Abstract
Colorectal cancer (CRC) is one of the leading causes of cancerrelated death worldwide. Despite progress in treatment of cancers, CRC with KRAS mutations are resistant towards anti-EGFR treatment. MicroRNAs have been discovered in an exponential manner within the last few years and have been known to exert either an onco-miRNA or tumor suppressive effect. Here, the various roles of microRNAs involved in the initiation and progression of KRAS-regulated CRC are summarized. A thorough understanding of the roles and functions of the plethora of microRNAs associated with KRAS in CRC will grant insights into the provision of other potential therapeutic targets as well as treatment. MicroRNAs may also serve as potential molecular classifier or early detection biomarkers for future treatment and diagnosis of CRC.
Key words: Cancer, K-ras, gene, protein, colorectal, microRNA, regulation
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the fourth leading cause of cancer death worldwide. 1 CRC is a multistep carcinogenesis caused by the accumulation of genetic mutations and alteration in signaling pathways. 35% of CRC is due to gene mutations,2 with KRAS gene mutations accounting for 40% of these CRC cases and NRAS accounting for nearly 5%.3 Lifestyle factors such as smoking, lack of exercise and fat-rich diet, increase the chances of having CRC. Aging itself can also result in epigenetic changes that contributes to cancer. Sedentary lifestyle has increased CRC incidence in younger population by approximately 2%.4
RAS proteins are GTPases that regulate the RAS signaling pathway that control cell proliferation and cell survival and are often mutated in human cancers. Human RAS genes are comprised of Kirsten RAS (KRAS), Neuroblastoma RAS (NRAS) and Harvey RAS. The first 85 amino acid residues at the N-termini of all three ras isoforms are highly related - possessing a role in GTPase activity.5 However, different isoforms of ras protein are involved in different types of cancer. For example, KRAS mutations are frequently found in solid tumors such as lung, colorectal and pancreatic cancers,6 whereas NRAS are found mostly in hematopoietic tumors and melanomas.6,7
85% of KRAS gene mutations occurs in codons 12 and 13 of exon 2, while the remaining 15% is found within codon 61 of exon 3.8 During carcinogenesis, activation of KRAS proteins was not required for tumor initiation, however the activation significantly increased tumor incidence and accelerates tumor growth.9 KRAS mutations have been detected in both early and late CRC, indicating that KRAS mutations might occur in the early stage of tumor development.6
microRNAs (miRNA)
MicroRNAs (miRNA) are a class of small, single stranded, non-coding regulatory RNA molecules, approximately 20 nucleotides in length. An endogenous miRNA regulates gene expression by binding to complementary 3’ untranslated region (UTR) of target gene resulting in the degradation of mRNA or a repression in translation.10 The biogenesis of miRNA consists of the cleavage of primary miRNA (pri-miRNA) into precursor miRNA (pre-miRNA) in the nucleus. These pre-miRNAs will then be exported out into the cytoplasm and further processed into mature miRNAs.11-14 Mature miRNA can be derived from either the 3’ or 5’ ends of the pre-miRNA and presented as miRNA-3p or -5p, respectively.
miRNA is involved in various biological processes such as cell proliferation, migration, invasion, epithelial-mesenchymal transition, tumor initiation and development.14-18 miRNA can function as either tumor suppressor or oncogene in the regulation pathway. For example, miR-143 has a tumor suppressor effect in CRC,19 whereas miR-21 exerts an oncogenic effect.20
microRNAs and colorectal cancer
miRNAs expression vary widely in different cancer types.21 A comprehensive review of miRNAs in CRC will be useful for clarifying and summarizing the roles of miRNAs in CRC. In this review, the involvement of various miRNAs towards KRAS regulation in the context of CRC is elaborated. Dysregulation of miRNAs can be seen in every stage of CRC initiation, progression and development. Let-7, miR-18a and miR-30 can be found in advanced stage CRC, whereas miR-193a is more frequently associated with early stage CRC.22-25 Recent studies reported that miRNAs in tissue are concordant to the expression of those in serum, blood and plasma. miR-193a-3p, miR-23a and miR-338-5p were found to be present in tissue and blood samples. Therefore, miRNAs may be a potential molecular classifier, early detection biomarkers and therapeutic targets for future treatment and diagnosis of CRC.
miRNAs and KRAS
Let-7 family
Lethal-7, or mostly termed as let-7, is one of the earliest discovered miRNAs. Let-7 is negatively regulated with KRAS expression in CRC.26 An overexpression of let-7 reduces KRAS and DNA damage repair genes, such as RAD51 and CDC25.27 There are currently thirteen known members in the let-7 family, locating at nine different loci. All the let-7 family members have highly similar sequence and share a seed region (GAGGUAG), which is a nucleotide motif, an important component for RNA-induced silencing complex target recognition.27
Of the 13 members, Let-7a-1-5p is most frequently downregulated in CRC. Administration of let-7a-1-5p precursor demonstrated a suppressive effect on growth and proliferation in human colon cancer cells, DLD-1 and SW480 cell lines.26 Overexpression of let-7a-1-5p reduces KRAS and c-myc protein expression, but not the KRAS and c-MYC mRNAs.26 Overexpression of let-7a decreases the radiosensitization of cells during therapy.27
Although let-7 is one of the first miRNAs discovered, the functional roles of let-7 family members have yet to be understood. Choo et al. demonstrated that let-7d-3p/5p are both co-expressed in colon cancers - Let-7d-3p specifically downregulates KRAS whereas let-7d-5p upregulates IGF1R and THBS1.11 In a study of 49 Stage II CRC patients, the upregulation of let-7b and let-7d were associated with microsatellite stability (MSS).28 Let-7d-3p targets KRAS protein in vascular smooth muscle cells and a transfection of let-7d-3p decreases the KRAS protein - cell growth were reduced, and the G1 cell cycle was induced when compared to the G2/M phase.29 A recent study by Gunel et al. showed that let-7d- 3p downregulates KRAS and HMGA2 in epithelial ovarian cancers.30 Further study on let-7d and its involvement in the cell cycle is required.
Let-7 complementary sites, LSCs
Polymorphisms, or heterogeneity, within miRNA binding site of target gene affects the ability of miRNA binding to its targeted gene and are associated with the development and progression of cancer.22 There are multiple let-7 complementary sites (LSCs) within the 3'UTR of KRAS mRNA. T to G base substitution on rs712 within the 3’UTR of KRAS mRNA weakens the binding to let-7, increasing the expression of constitutively active KRAS and thus activating the RAS-MAPK pathway which results in carcinogenesis. Let-7 complementary site 1 (LCS1), or also known as rs712, consists of a mismatch base substitution of T to G.31 Patients with SNP polymorphism of T to G substitution in rs712 were associated with an increase in CRC risk, advanced TNM stage with poor differentiation and node metastasis (Figure 1).22,32 Similarly, LCS6 is another intensely studied let-7 complementary site which also consists of a mismatched T to G base substitution. Polymorphism in LCS6, or rs61764370, alters the binding of let-7 to the KRAS mRNA, resulting in the increment of KRAS protein expression and therefore activating its downstream pathway, leading to cell proliferation and causes CRC.33
The evidence for the prognosis of CRC patients with regards to KRAS-LCS6 variants is currently conflicting. Smit et al. reported better prognosis and survival for CRC patients having KRAS-LCS6 variant, in both mutant and wild type KRAS in early stage CRC but not in advanced stage CRC.34 Correspondingly, Sclafani et al. demonstrated that a T-to-G base substitution in rs61764370 provides better prognosis in early stage CRC, with improved complete response, five years progression free survival (PFS) and overall survival (OS) of CRC and benefit from anti-EGFR monoclonal antibodies.35 However, in NORDIC VII cohort study, there was no significant association between KRAS-LCS6 variant with PFS and OS of CRC patients.36 Therefore, further studies on polymorphisms of let-7 in large population samples may yield more conclusive results.
