Abstract
Trimeric porins in the outer membrane (OM) of Gram-negative bacteria are the conduits by which nutrients and antibiotics diffuse passively into cells. The narrow gateways that porins form in the OM are also exploited by bacteriocins to translocate into cells by a poorly understood process. Here, using single-channel electrical recording in planar lipid bilayers in conjunction with protein engineering, we explicate the mechanism by which the intrinsically unstructured N-terminal translocation domain (IUTD) of the endonuclease bacteriocin ColE9 is imported passively across the Escherichia coli OM through OmpF. We show that the import is dominated by weak interactions of OmpF pores with binding epitopes within the IUTD that are orientationally biased and result in the threading of over 60 amino acids through 2 subunits of OmpF. Single-molecule kinetic analysis demonstrates that the IUTD enters from the extracellular side of OmpF and translocates to the periplasm where the polypeptide chain does an about turn in order to enter a neighboring subunit, only for some of these molecules to pop out of this second subunit before finally re-entering to form a stable complex. These intimately linked transport/binding processes generate an essentially irreversible, hook-like assembly that constrains an import activating peptide epitope between two subunits of the OmpF trimer.
Introduction
The asymmetric outer membrane (OM) of Gram-negative bacteria, comprising a lipopolysaccharide (LPS) outer leaflet and a phospholipid inner leaflet, provides a robust barrier against host defenses. In order for nutrients and metabolites to diffuse across this impervious membrane barrier, numerous β-barrel trimeric porins such as OmpF and OmpC pepper the OM. The same porins are also the major route into the cell for antibiotics.1 For essential nutrients larger than the porin molecular weight cutoff filter (<600 Da) such as large siderophore complexes and vitamins, active transporters known as TonB-dependent transporters (TBDTs) drive entry into the cell.2
Outer membrane proteins (OMPs) are exploited by bacteriophages,3 contact-dependent inhibitors,4 and bacteriocins, all of which use OMPs to enter bacterial cells.5 A common problem in the movement of large molecules across the OM is the absence of a direct energy source, which is often overcome by coupling transport to the proton motive force (PMF) across the inner membrane. Here, using single-channel recordings of OmpF pores, we elucidate the passive translocation steps that precede energized import of the bacteriocin ColE9 across the OM of E. coli.
Bacteriocins are classified into two groups based on which PMF-coupled system is exploited. Group B bacteriocins use TBDTs and PMF-coupled TonB.6−8 Group A bacteriocins also use TBDTs, but primarily as docking sites to localize to the cell surface. Thereafter, contact is made with the PMF-linked Tol-Pal system, which activates import into the cell. In order to contact Tol-Pal proteins, group A bacteriocins exploit OM porins, primarily OmpF and OmpC. Unlike TBDTs, porins are not directly linked to the PMF, necessitating the early steps of the group A bacteriocin import to be energy-independent.
Enzymatic E-colicins, exemplified by colicin E9 (ColE9), are the best characterized group A bacteriocins. The initial interaction of ColE9 with its target cell is through a high-affinity interaction with vitamin B12 receptor BtuB.9,10 Although BtuB is a TBDT, TonB plays no role in colicin E9 uptake. Instead, 83 intrinsically unstructured amino acids at the N-terminus of the colicin (the IUTD) thread through the trimeric porin OmpF11 to bind TolB, a component of the Tol-Pal system in the periplasm. Two OmpF binding sites (OBS1 and OBS2) in the IUTD simultaneously bind the same OmpF trimer as shown through protease protection, native MS, negative stain electron microscopy, and planar lipid bilayer (PLB) experiments.12 It has previously been proposed that the IUTD passes through one OmpF subunit before binding back into a second subunit of the same OmpF trimer from its periplasmic face.12 This is supported by a combination of fluorescence microscopy and PLB studies, showing the preferential binding of OBS1 to the periplasmic face of OmpF, thereby promoting a hooked conformation of the unstructured region with the TolB-binding epitope presented in the periplasm.13
While the interactions of OBS1 (1550 Da) and OBS2 (980 Da) peptides with OmpF have been observed in vitro, it remains unclear how these interactions facilitate cell entry. Previous work with biological nanopores has shown the versatile capabilities of PLBs, including the stochastic sensing of peptide substrates14 and the detection protein translocation.15,16 Single-channel recordings of OmpF in PLB have previously been used to define the orientation of OmpF within the membrane17 and to monitor the interaction of colicin fragments with OmpF.13,18 In the present work, we defined the roles of individual OBS1 and OBS2 peptides, demonstrating noncooperative and orientationally biased binding to the identical OmpF pores within the OmpF trimer. We also illuminate for the first time the two-step translocation process of the ColE9 IUTD through OmpF. We report kinetic rate constants for translocation steps under physiologically relevant conditions, including the low transmembrane potential that exists across the OM.19,20 Hence, our investigation demonstrates the kinetic basis of the steps involved in the initial passage of ColE9 across the OM, a prerequisite for subsequent import into the cell of this group A bacteriocin.
Results
Prior to single-molecule analysis, we performed bulk kinetic experiments to probe the association of OBS1, which associates first with OmpF in the OM. An analysis of the OBS1–OmpF interaction was performed by stopped-flow FRET between the tryptophans of OmpF (residues 61 and 214) and an AEDANS-labeled OBS1-fusion protein. (See Figure S1 for details.) The observed 0.33 ± 0.03 μM affinity measured in 20 mM potassium phosphate (pH 6.5, 1% (w/v) β-OG at 4 °C) is the product of rapid association (k1 = 9.80 ± 0.31 μM–1 s–1) and fast dissociation (k–1 = 3.22 ± 0.03 s–1) (Figure S1). Binding is electrostatically driven,21 as evidenced by the 10-fold decrease in k1 in the presence of 100 mM NaCl. Optimal binding occurred at acidic pH, with an 80-fold decrease in k1 upon increasing the pH from 6.2 to 7.7. This is consistent with previous work demonstrating that histidine residues in OBS1 (residues 9 and 14) attenuate OBS1–OmpF interactions when deprotonated upon increasing the pH.22 Equivalent experiments using an AEDANS-labeled OBS2-fusion failed to give a FRET signal, probably due to the lower affinity (Kd ≈ 24 μM) for this complex.22 When using FRET with detergent-solubilized OmpF, it is not possible to distinguish between binding events occurring at the extracellular surface of OmpF and those at its periplasmic face. As OBS1 is proposed to pass through OmpF from the extracellular environment into the periplasm, before binding back into an adjacent OmpF subunit, it is important to be able to track the translocation path and resolve these two binding processes.
