Abstract
SARS-CoV-2, a newly emerged pathogen in December 2019, marked as one of the highly pathogenic Coronavirus, and altogether this is the third coronavirus attack that crossed the species barrier. As of 1st July 2020, it is spreading around 216 countries, areas or territories, and a total of 10,185,374 and 503,862 confirmed cases and death reports, respectively. The SARS-CoV-2 virus entered into the target cells by binding with the hACE2 receptors. Spike glycoprotein promotes the entry of the virus into host target cells. Literature reported a significant mutation in receptor binding sites and membrane proteins of the previous SARS-CoV to turned as SARS-CoV-2 virus, responsible for most dreadful pandemic COVID-19. These modifications may be the probable reason for the extreme transmission and pathogenicity of the virus. A hasty spread of COVID-19 throughout the world is highly threatening, but still, scientists do not have a proper therapeutic measure to fight with it. Scientists are endeavoring across the world to find effective therapy to combat COVID 19. Several drugs such as Remdesivir, Hydroxychloroquine, Chloroquine, Ribavirin, Ritonavir, Lopinavir, Favipiravir, Interferons, Bevacizumab, Azithromycin, etc. are currently under clinical trials. Vaccine development from various pharmaceutical companies and research institutes is under progress, and more than ten vaccine candidates are in the various phases of clinical trials. This review work highlighted the origin, emergence, structural features, pathogenesis, and clinical features of COVID-19. We have also discussed the in-line treatment strategies, preventive measures, and vaccines to combat the emergence of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Virology, Pathogenesis, Clinical features, Treatment strategies
Graphical abstract
Highlights
-
•
The hACE2 receptors facilitate the entry of the SARS-CoV-2 virus.
-
•
USFDA announced emergency use approval for remdesivir to treat COVID-19.
-
•
Clinical manifestation of COVID-19.
-
•
Recent updates on vaccine developments.
1. Introduction
The Coronavirus (CoV) as believed to be originated from bats, was known for 800 years, confirmed by many works of literature. These are positive-sense single-strand RNA viruses with around 24 similar species from the family of coronaviridae. This family of coronaviridae is further categorized as α, β, λ, and δ based on its distinct genetic features. However, among these, only alpha (α) and beta (β) coronavirus genera are pathogenic to mammalian and humans (Chen et al., 2020b; Paules et al., 2020). The first isolated avian infectious bronchitis virus was noticed in the year 1937 and further known for annihilative infections in chicken. In this connection, Tyrrell and Bynoe et al. have propagated the human Coronavirus in 1965 on an in vitro ciliated embryonic cell culture of the human respiratory system (Berry et al., 2015; Su et al., 2016; Yang et al., 2020a). During the extensive research on Coronaviruses, a total of six coronaviruses were identified to cause respiratory disease, i.e., HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV (severe acute respiratory syndrome coronavirus), and MERS-CoV (middle east respiratory syndrome coronavirus). Interestingly, out of these HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1 were reported as less pathogenic compare to SARS-CoV and MERS-CoV (Bonilla-Aldana et al., 2020; Skariyachan et al., 2019; Walls et al., 2020b).
The origin of both SARS-CoV and MERS-CoV were from Guangdong, China (2002), and Arabian Peninsula (2012), respectively (Fig. 1 ) (Ghinai et al., 2020; Walls et al., 2020b). A vast population has been affected by respiratory disease due to these two Coronavirus outbreaks (Anderson et al., 2020; Liu et al., 2020b).
Fig. 1.
Comparative analysis of three different coronavirus outbreaks in the 21st century, including SARS-CoV, MERS-CoV and COVID-19.
In December 2019, a new outbreak was noticed after a massive admission of patients with common clinical symptoms of pneumonia in the local hospitals of Wuhan city, China. This incidence has dragged the attention of many physicians, followed by scientists and regulatory agencies across the world. Upon further investigations, the World Health Organization (WHO) confirms the novel Coronavirus named SARS-CoV-2 is responsible for these clinical symptoms, and further declared this diseased condition as COVID-19 (He et al., 2020; Yang et al., 2020b).
This disappointing outbreak of the COVID-19 (Coronavirus Disease 2019) situation spreading throughout the world was announced as a pandemic disease by WHO. As per the latest WHO situation report 162 released on 1st July 2020, 10,185,374 confirmed cases and 503,862 death cases reported throughout the world. For a better understanding of COVID-19 devasting effect, WHO describes the statistics by categorizing the whole world into six different continental regions. As on date, the region of America is reported as the worst affected region with 5,136,705 confirmed cases and 247,129 death reports. The second most terribly affected region is the European continent with 2,692,086 confirmed cases and 197,254 death reports. This data is followed by the Eastern-Mediterranean region at the third position (1,058,055 confirmed cases and 24,423 deaths) and South-East Asia at the forth position of COVID-19 disaster (784,931 cases and 21,593 deaths). While the Western Pacific region stands at the second least affected region (215,566 cases and 7440 death reports) and Africa is listed as a minimally affected region (297,290 cases and 6010 deaths) till date (WHO, 2020a). During this outbreak of COVID-19, the world is frightened with an unpredictable and hasty impact of the infection, and the data is changing day by day.
2. Virology of SARS-CoV-2
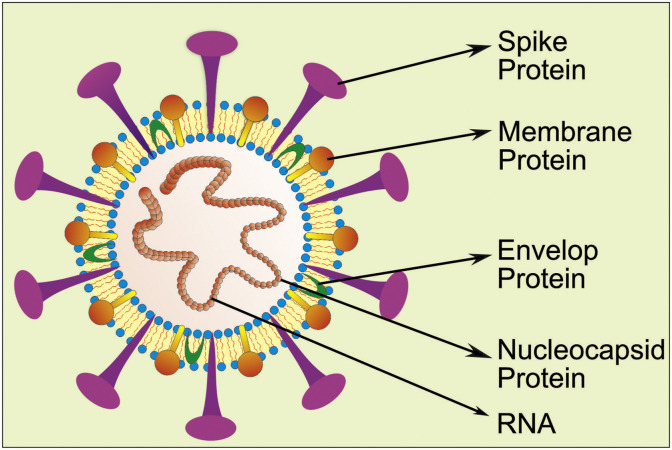
Coronaviruses are crown shape peplomers, positive-sense ssRNA (single strand RNA) virus, which was reported in the pleomorphic form with 80–160 nm size (Shang et al., 2020). It is a nonsegmented and RNA virus ranging from 26 to 32 kb. Coronavirus comes under the order-nidovirus, family-coronaviridae, subfamily-coronavineae which were further divided into α, β, ϒ and δ genus. Among these, α and β genus of Coronavirus mainly affect the human population. The α genus contains HCoV-229E (Human Coronavirus) and NL63, while the β genus consists of HKU1, 229E, OC43, MERS-CoV, SARS-CoV and latest outbreak SARS-CoV-2. Coronavirus has been mainly reported with single-strand RNA, nucleocapsid protein, envelop protein, membrane protein, spike glycoprotein (S), etc., as shown in Fig. 2 (Lei et al., 2018).
Fig. 2.
Schematic representation of the SARS-CoV-2 virus showing all its components, including a single strand RNA, envelop protein, nucleocapsid protein, spike protein, and membrane protein.
Spike (S) glycoprotein is responsible for the characteristic feature of the Coronavirus because it forms the crown-like structure on the outer surface of the virus. The S-protein divides into two subunits, namely, S1 and S2. The S1 subunit further classified into three domains, particularly A, B, and C (Angeletti et al.). Generally, domain A of the subunit S1 present on CoV-OC43 and CoV-HKU1 is binding with the host receptors (Hulswit et al., 2019). However, MERS-CoV uses both A and B domains to enter into the cell by binding to the DPP4 (Dipeptidyl peptidase-4) receptor (Park et al., 2019). While SARS-CoV-2 and SARS-CoV enter into the target cell through direct interaction with domain B. This, in turn, attaches to the human angiotensin converting enzyme-2 (hACE) receptor (Cui et al., 2019). Interestingly, the structure of the S protein in both SARS-CoV and novel SARS-CoV-2 viruses is almost similar to a few differences.
2.1. Unique features of the SARS-CoV-2 genome
The main genomic features of SARS-CoV-2 behind their effective binding with hACE 2 receptors in comparison with the SARS-CoV is not only mutations on the receptor-binding domain (RBD) of S protein but also the presence of O-linked glycans and polybasic furin cleavage site (Andersen et al., 2020; Walls et al., 2020a).
