Abstract
Alzheimer's disease (AD) is the most common neurodegenerative disorder caused by neuronal loss that results in cognitive and functional impairment. Formation of neurofibrillary tangles composed of abnormal hyperphosphorylation of tau protein is one of the major pathological hallmarks of AD. Importantly, several neurodegenerative disorders, including AD, are associated with abnormal protein phosphorylation events. However, little is known thus far on global protein phosphorylation changes in AD. We report a phosphoproteomics study examining the frontal gyrus of people with AD and age-matched cognitively normal subjects, using tandem mass tag (TMT) multiplexing technology along with immobilized metal affinity chromatography to enrich phosphopeptides. We identified 4631 phosphopeptides corresponding to 1821 proteins with liquid chromatography-mass spectrometry (MS)/MS analysis on an Orbitrap Fusion Lumos Tribrid mass spectrometer. Of these, 504 phosphopeptides corresponding to 350 proteins were significantly altered in the AD brain: 389 phosphopeptides increased whereas 115 phosphopeptides decreased phosphorylation. We observed significant changes in phosphorylation of known as well as novel molecules. Using targeted parallel reaction monitoring experiments, we validated the phosphorylation of microtubule-associated protein tau and myristoylated alanine-rich protein kinase C substrate (MARCKS) in control and AD (Control = 6, AD = 11) brain samples. In conclusion, our study provides new evidence on alteration of RNA processing and splicing, neurogenesis and neuronal development, and metabotropic glutamate receptor 5 (GRM5) calcium signaling pathways in the AD brain, and it thus offers new insights to accelerate diagnostics and therapeutics innovation in AD.
Keywords: Alzheimer's disease, proteomics, neuroscience, phosphorylation, biomarkers, diagnostics, multiplexing, dementia
Introduction
Post-translational modifications (PTMs) are key regulators of the protein function. There are more than 200 types of PTMs that are known to affect cellular function (Deribe et al., 2010). Protein phosphorylation at serine, threonine, and tyrosine residues is a common PTM that controls cellular signaling (Ardito et al., 2017). Phosphorylation of proteins is a reversible process that is regulated by kinases and phosphatases and their coordinated action results in a dynamic signaling cascade (Bononi et al., 2011). Protein phosphorylation controls numerous biological processes, including cell cycle homeostasis, metabolism, cell proliferation, and differentiation (Bononi et al., 2011).
To understand the complex regulatory circuits responsible for cellular response to external stimuli, global characterization of changes in protein phosphorylation is needed (Sathe et al., 2016). Current mass spectrometry (MS)-based approaches permit identification of thousands of phosphorylation sites, making them ideal for exploring signaling pathways mediated by phosphorylation (Zhong et al., 2012).
Alzheimer's disease (AD) is the most common neurodegenerative disorder leading to memory loss and subsequent functional impairment. Abnormal protein phosphorylation is detected in several neurodegenerative diseases, including AD. Aberrant phosphorylation of tau is correlated with disease progression and cognitive decline (Ghoshal et al., 2002). Analysis of postmortem brains from AD patients at autopsy has shown that the levels of phosphorylated tau are increased (Ksiezak-Reding et al., 1992).
In both AD patients and mouse models of tauopathy, increase in the phosphorylation levels of Tau Ser-324 (pSer-324) was observed. Lys-321/Ser-324 is also a known acetylation-phosphorylation switch that regulates tau polymerization and function (Carlomagno et al., 2017). The phosphorylation of microtubule-associated protein tau (MAPT) at serine 181 and threonine 231 in cerebrospinal fluid (CSF) is well known for the differentiation of AD from other forms of dementia (Hampel et al., 2010). Apart from tau, several other proteins are known to be hyperphosphorylated and involved in AD pathogenesis. These includes neurofilaments (Sternberger et al., 1985), microtubule-associated protein 1B (Ulloa et al., 1994), and CRMP2 (Cole et al., 2007).
Numerous MS-based studies attempted to detect the AD-related phosphorylation changes in human brain tissue. Zahid et al. (2012) carried out two-dimensional electrophoresis followed by Pro-Q diamond phosphoproteome staining and MS analysis for substantia nigra and cortex. They identified significant alteration in nine proteins, including GFAP, GAPDH, and AATC in the AD brain (Zahid et al., 2012). Similarly, another study was carried out by Triplett et al. (2016) on the inferior parietal lobule of patients with AD, mild cognitive impairment, and preclinical AD.
Significant alterations of phosphorylation were identified in 19 proteins involved in multiple signaling pathways, including neuronal plasticity, oxidative stress, and energy metabolism. Immobilized metal affinity chromatography (IMAC)-based phosphopeptide enrichment followed by label-free MS analysis from eight individual AD and eight age-matched controls identified hyperphosphorylation of heat-shock protein-beta-1 (HSPB1) and crystallin-alpha-B (CRYAB) in the brain of AD individuals (Dammer et al., 2015).
In this study, we employed isobaric tandem mass tag (TMT) multiplexing followed by IMAC-based phosphopeptide enrichment with high-resolution MS to detect changes in protein phosphorylation in the brain extract of confirmed AD cases with age-matched controls. Our analysis led to identification of 4631 phosphopeptides corresponding to 1821 proteins. Of these, 504 phosphopeptides correspond to 350 proteins that were significantly altered in the AD brains as compared with control brain samples.
Among the differentially phosphorylated proteins, we identified MAPT, which is a well-characterized protein known to be hyperphosphorylated in AD brains, validating the feasibility of our approach. Parallel reaction monitoring (PRM) assays were developed to quantify MAPT and myristoylated alanine-rich protein kinase C substrate (MARCKS) phosphorylation in control (n = 6) and AD (n = 11) brain samples to confirm the alterations that were observed in the discovery experiments.
Materials and Methods
Subjects
All brain tissue used in this analysis was derived from the autopsy collection of the Baltimore Longitudinal Study of Aging (BLSA). Human postmortem tissues were obtained under Johns Hopkins proper Institutional Review Board (IRB) protocols with consent from family under NA_00034084: The BLSA Autopsy Program: A Prospective Study of the Effects of Aging on Cognition and Brain Pathology. All case metadata, including disease status, neuropathological criteria, age, sex, postmortem interval, and APOE genotype, are provided in Table 1. These frontal brain tissues were pulverized in liquid nitrogen to a fine powder. The lysis was carried out in 4% sodium dodecyl sulfate (SDS) by using sonication. After tissue lysis, the proteins were precipitated by using ice-cold acetone.
Table 1.