miR-18a
miR-18a-3p is the first miRNA discovered to exclusively targets KRAS, but not HRAS and NRAS. miR-18a belongs to miR-17-92 clusters on the 13q31.1 region. The miR-17-92 cluster is a polycistron (an mRNA that can encode more than one polypeptide) which encodes six mature miRNAs belonging to four seed families, namely miR-17 family (miR-17 and miR-201), miR-18 family, miR-19 family (miR-19a and miR-19b-1) and miR-92 family.37 Expression of miR-18a-3p was inversely correlated with KRAS in HT29 CRC cell, squamous carcinoma A431 cells and fetal hepatic WRL-68 cells.38 Inhibition of miR-18a-3p increases KRAS expression, promotes cell proliferation and promotes the anchorage-independent growth in soft agar. These results were found via conducting experiments in the aforementioned three cell lines. Conversely, introduction of miR-18a-3p suppresses the cell proliferation and anchorage-independent growth.
Figure 1.
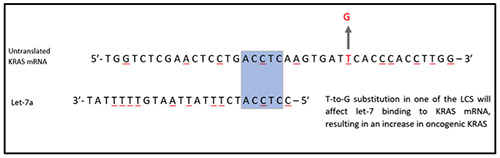
Let-7 Complementary Site (LCS). There are several LCS in KRAS mRNA. When there is a T to G substitution in the LCS, it will disrupt let-7 binding to KRAS mRNA thus resulting in an increase in KRAS as seen in CRC.
miR-18a expression is associated with tumor initiation and progression.23 In CRC clinical samples, miR-18a was found to be upregulated in serum and stool samples of advanced CRC.23,39 MiR-18a-3p may be a potential biomarker for screening and diagnosing CRC. Therefore, miR-18a-3p exerts a tumor suppressive effect on cancer cell lines while the upregulation of miR-18a is found in advanced CRC, proposing a biomarker role.
miR-29b
Another miRNA that has been suggested as a novel and useful prognostic marker for CRC is miR-29b. Putatively, miR-29b-3p is significantly reduced in CRC tumor samples by regulating apoptosis and cell cycle in CRC.40 This expression is an independent prognostic factor for disease-free survival, lymph node metastasis and T classification. For 5-year OS, similar association between lymph node metastasis, T classification, venous invasion and miR- 29b expression was found.41 In vitro, an administration of mimicmiR- 29b-3p showed low Ki-67, indicating a low proliferation of CRC cells. Moreover, miR-29b arrests cells at the G1/S cell transition in CRC cells.41
miR-29b expression was further applied as a treatment for KRAS mutant CRC. As an extension on the study of miR-29b for CRC treatment, Inoue et al. investigated the role of complementary strand of miR-29b-1-5p in CRC cell lines. In comparison to the expression levels of miR-29b-3p, miR-29b-1-5p was negligible in tumor samples. Remarkably, while miR-29b-1-5p lacks tumor inhibitory effects on KRAS mutant colorectal cancer cells, the complementary nucleic acid sequence illustrated antitumor effect on KRAS mutant CRC. This sequence was termed MIRTX.40
MIRTX significantly inhibits cell proliferation of KRAS mutant DLD1 and SW480 CRC cells, and KRAS-WT HT29 CRC cell lines.40 MIRTX induces apoptosis in DLD1 cells by downregulating protein expression of antiapoptotic proteins MCL1, BCL2 and BCL-xL, and increasing the expression of cleaved PARP and cleaved caspase-3. In addition, MIRTX directly binds to CXCR2 and PIK3R1, and suppresses the NFkB signaling pathway. In mouse xenograft models, systemic administration of MIRTX inhibits tumor growth without any reported side effects.40 In summary, miR-29b and the complementary strand of miR-29b-1-5p exerts anti-tumor and anti-apoptotic effect in both mutated and wild type KRAS CRC, by inducing targeting the apoptotic markers and the NFkb pathway.
Figure 2.
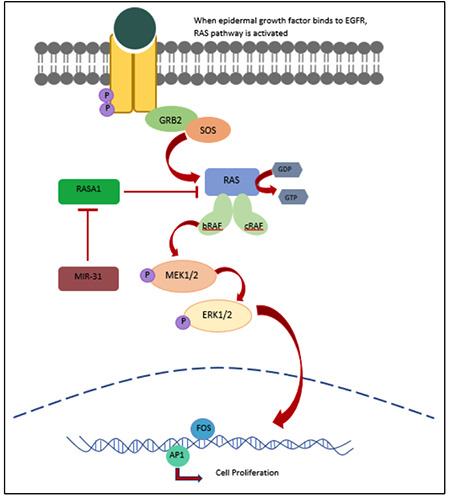
RAS-RAF pathway in relation to miR-31 and RASA1. When a growth factor ligand binds to epidermal growth factor receptor (EGFR), phosphorylation of the protein tyrosine kinase results in the recruitment of Grb2 adaptor protein and son of sevenless (SOS). This phosphorylation activates the RAS protein, which consequently bind to Raf kinase. Phosphorylated RAF in turn phosphorylates and activates subsequent downstream proteins MEK1/2 and ERK1/2, which result in nuclear transcription of genes responsible for cell proliferation. miR-31 directly inhibits RAS p21 GTPase Activating protein (RASA1) which affects the GTPase activity of RAS.
miR-30b
miR-30b-5p is often downregulated in CRC and its low expression is associated with poor differentiation, advanced TNM stage and poor prognosis.24 Tumor suppressive role of miR-30b-5p was demonstrated by Liao et al. when cell proliferation of SW620 and HCT116 colon cancer cell lines and tumor growth in mouse xenograft were reduced with an overexpression of miR-30b-5p. Similarly, inhibition of endogenous miR-30b-5p significantly increases the growth rate of SW480 and HCT15 colon cancer cell lines and caused an increase in colony number and size in soft agar assays.24 Additionally, overexpression of miR-30b-5p promotes G1 cell arrest and induces apoptosis.24 miR-30b-5p directly binds to the 3’UTR of KRAS mRNA, PIK3CD and BCL2. Ectopic expression of miR-30b-5p negatively regulates KRAS, PIK3CD and BCL2. Co-expression of KRAS, PIK3CD and BCL2 enhances the cell proliferation and apoptosis in CRC cells, remarkably, in comparison to single gene expression. Correspondingly, Kao et al. reported that an overexpression of miR-30b-5p significantly decreases the cell invasion and migration by using transwell assay.42,43 Further studies on miR-30b in CRC KRAS and its potential function in EMT await to be done.
miR-31
MiR-31, a high-profile miRNA, and its upregulation is correlated with advanced CRC with poor differentiation.44,45 Wild-type KRAS mCRC patients with high levels of miR-31-3p was significantly associated with shorter PFS.45-47 Moreover, when treated with Cetuximab, patients with high level of miR-31-3p have poor prognosis, with lower PFS and OS.45,48 Congruently, improved PFS and OS were seen in patients with low miR-31-3p when treated with Cetuximab. miR-31-3p may be a potential prognostic biomarker for anti-EGFR therapy against CRC.
