Summary
During development, cells undergo multiple, distinct morphogenetic processes to form a tissue or organ, but how their temporal order and time interval are determined remain poorly understood. Here we show that the nuclear receptors E75 and DHR3 regulate the temporal order and time interval between the collective migration and lumen formation of a coherent group of cells named border cells during Drosophila oogenesis. We show that E75, in response to ecdysone signaling, antagonizes the activity of DHR3 during border cell migration, and DHR3 is necessary and sufficient for the subsequent lumen formation that is critical for micropyle morphogenesis. DHR3's lumen-inducing function is mainly mediated through βFtz-f1, another nuclear receptor and transcription factor. Furthermore, both DHR3 and βFtz-f1 are required for chitin secretion into the lumen, whereas DHR3 is sufficient for chitin secretion. Lastly, DHR3 and βFtz-f1 suppress JNK signaling in the border cells to downregulate cell adhesion during lumen formation.
Subject Areas: Biological Sciences, Organizational Aspects of Cell Biology, Developmental Biology
Graphical Abstract

Highlights
-
•
E75 antagonizes DHR3's function in inducing lumen formation of border cells (BCs)
-
•
E75 and DHR3 temporally coordinate collective migration and lumen formation of BCs
-
•
DHR3 is required and sufficient for chitin secretion into the lumen
-
•
DHR3 and βFtz-f1 downregulate JNK signaling and cell adhesion in the BCs
Biological Sciences; Organizational Aspects of Cell Biology; Developmental Biology
Introduction
During development, a group or population of cells often has to undergo multiple, distinct morphogenetic processes in a certain temporal order (e.g. A, then B…) to form a tissue or organ (Webb and Oates, 2016). If the correct temporal order is not followed (e.g. process B occurring before process A), that tissue or organ would not form correctly (Rougvie, 2001; Thummel, 2001). Besides the correct order, the time interval between two processes is another important aspect of the temporal control for the morphogenetic processes. Making the interval too long or too short would also be detrimental to the formation of the organ or tissue. Despite their importance in development, our current understandings on how the temporal order and time intervals are regulated and determined are very limited.
The somatic follicle cells of the Drosophila egg chamber have served as an excellent model system to study multiple morphogenetic processes (Horne-Badovinac and Bilder, 2005). Specifically, during stage 9 of oogenesis, a group of about eight cells detaches from the anterior follicle epithelium and undergoes collective migration between the germ-line nurse cells in a posterior direction (Montell, 2003). By early stage 10A, this coherent cluster of cells would have migrated a distance of about 150 μm in 6 h, reaching the border between oocyte and nurse cells, hence the name border cells. About 6 h later, by stage 10B, the cluster of eight border cells would have migrated dorsally a short distance along the border, eventually stopping at the dorsal-most position of the border. Four hours later, by stage 12, this border cell cluster undergoes a second morphogenetic process to eventually form the tip of micropyle, a tubular structure required for sperm entry into the mature oocyte (Montell et al., 1992). Therefore, the formation of micropyle tip by border cells requires two distinct morphogenetic processes in a certain temporal order: first, the well-studied, stereotyped, collective migration process and then a largely uncharacterized morphogenetic process that transforms these border cells into the tip of the tubular structure. Furthermore, an interval of about 16 h exists between the beginning of collective migration and the start of the micropyle formation. However, whether and how the temporal order and the time interval between the two morphogenetic processes are regulated remain largely unknown.
Previous studies have shed light on the temporal regulation of border cell migration. The steroid hormone ecdysone, its receptor heterodimer ecdysone receptor (EcR) and ultraspiracle (USP), and their co-activator Taiman (Tai) had all been shown to be required for the initiation of border cell migration (Bai et al., 2000; Cherbas et al., 2003; Jang et al., 2009). Ecdysone and the EcR signaling had long been known to play important roles in coordination of growth and developmental timing during embryogenesis, larval molting, and metamorphosis in Drosophila (Jia et al., 2017; Kozlova and Thummel, 2003; Yamanaka et al., 2013). Active form of ecdysone is also made in the adult Drosophila ovaries to regulate progression of oogenesis (Ables et al., 2016; Buszczak et al., 1999; Carney and Bender, 2000). 20-Hydroxyecdysone, the active form of ecdysone, is locally synthesized by the follicle epithelium in individual egg chambers and reaches its highest levels around stages 9 and 10 (Domanitskaya et al., 2014; Margaret et al., 1989). Even small patches of wild-type follicle cells in mosaic stage 9 egg chambers were shown to produce a sufficient level of active ecdysone that allows the border cells to begin migration (Domanitskaya et al., 2014). The sufficiency of ecdysone/EcR signaling on initiation of border cell migration was further demonstrated by Jang and coworkers, in which early expression of the activated form of the co-activator Tai can precociously initiate border cell migration (Jang et al., 2009). However, what cellular processes in the border cells are directly regulated by EcR signaling and whether EcR also temporally regulates micropyle formation are currently unknown.
In this study, we show that E75 and DHR3, two nuclear receptors/transcription factors downstream of EcR signaling, regulate both the temporal order and time interval between border cell migration and micropyle formation. We find that EcR's migration-initiating ability is partially through E75's function. During border cell migration, EcR signaling activates the expression of both E75 and DHR3, with E75 repressing DHR3's function. Furthermore, de-repression of DHR3 function after completion of border cell migration switches on lumen formation, turning the cluster of border cells into the tip of micropyle. Such antagonist relationship between E75 and DHR3 (while both under the control of EcR signaling) provides the regulatory mechanism of temporal order and time interval between two distinct morphogenetic processes essential for the formation of a functional micropyle.
Results
RNAi Screen Identifies E75 Acting downstream of EcR Signaling
Ecdysone signaling was known to be critical for the temporal control of initiation of border cell migration (Bai et al., 2000; Jang et al., 2009), but the cellular processes directly regulated by ecdysone/EcR signaling are largely unknown. To identify them, we carried out a small-scale RNAi screen of candidate genes that were previously reported to be responsive to ecdysone in Drosophila larvae and pupae and in cell lines (Beckstead et al., 2005; Gauhar et al., 2009; Sap et al., 2015). We first screened through the well-established response genes of ecdysone signaling (Ashburner and Richards, 1976; Huet et al., 1995; Richards, 1995), including E74, E75, E93, Br-c, and DHR3. Two to three different RNAi lines were used to confirm that phenotypes were not due to off-target effects, and two RNAi lines for the EcR gene were used as positive controls. A border-cell-specific Gal4 driver, slbo-Gal4, was used to drive expression of various RNAi transgenes in border cells beginning at late stage 8 of oogenesis, before border cells initiate their migration at early stage 9. As expected, both EcR RNAi lines (9327 and v35078 lines) resulted in phenotypes of strong migration delay or block, consistent with the previous reported roles of EcR in initiating and promoting border cell migration (Figures 1A, 1B, and S1A–S1C) (Bai et al., 2000; Jang et al., 2009). In comparison, border cell clusters from the control (wild type) stage 10 egg chambers almost always reached the 100% migration position, with only 6% of clusters displaying moderate delay (stopping at the 75% position) (Figures 1B and S1A–S1C). Interestingly, of all the five ecdysone response genes tested, only E75 displayed strong migration defects. In fact, all three RNAi lines (v44851, 26717, Thu1738) consistently resulted in severe migration block and delay phenotypes as well as dramatic redistribution of F-Actin, when compared with the control (Figures 1A, 1B, and S1C). We then screened an additional collection of 20 genes that were considered ecdysone response genes or putative target genes of EcR/USP in previous reports (Beckstead et al., 2005; Gauhar et al., 2009; Li and White, 2003). However, none of the genes, when knocked down, displayed strong migration defects (Figures S1A–S1C). Only mild-to-moderate migration phenotypes were observed in a few of the RNAi experiments.
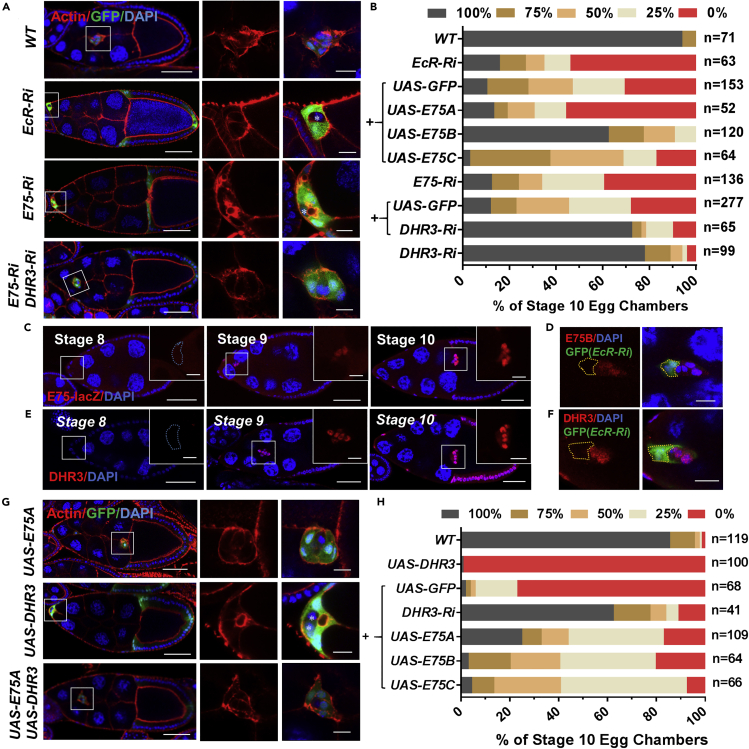
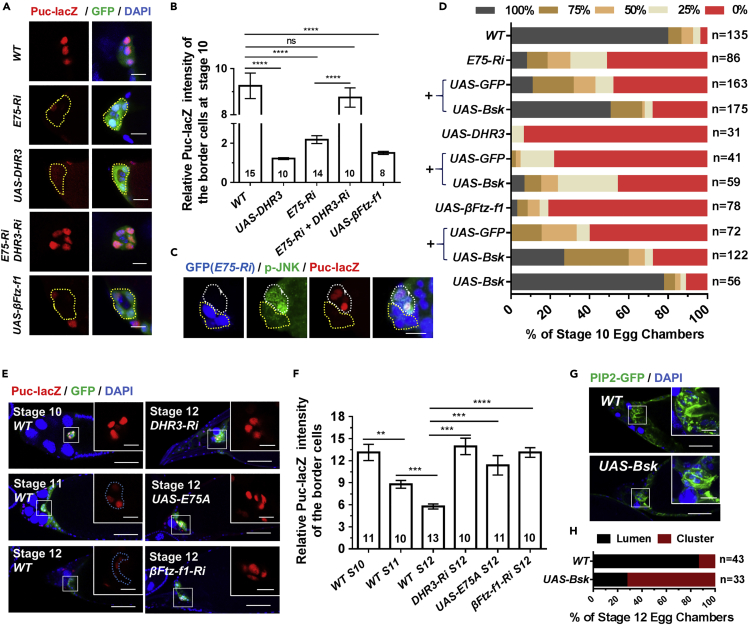
Figure 1.
