Abstract
Human milk oligosaccharides (HMOs) are a mixture of structurally diverse carbohydrates that contribute to shape a healthy gut microbiota composition. The great diversity of the HMOs structures does not allow the attribution of specific prebiotic characteristics to single milk oligosaccharides. We analyze here the utilization of four disaccharides, lacto-N-biose (LNB), galacto-N-biose (GNB), fucosyl-α1,3-GlcNAc (3FN) and fucosyl-α1,6-GlcNAc (6FN), that form part of HMOs and glycoprotein structures, by the infant fecal microbiota. LNB significantly increased the total levels of bifidobacteria and the species Bifidobacterium breve and Bifidobacterium bifidum. The Lactobacillus genus levels were increased by 3FN fermentation and B. breve by GNB and 3FN. There was a significant reduction of Blautia coccoides group with LNB and 3FN. In addition, 6FN significantly reduced the levels of Enterobacteriaceae family members. Significantly higher concentrations of lactate, formate and acetate were produced in cultures containing either LNB or GNB in comparison with control cultures. Additionally, after fermentation of the oligosaccharides by the fecal microbiota, several Bifidobacterium strains were isolated and identified. The results presented here indicated that each, LNB, GNB and 3FN disaccharide, might have a specific beneficial effect in the infant gut microbiota and they are potential prebiotics for application in infant foods.
Subject terms: Microbiology, Molecular biology
Introduction
Studies using in vitro analysis have shown that human milk unconjugated oligosaccharides (HMOs) and the glycan moiety of glycoproteins are effective in selectively promoting the growth of beneficial bacteria1–3. Bifidobacterium longum, Bifidobacterium bifidum, Bifidobacterium breve and Bifidobacterium catenulatum group are frequently isolated from breast-fed infant feces4,5. Some species of Lactobacillus, including Lactobacillus reuteri, Lactobacillus casei and Lactobacillus rhamnosus are also constituents of the infant gastrointestinal tract6,7. Among the bifidobacteria isolated from infant feces, there are important differences in the fermentation patterns of HMOs, which have more than two hundred distinct structures8–10, and this opens the possibility to accurately modulate the gut microbiota with specific HMOs11. To date only a few analyses have determined the physiological impact of individual HMOs on the infant gut microbiota. Thus, an in vitro fermentation system with stool samples from formula-fed babies demonstrated that lacto-N-biose (LNB), the main building block of type-1 HMOs, increased the total bifidobacterial population12. Especially B. bifidum numbers were highly induced in comparison to lactulose, raffinose, GOS or mannanoligosaccharides12. Lacto-N-neotetraose and 2′-fucosyllactose (2′FL) were fermented at a higher rate than 6′-sialyllactose in in vitro mixed culture systems by microbiota from breast or formula fed infants, but none of them showed an important effect in the number of bifidobacteria13. Total HMOs purified from human milk increased the number of Bifidobacterium spp. while the number of Escherichia spp. and Clostridium perfringens diminished during in vitro fermentation of infant microbiota1. In these cultures, most of the 2′FL, 3-fucosyllactose (3FL) and lactodifucotetraose (LDFT) from the HMOs supplement were consumed, suggesting that some specific oligosaccharides may be responsible of the prebiotic effect attributed to HMOs. A few studies using in vitro colon models14 and in vivo animal models also underline the ability of individual HMOs to modify the composition of the gastrointestinal microbiota15,16.
HMOs are present at high concentrations in human milk but not in infant formulas17. We have previously synthesized fucosyl- α-1,3-N-acetylglucosamine (3FN) and fucosyl-α-1,6-N-acetylglucosamine (6FN) using the transglycosylation activity of α-L-fucosidases18. 3FN forms part of HMOs2 and 6FN of N-glycosylated proteins, including mucins19. We have also produced LNB and galacto-N-biose (GNB), the core type-1 sugar from mucins, using the transgalactosylation activity of the β-galactosidase GnbG20. The synthesized fucosyl-N-GlcNAc disaccharides are fermented in vitro by pure cultures of L. casei, L. rhamnosus, B. breve and B. pseudocatenulatum species21. Regarding LNB, it was fermented in vitro for all the strains tested of B. bifidum, B. infantis, B. longum subsp. longum and B. breve, and for some strains of B. pseudocatenulatum, Bifidobacterium animalis subsp. animalis and Bifidobacterium pseudolongum22. In addition, we have seen that LNB and GNB are also fermented by pure cultures of Lactobacillus species20. In this work, we have investigated, in batch cultures using total microbiota isolated from stool samples of breastfed infants, the effects of LNB, GNB, 3FN and 6FN on infant microbiota composition and subsequent short chain fatty acids production. Our final aim is to compare the potential prebiotic effect of those four disaccharides.
Results
Total and specific bacterial levels modulation with human milk disaccharides
The disaccharides LNB, GNB, 3FN and 6FN were synthesized by transglycosylation reactions as previously described20,21, and they were tested independently in fermentation assays, using total microbiota isolated from stool samples of breastfed infants. The addition of any of those oligosaccharides decreased the culture pH (Table 1). However, while the cultures with LNB and GNB resulted in a significant pH reduction (more than 2.5 pH units), the cultures with 3FN and 6FN showed a change in pH (about 1.0 pH unit) similar to the control culture without added sugar. These results are in agreement with the amount of oligosaccharides metabolized by the cultured infant microbiota, being the percentages of LNB and GNB consumed higher than the ones of the fucosylated oligosaccharides consumed (Table 1). Microbiota modulation was first determined at group and genus level by analyzing the numbers of total bacteria, Bacteroides, Bifidobacterium, Blautia cocoides group, Enterobacteriaceae family and Lactobacillus spp. Total bacterial numbers were similar among all the cultures, including the control (Fig. 1). Since the disaccharides were not completely consumed, that result suggested that they can be utilized only by a few selected groups of bacteria present in the infant microbiota. The results presented here (p = 0.0964, Dunnett’s test) (Fig. 1a) and previous studies, using fecal bacteria from formula-fed infants, showed that the total bifidobacterial community incremented with LNB12. Contrary to Bifidobacterium genus, a significant (p = 0.0192, Dunnett’s test) decrease in the number of cells during fermentation of LNB was observed for Blautia coccoides group (Fig. 1a). Regarding the fucosyl-N-acetylglucosamine disaccharides, the Lactobacillus genus is significantly (p = 0.0540, Dunnett’s test) increased by 3FN (Fig. 1c). 6FN has no significant effect in the cell numbers of lactobacilli respect to the control (Fig. 1d). Significantly (p = 0.0822, Dunnett’s test) lower cell numbers were observed for Blautia coccoides group in cultures containing 3FN and for Enterobacteriaceae family (p = 0.0751, Dunnett’s test) in cultures with 6FN (Fig. 1c and d).
Table 1.