OBS Sequences Bind to OmpF in a Noncooperative Manner
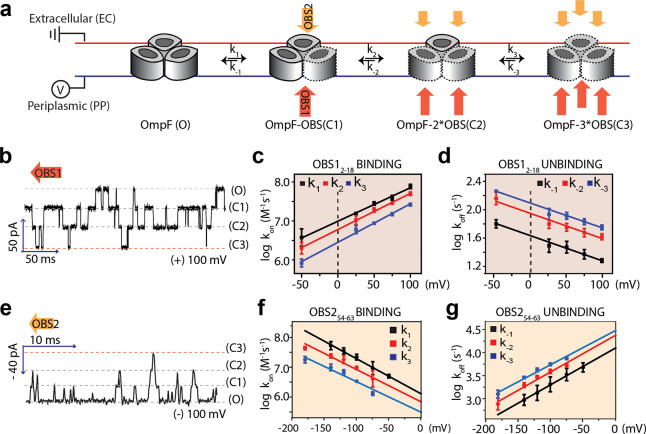
Single-molecule experiments were performed with an OmpF trimer inserted into a DPhPC planar lipid bilayer (PLB). The OmpF orientation was determined from the positive asymmetry of the I–V curve,17 allowing an analysis of OBS1 and OBS2 peptide binding to both the periplasmic side (trans, voltage applied) and the extracellular side (cis, at ground) of OmpF. Due to the net positive charge of OBS1 and OBS2 peptides, we applied positive potentials to promote the binding of OBS peptides to OmpF when added from the periplasmic side and negative potentials when added from the extracellular side. When OBS12–18 was added to the periplasmic side, stepwise and reversible binding was observed (Figure 1a), as shown by four conductance levels: all OmpF subunits unoccupied (O), one subunit occupied (C1), two subunits occupied (C2), and three subunits occupied (C3) (Figure 1a,b). More frequent binding events at higher applied positive potentials and at higher concentrations of peptide indicate a charge-dependent bimolecular interaction (Figure 1 and Figure S2). Rate constants (k1, k2, k3, k–1, k–2, and k–3) for the sequential binding and dissociation events were determined from the transition times among the four conductance levels of OmpF (Figure S2). Rate constants at 0 mV were determined by extrapolation from logarithmic plots of the rate constants against the applied potential (Figure 1c,d and Table 1). Ratios of association rate constants (3:2:1 k1/k2/k3) and dissociation rate constants (1:2:3 k–1/k–2/k–3) reflect the number of unoccupied and occupied sites, respectively, consistent with noncooperative binding. The Kd value of OBS12–18 for an OmpF monomer was calculated by using the average of three normalized rate constants (e.g., k1/3, k2/2, and k3), for both association and dissociation, extrapolated to 0 mV (13 ± 1.0 μM, Table 1). The difference in the Kd value obtained by isothermal calorimetry (ITC) (1.8 μM)22 and that obtained by PLB recording (13 μM) is likely due to the higher salt concentration in the bilayer recording buffer (100 mM KCl). When added from the extracellular side, OBS12–18 gave much weaker binding (Kd = 5 mM) with the transient occlusion of a single OmpF subunit (Figure S3), which is in agreement with simulations.13
Figure 1.
Stochastic binding of OBS peptides to OmpF. (a) Sequential binding and dissociation of either the OBS12–18 peptide (red) from the periplasmic side (PP) of OmpF or the OBS254–63 peptide (yellow) from the extracellular side (EC) of OmpF. (b) Current trace with 2 μM OBS12–18 on the periplasmic side at +100 mV. (c) Voltage dependences of the association rate constants of OBS12–18 (k1, black; k2, red; and k3, blue). (d) Voltage dependences of the dissociation rate constants of OBS12–18 (k–1, black; k–2, red; k–3, blue). (e) Current trace with 2 μM OBS254–63 on the extracellular side (EC) at −100 mV. (f) Voltage dependences of the association rate constants of OBS254–63 (k1, black; k2, red; and k3, blue). (g) Voltage dependences of the dissociation rate constants of OBS254–63 (k–1, black; k–2, red; and k–3, blue). R-squared values for the linear fittings were in the range 0.93 to 0.99. All rate constants (k values) at 0 mV are shown in Table 1.
Table 1. Rate Constants for OBS Peptide Binding to OmpFa.
| OBS12–18 from the periplasmic side | OBS254–63 from the extracellular side | |
|---|---|---|
| k1 (106 M–1 s–1) | 9.6 ± 0.60 | 4.0 ± 0.60 |
| k2 (106 M–1 s–1) | 6.1 ± 0.42 | 2.2 ± 0.64 |
| k3 (106 M–1 s–1) | 3.2 ± 0.25 | 0.97 ± 0.50b |
| k–1 (s–1) | 43 ± 1.5 | 1.3 ± 0.10 × 104 |
| k–2 (s–1) | 78 ± 3.9 | 2.5 ± 0.34 × 104 |
| k–3 (s–1) | 111 ± 6.6 | 3.2 ± 0.47 × 104 |
| Kd (μM)c | 13 ± 1.0 | 11 ± 2.0 × 103 |
The values are estimates at 0 mV obtained by extrapolation (Figure 1). Each rate constant and its standard deviation were determined from at least three independent experiments. Each experiment generated thousands of reversible binding events unless otherwise stated.
Due to fewer events in which all three subunits were occluded (<100) in the case of OBS254–63, the error in k3 is relatively large compared to those for the other rate constants.
Kd values for a single OmpF subunit were statistically corrected by using the mean association and dissociation rate constants.
The bimolecular association and dissociation of the OBS254–63 peptide with OmpF from the extracellular side also showed stochastic and noncooperative behavior as indicated by the ratios of rate constants (Figure 1e,g and Table 1). The Kd value extrapolated to 0 mV was 11 ± 2 mM (Table 1). The large discrepancy between the Kd value obtained in this study (11 mM) and the Kd value previously obtained by ITC (24 μM)22 may imply a different behavior of OBS2 depending on whether it binds to OmpF from the extracellular or the periplasmic side. This is further supported by the 4 orders of magnitude slower dissociation of OBS2 when OBS254–63 peptide was added to the periplasmic side of OmpF, resulting in a 200-times higher affinity (Kd = 60 μM, Figure S4).
OBS1 and OBS2 Display Distinct Orientational Bias in OmpF Binding
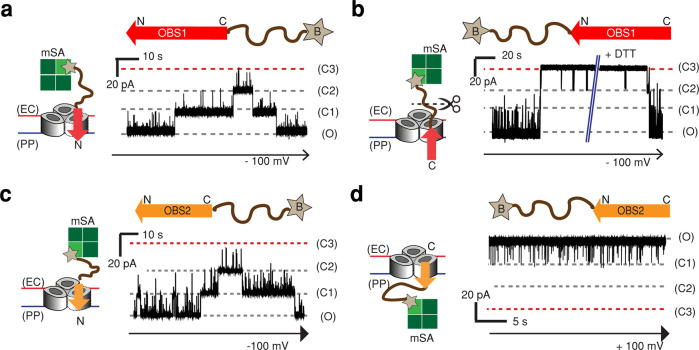
PLB experiments with OmpF in a defined orientation allow binding events at the extracellular and periplasmic faces to be differentiated, but ambiguity still exists with the potential for the peptide to insert either N-terminus or C-terminus first. To restrict the orientation of binding, OBS constructs were capped with monovalent streptavidin (mSA)23 at their C-terminus (OBS1–mSA, OBS2–mSA) or N-terminus (mSA–OBS1, mSA–OBS2). Using OBS1–mSA, we observed stepwise, reversible current blockades of OmpF from both the extracellular and periplasmic sides, indicating that the N-terminus of OBS1 can associate with OmpF from both the extracellular side (kon ≈ 1 μM–1 s–1, koff ≈ 1000 s–1) at −100 mV and the periplasmic side (kon ≈ 5 μM–1 s–1, koff ≈ 600 s–1) at +100 mV (Figure 2a and Figure S5a). Unlike the case of OBS1 peptide binding to OmpF from the periplasmic side (Figure 1b), all three OmpF subunits are not readily occluded by OBS1–mSA (Figure S5a). It is likely that the bulky mSA protein blocks the binding pathway to OmpF. This steric hindrance, observed only from the periplasmic side, reflects the structural and charge asymmetry of OmpF,1,11 likely affecting the OBS accessibility. In addition, threading events from the extracellular side were seen with relatively long residence times (kon ≈ 0.01 μM–1 s–1, koff ≈ 0.1 s–1) at −100 mV (Figure 2a). Such bidirectional binding of the OBS1 sequence led by its N-terminus indicates that OBS1 acts as a leader sequence for crossing the OM, before binding back into OmpF from the periplasmic face to attain the proposed hook-like conformation.12
Figure 2.