E75 Antagonizes DHR3 during Border Cell Migration
(A) Confocal images of egg chambers stained with phalloidin (red, for F-actin) and DAPI (blue, for nuclei) with indicated genotypes. The boxed regions are enlarged and shown to the right. Border cells expressing EcR RNAi displayed strong migration defects but exhibited a similar morphology and F-actin distribution pattern to that of wild-type (WT) border cells, whereas E75 RNAi border cells with migration defects displayed a very different morphology and F-actin distribution pattern to that of the EcR RNAi and WT border cells. Co-expression of DHR3 RNAi rescued E75 RNAi's morphology and F-actin defects. Ri is the abbreviation for RNAi for this and all subsequent figures. Posterior is to the right and anterior is to the left for this and all subsequent figures.
(B) Quantification of border cell migration with indicated genotypes. EcR-Ri denotes EcR RNAi, “+” indicates that these genotypes include both EcR RNAi and one of the denoted genotypes (UAS-GFP, UAS-E75A, UAS-E75B, and UAS-E75C). The “+” below E75 RNAi indicates that these genotypes include both E75 RNAi and one of the denoted genotypes (UAS-GFP, DHR3 RNAi). The x axis denotes the percentage of stage 10 egg chambers examined for each genotype that exhibited various degrees of migration, as represented by the five color-coded bars (see Figure S1 for details).
(C and E) Confocal images displaying β-galactosidase (β-gal) staining (C) and DHR3 staining (E) of stages 8, 9, and 10 egg chambers. Boxed region is enlarged to the right, showing a high-magnification view of the border cells.
(D and F) Confocal images showing antibody staining of E75B (D) and DHR3 (F) of individual stage 10 border cell clusters with flip-out clones expressing EcR RNAi (EcR-Ri). The flip-out clones (labeled by GFP and encircled by yellow dotted lines) clearly display marked reduction of E75B and DHR3, respectively. See Figure S1F for specificity of E75B antibody and Figure S1E for E75B staining of WT border cells.
(G) Border cells overexpressing DHR3 exhibit severe defects in migration and morphology, which could be rescued by co-expression of E75A. Border cells with E75A overexpression alone displayed wild-type phenotype. In (A) and (G) “∗” indicates polar cells that are labeled by absence of GFP.
(H) Quantification of rescue of border cell migration defects as resulted from DHR3 overexpression. “+” indicates that these genotypes include both UAS-DHR3 and one of the denoted genotypes (UAS-GFP, DHR3 RNAi, UAS-E75A, UAS-E75B, and UAS-E75C).
Scale bars: 50 μm in (A), (C), (E), and (G); 10 μm for high-magnification views in (A), (C–F), and (G). See also Figures S1 and S2.
We then proceeded to determine whether E75 acts downstream of EcR to initiate and promote border cell migration. Three distinct isoforms of E75 (A, B, C) were shown to be involved in different developmental and cellular processes (Huet et al., 1995; Li et al., 2016; Terashima and Bownes, 2006). Both E75A and E75C contain ligand-binding domain and DNA-binding domain, whereas E75B contains only the ligand-binding domain and no DNA-binding domain (Bialecki et al., 2002; Segraves and Hogness, 1990). E75B can heterodimer with DHR3 and inhibit its activity during metamorphosis (White et al., 1997). The sequences used in the three RNAi lines for E75 are all within the common region and would have knocked down all three isoforms. Therefore, we overexpressed each isoform to test its individual rescue ability on border cell migration defects as caused by EcR RNAi. We found that E75B overexpression markedly rescued EcR RNAi's migration defects, whereas E75C displayed a much weaker rescue effect and E75A showing no significant rescue (Figure 1B). Moreover, we found that E75's transcription levels (as represented by a previously used reporter E75-lacZ (Manning et al., 2017)) within border cells at stages 9 and 10 were much higher than those at stage 8 (Figure 1C), consistent with ecdysone signaling being significantly increased beginning at stage 9. And mosaic border cell clusters containing a clone of EcR RNAi-expressing cells demonstrate that E75B protein levels are drastically decreased when EcR function is reduced (Figure 1D), indicating that EcR activity is required for E75B expression during stage 9. Taken together, these results indicate that E75B is likely the major isoform and downstream factor to mediate EcR's temporal control on border cell migration. Other downstream factors besides E75B may also mediate ecdysone/EcR signaling because E75B can only partially rescue EcR's loss-of-function phenotype in border cells. Consistently, a recent study using microarray analysis also identified E75 as one of the target genes that are responsive to ecdysone signaling in the migratory border cells (Manning et al., 2017).
E75 Antagonizes DHR3's Function during Collective Migration of Border Cells
During metamorphosis, ecdysone-activated EcR turns on the expression of E75B, which then binds to DHR3 and antagonizes its activity (White et al., 1997). E75B and DHR3 are both nuclear receptors/transcription factors and are both induced by ecdysone, and E75B's inhibition of DHR3 function leads to suppression of DHR3's transcriptional activation of its target genes essential for metamorphosis (Caceres et al., 2011; Reinking et al., 2005; White et al., 1997). To determine whether antagonistic interaction also exists between E75 and DHR3 during border cell migration, we co-expressed DHR3 RNAi and E75 RNAi in border cells. We found that DHR3 reduction strongly rescued E75 RNAi's migration defects (Figures 1A and 1B), as well as the morphological defects of border cells (Figure 1A, also described in the section below). On the other hand, overexpression of DHR3 resulted in similar phenotypes of migration and morphology to those of E75 RNAi (Figures 1G and 1H), only with DHR3 overexpression's defects more severe than those of E75 RNAi (Figures S3A and S3B). Furthermore, E75 overexpression can in turn suppress DHR3 overexpression's severe defects (Figures 1G and 1H), with all three of its isoforms (E75A, E75B, E75C) displaying similar suppressing abilities. This is consistent with previous reports that both E75A and E75B isoforms can heterodimerize with DHR3 to inhibit DHR3's transcription activation ability (Sullivan and Thummel, 2003; White et al., 1997). Furthermore, all three isoforms contain the ligand-binding domain, which was previously shown to be involved in heterodimerization with DHR3 and inhibition of its activity (Reinking et al., 2005). Lastly, we showed that DHR3's levels were also increased in border cells beginning at stage 9 (Figure 1E), similar to E75's temporal expression pattern (Figure 1C), and its levels also depended on EcR's activity (Figure 1F). Together, these data demonstrate an antagonistic relationship between E75 and DHR3 during border cell migration, with both their expressions activated by EcR during the migratory process.
Conversely, we found that expressing E75 RNAi (B26717) or DHR3 in border cells significantly reduced the level of ecdysone response activity or EcR signaling (Figures S2A–S2C and S2F), which is represented by expression levels of EcRE-lacZ, a common reporter of EcR activity used in previous studies (Jang et al., 2009; Koelle et al., 1991). Moreover, expression of DHR3 RNAi could rescue E75 RNAi's EcRE-lacZ levels (Figures S2D–S2F). These data indicate that E75 can exert a positive feedback on EcR signaling by antagonizing DHR3's inhibitory effect on EcR signaling. This conclusion is consistent with previous studies that showed DHR3 physically interacted with EcR and suppressed its activity (Lam et al., 1997; White et al., 1997). It should be noted that DHR3 could also exert its inhibitory effect on EcR indirectly.