Decrease of pH values of culture supernatants and % of consumed oligosaccharides (mean ± standard deviation) from infant fecal microbiota fermentation assays.
| Control | LNB | GNB | 3FN | 6FN | |
|---|---|---|---|---|---|
|
pH decreasea |
0.83 ± 0.32 | 2.97 ± 0.26* | 2.89 ± 0.27* | 0.98 ± 0.34 | 0.95 ± 0.33 |
| % consumed disaccharides | - | 82.33 ± 15.94 | 78.33 ± 19.40 | 41.92 ± 9.49 | 23.51 ± 15.57 |
Control, without sugar added; LNB, lacto-N-biose; GNB, galacto-N-biose; 3FN, fucosyl-α-1,3-N-acetylglucosamine; 6FN, fucosyl-α-1,6-N-acetylglucosamine. aStatistical significant differences compared to control are indicated: *p < 0.05.
Figure 1.
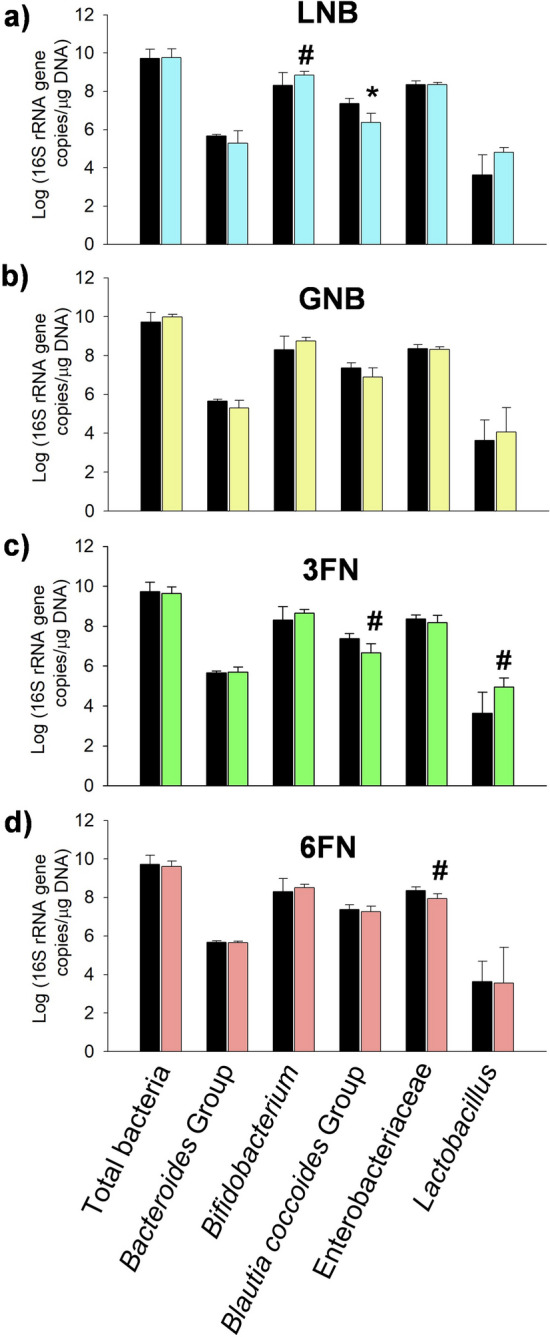
Total and specific bacterial levels measured with qPCR in cultures with fecal infant microbiota supplemented with oligosaccharides (clear bars). (a) lacto-N-biose (LNB); (b) galacto-N-biose (GNB); (c) fucosyl-α-1,3-N-acetylglucosamine (3FN); (d) fucosyl-α-1,6-N-acetylglucosamine (6FN). Control cultures are without carbohydrate added (dark bars). In all graphs, the data of the control is represented for better comparison. Data presented are mean values based on at least three replicates. Error bars indicate standard deviations. Statistically significant differences compared to control are indicated: #p < 0.1; *p < 0.05.
Bifidobacterium and Lactobacillus species modulation with human milk disaccharides
In addition to Bifidobacterium and Lactobacillus abundance at the genus level, Bifidobacterium species (Bifidobacterium breve, Bifidobacterium bifidum, Bifidobacterium catenulatum group, Bifidobacterium animalis subsp. lactis and Bifidobacterium longum group) and Lactobacillus species (Lactobacillus acidophilus, L. casei subgroup and Lactobacillus reuteri subgroup) were also analyzed (Fig. 2). B. lactis, L. reuteri subgroup and L. acidophilus were below detection limit levels. LNB (p < 0.0001, Dunnett’s test), GNB (p < 0.0001, Dunnett’s test) or 3FN (p = 0.0011, Dunnett’s test) supplementation significantly increased the number of cells of B. breve compared to control culture (Fig. 2). In addition, B. bifidum was also incremented (p = 0.0808, Dunnett’s test) with LNB (Fig. 2a). In agreement with this, LNB has been previously shown as a substrate supporting the growth of strains belonging to B. breve and B. bifidum22. Although LNB is also a substrate for B. longum species22 and they were in a high proportion compared with the other analyzed strains (Fig. 2a), there was not a significant cell number increase in the presence of that carbohydrate. In this respect, previous results showed that bifidobacteria increment in cultures with prebiotics is higher when these bacteria are at lower levels23. The 16S rRNA gene copy numbers of total bifidobacteria are higher than the sum of the numbers of the four species (B. breve, B. bifidum, B. catenulatum group and B. longum group) tested here (Figs. 1 and 2). Although these species are usually the dominant bifidobacteria detected in the infant feces, the literature showed that there is an extraordinary variation in the individual composition of Bifidobacterium species in the infant gut4. Therefore, the discrepancies observed between the 16S rRNA gene copy numbers might be due to the presence of other Bifidobacterium species that outcompete those generally infant gut-associated bifidobacterial species. Regarding 3FN, previous studies demonstrated that B. breve strain ATCC15700 was able to metabolize it21. The numbers of B. bifidum declined significantly (p = 0.0927, Dunnett’s test) in cultures with 3FN and they showed a tendency to decrease with 6FN (Fig. 2c,d). Two extracellular cell-anchored α-L-fucosidases, AfcA and AfcB, that hydrolyze α-1,2- and α-1,3/1,4-linked fucosyl residues, respectively, have been characterized in that species24,25. However, neither 3FN nor 6FN were catabolized by B. bifidum21, which is in agreement with the fact that those enzymes do not hydrolyze them26. Interestingly no differences were observed in the numbers of L. casei subgroup (Fig. 2) although as mentioned above these species contain the metabolic pathways to utilize LNB, GNB and 3FN as energy sources. Therefore, these results suggested a competition with other bacterial species with probably higher affinity for those substrates.
Figure 2.