Unlike other miRNAs, miR-31 does not directly target 3’UTR of KRAS mRNA. MiR-31-5p acts as a downstream target gene of MAPK signaling pathway.49 miR-31-5p directly targets RAS p21 GTPase Activating Protein 1 (RASA1), and the upregulation of miR-31-5p negatively regulates the expression of RASA1. The inhibition of RASA1 enhances the GTPase activating protein (GAP) for Ras, thus having a potential to prolong the half-life of GTP in KRAS.49 Additionally, phospho-ERK was significantly increased with an increase in miR-31-5p.50 Thus, an inhibition of RASA1 modulates the GTP binding by Ras and the constitutive phosphorylation of ERK1/2 in MAPK signaling pathway as illustrated in Figure 2. This inhibition promotes cell proliferation, suppresses apoptosis and deregulates the cell cycle which results in tumorigenesis.50 This was further corroborated when the size of tumor growth in immunodeficient mouse xenograft tumor model increased in vivo as RASA1 was inhibited.50
Figure 3.
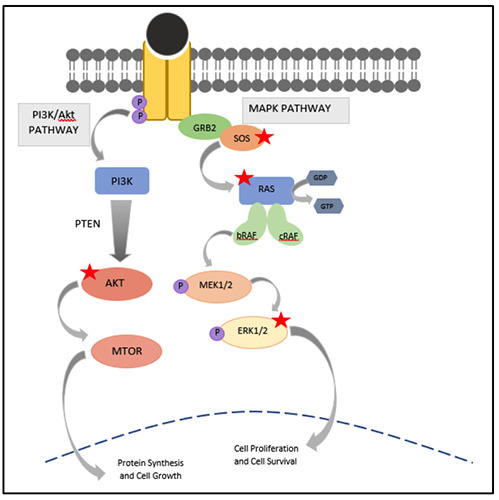
MAPK and PI3K pathways in relation to its interaction with syn-miR-143. Syn-miR-143 binds and inhibits KRAS mRNA and proteins SOS1, ERK and AKT. The red star denotes syn-miR-143.
miR-96
Another newly discovered miRNA in CRC is miR-96-5p. miR- 96-5p has been intensively studied in pancreatic and breast cancers, 51,52 but little is known in CRC. Recently, miR-96-5p was found to be upregulated in CRC samples and is associated with liver metastasis.53,54 Conversely, lower expression of miR-96-5p was significantly correlated with high grade tumor (G3), advanced tumor stage (Stage IV) and poor survival in CRC patients.53 Thus, miR-96-5p was validated to be an independent prognostic factor for colorectal cancer-specific survival.53
In vitro, an increase in miR-96-5p expression reduces the cellular growth of HCT-116 CRC cell lines and also reduces the colonies in soft agar.53 To support this, proliferation marker, cyclin D1, was also reduced, with an increase in cell cycle inhibitor p27 gene.
Administration of resveratrol in mutant KRAS-induced CRC mice showed improved conditions – minimal KRAS expression was found and tumor growth reduced. Mice that develops CRC also had delayed onset upon administration with resveratrol.9 Additionally, an increase in miR-96-5p expression was observed in tumor treated with resveratrol. These data show that the administration of resveratrol in KRAS mutated CRC may reduce the tumor growth and progression, by increasing the expression of miR-96-5p and inhibiting the translation of KRAS mRNA.9
miR-126
Reduced tumor expression of miR-126-5p was found in CRC tumor tissues.55,56 Introduction of miR-126-5p reduces the cell viability of HCT116, LoVo, SW403, SW116 and SW620 CRC cell lines. Low miR-126-5p expression is exclusively found in KRAS mutant cells and not in wild type KRAS cells. Therefore, Hara et al. conducted a study which focuses on the miR-126-5p expression in KRAS mutant cells. Results showed that an overexpression of miR- 126-5p increases the G1 compartment and inhibits colony formation. In the same study, a decreased tumor growth was observed in mouse xenografts formed in miR-126 transfected cells.57
miR-143
Another downregulated miRNA in human CRC is miR-143, and it directly targets KRAS protein.19,58,59 The gene of miR-143 is located on chromosome 5q33.1. Chen et al reported that as premiR-143-3p is transfected in LoVo and SW480 CRC cell, KRAS expression is significantly reduced by the inhibition of KRAS mRNA in the translational level. Conversely, an introduction of anti-miR-143-3p increase expression of K-ras proteins.19
Figure 4.
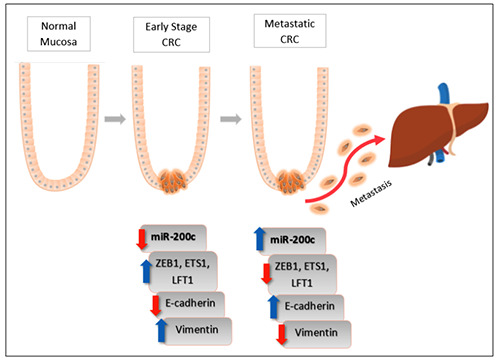
Epithelial-to-Mesenchymal Transition in CRC by an upregulation of miR-200c. In early stage CRC, lower expression of mIR- 200c stimulates the putative target genes Zeb1, ETS1 and LFT1 to suppress the epithelial marker E-cadherin. This results in epithelial cells losing its character and transition into mesenchymal cells, by the increase in vimentin, mesenchymal marker. As CRC progressed, the sudden increase in miR-200c expression decreases the putative target gene expression. This, in turn, increases the epithelial marker E-cadherin, establishing epithelial cells at the metastatic site.
Luciferase reporter assay confirmed the direct interaction between miR-143-3p and KRAS mRNA.19 The inhibition of miR- 143-3p showed an increased level in cell viability and cell proliferation by Ki-67 expression. To investigate the regulation of KRAS-miR-143-3p in MAPK signaling pathway, introduction of pre-miR-143-3p showed to inhibit the constitutive phosphorylation of ERK1/2, which is a downstream component in MAPK pathway, confirming the involvement of miR-143-3p in MAPK signaling pathway.19 Similar result was shown in a pancreatic ductal adenocarcinoma study, where an upregulation in miR-143-3p showed a downregulation in KRAS and ERK.60
Nuclease-resistance synthetic miR-143-3p (syn-miR-143-3p) has demonstrated a potent suppressive effect on human DLD-1 CRC cell harboring KRASG13D mutation, by inhibiting KRAS and its effector signal molecules. The transfection of syn-miR-143-3p on DLD-1 cells silences the expression level of both mRNA and protein levels of KRAS, ERK, AKT and SOS1 as in Figure 3.59 In a similar study, Akao et al. reported that a combination treatment between syn-miR-143-3p and a low dose of cetuximab significantly inhibit cell and tumor growth and inactivates both PI3K/Akt pathway and MAPK pathway, in vivo and in vitro. Conversely, the inactivation of KRAS by siR-KRAS alone was not effective, as the other KRAS effector signal molecules compensate the loss of KRAS. Syn-miR-143-3p affects the KRAS expression by silencing SOS1 expression resulting in the efficacy of the EGFR inhibitors. With more study, this remarkable finding may aid in the treatment of anti-EGFR therapy to KRAS mutant patients by combining synmiR- 143-3p and anti-EGFR antibodies.