Directional binding of OBS peptides to OmpF. Representative current traces of a single OmpF porin in the presence of OBS peptides capped either at (a, c) the C-terminus or (b, d) the N-terminus. The C-terminus-capped OBS1 and OBS2 constructs contain a 12-amino-acid SG linker (SSGGSSGGSSGG, brown curved line) and a single Cys residue with a cleavable linker (HPDP) downstream of the respective OBS sequence. (See the Methods section.) The N-terminus-capped OBS constructs have the same structures upstream of the OBS sequences. The Cys residue was biotinylated (star) and bound to monovalent streptavidin (mSA, green squares) in a 1:1 ratio to form OBS–mSA (C-terminus-capped) or mSA–OBS (N-terminus-capped). (a) Current trace of OmpF in the presence of OBS1–mSA (2.8 μM) at the extracellular surface at −100 mV. (b) Current trace of OmpF in the presence of mSA–OBS1 (2 μM) at the extracellular surface at −100 mV. Upon the addition of 10 mM DTT, the linker between the biotin and SG linker was cleaved, and OmpF returned to the fully open level. (c) Current trace of OmpF in the presence of OBS2–mSA (5 μM) at the extracellular surface at −100 mV. (d) Current trace of OmpF in the presence of mSA–OBS2 (1 μM) at the periplasmic surface at +100 mV.
When added to the extracellular side of OmpF, mSA–OBS1 inserted into OmpF with its C-terminus first and induced irreversible occlusions of all three OmpF subunits in a stepwise manner (kon = 2.4 ± 0.9 s–1) at −100 mV. The occluded OmpF regained the fully open state only when 10 mM DTT was added, which cleaves the disulfide linker between OBS1 and mSA and releases OBS1 most likely into the periplasmic side (Figure 2b). By contrast, no binding of mSA–OBS1 was detected from the periplasmic side (Figure S5b). Therefore, OBS1 can only insert from the periplasmic face of OmpF by leading with its N-terminus. The equivalent complex can be assembled with C-terminus-first insertion of mSA–OBS1 from the extracellular surface of OmpF, although dissociation back to the extracellular side is not possible in this case.
Similar to OBS1–mSA, OBS2–mSA (OBS2 blocked at its C-terminus) showed two types of reversible binding events when added from the extracellular side, with short (koff ≈ 100 s–1) and long (koff ≈ 0.5 s–1) residence times at −100 mV (Figure 2c), reflecting a transient association and long-lived binding, respectively. However, OBS2–mSA showed no entry from the periplasmic side (Figure S6a). With mSA–OBS2, no binding was observed from the extracellular side (Figure S6b). But reversible binding events were observed from the periplasmic side (Figure 2d), although all three subunits were not readily occluded, again likely due to the bulky mSA proteins. Therefore, OBS2 adopts a single orientation within the OmpF lumen, with its N-terminus directed toward the periplasm; OBS254–63 can approach this final orientation from either side of the membrane. The single orientation of OBS2 contrasts with the dual-binding behavior of OBS12–18, which must first cross the outer membrane into the periplasm with its N-terminus leading before binding back into its preferred conformation. Therefore, taken together with the 200-times higher affinity from the periplasmic side (60 μM, Figure S4), OBS2 is likely to be held in the OmpF lumen once adopting its final orientation.
ColE9-IUTD Threads through Multiple Monomers of the OmpF Trimer
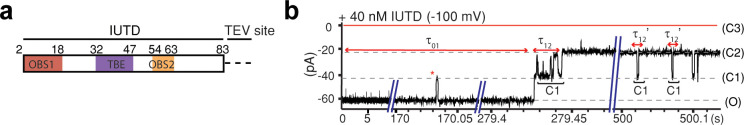
Within the context of intact ColE9 IUTD, the N-terminus of OBS1 is free to bind, but both ends of OBS2 are blocked with flanking sequences, which leaves questions of how the intact IUTD translocates through OmpF. To monitor ColE9–IUTD binding to OmpF, we used the IUTD sequence (residues 2–83) (Figure 3a), which was prepared by cleaving a TEV site between the IUTD and the fusion tag of an IUTD-fusion construct. The addition of 40 nM IUTD from the extracellular side (Figure 3b) resulted in a stepwise decrease in conductance to one-third of the open-pore level of homotrimer OmpF (O to C2), corresponding to the sequential occlusion of two OmpF subunits (Figure 3b). Initial association at the extracellular surface of OmpF blocked a single subunit, with the same current level (C1) maintained until OBS1 threaded into a second subunit (C2). During this process, OBS1 has presumably dissociated from the OmpF lumen into the periplasm and has been replaced by OBS2, translocating ∼60 amino acids across the outer membrane. However, as a full blockade of the first subunit is seen upon OBS1 binding, subsequent steps in this process were not observable. Once the IUTD had occluded two OmpF subunits, the current remained at one-third of the fully open state (C2) for the majority of the time, with occasional transient fluctuations between C2 and C1, which are likely due to the unbinding and rebinding of OBS1 to the second OmpF subunit (Figure 3b).
Figure 3.
Single-molecule observation of ColE9-IUTD translocation through OmpF. (a) The ColE9-IUTD sequence (residues 2–83) was produced by TEV protease, which cleaves the TEV site (ENLYFQ/GA) between the IUTD and the fusion tag. The locations of OBS1, TBE, and OBS2 are shown in red, purple, and orange with residue numbers. The residual amino acids from the TEV site (ENLYFQ) are shown as a dashed line. (b) Current trace of OmpF when 40 nM free IUTD (ColE92–83-ENLYFQ) was added to the extracellular side of OmpF at −100 mV. The IUTD first reversibly associates with a first OmpF subunit (red asterisk). The latency before the C1 state (τ01) indicates the time before translocation into the first OmpF subunit. The latency before the initial passage of OBS1 into the second subunit is τ12, and the latency before rebinding is τ12′.
In a continuous recording for up to an hour, the conductance level never returned to the fully open state (O), even when the applied potential was stepped from −100 to +100 mV (Figure S7), indicating that both OBS1 and OBS2 preferably remain bound to OmpF. The remaining OmpF subunit could be occluded by the addition of OBS12–26 peptide (1 μM) to the periplasmic side, as shown by additional current fluctuations between levels C2 and C3 (Figure S8).
A similar irreversible two-step decrease in conductance was observed with an IUTD-fusion construct (Figure S9a), which includes the DNase domain of ColE9 bound to its immunity protein (Im9), with additional short transitions observed between levels C2 and C3 most likely caused by the fusion domain transiently blocking the third OmpF subunit (Figure S9b). Despite this transient interaction of the fusion partner with OmpF, the fusion protein was the favored construct for further characterization due to its enhanced solubility (≥100 μM) compared to that of the IUTD alone (≤5 μM). The role of each OBS in OmpF binding was investigated by deleting either OBS12–32 or OBS254–62 from the IUTD-fusion. Only single-subunit occlusion was observed by the IUTD(ΔOBS1)-fusion (Figure S10a), confirming that the second occlusion occurs due to OBS1 binding from the periplasmic side after threading. The IUTD(ΔOBS2)-fusion retains the two-step reduction in conductance but reverses to the fully open state (O) (Figure S10b), showing that OBS2 plays a significant role in stabilizing the hooked conformation of the complex.
