Abstract
Background
Staphylococcus aureus (S. aureus), including methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA), is an eminent human pathogen that can colonize the human host and cause severe life-threatening infections. The development of a reliable, simple and rapid assay for detecting S. aureus and identifying MRSA is important for diagnosis and follow-up treatment.
Methods
A novel molecular diagnosis technique, named multiplex loop-mediated isothermal amplification linked to a nanoparticle-based lateral flow biosensor (m-LAMP-LFB), was applied to detect all S. aureus species and identify MRSA. Two sets of primers were designed based on the femA gene (S. aureus-specific gene) and the mecA gene (encoding penicillin-binding protein 2a), and the multiple-LAMP products were analyzed using LFB. The m-LAMP-LFB amplification conditions, including the target DNA concentration, reaction temperature and time, were optimized. The sensitivity and specificity of the m-LAMP-LFB method were tested in the current study, and the multiple-LAMP-LFB technology was applied to detect the MSSA and MRSA strains from clinical samples.
Results
The S. aureus- and MRSA-specific primers based on the femA and mecA genes allowed the multiple-LAMP technology to detect S. aureus and MRSA, respectively. The multiple-LAMP conditions were optimized at 63 °C for 40 min. The full process, including genomic DNA template preparation, LAMP, and product identification, could be achieved in 80 min. The limit of detection (LoD) of the multiple-LAMP assay for femA and mecA detection was 100 fg of genomic DNA template per reaction. The specificity of m-LAMP-LFB detection was 100 %, and no cross-reactions to non-S. aureus strains were observed.
Conclusion
The multiple-LAMP-LFB technique developed in the current study is a reliable, simple, rapid, specific and sensitive method to identify MSSA and MRSA infections for appropriate antibiotic therapy.
Keywords: Staphylococcus aureus, MRSA, Limit of detection, Loop-mediated isothermal amplification, Lateral flow biosensor
Background
Staphylococcus aureus, a gram-positive coccoid bacterium, is a common human pathogen that has the ability to cause a wide array of severe hospital and community-acquired infections, such as pneumonia, bacteremia, sepsis and toxic shock syndrome [1–3]. S. aureus infections have been typically treated with methicillin (a semisynthetic antibiotic), which was developed and applied to clinical practice in the late 1950s [4]. Unfortunately, a methicillin-resistant S. aureus (MRSA) strain was found among clinical isolates from patients hospitalized in 1960. By the 1980s, MRSA strains were globally epidemic in both community and healthcare settings [5–7]. According to a 2014 World Health Organization (WHO) report, MRSA was listed as one of the seven pathogens of international concern and has been associated with a high number of mortality and mortality [8, 9]. Thus, developing a reliable and rapid method of detection for the accurate differentiation of methicillin-susceptible S. aureus (MSSA) and MRSA isolates is necessary for the follow-up treatment and management of patients.
The traditional methods for the detection of MSSA and MRSA is based on cultivation [10] and include colony morphology, biochemical measurements and microdilution drug resistance testing; however, these methods are time-consuming and laborious. These methods require more than 2 days to identify MSSA or MRSA isolates. For this reason, the optimal treatment period is missed for many patients, or antibiotics are abused, resulting in multidrug resistance [11–13]. In recent decades, many molecular methods, such as polymerase chain reaction (PCR), multiplex PCR and real-time PCR, have been applied for detecting MSSA and MRSA isolates [10, 14, 15]. However, PCR-based methods require special experimental instruments and skilled personnel that may not be readily available in many resource-poor settings. Therefore, a cost-effective, simple, reliable, rapid, sensitive, and specific assay for the identification of MSSA and MRSA should be developed to improve treatment and prevent the spread and outbreak of these infections.
To overcome the drawbacks of PCR-based detection, a wide variety of isothermal amplification-based methods have been developed for use in molecular identification. Loop-mediated isothermal amplification (LAMP), as a reliable, low-cost, sensitive and rapid assay, has been widely applied to detect many bacterial pathogens, including Streptococcus pneumoniae, Salmonella and Brucella [16–18]. Unfortunately, the use of the multiplex LAMP (m-LAMP) method to distinguish MRSA from MSSA species has not been reported thus far. LAMP products have been analyzed by various methods, including agarose gel electrophoresis, visual inspection of color changes and turbidimetry changes [19–22]. However, these detection techniques are not specific for target genes and are likely to cause false positive results. To overcome this defect, a target-specific, visual and simple nanoparticle-based lateral flow biosensor (LFB) detection method was successfully designed and applied to analyze m-LAMP products in the current study. Hence, the multiplex-LAMP technique linked to a LFB detector (m-LAMP-LFB) was developed for the reliable, simple, specific, sensitive and visual identification of MSSA and MRSA strains by targeting the femA and mecA genes, respectively. femA is a S. aureus-specific gene that appears to be uniquely present in S. aureus as it shows no homology with other microbial genomes at GenBank by BLAST searches. mecA is an MRSA-specific gene that encodes penicillin-binding protein 2a. The optimal amplification conditions and feasibility of the m-LAMP-LFB assay were confirmed with pure cultures and clinical samples.
Materials and methods
Materials instruments
Bacterial genomic DNA extraction kits (QIAamp DNA minikits; Qiagen, Hilden, Germany) were purchased from Qiagen (Beijing, China). Universal isothermal amplification kits, a colorimetric indicator (malachite green, MG), and biotin-14-dCTP were obtained from Bei-Jing HaiTaiZhengYuan. Co., Ltd. (Beijing, China). The LFB materials, including the backing card, sample pad, absorbent pad, conjugate pad and nitrocellulose membrane (NC), were purchased from Jie-Yi Biotechnology. Co., Ltd. (Shanghai, China). Anti-FITC (rabbit anti-fluorescein antibody) and biotin-BSA (biotinylated bovine serum albumin) were purchased from Abcam. Co., Ltd. (Shanghai, China). Dye (Crimson red) streptavidin-coated polymer nanoparticles (129 nm, 10 mg ml− 1; 100 mM borate, pH 8.5, with 0.1% BSA, 0.05% Tween 20 and 10 mM EDTA) were purchased from Bangs Laboratories, Inc. (Indiana, USA).