E75 Antagonizes DHR3's Function in Lumen Formation during Border Cell Migration
We noted that border cell clusters with E75 knockdown or DHR3 overexpression displayed different morphology and F-actin staining pattern from border cells with reduced EcR function (Figure 1A), as indicated by our EcR RNAi result and previous reports (Hackney et al., 2007). The delayed border cell clusters with EcR RNAi often displayed a coherent and front-polarized morphology with F-actin enriched in the periphery of the cluster, similar to that of the wild-type clusters (Figure 1A). On the contrary, E75 RNAi- or DHR3-overexpressing border cells (driven by slbo-Gal4) lost the front-polarized cluster morphology that is characteristic of front-back polarity, and F-actin is instead enriched in the center of the cluster in a ring-like structure (Figures 1A and 1G), which is unique and never observed in any of the previously reported mutant phenotypes of border cells (to our knowledge). Closer examination revealed that this unique structure is not within individual border cell's cytoplasm but is instead composed of portions of outer border cells' inside membranes, which join together to form a continuous supra-cellular ring (Figures 2A–2E). And a similar but smaller ring can also form between two border cells in a flip-out clone that expresses E75 RNAi by Ay-Gal4 (Figure S3C). Moreover, such supra-cellular structure is also enriched with molecules that are typically associated with apical membranes (aPKC, Crb, Baz/Par3, PIP2-GFP reporter) (Figures 2A, 2C, 2E, and S3C) but not with lateral membranes (Dlg) (Figure 2A). In addition, E-cadherin relocalization was also observed, with E-cadherin sometimes ectopically enriched in a region adjacent to that of aPKC staining (Figure S8D). A typical supra-cellular ring encloses a space that resembles a lumen with significant depth (about 5 μm, Video S1) in the center of cluster, effectively displacing the two central polar cells to the side and underneath. To address whether polar cells are involved in the formation of the lumen, we closely followed the position of polar cells and found that lumens were mostly enclosed by border cells and that the ring-like lumen structures were not derived from the circumference of polar cells (Figures 1A, 1G, 2A, and 2B; polar cells marked by ∗). slbo-Gal4 expressed E75 RNAi or DHR3 only in border cells and not in polar cells, raising the possibility that differential expression at the interface of the border and polar cells could result in the unique lumen phenotype. We then used c306-Gal4 to drive E75 RNAi expression in both border cells and polar cells (Figure S3D), and we found similar lumen phenotype to that resulted from slbo-Gal4, suggesting differential expression of E75 RNAi or DHR3 in border cells and polar cells is not the cause of lumen formation.
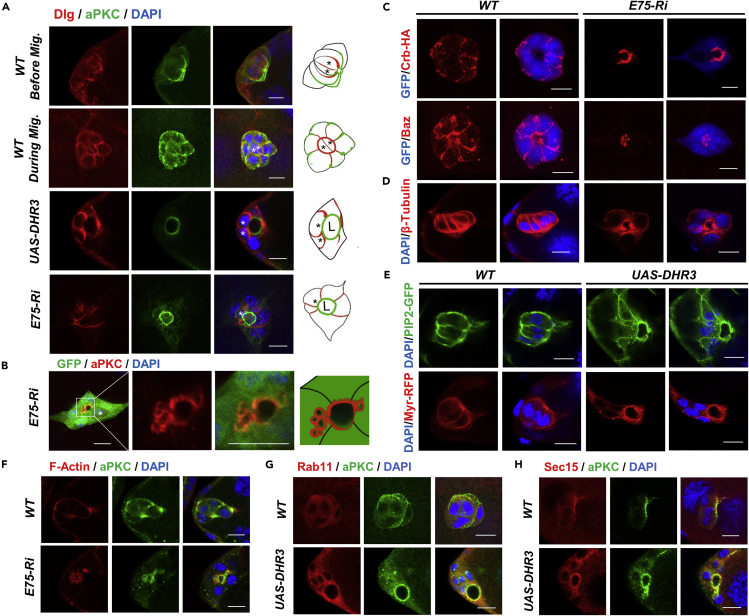
Figure 2.
E75 Loss of Function and DHR3 Overexpression Led to Precocious Lumen Formation of the Border Cells
(A) The first two rows show confocal images of wild-type border cells before migration (first row, early stage 9) and during migration (second row, mid stage 9), respectively. Before migration, the apical (stained with aPKC) and lateral (stained with Dlg) membranes of border cell cluster points to the posterior direction (to the right), with apical membrane more posterior than lateral membrane. During migration, the orientation of border cell cluster undergoes a 90° turn, resulting in the apical-lateral axis being perpendicular to the posterior direction (to the right). The two central polar cells are outlined by strong staining of Dlg and marked with “∗” in the diagrams to the right. The last two rows depict border cells with E75 RNAi or DHR3 overexpression that failed to migrate and instead formed lumen (marked with “L” in the diagrams) that is enclosed by aPKC-stained membrane. Displaced polar cells are marked with “∗”. Dlg staining is restricted to membranes between adjacent border cells. The first and last rows are resulted from projections of z-stacks of confocal sections, the others are single confocal sections.
(B–E) Images of border cells labeled with aPKC (B), Crb-HA and Baz (C), β-tubulin (D) staining, and PIP2-GFP and Myr-RFP (E) fluorescence, as resulted from E75 RNAi or DHR3 overexpression. DAPI labels all nuclei. PIP2-GFP serves as a reporter for PIP2-enriched membrane (PLCδ-PH-GFP, see Methods for details), and Myr-RFP (myristoylated RFP) serves as a general membrane marker.
(F–H) Images showing co-staining of aPKC with phalloidin (F-actin, F), Rab11 (recycling endosome marker, G), and Sec15 (exocyst component, H), as resulted from E75 RNAi or DHR3 overexpression. In this and other figures, all border cell clusters are oriented in such a way that their left side faces the anterior of egg chamber and their right side is toward the posterior of egg chamber.
Scale bars, 10 μm for all panels. See also Figure S3.
3D reconstruction of confocal z-stacks of wild-type (WT), E75 RNAi and DHR3 overexpressing border cells. The aPKC staining labels the apical membrane that surrounds the central lumen with significant depth (about 5 μm in depth) in the E75 RNAi or DHR3 overexpressing border cell clusters, which are the same clusters as those shown in fourth row and third row of Figure 2A respectively.
The strong and specific enrichment of apical markers such as aPKC in the membranes enclosing the luminal space suggests that the border cell cluster has undergone a lumen formation process to become a tubular structure with the apical membrane facing the central lumen. Interestingly, the E75 RNAi border cells displayed a range of lumen-like phenotypes. Half of them (50.0%) showed a clear lumen phenotype that is similar to that of the DHR3 overexpressing border cells, whereas majority of the rest (39.0%) exhibited little luminal space and discontinuous apical membrane patches as labeled by aPKC (Figures S3A and S3B), which resemble the previously reported structure of pre-apical patches (PAP) that are present during the intermediate stages of de novo lumen formation in several model systems (Bryant et al., 2010; Ferrari et al., 2008; Yang et al., 2013). These moderate phenotypes may reflect incomplete lumen formation or intermediate stages of lumen formation in the border cells, whereas the large lumen structure present in vast majority of the DHR3-overexpressing clusters and half of the E75 RNAi clusters may indicate complete lumen formation.
Formation of a tube and its enclosing lumen from non-epithelial cells is referred to as de novo lumen formation (Sigurbjornsdottir et al., 2014), which is a fundamental morphogenetic process central to animal development. Extensive studies in various in vitro and in vivo model systems have revealed that the initial stage of de novo lumen formation involves establishment of a new apical-basal polarity, which requires re-routing of multiple cellular processes and components including polarized intracellular trafficking, polarized actin and microtubule cytoskeleton, polarized distribution of apical markers, and newly synthesized membrane (Akhtar and Streuli, 2013; Datta et al., 2011; Sigurbjornsdottir et al., 2014). We found that in addition to the re-distribution of apical markers to the lumen-facing membrane, the intracellular traffic as well as cytoskeleton was also dramatically re-organized in the E75 RNAi- or DHR3-overexpressing border cells. Staining with Rab11 and Sec15 antibodies revealed that recycling endosome and exocyst were enriched in the cytoplasmic regions near the lumen-facing apical membrane, indicating a polarized transport toward the lumen (Figures 2G and 2H). Furthermore, F-actin and sometimes aPKC were observed localizing to large vacuole-like compartment adjacent to the lumen-facing membrane (Figures 2B and 2F), suggesting that these large vesicles could be in the process of fusing with the adjacent apical membrane. This phenomenon was similar to previous reports of VACs (vacuolar apical compartments) forming in the MDCK cells that are undergoing de novo lumen formation (Brignoni et al., 1993; Vega-Salas et al., 1988). In addition, the actin and microtubule cytoskeletons were re-organized in such a way that they are now mostly localized in and adjacent to the lumen-facing membrane. Interestingly, β-tubulin was re-organized into a distribution pattern that seems to radiate away from the central lumen (Figure 2D). Lastly, marked increase of intracellular membrane levels as indicated by Myr-RFP and PIP2-GFP was observed in the cytoplasm of DHR3 expressing border cells, suggesting that high levels of newly synthesized membrane are needed for formation and expansion of lumen-facing membrane (Figure 2E). Taken together, these results indicate that during border cell migration E75 acts to suppress DHR3's lumen formation function, which includes re-routing of endocytic recycling, re-distribution of apical markers, re-polarization of actin and microtubule cytoskeletons, and increased levels of membrane components.
DHR3 Is Later Required for the Formation of Micropyle Tip
We next sought to understand why E75 needs to suppress DHR3's lumen formation function during border cell migration. After border cells finish their anterior migration to the border at stage 10A, they would further migrate a short distance dorsally and finally stop at the dorsal border between nurse cells and oocyte at stage 10B. About four hours later, around stages 12, this cluster of border cells will undergo a morphogenetic transformation to form part of the micropyle, which is a tubular structure essential for sperm entry (Montell et al., 1992). Such a morphogenetic process is not well characterized and understood. Therefore, we wonder whether the lumen-forming phenotype from E75 knockdown or DHR3 over-activation represents the precocious occurrence of the morphogenetic event involved in micropyle formation. If that is the case, E75 may actually be preventing a late morphogenetic process from occurring earlier (i.e. before or during border cell migration). Therefore, E75 and DHR3 may function together to keep the correct temporal order between the two morphogenetic processes. To address this possibility, we first sought to characterize and understand the process that enables wild-type border cell cluster to be transformed into the tip of micropyle.