Single-Molecule Kinetic Analysis of the Two-Step Translocation Process
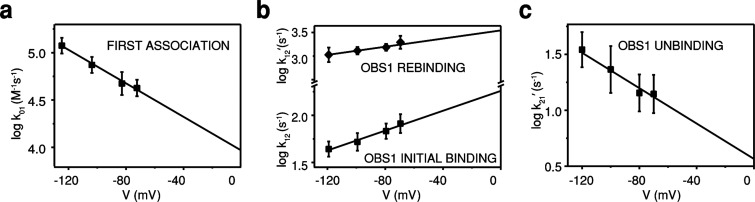
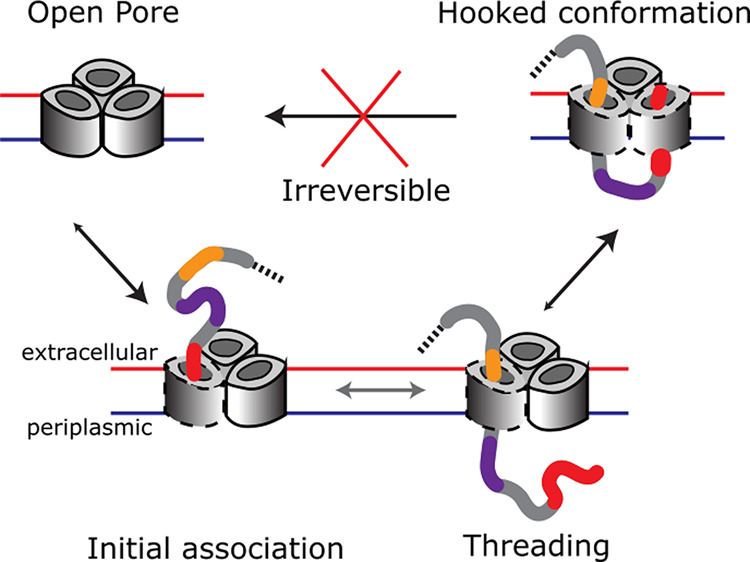
To quantitate the two-step translocation process, we examined the binding kinetics of the IUTD-fusion to OmpF at applied potentials of −70 to −120 mV over a concentration range of 0.1–1.0 μM (Figure S11). Rate constants obtained with the IUTD-fusion were in good agreement with the values obtained with the IUTD over a narrower range of applied potentials (Figure S12). The initial latency (τ01) became shorter at higher applied negative potentials (Figure S11a), indicating a charge-driven association. Also, the shorter latency (τ01) at higher concentrations of the IUTD-fusion (Figure S11b) indicates bimolecular association. Spike-like current fluctuations during the latency (τ01) appear to be failed attempts at threading (koff = 300 s–1 at −100 mV) (similar to Figure 3b), similar to the short-lived blockades seen when OBS1–mSA is added to the extracellular side (koff = 1000 s–1 at −100 mV) (Figure 2a). From the exponential dependence of the rate constant (k01) on the applied negative potential (Figure 4a), we obtained k01 = 9800 ± 1960 M–1 s–1 at 0 mV. After the initial association, the mean dwell time in C1 (τ12) before entry into the second subunit became longer at higher applied negative potentials (Figure S11c), in contrast to the effect of negative potentials on the initial association rate constant (k01). In this case, the rate constant (k12) was independent of the concentration of the IUTD-fusion on the extracellular side of OmpF, consistent with a unimolecular step (Figure S11d). This behavior implies that the translocated OBS1 binds to the second OmpF subunit from the periplasmic side against the force produced by the applied negative potential. From the exponential dependence on the applied potential (Figure 4b), we found k12 = 181 ± 20 s–1 at 0 mV. The two-subunit binding observed here and its voltage dependence support the mechanism previously proposed by Housden et al.12 in which the ColE9-IUTD becomes anchored to OmpF by penetration through one subunit from the extracellular side and then re-enters the porin through a second subunit, forming a hook-like conformation.
Figure 4.
Kinetic analysis of the two-step translocation process. (a) Voltage dependence of the logarithm of rate constant k01. (b) Voltage dependence of the logarithm of rate constants k12 and k12′. (c) Voltage dependence of the logarithm of rate constant k21′. Each data point plotted in panels a (first association) and b (initial binding) was obtained from the mean value of at least 10 independent measurements. Each data point plotted in panels b (rebinding) and c (unbinding) was generated from thousands of reversible binding events. The derived rate constants are provided in Table S1.
The second step, in which the OBS1 sequence binds from the periplasmic side of OmpF, is reversible, as shown by current fluctuations between levels C2 and C1 (Figure 3b). The rate constants for this transition and their voltage dependencies were obtained by analyzing thousands of events from extended current traces. From the exponential dependence on the applied potential (Figures 4b,c), rate constants at 0 mV were determined by extrapolation: k12′ (rebinding) = 5300 ± 500 s–1 and k21′ (unbinding) = 3.9 ± 0.7 s–1. The rate of recurring occlusion of the second subunit by binding of the OBS1 sequence (k12′) is 20 times faster than its initial rate of second subunit occlusion (k12). With no change in signal between the occlusion of the first OmpF subunit by OBS1 and its replacement by OBS2, it is likely that the initial rate observed for the second subunit occlusion is rate-limited by the transition within the first subunit from the initial OBS1 bound state to the OBS2 bound state.
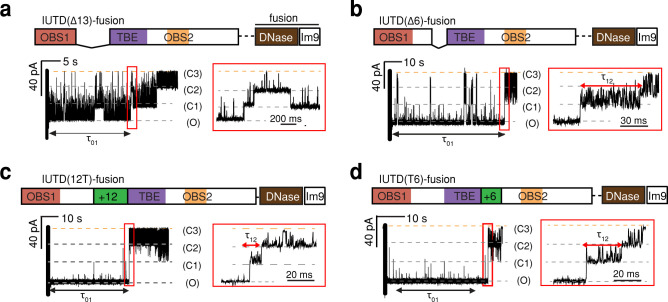
The IUTD is composed of OBS1 and OBS2 linked through the TolB binding epitope (TBE32–47). OBS1 and TBE as well as TBE and OBS2 are both connected by Ser-Gly-enriched linkers (residues 19–31 and 48–53). The impact of the length of the linker on IUTD·OmpF complex formation was assessed by using insertion and deletion mutants. Deletion of residues 19 to 31, the entire linker between OBS1 and the TBE (IUTD(Δ13)-fusion), retarded the binding of OBS1 to a second OmpF subunit, resulting mostly in one-step reversible binding and occasional second-subunit binding with a more than 10-fold reduction in the association rate constant (k12 = 3.0 ± 0.7 s–1 at −100 mV) (Figure 5a and Table S2). A more conservative deletion of six residues from within the same region, (IUTD(Δ6)-fusion), retained two-step binding with an approximately 3-fold reduction in both association rate constants, k01 and k12 (Figure 5b and Table S2). The insertion of 12 amino acids between OBS1 and TBE (IUTD(12T)-fusion) or 6 amino acids between TBE and OBS2 (IUTD(T6)-fusion) had little impact on the association rate constants of the two-step binding process (Figure 5c,d and Table S2). The linker length of the intact IUTD approaches the shortest functional linker length and is associated with the highest measured effective molarity (EM = 0.9 mM) among the tested insertion and deletion mutants (Table S3). The weak dependence of EM values on the linker length suggests that the linker region is flexible24 and does not dictate an optimal conformation for OBS1 binding back into OmpF.