2.2. Mutations on the RBD of S protein in SARS-CoV-2
Recent literature reveals that RBD of the S1 subunit of the “S” protein is responsible for binding with ACE2 (Angiotensin-converting enzyme) receptors (Letko et al., 2020). This RBD in coronaviruses is highly variable. Based on studies, six RBD amino acids that remain present on the S1 subunit are very critical for binding to the receptors. Interestingly, different structural and functional studies stated that five of these six residual proteins are distinct in SARS-CoV-2 compared to SARS-CoV. Accordingly, the coordinates based on the COVID-19 (L455, F486, Q493, S494, N501, and Y505) compared to those found in SARS-CoV (Y442, L472, N479, D480, T487, and Y4911) differ in only five distinct residues except Y4911 (Andersen et al., 2020). These differences may be the probable reason for the high affinity of SARS-CoV-2 with the receptors and therefore are optimal for receptor binding (Letko et al., 2020; Wan et al., 2020; Wrapp et al., 2020; Zhou et al., 2020).
2.3. Polybasic furin cleavage site and O linked glycans
A polybasic cleavage site (RRAR) is present at the union site of S1 and S2 subunit of Spike protein. In addition, proline residue is also introduced prior to this cleavage site, and the sequence in SARS-CoV-2 becomes PRRA (Unique protein sequencing in SARS-CoV-2). Insertion of the polybasic cleavage site facilitates an effective cleavage of S-protein by furin and other proteases, which plays a detrimental role in viral infectivity. In SARS-CoV-2, this feature leads to the addition of O-linked glycans to S673, T678, and S686, which flank the cleavage site. This cleavage site is not found in other β-coronavirus and thereore remains a unique characteristic feature of COVID-19 (Dhama et al., 2020). Similar experimentation in MERS-CoV showed that the insertion of the furin cleavage site in the 'S' protein enables the infection of human cells from MERS-CoV (Liu et al., 2015; Menachery et al., 2020). In the same series, in avian Influenza viruses not only the rapid replication and transcription upon insertion of the cleavage site has been seen but also the conversion of low pathogenicity virus into highly pathogenic forms is well observed (Alexander and Brown, 2009; Ito et al., 2001). However, the function of the predicated O-linked glycans is still not clear, but based on the literature, they may create a mucin-like domain that helps in immunoevasion (Bagdonaite and Wandall, 2018). In highly human pathogenic Coronaviruses, the ‘S’ trimers exist in a partially open state and remain in closed state in less pathogenic virus (Walls et al., 2020).
2.4. Entry and replication of Coronavirus in a host cell
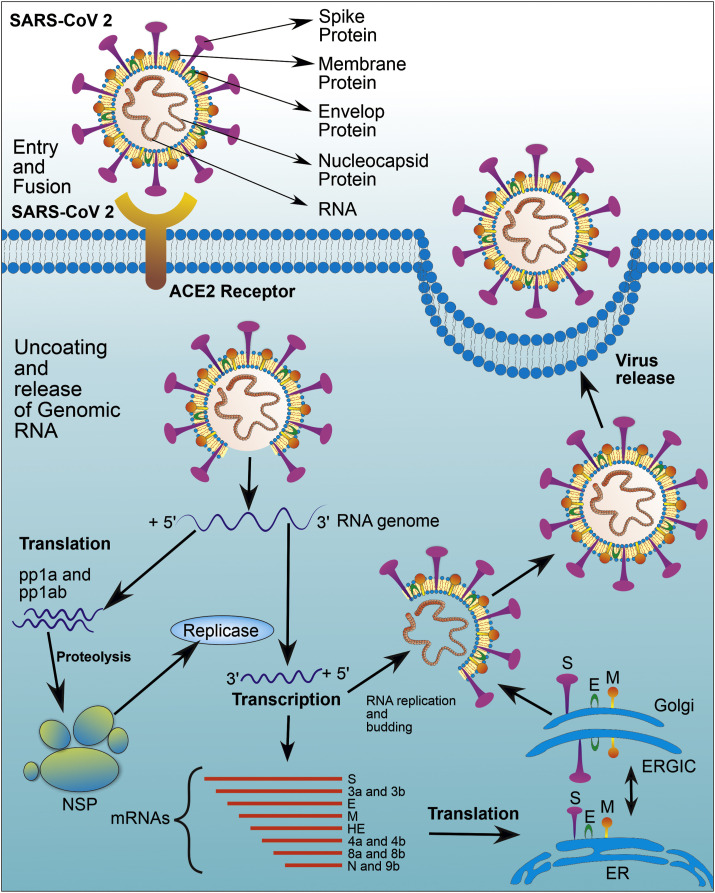
The Coronavirus enters into the host cell, particularly type-2 pneumocytes, by binding with specific cell surface receptors like a hACE-2 and CD90L (L-sign) in the case of SARS-CoV and SARS-CoV-2 while DPP4 receptor in case of MERS-CoV (Fig. 3 ) (Li et al., 2020). In the case of SARS-CoV-2, the RBD of the S1 subunit binds with the hACE2 receptor, while the S2 subunit facilitates fusion between the host and the viral cell membrane (Li et al., 2005).
Fig. 3.
Replication of the virus in host cell. SARS-CoV-2 enters into the host cell by binding with specific cell surface receptors like a human angiotensin-converting enzyme (hACE). S1 and S2 subunit of spike glycoproteins facilitate the process of entry and fusion between the host and the viral cell membrane. Followed by uncoating, Viral genomic mRNA is entered into the host cell cytoplasm. Two-third portion of the genomic RNA contains two ORFs mainly ORF1a and ORF1b which gets translated into two polypeptides namely pp1a and pp1ab which further gives rise to 16 no of NSPs through the proteolysis process. All these NSP proteins are involving in the replication and transcription process. One-third of the remaining viral genome transcribed into antisense RNA (3′ to 5′), further, it will replicate and formed to a full-length positive strand of genomic RNA with the help of replicase activity of viral RNA dependent RNA polymerase. On the other hand, antisense RNA is also able to synthesize several different small sizes nested (subgenomic) mRNA via discontinuous transcription and further translated into structural proteins like envelope protein (E), membrane protein (M), nucleocapsid (N) and spike proteins (S). Theses structural proteins are assembled into the nucleocapsid and viral envelope at the ER or ERGIC, followed by release of virus by exocytosis or by fusing with the plasma membrane.
The SARS-CoV-2 contains four distinct amino acids on its S1 and S2 subunits which results in the introduction of furin cleavage sites (Cui et al., 2019). The furin cleavage sites facilitate a very strong binding of the S-glycoprotein with the hACE-2 receptor of SARS-CoV-2 (Dhama et al., 2020). The cleavage of ‘S’ takes place at the boundary present between S1 and S2 subunits being bound into a perfusion confirmation form, which is non-covalent in nature present within the boundary of the membrane. Commonly protease enzyme lyse the protein present between the S1 and S2 subunits upon its binding with the hACE receptor (Liu et al., 2015). This lysis makes covalent conformation changes (irreversible), which increases the binding affinity of the protein and in turns extensively activates the protein for fusion (Belouzard et al., 2009). The furin cleavage sites and modified receptor binding sites of the S1 subunit may be the probable reasons behind the rapid transmission, replication, and infectivity of this virus (Zhou et al., 2020).
A viral genomic mRNA, along with the nucleocapsid, enters the host cell cytoplasm through either receptor-mediated endocytosis or directly through the host cell membrane after binding and fusion of ‘S’ protein. In general, the genomic RNA of the Coronavirus is very long and has nearly 30,000 nucleotides. Also, genomic RNA mainly consists of a 6–10 open reading frame (ORF) that encodes the replicase gene (Cowling and Leung, 2020). Both ORF1a (Open Reading Frame) and ORF1b present about a two-third portion of genomic RNA, and they translate this portion of genomic RNA into two large polyproteins, namely pp1a and pp1ab, respectively. Moreover, two protease enzymes, namely Papain like protease (PLpro) and 3C like protease (3CLpro) cleave these polyproteins into non-structural proteins (NSP) (Schoeman and Fielding, 2019). The polyproteins pp1ab and pp1b form NSP1-11 and NSP1-16, respectively. Since these proteins are involving in the replication and transcription process, these proteins are also called as replicase and transcriptase proteins. These NSPs now rearrange themselves within the membrane of the rough endoplasmic reticulum (RER) and make replicase-transcription complex (RTC) thereof. All these NSPs have specific functions like NSP1 (cellular mRNA degradation and inhibiting interferon signaling), NSP 12 (RNA dependent RNA polymerase activity), NSP13 (helicase activity), NSP 14 (exoribonuclease activity) and NSP 15 (endoribonuclease activity). One-third of the viral genome that is coded for remaining ORF transcribed into antisense RNA (3′ to 5′) with the help of RNA dependent RNA polymerase (Fehr and Perlman, 2015). This antisense RNA is further replicated and transformed to a full-length positive strand of genomic RNA with the help of replicase activity of viral RNA dependent RNA polymerase (Cui et al., 2019).