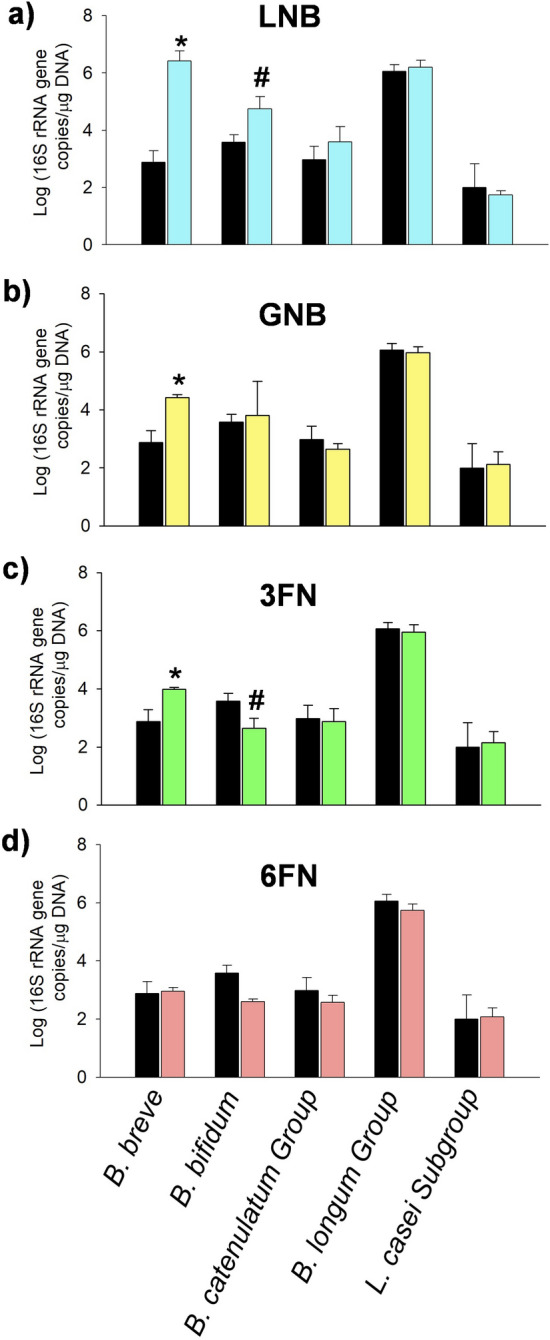
Bifidobacterium species/groups and Lactobacillus casei group levels measured with qPCR in cultures with fecal infant microbiota supplemented with oligosaccharides (clear bars). (a) lacto-N-biose (LNB); (b) galacto-N-biose (GNB); (c) fucosyl-α-1,3-N-acetylglucosamine (3FN); (d) fucosyl-α-1,6-N-acetylglucosamine (6FN). Control cultures are without carbohydrate added (dark bars). In all graphs, the data of the control is represented for better comparison. Data presented are mean values based on at least three replicates. Error bars indicate standard deviations. Statistically significant differences compared to control are indicated: #p < 0.1; *p < 0.05.
Lactate, formate and SCFA production changes with human milk disaccharides
Fermentation of non-digestible carbohydrates by the gut microbiota in the large intestine results in the accumulation of SCFA. About 90–95% of the SCFA present in the colon are acetate, propionate and butyrate27. Although lactate and formate are not SCFA, they are also major organic acids produced from the fermentation of carbohydrates by lactic acid bacteria28,29. Therefore, the concentration of lactate, formate and SCFA due to oligosaccharide fermentation by the infant fecal microbiota batch cultures were analyzed (Table 2). Lactate, formate and acetate increased significantly in the presence of LNB or GNB compared to the un-supplemented media control. Previous results with cultures containing LNB as carbon source and inoculated with fecal samples from formula-fed infants also showed an important acetate increment12. None of the tested sugars showed a butyrogenic effect in the cultures. Indeed, butyrate concentration significantly decreased with LNB and GNB fermentation (Table 2). Previous results demonstrated that oral administration of B. breve resulted in a decrease of fecal butyrate production in low birth weight infants30. As described above in the cultures supplemented with LNB or GNB significantly increased the cell counts of that species (Fig. 2), which might have a role lowering butyric acid levels. Additionally, Bacteroides and Blautia coccoides groups contain butyric acid producing genera31 that likely derive amino acids present in the culture media towards the synthesis of butyrate32. The availability of LNB or GNB in the cultures resulted in a tendency to decrease those bacterial groups with respect to the control (Fig. 1) and this could result in less production of butyric acid.
Table 2.
Lactate, formate and short-chain fatty acids concentration (mM) (mean ± standard deviation) in infant fecal microbiota fermentation with oligosaccharides.
| Control | LNB | GNB | 3FN | 6FN | |
|---|---|---|---|---|---|
| Lactate | 1.12 ± 0.14 | 4.93 ± 1.73* | 4.48 ± 3.71* | 1.06 ± 0.154 | 0.78 ± 0.36 |
| Formate | 2.43 ± 0.67 | 3.69 ± 0.38* | 4.62 ± 0.44* | 2.64 ± 0.91 | 2.77 ± 0.85 |
| Acetate | 3.67 ± 0.83 | 9.19 ± 5.44* | 7.55 ± 1.78# | 3.67 ± 1.07 | 3.61 ± 1.12 |
| Propionate | 1.45 ± 0.94 | 1.80 ± 0.86 | 3.07 ± 2.02 | 1.15 ± 0.62 | 0.87 ± 0.58 |
| Butyrate | 11.96 ± 3.80 | 6.84 ± 2.69# | 6.75 ± 4.85# | 10.62 ± 2.47 | 11.23 ± 1.61 |
Control, without sugar added; LNB, lacto-N-biose; GNB, galacto-N-biose; 3FN, fucosyl-α-1,3-N-acetylglucosamine; 6FN, fucosyl-α-1,6-N-acetylglucosamine. Statistical significant differences compared to control are indicated: #p < 0.1; *p < 0.05.
Identification of isolated Bifidobacterium species
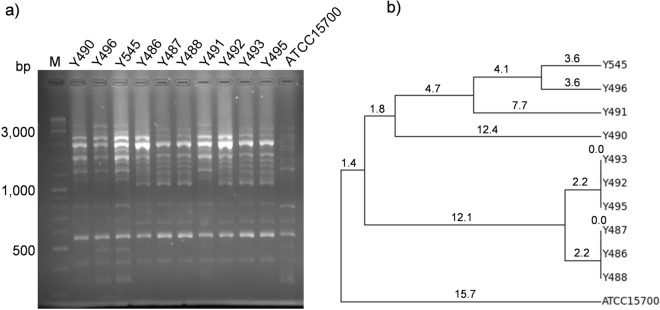
Samples from the disaccharide fermentation cultures were plated on MRS with mupirocin and cysteine in order to isolate Bifidobacterium species. Isolates were subjected to RAPD-PCR analysis and at least one representative of each band pattern was kept for subsequently species identification. Partial 16S rRNA gene sequencing led to the identification of one isolate as B. longum (Y485) and ten isolates as B. breve (Y486, Y487, Y488, Y490, Y491, Y492, Y493, Y495, Y496 and Y545). This result is in line with the fact that the latter species was stimulated by three out of the four disaccharides tested (Fig. 2). However, there are a number of cells of B. longum present in a high proportion in the cultures and therefore, other explanations for that result cannot be discarded33. Previous studies proved that RAPD-PCR analysis is an appropriate molecular tool to differentiate bifidobacteria at the strain level34. The RAPD-PCR profiles were almost similar in all isolated B. breve strains, suggesting that they were closely related (Fig. 3). However, the cluster analysis of RAPD-PCR profiles allowed differentiating the ten isolates to at least six different strains (Fig. 3). The B. longum strain Y485 and the B. breve strains Y486, Y490, Y491, Y492, Y496 and Y545 were selected to characterize their ability to utilize LNB, GNB, 3FN and 6FN. The B. breve type strain (ATCC 15700) was also included in this analysis. The ability of this strain to ferment 3FN was already demonstrated in our lab and it was used here as a positive control21. The results demonstrate that all the Bifidobacterium strains tested were able to ferment LNB and GNB as evidenced by the decreased pH of the culture media (Fig. 4). 3FN was not utilized by B. longum strain Y485 but it was fermented by all the B. breve strains tested. None of the strains fermented 6FN (Fig. 4).
