Abstract
Reactivation of the androgen receptor signaling pathway in the emasculated environment is the main reason for the occurrence of castration-resistant prostate cancer (CRPC). The immunophilin FKBP51, as a co-chaperone protein, together with Hsp90 help the correct folding of AR. Rapamycin is a known small-molecule inhibitor of FKBP51, but its effect on the FKBP51/AR signaling pathway is not clear. In this study, the interaction mechanism between FKBP51 and rapamycin was investigated using steady-state fluorescence quenching, X-ray crystallization, MTT assay, and qRT-PCR. Steady-state fluorescence quenching assay showed that rapamycin could interact with FKBP51. The crystal of the rapamycin-FKBP51 complex indicated that rapamycin occupies the hydrophobic binding pocket of FK1 domain which is vital for AR activity. The residues involving rapamycin binding are mainly hydrophobic and may overlap with the AR interaction site. Further assays showed that rapamycin could inhibit the androgen-dependent growth of human prostate cancer cells by down-regulating the expression levels of AR activated downstream genes. Taken together, our study demonstrates that rapamycin suppresses AR signaling pathway by interfering with the interaction between AR and FKBP51. The results of this study not only can provide useful information about the interaction mechanism between rapamycin and FKBP51, but also can provide new clues for the treatment of prostate cancer and castration-resistant prostate cancer.
Keywords: Prostate cancer, Rapamycin, Androgen receptor, FKBP51
Highlights
-
•
Rapamycin occupies the hydrophobic binding pocket of FK1 domain of FKBP51.
-
•
Rapamycin suppresses the AR signaling pathway by interacting with FKBP51.
-
•
Rapamycin inhibits the growth of prostate cancer cells via the AR signaling pathway.
1. Introduction
Prostate cancer (PCa) is the most common malignant tumor of the urinary system. The morbidity and mortality of PCa in male malignancies rank second and fifth in the world, respectively [1]. In 2015, there were about 60,300 new PCa patients in China and 26,600 deaths [2]. At present, the treatment of PCa, from surgical resection to standard endocrine therapy and radiotherapy, is only effective for early patients. However, the treatment for metastatic PCa and castration-resistant prostate cancer are not ideal [3]. The high recurrence rate after castration treatment is an important clinical feature of PCa which is also a hot but difficult issue to conquer [4,5].
Studies have shown that although CRPC appears to be resistant to androgen withdrawal, its progression is usually still dependent on AR [[6], [7], [8]]. The activation of AR after castration is associated with multiple various mechanisms, including the amplification of AR gene [9], the variable mutation of AR [10], co-activator and co-repressor modifications [11], as well as aberrant activation/post-translational modification [12]. In addition, altered steroidogenesis [13], AR splice variants, and signal transduction abnormalities of AR coactivators can also lead to CRPC progression [[14], [15], [16]]. How to inhibit or block the activation of AR in the androgen withdrawal environment is the key to the treatment of CRPC.
The three approved second-generation AR targeting agents, abiraterone, enzalutamide and apalutamide, effectively target AR signaling. Although they can prolong survival, they are ineffective for AR splice variants, acquired resistance to these three drugs will continue to develop mCRPC [[17], [18], [19]]. In addition, there are development of agents that target the AR axis [20,21], however, these new therapeutics only prolong median overall survival a few additional months. The FK506-binding protein 51 (FKBP51) is a key regulator of endocrine stress responses in mammals. Recently numerous interaction partners of FKBP51 have been described [22]. The immunophilin FKBP51 and FKBP52 act as synergistic chaperone (co-chaperone), together with Hsp90 help correct folding of AR, which are necessary for AR activation [[23], [24], [25]]. It has been reported that FKBP51 promoted the activity of AR and enhanced the ability of AR to bind androgens, leading to upregulation expression of downstream genes and finally promote proliferation of prostate cancer cells [26,27]. Moreover, Shang et al. [28] demonstrated that FKBP51 is regulated by AR-V7 and interacts with the AKT signaling pathway and NF-кB signaling to inhibit apoptosis of prostate cancer cells which finally promote the formation of CRPC. Therefore, inhibition of FKBP51 may reduce the activation of AR and the growth of prostate cancer cells, which is of great significance for the targeted treatment of CRPC.
Rapamycin is a macrolide isolated from the bacterium Streptomyces Hygroscopicus and is clinically used for the treatment of organ transplant rejection and autoimmune diseases. It can inhibit the intracellular receptor FKBP12 and the mammalian target of rapamycin (mTOR), which causes cell cycle to arrest in the G1 phase. The anti-cancer effects of rapamycin have been shown in several cancers including hematologic malignancies, breast cancer, rhabdomyosarcoma, and non-small cell lung cancer [[29], [30], [31], [32]]. Besides, rapamycin is effective in inhibiting the growth of hormone-dependent and hormone-independent prostate cancer cells [[33], [34], [35], [36], [37]]. The current research on rapamycin is mainly focused on the PI3K/AKT/mTOR signaling pathway [38,39], however the effect of rapamycin on the FKBP51/AR signaling pathway is still unknown.
The primary objective of this experiment is to investigate the effect of rapamycin on AR signaling pathway under androgen-dependent conditions and to determine whether the effect of rapamycin on AR is mediated by FKBP51 protein. The observations of this study will provide new mechanism for rapamycin in anti-tumor researches which would pave the way for further research on clinical research of CRPC.