Figure 5.
Effects of sequences between OBS1 and OBS2 on the occlusion of a second OmpF subunit. Current traces are shown after the addition of the 500 nM IUTD-fusion variant to the extracellular side of OmpF at an applied potential of −100 mV. The IUTD sequences are to scale, but the downstream-fusion tag has been compressed (color code in Figure 3). Expanded traces in the red boxes cover the second step in which the OBS1 sequence binds to the second OmpF subunit. (a) IUTD(Δ13)-fusion, lacking 13 residues, 19–31. (b) IUTD(Δ6)-fusion, lacking 6 residues, 26–31. (c) IUTD(12T)-fusion, containing an additional 12 residues (GSGSGG)2 (green) upstream of the TBE. (d) IUTD(T6)-fusion, containing an additional six residues (green) (GGSGSG) downstream of the TBE. The rate constants derived from these experiments are provided in Table S2.
Discussion
Following initial binding between ColE9 and BtuB on the E. coli cell surface with nanomolar affinity, the N-terminal IUTD, containing two OmpF binding sites (OBS1 and OBS2), crosses the OM through one subunit of an OmpF trimer and then reverses direction to enter a second OmpF subunit.12,13 While the previous studies with ColE311 and ColE912,13 provide a structural basis and thermodynamics for understanding the translocation process, a detailed mechanistic understanding has remained elusive. This would require not only monitoring the bidirectional interaction of OBS1 within the OmpF lumen but also deducing the kinetics of the ColE9-IUTD translocation through OmpF, which previous in vitro binding experiments have been unable to resolve. Here, using planar lipid bilayer recording, we dissect individual peptide interactions with OmpF in the context of their orientation and combine these individual events to elucidate the movement of ColE9-IUTD through OmpF in real time. We were able to observe the first passage of ColE9 translocating through OmpF and to determine kinetic rate constants of the two-step process.
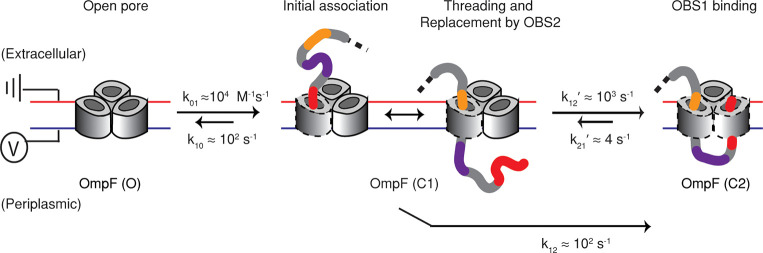
We found that ColE9–IUTD translocates through OmpF in two distinct steps to form an essentially irreversible complex (Figure 6). Furthermore, translocation occurs at 0 mV, which reflects the very low potential across the bacterial OM. ColE9 entry is initiated by the electrostatic association (k01 ≈ 104 M–1 s–1) of the IUTD on the extracellular side of OmpF (Figure 6, O to C1). Transient binding of OBS1 to the first OmpF subunit is followed by either dissociation into the extracellular milieu or transport into the periplasm, with OBS2 replacing OBS1 in the OmpF lumen (Figure 6, substeps at C1). In response to a strong internal electric field in the OmpF pore,25 OBS1 assumes a parallel orientation to the pore axis and thus the threading becomes feasible without external energy input.15 With OBS2 residing in the first OmpF subunit, OBS1 now located in the periplasm is oriented in the opposite direction with respect to the internal electric field and can reach a stable conformation in the OmpF lumen, as indicated by simulation studies.13 Hence, OBS1 binds back and occludes a second subunit of OmpF within the same trimer. Due to its flexibility, the linker between OBS1 and OBS2 is able to accommodate the movement required for the occlusion of the second OmpF subunit. The initial rate for the occlusion of the second OmpF subunit is limited by the transport of OBS1 into the periplasm (k12 ≈ 10 2 s–1, C1 to C2 in Figure 6). Once in the periplasm, rebinding of OBS1 to the second OmpF subunit occurs with a rate constant that is 1 order of magnitude faster than the initial binding (k12′ ≈ 103 s–1, C1 to C2 in Figure 6). This translocation process is completed by mutually independent OmpF subunits within the same trimer, which allow a polypeptide to thread and bind to OmpF in both directions.12,26 Individual associations of OBS1 and OBS2 sequences to OmpF with relatively weak affinities (∼μM)22 combine to stably present the TolB binding epitope (TBE), which lies between OBS1 and OBS2, in the periplasm, raising the probability of recruitment by TolB.27,28 The binding of TolB, a component of the energized Tol-Pal system located at the cell envelope, triggers binding with TolA in the inner membrane27 and results in translocation of the colicin across the outer membrane through a poorly understood mechanism, followed by subsequent translocation across the inner membrane such that the cytotoxic domain can degrade its genomic target.
Figure 6.
Proposed mechanism of ColE9-IUTD threading and binding to OmpF. First, the IUTD associates with the extracellular side of OmpF in a bimolecular fashion (O to C1). The association-induced threading results in the occupancy of the first OmpF subunit by the OBS2 sequence (C1 substep). The translocated OBS1 on the periplasmic side turns and binds to a second OmpF subunit (C1 to C2).
These findings are equally applicable to other enzymatic E-colicins (ColE2–ColE8), which employ the same colicin framework to deliver varied nuclease domains to the E. coli cytoplasm. Furthermore, OBS1-like sequences have been identified at the N-termini of nuclease bacteriocins throughout the enterobacteriaceae,13,29 indicating widespread exploitation of homologous porins through the hooked conformation mechanism. Trimeric porin OmpF, which is one of the most abundant β-barrel OM proteins on the surface of Gram-negative bacteria, may be useful in the design of porin-specific delivery vehicles for antibacterial reagents targeting bacteria without a dedicated transporter.30
Unresolved questions remain, notably, how the C-domain nuclease of ColE9 penetrates the OM and IM to cause cell death.31 Interaction of the TBE–TolB complex with the periplasmic Tol machinery is thought to initiate pmf-driven cellular import by providing enough force to release the colicin immunity protein at the cell surface, followed by translocation of the C-terminal nuclease domain.16,27,32,33 The lipid membrane environment may also play a role in facilitating the release of the colicin immunity protein34 and in altering the binding properties of porins.35 More elaborate in vitro reconstitution of the ColE9 translocation machinery may be required for a detailed elucidation of this intriguing process.
