Sir,
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has, as on March 31, 2020, spread to over 207 countries around the world1,2, with a total of 896,475 confirmed cases and 45,525 deaths2. The number of reported SARS-CoV-2 cases in India is also on an increase with 1,636 cases and 38 deaths2. In the current pandemic situation, the isolation of SARS-CoV-2 is important for developing and evaluating diagnostic reagents, for antiviral studies and for screening of vaccine candidates. Earlier studies showed that SARS-CoV-2 could not replicate in several cell lines, which are routinely used for isolation of respiratory viruses3. Human and animal cell lines that were found to support SARS-CoV-1 replication during the first outbreak of SARS in China, 20024, are currently being studied. The virus was first isolated in the human airway epithelial cells from clinical specimens as part of early attempts to identify the aetiologic agent of infection5. We describe here the successful isolation and characterization of SARS-CoV-2 from clinical samples in India using Vero CCL-81 cells by observing cytopathic effects (CPEs) and cycle threshold (Ct) values in real-time reverse transcription-polymerase chain reaction (RT-PCR), electron microscopy and next-generation sequencing (NGS).
The first three SARS-CoV-2 cases were reported from Kerala during January 27-31, 2020. Later during March 2020, cases were also reported from a group of Italian tourists (n=15) and their contacts in New Delhi, India. Simultaneously, cases were reported in Agra, Uttar Pradesh, which was the outcome of close contact of an infected Delhi-based individual who returned from Italy. The designated COVID-19 testing laboratories of Virus Research Diagnostic Laboratory network (All India Institute of Medical Sciences, New Delhi; Sawai Man Singh Medical College, Jaipur; and King George's Medical University, Lucknow) referred the specimens (throat swab/nasal swab, oropharyngeal swab/sputum) to the Indian Council of Medical Research-National Institute of Virology (ICMR-NIV), Pune, after screening for envelope (E) gene by real-time RT-PCR was done6. A total of 12 SARS-CoV-2 positive specimens having a Ct <30 for the E gene were included in the study. Of these, eight samples were from positive cases of Italian tourists and their contacts in New Delhi. The rest of the specimens were from four positive cases at Agra, Uttar Pradesh, and the close contact cases of an infected Delhi-based individual who returned from Italy.
The clinical specimens of the 12 cases were used for infecting Vero CCL-81 which was maintained in Eagle's minimum essential medium (MEM; Gibco, UK) supplemented with 10 per cent foetal bovine serum (FBS) (HiMedia, Mumbai), penicillin (100 U/ml) and streptomycin (100 mg/ml). Likewise, 100 μl was inoculated onto 24-well cell culture monolayers of Vero CCL-81, before growth medium was decanted. The cells were incubated for one hour at 37°C to allow virus adsorption, with rocking every 10 min for uniform virus distribution. After the incubation, the inoculum specimen was removed and the cells were washed with 1X phosphate-buffered saline (PBS). The MEM supplemented with two per cent FBS was added to each well. The cultures were incubated further in five per cent CO2 incubator at 37°C and observed daily for CPEs under an inverted microscope (Nikon, Eclipse Ti, Japan). Cellular morphological changes were recorded using a camera (Nikon, Japan). From each well of cell culture plate, on the third post-infection day (PID-3) of passage-1 (P-1), 50 μl of supernatant was taken and tested for SARS-CoV-2 using real-time RT-PCR for E and RNA-dependent RNA polymerase (RdRp) (2) genes as described earlier7,8. Similar testing was repeated on the cell supernatant of passage-2(P-2) at PID-4 for observing viral copy number. Cultures that showed CPE on PID-4 were centrifuged at 4815 × g for 10 min at 4°C; the supernatants were processed immediately or stored at −86°C. Further, those that showed CPE were grown in T-25 cm2 flasks at P-2 and titration was done after serial dilution. Tissue culture infective dose 50 per cent (TCID50) values were calculated by the Reed and Muench method9. CPEs were observed in 9 of 12 cultures in the P-1. The TCID50 values ranged from 105.5 to 106.4/ml for the different clinical specimens passaged in Vero CCL-81 at P-2. The cells were examined microscopically for cellular morphological changes following inoculation.