Further, this antisense RNA synthesize several different small sizes nested (subgenomic) mRNA via discontinuous transcription process by RNA dependent RNA polymerase (Chen et al., 2020b). Further translation of this subgenomic mRNA gives rise to viral structural proteins including envelope protein (E), membrane protein (M), nucleocapsid protein (N), and spike proteins (S) along with some accessory proteins (Knoops et al., 2008). The three structural proteins (E, M, and S) introduced at the endoplasmic reticulum (ER) membrane or Golgi membrane. The viral genomic RNA, combine with the nucleocapsid protein forms nucleocapsids. This viral particle buds into the ER or Golgi intermediate compartment complex (ERGIC) (Alsaadi and Jones, 2019). The mature virion/virus-containing vesicles are finally released from the cell either by the mechanism of exocytosis or by fusing with the plasma membrane. This cycle continues when the mature virion/virus infects a new host (Fig. 3) (Perlman and Netland, 2009).
3. Pathogenesis of COVID-19
Although the pathogenesis behind COVID-19 is not well understood, the pathogenesis of MERS-CoV and SARS-CoV still can be the best source of information regarding COVID-19 (Li et al., 2020b).
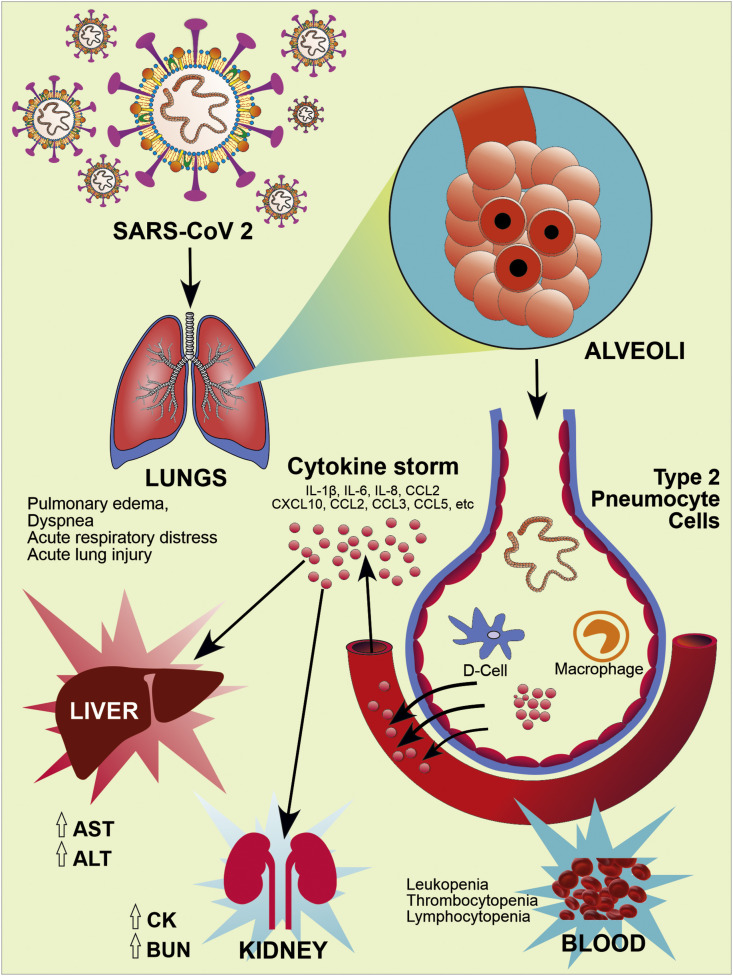
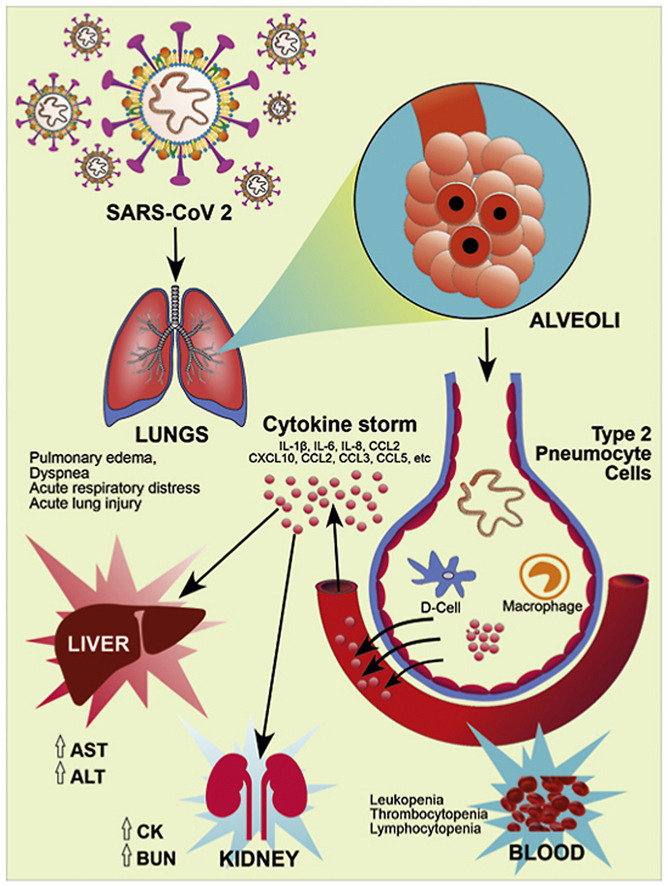
Recent literature suggests that the modified residues of RBD of S1 subunit, presence of RRAR and partially opened state of ‘S’ trimer may be a reason for high pathogenicity and transmission capacity of COVID-19. These RBD of S1 subunit on Spike proteins binds to the hACE2 receptor which are mainly present in the lungs, particularly type-2 pneumocytes. This leads to the subsequent downregulation of hACE2 receptors (Kuba et al., 2005; Xu et al., 2020). The downregulation of ACE2 receptors may lead to the increased production of angiotensin-2 (AT2) by the related enzyme ACE1. Increased production of AT2 potentially increases pulmonary vascular permeability and may cause lung injury (Imai et al., 2005). Further, SARS-CoV-2 contains antigen-presenting cells that attach to the dendritic cell of a host which activates macrophages and leads to the severe immunological reaction resulting in excessive release of pro-inflammatory cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.) called 'cytokines storm' (Cameron et al., 2008). These inflammatory mediators, further damage the epithelial cells lining and reach into the blood circulation where it causes damage to other organs (Li et al., 2020b; Rothan and Byrareddy, 2020).
4. Clinical features of COVID-19
The clinical symptoms of COVID-19 infection can be seen after 5–6 days of incubation which mainly differs from age and immune system of the person. Males are more susceptible to SARS-CoV-2 as compare to females. People with the age of more than 60 are more sensitive to SARS-CoV-2 (Li et al., 2020a).
The most common clinical features of SARS-CoV-2 infection are fever (more than 80% cases), cough (more than 60% cases), fatigue (more than 35% cases), sputum production (more than 30% cases), and shortness of breath (more than 15% cases). However, less common features include headache, muscle weakness, breathlessness, sore throat, and pleuritic pain (10–15%). Apart from this nausea, vomiting, chest tightness is the rare features of COVID-19 The primary cause of mortality and severity of COVID-19 are respiratory failure (69.5%), sepsis or multi-organ failure (28%), cardiac failure (14.6%), and renal failure (3.7%). (Heymann and Shindo, 2020; Zhang et al., 2020).