2. Materials and methods
2.1. Reagents and antibodies
Rapamycin was purchased from Selleck Ltd., which was dissolved in dimethyl sulfoxide (DMSO) to form a 5 mM stock solution, and aliquots were stored at −20 °C. 4, 5 α-dihydrotestosterone (DHT) (Sigma-Aldrich) was dissolved in ethanol. Fetal bovine serum (FBS), antibiotics and RPMI 1640 medium were purchased from Gibco (Carlsbad, CA, United States). RIPA lysis buffer and antibodies of AR, FKBP51, PSA, and GAPDH were purchased from Sigma Chemical Co (St. Louis, MO, USA). Fluorescein-conjugated goat anti-rabbit secondary antibody was produced from Abcam (ab50887, Abcam, Cambridge, UK).
2.2. Cell culture
The human prostate cancer cells of LNCaP and 22Rv1 were purchased from the American Type Culture Collection (ATCC) and cultured in RPMI 1640 medium supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin. All the cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
2.3. MTT assay
LNCaP and 22Rv1 cells were each planted at a density of 5 × 103 cells per well in a 96-well plate and cultured overnight for cell adhesion. 1 nM dihydrotestosterone (DHT) and different concentrations (5 nM, 10 nM, 20 nM, 50 nM and 100 nM) of rapamycin were applied as treatment for another 24 h and 48 h. DMSO treatment served as vehicle control. Every dosage was repeated three times and at least three independent experiments were performed. Subsequently, 0.5 mg/ml MTT reagent (Sigma-Aldrich) was added to each well and incubated at 37 °C for another 4 h, and then the dark blue crystals were dissolved with 100 ml DMSO. Absorbance was measured at a wavelength of 490 nm with the BioRad microplate reader. The percentages obtained from the absorbance of the treated cells divided by the absorbance of untreated cells were presented as the cell viabilities.
2.4. Endogenous expression of androgen-activated genes
The LNCaP cells grown to the logarithmic phase were subcultured into culture dishes and divided into four groups: blank control with DMSO added, DHT-added, 30 nM rapamycin+1 nM DHT, and 30 nM rapamycin +10 nM DHT. After attached, the cells were starved for 12 h in a serum-free RPMI 1640 medium. After rapamycin of 30 nM was added for 2 h, 1 nM DHT and 10 nM DHT were added respectively to the cells for 12 h. The same volume of culture medium was added to the blank control group, and 10 nM DHT was added to the DHT control group.
Total RNA was extracted using Trizol. cDNA was synthesized and amplified using the Transcript First-Strand cDNA Synthesis SuperMix (TaKaRa Biotechnology, Dalian, China) in accordance with the manufacturer's protocol. The real-time quantitative reverse transcription PCR (qRT-PCR) was performed by SYBR Premix Ex TapTM II (TaKaRa Biotechnology, Dalian, China) and the assay was carried out in triplicate on a CFX96™ Real-Time system (Bio-Rad, Hercules, CA). The designs of primer sequences referred our previous work [27]. All reactions were performed in triplicate.
2.5. Western blot
LNCaP cells were plated in 10-cm dishes at 1 × 105 cells/dish in RPMI 1640 medium. After 24 h, the cells were treated with 1.0 nM DHT and different concentrations (1 nM, 5 nM, 10 nM) of rapamycin for 24 h. Preparation of whole-cell protein lysates for western blot analysis was conducted as follows. After treatment, cells were lysed for at least 30 min and then centrifuged at 14,000×g for 10 min at 4 °C, and the supernatant was collected. Protein concentrations were determined by Bio-Rad Protein Assay kit (Bio-Rad, Philadelphia, PA, United States). Then, equal amounts of total protein of each sample was resuspended in loading buffer and denatured for 5 min at 100 °C. The total protein (30 μg) of each sample was separated by 10% SDS-PAGE and then transferred to PVDF membranes (Millipore, United States). After blocking with 5% non-fat dry milk in 1 × TBST for 1 h at room temperature, the membranes were incubated overnight with different primary antibodies [AR, FKBP51, PSA, and GAPDH] at 4 °C. After washing the membranes in TBST five times (5 min per time), horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1: 1000 dilution) were added at room temperature for 2 h. Proteins were detected using a ChemiDoc XRS imaging system and GAPDH was used as the loading control.
2.6. Cloning, expression, and purification of the FK1 domain of FKBP51
The FK1 domain of FKBP51 was expressed and purified as previously described [27]. Namely the sequence encoding FK1 domain was codon-optimized, synthesized and cloned into a pET28b-derived vector. Using heat-shock, the plasmid was transformed into Escherichia coli BL21(DE3) competent cells. Then the cells were allowed to grow to an optical density (OD600 nm) between 0.6 and 0.8 in LB medium containing 50 μg/ml kanamycin at 37 °C, 220 rpm. 0.2 mM isopropyl-1-thiogalactopyranoside (IPTG) was added to induce protein expression at 16 °C for 20 h. After centrifugation at 6000g for 30 min, the pellet was resuspended in 20 mM Tris–HCl pH 6.0 and the bacterial cells were disrupted through sonication. After centrifuged at 12,000 g for 30 min, the supernatant was loaded onto a Hitrap SP-Bestarose FF column (GE Healthcare) equilibrated with 20 mM Tris–HCl, pH 6.0. The FK1 protein was eluted with a NaCl gradient ranging from 20 mM to 500 mM and loaded onto a HiLoadTM 16/60 SuperdexTM 200 column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 8.0, 100 mM NaCl. The purified FK1 domain was finally concentrated to 50 mg/ml, flash-frozen in liquid nitrogen and stored −80 °C for further usage.