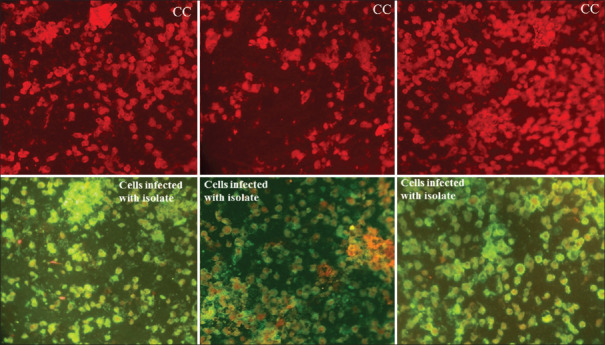
Vero CCL-81 cells infected with SARS-CoV-2 strain NIV-2020-770 and uninfected cells (CC) were transferred onto microcavity slides and fixed with acetone. Serum samples (1:25 dilution) from the confirmed COVID-19 cases (POD nCOV-S11, nCOV-S13 and nCOV-S7) and negative serum samples were added followed by incubation at 37°C for 1.5 h10. Antibody reactivity was visualized using anti-human immunoglobulin fluorescein-isothiocynate. In immunofluorescence assay of COVID-19 positive patients, three serum samples exhibited specific reactivity against SARS-CoV-2 virus isolate (Fig. 1).
Fig. 1.
Immunofluorescence images (red panel) showing uninfected Vero CCL-81 cells probed by positive patient serum samples after post infection day of 13th (left), 11th (middle) and seventh (right) and with SARS-CoV-2 strain NIV-2020-770 infected Vero CCL-81 cells probed by positive patients serum (green panel) showing the reactivity of virus and antibody.
Vero CCL-81 cells that were inoculated with the samples showed evidence of cell rounding and detachment from 9 of 12 clinical samples in P-1 at PID-4. Syncytial cells formed large cell masses that increased in size and number as the infection progressed. Enhanced CPE was noted in P-2 at PID-2. The cells were detached from the tissue culture plate surfaces by PID-3. Similar cellular morphological changes were observed after passaging of the above nine samples up to P-2. No cellular changes were observed in the cell control during both passages. Figure 2 depicts the day-wise changes during the passage of a representative clinical isolate (NIV-2020-770). Virus replication was confirmed using real-time RT-PCR with RNA extracted from the cell culture medium on PID-3. The Ct values ranged from 9.79 to 15.41 (in Vero CCL-81 cells) for the isolates at P-2, which were lower than the Ct values of 16-25.1 in the clinical samples (Table I). The number of virus copies in the isolates at P-1 in Vero CCL-81 cells ranged from 5.18×107 to 8.12×108 copy/ml and increased 1-26 fold to a range of 1.69×108 to 6.77×109 in the cell culture supernatants at P-2 (Table I).
Fig. 2.
Cytopathic effect of the SARS-CoV-2 isolate (NIV-2020-770) demonstrated in Vero CCL-81 cells on different post-infection days (PID).
Table I.
Cycle threshold (Ct) of SARS-CoV-2 positive clinical specimens and respective viral copy number in isolates in different passages for two different cell culture types using real-time reverse transcription-polymerase chain reaction (RT-PCR). E gene was targeted in all
| Serial number | Sample ID | Isolate ID | Ct (copy number) of viral RNA in real-time RT-PCR | ||
|---|---|---|---|---|---|
| Original (clinical) samples by qRTPCR (Ct) | Vero CCL-81 passage-1 Ct (copy number) | Vero CCL-81 passage-2 Ct (copy number) | |||
| 1 | nCoV-763 | NIV-2020-763 | 18.07 | 10.56 (4.08×109) | 11.14 (2.77×109) |
| 2 | nCoV-770 | NIV-2020-770 | 18 | 15.15 (1.96×108) | 11.62 (2.02×109) |
| 3 | nCoV-772 | NIV-2020-772 | 20.2 | 14.00 (4.18×108) | 10.93 (3.19×109) |
| 4 | nCoV-773 | NIV-2020-773 | 25.1 | 17.15 (5.18×107) | 15.41 (1.69×108) |
| 5 | nCoV-781 | NIV-2020-781 | 22.1 | 14.91 (2.27×108) | 10.0 (5.91×109) |
| 6 | nCoV-C132 | NIV-2020-C132 | 16 | 13.68 (5.12×108) | 10.73 (3.64×109) |
| 7 | nCoV-777 | NIV-2020-777 | 23.3 | 13.31 (6.57×108) | 9.99 (5.92×109) |
| 8 | nCoV-C31 | NIV-2020-C31 | 25 | 12.99 (8.12×108) | 9.79 (6.77×109) |
| 9 | nCoV-C32 | NIV-2020-C32 | 16 | 13.21 (7.01×108) | 10.25 (5.05×109) |
Serial numbers 1-7: Italian tourists who arrived in Delhi, India and an Indian contact of the cohort; Serial numbers 8-9: Close contacts in Agra, Uttar Pradesh, of an infected Delhi-based person who returned from Italy. qRT-PCR, quantitative RT-PCR
On PID-4, enhanced CPE was observed. The P-1 material was reinoculated in a new batch of cells, and it showed progressive enhancement of CPE as observed day-wise. Further, an aliquot of cell culture supernatant was harvested from infected Vero CCL-81 showing CPE and the supernatant used for negative staining as described elsewhere11,12. Distinct CoV particles with an average size of 95±10 nm having a distinct envelope fringe could be detected in the fields scanned (Fig. 3), as observed earlier13.