In a study of 77 CRC patients with wildtype KRAS, low expression levels of miR-143 was identified as an independent prognostic factor for CRC.61 Patients having low miR-143 expression showed high tumor grade, high tumor stage and elevated CEA tumor marker. Moreover, on EGFR-targeted therapy, patients with low miR-143 level was presented with PFS.
miR-155
miR-155, located within chromosome 21q21, is highly conserved from B-cell integration cluster, which is a non-coding transcript found in activated immune cells.62Although miR-155 is found upregulated in many cancers, there are however various contradicting findings. High miR-155-5p expression was significantly found in most CRC patients and is associated with tumor location, tumor grade, TNM staging and distant metastasis.16,63,64 High serum level of miR-155-5p was found in CRC concordant to tissue samples, and has a significant impact on OS and PFS.65 Parallel testing of serum miR-155-5p and postoperative CEA levels may provide more accurate diagnosis for CRC, as there is a positive correlation between miR-155-5p levels and postoperative CEA levels seen in recurrence and metastatic group of CRC patients.66 Thus, miR-155-5p may act as a tumor biomarker, useful in diagnosis and prognosis of CRC patients. On the other hand, Forzati et al. stated that an increase in miR-155-5p level significantly reduces protein level of KRAS.67 In HT-29 CRC cells, a transfection of mimic miR-155-5p stimulates cell proliferation and produces the highest invasion metastasis on transwell test when compared to anti-miR-155-5p.16 miR-155-5p is induced by factors that promote tumor inflammation, and currently miR-155-5p is known to target more than 100 genes, thus these target genes may affect the conflicting outcome. More studies in miR- 155-5p and its effect are needed.
miR-193
miR-193a-3p expression is downregulated in CRC tissue and is significantly correlated with early stage CRC, with an increase in polyp formation and mucosa perforation.25 Dual Luciferase Reporter Assay confirmed that miR-193a-3p directly targets the 3’UTR of KRAS mRNA.68 Downregulation of miR-193a-3p was found more in SW480 (Stage II) than SW48 (Stage III) CRC cell lines, when compared to the non-neoplastic colon epithelial FHC cells - This supports the downregulation of miR-193a-3p in early stage CRC.
When ectopic expression of miR-193a-3p was introduced, the translation of KRAS mRNA was reduced. miR-193a inhibited cell proliferation and migration significantly as shown by EMT marker TWIST.25,68 miR-193a is involved in cell cycle events - reduction of cells in the G0/G1 phase and an increase in the G2/M phase. Under fluorescence microscope, miR-193a showed apoptotic cell phenotype by showing either shrinkage or fragmentation of the nucleus or the disturbance of cell membrane.25
Significant increase in miR-193a-3p, miR-23a and miR-338-5p were detected in both tissue and blood levels of CRC. The combination of these triple miRNA classifier produced improved diagnostic value when compared to individual or two miRNAs combined. Thus, the triple miRNA classified has become a potential blood biomarker for early detection of CRC.69 Thus, miR-193a-3p exerts a tumor suppressive effect on CRC via reduction of KRAS expression.
miR-200c
Significant upregulation of miR-200c-3p was observed in CRCs and was associated with advanced stage, high grade tumor, lymphovascular invasion and lymph node metastasis.70,71 An increase level of miR-200c-3p showed worse OS and worse recurrence free survival in CRC.71
Incidentally, higher miR-200c-3p expression was exclusively found in CRC with KRAS mutation. Oncogenic KRAS level was higher in DLD-1 cells compared to DKO-4 (DLD-1 cells with reduced mutant KRAS).72 This result was corroborated by a similar study conducted by Tsunoda et al. using different CRC cells, whereby HCT116 cells has higher miR-200c expression vs HKe3 cells (HCT116 cells with deleted mutant KRAS).73 Conclusively, KRAS induces the miR-200c-3p expression in both DLD-1 and HCT116 colorectal cells.
miR-200c-3p is a crucial modulator of EMT by directly targeting the genes ZEB1 (zinc finger E-box-binding homeobox), ETS1 (v-ets erythroblastosis virus E26 oncogene homologue 1) and FLT1 (fms-related tyrosine kinase 1).71,74,75 miR-200c-3p expression was observed more frequently in liver metastasis compared to primary CRC.17 Transfection of miR-200c-3p in RKO and SW620 CRC cell lines stimulated cell proliferation, but reduced cell invasion and migration. The increase in miR-200c suppresses the target genes ZEB1, ETS1 and FLT1 and stimulates the epithelial marker, E-cadherin establishing cells in the distant site. Conversely, in primary CRC, lower expression of miR-200c increases the target genes which result in the loss of E-cadherin and drives infiltrating cells out into the system, which are summarized in Figure 4. Agreeably, Spaderna et al. reported that a loss in basal membrane is associated with distant metastasis and poor survival of CRC.76 Taken together, an increase in miR-200c-3p can be a potential biomarker for distant metastasis.
miR-217
Another novel gene expression regulator is miR-217 which is significantly downregulated in CRC. Early stage CRC showed higher expression of miR-217-5p compared to the advanced stage. Lower levels of miR-217-5p is associated with poorer OS and higher grade tumor.77
Table 1.
Summary of the miRNA and their functions in CRC.
