Abstract
Myiopsitta monachus is a small Neotropical parrot (Psittaciformes: Arini Tribe) from subtropical and temperate regions of South America. It has a diploid chromosome number 2n = 48, different from other members of the Arini Tribe that have usually 70 chromosomes. The species has the lowest 2n within the Arini Tribe. In this study, we combined comparative chromosome painting with probes generated from chromosomes of Gallus gallus and Leucopternis albicollis, and FISH with bacterial artificial chromosomes (BACs) selected from the genome library of G. gallus with the aim to shed light on the dynamics of genome reorganization in M. monachus in the phylogenetic context. The homology maps showed a great number of fissions in macrochromosomes, and many fusions between microchromosomes and fragments of macrochromosomes. Our phylogenetic analysis by Maximum Parsimony agree with molecular data, placing M. monachus in a basal position within the Arini Tribe, together with Amazona aestiva (short tailed species). In M. monachus many chromosome rearrangements were found to represent autopomorphic characters, indicating that after this species split as an independent branch, an intensive karyotype reorganization took place. In addition, our results show that M. monachus probes generated by flow cytometry provide novel cytogenetic tools for the detection of avian chromosome rearrangements, since this species presents breakpoints that have not been described in other species.
Keywords: Myiopsitta, Arini tribe, phylogenetic, karyotype, rearrangements, breakpoints
Introduction
The order Psittaciformes comprises some 330–350 species, grouped into three families according to a new classification: Strigopidae, Cacatuidae and Psittacidae (Rheindt et al., 2014; Winkler et al., 2015). Within the Psittaciformes, the earliest divergence was between the New Zealand-restricted Strigopidae family (Kakapo, Kea, and Kaka) and other parrots, followed by the divergence between the Australasian cockatoos (Cacatuidae) and remaining parrots (Tavares et al., 2006; Wright et al., 2008). Recently, some studies have focused on the phylogenetic reconstruction of this order based on ancient geographic considerations. Wright et al. (2008) supported the hypothesis of an Australasian origin, during the cretaceous in Gondwana and afterward the division of the African and India/Madagascar block, with subsequent diversification through vicariance and dispersal.
Some studies have proposed the division of Psittacidae family into Old World and New World parrots (Miyaki et al., 1998). According to Tavares et al. (2006), the New world Parrots share a much earlier common ancestor with Australian parrots (59 million years ago, Mya), prior to the separation of Australia from Antarctica and South America.
Concerning chromosomal data, New World parrots (Arini Tribe) show a karyotype homogeneity when compared to Old World species, with a constant diploid chromosome number (2n = 70) (Caparroz and Duarte, 2004). However, some species have atypical diploid chromosome numbers, as low as 2n = 48 in Myiopsitta monachus (Furo et al., 2017) and 2n = 64 in Graydidascalus brachyurus (Caparroz and Duarte, 2004), or higher, as in Forpus xanthopterygius and Brotogeris versicolurus with 2n = 86 and 82, respectively (de Lucca et al., 1991; de Lucca and Marco, 2014).
Advances in molecular cytogenetics have shown that ancestral macrochromosomes have been highly rearranged in Psittaciformes (Nanda et al., 2007; Furo et al., 2015a, 2018). Hence, although chromosome painting with probes derived from chicken Gallus gallus (GGA) revealed three associations shared by all species of Psittaciformes analyzed so far – GGA1/4, GGA6/7, and GGA8/9 (Seabury et al., 2013; Furo et al., 2015a, 2018) – there are few chromosomal similarities shared between New World and Old World Psittaciformes.
Nevertheless, the results of GGA probes are limited as they do not detect most intrachromosomal rearrangements useful for phylogenetic inferences (Furo et al., 2015a, b; Furo et al., 2018; Kretschmer et al., 2018a). The use of probes from the white hawk (Leucopternis albicollis, LAL-2n = 66), with a highly derived karyotype involving multiple fissions on the ancestral chromosome (GGA1-GGA5) (de Oliveira et al., 2010), have enabled the detection of key intrachromosomal rearrangements (inversions, fissions), which together with no-reciprocal translocations and tandem fusions represent the main types of mechanisms responsible for their karyotypical divergence (Furo et al., 2015a, 2018).
Myiopsitta monachus (MMO) is a South American species (Higdon, 1998) with an atypical karyotype, with a diploid chromosome number 2n = 48, which is the lowest known diploid chromosome number among the Arini Tribe (Neotropical Psittacidae) and, together with the African species Agapornis roseicollis, has the lowest chromosome complement in the order (Nanda et al., 2007). Furthermore, M. monachus also atypical sex chromosomes, with an enlarged W chromosome, similar in size and morphology to the Z. These features are uncommon among Neornithes birds, which generally present heteromorphic sex chromosomes. The enlargement of the W is due to the accumulation and amplification of repetitive DNA (Furo et al., 2017).
A study by Tavares et al. (2006) based on mitochondrial and nuclear sequences, placed M. monachus in a clade together with Amazona, Pionus, Pionopsitta, Triclaria, and Graydidascalus, despite the fact that the M. monachus karyotype has been characterized only by Giemsa staining and microsatellite mapping (Furo et al., 2017).
Hence, in view of the value of whole chromosome painting in the improved understanding of chromosomal evolution, and the fact that M. monachus has an atypical chromosome complement in which the homologies with other species is still unknown, the aim of this study has been (a) to investigate the mechanisms underlining the origin of the low 2n of M. monachus by using GGA BAC probes and whole chromosome painting probes from G. gallus and L. albicollis; (b) to compare the karyotypes of M. monachus and members of Psittaciformes, to better determine their phylogenetic relationships. Additionally, we show that the use of probes derived from species with highly derived karyotypes such as M. monachus provides greater resolution in detecting interchromosomal rearrangements.
Materials and Methods
Samples, Cell Culture, and Chromosome Preparation
The experiments followed ethical protocols and were approved by the ethics committee (CEUA-Universidade Federal do Pará) under no. 170/2013 and SISBIO 68443-1. Metaphases were obtained from fibroblast culture of skin biopsies and feather pulp of 1 (female) individual of Ara macao (AMA) and 5 (3 males and 2 females) of M. monachus (Table 1), following Sasaki et al. (1968) with modifications. The samples were firstly fractionated mechanically in a Petri dish and afterward were incubated in a solution of Collagenase type IV for dissociation. Cells were then cultivated in DMEM (GIBCO) supplemented with calf bovine serum 20%, AminiomaxTM-II 5% and Penicillin (PNS) 1%, and incubated at 37°C. Chromosome suspensions were obtained using colcemid (Gibco, 100 μl for 5 ml of complete medium) followed by a treatment in hypotonic solution (KCl 0,075 M) and fixation in Carnoy’s fixative methanol: acetic acid (3:1 v/v).
TABLE 1.
List of specimens collected in the present study.
| Species | Number of individuals/sex | City/state |
| Ara macao | 1 female | Belém/Pará-PA |
| Myiopsitta monachus | 3 males and 2 females | Sapucaia do Sul/Rio Grande do Sul-RS |
Flow Sorting and Generation of Chromosome-Specific Painting Probes
Chromosome preparations for flow cytometry were obtained from a fibroblast cell line of a male M. monachus. The flow sorting followed the method described by Nie et al. (2015). Chromosomes were stained with chromomycin A3 (40 μg/ml, Sigma) and Hoechst 33258 (2 μg/ml, Sigma). Sorting was performed using a dual-laser cell sorter (MoFlo, Beckman Coulter). The primary sorted chromosome material, i.e., macro- and microchromosomes of M. monachus (hereafter designated as MMO) was amplified by degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR) (Telenius et al., 1992) and the resulting products were then labeled with biotin-16-dUTP or digoxigenin-dUTPs during secondary DOP-PCR amplification. The identity of probes was validated by their hybridization back to the metaphases of the original species. Because many chromosomes of MMO have similar sizes (making their identification difficult) we used reciprocal chromosome painting between MMO and Ara macao (AMA) to resolve any ambiguity in the chromosomal assignment of each flow peak and, in particular, of the peaks that contained two chromosomes. As molecular karyotype characterization of Ara macao is known, the use of this species helped to clarify the assignment of some MMO peaks.