Fig. 3.
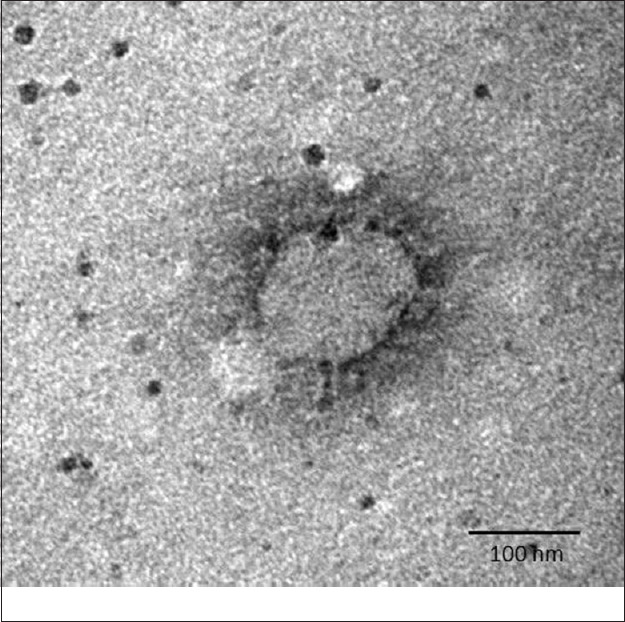
Transmission electron microscopy imaging of SARS-CoV-2. A negative-stained SARS-CoV-2 viral particle, demonstrating spike morphology of glycoprotein along with peplomeric projections, a feature typical to the family Coronaviridae, is seen.
Next-generation sequencing was performed on SARS-CoV-2 positive clinical samples (100 μl) included in the study and the tissue culture fluid (50 μl) of virus isolates at PID-3 as described earlier14,15. Reference-based mapping as implemented in the CLC genomics workbench 11.0 (CLC, Qiagen) was used to retrieve the sequence of the SARS-CoV-2. BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) identification of the viral genome sequences retrieved from the clinical samples and their isolates had 99.98 per cent identity with the SARS-CoV-2 isolate Wuhan-Hu-1 (Accession No. NC_045512). Details of the sequences obtained including the per cent of the reads mapped, total reads and the per cent of genome coverage recovered for the clinical samples and the isolates are provided in Table II. Partial sequences were retrieved from the clinical samples (nCoV-C 132 and nCoV-C 31) and were not included in the analysis.
Table II.