Methods
Plasmid Construction
pNGH25 for the IUTD-fusion (ColE91–83-TEV-E9 DNase·Im9His6), pNGH70 for the IUTD(ΔOBS2)-fusion, and pNGH72 for OmpF have been described elsewhere.22 The IUTD-fusion consists of the IUTD of ColE91–83, the TEV sequence (ENLYFQGA), and the C-terminal DNase domain, which is tightly bound to Im9-His6.
pSJL3, pSJL4, and pSJL5 encoding truncation variants IUTD(Δ6)-fusion and IUTD(Δ13)-fusion, and IUTD(ΔOBS1)-fusion were constructed by using pNGH25 as a template in a single PCR-mediated deletion reaction. A forward primer was designed to contain vector sequences downstream of the deleted fragments and a reverse primer was designed to bind the vector upstream of the deletion site. The primers used to construct pSJL3 are the following: forward, 5′-TGG TGC TTC TGA TGG TTC-3′; reverse, 5′-GTC GGG CCA CCA TTA ATG-3′. For pSJL4, the forward primer is 5′-GGT GCT TCT GAT GGT TCA G-3′, and the reverse primer is 5′-ACC ACT TGT GCT ATG CGC-3′.
pSJL6 and pSJL8 encoding insertion variants IUTD(12T)-fusion and IUTD(T6)-fusion were constructed by subcloning DNA fragments into the pUC57-Kan plasmid by using NdeI and NcoI restriction sites, which are at the beginning of the coding regions for the IUTD and DNase domains, respectively. Fragments including the IUTD insertion variants were ordered as synthetic genes (Genewiz). All of the constructs were sequenced before performing experiments.
Peptide Construction
OBS1 and OBS2 free peptides (NH2-OBS12–18-CONH2, NH2-OBS12–26-CONH2, NH2-OBS12–32-CONH2, and acetyl-OBS254–63-CONH2) were obtained from Peptide Synthetics. Purities were in excess of 90% as determined by HPLC analysis.
Cysteine-containing OBS peptides that were chemically biotinylated and then bound with mSA at the N or C terminus were obtained from Severn Biotech Ltd. Synthetic biotinylated peptides used in this study are listed in Table 2 (from the N-terminus to the C-terminus). Cleavable and noncleavable synthetic OBS peptides were used to monitor their binding and reversibility to OmpF.
Table 2. List of synthetic biotinylated OBS peptides.
| Cleavable | Noncleavable | |
|---|---|---|
| N-term-guided OBS1 | OBS12-18(SGGDGRGHNTGAHSTSG)-Linker(SSGGSSGGSSGG)-Cys-HPDP-Biotin | OBS12-18(SGGDGRGHNTGAHSTSG)-Linker(SSGGSSGGSSGG)-Cys-BMCC-Biotin |
| C-term-guided OBS1 | Biotin-HPDP-Cys-Linker(GGSSGGSSGGSS)-OBS12-18(SGGDGRGHNTGAHSTSG) | Biotin-BMCC-Cys-Linker(GGSSGGSSGGSS)-OBS12-18(SGGDGRGHNTGAHSTSG) |
| N-term-guided OBS2 | OBS254-63(IHWGGGSGRG)-Linker(SSGGSSGGSSGG)-Cys-HPDP-Biotin | OBS254-63(IHWGGGSGRG)-Linker(SSGGSSGGSSGG)-Cys-BMCC-Biotin |
| C-term-guided OBS2 | Biotin-HPDP-Cys-Linker(GGSSGGSSGGSS)-OBS254-63(IHWGGGSGRG) | Biotin-BMCC-Cys-Linker(GGSSGGSSGGSS)-OBS254-63(IHWGGGSGRG) |
The purities of biotinylated peptides were in excess of 90% as determined through HPLC analysis. The predicted masses of the biotinylated peptides were confirmed by MALDI as described elsewhere.36 The biotinylated OBS peptides were incubated with monovalent streptavidin (mSA) in a molar ratio of 1:1 and then added to either the extracellular or periplasmic side of OmpF. mSA was provided by Can Buldun in Mark Howarth’s group (Biochemistry Department, Oxford University).
Protein Expression and Purification
OmpF was expressed using the pNGH72 plasmid in E. coli BZB1107 cells (ompF–, ompC–)25 and purified using a protocol as described previously.12 In brief, transformants containing pNHG72 were grown in LB (5 L) containing ampicillin (100 μg mL–1) and induced by arabinose (0.2% w/v). Cell pellets were lysed by sonication, and membrane fractions were extracted through a series of ultracentrifugation processes. From the outer-membrane fraction, OmpF proteins were purified by three chromatography steps using Q-Sepharose, a 16/60 Sephacryl 300 size-exclusion column, and 4.6/100 mono-Q. All columns were equilibrated with 20 mM Tris-HCl (pH 8.0) buffer containing 5 mM EDTA and 1% (w/v) β-OG, and proteins were eluted using a 0–1 M LiCl gradient.
The IUTD-fusion encoded by pNGH25 was prepared as described elsewhere.22 The IUTD-fusion variants encoded by pSJL3, pSJL4, pSJL5, pSJL6, pSJL8, and pNGH70 were prepared by the same procedure with a slight modification. Transformants containing plasmids encoding the IUTD-fusion constructs were grown in LB (1 L) and induced by 1 mM IPTG for 2 h at 37 °C. His-tagged IUTD-fusion proteins were obtained in two steps, first via Ni-affinity chromatography (HisTrap 1 or 5 mL) and then with a size-exclusion column (HiLoad 26/60 Superdex 75 or HiLoad 16/60 Superdex 200). IUTD-fusion WT and variants were prepared in 20 mM phosphate buffer (pH 6.4) containing 100 mM KCl.
Untagged IUTD was obtained by using AcTEV protease (Invitrogen) according to the manufacturer’s protocol. The cleaved IUTD was collected by using Ni-NTA agarose (Qiagen); IUTD without the fusion tag passed through the column. The final product was examined by LC–MS (LCT–TOF, Waters), and the mass was determined using MassLynx software (8024 ± 44 Da). The calculated mass is 8048 Da.
Single-Channel Recordings in Planar Lipid Bilayers (PLB)
PLB recordings were carried out in potassium phosphate buffer (20 mM, pH 6.4) containing KCl (0.1 M) unless otherwise stated. A lipid bilayer of 1,2-diphytanoyl-sn-glycerol-3-phosphocholine (DPhPC; Avanti Polar Lipids, Alabaster, AL) dissolved in pentane was formed by the Montal–Mueller solvent-free method across a 100-μm-diameter aperture in a 25-μm-thick Teflon film (Goodfellow, Huntingdon, U.K).37 The film separated two 1.0 mL Delrin compartments designated as cis and trans. The cis compartment was connected to ground, and voltage was applied to the trans compartment. The current was amplified by using an Axopatch 200B amplifier, digitized with a Digidata 1440A A/D converter, and recorded with the pClamp 10.3 acquisition software (Molecular Devices) with a low-pass filter frequency of 2 kHz and a sampling frequency of 10 kHz. The data were analyzed and prepared for presentation with pClamp 10.3, QuB (https://qub.mandelics.com), and Origin 2017. All measurements were made at room temperature (20.5 ± 0.5 °C).
A single OmpF trimer was allowed to insert into the bilayer after the addition of a stock solution of OmpF to the cis compartment (<0.5 μL of stock concentration (5–66 μM) in Tris·HCl (20 mM, pH 8.0) containing LiCl (80 mM), EDTA (5 mM), and β-OG (1% w/v)); ±(200–300) mV was applied to induce protein insertion. After insertion had been detected, the buffer in the cis compartment was exchanged to prevent further insertion by replacing 30% of the compartment volume 5–10 times. A current–voltage plot (from −100 to +100 mV) was recorded for each OmpF trimer to define single OmpF insertion and its orientation in the bilayer.17
Kinetic Analysis of Single-Channel Recording Data
The rate constants (k01 and k12) for the two-step binding of the IUTD-fusion (or untagged IUTD) were determined by doing dwell-time analysis, and those for the bimolecular step were determined by measuring the concentration dependence (Figure S11). For the bimolecular first step, the rate constants (k01) were estimated by plotting the inverse of the mean dwell times (1/τ01) at the concentration of the IUTD-fusion. For the unimolecular second step, the rate constants (k12) were determined by taking the inverse of the mean dwell times (1/τ12). The rate constants (k01 and k12) obtained in this way were plotted on a log scale as a function of the applied potential (Figure 4), and the rate constants at 0 mV were estimated by extrapolating the linear regression (Figure 4). The error represents the deviation of linear fitting on the log scale.