Sample Details Used for the Analysis
| Patients | CERAD | Samples | Braak | Age | Sex | PMI (h) | ApoE | Detail sample break up |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Control | 2 | 80 | Male | 22 | 3/3 | Discovery/PRM |
| 2 | 0 | Control | 4 | 99 | Male | 24 | 3/3 | Discovery/PRM |
| 3 | 0 | Control | 3 | 82 | Male | 14.5 | 2/3 | Discovery/PRM |
| 4 | 0 | Control | 3 | 95 | Male | 17 | 2/3 | Discovery/PRM |
| 5 | 0 | Control | 2 | 84 | Male | 17 | 2/3 | PRM |
| 6 | 0 | Control | 4 | 86 | Male | 7 | 2/3 | PRM |
| 7 | 1 | AD | 3 | 94 | Male | 16 | 3/4 | PRM |
| 8 | 3 | AD | 4 | 83 | Female | 18 | 3/3 | PRM |
| 9 | 3 | AD | 5 | 81 | Male | 7.0 | 3/3 | PRM |
| 10 | 3 | AD | 6 | 82 | Male | 23.0 | 2/3 | Discovery/PRM |
| 11 | 3 | AD | 6 | 83 | Female | 11.0 | 3/4 | Discovery/PRM |
| 12 | 3 | AD | 6 | 94 | Female | 17.5 | 2/3 | Discovery/PRM |
| 13 | 3 | AD | 4 | 92 | Male | 7.0 | 3/3 | Discovery/PRM |
| 14 | 3 | AD | 5 | 88 | Male | 14 | 3/3 | Discovery/PRM |
| 15 | 3 | AD | 4 | 98 | Male | 20.0 | 3/3 | PRM |
| 16 | 3 | AD | 6 | 86 | Male | 8.0 | 4/4 | PRM |
| 17 | 3 | AD | 6 | 92 | Male | 12.0 | 3/4 | PRM |
AD, Alzheimer's disease; CERAD, Consortium to Establish a Registry for Alzheimer's Disease; PMI, post-mortem interval; PRM, parallel reaction monitoring.
In-solution trypsin digestion of proteins and TMT labelling
For discovery analysis, four brain samples from control and five from AD patients were used for the proteomic analysis. The details of the samples used for the discovery are provided in Table 1. From each brain sample, 500 μg of protein was reduced and alkylated with 5 mM dithiothreitol (DTT) for 20 min at 60°C and 10 mM iodoacetamide (IAA) for 20 min at room temperature, respectively. Protein digestion was carried out by using lys-C (1:100) for 4 h followed by trypsin (1:20) at 37°C for 12–16 h. The peptides were enriched by using Sep-Pak C18 material and labeled by using TMT as per the manufacturer's instructions (Catalog No. 90110; Thermo Fisher Scientific) (Sathe et al., 2019). Peptides derived from control brain samples were labeled with 126, 127N, 128N, and 128C TMT tags and patient samples were labeled with 129N, 129C, 130N, 130C, and 131 TMT tags.
IMAC enrichment
Initially, Fe-IMAC column (4 × 50 mm or 9 × 50 mm ProPac IMAC-10; Thermo Fisher Scientific) was connected to an Agilent 1260 high-pressure liquid chromatography (HPLC) system. After this, charging of the Fe-IMAC column was carried out as per manufacturer's instructions. Briefly, the columns were cleaned with three column volumes of 20 mM formic acid, followed by charging with three column volumes of 25 mM FeCl3, 100 mM acetic acid. To remove the unbound Fe ions, columns were again flushed with 20 mM formic acid. Deionized water was used to clean HPLC lines. The desalted peptides were reconstituted in 1 mL of Fe-IMAC solvent A (30% ACN, 0.07% [v/v] TFA) and loaded onto an Fe-IMAC column (0.1 mL/min over 10 min), and unbound peptides were removed out with Fe-IMAC solvent A (0.3 mL/min over 16 min).
Next, phosphopeptides were eluted by using a linear gradient from 0% to 45% Fe-IMAC solvent B (0.5% [v/v] NH4OH; 0.2 mL/min over 60 min). After gradient 100% solvent B was held for a 5-min period, the column was re-equilibrated with Fe-IMAC solvent A (0.5 mL/min over 30 min). The eluted peptides were collected according to the ultraviolet signal (280 nm) and dried.
Strong cation exchange chromatography fractionation
The TMT-labeled enriched phosphopeptides were subjected to stage tip-based strong cation exchange (SCX) chromatography fractionation to generate 6 fractions as previously described (Kulak et al., 2014). For packing, stage tip 3M™ Empore™ anion and cation exchange-SR extraction disks were used. These SCX chromatography resins were activated with acetonitrile followed by loading of phosphopeptides mixture, resuspended in the SCX chromatography solvent A (0.1% trifluoroacetic acid), and washed with 0.2% trifluoroacetic acid. After washing, the peptides were fractionated by different gradients of ammonium acetate (50 Mm to 300 mM) as described earlier (Kulak et al., 2014). A total of six fractions were collected and vacuum dried.
Liquid chromatography-MS/MS analysis
These phosphopeptides were analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, Bremen, Germany) that was equipped with an Easy-nLC 1200 nanoflow liquid chromatography (LC) system (Thermo Scientific). These IMAC-enriched phosphopeptides from six SCX chromatography fractions were analyzed on a mass spectrometer in triplicate. Vacuum-dried SCX chromatography fractionated phosphopeptides were reconstituted in 0.1% formic acid and loaded onto the enrichment column (100 μm × 2 cm, Nanoviper) at a flow rate of 3 μL/min. The peptides were resolved on an analytical column (75 μm × 50 cm, RSLC C18) at a flow rate of 300 nL/min by using a gradient of 5–30% solvent B (0.1% formic acid in 95% acetonitrile) for 80 min.
The total run time was set to 120 min. The mass spectrometer was operated in a data-dependent acquisition mode. A survey full scan MS (from m/z 350 to 1500) was acquired in the Orbitrap at a resolution of 120,000 (at 200 m/z). The automatic gain control (AGC) target for MS1 was set as 2 × 105, and ion filling time was set as 50 msec. The most abundant ions with charge state ≥2 were isolated in a 3-sec cycle, fragmented by using high-energy collision dissociation (HCD) fragmentation with 38% normalized collision energy, and detected at a mass resolution of 30,000 at 200 m/z. The AGC target for MS/MS was set as 5 × 104 and ion filling time was set at 100 msec, whereas dynamic exclusion was set to 30 sec with a 10-ppm (parts per million) mass window.
Data analysis
The acquired MS/MS database searches were carried out by using the SEQUEST search algorithm through Proteome Discoverer platform (version 2.1; Thermo Scientific) against Human RefSeq protein database (Version 73, containing protein entries with common contaminants).
The workflow included spectrum selector, SEQUEST search node, and percolator. Trypsin was specified as protease, and a maximum of two missed cleavages was allowed. The search parameters involved carbamidomethylation at cysteine; TMT 10-plex (+229.163) modification at N-terminus of peptide and lysine were set as fixed modifications, whereas oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. MS and MS/MS mass tolerances were set to 10 ppm and 0.1 Da, respectively. False discovery rate of 1% was set at the PSM level as well as at 1% at protein level. The MS data generated from this study have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via PRIDE partner repository with the dataset identifier PXD010807.
Sample preparation for targeted PRM analysis
From each brain sample, 500 μg protein lysate was used for targeted analysis. The protein was reconstituted with 100 mM triethylammonium bicarbonate (TEABC). The protein was reduced and alkylated by using 10 mM DTT followed by 20 mM IAA. Proteins were subjected to in-solution digestion by using Lys-C (1:00) for 4 h followed by trypsin (1:20) for 12–14 h. The peptides were cleaned by using Sep-Pak column C18 material, and samples were dried in a speed vac. These phosphopeptides were enriched by using IMAC enrichment.