Figure 3.
Evaluation of the genetic diversity among the Bifidobacterium breve strains isolated in this study using RAPD-PCR analysis. Agarose gel (2%) electrophoresis (a) and cluster analysis (b) of the DNA band patterns obtained from the B. breve strains Y486, Y487, Y488, Y490, Y491, Y492, Y493, Y495, Y496 and Y545. The type strain B. breve ATCC15700 was used as control. Cluster analysis was performed using the unweight pair group method with arithmetic mean (UPGMA) analysis. The numbers in the dendrogram provide a measure of genetic distances between isolates. Lane M, DNA molecular weight marker.
Figure 4.
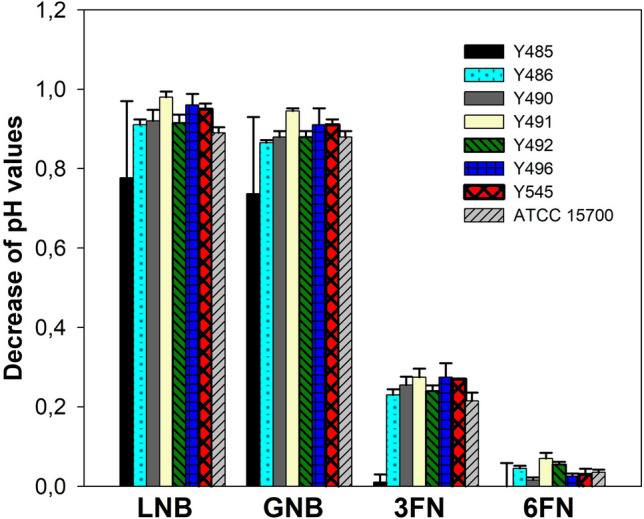
Decrease of pH values of culture supernatants from Bifidobacterium longum strain Y485 and Bifidobacterium breve strains Y486, Y490, Y491, Y492, Y496, Y545 and ATCC 15,700 cultured in MRS basal medium supplemented with 10 mM lacto-N-biose (LNB), galacto-N-biose (GNB), fucosyl- α-1,3-N-acetylglucosamine (3FN) or fucosyl-α-1,6-N-acetylglucosamine (6FN). Data presented are mean values based on at least three replicates. Error bars indicate standard deviations.
Discussion
Studies using in vitro analysis and animal models suggested that human milk oligosaccharides (HMOs) are crucial for modulating the infant gut microbiota and exerting important health benefits, including prevention of pathogen attachment and immunomodulation2,35–37. However, HMOs are not present or are present in low concentrations in infant formulas17. Therefore, there is a need to evaluate the influence of individual carbohydrates in the microbiota profile of newborns for their selection as prebiotic substrates. In this work, the disaccharides LNB, GNB, 3FN and 6FN, that form part of HMOs and mucosa-associated glycans, were tested individually in fermentation assays, using total microbiota isolated from stool samples of breastfed infants. Total levels of bifidobacteria were significantly increased by LNB. The effect of this disaccharide was previously analyzed in vitro on fecal bacteria from formula-fed infants and the total bifidobacterial community was also incremented12. These findings and our results confirmed LNB as an important growth-promoting factor for the genus Bifidobacterium. The transport and catabolism of LNB as well as GNB are mediated by the proteins encoded by the LNB/GNB gene cluster present in several Bifidobacterium species38–41. This gene cluster includes a key enzyme, the LNB/GNB phosphorylase (LNBP), which catalyzes the cleavage of the 1,3-β-glycosidic bond of LNB or GNB into galactose-1-phosphate and GlcNAc or GalNAc, respectively42. LNBP from Bifidobacterium species showed similar affinity for LNB and GNB, however, phosphorylases specific to GNB has also been characterized in intestinal bacteria43, which might explain the significant and nonsignificant differences found here between LNB and GNB, respectively, on Bifidobacterium cell counts (Fig. 1). Regarding lactobacilli, the operon gnb involved in the metabolism of LNB and GNB has been characterized in L. casei44. In this bacterium, those disaccharides are transported and phosphorylated by the PTSGnb and once inside the cell are hydrolyzed by the specific phospho-β-1,3-galactosidase GnbG. In addition to L. casei, the species Lactobacillus rhamnosus, Lactobacillus zeae, Lactobacillus gasseri and Lactobacillus johnsonii are also able to utilize LNB and GNB as a carbon source20. However, while L. rhamnosus and L. zeae contain a phospho-β-galactosidase homologous to GnbG, L gasseri and L. johnsonii do not have GnbG homologs, suggesting that the catabolism of both sugars in the latest species must depend on other glycosidases20.
Our results showed that the cell numbers of the Bacteroides and Blautia coccoides groups have a tendency to decrease during fermentation of LNB or GNB. BLAST searches using the amino acid sequence of LNBP from B. bifidum22 against the phylum Bacteroidetes did not show relevant homology with any protein. Analysis using GnbG from L. casei44 showed significant homology with proteins from several species belonging to Bacteroidetes, such as Sphingobacterium faecium45, Hymenobacter sp.46, Chryseobacterium sp.47 or Mucilaginibacter sp.48. However, the primers used to target Bacteroides group49 do not amplify the 16S rRNA gene of those genera and in addition, they are isolated from environments different from the intestine. These results suggested the absence of enzymes homologs to LNBP and GnbG in gut species of the Bacteroides group, however, other enzymatic strategies to metabolize those disaccharides cannot be discarded. Alternatively, the decrease of Bacteroides group counts might also be explained by the pH reduction determined in cultures with LNB or GNB since some Bacteroides species showed a poor growth at low pH50. Regarding B. coccoides group, some species contain homologs to LNBP and GnbG, although the possible role of these enzymes on LNB and GNB metabolism needs to be investigated.