Preparation of Chicken BAC Clones and FISH
The bacterial artificial chromosomes (BACs) containing fragments from chromosomes 17–28, were labeled by Nick translation, fluorescein isothiocyanate-12-UTP (FITC-dUTP) (p-arms) (Roche) and Texas Red-12-dUTP (q-arms) (Invitrogen) and used in fluorescent in situ hybridization experiments following O’Connor et al. (2018). The slides were analyzed with an Olympus BX-61 epifluorescence microscope equipped with a cooled CCD camera and appropriate filters. Images were captured using SmartCapture3 (Digital scientific United Kingdom).
Chromosome Painting With G. gallus and L. albicollis Probes
Sets of chromosome specific probes from G. gallus and L. albicollis were previously generated by flow-sorting of chromosomes in Cambridge Resource Center for Comparative Genomics (Cambridge, United Kingdom). We used a set of chicken chromosome probes with pairs 1–14. Comparative chromosome painting with L. albicollis probes used pairs homologous to chromosomes: GGA1 (LAL3, 6, 7, 15, and 18), GGA2 (LAL2, 4, and 20), GGA3 (LAL9, 13, 17, and 26), GGA4 (LAL1 and 16), GGA5 (LAL5) and GGA6 (LAL3) (de Oliveira et al., 2010). The protocols for hybridization followed de Oliveira et al. (2010). The slides were analyzed using a Zeiss Axioplan2 fluorescence microscope and ISIS software (Metasystems). Comparisons were based on the avian putative ancestral karyotype (PAK), in which pairs PAK 1–11 and PAK13-15 corresponded to GGA1-GGA3, GGA4q, GGA5-GGA9, GGA4p, GGA10, GGA12, GGA13, and GGA14, respectively (Griffin et al., 2007; Kretschmer et al., 2018a).
Phylogenetic Analysis Using Chromosomal Characters
A binary matrix was constructed showing the presence and absence of discrete characters through chromosomal rearrangements and chromosome homologies among MMO and other species previously reported: Amazona aestiva, Pyrrhura frontalis (PFR), Anodorhynchus hyacinthinus (AHY), Ara chloropterus (ACH) and Ara macao (AMA) from the Neotropical Psittacidae (Seabury et al., 2013; Furo et al., 2015a, 2018). As outgroup, we used G. gallus and Turdus rufiventris (TRU) (Supplementary Table S1) (Kretschmer et al., 2014). The matrix generated was used in a parsimony cladistic analysis, using PAUP 4.0b10 (Phylogenetic Analysis Using Parsimony). A heuristic search to find the most parsimonious tree(s) was performed using Tree Bisection Reconnection (TBR) branch-swapping; the posterior bootstrap probability was obtained with one thousand replicates. For homology analysis, we used the nomenclature of the Putative Avian Ancestral Karyotype (PAK) (Griffin et al., 2007). Conclusions concerning chromosomal rearrangements and divergence time of species were traced by comparing our phylogenetic data with previously published data from Tavares et al. (2006).
Results
Karyotype and Chromosome Mapping With G. gallus and L. albicollis Whole-Chromosome Probes
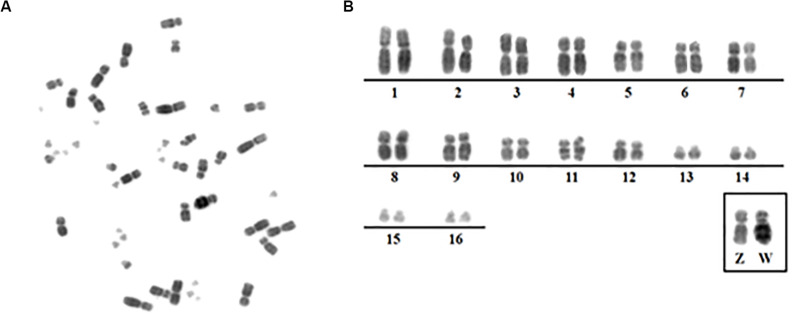
The chromosome number and morphology for the species analyzed here confirmed previous results: 2n = 68 in A. macao (AMA) and 2n = 48 in M. monachus (MMO) (Seabury et al., 2013; Furo et al., 2017; Figure 1).
FIGURE 1.
(A) Metaphase from the Myiospitta monachus female. (B) Karyotype from Myiopsitta monachus (2n = 48) showing only macrochromosomes 1–16 and ZW.
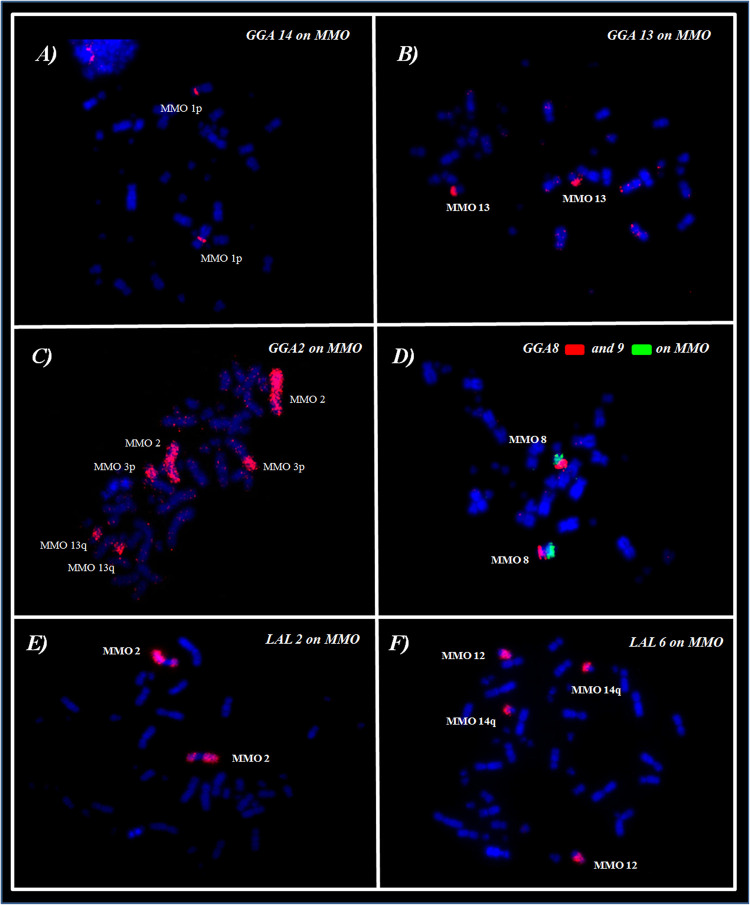
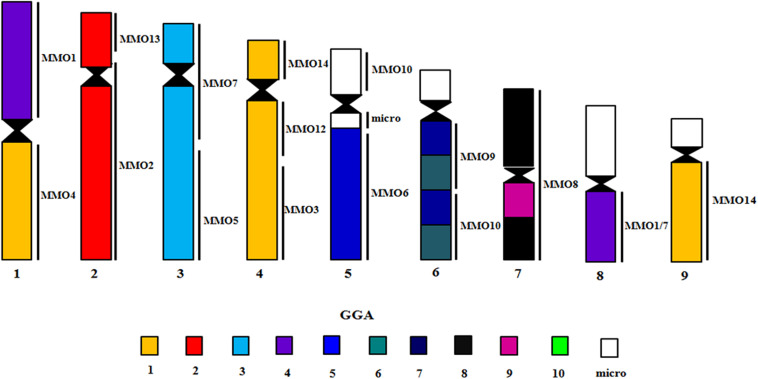
Gallus gallus whole chromosome probes corresponded to 21 homologous segments in the MMO genome, and revealed fissions in several ancestral pairs, and fusions involving the segments that were fissioned. The correspondences found were: GGA1 (MMO3q, 4p/q, 12, 14q), GGA2 (MMO 2, 3p, 13q), GGA3 (MMO5 and MMO7), GGA4 (MMO1q/p), GGA6 (MMO 9q, 10q), GGA7 (MMO 9q, 10q), GGA8 (MMO 8p), GGA9 (MMO 8q), GGA10 (MMO 9p, 11), GGA13 (MMO 13p), GGA14 (MMO1p).