2.7. Steady-state fluorescence quenching assay
The intrinsic fluorescence of the FK1 protein was monitored to investigate the interaction between the FK1 domain of FKBP51 and rapamycin. The FK1 protein with a final concentration of 20 μM was titrated with 100 μM rapamycin in the buffer of 100 mM NaCl, 20 mM Tris pH 8.0. At 295 nm excitation, the emission fluorescence in range of 310–400 nm was collected using a PerkinElmer LS 55 fluorescence spectrometer.
2.8. Crystallization and data collection
Protein crystallization was carried out by using the vapor-diffusion method at 16 °C. To set up the crystallization trials, 90 μl FKBP51 (30 mg/ml) was mixed with 10 μl rapamycin (10 mM), and the crystallization condition is 30% PEG 3350, 0.2 M NH4 acetate and 0.1 M HEPES pH 7.5. Crystals appeared in 1 day and grew to full size within 3–7 days. For data collection, the crystal was transferred to the cryoprotectant consisting of 50% saturated sucrose. X-ray data were collected using a Q315r CCD detector (ADSC) at the beamline BL17U1 at Shanghai Synchrotron Radiation Facility (SSRF) and processed using the HKL-2000 program [40].
2.9. Structure determination
The structure of FK1 was determined by the molecular replacement method with MOLREP [41] using the FKBP51 structure (PDB code 3O5R [42]) as the search model. The model was first refined by the REFMAC5 program [43] and rebuilt interactively in the program COOT [44] by using the σA-weighted electron density maps. The B-factors were then refined anisotropically by using the program phenix.refine [45]. One rapamycin molecule was added in the final model and the final model was validated with the programs MOLPROBITY [46] and PROCHECK [47]. The structure figures were prepared with the program PyMOL [48]. The sequence alignment was performed by using the programs MultAlin [49] and ESPript [50].
3. Results
3.1. Interaction between rapamycin and FKBP51
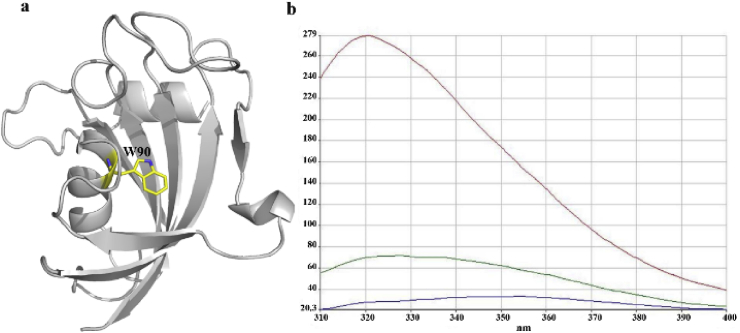
The major functional domain of FKBP51, namely the FK1 domain, was purified in our previous work [27]. To study whether rapamycin can interact with FKBP51, a steady-state fluorescence quenching assay was carried out and the result is displayed in Fig. 1. As structural analysis reveals there is only one tryptophan residue located at the bottom of the FK1 binding pocket (W90 in Fig. 1a), the excitation wavelength of 295 nm was used in this study to selectively excite the tryptophan residue only. As seen in Fig. 1b, the FK1 protein itself has strong fluorescence intensity (red curve). When 100 μM rapamycin was added to the FK1 solution, the initial fluorescence was strongly quenched (green curve) indicating a direct interaction between the binding pocket of FK1 and rapamycin. In the titration, the fluorescence of rapamycin alone was also monitored to eliminate interference come from the ligand (blue curve). The result indicates that rapamycin binds to the active pocket of FK1 which may interfere with the local chemical environment of W90.
Fig. 1.