Collective migration of border cells has been extensively studied, but the morphogenetic process that turns the border cells into micropyle tip is little studied. Previous work by Montell and coworkers first demonstrated that border cells develop into the tip of micropyle and contribute to the cellular process thought to maintain a functional opening, whereas the centripetal follicle cells form the bulk of the micropyle structure. Furthermore, in the absence of border cells, a slightly smaller micropyle structure could still form, but it lacks the functional opening required for sperm entry (Montell et al., 1992). We sought to describe and characterize such morphogenetic process in details, using markers of actin cytoskeleton, membrane, and apical polarity (Figures 3A–3D, Videos S2, S3, and S4). Similar to migratory border cells at stage 9, border cells at stage 10 (a period of about 10 h, Figure 3A) mostly retain the coherent cluster morphology as well as the distribution pattern of F-actin and apical polarity proteins. During stages 9 and 10, Par6-GFP was shown to localize between adjacent border cells in a thin section of junctional region (Figure 3B, Video S2), which was subsequently retracted and significantly shortened during stage 11 (a period of about 0.5 h). During stage 12, Par6-GFP localization is further remodeled, with its pattern shifted from junctional region between adjacent border cells to the membrane facing the lumen-like cavity (Figure 3B, Video S2). Consistently, Lifeact-GFP and a PIP2 membrane reporter (PIP2-GFP) both demonstrate a similar remodeling in their distribution patterns from stage 9 to stage 12, with Lifeact-GFP and PIP2-GFP highly enriched in the same membrane region surrounding the luminal space in wild-type border cells at stage 12 (Figures 3C and 3D, Videos S3 and S4).
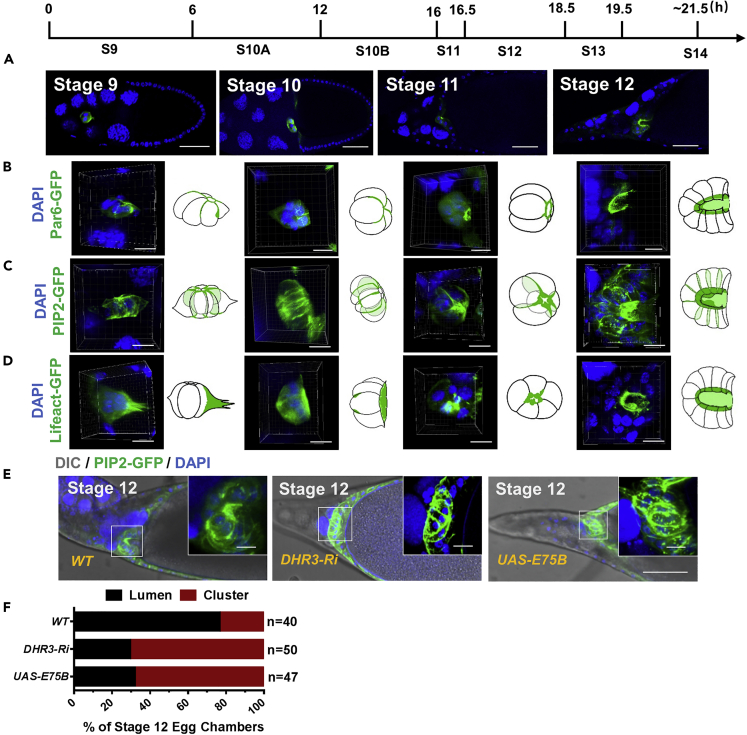
Figure 3.
DHR3 Is Required for Border Cells' Lumen Formation in the Micropyle at Stage 12
(A) A time course of developing wild type egg chambers at stages 9, 10, 11 and 12. (B–D) 3-D reconstruction of z-stacks of confocal section (see Methods for details) reveals the change of morphology from a cluster to the anterior part of the tubular micropyle (from stage 9 to stage 12). See also Videos S2, S3, and S4 that are generated from the 3-D reconstruction. Par6-GFP (B), Lifeact-GFP (C), and PIP2-GFP (D) fluorescence display a dynamic remodeling of apical polarity, F-actin and PIP2-enriched membrane in border cells during micropyle formation.
(E) DHR3 RNAi, E75B overexpression caused disruption of lumen formation, as compared to morphology of wild type border cells (outlined by PIP2-GFP) at stage 12. (F) The cluster or lumen morphology shown in (E) is quantified. As indicated by the diagram, 76.6% of stage 12 wild type border cells display obvious lumen morphology, whereas 70.4% of DHR3 RNAi and 68.0% of E75B display cluster morphology, which is characteristic of the wild type border cells at stage 10 (C).
Scale bars, 50 μm in (A) and (E); 10 μm in high-magnification views in (B–E). See also Figures S3 and S4 and Videos S2, S3, and S4.
3D reconstruction of confocal z-stacks of wild-type border cell clusters from stage 9 to stage 12, which are the same as those from Figure 3B and are labeled with Par6-GFP.
3D reconstruction of confocal z-stacks of wild-type border cell clusters from stage 9 to stage 12, which are the same as those from Figure 3C and are labeled with PIP2-GFP.
3D reconstruction of confocal z-stacks of wild-type border cell clusters from stage 9 to stage 12, which are the same as those from Figure 3D and are labeled with Lifeact-GFP.
A very small percentage of wild-type stage 11 or 12 egg chambers would contain border cells that failed to migrate properly and reach the oocyte border (Figures S4A-S4E). Interestingly, we found that those stages 11 and 12 border cells with migration defects also displayed lumen formation that was accompanied by the remodeling of apical markers, F-actin, and PIP2-enriched membrane and was similar to the DHR3-induced lumen formation process at stages 9 and 10 (Figures S4A–S4E). This result indicates that the remodeling process is autonomously initiated in border cells and is under strict temporal control. Finally, DHR3 knockdown or E75 overexpression each led to disruption of the remodeling process (Figures 3E and 3F). As shown by the PIP2-GFP marker (Figure 3E), most of DHR3 RNAi or E75 overexpressing border cell clusters at stages 12 and 13 displayed a cluster morphology that is characteristic of border cells at stages 9 and 10 (Figure 3C, Video S3), where the PIP2-GFP is broadly localized in membranes between adjacent border cells. Consequently, these border cells failed to develop into the anterior tip of micropyle that surrounds a lumen-like cavity.
Together, these results demonstrate that DHR3 activity is required for the morphogenetic process of lumen formation that is essential to micropyle formation. Interestingly, the morphogenetic remodeling process involved in micropyle formation is similar to the DHR3-induced lumen formation process occurred precociously in border cell cluster during stages 9 and 10, suggesting that DHR3 is not only required but also sufficient for all the remodeling events necessary for lumen formation.
Reduction of EcR Signaling and E75 Levels Causes De-repression of DHR3 Activity
We next sought to understand how DHR3 function is temporally regulated to limit its lumen-forming activity only to the period of micropyle formation and not to the period of collective migration. We reasoned that DHR3's activity in border cells had to be inhibited by E75 during stages 9 and 10, as shown by our results above (Figure 2). Afterward, DHR3's activity would need to be de-repressed beginning at stage 11 to start the morphogenetic process of lumen formation. We already showed that DHR3 function is antagonized by E75 and that both E75 and DHR3 are expressed by EcR during border cell migration at stage 9. We then examined the temporal expression patterns of E75 and DHR3 as well as the levels of EcR signaling. We found that EcR signaling, as reflected by its well-established reporter EcRE-lacZ, reaches its highest levels during stage 10B, and then dramatically declines from stage 11 to stage 13 (Figures 4A and 4E). Accordingly, both expression levels of the E75-lacZ reporter, which reflects the transcription levels of E75 (Figures 4B and 4F) (because E75B antibody cannot penetrate and stain stages 11–13 egg chambers very well), and the protein levels of DHR3 as detected by DHR3 antibody also decrease from stage 11 to stage 13 (Figures 4C and 4G). These results suggest that as ecdysone signaling decrease dramatically (beginning at stage 11) E75 levels should also decrease to a low level (at stages 11 and 12), which could be below the threshold level for inhibition of DHR3's activity (note: the real E75 protein level could be even lower than the E75-lacZ staining level due to β-galactosidase's long perdurance). To test the above idea, we need a good activity reporter for DHR3's function.
Figure 4.
Temporal Expression Patterns of EcRE-lacZ, E75-lacZ, DHR3, and βFtz-f1-lacZ from Stage 10B to Stage 13
(A–D) Confocal images showing antibody staining of β-gal that is expressed by the EcRE-lacZ reporter (A), E75-lacZ enhancer trap (B), and βFtz-f1-lacZ enhancer trap (D), as well as antibody staining of DHR3 (C), from stage 10B to stage 13. Boxed regions are enlarged and shown at the right of all panels. Areas encircled by yellow dotted lines (based on labeling of GFP as expressed by slbo-Gal4) highlight the border cell clusters (A–D) at stages 12 and 13. Scale bars, 10 μm.
(E–H) Quantification of antibody staining of border cell clusters in (A–D) from stage 10b to stage 13. (E, F, G, and H) corresponds to (A, B, C, and D) respectively. The number of border cell clusters examined (n) for each stage is given at the x-axis. Statistical analysis was performed using two-tailed Student's t test. Error bars indicate S.E.M.