The fusions detected were: GGA1/GGA2, in MMO3; GGA3/GGA4 in MMO7p, GGA8/GGA9 in MMO8 and GGA2/GGA13 in MMO13. In addition, we observed the fusion between GGA6/GGA7, as observed in all members of Arini Tribe (Seabury et al., 2013; Furo et al., 2015a, 2018). However, due to an additional rearrangement in M. monachus, this association originated two distinct pairs: MMO9 (micro/GGA7/GGA6) and MMO10 (micro/GGA6/GGA7) (Figures 2A–D and Table 2).
FIGURE 2.
FISH using whole-chromosome probes of (A–D) G. gallus and (E,F) L. albicollis on M. monachus.
TABLE 2.
Chromosomal correspondence, avian putative ancestral karyotype (PAK), G. gallus (GGA) and M. monachus (MMO) chromosomes.
| Ancestral chr. | Probes | Number of pairs | Pairs in MMO |
| PAK1 | GGA1 | 4 Pairs | 3q,4p/q,12,14q |
| PAK2 | GGA2 | 3 pairs | 2,3p,13q |
| PAK3 | GGA3 | 2 pairs | 5,7 |
| PAK4 | GGA4q | 2 pairs | 1q/7q |
| PAK5 | GGA5 | 1 pair | 6 |
| PAK6 | GGA6 | 2 pairs | 9q,10q |
| PAK7 | GGA7 | 2 pairs | 9q,10q |
| PAK8 | GGA8 | 1 pair | 8p |
| PAK9 | GGA9 | 1 pair | 8q |
| PAK10 | GGA4p | 2 pair | 1p, 7q |
| PAK11-13 | GGA10-12 | 2 pairs | 9p,11 |
| PAK12 | GGA11 | ||
| PAK14 | GGA13 | 1 pair | 13p |
| PAK15 | GGA14 | 1 pair | 1p |
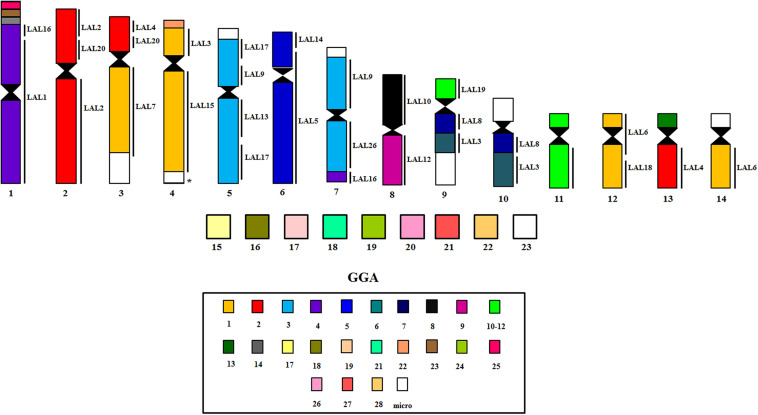
The experiments using LAL probes allowed the detection of inversions involving the ancestral chromosomes 1–5 (Figures 2E,F). In M. monachus an unusual rearrangement was detected with LAL probes, in this case, LAL16 (homologous to GGA4p), which has fissioned resulting in two segments, one of which fused with GGA4q and the other GGA3. The homology maps of M. monachus, G. gallus and L. albicollis by chromosome painting are shown in Figure 3 and Table 2.
FIGURE 3.
Homology maps with GGA, LAL probes and chicken BACs from microchromosomes on chromosomes of M. monachus.
Inferring Homeologies by BAC-FISH With GGA Probes Mapped Onto M. monachus Chromosomes
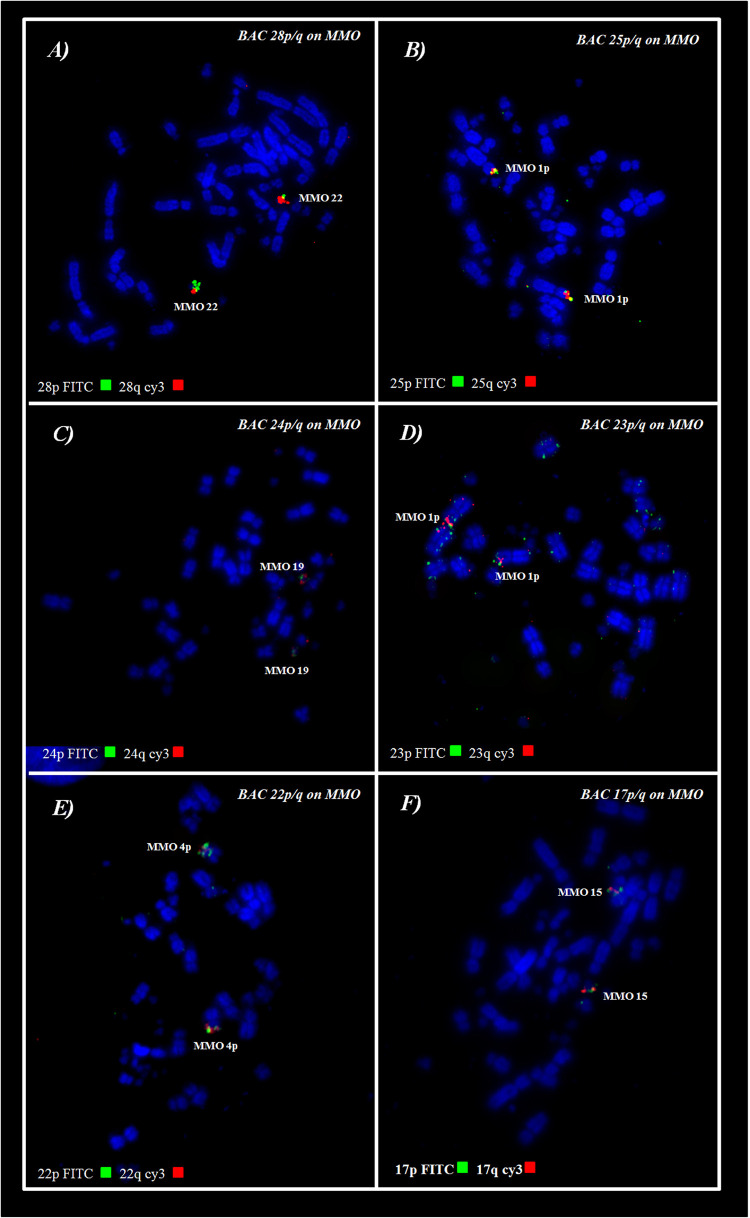
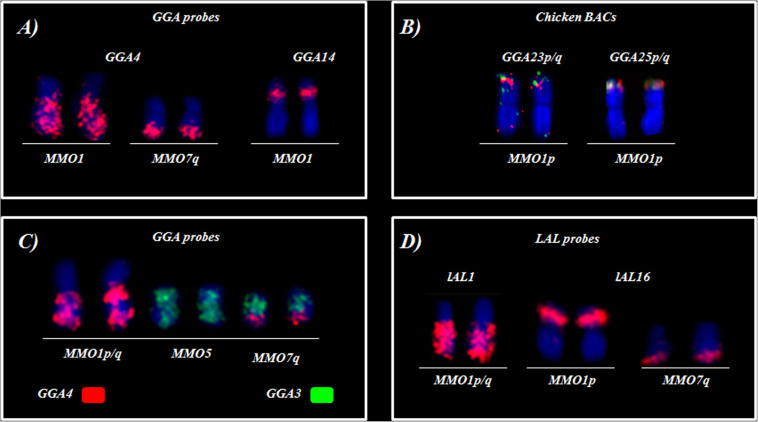
BAC probes corresponding to GGA chromosome pairs 17–28, except pair 20 (which did not produce reproducible results) revealed the occurrence of many tandem fusions between microchromosomes and also fusions between macrochromosomes and microchromosomes as revealed in MMO1 (GGA4p/q+GGA14+GGA23+GGA25) and MMO4 (GGA1+GGA22) (Figures 3, 4 and Table 3).
