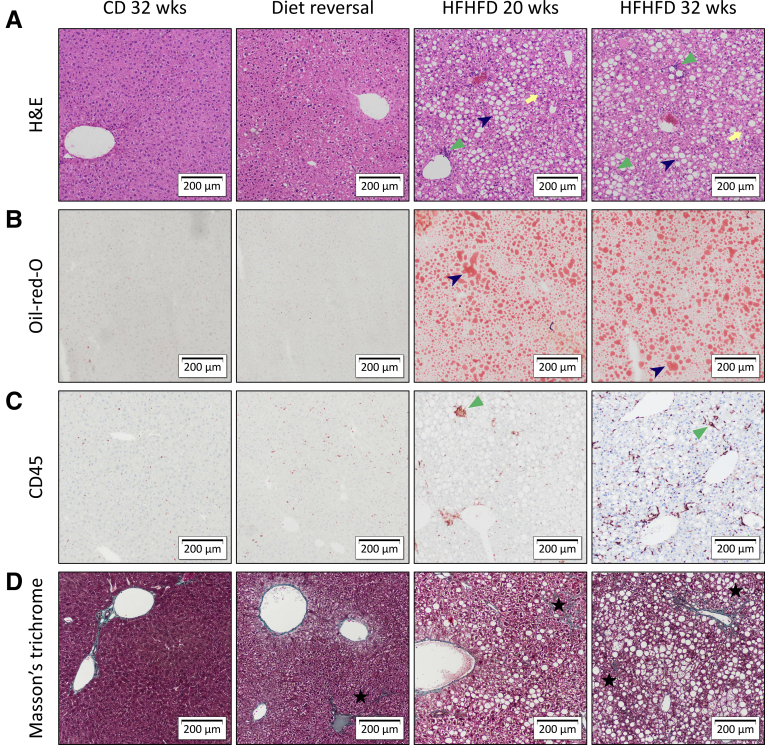
Figure 2.
Representative images of liver histology showing the effect of HFHFD feeding. Original magnification: ×10. Compared with CD-fed mice, HFHFD-fed mice showed increasing degrees of hepatic steatosis (dark blue arrowhead), ballooning (yellow arrow), lobular inflammation (green triangle), and fibrosis (black star). Staining was performed with (A) H&E, (B) Oil-red-O, (C) CD45 immunohistochemistry, and (D) MT.
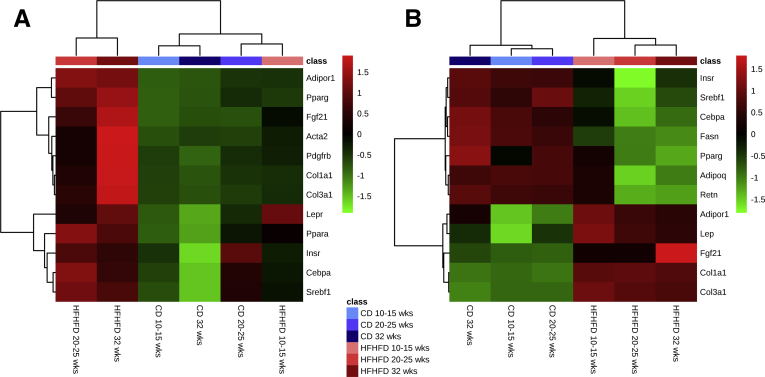
Figure 3.
Effect of HFHFD feeding on RNA expression of selected genes related to metabolism and fibrosis in (A) liver and (B) VAT. N = 5-6 per group. The top 12 most significant genes according to the Mann–Whitney U test are shown.
Figure 4.
Effect of HFHFD feeding on VAT histology. Representative images of VAT stained with (A) H&E and (B) CD45 immunohistochemistry. Original magnification: ×10. Compared with CD-fed mice, HFHFD-fed mice showed increasing degrees of adipocyte hypertrophy (red arrowhead) and adipose tissue inflammation (blue triangle). (C) Adipocyte diameter. (D) Quantification of VAT inflammation through CD45 immunohistochemistry, expressed as the number of inflammatory foci per mm2.
HFHFD Feeding Induced Concomitant Alterations in T-Cell Subsets in Liver and VAT
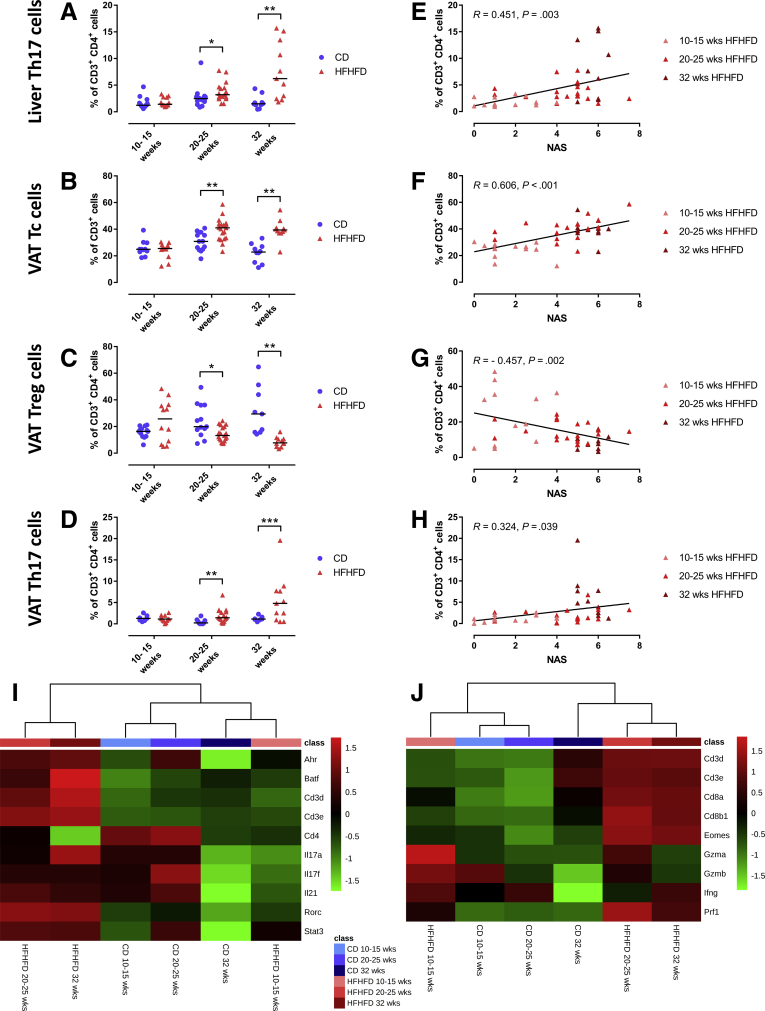
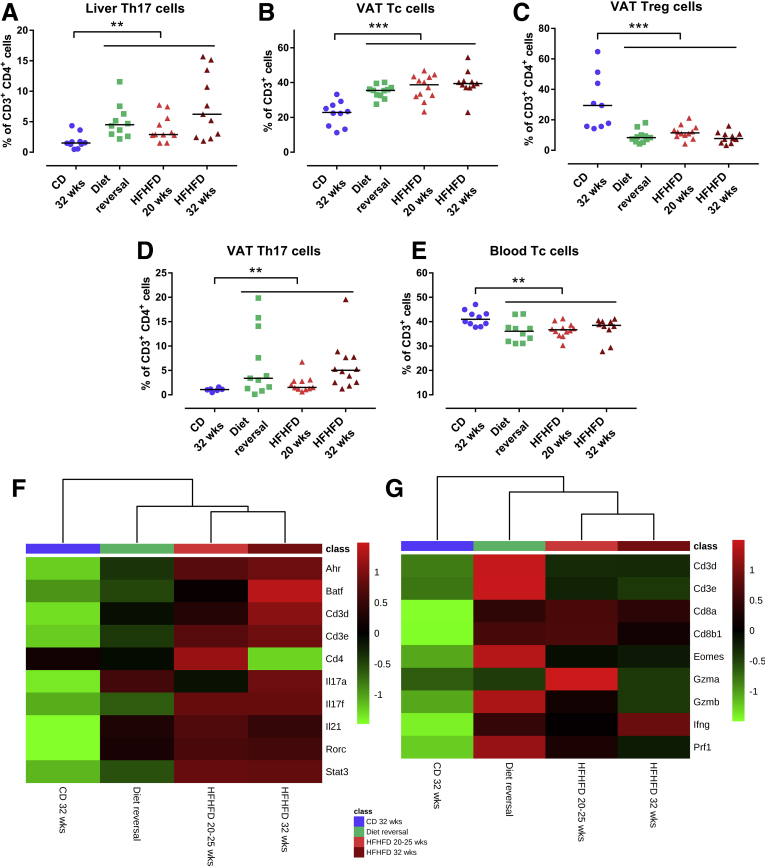
Tc, Th1, Th17, and Treg cells were quantified in liver, VAT, subcutaneous adipose tissue (SAT), blood, and spleen (Table 2). HFHFD feeding increased the proportion of hepatic Th17 cells compared with CD-fed mice from 20 to 25 weeks onward (Figure 5A). At the level of the VAT, HFHFD-fed mice showed increased proportions of Th17 and Tc cells compared with CD-fed mice, whereas the proportion of Treg cells decreased from 20 to 25 weeks onward (Figure 5B–D). Moreover, Tc cells were reduced in blood and spleen from 20 to 25 weeks and 32 weeks, respectively. Lastly, blood Th17 cells were expanded at 32 weeks of HFHFD feeding. No significant changes were observed at the level of the SAT. Furthermore, when exclusively considering the subgroup of HFHFD-fed mice, a strong positive correlation existed between the NAS and VAT Tc cells, as well as positive correlations between NAS and liver and VAT Th17 levels. In parallel, a strong negative correlation existed between VAT Treg cells and the NAS (Figure 5E–H). RNA clustering analysis of liver and VAT confirmed HFHFD-induced increase in liver Th17 cells, particularly through up-regulation of Rorc, coding for the canonical transcription factor RAR-related orphan receptor γt (RORγt), and VAT Tc cells, through up-regulation of Cd8a, Cd8b1, and Eomes (Figure 5I and J). Moreover, pathway analysis showed enrichment of the Th17 activation pathway in both liver (at 20–25 weeks and at 32 weeks, P < 10-12 and P < 10-18, respectively) and VAT (at 10–15 and 20–25 weeks, P < 10-9 and P < 10-4, respectively), as well as enrichment of the cytotoxic T-lymphocyte-associated protein 4 (CTLA4) signaling pathway in Tc cells in VAT (at 10–15 and 20–25 weeks, P < 10-4 and P < 10-5, respectively). Plasma cytokine analysis showed an increase in tumor necrosis factor α in HFHFD-fed mice at 32 weeks, but failed to show any systemic changes in other T-cell–associated cytokines (Table 3).