To obtain rate constants for the reversible rebinding of the OBS1 sequence (Figure 4 and Tables S1 and S2) after the initial two-step assembly, we measured the transition time from the C2 to C1 and the C2 to C3 states by using pClamp software and idealized current traces using the segmental k-means algorithm of QuB,38 according to a three-state linear model (C1 ↔ C2 ↔ C3). Given the kinetic model, rate constant estimation was performed using a maximum interval likelihood (MIL) function in QuB software.39,40 The MIL computation is carried out with 20 iterations over individual samples, consisting of 1000 transitions from one state to another using the mean dwell time in each state to produce rate constants for a proposed model with maximum probability.
Similar MIL analysis was performed with the same QuB software to determine rate constants for OBS peptide bindings to OmpF, according to a four-state linear model (Figure S2b). The MIL application yielded rates for each transition. By measuring the concentration dependency, rate constants for the bimolecular binding and dissociation events were determined (Figure S2). Since the binding and dissociation rate constants were exponentially proportional to the applied potential (Figure 1), the rate constants at 0 mV were estimated by extrapolating the linear regression to 0 mV on a log scale as a function of applied potential (Table 1).
Acknowledgments
We thank Can Buldun and Mark Howarth for monovalent streptavidin.
Glossary
Abbreviations
- ColE9
colicin E9
- OmpF
outer-membrane protein F
- FRET
fluorescence resonance energy transfer
- AEDANS
N-(acetaminoethyl)-1-naphthylamine-5-sulfonic acid
- β-OG
β-octylglucoside
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c02362.
Figures S1–S12 and Tables S1–S3 (PDF)
The research materials supporting this publication can be accessed by contacting the corresponding author.
Author Present Address
§ (M.H.Z.): Arthur Amos Noyes Laboratory of Chemical Physics, California Institute of Technology, Pasadena, California 91125, United States.
This research was supported by the BBSRC (BB/L021234/1) and a Wellcome Trust collaborative award to C.K. (201505/Z/16/Z).
The authors declare no competing financial interest.
Supplementary Material
References
- Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67 (4), 593–656. 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N.; Guillier M.; Barnard T. J.; Buchanan S. K. TonB-Dependent Transporters: Regulation, Structure, and Function. Annu. Rev. Microbiol. 2010, 64 (1), 43–60. 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi Silva J.; Storms Z.; Sauvageau D. Host Receptors for Bacteriophage Adsorption. FEMS Microbiol. Lett. 2016, 363 (4), fnw002. 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- Ruhe Z. C.; Low D. A.; Hayes C. S. Bacterial Contact-Dependent Growth Inhibition. Trends Microbiol. 2013, 21 (5), 230–237. 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E.; Buchanan S. K.; Duche D.; Kleanthous C.; Lloubes R.; Postle K.; Riley M.; Slatin S.; Cavard D. Colicin Biology. Microbiol. Mol. Biol. Rev. 2007, 71 (1), 158–229. 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilsenbeck J. L.; Park H.; Chen G.; Youn B.; Postle K.; Kang C. Crystal Structure of the Cytotoxic Bacterial Protein Colicin B at 2.5 Å Resolution. Mol. Microbiol. 2004, 51, 711–720. 10.1111/j.1365-2958.2003.03884.x. [DOI] [PubMed] [Google Scholar]
- White P.; Joshi A.; Rassam P.; Housden N. G.; Kaminska R.; Goult J. D.; Redfield C.; McCaughey L. C.; Walker D.; Mohammed S.; Kleanthous C. Exploitation of an Iron Transporter for Bacterial Protein Antibiotic Import. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (45), 12051–12056. 10.1073/pnas.1713741114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens H. M.; Lowe E. D.; Gault J.; Housden N. G.; Kaminska R.; Weber T. M.; Thompson C. M. A.; Mislin G. L. A.; Schalk I. J.; Walker D.; Robinson C. V.; Colin K.. Pyocin S5 Import into Pseudomonas aeruginosa Reveals a Generic Mode of Bacteriocin Transport. mBio 2020, 11, 10.1128/mBio.03230-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G.; Zakharov S. D.; Zhalnina M. V.; Bano S.; Eroukova V. Y.; Rokitskaya T. I.; Antonenko Y. N.; Wiener M. C.; Cramer W. A. The Structure of BtuB with Bound Colicin E3 R-Domain Implies a Translocon. Nat. Struct. Mol. Biol. 2003, 10 (11), 948–954. 10.1038/nsb997. [DOI] [PubMed] [Google Scholar]
- Housden N. G.; Loftus S. R.; Moore G. R.; James R.; Kleanthous C. Cell Entry Mechanism of Enzymatic Bacterial Colicins: Porin Recruitment and the Thermodynamics of Receptor Binding. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (39), 13849–13854. 10.1073/pnas.0503567102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita E.; Zhalnina M. V.; Zakharov S. D.; Sharma O.; Cramer W. A. Crystal Structures of the OmpF Porin: Function in a Colicin Translocon. EMBO J. 2008, 27 (15), 2171–2180. 10.1038/emboj.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden N. G.; Hopper J. T. S.; Lukoyanova N.; Rodriguez-Larrea D.; Wojdyla J. A.; Klein A.; Kaminska R.; Bayley H.; Saibil H. R.; Robinson C. V.; Kleanthous C. Intrinsically Disordered Protein Threads through the Bacterial Outer-Membrane Porin OmpF. Science (Washington, DC, U. S.) 2013, 340 (6140), 1570–1574. 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden N. G.; Rassam P.; Lee S.; Samsudin F.; Kaminska R.; Sharp C.; Goult J. D.; Francis M. L.; Khalid S.; Bayley H.; Kleanthous C. Directional Porin Binding of Intrinsically Disordered Protein Sequences Promotes Colicin Epitope Display in the Bacterial Periplasm. Biochemistry 2018, 57 (29), 4374–4381. 10.1021/acs.biochem.8b00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.; Cheley S.; Alexander L. T.; Knapp S.; Bayley H. Stochastic Detection of Pim Protein Kinases Reveals Electrostatically Enhanced Association of a Peptide Substrate. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E4417. 10.1073/pnas.1312739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J.; Mohammad M. M.; Cheley S.; Bayley H.; Movileanu L. Catalyzing the Translocation of Polypeptides through Attractive Interactions. J. Am. Chem. Soc. 2007, 129 (45), 14034–14041. 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Larrea D.; Bayley H. Multistep Protein Unfolding during Nanopore Translocation. Nat. Nanotechnol. 2013, 8 (4), 288–295. 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu S. A.; Lee S.; Housden N. G.; Kaminska R.; Kleanthous C.; Bayley H. Orientation of the OmpF Porin in Planar Lipid Bilayers. ChemBioChem 2017, 18 (6), 554–562. 10.1002/cbic.201600644. [DOI] [PubMed] [Google Scholar]