FIGURE 4.
FISH using Chicken BACs from microchromosomes on M. monachus. (A,C) Examples of conserved microchromosomes. (B,D–F) Examples of microchromosomes involved in fusions with macrochromosomes. Red signals represent probes labelled with Cy3; Green signals represent probes labelled with FTIC.
TABLE 3.
Chromosomal correspondence between chicken BACs from microchromosome (GGA) in M. monachus (MMO).
| GGA Chr. | BAC Name | MMO Chr. |
| 17q | CH261-42P16 | 15q |
| 17p | CH261-113A7 | 15q |
| 18p | CH261-60N6 | 16p |
| 18q | CH261-72B18 | 16q |
| 19p | CH261-10F1 | 17p |
| 19q | CH261-50H12 | 17p |
| 21q | CH261-122K8 | 18q |
| 21p | CH261-83I20 | 18p |
| 22q | CH261-18G17 | 4p |
| 22p | CH261-40J9 | 4q |
| 23p | CH261-191G17 | 1p |
| 23q | CH261-90K11 | 1p |
| 24p | CH261-103F4 | 19p |
| 24q | CH261-65O4 | 19q |
| 25q | CH261-127K7 | 1p |
| 25p | CH261-59C21 | 1p |
| 26q | CH261-170L23 | 20q |
| 26p | CH261-186M13 | 20p |
| 27q | CH261-28L10 | 21p |
| 27p | CH261-66M16 | 21q |
| 28p | CH261-64A15 | 22p |
| 28q | CH261-72A10 | 22q |
Flow Karyotype of M. monachus
The 48 chromosomes of M. monachus were resolved into 15 peaks by flow cytometry (Figure 5). The chromosomes in each peak of the flow karyotype were identified on MMO metaphases using FISH with labeled peak-specific DNA (Figures 6A–C). MMO chromosomes 1, 4, 9, 12, and 13 pairs formed a separate peak each. However, chromosomes 2+Z were found in the same peak as well as chromosomes 3+4, 5+6, 7+8, 10+11, and 14+15 and microchromosomes 15–23. However, some chromosomes were found in more than one peak, such as MMO 4, 7, and 8, and a possible explanation is the presence of repetitive DNA in some chromosomes of MMO, with a significant difference in the number and overall GC content of repeats between the homologs of some pairs, as described by Furo et al. (2017). These heteromorphisms would be sufficient to separate the homologs into different positions in the flow-karyotype (Figure 5).
FIGURE 5.
The bivariate flow karyotype of the Myiopsitta monachus. Chromosomes were sorted for DNA content and AT to GC base pair rations into 15 peaks after staining with Hoechst 22358 (vertical axis) and chromomycin-A (horizontal axis). Legend: **Different peaks contain same chromosomes.
FIGURE 6.
FISH using whole-chromosome probes [MMO (A–C)] onto M. monachus and [MMO (D,E)] M. monachus on A. macao.
Chromosome Painting Between M. monachus and A. macao
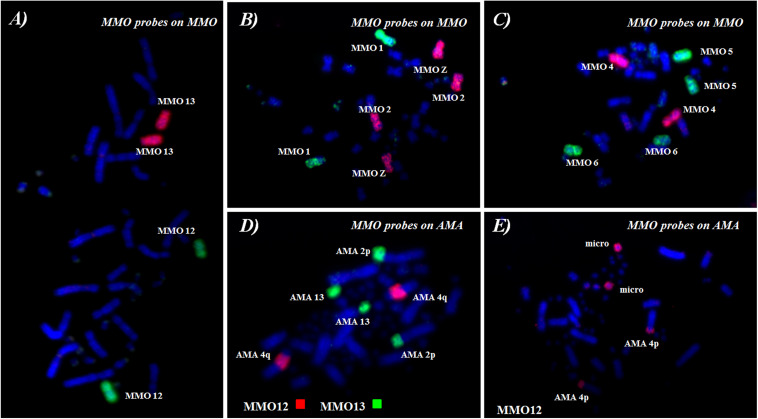
Reciprocal chromosome painting between MMO and AMA established chromosome homologies between these species and defined the chromosome content of MMO peaks. FISH examples are shown in Figures 6D,E, and results of reciprocal chromosome painting between MMO and AMA are summarized in the karyotype of AMA (Figure 7 and Table 4).
FIGURE 7.
Homology map with GGA and MMO probes on metaphase chromosomes of A. macao. The results with GGA probes were obtained from Seabury et al. (2013).
TABLE 4.
Chromosomal correspondence between M. monachus (MMO), G. gallus (GGA) and A. macao (AMA) chromosomes.