See also Figure S5.
Previous literatures indicate that DHR3's immediate downstream target gene during metamorphosis is βFtz-f1 (Jia et al., 2017; Kageyama et al., 1997; Lam et al., 1997), whose expression levels serve as a readout for DHR3 activity. We obtained an enhancer trap line for βFtz-f1, βFtz-f1-lacZ, which supposedly could reflect the transcription level of βFtz-f1. We found that its expression could serve as a bona fide reporter for DHR3 activity, based on the following results. First, βFtz-f1-lacZ expression is initially at non-detectable levels at stages 9 and 10A (Figure S5A), and at very low levels at stage 10B (Figures 4D and 4H), then it abruptly reaches much higher levels at stages 11, 12, and 13 (Figures 4D and 4H). Therefore, βFtz-f1-lacZ's temporal expression pattern is highly consistent with our above prediction about the temporal regulation of DHR3 activity. Second, DHR3 overexpression led to precocious expression of βFtz-f1-lacZ within border cells during stages 9 and 10, whereas coexpression of E75B and DHR3 suppressed such precocious expression (Figures 5A and 5C). Conversely, DHR3 RNAi, E75B overexpression, or E75B and DHR3 coexpression, each suppressed βFtz-f1-lacZ's normal expression in border cells during stage 11 (Figures 5B and 5D). Furthermore, DHR3 overexpression in the follicle cells at the stage 9, when βFtz-f1-lacZ is not normally expressed, ectopically turned on βFtz-f1-lacZ's expression in the follicle cells (Figure S5B). Third, expression of βFtz-f1-RNAi in the background of E75 RNAi partially rescues border cell's migration defects (Figures 5E and S6A), suggesting that βFtz-f1 functions downstream of DHR3. Lastly, expressing βFtz-f1-RNAi in the border cells resulted in the disruption in the formation of micropyle tip, similar to the loss-of-function defects of DHR3 RNAi (Figures S6B and S6C). On the other hand, overexpression of βFtz-f1 in stage 10 border cells resulted in actin-enriched patches that are similar to PAP from moderate E75 RNAi defects (Figures S3A, S3B, and S6A), suggesting an incomplete lumen forming phenotype. Taken together, these results support the conclusion that reduction in EcR signaling and E75 levels leads to de-repression of DHR3 activity (as represented by the βFtz-f1-lacZ reporter) beginning at stage 11, which serves to switch on lumen formation for micropyle formation (during stages 11–13). Moreover, βFtz-f1 acts downstream of DHR3 to mediate micropyle formation.
Figure 5.
βFtz-f1 Acts Downstream of DHR3 and βFtz-f1-lacZ Serves as a Reporter of DHR3 Activity
(A) βFtz-f1-lacZ levels in border cells at stage 10 as represented by β-gal antibody staining. Compared with wild-type (WT) control, E75 RNAi and DHR3 overexpression both result in significant increase of βFtz-f1-lacZ levels (quantified in C), whereas double knock down of E75 and DHR3 (E75 Ri + DHR3 Ri) and overexpression of both DHR3 and E75 (UAS-DHR3 + UAS-E75) abolish the increase (quantified in C).
(B) βFtz-f1-lacZ levels in border cells at stage 11 as represented by β-gal staining. Compared with wild-type (WT) control, DHR3 RNAi and E75 overexpression both result in significant reduction of βFtz-f1-lacZ levels (quantified in D). Yellowed dotted lines (A and B) outline individual border cell clusters, as labeled with GFP expressed by slbo-Gal4. Boxed regions (A, B) are enlarged and shown at the right of all panels. Scale bars, 50 μm for egg chambers; 10 μm for border cells.
(C and D) Quantification of βFtz-f1-lacZ levels. The number of border cell clusters examined for each genotype is indicated within its corresponding column. Statistical analysis was performed using two-tailed Student's t test. Error bars indicate S.E.M. ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns, not significant.
(E) Quantification of rescue of border cell migration defects of E75 RNAi by co-expression of βFtz-f1 RNAi. Represented images for the indicated genotypes are shown in Figure S6A.
See also Figure S6.
DHR3 Is Required and Sufficient for Chitin Secretion into the Lumen
An essential feature of de novo lumen formation in the vertebrates is the secretion of glycoprotein such as the negatively charged podocalyxin into the lumen to keep the lumen membranes apart and promote the expansion of luminal space (Bryant et al., 2014; Strilic et al., 2010). Although Drosophila does not possess a podocalyxin homolog, the tube formation during Drosophila tracheal development requires the secretion of chitin into the lumen (Devine et al., 2005). Chitin is a long-chain polymer of N-acetylglucosamine, which is also a primary component of the Drosophila exoskeleton (Moussian et al., 2005; Zhu et al., 2016).
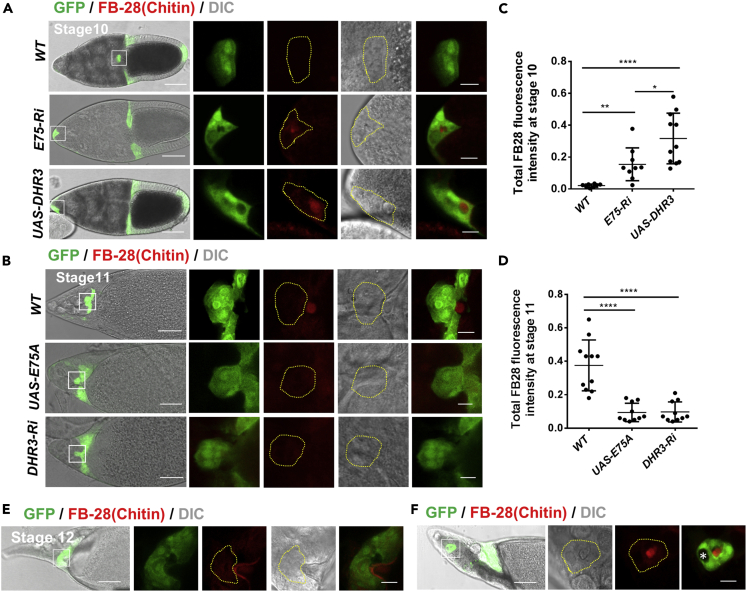
We then proceeded to determine whether chitin is present in the lumen enclosed by the border cells and whether DHR3 acts to promote secretion of chitin into the lumen. Interestingly, we found that chitin (labeled by FB-28) is only present in the extracellular space adjacent to wild-type border cells during and after stage 11 (Figures 6B, 6D, 6E, and S7E), whereas it is not present around the border cells before stage 11 (Figures 6A, 6C, and S7E). A very small percentage of wild-type stage 12 egg chambers would contain border cells that failed to migrate properly and reach the oocyte border, and we found that chitin is present within the lumen surrounded by those border cells (Figure 6F). Together, these results indicate that chitin is present in the lumen within the border cell cluster. In addition, the temporal and localization patterns of chitin suggest that it is secreted by border cells beginning at stage 11 during lumen formation for the micropyle tip. Furthermore, we found that expressing E75 RNAi or DHR3 specifically in the border cells (by slbo-Gal4) each resulted in chitin being precociously localized within the lumen of border cell clusters that failed to migrate to the oocyte border at stage 10 (Figures 6A and 6C), indicating that DHR3 activation is sufficient to induce chitin secretion. On the contrary, DHR3 knockdown or E75 overexpression in border cells led to loss of extracellular chitin adjacent to border cells (Figures 6B and 6D), indicating that DHR3 is required for chitin secretion by the border cells. Together, these results indicate that DHR3 activity is necessary and sufficient for chitin secretion by the border cells during lumen formation. To further test whether βFtz-f1 is also sufficient for chitin secretin, we examined and found no chitin secretion in βFtz-f1 overexpressing border cells at stage 10 (Figures S7A and S7C). This result could be due to the aforementioned fact that βFtz-f1 overexpression only resulted in PAP (incomplete lumen formation, Figure S6A). Hence, it is conceivable that chitin secretion may only occur after lumen formation progresses to a certain degree. On the other hand, we found that βFtz-f1 is required for chitin secretion by the border cells during micropyle formation (Figures S7B and S7D), similar to DHR3's role (Figures 6B and 6D). These results suggest that βFtz-f1 may act downstream of DHR3 to partially mediate DHR3's chitin secretion role.
Figure 6.
DHR3 Is Necessary and Sufficient for Chitin Secretion by the Border Cells during Lumen Formation
(A–D) (A and B) Confocal and DIC images showing chitin staining in stage 10 (A) and stage 11 (B) egg chambers. Chitin is labeled with the Fluorescent Brightener 28 (FB28) dye. (A) Chitin is not detected within or close to wild-type (WT) border cells at stage 10, whereas E75 RNAi and DHR3 overexpression in the border cells result in precocious secretion of chitin to the lumen (quantified in C). (B) Starting at stage 11, chitin is detectable adjacent to WT border cell cluster, but DHR3 RNAi or E75 overexpression abolishes chitin staining (quantified in D). Yellow dotted lines outline individual border cell clusters as labeled with GFP expressed by slbo-Gal4. Scale bars, 50 μm for egg chambers; 10 μm for border cells. (C and D) Quantification of chitin levels of border cells for the indicated genotypes.
(E and F) Chitin staining of normal stage 12 border cells (E) and delayed stage 12 border cells (F). “∗” marks polar cells that are labeled by absence of GFP. Statistical analysis was performed using two-tailed Student's t test. Error bars indicate S.E.M. ∗∗, p < 0.01; ∗∗∗∗, p < 0.0001.