PRM analysis of brain samples
For PRM analysis we have used 17 brain samples, including the samples used for the discovery analysis. The total nine samples used in discovery along with additional eight samples were included for the PRM analysis. The details of the samples are displayed in Table 1. The enriched phosphopeptides from each sample were reconstituted in 0.1% formic acid and analyzed on a Q Exactive HF mass spectrometer (Thermo Scientific, Bremen, Germany) as single mass spectrometer analysis. The mass spectrometer is connected with an RSLC nanoflow LC system (Thermo Scientific, San Jose, CA, USA) (Sathe et al., 2018). Peptide digests were loaded onto an enrichment column (100 μm × 2 cm, Nanoviper) at a flow rate of 3 μL/min.
For PRM analysis, enriched phosphopeptides from each sample were resolved on an analytical column (75 μm × 50 cm, RSLC C18). Linear gradient ranging from 4% to 35% buffer B over 90 min were used. Targeted phosphopeptides were monitored by using a targeted inclusion list. The parameters for PRM scan were set as: AGC target value of 5 × 105, Orbitrap resolution of 35,000, injection time of 100 msec, isolation window of 2 m/z, HCD normalized collision energy of 32, and starting mass of m/z 110. Skyline software package was used for the processing of PRM data. Peak integration was performed within the platform and reviewed manually.
Subsequently, results were exported for additional data and statistical analysis. Statistical analysis of phosphoproteomics data was performed with the Perseus software package (Tyanova et al., 2016). Statistical analysis of PRM data was carried out by using Graph Pad Prism version 5.04 (GraphPad Software, Inc., San Diego, CA, USA). Student's t-test was used to examine group comparisons in the validation studies described next. Proteins with p-value ≤0.05 and fold change cut-off of ≥1.5-fold were considered significantly different in AD.
Bioinformatics analysis
Molecular function and localization of phosphoproteins was obtained from the Human Protein Reference Database (HPRD) (Prasad et al., 2009). Biological process annotation of these phosphoproteins was also obtained from HPRD. Further protein–protein interaction network analysis was carried out by using String. Proteins with altered phosphorylation in the AD brain were used as input for the string analysis (Abdelmohsen and Gorospe, 2012). We also included the second shell for analysis, which adds additional proteins that are known interactors along with the input proteins.
Results and Discussion
Phosphoproteomic analysis of the brain
To detect AD-related changes in protein phosphorylation, phosphoproteome analysis of frontal gyrus of five AD patients and four cognitively normal postmortem human brain tissues was performed. For this analysis, we used a TMT-based quantitative proteomics approach along with IMAC-based phosphopeptide enrichment. Briefly, 500 μg of peptides was TMT labeled, subsequently pooled together, and subjected to IMAC enrichment. Enriched fractions were subjected to zip tip-based SCX chromatography fractionation. Each of the six fractions was analyzed on Orbitrap Fusion Lumos in replicates.
The workflow for the experiment is shown in Figure 1. LC-MS/MS analysis of these samples resulted in 4631 identified phosphopeptides that were derived from 1821 proteins (Supplementary Table S1). Out of the 504 differentially altered phosphopeptides identified (Fig. 2A, B), 389 peptides were hyperphosphorylated and 115 peptides were hypophosphorylated in AD (Supplementary Table S2). A partial list of the hyperphosphorylated peptides is provided in Table 2 and that of hypophosphorylated proteins is provided in Table 3.
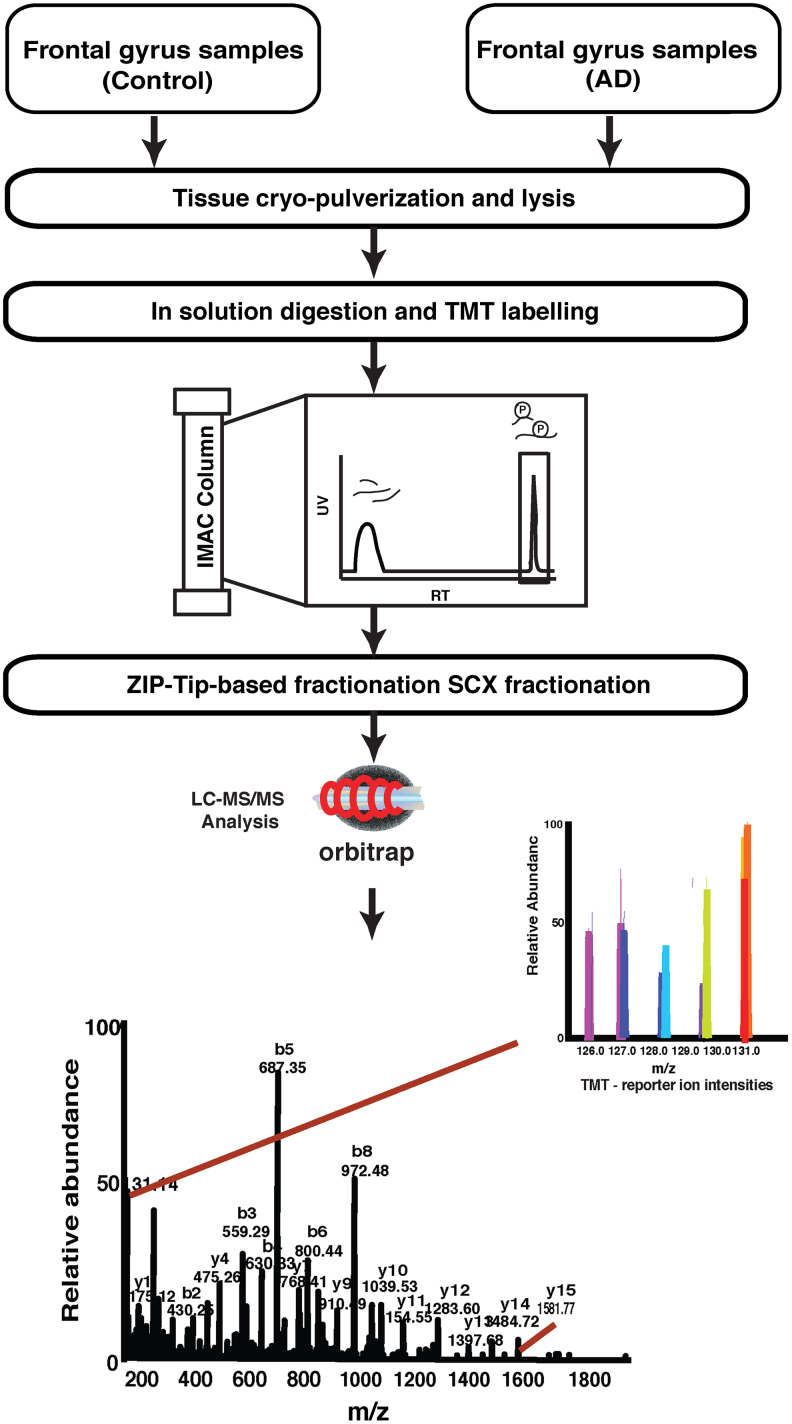
FIG. 1.
A schematic of the workflow used to study the phosphoproteomic changes in frontal gyrus of AD patients. The proteins harvested from AD and controls samples were subjected to in-solution digestion followed by TMT labeling. After TMT labeling, the samples were subjected to IMAC enrichment and the enriched phosphopeptides were fractionated by using stage tip-based SCX chromatography fractionation AD, Alzheimer's disease; IMAC, immobilized metal affinity chromatography; SCX, strong cation exchange; TMT, tandem mass tag.