Table 2.
Flow Cytometric Characterization of Tc, Treg, Th1, and Th17 Cells in Liver, VAT, SAT, Blood, and Spleen
| Liver |
VAT |
SAT |
Blood |
Spleen |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD | HFHFD | P | CD | HFHFD | P | CD | HFHFD | P | CD | HFHFD | P | CD | HFHFD | P | |
| Tc cells, % of XXX cells |
|||||||||||||||
| 10–15 wk | 41.4 | 46.6 | .021 | 27.4 | 25.5 | .569 | 37.1 | 39.4 | .120 | 37.2 | 34.7 | .217 | 34.1 | 32.9 | .061 |
| (36.8–44.9) | (38.0–52.2) | (24.0–30.1) | (19.3–28.3) | (27.6–40.0) | (33.9–45.6) | (34.8–39.6) | (33.7–39.9) | (32.6–38.2) | (29.7 - 35.8) | ||||||
| 20–25 wk | 46.9 | 42.5 | .336 | 26.3 | 41.2 | .002 | 46.6 | 41.1 | .172 | 40.8 | 37.2 | .006 | 33.0 | 34.1 | .509 |
| (36.5–52.9) | (40.2–46.6) | (24.5–35.4) | (33.3–44.1) | (45.3–48.0) | (37.2–44.4) | (38.8–44.2) | (35.9–40.4) | (31.1–39.1) | (29.7 - 36.5) | ||||||
| 32 wk | 49.2 | 49.5 | .809 | 18.7 | 40.1 | <.001 | 34.8 | 38.8 | .442 | 41.0 | 38.5 | .049 | 33.6 | 29.7 | .043 |
| (44.5–54.6) | (47.6–53.9) | (12.7–22.6) | (38.5–43.7) | (28.3–44.9) | (35.1–49.5) | (38.9–43.7) | (34.9–40.2) | (28.9–38.3) | (24.5 - 33.8) | ||||||
| Treg cells, % of XXX cells |
|||||||||||||||
| 10–15 wk | 1.6 | 1.6 | .899 | 16.6 | 18.9 | .270 | 8.9 | 5.7 | 1.000 | 5.3 | 4.3 | .611 | 5.7 | 6.0 | .830 |
| (0.8–2.3) | (1.1–2.9) | (13.2–20.5) | (6.4–36.4) | (5.0–10.9) | (4.6–11.3) | (3.0–6.7) | (3.5–6.5) | (3.8–13.7) | (4.3 - 8.2) | ||||||
| 20–25 wk | 2.3 | 2.2 | .790 | 19.9 | 12.7 | .022 | 3.1 | 7.9 | .432 | 6.3 | 7.3 | .083 | 3.1 | 3.6 | .478 |
| (1.8–3.2) | (1.7–3.5) | (13.7–36.2) | (9.7–19.1) | (2.8–3.4) | (4.1–10.0) | (0.9–7.2) | (4.0–8.2) | (2.3–5.7) | (2.7 - 5.4) | ||||||
| 32 wk | 1.9 | 4.4 | .055 | 30.9 | 7.8 | <.001 | 6.9 | 6.3 | .791 | 6.9 | 7.8 | .512 | 2.1 | 5.0 | .604 |
| (1.0–3.0) | (2.7–7.7) | (15.4–54.7) | (4.7–10.6) | (5.1–12.2) | (5.1–9.0) | (4.0–7.7) | (5.7–8.5) | (1.0–7.1) | (0.9 - 13.1) | ||||||
| Th1 cells, % of XXX cells |
|||||||||||||||
| 10–15 wk | 1.1 | 1.8 | .280 | 0.9 | 0.9 | .711 | 2.5 | 1.2 | .238 | 1.9 | 1.5 | .376 | 2.2 | 1.9 | .941 |
| (0.8–1.5) | (0.6–3.8) | (0.9–5.4) | (0.7–3.3) | (1.6–7.6) | (0.9–7.3) | (1.8–2.4) | (0.7–2.8) | (1.5–2.8) | (1.7 - 3.5) | ||||||
| 20–25 wk | 1.0 | 0.7 | .852 | 0.8 | 0.6 | .494 | 2.2 | 1.1 | .615 | 3.9 | 2.9 | .215 | 2.8 | 3.7 | .714 |
| (0.4–2.9) | (0.5–1.5) | (0.1–1.3) | (0.3–0.9) | (0.6–3.8) | (0.8–1.7) | (2.9–5.0) | (2.1–4.7) | (1.6–5.2) | (1.8 - 4.0) | ||||||
| 32 wk | 0.6 | 0.7 | .809 | 0.8 | 0.5 | .151 | 3.2 | 1.4 | .375 | 2.5 | 1.6 | .512 | 2.3 | 2.2 | 1.000 |
| (0.4–2.5) | (0.4–1.2) | (0.6–2.2) | (0.3–0.9) | (1.7–6.3) | (1.0–3.4) | (0.6–3.1) | (0.8–5.1) | (0.9–4.3) | (1.4 - 5.9) | ||||||
| Th17 cells, % of XXX cells |
|||||||||||||||
| 10–15 wk | 1.2 | 1.4 | .202 | 1.1 | 1.1 | .711 | 1.7 | 2.6 | .272 | 1.3 | 1.2 | .538 | 2.0 | 0.6 | .370 |
| (0.9–1.6) | (1.3–2.8) | (0.8–1.7) | (0.8–2.0) | (0.6–5.6) | (1.4–5.8) | (0.6–3.3) | (0.9–2.2) | (0.3–5.4) | (0.4 - 2.7) | ||||||
| 20–25 wk | 2.5 | 2.9 | .018 | 0.3 | 1.4 | .001 | 0.3 | 1.8 | .198 | 1.3 | 1.3 | .837 | 0.8 | 0.9 | .914 |
| (2.0–2.9) | (2.5–4.4) | (0.0–1.0) | (0.9–2.9) | (0.0–0.6) | (1.1–3.4) | (0.6–2.6) | (0.7–3.2) | (0.5–7.2) | (0.4 - 2.0) | ||||||
| 32 wk | 1.5 | 3.0 | <.001 | 1.1 | 3.9 | <.001 | 0.9 | 2.5 | .085 | 2.0 | 3.3 | .010 | 1.1 | 2.4 | 1.000 |
| (1.1–2.2) | (2.2–7.6) | (0.8–1.3) | (2.2–6.5) | (0.2–1.2) | (1.0–4.0) | (1.0–3.0) | (2.8–3.5) | (0.5–4.7) | (1.0 - 3.0) | ||||||
NOTE. Data are presented as medians (interquartile range). Statistical analysis was performed with a Mann–Whitney U test. Significant P values are in bold.
Figure 5.
HFHFD-induced T-cell alterations in liver and VAT. Comparisons of (A) liver Th17, (B) VAT Tc, (C) VAT Treg, and (D) VAT Th17 cells between CD-fed and HFHFD-fed mice at 10–15, 20–25, and 32 weeks. Correlations in HFHFD-fed mice between NAS and (E) liver Th17, (F) VAT Tc, (G) VAT Treg, and (H) VAT Th17 cells. T-cell subsets were characterized via flow cytometry. (I and J) Heat maps showing (I) hepatic RNA expression of Th17 cell–related genes and (J) VAT RNA expression of Tc cell–related genes. Medians are shown with a horizontal black line. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 (Mann–Whitney U), N = 10–20 per group.
Table 3.