Design of LAMP detection primers
Based on the reaction mechanism of LAMP, two sets of specific primers were designed according to the target genes femA (GenBank Accession No. NC 007795) and mecA (GenBank Accession No. X52593) to detect S. aureus and MRSA, respectively. The primers were designed with Primer Explorer V4 (http://primerexplorer.jp/e/; Eiken Chemical Co., Ltd., Tokyo, Japan) online primer design software and checked with the basic local alignment search tool (BLAST). The primer positions are shown in Fig. 1, and the primer sequences and modifications are shown in Table 1. All of the primers were synthesized by TsingKe Biotech Co., Ltd. (Beijing, China) with HPLC purification grade.
Fig. 1.
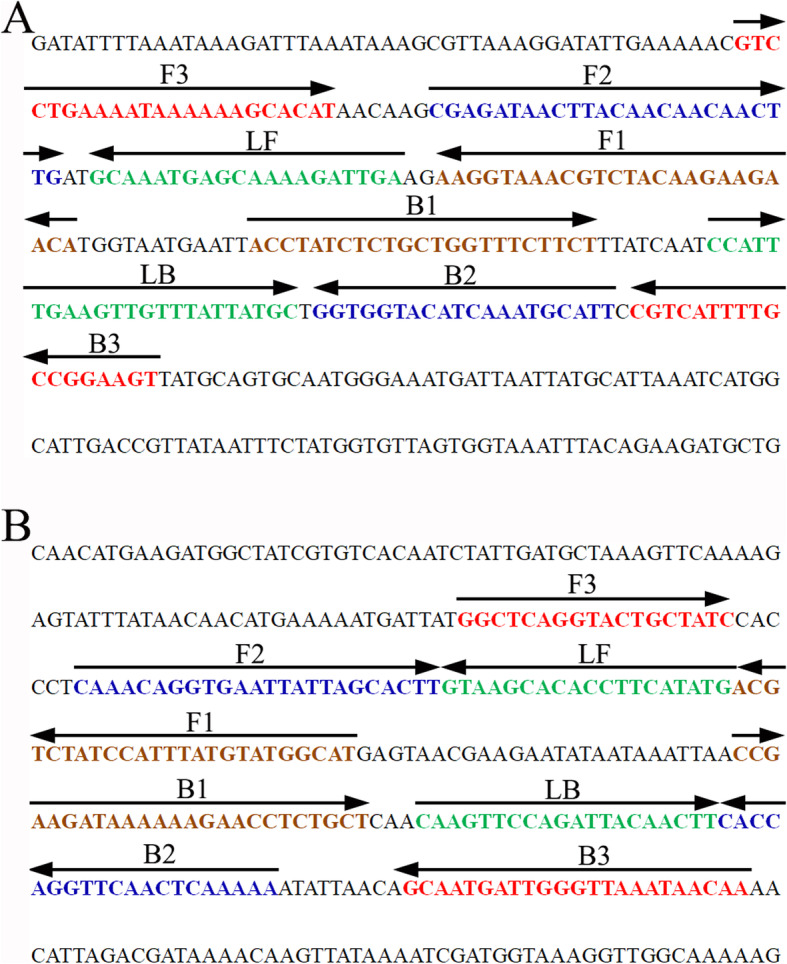
Sequence and location of the femA (a) and mecA (b) genes used to design m-LAMP primers. The nucleotide sequences of the sense strand of the femA and mecA genes are shown in the diagram. Right arrows and left arrows indicate sense and complementary sequences, respectively, which were used in the current study
Table 1.
The primers used in the present study
| Primer name | Sequence and modifications | Length | Gene |
|---|---|---|---|
| F3 | 5′-GTCCTGAAAATAAAAAAGCACAT-3′ | 23 nt | femA |
| B3 | 5′-ACTTCCGGCAAAATGACG-3′ | 18 nt | |
| FIP | 5′-TGTTCTTCTTGTAGACGTTTACCTT-CGAGATAACTTACAACAACAACTTG-3′ | 50 mer | |
| FIP* | 5′-Dig- TGTTCTTCTTGTAGACGTTTACCTT-CGAGATAACTTACAACAACAACTTG −3′ | 50 mer | |
| BIP | 5′-ACCTATCTCTGCTGGTTTCTTCT-AATGCATTTGATGTACCACC-3′ | 43 nt | |
| LF | 5′-TCAATCTTTTGCTCATTTGC-3′ | 20 nt | |
| LF* | 5′-Biotin-TCAATCTTTTGCTCATTTGC-3′ | 20 nt | |
| LB | 5′-CCATTTGAAGTTGTTTATTATGC-3′ | 23 nt | |
| F3 | 5′-GGCTCAGGTACTGCTATC-3′ | 18 nt | mecA |
| B3 | 5′-TTGTTATTTAACCCAATCATTGC-3′ | 23 nt | |
| FIP | 5′-ATGCCATACATAAATGGATAGACGT-CAAACAGGTGAATTATTAGCACTT-3′ | 49 nt | |
| FIP* | 5′-FAM-ATGCCATACATAAATGGATAGACGT-CAAACAGGTGAATTATTAGCACTT-3′ | 49 nt | |
| BIP | 5′-CCGAAGATAAAAAAGAACCTCTGCT-TTTTTGAGTTGAACCTGGTG-3′ | 45 nt | |
| LF | 5′-CATATGAAGGTGTGCTTAC-3′ | 19 nt | |
| LF* | 5′-Biotin-CATATGAAGGTGTGCTTAC-3′ | 19 nt | |
| LB | 5′-CAAGTTCCAGATTACAACTT-3′ | 20 nt |
Note: femA-FIP*, 5′-labeled with Dig when used in LAMP-LFB assay; femA-LF*, 5′-labeled with biotin when used in LAMP-LFB assay
mecA-FIP*, 5′-labeled with FAM when used in the LAMP-LFB assay; mecA-LF*, 5′-labeled with biotin when used in the LAMP-LFB assay;
Abbreviations: Dig digoxigenin, FAM carboxyfluorescein, nt nucleotide, mer monomeric unit
Bacterial strains and genomic DNA template preparation
In the current study, a total of 49 strains, including the S. aureus reference strain (ATCC 25923), 11 isolated MSSA strains, the methicillin resistant S. aureus reference strain (ATCC 43300), 16 MRSA isolated strains, and 20 non-S. aureus strains, were used for m-LAMP-LFB detection (Table 2). Genomic DNA templates were obtained using DNA extraction kits in accordance with the manufacturer’s instructions, and the concentration and purity were identified with a Nano drop ND-2000 (Beijing, China) at A260/280. The DNA templates were stored at − 20 °C before use. The genomic DNA of S. aureus (ATCC 25923) and MRSA (ATCC 43300) were serially diluted to concentrations ranging from 10 ng/μL to 100 ag/μL (10 ng/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL, 1 fg/μL, and 100 ag/μL), which were used to optimize the reaction temperature, reaction time, and sensitivity.