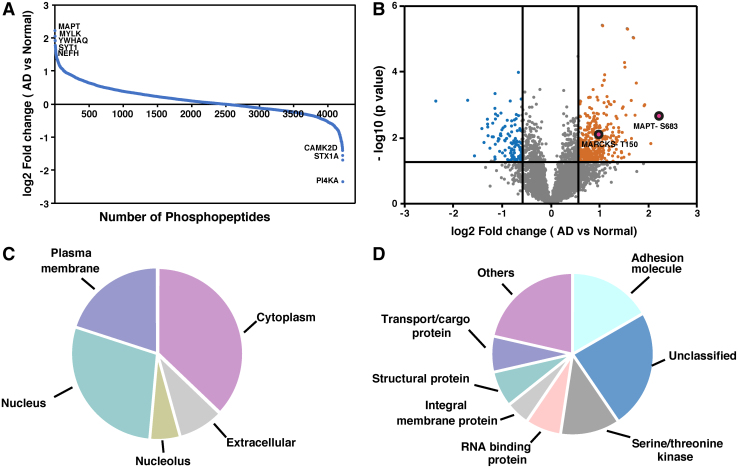
FIG. 2.
Summary of TMT-based discovery experiments. (A) Distribution of fold-change values (log2 ratios; AD/Controls) for phosphopeptides in the study. (B) Volcano plot of identified phosphopeptides. (C) Localization of proteins that are differentially phosphorylated in the brain of AD patients. (D) Molecular function of proteins that are differentially phosphorylated in the brain of AD patients.
Table 2.
Select Hyperphosphorylated Phosphosites in Alzheimer's Disease Identified in This Study
| Gene symbol | Protein | Phosphosite | Fold-change (AD/controls) |
|---|---|---|---|
| MAPT | Microtubule-associated protein tau | S683 | 4.5 |
| MYLK | Myosin light chain kinase | T1788, S1789 | 3.7 |
| SYT1 | Synaptotagmin 1 | T127 | 3.2 |
| NEFH | Neurofilament heavy | S763 | 3.2 |
| SPP1 | Secreted phosphoprotein 1 | S267 | 3.0 |
Table 3.
Select Hypophosphorylated Phosphosites in Alzheimer's Disease Identified in This Study
| Gene symbol | Protein | Phosphosite | Fold-change |
|---|---|---|---|
| CAMK2D | Calcium-/calmodulin-dependent protein kinase II delta | T287 | 0.4 |
| HSPA4L | Heat shock protein family A (Hsp70) | T576 | 0.4 |
| USP5 | Ubiquitin-specific peptidase 5 | S751 | 0.4 |
| NAA38 | N(alpha)-acetyltransferase 38, NatC auxiliary subunit | S28 | 0.3 |
| PI4KA | Phosphatidylinositol 4-kinase alpha | S1828 | 0.2 |
Bioinformatics analysis of proteins altered in AD brain
We observed alteration in the phosphorylation of several signaling molecules in the AD brain. Further, we carried out a bioinformatics analysis of the differentially phosphorylated proteins to categorize them based on their cellular localization and biological function. The categorization is based on HPRD annotations, a manually curated Gene Ontology compliant database (Prasad et al., 2009). We observed that most of the altered phosphorylated proteins were localized in the cytoplasm (37%), followed by nucleus (29%), plasma membrane (20%), and extracellular (8%) sites (Fig. 2C). The phosphorylated proteins that were differentially altered in AD samples were involved in a broad range of molecular functions. Most of the proteins were involved in adhesion, binding activity, and enzyme regulatory activity (Fig. 2D).
A major category (24%) of proteins altered in the AD brain was less well characterized in terms of their function. These include SAMD4A, TCEAL6, AFTPH, and NOP56. The other major categories of proteins identified were adhesion molecules (17%), serine/threonine kinases (12%), transport/cargo proteins (7%), and structural proteins. We also classified AD-affected phosphorylated proteins based on biological processes and found that most of the altered proteins were involved in cell growth and/or maintenance (24%), followed by cell communication (17%) and metabolism (14%).
Known AD biomarkers identified to be altered in their phosphorylation levels
Since we were interested in identifying altered phosphorylation events in the AD brain, we first studied the list of known biomarkers to determine whether their phosphorylation was also in agreement with other published studies. The MAPT plays a vital role in microtubule assembly and stabilizing microtubules. In AD and other tauopathies, abnormally hyperphosphorylated tau aggregates and accumulates in the neuropil and forms neurofibrillary tangles (Ma et al., 2017). This has been widely studied and, as expected, we identified hyperphosphorylation of MAPT in the brain of AD-affected individuals. The representative MS/MS spectra are shown in Figure 3A.
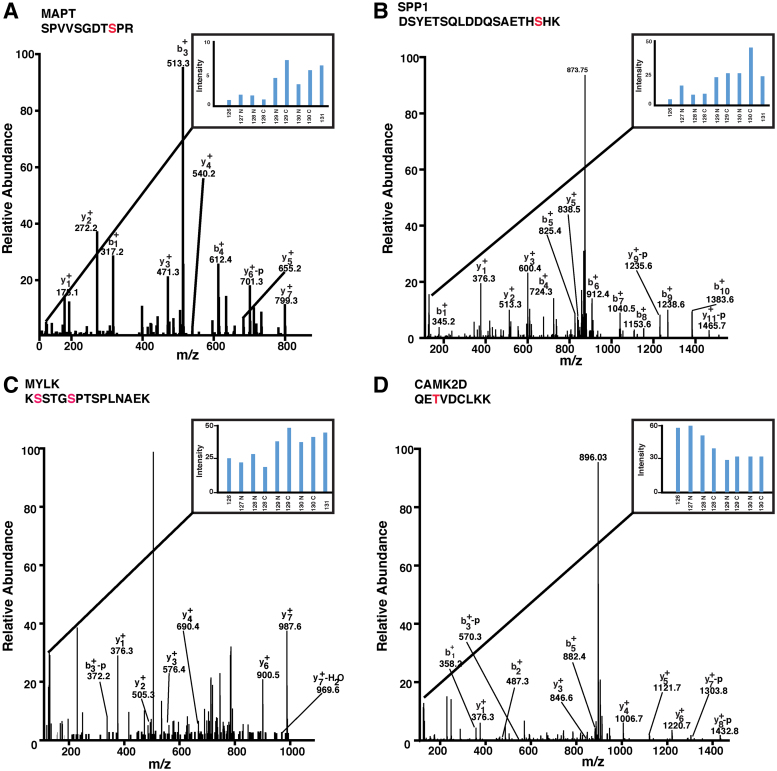
FIG. 3.
Representative MS/MS spectra of altered phosphopeptides identified in the AD brain. (A) MAPT; (B) SPP1; (C) MYLK; (D) CAMK2D. CAMK2D, calcium-/calmodulin-dependent protein kinase II delta; MS, mass spectrometry; MYLK, myosin light chain kinase; SPP1, secreted phosphoprotein 1.