Per cent of the reads mapped, total reads and the per cent of genome coverage recovered for the clinical samples and the isolates
| Sample type | Sample/isolate details | Total reads | Per cent of reads mapped | Per cent of genome recovered | Position of nucleotide in genome[17] | |
|---|---|---|---|---|---|---|
| 8782 | 28144 | |||||
| Isolate | NIV-2020-763 | 10,054,258 | 94.8 | 100 | C | T |
| NIV-2020-770 | 4,384,130 | 99.0 | 100 | C | T | |
| NIV-2020-772 | 3,482,648 | 98.4 | 99.9 | C | T | |
| NIV-2020-773 | 5,952,758 | 94.2 | 99.9 | C | T | |
| NIV-2020-777 | 3,949,748 | 98.7 | 100 | C | T | |
| NIV-2020-781 | 2,226,464 | 91.6 | 99.9 | C | T | |
| NIV-2020-C32 | 4,159,878 | 99.0 | 100 | C | T | |
| Clinical sample | nCoV-763 | 8,721,610 | 84.9 | 99.9 | T | T |
| nCoV-770 | 5,197,614 | 93.1 | 99.9 | T | T | |
| nCoV-772 | 4,222,912 | 81.7 | 99.8 | C | T | |
| nCoV-773 | 9,951,190 | 19.98 | 99.8 | C | T | |
| nCoV-777 | 8,808,756 | 26.93 | 99.8 | C | T | |
| nCoV-781 | 15,688,460 | 35.5 | 99.9 | C | T | |
| nCoV-C32 | 2,772,158 | 88.5 | 100 | C | T | |
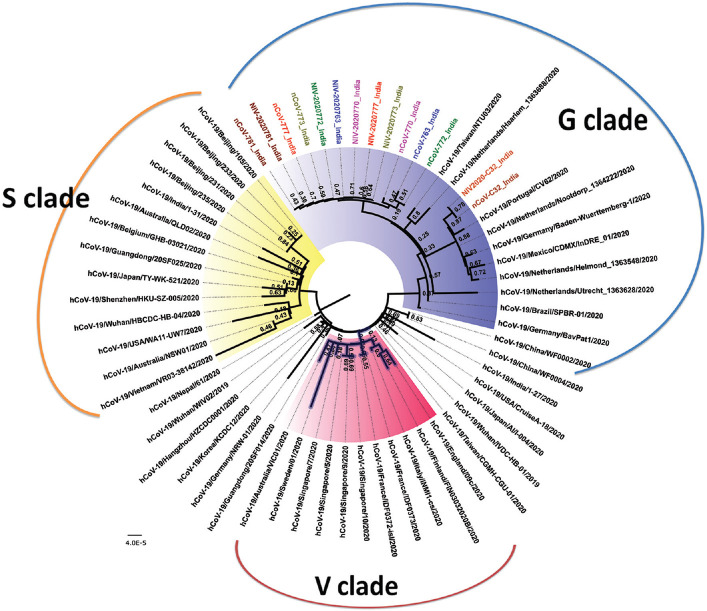
MEGA software version 7.0.1116 was used for the multiple alignments of the sequences retrieved in this study and the sequences from the Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org/) (Supplementary Table (1.6MB, pdf) (available from http://www.ijmr.org.in/articles/2020/151/2/images/IndianJMedRes_2020_151_2_244_282559_sm7.pdf)). A neighbour-joining tree was generated using the best substitution model (Kimura 2-parameter model) with a bootstrap of 1000 replicates. As per Tang et al17, the circulating SARS-CoV-2 can be grouped into two types (S and L type) based on the two different single-nucleotide polymorphisms (SNPs) at positions 8782 and 28144 in the genome. The S type possesses TC SNPs while the L type possesses CT SNPs at positions 8782 and 28144, respectively. In the present study, it was observed that two sequences from clinical samples (nCoV-763 and nCoV-770) had TT SNPs, while the other sequences had CT as the SNP (L type) (Table II). The TT SNPs have been observed in few of the GISAID sequences, including one of the Kerala genome sequences (nCoV-19/India/31 January 2020) submitted by us earlier. All the isolates of the clinical samples were of L type. Specific amino acid mutations in the nsp3 region, spike protein and ORF8, in general, lead to the formation of V, G and S genetic variants/clades, respectively, as per the recent classification followed by GISAID. It was observed that the clinical samples, as well as the isolates, had the mutation D614G in the spike protein, classifying the study samples and isolates into the G clade (Table II and Fig. 4). No specific substitutions were observed in any of the isolate sequences with respect to the corresponding clinical sample sequences, as these were sequences from a low passage. The sequences of the clinical samples and the isolate from the contact of the infected Delhi-based individual, who returned from Italy, further showed two mutations, R203K and G204R in the nucleocapsid protein (N). Although all strains demonstrated 99.6 per cent identity with the original Wuhan Hu-1 sequence, the role of unique SNPs and mutations in identifying the source of infection needs to be explored.
Fig. 4.
Neighbour-joining tree of SARS-CoV-2. The phylogenetic tree is generated using the best substitution model. A bootstrap of 1000 replicates was used to assess the statistical robustness of the tree. Same colours are used for sequences derived from a clinical sample and the respective isolate. Clinical samples are labelled with initials as nCoV while the isolates are labelled with initials as NIV. The clades are represented by different colours in the core region (S - yellow, V - pink, G - blue and unclassified - not coloured).