See also Figure S7.
DHR3 and βFtz-f1 Suppress JNK Signaling in the Border Cells
Lastly, we sought to explore what signaling pathways DHR3 regulates in border cells. We tested reporters for a number of signaling pathways previously known to play essential roles in the border cells, including JAK/STAT (Beccari et al., 2002; Silver et al., 2005), Notch (Wang et al., 2007), JNK (c-Jun N-terminal kinase) (Llense and Martin-Blanco, 2008; Melani et al., 2008), and Dpp (Luo et al., 2015). Among them, JNK was the only signaling found to be severely affected by E75 knockdown or DHR3 overexpression (Figure S8A). Staining for Puc-lacZ, a widely used reporter for JNK signaling, revealed that both E75 RNAi and DHR3 overexpression caused strong reduction of Puc-lacZ reporter activity in stage 10 (Figures 7A–7C), indicating that increased DHR3 activity suppresses JNK signaling. Indeed, knockdown of DHR3 in the background of E75 RNAi rescued the level of Puc-lacZ expression back to the wild-type level in stage 10 (Figures 7A and 7B). Furthermore, overexpressing bsk (encoding Drosophila JNK) in the background of E75 RNAi or DHR3 overexpression partially rescued the severe migration defects and precocious lumen formation of border cells that were resulted from E75 loss of function (Figures 7D and S8B). Together, these results demonstrate that reduction of E75 or increase of DHR3 activity leads to downregulation of JNK signaling in the migratory border cells at stages 9 and 10.
Figure 7.
DHR3 and βFTZ-f1 Downregulate JNK Signaling in the Border Cells
(A) Puc-lacZ expression levels in migratory border cells at stage 9 or 10 as indicated by representative confocal images of b-gal antibody staining. (B) Quantification indicates that E75 RNAi, DHR3 and bFTZ-f1 overexpression as shown in (A) each results in strong and significant decrease of Puc-lacZ levels as compared to wild type (WT) control, while coexpression of DHR3 RNAi and E75 RNAi returns the Puc-lacZ levels to that of WT.
(C) A mosaic border cell cluster containing a clone of E75 RNAi expression cells (marked by GFP, outlined with yellow dotted line), which exhibit reduction of Puc-lacZ and p-JNK levels as compared with those in the adjacent wild-type cells (no GFP, outlined with white dotted line).
(D) Quantification of partial rescue of border cell migration defects of E75 RNAi, DHR3 overexpression, and βFTZ-f1 overexpression by coexpression of Bsk.
(E) Representative confocal images show that Puc-lacZ expression levels in WT border cells are decreased from stage 10 to 12, while expression of DHR3 RNAi, E75A and bFTZ-f1 RNAi elevates Puc-lacZ levels in border cells at stage 12. (F) Quantification of the results as shown in (E).
(G) Bsk overexpression caused disruption of lumen formation, as compared to morphology of wild type border cells (outlined by PIP2-GFP) at stage 12. (H) Quantification of the cluster or lumen morphology as shown in (G). 86.0% of stage 12 wild type border cells display obvious lumen morphology, whereas 72.7% of Bsk overexpressing border cells display cluster morphology, which is similar to that of the wild type border cells at stage 10. (B and F) The number of border cell clusters examined for each genotype is indicated within its corresponding column. Statistical analysis was performed using unpaired two-tailed Student's t test.
Error bars indicate S.E.M. ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns, not significant. Scale bars, 50 μm for egg chambers, 10 μm for border cells. See also Figure S8.
JNK signaling pathway was previously reported to be required for cell-cell adhesion between adjacent border cells during their collective migration (Llense and Martin-Blanco, 2008; Melani et al., 2008). Downregulation of JNK signaling in border cells resulted in strong disruption of the coherent cluster morphology, with the most severe cases showing individual border cells dissociated from the cluster. We found that reduction of E75 or increase of DHR3 activity leads to similar but milder defects in cluster morphology. Twenty percent of E75 RNAi expressing border cell clusters (n = 64) at 29°C displayed an incoherent and outstretched cluster morphology (Figure S8C, second row). Furthermore, 27% of DHR3 expressing border cell clusters at 25°C (n = 46) exhibited a more severe incoherent and outstretched cluster phenotype (Figure S8C, third row). It should be noted here that our experiments involving Gal4 were normally carried out at 29°C to maximize the efficiency of UAS/Gal4 system and phenotypic severity. Almost all of the DHR3 expressing clusters at 29°C displayed severe migration block phenotype (no migration, Figure S3B), whereas a majority of DHR3 expressing clusters at 25°C could detach and migrate. Importantly, we only observed incoherent cluster morphology in those DHR3 clusters (25°C) and E75 RNAi clusters (29°C) that have detached and undergone migration, suggesting that strong cell-cell adhesion is especially needed for migratory border cells to maintain their coherent cluster morphology. Lastly, E75 RNAi expression in a small Flip-out clone of two border cells resulted in these two border cells losing cohesion and almost dissociating themselves from the rest of cluster (Figure S3C). Taken together, these results demonstrate that E75 RNAi or DHR3 expression could result in significant cell-cell adhesion defects, indicating that increased DHR3 activity could downregulate cell adhesion.
We next tested whether JNK signaling was negatively regulated by DHR3 and βFtz-f1 during micropyle formation. We showed that in the wild type the level of JNK signaling was reduced from stage 10 to stage 11 and then further reduced from stage 11 to stage 12 (Figures 7E and 7F). Knockdown of βFtz-f1 in the border cells at stage 12 increased the originally low JNK signaling to a much higher level, which is comparable with the level at stage 10 (Figures 7E and 7F). These results suggest that JNK signaling needs to be suppressed in order for lumen formation to proceed properly during stages 11 and 12. Indeed, overexpression of bsk and hence increase of JNK signaling resulted in disruption of lumen formation during formation of the micropyle tip at stage 12 (Figures 7G and 7H). Taken together, these results suggest that JNK-mediated cell adhesion between border cells is temporally and differentially regulated during two different morphogenetic processes—collective migration and micropyle formation—and that its downregulation by DHR3 is essential for lumen formation in the latter process.
Discussion
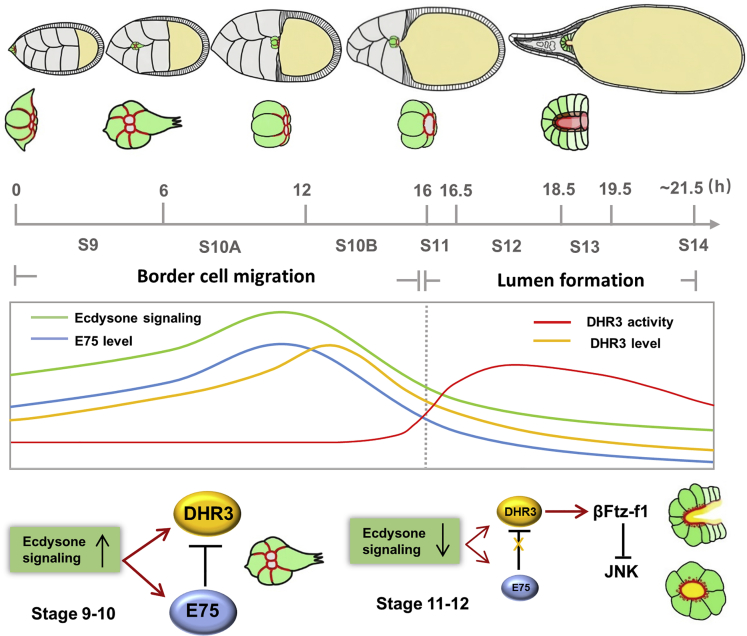
We demonstrate that two nuclear receptors, E75 and DHR3, are critical for temporal regulation and coordination between two very different morphogenetic processes of the border cell cluster, namely its collective migration and its lumen formation. First, our results revealed that the levels of E75 and DHR3 (in response to ecdysone) are the underlying control of the temporal order (Figure 8). Strong loss of function of E75 or DHR3 overexpression disrupts the temporal order and causes lumen formation to occur first. Consequently, collective migration could not take place afterward, because of the unique nature of the lumen structure, which precludes migration from occurring. Second, levels of E75 and DHR3 together with the antagonism between the two nuclear receptors underlie the mechanistic control of the time interval between the two morphogenetic processes (Figure 8). E75 acts as a molecular timer. Its expression level determines the length of interval between migration and lumen formation (Figure 8). Very little E75 (strong loss-of-function) causes lumen formation to occur before migration could take place, effectively resulting in no interval between the two morphogenetic processes. Moderate E75 loss-of-function phenotype demonstrates that collective migration could take place at early stage 9 (Figures S3A and S3B) but accompanied with a precocious occurrence of lumen formation at late stage 9 or stage 10, indicating a shortened interval. On the other hand, too much E75 (E75 overexpression) results in reduced occurrence of lumen formation at stage 12 or 13 (Figure 3F), suggesting an expanded interval. Finally, it is important to note that border cells that supposedly contain the wild-type levels of E75 and DHR3 but display migration defects exhibit a normal time interval (Figures S4A–S4F, and 6F). During tissue or organ formation, it is not uncommon for a certain cell population to undergo two vastly different morphogenetic processes. This study provides a novel mechanistic insight into the molecular machinery that coordinates both the order and time interval between morphological processes.
Figure 8.
Model that Shows How E75 and DHR3 Temporally Coordinate the Migration and Lumen Formation of Border Cells
See description in the Discussion section for details.