- Zakharov S. D.; Eroukova V. Y.; Rokitskaya T. I.; Zhalnina M. V.; Sharma O.; Loll P. J.; Zgurskaya H. I.; Antonenko Y. N.; Cramer W. A. Colicin Occlusion of OmpF and TolC Channels: Outer Membrane Translocons for Colicin Import. Biophys. J. 2004, 87 (6), 3901–3911. 10.1529/biophysj.104.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen K.; Hellman J.; Nikaido H. Porin Channels in Intact Cells of Escherichia Coli Are Not Affected by Donnan Potentials across the Outer Membrane. J. Biol. Chem. 1988, 263 (3), 1182–1187. [PubMed] [Google Scholar]
- Stock J. B.; Rauch B.; Roseman S. Periplasmic Space in Salmonella Typhimurium and Escherichia Coli. J. Biol. Chem. 1977, 252 (21), 7850–7861. [PubMed] [Google Scholar]
- Schreiber G.; Haran G.; Zhou H. X. Fundamental Aspects of Protein - Protein Association Kinetics. Chem. Rev. 2009, 109 (3), 839–860. 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden N. G.; Wojdyla J. a; Korczynska J.; Grishkovskaya I.; Kirkpatrick N.; Brzozowski a M.; Kleanthous C. Directed Epitope Delivery across the Escherichia Coli Outer Membrane through the Porin OmpF. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (50), 21412–21417. 10.1073/pnas.1010780107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M.; Chinnapen D. J. F.; Gerrow K.; Dorrestein P. C.; Grandy M. R.; Kelleher N. L.; El-Husseini A.; Ting A. Y. A Monovalent Streptavidin with a Single Femtomolar Biotin Binding Site. Nat. Methods 2006, 3 (4), 267–273. 10.1038/nmeth861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy V. M.; Semetey V.; Bracher P. J.; Shen N.; Whitesides G. M. Dependence of Effective Molarity on Linker Length for an Intramolecular Protein-Ligand System. J. Am. Chem. Soc. 2007, 129 (5), 1312–1320. 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur D.; Schirmer T.; Fourel D.; Simonet V.; Rummel G.; Widmer C.; Rosenbusch J. P.; Pattus F.; Pagès J. M. Structural and Functional Alterations of a Colicin-Resistant Mutant of OmpF Porin from Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A. 1994, 91 (22), 10675–10679. 10.1073/pnas.91.22.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehna G.; Zhang G.; Gong X.; Duszyk M.; Okon M.; McIntosh L. P.; Weiner J. H.; Strynadka N. C. J. A Protein Export Pathway Involving Escherichia Coli Porins. Structure 2012, 20 (7), 1154–1166. 10.1016/j.str.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Bonsor D. A.; Hecht O.; Vankemmelbeke M.; Sharma A.; Krachler A. M.; Housden N. G.; Lilly K. J.; James R.; Moore G. R.; Kleanthous C. Allosteric Beta-Propeller Signalling in TolB and Its Manipulation by Translocating Colicins. EMBO J. 2009, 28 (18), 2846–2857. 10.1038/emboj.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakos G.; Housden N. G.; Lilly K. J.; Kaminska R.; Kleanthous C. Kinetic Basis for the Competitive Recruitment of TolB by the Intrinsically Disordered Translocation Domain of Colicin E9. J. Mol. Biol. 2012, 418 (5), 269–280. 10.1016/j.jmb.2012.01.039. [DOI] [PubMed] [Google Scholar]
- Sharp C.; Bray J.; Housden N. G.; Maiden M. C. J.; Kleanthous C. Diversity and Distribution of Nuclease Bacteriocins in Bacterial Genomes Revealed Using Hidden Markov Models. PLoS Comput. Biol. 2017, 13 (7), e1005652. 10.1371/journal.pcbi.1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovich M. A. T.; Hansford K. A.; Gong Y.; Butler M. S.; Muldoon C.; Huang J. X.; Ramu S.; Silva A. B.; Cheng M.; Kavanagh A. M.; Ziora Z.; Premraj R.; Lindahl F.; Bradford T. A.; Lee J. C.; Karoli T.; Pelingon R.; Edwards D. J.; Amado M.; Elliott A. G.; Phetsang W.; Daud N. H.; Deecke J. E.; Sidjabat H. E.; Ramaologa S.; Zuegg J.; Betley J. R.; Beevers A. P. G.; Smith R. A. G.; Roberts J. A.; Paterson D. L.; Cooper M. A.. Protein-Inspired Antibiotics Active against Vancomycin- and Daptomycin-Resistant Bacteria. Nat. Commun. 2018, 9 ( (1), ). 10.1038/s41467-017-02123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer W. A.; Sharma O.; Zakharov S. D. On Mechanisms of Colicin Import: The Outer Membrane Quandary. Biochem. J. 2018, 475 (23), 3903–3915. 10.1042/BCJ20180477. [DOI] [PubMed] [Google Scholar]
- Hands S. L.; Holland L. E.; Vankemmelbeke M.; Fraser L.; Macdonald C. J.; Moore G. R.; James R.; Penfold C. N. Interactions of TolB with the Translocation Domain of Colicin E9 Require an Extended TolB Box. J. Bacteriol. 2005, 187 (19), 6733–6741. 10.1128/JB.187.19.6733-6741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankemmelbeke M.; Housden N. G.; James R.; Kleanthous C.; Penfold C. N. Immunity Protein Release from a Cell-Bound Nuclease Colicin Complex Requires Global Conformational Rearrangement. MicrobiologyOpen 2013, 2 (5), 853–861. 10.1002/mbo3.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. L.; Ridley H.; Marchetti R.; Silipo A.; Griffin D. C.; Crawford L.; Bonev B.; Molinaro A.; Lakey J. H. The Antibacterial Toxin Colicin N Binds to the Inner Core of Lipopolysaccharide and Close to Its Translocator Protein. Mol. Microbiol. 2014, 92 (3), 440–452. 10.1111/mmi.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko I.; Degiacomi M. T.; Lee S.; Newport T. D.; Gault J.; Reading E.; Hopper J. T. S.; Housden N. G.; White P.; Colledge M.; Sula A.; Wallace B. A.; Kleanthous C.; Stansfeld P. J.; Bayley H.; Benesch J. L. P.; Allison T. M.; Robinson C. V. Lipid Binding Attenuates Channel Closure of the Outer Membrane Protein OmpF. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (26), 6691–6696. 10.1073/pnas.1721152115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Bayley H. Semisynthetic Protein Nanoreactor for Single-Molecule Chemistry. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (45), 13768–13773. 10.1073/pnas.1510565112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsmann T.; Heimburg T.; Keyser U.; Mahendran K. R.; Winterhalter M. Protein Reconstitution into Freestanding Planar Lipid Membranes for Electrophysiological Characterization. Nat. Protoc. 2015, 10 (1), 188–198. 10.1038/nprot.2015.003. [DOI] [PubMed] [Google Scholar]
- Qin F. Restoration of Single-Channel Currents Using the Segmental k-Means Method Based on Hidden Markov Modeling. Biophys. J. 2004, 86 (3), 1488–1501. 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai C.; Sachs F. Solving Ion Channel Kinetics with the QuB Software. Biophys. Rev. Lett. 2013, 8 (3–4), 191–211. 10.1142/S1793048013300053. [DOI] [Google Scholar]
- Qin F.; Auerbach A.; Sachs F. Maximum Likelihood Estimation of Aggregated Markov Processes. Proc. R. Soc. London, Ser. B 1997, 264 (1380), 375–383. 10.1098/rspb.1997.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.