Microtubule-associated protein 1A (MAP1A) is another protein that belongs to the microtubule-associated protein family. We have identified hyperphosphorylation of MAP1A on several sites in the AD brain. Members of this family are involved in microtubule assembly, which is an essential step in neurogenesis. Mutations in the MAP1A gene cause degeneration in Purkinje cells (Liu et al., 2015). We also observed significant hyperphosphorylation of osteopontin in AD-affected patients. The representative MS/MS spectra are shown in Figure 3B. Osteopontin is a secreted glycoprotein expressed by cytotoxic T cells and is a known marker for neuroinflammation. It activates and recruits macrophages at the site of inflammation. Its expression in pyramidal neurons is known to be increased in AD patients (Wung et al., 2007). It is also upregulated in AD transgenic mouse models (Wirths et al., 2010).
Increased abundance of osteopontin was also reported in CSF of familial AD individuals (Begcevic et al., 2018). We also detected hyperphosphorylation of neurofilament heavy (NEFH) chain in AD patient samples. The NEFH chain plays a vital role in intracellular transport to axons and dendrites. It is also used as a biomarker of neuronal damage and amyotrophic lateral sclerosis (Gendron et al., 2017). Changes in CSF levels of neurofilament proteins are also useful in discriminating patients with neuroinflammation and normal controls or AD patients (Fialova et al., 2017). NCAM2 is a type I membrane protein that belongs to the immunoglobulin superfamily. It is mainly expressed in the brain and it is involved in neurite outgrowth to enable dendritic and axonal compartmentalization (Winther et al., 2012).
In a recent study, it was observed that the levels of NCAM2 increased in the hippocampus of AD patients but decreased in the synaptosomes isolated from this brain region (Leshchyns'ka et al., 2015). We have detected hyperphosphorylation of NCAM2 in AD-affected patients.
Alteration in phosphorylation of protein kinases in the brain of AD-affected individuals
Protein kinases perform a critical role in the regulation of cellular signaling networks. Phosphorylation results in a functional change in the target protein by altering enzyme activity, cellular location, or association with other proteins (Manning et al., 2002). We identified significant alterations in the phosphorylation of 12 kinases in brains of AD-affected patients compared with normal patients. MYLK (myosin light chain kinase) is a calcium-/calmodulin-dependent kinase. It phosphorylates myosin regulatory light chains to enable myosin interaction with actin filaments and produce contractile activity (Sanders et al., 1999). In our analysis of brain tissue, we observed 3.7-fold hyperphosphorylation of MYLK in the AD patients as compared with age-matched controls. The representative MS/MS spectra are shown in Figure 3C.
We also observed hyperphosphorylation of CAMK2D (calcium-/calmodulin-dependent protein kinase II delta), a serine/threonine protein kinase. The representative MS/MS spectra are shown in Figure 3D. Calcium signaling is critical for numerous aspects of plasticity at glutamatergic synapses (Blackwell and Jedrzejewska-Szmek, 2013). All these altered kinases, namely MYLK, PI4KA, and CAMK2D, identified in the AD brain are purported to play a role in calcium signaling. Aberrant regulation of calcium signaling has been implicated in numerous neurodegenerative diseases, including AD (Berridge, 2013). Alzheimer animal models and studies in neurons have shown that the expression of amyloid precursor protein and familial AD mutants of presenilin change calcium homeostasis and cause synaptic dysfunction and dendritic spine loss in neurons (Thinakaran and Sisodia, 2006).
Targeted PRM analysis
From our phosphoproteomic analysis of frontal cortex from AD patients and cognitively normal postmortem human brain tissue, we were able to identify several known as well as novel proteins that have significantly altered phosphorylation levels in AD. From this analysis, we identified hyperphosphorylation of MAPT at serine 654 and MARCKS at T-150. For confirmation in a larger set of individuals, we carried out targeted PRM on these phosphopeptides in a total of 17 brain samples (this included samples used in discovery analysis as well as additional brain samples).
The workflow for the PRM analysis is shown in Figure 4A. The MAPT protein plays a critical role in stabilization of microtubules. It is abundant in neurons of the central nervous system and also expressed at very low levels in CNS astrocytes and oligodendrocytes (Shin et al., 1991). Hyperphosphorylation of MAPT leads to the self-assembly of tangles of paired helical and straight filaments. This filament formation is involved in the pathogenesis of AD, frontotemporal dementia, and other tauopathies (Alonso et al., 2001). In AD, all 20 epitopes of p-Tau are hyperphosphorylated (Duka et al., 2013). Phosphorylation of tau at S683 in combination with phosphorylation at other sites causes neurodegeneration (Alonso et al., 2010). In the AD Drosophila model, phosphorylation of Tau S683 is important for Abeta42-induced toxicity (Iijima et al., 2010).
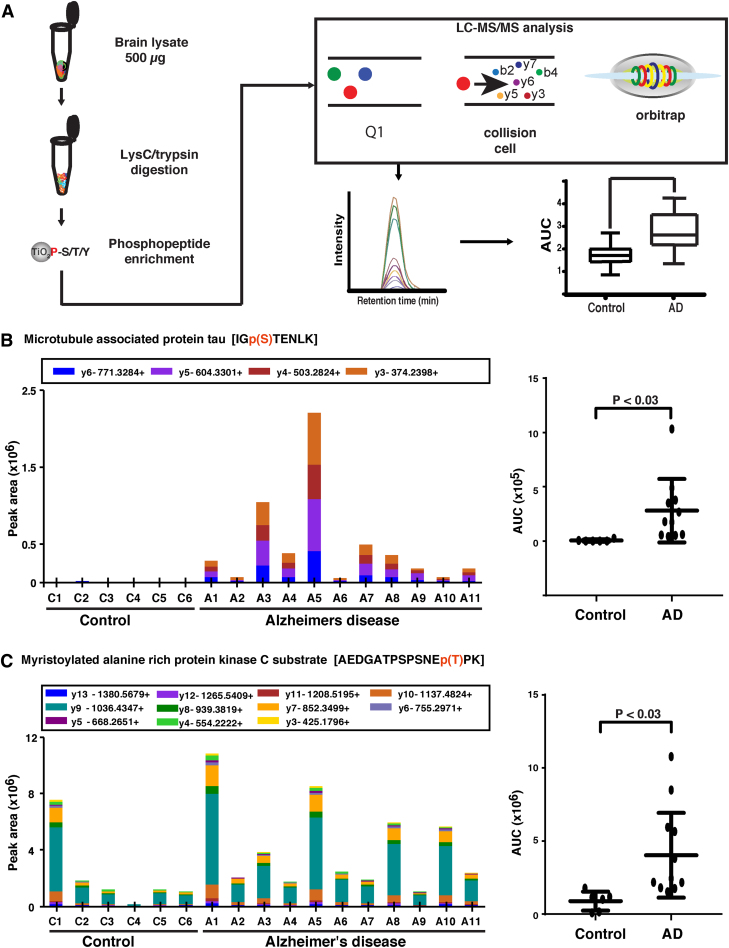
FIG. 4.
Validation of selected candidate phosphopeptides by PRM assays. (A) Workflow of the validation approach that was carried out by using PRM assays for a subset of altered phosphosites. (B) MAPT. (C) MARCKS. The peptide sequences and p-values are shown in each case. MARCKS, myristoylated alanine-rich protein kinase C substrate; PRM, parallel reaction monitoring.