Steady-state fluoresce quenching assay. a. The binding pocket of FK1 with the W90 residue shown in yellow sticks. b. Fluorescence emission spectra of FKBP51 and rapamycin resulting from excitation at 295 nm. The fluorescence intensity spectra of the FK1 domain (20 μM) and rapamycin (100 μM) alone are shown in red and purple, respectively. The green curve represents the fluorescence intensity spectra of 20 μM FK1 titrated with 100 μM rapamycin. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Protein-ligand interactions
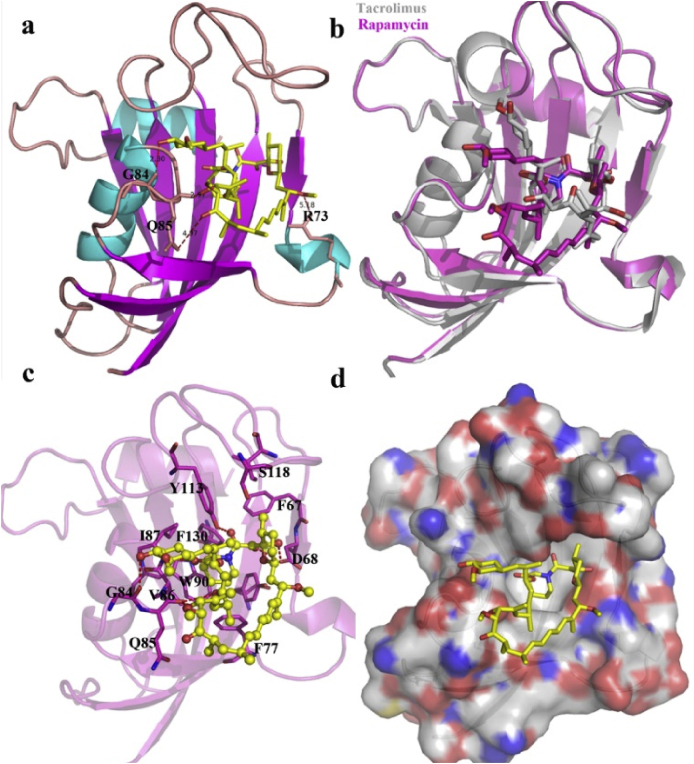
The structure of the determined FK1 domain is comprised of a twisted five-stranded anti-parallel β-sheet wrapping around a short α-helix (Fig. 2). The newly crystallized FK1 structure is aligned well with the pre-resolved FK1 structure (grey cartoon in Fig. 2b, PDB code: 3O5R). In addition, the binding partner rapamycin shares a very similar binding pose to that observed for the ligand FK506 (tacrolimus) embedding in the FK1 region and occupying the active sites (Fig. 2a and c). Remarkably, the macrolide moiety of rapamycin and FK506 locate in the same direction with the cyclohexane motif protruding outside the binding pocket. Binding of rapamycin to FK1 is rather rigid and doesn't induce significant conformational changes in the FK506 binding pocket (Figs. 2b and 3). The piperidine group of rapamycin sits face-to-face with the side chain of the residue W90 in the binding pocket. This is corroborated with the results of our fluorescence quenching assay that rapamycin joins the vicinity of the W90 and quenches the inherent fluorescence of the only tryptophan residue in the FK1 domain. Rapamycin binding to FK1 forms several hydrogen bond interactions with the backbone atoms of the surrounding D68, G84, Q85, I87 and Y113 residues which would contribute favorably to the binding process. Apart from the hydrogen bond interactions, hydrophobic interactions from residues such as F67, F77, V86, S118, F130 also contribute greatly to the components binding.
Fig. 2.
The interaction between FK1 domain of FKBP51 and rapamycin. a. Binding pocket of rapamycin (yellow sticks) on FK1 domain (cartoon representation). b. Superposition of the binding poses of rapamycin (magenta) and tacrolimus (grey) in the FK1 domain. c. The binding interaction between rapamycin and FK1 with those residues contributing to the binding shown in magenta sticks. d. Surface representation of the FK1 domain with rapamycin shown in yellow sticks. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
The chemical structures of rapamycin and tacrolimus.
3.3. Rapamycin inhibits AR signaling
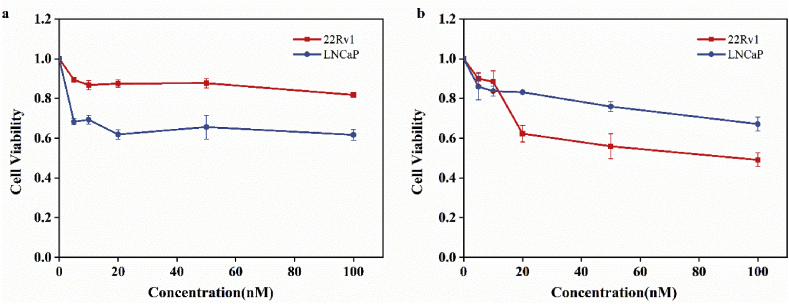
As AR-mediated signaling is essential for the proliferation of prostate cancer cells, androgen-dependent cells LNCaP and androgen responsive cells 22Rv1 were used as vitro models to investigate whether targeting FKBP51 could inhibit AR signaling. As can be seen in Fig. 4, both 22Rv1 and LNCaP cells growth were obviously inhibited when treated with rapamycin. The androgen-dependent (with 1 nM DHT supplied) proliferation of two cell lines was inhibited with a concentration-dependent manner.
Fig. 4.
Results of MTT assay of LNCaP and 22Rv1 cells treated with 1 nM dihydrotestosterone (DHT) and different concentrations (5 nM, 10 nM, 20 nM, 50 nM, 100 nM) of rapamycin for 24 h (a) and 48 h (b).
Treating LNCaP cells with 100 nM rapamycin for 24 h (Fig. 4a) and 48 h (Fig. 4b) resulted in about 40% and 50% growth inhibition, respectively. For 22Rv1 cells, the growth inhibition rates were ~15% and ~30% when treated for 24 h and 48 h, respectively. The MTT assay suggested that the occupation of the FK1 domain with rapamycin may suppress the androgen-induced growth of prostate cancer cells via inhibiting AR signaling.