| miRNA | Dysregulation in Function colorectal cancer | KRAS as a target gene (www.targetscan.org) | References | |
|---|---|---|---|---|
| Let-7 | Downregulated | Overexpression of Let-7a-1 in DLD1 and SW480: Inhibits growth and cell proliferation Reduces KRAS and C-myc protein àdecrease the radiosensitization of cells during therapy Overexpression of Let-7d in HCT15: Reduces KRAS mRNA and protein expression in smooth muscle cell Reduces cell growth Induced G1 cell cycle Increase in microsatellite stability (MSS) | KRAS is a target of: Let-7a-3p, Let-7a-2-3p Let-7b-3p Let-7c-3p Let-7f-1-3p Let-7g-3p | 11,26,27,29 |
| miR-18a-3p | Downregulated | Inhibition in HT29, A431 and WRL-68 cells: Increases KRAS expression Promotes cell proliferation Promotes Anchorage-independent growth in soft agar | KRAS is a target of miR-18a-3p, but not miR-18-5p. | 38 |
| miR-29b-3p | Downregulated | Overexpression in SW480 cells: Reduces ki-67 level which showed low cell proliferation, Stimulate apoptosis Arrest cells at G1/S Cell Cycle Overexpression of MIRTX (complementary strand of miR-29b-1-5p) in DLD1 and SW480: Anti-tumor effect on KRAS mutant CRC Inhibits cell proliferation Induces apoptosis - by regulating antiapoptotic and proapoptotic proteins - suppresses NF-kB pathway | KRAS is a target of miR-29b-1-5p, but not miR-29b-2-5p | 41 |
| miR-30b | Downregulated | Overexpression in SW620 and HCT116: Reduces cell proliferation, Reduced tumor growth in mouse xenograft, Increases G1 cell arrest Induces apoptosis | KRAS is a target of both miR-30-5p and miR-30-3p, but not miR-30b-3p | 24 |
| miR-31-3p | Upregulated | miR-31 inhibits RASA1, which enhances GTPase for RAS - Indirectly targets 3’ UTR of KRAS Inhibition of RASA1: Increases phosphorylation of ERK1/2 Promotes cell proliferation Suppresses apoptosis Deregulate cell cycle | KRAS is not listed as a target of miR-31-3p in TargetScan database. | 49 |
| miR-96 | Downregulated | Overexpression in HCT-116 Reduced cell growth Reduced colonies in soft agar Reduced in proliferation marker cyclin D1 Increase in cell cycle inhibitor p27 gene When treated with Resveratrol (in vivo) Reduced KRAS expression Reduced tumor progression and growth Delayed onset of tumor | KRAS is a target of miR-96-5p, but not miR-96-3p | 9,53 |
| miR-126 | Downregulated | Overexpression in HCT116, LoVo, SW403, SW116 and SW620: Reduces cell viability, but not miR-126-3p Increase G1 compartment Inhibit colony formation Increase apoptosis, Decreased tumor growth in mouse xenografts *Lower miR-126 expression is exclusively found in KRAS mutant only | miR-126-5p targets KRAS, | 55-57 |
| miR-143 | Downregulated | Overexpression in LoVo, SW480 and DLD-1 cells: Reduces KRAS expression by inhibiting KRAS mRNA in translational level Inhibits ERK1/2 phosphorylation that results in the Inhibition of tumor cell growth Inhibition in LoVo and SW480 cells: Increased in cell viability and cell proliferation | miR-143-3p targets KRAS, but miR-143-5p does not | 19,59 |
| miR-155 | Upregulated | Overexpression in HT-29 cells: Promotes proliferation, invasion and metastasis | miR-155-5p targets KRAS, but | 16 |
| (Downregulated) | *Transfection of miR-155 reduces KRAS protein level | miR-155-3p does not | 67 | |
| miR-193a-3p | Downregulated | Overexpression in SW480 and SW48 Reduced amount of K-ras proteins Reduces cell proliferation and cell migration by inhibiting EMT markers Involved in cell cycle event - reduction of cells in the G0/G1 phase and increase in G2-M phase Showed apoptotic cell structure by either shrinkage or fragmentation (under fluorescence) | miR-193-3p is a target for KRAS, but not miR-193a-5p | 25,68 |
| miR-200c | Upregulated | Overexpression in RKO and SW620 cells Increases cell proliferation Reduced cell invasion and migration KRAS level is higher in DLD-1 cells and HCT116 cells when compared with their respective knockdown cells. miR-200c is a modulator of EMT by inhibiting ZEB1 and ZEB2 in these cells. *High miR-200c is exclusively found in KRAS mutated CRC | miR-200bc-3p is a target for KRAS, but not miR-200c-5p | 17,71-75 |
| miR-217 | Downregulated | Overexpression in RKO and SW480 cells Inhibit tumor growth Reduces cell proliferation Enhances apoptosis by decreasing Bcl-xl and Bcl-2 miR-217 targets 3’UTR of MAPK1 and KRAS | miR-217 is a target for KRAS | 78,87 |
| miR-384 | Downregulated | Overexpression in SW480 and HCT116 cells Reduces cell viability Inhibits cell migration and invasion Reduction of metastatic nodule in liver (in vivo) Inhibition in LoVo and SW620 cells Stimulates cell viability Increase cell migration and invasion | miR-384 is a target for KRAS | 82 |
| miR-543 | Downregulated | Target gene: KRAS, MTA1 and HMGA2 Overexpression in SW620 and LoVo cells Inhibits KRAS mRNA Reduces expression levels of p-MEK and p-ERK Represses cell proliferation and metastasis Inhibit tumor growth, in vivo *Directly targets KRAS, MTA1 and HMGA2 | miR-543 is a target for KRAS | 84,85 |
| miR-4689 | Downregulated | Overexpression in DLD-1 cells Induces proapoptotic genes, BAX, BAD, BAK and Cyt C Suppresses antiapoptotic genes; BCL2 and BCLXL In vivo, miR-4689 inhibit tumor growth in mouse xenograft *Direct targets: KRAS and AKT1 at mRNA and protein level *Exclusively targets KRASG12V CRC | miR-4689 is not listed as a target for KRAS in TargetScan | 86 |
MiR-217-5p regulates the MAPK signaling pathway as there is a putative binding site of miR-217 within the 3’UTR of both MAPK1 and KRAS mRNA.77,78 Overexpression of miR-217-5p showed a downregulation of MAPK1 and KRAS in RKO and SW480, which are CRC malignant cells. MiR-217-5p was also demonstrated to target KRAS and is downregulated in pancreatic cancer.79,80 Furthermore, overexpression of miR-217-5p inhibits tumor growth and enhances apoptosis by decreasing the expression of Bcl-xl and Bcl-2.77 Conversely, a reduction in miR-217-5p expression increases the cell proliferation of CRC and tumor progression. As miR-217 demonstrated a tumor suppressive effect by inhibiting MAPK1 and KRAS, miR-217-5p could act as an inhibitor that suppresses tumor proliferation and enhances apoptosis by targeting the MAPK pathway.
miR-384
miR-384 was recently identified in CRC, breast cancer and Hepatitis B-related hepatocellular carcinoma.81-83 miR-384-3p is downregulated in CRC and targets the 3’UTR of KRAS. Low expression of miR-384-3p is correlated with invasive depth, lymph node and distant metastasis.82 In a study of SW480 and HCT116 CRC cells, a transfection of mimic-miR-384-3p reduces cell viability and inhibits cell migration and invasion. In vivo, mimic-miR- 384-3p in mice showed significant reduction of visible metastatic nodules in the liver compared to the control. Thus, overexpression of miR-384-3p increases the overall survival of the mice. Conversely, when endogenous miR-384-3p was inhibited in LoVo and SW620 CRC cells, cell viability increases, and it promotes cell migration and invasion. Similar to previous miR-217, miR-384-3p could be a potential therapeutic target by regenerating miR-384-3p to inhibit cell migration and invasion, and to reduce CRC tumorigenesis. 82
miR-543
Another novel miRNA that is downregulated in CRC is miR- 543, and it directly targets KRAS, MTA1 and HMGA2.84 Lower miR-543-3p expression is associated with distant metastatic status of CRC patients and high metastatic potential in CRC cell lines.84 A study by Fan et al. (2016) reported that miR-543-3p expression is downregulated in CRC tissue, APCmin mice and colitis-associated colon cancer mice. In SW620 and LoVo cells, overexpression of miR-543-3p inhibits KRAS mRNA and reduces the expression level of p-MEK and p-ERK, suggesting the involvement of miR- 543-3p in MAPK pathway. Moreover, overexpression of miR-543-3p suppresses cell proliferation and metastasis of CRC cell, and inhibits tumor growth and metastasis in vivo.84,85 Taken together, miR-543-3p illustrates tumor suppressive effect on CRC.