In our discovery analysis, we identified significant alteration of the S683 site of MAPT in the frontal cortex of AD-affected individuals. It is 4.5-fold higher in the brain of AD individuals as compared with normal individuals. We have further validated our results by using PRM on 17 frontal gyrus brain tissue samples. The phosphopeptide (MAPT-Ser683-IGpSTENLK) that we monitored has a significantly higher abundance (p < 0.003) in AD compared with controls, as shown in Figure 4B. MARCKS is an actin filament crosslinking protein localized to the plasma membrane. Its activity is inhibited by protein kinase C (PKC)-mediated phosphorylation and by binding to the calcium–calmodulin complex (Hartwig et al., 1992). MARCKS is also known to interact with Rab10 and is involved in Oligodendrocyte precursor cell maturation (Zhang et al., 2017).
In our analysis, we detected significant alteration of the T150 site of MARCKS in the brain of AD-affected individuals. It is 1.9-fold higher in the brain of AD individuals as compared with normal individuals. A previous study reported a reduction in MARCKS phosphorylation in neurons but enhanced phosphorylation in microglia and dystrophic neurites, suggesting that phosphorylated MARCKS could be an indicator of neuro-inflammation (Kimura et al., 2000). We further validated our results by using PRM on 17 frontal gyrus brain tissue samples. The phosphopeptide (MARCKS-Thr150-AEDGATPSPSNEpTPK) that we monitored is significantly more phosphorylated (p < 0.003) in AD compared with controls, as shown in Figure 4C.
Signaling pathways altered in AD pathogenesis
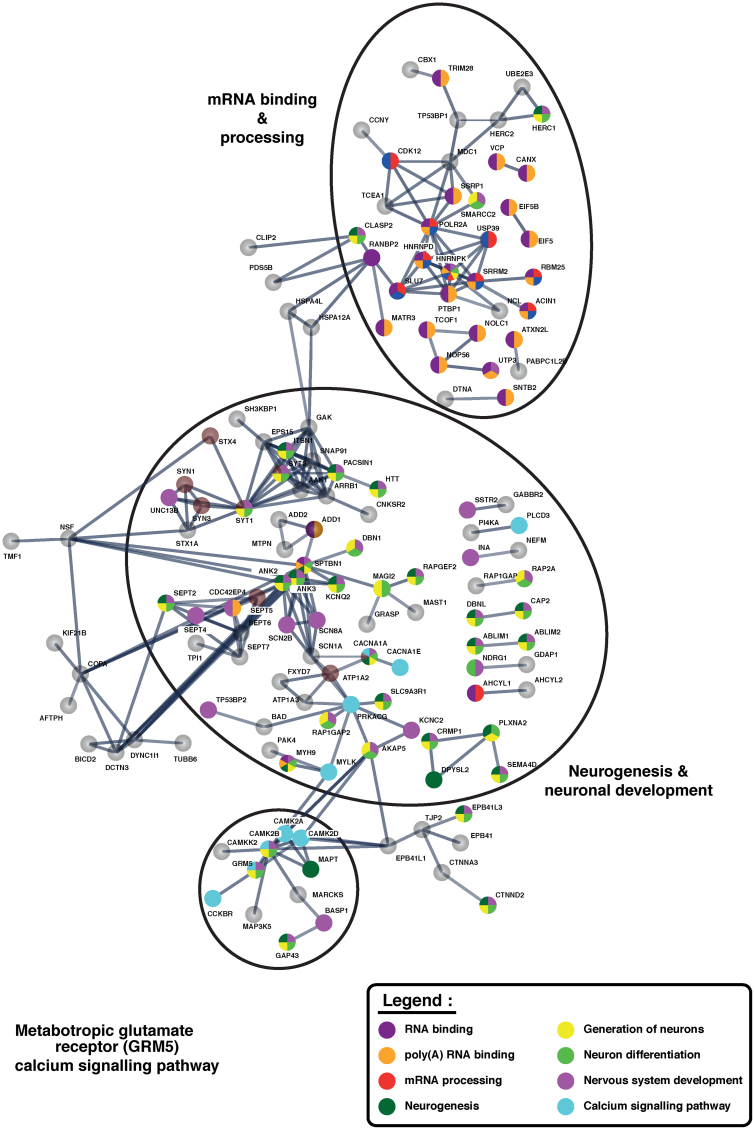
To understand biological networks that play a role in AD pathogenesis, protein–protein interaction network analysis of aberrantly phosphorylated proteins was carried out by using the STRING network analysis tool (Szklarczyk et al., 2015). From the enriched networks, we identified several molecules involved in the RNA processing and splicing, neurogenesis and neuronal development, and metabotropic glutamate receptor 5 (GRM5) calcium signaling pathway (Fig. 5). Several groups have previously reported alterations in RNA splicing in AD (Bai et al., 2013; Wong, 2013). We observed hypophosphorylation of N(alpha)-acetyltransferase 38 at Serine 29. N(alpha)-acetyltransferase 38 is a ribonucleoprotein that binds to the 3′-terminal U-tract of U6 snRNA (small nuclear RNA) and is involved in RNA splicing (Tomasevic and Peculis, 2002; Vindry et al., 2017).
FIG. 5.
Network analysis of significantly altered phosphorylated proteins in AD. Three major enriched pathways are circled. The color of the nodes indicates the biological process that the proteins are involved in.
We also identified NOP56, a nuclear protein hyperphosphorylated in the AD brain. Nop56 is essential for 60S ribosomal subunit assembly and it is also involved in pre-rRNA (ribosomal RNA) processing (Hayano et al., 2003). In addition, we observed hyperphosphorylation of nucleolin at S580 in the AD brain. Nucleolin is a nucleolar phosphoprotein and it plays an important role in the synthesis and maturation of ribosomes (Abdelmohsen and Gorospe, 2012). Another enriched network was neurogenesis and neuronal development. Neurogenesis occurs in the subventricular zone and the subgranular zone of the dentate gyrus of the hippocampus (Rodriguez and Verkhratsky, 2011). Hippocampal structure and function are known to be affected in early AD. As the hippocampus forms the neurogenic niches of the brain, this process is affected in AD (Taupin, 2006).
In a murine model of AD, enhanced neurogenesis is also associated with oligomeric Aβ (Jin et al., 2004). In another study, it has been reported that neurogenesis is disturbed in transgenic mice expressing mutated presenilins (Chevallier et al., 2005). The synaptotagmin protein is integral to the membrane of synaptic vesicles (Fernandez-Chacon et al., 2001). Synaptotagmin 1 (SYT1) expression also increases during neurogenesis (Morgado et al., 2015). In our phosphoproteomics analysis, we observed hyperphosphorylation of SYT1 at T127 in AD-affected brains.
Another important protein, CRMP1, expressed in the nervous system is also involved in neuronal development (Makihara et al., 2016). We observed hyperphosphorylation of CRMP1 in the AD brain. Another important network enriched in our analysis is the GRM5-calcium signaling pathway. Several studies have reported an interaction between mGluR5 and several molecules associated with AD pathogenesis (Renner et al., 2010). mGluR5 and CaMKIIα signaling are involved in learning and memory. We identified alteration in the phosphorylation of CAMK2A, CAMK2B, and CAMK2D in the AD brain. The association of the GRM5-calcium signaling pathway with AD pathogenesis makes components of this pathway potential pharmacological targets in AD (Raka et al., 2015).