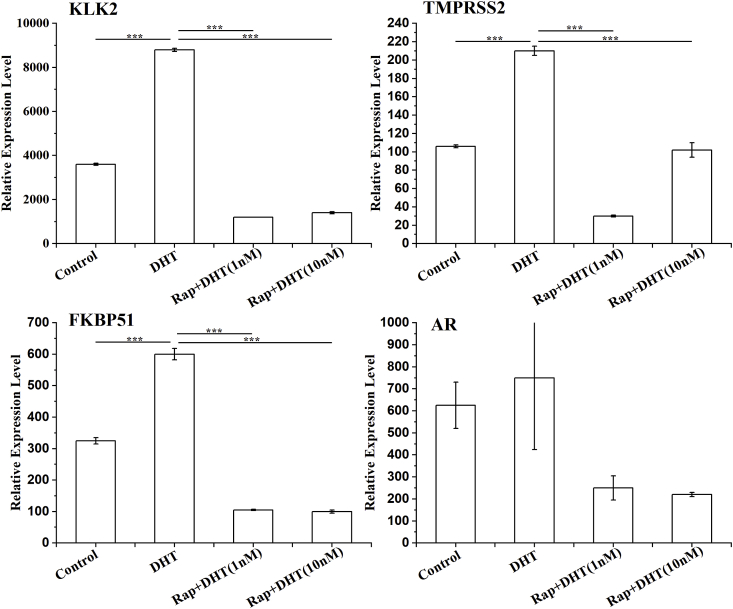
To determine the inhibition level of AR-activated gene transcription in the presence of rapamycin, a quantitative real-time PCR was performed. The results showed that in the presence of DHT, rapamycin inhibited the expression of AR and FKBP51 as well as the expression of downstream genes KLK2 and TMPRSS2, which are regulated by AR (Fig. 5). Among them, the mRNA expressions of KLK2 and TMPRSS2 were significantly down-regulated while the expression level of AR indicated no significance.
Fig. 5.
Rapamycin inhibits AR-regulated downstream gene mRNA expression. LNCaP cells were starved for 12 h, then pretreated with rapamycin at 30 nM for 2 h, and then incubated with DHT (1 nM or 10 nM) for 12 h. Relative expression levels of KLK2 (a), TMPRSS2 (b), FKBP51 (c) and AR (d) were assayed by qRT-PCR. All the experiments were repeated at least three times. GAPDH was used as a loading control. *** indicates that P < 0.001.
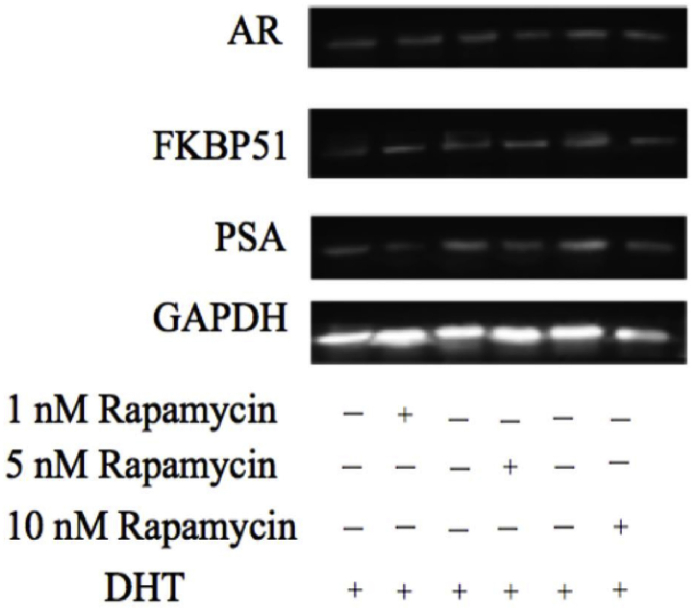
To further examine the inhibition effect of rapamycin on protein, we examined the protein expression of PSA, AR, and FKBP51 using western blot. LNCaP cells were incubated with different concentrations of rapamycin in the presence of 1 nM DHT. After 24 h, the proteins were extracted to detect with western blot analysis, GAPDH used as control. The results showed that different concentrations of rapamycin inhibited the expression of PSA protein under DHT conditions (Fig. 6). However, rapamycin did not induce influence on the protein expression of AR and FKBP51. Overall, we speculate that rapamycin may inhibit the activity of AR without influence the protein expression.
Fig. 6.
Western blot analysis of key protein (AR, FKBP51, and PSA) expression level after treatment with different concentrations of rapamycin and 1 nM DHT for 24 h.
4. Discussion
In this work, the interaction between rapamycin and the FK1 domain of FKBP51 was investigated. Rapamycin that could interact with the FK1 domain of FKBP51 was identified by fluorescence quenching assay. Structure of the rapamycin-FK1 complex revealed that rapamycin occupies a hydrophobic pocket of FKBP51 which is important for AR activity. Structural analysis indicated that rapamycin is stably bound to the FKBP51 pocket and the hotspot residues involved in rapamycin binding are mainly hydrophobic which may overlap with the AR interaction sites. Although there have been some reports on the role of FKBP51 and AR in the literature, there is no unified conclusion on whether FKBP51 has an enhancement effect on AR [[51], [52], [53]].
The structure of the binary complex we obtained is basically the same as that of Marz et al., which is a ternary complex of the FK1 domain of FKBP51, the FRB domain of mTOR, and rapamycin [54]. The rapamycin poses in the active pockets of the binary and ternary complexes were identical, but the loop region of the protein was different, and the two loop regions were closer together in the binary complex than in the ternary complex.
We further showed that rapamycin inhibited the androgen-dependent growth of the prostate cancer cells and down-regulated the expression levels of AR activated genes. This reveals another possible signaling pathway for rapamycin in prostate cancer cells, which inhibits the growth of prostate cancer cells via the AR signaling pathway. However, the role of FKBP51 in AR signalling appears controversially, as both positive and negative modulatory effects of FKBP51 on activity of AR have been reported [55,56]. Overall, our studies demonstrate that rapamycin suppresses the AR signaling pathway by interacting with FKBP51, providing a new mechanism of rapamycin in prostate cancer therapy. In addition, rapamycin may act as a bridge connecting the mTOR signaling pathway and the AR signaling pathway and further research is needed to verify whether these two signaling pathways have cross-interaction. This provides a new mechanism for rapamycin in anti-tumor, paving the way for further research on clinical research of castration-resistant prostate cancer.