Effect of HFHFD Feeding on Plasma Cytokine Concentrations
| Cytokine concentrations in plasma | CD | HFHFD | P |
|---|---|---|---|
| IFNγ, pg/mL | |||
| 10–15 wk | 2.24 (2.24–2.24) | 2.24 (2.00–7.98) | .429 |
| 20–25 wk | 2.24 (2.24–3.49) | 2.24 (2.24–2.24) | .662 |
| 32 wk | 2.24 (2.24–2.86) | 2.24 (2.24–2.24) | .537 |
| IL5, pg/mL | |||
| 10–15 wk | 3.05 (2.15–3.20) | 3.00 (3.00–3.23) | 1.000 |
| 20–25 wk | 2.43 (2.15–2.49) | 2.15 (2.15–5.50) | .931 |
| 32 wk | 2.15 (2.15–2.15) | 3.32 (2.38–4.36) | .082 |
| TNFα, pg/mL | |||
| 10–15 wk | 2.47 (2.17–2.50) | 2.72 (3.00–5.41) | .247 |
| 20–25 wk | 3.51 (3.33–3.65) | 4.28 (2.89–5.58) | .662 |
| 32 wk | 3.30 (2.11–3.51) | 8.38 (6.47–9.31) | .030 |
| IL2, pg/mL | |||
| 10–15 wk | 2.04 (1.75–2.17) | 1.47 (1.00–2.94) | .662 |
| 20–25 wk | 4.83 (2.98–5.65) | 1.35 (1.07–3.19) | .177 |
| 32 wk | 1.20 (1.02–1.46) | 1.99 (1.44–2.94) | .177 |
| IL6, pg/mL | |||
| 10–15 wk | 2.51 (2.51–2.83) | 3.51 (4.00–15.74) | .126 |
| 20–25 wk | 8.47 (4.45–26.02) | 5.11 (4.56–31.84) | .792 |
| 32 wk | 4.58 (2.51–6.15) | 4.93 (3.93–9.83) | 1.000 |
| IL4, pg/mL | |||
| 10–15 wk | 1.62 (1.62–1.62) | 1.62 (2.00–1.62) | .662 |
| 20–25 wk | 1.62 (1.62–1.62) | 1.62 (1.62–1.62) | .662 |
| 32 wk | 1.62 (1.62–1.62) | 1.62 (1.62–1.71) | .429 |
| IL10, pg/mL | |||
| 10–15 wk | 7.21 (7.21–7.21) | 7.21 (7.00–7.21) | .662 |
| 20–25 wk | 18.22 (7.21–33.77) | 7.21 (7.21–7.21) | .177 |
| 32 wk | 7.21 (7.21–24.68) | 19.48 (7.91–29.83) | .537 |
| IL9, pg/mL | |||
| 10–15 wk | 1.08 (1.08–2.89) | 2.37 (2.00–2.58) | .792 |
| 20–25 wk | 2.25 (1.62–2.91) | 1.09 (1.08–7.92) | .429 |
| 32 wk | 1.08 (1.08–1.12) | 1.56 (1.33–1.97) | .126 |
| IL17A, pg/mL | |||
| 10–15 wk | 2.66 (2.66–2.66) | 2.75 (3.00–3.31) | .537 |
| 20–25 wk | 4.13 (3.77–5.59) | 2.66 (2.66–3.67) | .329 |
| 32 wk | 2.66 (2.66–5.00) | 4.35 (2.88–6.74) | .429 |
| IL17F, pg/mL | |||
| 10–15 wk | 1.58 (1.58–1.58) | 1.58 (2.00–1.58) | .662 |
| 20–25 wk | 1.86 (1.58–2.11) | 1.58 (1.58–1.58) | .177 |
| 32 wk | 1.58 (1.58–2.93) | 1.58 (1.58–2.36) | .792 |
| IL21, pg/mL | |||
| 10–15 wk | 3.02 (3.02–3.02) | 3.02 (3.00–3.02) | .662 |
| 20–25 wk | 3.02 (3.02–3.02) | 3.02 (3.02–3.02) | 1.000 |
| 32 wk | 3.02 (3.02–3.02) | 3.02 (3.02–3.02) | .662 |
| IL22, pg/mL | |||
| 10–15 wk | 8.26 (4.44–14) | 4.44 (4.00–5.69) | .329 |
| 20–25 wk | 9.28 (4.44–14.74) | 4.44 (4.44–4.44) | .126 |
| 32 wk | 4.44 (4.44–16.5) | 4.88 (4.44–5.42) | .792 |
| IL13, pg/mL | |||
| 10–15 wk | 5.12 (5.12–5.12) | 5.12 (5.00–5.12) | .662 |
| 20–25 wk | 5.12 (5.12–5.12) | 5.12 (5.12–5.12) | 1.000 |
| 32 wk | 5.12 (5.12–5.12) | 5.12 (5.12–5.12) | 1.000 |
NOTE. Data are presented as medians (interquartile range). Statistical analysis was performed with the Mann–Whitney U test. Significant P values are in bold.
IFNγ, interferon γ; TNFα, tumor necrosis factor α.
Diet Reversal Reverted Most of the HFHFD-Induced Metabolic Changes Toward a CD-Like Phenotype
A DR was performed by substituting the HFHFD with CD after 20 weeks, subsequently continuing the CD for an additional 12 weeks (Figure 6A). Compared with mice that were fed the HFHFD for 20 and 32 weeks, DR mice showed a significantly lower body weight (Figure 6B), adipose tissue mass (Figure 6C), and plasma cholesterol levels (Figure 6D), returning to levels that were not significantly different from mice fed the CD for 32 weeks (Table 4). Plasma leptin levels seemed to decrease as well, although a level of significance was not reached (Figure 6E). Moreover, GTT and ITT showed improved glucose tolerance and insulin sensitivity in DR mice, again reverting to levels not significantly different from CD-fed mice (Figure 6F and G). Plasma ALT levels and liver weight were significantly lower in DR mice compared with mice fed the HFHFD for 32 weeks (Figure 6C and H). Furthermore, DR improved the histologic degree of severity as assessed by a decrease in NAS compared with mice fed the HFHFD for 20 and 32 weeks (Figures 6I–M and 7A–C). Importantly, the NAS was not significantly different between DR mice and mice fed the CD for 32 weeks. In addition, automated quantification on Masson’s trichrome (MT)-stained liver tissue showed a decrease in fibrosis, although this effect was not observed when the tissue was evaluated by means of the semiquantitative NASH-clinical research network grading system (Figures 6N and O and 7D). Moreover, hepatic RNA analysis of the fibrosis-related genes Acta2, Col1a1, and Col1a3 also reverted to normal (Figure 8A). Together, these findings suggest a normalization of the collagen content and production, despite the presence of some residual fibrotic strains. Hepatic RNA expression of most metabolic genes that were up-regulated by HFHFD feeding also decreased to normal levels, including Ppara, Pparg, and Fgf21 (Figure 8A). Histologic assessment of the VAT showed a resolution of adipocyte hypertrophy (Figure 9A and C). VAT RNA analysis showed a decrease in expression of most genes that were up-regulated by HFHFD feeding, including Lep and Fgf21. Interestingly, this normalization was not seen in the down-regulated genes, including Retn, Fasn, and Pparg (Figure 8B).
Figure 6.
Effect of diet reversal on phenotype. (A) Study design and legend. (B) Evolution total body weight. (C) Tissue weights. (D) Plasma cholesterol. (E) Plasma leptin. (F) GTT, absolute glycemia in mg/dL, and (G) ITT, percentage of glycemia at 0 minutes. The area under the curve was compared between the different groups. Medians with interquartile range are shown. (H) Plasma ALT. (I) NAS. N = 10–20 per group. (J) Quantification of hepatic steatosis through Oil-red-O staining, expressed as the percentage stained. (K) Morphometric quantification of macrovesicular steatosis on H&E-stained liver tissue, expressed as the percentage of macrovesicular steatosis. (L and M) Quantification of hepatic inflammation through (L) CD45 and (M) F4/80 immunohistochemistry, expressed as the number of inflammatory foci and crown-like structures per mm2 respectively. (N) Fibrosis grade according to NASH-clinical research network. (O) Quantification of fibrosis through MT staining, expressed as the percentage stained. Medians are depicted with a horizontal black line. #P < .001, HFHFD-fed mice (including the DR group, before DR) vs CD-fed mice. $P < .01, 32 weeks HFHFD group vs the CD and DR groups. ∗∗P < .01, ∗∗∗P < .001 (Kruskal–Wallis, homogenous subsets are shown with a horizontal line above the data points).
Table 4.
Effect of Diet Reversal on Plasma Biochemistry
| Biochemical plasma analysis | CD 32 wk (1) | Diet reversal (2) | HFHFD, 20 wk (3) | HFHFD, 32 wk (4) | P | P, (1) vs (2) | P, (2) vs (3) | P (2) vs (4) |
|---|---|---|---|---|---|---|---|---|
| AST, U/L | 110 (76–188) | 182 (113–259) | 245 (212–322) | 318 (266–457) | .002 | .165 | .228 | .013 |
| ALT, U/L | 15 (13–30) | 10 (6–14) | 19 (6–45) | 141 (121–191) | <.001 | .278 | .387 | <.001 |
| Fasted glycemia, mg/dL | 155 (135–172) | 178 (156–185) | 236 (220–257) | 197 (179–225) | <.001 | .159 | <.001 | .032 |
| Total cholesterol, mg/dL | 63 (61–70) | 64 (60–72) | 140 (133–157) | 178 (169–192) | <.001 | .853 | .004 | <.001 |
| LDL, mg/dL | 6 (5–8) | 8 (7–9) | 21 (17–32) | 26 (22–30) | <.001 | .123 | <.001 | <.001 |
| HDL, mg/dL | 59 (55–64) | 65 (51–80) | 112 (81–137) | 138 (116–154) | <.001 | .436 | .122 | .001 |
| Triglycerides, mg/dL | 107 (94–120) | 135 (98–149) | 61 (51–72) | 67 (64–76) | <.001 | .247 | <.001 | <.001 |
| Leptin, ng/mL | 20 (12–48) | 44 (18–69) | 70 (69–72) | 78 (77–79) | .019 | .247 | .643 | .093 |
| Adiponectin, μg/mL | 64 (49–65) | 56 (51–60) | 61 (60–61) | 58 (55–59) | .717 |
NOTE. Data are presented as medians (interquartile range). Statistical analysis was performed with the Kruskal–Wallis test according to a stepwise step-down method to determine homogenous subsets. Significant P values are in bold.