Limitations of the study
The study has several limitations, with the chief among them being the small sample sizes for discovery (9) and validation (17). This is due to a lack of well-curated postmortem brain samples as is common in most neurodegenerative brain disorders (Ping et al., 2018; Portelius et al., 2017). Another limitation is that the BLSA is a unique cohort of highly educated participants (the mean education is ∼17 years); thus, findings may not generalize to the population. This study is also limited in sampling of the medial frontal gyrus. Tau pathology and neuronal degeneration begin in the entorhinal cortex and hippocampus; whereas amyloid deposition, which is known to precede tau pathology, starts in the default mode network comprising the precuneus, medial orbitofrontal and posterior cingulate cortices (Palmqvist et al., 2017).
In future studies, we plan to analyze changes in the medial temporal lobes. This study also compared changes between cognitively normal and subjects with AD. This could bias biomarker findings toward finding changes that are downstream of more proximate events. In the future, we plan to look for similar changes in asymptomatic and prodromal AD subjects. Also, the insoluble brain fraction may harbor potentially important pathogenic proteins/peptides. Despite these limitations, we have recapitulated established biomarkers from other studies while also finding novel markers.
Conclusions
We have used brain samples from the well-characterized BLSA cohort combined with TMT multiplexing, IMAC enrichment to identify alterations in protein phosphorylation. This led to the identification 4631 phosphopeptides corresponding to 1821 proteins. Out of these, 504 phosphopeptides are altered in the AD-affected brain. We have validated known and novel sites in additional brain samples by using PRM. The alteration in protein phosphorylation will be useful for the understanding of the signaling associated with AD pathogenesis.
In all, this study provides new evidence on alteration of RNA processing and splicing, neurogenesis and neuronal development, and GRM5 calcium signaling pathways in the AD brain; thus, it offers new insights to accelerate diagnostics and therapeutics innovation in AD.
Supplementary Material
Abbreviations Used
- AD
Alzheimer's disease
- AGC
automatic gain control
- BLSA
Baltimore Longitudinal Study of Aging
- CSF
cerebrospinal fluid
- DTT
dithiothreitol
- GRM5
metabotropic glutamate receptor 5
- HCD
high-energy collision dissociation
- HPLC
high-pressure liquid chromatography
- HPRD
Human Protein Reference Database
- IAA
iodoacetamide
- IMAC
immobilized metal affinity chromatography
- LC
liquid chromatography
- MAP1A
microtubule-associated protein 1A
- MAPT
microtubule-associated protein tau
- MARCKS
myristoylated alanine-rich protein kinase C substrate
- MS
mass spectrometry
- MYLK
myosin light chain kinase
- NEFH
neurofilament heavy
- ppm
part per million
- PRM
parallel reaction monitoring
- PTM
post-translational modification
- SCX
strong cation exchange
- Syt1
synaptotagmin 1
- TMT
tandem mass tag
Author Disclosure Statement
The authors declare there are no conflicting financial interests.
Funding Information
This study was supported, in part, by the National Institute on Aging (NIA) grant U19AG03365 (M.A.), NINDS grant P50NS38377 (A.P.), and Wellcome Trust/DBT India Alliance Margdarshi Fellowship grant IA/M/15/1/502023 (A.P.).
Supplementary Material
References
- Abdelmohsen K, and Gorospe M (2012). RNA-binding protein nucleolin in disease. RNA Biol 9, 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Zaidi T, Novak M, Grundke-Iqbal I, and Iqbal K (2001). Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc Natl Acad Sci U S A 98, 6923–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso AD, Di Clerico J, Li B, et al. (2010). Phosphorylation of tau at Thr212, Thr231, and Ser262 combined causes neurodegeneration. J Biol Chem 285, 30851–30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardito F, Giuliani M, Perrone D, Troiano G, and Lo Muzio L (2017). The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int J Mol Med 40, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Hales CM, Chen PC, et al. (2013). U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc Natl Acad Sci U S A 110, 16562–16567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begcevic I, Brinc D, Brown M, et al. (2018). Brain-related proteins as potential CSF biomarkers of Alzheimer's disease: A targeted mass spectrometry approach. J Proteomics 182, 12–20 [DOI] [PubMed] [Google Scholar]
- Berridge MJ. (2013). Dysregulation of neural calcium signaling in Alzheimer disease, bipolar disorder and schizophrenia. Prion 7, 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell KT, and Jedrzejewska-Szmek J (2013). Molecular mechanisms underlying neuronal synaptic plasticity: Systems biology meets computational neuroscience in the wilds of synaptic plasticity. Wiley Interdiscip Rev Syst Biol Med 5, 717–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi A, Agnoletto C, De Marchi E, et al. (2011). Protein kinases and phosphatases in the control of cell fate. Enzyme Res 2011, 329098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno Y, Chung DC, Yue M, et al. (2017). An acetylation-phosphorylation switch that regulates tau aggregation propensity and function. J Biol Chem 292, 15277–15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier NL, Soriano S, Kang DE, et al. (2005). Perturbed neurogenesis in the adult hippocampus associated with presenilin-1 A246E mutation. Am J Pathol 167, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AR, Noble W, van Aalten L, et al. (2007). Collapsin response mediator protein-2 hyperphosphorylation is an early event in Alzheimer's disease progression. J Neurochem 103, 1132–1144 [DOI] [PubMed] [Google Scholar]
- Dammer EB, Lee AK, Duong DM, et al. (2015). Quantitative phosphoproteomics of Alzheimer's disease reveals cross-talk between kinases and small heat shock proteins. Proteomics 15, 508–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, and Dikic I (2010). Post-translational modifications in signal integration. Nat Struct Mol Biol 17, 666–672 [DOI] [PubMed] [Google Scholar]
- Duka V, Lee JH, Credle J, et al. (2013). Identification of the sites of tau hyperphosphorylation and activation of tau kinases in synucleinopathies and Alzheimer's diseases. PLoS One 8, e75025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fernandez-Chacon R, Konigstorfer A, Gerber SH, et al. (2001). Synaptotagmin I functions as a calcium regulator of release probability. Nature 410, 41–49 [DOI] [PubMed] [Google Scholar]
- Fialova L, Bartos A, and Svarcova J (2017). Neurofilaments and tau proteins in cerebrospinal fluid and serum in dementias and neuroinflammation. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 161, 286–295 [DOI] [PubMed] [Google Scholar]
- Gendron TF, C9ORF72 Neurofilament Study Group, Daughrity LM, et al. (2017). Phosphorylated neurofilament heavy chain: A biomarker of survival for C9ORF72-associated amyotrophic lateral sclerosis. Ann Neurol 82, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal N, Garcia-Sierra F, Wuu J, et al. (2002). Tau conformational changes correspond to impairments of episodic memory in mild cognitive impairment and Alzheimer's disease. Exp Neurol 177, 475–493 [DOI] [PubMed] [Google Scholar]
- Hampel H, Blennow K, Shaw LM, et al. (2010). Total and phosphorylated tau protein as biological markers of Alzheimer's disease. Exp Gerontol 45, 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH, Thelen M, Rosen A, et al. (1992). MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature 356, 618–622 [DOI] [PubMed] [Google Scholar]