The fucosyl-disaccharide 3FN is hydrolyzed by the α-L-fucosidase AlfB from L. casei51 and the ability of this bacterium to ferment 3FN rely upon this enzyme21,52. Homologs to AlfB are present in strains of the L. casei/L. paracasei/L. rhanmosus group and other lactobacilli species, including Lactobacillus sharpeae, Lactobacillus fabifermentans, Lactobacillus paralimentarius, Lactobacillus tucceti, Lactobacillus ghanensis, Lactobacillus apis, Lactobacillus vini, Lactobacillus gigeriorum, Lactobacillus panisapium and Lactobacillus helsingborgensis (https://www.ncbi.nih.gov/genome). In accordance with this, the Lactobacillus genus is significantly increased by 3FN (Fig. 1c), although whether the lactobacilli present in the fermentation cultures analyzed here contain homologs to AlfB would require further analysis. As well, other catabolic systems different to those already characterized for 3FN utilization might exist. Regarding 6FN, lactobacilli cell counts were not affected. Curiously, 6FN is cleaved by the α-L-fucosidase AlfC from L. casei strain BL2351, but unlike 3FN, it is not a metabolic substrate for this bacterium. The alfC gene forms part of the alf2 operon that is induced by the glycoamino acid 6FN-Asn, a constituent of the mammalian N-glycoproteins, but not by its glycan moiety 6FN53. L. casei strain BL87 and L. rhamnosus BL327 contain homologs to AlfC and they are able to metabolize 6FN although with low efficiency21. BLAST searches using the amino acid sequences of AlfB and AlfC from L. casei51 against Blautia coccoides group and Enterobacteriaceae, respectively, did not show relevant homology with any protein, suggesting the absence of enzymes homologs to those α-L-fucosidases. However, the presence on those bacterial groups of other enzymes able to catabolize 3FN and 6FN cannot be ruled out. The reduced numbers of Enterobacteriaceae in the presence of 6FN, might be relevant for specific infant gastrointestinal disorders such as the development of necrotizing enterocolitis, which has been associated with an increased relative abundance of Enterobacteriaceae strains54.
The major products of non-digestible carbohydrate metabolism by the gut microbiota are SCFA and organic acids such as formate and lactate. Both, organic acids and SCFA, resulted in a decrease of the luminal pH, which in turn resulted in inhibition of pathogens growth and increment of nutrients absorption55. SCFA are a source of energy for colonocytes56 and also for some members of the microbiota31. In addition, SCFA have important roles in colorectal cancer, immune system and inflammatory responses57. The fermentation of LNB and GNB by the infant microbiota resulted in an increment of the production of lactate, formate and acetate (Table 2). According to this, Bifidobacterium species produced both acetate and lactate through the “bifid-shunt” route during carbohydrate fermentation58. Stool pH of breastfed infants is lower than that of bottle-fed infants59 and it is due to the high lactate and acetate levels present in the feces of breastfed infants11,60. Interestingly, research using an animal model has demonstrated that increased production of acetate by bifidobacteria protects the host against enteropathogenic infection61. Lactate has also been shown to be important in maintaining intestinal barrier function through stimulation of enterocyte proliferation in a murine model62. Although the cell numbers of lactobacilli and B. breve increase after fermentation of 3FN by the infant microbiota, there is not an increment in the production of lactate. Previous studies have shown that in anaerobiosis the L-fucose metabolism by some bacteria produces more efficiently 1,2-propanediol than lactate63,64. However, this metabolite has not been detected in the supernatants from the 3FN-supplemented cultures (data not shown), suggesting that either it was not produced or if produced, it was further metabolized by other members of the microbiota present in the fermentation cultures. Some bacteria utilize 1,2-propanediol, via propionaldehyde, to produce propionate and/or propanol31. Since the concentration of propionate did not increase with 3FN, the possible 1,2-propanediol produced would be derived to propanol. Whether the lactobacilli and bifidobacteria present in the fermentation cultures analyzed here produce 1,2-propanediol, that is further derived to propanol by other bacteria also present in the cultures, is unknown. Additional studies will be needed to find out why the lactate levels do not change in the cultures with 3FN.
This study demonstrates that the human milk and mucosa-associated disaccharides LNB, GNB and 3FN and 6FN are fermented in vitro by the microbiota isolated from stool samples of breastfed infants. The results presented here allow a comparison among four individual disaccharides and they indicated that each disaccharide distinctly modulates the microbiota composition and activity. The growth of specific Bifidobacterium species is significantly promoted by LNB, GNB and 3FN, and the growth of lactobacilli by 3FN. Furthermore, LNB and GNB enhance the production of lactate, formate and acetate. Regarding 6FN, Enterobacteriaceae family counts are significantly reduced, although further work is needed to determine the relevance of this finding. Overall, our results highlight the prebiotic potential of each LNB, GNB and 3FN for application in functional infant foods.
Methods
Enzymatic synthesis and purification of human milk disaccharides
The disaccharides LNB, GNB, 3FN and 6FN were produced in our laboratory as previously described18,20,21. Briefly, transgalactosylation reactions containing 100 mM Tris–HCl buffer, pH 7.5, o-nitrophenyl β-D-galactopyranoside 40 mM, 0.17 U/ml of GnbG and N-acetylglucosamine (GlcNAc) 200 mM or N-acetylgalactosamine (GalNAc) 200 mM were carried out at 42 °C for 3 h to synthesize LNB and GNB, respectively. Transfucosylation reactions containing 100 mM Tris–HCl buffer, pH 7.0, p-nitrophenyl α-L-fucopyranoside 50 mM, N-acetylglucosamine (GlcNAc) 150 mM and 180 U/ml of AlfB or 800 U/ml AlfC, were carried out at 42 °C for 15 min (AlfB) and 10 min (AlfC) to synthesize 3FN and 6FN, respectively.
The transglycosylation reaction products were purified by high-performance liquid chromatography (HPLC) with a Jasco PU2080Plus system coupled to a refractive index detector (Jasco RI-2031 Plus) using a preparative Rezex RCM-Monosaccharide column (Phenomenex, Torrance, CA, USA), as described previously21. Based on HPLC analysis, the purity of the synthesized oligosaccharides was found to be more than 99%.
Batch culture fermentation assays
Stool samples from five healthy infants exclusively breast-fed and two months old were selected from a previous study65. A pooled fecal sample was used to isolate the microbiota as previously described66. Two grams of feces were homogenized in 18 ml of physiological serum (NaCl 0.9%), placed on top of 3.5 ml Nycodenz 80% solution (Alere Technologies AS) and centrifuged for 1 h at 3,327 xg (Hermle Z383K). The layer containing the microbiota was collected and stored in 20% glycerol at – 80 °C. This microbiota was used to inoculate two ml of basal medium containing bactopeptone (Difco), 2 g/L; yeast extract (Pronadisa), 2 g/L; NaCl 0.1 g/L; K2HPO4 0.04 g/L; KH2PO4 0.04 g/L; MgSO4·7H2O 0.01 g/L; CaCl2·H2O 0.01 g/L; NaHCO3 2 g/L; L-cysteine 0.5 g/L; bile salts 0.5 g/L; Tween 80 2 ml/L; haemin solution 0.05 g/L (Sigma); vitamin K1 (Sigma), 10 μl/L; and resazurin 0.025% solution, 4 ml/L. The pH of the medium was adjusted to 7.4. Four cultures were initiated with the purified disaccharides, LNB, GNB, 3FN or 6FN at 10 mM. A control culture without carbohydrate added was also included. Cultures were grown at 37 °C under anaerobic conditions using an anaerobic atmosphere generation system (Anaerogen, Oxoid), and at 48 h they were centrifuged, and pellets and supernatants were collected. Three independent cultures were carried out for each oligosaccharide.