After the first isolation of the virus in the human airway epithelial cells reported by China5, countries such as Australia18, Korea19, Germany20 and the USA21 have also isolated the SARS-CoV-2 strain. In India, initial attempts to isolate the virus from the first three cases did not succeed due to low titres in the clinical specimens. This is the first successful virus isolation of SARS-CoV-2 in the Vero CCL-81 cells in India from nasal and throat swabs of persons with a travel history from Italy and their contacts. Isolation of SARS-CoV-2 from clinical samples will be helpful to address key questions of correlating the differential cell line susceptibility and viral replication efficiency, especially important for clinical samples with low viral titres. Isolation of the virus in such a pandemic situation would help to develop indigenously designed reagents such as positive controls, virus antigen and antibodies, which could lead to the indigenous development of sero-diagnostic assays. These assays would be critical for conducting population-based serosurveys. Propagation in culture will also facilitate antiviral susceptibility studies and vaccine efforts in India.
Acknowledgement for the list of the sequences downloaded from GISAID database that were used in the study
Footnotes
Supplementary material available from http://www.ijmr.org.in/article.asp?issn=0971-5916;year=2020;volume=151;issue=2;spage=244;epage=250;aulast=Sarkale
Financial support & sponsorship: Financial support was provided by the Department of Health Research, Ministry of Health & Family Welfare, New Delhi, at ICMR-National Institute of Virology, Pune
Conflicts of Interest: None.
References
- 1.Worldometer. COVID-19 coronavirus pandemic. [accessed on March 29, 2020]. Available from: https://wwwworldometersinfo/coronavirus/
- 2.World Health Organization. Coronavirus disease (COVID-2019) situation reports. WHO; 2020. [accessed on March 29, 2020]. Available from: https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/situation-reports . [Google Scholar]
- 3.Kaye M, Druce J, Tran T, Kostecki R, Chibo D, Morris J, et al. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–33. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillim-Ross L, Taylor J, Scholl DR, Ridenour J, Masters PS, Wentworth DE. Discovery of novel human and animal cells infected by the severe acute respiratory syndrome coronavirus by replication-specific multiplex reverse transcription-PCR. J Clin Microbiol. 2004;42:3196–206. doi: 10.1128/JCM.42.7.3196-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. WHO; 2020. [accessed on February 9, 2020]. Available from: https://wwwwhoint/eme gencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance . [Google Scholar]
- 7.Yadav PD, Potdar VA, Choudhary ML, Nyayanit DA, Agrawal M, Jadhav SM, et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020 doi: 10.4103/ijmr.IJMR_663_20. doi: 104103/ijmrIJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary ML, Vipat V, Jadhav S, Basu A, Cherian S, Abraham P, et al. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020 doi: 10.4103/ijmr.IJMR_671_20. doi: 104103/ijmrIJMR_671_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epi emiol. 1938;27:493–7. [Google Scholar]
- 10.Haveri A, Smura T, Kuivanen S, Österlund P, Hepojoki J, Ikonen N, et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Euro Surveill. 2020;25:2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangodkar S, Jain P, Dixit N, Ghosh K, Basu A. Dengue virus-induced autophagosomes and changes in endomembrane ultrastructure imaged by electron tomography and whole-mount grid-cell culture techniques. J Electron Microsc (Tokyo) 2010;59:503–11. doi: 10.1093/jmicro/dfq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner S, Horne RW. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta. 1959;34:103–10. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- 13.Prasad S, Potdar V, Cherian S, Abraham P, Basu A. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020 doi: 10.4103/ijmr.IJMR_577_20. doi: 104103/ijmrIJMR_577_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav PD, Albariño CG, Nyayanit DA, Guerrero L, Jenks MH, Sarkale P, et al. Equine encephalosis virus in India, 2008. Emerg Infect Dis. 2018;24:898–901. doi: 10.3201/eid2405.171844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav PD, Nyayanit DA, Shete AM, Jain S, Majumdar TP, Chaubal GY, et al. Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick Borne Dis. 2019;10:23–33. doi: 10.1016/j.ttbdis.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020:nwaa036. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caly L, Druce J, Roberts J, Bond K, Tran T, Kostecki R, et al. Isolation and rapid sharing of the 2019 novel coronavirus (SAR-CoV-2) from the first diagnosis of COVID-19 in Australia. Med J Aust. 2020 doi: 10.5694/mja2.50569. doi: 105694/mja250569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Chung YS, Jo HJ, Lee NJ, Kim MS, Woo SH, et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, Bojkova D, et al. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020;382:1278–80. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Severe acute respiratory syndrome coronavirus 2 from patient with 2019 novel coronavirus disease. United States Emerg Infect Dis. 2020:26. doi: 10.3201/eid2606.200516. doi: 103201/eid2606200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Acknowledgement for the list of the sequences downloaded from GISAID database that were used in the study