It is well known in the metamorphosis literatures that E75 and DHR3 genes are sequentially activated by ecdysone (Huet et al., 1995; White et al., 1997). As a result, E75 is considered as an “early response gene,” whereas DHR3 is considered as an “early-late response gene” (Huet et al., 1995; White et al., 1997). A previous study reported that E75B and DHR3 reached their respective peak protein levels at different time points (about 2 h apart) immediately after pupariation (White et al., 1997). Because E75B level began to decrease a couple of hours before DHR3 did, DHR3 was eventually de-repressed, resulting in the gene activation of βFtz-f1 and appearance of βFtz-f1 protein about 2 h after the E75B decrease began, a time point where E75 was at a low level and DHR3 at a medium level (White et al., 1997). It had been suggested that the ecdysone response genes that control pupal transition were also used reiteratively during oogenesis (Ables et al., 2016). Therefore, it is conceivable that E75 and DHR3 genes could also be sequentially activated by ecdysone in the border cells during early stage 9, and their protein levels reach their respective peaks at different time points during stage 10 (1–2 h apart, Figure 8). It should be noted that such a small time difference (1–2 h) would not be readily detectable with our reporters and antibodies. Consequently, as the E75 level drops below a threshold level immediately before stage 11, the DHR3 protein level, although declining, is still at an intermediate level (Figure 8). As a result, a sufficient amount of DHR3 is de-repressed (activated) to activate βFtz-f1 expression and start the lumen formation process.
Our study also uncovers a surprising mechanism of how a nuclear receptor controls the process of de novo lumen formation. DHR3 seems to act as a master switch or potent inducer for lumen formation because it is necessary and sufficient for lumen formation of border cells both during stage 9 and during stages 11–13. Activation of DHR3 function in border cells seems to simultaneously induce multiple cellular processes that were previously demonstrated to be essential for de novo lumen formation in other systems (Sigurbjornsdottir et al., 2014). In addition, DHR3 is necessary and sufficient for the secretion of chitin into the lumen of border cells both at stage 9 and at stage 12. Chitin had been previously shown to be required for tube expansion and maturation during Drosophila tracheal morphogenesis (Devine et al., 2005). Its function seems to provide an extracellular matrix support (Moussian et al., 2006; Wang et al., 2006). The mechanism by which chitin affects tube morphogenesis remains poorly understood. How DHR3 induces chitin synthesis and secretion and whether chitin is required for lumen formation and tube maturation in micropyle remain to be further determined.
Furthermore, we demonstrate that DHR3's lumen-inducing function is mainly mediated through βFtz-f1, a nuclear receptor and transcription factor that has been well established to be DHR3's immediate target gene during metamorphosis. However, βFtz-f1 does not seem to mediate all of DHR3's functions because overexpression of βFtz-f1 could not induce complete lumen structure and chitin secretion, suggesting that other factors downstream of DHR3 may contribute to lumen formation. Lastly, we show that JNK signaling is downregulated by DHR3 and βFtz-f1, suggesting that cell adhesion between adjacent border cells needs to be reduced during lumen formation. This is consistent with the idea that remodeling of apical polarity, cytoskeleton, and membrane during lumen formation may require downregulation of cell-cell adhesion. Given the multiple functions as demonstrated for DHR3, it will be interesting to test whether these lumen inducing functions will be conserved in other developmental contexts in Drosophila and vertebrate. It is reported that E75 and DHR3 are expressed in tissues that form luminal structures including gut, trachea, and salivary gland (Wilk et al., 2013). Moreover, the ligand sensor of DHR3 displays its activity in gut, trachea, and Malpighian tubules (Palanker et al., 2006). Further study is needed to determine whether E75 and DHR3 are involved in lumen formation in those tissues. Interestingly, previous studies reported that the mammalian homolog of DHR3, RORα, was enriched in human mammary duct, and its inactivation impaired polarized acinar morphogenesis (Xiong et al., 2012; Xiong and Xu, 2014), raising the possibility of a similar lumen-forming role in vertebrate.
Although treated as an excellent model system for collective migration, border cells' physiological function during oogenesis is to make a functional opening within the micropyle for sperm entry. How the border cell cluster develops into the anterior tip of the tubular structure of micropyle is poorly understood. Our study reveals a dynamic remodeling of apical polarity molecules, F-actin, and PIP2-enriched membrane, which is consistent with the process of de novo lumen formation. The functional roles of DHR3, βFtz-f1, EcR, E75, and JNK during micropyle formation, as demonstrated by our study, provide the first detailed analysis of this morphogenetic process. We suggest that in addition to collective migration, border cells could also serve as a model system to study de novo lumen formation in Drosophila.
Limitations of the Study
In this study, we demonstrate that DHR3's lumen-inducing function is mainly mediated through βFtz-f1, a nuclear receptor and transcription factor that has been well established to be DHR3's immediate target gene during metamorphosis. However, what target genes are directly activated or repressed by βFtz-f1 are still unknown. Further studies will be needed to reveal those target genes, for their discovery will be critical to understanding the molecular mechanisms of lumen formation.
Resource Availability
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Jiong Chen (chenjiong@nju.edu.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Henry Krause and Oren Schuldiner for E75 related flies and antibodies, Carl Thummel for DHR3 antibody. We also thank Jacques Montagne, Juan Huang, Lei Xue, Xiaobo Wang, Zizhang Zhou, the Bloomington Drosophila Stock Center, Tsinghua University Fly Stock Center, National Institute of Genetics Stock Center (Japan), and Vienna Drosophila RNAi Center for other fly stocks. We thank Zhenji Gan for critical comments. This work is supported by grants from the National Natural Science Foundation of China (31970743, 31571435) and Natural Science Foundation of Jiangsu Province (BK20171337) to J.C., grants from the National Natural Science Foundation of China (31900563) and Natural Science Foundation of Jiangsu Province (BK20190303) to H.W. G.E. is supported by grants from the Canadian Institutes of Health Research (CIHR; MOP-148560) and the Natural Sciences and Engineering Research Council of Canada.
Author Contributions
Conceptualization, J.C., X.P.W., S.L., and G.E.; Methodology, X.P.W., J.C., and H.W.; Investigation, X.P.W., L.L.; Resources, S.L., G.E.; Visualization, X.P.W. and H.W.; Writing, J.C., X.P.W., H.W., and G.E.; Supervision, J.C. and H.W.
Declaration of Interests
The authors declare no competing financial interests.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101335.
Contributor Information
Heng Wang, Email: wangheng@nicemice.cn.
Jiong Chen, Email: chenjiong@nju.edu.cn.
Supplemental Information
References
- Ables E.T., Hwang G.H., Finger D.S., Hinnant T.D., Drummond-Barbosa D. A genetic mosaic screen reveals ecdysone-responsive genes regulating Drosophila oogenesis. G3 (Bethesda) 2016;6:2629–2642. doi: 10.1534/g3.116.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar N., Streuli C.H. An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium. Nat. Cell Biol. 2013;15:17–27. doi: 10.1038/ncb2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Richards G. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. Dev. Biol. 1976;54:241–255. doi: 10.1016/0012-1606(76)90302-x. [DOI] [PubMed] [Google Scholar]
- Bai J., Uehara Y., Montell D.J. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Beccari S., Teixeira L., Rorth P. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech. Dev. 2002;111:115–123. doi: 10.1016/s0925-4773(01)00615-3. [DOI] [PubMed] [Google Scholar]
- Beckstead R.B., Lam G., Thummel C.S. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialecki M., Shilton A., Fichtenberg C., Segraves W.A., Thummel C.S. Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev. Cell. 2002;3:209–220. doi: 10.1016/s1534-5807(02)00204-6. [DOI] [PubMed] [Google Scholar]
- Brignoni M., Podesta E.J., Mele P., Rodriguez M.L., Vega-Salas D.E., Salas P.J. Exocytosis of vacuolar apical compartment (VAC) in Madin-Darby canine kidney epithelial cells: cAMP is involved as second messenger. Exp. Cell Res. 1993;205:171–178. doi: 10.1006/excr.1993.1072. [DOI] [PubMed] [Google Scholar]
- Bryant D.M., Datta A., Rodriguez-Fraticelli A.E., Peranen J., Martin-Belmonte F., Mostov K.E. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.M., Roignot J., Datta A., Overeem A.W., Kim M., Yu W., Peng X., Eastburn D.J., Ewald A.J., Werb Z. A molecular switch for the orientation of epithelial cell polarization. Dev. Cell. 2014;31:171–187. doi: 10.1016/j.devcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., Freeman M.R., Carlson J.R., Bender M., Cooley L., Segraves W.A. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development. 1999;126:4581–4589. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- Caceres L., Necakov A.S., Schwartz C., Kimber S., Roberts I.J., Krause H.M. Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev. 2011;25:1476–1485. doi: 10.1101/gad.2064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney G.E., Bender M. The Drosophila ecdysone receptor (EcR) gene is required maternally for normal oogenesis. Genetics. 2000;154:1203–1211. doi: 10.1093/genetics/154.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L., Hu X., Zhimulev I., Belyaeva E., Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- Datta A., Bryant D.M., Mostov K.E. Molecular regulation of lumen morphogenesis. Curr. Biol. 2011;21:R126–R136. doi: 10.1016/j.cub.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine W.P., Lubarsky B., Shaw K., Luschnig S., Messina L., Krasnow M.A. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc. Natl. Acad. Sci. U S A. 2005;102:17014–17019. doi: 10.1073/pnas.0506676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanitskaya E., Anllo L., Schupbach T. Phantom, a cytochrome P450 enzyme essential for ecdysone biosynthesis, plays a critical role in the control of border cell migration in Drosophila. Dev. Biol. 2014;386:408–418. doi: 10.1016/j.ydbio.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A., Veligodskiy A., Berge U., Lucas M.S., Kroschewski R. ROCK-mediated contractility, tight junctions and channels contribute to the conversion of a preapical patch into apical surface during isochoric lumen initiation. J. Cell Sci. 2008;121:3649–3663. doi: 10.1242/jcs.018648. [DOI] [PubMed] [Google Scholar]