Recently, Guan et al. published a paper in the New England journal of medicine on clinical features of COVID-19 in China. They collected data from 1099 patients with conformed COVID-19 cases. As per that report, the most common symptoms were fever (88.7%), cough (67.8%), CT (Computer Tomography) scan abnormalities (86.2%). The most common pattern observed in CT scan were ground-glass opacity and patchy bilateral shadowing. Further laboratory findings revealed the presence of lymphocytopenia (83.2%), thrombocytopenia (36.2%), and leukopenia (33.7%). That study also reported that most of the patients were observed with high levels of C-reactive protein. Alanine transaminase (ALT), aspartate aminotransferase (AST), Creatinine kinase (CK), blood urea nitrogen (BUN), and D-dimer were elevated less commonly. In this study, out of 1099 patients, 5% were admitted in ICU, 2.3% underwent invasive mechanical ventilation, and 1.4% of patients died (Guan et al., 2020).
The excessive sputum production and cough like symptoms are due to the presence of ACE receptor on the alveoli and COVID-19 has a high affinity towards the hACE receptor (Liu et al., 2020b). Apart from this, SARS-CoV-2 is showing some unique symptoms like infection of the upper respiratory system, mainly rhinorrhea, pharyngitis, and sneezing makes it unique from other members of the corona family-like SARS-CoV and MERS-CoV (Huang et al., 2020a). From the 82 death samples, scientists found the main cause of mortality in COVID-19 is respiratory tract infection, they also found the high level of liver biomarkers ALT, AST which indicate that liver failure can also be one of the key players (Zhang et al., 2020).
New findings show that in COVID-19, patients have the chance to develop CNS (Central Nervous System) symptoms like dizziness, acute cerebrovascular disease, headaches, unconsciousness with some PNS (Peripheral Nervous System) symptoms like hypogeusia, hyposmia, and skeletal muscle injury are high (Mao et al., 2020). Patients with a condition like hypertension, COPD (Chronic Obstructive Pulmonary Disease), diabetes, and cardiovascular disease are more prone to cause SARS-CoV-2 infection. Respiratory distress syndrome, septic shock, metabolic acidosis, coagulation dysfunction, and multiple organ failure are noted as the main complications of COVID-19 (Fig. 4 ) (Wang et al., 2020b). Some of the important clinical features with detailed descriptions are mentioned below.
Fig. 4.
Pathogenesis and Clinical Manifestation of COVID-19. RBD of S1 subunit of spike glycoproteins on the envelope of SARS-CoV-2 binds to the human hACE2 receptor, particularly in type-2 pneumocytes of lung tissue, and causes lung injury. ACE2 receptors expressed on other tissues like oral mucosa, vascular endothelial cells, gastrointestinal tract, kidney, heart, blood vessels also prone to infection. Further, SARS-CoV-2 infection activates host innate and adaptive immune response. Uncontrolled dysregulation of the host immune response against infection may cause the releases of excessive pro-inflammatory mediators and lead to the cytokine storm. It causes harmful tissue damage at the systemic level.
4.1. Severe pneumonia
Severe fever, followed by shaking chills, breathlessness, and chest pain, are the main pathological symptoms of pneumonia, which were found to be in SARS-CoV-2 infection. Cyanosis also can occur, especially in children during the infection. Breathing rate >30/min and hypoxia SpO2 below 90% are the main clinical basis behind pneumonia (Peng et al., 2020).
4.2. Acute respiratory distress syndrome (ARD)
Pro-inflammatory cytokines are secreted by alveolar macrophages results in the recruitment of neutrophil and monocyte or macrophage along with activation of T-cells of epithelial cells, which further starts inflammation and tissue injury (Ritchie and Singanayagam, 2020). In medical term, ARD is called “diffuse alveolar damage,” or epithelial-cell hyperplasia. The diagnostic parameter for ARD is hypoxia, which mainly decides based on the ratio between the blood pressure of oxygen and the percentage of oxygen supply, formulated as PaO2/FiO2 mmHg. In severe cases, PaO2/FiO2 found to be ≤ 100 mmHg while in moderate cases, found to be ≤ 200. This ratio falls from 200 to 300 in mild cases of ARD due to SARS-CoV-2 (Wang et al., 2020b). Other parameters that can distinguish ARD are bilateral opacities followed by the collapse of lungs, which can be diagnosed through CT scan or ultrasound, where lung infiltration found to be >50% (Lai et al., 2020a).
5. Diagnosis for the COVID-19
To control COVID-19, an accurate line of diagnosis is required to confirm its presence. Interestingly, numerous approaches are available for the diagnosis of COVID-19. Once confirmed, it must be followed by the isolation of individuals from the population. Hence, preliminary prevention is limited to diagnostics. During the COVID-19 outbreak, proper tracking of travel history is highly advisable, as witnessed by a series of earlier incidences from China (Wuhan city) (Lai et al., 2020b). The suspected cases shall be confirmed by a preliminary thermal screening followed by the molecular tests as well as CT imaging as per the regulatory guidelines (Singhal, 2020). The recommended diagnostic techniques towards the screening of the COVID-19 are mention below.
5.1. Infrared sensors and thermal scanning
It is a reliable technique to isolate the affected population from the large population. It is also essential for initial screening for auxiliary diagnosis. A thermal camera was first introduced at hospital entrances and in the emergency department to recognize any persons with incremental body temperature. The scanning distance required for such cameras is 10 m. Thermal cameras captured and detect infrared energy in terms of heat and convert it into a visual image. The installation of such cameras at various public places, including airports, railway stations, academic institutions, and research centers, is useful to screen the body temperature of mass gatherings with the accuracy up to ±0.5 °C. Generally, the thermal cameras are operated at longer wavelength IR energy (Lee et al., 2020). Whereas IR scanners are used to scan individuals, but it needs more time to screen a large population. Hence, it may be concluded that thermal cameras are the better choice to screen at a large scale over IR scanners.
5.2. Nucleic acid amplification test (NAAT)
NAAT is being used to confirm COVID-19 disease through a nasal swab or blood sample using a real-time fluorescence polymerase chain reaction (RT-PCR) as per the WHO established protocol (Wu et al., 2020). As per recent literature, if a viral load is very less, a limited sensitivity is witnessed due to the low detection of virus nucleic acid. This low detection may lead to false-negative results thereof (Dai et al., 2020). Recently, US-FDA and various regulators have given emergency use authorization for diagnostic kits to detect COVID-19 based on RT-PCR technology (Devices, 2020). Cobas® SARS-CoV-2 was the first commercial diagnostic kit to meet the required emergency medical needs under Clinical Laboratory Improvement Amendments of 1988 (CLIA) to perform moderate to high complexity tests (Roche's, 2020).
However, numerous potential pre-analytical and analytical risks are associated with RT-PCR. Such risks are false () identification, sample preparation, specificity, sensitivity, stability issues, and also, it is a time-consuming process. Although calibration of the instruments is also a matter of great concern, as it may provide false or misleading results, therefore it is highly recommended to calibrate the RT-PCR regularly to avoid any misleading readings. Besides, the diagnostic kit should also be pre-validated in terms of the accuracy and reliability of the data (Lippi et al., 2020; SK Khaja et al., 2020).
5.3. Computerized tomography scan
Chinese researchers have recommended that CT imaging was the primary basis for the diagnosis of COVID-19 in the current situation due to the limitation of NAAT (Ai et al., 2020). However, this was a matter of case study after Huang et al. report, claiming a patient history of fever (37.8 °C) only. In the report, the patient was showing symptoms of sore throat and fatigue. Initially, it was observed negative results of COVID-19 with a fluorescent RT-PCR test of the sputum till 06 days. This patient had a travel history of Wuhan city, China. The examination of the suspected patient was done with chest CT and showed the first day multiple peripheral ground-glass opacities in both lungs with more involvement of the left upper lobe, and liner segment. This group reported the progression of ground-glass opacities in the lungs, which increased after three days of hospitalization due to COVID-19 (Huang et al., 2020b). Therefore, it is highly recommended by the researchers that chest CT imaging has a high rate of detection of viral pneumonia. However, other observations, including ground-glass opacities (GGOs) with patchy consolidation and posterior part or lower lobe involvement predilection in case of COVID-19 (Zu et al., 2020). Li et al. reported the development and clinical implementation of a rapid IgM-IgG combined antibody test for the diagnosis of SARS-CoV-2 infection. This group developed a rapid (within 15 min) and simple point-of-care lateral flow immunoassay. This developed immunoassay can detect IgM and IgG antibodies simultaneously against the SARS-CoV-2 virus at different stages of COVD-19. This research group validated test kit against clinical samples for rapid screening and this kit has passed for specificity tests also (Li et al., 2020c).