AST, aspartate aminotransferase.
Figure 7.
Representative images of liver histology showing the effect of diet reversal. Original magnification: ×10. Diet reversal induced resolution of hepatic steatosis (blue arrowhead), inflammation (green triangle), and ballooning (yellow arrow), whereas fibrosis persisted (black star). Staining was performed with (A) H&E, (B) Oil-red-O, (C) CD45 immunohistochemistry, and (D) MT.
Figure 8.
Effect of DR on RNA expression of selected genes related to metabolism and fibrosis in (A) liver and (B) VAT. N = 5-6 per group. The top 12 most significant genes according to the Mann–Whitney U test are shown.
Figure 9.
Effect of diet reversal on VAT histology. Representative images of VAT stained with (A) H&E and (B) CD45 immunohistochemistry. Original magnification: ×10. Although diet reversal reduced adipocyte hypertrophy (red arrowhead), adipose inflammation persisted (blue triangle). (C) Adipocyte diameter. (D) Quantification of VAT inflammation through CD45 immunohistochemistry, expressed as the number of inflammatory foci per mm2. Medians are shown with a horizontal black line. ∗∗P < .01 (Kruskal–Wallis, homogenous subsets are shown with a horizontal line above the data points).
The HFHFD-Induced T-Cell Alterations Were Not Affected by the DR-Induced Phenotypical Changes
Although DR induced distinct changes toward a CD-like phenotype, the earlier-described HFHFD-induced T-cell alterations persisted. Specifically, DR mice showed an increased proportion of hepatic and VAT Th17 cells, as well as VAT Tc cells, and a reduction of VAT Treg cells, compared with mice fed CD for 32 weeks. In addition, a reduction in blood Tc cells was observed (Figure 10A–E). Moreover, no significant difference existed between the DR mice and mice fed a HFHFD for 20 and 32 weeks. RNA clustering analysis confirmed the persistence of the increase in hepatic Th17 cells, especially the canonical gene Rorc (Figure 10F), and VAT Tc cells, especially the gene Eomes, coding for eomesodermin, which is highly expressed in Tc cells (Figure 10G). In addition, CD45 immunohistochemistry of the VAT showed a persistence of adipose tissue inflammation despite the resolution of adipocyte hypertrophy (Figure 9A-D).
Figure 10.
Effect of diet reversal on HFHFD-induced T-cell alterations in liver, VAT, and blood. (A–E) Comparisons of liver Th17, VAT Tc, VAT Treg, VAT Th17, and blood Tc cells. T-cell subsets were characterized via flow cytometry. Heat maps showing the (F) hepatic RNA expression of Th17 cell–related genes and (G) VAT RNA expression of Tc cell–related genes. N = 10–20 per group. Medians are depicted with a horizontal black line. ∗∗P < .01 and ∗∗∗P < .001 (Kruskal–Wallis, homogenous subsets are shown with a horizontal line above the data points).
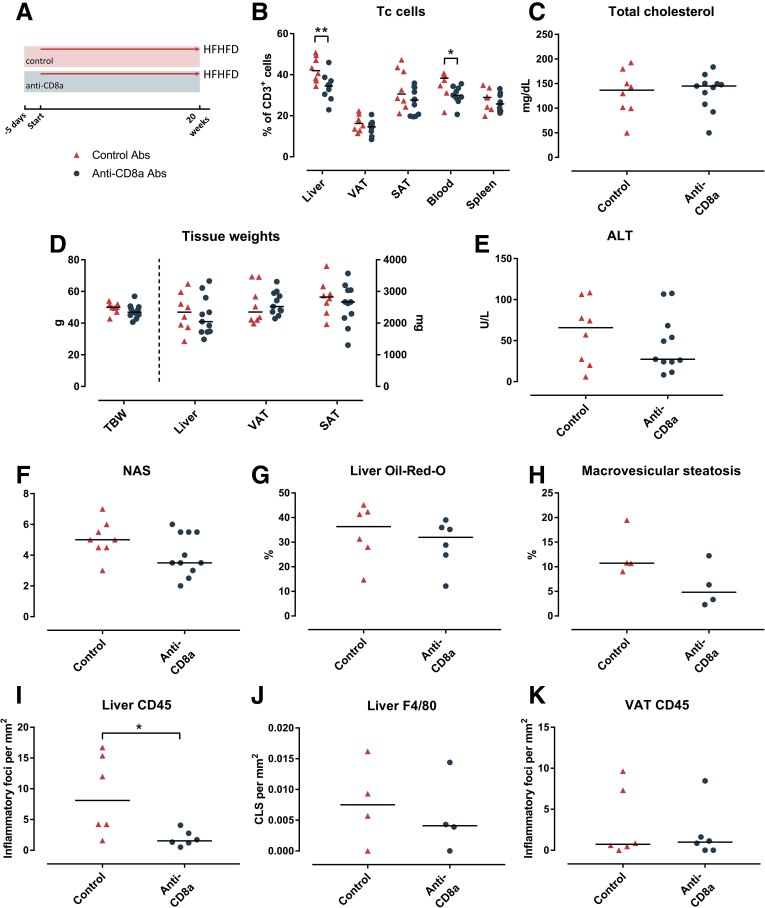
Anti-CD8a Antibodies Attenuated NASH, Whereas Anti–Interleukin 17A Antibodies Reduced Hepatic Inflammation
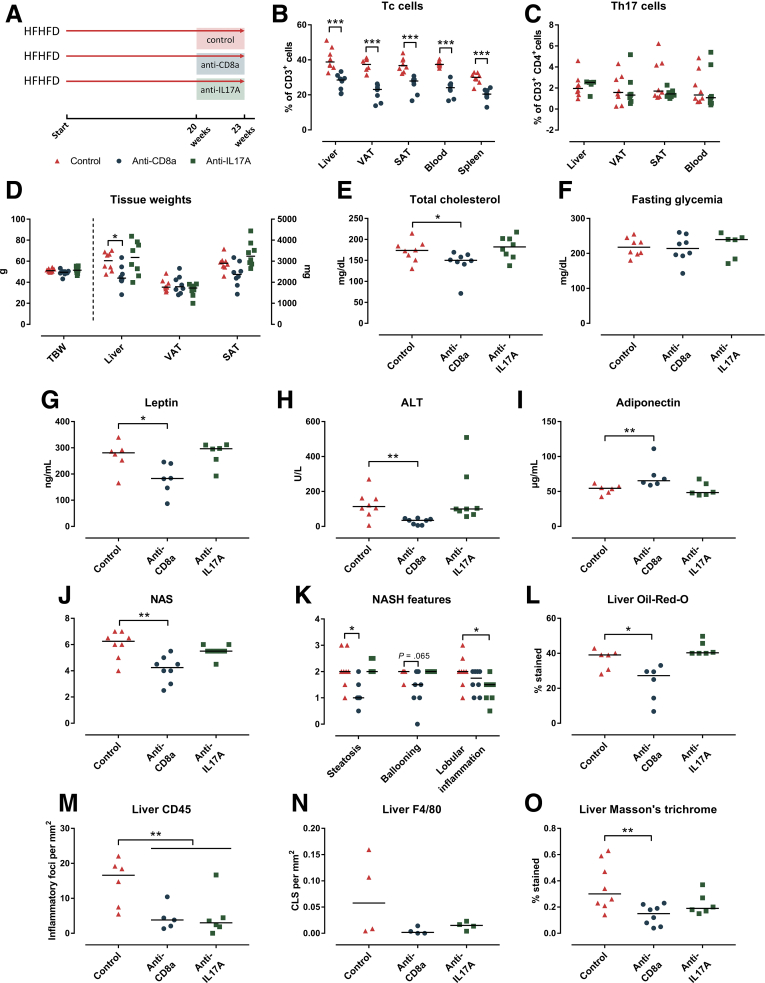
After being fed the HFHFD for 20 weeks, in a curative set-up, mice were treated for an additional 3 weeks once weekly via intraperitoneal injection with either neutralizing anti-CD8a antibodies, neutralizing interleukin (IL)17A antibodies, or inactive isotype control antibodies, while continuing HFHFD feeding (Figure 11A). Treatment with the anti-CD8a antibodies effectively reduced the percentage of Tc cells in VAT, as well as in the liver, SAT, blood, and spleen (Figure 11B), whereas it did not affect the proportions of Th1, Th17, or Treg cells in any of the investigated tissues. The decrease in Tc cells was associated with a reduction in liver weight and plasma ALT, cholesterol, and leptin levels, as well as an increase in adiponectin levels, while not affecting total body weight or fasting glycemia (Figure 11D–I). Furthermore, histologic assessment showed a decrease in NAS (Figure 11J), predominantly by reducing steatosis and, to a lesser extent, ballooning (Figures 11K and 12A). Automated quantification of Oil-red-O–stained liver tissue confirmed the decrease in steatosis (Figures 11L and 12B). Automated analysis of CD45- and F4/80-immunohistochemically stained liver tissue showed a reduction of hepatic inflammation (Figures 11M and N and 12C). Furthermore, automated analysis of MT-stained slides showed a significant reduction in fibrosis (Figures 11O and 12D). At the level of the VAT, no effect was observed on adipocyte diameter, adipose tissue inflammation (Figure 13A–D), or adipose tissue mass (Figure 11D). Interestingly, when mice were treated in a preventive set-up rather than a curative set-up by treating them with neutralizing anti-CD8a antibodies for 20 weeks with concomitant HFHFD feeding, the effects were less pronounced, possibly by an impaired effect of the antibodies at the level of the VAT (Figure 14).