| Chr MMO | GGA | Chr AMA |
| 2, Z | 2q, Z | 2q, Z |
| 1 | 4q, 4p,14,23,25,28 | 1p,8q + 4 micros |
| 2, 3, 4, Z | 2p/q,1p/q, Z, micros | 1q,2q/p,4q,Z, micros |
| 4 | 1p | 1q |
| 5,6 | 3p/q,5 | 3q,5q |
| 5,6,7,8 | 3p/q,5,8,9 | 3,5,6, |
| 8,9 | 7,6,8,9 | 6,7 |
| 7 | 3q | 6 |
| 10,11 | 6,7,10,micro | 7,micro |
| 12 | 1q | 4q |
| 13 | 2p + micro | 2p + micro |
| 14,15 | 1q (MMO14), micro | 1q (MMO14), micro |
Phylogenetic Analysis
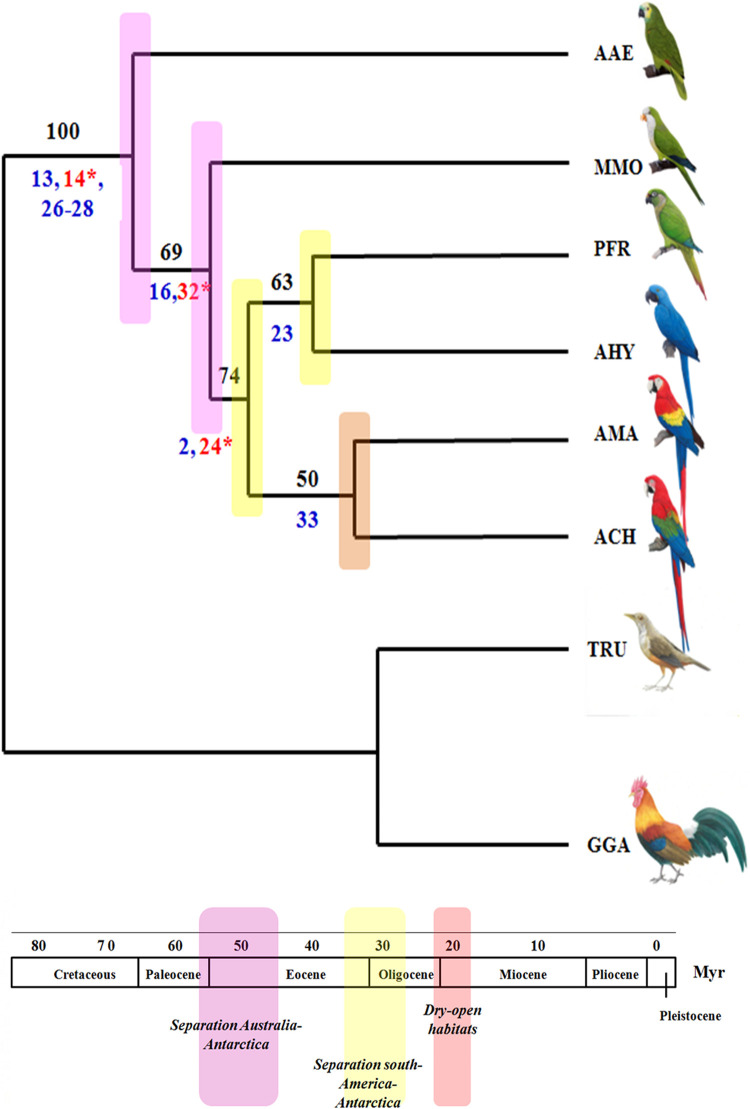
Two most parsimonious trees were obtained using PAUP (Figure 10). The consensus tree presented score 37, and was obtained from 34 characters, 12 of them being phylogenetically informative. The tree length was 37, consistency index and homoplasic index was 0,9189 and 0,0811, respectively. The Psittaciformes are well supported as a monphyletic group, with high bootstrap support (100). In the basal branch is A. aestiva, followed by a branch including the other membership of Arini tribe (bootstrap of 69), the first species to split within this clade was MMO, followed by a branch (bootstrap 74) that contain the sister groups AHY/PFR and AMA/ACH, with bootstrap support of 63 and 50, respectively. The low bootstrap support in some branches were due to the low number of informative characters (numbers below of the branches), as most chromosome characters were autopomorphies, mainly due to intense chromosomal reshuffling observed in some species, such as M. monachus. In addition, some chromosome rearrangements could not be included because there is no information concerning chromosome homology involving the microchromosomes, with the exception of MMO.
FIGURE 10.
Phylogenetic analysis of Maximum Parsimony made using PAUP based on chromosome rearrangements present in Neotropical Psittacidae, according to results obtained by FISH from G. gallus probes (Legend: AMA, Ara macao, ACH, Ara chloropterus, AHY, Anodorhynchus hyacinthinus, PFR, Pyrrhura frontalis, AAE, Amazona aestiva, GGA, Gallus gallus, TRU, Turdus rufiventris, MMO, Myiopsitta monachus, Myr, millions of years). The red numbers correspond to features that appear in several clades, however, the blue numbers correspond to the characteristics that place the clades together; the bootstrap numbers are in black. The estimation of geological events was based on the literature review by Tavares et al. (2006).
Discussion
Chromosome Evolution in Psittaciformes
The advances in the last decades in comparative cytogenetics have clarified many aspects of the dynamic organization of avian genomes, mainly thanks to the use of G. gallus probes in many species belonging to different orders. For instance, it was possible to identify that their ancestral karyotype structure was kept stable over millions of years of evolution in many branches (Kretschmer et al., 2018a). This conservatism seems to count not only for macrochromosomes but also microchromosomes, as revealed by recent studies with chicken BACs from microchromosomes (O’Connor et al., 2018). Despite this, some avian groups such as Charadriiformes, Accipitriformes and Psittaciformes, exhibit highly reorganized karyotypes (de Oliveira et al., 2005, 2010; Nie et al., 2009; Furo et al., 2015a, 2018).
Important insights into chromosomal diversification in Psittaciformes have been highlighted by earlier studies with sets of G. gallus and L. albicollis whole-chromosome probes. According to a recent study by Furo et al. (2018), the chromosomal synapomorphies found in Psittaciformes, such as the associations between different pairs of the putative ancestral karyotype (PAK), such as PAK1/PAK4 (GGA1/GGA4q), PAK6/PAK7 (GGA6/GGA7) and PAK8/PAK9 (GGA8/GGA9) demonstrate a closer phylogenetic relationship among some genera of Neotropical Psittacidae. In addition, the data from the present study do not support tail size as a relevant taxonomic criterion for classifying this group, as proposed by some authors (Montón, 1977; Miyaki et al., 1997, 1998; Sick, 1997; Francisco and Galetti, 2001).
So far, all species of the Neotropical Psittacidae analyzed by chromosome painting showed homogeneity in the diploid chromosome number, corresponding generally to 2n = 70, however, M. monachus 2n = 48, represents an exception within this group. In fact, M. monachus shares the lowest 2n among Psittaciformes together with the African species A. roseicollis (ARO) (African Psittacidae) (Nanda et al., 2007). Nevertheless, phylogenetic studies, including our chromosome painting data, reveal that this similarity was limited to the chromosome number only. This happened because of the high rate of karyotype repatterning that occurred independently during their evolution, as observed in this study using G. gallus probes. Hence, the chromosome count is not always indicative of phylogenic proximity as in, for instance, Agapornis which is related to the Loriini (represented by Lorius) from Indonesia (Rheindt et al., 2014).
Likewise, the utilization of L. albicollis probes in M. monachus allowed us to identify a great number of pericentric and paracentric inversions. This is in agreement with the view that chromosomal rearrangements (inversions, fissions), which together with no-reciprocal translocations and tandem fusions represent the main rearrangements associated with karyotype evolution in Psittaciformes (Seabury et al., 2013). The methods used, however, are unable to distinguish inversions from centromeric repositioning (Rocchi et al., 2012).
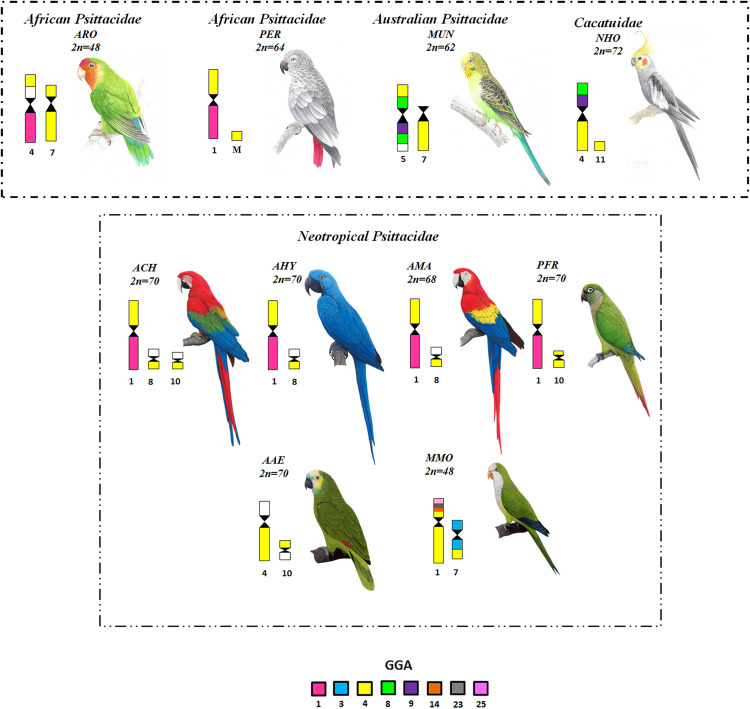
In general, the ancestral macrochromosomes are involved in many fissions and fusions in M. monachus, several representing autapomorphies. For instance, an unusual rearrangement in MMO is the fusion between a small segment of PAK10 (GGA4p) with PAK4 (GGA4q). Generally, GGA4 (PAK4 and PAK10) corresponds to two pairs in most species of birds, belonging to different orders, such as Galliformes, Anseriformes, Passeriformes, Falconiformes, Strigiformes and Struthioniformes (Guttenbach et al., 2003; Kretschmer et al., 2014, 2015, 2018b). However, in M. monachus and other species of this order, these chromosomes are fused with other elements as shown in Figure 9 (Nanda et al., 2007; Seabury et al., 2013; Furo et al., 2015a, 2018). In M. monachus a fusion between these segments followed by fission in a region of PAK10 (GGA4p) was observed, nevertheless, the breakpoints of the segments corresponding to PAK10 (GGA4p) were not found in other species so far, because majority of molecular cytogenetic studies in birds has been limited to the use of G. gallus probes. It is thus not possible to know whether this peculiarity is exclusive to this species (Figure 8).