miR-4689
miR-4689 is a novel miRNA that is downregulated in exclusively mutant KRASG12V CRC.86 Lower expression of miR-4689-5p was found in KRAS mutated CRC patients when compared to the normal adjacent mucosa. Transfection of miR-4689-5p in DLD1 cells exhibits apoptotic effect by stimulating the expression of proapoptotic genes, BAX, BAD, BAK and Cyt C, while simultaneously suppressing the expression of antiapoptotic genes, BCL2 and BCLXL. In addition, systemic administration of miR-4689-5p drastically inhibited tumor growth in mouse xenograft.86 Direct interaction between KRAS and AKT1 to miR-4689-5p was detected by luciferase reporter activity, both at the mRNA and protein level.86 This suggests that miR-4689-5p could have a suppressive and apoptotic role in both MAPK and PI3K/AKT pathway which is crucial in EGFR therapy against KRASG12V mutant CRC (Table 1).87
miRNA as a potential treatment
The ability of miRNAs to regulate cell growth and proliferation has opened a new avenue of cancer treatment. miRNA-based therapy against CRC was demonstrated by the study of complementary strand of miR-29b-1-5p, or also termed as MIRTX.41 MIRTX significantly inhibits cell proliferation of KRAS mutant CRC cell lines and suppresses tumor growth in mouse xenografts. Additionally, MIRTX promotes apoptosis by targeting CXCRs and PIK3R1 mRNA which is involved in NF-kB signaling pathway. In the same study, MIRTX regulates the antiapoptotic BCL2, BCL-xL and MCL1 and proapoptotic factors caspase-3 and PARP showing that MIRTX is involved in apoptosis of CRC.41 Akao et al. (2018) constructed a nuclease-resistance synthetic miR-143 (syn-miR- 143) which is a potent suppressive effect that targets the MAPK pathway in KRAS mutated CRC by inhibiting KRAS, ERK, AKT and SOS1, as in Figure 2. A combination treatment of syn-miR-143 and low dose of Cetuximab significantly inhibit cell and tumor growth.59 From these findings, miRNA-based therapy could potentially be attainable against refractory KRAS mutant CRC by inhibiting cell proliferation and inducing apoptosis.
miRNA and its effect on chemotherapy
Cetuximab is an anti-EGFR antibody that has successfully treated mCRC patients. However, CRC patients with KRAS mutation are often resistant to anti-EGFR, thus KRAS mutation status has become the negative predictor prior in receiving anti-EGFR therapy. Recently, administration of chemotherapy has been monitored in relation to the miRNA status.
In the new EPOC (eloxatin peri-operative chemotherapy) study where patients with resectable metastases underwent surgery alone, or surgery with the administration of neoadjuvant chemotherapy, miR-31-3p was evaluated. Patients with high miR- 31-3p tumor showed poor survival and PFS when treated with cetuximab and chemotherapy, thus they do not benefit from the combination therapy prior to surgery.45 When treated with Bevacizumab, patients with low miR-126-3p expression was significantly associated with shorter PFS and OS.55 Whereas, in patients exhibiting high levels of let-7a, a combination therapy of Cetuximab and Irinotecan increases the survival rate and PFS of the patients that harbors KRAS mutation.88 With more study, miRNA may be a potential prognostic marker especially in the effectiveness of a treatment.
miRNA as potential biomarkers
Recent studies have shown that miRNA is present in body fluids, such as blood plasma, serum, urine, saliva and semen.14,39,44 Circulating miRNA are chemically stable and are potentially used for a non-invasive detection tool for CRC. Expression levels of miR-18a, miR-21, miR-92a and miR-221 are upregulated in CRC and is concordant to the high expression in tissue samples.23,89 Moreover, stool-based miR-92a and miR-21 may differentiate proximal CRC from distal CRC.89 From this, miRNA can be a potential biomarker used to diagnose CRC. To increase sensitivity and specificity, miRNAs are also used - a combination of miR-221, miR-135b and miR-18a produces better result when compared with miR-18a alone.23
CRC can also be screened by drawing blood of CRC samples. Concordant expression of miR-23a, miR-193a-3p and miR-338-5p were found in both tissue and blood of CRC samples samples.69 Blood testing for miRNA may be a potent predictor for CRC staging as high levels of miR-18a and miR-29a were found in Stage III CRC patients.39 Additionally, Carcinoembryonic Antigen (CEA) is significantly increased in cancers, mainly in CRC. Parallel testing of miR-155 and serum CEA level preoperatively can provide more accurate diagnosis for CRC.66 Thus, with intense research and clinical trial, a non-invasive procedure such as drawing blood and serum can be an alternative method for screening CRC in the future.
Conclusions
This review outlines the role of different miRNAs in KRASregulated CRC. miRNA is a short stranded, non-coding RNA molecule that is involved in several biological processes such as cell proliferation, migration and invasion and EMT. Different miRNA can act as either tumor suppressor or oncogene.
KRAS mutated CRC are known to be ineffective against anti- EGFR treatment, thus exploring miRNA could open up various avenues to treatment of CRC. Interestingly, some miRNAs are found to interact directly with mutated KRAS in CRC and has produced positive outcome. MiR-200 and miR-126 are found exclusively in KRAS mutated CRC and not in the wild type variant,57,71 whereas complementary strand of miR-29b-1-5p exhibits antitumor effect on KRAS mutated CRC.40 These showed that miRNA could potentially be use for treatment against mutated KRAS with further studies needed.
Certain miRNAs have the potential to become early detection marker through a non-invasive method such as in the blood, stool and serum. For instance, miR-18a was found upregulated in serum and stool samples of advanced stage CRC.23,39 Nevertheless, further validation of miRNA as biomarkers is necessary prior to implementation of the miRNA as biomarkers.
In summary, miRNAs are potential biomarkers and may give insights into CRC prognosis. With the plethora of miRNAs known and more to be discovered, the outlining of the various functions of miRNAs is therefore important to further elucidate CRC and the discovery for potential biomarkers and therapeutic targets.
Acknowledgments
We would like to thank Universiti Brunei Darussalam for financial support and scholarship (UBD/OAVCRI/ CRGWG(020)/180101 and UBD/RSCH/1.6/FICBF(b)/2019/005).