Figure 11.
Effect of curative anti-CD8a and anti-IL17A treatment on HFHFD-induced metabolic alterations and NASH. (A) Study design. (B) Tc cell numbers. (C) Th17 cell numbers. (D) Total body weight and tissue weights. (E) Plasma cholesterol. (F) Fasted glycemia. (G) Plasma leptin. (H) Plasma adiponectin. (I) ALT. (L) (J) NAS. (K) NASH features graded according to NAS. (L) Quantification of steatosis through Oil-red-O staining. (M and N) Quantification of liver inflammation through (M) CD45 and (N) F4/80 immunohistochemistry, respectively, expressed as the number of inflammatory foci and crown-like structures per mm2. (O) Quantification of fibrosis through Masson’s trichrome staining. N = 8 per group. Medians are shown with a horizontal black line. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 (Mann–Whitney U).
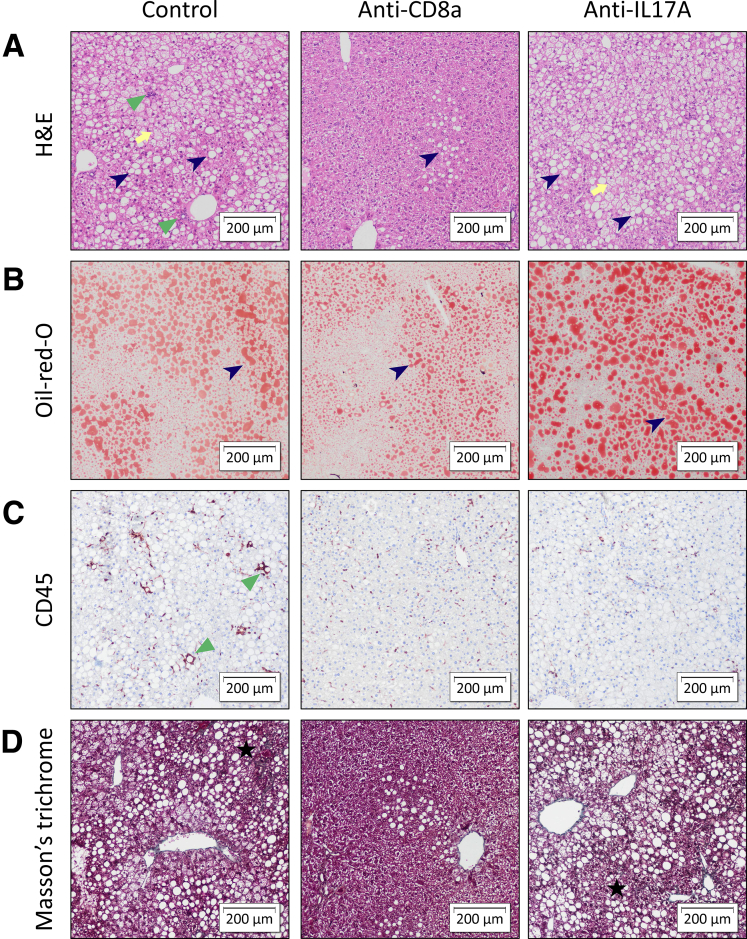
Figure 12.
Representative images of liver histology showing the effect of curative anti-CD8a and anti-IL17A treatment on HFHFD-induced NASH. Original magnification: ×10. Histologic staining was performed with (A) H&E, (B) Oil-red-O, (C) CD45 immunohistochemistry, and (D) MT. Anti-CD8a antibodies reduced steatosis (dark blue arrowhead), hepatic inflammation (green triangle), and fibrosis (black star). Anti-IL17A antibodies reduced hepatic inflammation, while not affecting steatosis or fibrosis. Ballooning is marked with a yellow arrow.
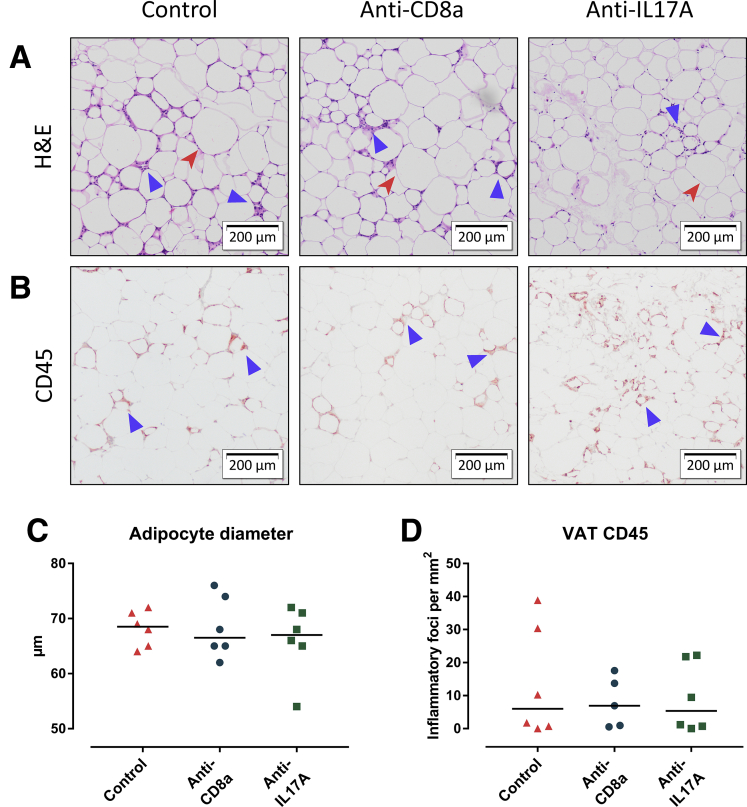
Figure 13.
The effect of curative anti-CD8a and anti-IL17A treatment on VAT. Representative images of VAT stained with (A) H&E and (B) CD45 immunohistochemistry. Original magnification: ×10. Neither anti-CD8a nor anti-IL17A treatment affected adipocyte hypertrophy (red arrowhead), or adipose tissue inflammation (blue triangle). (C) Adipocyte diameter. (D) Quantification of VAT inflammation through CD45 immunohistochemistry, expressed as the number of inflammatory foci per mm2. Medians are shown with a horizontal black line.
Figure 14.
Effect of preventive anti-CD8a treatment on HFHFD-induced metabolic alterations and NASH. (A) Study design and legend. (B) Tc cell numbers in liver, VAT, SAT, blood, and spleen. (C) Plasma cholesterol. (D) Total body weight and tissue weights. (E) Plasma ALT. (F) NAS. (G) Quantification of Oil-red-O–stained liver tissue, expressed as the percentage stained. (H) Morphometric quantification of macrovesicular steatosis on H&E-stained liver tissue, expressed as the percentage of macrovesicular steatosis. (I and J) Quantification of liver inflammation through (I) CD45 and (J) F4/80 immunohistochemistry expressed as the number of inflammatory foci and crown-like structures per mm2, respectively. (K) Quantification of VAT inflammation through CD45 immunohistochemistry, expressed as the number of inflammatory foci per mm2. N = 8–11 per group. Medians are shown with a horizontal black line. ∗P < .05, ∗∗P < .01 (Mann–Whitney U).
In contrast to treatment with anti-CD8a antibodies, curative treatment with neutralizing anti-IL17A antibodies failed to induce any phenotypical changes, or a reduction in the NAS, steatosis, or fibrosis (Figures 11D–L and O and 12A, B, and D). Nevertheless, when considering the individual components of the NAS, treatment with anti-IL17A antibodies did induce a reduction in lobular inflammation (Figure 11K). This was confirmed through quantification of CD45 and F4/80 immunohistochemistry (Figures 11M and N and 12C). This anti-inflammatory effect was not observed at the level of the VAT, and there was no effect on adipocyte diameter or adipose tissue mass (Figures 11D and 13A–D). As expected, treatment with anti-IL17A antibodies did not affect Th17 cell numbers because IL17A is not involved in the Th17 differentiation pathway (Figure 13C).
Discussion
In this study, we have shown a key role for differentially active T-cell subsets in the pathogenesis of NASH at the level of the liver and the VAT, showing not only persisting disruptions in the T-cell subset profile, even upon resolution of the metabolic alterations and NASH, but also mechanistically establishing the importance of these disruptions by inhibiting Tc cell and Th17 cell functionality.
First, prolonged HFHFD feeding induced NASH accompanied by several important metabolic disruptions, including obesity, dyslipidemia, and insulin resistance, thereby closely mimicking the human pathology, both phenotypically and pathophysiologically. In addition, HFHFD feeding resulted in the altered expression of several essential genes involved in the regulation of metabolic processes, including Ppara and Fgf21 in the liver and Retn, Fasn, and Pparg in the VAT. Furthermore, we showed that the development of NASH was accompanied by marked alterations in T-cell numbers, both at the level of the liver and the VAT. Importantly, these changes were confirmed by 2 independent techniques (ie, flow cytometry and RNA analysis), and at 2 separate time points. Moreover, the experiment’s longitudinal approach allows hypothesizing on the precise timing and relevance of the observed changes: although HFHFD feeding resulted in early metabolic alterations at 10–15 weeks, including adiposity, dyslipidemia, and hyperleptinemia, and the development of hepatic steatosis, the emergence of the earlier-described T-cell alterations occurred at a later stage (ie, from 20–25 weeks onward), and coincided with the development of major obesity-related disruptions, including insulin resistance, overt hepatic and VAT inflammation, and NASH. This suggests a close interaction between the T-cell subsets present at the level of the VAT and the liver specifically in the pathogenesis of NASH, rather than in the earlier processes involved in the development of steatosis.