Table 2.
Bacterial strains used in the current study
| No. | Bacteria | Strain no. (source of strains)a | No. of strains | m-LAMP-LFB resultb | |
|---|---|---|---|---|---|
| femA | mecA | ||||
| 1 | S. aureus (MSSA) | ATCC 25923 | 1 | P | N |
| 2 | S. aureus (MSSA) | Isolated strains (2nd GZUTCM) | 11 | P | N |
| 3 | S. aureus (MRSA) | ATCC 43300 | 1 | P | P |
| 4 | S. aureus (MRSA) | Isolated strains (2nd GZUTCM) | 17 | P | P |
| 5 | Enterococcus faecalis | ATCC 29212 | 1 | N | N |
| 6 | Pseudomonas aeruginosa | ATCC 27853 | 1 | N | N |
| 7 | Shigella flexneri | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 8 | Listeria monocytogenes | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 9 | Mycobacterium tuberculosis | Isolated strains (GZCDC) | 1 | N | N |
| 10 | Acinetobacter baumannii | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 11 | Bacillus cereus | Isolated strains (GZCDC) | 1 | N | N |
| 12 | Vibrio parahaemolyticus | Isolated strains (GZCDC) | 1 | N | N |
| 13 | Leptospira interrogans | Isolated strains (GZCDC) | 1 | N | N |
| 14 | Enterohemorrhagic Escherichia coli | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 15 | Enteroaggregative Escherichia coli | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 16 | Streptococcus pneumoniae | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 17 | Staphylococcus saprophyticus | Isolated strains (GZCDC) | 1 | N | N |
| 18 | Enterotoxigenic Escherichia coli | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 19 | Invasive Escherichia coli | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 20 | Haemophilus parainfluenzae | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 21 | Shigella boydii | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 22 | Enteropathogenic Escherichia coli | Isolated strains (GZCDC) | 1 | N | N |
| 23 | Klebsiella pneumoniae | Isolated strains (2nd GZUTCM) | 1 | N | N |
| 24 | Bordetella parapertussis | Isolated strains (GZCDC) | 1 | N | N |
aATCC American Type Culture Collection, 2nd GZUTCM Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine, GZCDC Guizhou Provincial Center for Disease Control and Prevention
bP Positive, N Negative
Gold nanoparticle-based lateral flow biosensor preparation
The LFB platform was prepared according to a previous report [23]. Briefly, the LFB contained four components: an absorbent pad, NC membrane, sample pad, and conjugate pad. The components were assembled orderly on a backing card. The capture reagents, including anti-Dig, anti-FAM, and biotin-BSA, were immobilized by physical adsorption on the reaction regions. Then, anti-Dig was immobilized at test line 1 (TL1) (MSSA), and anti-FAM was immobilized at test line 2 (TL2) (MRSA), while biotin-BSA was immobilized at the control line (CL); each line was separated by 5 mm. SA-PNPs (dye streptavidin-coated polymer nanoparticles) were gathered on the conjugate pad. The prepared biosensors were preserved in a plastic box with a desiccant gel at room temperature before use.
The standard LAMP reaction
The single LAMP reactions for MSSA or MRSA were performed in 25 μl reaction systems as previously described [18]. Briefly, 0.4 μM each outer primer, F3 and B3, 0.8 μM each loop primer, LF* and LB, 1.6 μM each inner primer, FIP* and BIP, 0.4 mM biotin-14-dCTP, 1 μl (8 U) of Bst DNA polymerase, 12.5 μl of 2× reaction buffer, and 1 μl of DNA template were added to a tube. The mixtures were heated at 64 °C for 1 h. Genomic DNA from non-S. aureus strains, including Pseudomonas aeruginosa and Enterococcus faecalis, was used as a negative control (NC), and double distilled water (DW) was used as the template in the blank control (BC).
The m-LAMP reaction was performed in a one-step reaction in a 25 μl reaction system containing 12.5 μl of 2× reaction buffer; 0.2 μM each outer primer, femA-F3, femA-B3, mecA-F3 and mecA-B3; 0.4 μM each loop primer, femA-LF*, femA-LB, mecA-LF* and mecA-LB; 0.8 μM each inner primer, femA-FIP*, femA-BIP, mecA-FIP* and mecA-BIP; 0.4 mM biotin-14-dCTP; 1 μl (8 U) of Bst DNA polymerase; and 1 μl of DNA template. The reaction conditions were as described above.
Detection of LAMP products
Colorimetric indicator (malachite green, MG) and lateral flow biosensor (LFB) methods were applied for the determination and verification of the femA-LAMP, mecA-LAMP, and m-LAMP products. For the products amplified effectively, the color changed from colorless to light green in the MG assay. However, the color of the negative and blank controls remained colorless. The strategy of visualization of LAMP products with LFB was as previously described [24].
Temperature optimization of the LAMP assays
The effect of temperature on each LAMP reaction (femA-LAMP and mecA-LAMP) was tested during the amplification stage. Reaction temperatures ranging from 60 to 67 °C with 1 °C intervals were tested. Reaction mixtures with 1 μL of genomic DNA from P. aeruginosa and E. faecalis were used as negative controls (NCs), and 1 μL of distilled water (DW) was used as a blank control. The LAMP amplicons were analyzed by examining the turbidity of the products. The curves of the DNA concentrations of each amplified product are shown in the graph. Turbidity > 0.1 was considered positive.