6. Current inline treatments
As per the latest report of WHO, FDA and CDC (Centre for Disease Control and Prevention), no appropriate therapy, medicine or vaccine are developed and approved till date for the prevention and treatment of SARS-CoV-2 infection (CDC, 2020; FDA; Organization, 2020). However, scientific fraternity throughout the globe are aggressively working to find out a promising solution to this epidemic outbreak originated from China. Many preclinical and clinical investigations by different Institutions, Government bodies, research centers and pharmaceutical industries focused on testing the efficiency of various existing drug moieties based on their previous history to treat viral infections.
In this sequence, Wang and team in February 2020, estimated the antiviral efficacy of five different FDA approved drugs, including chloroquine, ribavirin, nitazoxanide, penciclovir, nafamostat along with two commonly used wide spectrum antiviral agents favipiravir and remdesivir against COVID-19 infection. The drugs were tested in vitro on the clinically isolated sample of COVID-19 strain. They have performed cytotoxicity assessment and evaluated the drug effect on yield of virus and rate of infection on the in vitro culture of COVID-19. The study revealed two drugs, named chloroquine and remdesivir, effectively controlled the COVID-19 infection in in vitro cell culture. Based on their safety profile and efficacy in other viral infections, the author suggested the drug should be tried in human patients suffering from COVID-19 infection (Wang et al., 2020a).
6.1. Remdesivir
During the outbreak, remdesivir was tested on three COVID-19 infected patients in the U.S., which showed improvement of symptoms with no significant side effects, and also FDA has given permission about 250 patients to use this product. As far as safety is a major concern, Gilead Sciences announced phase III clinical trial of remdesivir to prove its safety and efficacy in COVID-19 infection. After a preliminary investigation, Gilead announced the results of the initial Phase 3 clinical trial. The drug significantly reduced the mortality rate upon 14 days of treatment and showed improvement in 64% of cases (Gilead, 2020). Based on these results, the FDA issued Emergency Use Authorizations for the use of remdesivir against COVID-19; however, full approval is still not provided anywhere in the world (FDA, 2020).
6.2. Chloroquine
The FDA already approves the drug chloroquine for treating malaria, arthritis and lupus (Solomon and Lee, 2009). Recent studies also showed chloroquine as a promising drug candidate for the treatment of SARS-CoV-2 virus infection (Touret and de Lamballerie, 2020). More than 35 different clinical trials registered in China and the USA for evaluating the application of chloroquine against COVID-19 infection. The preliminary investigations, past success in treating viral infections and safety profile will give a hope that chloroquine possibly provides effective therapy for this pandemic situation (Gao et al., 2020; Touret and de Lamballerie, 2020; Wang et al., 2020a). The National Health Commission of the People's Republic of China reported that chloroquine had been included as trial drugs in the Guideline of Diagnosis and Treatment of COVID-19 (6th edition) due to positive results in preliminary clinical studies. Chloroquine, thus considered a safer option of treatment than the other drugs, but its overdose could cause toxicity, which may lead to death (Liu et al., 2020a).
6.3. Hydroxychloroquine
Recent literature suggests that a hydroxyl derivative of chloroquine, Hydroxychloroquine, can be used as a promising and safer alternative of chloroquine to treat SARS-CoV-2 infection. Besides, it is found less toxic than chloroquine in animal studies (Liu et al., 2020a). To assess the efficiency of Hydroxychloroquine against SARS-CoV-2 infection, Liu et al. studied its antiviral activity as compared to chloroquine in an in vitro studies. The study showed that Hydroxychloroquine effectively reduces SARS-CoV-2 infection in in vitro model. Also, a higher cytokinin level were observed in the plasma of COVID-19 infected patients. This, in turn, increases the severity of the disease. As per this study, Hydroxychloroquine can reduce the cytokines in serum. Thus, the author proposed that Hydroxychloroquine could be a potential candidate to combat the infection. However, there is no sufficient evidence proving the efficacy of Hydroxychloroquine in SARS-CoV-2 in clinical models relative to chloroquine (Liu et al., 2020a). Several countries suggested the inclusion of Hydroxychloroquine in the clinical guidelines to treat COVID 19 hospitalized patients and also to the high-risk exposure population like health care professionals. Based on encouraging preclinical data and initial clinical data on a small population, many companies are registered more than 300 clinical trials across the globe to investigate the efficacy and safety of this drug in the treatment of COVID 19.
An open-label nonrandomized clinical trial was conducted to evaluate the role of Hydroxychloroquine on respiratory viral loads of COVID 19 patients. 26 COVID 19 patients were treated with Hydroxychloroquine at 600 mg/day for 10 days. As per their findings, it is efficient in clearing the viral load in most of the patients by day 5. However, these findings are difficult to interpret, as they have done on a very small population, the lack of a randomized control group, and the omission of 6 patients from the analysis (Gautret et al., 2020b).
A small randomized trial from Shanghai, China, have been reported on Hydroxychloroquine. They enrolled 30 patients to conduct this pilot study, and the patients were randomized 1:1 to treatment and control group. In the treatment group, given 400 mg of the drug daily for 5 days and in control group patients received conventional treatment only. This report showed that 86% and 93% of the patients had COVID 19 negative results by day 7 in Hydroxychloroquine treated group and control group, respectively (Chen et al., 2020a).
These two results are promising with benefits of Hydroxychloroquine but should be supplemented by more extensive studies as Some observational studies reported use of Hydroxychloroquine not associated with significant benefits in COVID 19 hospitalized patients (Geleris et al., 2020).
An observational study of Hydroxychloroquine in hospitalized patients with COVID 19 was conducted on 1446 adult patients who were admitted with COVID 19 in New York-Presbyterian Hospital (NYP)-Columbia University Irving Medical Center (CUIMC) in northern Manhattan. As per the study protocol, the suggested hydroxychloroquine dose was 600 mg twice on day one, and the next four days, they have given at the dose of 400 mg per day in combination with Azithromycin. The primary endpoint of this study was baseline to intubation or death. In this report, as per their findings, the primary endpoint event is slightly higher in the Hydroxychloroquine treatment group when compared with patients who did not take this drug. However, they also claimed that there was no significant association between the use of Hydroxychloroquine and the composite primary endpoint but the results do not support the use of Hydroxychloroquine at present, outside randomized clinical trials testing its efficacy (Gautret et al., 2020a).
Drugs for the treatment of infectious disease must have an excellent safety profile. However, few studies with Hydroxychloroquine also described mixed results. Importantly, treatment regimens which are also included Azithromycin in combination may have severe side-effects, including cardiac toxicity like QT interval prolongation, torsades de pointes, and drug-induced sudden cardiac death (Alexander et al., 2011; Huang et al., 2007; Kezerashvili et al., 2007).
Ehud Chorin et al. studied QT interval in COVID 19 patients who are treated with Hydroxychloroquine and Azithromycin. They reviewed the data of 84 patients, they treated with Hydroxychloroquine on the first day at a dose of 400 mg twice a day, followed by 200 mg twice daily. Azithromycin was given at a dose of 500 mg per day. They observed prolongation of QT interval from day 4 of therapy. Very importantly, severely prolonged QT interval was observed in a subset of nine patients with more than 500 mg against the baseline average of 435 ± 24 mg (Mean ± SD). QT interval prolongation is a known marker for the high risk of sudden cardiac death (Chorin et al., 2020).
6.4. Favipiravir
In the latest news, on 18th March 2020, reported that Favipiravir marketed as Avigan found effective against COVID-19. Xinmin Zhang from the Ministry of Science and Technology, China, said that Favipiravir gives positive results in a clinical trial on 340 patients in Shenzhen and Wuhan city. The drug was developed by Toyama Chemicals and prescribed for influenza infection. In February 2020, it got approval for the experimental treatment of novel coronavirus infection. In the Wuhan trial, the treatment with favipiravir improved the symptoms of infection and reduced the duration of fever. According to the Guardian Report, some patients with early COVID-19 symptoms showed recovery; however, the patients with severe or advanced symptoms do not show a similar response ().