References
- 1.Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2016;1-9. [DOI] [PubMed] [Google Scholar]
- 2.Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Epidemiol Genet 2016;7:105-14. [PMC free article] [PubMed] [Google Scholar]
- 3.Zenonos K, Kyprianou K. RAS signaling pathways, mutations and their role in colorectal cancer. World J Gastrointest Oncol 2013;5:97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahnen DJ, Wade SW, Jones WF, et al. The Increasing Incidence of Young-Onset Colorectal Cancer: A Call to Action. Mayo Clin Proc [Internet] 2014;89:216-24. [DOI] [PubMed] [Google Scholar]
- 5.Jancik S, Drabek J, Radzioch D, Hajduch M. Clinical Relevance of KRAS in Human Cancers. J Biomedcine Biotechnol 2010;2010:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-medarde A, Santos E. Ras in Cancer and Developmental Diseases. Genes Cancer 2011;2:344-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prior IA, Lewis PD, Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res 2012;72:2457-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vakiani E, Solit DB. KRAS and BRAF : drug targets and predictive biomarkers. J Pathol 2011;223:219-29. [DOI] [PubMed] [Google Scholar]
- 9.Saud SM, Li W, Morris NL, et al. Resveratrol prevents tumorigenesis in mouse model of KRAS activated sporadic colorectal cancer by suppressing oncogenic KRAS expression. Carcinogenesis 2014;35:2778-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Z, Qiu X, Wang D, et al. MiR-181a-5p inhibits cell proliferation and migration by targeting KRAS in non-small cell lung cancer A549 cells. Acta Biochim Biophys Sin (Shanghai) 2015;47:630-8. [DOI] [PubMed] [Google Scholar]
- 11.Choo KB, Soon YL, Nguyen PNN, et al. MicroRNA-5p and - 3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci 2014;21:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S, Gregory RI. MicroRNA biogenesis pathway in cancer. Nat Rev Cancer 2015;15:321-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol 2009;27:5848-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baran B, Ozupek N, Calibasi-Kocal G, Basbinar Y. MicroRNA (miRNAs) in Colorectal Cancer. IntechOpen 2018. [Epub ahead of print]. [Google Scholar]
- 15.Guo Y, Bao Y, Yang W. Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis. Int J Mol Sci 2017;18:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Y, Wang H, Sun Z, et al. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int J Exp Pathol 2015;8:6988-94. [PMC free article] [PubMed] [Google Scholar]
- 17.Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-tomesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013;62:1315-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol 2012;59:467-74. [PubMed] [Google Scholar]
- 19.Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009;28:1385-92. [DOI] [PubMed] [Google Scholar]
- 20.Schetter AJ, Okayama H, Harris CC. Role of microRNA in colorectal cancer. Cancer J 2013;18:244-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Slack FJ. MicroRNA-mediated regulation of KRAS in cancer. J Hematol Oncol 2014;7:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X-M, Sun R-F, Li Z-H, et al. A let-7 KRAS rs712 polymorphism increases colorectal cancer risk. Tumor Biol [Internet] 2014;35:831-5. [DOI] [PubMed] [Google Scholar]
- 23.Yau TO, Wu CW, Dong Y, et al. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer 2014;111:1765-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao WT, Ye YP, Zhang NJ, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol 2014;232:415-27. [DOI] [PubMed] [Google Scholar]
- 25.Mamoori A, Wahab R, Islam F, et al. Clinical and biological significance of miR-193a-3p targeted KRAS in colorectal cancer pathogenesis. Hum Pathol 2018;71:145-56. [DOI] [PubMed] [Google Scholar]
- 26.Akao Y, Nakagawa Y, Naoe T. let-7 MicroRNA Functions as a Potential Growth Suppressor in Human Colon Cancer Cells. Biol Pharm Bull 2006;29:903-6. [DOI] [PubMed] [Google Scholar]
- 27.Kolenda T, Przybyla W, Teresiak A, et al. The mystery of let- 7d - A small RNA with great power. Contemp Oncol 2014;18:293-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margetis N, Mariolis-sapsakos T. Let-7 microRNA as a Genetic Regulator of Colorectal Carcinogenesis Process : Principles and Queries. JSM Gastroenterol Hepatol 2017;5:1086. [Google Scholar]
- 29.Yu M-L, Wang J-F, Wang G-K, et al. Vascular Smooth Muscle Cell Proliferation Is Influenced by let-7d MicroRNA and Its Interaction With KRAS. Circ J [Internet] 2011;75:703-9. [DOI] [PubMed] [Google Scholar]
- 30.Gunel T, Dogan B, Gumusoglu E, et al. Regulation of HMGA2 and KRAS gene in epithelial ovarian cancer by miRNA hsalet- 7d-3p. J Cancer Res Ther 2019;15:1321-7. [DOI] [PubMed] [Google Scholar]
- 31.Li Z-H, Pan X-M, Han B-W, et al. A let-7 binding site polymorphism rs712 in the KRAS 3’ UTR is associated with an increased risk of gastric cancer. Tumor Biol [Internet] 2013;34:3159-63. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Q-H, Peng H-X, Zhang Y, et al. rs712 polymorphism within let-7 microRNA-binding site might be involved in the initiation and progression of colorectal cancer in Chinese population. Onco Targets Ther 2015;8:3041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saridaki Z, Weidhaas JB, Lenz H-J, et al. A let-7 microRNAbinding site polymorphism in KRAS predicts improved outcome in metastatic colorectal cancer (mCRC) patients treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res 2014;29:4499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smits KM, Paranjape T, Nallur S, et al. A Let-7 MicroRNA SNP in the KRAS 3′UTR Is Prognostic in Early-Stage Colorectal Cancer. Clin Cancer Res 2011;17:7723-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sclafani F, Chau I, Cunningham D, et al. Prognostic role of the LCS6 KRAS variant in locally advanced rectal cancer: Results of the EXPERT-C trial. Ann Oncol 2015;26:1936-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjersem JB, Ikdahl T, Guren T, et al. Let-7 miRNA-binding site polymorphism in the KRAS 3 0 UTR ; colorectal cancer screening population prevalence and influence on clinical outcome in patients with metastatic colorectal cancer treated with 5-fluorouracil and oxaliplatin + / − cetuximab. BMC Cancer [Internet] 2012;12:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong L, Lai M, Chen M, et al. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogeneinduced senescence. Cancer Res 2010;70:8547-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsang WP, à TTK. The miR-18a à microRNA functions as a potential tumor suppressor by targeting on. Carcinogenesis 2009;30:953-9. [DOI] [PubMed] [Google Scholar]
- 39.Vega AB, Pericay C, Moya I, et al. microRNA expression profile in stage III colorectal cancer: Circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep 2013;30:320-6. [DOI] [PubMed] [Google Scholar]
- 40.Inoue A, Mizushima T, Wu X, et al. A miR-29b Byproduct Sequence Exhibits Potent Tumor-Suppressive Activities via Inhibition of NF-κB Signaling in KRAS -Mutant Colon Cancer Cells. Mol Cancer Ther [Internet] 2018;17:1-11. [DOI] [PubMed] [Google Scholar]
- 41.Inoue A, Yamamoto H, Uemura M, et al. MicroRNA-29b is a Novel Prognostic Marker in Colorectal Cancer. Ann Surg Oncol [Internet] 2015;22:1410-8. [DOI] [PubMed] [Google Scholar]
- 42.Kao CJ, Martiniez A, Shi XB, et al. MiR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014;33:2495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valentino A, Reclusa P, Sirera R, et al. Exosomal microRNAs in liquid biopsies: future biomarkers for prostate cancer. Clin Transl Oncol 2017;19:651-7. [DOI] [PubMed] [Google Scholar]
- 44.Schee K, Boye K, Abrahamsen TW, et al. Clinical relevance of microRNA miR-21, miR-31. Colorectal Cancer 2012;12:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pugh S, Thiébaut R, Bridgewater J, et al. Association between miR-31-3p expression and cetuximab efficacy in patients with KRAS wild-type metastatic colorectal cancer: a post-hoc analysis of the New EPOC trial. Oncotarget [Internet] 2017;8:93856-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramon L, David C, Fontaine K, et al. Technical Validation of a Reverse-Transcription Quantitative Polymerase Chain Reaction In vitro Diagnostic Test for the Determination of MiR-31-3p Expression Levels in Formalin-Fixed Paraffin- Embedded Metastatic Colorectal Cancer Tumor Specimens. Biomark Insights 2018;13:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manceau G, Imbeaud S, Thiébaut R, et al. Hsa-miR-31-3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res 2014;20:3338-47. [DOI] [PubMed] [Google Scholar]