Analytical sensitivity of LAMP-LFB assays
The sensitivity of each LAMP-LFB reaction (femA-LAMP-LFB, mecA-LAMP-LFB, and m-LAMP-LFB) was determined using 1 μl of the serial dilution (10 ng, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg and 100 ag per microliter) of the extracted genomic DNA. The LAMP-LFB reactions were carried out as described above, and the results were tested using a colorimetric indicator (MG) and LFB. The limit of detection (LoD) of single and multiplex reactions was verified as the last dilution of each positive test. Three replicates were tested for each dilution.
Optimization of the amplification time for the multiplex-LAMP-LFB assay
To optimize the reaction time of m-LAMP-LFB, four amplification times (20, 30, 40 and 50 min) were evaluated. The LAMP-LFB reactions were carried out as described above, and the results were tested using LFB. Each amplification time was tested at least three times.
Specificity analysis of m-LAMP-LFB detection
To determine the specificity of the m-LAMP-LFB assay, genomic DNA (at least 10 ng per microliter) from 12 MSSA strains, 17 MRSA strains, and 20 non-S. aureus strains (Table 2) was used for m-LAMP, and all of the results were tested using the LFB method. All examinations were confirmed at least three times.
Application of the m-LAMP-LFB method to analyze the clinical samples
To verify the applicability of the m-LAMP-LFB method for detecting S. aureus and identifying MRSA strains, a total of 63 whole blood samples, which were suspected of S. aureus infection, were collected from the Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine. The clinical samples were detected for MSSA and MRSA using traditional culture, PCR, and m-LAMP-LFB methods. Traditional culture methods included blood culture, colony morphology, Gram staining, biochemical identification, and methicillin susceptibility testing. PCR diagnosis was carried out using S. aureus- and MRSA-specific primers targeting the femA and mecA genes, respectively. The m-LAMP-LFB detection was performed as described above.
Results
Verification and analysis of femA- and mecA-LAMP products
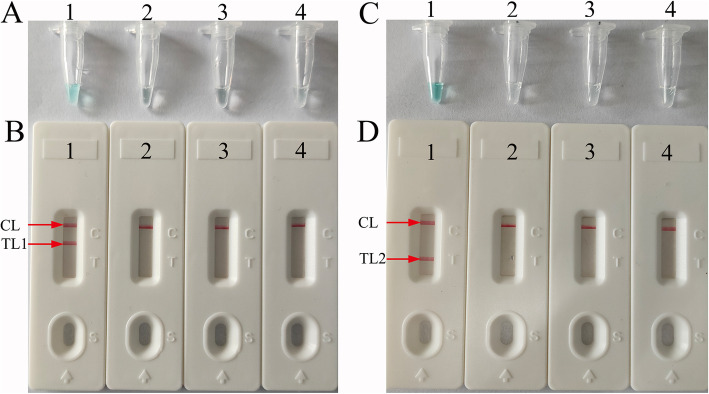
To confirm the amplification with the two sets of LAMP primers, the femA-, mecA-, and m-LAMP mixtures were incubated at a constant temperature of 63 °C for 1 h. Then, the femA-, mecA-, and m-LAMP products were analyzed with colorimetric indicator (MG) and lateral flow biosensor (LFB) methods, respectively. The color of the positive results in the femA-, mecA-, and m-LAMP reactions changed from colorlessness to bright green, while the negative and blank control reactions remained colorless (Fig. 2a, c). LFB was used for further confirmation of femA-, mecA-, and m-LAMP. For femA-LAMP detection, two crimson red bands (CL and TL1) appeared, indicating positive results, and CL and TL2 were visible for mecA-LAMP, indicating successful amplification, while the negative and blank controls only appeared as a crimson red line (CL) in the biosensor (Fig. 2b, d). Therefore, the results suggested that the two sets of LAMP primers for femA and mecA detection were valid for the development of the m-LAMP techniques.
Fig. 2.
Detection and verification of femA-and mecA-LAMP products. a, b Color and lateral flow biosensor detection of femA-LAMP products. Tube A1/Biosensor B1, positive amplification of the femA gene (S. aureus, ATCC25923); Tube A2 /Biosensor B2, negative amplification (Pseudomonas aeruginosa); Tube A3/Biosensor B3, negative amplification (Enterococcus faecalis); Tube 4/Biosensor 4, blank control (DW). c, d Color and lateral flow biosensor detection of mecA-LAMP products. Tube A1/Biosensor B1, positive amplification of the mecA gene (methicillin-resistant S. aureus, ATCC43300); Tube A2/Biosensor B2, negative amplification (Pseudomonas aeruginosa); Tube A3/Biosensor B3, negative amplification (Enterococcus faecalis); Tube 4/Biosensor 4, blank control (DW)
Optimal amplification temperature for femA- and mecA-LAMP
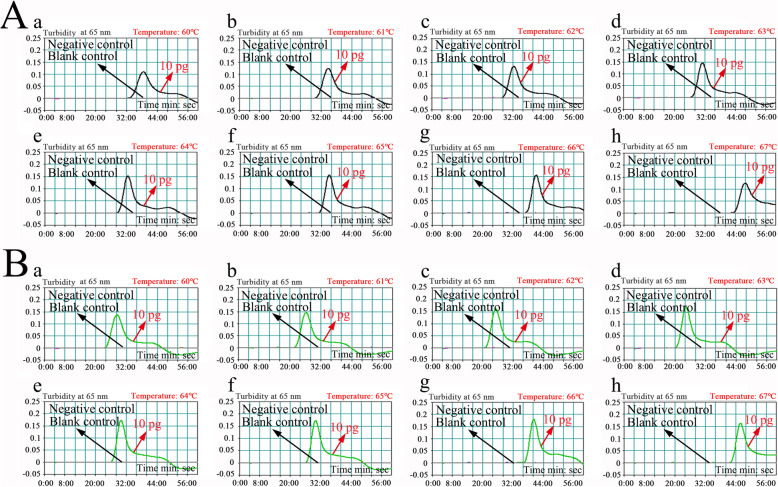
The reaction temperature is crucial for LAMP. In this study, the reaction temperature of femA- and mecA-LAMP was tested at different temperatures (from 60 to 67 °C with 1 °C intervals) with genomic template (10 pg/μl) extracted from purified cultures (ATCC 43300). The LAMP protocol was performed as described above, and the femA- and mecA-LAMP reactions were monitored by means of real-time turbidity measurements. Kinetic graphs were recorded at all temperatures. The results showed that femA-LAMP was amplified faster in the temperature range from 63 to 66 °C, and mecA-LAMP was amplified faster at 61 to 67 °C (Fig. 3). Hence, the amplification temperature of 63 °C was considered the optimal temperature for the rest of the m-LAMP reactions in the present study.