Although preclinical studies have shown that rapamycin analogues can inhibit the formation of prostatic epithelioma in mice [57], with good efficacy in the treatment of metastatic renal cell carcinoma and pancreatic neuroendocrine tumors at cell level, the application of rapamycin analogues in the treatment of men with CRPC did not cause significant improvements in clinical symptoms, serum PSA levels, and time to disease progression [[58], [59], [60]]. We will continue to study the relevant mechanisms to understand why the clinical application of rapamycin is not effective as that at the cell level in our future study.
For androgen-independent AR activation, enzalutamide and abiraterone may become ineffective. However, the clinical application of two more beneficial drugs, docetaxel and cabazitaxel, are limited due to severe adverse effects and drug resistance. In the last few years, several new agents for treatment of mCRPC have shown survival benefits in the clinical, including sipuleucel-T, Radium-223, denosumab, and bisphosphonates. Currently, novel treatments such as immunotherapeutics, therapies targeting other oncogenic and genomic pathways, particularly PARP inhibitors and PD-1 inhibitors, are under clinical investigation [61]. It may be a good choice of prostate cancer treatment to develop the appropriate individualized therapy, which will improve clinical outcomes, and prevent unfavorable side effects and costly therapies. In-depth understanding of the action mechanism of FKBP51 inhibitor on AR pathway will help improve the treatment efficacy of AR pathway.
Author statement
I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere. All the authors transfer all copyright ownership of this manuscript to Biochemistry and Biophysics Reports, in the event the work is published. The authors confirm that they have reviewed and approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility for their assistance in the data collection. This work was supported by the grants from the National Natural Science Foundation of China (Grant Nos. 81874088), Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital (CY2017-MS01) and Special Fund for Doctoral Students of Lanzhou University Second Hospital (YJS-BD-20).
References
- 1.Qi D., Wu C., Liu F., Gu K., Shi Z., Lin X., Tao S., Xu W., Brendler C.B., Zheng Y., Xu J. Trends of prostate cancer incidence and mortality in Shanghai, China from 1973 to 2009. Prostate. 2015;75:1662–1668. doi: 10.1002/pros.23046. [DOI] [PubMed] [Google Scholar]
- 2.Chen W. Cancer statistics: updated cancer burden in China. Chin. J. Canc. Res. 2015;27:1. doi: 10.3978/j.issn.1000-9604.2015.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westdorp H., Skold A.E., Snijer B.A., Franik S., Mulder S.F., Major P.P., Foley R., Gerritsen W.R., de Vries I.J. Immunotherapy for prostate cancer: lessons from responses to tumor-associated antigens. Front. Immunol. 2014;5:191. doi: 10.3389/fimmu.2014.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acar O., Esen T., Lack N.A. New therapeutics to treat castrate-resistant prostate cancer. Sci. World J. 2013;2013 doi: 10.1155/2013/379641. 379641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantoff P.W., Higano C.S., Shore N.D., Berger E.R., Small E.J., Penson D.F., Redfern C.H., Ferrari A.C., Dreicer R., Sims R.B., Xu Y., Frohlich M.W., Schellhammer P.F., Investigators I.S. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 6.Cai C., He H.H., Chen S., Coleman I., Wang H., Fang Z., Chen S., Nelson P.S., Liu X.S., Brown M.J. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Canc. Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knudsen K.E., Penning T.M. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol. Metabol. 2010;21:315–324. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peter S N. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. J. Clinc. Oncol.: Off. J. Am. Soc. Clinc. Oncol. 2012;6 doi: 10.1200/JCO.2011.39.1300. [DOI] [PubMed] [Google Scholar]
- 9.Eick G.N., Thornton J.W.J., endocrinology c. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Molecular. 2011;334:31–38. doi: 10.1016/j.mce.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 10.van de Wijngaart D.J., Molier M., Lusher S.J., Hersmus R., Jenster G., Trapman J., Dubbink H.J.J. Systematic structure-function analysis of androgen receptor Leu701 mutants explains the properties of the prostate cancer mutant L701H. J. Biol. Chem. 2010;285:5097–5105. doi: 10.1074/jbc.M109.039958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heemers H.V., Tindall D.J.J. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr. Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M.J. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yepuru M., Wu Z., Kulkarni A., Yin F., Barrett C.M., Kim J., Steiner M.S., Miller D.D., Dalton J.T., Narayanan R.J. Steroidogenic enzyme AKR1C3 is a novel androgen receptor-selective coactivator that promotes prostate cancer growth. Clin. Canc. Res. 2013;19:5613–5625. doi: 10.1158/1078-0432.CCR-13-1151. [DOI] [PubMed] [Google Scholar]
- 14.Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Canc. Res. 2013;73:4599–4605. doi: 10.1158/0008-5472.CAN-12-4414. [DOI] [PubMed] [Google Scholar]