The earlier-described T-cell alterations are in line with previous reports by both our own research group and other investigators. Specifically, Th17 cells were shown previously to be increased in the liver2, 3, 4, 5, 6 and VAT,6, 7, 8 Tc cells were shown to be increased in VAT,9, 10, 11 and Treg cells were shown to be decreased in VAT.12, 13, 14 Conversely, a number of other reported alterations were not confirmed in our model, including an increase in hepatic Tc cells15, 16, 17 and Th1 cells,4,18 a decrease in hepatic Treg cells,3,19 and an increase in VAT Th1 cells.8,13,20,21 Moreover, although some investigators have described differential immune disturbances in the SAT,6,13,22 this was not observed in our model. Unfortunately, when considering the available literature, these discrepancies seem inherent to the research field. Although, in part, they might be owing to the use of different models and time points, they also stress the dynamic nature of NASH and the associated immunologic alterations. Importantly, we characterized the different T-cell subsets at various time points and in multiple tissues in one and the same model, whereas previous research often focused on 1 subset and/or tissue. This allowed us to map the dynamic nature of these alterations in relation to the onset and progression of NASH, both at the hepatic and extrahepatic levels.
Second, we show that, although the DR normalized most HFHFD-induced metabolic alterations, including NASH, none of the observed T-cell alterations were affected, underlining their relevance in NASH pathogenesis. We report on the persisting immune disruption at the level of multiple T-cell subsets in multiple affected tissues, most importantly the liver and VAT. Interestingly, at the level of the VAT, the DR-induced normalization of down-regulated RNA expression of the metabolic genes Fasn, Pparg, and Retn was somewhat incomplete, an observation that might be associated to the persisting immune disruption.
Concerning the effect of the DR, other investigators have reported similar findings in the context of the NLRP3 inflammasome,23 as well as in the obesity-associated impaired memory T-cell response to influenza.24 Alternatively, some studies reported a normalization of the investigated, nonimmunologic, alterations upon DR.25,26 Furthermore, Rau et al27 showed in NASH patients that the initially observed increase in the Th17/Treg ratio in peripheral blood normalized 1 year after bariatric surgery. It is important to note, however, that no data are available concerning the effect of this intervention at the level of the liver. Collectively, these findings challenge our current understanding of the reversibility of NASH on weight loss and raise the question of whether rechallenge with the HFHFD might elicit an enhanced metabolic response, including a rapid recurrence of NASH. This hypothesis could be very relevant considering the weight cycling effect that often is present in patients pursuing weight loss.
Next, to further investigate the mechanistic involvement of the observed alterations in Tc cells and Th17 cells in the pathogenesis of NASH, we sought to correct these disruptions in 2 ways. On the one hand, we administered anti-CD8a antibodies to generally reduce Tc cell numbers, while on the other hand, we administered anti-IL17A antibodies to inhibit Th17 functionality. Neutralizing anti-CD8a antibodies effectively decreased Tc cells in VAT and all other investigated tissues, thereby decreasing plasma ALT, cholesterol, and leptin levels, as well as histologically attenuating hepatic steatosis, inflammation, and fibrosis, thus improving NASH. Remarkably, no effect was observed with respect to adipose tissue hypertrophy and inflammation, total body weight, or fasting glycemia. This is in line with previous reports,15,28 although these did not include an examination of the involvement of the adipose tissue. In parallel, the anti-IL17A antibodies reduced hepatic inflammation, corresponding to the HFHFD-induced abundance in hepatic Th17 cells. Nevertheless, no effect was observed on plasma ALT, cholesterol, and leptin levels, total body weight, adipose tissue hypertrophy and inflammation, or fasting glycemia. The fact that the most pronounced effect on NASH was achieved by correcting an immune disruption at the level of the VAT again underlines the immense importance of adipose tissue inflammation in the pathogenesis of NASH and should encourage further exploration of the modulation of adipose tissue inflammation in the treatment of obesity-related metabolic disease, including NASH.
In conclusion, this study presents data that clearly support a role for hepatic and VAT Tc, Treg, and Th17 cells in NASH pathogenesis, with an emphasis on the critical involvement of VAT Tc cells. Importantly, these findings were shown using a holistic approach, investigating multiple T-cell subsets in multiple tissues at multiple time points, both at the cellular and the messenger RNA level, and confirming their relevance with loss-of-function experiments. Moreover, we report a persistence of these hepatic and VAT inflammatory changes despite metabolic normalization upon diet reversal, challenging our current understanding of the reversibility of NASH and other obesity-related conditions. Further exploration of the involved mechanisms constitutes an important future research goal in the search for a medical treatment for NASH.
Methods
Mice and Diet
Male 8-week-old C57BL/6J mice (Janvier Labs, Le Genest-Saint-Isle, France) were kept on a 12:12-hour light/dark cycle with controlled temperature and humidity and housed in enriched cages with a stainless-steel grid, filter top, and free access to tap water. All interventions were performed during the light cycle. Depending on the experimental group, mice had free access to standard chow as a CD (Carfil Quality, Oud-Turnhout, Belgium) or a HFHFD (D16042610; Research Diets, New Brunswick, NJ), containing 55 kcal% fat and 14 kcal% fructose. This study was approved by the Ethical Committee for Animal Testing at the University of Antwerp (2015-16). Laboratory animal-specific information was reported in accordance with the Animal Research: Reporting of In Vivo Experiments guidelines.
Experimental Set-Ups
In the first experiment, mice were fed either the HFHFD or the CD and killed at 10, 15, 20, 25, or 32 weeks after the start of the experiment (N = 6–12 per time point, per group). Based on their similar phenotype, mice were pooled as follows: 10–15 weeks, 20–25 weeks, and 32 weeks. In a second experiment, a DR was performed by feeding mice the HFHFD for 20 weeks, subsequently substituting the HFHFD for CD, which then was continued for another 12 weeks (N = 12). In a third experiment, HFHFD-fed mice (N = 8–11 per group) were injected intraperitoneally once weekly with neutralizing anti-CD8a antibodies (3 μg/g of body weight), neutralizing anti-IL17A antibodies (8 μg/g of body weight), or inactive isotype control antibodies (3 μg/g of body weight)10,29 (Table 5). Treatment was performed according to 2 experimental protocols: a preventive and curative set-up. Mice in the preventive set-up were administered the first dose 5 days before start of the HFHFD. Subsequently, they were treated once weekly for 20 weeks with concomitant HFHFD feeding. Mice in the curative set-up were fed the HFHFD for 20 weeks, after which they were treated once weekly for a total of 3 weeks with concomitant HFHFD feeding. Treatment with neutralizing anti-IL17A antibodies was performed only in the curative set-up.
Table 5.
Overview of Antibodies Used in This Study
| Use | Antigen | Fluorochrome | Clone | Supplier |
|---|---|---|---|---|
| Flow cytometry | CD45 | PE | 30-F11 | BioLegend |
| Flow cytometry | CD3 | FITC | 17A2 | BioLegend |
| Flow cytometry | CD4 | PerCP/Cy5.5 | GK15 | BioLegend |
| Flow cytometry | CD8a | APC | 53-6.7 | BioLegend |
| Flow cytometry | CD25 | PE | PC61 | BioLegend |
| Flow cytometry | Foxp3 | APC | FJK-16s | Thermo Fisher Scientific |
| Flow cytometry | T-bet | APC | 4B10 | Thermo Fisher Scientific |
| Flow cytometry | RORγt | PE | B2D | Thermo Fisher Scientific |
| In vivo | CD8a | 53-6.7 | BioLegend | |
| In vivo | IL17A | TC11-18H10.1 | BioLegend | |
| In vivo | IgG2a, κ control | RTK2758 | BioLegend |
Glucose and Insulin Tolerance Testing
GTT and ITT were performed by injecting mice intraperitoneally with a 20% glucose solution (Baxter, Deerfield, IL) at 2 mg glucose per gram of body weight or a 0.1U/mL insulin solution (Novo Nordisk, Bagsvaerd, Denmark) at 0.75 mU per gram of body weight, after being fasted for 6 hours. Glucose levels were determined with a portable glucometer (GlucoMen LX; A. Menarini Diagnostics, Florence, Italy) at 15, 30, 60, 90, and 120 minutes after the respective challenge and the area under the curve was calculated.