Fig. 3.
Optimization of amplification temperature for the femA-LAMP (a) and mecA-LAMP (b) primer sets. LAMP for the detection of femA (a) and mecA (b) was monitored through real-time turbidity, and the corresponding curves of DNA concentrations are displayed in the graphs. The threshold value was 0.1, and turbidity> 0.1 was considered positive. Mixtures with 10 pg of genomic DNA from P. aeruginosa (ATCC 27853) and E. faecalis (ATCC 29212) were used as negative controls (NCs), and 1 μl of double distilled water (DW) was used as a blank control (BC). Eight kinetic graphs were obtained at different temperatures (60–67 °C, 1 °C intervals) with 10 pg of target genomic DNA per reaction. a, The graphs from d (63 °C) to g (66 °C) showed robust amplification; b, the graphs from b (61 °C) to h (67 °C) showing robust amplification
Sensitivity of femA- and mecA-LAMP detection
The sensitivity of femA- and mecA-LAMP detection was evaluated with serially diluted genomic DNA at concentrations ranging from 10 ng to 100 ag per microliter. The LAMP amplicons were analyzed by visual inspection with MG reagents and lateral flow biosensors. The CL and TL1 lines appeared on the biosensor, showing positive results for the femA-LAMP assay, and two crimson lines (CL and TL2) were observed on the biosensor, indicating positive results for mecA-LAMP detection. For the negative controls, only the CL line appeared on the biosensor, indicating negative results. The results showed that the LoD of mecA-LAMP was 100 fg per reaction, which was the same as the LoD of the femA-LAMP assay (Fig. 4).
Fig. 4.
Sensitivity analysis of femA-LAMP (a, b) and mecA-LAMP (c, d) detection with serial dilutions of genomic DNA extracted from MSSA and MRSA strains, respectively
Two detection methods involving a colorimetric indictor (MG; a, c) or lateral flow biosensor (b, d) were used to analyze the amplification products. The genomic DNA was serially diluted (10 ng, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg and 100 ag per microliter) and subjected to standard LAMP. Tubes A1-A7 (Biosensors B1-B7), S. aureus (ATCC25923) genomic templates (10 ng-100 ag); Tube A8 (Biosensor B8), negative control (DW). The LoD of femA-LAMP detection was 100 fg of genomic template per reaction. Tubes C1-C7 (Biosensors D1-D7), methicillin-resistant S. aureus (ATCC43300) genomic templates (10 ng-100 ag); Tube C8 (Biosensor D8), blank control (DW). The LoD of mecA-LAMP detection was 100 fg of genomic template per reaction.
Sensitivity of m-LAMP detection
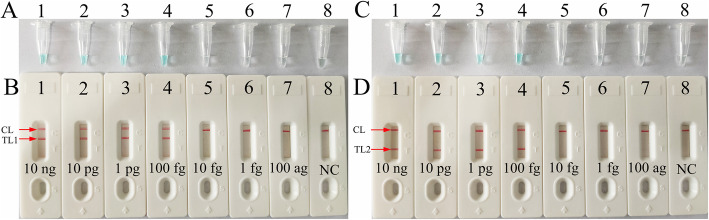
After m-LAMP was performed, the products were directly analyzed using LFB. The CL, TL1, and TL2 bands became crimson on the biosensor, reporting positive results for the femA and mecA genes. Only the CL line appeared on the biosensor, indicating negative results. The results showed that the LoD of m-LAMP-LFB for simultaneously assaying femA and mecA genes was also 100 fg of DNA per reaction (Fig. 5), which was consistent with single LAMP detection (Figs. 4 and 5).
Fig. 5.
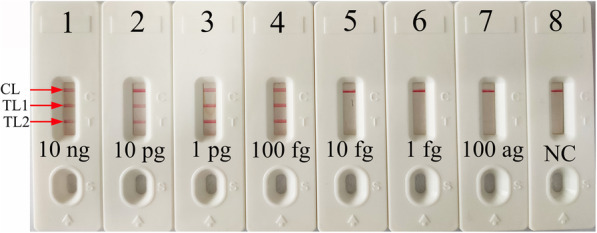
Sensitivity analysis of m-LAMP detection with serial dilutions of genomic DNA extracted from the MRSA strain. Two sets of primers targeting the femA and mecA genes were simultaneously added to a reaction vessel, and the LoD of m-LAMP for detecting S. aureus and identifying MRSA was analyzed with a lateral flow biosensor. Biosensors 1–8 represent the genomic DNA (methicillin-resistant S. aureus, ATCC43300) amounts of 10 ng, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg and 100 ag per reaction and blank control (DW), respectively. The LoD of the m-LAMP assay for femA and mecA detection was 100 fg of genomic template per reaction
Optimization of amplification time for m-LAMP-LFB detection
To obtain an optimal reaction time for m-LAMP, four reaction times (20, 30, 40, and 50 min) were tested at the optimal amplification temperature (63 °C). The results showed that the LoD of the genomic DNA template (100 fg of MRSA per reaction) was detected (displayed TL1, TL2 and CL) when the m-LAMP lasted 40 min (Fig. 6). Hence, a reaction time of 40 min was considered the optimal reaction time for m-LAMP detection. In summary, the whole detection procedure, including target genomic DNA preparation (30 min), m-LAMP (40 min) and analysis of results (2 min), could be completed within 80 min.
Fig. 6.