- 48.Mlcochova J, Faltejskova-vychytilova P, Ferracin M, et al. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget [Internet] 2015;6:38695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent OA, Mendell JT, Rottapel R. Transcriptional regulation of miR-31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res 2016;14:267-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D, Yu F, Ma Y, et al. MicroRNA-31 Activates the RAS Pathway and Functions as an Oncogenic MicroRNA in Human Colorectal Cancer by Repressing RAS p21 GTPase Activating Protein 1 (RASA1)*. J Biol Chem 2013;288:9508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z, Liu K, Wang Y, et al. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer Cell Int [Internet] 2015;15:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, Lv W, Zhang J-H, Lu D-L. miR96 functions as a tumor suppressor gene by targeting NUAK1 in pancreatic cancer. Int J Mol Med 2014;34:1599-605. [DOI] [PubMed] [Google Scholar]
- 53.Lena RA, Verena S, Elke W, et al. MiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol Carcinog [Internet] 2014;54:1442-50. [DOI] [PubMed] [Google Scholar]
- 54.Xu XM, Qian JC, Deng ZL, et al. Expression of miR-21, miR- 31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett 2012;4:339-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiala O, Pitule P, Hosek P, et al. The association of miR-126-3p, miR-126-5p and miR-664-3p expression profiles with outcomes of patients with metastatic colorectal cancer treated with bevacizumab. Tumor Biol 2017;39:1-9. [DOI] [PubMed] [Google Scholar]
- 56.Ebrahimi F, Gopalan V, Wahab R, et al. Deregulation of miR- 126 expression in colorectal cancer pathogenesis and its clinical significance. Exp Cell Res [Internet] 2015;339:333-41. [DOI] [PubMed] [Google Scholar]
- 57.Hara T, Jones MF, Subramanian M, et al. Selective targeting of KRAS-Mutant cells by miR-126 through repression of multiple genes essential for the survival of KRAS-Mutant cells. Oncotarget [Internet] 2014;5:7635-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mosakhani N, Sarhadi V, Borze I, et al. MicroRNA Profiling Differentiates Colorectal Cancer According to KRAS Status. Genes Chromosomes Cancer 2012;51:1-9. [DOI] [PubMed] [Google Scholar]
- 59.Akao Y, Kumazaki M, Shinohara H, et al. Impairment of KRas signaling networks and increased efficacy of epidermal growth factor receptor inhibitors by a novel synthetic miR- 143. Cancer Sci 2018;1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie F, Li C, Zhang X, et al. MiR-143-3p suppresses tumorigenesis in pancreatic ductal adenocarcinoma by targeting KRAS. Biomed Pharmacother [Internet] 2019;119:109424. [DOI] [PubMed] [Google Scholar]
- 61.Pichler M, Winter E, Stotz M, et al. Down-regulation of KRAS-interacting miRNA-143 predicts poor prognosis but not response to EGFR-targeted agents in colorectal cancer. Br J Cancer [Internet] 2012;106:1826-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higgs G, Slack F. The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinforma [Internet] 2013;3:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao H, Huang S, Liu A, Chen Z. Up-regulated expression of miR-155 in human colonic cancer. J Cancer Res Ther 2018;14:604-7. [DOI] [PubMed] [Google Scholar]
- 64.Shibuya H, Iinuma H, Shimada R, et al. Clinicopathological and Prognostic Value of MicroRNA-21 and MicroRNA-155 in Colorectal Cancer. Oncology 2011;79:313-20. [DOI] [PubMed] [Google Scholar]
- 65.Lv Z, Fan Y, Chen H, Zhao D. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumor Biol [Internet] 2015;36:1619-25. [DOI] [PubMed] [Google Scholar]
- 66.Hongliang C, Shaojun H, Aihua L, Hua J. Correlation between expression of miR-155 in colon cancer and serum carcinoembryonic antigen level and its contribution to recurrence and metastasis forecast. Saudi Med J 2014;35:547-53. [PubMed] [Google Scholar]
- 67.Forzati F, De Martino M, Esposito F, et al. miR-155 is positively regulated by CBX7 in mouse embryonic fibroblasts and colon carcinomas, and targets the KRAS oncogene. BMC Cancer 2017;17:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan Q, Hu X, Zhang H, et al. MiR-193a-3p is an Important Tumour Suppressor in Lung Cancer and Directly Targets KRAS. Cell Physiol Biochem 2017;44:1311-24. [DOI] [PubMed] [Google Scholar]
- 69.Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier : early detection of colorectal cancer. BMC Cancer [Internet] 2013;13:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paterson EL, Kazenwadel J, Bert AG, et al. Down-Regulation of the miRNA-200 Family at the Invasive Front of Colorectal Cancers with Degraded Basement Membrane Indicates EMT Is Involved in Cancer Progression. Neoplasia [Internet] 2013;15:180-IN22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roh MS, Lee HW, Jung SB, et al. Expression of miR-200c and its clinicopathological significance in patients with colorectal cancer. Pathol Res Pract 2018;214:350-5. [DOI] [PubMed] [Google Scholar]
- 72.Ota T, Doi K, Fujimoto T, et al. KRAS up-regulates the expression of miR-181a, miR-200c and miR-210 in a three-dimensional- specific manner in DLD-1 colorectal cancer cells. Anticancer Res 2012;32:2271-6. [PubMed] [Google Scholar]
- 73.Tsunoda T, Takashima Y, Yoshida Y, et al. Oncogenic KRAS Regulates miR-200c and miR-221 / 222 in a 3D-Specific Manner in Colorectal Cancer Cells. Anticancer Res 2011;31:2453-60. [PubMed] [Google Scholar]
- 74.Kopp F, Wagner E, Roidl A. The proto-oncogene KRAS is targeted by miR-200c. Oncotarget [Internet] 2014;5:185-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev [Internet] 2009;23:2140-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spaderna S, Schmalhofer O, Hlubek F, et al. A Transient, EMTLinked Loss of Basement Membranes Indicates Metastasis and Poor Survival in Colorectal Cancer. Gastroenterology 2006;131:830-40. [DOI] [PubMed] [Google Scholar]
- 77.Zhang N, Lu C, Chen L. MiR-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancer. Oncol Lett 2016;12:4589-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao WG, Yu SN, Lu ZH, et al. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis 2010;31:1726-33. [DOI] [PubMed] [Google Scholar]
- 79.Gilles ME, Hao L, Brown K, et al. Tumor penetrating nanomedicine targeting both an oncomiR and an oncogene in pancreatic cancer. Oncotarget 2019;10:5349-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kasinski AL. Combatting PDAC with two tumor-targeting small RNAs. Oncotarget 2019;10:5892-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bai PS, Xia N, Sun H, Kong Y. Pleiotrophin, a target of miR- 384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma. J Cell Mol Med 2017;21:3023-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y-X, Chen Y-R, Liu S-S, et al. MiR-384 inhibits human colorectal cancer metastasis by targeting KRAS and CDC42. Oncotarget [Internet] 2016;7:84826-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y, Zhang Z, Wang J. MicroRNA-384 inhibits the progression of breast cancer by targeting ACVR1. Oncol Rep 2018;39:2563-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan C, Lin Y, Mao Y, et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget [Internet] 2016;7:21825-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun J, Zhou J, Dong M, Sheng W. Dysregulation of MicroRNA-543 expression in colorectal cancer promotes tumor migration and invasion. Mol Carcinog [Internet] 2016;56:250-7. [DOI] [PubMed] [Google Scholar]
- 86.Hiraki M, Nishimura J, Takahashi H, et al. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic Acids 2015;4:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, Wang J. MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol Chem 2017;398:929-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruzzo A, Graziano F, Vincenzi B, et al. High Let-7a MicroRNA Levels in KRAS-Mutated Colorectal Carcinomas May Rescue Anti-EGFR Therapy Effects in Patients with Chemotherapy-Refractory Metastatic Disease. Oncologist 2012;17:823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu CW, Ng SSM, Dong YJ, et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut [Internet] 2012;61:739 LP- 745. [DOI] [PubMed] [Google Scholar]