Quantification of bacterial groups, genera and species by specific real-time PCR
Bacterial DNA was extracted from each pellet of the cultures using the MasterPure DNA extraction Kit (Epicentre) according to the manufacturer’s instructions with some modifications. These include a previous treatment with lysozyme and mutanolysin for 60 min at 37 °C followed by cell disruption with 0.1 mm diameter glass beads. DNA concentration was measured using a Qubit 2.0 Fluorometer (Life Technology).
Quantitative real-time PCR (qPCR) assays were performed as previously described67 and using a series of bacterial group, genus and species-specific 16S rRNA gene primer pairs49,67–74 listed in supplementary Table S1. The qPCR amplification and detection were performed in a LightCycler 480 Real-Time PCR System (Roche Technologies). Each reaction mixture (10 μl) contained SYBR Green PCR Master Mix (Roche), 0.25 μl of each primer (10 μM) and 1 μl of template DNA. All samples were analyzed in duplicate. Standard curves obtained by using serial tenfold dilutions of specific DNA fragments were used to calculate bacterial concentration in each sample. A detection limit of 100 copies per reaction was established.
Analysis of disaccharide consumption
To determine the carbohydrates present in the supernatants from the infant fecal microbiota fermentation assays, filtered culture supernatants were analyzed by high-pH anion-exchange chromatography with pulsed amperometric detection in a ICS3000 chromatographic system (Dionex) using a CarboPac PA100 column. A gradient of 10 to 100 mM NaOH was used at 27 °C for 15 min at a flow rate of 1 ml/min. Disaccharides were confirmed by comparison of their retention times with those of standards. The obtained calibration curves for LNB, GNB, 3FN and 6FN were linear in the range tested, 0.05–0.25 mM.
Lactate, short-chain fatty acids and 1,2-propanediol analysis
Filtered culture supernatants were analyzed by HPLC with a Jasco PU2080Plus system coupled to a UV (210 nm) or refractive index (Jasco RI-2031Plus) detectors and using a Rezex ROA-Organic Acid or Rezex RCM-Monosaccharide columns (Phenomenex, Torrance, CA, USA). Lactic, formic, acetic, propionic, butyric acids (Rezex ROA-Organic Acid column) and 1,2-propanediol (Rezex RCM-Monosaccharide column) were determined as previously described63,75. The column temperatures were kept at 40 °C and 80 °C, respectively, in phosphoric acid 0.1% or water.
Isolation of Bifidobacterium species
A dilution series was made from the LNB, GNB, 3FN and 6FN fermentation cultures, and 0.1 ml aliquots were plated on MRS agar with mupirocin 50 mg/L and cysteine 0.1%. These plates were incubated in anaerobic jars at 37 °C for 72 h in order to isolate Bifidobacterium species. More than ten colonies from each of the four disaccharide-supplemented cultures were selected until reaching a total number of fifty colonies. These were subsequently subjected to randomly amplified polymorphic DNA (RAPD) analysis. Each RAPD-PCR reaction (25 μl) contained bacterial cells from the colonies as source of DNA template, random primer MCV76 described in Table S1 at a final concentration of 2 μM, 400 μM of each dNTP, MgCl2 4 mM, dimethyl sulfoxide (DMSO) 4%, 2 U of Taq DNA polymerase and PCR buffer (NZYtech). Amplifications were performed at 96 °C for 7 min, followed by 35 cycles of 30 s at 95 °C, 45 s at 30 °C and 1 min at 72 °C and extension for 7 min at 72 °C. Samples were analyzed by agarose gel electrophoresis and one representative isolate of each band pattern was kept for further analysis. Partial 16S rRNA gene of these bacterial isolates was amplified by PCR using cells from the colonies as the template and the primers 27F77 and 924R78 (Table S1). The PCR products were sequenced by the Central Service of Research Support of the University of Valencia (Spain). The sequences were used in BLAST searches to identify each isolated.
Genetic distance between the B. breve isolates was calculated using the Dice coefficient of similarity and the strains were clustered in a dendrogram using the unweight pair group method with arithmetic mean (UPGMA) analysis with PyElph 1.4 software tool79.
The growth of individual B. longum and B. breve strains in the presence of each disaccharide at 10 mM was tested in MRS basal medium using the culture conditions previously described for the Bifidobacterium strains21.
Statistical analysis
One-way ANOVA with Dunnett’s multiple comparisons test was performed using GraphPad Prism, version 6.07 (GraphPad Software Inc., San Diego, CA, USA) and it was used to detect statistically significant differences between the control and the disaccharide-supplemented groups (LNB, GNB, 3FN and 6FN) on the following: numbers of total and specific bacterial groups, genera and species; pH decrease of culture supernatants; lactate, formate and SCFA. Statistical significance was accepted at two levels #p < 0.1; *p < 0.05.
Nucleotide sequence accession numbers
The partial nucleotide sequence of the 16S rRNA gene amplicons have been deposited at the GenBank database under the accession numbers MN650609 to MN650619.
Ethical statement
All applicable international, national, and/or institutional guidelines for the use of human samples were followed. Samples were selected from the project “The power of maternal microbes on infant health (MAMI)”. The study protocol with the registration number 2015/0024 was approved by the Ethics Committee of Spanish National Research Council (CSIC). Written informed consent is obtained from a parent and/or legal guardian.
Data availability statement
All data generated or analyzed during this study are included in this published article.
Supplementary information
Acknowledgements
This work was supported by the Spanish Ministry for Economy and Competitiveness (MINECO)/FEDER through the Project AGL2017-84165-C2 (1-R and 2-R). M.C.C. would like to acknowledge the support from European Research Council under the European Union's Horizon 2020 research and innovation programme (ERC starting grant, n° 639226). J.R-D. was supported by a Ramon y Cajal Contract RYC-2013-12442 by the Spanish Ministry for Economy and Competitiveness.
Author contributions
M.C.C., J.R.D. and M.J.Y. conceived and designed research. A.R.C. and C.A. conducted experiments. M.J.Y. drafted the manuscript. A.R.C., C.A., M.C.C. and J.R.D. helped improving the draft. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jesús Rodríguez-Díaz, Email: jesus.rodriguez@uv.es.
María J. Yebra, Email: yebra@iata.csic.es
Supplementary information
is available for this paper at 10.1038/s41598-020-68718-4.