- 15.Mitsiades N., Sung C.C., Schultz N., Danila D.C., He B., Eedunuri V.K., Fleisher M., Sander C., Sawyers C.L., Scher H.I. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Canc. Res. 2012;72:6142–6152. doi: 10.1158/0008-5472.CAN-12-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karantanos T., Corn P.G., Thompson T.C. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sternberg C., De Bono J., Chi K., Fizazi K., Mulders P., Cerbone L., Hirmand M., Forer D., Scher H.J. Improved outcomes in elderly patients with metastatic castration-resistant prostate cancer treated with the androgen receptor inhibitor enzalutamide: results from the phase III AFFIRM trial. Ann. Oncol. 2014;25:429–434. doi: 10.1093/annonc/mdt571. [DOI] [PubMed] [Google Scholar]
- 18.Efstathiou E., Titus M., Wen S., Hoang A., Karlou M., Ashe R., Tu S.M., Aparicio A., Troncoso P., Mohler J., Logothetis C.J. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur. Urol. 2015;67:53–60. doi: 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C., Chen Y., Mohammad T.A., Chen Y., Fedor H.L., Lotan T.L., Zheng Q., De Marzo A.M., Isaacs J.T., Isaacs W.B., Nadal R., Paller C.J., Denmeade S.R., Carducci M.A., Eisenberger M.A., Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bambury R.M., Rathkopf D.E. Elsevier; 2016. Novel and Next-Generation Androgen Receptor–Directed Therapies for Prostate Cancer: beyond Abiraterone and Enzalutamide, Urologic Oncology: Seminars and Original Investigations; pp. 348–355. [DOI] [PubMed] [Google Scholar]
- 21.De Maeseneer D.J., Van Praet C., Lumen N., Rottey S. Elsevier; 2015. Battling Resistance Mechanisms in Antihormonal Prostate Cancer Treatment: Novel Agents and Combinations, Urologic Oncology: Seminars and Original Investigations; pp. 310–321. [DOI] [PubMed] [Google Scholar]
- 22.Hähle Andreas, Merz Stephanie, Meyners Christian, Hausch Felix. The many faces of FKBP51. Biomolecules. 2019;9 doi: 10.3390/biom9010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storer C.L., Dickey C.A., Galigniana M.D., Rein T., Cox M.B. FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol. Metabol. 2011;22:481–490. doi: 10.1016/j.tem.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni L., Yang C.S., Gioeli D., Frierson H., Toft D.O., Paschal B.M. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol. Cell Biol. 2010;30:1243–1253. doi: 10.1128/MCB.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guy N.C., Garcia Y.A., B Cox M.J. Therapeutic targeting of the FKBP52 co-chaperone in steroid hormone receptor-regulated physiology and disease. Curr. Mol. Pharmacol. 2016;9:109–125. doi: 10.2174/1874467208666150519114115. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt M.V., Paez‐Pereda M., Holsboer F., Hausch F.J. The prospect of FKBP51 as a drug target. ChemMedChem. 2012;7:1351–1359. doi: 10.1002/cmdc.201200137. [DOI] [PubMed] [Google Scholar]
- 27.Wu D., Tao X., Chen Z.-P., Han J.-T., Jia W.-J., Zhu N., Li X., Wang Z., He Y.-X.J. The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells. J. Hazard Mater. 2016;307:193–201. doi: 10.1016/j.jhazmat.2015.12.045. [DOI] [PubMed] [Google Scholar]
- 28.Yu J., Sun L., Hao T., Zhang B., Chen X., Li H., Zhang Z., Zhu S., Quan C., Niu Y. Restoration of FKBP51 protein promotes the progression of castration resistant prostate cancer. Ann. Transl. Med. 2019;7 doi: 10.21037/atm.2019.11.127. 729-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panwalkar A., Verstovsek S., Giles F.J.J. Mammalian target of rapamycin inhibition as therapy for hematologic malignancies. Canc. Res. 2004;100:657–666. doi: 10.1002/cncr.20026. [DOI] [PubMed] [Google Scholar]
- 30.Noh W.-C., Mondesire W.H., Peng J., Jian W., Zhang H., Dong J., Mills G.B., Hung M.-C., Meric-Bernstam F. Determinants of rapamycin sensitivity in breast cancer cells. Clin. Canc. Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- 31.Hosoi H., Dilling M.B., Shikata T., Liu L.N., Shu L., Ashmun R.A., Germain G.S., Abraham R.T., Houghton P.J.J. Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Canc. Res. 1999;59:886–894. [PubMed] [Google Scholar]
- 32.Miyake N., Chikumi H., Takata M., Nakamoto M., Igishi T., Shimizu E.J. Rapamycin induces p53-independent apoptosis through the mitochondrial pathway in non-small cell lung cancer cells. Oncol. Rep. 2012;28:848–854. doi: 10.3892/or.2012.1855. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Mikhailova M., Bose S., Pan C.X., deVere White R.W., Ghosh P.M. Regulation of androgen receptor transcriptional activity by rapamycin in prostate cancer cell proliferation and survival. Oncogene. 2008;27:7106–7117. doi: 10.1038/onc.2008.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morikawa Y., Koike H., Sekine Y., Matsui H., Shibata Y., Ito K., Suzuki K.J., communications b.r. Rapamycin enhances docetaxel-induced cytotoxicity in a androgen-independent prostate cancer xenograft model by survivin downregulation. Biochemical. 2012;419:584–589. doi: 10.1016/j.bbrc.2012.02.089. [DOI] [PubMed] [Google Scholar]