Procurement of Target Tissues, Immune Cell Isolation, and Flow Cytometry
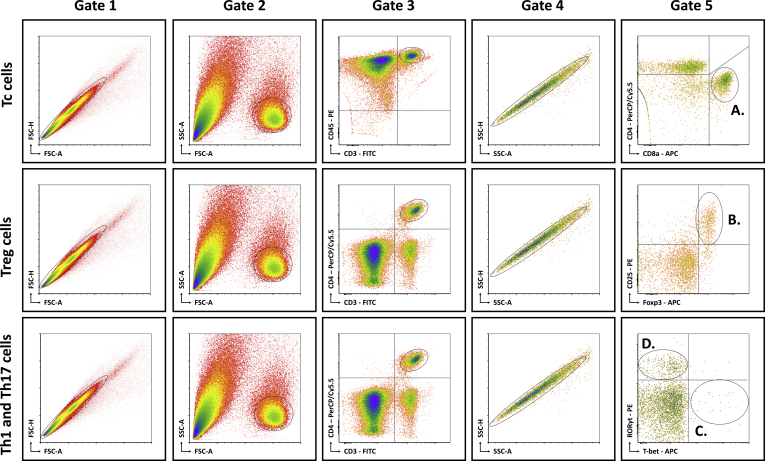
Mice were anesthetized with pentobarbital (0.12 mg/g body weight, Nembutal; Ceva, Libourne, France) at 8–9 AM, without being fasted. Blood was collected on EDTA by cardiac puncture. Liver, VAT, SAT, and spleen were excised and processed as described previously.6 The resulting single-cell suspensions were incubated at 4°C at approximately 106 cells per 100 μL with fluorochrome-conjugated antibodies (Table 5). Flow cytometry was performed on an Accuri C6 (BD Biosciences, Franklin Lakes, NJ). Data analysis was performed with FCS Express 4 (De Novo Software, Glendale, CA). Lymphocyte populations were determined by forward scatter and side scatter. Doublets and triplets were filtered out twice by plotting area to height for both forward scatter and side scatter. Concerning the fluorochrome-stained cells, positive populations were defined as being above the 99th percentile of fluorescence-minus-one staining. The gating strategy is shown in Figure 15.
Figure 15.
Schematic overview of flow cytometric gating strategy for the identification of (A) Tc cells, (B) Treg cells, (C) Th1 cells, and (D) Th17 cells in liver, VAT, SAT, spleen, and blood. Lymphocyte populations were determined by FSC-A and SSC-A. Doublets and triplets were filtered out twice by plotting the area to height for both FSC and SSC. Concerning the fluorochrome-stained cells, positive populations were defined as being above the 99th percentile of FMO staining, indicated by the quadrants. -A, area; FSC, forward scatter; FMO, fluorescence minus one; -H; height; RORγt, RAR-related orphan receptor γt; SSC, side scatter.
Biochemical Analysis
Whole blood samples were centrifuged for 10 minutes at 1500 × g and plasma was collected. Biochemical analysis was performed with an automated Dimension Vista 1500 System (Siemens Healthineers, Erlangen, Germany) and included aspartate aminotransferase, ALT, cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and triglycerides. Plasma leptin and adiponectin concentrations were determined using the Mouse Leptin and Adiponectin Enzyme-Linked Immunosorbent Assay Kit (Thermo Fisher Scientific, Waltham, MA), respectively (N = 6 per group). Plasma cytokine concentrations were determined with the LEGENDplex Mouse Th Cytokine Panel (N = 6 per group; BioLegend, San Diego, CA). Analysis was performed on an Accuri C6 (BD Biosciences) and data were processed using the LEGENDplex Software v8.0 (BioLegend).
Histology
Liver and VAT were prepared for histologic analysis and stained with H&E, MT, and Oil-red-O as previously described.30 Immunohistochemistry was performed using anti-CD45 antibodies (ab10558; Abcam, Cambridge, UK) and anti-F4/80 antibodies (MCA497G; Bio-Rad Laboratories, Hercules, CA). Secondary antibodies were species-appropriate horseradish-peroxidase conjugates (Vectastain ABC; Vector Laboratories, Burlingame, CA).
The histologic hallmarks of NASH in the liver were assessed on H&E-stained slides by means of the NAS.31 Steatosis was quantified on Oil-red-O–stained slides with ImageJ (version 1.52p, National Institutes of Health, Bethesda, MD) and expressed as the percentage stained. In addition, macrovesicular steatosis was quantified morphometrically with Biocellvia software (Biocellvia, Marseille, France) and expressed as a percentage of the total area. Hepatic inflammation was quantified immunohistochemically by quantification of the amount of CD45+ inflammatory foci per mm2 (ImageJ) and the amount of F4/80+ crown-like structures per mm2 (Biocellvia). Fibrosis was graded according to the accepted NASH-clinical research network definitions31 and quantified with ImageJ, both on MT-stained slides. Adipocyte diameters in VAT were determined with the ImageJ plugin Adiposoft,32 and adipose tissue inflammation was quantified immunohistochemically as the number of CD45+ inflammatory foci per mm2 with ImageJ.
Multiplex RNA Analysis
At death, liver tissue and VAT were snap-frozen in liquid nitrogen and stored at -80°C. A random selection was made of 5-6 samples from CD-fed and HFHFD-fed mice at every grouped time point (10–15 weeks, 20–25 weeks, and 32 weeks), as well as from the DR group. This resulted in the analysis of 80 samples. Subsequently, the tissues were homogenized (Precellys Lysing Kit; Bertin Instruments, Montigny-le-Bretonneux, France) and total RNA was extracted from liver (NucleoSpin RNA Plus; Macherey-Nagel, Düren, Germany) and VAT (RNeasy Lipid Tissue Mini Kit; Qiagen, Hilden, Germany). RNA expression of 581 genes was determined (nCounter Mouse Immunology panel and 20 Panel Plus genes; NanoString Technologies, Seattle, WA). After hybridization, transcripts were quantified (nCounter; NanoString Technologies). Results were normalized in the nSolver Analysis Software (NanoString Technologies) by the geometric mean of 14 housekeeping genes and 6 positive controls.
Statistics
For statistical analysis, the Mann–Whitney U test, the Kruskal–Wallis test according to a stepwise step-down protocol to identify homogenous subsets, or Pearson correlation was used in SPSS 25 (IBM, Armonk, NY) as appropriate. Significance was assumed at P < .05. Graphs were designed in GraphPad Prism 7 (GraphPad Software, San Diego, CA). Concerning the RNA analysis, a selection of 47 relevant genes was made based on their expression in the T-cell subsets of interest or their involvement in metabolism or fibrogenesis. Heat maps were generated using MetaboAnalyst 4.0 (Xia Lab, McGill University, Montreal, Canada), clustering was performed using Ward’s method. Differentially expressed genes were identified with the nSolver Analysis Software (NanoString Technologies). Pathway analysis was performed on the entire set of 581 genes with Ingenuity Pathway Analysis (Qiagen) with the following criteria: P < .0001 and overlap greater than 5%.
All authors had access to the study data and have reviewed and approved the final manuscript.
CRediT Authorship Contributions
Mikhaïl A. Van Herck, MD (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Equal; Visualization: Lead; Writing – original draft: Lead); Luisa Vonghia, MD, PhD (Methodology: Equal; Supervision: Equal; Writing – review & editing: Equal); Wilhelmus J Kwanten, MD, PhD (Writing – review & editing: Equal); Yvon Julé, PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Equal); Thomas Vanwolleghem, MD PhD (Writing – review & editing: Equal); Didier Ebo, MD, PhD (Writing – review & editing: Equal); Peter P. Michielsen, MD, PhD (Writing – review & editing: Equal); Joris De Man, MSc (Writing – review & editing: Equal); Lucio Gama, PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Equal); Benedicte Y. De Winter, MD PhD (Conceptualization: Equal; Methodology: Equal; Supervision: Equal; Writing – review & editing: Equal); Sven Francque, MD, PhD (Conceptualization: Equal; Funding acquisition: Lead; Methodology: Equal; Supervision: Lead; Writing – review & editing: Equal).
The authors thank Petra Aerts, Ilse Goolaerts, Angelika Jürgens, Rita Van Den Bossche, Marleen Vinckx, and Lieve Vits for their technical support, Stijn Van Hees for his scientific input, and Stijn Brusselmans and Louise Mees for proofreading the manuscript.
Footnotes
Conflicts of interest This author discloses the following: Yvon Julé is the chief science officer at Biocellvia, which performed some of the histologic analyses. The remaining authors disclose no conflicts.
Funding Supported by the University Research Fund (BOF), University of Antwerp (DOCPRO4, 31879) (M.A.V.H. and S.M.F.), and a senior clinical research fellowship from Research Foundation Flanders (1802154N) (S.M.F.).
Contributor Information
Mikhaïl A. Van Herck, Email: mikhail.vanherck@uantwerpen.be.
Luisa Vonghia, Email: luisa.vonghia@uza.be.