References
- 1.Yu ZT, et al. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology. 2013;23:169–177. doi: 10.1093/glycob/cws138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuñiga M, Monedero V, Yebra MJ. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front. Microbiol. 2018;9:1917. doi: 10.3389/fmicb.2018.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido D, et al. Endo-beta-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol. Cell Proteomics. 2012;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turroni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avershina E, et al. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl. Environ. Microbiol. 2013;79:497–507. doi: 10.1128/AEM.02359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin R, Heilig GH, Zoetendal EG, Smidt H, Rodriguez JM. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J. Appl. Microbiol. 2007;103:2638–2644. doi: 10.1111/j.1365-2672.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- 7.Albesharat R, Ehrmann MA, Korakli M, Yazaji S, Vogel RF. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011;34:148–155. doi: 10.1016/j.syapm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Kobata A. Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. Phys. Biol. Sci. 2010;86:731–747. doi: 10.2183/pjab.86.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locascio RG, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb. Biotechnol. 2009;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Moyano S, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl. Environ. Microbiol. 2013;79:6040–6049. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuki T, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satoh T, et al. In vitro comparative evaluation of the impact of lacto-N-biose I, a major building block of human milk oligosaccharides, on the fecal microbiota of infants. Anaerobe. 2013;19:50–57. doi: 10.1016/j.anaerobe.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Vester Boler BM, et al. In vitro fermentation characteristics of select nondigestible oligosaccharides by infant fecal inocula. J. Agric. Food Chem. 2013;61:2109–2119. doi: 10.1021/jf305056f. [DOI] [PubMed] [Google Scholar]
- 14.Wiese M, et al. CoMiniGut-a small volume in vitro colon model for the screening of gut microbial fermentation processes. PeerJ. 2018;6:e4268. doi: 10.7717/peerj.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss GA, Chassard C, Hennet T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Brit. J. Nutr. 2014;111:1602–1610. doi: 10.1017/S0007114513004200. [DOI] [PubMed] [Google Scholar]
- 16.Tarr AJ, et al. The prebiotics 3'Sialyllactose and 6'Sialyllactose diminish stressor-induced anxiety-like behavior and colonic microbiota alterations: evidence for effects on the gut-brain axis. Brain Behav. Immun. 2015;50:166–177. doi: 10.1016/j.bbi.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urashima T, Taufik E, Fukuda K, Asakuma S. Recent advances in studies on milk oligosaccharides of cows and other domestic farm animals. Biosci. Biotechnol. Biochem. 2013;77:455–466. doi: 10.1271/bbb.120810. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Diaz J, Carbajo RJ, Pineda-Lucena A, Monedero V, Yebra MJ. Synthesis of fucosyl-N-acetylglucosamine disaccharides by transfucosylation using alpha-L-fucosidases from Lactobacillus casei. Appl. Environ. Microbiol. 2013;79:3847–3850. doi: 10.1128/AEM.00229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parry S, et al. N-Glycosylation of the MUC1 mucin in epithelial cells and secretions. Glycobiology. 2006;16:623–634. doi: 10.1093/glycob/cwj110. [DOI] [PubMed] [Google Scholar]
- 20.Bidart GN, Rodriguez-Diaz J, Palomino-Schatzlein M, Monedero V, Yebra MJ. Human milk and mucosal lacto- and galacto-N-biose synthesis by transgalactosylation and their prebiotic potential in Lactobacillus species. Appl. Microbiol. Biotechnol. 2017;101:205–215. doi: 10.1007/s00253-016-7882-0. [DOI] [PubMed] [Google Scholar]
- 21.Becerra JE, Coll-Marques JM, Rodriguez-Diaz J, Monedero V, Yebra MJ. Preparative scale purification of fucosyl-N-acetylglucosamine disaccharides and their evaluation as potential prebiotics and antiadhesins. Appl. Microbiol. Biotechnol. 2015;99:7165–7176. doi: 10.1007/s00253-015-6666-2. [DOI] [PubMed] [Google Scholar]
- 22.Xiao JZ, et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl. Environ. Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Preter V, et al. Baseline microbiota activity and initial bifidobacteria counts influence responses to prebiotic dosing in healthy subjects. Aliment. Pharmacol. Ther. 2008;27:504–513. doi: 10.1111/j.1365-2036.2007.03588.x. [DOI] [PubMed] [Google Scholar]
- 24.Katayama T, et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95) J. Bacteriol. 2004;186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashida H, et al. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 26.Sakurama H, et al. 1,3–1,4-α-L-Fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J. Biol. Chem. 2012;287:16709–16719. doi: 10.1074/jbc.M111.333781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rios-Covian D, et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hove H, Nordgaard-Andersen I, Mortensen PB. Faecal DL-lactate concentration in 100 gastrointestinal patients. Scand. J. Gastroenterol. 1994;29:255–259. doi: 10.3109/00365529409090473. [DOI] [PubMed] [Google Scholar]
- 29.Liong MT, Shah NP. Production of organic acids from fermentation of mannitol, fructooligosaccharide and inulin by a cholesterol removing Lactobacillus acidophilus strain. J. Appl. Microbiol. 2005;99:783–793. doi: 10.1111/j.1365-2672.2005.02677.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, et al. Effects of oral administration of Bifidobacterium breve on fecal lactic acid and short-chain fatty acids in low birth weight infants. J. Pediatr. Gastroenterol. Nutr. 2007;44:252–257. doi: 10.1097/01.mpg.0000252184.89922.5f. [DOI] [PubMed] [Google Scholar]
- 31.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 32.Kumari M, Kozyrskyj AL. Gut microbial metabolism defines host metabolism: an emerging perspective in obesity and allergic inflammation. Obes. Rev. 2017;18:18–31. doi: 10.1111/obr.12484. [DOI] [PubMed] [Google Scholar]
- 33.Serafini F, et al. Insights into physiological and genetic mupirocin susceptibility in bifidobacteria. Appl. Environ. Microbiol. 2011;77:3141–3146. doi: 10.1128/AEM.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarocki P, et al. Comparison of various molecular methods for rapid differentiation of intestinal bifidobacteria at the species, subspecies and strain level. BMC Microbiol. 2016;16:159. doi: 10.1186/s12866-016-0779-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 36.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Nat. Acad. Sci. USA. 2011;108:4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–664. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl. Environ. Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James K, Motherway MO, Bottacini F, van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci. Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki R, et al. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 2008;283:13165–13173. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 41.Fushinobu S. Unique sugar metabolic pathways of bifidobacteria. Biosci. Biotechnol. Biochem. 2010;74:2374–2384. doi: 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- 42.Derensy-Dron D, Krzewinski F, Brassart C, Bouquelet S. Beta-1,3-galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol. Appl. Biochem. 1999;29:3–10. [PubMed] [Google Scholar]