6.5. Bevacizumab
Bevacizumab, a recombinant humanized monoclonal antibody against VEGF, was first approved by USFDA on 26th February 2004 for the first-line treatment for metastatic colorectal cancer (Cowling and Leung, 2020). Subsequently, the FDA approved this product along with chemotherapy to treat many cancers like lung cancer, renal cancer, cervical cancer, ovarian cancer, etc. (Zhang and Zhou, 2018). In addition, recent studies suggest that higher levels of blood VEGF in COVID-19 patients compared with normal and also pulmonary edema, dyspnea, acute respiratory distress and acute lung injury are the most detrimental symptoms of COVID-19. Numerous studies reported that VEGF was a critical factor in pulmonary edema, acute respiratory distress and acute lung injury (Schoeman and Fielding, 2019). Based on this evidences, Qilu Hospital of Shandong University has also initiated clinical trials on this product for treating the COVID-19 (Clinical trials.gov).
6.6. Protease inhibitors
Protease inhibitors are approved by USFDA to treat HIV, and also, these drugs previously tried for the treatment of SARS-CoV patients. However, their clinical efficacy was inconclusive. Protease enzymes are mainly responsible for the synthesis of structural and functional proteins from polyproteins such as pp1ab and pp1a, which further helps for the replication of viruses. In in vitro studies against coronaviruses, the protease inhibitors like lopinavir/ritonavir have shown inhibitory activity on 3-chymotrypsin like protease enzyme, which plays a vital role in the replication and synthesis of critical structural proteins (Chu et al., 2004; de Wilde et al., 2014).
Currently, many countries are included lopinavir/ritonavir in their clinical guidelines to treat COVID 19, and 48 clinical trials are registered around the world till 14th June 2020 (Clinicaltrial.gov.in). However, clinical trial data on the safety and efficacy of these drugs for treating COVID 19 infection is very limited as only one result of clinical trial data (Cao et al. and Ivan fan-Ngai Hung et al.) is published to date and results of remaining yet to be released.
Cao and his team conducted an open-label, randomized and controlled clinical trial to find the effectiveness of a combination of Lopinavir and Ritonavir for the treatment of COVID-19 from 18th January 2020 to 3rd February 2020 in a total of 199 patients who have confirmed with COVID-19 and having a relatively same viral load. In this trial, they found that the proposed combination, along with standard care, did not show significant clinical improvement and also not reduced the mortality when compared with alone stand care treatment group. Based on these results, they suggested that future studies are required with more combination drugs to assess the effectiveness of Lopinavir and Ritonavir for COVID-19 infection (Belouzard et al., 2009).
Presently, the physicians are trying various relevant drugs and combined dosage regimens based on the preliminary symptoms to treat the suspected or infected patients throughout the world. No drug is approved by FDA or WHO for COVID-19 but are under investigations and hopefully will soon give a satisfactory answer.
Current clinical studies on SARS-CoV-2 patients reported remdesivir and Hydroxychloroquine to be an effective drug candidate towards the treatment of SARS-CoV-2 infection. However, robust data from clinical trials are needed to support the efficacy and safety profile of these drugs since very few such studies have been reported. Further, these studies are also required to answer several questions like a dose of the drug, duration of the treatment, defined parameters during treatment management/monitoring, to identify the high-risk population and severe adverse effects.
7. Progress on COVID-19 vaccines development
Besides, clinical trials have also initiated to estimate some experimental COVID-19 vaccines on humans. The trials focusing on testing the efficacy of a vaccine to boost the immunity against COVID-19 and its safety profile. Dr. Fauci, Director, National Institute of Allergy and Infectious Diseases said that, if all goes well the vaccines will be launched for general use in the next 12–18 months. Vaccine development for any infectious disease is a systematic, tedious and long-run process. The first vaccine for the Ebola virus was approved by FDA in the year 2019, after 43 years of the viral outbreak. After all the gigantic investigations and investments, the scientists only get a few strides towards developing vaccines for RSV (Respiratory Syncytial Virus) or HIV (Human Immunodeficiency Virus) (Pronker et al., 2013). Usually, successful development of a vaccine takes around ten years. In this pandemic crisis of COVID-19, the world is sorely needed a potential vaccine to combat this threatful situation. As per the available data, around ten vaccines are presently under clinical trial against SARS-CoV-2 from which the scientists from Oxford University and AstraZeneca expected to have the data of phase III trial by this summer (as shown in Table 1 ). However, many experts around the globe suspect the credibility of this work as it takes a minimum of 18 months to develop the first vaccine even after tremendous efforts. While some presume that a huge dose of vaccines (around hundreds of million) will be ready to launch by the end of this year (Iserson, 2020).
Table 1.
Current clinical development of COVID-19 vaccines (Adopted form (Pronker et al., 2013)).
| S. No. | Therapeutic agents | Properties | Organization/Company | Status |
|---|---|---|---|---|
| mRNA-1273 | mRNA vaccine | Moderna and NIAID | Phase 2 | |
| BNT162 | mRNA vaccine | BioNTech and Pfizer | Phase 1/2 | |
| INO-4800 | DNA vaccine | Inovio Pharmaceuticals | Phase 1 | |
| AZD1222 | Adenovirus vaccine | University of Oxford and AstraZeneca | Phase 2b/3 | |
| Ad5-nCoV | Adenovirus vaccine | CanSino Biologics | Phase 2 | |
| Unnamed | Inactivated virus | Wuhan Institute of Biological Products and Sinopharm | Phase 1/2 | |
| Unnamed | Inactivated virus | Beijing Institute of Biological Products and Sinopharm | Phase 1/2 | |
| PiCoVacc | Inactivated virus with adjuvant | Sinovac | Phase 1/2 | |
| Unnamed | Inactivated virus | Institute of Medical Biology and Chinese Academy of Medical Sciences |
Phase 1 | |
| NVX-CoV2373 | Protein subunit | Novavax | Phase 1/2 |
Various emerging technologies have been implemented to design the advanced vaccine by the pharmaceutical and biotech industries and organizations. One such example is the mRNA-1273 vaccine candidate by Moderna which undergoes clinical trial just after 66 days of SARS-CoV-2 genetic sequencing, demonstrates the potency of the nucleotide-based vaccine. Unlike the traditional vaccination approach of using a weakened strain of the virus, Moderna adopted a lipid nanocarrier to deliver the mRNA template to the host cell. Although, this vaccine delivers a viral genetic sequence into the host cell and activates the immune system by expressing the respective antigen. Just like other approaches, Moderna also attempted to train the immune system to distinguish the spike protein of SARS-CoV-2 which is responsible for the entry of SARS-CoV-2 virus into the host cell (Thanh Le et al., 2020).
On the other hand, the University of Oxford and AstraZeneca embodies AZD1222 (a recombinant vaccine) to obtain a similar result. They have engineered a chimpanzee adenovirus for carrying a spike DNA antigen. This approach supposes to produce a firm memory B cells and T cells due to the combined immunogenic effect of the spike DNA antigen and adenovirus which might produce a better prophylactic effect in a lesser dose (Lv et al., 2020; Pronker et al., 2013). Similarly, various other advanced strategies have been adopted to develop a vaccine instead of the traditional one which gives a positive hope to find a promising solution in a minimum possible time. However, the researchers and developers are facing many challenges in distinct fronts, from the regulatory to the organizational aspects. Time and worsening of disease are other crucial factors that seem difficult to be met. Even after the clinical success of a vaccine, it brings a different line of trouble of producing a huge amount of dose in a large scale and its distribution amongst the worldwide population (Lv et al., 2020). So, in present scenario authorities, themselves do not have a precise answer and the world has to just wait and watch with precautions to protect ourselves and loved ones.
8. Preventive measures against transmission of COVID-19
Transmission between humans to humans and through various surfaces like metal, cardboard, plastic is very high in the COVID-19 disease (Bedford et al., 2020). Therefore, to prevent the spread of Coronavirus (SARS-CoV-2), various preventive actions have been advised by WHO and other health ministries across various countries. Such preventive actions include social distancing, use of personal protective equipment (PPE), face masks/shields, and hand sanitizers.
8.1. Social distancing
As per WHO recommendation, people should keep at least a 3-m distance from each other to reduce the chances of human to human transmission of COVID-19. Besides this, mass gatherings should also be avoided or prohibited to reach the disease in the community transmission stage. People who came in direct contact with an infected person should be isolated and quarantined for at least 14 days to stop the transmission. The healthcare workers like doctors, nurses, and other paramedical staff, fighting against coronavirus disease should also remain isolated from their family members (Anderson et al., 2020).