- Hayano T, Yanagida M, Yamauchi Y, et al. (2003). Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J Biol Chem 278, 34309–34319 [DOI] [PubMed] [Google Scholar]
- Iijima K, Gatt A, and Iijima-Ando K (2010). Tau Ser262 phosphorylation is critical for Abeta42-induced tau toxicity in a transgenic Drosophila model of Alzheimer's disease. Hum Mol Genet 19, 2947–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Galvan V, Xie L, et al. (2004). Enhanced neurogenesis in Alzheimer's disease transgenic (PDGF-APPSw,Ind) mice. Proc Natl Acad Sci U S A 101, 13363–13367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Yamamoto H, Takamatsu J, et al. (2000). Phosphorylation of MARCKS in Alzheimer disease brains. Neuroreport 11, 869–873 [DOI] [PubMed] [Google Scholar]
- Ksiezak-Reding H, Liu WK, and Yen SH (1992). Phosphate analysis and dephosphorylation of modified tau associated with paired helical filaments. Brain Res 597, 209–219 [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, and Mann M (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- Leshchyns'ka I, Liew HT, Shepherd C, et al. (2015). Abeta-dependent reduction of NCAM2-mediated synaptic adhesion contributes to synapse loss in Alzheimer's disease. Nat Commun 6, 8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lee JW, and Ackerman SL (2015). Mutations in the microtubule-associated protein 1A (Map1a) gene cause Purkinje cell degeneration. J Neurosci 35, 4587–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma RH, Zhang Y, Hong XY, et al. (2017). Role of microtubule-associated protein tau phosphorylation in Alzheimer's disease. J Huazhong Univ Sci Technolog Med Sci 37, 307–312 [DOI] [PubMed] [Google Scholar]
- Makihara H, Nakai S, Ohkubo W, et al. (2016). CRMP1 and CRMP2 have synergistic but distinct roles in dendritic development. Genes Cells 21, 994–1005 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, and Sudarsanam S (2002). The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- Morgado AL, Xavier JM, Dionisio PA, et al. (2015). MicroRNA-34a modulates neural stem cell differentiation by regulating expression of synaptic and autophagic proteins. Mol Neurobiol 51, 1168–1183 [DOI] [PubMed] [Google Scholar]
- Palmqvist S, Scholl M, Strandberg O, et al. (2017). Earliest accumulation of beta-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat Commun 8, 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping L, Duong DM, Yin L, et al. (2018). Global quantitative analysis of the human brain proteome in Alzheimer's and Parkinson's Disease. Sci Data 5, 180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelius E, Brinkmalm G, Pannee J, et al. (2017). Proteomic studies of cerebrospinal fluid biomarkers of Alzheimer's disease: An update. Expert Rev Proteomics 14, 1007–1020 [DOI] [PubMed] [Google Scholar]
- Prasad TS, Kandasamy K, and Pandey A (2009). Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol 577, 67–79 [DOI] [PubMed] [Google Scholar]
- Raka F, Di Sebastiano AR, Kulhawy SC, et al. (2015). Ca(2+)/calmodulin-dependent protein kinase II interacts with group I metabotropic glutamate and facilitates receptor endocytosis and ERK1/2 signaling: Role of beta-amyloid. Mol Brain 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M, Lacor PN, Velasco PT, et al. (2010). Deleterious effects of amyloid beta oligomers acting as an extracellular scaffold for mGluR5. Neuron 66, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, and Verkhratsky A (2011). Neurogenesis in Alzheimer's disease. J Anat 219, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LC, Matsumura F, Bokoch GM, and de Lanerolle P (1999). Inhibition of myosin light chain kinase by p21-activated kinase. Science 283, 2083–2085 [DOI] [PubMed] [Google Scholar]
- Sathe G, Na CH, Renuse S, et al. (2018). Phosphotyrosine profiling of human cerebrospinal fluid. Clin Proteomics 15, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G, Na CH, Renuse S, et al. (2019). Quantitative proteomic profiling of cerebrospinal fluid to identify candidate biomarkers for Alzheimer's disease. Proteomics Clin Appl 13, e1800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G, Pinto SM, Syed N, et al. (2016). Phosphotyrosine profiling of curcumin-induced signaling. Clin Proteomics 13, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin RW, Iwaki T, Kitamoto T, and Tateishi J (1991). Hydrated autoclave pretreatment enhances tau immunoreactivity in formalin-fixed normal and Alzheimer's disease brain tissues. Lab Invest 64, 693–702 [PubMed] [Google Scholar]
- Sternberger NH, Sternberger LA, and Ulrich J (1985). Aberrant neurofilament phosphorylation in Alzheimer disease. Proc Natl Acad Sci U S A 82, 4274–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, et al. (2015). STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupin P. (2006). Neurogenesis in the adult central nervous system. C R Biol 329, 465–475 [DOI] [PubMed] [Google Scholar]
- Thinakaran G, and Sisodia SS (2006). Presenilins and Alzheimer disease: The calcium conspiracy. Nat Neurosci 9, 1354–1355 [DOI] [PubMed] [Google Scholar]
- Tomasevic N, and Peculis BA (2002). Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol Cell Biol 22, 4101–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JC, Swomley AM, Cai J, Klein JB, and Butterfield DA (2016). Quantitative phosphoproteomic analyses of the inferior parietal lobule from three different pathological stages of Alzheimer's disease. J Alzheimers Dis 49, 45–62 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- Ulloa L, Montejo de Garcini E, Gomez-Ramos P, Moran MA, and Avila J (1994). Microtubule-associated protein MAP1B showing a fetal phosphorylation pattern is present in sites of neurofibrillary degeneration in brains of Alzheimer's disease patients. Brain Res Mol Brain Res 26, 113–122 [DOI] [PubMed] [Google Scholar]
- Vindry C, Marnef A, Broomhead H, et al. (2017). Dual RNA processing roles of Pat1b via cytoplasmic Lsm1–Lsm7 and Nuclear Lsm2–Lsm8 complexes. Cell Rep 20, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther M, Berezin V, and Walmod PS (2012). NCAM2/OCAM/RNCAM: Cell adhesion molecule with a role in neuronal compartmentalization. Int J Biochem Cell Biol 44, 441–446 [DOI] [PubMed] [Google Scholar]
- Wirths O, Breyhan H, Marcello A, et al. (2010). Inflammatory changes are tightly associated with neurodegeneration in the brain and spinal cord of the APP/PS1KI mouse model of Alzheimer's disease. Neurobiol Aging 31, 747–757 [DOI] [PubMed] [Google Scholar]
- Wong J. (2013). Altered expression of RNA splicing proteins in Alzheimer's disease patients: Evidence from two microarray studies. Dement Geriatr Cogn Dis Extra 3, 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wung JK, Perry G, Kowalski A, et al. (2007). Increased expression of the remodeling- and tumorigenic-associated factor osteopontin in pyramidal neurons of the Alzheimer's disease brain. Curr Alzheimer Res 4, 67–72 [DOI] [PubMed] [Google Scholar]
- Zahid S, Oellerich M, Asif AR, and Ahmed N (2012). Phosphoproteome profiling of substantia nigra and cortex regions of Alzheimer's disease patients. J Neurochem 121, 954–963 [DOI] [PubMed] [Google Scholar]
- Zhang ZH, Ma FF, Zhang H, and Xu XH (2017). MARCKS is necessary for oligodendrocyte precursor cell maturation. Neurochem Res 42, 2933–2939 [DOI] [PubMed] [Google Scholar]
- Zhong J, Kim MS, Chaerkady R, et al. (2012). TSLP signaling network revealed by SILAC-based phosphoproteomics. Mol Cell Proteomics 11, M112.017764 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.