- 43.Nakajima M, Nihira T, Nishimoto M, Kitaoka M. Identification of galacto-N-biose phosphorylase from Clostridium perfringens ATCC13124. Appl. Microbiol. Biotechnol. 2008;78:465–471. doi: 10.1007/s00253-007-1319-8. [DOI] [PubMed] [Google Scholar]
- 44.Bidart GN, Rodriguez-Diaz J, Monedero V, Yebra MJ. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in Lactobacillus casei. Mol. Microbiol. 2014;93:521–538. doi: 10.1111/mmi.12678. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Chen W, Ke W, Chen H. Screening of a glucoside 3-dehydrogenase-producing strain, Sphingobacterium faecium, based on a high-throughput screening method and optimization of the culture conditions for enzyme production. Appl. Microbiol. Biotechnol. 2014;172:3448–3460. doi: 10.1007/s12010-014-0773-x. [DOI] [PubMed] [Google Scholar]
- 46.Buczolits S, Denner EB, Kampfer P, Busse HJ. Proposal of Hymenobacter norwichensis sp. nov., classification of 'Taxeobacter ocellatus', 'Taxeobacter gelupurpurascens' and 'Taxeobacter chitinovorans' as Hymenobacter ocellatus sp. nov., Hymenobacter gelipurpurascens sp. nov. and Hymenobacter chitinivorans sp. nov., respectively, and emended description of the genus Hymenobacter Hirsch et al. 1999. Int. J. Syst. Evol. Microbiol. 2006;56:2071–2078. doi: 10.1099/ijs.0.64371-0. [DOI] [PubMed] [Google Scholar]
- 47.Pal M, Swarnkar MK, Dhar H, Chhibber S, Gulati A. Genome assembly of Chryseobacterium sp. strain IHBB 10212 from glacier top-surface soil in the Indian trans-Himalayas with potential for hydrolytic enzymes. Genom. Data. 2017;13:46–49. doi: 10.1016/j.gdata.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh TJ, Han SR, Kang S, Park H, Kim AY. Complete genome sequence of the xylan-degrading Mucilaginibacter sp. strain PAMC26640 isolated from an Arctic lichen. J. Biotechnol. 2016;227:23–24. doi: 10.1016/j.jbiotec.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 49.Echarri PP, et al. Assessment of intestinal microbiota of full-term breast-fed infants from two different geographical locations. Early Hum. Dev. 2011;87:511–513. doi: 10.1016/j.earlhumdev.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 2005;71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez-Diaz J, Monedero V, Yebra MJ. Utilization of natural fucosylated oligosaccharides by three novel alpha-L-fucosidases from a probiotic Lactobacillus casei strain. Appl. Environ. Microbiol. 2011;77:703–705. doi: 10.1128/AEM.01906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Diaz J, Rubio-del-Campo A, Yebra MJ. Lactobacillus casei ferments the N-Acetylglucosamine moiety of fucosyl-alpha-1,3-N-acetylglucosamine and excretes L-fucose. Appl. Environ. Microbiol. 2012;78:4613–4619. doi: 10.1128/AEM.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becerra JE, et al. Unique microbial catabolic pathway for the human core N-glycan constituent fucosyl-alpha-1,6-N-acetylglucosamine-asparagine. mBio. 2020 doi: 10.1128/mBio.02804-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pammi M, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 56.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 57.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microb. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogawa K, Ben RA, Pons S, de Paolo MI, Bustos Fernandez L. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J. Pediatr. Gastroenterol. Nutr. 1992;15:248–252. doi: 10.1097/00005176-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 60.Bridgman SL, et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: differences in relative versus absolute concentrations. Front. Nutr. 2017;4:11. doi: 10.3389/fnut.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 62.Garrote GL, Abraham AG, Rumbo M. Is lactate an undervalued functional component of fermented food products? Front. Microbiol. 2015;6:629. doi: 10.3389/fmicb.2015.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Becerra JE, Yebra MJ, Monedero V. An L-fucose operon in the probiotic Lactobacillus rhamnosus GG is involved in adaptation to gastrointestinal conditions. Appl. Environ. Microbiol. 2015;81:3880–3888. doi: 10.1128/AEM.00260-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bunesova V, Lacroix C, Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16:248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia-Mantrana I, et al. MAMI: a birth cohort focused on maternal-infant microbiota during early life. BMC Pediatr. 2019;19:140. doi: 10.1186/s12887-019-1502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hevia A, Delgado S, Margolles A, Sanchez B. Application of density gradient for the isolation of the fecal microbial stool component and the potential use thereof. Sci. Rep. 2015;5:16807. doi: 10.1038/srep16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mira-Pascual L, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 68.Gueimonde M, Tolkko S, Korpimaki T, Salminen S. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 2004;70:4165–4169. doi: 10.1128/AEM.70.7.4165-4169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuki T, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 2002;68:5445–5451. doi: 10.1128/aem.68.11.5445-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartosch S, Fite A, Macfarlane GT, McMurdo ME. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004;70:3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gueimonde M, Debor L, Tolkko S, Jokisalo E, Salminen S. Quantitative assessment of faecal bifidobacterial populations by real-time PCR using lanthanide probes. J. Appl. Microbiol. 2007;102:1116–1122. doi: 10.1111/j.1365-2672.2006.03145.x. [DOI] [PubMed] [Google Scholar]
- 72.Sheu SJ, et al. Use of tuf gene-based primers for the PCR detection of probiotic Bifidobacterium species and enumeration of bifidobacteria in fermented milk by cultural and quantitative real-time PCR methods. J. Food Sci. 2010;75:M521–527. doi: 10.1111/j.1750-3841.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 73.Larsen N, et al. Predominant genera of fecal microbiota in children with atopic dermatitis are not altered by intake of probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium animalis subsp lactis Bi-07. FEMS Microbiol. Ecol. 2011;75:482–496. doi: 10.1111/j.1574-6941.2010.01024.x. [DOI] [PubMed] [Google Scholar]
- 74.Matsuda K, et al. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl. Environ. Microbiol. 2009;75:1961–1969. doi: 10.1128/AEM.01843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarmiento-Rubiano LA, Zuñiga M, Perez-Martinez G, Yebra MJ. Dietary supplementation with sorbitol results in selective enrichment of lactobacilli in rat intestine. Res. Microbiol. 2007;158:694–701. doi: 10.1016/j.resmic.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 76.Tynkkynen S, Satokari R, Saarela M, Mattila-Sandholm T, Saxelin M. Comparison of ribotyping, randomly amplified polymorphic DNA analysis, and pulsed-field gel electrophoresis in typing of Lactobacillus rhamnosus and L. casei strains. Appl. Environ. Microbiol. 1999;65:3908–3914. doi: 10.1128/AEM.65.9.3908-3914.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rudi K, Skulberg OM, Larsen F, Jakobsen KS. Strain characterization and classification of oxyphotobacteria in clone cultures on the basis of 16S rRNA sequences from the variable regions V6, V7, and V8. Appl. Environ. Microbiol. 1997;63:2593–2599. doi: 10.1128/AEM.63.7.2593-2599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavel AB, Vasile CI. PyElph - a software tool for gel images analysis and phylogenetics. BMC Bioinform. 2012;13:9. doi: 10.1186/1471-2105-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.