8.2. Personal protective equipment (PPE)
PPEs are being used by health care professionals to protect from the infection of SAR-CoV-2 to avoid secondary transmission in hospitals. PPE should cover the whole body to prevent the chances of infection through the patient. Health care professionals should also wear respirator masks such as N95 (filters >95% of airborne particles), FFP2 (filters > 94% of airborne particles), and FFP3 (filters > 99% of airborne particles) to avoid infection. Adequate training of PPEs handling should be given to health care workers to avoid careless handling. Also, proper care should be taken for the disposal of used PPEs (Kraemer et al., 2020).
8.3. Masks
Many health advisory bodies have compelled the use of masks across the world as a preventive measure to avoid the spread of COVID-19. Mask control the virus spread through the droplets of an infected person by acting as a shield/barrier to prevent the exposure of the critical body parts responsible for the spreading of the COVID-19. However, in the absence of a mask, it is advisable to cover the face with bending elbow over the face or to cover with a handkerchief/tissue paper while sneezing or coughing (Adhikari et al., 2020; WHO, 2020b).
8.4. Hand sanitizers
Cleaning of hands from time to time with alcohol-based sanitizers is advisable to protect individuals against coronavirus disease. Washing hands with soap and water before and after meals must be practiced as a preventive measure. As suggested, the content of alcohol should be more than 70% in hand sanitizes, and therefore children should be kept away from alcohol-based sanitizers (WHO, 2020b).
8.5. Management of stress during COVID-19
Throughout the world, people are facing different kinds of challenges in their lifestyles due to increased mental stress in the current scenario of the COVID-19 pandemic. This increased stress can be well acquainted with a small change in daily lifestyle like practicing of Indian/western classical yoga, a balanced diet containing low calories and more proteins, lightweight indoor exercises, working on a new hobby or skill development, etc. (Melbourne, 2020).
9. Conclusion
In history, mankind has already experienced two major coronavirus outbreaks in 21st century itself including SARS-CoV (originated and affected Guangdong province China in 2002) and MERS-CoV (originated and affected middle-east countries in 2012). Both of these are highly contagious, affected millions of people by human to human transmission and declared as an epidemic at that time. From the past three months, we are again going through a highly crucial time which even put a question mark for the existence of mankind due to a highly contagious strain of another coronavirus known as SARS-CoV-2. The latest version of coronavirus species has some distinct features due to genetic mutation in the previous species including mutated RBD protein with high affinity to host cell receptors and polybasic furin cleavage and O-linked glycan which may result in immune evasion. Such features increase the pathogenicity and complexities of the virus. Upon three months of COVID-19 outbreak, we do not have a promising diagnostic tool and therapeutic approach to treat the infection. However, various approaches are presently under investigation and various clinical trials were registered in the Chinese clinical trial registry and U.S. Govt. clinical trial registry. Some of the already approved molecules by FDA with previous success in different antiviral and inflammatory diseases may give a positive response in the treatment of COVID-19. Although, there is a great need to produce sufficient evidence by successful clinical studies to assure the safety and efficacy of these drug substances. The development of a vaccine for the prevention and cure of COVID-19 and a promising diagnostic tool is also essential. Hopefully, the constant scientific efforts and thorough understanding of SARS-CoV-2 pathogenesis and life cycle will soon find out the solution for this.
Author statement
Pavan Kumar Samudrala: Writing-Review editing. Pramod Kumar: Idea creation, Writing-Review editing. Kamlesh Choudhary: Writing-Original Draft. Nagender Thakur: Writing-Original Draft. Gaurav Suresh Wadekar: Writing-Original Draft. Richa Dayaramani: Formal analysis. Mukta Agrawal: Writing-Original Draft. Amit Alexander: Conceptualization, Graphical Designing, Visualization, Supervision.
Declaration of competing interest
None.
Acknowledgment
The author wants to show his gratitude to National Institute of Pharmaceutical Education and Research (NIPER GUWAHATI), Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Govt. of India, Changsari, Kamrup-781101, Guwahati, Assam, India for providing necessary facilities for the compilation of this review work and support.
References
- Adhikari S.P., Meng S., Wu Y.J., Mao Y.P., Ye R.X., Wang Q.Z., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander A., Ajaz A., Swarna, Sharma M., Tripathi D.K. Polymers and permeation enhancers: specialized components of mucoadhesives. Stamford J. Pharm. Sci. 2011;4:91–95. [Google Scholar]
- Alexander D.J., Brown I.H. Revue scientifique et technique (International Office of Epizootics) Vol. 28. 2009. History of highly pathogenic avian influenza; pp. 19–38. [DOI] [PubMed] [Google Scholar]
- Alsaadi E.A.J., Jones I.M. vol. 14. 2019. pp. 275–286. (Membrane Binding Proteins of Coronaviruses). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti, S., Benvenuto, D., Bianchi, M., Giovanetti, M., Pascarella, S., Ciccozzi, M., COVID-2019: The Role of the nsp2 and nsp3 in its Pathogenesis. (n/a). [DOI] [PMC free article] [PubMed]
- Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M.-d., Sall A.A., Schuchat A., Ungchusak K., Wieler L.H. COVID-19: towards controlling of a pandemic. Lancet. 2020;395:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M., Gamieldien J., Fielding B.C. Identification of new respiratory viruses in the new millennium. Viruses. 2015;7:996–1019. doi: 10.3390/v7030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Aldana D.K., Holguin-Rivera Y., Cortes-Bonilla I., Cardona-Trujillo M.C., Garcia-Barco A., Bedoya-Arias H.A., Rabaan A.A., Sah R., Rodriguez-Morales A.J. Coronavirus infections reported by ProMED, February 2000-January 2020. Trav. Med. Infect. Dis. 2020:101575. doi: 10.1016/j.tmaid.2020.101575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, 2020.
- Chen J., Liu D., Liu L., Liu P., Xu Q., Xia L., Ling Y., Huang D., Song S., Zhang D., Qian Z., Li T., Shen Y., Lu H. [A pilot study of hydroxychloroquine in treatment of patients with moderate COVID-19] J. Zhejiang Univ. Med. Sci. 2020;49:215–219. doi: 10.3785/j.issn.1008-9292.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorin E., Dai M., Shulman E., Wadhwani L., Bar-Cohen R., Barbhaiya C., Aizer A., Holmes D., Bernstein S., Spinelli M., Park D.S., Chinitz L.A., Jankelson L. The QT interval in patients with COVID-19 treated with hydroxychloroquine and azithromycin. Nat. Med. 2020;26:808–809. doi: 10.1038/s41591-020-0888-2. [DOI] [PubMed] [Google Scholar]
- Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., Kao R.Y., Poon L.L., Wong C.L., Guan Y., Peiris J.S., Yuen K.Y. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling B.J., Leung G.M. Epidemiological research priorities for public health control of the ongoing global novel coronavirus (2019-nCoV) outbreak. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.6.2000110. bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.C., Zhang H.W., Yu J., Xu H.J., Chen H., Luo S.P., Zhang H., Liang L.H., Wu X.L., Lei Y., Lin F. CT imaging and differential diagnosis of COVID-19. Can. Assoc. Radiol. J. 2020;71(2):195–200. doi: 10.1177/0846537120913033. 846537120913033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devices V.M. Verdict Media Limited; 2020. Roche Coronavirus Test Receives Emergency Use Authorisation from FDA. [Google Scholar]
- Dhama K., Khan S., Tiwari R., Sircar S., Bhat S., Malik Y., Singh K., Chaicumpa W., Bonilla-Aldana D., Rodriguez-Morales A. 2020. Coronavirus Disease 2019 – COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DA.
- FDA, 2020. Gilead Sciences, Inc.