FIGURE 9.
Representation of chromosome organization of four ancestral chromosomes in Psittaciformes, obtained by chromosome painting with G. gallus probes and chicken BACs from microchromosomes. Legend: GGA (Gallus gallus), ARO (Agapornis roseicollis), PER (Psittacus erithacus), MUN (Melopsittacus undulatus), NHO (Nymphicus hollandicus), ACH (Ara chloropterus), AHY (Anodorhynchus hyacinthinus), AMA (Ara macao), PRF (Pyrrhura frontalis), AAE (Amazona aestiva), MMO (Myiopsitta monachus).
FIGURE 8.
Summary of chromosome rearrangements involving GGA4 chromosome in M. monachus. (A,C) G. gallus probes (B) Chicken BACs (D) L. albicollis probes.
In addition, although we found the fusion of PAK6 (GGA6)/PAK7 (GGA7) in the karyotype of M. monachus, it does not show the same pattern as observed in most species of Neotropical Psittacidae (Seabury et al., 2013; Furo et al., 2015a, 2018). Moreover, it was possible to confirm only one chromosome signature of Psittaciformes in the karyotype of this species, in this case, the fusion between PAK8(GGA8)/PAK9 (GGA9), a feature present in most species of this group, except for two species described recently: A. aestiva and Pyrrhura frontalis (Furo et al., 2018).
Thus, based on the chromosome painting data, Furo et al. (2018), considered the existence of two groups of Neotropical Psittacidae, according to the presence or absence of specific chromosomal synapomorphies, such as the association between PAK1/PAK4 (GGA1p/GGA4q), PAK6/PAK7 (GGA6/GGA7) and PAK8/PAK9 (GGA8/GGA9). In this case, M. monachus shows chromosomal characteristics found in both groups.
The role of microchromosomes in the karyotype evolution of birds has proven to be one of the greatest enigmas in the comparative chromosome analyses of birds (O’Connor et al., 2018). In general, the available chromosome painting probes correspond only to the first ten pairs of chicken chromosomes, which can be sorted by flow cytometry due to their differences in size and GC content (Yang et al., 1999). However, the analyses of microchromosomes are essential to draw a more accurate phylogenetic picture, especially in groups with low diploid numbers and atypical karyotypes, such as the Falconiformes and Psittaciformes. The introduction of comparative chromosome mapping of chicken BACs has helped to resolve this difficulty. Despite the constant chromosome complement (2n = 70) in most members of the Arini Tribe, it has been observed that several macrochromosome regions were not hybridized by any of the GGA whole-chromosome probes, which could indicate cryptic fusions with microchromosomes (Furo et al., 2015a, 2018; O’Connor et al., 2018). This conclusion is confirmed in M. monachus where many tandem fusions between microchromosomes and macrochromosomes were observed, reducing the diploid chromosome number of this species to 2n = 48.
Phylogenetic Relationship in Neotropical Psittacidae
Recent analyses of mDNA and nuclear genes have started to resolve the phylogenetic relationships in the Neotropical Psittacids, and comparative chromosome painting has helped also in the understanding of karyotype evolution in this group.
Tavares et al. (2006) in their study with nuclear and mitochondrial markers in 25 of the 30 genera of Neotropical Psittacidae suggested the division of this group into three large clades, which did not correspond to the grouping by tail size (short and long tail) as proposed by Sick (1997). In the proposal of Tavares et al. (2006), M. monachus is placed in a basal position in the phylogenetic tree, together with Amazonian species and allies of the genera: Pionus, Graydidascalus, Pionopsitta, Tricalia and Botogeris. These results are consistent with studies by Wright et al. (2008) who used multilocus molecular character sampling (3,941 bp from mitochondrial DNA (mtDNA) genes, cytochrome oxidase I and NADH dehydrogenase 2 and nuclear introns of rhodopsin intron 1, tropomyosin alpha-subunit intron 5, and transforming growth factor ss-2).
In our analysis of maximum parsimony, it was possible to clarify doubts about the phylogenetic position of M. monachus. Although this species has a long tail and a karyotype consisting mainly of metacentric and submetacentric chromosomes, as well as the association of PAK8/PAK9, our analysis placed this species in a basal position, corresponding to a second radiation, after A. aestiva. This result agrees with previous analysis performed with mDNA and nuclear sequences (Tavares et al., 2006; Wright et al., 2008). In this analysis, we also found that the main characters that united the macaws to a common ancestor, is the association of PAK4q/PAK1p (GGA4q/GGA1p), in addition to a fusion between PAK5 (GGA5) with a microchromosome (Characters 2, 24). As proposed by us previously (Furo et al., 2018), the fusion between PAK4q/PAK1p (GGA4q/GGA1p) was already present in the common ancestor of the macaws (Table 5 and Figure 10).
TABLE 5.
Correspondence between syntenic groups of Psittaciformes species analyzed by FISH and the putative ancestral avian karyotype (PAK) and Gallus gallus chromosomes (GGA).
| Species | Chromosomes | 2n | Distribution | References | ||||||||||
| GGA | 1 | 2 | 3 | 4q | 5 | 6 | 7 | 8 | 9 | 4p | 10 | 78 | Griffin et al., 2007 | |
| PAK | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 80 | Griffin et al., 2007 | |
| AHY | 1q/4 | 2 | 3 | 1p | 5 | 6q | 6q | 7p | 7q | 8q | 9q | 70 | Neotropical | Furo et al., 2015a |
| ACH | 1q/4 | 2/11 | 3 | 1p | 5q | 6q | 6q | 7pq | 7q | 8q/10q | 9 | 70 | Neotropical | Furo et al., 2015a |
| PFR | 1q/4 | 2 | 3 | 1p | 5q | 6q | 6q | 7 | 8 | 10 | 9 | 70 | Neotropical | Furo et al., 2018 |
| AMA | 1q/4/9q | 2 | 3 | 1p | 5q | 6q | 6q | 7pq | 7q | 8q | 70 | Neotropical | Seabury et al., 2013 | |
| MMO | 3q,4p/q,12,14q | 2,3p,13q | 5,7 | 1q | 6 | 9q,10q | 9q,10q | 8p | 8q | 1p/7q | 9p,11 | 48 | Neotropical | Present work |
| AAE | 2/5q | 1/12 | 3 | 4q | 6 | 7q | 7q | 11 | 8 | 10 | 9p | 70 | Neotropical | Furo et al., 2018 |
| PER | 1q/4 | 2 | 3 | 4q | 5q | 6q | 6q | 7q | 7q | micro | 70 | African | Furo et al., 2018 | |
| ARO | 3/4q | 2/9q | 1 | 7 | 8q | 6q | 6q | 5q | 5q/9q | 4p | 10 | 48 | African | Nanda et al., 2007 |
| MUN | 3/6 | 1 | 2 | 7 | 4q | 4p/8p | 4p | 5pq | 5q | 5p | 9q | 62 | Australia | Nanda et al., 2007 |
| NHO | 3/6 | 1 | 2 | 4 | 7q | 5 | 5 | 4p | 4p/10 | 11 | 9 | 72 | Australia | Nanda et al., 2007 |
GGA (Gallus gallus), PAK (Putative ancestral Karyotype), AHY (Anodorhynchus hyacinthinus), ACH (Ara chloropterus), PRF (Pyrrhura frontalis), AMA (Ara macao), MMO (Myiopsitta monachus), AAE (Amazona aestiva), PER (Psittacus erithacus), ARO (Agapornis roseicollis), MUN (Melopsittacus undulatus), NHO (Nymphicus hollandicus).