- Gauhar Z., Sun L.V., Hua S., Mason C.E., Fuchs F., Li T.R., Boutros M., White K.P. Genomic mapping of binding regions for the Ecdysone receptor protein complex. Genome Res. 2009;19:1006–1013. doi: 10.1101/gr.081349.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney J.F., Pucci C., Naes E., Dobens L. Ras signaling modulates activity of the ecdysone receptor EcR during cell migration in the Drosophila ovary. Dev. Dyn. 2007;236:1213–1226. doi: 10.1002/dvdy.21140. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S., Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev. Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Huet F., Ruiz C., Richards G. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121:1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- Jang A.C., Chang Y.C., Bai J., Montell D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat. Cell Biol. 2009;11:569–579. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q., Liu S., Wen D., Cheng Y., Bendena W.G., Wang J., Li S. Juvenile hormone and 20-hydroxyecdysone coordinately control the developmental timing of matrix metalloproteinase-induced fat body cell dissociation. J. Biol. Chem. 2017;292:21504–21516. doi: 10.1074/jbc.M117.818880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y., Masuda S., Hirose S., Ueda H. Temporal regulation of the mid-prepupal gene FTZ-F1: DHR3 early late gene product is one of the plural positive regulators. Genes Cell. 1997;2:559–569. doi: 10.1046/j.1365-2443.1997.1460344.x. [DOI] [PubMed] [Google Scholar]
- Koelle M.R., Talbot W.S., Segraves W.A., Bender M.T., Cherbas P., Hogness D.S. The drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- Kozlova T., Thummel C.S. Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science. 2003;301:1911–1914. doi: 10.1126/science.1087419. [DOI] [PubMed] [Google Scholar]
- Lam G.T., Jiang C., Thummel C.S. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- Li K., Tian L., Guo Z., Guo S., Zhang J., Gu S.H., Palli S.R., Cao Y., Li S. 20-Hydroxyecdysone (20E) primary response gene E75 isoforms mediate steroidogenesis autoregulation and regulate developmental timing in Bombyx. J. Biol. Chem. 2016;291:18163–18175. doi: 10.1074/jbc.M116.737072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.-R., White K.P. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev. Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Llense F., Martin-Blanco E. JNK signaling controls border cell cluster integrity and collective cell migration. Curr. Biol. 2008;18:538–544. doi: 10.1016/j.cub.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Luo J., Zuo J., Wu J., Wan P., Kang D., Xiang C., Zhu H., Chen J. In vivo RNAi screen identifies candidate signaling genes required for collective cell migration in Drosophila ovary. Sci. China Life Sci. 2015;58:379–389. doi: 10.1007/s11427-014-4786-z. [DOI] [PubMed] [Google Scholar]
- Manning L., Sheth J., Bridges S., Saadin A., Odinammadu K., Andrew D., Spencer S., Montell D., Starz-Gaiano M. A hormonal cue promotes timely follicle cell migration by modulating transcription profiles. Mech. Dev. 2017;148:56–68. doi: 10.1016/j.mod.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaret B.S., Thomas J.K., Charles W.W., Richard B.I. Ecdysteroid fluctuations in adult Drosophila melanogaster caused by elimination of pupal reserves and synthesis by early vitellogenic ovarian follicles. Insect Biochem. 1989;19:243–249. [Google Scholar]
- Melani M., Simpson K.J., Brugge J.S., Montell D. Regulation of cell adhesion and collective cell migration by hindsight and its human homolog RREB1. Curr. Biol. 2008;18:532–537. doi: 10.1016/j.cub.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Montell D.J. Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- Montell D.J., Rorth P., Spradling A.C. Slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Moussian B., Schwarz H., Bartoszewski S., Nusslein-Volhard C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster. J. Morphol. 2005;264:117–130. doi: 10.1002/jmor.10324. [DOI] [PubMed] [Google Scholar]
- Moussian B., Tang E., Tonning A., Helms S., Schwarz H., Nusslein-Volhard C., Uv A.E. Drosophila Knickkopf and Retroactive are needed for epithelial tube growth and cuticle differentiation through their specific requirement for chitin filament organization. Development. 2006;133:163–171. doi: 10.1242/dev.02177. [DOI] [PubMed] [Google Scholar]
- Palanker L., Necakov A.S., Sampson H.M., Ni R., Hu C., Thummel C.S., Krause H.M. Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development. 2006;133:3549–3562. doi: 10.1242/dev.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinking J., Lam M.M., Pardee K., Sampson H.M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Richards G.W.R.A. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121:1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- Rougvie A. Control of developmental timing in animals. Nat. Rev. Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- Sap K.A., Bezstarosti K., Dekkers D.H.W., van den Hout M., van Ijcken W., Rijkers E., Demmers J.A.A. Global quantitative proteomics reveals novel factors in the ecdysone signaling pathway in Drosophila melanogaster. Proteomics. 2015;15:725–738. doi: 10.1002/pmic.201400308. [DOI] [PubMed] [Google Scholar]
- Segraves W.A., Hogness D.S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Sigurbjornsdottir S., Mathew R., Leptin M. Molecular mechanisms of de novo lumen formation. Nat. Rev. Mol. Cell Biol. 2014;15:665–676. doi: 10.1038/nrm3871. [DOI] [PubMed] [Google Scholar]
- Silver D.L., Geisbrecht E.R., Montell D.J. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- Strilic B., Eglinger J., Krieg M., Zeeb M., Axnick J., Babal P., Muller D.J., Lammert E. Electrostatic cell-surface repulsion initiates lumen formation in developing blood vessels. Curr. Biol. 2010;20:2003–2009. doi: 10.1016/j.cub.2010.09.061. [DOI] [PubMed] [Google Scholar]
- Sullivan A.A., Thummel C.S. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol. Endocrinol. 2003;17:2125–2137. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- Terashima J., Bownes M. E75A and E75B have opposite effects on the apoptosis/development choice of the Drosophila egg chamber. Cell Death Differ. 2006;13:454–464. doi: 10.1038/sj.cdd.4401745. [DOI] [PubMed] [Google Scholar]
- Thummel C.S. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Vega-Salas D.E., Salas P.J., Rodriguez-Boulan E. Exocytosis of vacuolar apical compartment (VAC): a cell-cell contact controlled mechanism for the establishment of the apical plasma membrane domain in epithelial cells. J. Cell Biol. 1988;107:1717–1728. doi: 10.1083/jcb.107.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Jayaram S.A., Hemphala J., Senti K.A., Tsarouhas V., Jin H., Samakovlis C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 2006;16:180–185. doi: 10.1016/j.cub.2005.11.074. [DOI] [PubMed] [Google Scholar]
- Wang X., Adam J.C., Montell D. Spatially localized Kuzbanian required for specific activation of Notch during border cell migration. Dev. Biol. 2007;301:532–540. doi: 10.1016/j.ydbio.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Webb A.B., Oates A.C. Timing by rhythms: daily clocks and developmental rulers. Dev. Growth Differ. 2016;58:43–58. doi: 10.1111/dgd.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.P., Hurban P., Watanabe T., Hogness D.S. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Wilk R., Hu J., Krause H.M. Spatial profiling of nuclear receptor transcription patterns over the course of Drosophila development. G3 (Bethesda) 2013;3:1177–1189. doi: 10.1534/g3.113.006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Wang C., Evers B.M., Zhou B.P., Xu R. RORalpha suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012;72:1728–1739. doi: 10.1158/0008-5472.CAN-11-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Xu R. RORalpha binds to E2F1 to inhibit cell proliferation and regulate mammary gland branching morphogenesis. Mol. Cell. Biol. 2014;34:3066–3075. doi: 10.1128/MCB.00279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Rewitz K.F., O'Connor M.B. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zimmerman S., Brakeman P.R., Beaudoin G.M., 3rd, Reichardt L.F., Marciano D.K. De novo lumen formation and elongation in the developing nephron: a central role for afadin in apical polarity. Development. 2013;140:1774–1784. doi: 10.1242/dev.087957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K.Y., Merzendorfer H., Zhang W., Zhang J., Muthukrishnan S. Biosynthesis, turnover, and functions of chitin in insects. Annu. Rev. Entomol. 2016;61:177–196. doi: 10.1146/annurev-ento-010715-023933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D reconstruction of confocal z-stacks of wild-type (WT), E75 RNAi and DHR3 overexpressing border cells. The aPKC staining labels the apical membrane that surrounds the central lumen with significant depth (about 5 μm in depth) in the E75 RNAi or DHR3 overexpressing border cell clusters, which are the same clusters as those shown in fourth row and third row of Figure 2A respectively.
3D reconstruction of confocal z-stacks of wild-type border cell clusters from stage 9 to stage 12, which are the same as those from Figure 3B and are labeled with Par6-GFP.
3D reconstruction of confocal z-stacks of wild-type border cell clusters from stage 9 to stage 12, which are the same as those from Figure 3C and are labeled with PIP2-GFP.
3D reconstruction of confocal z-stacks of wild-type border cell clusters from stage 9 to stage 12, which are the same as those from Figure 3D and are labeled with Lifeact-GFP.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.