- 35.Imrali A., Mao X., Yeste-Velasco M., Shamash J., Lu Y.J. Rapamycin inhibits prostate cancer cell growth through cyclin D1 and enhances the cytotoxic efficacy of cisplatin. Am. J. Canc. Res. 2016;6:1772. [PMC free article] [PubMed] [Google Scholar]
- 36.Amato R.J., Jac J., Mohammad T., Saxena S.J. Pilot study of rapamycin in patients with hormone-refractory prostate cancer. Clin. Genitourin. Canc. 2008;6:97–102. doi: 10.3816/CGC.2008.n.015. [DOI] [PubMed] [Google Scholar]
- 37.Audetwalsh E., Dufour C.R., Yee T., Zouanat F.Z., Yan M., Kalloghlian G., Vernier M., Caron M., Bourque G., Scarlata E. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev. 2017;31:1228–1242. doi: 10.1101/gad.299958.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filippone S.M., Samidurai A., Roh S.K., Cain C.K., He J., Salloum F.N., Kukreja R.C., Das A., longevity c. Reperfusion therapy with rapamycin attenuates myocardial infarction through activation of AKT and ERK. Oxidative medicine. 2017:2017. doi: 10.1155/2017/4619720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y.Q., Guerra‐Librero A., Fernandez‐Gil B.I., Florido J., García‐López S., Martinez‐Ruiz L., Mendivil‐Perez M., Soto‐Mercado V., Acuña‐Castroviejo D., Ortega‐Arellano H. Combination of melatonin and rapamycin for head and neck cancer therapy: suppression of AKT/mTOR pathway activation, and activation of mitophagy and apoptosis via mitochondrial function regulation. J. Pineal Res. 2018;64 doi: 10.1111/jpi.12461. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 42.Bracher A., Kozany C., Thost A., Hausch F. Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:549–559. doi: 10.1107/S0907444911013862. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams P.D., Grosse-Kunstleve R.W., Hung L.-W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 46.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., III, Snoeyink J., Richardson J.S. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 48.DeLano W.L. The PyMOL molecular graphics system. 2002. http://www.pymol.org
- 49.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gouet P., Robert X., Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni L., Yang C., Gioeli D., Frierson H.F., Toft D.O., Paschal B.M. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol. Cell Biol. 2010;30:1243–1253. doi: 10.1128/MCB.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barent R.L., Nair S.C., Carr D.C., Ruan Y., Rimerman R.A., Fulton J., Zhang Y., Smith D.F. Analysis OF FKBP51/FKBP52 chimeras and mutants for HSP90 binding and association with progesterone receptor complexes. Mol. Endocrinol. 1998;12:342–354. doi: 10.1210/mend.12.3.0075. [DOI] [PubMed] [Google Scholar]
- 53.Riggs D.L., Cox M.B., Tardif H.L., Hessling M., Buchner J., Smith D.F. Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol. Cell Biol. 2007;27:8658–8669. doi: 10.1128/MCB.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marz A., Fabian A., Kozany C., Bracher A., Hausch F. Large FK506-binding proteins shape the pharmacology of rapamycin. Mol. Cell Biol. 2013;33:1357–1367. doi: 10.1128/MCB.00678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Periyasamy S., Hinds T.J., Shemshedini L., Shou W., Sanchez E.R. FKBP51 and Cyp40 are positive regulators of androgen-dependent prostate cancer cell growth and the targets of FK506 and cyclosporin A. Oncogene. 2009;29:1691–1701. doi: 10.1038/onc.2009.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulke J., Wochnik G.M., Langrollin I., Gassen N.C., Knapp R.T., Berning B., Yassouridis A., Rein T. Differential impact of tetratricopeptide repeat proteins on the steroid hormone receptors. PloS One. 2010;5 doi: 10.1371/journal.pone.0011717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majumder P.K., Febbo P.G., Bikoff R., Berger R., Xue Q., Mcmahon L., Manola J., Brugarolas J., Mcdonnell T.J., Golub T.R. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat. Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 58.Hudes G.R., Carducci M.A., Tomczak P., Dutcher J.P., Figlin R.A., Kapoor A., Staroslawska E., Sosman J.A., Mcdermott D.F., Bodrogi I. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N. Engl. J. Med. 2007;356:2271–2281. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 59.Yao J.C., Shah M.H., Ito T., Bohas C.L., Wolin E.M., Van Cutsem E., Hobday T.J., Okusaka T., Capdevila J., De Vries E.G.E. Everolimus for advanced pancreatic neuroendocrine tumors. N. Engl. J. Med. 2011;364:514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.George D.J., Halabi S., Healy P., Jonasch D., Anand M., Rasmussen J., Wood S.Y., Spritzer C.E., Madden J.F., Armstrong A.J. Phase 2 clinical trial of TORC1 inhibition with everolimus in men with metastatic castration-resistant prostate cancer. Urol. Oncol. seminars Original Invest. 2020;38 doi: 10.1016/j.urolonc.2019.08.015. [DOI] [PubMed] [Google Scholar]
- 61.Nuhn P., De Bono J.S., Fizazi K., Freedland S.J., Grilli M., Kantoff P.W., Sonpavde G., Sternberg C.N., Yegnasubramanian S., Antonarakis E.S. Update on systemic prostate cancer therapies: management of metastatic castration-resistant prostate cancer in the era of precision oncology. Eur. Urol. 2019;75:88–99. doi: 10.1016/j.eururo.2018.03.028. [DOI] [PubMed] [Google Scholar]