Optimization of the amplification time for m-LAMP detection. Different amplification times (a, 20 min; b, 30 min; c, 40 min; d, 50 min) were tested at 63 °C. Biosensors 1, 2, 3, 4, 5, 6, 7, and 8 represent genomic DNA (methicillin-resistant S. aureus, ATCC43300) levels of 10 ng, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg and 100 ag target template per reaction and negative control (DW), respectively. The best sensitivity was observed when the amplification lasted for 40 min (c)
Specificity of the m-LAMP assay
The specificity of m-LAMP detection was confirmed with MSSA, MRSA, and non-S. aureus isolates (Table 2). The genomic DNA extracted from MSSA and MRSA strains presented positive results. Three crimson lines (TL1, TL2 and CL) were displayed on the LFB, indicating positive results for the MRSA isolates. TL1 and CL appeared on the LFB, indicating positive results for the MSSA isolates. Other non-S. aureus strains and the blank control showed negative results (Fig. 7). Hence, the results confirmed that the m-LAMP-LFB method could accurately identify S. aureus and differentiate MRSA from all S. aureus strains.
Fig. 7.

Specificity analysis of m-LAMP-LFB detection using different strains. The m-LAMP reactions were carried out using different genomic DNA as templates, and each of the amplification products was determined by means of the visual LFB method. Biosensor 1, MSSA (ATCC 25923); Biosensors 2–12; eleven isolated strains of MSSA from the Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine; Biosensor 13, MRSA (ATCC 43300); Biosensors 14–29, sixteen isolated strains of MRSA from the Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine; Biosensors 30–49, Enterococcus faecalis, Pseudomonas aeruginosa, Shigella Flexneri, Listeria monocytogenes, Mycobacterium tuberculosis, Acinetobacter baumannii, Bacillus cereus, Vibrio parahaemolyticus, Leptospira interrogans, enterohemorrhagic Escherichia coli, enteroaggregative Escherichia coli, Streptococcus pneumonia, Staphylococcus saprophyticus, enterotoxigenic Escherichia coli, invasive Escherichia coli, Hemophilus parainfluenza, Shigella boydii, enteropathogenic Escherichia coli, Klebsiella pneumonia, Bordetella parapertussis, respectively; biosensor 50, blank control (DW)
Feasibility of the m-LAMP-LFB method using whole blood samples
To further demonstrate the feasibility of m-LAMP-LFB as a valuable method for the detection of MRSA and MRSA, 63 whole blood samples of suspected S. aureus-infected patients were collected from the Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine and tested by conventional culture-biotechnical methods, PCR detection, and m-LAMP-LFB assays. The results showed that 16 of 63 samples had been verified as MSSA-positive, and 12 of 63 samples had been verified as MRSA-positive through traditional culture techniques. The m-LAMP-LFB assay results were consistent with the traditional cultivation detection results. However, using PCR, only 14 and 9 samples were confirmed as MSSA- and MRSA-positive results, respectively (Table 3). These results suggested that the m-LAMP-LFB assay established in the current study could be used as an advanced tool to detect all S. aureus strains and separate MRSA from MSSA.
Table 3.
Comparison of conventional culture, PCR and m-LAMP-LFB methods to identify MSSA and MRSA in clinical samples
| Detection method | Clinical samples (n = 63) | ||
|---|---|---|---|
| MSSA-Positive | MRSA-Positive | Negative | |
| Culture | 16 | 12 | 35 |
| PCR | 14 | 9 | 40 |
| m-LAMP-FB | 16 | 12 | 35 |
Discussion
S. aureus, a ‘Janus-faced’ bacterium, is a commensal species and a pathogenic microorganism [25–27]. It is estimated that 20–30% of adult populations carry S. aureus in their nares; although this bacterium is commonly found, it only causes invasive infection when the host immune system is weakened [28–30]. In nosocomial settings, S. aureus is widely distributed in the environment, on surgical instrument surfaces, and on implanted medical devices, including prosthetic joints, catheters, and artificial heart valves [11, 31]. After S. aureus invades the human body, the pathogen rapidly enters the bloodstream and then diffuses into vital organs, causing pathological injury, such as osteomyelitis, endocarditis, and descending urinary tract infections [28]. Importantly, since methicillin-resistant S. aureus has emerged and spread worldwide, S. aureus has disrupted both the healthcare setting and community [4, 30], resulting in a huge socioeconomic burden in both developed and developing areas. The detection and identification of MSSA and MRSA is essential in cases of suspected S. aureus infections. However, traditional methods, including culture-based techniques, colony morphology, Gram staining, biochemical identification, methicillin susceptibility testing, and PCR-based detection (traditional PCR, multiple PCR and real-time PCR approaches), are time consuming and require expensive instruments. Moreover, the accurate interpretation of the results requires trained experts [14, 32]. Herein, developing a reliable, rapid, low-cost, simple, specific and sensitive detection method to accurately differentiate MSSA and MRSA is essential for disease diagnosis and therapy.
In the current study, m-LAMP-LFB detection targeting the femA and mecA genes was successfully established to assay all S. aureus species and identify MRSA. The sequences of S. aureus-LAMP primers were designed using the femA gene, which appears to be a unique feature of S. aureus and is not found in other Staphylococcus species [10]. Moreover, the MRSA-LAMP primers were designed with the mecA gene, encoding the low-affinity penicillin-binding protein 2a (PBP2a), which is associated with the methicillin resistance of S. aureus species [33]. The specificity of the m-LAMP assay was confirmed with genomic DNA from 12 MSSA, 17 MRSA, and 20 non-S. aureus isolates. m-LAMP detection of the femA gene identified S. aureus with 100% specificity, and the mecA gene identified MRSA with 100% specificity (Fig. 7).
In the present study, a nanoparticle-based lateral flow biosensor (LFB) was applied to analyze LAMP products. Although the amplification results could be detected equally with the turbidity and MG methods used in the current study, the LFB was deemed the preferred method for analyzing LAMP products; the LFB provides more visualization and does not require special instruments, regents and processes [34, 35]. In particular, the LFB applied in this study can simultaneously and visually detect two target genes (femA and mecA) in a single test. Compared with conventional culture and PCR-based methods, the m-LAMP-LFB technique is more sensitive, time-saving and cost-saving. The newly developed m-LAMP-LFB method was able to detect 100 fg of genomic DNA (Figs. 4 and 5). The entire detection process, including template preparation (approximately 30 min), isothermal amplification (40 min) and LFB reading (approximately 2 min), could be accomplished within 80 min. The total cost of one test, including genomic DNA extraction (approximately $1 USD), LAMP reaction (approximately $3.5 USD) and LFB reading (approximately $2 USD), is estimated to be $6.5 USD, which is cheaper than normal PCR-based methods. In addition, the advanced technique can decrease labor costs because performing the LAMP-LFB assay does not require skilled technical personnel. In conclusion, the newly developed m-LAMP-LFB technique in this study is a rapid, reliable and low-cost assay for the identification of MSSA and MRSA, and this technique can save detection time and help determine the optimal treatments for patients in a timely manner. In addition, accurate, timely and low-cost testing can reduce the patient’s financial burden, especially in resource-constrained regions of the world.