In an attempt to improve the phylogenetic tree, the conflicting character corresponding to the fusions between PAK4 (GGA4p) with microchomosomes – shared by ACH, AMA, and AAE – was removed from the binary matrix. This feature could support the relationship between AMA and ACH, however, only analysis with chicken BACs from microchomosomes could clarify whether these fusions involve the same microchromosomes. The use of chicken BACs in different species would help to improve support of the different branches in the phylogenetic tree.
Despite the high number of chromosomal rearrangements, most of them corresponded to autapomorphisms (Supplementary Table S1, Characters – 8–12, 17–20, 30,31). Because of the low number of informative chromosomal characters, chromosome painting data did not throw any light on the time of divergence of the species analyzed. However, consideration of our phylogenetic proposal and comparisons with previously published data, led us to assume that the common ancestor of the clade of M. monachus and phylogenetically related genera (for example, Amazona) could have been originated in the Eocene around 48 Mya. This was a period of major changes in South America, including drastic changes in temperature due to the separation of Australia-Antarctica caused by continental drift, which changed the fauna and flora scenario around 32 Mya. The ancestor of M. monachus would have appeared during the separation of South America and Antarctica (Tavares et al., 2006).
Significance of Whole Chromosome Painting With MMO Probes
Interchromosomal rearrangements have played an important role during the karyotype evolution of the Psittaciformes (Nanda et al., 2007; Seabury et al., 2013; Furo et al., 2015a, 2018). As mentioned before, M. monachus showed several fissions involving pairs corresponding to PAK1 (GGA1), PAK2 (GGA2), PAK3 (GGA3), PAK6 (GGA6), PAK7 (GGA7), e PAK10 (GGA4p). In this context, this species becomes interesting from the cytogenetic point of view. Hence, with the analysis of LAL probes in the metaphases of MMO, it was possible to check many breakpoints that were not recurrent. Furthermore, the utilization of these probes onto other species of Psittaciformes will be very useful for genomic comparisons, because the signals of hybridization are more evident due to phylogenetic proximity.
MMO probes were used in metaphases of A.macao and produced good signals which showed a fusion between MMO10 (microchromosome) with PAK5 (GGA5). Generally, the GGA5 probe does not hybridize to the entire length of particular chromosome pair in macaws and parrots (Furo et al., 2015a, 2018) and a gap was always observed, leading authors to suppose it corresponded to a fusion with a microchromosome. It would be interesting to use the same probes in other species of Neotropical Psittacidae to check for the same association.
Conclusion
The description of the M. monachus karyotype using different sets of chromosome probes showed that, although most species of Neotropical Psittacidae have a constant diploid chromosome number of 2n = 70 and a similar karyotype in terms of uni- and biarmed elements, this species shows high rate of karyotype repatterning, with 2n being reduced to 48 chromosomes. M. monachus is the first to have its karyotype described in detail, with probes corresponding to macro and microchromosomes, revealing many inversions and fissions, which together with no-reciprocal translocations and tandem fusions represent the main karyotype rearrangements in Psittaciformes. Additionally, although this species has a long tail, our phylogenetic analysis placed it in a basal position together with A. aestiva (short tail), mainly due the low number of chromosome synapomorphies, as most of the numerous rearrangements corresponded to autapomorphies. Thus, our results corroborate previous studies performed by mDNA and nuclear sequences. Moreover, we have shown that the MMO probes are useful tools in the analysis of evolutionary chromosome rearrangements, because they reveal novel breakpoints previously undescribed in the literature.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by CEUA-Universidade Federal do Pará, no. 170/2013.
Author Contributions
IF and EO: conceptualization, validation, and writing (original draft). IF, RK, AG, and EO: data curation and formal analysis. IF, RK, AG, RO’C, AG, and RG: investigation. IF, PO’B, and JP: methodology. EO: project administration. MF-S, RG, AG, and EO: funding acquisition. PO’B and MF-S: writing (review and editing). All authors corrected, revised, and discussed the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Parque Zoobotânico do Museu Paraense Emilio Goeldi (Belém, PA, Brazil) and Parque Zoológico do Rio Grande do Sul (Sapucaia do Sul, RS, Brazil) for samples used in this study. We are very grateful to CNPq (309699/2015-0), PROPESP-UFPA, and CAPES for technical and financial support. Finally, we would like to thank Mr. Alex Araújo and Rafael Nascimento for artwork in Figures 9, 10. We are thankful to PROPESP-UFPA (Edital 1/2020-PAPQ) for financial support.
Footnotes
Funding. This research was partially funded by a grant to EO from CNPq (307382/2019-2) and to MF-S from the Welcome Trust in support of the Cambridge Resource Center for Comparative Genomics, by the Biotechnology and Biological Sciences Research Council (BB/K008161/1) to the University of Kent, and by the project UIDB/CVT/00772/2020 funded by the Fundação para a Ciência e Tecnologia (FCT).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00721/full#supplementary-material
References
- Caparroz R., Duarte J. M. B. (2004). Chromosomal similarity between the scaly-headed parrot (Pionus maximiliani), the Short-tailed parrot (Graydidascalus brachyurus) and the yellow-faced parrot (Salvatoria xanthops) (Psittaciformes: Aves): a cytotaxonomic analysis. Genet. Mol. Biol. 27 522–528. 10.1590/S1415-47572004000400010 [DOI] [Google Scholar]
- de Lucca E. J., Marco D. A. (2014). Chromosomal polymorphism in Forpus Xanthopterygius (Psittaciformes: Aves). Caryologia 36 355–361. 10.1080/00087114.1983.10797676 [DOI] [Google Scholar]
- de Lucca E. J., Shirley L. R., Lanier C. (1991). Karyotype studies in twenty-two species of parrots (Psittaciformes: Aves). Brazil. J. Genet. 14 73–98. [Google Scholar]
- de Oliveira E. H. C., Habermann F., Lacerda O., Sbalqueiro I. J., Wienberg J., Muller E. S. (2005). Chromosome reshuffling in birds of prey: the karyotypes of the world’s largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus). Chromosoma 114 338–343. 10.1007/s00412-005-0009-5 [DOI] [PubMed] [Google Scholar]
- de Oliveira E. H. C., Tagliarini M. M., Rissino J. D., Pieczarka J. C., Nagamachi C. Y., O’Brien P. C. M., et al. (2010). Reciprocal chromosome painting between white hawk (Leucopternis albicollis) and chicken reveals extensive fusions and fissions during karyotype evolution of Accipitridae (Aves, Falconiformes). Chromosom. Res. 18 349–355. 10.1007/s10577-010-9117-z [DOI] [PubMed] [Google Scholar]
- Francisco M. R., Galetti J. P. M. (2001). Cytotaxonomic considerations on neotropical psittacidae birds and description of three new karyotypes. Hereditas 134 225–228. 10.1111/j.1601-5223.2001.00225.x [DOI] [PubMed] [Google Scholar]