References
- 1.Van Herck M.A., Weyler J., Kwanten W.J., Dirinck E.L., De Winter B.Y., Francque S.M., Vonghia L. The differential roles of T cells in non-alcoholic fatty liver disease and obesity. Front Immunol. 2019;10:82. doi: 10.3389/fimmu.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giles D.A., Moreno-Fernandez M.E., Stankiewicz T.E., Cappelletti M., Huppert S.S., Iwakura Y., Dong C., Shanmukhappa S.K., Divanovic S. Regulation of Inflammation by IL-17A and IL-17F modulates non-alcoholic fatty liver disease pathogenesis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He B., Wu L., Xie W., Shao Y., Jiang J., Zhao Z., Yan M., Chen Z., Cui D. The imbalance of Th17/Treg cells is involved in the progression of nonalcoholic fatty liver disease in mice. BMC Immunol. 2017;18:33. doi: 10.1186/s12865-017-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolla S., Alchera E., Imarisio C., Bardina V., Valente G., Cappello P., Mombello C., Follenzi A., Novelli F., Carini R. The balance between IL-17 and IL-22 produced by liver-infiltrating T-helper cells critically controls NASH development in mice. Clin Sci. 2015;130:193–203. doi: 10.1042/CS20150405. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y., Bian Z., Zhao L., Liu Y., Liang S., Wang Q., Han X., Peng Y., Chen X., Shen L., Qiu D., Li Z., Ma X. Interleukin-17 exacerbates hepatic steatosis and inflammation in non-alcoholic fatty liver disease. Clin Exp Immunol. 2011;166:281–290. doi: 10.1111/j.1365-2249.2011.04471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vonghia L., Ruyssers N., Schrijvers D., Pelckmans P., Michielsen P., Clerck L.D., Ramon A., Jirillo E., Ebo D., Winter B.D., Bridts C., Francque S. CD4+RORγt ++ and Tregs in a mouse model of diet-induced nonalcoholic steatohepatitis. Mediat Inflamm. 2015;2015:239623. doi: 10.1155/2015/239623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertola A., Ciucci T., Rousseau D., Bourlier V., Duffaut C., Bonnafous S., Blin-Wakkach C., Anty R., Iannelli A., Gugenheim J., Tran A., Bouloumie A., Gual P., Wakkach A. Identification of adipose tissue dendritic cells correlated with obesity-associated insulin-resistance and inducing Th17 responses in mice and patients. Diabetes. 2012;61:2238–2247. doi: 10.2337/db11-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong C.P., Park A., Yang B.G., Yun C.H., Kwak M.J., Lee G.W., Kim J.H., Jang M.S., Lee E.J., Jeun E.J., You G., Kim K.S., Choi Y., Park J.H., Hwang D., Im S.H., Kim J.F., Kim Y.K., Seoh J.Y., Surh C.D., Kim Y.M., Jang M.H. Gut-specific delivery of T-helper 17 cells reduces obesity and insulin resistance in mice. Gastroenterology. 2017;152:1998–2010. doi: 10.1053/j.gastro.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Kawanishi N., Mizokami T., Yano H., Suzuki K. Exercise attenuates M1 macrophages and CD8+ T cells in the adipose tissue of obese mice. Med Sci Sports Exerc. 2013;45:1684–1693. doi: 10.1249/MSS.0b013e31828ff9c6. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura S., Manabe I., Nagasaki M., Eto K., Yamashita H., Ohsugi M., Otsu M., Hara K., Ueki K., Sugiura S., Yoshimura K., Kadowaki T., Nagai R. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 11.Rausch M.E., Weisberg S., Vardhana P., Tortoriello D.V. Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. Int J Obes (Lond) 2008;32:451–463. doi: 10.1038/sj.ijo.0803744. [DOI] [PubMed] [Google Scholar]
- 12.Feuerer M., Herrero L., Cipolletta D., Naaz A., Wong J., Nayer A., Lee J., Goldfine A.B., Benoist C., Shoelson S., Mathis D. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winer S., Chan Y., Paltser G., Truong D., Tsui H., Bahrami J., Dorfman R., Wang Y., Zielenski J., Mastronardi F., Maezawa Y., Drucker D.J., Engleman E., Winer D., Dosch H.M. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15:921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z., Wang G., Zhu Y., Liu R., Song J., Ni Y., Sun H., Yang B., Hou M., Chen L., Ji M., Fu Z. PPAR-gamma agonist ameliorates liver pathology accompanied by increasing regulatory B and T cells in high-fat-diet mice. Obesity (Silver Spring) 2017;25:581–590. doi: 10.1002/oby.21769. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharjee J., Kirby M., Softic S., Miles L., Salazar-Gonzalez R.M., Shivakumar P., Kohli R. Hepatic natural killer T-cell and CD8+ T-cell signatures in mice with nonalcoholic steatohepatitis. Hepatol Commun. 2017;1:299–310. doi: 10.1002/hep4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazarian M., Revelo X.S., Nohr M.K., Luck H., Zeng K., Lei H., Tsai S., Schroer S.A., Park Y.J., Chng M.H.Y., Shen L., D'Angelo J.A., Horton P., Chapman W.C., Brockmeier D., Woo M., Engleman E.G., Adeyi O., Hirano N., Jin T., Gehring A.J., Winer S., Winer D.A. Type I interferon responses drive intrahepatic T cells to promote metabolic syndrome. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aai7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas J.T., Vonghia L., Mogilenko D.A., Verrijken A., Molendi-Coste O., Fleury S., Deprince A., Nikitin A., Woitrain E., Ducrocq-Geoffroy L., Pic S., Derudas B., Dehondt H., Gheeraert C., Van Gaal L., Driessen A., Lefebvre P., Staels B., Francque S., Dombrowicz D. Transcriptional network analysis implicates altered hepatic immune function in NASH development and resolution. Nat Metab. 2019;1:604–614. doi: 10.1038/s42255-019-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer M., Hines I.N., Milton R.J., Wheeler M.D. Favored T helper 1 response in a mouse model of hepatosteatosis is associated with enhanced T cell-mediated hepatitis. Hepatology. 2006;44:216–227. doi: 10.1002/hep.21221. [DOI] [PubMed] [Google Scholar]
- 19.Ma X., Hua J., Mohamood A.R., Hamad A.R., Ravi R., Li Z. A high-fat diet and regulatory T cells influence susceptibility to endotoxin-induced liver injury. Hepatology. 2007;46:1519–1529. doi: 10.1002/hep.21823. [DOI] [PubMed] [Google Scholar]
- 20.Rocha V.Z., Folco E.J., Sukhova G., Shimizu K., Gotsman I., Vernon A.H., Libby P. Interferon-gamma, a Th1 cytokine, regulates fat inflammation: a role for adaptive immunity in obesity. Circ Res. 2008;103:467–476. doi: 10.1161/CIRCRESAHA.108.177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan I.M., Dai Perrard X.Y., Perrard J.L., Mansoori A., Smith C.W., Wu H., Ballantyne C.M. Attenuated adipose tissue and skeletal muscle inflammation in obese mice with combined CD4+ and CD8+ T cell deficiency. Atherosclerosis. 2014;233:419–428. doi: 10.1016/j.atherosclerosis.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuniga L.A., Shen W.J., Joyce-Shaikh B., Pyatnova E.A., Richards A.G., Thom C., Andrade S.M., Cua D.J., Kraemer F.B., Butcher E.C. IL-17 regulates adipogenesis, glucose homeostasis, and obesity. J Immunol. 2010;185:6947–6959. doi: 10.4049/jimmunol.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christ A., Gunther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Bassler K., Klee K., Schulte-Schrepping J., Ulas T., Moorlag S., Kumar V., Park M.H., Joosten L.A.B., Groh L.A., Riksen N.P., Espevik T., Schlitzer A., Li Y., Fitzgerald M.L., Netea M.G., Schultze J.L., Latz E. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172:162–175 e14. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebeles J., Green W.D., Alwarawrah Y., Nichols A.G., Eisner W., Danzaki K., MacIver N.J., Beck M.A. Obesity-induced changes in T-cell metabolism are associated with impaired memory T-cell response to influenza and are not reversed with weight loss. J Infect Dis. 2019;219:1652–1661. doi: 10.1093/infdis/jiy700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radonjic M., Wielinga P.Y., Wopereis S., Kelder T., Goelela V.S., Verschuren L., Toet K., van Duyvenvoorde W., van der Werff van der Vat B., Stroeve J.H., Cnubben N., Kooistra T., van Ommen B., Kleemann R. Differential effects of drug interventions and dietary lifestyle in developing type 2 diabetes and complications: a systems biology analysis in LDLr-/- mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siersbaek M., Varticovski L., Yang S., Baek S., Nielsen R., Mandrup S., Hager G.L., Chung J.H., Grontved L. High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Sci Rep. 2017;7:40220. doi: 10.1038/srep40220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rau M., Schilling A.-K., Meertens J., Hering I., Weiss J., Jurowich C., Kudlich T., Hermanns H.M., Bantel H., Beyersdorf N., Geier A. Progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis is marked by a higher frequency of Th17 cells in the liver and an increased Th17/resting regulatory T cell ratio in peripheral blood and in the liver. J Immunol. 2016;196:97–105. doi: 10.4049/jimmunol.1501175. [DOI] [PubMed] [Google Scholar]
- 28.Wolf M.J., Adili A., Piotrowitz K., Abdullah Z., Boege Y., Stemmer K., Ringelhan M., Simonavicius N., Egger M., Wohlleber D., Lorentzen A., Einer C., Schulz S., Clavel T., Protzer U., Thiele C., Zischka H., Moch H., Tschop M., Tumanov A.V., Haller D., Unger K., Karin M., Kopf M., Knolle P., Weber A., Heikenwalder M. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Harley I.T.W., Stankiewicz T.E., Giles D.A., Softic S., Flick L.M., Cappelletti M., Sheridan R., Xanthakos S.A., Steinbrecher K.A., Sartor R.B., Kohli R., Karp C.L., Divanovic S. IL-17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology. 2014;59:1830–1839. doi: 10.1002/hep.26746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwanten W.J., Vandewynckel Y.P., Martinet W., De Winter B.Y., Michielsen P.P., Van Hoof V.O., Driessen A., Timmermans J.P., Bedossa P., Van Vlierberghe H., Francque S.M. Hepatocellular autophagy modulates the unfolded protein response and fasting-induced steatosis in mice. Am J Physiol Gastrointest Liver Physiol. 2016;311:G599–G609. doi: 10.1152/ajpgi.00418.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 32.Galarraga M., Campion J., Munoz-Barrutia A., Boque N., Moreno H., Martinez J.A., Milagro F., Ortiz-de-Solorzano C. Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. J Lipid Res. 2012;53:2791–2796. doi: 10.1194/jlr.D023788. [DOI] [PMC free article] [PubMed] [Google Scholar]