Conclusions
In the current study, a reliable, rapid and simple m-LAMP-LFB technique based on the femA and mecA genes was successfully developed for assaying S. aureus and identifying MRSA. This method could reliably, specifically, sensitively and rapidly detect all S. aureus species and identify MRSA isolates in samples. The amplification products were analyzed with LFB, which was objective, rapid, and easily interpretable. Hence, the m-LAMP-LFB assay could be considered a useful method for the reliable and rapid detection of S. aureus and identification of MRSA in clinical samples, particularly in resource-constrained regions of the world.
Acknowledgments
We thank all the medical workers in both the 2nd GZUTCM and GZCDC for their cooperation in this study.
Abbreviations
- MSSA
Methicillin susceptible Staphylococcus aureus
- MRSA
Methicillin resistant Staphylococcus aureus
- LAMP
Loop-mediated isothermal amplification
- LFB
Lateral flow biosensor
- m-LAMP-LFB
multiplex loop-mediated isothermal amplification linked to a nanoparticle-based lateral flow biosensor
- LoD
Limit of detection
- WHO
WORLD Health Organization
- PCR
Polymerase chain reaction
- MG
Malachite green
- ATCC
American type culture collection
- 2nd GZUTCM
Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine
- GZCDC
Guizhou Provincial Center for Disease Control and Prevention
- Dig
Digoxigenin
- FAM
Carboxyfluorescein
- nt
nucleotide
- mer
monomeric unit
- TL1
Test line 1
- TL2
Test line 2
- CL
Control line
- NC
Negative control
- BC
Blank control
- DW
Distilled water
- PBP2a
Penicillin-binding protein 2a
Authors’ contributions
XC, KM, and SL conceived and designed the study. XC and SL participated in primer design. XC, KM, XY, LX and YW contributed to all the laboratory work. KM, and LX contributed to the data collection. XC, XY and SL performed the statistical analysis. XC and SL wrote the initial draft of the manuscript. XC, YW, and SL revised the manuscript. All the authors read and approved the final manuscript.
Funding
This study was supported by the Program of the Scientific and Technological in Guiyang City (Grant No.Zhu Ke He [2020]1001), the High-Level Creative Talents Cultivation in Guizhou Province (Qian Ke He [2016]4021), the Program of Scientific and Technological Innovation Team of Guizhou Province (Grant No. Qian Ke He Platform talent [2018]5606), the Guizhou Provincial Special Funds for Science and Technology Supporting Program ([2019]1186, [2018]5767), and the Program of Science and Technology of Guizhou Provincial Health Commission (gzwjkj2018–1-012).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by the Human Ethics Committee of the Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine and complied with the Declaration of Helsinki. All data/isolates were analyzed anonymously.
Consent for publication
Not applicable.
Competing interests
All of the authors declare that there are no competing interests in this article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xu Chen, Email: xuchen1220@126.com.
Kai Ma, Email: makai1981@sina.com.
Xu Yi, Email: yixu2013@yeah.net.
Lijuan Xiong, Email: 279811189@qq.com.
Yu Wang, Email: wangzhongyuwy@163.com.
Shijun Li, Email: zjumedjun@163.com.
References
- 1.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VJ. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrel M, Perencevich EN, David MZ. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000-2013. Emerg Infect Dis. 2015;2(11):1973–1980. doi: 10.3201/eid2111.150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll DE, Marr I, Huang GKL, Holt DC, Tong SYC, Boutlis CS. Staphylococcus aureus prostatic abscess: a clinical case report and a review of the literature. BMC Infect Dis. 2017;17(1):509–507. doi: 10.1186/s12879-017-2605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson LR, Schora DM. Methicillin-resistant Staphylococcus aureus control in the 21st century: laboratory involvement affecting disease impact and economic benefit from large population studies. J Clin Microbiol. 2016;54(11):2647–2654. doi: 10.1128/JCM.00698-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okwu MU, Olley M, Akpoka AO, Izevbuwa OE. Methicillin-resistant Staphylococcus aureus (MRSA) and anti-MRSA activities of extracts of some medicinal plants: a brief review. AIMS Microbiolol. 2019;5(2):117. doi: 10.3934/microbiol.2019.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naimi HM, Rasekh H, Noori AZ, Bahaduri MA. Determination of antimicrobial susceptibility patterns in Staphylococcus aureus strains recovered from patients at two main health facilities in Kabul, Afghanistan. BMC Infect Dis. 2017;17(1):737. doi: 10.1186/s12879-017-2844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy H, Rudkin JK, Black NS, Gallagher L, O'Neill E, O'Gara JP. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Mi. 2015;5:1. doi: 10.3389/fcimb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong EF, Johnson JK, Jabra-Rizk MA. Community-associated methicillin-resistant Staphylococcus aureus: an enemy amidst us. PLoS Pathog. 2016;12(10):e1005837. doi: 10.1371/journal.ppat.1005837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Che Hamzah AM, Yeo CC, Puah SM, Chua KH, Chew CH. Staphylococcus aureus infections in Malaysia: a review of antimicrobial resistance and characteristics of the clinical isolates, 1990-2017. Antibiotics. 2019;8(3):128–156. doi: 10.3390/antibiotics8030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38(3):1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):1–103. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dweba CC, Zishiri O, El Zowalaty M. Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infect Drug Resist. 2018;11:2497–2509. doi: 10.2147/IDR.S175967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz R, Afreixo V, Ramalheira E, Rodrigues C, Gago B. Evaluation of vancomycin MIC creep in methicillin-resistant Staphylococcus aureus infections-a systematic review and meta-analysis. Clin Microbiol Infect. 2018;24(2):97–104. doi: 10.1016/j.cmi.