- Furo I. O., Kretschmer R., dos Santos M. S., Carvalho C. A. L., Gunski R. J., O’Brien P. C. M., et al. (2017). Chromosomal mapping of repetitive DNAs in Myiopsitta monachus and Amazona aestiva (Psittaciformes, Psittacidae: Psittaciformes), with emphasis on the sex chromosomes. Cytogenet. Genom. Res. 151 151–160. 10.1159/000464458 [DOI] [PubMed] [Google Scholar]
- Furo I. O., Kretschmer R., O’Brien P. C. M., Ferguson-Smith M. A., de Oliveira E. H. C. (2015a). Chromosomal diversity and karyotype evolution in South American macaws (Psittaciformes, Psittacidae). PLoS One 10:e0130157. 10.1371/journal.pone.0130157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furo I. O., Monte A. A., dos Santos M. S., Tagliarini M. M., O’Brien P. C. M., Ferguson-Smith M. A., et al. (2015b). Cytotaxonomy of Eurypyga helias (Gruiformes, Eurypygidae): first karyotypic description and phylogenetic proximity with Rynochetidae. PLoS One 10:e0143982. 10.1371/journal.pone.0143982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furo I. O., Kretschmer R., O’Brien P. C. M., Pereira J. C., Garnero A. D. V., Gunski R. J., et al. (2018). Chromosome painting in neotropical long- and short-tailed parrots (Aves, Psittaciformes): phylogeny and proposal for a putative ancestral karyotype for tribe Arini. Genes 9:491. 10.3390/genes9100491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. K., Robertson L. B., Tempest H. G., Skinner B. M. (2007). The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genom. Res. 117 64–77. 10.1159/000103166 [DOI] [PubMed] [Google Scholar]
- Guttenbach M., Nanda I., Feichtinger W., Masabanda J. S., Griffin D. K., Schmid M. (2003). Comparative chromosome painting of chicken autosomal paints 1-9 in nine different bird species. Cytogenet Genom. Res. 103 173–184. 10.1159/000076309 [DOI] [PubMed] [Google Scholar]
- Higdon P. (1998). The Quaker Parrot: An Owners Guide To A Healthy Pet. Boston, MA: IDG. [Google Scholar]
- Kretschmer R., de Oliveira E. H. C., dos Santos M. S., Furo I. O., O’Brien P. C. M., Ferguson-Smith M. A., et al. (2015). Chromosome mapping of the large elaenia (Elaenia spectabilis): evidence for a cytogenetic signature for passeriform birds? Biol. J. Linn. Soc. 115 391–398. 10.1111/bij.12504 [DOI] [Google Scholar]
- Kretschmer R., Ferguson-Smith M. A., de Oliveira E. H. C. (2018a). Karyotype evolution in birds: from conventional staining to chromosome painting. Genes 9:181. 10.3390/genes9040181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer R., Furo I. O., Gunski R. J., Garnero A. D. V., Pereira J., O’Brien P. C. M., et al. (2018b). Comparative chromosome painting in Columbidae (Columbiformes) reinforces divergence in Passerea and Columbea. Chromosom. Res. 26 211–223. 10.1007/s10577-018-9580-5 [DOI] [PubMed] [Google Scholar]
- Kretschmer R., Gunski R. J., Garnero A. D. V., Furo I. O., O’Brien P. C. M., Ferguson-Smith M. A., et al. (2014). Molecular cytogenetic characterization of multiple intrachromosomal rearrangements in two representatives of the genus Turdus (Turdidae, Passeriformes). PLoS One 9:e103338. 10.1371/journal.pone.0103338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki C. Y., Duarte J. M. B., Caparroz R., Nunes A. L. V., Wajntal A. (1997). Sex identification of new world parrots (Psittacidae, Aves) using the human minisatellite probe 33.15. Auk 114 516–520. 10.2307/4089258 [DOI] [Google Scholar]
- Miyaki C. Y., Matioli S. R., Burke T., Wajntal A. (1998). Parrot evolution and paleogeographical events: mitochondrial DNA evidence. Mol. Biol. Evol. 15 544–551. 10.1093/oxfordjournals.molbev.a025954 [DOI] [Google Scholar]
- Montón M. (1977). Algunos aspectos significativos en la adaptación de los psitácidos de Sudamérica. Rev. Parque Zool. Barc. 30 19–22. [Google Scholar]
- Nanda I., Karl E., Griffin D. K., Schartl M., Schmid M. (2007). Chromosome repatterning in three representative parrots (Psittaciformes) inferred from comparative chromosome painting. Cytogenet. Genom. Res. 117 43–53. 10.1159/000103164 [DOI] [PubMed] [Google Scholar]
- Nie W., O’Brien P. C. M., Fu B., Wang J., Su W., He K., et al. (2015). Multidirectional chromosome painting substantiates the occurrence of extensive genomic reshuffling within Accipitriformes. BMC Evol. Biol. 15:205. 10.1186/s12862-015-0484-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W., O’Brien P. C. M., Ng B. L., Fu B., Volobouev V., Carter N. P., et al. (2009). Avian comparative genomics: reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes) - An atypical species with low diploid number. Chromosom. Res. 17 99–113. 10.1007/s10577-009-9021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor R. E., Kiazin L., Skinner B., Fonseka G., Joseph S., Jennings R., et al. (2018). Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 128 21–29. 10.1007/s00412-018-0685-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheindt F. E., Christidis L., Kuhn S., De Kloet S., Norman J. A., Fidler A. (2014). The timing of diversi? cation within the most divergent parrot clade. J. Avian Biol. 45 140–148. 10.1111/j.1600-048X.2013.00200.x [DOI] [Google Scholar]
- Rocchi M., Archidiacono N., Schempp W., Capozzi O., Stanyon R. (2012). Centromere repositioning in mammals. Heredity 108 59–67. 10.1038/hdy.2011.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Ikeuchi T., Maino S. (1968). A feather pulp culture for avian chromosomes with notes on the chromosomes of the peafowl and the ostrich. Experientia 24 1923–1929. 10.1007/BF02146680 [DOI] [PubMed] [Google Scholar]
- Seabury C. M., Dowd S. E., Seabury P. M., Raudsepp T., Brightsmith D. J., Liboriussen P., et al. (2013). A multi-platform draft de novo genome assembly and comparative analysis for the scarlet macaw (Ara macao). PLoS One 8:e62415. 10.1371/journal.pone.0062415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sick H. (1997). Ornitologia Brasileira. Rio de Janeiro: Editora Nova Fronteira. [Google Scholar]
- Tavares E. S., Baker A. J., Pereira S. L., Miyaki C. Y. (2006). Phylogenetic relationships and historical biogeography of neotropical parrots (Psittaciformes: Psittacidae: Arini) inferred from mitochondrial and nuclear DNA sequences. Soc. Syst. Biol. 55 454–470. 10.1080/10635150600697390 [DOI] [PubMed] [Google Scholar]
- Telenius H., Pelmear A. H., Tunnacliffe A., Carter N. P., Behmel A., Ferguson-Smith M. A., et al. (1992). Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosom. Cancer 4 257–263. 10.1002/gcc.2870040311 [DOI] [PubMed] [Google Scholar]
- Winkler D. W., Billerman S. W., Lovette I. J. (2015). Bird Families of the World. : An invitation to the Spectacular Diversity of Birds. Barcelona: Linx Edicions. [Google Scholar]
- Wright T. F., Schirtzinger E. E., Matsumoto T., Eberhard J. R., Graves G. R., Sanchez J. J., et al. (2008). A multilocus molecular phylogeny of the parrots (Psittaciformes): support for a gondwanan origin during the cretaceous. Mol. Biol. Evol. 25 2141–2156. 10.1093/molbev/msn160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., O’Brien P. C. M., Milne B. S., Graphodatsky A. S., Solanky N., Trifonov V., et al. (1999). A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62 189–202. 10.1006/geno.1999.5989 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.