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;382(25):2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghinai I., McPherson T.D., Hunter J.C., Kirking H.L., Christiansen D., Joshi K., Rubin R., Morales-Estrada S., Black S.R., Pacilli M., Fricchione M.J., Chugh R.K., Walblay K.A., Ahmed N.S., Stoecker W.C., Hasan N.F., Burdsall D.P., Reese H.E., Wallace M., Wang C., Moeller D., Korpics J., Novosad S.A., Benowitz I., Jacobs M.W., Dasari V.S., Patel M.T., Kauerauf J., Charles E.M., Ezike N.O., Chu V., Midgley C.M., Rolfes M.A., Gerber S.I., Lu X., Lindstrom S., Verani J.R., Layden J.E. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395(10320):1137–1144. doi: 10.1016/S0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilead . Gilead Sciences, Inc; 2020. Gilead Announces Results from Phase 3 Trial of Investigational Antiviral Remdesivir in Patients with Severe COVID-19. [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S.C., Du B., Li L.-j., Zeng G., Yuen K.-Y., Chen R.-c., Tang C.-l., Wang T., Chen P.-y., Xiang J., Li S.-y., Wang J.-l., Liang Z.-j., Peng Y.-x., Wei L., Liu Y., Hu Y.-h., Peng P., Wang J.-m., Liu J.-y., Chen Z., Li G., Zheng Z.-j., Qiu S.-q., Luo J., Ye C.-j., Zhu S.-y., Zhong N.-s. 2020. Clinical Characteristics of Coronavirus Disease 2019 in China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Deng Y., Li W. Coronavirus Disease 2019 (COVID-19): what we know? J. Med. Virol. 2020;92(7):719–725. doi: 10.1002/jmv.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D.L., Shindo N. COVID-19: what is next for public health? Lancet. 2020;395:542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.H., Wu C.H., Hsia C.P., Yin Chen C. Azithromycin-induced torsade de pointes. Pacing Clin. Electrophysiol. : PACE. 2007;30:1579–1582. doi: 10.1111/j.1540-8159.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., Hu X., Chen J., Liu B. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295:22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J.G., Lang Y., Bakkers M.J.G., Li W., Li Z., Schouten A., Ophorst B., van Kuppeveld F.J.M., Boons G.-J., Bosch B.-J., Huizinga E.G., de Groot R.J. Human coronaviruses OC43 and HKU1 bind to 9-<em>O</em>-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserson K.V. SARS-CoV-2 (COVID-19) vaccine development and production: an ethical way forward. Camb. Q. Healthc. Ethics. 2020:1–12. doi: 10.1017/S096318012000047X. CQ : the international journal of healthcare ethics committees. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Goto H., Yamamoto E., Tanaka H., Takeuchi M., Kuwayama M., Kawaoka Y., Otsuki K. Generation of a highly pathogenic avian influenza A virus from an avirulent field isolate by passaging in chickens. J. Virol. 2001;75:4439–4443. doi: 10.1128/JVI.75.9.4439-4443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kezerashvili A., Khattak H., Barsky A., Nazari R., Fisher J.D. Azithromycin as a cause of QT-interval prolongation and torsade de pointes in the absence of other known precipitating factors. J. Intervent. Card Electrophysiol. 2007;18:243–246. doi: 10.1007/s10840-007-9124-y. an international journal of arrhythmias and pacing. [DOI] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M.U.G., Yang C.H., Gutierrez B., Wu C.H., Klein B., Pigott D.M., du Plessis L., Faria N.R., Li R., Hanage W.P., Brownstein J.S., Layan M., Vespignani A., Tian H., Dye C., Pybus O.G., Scarpino S.V. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science (New York, N.Y.) 2020;368:493–497. doi: 10.1126/science.abb4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Liu Y.H., Wang C.Y., Wang Y.H., Hsueh S.C., Yen M.Y., Ko W.C., Hsueh P.R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J. Microbiol. Immunol. Infect. 2020;53(3):404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.K., Wang C.C., Lin M.C., Kung C.T., Lan K.C., Lee C.T. Effective strategies to prevent coronavirus disease-2019 (COVID-19) outbreak in hospital. J. Hosp. Infect. 2020;105(1):102–103. doi: 10.1016/j.jhin.2020.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharmaceut. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.) 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of A rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang Y., Wang F., Liu B., Li H., Tang G., Chang Z., Liu A., Fu C., Gao J. medRxiv; 2020. Sex Differences in Clinical Findings Among Patients with Coronavirus Disease 2019 (COVID-19) and Severe Condition. In press. [Google Scholar]
- Lippi G., Simundic A.-M., Plebani M. 7th. Vol. 58. Clinical Chemistry and Laboratory Medicine (CCLM); 2020. Potential Preanalytical and Analytical Vulnerabilities in the Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) pp. 1070–1076. [DOI] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., Yang D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Wu N.C., Mok C.K.P. COVID-19 vaccines: knowing the unknown. Eur. J. Immunol. 2020;50(7):939–943. doi: 10.1002/eji.202048663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Wang M., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Li Y., Jin H., Hu B. 2020. Neurological Manifestations of Hospitalized Patients with COVID-19 in Wuhan, China: a Retrospective Case Series Study. 2020.2002.2022.20026500. [Google Scholar]
- Melbourne U.o. University of Melbourne; 2020. Coronavirus (COVID-19): Managing Stress and Anxiety. [Google Scholar]
- Menachery V.D., Dinnon K.H., Yount B.L., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L., Graham B., Randell S.H., Lipkin W.I., Baric R.S. Trypsin treatment unlocks barrier for zoonotic bat. Coronavirus Infect. 2020;94 doi: 10.1128/JVI.01774-19. e01774-01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W.H. 2020. Clinical Management of Severe Acute Respiratory Infection when Novel Coronavirus (nCoV) Infection is Suspected. [Google Scholar]
- Park Y.-J., Walls A.C., Wang Z., Sauer M.M., Li W., Tortorici M.A., Bosch B.-J., DiMaio F., Veesler D. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. JAMA. 2020 doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Peng Q.Y., Wang X.T., Zhang L.N. Findings of lung ultrasonography of novel corona virus pneumonia during the 2019-2020 epidemic. Intensive Care Med. 2020;46(5):849–850. doi: 10.1007/s00134-020-05996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronker E.S., Weenen T.C., Commandeur H., Claassen E.H., Osterhaus A.D. Risk in vaccine research and development quantified. PloS One. 2013;8 doi: 10.1371/journal.pone.0057755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie A.I., Singanayagam A. Immunosuppression for hyperinflammation in COVID-19: a double-edged sword? Lancet. 2020;395(10230):1111. doi: 10.1016/S0140-6736(20)30691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche's . 2020. Roche's Cobas SARS-CoV-2 Test to Detect Novel Coronavirus Receives FDA Emergency Use Authorization and is Available in Markets Accepting the CE Mark. [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SK Khaja M., Bhaskar S., Gaurav S.W., Pramod k. Regulatory updates and analytical methodologies for nitrosamine impurities detection in sartans, ranitidine, nizatidine and metformin along with sample preparation techniques. Crit. Rev. Anal. Chem. 2020 doi: 10.1080/10408347.2020.1788375. In press. [DOI] [PubMed] [Google Scholar]
- Skariyachan S., Challapilli S.B., Packirisamy S., Kumargowda S.T., Sridhar V.S. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for Middle East respiratory syndrome coronavirus infections. Front. Microbiol. 2019;10:569. doi: 10.3389/fmicb.2019.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon V.R., Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur. J. Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antivir. Res. 2020;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. 2020;94 doi: 10.1128/JVI.00127-20. e00127-00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2020. Coronavirus Disease 2019 (COVID-19): Situation Report – 144, 12 June 2020. [Google Scholar]
- WHO . World Health Organization; 2020. Coronavirus Disease (COVID-19) Advice for the Public, Advice for Public. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.C., Chen C.S., Chan Y.J. Overview of the 2019 novel coronavirus (2019-nCoV): the pathogen of severe specific contagious pneumonia (SSCP) J. Chin. Med. Assoc. : J. Chin. Med. Assoc. 2020;83(3):217–220. [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhou X., Qiu Y., Feng F., Feng J., Jia Y., Zhu H., Hu K., Liu J., Liu Z., Wang S., Gong Y., Zhou C., Zhu T., Cheng Y., Liu Z., Deng H., Tao F., Ren Y., Cheng B., Gao L., Wu X., Yu L., Huang Z., Mao Z., Song Q., Zhu B., Wang J. 2020. Clinical Characteristics of 82 Death Cases with COVID-19. 2020.2002.2026.20028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou Q. Bevacizumab with dose-dense paclitaxel/carboplatin as first-line chemotherapy for advanced ovarian cancer. Eur. J. Pharmacol. 2018;837:64–71. doi: 10.1016/j.ejphar.2018.07.049. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020:200490. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]