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 14.Chung Y, Kim TS, Min YG, Hong YJ, Park JS, Hwang SM, Song K, Kim ES, Park KU, Kim HB, et al. Usefulness of multiplex real-time PCR for simultaneous pathogen detection and resistance profiling of Staphylococcal bacteremia. Biomed Res Int. 2016;2016:6913860–6913867. doi: 10.1155/2016/6913860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira LA, Harnett GB, Hodge MM, Cattell JA, Speers DJ. Real-time PCR assay for detection of blaZ genes in Staphylococcus aureus clinical isolates. J Clin Microbiol. 2014;52(4):1259–1261. doi: 10.1128/JCM.03413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DW, Kilgore PE, Kim EJ, Kim SA, Anh DD, Dong BQ, Kim JS, Seki M. The enhanced pneumococcal LAMP assay: a clinical tool for the diagnosis of meningitis due to Streptococcus pneumoniae. PLoS One. 2012;7(8):e42954. doi: 10.1371/journal.pone.0042954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge B, Domesle KJ, Yang Q, Hammack TS, Wang SS, Deng X, Hu L, Zhang G, Hu Y, Lai X, et al. Multi-laboratory validation of a loop-mediated isothermal amplification method for screening Salmonella in animal food. Front Microbiol. 2019;10:1–13. doi: 10.3389/fmicb.2019.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Liu Y, Wang Y, Chen H, Liu C, Wang Y. Lateral flow biosensor combined with loop-mediated isothermal amplification for simple, rapid, sensitive, and reliable detection of Brucella spp. Infect Drug Resist. 2019;12:2343–2353. doi: 10.2147/IDR.S211644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waterfield T, Fairley D, Lynn F, Blackwood B, Shields MD. A protocol for a systematic review of the diagnostic accuracy of loop-mediated-isothermal AMPlification (LAMP) in diagnosis of invasive meningococcal disease in children. Systematic Reviews. 2018;7(1):86–90. doi: 10.1186/s13643-018-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M, Kilgore PE, Kim EJ, Ohnishi M, Hayakawa S, Kim DW. Loop-mediated isothermal amplification methods for diagnosis of bacterial meningitis. Front Pediatr. 2018;6:57. doi: 10.3389/fped.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Q, Domesle KJ, Wang F, Ge B. Rapid detection of Salmonella in food and feed by coupling loop-mediated isothermal amplification with bioluminescent assay in real-time. BMC Microbiol. 2016;16:112. doi: 10.1186/s12866-016-0730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Wang Y, Ma A, Li D, Luo L, Liu D, Jin D, Liu K, Ye C. Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci Rep. 2015;5:11902. doi: 10.1038/srep11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao W, Wang Y, Wang G, Wang Y, Xiao J, Sun L, Li J, Wen S, Zhang T, Ma Q, et al. Development and clinical validation of multiple cross displacement amplification combined with nanoparticles-based biosensor for detection of Mycobacterium tuberculosis: preliminary results. Front Microbiol. 2019;10:2135. doi: 10.3389/fmicb.2019.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Li H, Wang Y, Zhang L, Xu J, Ye C. Loop-mediated isothermal amplification label-based gold nanoparticles lateral flow biosensor for detection of Enterococcus faecalis and Staphylococcus aureus. Front Microbiol. 2017;8:192. doi: 10.3389/fmicb.2017.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grousd JA, Rich HE, Alcorn JF. Host-pathogen interactions in gram-positive bacterial pneumonia. Clin Microbiol Rev. 2019;32(3):e00107. doi: 10.1128/CMR.00107-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galar A, Weil AA, Dudzinski DM, Muñoz P, Siedner MJ. Methicillin-resistant Staphylococcus aureus prosthetic valve endocarditis: pathophysiology, epidemiology, clinical presentation, diagnosis, and management. Clin Microbiol Rev. 2019;32(2):e00041. doi: 10.1128/CMR.00041-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassoun A, Linden PK, Friedman B. Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. Crit Care. 2017;21:211. doi: 10.1186/s13054-017-1801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balasubramanian D, Harper L, Shopsin B, Torres VJ. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis. 2017;75(1):ftx005. doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosimi RA, Beik N, Kubiak DW, Johnson JA. Ceftaroline for severe methicillin-resistant Staphylococcus aureus infections: a systematic review. Open Forum Infect Dis. 2017;4(2):ofx084. doi: 10.1093/ofid/ofx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aires-de-Sousa M. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. 2017;23(6):373–380. doi: 10.1016/j.cmi.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Holland TL, Arnold C, Fowler VG. Clinical management of Staphylococcus aureus bacteremia. JAMA. 2014;312(13):1330. doi: 10.1001/jama.2014.9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallin M, Maes N, Byl B, Jacobs F, De Gheldre Y, Struelens MJ. Clinical impact of a PCR assay for identification of Staphylococcus aureus and determination of methicillin resistance directly from blood cultures. J Clin Microbiol. 2003;41(8):3942–3944. doi: 10.1128/JCM.41.8.3942-3944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Talib H, Yean CY, Al-Khateeb A, Hasan H, Ravichandran M. Rapid detection of methicillin-resistant Staphylococcus aureus by a newly developed dry reagent-based polymerase chain reaction assay. J Microbiol Immunol Infect. 2014;47(6):484–490. doi: 10.1016/j.jmii.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Hu S, Niu L, Zhao F, Yan L, Nong J, Wang C, Gao N, Zhu X, Wu L, Bo T, et al. Identification of Acinetobacter baumannii and its carbapenem-resistant gene blaOXA-23-like by multiple cross displacement amplification combined with lateral flow biosensor. Sci Rep. 2019;9:17888. doi: 10.1038/s41598-019-54465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng X, Yang J, Wang M, Wu P, Du Q, He J, Tang Y. Visual and rapid detection of Acinetobacter baumannii by a multiple cross displacement amplification combined with nanoparticles-based biosensor assay. AMB Express. 2019;9(1):30. doi: 10.1186/s13568-019-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.